Abstract
The immense spread of coronavirus disease 2019 (COVID-19) has left healthcare systems incapable to diagnose and test patients at the required rate. Given the effects of COVID-19 on pulmonary tissues, chest radiographic imaging has become a necessity for screening and monitoring the disease. Numerous studies have proposed Deep Learning approaches for the automatic diagnosis of COVID-19. Although these methods achieved outstanding performance in detection, they have used limited chest X-ray (CXR) repositories for evaluation, usually with a few hundred COVID-19 CXR images only. Thus, such data scarcity prevents reliable evaluation of Deep Learning models with the potential of overfitting. In addition, most studies showed no or limited capability in infection localization and severity grading of COVID-19 pneumonia. In this study, we address this urgent need by proposing a systematic and unified approach for lung segmentation and COVID-19 localization with infection quantification from CXR images. To accomplish this, we have constructed the largest benchmark dataset with 33,920 CXR images, including 11,956 COVID-19 samples, where the annotation of ground-truth lung segmentation masks is performed on CXRs by an elegant human-machine collaborative approach. An extensive set of experiments was performed using the state-of-the-art segmentation networks, U-Net, U-Net++, and Feature Pyramid Networks (FPN). The developed network, after an iterative process, reached a superior performance for lung region segmentation with Intersection over Union (IoU) of 96.11% and Dice Similarity Coefficient (DSC) of 97.99%. Furthermore, COVID-19 infections of various shapes and types were reliably localized with 83.05% IoU and 88.21% DSC. Finally, the proposed approach has achieved an outstanding COVID-19 detection performance with both sensitivity and specificity values above 99%.
Keywords: COVID-19, Chest X-ray, Lung Segmentation, Infection Segmentation, Convolutional Neural Networks, Deep Learning
1. Introduction
The novel coronavirus 2019 (COVID-19) is an acute respiratory syndrome that has already caused over 4.9 million causalities and infected more than 243 million people, as of October 27, 2021 [1]. The business, economic, and social dynamics of the whole world have been affected due to this pandemic. Governments have imposed flight restrictions, social distancing, and taken measures to increase awareness of hygiene. Several studies have been done to forecast the future conditions of the virus and to recede its impact [2,3]. However, COVID-19 is still spreading at a very rapid rate. The common symptoms of coronavirus include fever, cough, shortness of breath, and pneumonia [4]. Severe cases of coronavirus disease result in acute respiratory distress syndrome (ARDS) or complete respiratory failure, which requires support from mechanical ventilation and an intensive-care unit (ICU). People with a compromised immune system or elderly people are more likely to develop serious illnesses, including heart and kidney failures and septic shock [4].
Reliable detection of COVID-19 is crucial. However, the diagnosis procedures thereof, particularly through clinical diagnosis, are not straightforward as the common symptoms of COVID-19 are generally indistinguishable from other viral infections [5,6]. Currently, the primary diagnostic tool to detect COVID-19 is reverse-transcription polymerase chain reaction (RT-PCR) arrays, where the presence of Severe Acute Respiratory Syndrome Related Coronavirus 2 (SARS-CoV-2) Ribonucleic acid (RNA) is tested on collected respiratory specimens from the suspected cases [7,8]. However, RT-PCR arrays have a high false alarm rate caused by sample contamination, and damage through the virus mutations in the COVID-19 genome [9,10]. Therefore, several studies have suggested using chest computed tomography (CT) imaging as a primary diagnostic tool since it has shown higher sensitivity compared to RT-PCR [11,12]. Besides, several studies [[11], [12], [13]] have suggested performing CT scans as a secondary test if the suspected patients show shortness of breath or other respiratory symptoms but the RT-PCR result comes negative. Despite the superior performance, CT scans do pose some difficulties and certain limitations. For example, their sensitivity is limited to early COVID-19 cases with no or minimum pneumonia symptoms, the corresponding image acquisition process is slow, and the whole process is costly. On the other hand, X-ray imaging is a cheaper, faster, and readily available method, where the body gets exposed to a much smaller amount of harmful radiation compared to CT [14]. Chest X-ray (CXR) imaging is widely used as an assistive diagnostic tool in COVID-19 screening, and it is reported to have high potential prognostic capabilities [15].
The majority of early COVID-19 cases have exhibited similar features on radiographic images, including bilateral, multi-focal, ground-glass opacities with posterior or peripheral distribution, mainly in the lower lung lobes, while it develops to pulmonary consolidation in the late stage [16,17]. Even though chest radiographs can help in the early screening of the suspected case, the images of several other types of viral pneumonia are similar. They show a high similarity with other inflammatory lung diseases as well. Therefore, it is difficult for medical doctors to distinguish COVID-19 infections from other viral pneumonia using only a chest X-ray. Hence, this symptom similarity can lead to a wrong diagnosis under the current situation, which may cause mistreatment leading to human causalities.
The tremendous development in Deep Learning techniques in recent years has led to many state-of-the-art performances in several Computer Vision tasks, such as image classification, object detection, and image segmentation. This breakthrough led to increased utilization of AI-based solutions in various life sciences fields, including the domain of biomedical health problems and complications. Specifically, Convolutional Neural Network (CNN) has been proven extremely beneficial in several biomedical imaging applications, such as skin lesion classification [18], brain tumor detection [19], breast cancer detection [20], and lung pathology screening [21,22]. Deep Learning techniques on chest X-ray images are gaining popularity with the availability of deep CNNs, showing promising results in various applications. Rajpurkar et al. [23] proposed the CheXNet network, one of the top-performing architectures for CXR, by training Densenet121 on the ChestX-ray14 dataset [24], one of the largest public CXR datasets with over 100 thousand X-ray images for 14 different pathologies. Rahman et al. [25] investigated several pre-trained CNNs to classify the CXR images as either healthy or having manifestations of pulmonary tuberculosis (TB). The proposed model was trained over a dataset of 3500 infected and 3500 Normal CXR images. The best performing model, DenseNet201, performed very well achieving 98.57% sensitivity and 98.56% specificity.
1.1. Related works
Recently, many studies have proposed Deep Learning approaches to automate COVID-19 detection from chest X-ray images [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35]] with high performance. Ozturk et al. [26] presented a modified version of DarkNet for binary classification (COVID-19 vs Normal) and multi-class classification (COVID-19 vs Non-COVID pneumonia vs Normal). They reported 90.65% sensitivity for the binary scheme and 85.35% sensitivity for the multi-class scheme on a dataset that includes 114 COVID-19 CXRs. Apostolopoulos et al. [27] evaluated MobileNetV2 on a dataset with 224 COVID-19 cases achieving high discrimination performance with 98.7% sensitivity. Wang et al. [28] introduced COVID-Net, a CNN architecture tailored for COVID-19 recognition. The network exhibited 91% sensitivity over a dataset with 358 COVID-19 CXRs. Waheed et al. [29] proposed a synthetic data augmentation technique to alleviate the scarcity of public COVID-19 CXR data using Auxiliary Classifier Generative Adversarial Network (ACGAN). Chowdhury et al. [30] investigated several deep CNNs (SqueezeNet, ResNet18, ResNet101, MobileNetV2, DenseNet201, and CheXNet) for both binary and multi-class schemes on a dataset that contains 423 COVID-19 CXR images. DenseNet201 showed the best classification performance with 99.7% and 97.9% sensitivity values for binary and multi-class schemes, respectively. Yamac et al. [31] utilized CheXNet as a feature extractor while a proposed classifier, Convolution Support Estimation Network (CSEN), discriminates the target CXR as COVID-19, Bacterial pneumonia, Viral Pneumonia, or Normal. The network produced satisfactory results with 98% sensitivity over the benchmark QaTa-COV19 dataset that includes 462 COVID-19 CXR images. Fan et al. [32] investigated the role of attention mechanism on the COVID-19 recognition scheme by introducing Multi-Kernel-Size Spatial-Channel Attention Network. The proposed network achieved 98.1% sensitivity and 98.3% specificity on a dataset that comprises 500 COVID-19 and 500 Non-COVID CXR images.
Oh et al. [33] proposed a patch-based deep CNN architecture for COVID-19 recognition. First, lung areas were extracted using a fully connected (FC)-DenseNet103 followed by patch-based classification using ResNet50, where a majority voting was utilized to make the final decision. The proposed pipeline achieved 95.5% Intersection over Union (IoU) for the lung segmentation task while it exhibited 96.9% sensitivity for the COVID-19 recognition task. In recent work [34], we investigated the ability of deep networks to distinguish between different Coronavirus family members (COVID-19, MERS-CoV, and SARS-CoV) using CXR images which is an extremely challenging task for medical doctors without the aid of clinical data. A cascaded system was proposed where first lung regions are segmented using U-Net model and then classified using a deep CNN classifier (SqueezeNet, ResNet18, InceptionV3, or DenseNet201). Our proposed pipeline achieved 93.1% IoU and 96.4% Dice Similarity Coefficient (DSC) for the segmentation task while it achieved 96.9% sensitivity for the recognition task. Motamed et al. [35] utilized a semi-supervised learning approach that only requires partial labels for the training data without the need for a single label from the positive class (COVID-19). The lung regions were first segmented using the U-Net model and then feed to the proposed randomized generative adversarial network (RANDGAN) for classification. Poor classification performance was achieved with 57% sensitivity and 80% specificity. Therefore, the introduced pipeline can have significant value in the very early stages of the emergence of a certain disease/pandemic where annotated data are scarce. However, supervised approaches are still a preferable choice as soon as enough annotated data are created to train the deep CNN models. Despite the high classification performance achieved in most of the recent studies, they also have highlighted certain issues and drawbacks thereof as follows. First of all, all of these studies suffer from the issue of a small dataset, while the largest one has only a few hundred CXR samples. This makes their performance evaluation questionable and it is difficult to generalize their results in practice. Secondly, they only aimed for COVID-19 detection and/or classification among other types without further assessment and localization. These issues limit their usability, particularly in a real clinical setting.
On the other hand, few studies [36,37] considered lung segmentation as the first stage in their detection system. This ensures reliable decision-making in the classification phase and guards the network against irrelevant features from non-lung areas, such as heart, bones, background, or text. However, the previous segmentation approaches were trained on a mixture of medium and high-quality CXR images comprising a total of 704 X-ray images for Normal and TB cases, mainly collected from Montgomery [38] and Shenzhen [39] CXR lung mask datasets. Therefore, the segmentation performance degrades in unseen scenarios such as severe COVID-19 cases or low-quality images with poor signal-to-noise (SNR) levels. The lung areas can be partially or incompletely segmented for severe COVID-19 infections, such as, bilateral consolidation or fluid accumulation at lower-lung lobes, which degrades the classification performance. Therefore, creating a large benchmark CXR dataset with ground-truth lung segmentation masks is extremely important, and will help the research community to provide a more reliable detection system for COVID-19 and other lung pathologies.
Along with COVID-19 detection, infection localization is another crucial task that helps in evaluating the status of the patient and deciding on the treatment plan [40]. Therefore, several studies utilized class activation maps which are generated from Deep Learning models trained for COVID-19 classification tasks to localize infected lung regions. Those localized regions are potential signatures for COVID-19. However, more precise and reliable localization can be provided by ground-truth infection masks from expert radiologists. Therefore, Degerli et al. [41] proposed a novel approach for COVID-19 infection map generation by compiling a COVID-19 dataset consisting of 2951 CXR images with annotated ground-truth infection segmentation masks. Several encode-decoder (E-D) CNNs were trained and evaluated on the generated dataset, where the best performing network achieved an F1-score of 85.81% for infection localization. However, their proposed approach is limited only to COVID-19 infection localization. Therefore, there is certainly room for improvement particularly in the context of both localizing and quantifying infection regions by computing the overall percentage of infected area in the lungs. This can help medical doctors to quantify the severity and track the progression of COVID-19 pneumonia.
With the above backdrop, in this work, we attempt to overcome the aforementioned limitations and challenges. This paper makes the following key contributions:
-
-
We present the largest COVID-19 benchmark dataset, namely, COVID-QU-Ex [65], having 11,956 COVID-19, 11,263 Non-COVID (but diseased), and 10,701 Normal (healthy) CXR images. It is expected that COVID-QU-Ex will be regarded as the most reliable benchmark hitherto available for reliable evaluation for COVID-19 detection, localization, and quantification models, particularly the ones involving state-of-the-art deep network architectures.
-
-
We have prepared the ground-truth lung segmentation masks for the entire COVID-QU-Ex dataset applying an elegant human-machine collaborative approach that significantly reduces human labour to annotate the images. This is the first-ever attempt to provide ground-truth lung segmentation masks at such a large scale. Both the dataset and the ground-truth masks will be released along with this study as a public benchmark dataset. We believe that COVID-QU-Ex will be extremely beneficial for researchers, doctors, and engineers around the world to come up with innovative solutions for the early detection of COVID-19 with the help of the large benchmark COVID-19 CXR images with their ground-truth lung masks.
-
-
Furthermore, we have experimented with three state-of-the-art image segmentation architectures, namely, U-Net [42], U-Net++ [43], and Feature Pyramid Networks (FPN) [44] with different backbone encoder structures for both lung and infection segmentation tasks thereby identifying which model is better suited for which task. As the backbone encoder, we started with shallow structures and went on to deeper ones thereby covering ResNet18, ResNet50 [45], DenseNet121, DenseNet161 [46], and InceptionV4 [47].
-
-
Finally, we have proposed a novel and robust system for lung segmentation and COVID-19 localization with infection quantification from CXR images. This is a crucial accomplishment for a reliable diagnosis and assessment of the disease with the highest accuracy ever reached.
2. The benchmark COVID-QU-Ex dataset
In this section, we will first show the data compilation process; then, we will present the proposed approach for ground-truth lung mask generation.
2.1. Data compilation
Due to the emerging nature of the pandemic, initially, only limited efforts were being made by the highly infected countries on sharing clinical and radiography data publicly. Therefore, a group of researchers from Qatar University (QU) and Tampere University (TU), created two datasets, COVID-QU [48] and QaTa-Cov19 datasets [41]. The COVID-QU dataset consists of 3616 COVID-19, 8851 Non-COVID cases, and 6012 Normal cases, whereas the QaTa-Cov19 dataset comprises 2951 COVID-19 CXR along with their ground-truth infection masks. Gradually, more X-rays have become publicly available. Hence, we extended those datasets creating COVID-QU-Ex [65], which include over 33,000 CXR images, from three different classes:
-
1)
11,956 COVID-19 cases
-
2)
11,263 Non-COVID infections (viral or bacterial pneumonia) cases
-
3)
10,701 Normal (healthy) cases
In this study, only posterior-to-anterior (PA) or anterior-to-posterior (AP) chest X-rays were considered as this view of radiography is preferred and widely used by the radiologist, whereas a lateral image is usually taken to complement the frontal view. Besides, a very small portion of the compiled dataset were lateral X-rays. Thus, they were excluded from this study [49]. This dataset was created by utilizing numerous publicly available datasets and repositories, all of which are scattered, and with varying formats. The quality of the dataset was ensured through a rigorous quality control process where duplicates, extremely low-quality, and over-exposed images were identified and removed. The resulting dataset thus comprises images of high interclass dissimilarity with few varying resolutions, quality, and SNR levels (See Fig. 1 for some representative samples).
Fig. 1.
Sample chest X-ray images from the COVID-QU-Ex dataset for Normal, Non-COVID, and COVID-19 classes. All images are rescaled with the same factor to illustrate the diversity of the dataset.
Details of different data sources are given below:
COVID-19 CXR dataset: This dataset contains 11,956 positive COVID-19 CXR images among which 10,814 images are collected from the BIMCV-COVID19+ dataset [50], 183 CXR images from a German medical school [51], 559 CXR images from SIRM, Github, Kaggle, and Tweeter [[52], [53], [54], [55]], and 400 CXR images from another COVID-19 CXR repository [56].
RSNA CXR dataset (Non-COVID infections and Normal CXR): RSNA pneumonia detection challenge dataset [57] consists of 26,684 CXR images, where 8,851 images are Normal, 11,821 are abnormal, and 6,012 are lung opacity images. All images are in DICOM format. We have included 8,851 Normal and 6,012 lung opacity CXR images from this dataset in our COVID-QU-Ex dataset, where the latter is considered as Non-COVID images.
Chest-Xray-Pneumonia dataset: This is a Kaggle dataset [58] that comprises 1,300 viral pneumonia, 1,700 bacterial pneumonia, and 1,000 Normal CXR images. The viral and bacterial pneumonia images of this dataset are added as Non-COVID (diseased) images in our COVID-QU-Ex dataset.
PadChest dataset: PadChest [59] dataset comprises more than 160,000 CXR images from 67,000 patients that were collected and reported by radiologists at Hospital San Juan (Spain) from 2009 to 2017. We included 4,000 Normal, and 4,000 pneumonia/infiltrate (Non-COVID) cases from this dataset in our COVID-QU-Ex dataset.
Montgomery and Shenzhen CXR lung masks dataset: This dataset consists of 704 CXR images with their corresponding lung segmentation masks. In the first stage of the proposed human-machine collaborative approach, the lung masks from this dataset were used as the initial ground truth masks to train the lung segmentation models. The dataset was acquired by Shenzhen Hospital in China [39], and the tuberculosis control program of the Department of Health and Human Services of Montgomery County, MD, USA [38]. Montgomery dataset consists of 80 Normal and 58 tuberculosis CXR with lung segmentation masks. On the other hand, the Shenzhen dataset comprises 326 Normal and 336 tuberculosis CXR, where 566 out of 662 CXR are provided with their corresponding masks.
QaTa-Cov19 CXR infection mask dataset [60]: This dataset was created by a research group from Qatar University and Tampere University. It consists of nearly 120,000 CXR images, including 2913 COVID-19 images with their corresponding ground-truth infection masks, but no ground-truth lung masks are provided. Thus, these ground-truth infection masks were used to train and evaluate the infection segmentation models.
2.2. Collaborative human-machine segmentation approach for lung ground-truth mask generation
Recent advancements in Deep Learning techniques have brought about remarkable success. However, supervised Deep Learning approaches require large and annotated data for training. Lack of adequate and quality data (including ground truth masks) often degrades the performance of the models, resulting in poor generalization capabilities. On the other hand, the process of producing ground truth segmentation masks is an exhaustive task, where human experts need to delineate pixel-wise masks. This process is bound to suffer from the varying subjectivity and hand-crafting levels of the human annotators. To overcome these issues, here, a collaborative human-machine segmentation approach is proposed to accurately produce the ground-truth lung segmentation masks for CXR images. The majority of the manual annotation process was assigned to biomedical engineering researchers from Qatar University (QU) team to reduce the load on medical collaborators from Hamad Medical Corporation (HMC). All researchers attended several training sessions conducted by MDs to grasp a general understating of Chest X-ray imaging and get exposed to a variety of cases with mild, moderate, or severe infections. This human-machine collaborative approach is performed in four main stages as follows.
Stage Ⅰ (Initial Training):
In the first stage, three variants of the U-Net [42] segmentation model, are trained on 704 CXR images and ground-truth lung masks publicly available from Montgomery and Shenzhen dataset mentioned previously. The ground-truth CXR lung masks are referred to as the CXR-lung-mask-repository in Fig. 2 , and it is enlarged throughout the mask creation process. Next, the best performing network in terms of Dice Similarity Coefficient (DSC) is selected as the main network for Stage Ⅱ, which is referred to as the CXR-Segmentation network in Fig. 2.
Stage Ⅱ (Collaborative Evaluation):
Fig. 2.
Collaborative human-machine approach to create ground-truth lung segmentation masks for COVID-QU-Ex CXR dataset. Stage Ⅰ: Three segmentation networks are trained on a repository of 704 CXR lung segmentation masks, and the best network in terms of DSC is selected for the subsequent stages. Stage Ⅱ: An iterative training is utilized to create lung masks for a subset of 3000 CXR samples from the COVID-QU-Ex dataset. Firstly, A subset of 500 samples is inferred by the CXR segmentation model and the outputs are evaluated manually as accept, reject, modify, or exclude. Next, the modified masks are added to the lung repository and the network is re-trained on the extended dataset. These steps are repeated until generating ground-truth masks for the 3000 CXR samples is completed. Stage Ⅲ: six deep segmentation networks are trained using the 3000 ground-truth masks generated in the previous stage. The trained networks are used to predict segmentation masks for the rest of the COVID-QU-Ex dataset (30,920 images). Stage Ⅳ: a final verification is performed by MDs on randomly selected 6788 CXR samples (20% of the full dataset) that well presents the diversity of the COVID-QU-Ex dataset.
In the second stage, an iterative training is utilized to create lung masks for a subset of 3000 CXR samples (∼10% of the full dataset) that well represent the diversity of the COVID-QU-Ex dataset. Firstly, a subset of 500 samples is selected and inferred using the CXR-Segmentation network. The predicted lung masks are then evaluated by researchers as “accept”, “reject”, “unsure”, or “exclude”. Accepted masks that accurately cover the lung areas are added to the CXR-lung-mask-repository. Rejected masks either miss certain parts of the lung or include irrelevant parts. These rejected masks are then manually examined by the researchers, and the corrected masks are finally added to the CXR-lung-mask-repository. The “unsure” masks are the severe cases with highly infected areas. These are usually consolidations or fluid accumulation at the lower lung lobes with a whitish color, which makes them indistinguishable from neighboring organs. The unsure masks are first assessed by MDs; then, researchers adjust the masks based on their recommendations. Finally, the “excluded” masks are the ones where the quality is extremely bad for proper lung segmentation. Eventually, the CXR-Segmentation network is re-trained on the extended mask dataset (extended through the above-mentioned protocol). Then the second subset of 500 samples is selected, and the steps of Stage Ⅱ are repeated. This process is repeated until generating ground-truth masks for 3000 CXR samples is completed.
Stage Ⅲ (Collaborative Selection):
In the third stage, six deep segmentation networks from the models of U-Net [42], U-Net++ [43], and FPN [44] are trained using the 3000 ground-truth masks generated in Stage Ⅱ by the proposed approach. The trained networks are used to predict segmentation masks for the rest of the COVID-QU-Ex dataset, which is 30,920 unannotated samples (∼90% of the full dataset). Among the six predictions, researchers selected the best one as the ground truth or discarded the sample for now if none of the masks segments the lung properly. The latter is a minority case that included less than 5% of the unannotated data. The network that registered the highest number of selection (as above) is considered as the best-performing network and used for a new training with the CXR-lung-masks-repository.
The discarded cases are then inferred by the best-performing segmentation network and evaluated manually following the steps in Stage Ⅱ. As a result, the ground-truth masks for 33,920 CXR images are gathered to construct the benchmark COVID-QU-Ex lung masks dataset.
The proposed systematic collaboration ensured a good compromise between human intervention and machine training throughout the entire process. In Stage Ⅱ, a smaller subset (∼10%) of the dataset was annotated where manual modification was performed by RAs. On the other hand, a larger subset (∼90%) of the dataset was annotated in Stage Ⅲ, where the performance of the segmentation models has been enhanced. Thus, the load was reduced on the RAs, and they had to select among different network predictions rather than manually modifying the predicted masks. This approach saved valuable human labor time. Also, it enhanced the quality and reliability of the generated masks and reduced subjectivity.
Stage Ⅳ (Final Verification):
In the final stage, a final verification is performed by two radiologists on randomly selected 6788 CXR samples (20% of the full dataset). To ensure that the diversity of the COVID-QU-Ex dataset is well-captured during this verification, the samples are selected from COVID, Non-COVID, and Normal classes, with different resolution, quality, and SNR levels. Both radiologists accepted >97% of the annotated subset, while the rejected masks were modified by the radiologists then added to the dataset. Considering the noisy nature of the radiographic imaging and the subjectivity in the annotation process it is acceptable to have such a small rejection rate (∼3%). Thus, the constructed COVID-QU-Ex dataset can be used as a reliable ground-truth lung segmentation masks dataset. In this study, the verified subset (20%) was considered as a test set for all the experimental evaluations, while the remaining data (80%) were used for training and validation.
3. Methods
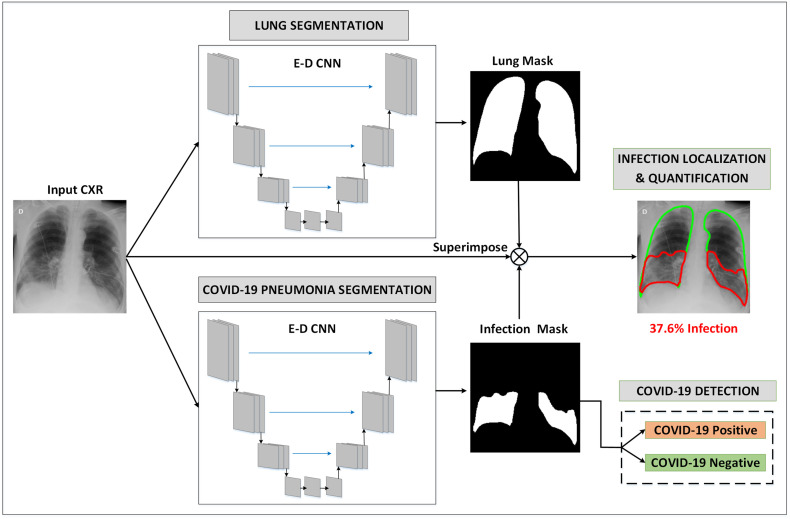
In this section, we describe the proposed unified approach for lung segmentation and COVID-19 localization with infection quantification from the CXR images. The schematic representation of the pipeline of the proposed COVID-19 recognition system is shown in Fig. 3 . A binary lung mask is first generated from the input CXR image using the 1st encoder-decoder (E-D) CNN. In parallel, the input CXR is fed to the 2nd E-D CNN to generate COVID-19 infection masks. Then, the generated lung and infection masks are superimposed with the CXR image to localize and quantify COVID-19 infected lung regions. Finally, the generated infection mask is used to detect COVID-19 positive cases from COVID-19 negative cases. In what follows, we will describe these steps in detail.
Fig. 3.
Schematic representation of the pipeline of the proposed system. The input CXR image is fed to two ED-CNNs in parallel, to generate two binary masks: lung, and COVID-19 infection masks. Next, the generated masks are superimposed with the CXR image to localize and quantify COVID-19 infected lung regions. Finally, the generated infection mask is used to detect COVID-19 positive cases from COVID-19 negative cases.
The pseudo-code for training and evaluating the proposed COVID-19 recognition system is shown in Algorithm 1 and Algorithm 2, respectively.
3.1. Network models for lung and COVID-19 infection segmentation
Lung and COVID-19 pneumonia (infection) segmentation were performed on CXR images using three state-of-the-art deep E-D CNNs: U-Net [42], U-Net++ [43], and FPN [44], with different backbone (encoder) models using the variants of ResNet [45], DenseNet [46], and InceptionV4 [47] networks. Five variants of the backbone models were considered starting from shallow to deep structures: ResNet18, ResNet50, DenseNet121, DenseNet161, and InceptionV4.
The deployed encoder-decoder blocks provide a firm segmentation model that captures the context in the contracting path and empowers precise localization by the expanding path. The U-Net architecture has a classical decoder part that is symmetric to the encoder part, where max-pooling operations are replaced with up-sampling operations. Besides, high-resolution features from the encoder path are merged with the up-sampled output from the corresponding decoder path through skip connection. On the other hand, the U-Net++ is a recent implementation that has further developed the decoder block. The encoder and decoder blocks are connected through a series of nested dense convolutional blocks. This ensures a firm bridge between the encoder and decoder parts of the network, where information can be transferred to the final layers more intensively compared to the conventional U-Net. Both U-Net and U-Net++ architectures utilize 1 × 1 convolution to map the output from the last decoding block to two-channel feature maps, where a pixel-wise SoftMax activation function is applied to map each pixel into a binary class of background or lung for Lung segmentation task, and background or lesion for infection segmentation task. In contrast, the FPN employs the encoder-decoder as a pyramidal hierarchy by generating prediction masks at each spatial level of the decoder path. All predicted feature maps are up-sampled to the same size, concatenated, convolved with a 3 × 3 convolutional filter, and then SoftMax activation is applied to generate the final prediction mask.
To ensure efficient training and faster convergence, transfer learning was leveraged on the encoder side of the segmentation networks by initializing the convolutional layers with ImageNet [61] weights.
3.1.1. Segmentation loss function
The cross-entropy (CE) loss is used as the cost function for the segmentation networks:
| (1) |
Here, denotes the kth pixel in the predicted segmentation mask, denotes its SoftMax probability, is a binary random variable getting 1 if , otherwise 0, and denotes the class category, i.e., for the lung segmentation task, and for the infection segmentation.
3.2. Post-processing
The predicted segmentation masks, , by the segmentation models are defined as , where and represent the size of the image. In the post-processing step, binary segmentation masks are first generated by thresholding with a fixed value of 0.5. The predicted pixels are classified as lung if for the lung segmentation task, while classified as COVID-19 infection if for the infection segmentation task. The binary lung masks are further processed by hole filling and removal of small regions, <5% of the total positive predicted pixels. As a result, we increase the true-positives while minimizing the false-positives, i.e., non-lung regions that are falsely predicted as a lung. In contrast, infection masks are masked with post-processed lung masks to ensure that the infection region falls within the lung area and remove the false positives outside the lung region.
3.3. COVID-19 detection and quantification
The detection of COVID-19 is performed based on the prediction maps generated by the infection segmentation network. Accordingly, a CXR image is classified as COVID-19 positive if at least one pixel of lung areas is predicted as COVID-19 infection, i.e., . Otherwise, the image is considered as COVID-19 negative, i.e., it could be an image of a healthy person or a patient with Non-COVID pneumonia. Furthermore, COVID-19 infection is quantified by computing the overall percentage of infected lungs by dividing the sum of predicted infection pixels over the sum of predicted lung pixels. In addition, the infection percentage of each lung is computed in a similar manner, enabling doctors to assess the progression of COVID-19 for each lung individually.
3.4. Experimental setup
The lung segmentation task was conducted over the COVID-QU-Ex dataset. In contrast, the infection segmentation and COVID-19 detection tasks were conducted over a subset of the COVID-QU-Ex dataset comprising 2913 CXR samples with corresponding infection masks from the QaTa-Cov19 dataset [60]. The CXR images were resized to have a fixed dimension of 256 × 256 pixels to be used as the input for the deep networks. In all our experiments, we assumed an 80-20 split for train and test purposes respectively. Besides, 20% of training data was used as a validation set for model selection and to avoid overfitting. Table 1 summarizes the number of images per class used for training, validation, and testing.
Table 1.
Number of mages per class per train, validation, and test sets for each of the 5 folds used for lung segmentation, infection segmentation, and COVID-19 detection tasks.
| Dataset Name | Task | Class | # of Samples | Training Samples | Validation Samples | Test Samples | |
|---|---|---|---|---|---|---|---|
| COVID-QU-Ex dataset | Lung Segmentation |
COVID-19 | 11,956 | 7658 | 1903 | 2395 | |
| Non-COVID | 11,263 | 7208 | 1802 | 2253 | |||
| Normal | 10,701 | 6849 | 1712 | 2140 | |||
| Total | 33,920 | 21,715 | 5417 | 6788 | |||
| COVID-QU-Ex and QaTa-Cov19 [60] datasets | Infection Segmentation and COVID-19 Detection |
COVID-19 positive | 2913 | 1864 | 466 | 583 | |
| COVID-19 negative | Non-COVID | 1457 | 932 | 233 | 292 | ||
| Normal | 1456 | 932 | 233 | 291 | |||
| Total | 5826 | 3728 | 932 | 1166 | |||
Adam optimizer was used, with the initial learning rate, , momentum updates, and an adaptive learning rate that decreases the learning parameter by a factor of if validation loss did not improve for consecutive epochs, early stopping criterion of epochs, where training stops if validation loss did not improve for consecutive epochs, and mini-batch size of images with backpropagation epochs.
3.5. Evaluation metrics
We evaluate our approach as follows. The segmentation tasks are evaluated at the pixel level, where the foreground (lung or infected region) is considered as the positive class and the background as the negative class. For the COVID-19 detection task, the performance metric is computed per CXR sample, where X-rays with COVID-19 infection are considered as the positive class and X-rays of healthy people or patients with Non-COVID pneumonia are considered as the negative class.
The performance of deep CNNs is assessed using different evaluation metrics with a 95% confidence interval (CI). Notably, the CI (r) for each evaluation metric is computed as follows:
| (2) |
Here, N is the number of test samples, and is the level of significance that is 1.96 for 95% CI.
3.5.1. Segmentation evaluation metrics
The performance of the lung and lesion segmentation networks is evaluated using three evaluation metrics, namely, Accuracy, Intersection over Union (IoU), and Dice Similarity Coefficient (DSC) as per the following equations.
| (3) |
Here, is the ratio of the correctly classified pixels among the image pixels. TP, TN, FP, FN represent the true positive, true negative, false positive, and false negative, respectively.
| (4) |
| (5) |
Here, both are statistical measures of spatial overlap between the binary ground-truth and the predicted segmentation masks, where the main difference is that the latter considers double weight for pixels (true lung/lesion predictions) compared to the former.
3.5.2. COVID-19 detection evaluation metrics
The performance of the COVID-19 detection scheme is assessed using five evaluation metrics, namely, Accuracy, Precision, Sensitivity, F1-score, and Specificity as per the following equations.
| (6) |
Here, is the rate of correctly classified positive class CXR samples among all the samples classified as positive samples.
| (7) |
Here, is the rate of correctly predicted positive samples from among the positive class samples.
| (8) |
Here, (i.e., F1-score) is the harmonic average of precision and sensitivity.
| (9) |
Here, specificity is the sensitivity of the negative class samples.
PyTorch [62] library with Python 3.7 was used to train and evaluate the deep CNN networks, running on a PC with Intel® Core™ i9-9900K CPU at 3.6 GHz, with 32 GB RAM, and with an 8-GB NVIDIA GeForce GTX 1080 GPU card.
4. Results
In this section, both quantitative and qualitative results are reported with an extensive set of comparative evaluations for lung segmentation, infection segmentation, and COVID-19 detection tasks.
4.1. Lung segmentation results
The performance of the lung segmentation models over the test (unseen) set is tabulated in Table 2 . Recall that, each model was evaluated with five different encoder structures. For all models, it was observed that DenseNet encoders exhibit the top segmentation performance as they can share pieces of collective knowledge by densely connecting convolutional layers to their subsequent layers, thereby preserving the information coming from the earlier layer through the output layer. The FPN model with DenseNet121 encoder holds the leading position with 96.11% IoU, and 97.99% DSC.
Table 2.
Performance metrics (%) for lung region and COVID-19 infected region segmentation computed over test (unseen) set with three network models and five encoder architectures. x ± y means that the achieved metric value is x with standard deviation y.
| Task | Model | Encoder | Accuracy | IoU | DSC |
|---|---|---|---|---|---|
| Lung Segmentation |
U-Net | ResNet18 | 99.07 ± 0.23 | 95.91 ± 0.47 | 97.88 ± 0.34 |
| ResNet50 | 99.08 ± 0.23 | 95.93 ± 0.47 | 97.89 ± 0.34 | ||
| DenseNet121 | 99.1 ± 0.22 | 96.06 ± 0.46 | 97.96 ± 0.34 | ||
| DenseNet161 | 99.1 ± 0.22 | 96.02 ± 0.47 | 97.94 ± 0.34 | ||
| InceptionV4 | 99.07 ± 0.23 | 95.9 ± 0.47 | 97.88 ± 0.34 | ||
| U-Net ++ | ResNet18 | 99.07 ± 0.23 | 95.9 ± 0.47 | 97.88 ± 0.34 | |
| ResNet50 | 99.1 ± 0.22 | 96.04 ± 0.46 | 97.95 ± 0.34 | ||
| DenseNet121 | 99.11 ± 0.22 | 96.1 ± 0.46 | 97.98 ± 0.33 | ||
| DenseNet161 | 99.09 ± 0.23 | 95.98 ± 0.47 | 97.92 ± 0.34 | ||
| InceptionV4 | 99.08 ± 0.23 | 95.96 ± 0.47 | 97.91 ± 0.34 | ||
| FPN | ResNet18 | 99.06 ± 0.23 | 95.86 ± 0.47 | 97.86 ± 0.34 | |
| ResNet50 | 99.07 ± 0.23 | 95.91 ± 0.47 | 97.88 ± 0.34 | ||
| DenseNet121 | 99.12 ± 0.22 | 96.11 ± 0.46 | 97.99 ± 0.33 | ||
| DenseNet161 | 99.09 ± 0.23 | 96.01 ± 0.47 | 97.94 ± 0.34 | ||
| InceptionV4 | 99.07 ± 0.23 | 95.92 ± 0.47 | 97.89 ± 0.34 | ||
| Infection Segmentation |
U-Net | ResNet18 | 98.02 ± 0.8 | 82.92 ± 2.16 | 88.1 ± 1.86 |
| ResNet50 | 97.84 ± 0.83 | 81.73 ± 2.22 | 87.02 ± 1.93 | ||
| DenseNet121 | 97.98 ± 0.81 | 82.53 ± 2.18 | 87.74 ± 1.88 | ||
| DenseNet161 | 97.86 ± 0.83 | 81.95 ± 2.21 | 87.19 ± 1.92 | ||
| InceptionV4 | 97.98 ± 0.81 | 82.03 ± 2.2 | 87.11 ± 1.92 | ||
| U-Net ++ | ResNet18 | 97.9 ± 0.82 | 82.9 ± 2.16 | 88.06 ± 1.86 | |
| ResNet50 | 97.93 ± 0.82 | 82.59 ± 2.18 | 87.78 ± 1.88 | ||
| DenseNet121 | 97.97 ± 0.81 | 83.05 ± 2.15 | 88.21 ± 1.85 | ||
| DenseNet161 | 97.95 ± 0.81 | 81.55 ± 2.23 | 86.66 ± 1.95 | ||
| InceptionV4 | 97.9 ± 0.82 | 81.13 ± 2.25 | 86.22 ± 1.98 | ||
| FPN | ResNet18 | 97.84 ± 0.83 | 81.9 ± 2.21 | 87.25 ± 1.91 | |
| ResNet50 | 97.84 ± 0.83 | 80.83 ± 2.26 | 86.25 ± 1.98 | ||
| DenseNet121 | 97.99 ± 0.81 | 82.55 ± 2.18 | 87.71 ± 1.88 | ||
| DenseNet161 | 97.95 ± 0.81 | 81.89 ± 2.21 | 87.08 ± 1.93 | ||
| InceptionV4 | 97.99 ± 0.81 | 83.08 ± 2.15 | 88.13 ± 1.86 |
The outputs of the three top-performing networks compared with the ground-truth are shown in Fig. 4 . An interesting observation is that the three networks can reliably segment lung regions not only for COVID-19 cases, but for Non-COVID-19 pneumonia as well with different severity levels, i.e., mild, moderate, or severe. This elegant performance may be attributed to the large and diverse COVID-QU-Ex dataset (33,920 samples) comprising CXR samples with different quality, resolution, and SNR levels from COVID-19, Non-COVID-19, and Normal classes. Thus, our benchmark dataset is expected to help researchers to overcome the challenges and limitations faced, mainly in the lung segmentation phase for COVID-19 or other lung pathology problems. As most of the previous approaches were trained over Montgomery [38] and Shenzhen [39] CXR lung mask datasets that comprise medium and high-quality X-ray images from Normal and TB classes, the previous segmentation approaches were falling in unseen scenarios, such as, severe infection or low-quality images [37].
Fig. 4.
Sample qualitative evaluation of generated lung masks by the three top-performing networks. Column 1 shows the CXR image, Column 2 shows ground truths, and the lung masks of the top three networks are shown in Columns 3–5, respectively.
4.2. Infection segmentation results
The infection segmentation model has been first evaluated over two different configurations: cascaded and parallel segmentation. For the cascaded scheme, the lung region was first segmented using the lung segmentation model; then the segmented CXR was fed to the infection segmentation model whereas the plain CXR was fed to both models independently for the parallel scheme.
FPN model with DenseNet161 encoder was trained and evaluated on both schemes. The parallel scheme showed slightly better results with 87.08% DSC compared to 86.84% DSC for the cascaded scheme. Therefore, the parallel scheme was used as the main configuration for the remaining experiments. The performance of the infection segmentation models is presented in Table 2. U-Net++ model with DenseNet121 encoder showed the best performance with IoU and DSC values of 83.05% and 88.21%, respectively. Besides, the InceptionV4 encoder showed the best performance among FPN models with 83.08% IoU and 88.13% DSC. In contrast, the shallowest encoder, ResNet18 did better among U-Net models with IoU and DSC values of 82.92% and 88.1%, respectively.
Fig. 5(a) shows the robustness of three top-performing networks to reliably segment COVID-19 infections of various shapes (small, medium, or large) with different severity levels (mild, moderate, severe, or critical). In general, the FPN models produced smoother masks with better localization of infected regions compared to U-Net and U-Net++ models. This can be inspired by the hierarchy architecture of FPN where predictions are made on each spatial level of the decoder path, then merged to produce the final prediction mask, whereas only the final decoder block is used to generate the prediction mask in U-Net and U-Net ++ models. Fig. 5(b) shows infection localization and severity grading of COVID-19 pneumonia for a 42-year female patient on the 1st day (of hospital admission), 2nd day, and 3rd day using the proposed COVID-19 recognition system, where two parallel FPN with DenseNet121 encoders models were used for the lung and the infection segmentation tasks.
Fig. 5.
(a) Sample qualitative evaluation of generated infection masks by the three top-performing networks. Column 1 shows the CXR image, Column 2 shows ground truths, and the lung masks of the top three networks are shown in Columns 3–5, respectively. (b) Infection localization and severity grading of COVID-19 pneumonia for a 42-year female patient on the 1st, 2nd, and 3rd days of admission using the proposed system.
4.3. COVID-19 detection results
The performance of infection segmentation networks for COVID-19 detection from the CXR images is presented in Table 3 . The sensitivity was considered as the primary metric for the detection task, as missing any COVID-19 positive case is critical. All the networks achieved high sensitivity values (>97%), where U-Net with DenseNet121 backbone and FPN with ResNet18 backbone achieved the best performance with a sensitivity of 99.66%. Similarly, all models showed high specificity values (>97%), where U-Net++ with ResNet18 backbone exhibited the best performance with 100% specificity, indicating the absence of any false alarm.
Table 3.
COVID-19 detection performance results (%) computed over test (unseen) set with three network models, and five encoder architectures. x ± y means that the achieved metric value is x with standard deviation y.
| Model | Encoder | Accuracy | Precision | Sensitivity | F1-score | Specificity |
|---|---|---|---|---|---|---|
| U-Net | ResNet18 | 98.89 ± 0.6 | 99.14 ± 0.53 | 98.63 ± 0.67 | 98.88 ± 0.6 | 99.14 ± 0.53 |
| ResNet50 | 98.89 ± 0.6 | 98.47 ± 0.7 | 99.31 ± 0.48 | 98.89 ± 0.6 | 98.46 ± 0.71 | |
| DenseNet121 | 98.8 ± 0.62 | 97.98 ± 0.81 | 99.66 ± 0.33 | 98.81 ± 0.62 | 97.94 ± 0.82 | |
| DenseNet161 | 98.71 ± 0.65 | 97.97 ± 0.81 | 99.49 ± 0.41 | 98.72 ± 0.65 | 97.94 ± 0.82 | |
| InceptionV4 | 98.03 ± 0.8 | 98.28 ± 0.75 | 97.77 ± 0.85 | 98.02 ± 0.8 | 98.28 ± 0.75 | |
| U-Net ++ | ResNet18 | 99.23 ± 0.5 | 100 ± 0 | 98.46 ± 0.71 | 99.22 ± 0.5 | 100 ± 0 |
| ResNet50 | 99.14 ± 0.53 | 99.83 ± 0.24 | 98.46 ± 0.71 | 99.14 ± 0.53 | 99.83 ± 0.24 | |
| DenseNet121 | 99.23 ± 0.5 | 99.14 ± 0.53 | 99.31 ± 0.48 | 99.22 ± 0.5 | 99.14 ± 0.53 | |
| DenseNet161 | 98.2 ± 0.76 | 97.95 ± 0.81 | 98.46 ± 0.71 | 98.2 ± 0.76 | 97.94 ± 0.82 | |
| InceptionV4 | 98.2 ± 0.76 | 98.45 ± 0.71 | 97.94 ± 0.82 | 98.19 ± 0.77 | 98.46 ± 0.71 | |
| FPN | ResNet18 | 98.54 ± 0.69 | 97.48 ± 0.9 | 99.66 ± 0.33 | 98.56 ± 0.68 | 97.43 ± 0.91 |
| ResNet50 | 98.46 ± 0.71 | 98.46 ± 0.71 | 98.46 ± 0.71 | 98.46 ± 0.71 | 98.46 ± 0.71 | |
| DenseNet121 | 98.97 ± 0.58 | 99.65 ± 0.34 | 98.28 ± 0.75 | 98.96 ± 0.58 | 99.66 ± 0.33 | |
| DenseNet161 | 98.11 ± 0.78 | 97.3 ± 0.93 | 98.97 ± 0.58 | 98.13 ± 0.78 | 97.26 ± 0.94 | |
| InceptionV4 | 99.23 ± 0.5 | 99.31 ± 0.48 | 99.14 ± 0.53 | 99.22 ± 0.5 | 99.31 ± 0.48 |
4.4. Computational complexity analysis
Table 4 compares the segmentation models in terms of inference time and the number of trainable parameters. The results present the inference time per CXR sample. It can be noticed that, due to their shallow and close structures, FPN and U-Net models are faster than U-Net ++ models. FPN with ResNet18 encoder is the fastest network taking up to 5.74 ms per image. In contrast, the U-Net++ model is the slowest with the highest number of trainable parameters. The most computationally demanding model is UNet++ with InceptionV4 encoder having a staggering 59.35 M trainable parameters. However, UNet++ with DenseNet161 encoder is the slowest, with an inference time of 48.62 ms as it is the deepest model with 161 layers. Note that, for systems with limited computational capabilities, where both lung and infection segmentation cannot be used in parallel, the two models can be used consecutively. This will double (×2) the inference time. However, we can still say that the full system can be used for real-time clinical applications as the overall inference time is still less than 100 ms in the worst case, which means that multiple images can be processed within a second.
Table 4.
The number of trainable parameters of the models with their inference time (ms) per CXR sample.
| Model | Encoder | Trainable parameters | Inference Time (ms) |
|---|---|---|---|
| U-Net | ResNet18 | 14.32 M | 5.78 |
| ResNet50 | 32.5 M | 10.44 | |
| DenseNet121 | 13.60 M | 22.86 | |
| DenseNet161 | 38.73 M | 29.74 | |
| InceptionV4 | 48.79 M | 26.53 | |
| U-Net ++ | ResNet18 | 15.96 M | 8.30 |
| ResNet50 | 48.97 M | 19.90 | |
| DenseNet121 | 30.06 M | 25.13 | |
| DenseNet161 | 79.04 M | 48.62 | |
| InceptionV4 | 59.35 M | 32.53 | |
| FPN | ResNet18 | 13.04 M | 5.74 |
| ResNet50 | 26.11 M | 10.34 | |
| DenseNet121 | 9.29 M | 22.68 | |
| DenseNet161 | 29.49 M | 29.62 | |
| InceptionV4 | 43.57 M | 26.08 |
4.5. Comparison with related work
Table 5 compares the proposed work with recent literature about automatic COVID-19 pneumonia diagnosis from CXR images for three main tasks: classification, localization, and quantification. First, despite the superior classification performance achieved in most of the studies, small datasets have been used, with few hundred samples only, except [41] where they used 2951 COVID-19 CXRs. On the other hand, we evaluated our pipeline on four times larger cohort datasets with 11,956 COVID-19 CXRs, where >97% sensitivity and specificity values were achieved. This elegant performance is exhibited by the high diversity in the COVID-QU-Ex dataset which ensured good generalization capabilities by the deep CNN models. In addition, we provided a robust lung segmentation model which guards the detection and localization schemes against irrelevant features from non-lung areas. Therefore, empowered by the largest ever ground-truth lung segmentation mask dataset (33,920 samples), an outstanding performance was achieved with 97.9% DSC. Finally, only a single study [41] provided precise and reliable localization of COVID-19 infected lung regions based on ground-truth annotation from medical experts, where the proposed model achieved 83.2% DSC for localizing infected regions. In contrast, our model showed higher localization performance with 88.1% DSC. Moreover, our deployment of lung and infection segmentation models enabled both localization and quantification of infected regions. Therefore, our system could facilitate early intervention and provide a unified solution that helps doctors to access the severity and track the progression of the disease.
Table 5.
Comparing the proposed work with recent literature about automatic COVID-19 diagnosis using CXR images, in terms of utilized Dataset, whether Lung and/or Infection Segmentation models are used, deployed Networks, and achieved Results.
| Ref. | Dataset (# of subjects) | Lung Seg. |
Infection Seg. | Network | Results |
|---|---|---|---|---|---|
| [27] | COVID19 (224) Non-COVID (714) Healthy (504) |
Classification model MobileNetV2 |
Class. Sens. 98.7% Spe. 96.5% |
||
| [28] | COVID19 (358) Non-COVID (8,066) Healthy (5,538) |
Classification model COVID-Net |
Class. Sens. 91.0% Spe. 99.5% |
||
| [29] | COVID-19 (403) Healthy (721) |
Augmentation model CovidGAN Classification model VGG16 |
Class. Sens. 90.0% Spe. 97.0% |
||
| [30] | COVID-19 (423) Non-COVID (1,485) Healthy (1,579) |
Classification model DenseNet201 |
Class. Sens. 99.7% Spe. 99.6% |
||
| [31] | COVID-19 (462) Non-COVID (2,485) Healthy (1,579) |
Feature extraction model CheXNet Classification model Convolutional Support Estimation Network |
Class. Sens. 98.5% Spe. 94.7% |
||
| [32] | COVID-19 (500) Non-COVID (500) |
Classification model Multi-Kernel-Size Spatial-Channel Attention Network |
Class. Sens. 98.1% Spe. 98.3% |
||
| [33] | COVID-19 (180) Non-COVID (74) TB (57) Healthy (191) |
✓ |
Segmentation model FC-DenseNet103 Patch-based classification model ResNet50 |
Lung Seg. IoU 95.5% Class. Sens. 85.9% Spe. 96.4% |
|
| [34] | COVID-19 (423) MERS-CoV (144) SARS-CoV (134) |
✓ |
Segmentation model U-Net Classification model InceptionV3 |
Lung Seg. IoU 93.1% DSC 96.4% Class. Sens. 96.9% Spe. 91.7% |
|
| [35] | COVID-19 (573) Non-COVID (5,559) Healthy (8,066) |
✓ |
Classification model RAND-GAN |
Lung Seg. DSC 83.0% Class. Sens. 57.0% Spe. 80.0% |
|
| [41] | COVID-19 (2951) Non-COVID (116,365) |
✓ |
Segmentation models U-Net, U-Net++, and DLA Backbone Encoders: CheXNet, DenseNet121, InceptionV3, and ResNet50 |
Infection Seg. DSC 83.2% Class. Sens. 94.9% Spe. 99.9% |
|
| This work | COVID-19 (11,956) Non-COVID (11,263) Healthy (10,701) |
✓ | ✓ |
Segmentation models U-Net, U-Net++, and FPN Backbone Encoders ResNet18, ResNet50, DenseNet121, DenseNet161, InceptionV4 |
Lung Seg. IoU 96.1% DSC 97.9% Class. Sens. 99.6% Spe. 97.4% Infection Seg. IoU 83.1% DSC 88.1% |
5. Conclusion
Early identification and isolation of highly infectious COVID-19 cases play a vital role in treatment as well as preventing the spread of the virus. X-ray imaging is a low-cost, easily accessible, and fast method that can be an excellent alternative for conventional diagnostic methods such as RT-PCR and CT scans. Therefore, numerous studies proposed AI-based solutions for automatic and real-time detection of COVID-19. In general, these methods showed outstanding performance for early detection and diagnosis. However, they have used limited CXR repositories for evaluation with a small number, a few hundreds, of COVID-19 samples. Thus, the generalization of the achieved results on a large cohort dataset is not guaranteed. In addition, they showed limited performance in infection localization and severity grading of COVID-19 pneumonia. In this study, we proposed a robust and comprehensive system to segment the lung, detect, localize, and quantify COVID-19 infections from the CXR images. To accomplish this, we compiled the largest CXR dataset hitherto known, namely, COVID-QU-Ex [65], which consists of 11,956 COVID-19, 11,263 Non-COVID pneumonia, and 10,701 Normal CXR images. Moreover, we constructed ground-truth lung segmentation masks for the benchmark dataset using an elegant collaborative human-machine approach, which saved valuable human labour time and minimized subjectivity in the annotation process. The publicly shared dataset will help researchers to investigate deep CNN models on a comparatively larger dataset, which can provide more reliable solutions for COVID-19 and other lung pathology problems. Extensive experiments on COVID-QU-Ex showed superior lung segmentation performance with 96.11% IoU and 97.99% DSC. Moreover, the proposed system proved reliable in localizing COVID-19 infection of various severity, achieving IoU and DSC values of 83.05% and 88.21%, respectively. Furthermore, unprecedented COVID-19 detection performance was achieved with sensitivity and specificity values > 99%. To the best of our knowledge, this is the first study that utilizes both lung and infection segmentation to detect, localize and quantify COVID-19 infection from X-ray images. Therefore, it can assist the medical doctors to better diagnose the severity of COVID-19 pneumonia and follow up the progression of the disease easily.
In the future, we plan to explore robust quantization and model compression techniques to further reduce the model complexity and accelerate the inference process, using the new generation of heterogeneous network models such as Self-Organized Operational Neural Networks [63,64].
Data availability
The COVID-QU-Ex chest X-ray datasets and corresponding lung mask created during the current study are available in the following Kaggle repository: www.kaggle.com/dataset/cf77495622971312010dd5934ee91f07ccbcfdea8e2f7778977ea8485c1914df.
Author contributions
Experiments were designed by AMT, MEHC, and SK. Experiments were performed by AMT, AK, TR, YQ, and UK. Data were compiled and created by AMT, AK, TR, YQ, UK, NI, SM, ME, KH, and TH. Results were analyzed by AMT, MEHC, SK, MSR, SAM, KH, and TH. The project is supervised by MEHC and SK. All the authors were involved in the interpretation of data and paper writing and revision of the article.
Funding
Qatar University COVID19 Emergency Response Grant (QUERG-CENG-2020-1) from Qatar University, and UREP28-144-3-046 grant from Qatar National Research Fund provided the support for the work and the claims made herein are solely the responsibility of the authors. Open access publication is supported by Qatar National Library (QNL).
Conflicts of interest
The authors report no declarations of interest.
References
- 1.World Health Organization WHO coronavirus disease (COVID-19) Dashboard. 2020. https://covid19.who.int/?gclid=Cj0KCQjwtZH7BRDzARIsAGjbK2ZXWRpJROEl97HGmSOx0_ydkVbc02Ka1FlcysGjEI7hnaIeR6xWhr4aAu57EALw_wcB Available.
- 2.Shastri S., Singh K., Kumar S., Kour P., Mansotra V. Time series forecasting of Covid-19 using deep learning models: India-USA comparative case study. Chaos, Solit. Fractals. 2020;140:110227. doi: 10.1016/j.chaos.2020.110227. 2020/11/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shastri S., Singh K., Deswal M., Kumar S., Mansotra V. Spatial Information Research, 2021/06/12. 2021. CoBiD-net: a tailored deep learning ensemble model for time series forecasting of covid-19. [Google Scholar]
- 4.Pormohammad A., et al. Comparison of confirmed COVID-19 with SARS and MERS cases - clinical characteristics, laboratory findings, radiographic signs and outcomes: a systematic review and meta-analysis. Rev. Med. Virol. 2020;30(4) doi: 10.1002/rmv.2112. p. e2112, 2020/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singhal T. 2020. A Review of Coronavirus Disease-2019 (COVID-19) pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohrabi C., et al. 2020. World Health Organization Declares Global Emergency: A Review of the 2019 Novel Coronavirus (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kakodkar P., Kaka N., Baig M.J.C. vol. 12. 2020. (A Comprehensive Literature Review on the Clinical Presentation, and Management of the Pandemic Coronavirus Disease 2019 (COVID-19)). no. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., et al. 2020. Stability Issues of RT-PCR Testing of SARS-CoV-2 for Hospitalized Patients Clinically Diagnosed with COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 Detection: Issues Affecting the Results. Taylor & Francis; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia J., Tong J., Liu M., Shen Y., Guo D.J. vol. 92. 2020. pp. 589–594. (Evaluation of Coronavirus in Tears and Conjunctival Secretions of Patients with SARS-CoV-2 Infection). no. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ai T., et al. 2020. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: a Report of 1014 Cases; p. 200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A.J. 2020. Coronavirus Disease 2019 (COVID-19): a Systematic Review of Imaging Findings in 919 Patients; pp. 1–7. [DOI] [PubMed] [Google Scholar]
- 13.Fang Y., et al. 2020. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR; p. 200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner D.J., Hall E.J. vol. 357. 2007. pp. 2277–2284. (Computed Tomography—An Increasing Source of Radiation Exposure). no. 22. [DOI] [PubMed] [Google Scholar]
- 15.Shi F., et al. 2020. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for Covid-19. [DOI] [PubMed] [Google Scholar]
- 16.Huang C., et al. vol. 395. 2020. pp. 497–506. (Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China). no. 10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosseiny M., Kooraki S., Gholamrezanezhad A., Reddy S., Myers L.J. vol. 214. 2020. pp. 1078–1082. (Radiology Perspective of Coronavirus Disease 2019 (COVID-19): Lessons from Severe Acute Respiratory Syndrome and Middle East Respiratory Syndrome). no. 5. [DOI] [PubMed] [Google Scholar]
- 18.Esteva A., et al. vol. 542. 2017. pp. 115–118. (Dermatologist-level Classification of Skin Cancer with Deep Neural Networks). no. 7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong H., Yang G., Liu F., Mo Y., Guo Y. Annual Conference on Medical Image Understanding and Analysis. Springer; 2017. Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks; pp. 506–517. [Google Scholar]
- 20.Shen L., Margolies L.R., Rothstein J.H., Fluder E., McBride R., Sieh W. J. S. r. vol. 9. 2019. pp. 1–12. (Deep Learning to Improve Breast Cancer Detection on Screening Mammography). no. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardila D., et al. vol. 25. 2019. pp. 954–961. (End-to-end Lung Cancer Screening with Three-Dimensional Deep Learning on Low-Dose Chest Computed Tomography). no. 6. [DOI] [PubMed] [Google Scholar]
- 22.Tahir A., et al. 2020. Coronavirus: Comparing COVID-19, SARS and MERS in the Eyes of AI. arXiv preprint arXiv:1706.05587. [Google Scholar]
- 23.Rajpurkar P., et al. vol. 15. 2018. (Deep Learning for Chest Radiograph Diagnosis: A Retrospective Comparison of the CheXNeXt Algorithm to Practicing Radiologists). no. 11, p. e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanford ML Group CheXpert: A Large Dataset of Chest X-Rays and Competition for Automated Chest X-Ray Interpretation. https://stanfordmlgroup.github.io/competitions/chexpert/ Available.
- 25.Rahman T., et al. vol. 8. 2020. pp. 191586–191601. (Reliable Tuberculosis Detection Using Chest X-Ray with Deep Learning, Segmentation and Visualization). [Google Scholar]
- 26.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. 2020/06/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. 2020/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):19549. doi: 10.1038/s41598-020-76550-z. 2020/11/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waheed A., Goyal M., Gupta D., Khanna A., Al-Turjman F., Pinheiro P.R. CovidGAN: data augmentation using auxiliary classifier GAN for improved covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowdhury M.E.H., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 31.Yamaç M., Ahishali M., Degerli A., Kiranyaz S., Chowdhury M.E.H., Gabbouj M. Convolutional sparse support estimator-based COVID-19 recognition from X-ray images. IEEE Trans. Neural Netw. Learn. Syst. 2021;32(5):1810–1820. doi: 10.1109/TNNLS.2021.3070467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan Y., Liu J., Yao R., Yuan X. COVID-19 detection from X-ray images using multi-kernel-size Spatial-Channel attention network. Pattern Recogn. 2021;119:108055. doi: 10.1016/j.patcog.2021.108055. 2021/11/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imag. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 34.Tahir A., et al. Deep learning for reliable classification of COVID-19, MERS, and SARS from chest X-ray images. Cogn. Comput. 2021 doi: 10.1007/s12559-021-09955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motamed S., Rogalla P., Khalvati F. RANDGAN: randomized generative adversarial network for detection of COVID-19 in chest X-ray. Sci. Rep. 2021;11(1):8602. doi: 10.1038/s41598-021-87994-2. 2021/04/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. "Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh Y., Park S., Ye J.C. 2020. Deep Learning Covid-19 Features on Cxr Using Limited Training Data Sets. [DOI] [PubMed] [Google Scholar]
- 38.Jaeger S., et al. vol. 33. 2013. pp. 233–245. (Automatic Tuberculosis Screening Using Chest Radiographs). no. 2. [DOI] [PubMed] [Google Scholar]
- 39.Candemir S., et al. vol. 33. 2013. pp. 577–590. (Lung Segmentation in Chest Radiographs Using Anatomical Atlases with Nonrigid Registration). no. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi F., et al. 2020. Large-scale Screening of Covid-19 from Community Acquired Pneumonia Using Infection Size-Aware Classification. [DOI] [PubMed] [Google Scholar]
- 41.Degerli A., et al. COVID-19 infection map generation and detection from chest X-ray images. Health Inf. Sci. Syst. 2021;9(1):15. doi: 10.1007/s13755-021-00146-8. 2021/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 43.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. Unet++: a nested u-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin T.-Y., Dollár P., Girshick R., He K., Hariharan B., Belongie S. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Feature pyramid networks for object detection; pp. 2117–2125. [Google Scholar]
- 45.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 46.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 47.Szegedy C., Ioffe S., Vanhoucke V., Alemi A. vol. 31. 2017. Inception-v4, inception-resnet and the impact of residual connections on learning. (Proceedings of the AAAI Conference on Artificial Intelligence). no. 1. [Google Scholar]
- 48.Rahman T., et al. 2020. Exploring the Effect of Image Enhancement Techniques on COVID-19 Detection Using Chest X-Rays Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corne J. Elsevier Health Sciences; 2015. Chest X-Ray Made Easy E-Book. [Google Scholar]
- 50.Medical Imaging Databank of the Valencia Region BIMCV-COVID19+: a large annotated dataset of RX and CT images of COVID19 patients. https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/#1590858128006-9e640421-6711 Available.
- 51.GitHub covid-19-image-repository. 2020. https://github.com/ml-workgroup/covid-19-image-repository/tree/master/png Available.
- 52.Eurorad https://www.eurorad.org/ Available.
- 53.GitHub covid-chestxray-dataset. 2020. https://github.com/ieee8023/covid-chestxray-dataset Available.
- 54.SIRM COVID-19 DATABASE. 2020. https://www.sirm.org/category/senza-categoria/covid-19/ Available.
- 55.Kaggle COVID-19 radiography Database. 2020. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database Available.
- 56.GitHub COVID-CXNet. 2020. https://github.com/armiro/COVID-CXNet Available.
- 57.Kaggle RSNA pneumonia detection challenge. 2018. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data Available.
- 58.Kaggle Chest X-ray images (pneumonia) 2018. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia Available.
- 59.Medical Imaging Databank of the Valencia Region PadChest: A large chest x-ray image dataset with multi-label annotated reports. https://bimcv.cipf.es/bimcv-projects/padchest/ Available. [DOI] [PubMed]
- 60.Degerli A., et al. 2020. COVID-19 Infection Map Generation and Detection from Chest X-Ray Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Image-net.org ImageNet. http://www.image-net.org/ Available.
- 62.Pytorch.org PyTorch. https://pytorch.org/ Available.
- 63.Malik J., Kiranyaz S., Gabbouj M.J.N.N. vol. 135. 2021. pp. 201–211. (Self-organized Operational Neural Networks for Severe Image Restoration Problems). [DOI] [PubMed] [Google Scholar]
- 64.Kiranyaz S., Malik J., Abdallah H.B., Ince T., Iosifidis A., Gabbouj M.J. 2020. Self-Organized Operational Neural Networks with Generative Neurons. [DOI] [PubMed] [Google Scholar]
- 65.Tahir A.M., Chowdhury M.E.H., Qiblawey Y., Khandakar A., Rahman T., Kiranyaz S. Kaggle; 2021. COVID-QU-Ex. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The COVID-QU-Ex chest X-ray datasets and corresponding lung mask created during the current study are available in the following Kaggle repository: www.kaggle.com/dataset/cf77495622971312010dd5934ee91f07ccbcfdea8e2f7778977ea8485c1914df.