Abstract
Two new complexes of Co(II) and Zn(II) 2-chlorobenzoate (2-ClBA) with 3-cyanopyridine (CNP) of the general formula [Co(2-ClBA)2(CNP)2(H2O)2] and [Zn(2-ClBA)2(CNP)2(H2O)2] were synthesized. The structures of the complexes were characterized by single crystal XRD and FT-IR and NMR spectroscopy and Mass Spectrometry (MALDI-TOF MS) methods. Mononuclear complexes exhibit octahedral coordination. In addition, Hirshfeld surface analysis was performed to determine non-covalent interactions in crystal packing. The geometry optimization of the molecules was carried out using the LANL2DZ level of theory of the DFT method and the obtained findings were confirmed by comparing with the data obtained from the single crystal X-ray diffraction method. The theoretical and experimental bond angles and lengths are very close to each other. The effectiveness of the complexes against SARS-CoV-2 enzymes was investigated in silico using the molecular docking method, and a binding score of -8.0 kcal/mol on NSP16 of complex 1 as an inhibitor was obtained. To investigate the drug potential of the complexes, their pharmacokinetic and toxicokinetic properties were estimated by ADMET calculations.
Keywords: Transition metal complex, Arylcarboxylates, SARS-CoV-2, Molecular docking, DFT, ADMET
Graphical abstract

1. Introduction
Metal(II) aryl carboxylate complexes have been widely studied for many years because of their many potential applications in fields such as biology, pharmacology, catalysis, sensors, and magnetism. Most of these carboxylate-containing ligands coordinate with metal ions in many different types such as monodentate, bidentate, bridging, chelating-bidentate, bridging bidentate. Different coordination types ensure the diversification of their properties in application areas [1], [2], [3], [4]. The auxiliary ligand in the structure has a great role in the exhibit of all these different coordination modes of benzoic acid and its derivatives [5]. Arylcarboxylic acids and N-donor ligands are widely used in the synthesis of supramolecular structures [6]. Supramolecular structures are formed through different components interacting with non-covalent interactions such as hydrogen bonding [7], [8], [9], [10], dipole-dipole interactions, steric repulsions, van der Waals interactions, and London dispersion forces. These non-covalent interactions are important for biological systems as well as contribute to the stability of the crystal structure [11], [12], [13], [14], [15].
Cobalt is a bio-metal found in plants, human and animal metabolisms. In humans, cobalt is at the center of cobalamin (vitamin B12), which is necessary for many physiological processes such as DNA synthesis and the proper functioning of the nervous system. Cobalt complexes are known for their anticancer and antimicrobial properties. Recent research has focused on the synthesis and evaluation of complexes with biologically interesting ligands to increase complex activity [16], [17], [18]. Zinc is the second most abundant trace element in the human body. Approximately 95% of the zinc in the human body is in the cell. It is essential for growth and development in all forms of life. In case of both deficiency and excess, it causes some health problems. It has been suggested to have beneficial therapeutic and preventive effects on infectious diseases, including shortening the duration of the common cold for humans [17,[19], [20], [21]]. Besides its essential role in living systems, the zinc (II) ion is a component of compounds used medicinally in the treatment of skin diseases and as a nutritional supplement. Zinc(II) complexes are known to be used as antimicrobial [22], antifungal [23], anticancer [24], anti-inflammatory [25], antioxidant [26] and antidiabetic [27] agents.
The disease COVID19, which emerged with the spread of a new Coronavirus called SARS-Cov-2 in late 2019, caused a worldwide pandemic and brought most activities in the world to a standstill. With the pandemic, people had to take some disposable precautions, such as masks, visors, overalls, with the guidance of some authorities. Many researchers continue to work for both treatment and permanent prevention [28], [29], [30]. Today, drug materials synthesized for use in drug therapy for many viral diseases such as Herpes and HIV are limited. This is also true for SARS-CoV-2 [31], [32], [33]. While some of the known drugs used in the treatment of SARS-CoV-2 since the beginning of the pandemic are still in use, the use of some of them has been stopped [34]. We wanted to support with an in silico study whether two compounds synthesized for the first time in this study can be antiviral agent candidates for use in the treatment of SARS-CoV-2.
2. Materials and method
2.1. Chemicals
All chemicals and solvents were purchased commercially and used without further purification.
Sodium bicarbonate, 2-chlorobenzoic acid, 3-cyanopyridine, ethanol, and the metal salts CoSO4·6H2O and ZnSO4·7H2O were obtained from Sigma Aldrich, Germany.
2.2. Measurements
FT-IR spectra were recorded in the range of 600–4000 cm−1 with a Perkin Elmer Frontier™ spectrometer with an ATR detector using solid samples. 1H NMR spectrum of the complex 2 was recorded on Bruker Avance III 500 spectrometer in deuterated DMSO (d6-DMSO). Mass spectra of the complexes were obtained by Bruker Micro flex LT MALDI-TOF MS spectrometer using dithranol (DIT) as MALDI matrix.
2.3. Preparation of the complex 1
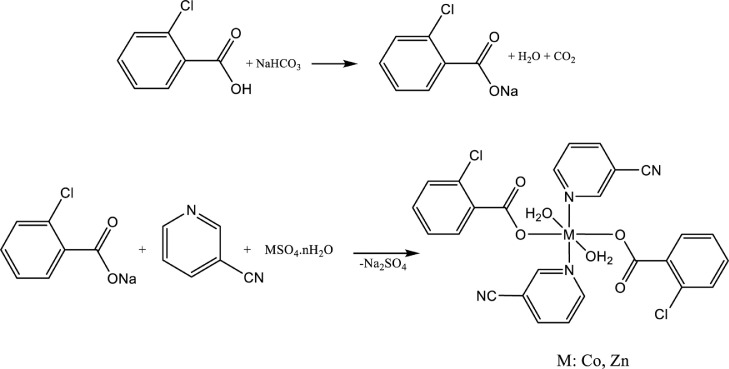
Sodium bicarbonate (0.84 g, 10 mmol) and 2-chlorobenzoic acid (1.56 g, 10 mmol) were mixed in 100 mL of distilled water and heated and stirred at 60 °C until the CO2 gas was completely removed. 3-cyanopyridine (1.04 g, 10 mmol) in 20 mL of ethanol was added to a 50 mL aqueous solution of CoSO4·7H2O (1.40 g, 5 mmol), followed by the addition of a previously prepared solution of sodium 2-chlorobenzoate (1.78 g, 10 mmol) in 100 mL of distilled water. The synthesis is given in Scheme 1. The pink and colorless solutions obtained were slowly evaporated at room temperature and pink and colorless single crystals suitable for X-ray structure determination were obtained in two weeks. The crystals obtained were filtered, washed with distilled water, and left to dry at room temperature. The purity of the complex was checked by the thin-layer chromatography (TLC) technique. Yield 2.30 g (74.92%). Selected IR bands (cm−1): ν(OH)H2O 3375, ν(C—H)as 2973, ν(C—N)s 2237, v(COO)as 1596, v(COO−)s 1408, ν(C—C)phen 1438, δ(C—Cl) 810. m/z: [M-CNP]+ = 510.82; [M-2CN +5H]+ = 568.15; [M+DIT-2H2O—CN+Na]+ = 793.72;[M+DIT-2H2O—Na]+ = 851.24 (Fig. S1).
Scheme 1.
The synthesis procedure of the complexes.
2.4. Preparation of the complex 2
Unlike the synthesis of Complex 1, ZnSO4·7H2O (1.43 g, 5 mmol) was used as the metal salt. Colorless crystals were obtained from the colorless clear solution after four weeks, filtered and washed with distilled water. The purity of the complex was checked by thin layer chromatography (TLC) technique. The synthesis is given in Scheme 1. Yield 2.51 g (81.29%). Selected IR bands (cm−1): ν(OH)H2O 3372, ν(C—H)as 2980, ν(C—N)s 2237, v(COO)as 1596, v(COO−)s 1407, ν(C—C)phen 1438, δ(C—Cl) 810. 1H NMR (400 MHz, DMSO‑d 6) δ 7.33–7.44 (m, 6H, ArH), 7.64–7.69 (m, 4H, ArH), 8.33 (d, 2H, ArH; J = 8.00 Hz), 8.87 (m, 2H, ArH), 9.04 (s, 2H, ArH), 3.33 (s, 4H, 2H2O). m/z: [M-CNP-H2O]+ = 371.95; [M-2CN +Na]+ = 590.87; [M + 2Na]+ = 665.52; [M+DIT-CN—H2O+4H]+ = 806.92 (Fig. S2).
2.5. X-ray crystallography
Single-crystal X-ray diffraction analyses of compounds (1 and 2) were performed on a Bruker APEX-II CCD diffractometer using Mo Kα (λ= 0.71073 Å) radiation at a temperature of 296(2) K. Structures were solved by direct methods [35] and refined by full-matrix least-squares against F2 using all data [35]. All non-H atoms were refined anisotropically. The water H atoms were located in different Fourier maps and refined freely, while the C-bound H atoms were positioned geometrically at distances of 0.93 Å (for aromatic CH) from the parent C atoms; a riding model was used during the refinement processes and the Uiso(H) values were constrained to be Uiso(H) = 1.2 X Ueq(C). Experimental data are given in Table 1 .
Table 1.
Experimental details for complexes (1 and 2).
|
Complex |
||
|---|---|---|
| 1 | 2 | |
| Empirical Formula | C26H20Cl2CoN4O6 | C26H20Cl2N4O6Zn |
| formula weight | 614.31 | 620.75 |
| color/shape | pink/prism | colourless/prism |
| Crystal System | monoclinic | monoclinic |
| Space Group | P 21/c | P 21/c |
| a (Å) | 7.0846(2) | 7.1077(2) |
| b (Å) | 13.6645(3) | 13.6178(4) |
| c (Å) | 14.1157(3) | 14.1456(4) |
| α (°) | 90 | 90 |
| β (°) | 101.42 (2) | 101.714(2) |
| γ (°) | 90 | 90 |
| V (Å3) | 1339.43(6) | 1340.65(7) |
| Z | 2 | 2 |
| μ (Mo Kα) (mm−1) | 0.889 | 1.164 |
| ρ (calcd) (mg m−3) | 1.523 | 1.538 |
| Number of Reflections Total | 25,938 | 22,198 |
| Number of Reflections Unique | 3354 | 3341 |
| Rint | 0.0288 | 0.0248 |
| 2θmax (°) | 56.82 | 56.64 |
| Tmin / Tmax | 0.64 / 0.79 | 0.56 / 0.70 |
| Number of Parameters | 186 | 186 |
| GOF | 1.047 | 1.064 |
| R [F2 >2σ(F2)] | 0.0283 | 0.0283 |
| wR | 0.0774 | 0.0772 |
| (Δρ)max (e Å−3) | 0.393 | 0.418 |
| (Δρ)min (e Å−3) | −0.174 | −0.411 |
2.6. Hirshfeld surface analysis
Hirshfeld surface analysis was used to visualize the intermolecular interactions of the synthesized complexes [36,37]. CrystalExplorer Version 21.5 [38] was used to generate the Hirshfeld surface [39] and 2D fingerprint plots [37].
2.7. Density functional theory calculations
Gaussian 09 [40] was preferred for theoretical calculations and Gaussview 5.0 [41] was used to visualize. The Avogadro software [42] was partially used to visualize bond length and bond angle. Geometry optimizations of complexes 1 and 2 were calculated with Becke-3-Lee-Yang-Parr's functional correlation (B3LYP) [43], [44], [45] of LANL2DZ level of theory in the DFT method. LANL2DZ (Los Alamos National Laboratory 2 double ζ) is mostly used for calculations involving heavy elements [46]. The geometry optimization of the complex was chosen as a stable form with C1 symmetry. In addition to geometry optimization, frequency analyses were also performed and compared with experimental data. Total electron density surface was calculated according to electrostatic potential values. The total electron surface was visualized using SCF/ESP as the density matrix.
2.8. Molecular docking
Autodock Vina [47] was used to calculate binding affinity for synthesized cobalt and zinc complexes 1 and 2 The X-ray crystal structure of SARS-CoV-2 non-structural protein (PDB code: 6M0J [48], 7BV2 [49], 6WXC [50], 7BQY [51], 6WKQ [52], 7MEQ [53]) was resolved using X-ray diffraction method with a resolution factor of 2.50 Å was retrieved from the RCSB Protein Data Bank (https://www.rcsb.org). In the protein, the presence of water molecules from the protein structure was removed and polar hydrogens and Kollman charges were added. Automatically the root of each ligand molecule is detected, and torsions were selected. The ligand's torsions were allowed to rotate, and the selected residues were tested. The ligands were docked blindly to see where they would preferentially bind. The amino acids in the active site of NSP-12 and NSP-12 with RNA were determined using the BIOVA Discovery Studio Visualizer 2021 [54]. Pre-calculated grid maps were required for running the program, which was calculated using the AutoGrid program. The energy scoring grid box was set to 32 × 32 × 32 dimension (x, y, and z) centered at X = 91.692; Y = 92.471; and Z = 103.743 with 0.325 Angstroms grid points spacing assigned with default atomic salvation parameters. As a docking engine, the Lamarckian Genetic Algorithm was used, with all docking parameters set to default. The visualization processes were carried out using the BIOVA Discovery Studio Visualizer 2021.
2.9. ADMET predictions
In drug design, determining the pharmacokinetic and toxicokinetic properties of drug candidate molecules prevents multiple experiments, saving both time and cost and making the success rate higher. Absorption, distribution, metabolism, excretion, and toxicity parameters (ADMET) describe the properties that a drug molecule must carry. The SwissADME online database [55] was used to estimate the absorption, distribution, metabolism, and excretion values of the synthesized complexes. The ProTox-II online database [56] was used to estimate toxicity values.
3. Results and discussion
3.1. Description of the structure
The single crystal X-ray analysis results and spectroscopic data of complexes 1 and 2 are compatible with each other. Selected bond lengths and angles are given in Table 2 . Hydrogen bond geometries are given in Table 3 . Fig. 1 a and 1b show the molecular structures and atom-numbering schemes, whereas Fig. 2 a and 2b show the partial packing diagrams.
Table 2.
Selected bond lengths (Å) and angles (°) for complexes (1 and 2).
|
Complex 1 |
Complex 2 |
||||
|---|---|---|---|---|---|
| Bond lengths | X-Ray | DFT | X-Ray | DFT | |
| Co1—O1 | 2.0749(9) | 2.0165 | Zn1—O1 | 2.0832(10) | 2.1112 |
| Co1—O3 | 2.0976(11) | 2.1868 | Zn1—O3 | 2.1118(12) | 2.0903 |
| Co1—N1 | 2.1815(12) | 1.9762 | Zn1—N1 | 2.1906(12) | 2.2430 |
| O1—C1 | 1.2569(16) | 1.3029 | O1—C1 | 1.2536(17) | 1.2999 |
| O2—C1 | 1.2526(17) | 1.2876 | O2—C1 | 1.2509(18) | 1.2925 |
| C11—C13 | 1.441(2) | 1.4381 | C9—C13 | 1.442(2) | 1.4377 |
| N2—C13 | 1.134(2) | 1.1820 | N2—C13 | 1.134(2) | 1.1822 |
| Bond angles | |||||
| O1—Co1—O3 | 88.20 (4) | 88.18 | O1—Zn1—O3 | 87.77 (5) | 87.95 |
| O1—Co1—O3i | 91.80 (4) | 91.82 | O2—Zn1—O3ii | 92.23 (5) | 92.06 |
| O1—Co1—N1 | 88.71 (4) | 88.71 | O1—Zn1—N1 | 90.76 (5) | 90.27 |
| O1—Co1—N1i | 91.29 (4) | 90.29 | O1—Zn1—N1ii | 89.24 (5) | 89.73 |
| O3—Co1—N1 | 93.42 (5) | 93.53 | O3—Zn1—N1 | 86.77 (5) | 89.57 |
| O3—Co1—N1i | 86.58 (9) | 86.47 | O3i—Zn1—N1ii | 93.23 (5) | 90.43 |
| O1—C1—O2 | 125.56 (13) | 124.26 | O1—C1—O2 | 125.63 (13) | 123.78 |
Symmetry codes: (i) −x, −y, −z, (ii) −x, −y + 1, −z.
Table 3.
Hydrogen-bond geometry (Å, o) for complexes (1 and 2).
| Complex | D-H···A | D-H | H···A | D···A | D-H···A |
|---|---|---|---|---|---|
| 1 | O3—H31···O2i | 0.76 (2) | 2.07 (2) | 2.7690 (17) | 153 (2) |
| O3—H32···O2ii | 0.81 (2) | 1.85 (2) | 2.6499 (17) | 166 (2) | |
| C4—H4···O2iii | 0.93 | 2.48 | 3.393 (2) | 167 | |
| 2 | O3—H31···O2iv | 0.79 (2) | 1.88 (2) | 2.6483 (19) | 164 (3) |
| O3—H32···O2v | 0.75 (2) | 2.08 (2) | 2.7640 (18) | 151 (2) |
Symmetry codes: (i) x + 1, y, z; (ii) − x, − y, − z; (iii) x, − y + ½, z – ½; (iv) − x, − y + 1, − z; (v) x – 1, y, z.
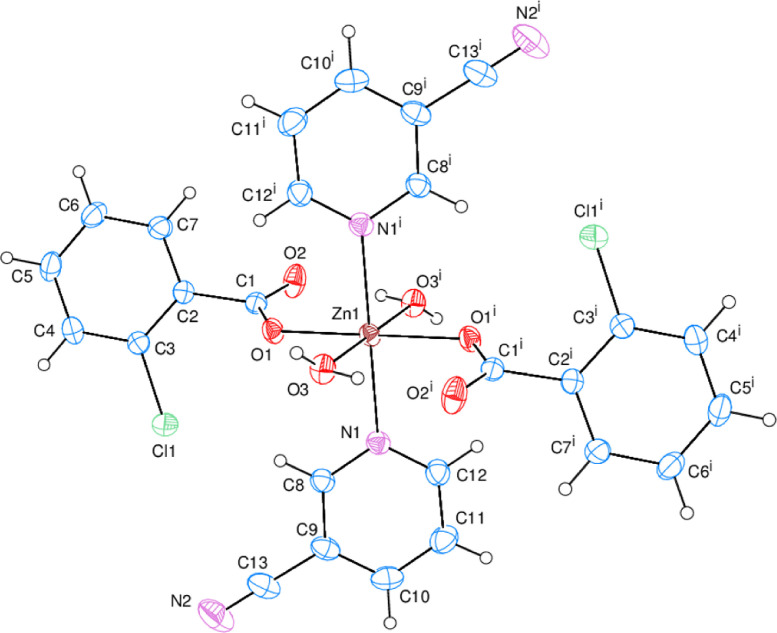
Fig. 1.
(a) An ORTEP-3 [58] view of complex 1. The thermal ellipsoids are drawn at the 50% probability level (Symmetry code: −x, −y, −z). (b) An ORTEP-3 [58] view of complex 2. The thermal ellipsoids are drawn at the 50% probability level (Symmetry code: −x, −y + 1, −z).
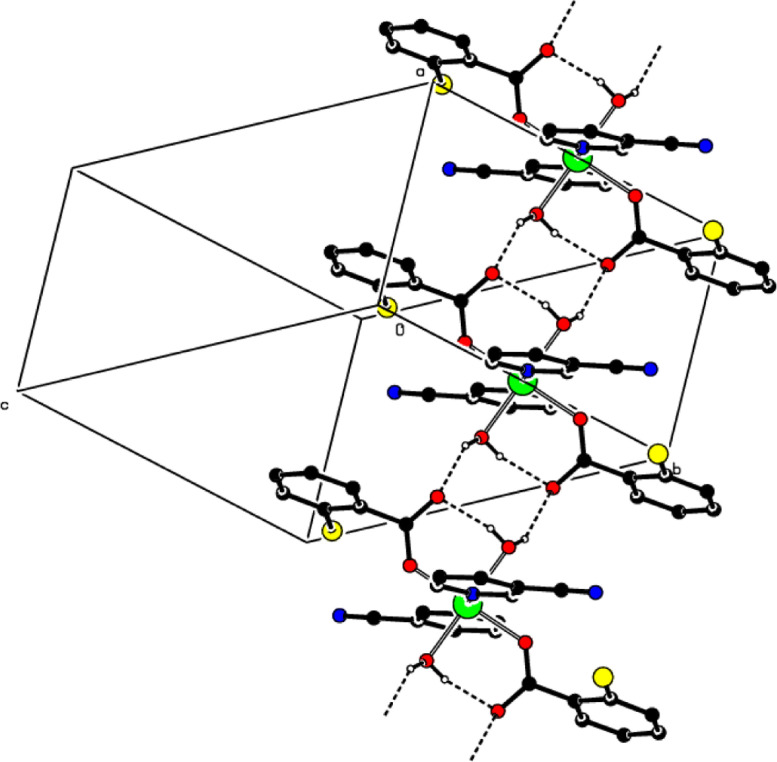
Fig. 2.
(a) A partial packing diagram of complex 1. Intermolecular O—Hw···Oc and C—Hbnz···Oc (w = water, bnz = benzene, c = non-coordinating carboxylate O atom) hydrogen bonds are shown as dashed lines. Non-bonding H atoms have been omitted for clarity. (b) A partial packing diagram of complex 2. Intermolecular O—Hw···Oc (w = water, c = non-coordinating carboxylate O atom) hydrogen bonds are shown as dashed lines. Non-bonding H atoms have been omitted for clarity.
The asymmetric units of the mononuclear complexes (1 and 2) include metal (Co1 for 1 and Zn1 for 2) cations, positioned on symmetry centers, one 3-cyanopyridine (CNP) ligand, one 2-chlorobenzoate (2-ClBA) anion, and one water molecule. All ligands are monodentately coordinated to Metal cations (Fig. 1a and 1b). (Fig. 1a and 1b).
The MII cations (CoII and ZnII) are coordinated monodentately through the two symmetry-related 2-ClBA anions’ the two carboxylate oxygen atoms (O1 and O1i) and two oxygen atoms (O3 and O3i) of the two symmetry-related water at distances of [2.0749 (9) and 2.0976 (11) Å (for 1) and [2.0832 (10) and 2.1118 (12) Å (for 2)], respectively, to form slightly distorted square-planar environments. The slightly distorted octahedral coordination geometry has been completed via the two symmetry-related N atoms (N1 and N1i) of the two symmetry-related CNP ligands in the axial positions. M—N1 and M—N1i distances were found as 2.1815 (12) Å (for 1) and 2.1906 (12) Å (for 2)], respectively [symmetry codes: (i) – x, – y, – z (for 1) and (i) −x, −y + 1, −z (for 2)] (Fig. 1a and 1b). The close bong lengths of the C1—O1 [1.2569 (16) Å (for 1) and 1.2536 (17) Å (for 2)] and C1—O2 [1.2526 (17) Å (for 1) and 1.2509 (18) Å (for 2)] in the carboxylate groups are more suitable for delocalized bonding arrangements, rather than localized single and double bonds. The M—O bond lengths [2.0976 (11) Å (for 1) and 2.1118 (12) Å (for 2)] for water O atoms are about 0.023 Å (for 1) and 0.029 Å (for 2) longer than those the 2-chlorobenzoate O atoms [2.0749 (9) Å (for 1) and 2.0832 (10) Å (for 2)]. In the MO4N2 octahedrons, the M—N bond lengths [2.1815 (12) Å (for 1) and 2.1906 (12) Å (for 2)] are the longest. The Co and Zn atoms are located at 0.3355(1) Å (for 1) and 0.3276(1) Å (for 2) below and above the planar (O1/O2/C1) carboxylate groups, respectively. The O1—C1—O2 bond angles [125.56 (13)° (for 1) and 125.56 (13)° (for 2)] appear to be substantially greater than in a free acid [122.2°]. The O—M—O and O—M—N bond angles [range 86.58 (5) to 93.42 (5)° (for 1) and 86.77 (5) to 93.23 (5)° (for 2) for cis angles; all-trans angles are 180° due to symmetry] are slightly different from ideal, with same average values of 90.00(4)° (for 1) and 89.89 (5)° (for 2).
The dihedral angles between the planar carboxylate groups (O1/O2/C1) and the neighboring benzene A (C2—C7) rings of complexes 1 and 2 are 62.81 (6)° and 64.19 (7)°, respectively. while the benzene (A) and pyridine (B) (N1/C9—C13) rings are oriented at dihedral angles of A/B = 12.62 (5)° (for 1) and 13.04 (6)° (for 2).
In complex 1, the water ligands to the 2-ClBA anions are connected by the intermolecular O—Hw···Oc (w = water, c = non-coordinating carboxylate oxygen atom) hydrogen bonds (Table 1), through the R4 4(8) ring motifs [57], leading to the formations of layers along the a-axis direction. These layers are further connected by the weak intermolecular C—Hbnz···Oc (bnz = benzene) hydrogen bonds (Table 1), resulting in the construction of a three-dimensional network structure (Fig. 2a). In complex 2, the water ligands to the 2-ClBA anions are connected by the intermolecular O—Hw···Oc (w = water, c = non-coordinating carboxylate O atom) hydrogen bonds (Table 1), through the R4 4(8) ring motifs [57], resulting to the formations of layers along the a-axis direction (Fig. 2b). The π···π interactions between the benzene A (C2—C7) and pyridine B (N1/C8—C12) rings, Cg1— Cg2i (for 1) and Cg1— Cg2ii (for 2) [Symmetry codes: (i) 1 + x, ½ - y, ½ + z, (ii) 1 - x, ½ + y, ½ - z, where Cg1 and Cg2 are the centroids of rings A and B, respectively] may help to stabilize the crystal structures with the centroid-centroid distances of 3.7940(9) Å (for 1) and 3.7857(10) Å (for 2).
3.2. Hirshfeld surface analysis
Intermolecular interactions can be represented with varied colors and color intensities on Hirshfeld surfaces, indicating short or long contacts and the strength of interactions [59]. Fig. 3 a and 3c show the complexes' dnorm maps. On the dnorm maps, Contacts with distances equal to the total of van der Waals radii are indicated by the white surface. The colors red and blue show distances that are shorter (in close contact) or longer (distinct touch) than the van der Waals radii. In the dnorm maps of the complexes, dark red spots are seen near the hydrogen bond donor and acceptor atoms that form hydrogen bonds. Other intermolecular interactions are seen as faint red [60], [61], [62]. Convex blue regions and concave red regions on the shape-index for complexes 1 and 2 indicate donor and acceptor groups, respectively (Fig. 3b and 3d). The existence of adjacent red and blue triangles in the crystal structures of complexes 1 and 2 confirms π-π stacking interactions.
Fig. 3.
The Hirshfeld surface mapped for complexes 1 and 2 over (a and c) dnorm in the range −0,5655 to 1,2178 a.u., and −0,5612 to 1,2270 a.u., respectively. A view of the Hirshfeld surface mapped for complexes 1 and 2 over (b and d) the shape-index.
The contribution percentage values for each interaction in 2D fingerprint plots are given in Fig. 4 . The contribution values for each interaction of the two isostructure complexes are almost the same. 2D fingerprint graphs of important intermolecular contacts are given in Figs. S3 and S4 in the Supporting Material file. On the Hirshfeld surfaces of complexes 1 and 2, the H…H interactions were shown in the largest region of the fingerprint plot at de + di= 2.46 Å (27.3% contribution for complex 1 and 27.4% contribution for complex 2). Because of the abundance of hydrogen in the structure, H…H interactions make the most significant contribution to the structure. For complexes 1 and 2, H…N/N…H interactions, which are the second significant contribution, contributed 19.7% to the fingerprint plot to the total Hirshfeld surface with the tips at de + di = 2.64 Å. C…C interactions which have bullet shape cover 14.9% contributions in Hirshfeld surface of complexes 1 and 2. C—H…π and π-π stacking interactions are responsible for these interactions. These weak interactions greatly contribute to the crystal packing of the complexes. For complexes 1 and 2, the contribution of H…F/F…H contacts in the fingerprint plot to the total Hirshfeld surface is 8.7%, 10.3%, and 12.2%, respectively. H⋯Cl/Cl⋯H contacts have been observed the symmetrical two wings in the fingerprint plot delineated and they have been contributed with 12.0% to Hirshfeld surface with the tips at de + di= 2.94 Å. H…O/O…H interactions 12.5% to the overall crystal packing were identified as the complex 1 and 2′s fifth significant intermolecular interactions with tips at de + di = 1.1 + 0.78 Å =1.88 Å. These interactions are also represented by two spike wings. These interactions are due to O—H…O hydrogen bonds in the crystal structure. The contribution of H…C/C…H interactions to the Hirshfeld surface of complexes 1 and 2 are 9.2%. These interactions occurred symmetrically and over a wide area, with a high concentration at the fingerprint plot's edges with the tips at de + di = 2.02 Å. C—H.... π and π-π stacking interactions are responsible for these interactions. These weak interactions greatly contribute to the crystal packing of the complexes. In addition, C…Cl/Cl…C (1.4%), C…N/N…C (0.8%), N…N (0.7%) and N…Cl/Cl…N (0.3%) interactions were also observed, these interactions made a negligible contribution to the Hirshfeld surface.
Fig. 4.
The percentages of the intermolecular interactions from obtained Hirshfeld surface analysis of complexes 1 and 2.
3.3. FT-IR spectroscopy
Experimental and theoretical FT-IR spectra of the complexes are given in Figs S5 and S6 in the Supporting Material file. The FT-IR spectra of [Co(2-ClBA)2(CNP)2(H2O)2] and [Zn(2-ClBA)2(CNP)2(H2O)2] show bands with wavenumbers close to those reported in the literature for similar structures. When the FT-IR spectra of the complexes are examined, the absorption bands observed in the range of 3550–3300 cm−1 are due to stretching vibrations of water molecules. These vibrations were found in the different regions in FT-IR spectra calculated using the B3LYP/LANL2DZ level of theory. In the theoretical study, O—H bands of both complexes were observed around 2850 cm−1. Reviewing previous studies, this large chemical shift is attributed to anharmonic contributions. Due to the anharmonic effects, the vibrations of hydrogen bond formation affected by the H-bond are observed in a different region [63], [64], [65]. The stretching vibrations related to the aromatic C—H stretching vibrations were observed around 3000 cm–1 in the FT-IR Spectra of complexes 1 and 2. The mentioned vibrations for FT-IR Spectra were computed around 3200–3100 cm−1. The υ(C—N) stretching vibration of 3-cyanopyridine is reported at 2230 cm−1 in the FT-IR spectra. In the FT-IR spectra, while the experimentally observed and theoretically calculated values for complex 1 are 2237 cm−1 and 2272 cm−1, respectively and for complex 2, these values are 2237 cm−1 and 2272 cm−1, respectively. The fact that these vibrations occur in the same region without chemical shifts also shows that the metal atom is not coordinated with the cyano group. The FT-IR spectroscopy gives also important information regarding the carboxylate ion's coordination type [66]. This information is determined by the Δν value calculated from the difference between asymmetric and symmetric carboxylate vibrations. If this value is greater than the Δν value of the sodium salt of the acid, it is suggested that the coordination type of the carboxylate group be monodentate. Asymmetric and symmetrical vibrations of the COO− moiety for complexes 1 and 2 were observed at 1596 cm−1 and 1375 cm−1 and, respectively. The Δν(COO−) values for complexes 1 and 2 were calculated as 210 and 220 cm−1, respectively. Theoretically asymmetric and symmetrical vibrations of the COO− moiety were calculated at 1514 cm−1 and 1376 cm−1 for complex 1 and 1511 and 1367 cm−1 for complex 2, respectively. When the synthesized complexes' (COO−) values are compared to the sodium salt of 2-chlorobenzoic acids, it is evident that the 2-chlorobenzoate anions coordinate monodentate. The υ(C—N) stretching vibration of the pyridine ring was observed at 1048 cm−1 for complex 1 and 1047 cm−1 for complex 2. For complexes 1 and 2, these vibrations were calculated theoretically at 1061 cm−1 and 1081 cm−1, respectively. The complexes' and 2-chlorobenzoate's ν(C—Cl) vibrations were recorded at the same frequency (about 810 cm−1). This result supports that the 2-chlorobenzoate anion of the metal atom is not coordinated with the chloro groups [67,68].
3.4. 1H NMR spectra
Because Co(II) is paramagnetic, no signal was observed in 1H NMR for complex 1. Resonances of the aromatic protons of benzene rings of 2-chlorobenzoic acid and pyridine ring of 3-cyanopyridine showed at 7.33–9.04 ppm for complex 2. The signal related to the proton of coordinated water molecules for complex 2 was observed at δ 3.33 ppm (Fig. S7) [60].
3.4. DFT results
Density functional theory was used to study the chemical properties of complexes 1–2 and calculated using the LANL2DZ level of theory of the B3LYP basis set. The geometries of the crystal structures were optimized in the gas phase and some computational parameters were evaluated. The Koopmans theorem shows the relationship between ionization potential and electron affinity with HOMO and LUMO orbital energies: electron affinity is the inverse of LUMO energy value, while ionization potential is the inverse of HOMO value. From these energy data, ionization potential (I.P.), electron affinity (E.A.), electronegativity (χ), electrophilicity index (ω), global softness (σ) and chemical hardness (η) values of complexes 1–2 were calculated according to the formulas specified in Table 4 [69], [70], [71], [72], [73], [74].
Table 4.
Some calculated chemical parameters of complexes 1 and 2 by the DFT method.
| Unit | Complex 1 | Complex 2 | ||
|---|---|---|---|---|
| Polarizability (α) | cm−3 | 9890.59 | 9307.57 | |
| Total Energy (ETotal) | eV | −50,287.29 | −48,124.90 | |
| LUMO +1 Energy (ELUMO+1) | eV | −2.31 | −2.28 | |
| LUMO Energy (ELUMO) | eV | −2.35 | −2.29 | |
| HOMO Energy (EHOMO) | eV | −5.95 | −7.01 | |
| HOMO −1 Energy (EHOMO-1) | eV | −6.69 | −7.04 | |
| Energy Gap (EGAP) | eV | 3.60 | 4.72 | |
| Dipole Moment (D) | eV | 0.00 | 0.00 | |
| Ionization Potential (IP) | eV | 5.95 | 7.01 | |
| Electron Affinity (EA) | eV | 2.35 | 2.29 | |
| Electronegativity (χ) | eV | 4.15 | 4.65 | |
| Chemical Hardness (η) | eV | 1.80 | 2.36 | |
| Global Softness (σ) | eV−1 | 0.28 | 0.21 | |
| Electrophilicity index (ω) | eV | 4.78 | 4.58 | |
| EGAP = EHOMO–ELUMO | IP = –EHOMO | EA = –ELUMO | χ = (IP + EA)/2 | |
| η = (IP – EA)/2 | σ = 1/2η | ω = χ2/2η | ||
The calculated bond lengths and angles of complexes 1 and 2 optimized with DFT indicate that the X-ray values for complex 1 bond lengths are numerically close to each other compared to experimental values. The computational bond lengths for Co1—O1 and Co1—O3 are 2.0165 and 2.1868 Å, respectively. For Co1—N1, the calculated bond length was 1.9762 Å and lower than the experimental value (2.1815 Å). The computational bond lengths for Zn1—O1 and Zn1—O3 are 2.1112 and 2.0903 Å, respectively. For Zn1—N1, the computational bond length was 2.2430 Å and greater than the experimental value (2.1906 Å). The bond angles for both Co and Zn complexes are the same as experimental values (Table 2).
Electron migration is easier in molecules with a high polarizability. The cobalt complex can be more polarized than the zinc complex. The electronic energy of the cobalt complex is lower, i.e., more stable, than the energy of the zinc complex. This situation is in correlation with the band gap and the bandgap of complex 1 (3.60 eV) is narrower than the bandgap of complex 2 (4.72 eV) as seen in Fig. 5 . There is a positive correlation between molecular docking results and bandgap values. Reactive complex 1, which has a narrower bandgap and easier electron transitions, is more effective compared to complex 2, which has fewer values.
Fig. 5.
The calculated molecular orbital diagram of complexes 1 and 2 by the DFT method.
3.5. Molecular docking results
The Coronavirus consists of Envelope (E), Membrane (M), Spike (S), Nucleocapsid (N), and genomic RNA and nonstructural proteins (NSP1–16). Inhibition of one or more of these proteins will stop or slow the effects of the Coronavirus. There are some model inhibitors for enzyme inhibition, but their efficacy is also insufficient. N3 [K], Remdesivir nucleoside monophosphate (K), Tipiracil [K], Sinefungin [K] and N-Acetyl-beta-d-glucosamine [K] are model inhibitors. Despite being a small molecule, favipiravir is a highly effective antiviral because it exhibits covalent interactions with Coronavirus proteins. By taking all these model inhibitors as a reference, it is possible to discover new inhibitors that are more effective and have lower toxicity.
Complexes 1 and 2 were inserted by molecular docking study on five important proteins of SARS-CoV-2 (Spike, Main protease, NSP12, NSP15, and NSP16) and ACE2 and Transmembrane protease, serine 2 on the cell membrane, and their binding affinities and ligand efficiencies were computed (Table 5 ). Complex 1 has the most effective binding score for NSP16 (−8.00 kcal/mol). NSP16 plays an important role in viral transcription by stimulating 2′-O-methyltransferase activities [75]. Thus, complex 1 being a specific inhibitor candidate for NSP16 may inhibit viral transcription. In addition, the binding score for the spike protein of complex 1, Coronavirus is −7.90 kcal/mol. The spike protein enters the cell by interacting with ACE2 in the cell membrane. Complex 1 has a high docking score for both spike protein and ACE2. Therefore, complex 1 placed in the catalytic region between spike + ACE2 can act as an antagonist and prevent it from penetrating the cell. Complex 1 has a binding value of −7.70 kcal/mol for the main protease, which is essential for viral replication and feeds non-structural proteins [76]. For the docked NSP12, NSP15, and TMPRSS2 proteins, the complex 1 model inhibitor had slightly lower scores and ligand efficiencies (Fig. 6 and Table 5).
Table 5.
The docking results of complexes 1 and 2 on SARS-CoV-2 proteins.
 |
Fig. 6.
The Molecular docking results of complexes 1 and 2 on the NSP12 protein of Coronavirus. (a) Docking result of complex 1 and the spike protein of the Coronavirus. (b) 2D interactions of the complex 1 with amino acids in the active site of the spike protein. (c) Docking result of complex 1 and the NSP16 protein of the Coronavirus. (d) 2D interactions of the complex 1 with amino acids in the active site of the NSP16 protein.
The binding scores of complex 2 correlate with those of complex 1, the main protease and ACE2 docking scores are the same. The docking score of zinc complex for main protease and ACE2 is −7.70 kcal/mol. In other proteins, the zinc complex has relatively lower scores and ligand efficiencies than the cobalt complex. This shows that ligands rather than the central metal atom are effective on the enzyme. It was determined that there are conventional hydrogen, carbon-hydrogen, electrostatic salt bridge-attractive charge, hydrophobic π-π stacked or T-shaped, hydrophobic π-alkyl, π-sigma, π-sulfur, and halogen bonds non-covalent interactions between candidate inhibitors and amino acids. Non-covalent interactions of candidate inhibitors with amino acids are given in Table 5. Considering all these results, it is predicted that the complexes are very suitable for non-covalent interaction due to their structure and will exhibit a positive inhibitory effect when interacting with Coronavirus enzymes.
3.6. Estimated pharmacokinetic and toxicokinetic properties
Predicting the pharmacokinetic and toxicokinetic properties of drug candidate compounds increases the success of reaching the target in drug discovery. Lipinski et al. proposed five rules for a compound to be a drug [77]. Complexes 1 and 2 conform to Lipinski's rules, except for one (both compounds have a molecular weight above 500 g/mol). There is no difference between the predicted values of cobalt and zinc complexes. Due to the bulky nature of the compounds, their gastrointestinal absorption is low, they cannot cross the blood-brain barrier (BBB), and they cannot be used as substrates of P-glycoprotein. The complexes' solubility in water/octanol is low due to their rigid structure. The complexes can induce or inhibit cytochrome P450 enzymes (CYPs) CYP2C19 and CYP2C9. In addition, it does not interact with CYP1A2, CYP2D6, and CYP3A4. The lack of lipophilic groups in the structures of the complexes also reduces skin permeation or lipid permeability. The estimated toxicity values were examined and the compounds were determined to be in the nontoxic class. The similarity rate to the compounds used in toxicity estimation is 38%. The estimated lethal dose amounts are similar for complexes 1 and 2, being 2190 mg/kg. Complexes 1 and 2 are inactive, ie nontoxic, as hepatotoxicity, carcinogenicity, immunotoxicity, mutagenicity, cytotoxicity, and mitochondrial membrane potential (MMP). Considering all these advantages and disadvantages, it is predicted that our compounds can be candidate drugs, drug formulation studies can be conducted and they will be a guide for those who study similar molecules. The estimated pharmacokinetic properties for complexes 1–2 are given in Table 6 .
Table 6.
The pharmacokinetic properties of the complexes 1–2.
| Properties |
Complex |
|
|---|---|---|
| (1) | (2) | |
| Molecular weighta | 614.30 | 620.75 |
| Number of atoms | 59 | 59 |
| Heavy atoms | 39 | 39 |
| Rotatable bonds | 8 | 8 |
| H-Bond acceptors | 8 | 8 |
| H-Bond donors | 2 | 2 |
| Molar refractivity | 137.60 | 137.60 |
| TPSA (Å2) | 126.40 | 126.40 |
| Log Po/w | 2.53 | 2.55 |
| GI absorption | Low | Low |
| BBB permeant | No | No |
| P-gp substrate | No | No |
| CYP1A2 inhibitor | No | No |
| CYP2C19 inhibitor | Yes | Yes |
| CYP2C9 inhibitor | Yes | Yes |
| CYP2D6 inhibitor | No | No |
| CYP3A4 inhibitor | No | No |
| Log Kp (cm/s) | −6.21 | −6.25 |
| Lipinski | Yes | Yes |
| Toxicity classb | 5 | 5 |
| Predicted LD50c | 2190 | 2190 |
| Hepatotoxicity | Inactive | Inactive |
| Carcinogenicity | Inactive | Inactive |
| Immunotoxicity | Inactive | Inactive |
| Mutagenicity | Inactive | Inactive |
| Cytotoxicity | Inactive | Inactive |
| MMPd | Inactive | Inactive |
Molecular weight unit is g/mol.
The toxicity class consists of six numbers. Number 1 means toxic; number 6 means nontoxic.
Predicted LD50 unit is mg/kg.
MMP: Mitochondrial Membrane Potential.
4. Conclusion
The new cobalt(III) and zinc(II) 2-chlorobenzoate with 3-cyanopyridine complexes were synthesized and determined the crystal structure. The M2+ cations have octahedrally coordinated by two 2-chlorobenzoate anions, two 3-cyanopyridine ligands, and two water molecules leading to an overall MN2O4 coordination environment. It was determined by Hirshfeld surface analysis that the functional groups in the crystal structure of the complexes formed non-covalent interactions. We optimized the crystal structures of complexes 1–2 and calculated their physiological properties to understand their activities. The compounds were evaluated in terms of the structure-activity relationship and there were no major differences between the results. The substrate effect of our complexes was investigated as in silico against the proteins of SARS-CoV-2, which caused the global pandemic, and satisfactory results were found. Based on the data obtained, it is believed that in vitro/in vivo experiments of complexes can be conducted and will be promising for further research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The numerical calculations reported in this paper were fully performed at TUBITAK ULAKBIM, High Performance, and Grid Computing Center (TRUBA resources). The authors thank Murat Beytur for his supports.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.131825.
Appendix. Supplementary materials
Appendix
Crystallographic data for the structural analysis have been deposited with the Cambridge Crystallographic Data centre, CCDC 2,099,779 for complex 1 and 2,099,780 for complex 2. Copies of this information may be obtained free of charge from The Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax: 44–1223–336,033; e-mail: deposit@ ccdc.cam.ac.uk or www: http://www.ccdc.cam.ac.uk).
References
- 1.Hao J., Yu B., Van Hecke K., Cui G. A series of d 10 metal coordination polymers based on a flexible bis(2-methylbenzimidazole) ligand and different carboxylates: synthesis, structures, photoluminescence and catalytic properties. CrystEngComm. 2015;17:2279–2293. doi: 10.1039/C4CE02090A. [DOI] [Google Scholar]
- 2.Palanisami N., Murugavel R. Synthesis, spectral characterization, and single crystal X-ray structures of a series of manganese-2,2′-bipyridine complexes derived from substituted aromatic carboxylic acids. Inorg. Chim. Acta. 2011;365:430–438. doi: 10.1016/j.ica.2010.09.040. [DOI] [Google Scholar]
- 3.Rueff J.-.M., Masciocchi N., Rabu P., Sironi A., Skoulios A. Structure and magnetism of a polycrystalline transition metal soap-CoII[OOC(CH2)10COO](H2O)2. Eur. J. Inorg. Chem. 2001;2001:2843–2848. doi: 10.1002/1099-0682(200111)2001:11<2843::AID-EJIC2843>3.0.CO;2-T. [DOI] [Google Scholar]
- 4.Wang Q., Yu Z., Wang Q., Li W., Gao F., Li S. Synthesis, crystal structure and DNA-binding properties of a mononuclear copper complex with pyridine-2-carboxylate ligand. Inorg. Chim. Acta. 2012;383:230–234. doi: 10.1016/j.ica.2011.11.013. [DOI] [Google Scholar]
- 5.Ozarowski A., Calzado C.J., Sharma R.P., Kumar S., Jezierska J., Angeli C., Spizzo F., Ferretti V. Metal–metal interactions in trinuclear copper(II) complexes [Cu 3 (RCOO) 4 (H 2 TEA) 2] and binuclear [Cu 2 (RCOO) 2 (H 2 TEA) 2]. syntheses and combined structural, magnetic, high-field electron paramagnetic resonance, and theoretical studies. Inorg. Chem. 2015;54:11916–11934. doi: 10.1021/acs.inorgchem.5b02199. [DOI] [PubMed] [Google Scholar]
- 6.Gómez V., Corbella M. Versatility in the coordination modes of n -chlorobenzoato ligands: synthesis, structure and magnetic properties of three types of polynuclear Mn II compounds. Eur. J. Inorg. Chem. 2009;2009:4471–4482. doi: 10.1002/ejic.200900532. [DOI] [Google Scholar]
- 7.Siva V., Murugan A., samad Shameem A., Athimoolam S., Bahadur S.A. A new metal-organic hybrid material: synthesis, structural, electro-optical properties and quantum chemical investigation. Opt. Mater. 2021;121 doi: 10.1016/j.optmat.2021.111616. (Amst) [DOI] [Google Scholar]
- 8.Siva V., Shameem A., Murugan A., Athimoolam S., Vinitha G., Bahadur S.A. Structural, thermal and electro-optical properties of guanidine based metal-organic framework (MOF) Chin. J. Phys. 2020;68:764–777. doi: 10.1016/j.cjph.2020.09.036. [DOI] [Google Scholar]
- 9.Siva V., Shameem A., Murugan A., Athimoolam S., Suresh M., Asath Bahadur S. A promising guanidinium based metal-organic single crystal for optical power limiting applications. Chin. J. Phys. 2020;64:103–114. doi: 10.1016/j.cjph.2020.01.001. [DOI] [Google Scholar]
- 10.Siva V., Bahadur S.A., Shameem A., Athimoolam S., Lakshmi K.U., Vinitha G. Synthesis, structural, vibrational, thermal, dielectric and optical properties of third order nonlinear optical single crystal for optical power limiting applications. J. Mol. Struct. 2019;1191:110–117. doi: 10.1016/j.molstruc.2019.04.091. [DOI] [Google Scholar]
- 11.Kubik S. De Gruyter; 2020. Supramolecular Chemistry: From Concepts to Applications. [DOI] [Google Scholar]
- 12.Lehn J.M. VCH; Weinheim: 2006. Supramolecular Chemistry Concepts and Perspectives.http://onlinelibrary.wiley.com/book/10.1002/3527607439 (accessed July 27, 2021) [Google Scholar]
- 13.Steed J.W., Atwood J.L. 2nd ed. Wiley; Chichester: 2009. Supramolecular Chemistry. [Google Scholar]
- 14.Chitra Devi A., Siva V., Thangarasu S., Athimoolam S., Asath Bahadur S. Supramolecular architecture, thermal, quantum chemical analysis and in vitro biological properties on sulfate salt of 4-aminoantipyrine. J. Mol. Struct. 2021;1245 doi: 10.1016/j.molstruc.2021.131033. [DOI] [Google Scholar]
- 15.Patil S.S., Tadavi S.K., Dikundwar A., Bendre R.S. The transition metal complexes of Fe(II), Ni(II) and Cu(II) derived from phthalazine based ligands: synthesis, crystal structures and biological activities. J. Mol. Struct. 2022;1247 doi: 10.1016/j.molstruc.2021.131293. [DOI] [Google Scholar]
- 16.Akcha S., Gómez-Ruiz S., Kellou-Tairi S., Lezama L., Pérez F.B., Benali-Baitich O. Synthesis, characterization, solution equilibria, DFT study, DNA binding affinity and cytotoxic properties of a cobalt(II) complex with a 5-pyrazolone ligand. Inorg. Chim. Acta. 2018;482:738–748. doi: 10.1016/j.ica.2018.06.051. [DOI] [Google Scholar]
- 17.Crichton R.R. 1st ed. Elsevier; Amsterdam: 2008. Biological Inorganic Chemistry: An Introduction. Repr. [Google Scholar]
- 18.Talwar G.P., Srivastava L.M. Prentice Hall Of India; New Delhi: 2006. Textbook of Biochemistry and Human Biology. [Google Scholar]
- 19.Andrejević T.P., Warżajtis B., Glišić B.Đ., Vojnovic S., Mojicevic M., Stevanović N.Lj., Nikodinovic-Runic J., Rychlewska U., Djuran M.I. Zinc(II) complexes with aromatic nitrogen-containing heterocycles as antifungal agents: synergistic activity with clinically used drug nystatin. J. Inorg. Biochem. 2020;208 doi: 10.1016/j.jinorgbio.2020.111089. [DOI] [PubMed] [Google Scholar]
- 20.Larson C.P., Saha U.R., Nazrul H. Impact monitoring of the national scale up of zinc treatment for childhood diarrhea in Bangladesh: repeat ecologic surveys. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vahrenkamp H. Why does nature use zinc-a personal view. Dalton Trans. 2007:4751–4759. doi: 10.1039/B712138E. [DOI] [PubMed] [Google Scholar]
- 22.Tarushi A., Lafazanis K., Kljun J., Turel I., Pantazaki A.A., Psomas G., Kessissoglou D.P. First- and second-generation quinolone antibacterial drugs interacting with zinc(II): structure and biological perspectives. J. Inorg. Biochem. 2013;121:53–65. doi: 10.1016/j.jinorgbio.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Xie J., Zhu L., Zhu T., Jian Y., Ding Y., Zhou M., Feng X. Zinc supplementation reduces Candida infections in pediatric intensive care unit: a randomized placebo-controlled clinical trial. J. Clin. Biochem. Nutr. 2019;64:170–173. doi: 10.3164/jcbn.18-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anjum R., Palanimuthu D., Kalinowski D.S., Lewis W., Park K.C., Kovacevic Z., Khan I.U., Richardson D.R. Synthesis, characterization, and in vitro anticancer activity of copper and zinc bis(thiosemicarbazone) complexes. Inorg. Chem. 2019;58:13709–13723. doi: 10.1021/acs.inorgchem.9b01281. [DOI] [PubMed] [Google Scholar]
- 25.Peter A.S. Oral zinc sulphate in rheumatoid arthritis. Lancet N. Am. Ed. 1976;308:539–542. doi: 10.1016/S0140-6736(76)91793-1. [DOI] [PubMed] [Google Scholar]
- 26.Prasad A.S. Zinc is an antioxidant and anti-inflammatory agent: its role in human health. Front. Nutr. 2014:1. doi: 10.3389/fnut.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakurai H., Kojima Y., Yoshikawa Y., Kawabe K., Yasui H. Antidiabetic vanadium(IV) and zinc(II) complexes. Coord. Chem. Rev. 2002;226:187–198. doi: 10.1016/S0010-8545(01)00447-7. [DOI] [Google Scholar]
- 28.Jain S., Potschka H., Chandra P.P., Tripathi M., Vohora D. Management of COVID-19 in patients with seizures: mechanisms of action of potential COVID-19 drug treatments and consideration for potential drug-drug interactions with anti-seizure medications. Epilepsy Res. 2021;174 doi: 10.1016/j.eplepsyres.2021.106675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of Coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verma T., Sinha M., Nitin B., Yadav S., Shah K., Chauhan N. A review on Coronavirus disease and potentially active drugs targeting Coronavirus. Biomed. Biotechnol. Res. J. 2021;5:110. doi: 10.4103/bbrj.bbrj_14_21. [DOI] [Google Scholar]
- 31.Prusoff W.H., Lin T.S., August E.M., Wood T.G., Marongiu M.E. Approaches to antiviral drug development. Yale J. Biol. Med. 1989;62:215–225. [PMC free article] [PubMed] [Google Scholar]
- 32.A.S. Dousari, M.T. Moghadam, N. Satarzadeh, COVID-19 (Coronavirus disease 2019): A new Coronavirus disease, Infect. Drug. Resist. 13 (2020) 2819–2828. doi: 10.2147/IDR.S259279. [DOI] [PMC free article] [PubMed]
- 33.Wang X.J., Li J.X., Wang M.R., Zhou Z.Y., Zhu B.C., Zhang X.X., Zhang R., Tang W., Wu Y.F., Zhang W.T., Li Q.Y. Antiviral properties of traditional chinese medicine against Coronavirus: research clues for Coronavirus disease-2019. World J. Tradit. Chin. Med. 2020;6:132. doi: 10.4103/wjtcm.wjtcm_15_20. [DOI] [Google Scholar]
- 34.Fakhar Z., Khan S., AlOmar S.Y., Alkhuriji A., Ahmad A. ABBV-744 as a potential inhibitor of SARS-CoV-2 main protease enzyme against COVID-19. Sci. Rep. 2021;11:234. doi: 10.1038/s41598-020-79918-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheldrick G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 36.Hirshfeld F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta. 1977;44:129–138. doi: 10.1007/BF00549096. [DOI] [Google Scholar]
- 37.Spackman M.A., Jayatilaka D. Hirshfeld surface analysis. CrystEngComm. 2009;11:19–32. doi: 10.1039/B818330A. [DOI] [Google Scholar]
- 38.Spackman P.R., Turner M.J., McKinnon J.J., Wolff S.K., Grimwood D.J., Jayatilaka D., Spackman M.A. CrystalExplorer : a program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021;54:1006–1011. doi: 10.1107/S1600576721002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKinnon J.J., Jayatilaka D., Spackman M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007:3814–3816. doi: 10.1039/b704980c. [DOI] [PubMed] [Google Scholar]
- 40.Frisch M., Trucks G., Schlegel H., Scuseria G., Robb M., Cheeseman J., Scalmani G., Barone V., Mennucci B., Petersson G., Nakatsuji H., Caricato M., Li X., Hratchian H., Izmaylov A., Bloino J., Zheng G., Sonnenberg J., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J., Peralta J., Ogliaro F., Bearpark M., Heyd J., Brothers E., Kudin K., Staroverov V., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J., Iyengar S., Tomasi J., Cossi M., Rega N., Millam J., Klene M., Knox J., Cross J., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R., Yazyev O., Austin A., Cammi R., Pomelli C., Ochterski J., Martin R., Morokuma K., Zakrzewski V., Voth G., Salvador P., Dannenberg J., Dapprich S., Daniels A., Farkas J., Foresman J., Ortiz J., Cioslowski D.Fox. Gaussian, Inc.; Wallingford CT: 2009. Gaussian 09, Revision B.01, Gaussian 09, Revision B.01. [Google Scholar]
- 41.Dennington R., Keith T., Millam J. Shawnee Mission KS; 2009. Semichem Inc., GaussView, Version 5. [Google Scholar]
- 42.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becke A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/PhysRevA.38.3098. [DOI] [PubMed] [Google Scholar]
- 44.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. doi: 10.1063/1.464913. [DOI] [Google Scholar]
- 45.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y., Weaver M.N., Merz K.M. Assessment of the “6-31+G** + LANL2DZ” mixed basis set coupled with density functional theory methods and the effective core potential: prediction of heats of formation and ionization potentials for first-row-transition-metal complexes. J. Phys. Chem. A. 2009;113:9843–9851. doi: 10.1021/jp807643p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009 doi: 10.1002/jcc.21334. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 49.Yin W., Mao C., Luan X., Shen D.-.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y., Wower J., Maltseva N., Chang C., Jedrzejczak R., Wilamowski M., Kang S., Nicolaescu V., Randall G., Michalska K., Joachimiak A. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun. Biol. 2021;4:193. doi: 10.1038/s42003-021-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 52.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Brunzelle J., Satchell K.J.F. High-resolution structures of the SARS-CoV-2 2′- O -methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 2020;13:eabe1202. doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraser B., Beldar S., Hutchinson A., Li Y., Seitova A., Edwards A.M., Benard F., Arrowsmith C.H., Halabelian L. Crystal structure of human TMPRSS2 in complex with Nafamostat: protein. data bank. 2021 [Google Scholar]
- 54.BIOVIA . Dassault Systèmes; San Diego: 2021. Dassault Systèmes, BIOVA Discovery Studio Visualizer 2021. v21.1.0.20298. [Google Scholar]
- 55.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257–W263. doi: 10.1093/nar/gky318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein J., Davis R.E., Shimoni L., Chang N.L. Patterns in hydrogen bonding: functionality and graph set analysis in crystals. Angew. Chem. Int. Ed. Engl. 1995;34:1555–1573. doi: 10.1002/anie.199515551. [DOI] [Google Scholar]
- 58.Farrugia L.J. WinGX and ORTEP for Windows : an update. J. Appl. Crystallogr. 2012;45:849–854. doi: 10.1107/S0021889812029111. [DOI] [Google Scholar]
- 59.D. Jayatilaka, D.J. Grimwood, A. Lee, A. Lemay, A.J. Russel, C. Taylor, S.K. Wolff, P. Cassam-Chenai, A. Whitton, A. (2005). TONTO-a system for computational chemistry. Available at: http://hirshfeldsurface.net/
- 60.Spackman M.A., McKinnon J.J., Jayatilaka D. Electrostatic potentials mapped on Hirshfeld surfaces provide direct insight into intermolecular interactions in crystals. CrystEngComm. 2008 doi: 10.1039/b715227b. 10.1039.b715227b. [DOI] [Google Scholar]
- 61.Venkatesan P., Thamotharan S., Ilangovan A., Liang H., Sundius T. Crystal structure, Hirshfeld surfaces and DFT computation of NLO active (2E)-2-(ethoxycarbonyl)-3-[(1-methoxy-1-oxo-3-phenylpropan-2-yl)amino] prop-2-enoic acid. Spectrochim. Acta Part A. 2016;153:625–636. doi: 10.1016/j.saa.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Kaiba A., Geesi M.H., Riadi Y., Ibnouf E.O., Aljohani T.A., Guionneau P. A new Organic–Inorganic hybrid compound (NH3(CH2)2C6H5)2[SnCl6]: crystal structure, characterization, Hirshfeld surface analysis, DFT calculation, vibrational properties and biological evaluation. J. Solid State Chem. 2021;304 doi: 10.1016/j.jssc.2021.122587. [DOI] [Google Scholar]
- 63.Sandorfy C. Anharmonicity and hydrogen bonding. J. Mol. Struct. 2002;614:365–366. doi: 10.1016/S0022-2860(02)00277-6. [DOI] [Google Scholar]
- 64.Smith Z., Wilson E.B., Duerst R.W. The infrared spectrum of gaseous malonaldehyde (3-hydroxy-2-propenal) Spectrochim. Acta Part A Mol. Spectrosc. 1983;39:1117–1129. doi: 10.1016/0584-8539(83)80135-4. [DOI] [Google Scholar]
- 65.Hansen P.E., Spanget-Larsen J. NMR and IR investigations of strong intramolecular hydrogen bonds. Molecules. 2017;22:552. doi: 10.3390/molecules22040552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakamoto K. 4th ed. Wiley; New York: 1986. Infrared and Raman spectra of Inorganic and Coordination Compounds. [Google Scholar]
- 67.Bellamy L.J. Springer; Netherlands: 1980. The Infrared Spectra of Complex Molecules: Volume Two Advances in Infrared Group Frequencies. [DOI] [Google Scholar]
- 68.Pavia D.L., Lampman G.M., Kriz G.S. 3rd ed. Brooks/Cole; South Melbourne: 2001. Introduction to spectroscopy: a Guide For Students of Organic Chemistry. [Google Scholar]
- 69.Mulliken R.S. A new electroaffinity scale; together with data on valence states and on valence ionization potentials and electron affinities. J. Chem. Phys. 1934;2:782–793. doi: 10.1063/1.1749394. [DOI] [Google Scholar]
- 70.Pearson R.G. Recent advances in the concept of hard and soft acids and bases. J. Chem. Educ. 1987;64:561. doi: 10.1021/ed064p561. [DOI] [Google Scholar]
- 71.Parr R.G., Pearson R.G. Absolute hardness: companion parameter to absolute electronegativity. J. Am. Chem. Soc. 1983;105:7512–7516. doi: 10.1021/ja00364a005. [DOI] [Google Scholar]
- 72.Parr R.G., Chattaraj P.K. Principle of maximum hardness. J. Am. Chem. Soc. 1991;113:1854–1855. doi: 10.1021/ja00005a072. [DOI] [Google Scholar]
- 73.Parr R.G., Szentpály L.v., Liu S. Electrophilicity index. J. Am. Chem. Soc. 1999;121:1922–1924. doi: 10.1021/ja983494x. [DOI] [Google Scholar]
- 74.Padmanabhan J., Parthasarathi R., Subramanian V., Chattaraj P.K. Electrophilicity-based charge transfer descriptor. J. Phys. Chem. A. 2007;111:1358–1361. doi: 10.1021/jp0649549. [DOI] [PubMed] [Google Scholar]
- 75.Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hussein R.K., Elkhair H.M. Molecular docking identification for the efficacy of some zinc complexes with chloroquine and hydroxychloroquine against main protease of COVID-19. J. Mol. Struct. 2021;1231 doi: 10.1016/j.molstruc.2021.129979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Crystallographic data for the structural analysis have been deposited with the Cambridge Crystallographic Data centre, CCDC 2,099,779 for complex 1 and 2,099,780 for complex 2. Copies of this information may be obtained free of charge from The Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax: 44–1223–336,033; e-mail: deposit@ ccdc.cam.ac.uk or www: http://www.ccdc.cam.ac.uk).