Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) that have become dominant as the pandemic progresses bear the ORF8 mutation together with multiple spike mutations. A 382-nucleotide deletion (Δ382) in the ORF7b and ORF8 regions has been associated with milder disease phenotype and less systemic inflammation in COVID-19 patients. However, its impact on host immunity against SARS-CoV-2 remains undefined. Here, RNA-sequencing was performed to elucidate whole blood transcriptomic profiles and identify contrasting immune signatures between patients infected with either wildtype or Δ382 SARS-CoV-2 variant. Interestingly, the immune landscape of Δ382 SARS-CoV-2 infected patients featured an increased adaptive immune response, evidenced by enrichment of genes related to T cell functionality, a more robust SARS-CoV-2-specific T cell immunity, as well as a more rapid antibody response. At the molecular level, eukaryotic initiation factor 2 signaling was found to be upregulated in patients bearing Δ382, and its associated genes were correlated with systemic levels of T cell-associated and pro-inflammatory cytokines. This study provides more in-depth insight into the host–pathogen interactions of ORF8 with great promise as a therapeutic target to combat SARS-CoV-2 infection.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-021-01142-z.
Keywords: COVID-19, SARS-CoV-2, Transcriptome, ORF8, Adaptive immune response, CD4+ T cell response, CD8+ T cell response, Antibody response
Introduction
Genetic mutation events might enable viruses to cross the species barrier to infect a new host and subsequently allow virus adaptation to the host [1–3]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has mutated throughout the pandemic, and multiple variants of concern (VOCs) continue to spread globally. The B.1.1.7 (alpha) VOC has become one of the SARS-CoV-2 strains in the United Kingdom since its emergence in September 2020 due to its increased transmissibility [4]. The P.1 (gamma) and B.1.351 (beta) variants are listed as VOCs as they are associated with enhanced transmissibility and decreased effectiveness of available therapeutics against coronavirus disease 2019 (COVID-19) [5]. The B.1.617.2 (delta) variant discovered initially in India has become the most dominant strain that largely drives the second or third wave of infections in multiple countries [6, 7]. It is thus important to assess the possible effects of various mutations present in these VOCs on the host immune landscape. Intriguingly, VOCs Alpha, Gamma and Delta variants have a mutation that potentially truncates the ORF8 protein or renders it inactive.
Associated with host adaptation and viral replication [8–10], the open reading frame (ORF) 8 region was identified as a mutation hotspot in SARS-CoV during the 2002–2003 outbreak [9]. Deletion events in the SARS-CoV-2 ORF8 region have been reported in several countries, including Singapore and Taiwan (382-nucleotide (nt) deletion, termed Δ382 SARS-CoV-2), Bangladesh (345-nt deletion), Australia (138-nt deletion), and Spain (52-nt deletion) [11, 12]. Several mutations in SARS-CoV-2 ORF8 protein have also been observed in multiple virus strains [13–15] and these variants account for around 5% of infections worldwide [16]. Although truncated ORF8 variants contribute to milder infections [17], they were the major variants in Asia and North America during the early pandemic [18]. Understanding the natural biology of host immune response following infection with a SARS-CoV-2 variant that causes a milder disease phenotype will provide important insights into preventive and therapeutic strategies for patient management and improve COVID-19 prognosis. Moreover, the findings could provide clues on the high transmissibility of VOCs that are currently in circulation.
While in vitro studies have suggested that SARS-CoV-2 ORF8 downregulates major histocompatibility complex (MHC)-I molecules and inhibits type I interferon signaling pathway [19, 20], the functional impacts of ORF8 deletion on the cellular host immune response against SARS-CoV-2 is unknown [17]. To decipher the underlying molecular mechanisms of this natural genetic deletion, a comprehensive characterization of the whole blood transcriptomic profiles between coronavirus disease 2019 (COVID-19) patients infected with wildtype (WT) and Δ382 SARS-CoV-2 was performed in this study. High-density RNA-sequencing (RNA-seq) revealed upregulated eIF2 signaling and cellular stress responses, with an under-expression of neutrophil activation-associated signature in Δ382 SARS-CoV-2 infected patients. More robust T and B cell responses were observed, evidenced by enrichment of effector cytotoxic genes and upregulation of SARS-CoV-2 specific T cell immunity and antibody responses.
Methods
COVID-19 Patients
A total of 66 patients (WT, n = 36 and Δ382, n = 30) who tested PCR-positive for SARS-CoV-2 in nasopharyngeal swab samples was recruited into the study from February to April 2020 (Supplemental Table 1). Demographic data, clinical manifestations, and gene deletion status were obtained from patient records throughout hospitalization (Supplemental Table 1). Blood was collected in Cell Preparation Tubes (CPT; BD) from COVID-19 patients at acute and convalescent timepoints. Plasma fraction was extracted from CPT tubes for serology and multiplex microbead-based immunoassay while isolated peripheral blood mononuclear cells (PBMCs) were then used for T cell restimulation analysis. Whole blood samples of COVID-19 patients and healthy controls (Supplemental Table 2) were also collected into Tempus™ Blood RNA Tubes (Applied Biosystems) and stored at − 80 °C for transcriptomic profiling.
Detection of 382-nt Deletion in SARS-CoV-2 ORF8
Detection of the 382-nt deletion in COVID-19 patients was performed as previously described [11]. Briefly, confirmation of 382-nt deletion in SARS-CoV-2 ORF8 was performed by designing two specific PCR primers flanking the deleted region (F1 primer: 5′-TGTTAGAGGTACAACAGTACTTT-3′, and R1 primer: 5′-GGTAGTAGAAATACCATCTTGGA-3′). A hemi-nested PCR was performed with a different forward primer (F2 primer: 5′-TGTTTATAACACTTTGCTTCACA-3′) and R1 primer for samples with low cycle threshold (Ct) values. PCR products were visualized by gel electrophoresis, and 382-nt deletions were verified by Sanger sequencing.
RNA Extraction
Whole blood samples of 25 COVID-19 patients and six healthy controls were collected into Tempus™ Blood RNA Tubes (Applied Biosystems) and stored at − 80 °C. Patient samples at acute (SARS-CoV-2 PCR-positive) and recovered (SARS-CoV-2 PCR-negative) stages were selected for RNA extraction. Tempus™ Blood RNA Tubes were heat-inactivated at 60 °C for 30 min according to regulatory requirements, followed by RNA extraction using MagMAX™ for Stabilized Blood Tubes RNA Isolation Kit (Invitrogen) as per manufacturer’s instructions.
RNA-Sequencing (RNA-Seq)
Purified RNA was analyzed on Bioanalyser (Agilent) for quality assessment. RNA samples with RNA Integrity Number (RIN) of more than 6 were selected for the study (RIN ranging from 6.3 to 9.1 and with a median of 7.5) (Supplemental Table 1). cDNA libraries were prepared by Smart-Seq v2 [21], using a modification of the GlobinLock (GL) method [22] to block transcription of globin mRNA. Human “DNA 3 long A” and “DNA 3 long B” oligonucleotides (0.6 pmol each) were added to 2 ng of total blood RNA in 2.3 μL, denatured at 95 °C for 30 s, incubated at 60 °C for 10 min for GL oligo hybridization, and held at 42 °C for the loading of the reverse transcriptase (RT) mixture. RT and subsequent steps were according to Smart-Seq v2 with the following modifications: (1) addition of 20 μM template switching oligos (TSO) and (2) use of 200 pg cDNA with 1/5 reaction of Nextera XT Kit (Illumina). The length distribution of the cDNA libraries was monitored using a DNA High Sensitivity Reagent Kit on the LabChip (Perkin Elmer). All samples were subjected to an indexed paired-end sequencing run of 2 × 151 cycles on a HiSeq 4000 system (19 samples/lane; Illumina).
Bioinformatics and Differential Gene Expression Analysis
STAR aligner [23] was used to map paired-end raw reads to human genome build GRCh38 and counted for genes using featureCounts [24] based on GENCODE v31 gene annotation [25]. Log2 transformed counts per million mapped reads (log2CPM), and log2 transformed reads per kilobase per million mapped reads (log2RPKM) were computed using the edgeR Bioconductor package [26]. Sequencing coverage statistics are listed in Supplemental Table 3. Data are accessible at NCBI’s Gene Expression Omnibus (GEO) database (GSE155454). Genes with log2CPM inter-quartile range (IQR) of less than 0.5 across all samples were filtered out from subsequent differential expression gene (DEG) analysis. Respective DEG analyses for PCR (positive vs. negative), mutation (wildtype (WT) vs. Δ382), and disease severity profiles were done using edgeR [26]. DEG comparison analysis between PCR profiles was done using sample blocking model design by comparing paired samples from the same individual. Multiple testing correction was performed by using a false-discovery rate approach with the Benjamini–Hochberg method [27]. Principal Component Analysis (PCA) was performed on log2RPKM values using R function “prcomp.” All DEG analyses and PCA were done in the R statistical language (version 3.3.3) [28].
Integrative Pathway and Network Analysis
Biological processes, canonical pathways and upstream regulators were predicted from the DEGs with Ingenuity Pathway Analysis (IPA; Qiagen). Gene Ontology (GO) enrichment analysis (including biological processes, cellular component and molecular function categories) for DEGs was performed using Enrichr functional annotation tool [29], with the Fisher’s exact p-value set to < 0.01. The smallest p-value indicates the highest degree of enrichment. ClueGO (version 2.5.7), a plug-in app of Cytoscape (version 3.8.0; NIGMS; http://www.cytoscape.org/), was used to visualize and explore enriched pathways and biological terms related to DEGs. Heatmaps of log2RPKM values for DEGs were generated using ClustVis [30] and the rows are clustered using correlation distance and average linkage.
Multiplex Microbead-Based Immunoassay
Patient plasma samples were inactivated with Triton™ X-100 (Thermo Fisher Scientific) to a final concentration of 1% for 2 h in the dark. Measurement of immune mediators was done using the Cytokine/Chemokine/Growth Factor 45-plex Human ProcartaPlex™ (Thermo Fisher Scientific) with the Luminex™ assay [17]. Briefly, standards and plasma from COVID-19 patients and healthy controls were incubated with fluorescent-coded magnetic beads pre-coated with respective antibodies in a black 96-well clear-bottom plate overnight at 4 °C. After incubation, plates were washed five times with wash buffer (PBS with 1% BSA (Capricorn Scientific) and 0.01% Tween (Promega)). Sample-antibody-bead complexes were incubated with biotinylated detection antibodies for one hour and washed five times with wash buffer. Subsequently, streptavidin-PE was added and incubated for another 30 min. Plates were washed five times before sample-antibody-bead complexes were re-suspended in sheath fluid for acquisition on the FLEXMAP® 3D (Luminex) using xPONENT® 4.0 (Luminex) software. Data analysis was done on Bio-Plex Manager™ 6.1.1 (Bio-Rad). Standard curves were generated with a 5-PL (5-parameter logistic) algorithm; reporting mean fluorescence intensity (MFI) and concentration data values. The concentrations were logarithmically transformed to ensure normality. The logarithmically transformed value of Limit of Quantification (LOQ) was assigned to samples with concentrations out of the measurement range.
SARS-CoV-2-Specific T cells by Intracellular Cytokine Staining (ICS)
To profile the SARS-CoV-2 specific T effector subsets in COVID-19 patients, frozen PBMCs from the first convalescent timepoint (median 19.5 days PIO, IQR 16–26) were thawed and rested overnight at 37 °C in RPMI 1640 (Hyclone) supplemented with 5% human serum (Innovative Research), followed by stimulation with phorbol 12-myristate 13-acetate (PMA 100 ng/mL, Sigma Aldrich) and ionomycin (1 µg/mL, Sigma Aldrich), or pooled SARS-CoV-2 PepTivator S, S1, M and N peptides (0.6 nmol/mL each) (Miltenyi Biotec) for 6 h. Brefeldin A and Monesin (1 × , ThermoFisher Scientific) were added at 2 h post-stimulation. Cells were stained for surface markers in the dark at room temperature for 30 min (Supplemental Table 4), followed by fixation and permeabilization for 30 min with Foxp3/Transcription Factor Staining Buffer Set (ThermoFisher Scientific). Permeabilized cells were then stained for intracellular cytokines in the dark at room temperature for 30 min (Supplemental Table 4). Cells were then washed with PBS and centrifuged at 800 × g for 5 min before transferring to respective polystyrene FACS tubes containing 5 μL (5.4 × 103 beads) of CountBright Absolute Counting Beads (Invitrogen). Cells were acquired with the Cytek Aurora cytometer (Cytek Biosciences) and analyzed using FlowJo (Tree Star).
Serology Profiling
Antibody response against the full-length SARS-CoV-2 spike protein was examined using an S protein flow-based (SFB) assay [31]. Cells expressing full-length SARS-CoV-2 spike protein were seeded at 1.5 × 105 cells per well in 96 well plates (Thermo Fisher Scientific). The cells were first incubated with plasma samples from COVID-19 patients and healthy controls (1:100 dilution in 10% fetal bovine serum, FBS), followed by secondary incubation with a double stain, consisting of Alexa Fluor 647-conjugated anti-human IgM or IgG (Thermo Fisher Scientific; 1:500 dilution in 10% FBS) and propidium iodide (Sigma-Aldrich; 1:2500 dilution). Cells were acquired on LSRII 4 Laser flow cytometer (BD Biosciences) and analyzed using FlowJo (Tree Star). A positive antibody response cut-off is defined as the healthy controls’ mean + 3 standard deviations (SD).
To further define the antibody response against the SARS-CoV-2 spike protein, a peptide-based ELISA was conducted against two epitopes, S14P5 and S21P2, to determine patients’ IgG antibody response against SARS-CoV-2 [32, 33]. Briefly, 50 μL of 0.5 μg/mL of NeutrAvidin protein (Thermo Fisher Scientific) was coated on Nunc Maxisorp flat-bottom 96-well plates (Thermo Fisher Scientific) overnight at 4 °C. Blocking was done with 0.01% polyvinyl alcohol (PVA; Sigma-Aldrich) in PBS containing 0.1% Tween-20 (0.1% PBST; Sigma-Aldrich). Biotinylated peptides (GenScript; 1:2000 dilution in 0.1% PBST) were added to wells, followed by 1% Triton X-100-inactivated plasma samples from COVID-19 patients and healthy controls (1:1000 dilution in 0.1% PBST), and horseradish peroxidase (HRP)-conjugated goat anti-human IgG (H + L) (Jackson ImmunoResearch) in blocking buffer. For color development, 3,3′,5,5′-tetramethylbenzidine (TMB; Sigma-Aldrich) was added for 5 min, and the reaction stopped with 0.16 M sulfuric acid (Merck). Absorbance was measured on an Infinite M200 plate reader (Tecan) at 450 nm and a reference wavelength of 690 nm. Before subtracting background signals, raw optical density (OD) values were normalized to a positive control to account for plate-to-plate variations. Incubation was done at 37 °C for one hour in between steps.
Data and Statistical Analyses
Data analyses were done using GraphPad Prism (GraphPad Software, version 8.4.3). Mann–Whitney U tests were conducted on the logarithmically transformed concentration of immune mediators. Comparison of SARS-CoV-2 specific T cell responses and serological profiles between WT- and Δ382-infected patients were analyzed by Mann–Whitney U tests. A cut-off value of mean + 3SD of healthy controls was used as a baseline to classify the serological profile of COVID-19 patients as positive or negative [32, 33].
Results
Wildtype and Δ382 SARS-CoV-2 Infections Activate TLR and PRR Pathways and Antiviral Interferon Responses in COVID-19 Patients
We herein studied 30 Δ382 SARS-CoV-2 infected patients and compared their transcriptomic signatures, systemic soluble immune mediator levels, and adaptive immune responses against 36 WT infected patients (Supplemental Table 1). To uncover the molecular mechanisms underlying the milder disease phenotype in Δ382 SARS-CoV-2 infections [17], RNA-seq of whole blood from 25 COVID-19 patients was performed (WT, n = 14 and Δ382, n = 11) in this study (Supplemental Table 1).
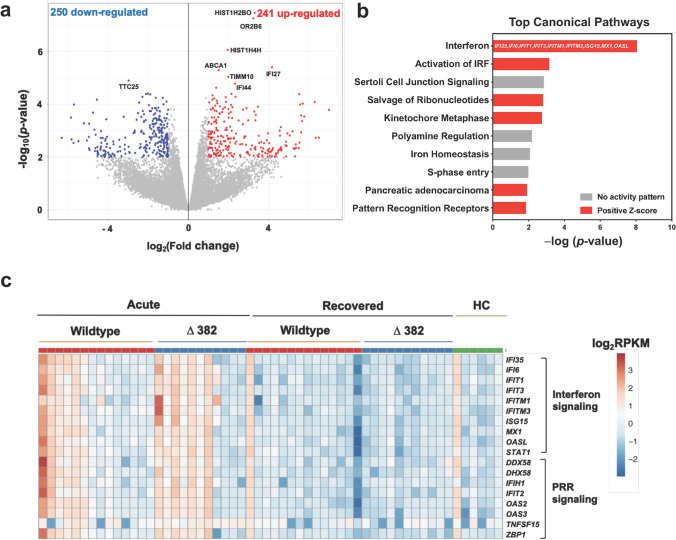
Firstly, a comparison of transcriptomic profiles during the acute (SARS-CoV-2 PCR-positive, median 8 days post-illness onset [PIO]) and recovered (SARS-CoV-2 PCR-negative, median 21 days PIO) phases of disease was performed on 13 WT SARS-CoV-2 infected patients with paired samples to identify transcriptomic changes specific to virus infection. Following correction for multiple testing (q-value ≤ 0.05 and |FC| > 2), only eight genes remained significant, as highlighted in the volcano plot (Fig. 1a). To gain an understanding of the underlying mechanisms, less stringent criteria of p < 0.01 and |FC|> 2 was applied to pathway and Gene Ontology (GO) analyses [34]. This yielded 491 significant transcripts, in which 241 differentially expressed genes (DEGs) were found to be enriched, whereas 250 were downregulated (Fig. 1a, Supplemental Table 5) during the acute phase of infection.
Fig. 1.
Whole blood transcriptome analysis in COVID-19 patients. RNA-seq of whole blood from COVID-19 patients (n = 25) at acute (SARS-CoV-2 PCR-positive, median 8 days PIO) and recovered (SARS-CoV-2 PCR-negative, median 21 days PIO) phases and healthy controls (n = 6) was performed. a Volcano plot indicating DEGs between blood samples collected at acute and recovered phases in patients with WT SARS-CoV-2 infection (n = 13), with thresholds of p-value < 0.01 and |FC|> 2. Numbers of over-expressed and under-expressed genes are indicated. b Top IPA canonical pathways showing differential expression of genes related to IFN and PRR signaling in WT SARS-CoV-2 infection. Pathways are ranked by − log(p-value), and the color scheme is based on predicted activation Z-scores, with activation in red and undetermined directionality in gray. DEGs related to the IFN pathway are indicated on the bar graph. c Heatmap of DEGs related to IFN and PRR signaling between COVID-19 patients infected with WT (n = 14) and Δ382 SARS-CoV-2 (n = 11) at acute and recovered phases and healthy controls. Heatmap is scaled based on log2RPKM values, with blue and red indicating low and high expressions, respectively. WT, wildtype; DEGs, differentially expressed genes; FC, fold change; FDR, false discovery rate; PCR, polymerase chain reaction; PIO, post-illness onset; HC, healthy controls; RPKM, reads per kilobase per million reads mapped; IFN, interferon; PRR, pattern recognition receptor
To elucidate the biological processes and pathways of the involved DEGs, further analyses were performed using GO enrichment and Ingenuity Pathway Analysis (IPA) on the dataset of 491 DEGs. Gene functional enrichment analysis revealed that robust type I IFN, classical complement, and humoral and cellular immune responses were induced, whereas biological processes such as “target of rapamycin (TOR) signaling,” “protein catabolic process,” and “positive regulation of autophagy” were shown to be downregulated in the acute phase of WT SARS-CoV-2 infection (Supplemental Fig. 1). IPA analysis revealed IFN signaling, IFN regulatory factor (IRF) activation, pattern recognition receptors (PRR)-induced signaling and salvage pathway of ribonucleotides as the top canonical pathways induced upon SARS-CoV-2 infection (Fig. 1b). Overall, the results showed that antiviral IFN responses and virus sensing PRR signaling pathways were activated following WT SARS-CoV-2 infection (Fig. 1c). A differential response observed among patients in both WT and Δ382 SARS-CoV-2 groups might be explained by the heterogeneity of disease progression in these patients, whereby those with a more severe disease outcome demonstrated more robust type I interferon responses [35, 36].
Δ382 SARS-CoV-2 Infection Upregulates eIF2 Signaling and Cellular Stress Responses in COVID-19 Patients
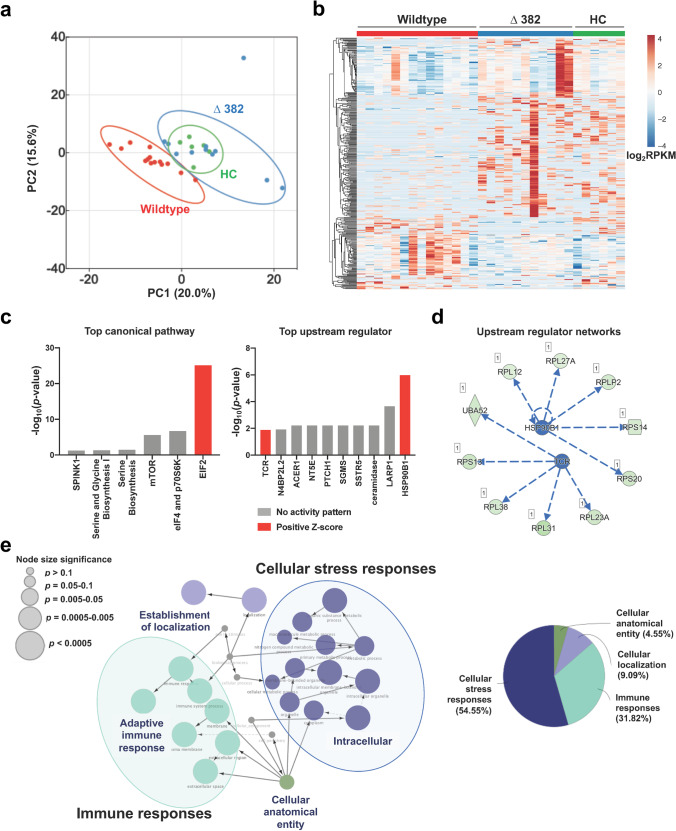
A comparative analysis was next carried out to identify specific differences in the whole blood transcriptomes of patients during the acute phase of WT (median 9.5 days PIO, n = 14) and Δ382 SARS-CoV-2 infection (median 6 days PIO, n = 11), as well as healthy controls (n = 6; Supplemental Table 2). With the cut-off criteria of p-value < 0.01 and |FC|> 2 [34], 358 DEGs were identified, in which 259 were over-expressed, and 99 were under-expressed in Δ382 SARS-CoV-2 infected patients (Supplemental Table 6). Principal component analysis (PCA) showed a clear distinction between the two groups of patients, while Δ382 SARS-CoV-2 infected patients overlapped with healthy controls (Fig. 2a). Notably, gene expression signatures comprising significant DEGs illustrated contrasting whole blood transcriptomic responses between WT and Δ382 SARS-CoV-2 infections, with the profile of Δ382 SARS-CoV-2 being closer to that of the healthy uninfected controls (Fig. 2b). Further analysis of the 358 DEGs revealed eight significant canonical signaling pathways, with the eukaryotic initiation factor 2 (eIF2) signaling pathway being the most significantly enriched in Δ382 SARS-CoV-2 infection (Fig. 2c).
Fig. 2.
Effects of 382-nt deletion in SARS-CoV-2 ORF8 genome (Δ382) on whole blood transcriptome of COVID-19 patients. RNA-seq of whole blood from COVID-19 patients infected with WT (n = 14) and Δ382 SARS-CoV-2 (n = 11) at the acute phase of infection (SARS-CoV-2 PCR-positive; median 8 days PIO) was performed. Only samples with RNA integrity number > 6 were sent for sequencing and included in the analysis a PCA of COVID-19 patients and healthy controls based on DEGs, with p-value < 0.01 and |FC|> 2. b Heatmap of 358 DEGs, scaled based on log2RPKM values, with blue and red colors indicating low and high expressions, respectively. c Top canonical pathways and upstream regulators identified by IPA based on the DEGs. Bar graphs are ranked by significance, with red indicating positive predicted activation Z-scores and gray indicating undetermined directionality. d An integrated network of HSP90B1 and TCR and their targeted genes. Stimulation of HSP90B1 and TCR leads to overexpression of the downstream genes. e GO pathway term enrichment networks of DEGs using Cytoscape add-on ClueGO. Each of the GO terms is statistically significant (Benjamini–Hochberg correction < 0.05). The filled colored circles (nodes) represent a statistically significant enriched parent GO term. The lines (edges) between nodes show overlapping genes within terms, with node size representing the term enrichment significance. The overview chart shows the distribution of the functionally grouped GO terms. The cut-off for terms in the functionally grouped networks was set at p-value < 0.05. WT, wildtype; PCA, principal component analysis; DEGs, differentially expressed genes; FC, fold change; RPKM, reads per kilobase per million reads mapped; PCR, polymerase chain reaction; IPA, Ingenuity Pathway Analysis; HSP90B1, heat shock protein 90 kDa beta member 1; TCR, T cell receptor; GO, gene ontology
In addition, Heat Shock Protein 90 Beta Family Member 1 (HSP90B1) and T Cell Receptor (TCR) were predicted to be highly stimulated in Δ382 SARS-CoV-2 infected patients compared to WT patients (Fig. 2c). Stimulation of both HSP90B1 and TCR formed a regulatory network involving ribosomal stress-induced related genes (Fig. 2d). Moreover, a network‑based analysis of GO terms (biological, cellular, and molecular processes) using ClueGO (plugged into Cytoscape) showed that cellular stress and immune responses (54.55% and 31.82%, respectively) were significantly enriched in the transcriptomic profiles of Δ382 SARS-CoV-2 infected patients, as compared to WT patients (Fig. 2e).
Infection with Δ382 SARS-CoV-2 Is Associated with Lower Activation of Neutrophils and More Robust T cell Immunity
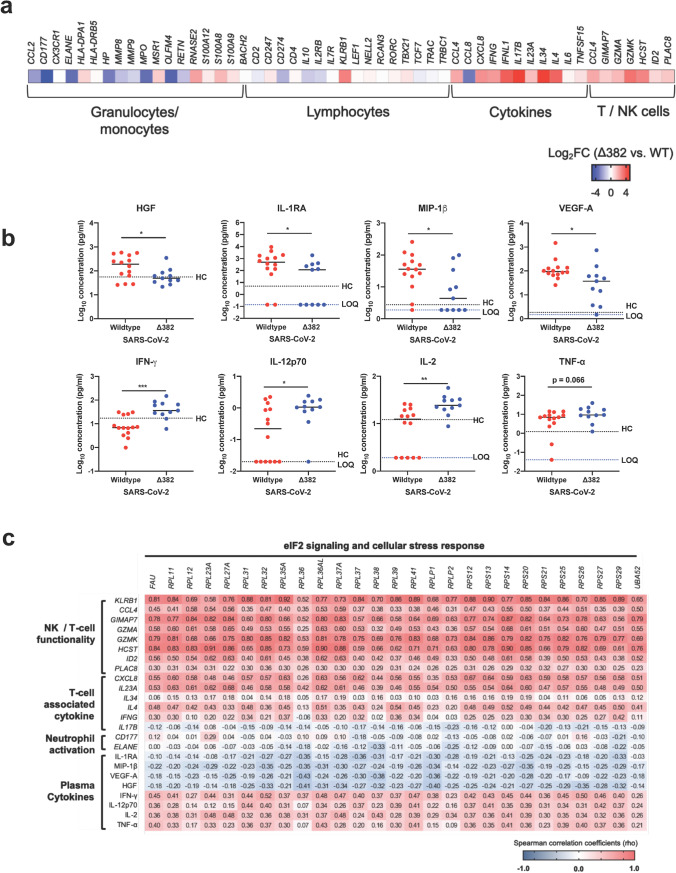
Next, previously reported DEGs in the whole blood transcriptomes of COVID-19 patients [37, 38] were compared against the WT (n = 14) and Δ382 SARS-CoV-2 (n = 11) infected patients in our study. Increased gene expression was found for granulocyte- and monocyte-associated molecules (eosinophil-derived neurotoxin; RNASE2) and lymphocyte-associated molecules (natural killer cell surface protein P1A; KLRB1) in Δ382 SARS-CoV-2 infected patients (Fig. 3a). Notably, lower expressions of neutrophil activation-associated CD177 and neutrophil elastase (ELANE) were observed in Δ382 SARS-CoV-2 infected patients (Fig. 3a). Moreover, a higher expression of T cell cytokine genes (CCL4, CXCL8, IFNG, IL17B, IL23A, IL34) and genes associated with T/NK cell functionality (CCL4, CIMAP7, GZMA, GZMK, HCST, ID2, PLAC8) was also observed in Δ382 SARS-CoV-2 infected patients (Fig. 3a).
Fig. 3.
Effects of 382-nt deletion in SARS-CoV-2 ORF8 genome on immune responses in COVID-19 patients. Transcriptomic and cytokine profiles of COVID-19 patients infected with WT (n = 14) or Δ382 SARS-CoV-2 (n = 11) at the acute phase of infection (SARS-CoV-2 PCR-positive; median 8 days PIO) were analyzed. a Expressions of genes associated with granulocytes, monocytes, lymphocytes, cytokines and T/NK cell functionality were compared between COVID-19 patients infected with WT or Δ382 SARS-CoV-2. Heatmaps of the DEGs, scaled based on log2FC values, with blue and red colors indicating low and high expressions, respectively. b Plasma immune mediator levels of COVID-19 patients infected with WT (n = 14) or Δ382 SARS-CoV-2 (n = 11) at the acute phase of infection (SARS-CoV-2 PCR-positive; median 8 days PIO) and profiles of significant immune mediators are illustrated as scatter plots and shown as mean. Mann–Whitney U tests were conducted on the logarithmically transformed concentration values (*p < 0.05; **p < 0.01; ***p < 0.001). Respective mean concentrations of immune mediators from healthy controls (HC; n = 23) are indicated as black dotted lines. Patient samples with concentrations out of measurement range are presented as the logarithmically transformed value of LOQ and indicated as blue dotted lines. c Association between elF2 signaling and immune signatures in Δ382 SARS-CoV-2 infection. Spearman’s correlation matrix for the genes associated with eIF2 signaling, T cell functionality, neutrophil activation and plasma cytokines. Colors represent the Spearman correlation coefficients (rho) between the expression of genes related to eIF2 signaling and genes or immune mediators associated with different immune phenotypes. FC, fold change, WT, wildtype; PCR, polymerase chain reaction; PIO, post-illness onset; DEGs, differentially expressed genes; LOQ, limit of quantification
In addition to the increased expression of T cell activation associated genes, quantified acute plasma immune mediator levels in patients also showed higher levels of T cell-associated cytokines (IFN-γ, IL-12p70, IL-2, and TNF-α), with lower levels of chemokines (MIP-1β) and disease severity associated cytokines (HGF, IL-1RA, and VEGF-A) [39] (Fig. 3b and Supplemental Table 7). Interestingly, correlation analysis also showed that the levels of genes related to T cell functionality and neutrophil activation, together with T cell-associated and pro-inflammatory immune mediators, are associated with expression levels of genes related to cellular stress responses and eIF2 signaling (Fig. 3c). Increased activation of eIF2 signaling is associated with enhanced T cell function and reduced inflammatory response, and neutrophil activation.
Δ382 SARS-CoV-2 Infected Patients Have more Robust Virus-Specific Adaptive Immune Responses
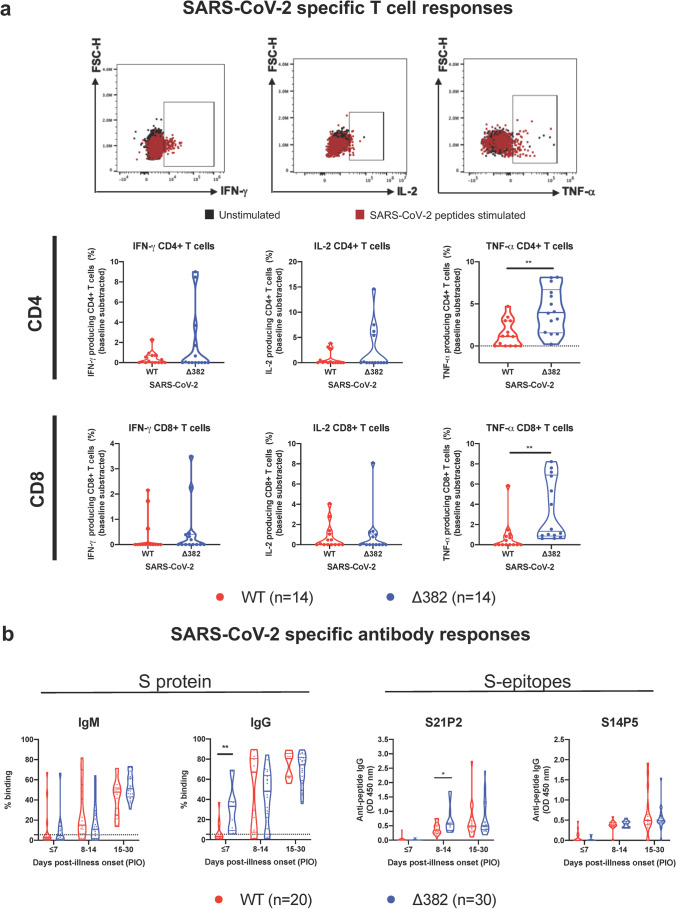
To further assess the difference in T cell responses, PBMCs of 28 patients collected shortly after acute infection (median 19.5 days PIO, IQR 16–26) were stimulated with pooled pan-SARS-CoV-2 peptides or with PMA/ionomycin [40]. Intracellular cytokine staining demonstrated virus-specific CD4+ and CD8+ T cells responses in these patients. Δ382 SARS-CoV-2 infected patients had higher CD4+ and CD8+ T cells expressing TNF-α than WT infected patients following peptide stimulation (Fig. 4a, Supplemental Fig. 2), indicating a more robust SARS-CoV-2 specific T cell response in patients with mutant virus infection. Although not significant, they also had higher IL-2 and IFN-γ expressing virus-specific CD4+ and CD8+ T cells. Intriguingly, the general Th1 responses following PMA/ionomycin stimulation were higher in WT infection compared to Δ382 SARS-CoV-2 infection (Supplemental Fig. 3). WT infected patients also showed higher IL-17+ CD4+ T cells but with lower IL-4+/IL-6+ CD4+ T cells compared to patients with mutant virus infection.
Fig. 4.
Effects of 382-nt deletion in SARS-CoV-2 ORF8 genome on adaptive immune responses to COVID-19. a SARS-CoV-2 specific CD4+ and CD8+ non-T follicular helper (TFH) cells were characterized with flow cytometry-based on the expression of IFN-γ, IL-2, and TNF-⍺ upon SARS-CoV-2 peptide stimulation in WT (n = 14) or Δ382 SARS-CoV-2 (n = 14) infected patients. Statistical analyses were performed with the Mann–Whitney U test (**p < 0.01). b Antibody responses in Δ382 SARS-CoV-2 infected patients. Spike protein-specific antibody response was characterized using an S-flow assay. Plasma samples of COVID-19 patients infected with either WT (n = 20) or Δ382 SARS-CoV-2 (n = 30) were screened at 1:100 dilution against cells expressing the full-length SARS-CoV-2 spike protein, with healthy donors (n = 22) screened in parallel. IgM and IgG profiles of COVID-19 patients at timepoints ≤ 7, 8 to 14, and 15 to 30 days PIO are illustrated as violin plots. The dotted line indicates the mean + 3SD of healthy donors. Data are shown as mean ± SD of two independent experiments. For determination of anti-peptide IgG responses, plasma samples o COVID-19 patients infected with either WT (n = 20) or Δ382 SARS-CoV-2 (n = 30) were tested at 1:1000 dilution on an IgG ELISA against SARS-CoV-2 spike epitopes S21P2 and S14P5. Healthy controls (n = 22) were screened in parallel. Antibody profiles of COVID-19 patients at timepoints ≤ 7, 8 to 14, 15 to 30 days PIO are illustrated as violin plots. The dotted line indicates the mean + 3SD of healthy controls. Data are shown as mean ± SD of two independent experiments. Statistical analysis was carried out using Mann–Whitney U tests (*p < 0.05). WT, wildtype
In concordance with robust SARS-CoV-2 specific T cell response, further characterization of humoral response with the S-flow assay [41] revealed a higher level of detectable IgG response against the SARS-CoV-2 spike protein in Δ382 SARS-CoV-2 patients (n = 30) compared to WT infected patients (n = 20) at the acute phase of infection (< 7 days PIO). However, this phenomenon was not observed at later time points (Fig. 4b). In addition, a peptide-based ELISA against two previously identified immunodominant B-cell linear epitopes, S21P2 and S14P5 on the spike glycoprotein [32, 33], revealed that the IgG levels against S21P2 were significantly higher in Δ382 SARS-CoV-2 infected patients at the later PIO) (Fig. 4b).
Discussion
Understanding the impact of virus mutation on the pathophysiology of COVID-19 is imperative to finding new treatments and providing essential clues on its potential impact on transmission rates and disease severity. Here, we analyzed in detail the peripheral blood transcriptomes and adaptive immune responses of COVID-19 patients with Δ382 SARS-CoV-2 infection and elucidated several key immune responses potentially associated with the deletion of SARS-CoV-2 ORF8.
This study reveals distinctly different transcriptomic profiles between WT and Δ382 SARS-CoV-2 infections, with the Δ382 eliciting a more active cellular stress response and an upregulated eIF2 signaling in the infected patients. Studies have demonstrated that coronaviruses induce cellular stress responses following infection [42–44] by targeting unfolded protein response (UPR) pathways to cause an imbalance in cellular homeostasis, which subsequently induces endoplasmic reticulum (ER) stress that favors viral replication [45]. While SARS-CoV ORF8 protein has been previously reported to induce ER stress by specifically targeting activating transcription factor 6 (ATF6) [46], our results suggest that the SARS-CoV-2 ORF8 protein potentially triggers protein kinase RNA-like ER kinase (PERK) and eIF2 signaling mechanisms, further strengthening an earlier report showing interactions between SARS-CoV-2 ORF8 protein and human proteins involved in ER quality control [47]. ER stress is known to propagate inflammatory responses [48, 49], which is consistent with our findings showing reduced systemic inflammation and ER stress levels in Δ382 SARS-CoV-2 infected patients (Fig. 5). Notably, there is still controversy amongst the literature about the susceptibility of human PBMCs to SARS-CoV-2 [50, 51]. Although SARS-CoV-2 was detected in immune cells of COVID-19 patients [51–53], it remains to be determined whether enhanced activation of the eIF2 pathway in Δ382 SARS-CoV-2 infected patients is directly caused by a loss of ORF8’s impact on ER quality control or due to secondary peripheral immune responses to the mutant virus infection, mediated by soluble factors.
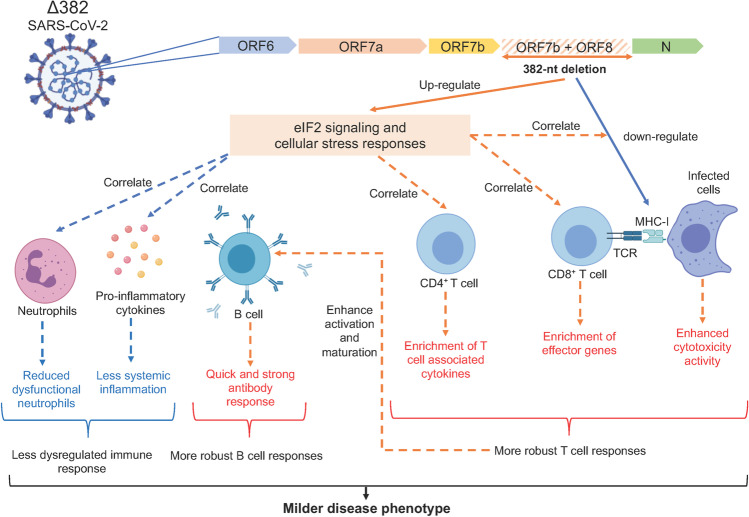
Fig. 5.
Molecular mechanisms underlying the milder disease phenotype in Δ382 SARS-CoV-2 infections. A SARS-CoV-2 variant with a 382-nucleotide deletion (Δ382) truncates ORF7b and removes the ORF8 transcription-regulatory sequence, eliminating ORF8 transcription. The ORF8 382-nt deletion has recently been associated with a milder disease phenotype. The attenuation of SARS-CoV-2 ORF8 upregulates eIF2 signaling and cellular stress responses at the acute phase of infection, potentially interrupting the downregulation of MHC-I molecules by ORF8 and also enhances the activation of both CD4+ and CD8+ T cells, evidenced by enrichment of effector cytotoxic genes and upregulation of SARS-CoV-2 specific T cell responses in Δ382 SARS-CoV-2 infected patients. Enhanced T cell responses may in turn mediate rapid and effective antibody responses in Δ382 SARS-CoV-2 infection. More pronounced cellular stress responses may further reduce systemic inflammation and dysfunctional neutrophils in Δ382 SARS-CoV-2 infected patients. Overall, the attenuation of SARS-CoV-2 ORF8 produced a molecular phenotype characterized by more pronounced cellular stress responses and a less dysregulated immune phenotype with more robust T and B cell responses. ORF, open reading frame; eIF2, eukaryotic initiation factor 2; MHC-I, major histocompatibility complex 1; CD, cluster of differentiation
SARS-CoV-2 ORF8 has also been reported to interact with MHC-I molecules [20] and subsequently downregulate cytotoxic functions of T lymphocytes. Cytotoxic effector genes (GZMA, GZMB, ID2, and PLAC8) were enriched in Δ382 SARS-CoV-2 infected patients, coupled with high plasma levels of IFN-γ, TNF-α, and IL-2 during the acute phase of virus infection. In addition to robust cytotoxic T cell functions, enrichment of KLRB1, GZMA, and GZMK transcripts may indicate enhanced NK cell cytotoxic activity in Δ382 SARS-CoV-2 infected patients, in which the function is impaired in severe COVID-19 patients [54]. Our findings are in agreement with other single-cell studies reporting an enrichment of effector populations with a cytotoxic phenotype (effector CD8+, MAIT and NK T cells) in COVID-19 individuals with milder disease phenotype [55, 56] and further highlight the impact of SARS-CoV-2 ORF8 on cytotoxic cellular responses in COVID-19 (Fig. 5). A higher magnitude of virus-specific T cells was induced following Δ382 infection, and this is in line with other studies in which highly functional virus-specific cellular immune response resulted in better disease outcomes in COVID-19 [57, 58]. It is important to note that genetic variability of the cohort could also explain some of the differences in the T cell response since a few human leukocyte antigen (HLA) alleles, which are predominant in Asia, are associated with COVID-19 severity [59, 60]. Additionally, enhanced effector functions of virus-specific T cells may in turn mediate rapid and protective antibody responses against SARS-CoV-2 infection [61]. Concordantly, higher IgG responses during the early phase of disease were observed in Δ382 SARS-CoV-2 infected patients, which could indicate a more robust CD4+ T cell response driving B cell activation and maturation in these patients [62, 63] (Fig. 5). Thus, deletion of ORF8 could result in increased immunogenicity against SARS-CoV-2. Intriguingly, while IgG levels at the later phase of infection have been associated with severe COVID-19 [33, 64], Δ382-infected patients with a milder disease phenotype in this report had higher IgG levels at the early acute phase of infection. Our observations are consistent with the findings, which found that S-specific antibody responses were elevated early in COVID-19 individuals who recovered from the disease compared to deceased patients [65]. Further work to fully define the exact roles of IgG in SARS-CoV-2 infection will bring additional insights into this phenomenon.
The increased effectiveness of the virus-specific adaptive T and B cell responses may explain the reduced need for sustained, pathogenic pro-inflammatory responses. Δ382 SARS-CoV-2 infected patients had lower pro-inflammatory cytokines, chemokines and growth factors strongly associated with severe COVID-19 [66, 67]. The N protein of SARS-CoV-2 was reported to promote inflammation by increasing IL-6 levels following virus infection [68]. Meanwhile, we did not observe any significant difference in the IL-6 levels between the WT and Δ382 infected patients [17], suggesting differential roles of ORF8 in inducing hyperinflammation in COVID-19. More interestingly, general pro-inflammatory Th1 responses were more robust in WT infected patients. The non-specific and uncontrolled activation of CD4+ T cells maybe the cause and effect of heightened inflammation observed in WT infection. Lymphopenia and dysregulated myelopoiesis with immature and dysfunctional neutrophils have also been associated with COVID-19 severity [66, 69–72]. While lymphocyte and neutrophil counts of Δ382 SARS-CoV-2 infected patients did not significantly differ from WT infected patients [17] (Supplemental Table 1), Δ382 infected patients demonstrated molecular signatures characterized by reduced dysfunctional neutrophils and more robust T cell responses (Fig. 5).
Similar to other coronaviruses [73, 74], SARS-CoV-2 activates cellular viral sensors, host defenses and IFN responses during an active virus infection. Despite previous reports showing SARS-CoV-2 ORF8 as a potent IFN antagonist [75], there were no significant differences in the host innate antiviral responses between WT and Δ382 groups in this study (Fig. 1c). This indicates potential functional compensation for the loss of ORF8 by other potent interferon antagonists such ORF3b and ORF6 [75] in the Δ382 SARS-CoV-2 infections.
While mutations on spike protein in SARS-CoV-2 resulted in more efficient virus transmission and severe disease outcomes [76–78], the ORF8 deletion does not appear to increase viral load in patients [11] and is associated with milder disease [17]. While in vitro infection of Δ382 in human nasal epithelial cells induced similar inflammatory signatures [79], it was not clear how the immune profiles in patients would be affected with the absence of ORF8. Nonetheless, putting all the data together, it is clear that the disease phenotypic difference is likely due to the functional implication of ORF8 on the host immune system. Δ382 SARS-CoV-2 viruses likely stimulate the activation of common host response signaling mechanisms similar to WT viruses, but with different degrees of magnitude. Our key findings of the functional implication of ORF8 on host immune surveillance further define the relevance of inhibiting ORF8 function as a possible target for therapeutic intervention in COVID-19. Antiviral drugs can be developed against the SARS-CoV-2 ORF8 protein. However, the hypervariable nature of the ORF8 gene and the rapid evolution it undergoes can compromise the suitability of the ORF8 protein as an antiviral target. Alternatively, host-directed strategies [47] can be developed to target the host factors to which ORF8 establishes critical interactions. In this case, a drug that can modulate ER stress responses may benefit the outcome of COVID-19 [80]. Interestingly, the VOCs that have become dominant as the pandemic progresses bear the ORF8 mutation together with multiple spike mutations. The asymptomatic or mild disease phenotype attributed to the absence of ORF8 and the enhanced infectivity caused by the spike mutations may explain the high transmissibility of these VOCs. Monitoring ORF8 mutation throughout the progression of the COVID-19 pandemic is therefore important, particularly when associated with relevant spike mutations.
Our study has caveats that should be noted. Due to Singapore’s strict and efficient public health measures during the pandemic in 2020, the cases of Δ382 SARS-CoV-2 infections were kept to a low number. As such, we only had a limited number of patients with the mutant virus infection in this study. In addition, host genetic background and differences in variables such as sex and comorbidities between the WT and Δ382 groups, although not significant (Supplemental Table 1), may be confounding in this study. Nevertheless, our observations provide exciting insights into the potential impacts of ORF8 deletion on human immune responses during SARS-CoV-2 infection, and this well-characterized dataset would value-add to the current COVID-19 resource. Future studies with engineered viruses and animal models could further address the interactions and mechanisms between ORF8 and ER stress or T cell responses during COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Supplemental Fig. 1 GO-term enrichment of DEGs in whole blood of COVID-19 patients infected with WT SARS-CoV-2 during the acute phase of infection (SARS-CoV-2 PCR-positive; median 8 days PIO, n=13). GO-term functional enrichments for biological process, molecular function, and cellular component were performed for both up-regulated and down-regulated genes. Pathways are ranked by –log10(p-value). GO, gene ontology; DEGs, differentially expressed genes; WT, wildtype; PCR, polymerase chain reaction; PIO, post-illness onset. Supplemental Fig. 2 Representative gating strategy for the characterization of IFN-γ, IL-2, TNF-⍺ expression of CD4+ and CD8+ T cells in isolated PBMCs of COVID-19 patients by flow cytometry upon peptide stimulation. Supplemental Fig. 3 WT SARS-CoV-2 infected patients exhibit a higher systemic non-specific Th1 response, compared to Δ382 SARS-CoV-2 infected patients. CD4+ non T follicular helper (TFH) cells were characterized based on the expression of IFN-γ, IL-2, TNF-⍺, IL-17A, IL-4, IL-6 and IL-10 upon PMA/ Ionomycin stimulation. Statistical analyses were performed with unpaired test (*p < 0.05; **p < 0.01; ***p < 0.001).WT, wildtype. (PDF 2108 KB)
Acknowledgements
The authors would like to thank the participants who donated blood samples to this study and the healthcare workers caring for COVID-19 patients. The authors also wish to thank the team at NCID, National University Hospital and Singapore General Hospital, for their help in patient recruitment, and Dr. Danielle Anderson and her team at Duke-NUS Medical School for their technical assistance. The authors are also grateful to Dr. Olaf Rötzschke, Wilson How, and Norman Leo Fernandez from the Singapore Immunology Network (SIgN) Multiplex Analysis of Proteins (MAP) platform, for their assistance in running multiplex microbead-based immunoassay.
Author Contribution
SWF and NKWY conceptualized, acquired, analyzed, interpreted the data, and wrote the manuscript. YHC, YSG, and SNA acquired, analyzed, interpreted the data, and wrote the manuscript. NA, MPR, JL, SHF, CYPL, GC, RSLC, ATR, MZT, ZWC, CMP, and BL acquired and analyzed the data. BEY, PAT, SK, YSL, and DCL designed and supervised sample collection. SB, SWH, LR, and LFPN conceptualized, designed, analyzed the data, and wrote the manuscript. All authors revised and approved the final version of the manuscript.
Funding
The study was supported by core and COVID-19 (project number H20/04/g1/006) research grants provided to Singapore Immunology Network by the Biomedical Research Council (BMRC) and A*ccelerate GAP-funded project (ACCL/20-GAP001-C20H-E) from the Singapore Ministry of Health’s National Medical Research Council. This study was also funded by the National Medical Research Council (NMRC) COVID-19 Research fund (COVID19RF-001, COVID19RF-007 and COVID19RF-060). SIgN Immunomonitoring Platform is supported by a BMRC IAF 311006 grant and BMRC transition funds #H16/99/b0/011. ATR is supported by the Singapore International Graduate Award (SINGA), A*STAR.
Data Availability
RNA-seq data are accessible at NCBI’s Gene Expression Omnibus (GEO) database (GSE155454). All data are available upon request: Lisa F.P. Ng, lisa_ng@IDLabs.a-star.edu.sg.
Code Availability
Not applicable.
Declarations
Ethics Approval and Consent to Participate
The study design and protocols for the COVID-19 PROTECT study group were evaluated by National Healthcare Group (NHG) Domain Specific Review Board (DSRB) and approved under study number 2012/00917. Collection of healthy donor samples was approved by SingHealth Centralised Institutional Review Board (CIRB) under study number 2017/2806 and NUS IRB 04–140. Written informed consent was obtained from participants in accordance with the Declaration of Helsinki for Human Research.
Consent for Publication
The authors affirm that human research participants provided informed consent for publication of the data.
Conflict of Interest
A patent application for the SFB assay has been filed (Singapore patent 10202009679P: A Method Of Detecting Antibodies And Related Products. YSG, LFPN, and LR). A patent application on the identified linear epitopes S14P5 and S20P2 has also been filed (PCT/SG2021/050178: Antibody-binding linear B cell epitopes of SARS-CoV and SARS-CoV-2. NKWY, SNA, GC, CMP, CYPL, RSLC, LR and LFPN). All other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Siew-Wai Fong and Nicholas Kim-Wah Yeo contributed equally to this work.
Change history
11/27/2021
A Correction to this paper has been published: 10.1007/s10875-021-01184-3
References
- 1.Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proceedings of the National Academy of Sciences of the United States of America. 2005. [DOI] [PMC free article] [PubMed]
- 2.Zhang CY, Wei JF, He SN. Adaptive evolution of the spike gene of SARS coronavirus: changes in positively selected sites in different epidemic groups. BMC Microbiology. 2006. [DOI] [PMC free article] [PubMed]
- 3.Webby R, Hoffmann E, Webster R. Molecular constraints to interspecies transmission of viral pathogens. Nat Med. 2004;10(12 Suppl):S77–81. doi: 10.1038/nm1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage-United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(3):95–9. [DOI] [PMC free article] [PubMed]
- 5.Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–51 e4. [DOI] [PMC free article] [PubMed]
- 6.Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7). [DOI] [PMC free article] [PubMed]
- 7.Dougherty K, Mannell M, Naqvi O, Matson D, Stone J. SARS-CoV-2 B.1.617.2 (Delta) Variant COVID-19 outbreak associated with a gymnastics facility-Oklahoma, April-May 2021. MMWR Morb Mortal Wkly Rep. 2021;70(28):1004–7. [DOI] [PMC free article] [PubMed]
- 8.Muth D, Corman VM, Roth H, Binger T, Dijkman R, Gottula LT, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Scientific Reports. 2018. [DOI] [PMC free article] [PubMed]
- 9.He JF, Peng GW, Min J, Yu DW, Liang WJ, Zhang SY, et al. Molecular evolution of the SARS coronavirus, during the course of the SARS epidemic in China. Science. 2004. [DOI] [PubMed]
- 10.Lau SKP, Feng Y, Chen H, Luk HKH, Yang W-H, Li KSM, et al. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. Journal of Virology. 2015. [DOI] [PMC free article] [PubMed]
- 11.Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11(4):e01610–20. [DOI] [PMC free article] [PubMed]
- 12.Gong Y-N, Tsao K-C, Hsiao M-J, Huang C-G, Huang P-N, Huang P-W, et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerging Microbes & Infections. 2020. [DOI] [PMC free article] [PubMed]
- 13.Laha S, Chakraborty J, Das S, Manna SK, Biswas S, Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol. 2020;85:104445. [DOI] [PMC free article] [PubMed]
- 14.Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92(5):522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Chen J, Gao K, Hozumi Y, Yin C, Wei GW. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun Biol. 2021;4(1):228. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamed SM, Elkhatib WF, Khairalla AS, Noreddin AM. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci Rep. 2021;11(1):8435. doi: 10.1038/s41598-021-87713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020. [DOI] [PMC free article] [PubMed]
- 18.Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 Mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Research. 2020. [DOI] [PMC free article] [PubMed]
- 20.Zhang Y, Zhang J, Chen Y, Luo B, Yuan Y, Huang F, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv. 2020.
- 21.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 22.Krjutskov K, Koel M, Roost AM, Katayama S, Einarsdottir E, Jouhilahti EM, et al. Globin mRNA reduction for whole-blood transcriptome sequencing. Sci Rep. 2016;6:31584. doi: 10.1038/srep31584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013. [DOI] [PMC free article] [PubMed]
- 24.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014. [DOI] [PubMed]
- 25.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Research. 2012. [DOI] [PMC free article] [PubMed]
- 26.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009. [DOI] [PMC free article] [PubMed]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological). 1995.
- 28.Team RDC. R: a language and environment for statistical computing. 2017.
- 29.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016. [DOI] [PMC free article] [PubMed]
- 30.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Research. 2015. [DOI] [PMC free article] [PubMed]
- 31.Goh YS, Chavatte J-M, Lim A, Amrun SN, Lee CY-P, Chee RS-L, et al. A highly sensitive FACS assay for serological detection and analysis of symptomatic and asymptomatic COVID-19 patients. 2020.
- 32.Poh CM, Carissimo G, Wang B, Amrun SN, Lee CYP, Chee RSL, et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nature Communications. 2020;11(1). [DOI] [PMC free article] [PubMed]
- 33.Naqiah Amrun S, Lee CY-P, Lee B, Fong S-W, Young BE, Sin-Ling Chee R, et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. SSRN Electronic Journal. 2020. [DOI] [PMC free article] [PubMed]
- 34.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–90 e2. [DOI] [PMC free article] [PubMed]
- 36.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martinez-Colon GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aschenbrenner AC, Mouktaroudi M, Kraemer B, Antonakos N, Oestreich M, Gkizeli K, et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. medRxiv. 2020:2020.07.07.20148395–2020.07.07. [DOI] [PMC free article] [PubMed]
- 38.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nature Medicine. 2020. [DOI] [PMC free article] [PubMed]
- 39.Young BE, Ong SWX, Ng LFP, Anderson DE, Chia WN, Chia PY, et al. Immunological and viral correlates of COVID-19 disease severity: a prospective cohort study of the first 100 patients in Singapore. SSRN Electronic Journal. 2020.
- 40.Chan YH, Fong SW, Poh CM, Carissimo G, Yeo NK, Amrun SN, et al. Asymptomatic COVID-19: disease tolerance with efficient anti-viral immunity against SARS-CoV-2. EMBO Mol Med. 2021;13(6):e14045. [DOI] [PMC free article] [PubMed]
- 41.Goh YS, Chavatte JM, Jieling AL, Lee B, Hor PX, Amrun SN, et al. Sensitive detection of total anti-Spike antibodies and isotype switching in asymptomatic and symptomatic COVID-19 patients. Cell Rep Med. 2021:100193. [DOI] [PMC free article] [PubMed]
- 42.Tang BSF, Chan KH, Cheng VCC, Yuen KY. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by SARS coronavirus and human coronavirus 229E. Hong Kong Medical Journal. 2009. [PubMed]
- 43.Yeung YS, Yip CW, Hon CC, Chow KYC, Ma ICM, Zeng F, et al. Transcriptional profiling of Vero E6 cells over-expressing SARS-CoV S2 subunit: Insights on viral regulation of apoptosis and proliferation. Virology. 2008. [DOI] [PMC free article] [PubMed]
- 44.Versteeg GA, van de Nes PS, Bredenbeek PJ, Spaan WJM. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. Journal of Virology. 2007. [DOI] [PMC free article] [PubMed]
- 45.Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discovery. 2019. [DOI] [PMC free article] [PubMed]
- 46.Sung SC, Chao CY, Jeng KS, Yang JY, Lai MMC. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 2009. [DOI] [PMC free article] [PubMed]
- 47.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020. [DOI] [PMC free article] [PubMed]
- 48.Nabar NR, Kehrl JH. The transcription factor EB links cellular stress to the immune response. Yale J Biol Med. 2017;90(2):301–315. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banerjee A, Nasir JA, Budylowski P, Yip L, Aftanas P, Christie N, et al. Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(9):2054–2063. doi: 10.3201/eid2609.201495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pontelli MC, Castro IA, Martins RB, Veras FP, Serra LL, Nascimento DC, et al. Infection of human lymphomononuclear 1 cells by SARS-CoV-2. 2020.
- 52.Pagano N, Laurent-Rolle M, Hsu JC-C, Team TYIR, Vogels CB, Grubaugh ND, et al. Long SARS-CoV-2 nucleocapsid sequences in blood monocytes collected soon after hospital admission. 2021.
- 53.Liu T, Jia P, Fang B, Zhao Z. Differential expression of viral transcripts from single-cell RNA sequencing of moderate and severe COVID-19 patients and its implications for case severity. Front Microbiol. 2020;11:603509. [DOI] [PMC free article] [PubMed]
- 54.Osman M, Faridi RM, Sligl W, Shabani-Rad MT, Dharmani-Khan P, Parker A, et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020;4(20):5035–5039. doi: 10.1182/bloodadvances.2020002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med. 2021;27(5):904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren X, Wen W, Fan X, Hou W, Su B, Cai P, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184(7):1895–913 e19. [DOI] [PMC free article] [PubMed]
- 57.Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34(6):108728. [DOI] [PMC free article] [PubMed]
- 58.Le Bert N, Clapham HE, Tan AT, Chia WN, Tham CYL, Lim JM, et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J Exp Med. 2021;218(5). [DOI] [PMC free article] [PubMed]
- 59.Wang F, Huang S, Gao R, Zhou Y, Lai C, Li Z, et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6(1):83. doi: 10.1038/s41421-020-00231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khor SS, Omae Y, Nishida N, Sugiyama M, Kinoshita N, Suzuki T, et al. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, age and sex are associated with severity of Japanese COVID-19 with respiratory failure. Front Immunol. 2021;12:658570. [DOI] [PMC free article] [PubMed]
- 61.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keightley RG, Cooper MD, Lawton AR. The T cell dependence of B cell differentiation induced by pokeweed mitogen. The Journal of Immunology. 1976;117(5 Part 1):1538 LP-44. [PubMed]
- 63.Hirohata S, Jelinek DF, Lipsky PE. T cell-dependent activation of B cell proliferation and differentiation by immobilized monoclonal antibodies to CD3. The Journal of Immunology. 1988;140(11):3736 LP-44. [PubMed]
- 64.Qu J, Wu C, Li X, Zhang G, Jiang Z, Li X, et al. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2020. [DOI] [PMC free article] [PubMed]
- 65.Atyeo C, Fischinger S, Zohar T, Slein MD, Burke J, Loos C, et al. Distinct Early Serological Signatures Track with SARS-CoV-2 Survival. Immunity. 2020;53(3):524–32 e4. [DOI] [PMC free article] [PubMed]
- 66.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020. [DOI] [PMC free article] [PubMed]
- 67.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020. [DOI] [PMC free article] [PubMed]
- 68.Pan P, Shen M, Yu Z, Ge W, Chen K, Tian M, et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat Commun. 2021;12(1):4664. doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. New England Journal of Medicine. 2020. [DOI] [PMC free article] [PubMed]
- 70.Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. Journal of Translational Medicine. 2020. [DOI] [PMC free article] [PubMed]
- 71.Carissimo G, Xu W, Kwok I, Abdad MY, Chan YH, Fong SW, et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early prognostic marker for severe COVID-19. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 72.Schulte-Schrepping J, Reusch N, Paclik D, Baßler K, Schlickeiser S, Zhang B, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020. [DOI] [PMC free article] [PubMed]
- 73.Scheuplein VA, Seifried J, Malczyk AH, Miller L, Höcker L, Vergara-Alert J, et al. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. Journal of Virology. 2015. [DOI] [PMC free article] [PubMed]
- 74.Wang Y, Liu L. The membrane protein of severe acute respiratory syndrome coronavirus functions as a novel cytosolic pathogen-associated molecular pattern to promote beta interferon induction via a toll-like-receptor-related TRAF3-independent mechanism. mBio. 2016. [DOI] [PMC free article] [PubMed]
- 75.Li J-Y, Liao C-H, Wang Q, Tan Y-J, Luo R, Qiu Y, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Research. 2020. [DOI] [PMC free article] [PubMed]
- 76.Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, Izard T, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 77.Yurkovetskiy L, Pascal KE, Tomkins-Tinch C, Nyalile T, Wang Y, Baum A, et al. SARS-CoV-2 Spike protein variant D614G increases infectivity and retains sensitivity to antibodies that target the receptor binding domain. bioRxiv. 2020:2020.07.04.187757–2020.07.04.
- 78.Eaaswarkhanth M, Al Madhoun A, Al-Mulla F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? International Journal of Infectious Diseases. 2020. [DOI] [PMC free article] [PubMed]
- 79.Gamage AM, Tan KS, Chan WOY, Liu J, Tan CW, Ong YK, et al. Infection of human nasal epithelial cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020;16(12):e1009130. [DOI] [PMC free article] [PubMed]
- 80.Vela JM. Repurposing sigma-1 receptor ligands for COVID-19 therapy? Front Pharmacol. 2020;11:582310. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Supplemental Fig. 1 GO-term enrichment of DEGs in whole blood of COVID-19 patients infected with WT SARS-CoV-2 during the acute phase of infection (SARS-CoV-2 PCR-positive; median 8 days PIO, n=13). GO-term functional enrichments for biological process, molecular function, and cellular component were performed for both up-regulated and down-regulated genes. Pathways are ranked by –log10(p-value). GO, gene ontology; DEGs, differentially expressed genes; WT, wildtype; PCR, polymerase chain reaction; PIO, post-illness onset. Supplemental Fig. 2 Representative gating strategy for the characterization of IFN-γ, IL-2, TNF-⍺ expression of CD4+ and CD8+ T cells in isolated PBMCs of COVID-19 patients by flow cytometry upon peptide stimulation. Supplemental Fig. 3 WT SARS-CoV-2 infected patients exhibit a higher systemic non-specific Th1 response, compared to Δ382 SARS-CoV-2 infected patients. CD4+ non T follicular helper (TFH) cells were characterized based on the expression of IFN-γ, IL-2, TNF-⍺, IL-17A, IL-4, IL-6 and IL-10 upon PMA/ Ionomycin stimulation. Statistical analyses were performed with unpaired test (*p < 0.05; **p < 0.01; ***p < 0.001).WT, wildtype. (PDF 2108 KB)
Data Availability Statement
RNA-seq data are accessible at NCBI’s Gene Expression Omnibus (GEO) database (GSE155454). All data are available upon request: Lisa F.P. Ng, lisa_ng@IDLabs.a-star.edu.sg.
Not applicable.