Abstract
It is known that nitrate inhibits ruminal methanogenesis, mainly through competition with hydrogenotrophic methanogens for available hydrogen (H2) and also through toxic effects on the methanogens. However, there is limited knowledge about its effects on the others members of ruminal microbiota and their metabolites. In this study, we investigated the effects of dietary nitrate inclusion on enteric methane (CH4) emission, temporal changes in ruminal microbiota, and fermentation in Holstein calves. Eighteen animals were maintained in individual pens for 45 d. Animals were randomly allocated to either a control (CTR) or nitrate (NIT, containing 15 g of calcium nitrate/kg dry matter) diets. Methane emissions were estimated using the sulfur hexafluoride (SF6) tracer method. Ruminal microbiota changes and ruminal fermentation were evaluated at 0, 4, and 8 h post-feeding. In this study, feed dry matter intake (DMI) did not differ between dietary treatments (P > 0.05). Diets containing NIT reduced CH4 emissions by 27% (g/d) and yield by 21% (g/kg DMI) compared to the CTR (P < 0.05). The pH values and total volatile fatty acids (VFA) concentration did not differ between dietary treatments (P > 0.05) but differed with time, and post-feeding (P < 0.05). Increases in the concentrations of ruminal ammonia nitrogen (NH3–N) and acetate were observed, whereas propionate decreased at 4 h post-feeding with the NIT diet (P < 0.05). Feeding the NIT diet reduced the populations of total bacteria, total methanogens, Ruminococcus albus and Ruminococcus flavefaciens, and the abundance of Succiniclasticum, Coprococcus, Treponema, Shuttlewortia, Succinivibrio, Sharpea, Pseudobutyrivibrio, and Selenomona (P < 0.05); whereas, the population of total fungi, protozoa, Fibrobacter succinogenes, Atopobium and Erysipelotrichaceae L7A_E11 increased (P < 0.05). In conclusion, feeding nitrate reduces enteric CH4 emissions and the methanogens population, whereas it decreases the propionate concentration and the abundance of bacteria involved in the succinate and acrylate pathways. Despite the altered fermentation profile and ruminal microbiota, DMI was not influenced by dietary nitrate. These findings suggest that nitrate has a predominantly direct effect on the reduction of methanogenesis and propionate synthesis.
Keywords: Methane emission, Nitrate, Microbiota, Ruminal fermentation
1. Introduction
The livestock sector contributes significantly to global food security, accounting for 17% and 33% of the world consumption of kilojoules and proteins (Rosegrant et al., 2009). An increase in global demands of meat (73%) and milk (58%) is projected for the year 2050, compared to 2010; due to projected increases in world population, urbanization, and income in developing countries (FAOSTAT, 2020; Gerber et al., 2013a). To meet this demand, the main challenge for the livestock sector is to increase production efficiency while reducing the impact on the environment.
Ruminal fermentation plays a crucial role in the digestion and transformation of structural and non-structural carbohydrates into useful products and metabolites beneficial to ruminant animals (Armstrong and Blaxter, 1957), a process that also generates methane (CH4) which is then released into the atmosphere. This CH4 source accounts for approximately 6% of global greenhouse gas (GHG) emissions (Gerber et al., 2013b). Methane is produced during ruminal methanogenesis by the methanogens that are closely associated with ciliated protozoa and hydrogen (H2) producing bacteria (Morgavi et al., 2010).
Studies have shown that CH4 emission is strongly related to feed intake and diet composition (Hellwing et al., 2018; Hristov et al., 2018), hence dietary interventions are among the most preferred GHG mitigation avenues leading to concurrent reduction of emission intensities by increasing animal performance (Beauchemin et al., 2020). One of these strategies is the supplementation of diets with nitrate (NO3─). The mitigation of CH4 emissions in vivo through the use of NO3─ has proven to be effective, with an observed reduction potential ranging from 6.8% to 12.5% for each 1% of NO3─ added daily on a dry matter (DM) basis (Lund et al., 2014; van Zijderveld et al., 2010; Veneman et al., 2015). It is known that NO3─ in the rumen acts as an electron acceptor, thereby competing for dissolved H2 between nitrate-reducing microorganisms and methanogenic archaea (Latham et al., 2016). This mode of action is thermodynamically explained, given that reduction of NO3─ to NH3 is more favorable (delta Gibbs [ΔG] = −599.6 kJ/mol) than methanogenesis (ΔG = −136 kJ/mol) (Thauer et al., 1977).
To date, there are no conclusive research results on the effects of NO3─ addition on ruminal fermentation and the ruminal microbiota, mainly due to the scarcity of research results. While some studies report a decrease in propionate production and an increase in acetate production (Olijhoek et al., 2016; Troy et al., 2015), another study reported an opposite effect (Wang et al., 2018), and others did not reveal significant changes (Popova et al., 2017; Zhang et al., 2019). It has been also reported that although methanogenesis was inhibited with the inclusion of NO3─, the concentration of H2 (gaseous and dissolved) increased (Guyader et al., 2015; Olijhoek et al., 2016). All these reports suggest that competition for available H2 cannot be regarded as the only mode of action of NO3─ to reduce CH4 emissions. A possible explanation for the inhibition of methanogenesis could be the toxic effects of compounds derived from nitrate-reduction causing a reduction in the relative abundance and activity of methanogens (Granja-Salcedo et al., 2019; Iwamoto et al., 2002; Popova et al., 2019).
The production of volatile fatty acids (VFA), H2, and CH4 peak shortly following feeding (van Lingen et al., 2017). Also, the composition of ruminal microbiota varies widely during the day due to the different fermentation stages of feeds (Shaani et al., 2018). However, several studies that evaluated the effects of NO3─ on ruminal microbiota focused their analysis on a single follow-up time and did not report significant changes in the bacterial, protozoan, methanogenic and fungal communities (Granja-Salcedo et al., 2019; Popova et al., 2017). Therefore, it is important to consider the variation in the dynamics of the ruminal microbiota and its metabolites in response to the inclusion of NO3─ in the diet. Thus, the aim of this work was to simultaneously address the temporal changes in the population of ruminal microbiota (methanogens, bacteria, protozoa, and fungi), the concentration of dissolved metabolites (ruminal ammonia nitrogen [NH3–N] and VFA), and the emissions of CH4 in response to the inclusion of NO3─ in the diet of calves, with the purpose of understanding the effect in greater detail. In order to safeguard against unwanted effects on animal health and performance, an intermediate level of NO3─ inclusion (15 g/kg DM) was chosen based on the finding of previous studies (van Zijderveld et al., 2011a; Olijhoek et al., 2016), and a gradual acclimatization to increasing levels was implemented. We hypothesized that nitrate feeding would induce changes in ruminal microbiota composition, resulting in changes in ruminal fermentation profiles and lower CH4 emission.
2. Materials and methods
2.1. Experimental design and animal procedures
The protocols, procedures, and the care of the animals were approved by the Institutional Committee for the Care and Use of Animals (CICUAE File No. 2017/124, approval date September 12, 2017) of the National Institute of Agricultural Technology (INTA), Argentina.
The experiment was conducted in the Experimental Dairy Centre of the Balcarce Experimental Station at INTA, Argentina. Eighteen Holstein calves (7 heifers and 11 steers) of 8.1 ± 0.5 months of age and 214 ± 13.5 kg of live weight (LW) were randomly allocated to either a control diet (CTR; including 5 steers and 4 heifers) or a nitrate diet (NIT; including 6 steers and 3 heifers). The CTR group received a total mix ration (on DM basis), containing ground corn (69.3%), soybean expeller (8.4%), urea (0.8%), vitamin-mineral premix (1.1%), and grass hay (20.4%); whereas the NIT group received a total mix ration (on DM basis), containing ground corn (68.4%), soybean expeller (8.4%), urea (0.2%), vitamin-mineral premix (1.1%), grass hay (20.4%), and 1.5% of nitrate (as calcium nitrate, Calcinit YARALIVA, Olso, Norway), (Appendix Table 1).
The trial included 30 d of acclimatization to diet, followed by a 15-d period of measurements (on d 31 to 45). To reduce the risks of toxicity from nitrites (ruminal microorganism convert NO3─ to nitrite [NO2─]), the amount of NO3─ in the diet was gradually increased as previously described (Ortiz-Chura et al., 2021). Throughout the experiment, the animals were kept in individual pens of 36 m2 (9 m length × 4 m width), which were constructed outdoors using electric fences. The experimental animals were fed ad libitum twice a day (08:00 and 16:00; equal rations), and drinking water was freely available to them.
2.2. Diet chemical composition and feed intake
The experimental animals were fed at 100% of their ad libitum consumption observed during the last week of the acclimatization period. Feed dry matter intakes (DMI, in kilograms per day) by individual animals were measured daily as the difference between the amount offered and the amount refused. Refused feed was collected from the feeding bins immediately before feeding. Dry matter intake was also measured during the days of CH4 emission measurements (on d 36 to 40), when the feeding level was set to 95% of the ad libitum feed intake measurement period to ensure the complete intake of the diet. The experimental animals were weighed on d 1 and 45 (initial and final weight, respectively), following a fasting period of 24 h before initial and final weighing, when feed and water were removed.
The dry matter concentration of feed offered was determined by oven drying at 105 °C for 24 h. Ash concentrations were determined by incineration at 550 °C for 4 h (AOAC, 1990; method 942.05). The total nitrogen (TN) concentration was determined using a combustion type auto-analyzer (Leco FP-2000, Leco Corp., St. Joseph, MI) according to AOAC Official Methods (1990; method 990.03) and the crude protein (CP) was calculated as TN × 6.25. The ether extract (EE) concentration was determined after extraction with petroleum ether using a Soxhlet System Apparatus (Electromantle ME1000, UK) according to AOAC (1990; method 920.39). The concentration of the neutral detergent fiber (NDF) was determined using the Ankom 220 fiber analyzer (ANKOM Technology, Macedonia NY–USA) according to Van Soest et al. (1991). The starch concentration was analyzed using the enzymatic method (MacRae and Armstrong, 1968).
2.3. Enteric methane emission
Methane emissions from individual animals were estimated using the principles of the sulfur hexafluoride (SF6) tracer technique (Zimmerman, 1993; Johnson et al., 1994), with a slight modification as described by Pinares-Patiño et al. (2012). This modification involved extending the duration of breathed air sample collection from every 24 h (traditional technique) to 5 consecutive days (adapted technique). Briefly, at the beginning of the acclimatization period, the calves were orally dosed with permeation tubes containing the tracer. The tubes were chosen from a larger number based on their high linearity of mass loss (R2 > 0.99) and narrow range of pre-calibrated rates of SF6 permeation (4.57 ± 0.64 mg/d; range 3.85 to 5.99 mg/d). The sample collection system consisted of polyvinyl chloride yoke-shaped devices (2.5 L volume) and the sample flow regulator was a metal capillary (10 cm length), with a small section (5 mm) pressed until the desired flow was achieved (initial flow rate 0.25 mL/min). Approximately, 500 mbar of internal pressure in the collection device at the end of sample collection was aimed (Pinares-Patiño and Clark, 2008). The sampling period was 5 consecutive days as proposed by Gere and Gratton (2010) and validated by Pinares-Patiño et al. (2012). Parallel to the animal breath sample collections, background air samples were also collected at a surrounding site, away from the animal pens to determine the baseline atmospheric concentrations of CH4 and SF6. These background samples were collected in duplicate, facing the wind direction. Given the disposition of the animal pens, it was assumed that concentrations of CH4 and SF6 in the inspired air by animals would be similar to those at the background samplers. Sampling for CH4 emission started at 08:30 on d 36 and ended at 08:00 on d 40. The concentrations of CH4 and SF6 were analyzed using a gas chromatograph (PerkinElmer 600, USA) as described by Gere et al. (2019).
Methane emissions were calculated using the permeation rate of each SF6 permeation tube and the concentration of CH4 and SF6 measured in each sample, according to the equation described by Williams et al. (2011):
where PR SF6 is the permeation rate of the SF6 tubes; [CH4] and [SF6] are the concentrations of these gases in the samplers; [BG] is the baseline atmospheric concentrations (baseline values were 2.8 ± 0.3 parts per million and 6.3 ± 0.7 parts per trillion for CH4 and SF6, respectively); MWCH4 and MWSF6 are the molecular mass of CH4 (16.04 g/mol) and SF6 (146.06 g/mol), respectively.
2.4. Sampling and processing of ruminal fluid
The ruminal fluid of the 18 animals in study was sampled at 3 evaluation times: 0 h pre-feeding (08:00), 4 h post-feeding (12:00), and 8 h post-feeding (16:00; immediately before the afternoon feeding) on d 41, 42 and 43, respectively. Samples of ruminal fluid (approximately 200 mL per animal) were obtained by esophageal tubing using a flexible PVC tube (2 mm of wall thickness and 8 mm of internal diameter; Tecnocom, Buenos Aires, Argentina) with about 25 holes of 3 mm diameter in the 15 cm-probe head, and an electric vacuum pump (down to 7 mbar; PXC-100, BOMBAS PASCAL S.A., Argentina). Then, the samples were filtered with a 3-cloth gauze, and immediately afterward the pH of each sample was measured. Finally, 50 mL was subsampled for microbial studies, 10 mL for determination of VFA concentration, 10 mL for ammonia concentration, and 5 mL for the count of ciliated protozoa. The samples for microbial studies were lyophilized and stored at −70 °C, and the samples for ruminal fermentation were acidified by the addition of 100 μL of 99% sulfuric acid and kept at −20 °C until their use. Samples for protozoa count were conserved in a solution of 10% formaldehyde (vol/vol), at room temperature.
2.5. Determinations of the ruminal fermentation parameters
Concentrations of VFA were determined by gas chromatography using Konik 5000B equipment (KONIK Group. Miami, USA) and according to the procedures described by Friggens et al. (1998). The concentration of total VFA was expressed in mM, and the fermentation profiles of acetate, propionate, iso-butyrate, butyrate, iso-valerate, and valerate were expressed in percentage of total VFA. Ruminal NH3–N concentration was determined using the colorimetry technique using the uremia kit (Lab Wiener, Rosario-Argentina) according to the manufacturer's instructions. The pH of the ruminal samples was analyzed with a potentiometer (Corning Ltd, Halstead, Essex, UK).
2.6. DNA extraction, microbial quantification of specific populations and counting of protozoa
Total genomic DNA was extracted from 100 mg of the lyophilized ruminal sample using the commercial Mini Kit DNA Stool QIAamp extraction kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's instructions. The DNA concentration was analyzed qualitatively and quantitatively using electrophoretic runs on 0.8% agarose gels and fluorometry (Qubit 2.0, Qubit dsDNA Broad Range Assay Kit, Life Technologies, Oregon, USA).
The absolute quantification of specific microbial populations was determined using real-time PCR (qPCR) by comparison with serial dilutions (108 to 101) of specific plasmid DNA standards. The quantitative qPCR was performed using the StepOnePlus Real-Time System (TermoFisher Scientific, USA). A total of 2 μL of genomic DNA (10 ng/μL) was added to the amplification reaction containing 20 pmol of each primer, 4 μL of 5X HOT FIREPol EvaGreen qPCR Mix Plus (Solis BioDyne, Estonia), and DNA/RNA free water adjusting to the total volume (20 μL) in duplicate for each sample. The primers used for the amplification of the 16S rRNA and methyl coenzyme-M reductase (mcrA) gene region were obtained from the bibliographic reference (Appendix Table 2).
The amplification of the fragments was previously fine-tuned (Ortiz-Chura et al., 2018). The procedure briefly consisted of a cycle of 95 °C for 15 min, followed by 40 cycles of 95 °C for 30 s for denaturation; hybridization at 60 °C for 30 s, but which varied according to the primer, and 72 °C for 1 min for the extension. The quantification was based on the construction of regression curves of the standard plasmid with already known concentrations. In each reaction, the linear regression values of the standard curve were within normal limits (R2 = 0.99, slope = −3.2 to −3.6 and efficiency = 95% to 110%).
The ruminal ciliated protozoa count was performed according to the methodology described by Dehority (1993). This procedure consisted of mixing the previously filtered fluid samples with a formalin saline solution. Then, the samples were stained with methyl green and the count of the number of ciliated protozoa was performed using an optical microscope (Nikon eclipse E200MV, Nikon Tokyo, Japan).
2.7. Microbial analysis by 16S rRNA amplicon sequencing
In order to analyze the meta-taxonomic temporal changes of ruminal bacteria, a total of 30 samples of genomic DNA of ruminal content were evaluated, consisting of 5 samples randomly chosen from each treatment group, at 3 separate evaluation times. The samples for 16S rRNA amplicon sequencing were sent to the Genomics and Sequencing Service of the Research Center in Veterinary and Agronomic Sciences of INTA, Argentina. The processing of samples for sequencing is briefly described below. A PCR was used to amplify the V3 to V4 regions of the 16S rRNA gene for bacteria. For this purpose, 4 primers were used: 2 specific −341f/805r (341F: CCTACGGGNGGCWGCAG and 805R: GACTACHVGGGTATCTAATCC) + 2 that contain the adapter sequences P5/P7 for the indexed flow cells (Klindworth et al., 2012). The reactions were kept at 95 °C for 3 min to denature the DNA, proceeding to amplification for 30 cycles at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 45 s; a final extension of 10 min at 72 °C was added to ensure complete amplification.
The expected fragment length of the PCR products was verified with agarose gel electrophoresis (1%) with ethidium bromide, and the size of the amplicon was estimated comparing it with a scale of 1 kb plus DNA (1 kb plus the scale of DNA, Invitrogen, Carlsbad, CA, USA). In addition, the PCR fragments were purified using the Agencourt AMPure XP kit (Beckman Coulter, USA) following the manufacturer's instructions. Finally, to validate the library, the size of the DNA fragment was verified using the 5200 Fragment Analyzer System (Agilent Technology Inc. CA. USA). The amplicons were sequenced with the Illumina MiSeq platform (Illumina Inc., San Diego, USA). Raw data of the sequencing from our samples are available upon request.
2.8. Bioinformatic analysis
Sequencing of the bacterial 16S rRNA amplicon yielded 3,178,366 paired-end reads with a mean length of 464 pb, which merged using Mothur's Make. Contig command (Schloss et al., 2009). Further, sequences were quality trimmed and those that did not meet the following criteria were excluded from analysis: length 200 to 600 bp, ambiguous bases > 6, Phred score > 25, homopolymer < 8, mismatches in primers < 4. Operational taxonomic units (OTU) picking was performed using the Uclust algorithm with GreenGenes v13.8 database as a reference and 97% of similarity threshold (Seedorf et al., 2014). Representative sequences were aligned using PyNAST (Caporaso et al., 2009), and chimeric sequences were searched using the Chimera. Uchime command from the Mothur package. The taxonomy was assigned using the GreenGenes v13.8 database.
2.9. Statistical analysis
Data for feed intake and CH4 emission were analyzed using a general linear model (GLM) SAS Software (13.1, SAS Institute SAS, 2013, Cary, NC, USA) as a randomized complete block design (Block = Sex), according to the following model I:
| Yij = μ + Sexi + Treatj + еij, |
where: Yij = response variable; μ = general mean of the experiment; Sexi = sex (i = 2); Treatj = dietary treatments, CTR versus NIT (j = 2); еij = experimental error. Data for pH, NH3–N, VFA, ciliated protozoa, the concentration of bacteria, methanogens and fungi by qPCR, and the alpha-diversity indices (Chao 1 and ACE estimators and Shannon and Simpson diversity indices) were compared between dietary treatments and sampling times by a repeated measures ANOVA using the PROC MIXED of the SAS Software, according to the model II:
| Yijkl = μ + Sexi + Treatj + Animk (Treat) + Timel + Treat × Time + еijkl, |
where: Yijkl = response variable; μ = general mean of the experiment; Sexi = sex (i = 2; block); Treatj = dietary treatments, CTR versus NIT (j = 2); Anim (Treat)k = is the repeated effect of the kth calf within the jth dietary treatments (k = 18); Timel = sampling times (l = 3); Treat × Time = interaction; еijkl = experimental error.
The OTU tables and raw counts per taxonomic level were used to compute the relative abundance of bacterial phyla and genera. The data of abundance were analyzed according to model II using PROC MIXED of SAS Software, either for data with normal distribution or for those data that were transformed to normalize them using the square root function. Prior to the alpha and beta-diversity analysis, the data from the OTU table of the samples were normalized using the rarefy function of the vegan package in R Studio 3.6.1 (R Core Team, 2019), which is applied as an ad hoc medium to normalize the microbiome counts that have resulted from libraries of very different sizes.
For the beta-diversity analysis, the ordering graphs of the non-metric dimensional scale (NMDS) were applied using the Bray–Curtis dissimilarity distance in the R ggplot2 package (Wickham, 2016) and the meta MDS function in the R vegan statistical package (Oksanen et al., 2019), in which the OTU were rarefied to the lowest sequence number. For statistical analysis of beta-diversity, we performed the permutational multivariate analysis of variance (PERMANOVA) using Adonis function in R. In addition, for each set of beta-diversity data, the differences were compared using the group dispersion homogeneity test using the betadisper function in the R vegan package (Anderson, 2006). All differences were significant when P < 0.05.
Spearman's correlation analysis was used to determine the degree of association between enteric CH4 emission, ruminal fermentation, and the relative abundance of bacterial communities from sequencing results. The analyses were performed using the corrplot function in R.
3. Results
3.1. Feed intake and CH4 emission
Feed intake, initial and final weights did not differ (P > 0.05) between dietary treatments. Compared to the CTR diet, feeding the NIT diet decreased both CH4 emission (g/d; P = 0.009) and CH4 yield (g/kg DMI; P = 0.041), by 27% and 21%, respectively (Table 1).
Table 1.
Feed dry matter intake (DMI), average liveweight (LW) and methane emissions from Holstein calves fed a control diet (CTR) and a nitrate-containing diet (NIT).
| Item | Diet |
SEM | P-value | |
|---|---|---|---|---|
| CTR | NIT | |||
| DMI1, kg of DM/d | 8.8 | 8.2 | 0.23 | 0.117 |
| Initial LW, kg | 213.9 | 214.2 | 4.76 | 0.960 |
| Final LW, kg | 267.6 | 265.8 | 5.81 | 0.832 |
| DMI2, kg of DM/d | 8.0 | 7.3 | 0.37 | 0.086 |
| CH4 emission, g/d | 242.7 | 178.0 | 15.33 | 0.009 |
| CH4 yield, g/kg of DMI | 30.8 | 24.4 | 2.16 | 0.041 |
DM = dry matter.
DMI ad libitum.
DMI restricted (95%) during the methane measurement period.
3.2. Temporal changes of ruminal fermentation parameters
Ruminal pH and total VFA concentration did not differ (P > 0.05; Table 2) between dietary treatments. There was no Diet × Time (i.e., sampling time) interaction effect on pH (P = 0.136). However, pH changed as a function of sampling time (P = 0.001). The initial pH was close to neutral, but 4 h post-feeding, this value decreased significantly in both dietary treatments; then the pH value tended to increase until reaching an average value of 6.8 in both treatments. There were Diet, Time, and Diet × Time interaction effects (P < 0.05) on the concentration of NH3–N. The concentrations of NH3–N did not differ (P > 0.05) at 8 h post-feeding period, but for the pre-feeding period it was higher for the CTR than for the NIT diet; whereas at 4 h post-feeding it was higher for the NIT than for the CTR diet (11.8 vs. 5.2 mg/dL, respectively) (Fig. 1A).
Table 2.
Effects of diet, time of collection of sample and their interaction on ruminal pH and fermentation parameters in ruminal fluid collected from Holstein calves fed a control diet (CTR) and a nitrate-containing diet (NIT).
| Item | Diet |
SEM |
P-value |
|||
|---|---|---|---|---|---|---|
| CTR | NIT | Diet | Time | D × T | ||
| pH | 6.8 | 6.8 | 0.03 | 0.753 | 0.001 | 0.136 |
| NH3–N, mg/dL | 8.5 | 10.2 | 0.35 | 0.047 | 0.002 | 0.001 |
| Total VFA, mmol/L | 236.2 | 228.4 | 8.23 | 0.253 | 0.036 | 0.092 |
| Fermentation profile, % | ||||||
| Acetate | 69.6 | 75.3 | 0.55 | 0.009 | 0.279 | 0.001 |
| Propionate | 20.9 | 14.9 | 0.49 | 0.010 | 0.245 | 0.001 |
| Butyrate | 6.2 | 6.9 | 0.19 | 0.198 | 0.817 | 0.149 |
| Iso-butyrate | 1.3 | 0.8 | 0.13 | 0.115 | 0.255 | 0.308 |
| Valerate | 0.6 | 0.4 | 0.02 | 0.037 | 0.917 | 0.165 |
| Iso-valerate | 1.3 | 1.4 | 0.05 | 0.748 | 0.411 | 0.098 |
| A/P ratio | 3.7 | 5.4 | 0.14 | 0.005 | 0.196 | 0.001 |
D × T = Diet × Time interaction; NH3–N = ammonia nitrogen; VFA = Volatile fatty acids; A/P = acetate to propionate ratio.
Fig. 1.
Temporal changes of fermentation parameters (mean ± standard error) in ruminal fluid collected from Holstein calves fed a control diet (CTR; grey line) and a nitrate-containing diet (NIT; black line). (A) Ruminal ammonia (NH3–N); (B) total volatile fatty acids; (C) molar proportion of acetate; (D) molar proportion of propionate. 0 h: pre-feeding; 4 h: 4 h post-feeding; 8 h: 8 h post-feeding. Significance level: ∗, P ≤ 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
Total VFA concentration showed no Diet × Time interaction (P = 0.092). However, it changed with sampling time (P = 0.036), concentrations being higher at 4 h post-feeding than pre-feeding and 8 h post-feeding periods (Fig. 1B). The fermentation profile analysis showed Diet × Time interaction (P < 0.05) for the molar proportion of acetate, propionate, and for the acetate to propionate (A/P) ratio, whereas the molar proportions of the other VFA showed no Diet × Time interaction (P > 0.05) (Table 2). No effects of sampling time on the molar proportions of VFA were observed (P > 0.05). Moreover, there were diet effects on the molar proportion of acetate, propionate, valerate, and on the A/P ratio (P < 0.05; Table 2). The molar proportion of acetate increased between 0 h and 4 h for the NIT diet, whereas in the same period it decreased for CTR diet, hence at 4 h post-feeding, the acetate molar proportion was much higher for the NIT than for the CTR diet (Fig. 1C). For the same period of sampling, the molar proportion of propionate showed opposite changes than those for acetate, hence at 4 h post-feeding, propionate molar proportion was higher for the CTR than the NIT diet (Fig. 1D). The A/P ratio in the NIT diet increased from 0 h to 4 h post-feeding, hence, at 4 h post-feeding it was much higher than for the CTR diet (P < 0.05).
3.3. Temporal changes of ruminal microbiota using qPCR
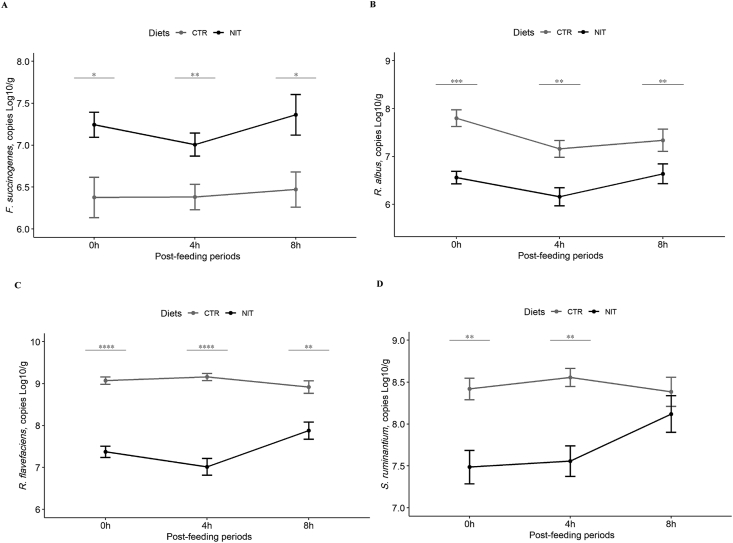
Feeding the NIT diet led to a significant decrease in the bacterial population (P = 0.001; Fig. 2A). Likewise, a time sampling effect was observed (P = 0.002), but no Diet × Time interaction effect was observed (P = 0.353) (Table 3). The methanogens population showed a Diet × Time interaction (P < 0.013) (Table 3). Compared to the CTR diet, the NIT feeding decreased the methanogens population, by 0.31 and 0.42 log10/g at 4 and 8 h post-feeding, respectively (Fig. 2B). The total fungal population showed a Diet × Time interaction (P = 0.003) (Table 3). The fungal population increased at 4 h and 8 h post-feeding of the NIT diet in comparison to the CTR diet (Fig. 2C). In addition, feeding the NIT diet increased significantly the population of ciliated protozoa (P = 0.001; Fig. 2D) and Fibrobacter succinogenes (P = 0.001; Fig. 3A) compared to the CTR diet. However, neither Diet × Time interaction nor time sampling effects were observed (P > 0.05).
Fig. 2.
Temporal changes of ruminal microbial populations (mean ± standard error) in ruminal fluid collected from Holstein calves fed a control diet (CTR; grey line) and a nitrate-containing diet (NIT; black line). (A) Total bacteria; (B) total methanogens; C) total fungi; D) total ciliate protozoa. 0 h: Pre-feeding; 4 h: 4 h post-feeding; 8 h: 8 h post-feeding. Significance level: ∗, P ≤ 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001.
Table 3.
Effect of diet, time collection and their interaction on ruminal microbiota (copies log10/g) in ruminal fluid collected from Holstein calves fed a control diet (CTR) and a nitrate-containing diet (NIT).
| Item | Diet |
SEM |
P-value |
|||
|---|---|---|---|---|---|---|
| CTR | NIT | Diet | Time | D × T | ||
| Total bacteria | 11.8 | 11.4 | 0.03 | 0.001 | 0.002 | 0.353 |
| Methanogens | 8.4 | 8.2 | 0.03 | 0.048 | 0.001 | 0.013 |
| Fungi | 4.8 | 6.5 | 0.12 | 0.001 | 0.122 | 0.003 |
| Protozoa, log10/mL | 4.8 | 6.0 | 0.04 | 0.001 | 0.057 | 0.140 |
| Fibrobacter succinogenes | 6.4 | 7.2 | 0.07 | 0.001 | 0.460 | 0.747 |
| Ruminococcus albus | 7.4 | 6.5 | 0.06 | 0.001 | 0.003 | 0.182 |
| Ruminococcus flavefaciens | 9.1 | 7.4 | 0.05 | 0.001 | 0.051 | 0.061 |
| Selenomonasruminatium | 8.5 | 7.7 | 0.06 | 0.001 | 0.160 | 0.046 |
| Veillonellaparvula | 5.3 | 5.2 | 0.08 | 0.349 | 0.110 | 0.800 |
D × T = Diet × Time interaction.
Fig. 3.
Temporal changes of ruminal microbial populations (mean ± standard error) in ruminal fluid collected from Holstein calves fed a control diet (CTR; grey line) and a nitrate-containing diet (NIT; black line). (A) Fibrobacter succinogenes; (B) Ruminococcus albus; (C) Ruminococcus flavefaciens; (D) Selenomonas ruminantium. 0 h: Pre-feeding; 4 h: 4 h post-feeding; 8 h: 8 h post-feeding. Significance level: ∗, P ≤ 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; ∗∗∗∗, P < 0.0001.
The populations of Ruminococcus albus, Ruminococcus flavefaciens, and Veillonella parvula showed no Diet × Time interaction (P > 0.05). However, feeding the NIT diet decreased the populations of R. albus and R. flavefaciens compared to the CTR diet (P < 0.05; Fig. 3B and C). No differences were observed between dietary treatments for the population of V. parvula (P = 0.349) and no sampling time effect was observed either (P = 0.110). The population of Selenomonas ruminantium showed a Diet × Time interaction (P = 0.046), the population was much higher for the CTR than for the NIT diet at the pre-feeding and 4 h post-feeding periods (Fig. 3D).
3.4. Diversity and composition of bacterial communities
A total of 2,783,124 bacterial and archaeal sequences were obtained as a result of the filtering analysis from 30 samples. The results revealed that the richness indices (Chao1 and ACE) and alpha-diversity estimators (Shannon and Simpson) of the ruminal bacteria did not differ between the dietary treatments (P > 0.05) or during the 3 evaluation times (P > 0.05) (Appendix Table 3). The beta-dispersion analysis showed differences between dietary treatments (Appendix Fig. A). However, the beta-diversity analysis of bacterial communities revealed significant differences between the dietary treatments (P < 0.001); and showed a distinct separation in the analysis of NMDS plot between the dietary treatments (Fig. 4). As regards the time factor, no separation was observed between the 3 levels, and no significant differences were found (P = 0.996).
Fig. 4.
Comparison of bacterial community structure and composition between dietary treatments, control (CTR; ●) and nitrate (NIT; ▲). During the sampling times, 0 h: pre-feeding (red round symbol), 4 h: 4 h post-feeding (green round symbol), and 8 h: 8 h post-feeding (blue round symbol). Data were examined by multivariate analysis, non-metric multidimensional scaling (NMDS) plots derived from Bray Curtis dissimilarity analysis between animals. Samples were plotted along the first 2 component axes. The microbial composition was compared using the Adonis Test. Diets effect, P-value < 0.001, Time effect, P-value = 0.996.
A total of 24 bacterial and archaeal phyla were identified. The 12 most abundant bacterial phyla found in both treatments and in the 3 evaluation times were those summarized in Table 4 and Fig. 5. The Euryarchaeota phyla was the only one identified within the archaeal community, and the sequences that were not classified under any phylum (other) in the dietary treatments and during the 3 evaluation times were on average (7.6 ± 0.96)% and (7.4 ± 1.54)% (mean ± standard deviation) in the CTR diet and the NIT diet, respectively. The Firmicutes and Bacteroidetes phyla were the most abundant ones, accounting for 84.1% and 85.7% of the total bacterial phyla in the CTR diet and the NIT diet, respectively. Likewise, changes in the relative abundance of Firmicutes and Bacteroidetes phyla were variable among animals within each dietary treatment, i.e., the coefficient of variation (CV) in the pre-feeding period ranged from 5.9% to 13.9%. In contrast, in the 4 h and 8 h post-feeding periods, the CV ranged from 6.1% to 26.3% (Fig. 5). However, the relative abundance of these dominant phyla, that of the Tenericutes, WPS-2, Cyanobacteria, Euryarchaeota phyla, and the Firmicutes to Bacteroidetes ratio (F/B) did not result in a Diet × Time interaction and neither did they differ between dietary treatments (P > 0.05).
Table 4.
Effects of diet, time collection of sample and their interaction on the relative abundance of ruminal bacteria at phylum level (% of the total) in ruminal fluid collected from Holstein calves fed a control diet (CTR) and a nitrate-containing diet (NIT).
| Item | Diet |
Time1 |
SEM |
P-value |
|||||
|---|---|---|---|---|---|---|---|---|---|
| CTR | NIT | 0 h | 4 h | 8 h | Diet | Time | D × T | ||
| Firmicutes | 47.74 | 50.31 | 49.02 | 50.64 | 47.42 | 0.952 | 0.358 | 0.406 | 0.214 |
| Bacteroidetes | 36.38 | 35.39 | 35.15 | 34.85 | 37.66 | 0.950 | 0.790 | 0.432 | 0.177 |
| Proteobacteria | 2.34 | 1.07 | 1.64 | 2.02 | 1.46 | 0.199 | 0.014 | 0.514 | 0.229 |
| Spirochaetes | 2.21 | 0.81 | 1.70 | 1.26 | 1.58 | 0.003 | 0.038 | 0.732 | 0.724 |
| Tenericutes | 1.51 | 1.86 | 1.55 | 1.61 | 1.90 | 0.117 | 0.381 | 0.451 | 0.178 |
| Actinobacteria | 1.31 | 1.28 | 1.57a | 1.46a | 0.87b | 0.096 | 0.964 | 0.019 | 0.003 |
| TM7 | 0.43 | 1.09 | 0.63b | 1.05a | 0.60b | 0.076 | 0.016 | 0.047 | 0.211 |
| Cyanobacteria | 0.12 | 0.06 | 0.06b | 0.12a | 0.09ab | 0.008 | 0.218 | 0.005 | 0.661 |
| WPS-2 | 0.08 | 0.01 | 0.10 | 0.01 | 0.02 | 0.005 | 0.274 | 0.053 | 0.434 |
| Verrucomicrobia | 0.07 | 0.27 | 0.15 | 0.17 | 0.21 | 0.013 | 0.015 | 0.216 | 0.031 |
| Synergistetes | 0.03 | <0.01 | 0.02 | 0.01 | 0.02 | 0.003 | 0.013 | 0.415 | 0.322 |
| Fibrobacteres | <0.01 | 0.08 | 0.04 | 0.02 | 0.06 | 0.009 | 0.001 | 0.349 | 0.321 |
| Euryarchaeota | 0.12 | 0.33 | 0.27a | 0.27a | 0.13b | 0.029 | 0.249 | 0.037 | 0.249 |
| F/B ratio | 1.38 | 1.52 | 1.45 | 1.59 | 1.32 | 0.070 | 0.574 | 0.313 | 0.234 |
D × T = Diet × Time interaction; F/B ratio = Firmicutes to Bacteroidetes ratio.
a, b Within rows, means with different superscripts differ at P ≤ 0.05.
0 h: pre-feeding period; 4 and 8 h: post-feeding periods.
Fig. 5.
Ruminal relative abundance of the most abundant phyla in individual ruminal fluid collected from Holstein calves fed a control diet (CTR; n = 5) and a nitrate-containing (NIT; n = 5) diet. During 3 sampling times; 0 h: pre-feeding; 4 h: 4 h post-feeding; 8 h: 8 h post-feeding.
In turn, no interaction was observed for the Proteobacteria, Spirochaetes, and Synergistetes phyla (P > 0.05), but their abundance decreased as a result of feeding the NIT diet (P < 0.05). The TM7 and Fibrobacteres phyla did not show any interaction either (P < 0.05), but their abundance increased as a result of feeding the NIT diet compared to the CTR diet. Moreover, a significant effect of sampling times was observed on the relative abundance of TM7, Cyanobacteria, and Euryarchaeota (P < 0.05). However, the Firmicutes, Bacteroidetes, Proteobacteria, Spirochaetes, Tenericutes, WPS-2, Synergistetes, and Fibrobacteres phyla and the F/B ratio did not differ among the sampling times (P > 0.05). An interaction of Diet × Time (P < 0.05) was shown for Actinobacteria and Verrucomicrobia. Compared to the CTR diet, feeding the NIT diet increased the relative abundance of Actinobacteria at 0 h (P < 0.05), whereas, the relative abundance of Verrucomicrobia was increased at 4 and 8 h post-feeding (P < 0.05) (Table 4).
Feeding the NIT diet reduced the ruminal abundance of Succiniclasticum, Coprococcus, Treponema, Shuttlewortia, Succinivibrio, Pseudobutyrivibrio, Sharpea, and Selenomona, compared to the CTR diet (P < 0.05). This contrasts with the abundance of Atopobium, Erysipelotrichaceae_L7A_E11, and Fibrobacter, which increased in the NIT diet (P < 0.05). However, no differences were found between dietary treatments for the Prevotella, Ruminococcus, Butyrivibrio, Clostridium, Oscillospira, Bulleidia, Moryella, Streptococcus, Schwartia, Lactobacillus, and Mogibacterium genera (P > 0.05) (Table 5). On the other hand, significant changes were observed as a result of the sampling times, as the abundance of the genera Pseudobutyrivibrio, Butyrivibrio, Clostridium, and Mogibacterium decreased and then recovered. In contrast, the abundance of the genus Ruminococcus increased at 4 h post-feeding (P < 0.05). Furthermore, a Diet × Time interaction was observed (P < 0.05) for the Atopobium and Erysipelotrichaceae L7A_E11 genera. The abundance of Atopobium was higher for the NIT diet than for the CTR diet only in the pre-feeding period (P < 0.05); whereas, the abundance of genus L7A_E11 from the Erysipelotrichaceae family was higher at 0 and 4 h post-feeding periods for the NIT diet than for the CTR diet (P < 0.05).
Table 5.
Effects of diet, time collection of sample and their interaction on the relative abundance of ruminal bacteria at genus level (% of the total) in ruminal fluid collected from Holstein calves fed a control diet (CTR) and a nitrate-containing diet (NIT).
| Item | Diet |
Time1 |
SEM |
P-value |
|||||
|---|---|---|---|---|---|---|---|---|---|
| CTR | NIT | 0 h | 4 h | 8 h | Diet | Time | D × T | ||
| Group of diminished bacteria by NIT diet | |||||||||
| Succiniclasticum | 1.90 | 0.59 | 1.19 | 1.16 | 1.39 | 0.069 | 0.004 | 0.354 | 0.132 |
| Coprococcus | 1.87 | 0.44 | 0.87 | 1.67 | 0.92 | 0.225 | 0.015 | 0.167 | 0.848 |
| Treponema | 1.31 | 0.66 | 1.10 | 0.89 | 0.97 | 0.315 | 0.033 | 0.062 | 0.585 |
| Shuttlewortia | 1.07 | 0.20 | 0.68 | 0.56 | 0.65 | 0.117 | 0.007 | 0.052 | 0.591 |
| Succinivibrio | 0.84 | 0.01 | 0.34 | 0.57 | 0.37 | 0.106 | 0.006 | 0.603 | 0.279 |
| Pseudobutyrivibrio | 0.58 | 0.04 | 0.41a | 0.16b | 0.36ab | 0.036 | 0.002 | 0.016 | 0.056 |
| Sharpea | 0.32 | 0.04 | 0.14 | 0.22 | 0.18 | 0.032 | 0.010 | 0.642 | 0.185 |
|
Selenomona |
0.22 |
0.10 |
0.13 |
0.13 |
0.21 |
0.023 |
0.024 |
0.270 |
0.271 |
| Group of increased bacteria by NIT diet | |||||||||
| Atopobium | 0.18 | 0.52 | 0.55a | 0.36ab | 0.13b | 0.052 | 0.023 | 0.015 | 0.008 |
| Erysipelotrichaceae_L7A_E11 | 0.08 | 0.21 | 0.20a | 0.10b | 0.13b | 0.011 | 0.010 | 0.005 | 0.010 |
|
Fibrobacter |
<0.01 |
0.08 |
0.04 |
0.02 |
0.06 |
0.009 |
0.003 |
0.240 |
0.192 |
| Group of bacteria unchanged by NIT diet | |||||||||
| Prevotella | 17.16 | 16.68 | 15.79 | 17.40 | 17.58 | 0.583 | 0.856 | 0.409 | 0.778 |
| Ruminococcus | 6.18 | 7.34 | 4.71b | 9.66a | 5.90b | 0.530 | 0.483 | 0.004 | 0.511 |
| Butyrivibrio | 3.55 | 4.59 | 5.08a | 3.42b | 3.72ab | 0.254 | 0.207 | 0.037 | 0.065 |
| Ruminococeae_Clostridium | 0.92 | 0.99 | 0.96b | 0.89b | 1.02a | 0.110 | 0.804 | 0.011 | 0.823 |
| Oscillospira | 0.79 | 0.80 | 0.74 | 0.80 | 0.84 | 0.093 | 0.971 | 0.920 | 0.828 |
| Bulleidia | 0.65 | 0.42 | 0.49 | 0.77 | 0.35 | 0.143 | 0.886 | 0.133 | 0.164 |
| Moryella | 0.13 | 0.19 | 0.19 | 0.12 | 0.18 | 0.018 | 0.132 | 0.277 | 0.572 |
| Streptococcus | 0.07 | 0.07 | 0.07 | 0.08 | 0.06 | 0.005 | 0.642 | 0.185 | 0.262 |
| Schwartia | 0.07 | 0.02 | 0.05 | 0.03 | 0.06 | 0.004 | 0.065 | 0.088 | 0.417 |
| Lactobacilus | 0.07 | 0.02 | 0.05 | 0.04 | 0.04 | 0.016 | 0.090 | 0.535 | 0.067 |
| Mogibacterium | 0.04 | 0.06 | 0.07a | 0.05ab | 0.03b | 0.005 | 0.256 | 0.020 | 0.184 |
D × T = Diet × Time interaction.
a, b Within a row, means with different superscripts differ at P ≤ 0.05.
10 h: pre-feeding period; 4 and 8 h: post-feeding periods.
3.5. Correlation between methane emission, ruminal fermentation, and ruminal microbiota
In order to estimate the degree of association between enteric CH4 emission, ruminal fermentation and ruminal microbiota, a correlation analysis was performed for both dietary treatments at 0 h (pre-feeding period). In the CTR diet (Fig. 6A), the CH4 emission and yield were not significantly correlated with any member of the ruminal microbiota. The proportion of acetate was positively correlated with the Tenericutes phyla (r = 0.90, P < 0.05) and negatively correlated with the F/B ratio (r = −0.90, P < 0.05), Shuttlewortia (r = −0.90, P < 0.05) and Selenomona (r ≥ −0.90, P < 0.001). Propionate was negatively correlated with Cyanobacteria (r = −0.90, P < 0.05) and Erysipelotrichaceae L7A_E11 (r = −0.90, P < 0.05), and positively correlated with Actinobacteria (r = 0.90, P < 0.05), Fibrobacteres (r = 0.90, P < 0.05), Succiniclasticum (r = 0.90, P < 0.05), Shuttlewortia (r ≥ 0.90, P < 0.001), Selenomona (r = 0.90, P < 0.05), Lactobacillus (r = 0.90, P < 0.05) and Fibrobacter (r = 0.90, P < 0.05). NH3–N was negatively correlated with Bacteroidetes (r = −0.90, P < 0.05) and Moryella (r = −0.90, P < 0.05) and pH was positively correlated with the relative abundance of the Butyrivibrio genus (r = 0.90, P < 0.05).
Fig. 6.
Spearman's correlation analysis between methane (CH4) emission, ruminal fermentation and ruminal microbiota by 16S rRNA amplicons sequencing for the pre-feeding period (0 h), for a control diet (A) and a nitrate containing diet (B). Significance level: ∗: P ≤ 0.05; ∗∗: P < 0.01; ∗∗∗: P < 0.001. CH4 total: expressed in grams of CH4 per animal; CH4 yield: expressed in grams of CH4 per kilogram of dry matter intake.
On the other hand, in the NIT diet (Fig. 6B), the CH4 emission was positively correlated with Succinivibrio (r ≥ 0.90, P < 0.001), and negatively correlated with Cyanobacteria (r ≥ −0.90, P < 0.001) and Pseudobutyrivibrio (r = −0.90, P < 0.05). Methane yield was positively correlated with TM7 (r = 0.90, P < 0.05), Bulleidia (r = 0.90, P < 0.05), and negatively correlated with Tenericutes (r = −0.90, P < 0.05), Fibrobacteres (r = −0.90, P < 0.05) and Fibrobacter (r = −0.90, P < 0.05). The proportion of acetate was positively correlated with Cyanobacteria (r = 0.90, P < 0.05) and negatively correlated with Prevotella (r = −0.90, P < 0.05) and Succinivibrio (r = −0.90, P < 0.05). In contrast, propionate was positively correlated with Succinivibrio (r ≥ 0.90, P < 0.001) and negatively with Cyanobacteria (r = −0.96, P < 0.001) and Pseudobutyrivibrio (r = −0.90, P < 0.05). Butyrate was negatively correlated with Fibrobacteres (r = −0.90, P < 0.05) and Fibrobacter (r = −0.90, P < 0.05). The total VFA concentration was positively correlated with Succiniclasticum (r = 0.90, P < 0.05), and negatively correlated with Shuttlewortia (r = −0.90, P < 0.05). The NH3–N concentration was positively correlated with Spirochaetes (r = 0.90, P < 0.05) and Treponema (r = 0.90, P < 0.05). Ruminal pH was positively correlated with Bacteroidetes (r ≥ 0.90, P < 0.001) and Prevotella (r = 0.90, P < 0.05), and negatively correlated with the F/B ratio (r = −0.90, P < 0.05), Coprococcus (r ≥ −0.90, P < 0.001) and Erysipelotrichaceae L7A E11 (r = −0.90, P < 0.05).
4. Discussion
4.1. Feed intake, methane emission, and methanogen population
Dry matter intake was not affected when nitrate was included in the diet of dairy calves at 15 g/kg of DM. This is in agreement with findings from previous studies (Hulshof et al., 2012; Olijhoek et al., 2016). However, recent studies by Meller et al. (2019) and Rebelo et al. (2019) reported a decrease of around 8.0% in DMI for inclusions of nitrate at similar levels of inclusion. This decrease in DMI was attributed, initially, to nitrate toxicity (evidenced by increased blood methemoglobin level [>20% of total hemoglobin]), when animals received diets >1% (on DM basis) of nitrate and without considering a scheme of dietary acclimatization (Bruning-Fann and Kaneene, 1993). Likewise, when the methemoglobinemia incidence was controlled, by gradual acclimatization or by using protected nitrate, the reduction in DMI was explained mainly due to the organoleptic properties of nitrate (bitter taste), both in cattle (Newbold et al., 2014; Lee et al., 2015) and in sheep (Li et al., 2012). Therefore, it seems that a gradual acclimatization scheme to nitrate is key to maintain DMI levels without affecting animal performance (van Zijderveld et al., 2011b) and without compromising animal health (Ortiz-Chura et al., 2021).
Methane yield was reduced by 21% when nitrate was included in the diet of dairy calves. This result agrees with findings from previous studies showing a reduction of between 16% and 25% with inclusion levels between 13 and 21 g of NO3─/kg of DM (Klop et al., 2016; Lund et al., 2014; Olijhoek et al., 2016; van Zijderveld et al., 2011a). In contrast, Meller et al. (2019) and Rebelo et al. (2019) reported that although the CH4 emission was decreased as a result of dietary nitrate, no differences in CH4 yield were observed. This lower CH4 emission and null effect on CH4 yield were mainly attributed to lowered DMI by nitrate effect because feed intake is positively associated with enteric CH4 emissions (Hristov et al., 2018). However, in the present study, the lower CH4 emission is not related to DMI variation, as it did not differ significantly among dietary treatments. Therefore, the lower CH4 emission would be mainly related to the mode of action of nitrate at the ruminal level.
In the rumen, the reduction of NO3─ to NH3 is energetically more favorable than carbon dioxide reduction. Stoichiometrically, the addition of 1 mol of NO3─ reduces by 1 mol the CH4 production, which is equivalent to 28.4 g of CH4/100 g of NO3─ (Latham et al., 2016). Thus, assuming a complete reduction of NO3─, the addition of 15 g of calcium nitrate (equivalent to 11.3 g NO3─/kg of DM) would have a reduction potential of 2.9 g CH4/kg of DM. However, the CH4 reduction observed in the present study (6.4 g/kg of DM) was much higher than the stoichiometric expectation. Reported CH4 reduction efficiencies greater than 100% of those from the stoichiometric expected value (Zhang et al., 2019), and increasing H2 concentration (gaseous and dissolved) after feeding dietary nitrate (Guyader et al., 2015; Olijhoek et al., 2016), suggest that nitrate may not act only as an effective H2 sink. In this sense, the toxic effects of nitrate-reducing metabolites (e.g., nitrite, nitrate esters, nitric oxide, and nitrous oxide) on ruminal methanogen growth seem to be more important because this inhibitory effect was previously reported in in vitro (Iwamoto et al., 2002) and in vivo studies (Asanuma et al., 2015). In addition, some studies suggest that hydrogenotrophic methanogens such as Methanobrevibacter spp. would be very sensitive to dietary nitrate (Bowen et al., 2020). Although the direct and indirect effects on methanogens and other members of the ruminal microbiota remain unclear, these inhibitory effects may result from the oxidizing nature of nitrite in relation to its antimicrobial properties (Cammack et al., 1999; Marais et al., 1988).
In the present study, the decrease in CH4 emissions and CH4 yield as a result of NIT diet feeding seems not only due to thermodynamic competition for H2 but also to the inhibition of the methanogen population (mcrA; a conserved gene involved in methanogenesis) observed at 4 and 8 h post-feeding. Similar results in mcrA copy reduction were reported in dairy cows fed nitrate (Veneman et al., 2015). In the same line, Granja-Salcedo et al. (2019) and Bowen et al. (2020) revealed that lower CH4 emissions in beef cattle fed nitrate were associated with a smaller Euryarchaeota to bacteria ratio and a lower abundance of M. spp.
4.2. Ruminal fermentation and microbiota
Ruminal pH usually decreases after feeding and then gradually increases due to VFA absorption, rumination, and salivation (Aschenbach et al., 2011). This pattern of change in pH was also observed in the present study, for a diet low in forage to concentrate ratio (20:80). It is well known (e.g., van Lingen et al., 2017) that following feeding, the molar proportion of acetate decreases and then recovers, whereas the molar proportion of propionate increases and then decreases. In this study, the described patterns of change in molar proportions of VFA were observed in the CTR diet. On the other hand, feeding the NIT diet altered the fermentation profile in a contrasting pattern; i.e., an increase in the molar proportion of acetate and a decrease in propionate molar proportion (at 4 h post-feeding period). This latter profile was also observed in in vitro (Lee et al., 2017) and in vivo (Hulshof et al., 2012; Olijhoek et al., 2016; Troy et al., 2015) studies, when nitrate was included in the diet.
Propionate synthesis is thermodynamically considered a more favorable pathway under high H2 pressure conditions (typically after feeding) (Janssen, 2010), and can sometimes be stimulated when methanogenesis is inhibited (Wang et al., 2018). In this study, although the concentration of dissolved H2 was not evaluated, the results suggest that the available H2 was not incorporated into the propiogenesis either, as the proportion of propionate decreased when fed the NIT diet. Therefore, it might be suggested that changes in the fermentation profile might be more related to the toxic effects of the nitrate-reduction metabolites on bacterial populations involved in propionate synthesis than on changes in metabolic hydrogen flow.
Relative abundances of Succiniclasticum, Coprococcus, Treponema, Shutlewortia, Succinivibrio, Pseudobutyrivibrio, Sharpea, and Selenomona were decreased when nitrate was included in the diet of dairy calves. These findings are unprecedented because, on the one hand, previous studies that reported a reduction in propionate synthesis did not evaluate the effects of nitrate feeding on the ruminal microbiota. On the other hand, studies that evaluated the effects of nitrate on the ruminal microbiota did not show changes in propionate synthesis. Furthermore, our results suggest that this detrimental effect on bacterial abundance could be associated with the lower propionate production because the conversion of pyruvate to propionate involves the acrylate and the succinate pathways. For instance, genes encoding these metabolic pathways were identified in the Succinivibrio dextrinosolvens genome (Hackmann et al., 2017; Hailemariam et al., 2020), which participates in the active synthesis of succinate (O'Herrin and Kenealy, 1993). In addition, this study and another study conducted by Ren et al. (2019) observed that Succinivibrio was positively correlated with propionate.
Another bacteria which could participate in the succinate pathway through the production of succinate, formate, and acetate is Treponema briyantti (Stanton, 1984), but in this study, the Treponema genus was not correlated to the production of propionate. Concomitantly, genes that participate in the synthesis of propionate through the succinate pathway were identified in the genome of some species of the Selenomona genus (Hackmann et al., 2017; Wang et al., 2020). In turn, Succiniclasticum ruminis has been identified as an active participant in the conversion of succinate to propionate (van Gylswyk, 1995). In addition, this study revealed a strong correlation between the abundance of Selenomona and Succiniclasticum and the production of propionate. Furthermore, it was determined that the Sharpea spp. (Kumar et al., 2018) and Pseudobutyrivibrio xylanovorans bacteria (Palevich et al., 2020) produce lactate, formate, and butyrate as a final product of glucose fermentation. It was also determined that Coprococcus participates in the production of propionate through the acrylate pathway using lactate (Reichardt et al., 2014). However, in the present study, no strong correlations were found between Coprococcus and propionate production.
On the other hand, the relative abundance of bacteria of the genus Atopobium (from Actinobacteria phyla) increased in response to the feeding the NIT diet. Also, the genus Atopobium was found to be a prevalent member of the human gut microbiota or ruminal microbiota (Harmsen et al., 2000). In the rumen, the genus Atopobium was described as a member of the epimural community (Chen et al., 2011), and their abundance increase under high-concentrate diets (>70%) (Mao et al., 2013; Petri et al., 2013). In turn, the L7A_E11 genus was phylogenetically categorized within the Erysipelotrichaceae family. The genome of this bacterium was recently sequenced from the metagenome of beef cattle (Stewart et al., 2018). Although very little is known about these bacteria, the relative abundance of the Erysipelotrichaceae family has been associated with the energy and protein metabolism of animal gut microbiota (Bermingham et al., 2017). Some previous studies on beef cattle showed a positive correlation between the abundance of L7A_E11 and total VFA concentration (Bi et al., 2018), and with the level of muscle marbling (Kim et al., 2020). However, their role in the rumen remains unknown.
The ruminal population of R. albus and R. flavefaciens decreased, whereas the population of F. succinogenes increased when nitrate was included in the diet of dairy calves. However, these results were contradictory to the findings of previous studies (Iwamoto et al., 2002; Asanuma et al., 2015) because these studies revealed a reduction in the populations of these 3 fibrolytic bacteria, whereas other studies showed no change, except for F. succinogenes, which increased (Granja-Salcedo et al., 2019). The reason for these contradictory findings could be related to the technique used in the different studies (in vitro vs. in vivo), sampling times (pre- and post-feeding periods), ruminal sampling technique (ruminal cannula vs. stomach tubing), DNA extraction method, ruminal fraction used (liquid, solid or mixture), among others. Similarly, the basal diet used in previous studies seems to play a crucial role because, in recent studies, when nitrate was used as a non-protein nitrogen source in low-quality diets, it was shown that higher fiber digestibility was associated with higher R. albus and total protozoa population (Zhang et al., 2019). However, our findings could partly explain the lower CH4 emissions for the NIT diet because the genus Ruminococcus has been considered an H2 producer, but this is not the case for F. succinogenes (Chaucheyras-Durand et al., 2010). Furthermore, F. succinogenes was found in higher density in low CH4-emitting beef cattle phenotypes (Tapio et al., 2017) and, in the present study, it was negatively correlated with CH4 production, suggesting an indirect effect on CH4 emission for the NIT diet.
The population of the genus Selenomona and the species S. ruminantium decreased, but there was no change in the population of V. parvula in response to the NIT diet. The results were contradictory with findings from previous studies (Granja-Salcedo et al., 2019; Iwamoto et al., 2002; Zhao et al., 2018), although another study showed no changes in the populations of nitrate-reducing bacteria when nitrate was included in the diet (Popova et al., 2017). The reason for these differences might be related to the methodology applied in the different studies (in vitro vs. in vivo), and sampling times (pre- and post-feeding), among others. We recommend furthering these studies using meta-transcriptomic tools to more deeply understand the nitrate- and nitrite-reduction process in the rumen.
Ciliated protozoa populations increased significantly as a result of feeding the NIT diet, which is in line with the results obtained in goats (Zhang et al., 2019) and in vitro studies (Lin et al., 2011). Furthermore, in a recent in vivo experiment with defaunated and faunated sheep supplemented with 1.8% of nitrate (on a DM basis), it was reported that nitrate metabolizes more rapidly in the presence of ruminal protozoa, and defaunated sheep may have an increased risk of poisoning due to nitrite accumulation in the rumen (Villar et al., 2020). In contrast, Iwamoto et al. (2001) observed that nitrite inhibited the growth of protozoa, and recently it was determined that nitrite interfered with the motility and chemotaxis of protozoa (mainly on isotrichids) in in vitro cultures (Roman-Garcia et al., 2019). In other studies, in sheep (van Zijderveld et al., 2010) and cattle (Popova et al., 2017) the protozoan population was not influenced by nitrate supplementation.
Previous studies suggested a smaller relationship between the fungi population and nitrate metabolism (Lin et al., 2011; Zhang et al., 2019). In contrast, the present study found that the fungi population increased as a result of feeding the NIT diet, suggesting that they might be involved in nitrate reduction. However, more studies are required to confirm the role of fungi in nitrate metabolism in the rumen. Finally, the variation observed among animals for both relative abundances at the phylum level and beta dispersion analysis of bacterial communities is mainly due to the age of the animals in this experiment, because in general, a higher variation of microbial composition was observed in young animals than in adults due to the status of rumen maturity, and the development and establishment of ruminal microbiota (Jami et al., 2013).
The results confirmed the hypothesis that nitrate feeding induced changes in the populations of total bacteria, methanogens, ciliated protozoa, total fungi, fibrolytic bacteria, and bacteria involved in the succinate and acrylate pathways, thus, these changes were associated with decreased enteric CH4 emission and altered ruminal fermentation profiles.
5. Conclusions
This study, in dairy calves, confirmed the ability of inclusion of nitrate in the diet to reduce enteric CH4 emission and CH4 yield without significantly affecting feed intake. The lower enteric CH4 emission in response to nitrate feeding was over the stoichiometrically predicted reduction potential and also associated with a lower density of total methanogen population, suggesting a predominantly direct effect on ruminal methanogenesis. The decrease in the molar proportion of propionate in ruminal fluid in response to nitrate was associated with a reduction in the relative abundance of the genera Succiniclasticum, Coprococcus, Treponema, Shutlewortia, Succinivibrio, Pseudobutyrivibrio, Sharpea, and Selenomona genera.
Author contributions
Abimael Ortiz-Chura: Conceptualization, Methodology, Investigation, Software, and Writing- Original draft preparation. Gisela Marcoppido: Investigation, Resources. Gustavo Depetris: Investigation, Methodology. José Gere: Formal analysis, Data curation, Funding acquisition. Claudia Faverín: Investigation, Software, Cesar Pinares-Patiño: Writing, Review, and Editing. Silvio Cravero: Resources, Visualization. Angel Cataldi: Supervision, Validation. María E. Cerón-Cucchi: Investigation, Funding acquisition, Writing, Review, and Editing.
Conflicts of interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This study was supported by a grant from the FonCyT-Argentina: PICT 2015-294, PID MS-UTNBA-0004540 and INTA I058. We wish to thank Dra Milka Popova (from INRAE-France) for the support in the bioinformatics analysis. We also thank Eng. Ricardo Bualo and Ricardo Arias for their help and technical support in methane emission measurements.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2021.07.005.
Appendix.
The following is the Supplementary data to this article:
References
- Anderson M.J. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62(1):245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- AOAC . 15 ed. 1990. Official methods of analysis. Washington, USA. [Google Scholar]
- Armstrong D.G., Blaxter K.L. The utilization of acetic, propionic and butyric acids by fattening sheep. Br J Nutr. 1957;11(4):413–425. doi: 10.1079/bjn19570063. [DOI] [PubMed] [Google Scholar]
- Asanuma N., Yokoyama S., Hino T. Effects of nitrate addition to a diet on fermentation and microbial populations in the rumen of goats, with special reference to Selenomonas ruminantium having the ability to reduce nitrate and nitrite. Anim Sci J. 2015;86(4) doi: 10.1111/asj.12307. 384-378. [DOI] [PubMed] [Google Scholar]
- Aschenbach J.R., Penner G.B., Stumpff F., Gäbel G. Ruminant Nutrition Symposium: role of fermentation acid absorption in the regulation of ruminal pH. J Anim Sci. 2011;89(4):1092–1107. doi: 10.2527/jas.2010-3301. [DOI] [PubMed] [Google Scholar]
- Beauchemin K.A., Ungerfeld E.M., Eckard R.J., Wang M. Review: fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal. 2020 doi: 10.1017/S1751731119003100. [DOI] [PubMed] [Google Scholar]
- Bermingham E.N., Maclean P., Thomas D.G., Cave N.J., Young W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ. 2017 doi: 10.7717/peerj.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y., Zeng S., Zhang R., Diao Q., Tu Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018;18(1):69. doi: 10.1186/s12866-018-1213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.M., Cormican P., Lister S.J., McCabe M.S., Duthie C.A., Roehe R. Links between the rumen microbiota, methane emissions and feed efficiency of finishing steers offered dietary lipid and nitrate supplementation. PLoS One. 2020 doi: 10.1371/journal.pone.0231759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning-Fann C.S., Kaneene J.B. The effects of nitrate, nitrite, and N-nitroso compounds on animal health. Vet Hum Toxicol. 1993;35(3):237–253. [PubMed] [Google Scholar]
- Cammack R., Joannou C.L., Cui X.Y., Torres Martinez C., Maraj S.R., Hughes M.N. Nitrite and nitrosyl compounds in food preservation. Biochim Biophys Acta Bioenerg. 1999;1411(2):475–488. doi: 10.1016/s0005-2728(99)00033-x. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Bittinger K., Bushman F.D., DeSantis T.Z., Andersen G.L., Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2009;26(2):266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaucheyras-Durand F., Masséglia S., Fonty G., Forano E. Influence of the composition of the cellulolytic flora on the development of hydrogenotrophic microorganisms, hydrogen utilization, and methane production in the rumens of gnotobiotically reared lambs. Appl Environ Microbiol. 2010;76(24) doi: 10.1128/AEM.01784-10. 7937-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Penner G.B., Li M., Oba M., Guan L.L. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl Environ Microbiol. 2011;77(16) doi: 10.1128/AEM.00375-11. 5781/5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehority B.A. 1st ed. CRC Press; 1993. Laboratory manual for classification and morphology of rumen ciliate Protozoa. [Google Scholar]
- FAOSTAT . 2020. Food and agriculture organization of the united nations (FAO)http://www.fao.org/faostat [Google Scholar]
- Friggens N.C., Oldham J.D., Dewhurst R.J., Horgan G. Proportions of volatile fatty acids in relation to the chemical composition of feeds based on grass silage. J Dairy Sci. 1998;81(5):1331–1344. doi: 10.3168/jds.S0022-0302(98)75696-6. [DOI] [PubMed] [Google Scholar]
- Gerber P.J., Steinfeld H., Henderson B., Mottet A., Opio C., Dijkman J. Food and Agriculture Organization of the United Nations (FAO); Rome: 2013. Tackling climate change through livestock: a global assessment of emissions and mitigation opportunities. [Google Scholar]
- Gerber P.J., Hristov A.N., Henderson B., Makkar H., Oh J., Lee C. Technical options for the mitigation of direct methane and nitrous oxide emissions from livestock: a review. Animal. 2013;7(s2):220–234. doi: 10.1017/S1751731113000876. [DOI] [PubMed] [Google Scholar]
- Gere J., Gratton R. Simple, low-cost flow controllers for time averaged atmospheric sampling and other applications. Latin American Applied Research Pesquisa Aplicada Latino Americana Investigación Aplicada Latinoamericana. 2010;40:377–381. [Google Scholar]
- Gere J.I., Bualó R.A., Perini A.L., Arias R.D., Ortega F.M., Wulff A.E. Methane emission factors for beef cows in Argentina: effect of diet quality. N Z J Agric Res. 2019 9-1. [Google Scholar]
- Granja-Salcedo Y.T., Fernandes R.M., de Araujo R.C., Kishi L.T., Berchielli T.T., de Resende F.D. Long-term encapsulated nitrate supplementation modulates rumen microbial diversity and rumen fermentation to reduce methane emission in grazing steers. Front Microbiol. 2019;10:614. doi: 10.3389/fmicb.2019.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyader J., Eugène M., Meunier B., Doreau M., Morgavi D.P., Silberberg M. Additive methane-mitigating effect between linseed oil and nitrate fed to cattle 1. J Anim Sci. 2015;93(7):3564–3577. doi: 10.2527/jas.2014-8196. [DOI] [PubMed] [Google Scholar]
- Hackmann T.J., Ngugi D.K., Firkins J.L., Tao J. Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ Microbiol. 2017;19(11):4670–4683. doi: 10.1111/1462-2920.13929. [DOI] [PubMed] [Google Scholar]
- Hailemariam S., Zhao S., Wang J. Complete genome sequencing and transcriptome analysis of nitrogen metabolism of Succinivibrio dextrinosolvens strain Z6 isolated from dairy cow rumen. Front Microbiol. 2020 doi: 10.3389/fmicb.2020.01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen H.J., Wildeboer-Veloo A.C., Grijpstra J., Knol J., Degener J.E., Welling G.W. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microbiol. 2000;66(10):4523–4527. doi: 10.1128/aem.66.10.4523-4527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwing A.L.F., Lund P., Mogensen L., Vestergaard M. Growth, feed intake, methane emissions and carbon footprint from Holstein bull calves fed four different rations. Livest Sci. 2018;214:51–61. [Google Scholar]
- Hristov A.N., Kebreab E., Niu M., Oh J., Bannink A., Bayat A.R. Symposium review: uncertainties in enteric methane inventories, measurement techniques, and prediction models. J Dairy Sci. 2018;101(7):6655–6674. doi: 10.3168/jds.2017-13536. [DOI] [PubMed] [Google Scholar]
- Hulshof R.B.A., Berndt A., Gerrits W.J.J., Dijkstra J., van Zijderveld S.M., Newbold J.R. Dietary nitrate supplementation reduces methane emission in beef cattle fed sugarcane-based diets 1. J Anim Sci. 2012;90(7):2317–2323. doi: 10.2527/jas.2011-4209. [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Asanuma N., Hino T. Ability of Selenomonas ruminantium, veillonella parvula, and wolinella succinogenes to reduce nitrate and nitrite with special reference to the suppression of ruminal methanogenesis. Anaerobe. 2002;8(4):209–215. [Google Scholar]
- Iwamoto M., Asanuma N., Hino T. Effect of protozoa on nitrate and nitrite reduction in ruminal microbiota. Kanto J Anim Sci. 2001;51:9–25. [Google Scholar]
- Jami E., Israel A., Kotser A., Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013;7(6):1069–1079. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P.H. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim Feed Sci Technol. 2010;160(1):1–22. [Google Scholar]
- Johnson K., Huyler M., Westberg H., Lamb B., Zimmerman P. Measurement of methane emissions from ruminant livestock using a sulfur hexafluoride tracer technique. Environ Sci Technol. 1994;28(2):359–362. doi: 10.1021/es00051a025. [DOI] [PubMed] [Google Scholar]
- Kim M., Park T., Jeong J.Y., Baek Y., Lee H.J. Association between rumen microbiota and marbling score in Korean native beef cattle. Animals. 2020;10(4):712. doi: 10.3390/ani10040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012;41(1):e1–e. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klop G., Hatew B., Bannink A., Dijkstra J. Feeding nitrate and docosahexaenoic acid affects enteric methane production and milk fatty acid composition in lactating dairy cows. J Dairy Sci. 2016;99(2):1161–1172. doi: 10.3168/jds.2015-10214. [DOI] [PubMed] [Google Scholar]
- Kumar S., Treloar B.P., The K.H., McKenzie C.M., Henderson G., Attwood G.T. Sharpea and Kandleria are lactic acid producing rumen bacteria that do not change their fermentation products when co-cultured with a methanogen. Anaerobe. 2018;54:31–38. doi: 10.1016/j.anaerobe.2018.07.008. [DOI] [PubMed] [Google Scholar]
- Latham E.A., Anderson R.C., Pinchak W.E., Nisbet D.J. Insights on alterations to the rumen ecosystem by nitrate and nitrocompounds. Front Microbiol. 2016;7:228. doi: 10.3389/fmicb.2016.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Araujo R.C., Koenig K.M., Beauchemin K.A. In situ and in vitro evaluations of a slow release form of nitrate for ruminants: nitrate release rate, rumen nitrate metabolism and the production of methane, hydrogen, and nitrous oxide. Anim Feed Sci Technol. 2017;231:97–106. [Google Scholar]
- Lee C., Araujo R.C., Koenig K.M., Beauchemin K.A. Effects of feed consumption rate of beef cattle offered a diet supplemented with nitrate ad libitum or restrictively on potential toxicity of nitrate. J Anim Sci. 2015;93(10):4956–4966. doi: 10.2527/jas.2015-9435. [DOI] [PubMed] [Google Scholar]
- Li L., Davis J., Nolan J., Hegarty R. An initial investigation on rumen fermentation pattern and methane emission of sheep offered diets containing urea or nitrate as the nitrogen source. Anim Prod Sci. 2012;52(7):653–658. [Google Scholar]
- Lin M., Schaefer D.M., Guo W.S., Ren L.P., Meng Q.X. Comparisons of in vitro nitrate reduction, methanogenesis, and fermentation acid profile among rumen bacterial, Protozoal and fungal fractions. Asian-Australas J Anim Sci. 2011;24(4):471–478. [Google Scholar]
- Lund P., Dahl R., Yang H.J., Hellwing A.L.F., Cao B.B., Weisbjerg M.R. The acute effect of addition of nitrate on in vitro and in vivo methane emission in dairy cows. Anim Prod Sci. 2014;54(9):1432–1435. [Google Scholar]
- MacRae J.C., Armstrong D.G. Enzyme method for determination of α-linked glucose polymers in biological materials. J Sci Food Agric. 1968;19(10):578–581. [Google Scholar]
- Mao S.Y., Zhang R.Y., Wang D.S., Zhu W.Y. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. 2013;24:12–19. doi: 10.1016/j.anaerobe.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Marais J.P., Therion J.J., Mackie R.I., Kistner A., Dennison C. Effect of nitrate and its reduction products on the growth and activity of the rumen microbial population. Br J Nutr. 1988;59(2):301–313. doi: 10.1079/bjn19880037. [DOI] [PubMed] [Google Scholar]
- Meller R.A., Wenner B.A., Ashworth J., Gehman A.M., Lakritz J., Firkins J.L. Potential roles of nitrate and live yeast culture in suppressing methane emission and influencing ruminal fermentation, digestibility, and milk production in lactating Jersey cows. J Dairy Sci. 2019;102(7):6144–6156. doi: 10.3168/jds.2018-16008. [DOI] [PubMed] [Google Scholar]
- Morgavi D.P., Forano E., Martin C., Newbold C.J. Microbial ecosystem and methanogenesis in ruminants. Animal. 2010;4(7):1024–1036. doi: 10.1017/S1751731110000546. [DOI] [PubMed] [Google Scholar]
- Newbold J.R., van Zijderveld S.M., Hulshof R.B.A., Fokkink W.B., Leng R.A., Terencio P. The effect of incremental levels of dietary nitrate on methane emissions in Holstein steers and performance in Nelore bulls 1. J Anim Sci. 2014;92(11):5032–5040. doi: 10.2527/jas.2014-7677. [DOI] [PubMed] [Google Scholar]
- O'Herrin S.M., Kenealy W.R. Glucose and carbon dioxide metabolism by Succinivibrio dextrinosolvens. Appl Environ Microbiol. 1993;59(3):748–755. doi: 10.1128/aem.59.3.748-755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Guillaume Blanchet F., Friendly F., Roeland Kindt L., Legendre P., McGlinn D. 2019. Vegan: community ecology package. [Google Scholar]
- Olijhoek D.W., Hellwing A.L.F., Brask M., Weisbjerg M.R., Højberg O., Larsen M.K. Effect of dietary nitrate level on enteric methane production, hydrogen emission, rumen fermentation, and nutrient digestibility in dairy cows. J Dairy Sci. 2016;99(8):6191–6205. doi: 10.3168/jds.2015-10691. [DOI] [PubMed] [Google Scholar]
- Ortiz-Chura A., Fernández Pepi M.G., Wawrzkiewicz M., Cerón Cucchi M.E., Cravero S., Jaurena G. Microbial populations and ruminal fermentation of sheep and llamas fed low quality forages. Small Rumin Res. 2018;168:47–51. [Google Scholar]
- Ortiz-Chura A., Marcoppido G., Gere J., Depetris D., Stefañuk F., Trangoni M.D. Changes in hematological, biochemical and blood gases parameters in response to progressive inclusion of nitrate in the diet of Holstein calves. Vet World. 2021;14(1):61–69. doi: 10.14202/vetworld.2021.61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevich N., Maclean P.H., Kelly W.J., Leahy S.C., Rakonjac J., Attwood G.T. Complete genome sequence of the polysaccharide-degrading rumen bacterium Pseudobutyrivibrio xylanivorans MA3014 reveals an incomplete glycolytic pathway. Genome Biol Evol. 2020;12(9):1566–1572. doi: 10.1093/gbe/evaa165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R.M., Schwaiger T., Penner G.B., Beauchemin K.A., Forster R.J., McKinnon J.J. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl Environ Microbiol. 2013;79(12):3744–3755. doi: 10.1128/AEM.03983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinares-Patiño C., Gere J., Williams K., Gratton R., Juliarena P., Molano G. Extending the collection duration of breath samples for enteric methane emission estimation using the SF6 tracer technique. Animals. 2012;2(2):275–287. doi: 10.3390/ani2020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinares-Patiño C.S., Clark H. Reliability of the sulfur hexafluoride tracer technique for methane emission measurement from individual animals: an overview. Aust J Exp Agric. 2008;48(2):223–229. [Google Scholar]
- Popova M., Guyader J., Silberberg M., Seradj A.R., Saro C., Bernard A. Changes in the rumen microbiota of cows in response to dietary supplementation with nitrate, linseed, and saponin alone or in combination. Appl Environ Microbiol. 2019 doi: 10.1128/AEM.02657-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova M., McGovern E., McCabe M.S., Martin C., Doreau M., Arbre M. The structural and functional capacity of ruminal and cecal microbiota in growing cattle was unaffected by dietary supplementation of linseed oil and nitrate. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: a language and environment for statistical computing. [Google Scholar]
- Rebelo L.R., Luna I.C., Messana J.D., Araujo R.C., Simioni T.A., Granja-Salcedo Y.T. Effect of replacing soybean meal with urea or encapsulated nitrate with or without elemental sulfur on nitrogen digestion and methane emissions in feedlot cattle. Anim Feed Sci Technol. 2019 doi: 10.1016/j.anifeedsci.2019.114293. [DOI] [Google Scholar]
- Reichardt N., Duncan S.H., Young P., Belenguer A., McWilliam Leitch C., Scott K.P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8(6):1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Su X., Bai H., Yang Y., Wang H., Dan Z. Specific enrichment of microbes and increased ruminal propionate production: the potential mechanism underlying the high energy efficiency of Holstein heifers fed steam-flaked corn. Amb Express. 2019;9(1):209. doi: 10.1186/s13568-019-0937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Garcia Y., Wenner B.A., Welty C.M., Wagner B.K., Plank J.E., Meller R.A. Rumen microbial responses to supplemental nitrate. I. Yeast growth and protozoal chemotaxis in vitro as affected by nitrate and nitrite concentrations. J Dairy Sci. 2019;102(3):2207–2216. doi: 10.3168/jds.2018-15274. [DOI] [PubMed] [Google Scholar]
- Rosegrant M.W., Fernandez M., Sinha A., Alder J., Ahammad H., Fraiture C.D. IFPRI; 2009. Looking into the future for agriculture and AKST.http://www.agassessment-watch.org/report/North%20America%20and%20Europe_Subglobal%20Report%20(English).pdf [Google Scholar]
- SAS . SAS Institute Inc.; Cary, NC: 2013. SAS/STAT 13.1 user's guide. [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf H., Kittelmann S., Henderson G., Janssen P.H. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ. 2014 doi: 10.7717/peerj.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaani Y., Zehavi T., Eyal S., Miron J., Mizrahi I. Microbiome niche modification drives diurnal rumen community assembly, overpowering individual variability and diet effects. ISME J. 2018;12(10):2446–2457. doi: 10.1038/s41396-018-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T.B. Glucose metabolism of Treponema bryantii, an anaerobic rumen spirochete. Can J Microbiol. 1984;30(5):526–531. doi: 10.1139/m84-080. [DOI] [PubMed] [Google Scholar]
- Stewart R.D., Auffret M.D., Warr A., Wiser A.H., Press M.O., Langford K.W. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun. 2018;9(1):870. doi: 10.1038/s41467-018-03317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapio I., Snelling T.J., Strozzi F., Wallace R.J. The ruminal microbiome associated with methane emissions from ruminant livestock. J Anim Sci Biotechnol. 2017;8:7. doi: 10.1186/s40104-017-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R.K., Jungermann K., Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy S.M., Duthie C.A., Hyslop J.J., Roehe R., Ross D.W., Wallace R.J. Effectiveness of nitrate addition and increased oil content as methane mitigation strategies for beef cattle fed two contrasting basal diets 1. J Anim Sci. 2015;93(4):1815–1823. doi: 10.2527/jas.2014-8688. [DOI] [PubMed] [Google Scholar]
- van Gylswyk N.O. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int J Syst Bacteriol. 1995;45(2):297–300. doi: 10.1099/00207713-45-2-297. [DOI] [PubMed] [Google Scholar]
- van Lingen H.J., Edwards J.E., Vaidya J.D., van Gastelen S., Saccenti E., van den Bogert B. Diurnal dynamics of gaseous and dissolved metabolites and microbiota composition in the bovine rumen. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P.J., Robertson J.B., Lewis B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74(10):3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- van Zijderveld S.M., Gerrits W.J.J., Apajalahti J.A., Newbold J.R., Dijkstra J., Leng R.A. Nitrate and sulfate: effective alternative hydrogen sinks for mitigation of ruminal methane production in sheep. J Dairy Sci. 2010;93(12):5856–5866. doi: 10.3168/jds.2010-3281. [DOI] [PubMed] [Google Scholar]
- van Zijderveld S.M., Gerrits W.J.J., Dijkstra J., Newbold J.R., Hulshof R.B.A., Perdok H.B. Persistency of methane mitigation by dietary nitrate supplementation in dairy cows. J Dairy Sci. 2011;94(8):4028–4038. doi: 10.3168/jds.2011-4236. [DOI] [PubMed] [Google Scholar]
- van Zijderveld S.M., Dijkstra J., Perdok H.B., Newbold J.R., Gerrits W.J. Dietary inclusion of diallyl disulfide, yucca powder, calcium fumarate, an extruded linseed product, or medium-chain fatty acids does not affect methane production in lactating dairy cows. J Dairy Sci. 2011;94(6):3094–3104. doi: 10.3168/jds.2010-4042. [DOI] [PubMed] [Google Scholar]
- Veneman J.B., Muetzel S., Hart K.J., Faulkner C.L., Moorby J.M., Perdok H.B. Does dietary mitigation of enteric methane production affect rumen function and animal productivity in dairy cows? PLoS One. 2015 doi: 10.1371/journal.pone.0140282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar M.L., Hegarty R.S., Clay J.W., Smith K.A., Godwin I.R., Nolan J.V. Dietary nitrate and presence of protozoa increase nitrate and nitrite reduction in the rumen of sheep. 2020. [DOI] [PubMed]
- Wang L., Zhang G., Li Y., Zhang Y. Effects of high forage/concentrate diet on volatile fatty acid production and the microorganisms involved in VFA production in cow rumen. Animals. 2020 doi: 10.3390/ani10020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Wang M., Ungerfeld E.M., Zhang X.M., Long D.L., Mao H.X. Nitrate improves ammonia incorporation into rumen microbial protein in lactating dairy cows fed a low-protein diet. J Dairy Sci. 2018;101(11):9789–9799. doi: 10.3168/jds.2018-14904. [DOI] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York: 2016. ggplot 2: elegant graphics for data analysis. [Google Scholar]
- Williams S.R.O., Moate P.J., Hannah M.C., Ribaux B.E., Wales W.J., Eckard R.J. Background matters with the SF6 tracer method for estimating enteric methane emissions from dairy cows: a critical evaluation of the SF6 procedure. Anim Feed Sci Technol. 2011;170(3):265–276. [Google Scholar]
- Zhang X., Medrano R.F., Wang M., Beauchemin K.A., Ma Z., Wang R. Effects of urea plus nitrate pretreated rice straw and corn oil supplementation on fiber digestibility, nitrogen balance, rumen fermentation, microbiota and methane emissions in goats. J Anim Sci Biotechnol. 2019 doi: 10.1186/s40104-019-0312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Meng Q., Li Y., Wu H., Huo Y., Zhang X. Nitrate decreases ruminal methane production with slight changes to ruminal methanogen composition of nitrate-adapted steers. BMC Microbiol. 2018 doi: 10.1186/s12866-018-1164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman P.R. United States Patent and Trademark Office; Alexandria, VA: 1993. System for measuring metabolic gas emissions from animals. Patent no. 5 265 618. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.