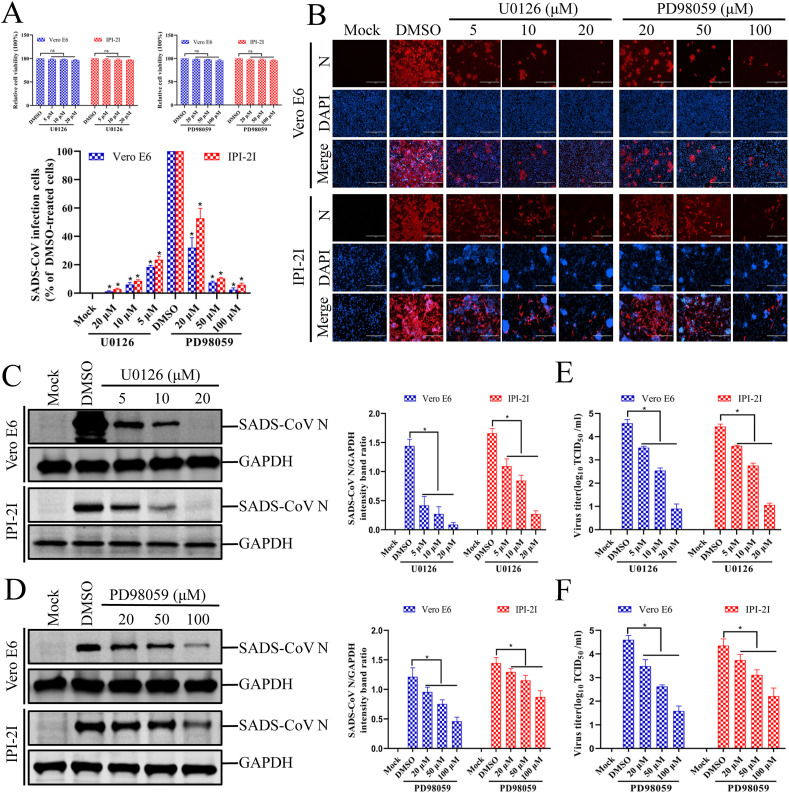
Fig. 3.
Inhibition of ERK1/2 activation impairs SADS-CoV infection. (A) U0126 and PD98059 treatments do not affect cell viability. Vero E6 and IPI-2I cells were treated with U0126 or PD98059 at different concentrations or with the vehicle control DMSO for 36 h. Cell cytotoxicity was analyzed with the CCK-8 kit, as described in the Materials and Methods. (B) SADS-CoV propagation in the presence of U0126 or PD98059. Vero E6 cells were pretreated with either inhibitor at the indicated concentrations for 1 h and then infected with SADS-CoV. SADS-CoV-infected cells were further incubated for 36 h in the presence of DMSO, U0126, or PD98059. At 36 hpi, the virus-infected cells were subjected to an immunofluorescence assay with an anti-SADS-CoV-N antibody, followed by DAPI counterstaining and examination under an inverted fluorescence microscope. The percentage of SADS-CoV-infected cells per view is expressed as the mean ± SD of three independent experiments. (C and D) Viral N protein expression in the presence of U0126 or PD98059. Vero E6 and IPI-2I cells were treated with DMSO, U0126, or PD98059 at the indicated concentration for 1 h before infection with SADS-CoV. The SADS-CoV-infected cells were then incubated for a further 36 h in the presence of DMSO, U0126, or PD98059. At 36 hpi, the cell lysates were examined with western blotting, probed with an antibody directed against SADS-CoV N protein. The blot was also reacted with a mouse monoclonal antibody directed against GAPDH to verify equal protein loading. Densitometric data for SADS-CoV N/GAPDH are expressed as the means ± SD of three independent experiments. (E and F) U0126 or PD98059 treatment suppressed SADS-CoV replication. Treatment and infection conditions were as described for panels C and D. The viral titers in the supernatants collected at 36 hpi were determined with the Spearman–Kärber method. Error bars represent the standard errors of the means of three independent experiments.