Abstract
Background
Early recurrence is the major cause of poor prognosis in hepatocellular carcinoma (HCC). Long non-coding RNAs (lncRNAs) are deeply involved in HCC prognosis. In this study, we aimed to establish a prognostic lncRNA signature for HCC early recurrence.
Methods
The lncRNA expression profile and corresponding clinical data were retrieved from total 299 HCC patients in TCGA database. LncRNA candidates correlated to early recurrence were selected by differentially expressed gene (DEG), univariate Cox regression and least absolute shrinkage and selection operator (LASSO) regression analyses. A 25-lncRNA prognostic signature was constructed according to receiver operating characteristic curve (ROC). Kaplan-Meier and multivariate Cox regression analyses were used to evaluate the performance of this signature. ROC and nomogram were used to evaluate the integrated models based on this signature with other independent clinical risk factors. Gene set enrichment analysis (GSEA) was used to reveal enriched gene sets in the high-risk group. Tumor infiltrating lymphocytes (TILs) levels were analyzed with single sample Gene Set Enrichment Analysis (ssGSEA). Immune therapy response prediction was performed with TIDE and SubMap. Chemotherapeutic response prediction was conducted by using Genomics of Drug Sensitivity in Cancer (GDSC) pharmacogenomics database.
Results
Compared to low-risk group, patients in high-risk group showed reduced disease-free survival (DFS) in the training (p < 0.0001) and validation cohort (p = 0.0132). The 25-lncRNA signature, AFP, TNM and vascular invasion could serve as independent risk factors for HCC early recurrence. Among them, the 25-lncRNA signature had the best predictive performance, and combination of those four risk factors further improves the prognostic potential. Moreover, GSEA showed significant enrichment of “E2F TARGETS”, “G2M CHECKPOINT”, “MYC TARGETS V1” and “DNA REPAIR” pathways in the high-risk group. In addition, increased TILs were observed in the low-risk group compared to the high-risk group. The 25-lncRNA signature negatively associates with the levels of some types of antitumor immune cells. Immunotherapies and chemotherapies prediction revealed differential responses to PD-1 inhibitor and several chemotherapeutic drugs in the low- and high-risk group.
Conclusions
Our study proposed a 25-lncRNA prognostic signature for predicting HCC early recurrence, which may guide postoperative treatment and recurrence surveillance in HCC patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-08827-z.
Keywords: Long non-coding RNA signature, Hepatocellular carcinoma, Early recurrence, Tumor infiltrating lymphocytes
Background
The very recent epidemiologic study has shown that liver cancer ranks the sixth commonly diagnosed cancer and the fourth leading cause of cancer death in the world. An estimated 84,100 liver cancer cases occurred and 78,200 liver cancer cases died in 2018 [1]. Hepatocellular carcinoma (HCC) compromises 75–85% of primary liver cancer [1]. The main clinical curative treatments for HCC include liver transplantation, percutaneous radiofrequency ablation and liver resection, among which liver resection is the most employed treatment [2]. Although 5-year overall survival rate reaches up to 50%, recurrence occurs in more than 70% HCC patients after curative surgery [3]. Clinically, the recurrence within 2-year after resection is defined as early recurrence, whereas the recurrence > 2-year is defined as late recurrence. Compared to late recurrence, HCC patients with early recurrence usually showed poorer prognosis [4].
Currently, many approaches, such as the TNM staging system of the American Joint Committee on Cancer (AJCC), the Barcelona Clinic Liver Cancer (BCLC) classification, and the Cancer of the Liver Italian Program (CLIP) staging system, have been employed to evaluate the prognosis of HCC patients [5]. However, their assessment criteria mainly rely on the clinicopathological features of HCC patients but do not take into account the critical and complicated molecular pathogenesis, an important factor in determining the outcome of HCC. Therefore, their prognostic predictive performance was unsatisfactory [6]. Meanwhile, serum alpha-fetoprotein (AFP) detection and medical imaging techniques are clinically used for post-surgery surveillance of recurrence in HCC patients, but with limited effectiveness due to the low specificity and sensitivity of those surveillance means [7].
The advent of high throughput array/sequencing and high-efficiency big data analysis in past decades makes it possible and reliable to construct multi-gene signatures to evaluate prognosis and predict therapeutic response in cancer patients. For example, a 70-gene signature had been established to aid decision making of adjuvant chemotherapy in patients with estrogen receptor-positive early breast cancer [8, 9]. More importantly, this 70-gene-signature based diagnostic test known as “MammaPrint” (Agendia, Amsterdam, The Netherlands) has been approved by the Food and Drug Administration (FDA) to predict breast cancer recurrence [10], and been validated in several retrospective studies [11, 12]. Additionally, an 18-gene signature ColoPrint (Agendia, Amsterdam, The Netherlands) was developed to predict disease relapse in patients with early-stage colorectal cancer (CRC) [13], and had been validated in other independent studies [14, 15]. Several multi-gene signatures have been constructed in HCC for prognosis evaluation. For example, Wei et al. developed a 20-miRNA signature to predict post-surgery survival in HCC patients [16]; Nault et al. constructed a 5-gene signature to evaluate the overall survival in HCC patients [17]; Kim et al. established a 233-gene signature to predict late recurrence in HCC patients [18]. Recently, prognostic signatures based on specific groups of genes such as glycolyis-related genes, metabolic-related genes and autophagy-related genes were also reported [19–21]. However, those multi-gene signatures of HCC mainly focus on overall survival and later recurrence, and few multi-gene signatures have been established to predict early recurrence in HCCs.
Long non-coding RNAs (lncRNAs) are a class of transcripts that are longer than 200 nucleotides (nt) and do not encode proteins [22]. Accumulating evidence has indicated the involvement of lncRNAs in diverse biological processes and disease pathogenesis [23]. Moreover, some lncRNAs have been reported to contribute to the initiation and progression in HCCs. For example, lncRNA-ANRIL has been reported to promote hepatocarcinoma cell proliferation [24]; and lncRNA-MALAT1 could function as a proto-oncogene to transform hepatocytes and enhance hepatocarcinoma cell growth [25]. In addition, some lncRNAs have been demonstrated to associate with HCC prognosis. For example, the overexpression of lncRNA-MVIH was associated with poor recurrence-free survival and overall survival in HCC patients [26]; LncRNA-PTTG3P expression was positively associated with tumor size, TNM stage and poor survival in HCC patients [27]. Although lncRNAs are involved in the progression and associated with prognosis in HCCs, lncRNA-based gene signatures for HCC prognostic evaluation, especially for early recurrence, are limited.
In this study, we analyzed the expression profile of lncRNAs and their association with early recurrence in the Liver Hepatocellular Carcinoma (LIHC) project from The Cancer Genome Atlas (TCGA) database (TCGA-LIHC). We constructed a 25-lncRNA signature significantly associated with HCC early recurrence. Based on this multi-lncRNA signature, HCC patients can be classified into low- and high-risk groups according to their risk scores. The early recurrence rate was significantly higher in the high-risk group than in the low-risk group. Moreover, the risk score negatively correlated with recurrence-free survival in HCC patients. Multivariate Cox regression analysis demonstrated that the 25-lncRNA signature, serum AFP, TNM stage and vascular invasion were 4 independent risk factors of HCC early recurrence. Compared with the other 3 risk factors, the 25-lncRNA signature had the best predictive performance for HCC early recurrence. Furthermore, the 25-lncRNA signature could synergize with serum AFP, TNM stage and vascular invasion to improve the prognosis evaluation for HCC early recurrence. In addition, in the context of this 25-lncRNA risk signature, we demonstrated that the “E2F TARGETS”, “G2M CHECKPOINT”, “MYC TARGETS V1” and “DNA REPAIR” were the most significantly enriched gene sets in the high-risk group. Moreover, the low-risk group showed greater tumor-infiltrating lymphocytes (TILs) compared to the high-risk group, and the 25-lncRNA prognostic signature was significantly negatively associated with the potent antitumor immune cells (i.e. type 1 T helper cell, effector memory CD8 T cell and activated CD8 T cell). Finally, the low-risk group was predicted to be more sensitive to immunotherapy like anti-PD-1 and chemotherapies like docetaxel, gefitinib and vinblastine, while the high-risk group was predicted to be more sensitive to doxorubicin, mitomycin C and paclitaxel. In conclusion, our findings may provide some insight into lncRNA-based personalized treatment and improve the strategy of post-surgery recurrence surveillance in HCC patients.
Methods
TCGA-LIHC database preparation and lncRNA profile mining
Gene expression profile of HCC and corresponding clinical information were downloaded from TCGA-LIHC (http://cancergenome.nih.gov/). Total 314 out of all 374 HCC samples with complete follow-up information (overall survival (OS) time, OS status, disease free survival (DFS) time and status) were retained. Among these 314 patients, some patients’ follow-up time was less than 1 month, and their OS and DFS status were labeled as “alive” and “recurrence free”. Therefore, these patients were not suitable for early recurrence analysis and they were excluded. Thus, we used 299 patients for signature construction in this study. The 299 HCC patients were then randomly divided into a training cohort (N = 150) and a validation cohort (N = 149). Based on the information of annotated lncRNAs in GENCODE V30, 14,847 human lncRNAs with Ensembl gene ID were obtained and their corresponding expression profile was extracted from the TCGA-LIHC.
Construction and validation of lncRNA-based risk signature
Most bioinformatics analyses were conducted using R software. DEG analysis was performed between the 150 HCC samples in the training cohort and 50 normal tissue samples from TCGA-LIHC project by using R package “edgeR” [28, 29]. Univariate Cox regression analysis was performed to select early recurrence related lncRNAs by using R package “survival” [30]. FunRich (version 3) was used to draw Venn diagram between differentially expressed lncRNAs and early recurrence related lncRNAs to obtain candidate lncRNAs for signature construction [31]. Candidate lncRNAs were then further analyzed in LASSO regression analysis by running R package “glmnet” for 1000 times [32], and the most powerful prognostic lncRNAs were selected through 10-fold cross-validation with lambda.min as the optimized cut-off [33]. Risk score of each patient was calculated in a linear combination of lncRNAs weighted by their corresponding regression coefficients and expression levels in indicated HCC patients by formula (risk score = ∑ coefficient × expression(gene)). Receiver operating characteristic curve (ROC) analysis was conducted by using R package “pROC” [34], and the predictive performance was assessed by calculating the area under curve (AUC). Finally, a combination of 25 lncRNAs was chosen for establishing risk signature because this 25-lcnRNA risk signature gave the largest AUC in ROC analysis. The 150 HCC patients were divided into the low-risk group (N = 75) and the high-risk group (N = 75) by using the median risk score as cut-off. A correlation analysis was performed between the risk score and early recurrence. Kaplan-Meier analysis, cumulative hazard and cumulative events analyses were conducted by using R package “survival” in the training cohort, the validation cohort and the total 299 HCC patients to investigate the early recurrence survival between low risk patients and high risk patients. Univariate and multivariate Cox analysis were done in the total 299 HCC patients with R package “survival” to evaluate whether the risk score could serve as an independent factor for early recurrence prediction in HCCs. Nomogram was constructed by using the 25-lncRNA signature, AFP, vascular invasion, TNM stages and their corresponding multivariate Cox regression coefficients, and calibration plots were generated with R package “regplot” [35]. C-index was used to evaluate the model performance for predicting early recurrence.
Gene set enrichment analysis (GSEA)
GSEA was conducted by using GSEA JAVA program (version 4.0.3) downloaded from official website (http://software.broadinstitute.org/gsea/index.jsp) to find out enriched gene sets. MsigDB h.all.v7.1.symbols.gmt gene set collection was chosen for identifying hallmarks of HCC early recurrence. The random sample permutations were set to be 1000 with the significance set as |NES| > 1, FDR q < 0.25 and nominal P < 0.05.
Analysis of the levels of tumor-infiltrating lymphocytes and immune therapy response prediction
Immune infiltration analysis was performed with single sample Gene Set Enrichment Analysis (ssGSEA) by using “GSVA” package in R [36]. A group of 28 cellmarker sets were used for calculating normalized enrichment score (NES) for each cell type in every 299 HCC samples [37]. Correlation analysis between risk scores and NES of immune cells was performed by function “cor.test” in R. TIDE (Tumor Immune Dysfunction and Exclusion) algorithm and SubMap modules from GenePattern were used to predict the response to immune checkpoint blockade for all 299 HCC samples [38–40].
Analysis of chemotherapeutic response prediction
Chemotherapeutic response prediction for every 299 HCC samples was conducted in R by using “pRRophetic” package based on the Genomics of Drug Sensitivity in Cancer (GDSC) pharmacogenomics database. The half maximal inhibitory concentration (IC50) was estimated by ridge regression and the prediction accuracy was evaluated by 10-fold cross-validation [41].
Real time quantitative RT-PCR
To validate the 25-lncRNA signature in clinical samples, 3 lncRNAs from the signature were selected and their relative expressions in HCC samples were detected by RT-qPCR. Total RNA from 36 paired HCC tumor and adjacent tissues provided by Xinhua Hospital were extracted by using TRIzol (Invitrogen, 15596026) according to the manufacturer’s instructions. cDNA was synthesized by using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, FSQ-301) in a SimpliAmp Thermal Cycler (Applied Biosystems). The 20 μL PCR reaction system consist of 2 μL cDNA, 0.8 μL forward primer, 0.8 μL reverse primer, 10 μL CYBR Premix Ex TaqII, 0.4 μL ROX Reference Dye II and 6 μL deionized water (Takara CYBR Premix Ex TaqII, RR820A). RT-PCR was performed in ABI Biosystems™ 7500 Real-Time qPCR System (Applied Biosystems). 18s was used as a housekeeping gene for normalization and the relative expression of selected genes was calculated by using 2−ΔΔCT method. Primers used were synthesized by GENEWIZ and the sequences of primers were ENSG00000231918 (GTGGCTCTGCCTTGGGTAAT, TTCCAGAACAACCTTGTCAGA), ENSG00000248596 (GCCAGAATTGGCGGTTTCTC, ATCGCTGAGTGTGTCGAGTG), and ENSG00000223392 (ATCCTTACCCTGCATTGCCC, ATGATCCAACCATCTGCAGGG).
Statistical analysis
DeLong’s test was used to compare the sensitivity and specificity of two ROC curves. Chi-square test was used to evaluate the impact of risk score group distribution on recurrence cases between 1 year and 2 years. The correlation of risk scores with disease free survival (DFS), NES of tumor-infiltrating lymphocytes and levels of immune checkpoints was analyzed by nonparametric Spearman’s rank correlation analysis. The log-rank test was used for Kaplan-Meier survival analyses, cumulative hazard and cumulative events analyses. The Cox proportional hazards regression model was used for univariate and multivariate analyses. Wilcoxon test was used for comparing NES of immune cells and IC50 of drugs between the low-risk and high-risk group. The difference was considered statistically significant when P < 0.05 in all statistical analysis.
Results
HCC dataset preparation and identification of candidate lncRNAs from the training cohort
HCC RNA-seq data and corresponding clinical information were downloaded from the TCGA-LIHC (Liver Hepatocellular Carcinoma) project. After removal of samples without complete survival information, total 299 out of all 374 HCC samples were enrolled in this study for further analysis. Table S1 shows the clinical characteristics of the 299 HCC samples, in which more than 50% HCC patients had recurrence. Because there are no suitable GEO datasets which are comparable to the TCGA-LIHC project containing comprehensive data on both lncRNA expression profile and patients’ clinical characteristics, we then randomly divided the 299 HCC patients into a training cohort (n = 150) and a validation cohort (n = 149) by using “split” function in R software instead of setting an external validation cohort. Bioinformatics analyses were first performed in the training cohort and further validated in the validation cohort (Fig. 1A).
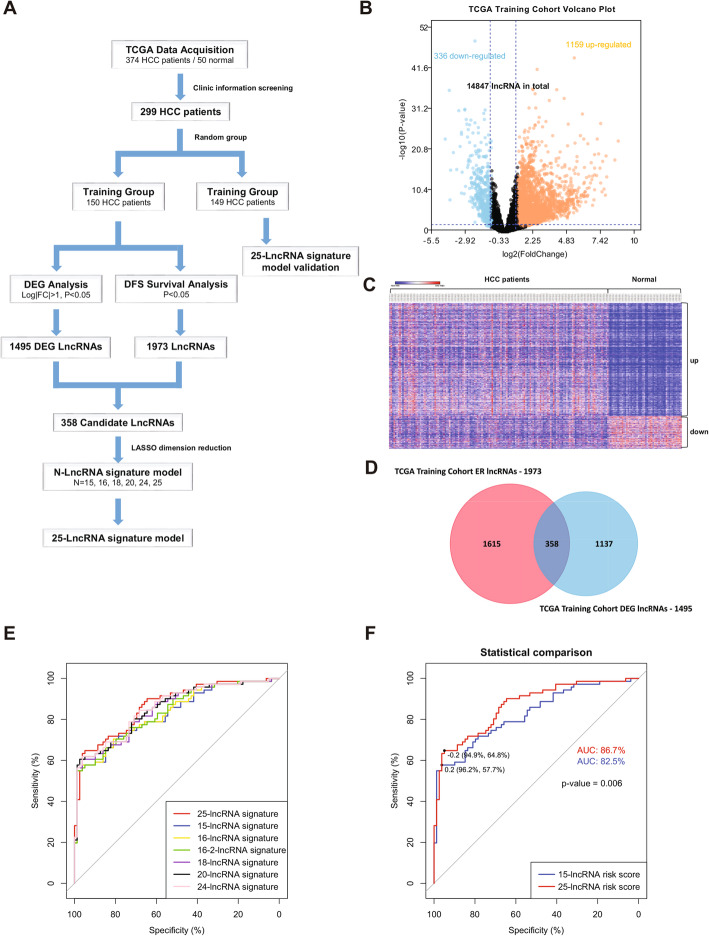
Fig. 1.
Data processing and lncRNA-based early risk signature construction from candidate lncRNAs. A) Schematic diagram of data processing and construction of lncRNA-based signature; B) Volcano plot of lncRNAs expression in the TCGA training cohort. Differentially expressed gene (DEG) analysis shows 1159 up-regulated lncRNAs and 336 down-regulated lncRNAs; C) Heatmap of 1495 DEG lncRNAs in 150 HCC samples and 50 normal tissues; D) Venn plot of DEG lncRNAs and early recurrence related lncRNAs (ER lncRNAs) in the TCGA training cohort. 358 DEG lncRNAs with potential prognostic value for HCC early recurrence were identified; E) ROC plot of 7 lncRNA signatures; F) ROC plot comparison between the 15-lncRNA risk signature and the 25-lncRNA risk signature, P = 0.006
To establish a lncRNA-based risk signature, differentially expressed gene (DEG) analysis on lncRNAs was performed between the training cohort and 50 normal controls from the TCGA-LIHC project. A total of 1495 DEG lncRNAs were found significantly dysregulated in HCC samples (1159 up-regulated and 336 down-regulated, log2|FC| > 1, FDR < 0.05) (Fig. 1B and C). Meanwhile, a univariate Cox regression analysis revealed that total 1973 lncRNAs were associated with HCC early recurrence (ER lncRNAs) (P < 0.05) (Fig. 1D). Finally, a Venn diagram between the 1495 DEG lncRNAs and the 1973 ER lncRNAs identified 358 lncRNA candidates which may have potential prognostic value for HCC early recurrence (Fig. 1D).
Pilot construction of multi-lncRNA signatures for HCC early recurrence
The least absolute shrinkage and selection operator (LASSO) Logistic Regression is a selection and shrinkage technique designed for regression model initially applied to Ordinary Logistic Regression [42]. LASSO can better identify those risk factors strongly linked to the outcome and is widely employed in signature construction [43]. To identify key lncRNAs suitable for establishing a risk signature for predicting HCC early recurrence, those 358 candidate lncRNAs (Fig. 1D) were further analyzed in LASSO regression. A total of 1000 LASSO regression iterations were performed by using the R package “glmnet”. Lambda.min was chosen as the optimized cut-off to select key lncRNAs for risk model (Fig. S1) [44]. Consequently, 7 lncRNA combinations were obtained after LASSO analysis (Table S2 and Fig. S1). Thus, 7 lncRNA risk signatures were individually constructed based on these combinations. The risk score of each HCC patient was calculated in a linear formula risk score = ∑ coefficient × expression(gene) (expression: lncRNA expression in individual HCC patients; coefficient: regression coefficients of indicated lncRNAs). To determine which lncRNA risk signature gives the best predictive performance on early recurrence, receiver operating characteristics (ROC) analysis was conducted between the 7 lncRNA risk signatures. As shown in Fig. 1E, all the 7 lncRNA risk signatures gave high area under the ROC curve (AUC, AUC > 80%), suggesting the reliability of our LASSO analysis. Among them, the 25-lncRNA risk signature gave the highest AUC (AUC = 86.70%) (Fig. 1F), suggesting the 25-lncRNA risk signature has the best predictive performance for HCC early recurrence.
Risk score calculation of the 25-lncRNA risk signature
Since the 25-lncRNA risk signature gave the best predictive performance, we then selected this signature to establish a risk model for HCC early recurrence. The detailed information of the 25 lncRNAs, including Ensembl gene ID, gene symbol, hazard ratio and coefficients, was summarized in Table 1. Among them, 19 lncRNAs (ENSG00000253417, ENSG00000272205, ENSG00000269894, ENSG00000275437, ENSG00000223392, ENSG00000248596, ENSG00000268201, ENSG00000247675, ENSG00000231918, ENSG00000234129, ENSG00000269974, ENSG00000236366, ENSG00000275223, ENSG00000253406, ENSG00000232079, ENSG00000255980, ENSG00000267905, ENSG00000176912, ENSG00000254333) had positive coefficients and were negatively associated with disease free survival (DFS), and the remainder 6 lncRNAs (ENSG00000259834, ENSG00000254887, ENSG00000259974, ENSG00000273837, ENSG00000231246, ENSG00000234283) had negative coefficients and were positively associated with DFS (Table 1). Here, we named those lncRNAs with positive coefficients as risk lncRNAs and those with negative coefficients as protective lncRNAs. The risk score could be calculated according to the coefficients of individual lncRNAs and their expression in corresponding HCC patients.
Table 1.
LncRNAs significantly associated with the disease free survival in the training cohort patients (N = 150)
| Ensembl | Gene Symbol | P value a | Hazard Ratio a | Coefficient b | associated diseases | Description | Reference |
|---|---|---|---|---|---|---|---|
| ENSG00000253417 | LINC02159 | < 0.001 | 2.85 | 0.069 | colorectal cancer, melanoma, head and neck squamous cell carcinoma | long intergenic non-protein coding RNA 2159 | [1–3] |
| ENSG00000272205 | < 0.001 | 2.47 | 0.233 | NR | NR | ||
| ENSG00000269894 | < 0.001 | 2.48 | 0.092 | NR | NR | ||
| ENSG00000275437 | < 0.001 | 4.78 | 0.083 | NR | NR | ||
| ENSG00000223392 | CLDN10-AS1 | < 0.001 | 1.82 | 0.278 | atherogenesis, lung adenocarcinoma, colorectal cancer, thyroid cancer, cholangiocarcinoma, colon adenocarcinoma | CLDN10 antisense RNA 1 | [4–6, 20] |
| ENSG00000248596 | LOC643201 | < 0.001 | 1.91 | 0.335 | colorectal cancer | centrosomal protein 192 kDa pseudogene | [5] |
| ENSG00000268201 | < 0.001 | 1.98 | 0.330 | NR | NR | ||
| ENSG00000247675 | LRP4-AS1 | < 0.001 | 7.36 | 0.567 | breast cancer, pancreatic neuroendocrine tumour | LRP4 antisense RNA 1 | [7] |
| ENSG00000231918 | LOC730100 | < 0.001 | 2.19 | 0.339 | glioma | uncharacterized LOC730100 | [8] |
| ENSG00000259834 | < 0.001 | 0.28 | −0.721 | NR | NR | ||
| ENSG00000234129 | 0.001 | 9.68 | 0.272 | NR | NR | ||
| ENSG00000269974 | 0.001 | 2.24 | 0.007 | NR | NR | ||
| ENSG00000254887 | LOC100505622 | 0.002 | 0.05 | −0.735 | gastric cancer | uncharacterized LOC100505622 | [12] |
| ENSG00000259974 | LINC00261 | 0.003 | 0.69 | − 0.187 | hepatocellular carcinoma, endometrial carcinoma, non-small cell lung cancer, colon cancer, esiohageal cancer, endometriosis, choriocarcinoma, gastric cancer, esophageal cancer, lung epithelial homeostasis, endoderm differentiation | long intergenic non-protein coding RNA 261 | [13–19, 25–27] |
| ENSG00000236366 | LOC153910 | 0.003 | 1.65 | 0.232 | lung function development, chronic obstructive pulmonary disease (COPD) and cardiovascular diseases (CVD) | uncharacterized LOC153910 | [3, 21–24] |
| ENSG00000275223 | 0.003 | 1.56 | 0.024 | NR | NR | ||
| ENSG00000253406 | 0.004 | 967.38 | 1.878 | NR | NR | ||
| ENSG00000232079 | LINC01697 | 0.009 | 1.53 | 0.142 | lung squamous cell carcinoma, gastric cancer, gastric adenocarcinoma, breast cancer | long intergenic non-protein coding RNA 1697 | [9–11] |
| ENSG00000273837 | 0.012 | 0.59 | −0.025 | NR | NR | ||
| ENSG00000255980 | LOC102724265 | 0.017 | 10.02 | 0.125 | NR | uncharacterized LOC102724265 | |
| ENSG00000267905 | 0.018 | 1.5 | 0.032 | NR | NR | ||
| ENSG00000176912 | TYMSOS | 0.023 | 1.28 | 0.038 | NR | TYMS opposite strand | |
| ENSG00000254333 | NDST1-AS1 | 0.032 | 1.21 | 0.044 | NR | NDST1 antisense RNA 1 | |
| ENSG00000231246 | LINC02884 | 0.033 | 0.17 | −0.041 | NR | long intergenic non-protein coding RNA 2884 | |
| ENSG00000234283 | 0.047 | 0.39 | −0.459 | NR | NR |
NR not reported
a Derived from the univariable Cox proportional hazards regression analysis in the 150 training cohort patients
b Derived from the LASSO regression analysis in the 150 training cohort patients
Please refer to the supplementary material for references citation
The 25-lncRNA risk signature correlates with HCC early recurrence
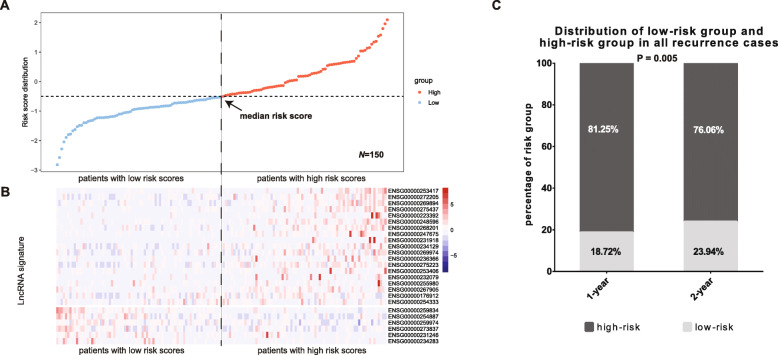
To determine whether the 25-lncRNA risk signature could predict HCC early recurrence, we first calculated the risk scores of the 150 HCC patients in the training cohort and then distributed them according to their risk scores from low to high (Fig. 2A). The median risk score was set as the cut-off to separate those patients into low-risk group (n = 75, patients’ risk scores < the median risk score) and high-risk group (n = 75, patients’ risk scores > the median risk score) (Fig. 2A). As shown in Fig. 2B, the 19 risk lncRNAs were mostly enriched in the high-risk group whereas the 6 protective lncRNAs were mainly enriched in the low-risk group. Moreover, 81.25% of recurrence cases in 1-year and 76.06% in 2-year came from the high-risk group, while the percentages were respectively 18.75 and 23.94% in the low-risk group (Fig. 2C). These results indicate that the 25-lncRNA risk signature have satisfying predictive potential for HCC early recurrence in the training cohort.
Fig. 2.
Correlation analysis of the 25-lncRNA risk signature with HCC early recurrence in the training cohort. A) The 150 HCC patients in the training cohort was ranked according to their risk scores from low to high, and the median risk score was set as the cut-off to divide the 150 HCC patients into low-risk group (n = 75) and high-risk group (n = 75); B) The 25-lncRNA expression profile in the 150 HCC patients. The 19 risk lncRNAs were enriched in the high-risk group and the 6 protective lncRNAs were enriched in the low-risk group; C) 81.25% and 76.06% HCC patients with recurrence in 1-year and 2-year respectively were classified in the high-risk group, and 18.72% and 23.94% HCC patients with recurrence in 1-year and 2-year respectively were classified in the low-risk group (P = 0.005)
Validation of the 25-lncRNA risk signature
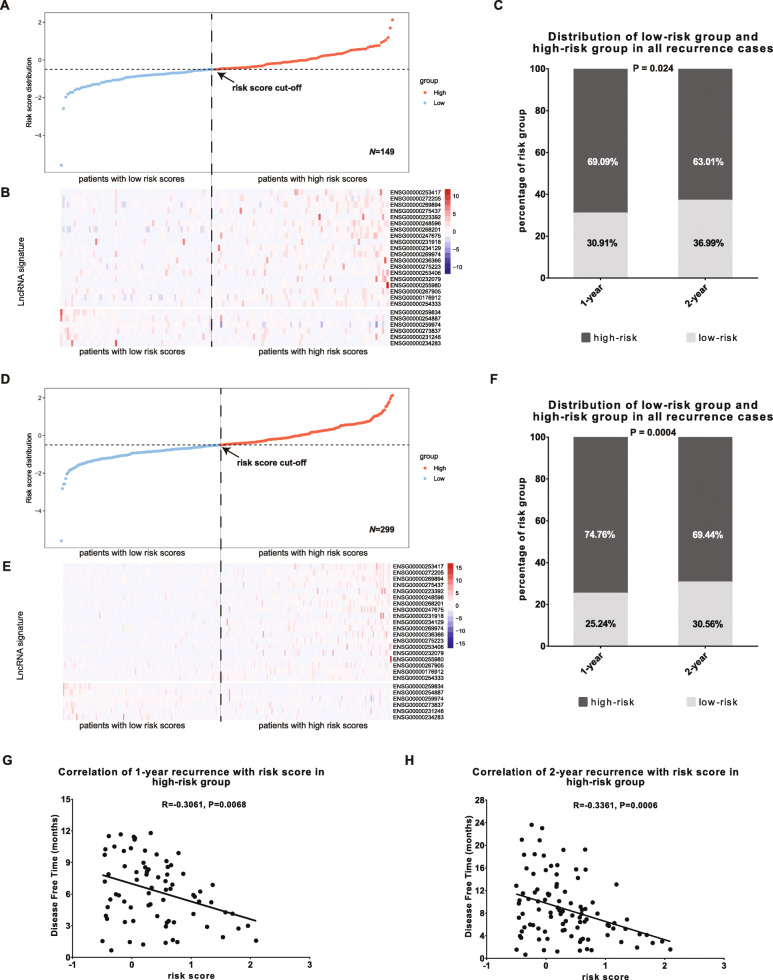
To validate the predictive potential of the 25-lncRNA risk signature, we evaluated it in the validation cohort. According to the median cut-off in the training cohort, the validation cohort (n = 149) was separated into the low-risk group (n = 69) and the high-risk group (n = 80) (Fig. 3A). In line with the finding in the training cohort, the risk lncRNAs were mainly enriched in the high-risk group whereas the protective ones were mainly enriched in the low-risk group (Fig. 3B). Meanwhile, 69.09% of 1-year recurrence cases and 63.01% of 2-year recurrence cases came from the high-risk group (Fig. 3C). Moreover, the predictive potential of the 25-lncRNA risk signature was also evaluated in the total 299 recruited HCC patients. Similarly, 299 HCC patients were separated into low-risk group (n = 144) and high-risk group (n = 155) according to the median cut-off in the training cohort (Fig. 3D). Most of the risk lncRNAs were enriched in the high-risk group and most of the protective lncRNAs were enriched in the low-risk group (Fig. 3E). Consistent with this finding, non-pair Wilcoxon test confirmed enrichment of the risk lncRNAs and the protective lncRNAs in the high-risk and low-risk groups respectively, except for the lncRNAs (ENSG00000255980, ENSG00000253406, ENSG00000232079, and ENSG00000234283) whose expression showed no significant changes between the low- and high-risk groups (Fig. S2). Patients in the high-risk group contributed 74.76% of 1-year recurrence cases and 69.44% of 2-year recurrence cases (Fig. 3F). More importantly, correlation assays showed significantly negative correlation of risk score with 1-year (Fig. 3G) or 2-year DFS (Fig. 3H) in the recurrent HCC patients in the high-risk group. No correlation was observed between risk score and DFS in recurrent HCC patients in the low-risk group (Fig. S3). These findings further validate the correlation of the 25-lncRNA risk signature with HCC early recurrence and indicate the great predictive potential of the risk signature on HCC early recurrence.
Fig. 3.
Correlation analysis of the 25-lncRNA risk signature with HCC early recurrence in the validation and entire TCGA cohort. A) The 149 HCC patients in the validation cohort were ranked according to their risk scores from low to high, and divided into the low-risk group (n = 69) and the high-risk group (n = 80) by using the same risk score cut-off in the training cohort; B) The 25-lncRNA expression profile in the 149 HCC patients. The 19 risk lncRNAs were enriched in the high-risk group and the 6 protective lncRNAs were enriched in the low-risk group; C) 69.09% and 63.01% HCC patients with recurrence in 1-year and 2-year respectively were classified in the high-risk group, and 30.91% and 36.99% HCC patients with recurrence in 1-year and 2-year respectively were assigned in the low-risk group (P = 0.024); D) The 299 HCC patients in the entire TCGA cohort were ranked according to their risk scores from low to high, and divided into the low-risk group (n = 144) and the high-risk group (n = 155) by using the same risk score cut-off in the training cohort; E) The 25-lncRNA expression profile in the 299 HCC patients. The 19 risk lncRNAs were enriched in the high-risk group and the 6 protective lncRNAs were enriched in the low-risk group; F) 74.46% and 69.44% HCC patients with recurrence in 1-year and 2-year respectively were assigned in the high-risk group, and 25.24% and 30.56% HCC patients with recurrence in 1-year and 2-year respectively were assigned in the low-risk group (P = 0.0004); G and H) Correlation of risk score with 1-year (G) or 2-year DFS (H) in the recurrent HCC patients in the high-risk group of the entire TCGA cohort
The primary purpose for the signature construction study is to accurately discriminate low- and high-risk patients. Therefore, the cut-off selection is critical for the accuracy of the prediction signature. In this study, we adopted the median risk score as cut-off which has been widely employed by many other groups [16, 45–48]. To investigate whether there are other cut-offs which could distinguish low- and high-risk of early recurrence better than the median cut-off, we adopted the cut-off derived from Youden index [49]. Although the Youden index/cut-off could separate patients into the low- and high-risk groups (Fig. S4A-C), the prediction performance for early recurrence is much poorer than that by using median cut-off (Fig. S4D-F). Therefore, the median risk score used in this study is an appropriate cut-off to accurately distinguish HCC patients with low or high early recurrence risk.
The 25-lncRNA risk signature precisely predicts early recurrence in HCC patients
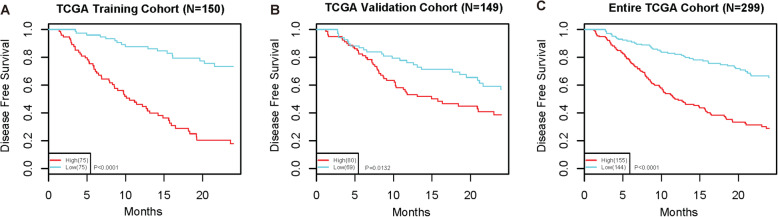
To further investigate the prognostic value of the 25-lncRNA risk signature for early recurrence, we analyzed cumulative hazard and event in HCC patients. Both cumulative hazards and cumulative events were significantly higher in the high-risk group than those in the low-risk group in either the training cohort (Fig. S5A and B), validation cohort (Fig. S5C and D) or total 299 HCC patients (Fig. S5E and F). Meanwhile, Kaplan-Meier analyses, in the training cohort (Fig. 4A), validation cohort (Fig. 4B) and 299 enrolled HCC patients (Fig. 4C), showed that the patients in the high-risk group had lower 2-year DFS than those in the low-risk group. These findings further indicate the prognostic value of the 25-lncRNA risk signature for HCC early recurrence.
Fig. 4.
Kaplan-Meier analysis of the association of the 25-lncRNA risk signature with early recurrence risk in HCCs. A-C) The association of the 25-lncRNA risk signature with 2-year DFS was analyzed in the training cohort (N = 150, P < 0.0001) (A), validation cohort (N = 149, P = 0.0132) (B), and the entire TCGA cohort (N = 299, P < 0.0001) (C). The statistical significance was determined by the log-rank test. The patients in each cohort were stratified into the high-risk and low risk groups based on the cut-off risk score in the training cohort
The 25-lncRNA risk signature is an independent prognostic factor for early recurrence in HCCs
To determine whether the 25-lncRNA risk signature is an independent prognostic factor for HCC early recurrence, we performed univariate and multivariate Cox regression analyses in the enrolled 299 HCC patients. The 25-lncRNA risk score and other clinicopathological factors, including gender, age, race, cirrhosis, vascular invasion, serum AFP level and TNM stage, were used as covariates. As shown in Table 2, the vascular invasion, serum AFP and 25-lncRNA risk score were significantly associated with 1-year and 2-year recurrence in HCC patients, while the TNM stage was significantly associated with 2-year recurrence but not with 1-year recurrence. These findings are consistent with previous studies showing that serum AFP [50], TNM stage [51] and vascular invasion [2] are independent risk factors for HCC early recurrence, and indicate that the 25-lncRNA risk signature could serve as an independent prognostic factor for HCC early recurrence.
Table 2.
Univariate and multivariate Cox analysis of risk factors in the TCGA entire group (N = 299)
| Characteristics | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | CI 95 | P Value | Hazard Ratio | CI 95 | P Value | |
| 1-year DFS | ||||||
| risk score of 25-lncRNA signature | 2.38 | 1.88–3.01 | < 0.001 | 2 | 1.48–2.71 | < 0.001 |
| TNM Stages | 1.96 | 1.55–2.47 | < 0.001 | 1.28 | 0.94–1.75 | 0.124 |
| Vascular Invasion | 1.88 | 1.35–2.61 | < 0.001 | 1.66 | 1.12–2.45 | 0.011 |
| AFP | 2.52 | 1.56–4.07 | < 0.001 | 2.23 | 1.34–3.74 | 0.002 |
| Cirrhosis | 1.11 | 0.65–1.9 | 0.709 | |||
| Gender | 1.17 | 0.78–1.77 | 0.444 | |||
| Age | 0.77 | 0.49–1.22 | 0.269 | |||
| Race | 1.15 | 0.83–1.59 | 0.396 | |||
| 2-year DFS | ||||||
| risk score of 25-lncRNA signature | 2.19 | 1.8–2.66 | < 0.001 | 1.92 | 1.48–2.49 | < 0.001 |
| TNM Stages | 1.96 | 1.61–2.39 | < 0.001 | 1.46 | 1.12–1.89 | 0.005 |
| Vascular Invasion | 1.8 | 1.36–2.38 | < 0.001 | 1.5 | 1.05–2.12 | 0.024 |
| AFP | 2.03 | 1.37–2.99 | < 0.001 | 1.77 | 1.16–2.71 | 0.008 |
| Cirrhosis | 1.25 | 0.81–1.93 | 0.305 | |||
| Gender | 1.21 | 0.86–1.71 | 0.27 | |||
| Age | 0.91 | 0.6–1.36 | 0.635 | |||
| Race | 0.84 | 0.62–1.12 | 0.229 | |||
In univariate and multivariate Cox analysis, risk score, TNM stages and vascular invasion were evaluated as continuous variables, and AFP, cirrhosis, gender, age and race were evaluated as category variables. Age category and AFP category were defined by 50 and 20 ng/ml as cut-off value, respectively
The combination of the 25-lncRNA risk signature, AFP, TNM stage and vascular invasion improves the prognosis evaluation and the construction of nomogram
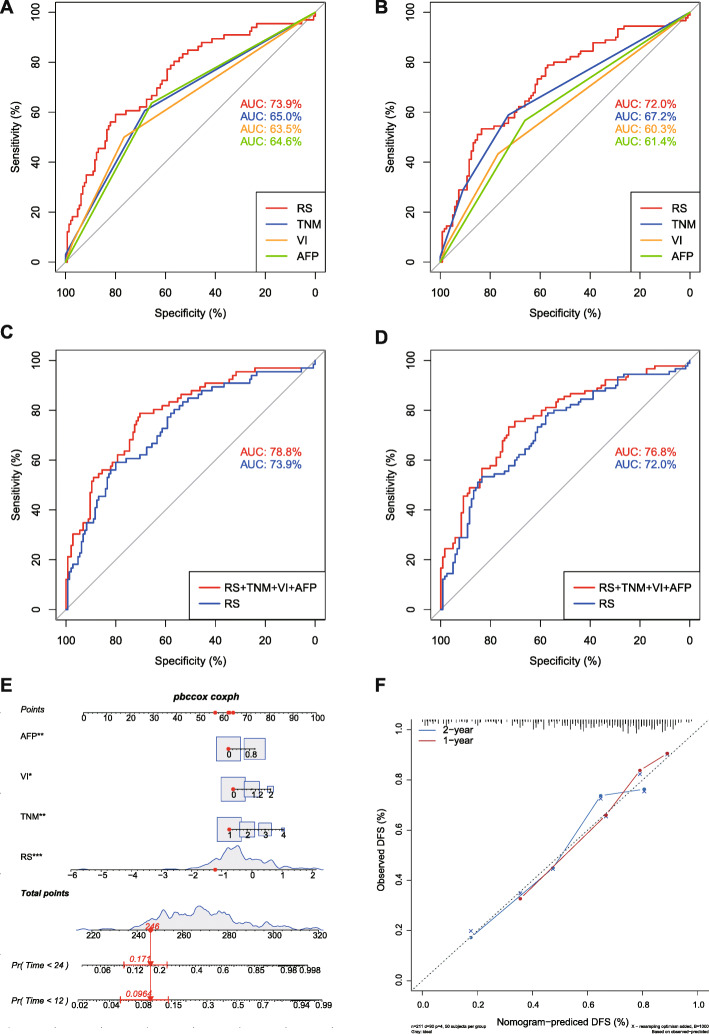
To investigate which independent risk factor gives the best predictive performance for HCC early recurrence, ROC analyses were performed by using “pROC”. As shown in Fig. 5A and B, the AUC of risk score for 1-year recurrence (73.86%) and 2-year recurrence (71.98%) were better those of AFP (64.58% for 1-year, 61.39% for 2-year recurrence), TNM (64.99% for 1-year, 67.17% for 2-year recurrence) and vascular invasion (VI) (63.47% for 1-year, 60.33% for 2-year recurrence). Moreover, compared to risk score alone, combining the risk score with AFP, TNM and VI further increased the predictive performance for 1-year recurrence (AUC: 78.79% vs. 73.86%) and 2-year recurrence (AUC: 76.82% vs. 71.98%) (Fig. 5C and D). The 95% confidence interval of AUC and C-index of above signatures were summarized in Table S3. An integrated Nomogram was further constructed by combining the 25-lncRNA signature, AFP, VI and TNM with a C-index 0.739 (Fig. 5E), and the calibration curves of the integrated nomogram for 1-year and 2-year DFS were presented in Fig. 5F. Therefore, the combination of the 25-lncRNA risk signature with AFP, TNM and VI could improve the prognosis evaluation for HCC early recurrence.
Fig. 5.
ROC analysis of the predictive performance and nomogram construction for early recurrence of the 25-lncRNA risk signature, TNM stage, vascular invasion and AFP. A-B) ROC analysis of the predictive performance of the 25-lncRNA risk signature, TNM stages, vascular invasion and AFP for 1-year DFS (A) and 2-year DFS (B) in the entire TCGA cohort; C-D) ROC analysis of the predictive performance of the combination of the 25-lncRNA risk signature, TNM stages, vascular invasion and AFP and risk score alone for 1-year DFS (C) and 2-year DFS (D) in the entire TCGA cohort. E) Nomogram of the 25-lncRNA signature risk score combined with AFP, vascular invasion and TNM stages; F) Calibration curves for the 25-lncRNA-signature-integrated nomogram for 1-year DFS and 2-year DFS. RS: the risk score of the 25-lncRNA signature, VI: vascular invasion
Biological processes associated with HCC early recurrence
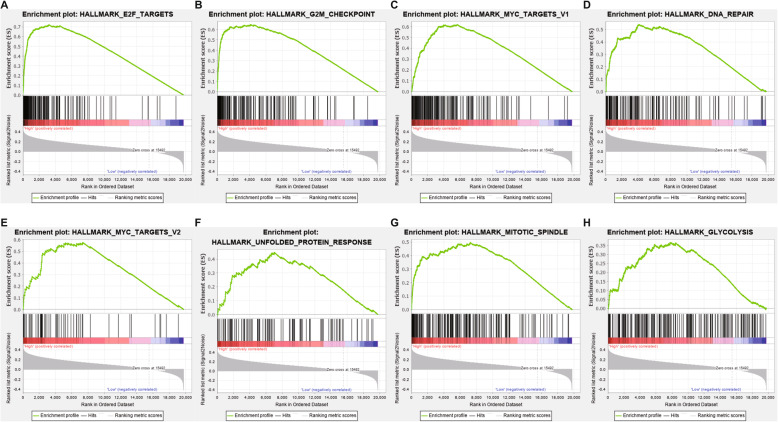
Previous studies have shown that lncRNAs function as key regulators of critical biological processes including cell differentiation, development, and apoptosis [52]. To investigate the biological processes associated with HCC early recurrence, gene set enrichment analysis (GSEA) was performed with hallmark pathways based on the gene expression profiling data from HCC patients in the high-risk and the low-risk groups. Eight gene sets were significantly enriched in the high-risk group while no significant gene set enrichment was observed in the low-risk group (|NES| > 1, FDR q-val < 0.25, NOM p-val < 0.05) (Table 3). Among them, the enrichment of gene sets of “E2F TARGETS”, “G2M CHECKPOINT”, “MYC TARGETS V1” and “DNA REPAIR” showed higher significance (|NES| > 1.5, FDR q-val < 0.10, NOM p-val < 0.01) (Table 3). The snapshots of enrichment results were displayed in Fig. 6 and the heatmaps for enriched gene sets were displayed in Fig. S6. These findings suggest that the 25 lncRNAs may affect HCC early recurrence through E2F, Myc, G2M and DNA repair pathways.
Table 3.
GSEA pathways up-regulated in high-risk group
| Gene Sets | SIZE | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|
| HALLMARK_E2F_TARGETS | 195 | 1.991 | 0.000 | 0.045 |
| HALLMARK_G2M_CHECKPOINT | 189 | 1.845 | 0.002 | 0.077 |
| HALLMARK_MYC_TARGETS_V1 | 194 | 1.844 | 0.004 | 0.052 |
| HALLMARK_DNA_REPAIR | 148 | 1.826 | 0.004 | 0.045 |
| HALLMARK_MYC_TARGETS_V2 | 58 | 1.641 | 0.040 | 0.153 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 107 | 1.562 | 0.043 | 0.207 |
| HALLMARK_MITOTIC_SPINDLE | 198 | 1.542 | 0.019 | 0.199 |
| HALLMARK_GLYCOLYSIS | 198 | 1.516 | 0.022 | 0.205 |
Fig. 6.
Gene set enrichment analysis illustrated upregulated gene sets in the high-risk group. A) Enrichment plot: HALLMARK_E2F_TARGETS; B) Enrichment plot: HALLMARK_G2M_CHECKPOINT; C) Enrichment plot: HALLMARK_MYC_TARGETS_V1; D) Enrichment plot: HALLMARK_DNA_REPAIR; E) Enrichment plot: HALLMARK_MYC_TARGETS_V2; F) Enrichment plot: HALLMARK_UNFOLDED_PROTEIN_RESPONSE; G) Enrichment plot: HALLMARK_MITOTIC_SPINDLE; H) Enrichment plot: HALLMARK_GLYCOLYSIS. |NES| > 1, FDR < 0.25, P < 0.05
The 25-lncRNA signature negatively associates with tumor infiltrating lymphocytes
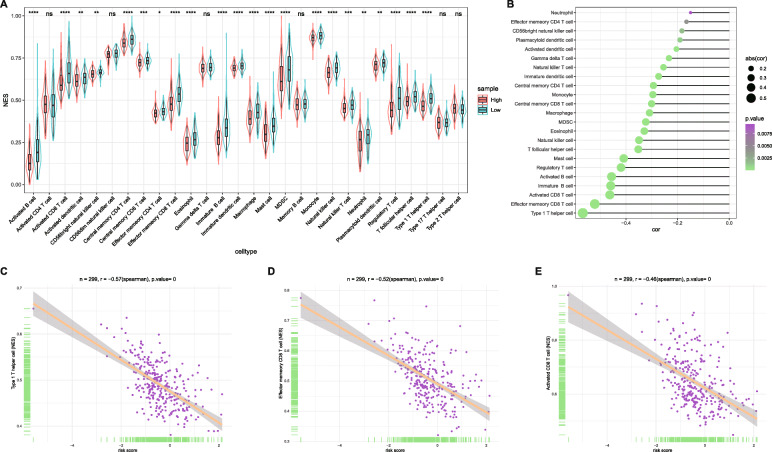
Tumor infiltrating lymphocytes (TILs) have been recognized as a prognostic factor in various types of cancers, and accumulation of TILs has been established as a positive prognostic factor in a number of solid cancers including melanoma [53], colon cancer [54] and ovarian cancer [55]. Previous studies have demonstrated that HCC patients with prominent TILs showed reduced recurrence and better prognosis compared with those without prominent TILs [56, 57]. Although the TILs are minority in the tumor bulk, the immune checkpoint molecules specifically express on T cells and antigen presenting cells but not tumor cells or other stromal cells in tumor bulk. Thus, it is a commonly accepted approach to evaluate TILs by using the expression levels of immune checkpoint molecules from bulk-tumor data [45, 58, 59]. To investigate whether the 25-lncRNA prognostic signature could reflect the levels of TILs, comparison of TILs was performed between the low- and high-risk groups. As shown in Fig. 7A, 22 out of 28 TILs showed significant enrichment in the low-risk group compared to the high-risk group (P < 0.05). Correlation analysis between risk scores and normalized enrichment scores (NES) of TILs revealed that the intratumor accumulation of 23 TILs was negatively associated with risk scores (P < 0.05, Fig. 7B). Among them, the type 1 T helper cell, effector memory CD8 T cell and activated CD8 T cell, which are well-known antitumor immune cells, ranked as the top 3 TILs negatively associated with the risk scores (|NES| > 0.4, Fig. 7C-E). These findings suggested that the 25-lncRNA prognostic signature may reflect the levels of TILs and predict the post-surgery prognosis in HCCs.
Fig. 7.
Association of the 25-lncRNA signature risk score with immune infiltration of 299 HCC samples. A) Comparisons of NES of immune cells between the low-risk and high-risk group, 22 immune cells showed higher NES in the low-risk group (P < 0.05); B) Correlation between risk scores and NES of immune cells, 23 immune cells were negatively associated with risk scores (P < 0.05); C)-E) Representative correlations between risk scores and type 1 T helper cell (C), effector memory CD8 T cell (D), activated CD8 T cell (E), |NES| > 0.4
The low-risk group patients showed more sensitivity in immunotherapies and the low- and high-risk group patients showed different chemotherapies responses
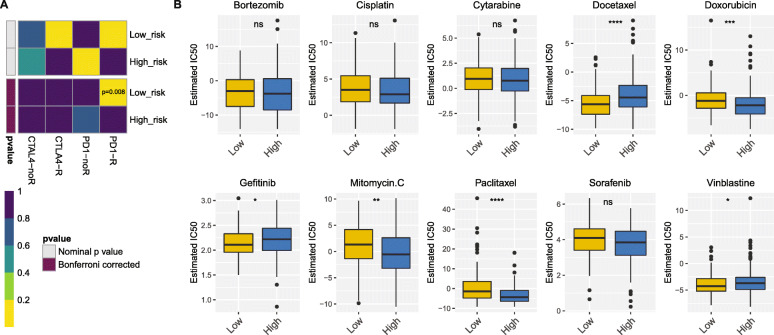
Since more TILs significantly enriched in the low-risk group patients, we attempted to further investigate whether the immunotherapies response are different in the low- and high-risk group. TIDE prediction suggest that there was no significantly difference in immunotherapies response between the low- (48.61%, 70/144) and high-risk (42.58%, 66/155) group (P = 0.297). However, by mapping the expression profile of the low- and high-risk group with a public dataset of 47 melanoma patients responded to immunotherapies in SubMap modules of GenePattern [60], the low-risk group showed prospective response to anti-PD-1 (programmed cell death protein 1) therapy (Bonferroni-corrected P = 0.008, Fig. 8A). Besides immunotherapies, we attempted to identify whether the 25-lncRNA prognostic signature could be applied to chemotherapies prediction. The results showed that the low-risk group had a lower half maximal inhibitory concentration of docetaxel, gefitinib and vinblastine, while the high-risk group had a lower half maximal inhibitory concentration of doxorubicin, mitomycin C and paclitaxel (Fig. 8B, P < 0.05). Thus, the 25-lncRNA prognostic signature could act as a potential predictor for immunotherapies and chemotherapies.
Fig. 8.
The prediction of immunotherapeutic and chemotherapeutic responses. A) SubMap analysis revealed that the low-risk group was more sensitive to PD-1 inhibitor (Bonferroni-corrected P = 0.008); B) The predicted IC50 for chemotherapeutic drugs in the low- and high-risk group. The low-risk group was related to a lower IC50 in docetaxel, gefitinib and vinblastine, while the high-risk group was related to a lower IC50 in doxorubicin, mitomycin C and paclitaxel (P < 0.05 by Wilcoxon test)
Discussion
As a class of non-coding transcripts, lncRNAs have been identified in all model organisms. So far, over 56,000 human lncRNAs have been reported in recent lncRNA annotations and the number of lncRNAs keeps growing [61]. Unlike protein-coding genes, most lncRNAs are less conserved, which leads to neglect of the function of lncRNAs [62]. However, accumulating evidence mainly at cellular level has indicated the involvement of lncRNAs in various biological processes such cell proliferation, apoptosis and nutrient sensing to cell differentiation [63]. Moreover, dysregulation of lncRNAs has been implicated in the pathogenesis of various diseases including cancers [64]. Many lncRNAs have shown their prognostic value in many types of cancers [26, 27]. In this study, we established a 25-lncRNA risk signature to predict HCC early recurrence. We demonstrated that, compared to AFP, TNM and VI, this 25-lncRNA risk signature possesses the best prognostic potential for HCC early recurrence. Moreover, the combination of the lncRNA risk signature with AFP, TNM and VI could further improve the predictive performance.
In this study, we define the recurrence in 2-year post-surgery as HCC early recurrence. This is in agreement with a previous study showing that the slopes of early recurrence curve and late recurrence curve are different and the intercept time point of the two curves is defined as the cut-off to separate early and late recurrence [4]. This separation criterion is widely adopted by many other studies [65]. We also noticed that some studies define the recurrence in 1-year post-surgery as HCC early recurrence [66, 67]. Therefore, we analyzed the association of the 25-lncRNA risk signature with both 1-year and 2-year recurrence in most of our analyses and found that this risk signature has great prognostic potential for both of them.
The 25-lncRNA risk signature includes 19 risk lncRNAs (coefficient > 0) and 6 protective lncRNAs (coefficient < 0) (Table 1). Among these lncRNAs, dysregulation of LINC02159, CLDN10-AS1, LOC643201, LRP4-AS1, LOC730100, LINC01697, LOC100505622, and LINC00261 has been reported in several types of cancers (Table 1). In addition, CLDN10-AS1 was reported to be involved in endothelial dysfunction in atherogenesis (Table 1). Some previous studies have suggested the association of LOC153910 with lung function development, risk of chronic obstructive pulmonary disease (COPD) and cardiovascular diseases (CVD) (Table 1). LINC00261 has shown to regulate endoderm differentiation, lung epithelial homeostasis and endometriosis (Table 1). Given the fact that most of lncRNAs demonstrate a tissue-specific expression pattern [68], further investigation of the role of those 25 lncRNAs in HCCs is warranted.
To investigate the biological processes or pathways related to HCC early recurrence, we performed GSEA to explore the hallmarks of gene sets in the high-risk group. Total 8 gene sets were significantly enriched in the high-risk group. Among them, the gene sets of “E2F TARGETS”, “G2M CHECKPOINT”, “MYC TARGETS V1” and “DNA REPAIR” showed higher significance in enrichment. In fact, members of those four gene sets have been reported to associate with poor prognosis in many types of cancers including HCC [69–72].
Accumulation of TILs is commonly related to an improved prognosis in many types of cancers. In the present study, greater intratumor accumulation of TILs was observed in the low-risk group compared to high-risk group. We demonstrated that the 25-signature risk score significantly and negatively associate with intratumor accumulation of type 1 T helper cell, effector memory CD8 T cell and activated CD8 T cell, which are well-known antitumor immune cells involved in cancer immune therapy [73–75], further suggesting that this 25-lncRNA signature has potential to predict the post-surgery prognosis in HCC patients. In addition, the immunotherapies prediction based on this 25-lncRNA signature suggested that the low-risk group had more effective response to PD-1 inhibitor. Moreover, chemotherapies prediction indicated that the low- and high-risk showed different sensitivity to drugs such as docetaxel and paclitaxel, but not cisplatin and sorafenib. Thus, different therapies might be adapted to HCC patient in the low- and high risk group according to the 25-lncRNA signature.
Although the 25-lncRNA risk signature was validated in the TCGA internal validation cohort and displayed good prognostic potential in the enrolled 299 HCC patients, an external validation cohort is missing in this study. This is because we failed to find any suitable GEO datasets or International Cancer Genome Consortium (ICGC) database which could apply sufficient information on both lncRNA expression profile and clinical survival. For example, there are two GEO datasets, GSE67260 and GSE113850, possess satisfied data on lncRNA expression profile but without clinical records. Moreover, expression profiles of 22 lncRNAs in the 25-lncRNA signature could be extracted from two ICGC datasets including LIRI-JP and LICA-FR, but disease free survival information is missing. The Cancer Genome Atlas (TCGA) is a multi-institutional, cross-discipline effort led by the National Cancer Institute recruiting cancer samples from different countries. For example, the HCC samples recruited in the TCGA-LIHC database were derived from Vietnam, United States, Canada, South Korea, Russia [76]. Therefore, those samples are actually derived from multi-centers and at certain level support the approach we used in this study by splitting them into a training cohort and a validation cohort. However, validation of this 25-lncRNA risk signature in an external cohort will be warranted as long as suitable data are available. Meanwhile, the validation of individual lncRNA included in this 25-lncRNA signature in clinical HCC tumor and paracancerous tissues were in processing but have not completed yet. LncRNAs ENSG00000231918, ENSG00000248596, and ENSG00000223392 were found to be upregulated in 36 HCC tumor tissues compared with paracancerous tissues (Fig. S7, Table S4).
Conclusions
In this study, we established a 25-lncRNA risk signature for HCC early recurrence. According to this risk signature, HCC patients could be accurately separated into the low- and high-risk groups. 1-year and 2-year recurrence rates were significantly higher in the high-risk group than those in the low-risk group. More importantly, the risk score significantly and negatively correlates with DFS in recurrent HCC patients in the high-risk group. Univariate and multivariate Cox regression analyses showed that the 25-lncRNA risk score, serum AFP, TNM stage and vascular invasion (VI) were independent prognostic factors for HCC early recurrence. Moreover, compared to serum AFP, TNM stage and VI, the 25-lncRNA risk signature showed better prognostic potential for HCC early recurrence. In addition, the combination of the 25-lncRNA risk signature with serum AFP, TNM stage and VI could further improve the prognostic potential for HCC early recurrence. Meanwhile, GSEA showed that several gene sets related to malignancy, such as “E2F TARGETS”, “G2M CHECKPOINT”, “MYC TARGETS V1” and “DNA REPAIR”, have significantly enriched in the high-risk group, suggesting that lncRNAs included in this risk signature may affect HCC progression through those biological pathways. Moreover, ssGSEA revealed greater TILs in the low-risk group compared to the high-risk group and the negative association between the 25-lncRNA risk signature score and the intratumor population of several key antitumor TILs such as type 1 T helper cell, effector memory CD8 T cell and activated CD8 T cell, and SubMap algorithm predicted that the low-risk group was more sensitive to anti-PD-1 therapy. Finally, Chemotherapies prediction revealed that the low risk was associated with sensitivity to docetaxel, gefitinib and vinblastine, while high risk was associated with sensitivity to doxorubicin, mitomycin C and paclitaxel.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- HCC
Hepatocellular carcinoma
- lncRNA
long non-coding RNA
- TCGA
The Cancer Genome Atlas
- DEG
Differentially expressed gene
- LASSO
Least absolute shrinkage and selection operator
- ROC
Receiver operating characteristic curve
- GSEA
Gene set enrichment analysis
- TILs
Tumor infiltrating lymphocytes
- ssGSEA
single sample Gene Set Enrichment Analysis
- GDSC
Genomics of Drug Sensitivity in Cancer
- AJCC
American Joint Committee on Cancer
- BCLC
Barcelona Clinic Liver Cancer
- CLIP
Cancer of the Liver Italian Program
- AFP
Alpha-fetoprotein
- FDA
Food and Drug Administration
- CRC
Colorectal cancer
- LIHC
Liver Hepatocellular Carcinoma
- OS
Overall survival
- DFS
Disease free survival
- ER lncRNAs
lncRNAs associated with HCC early recurrence
- VI
Vascular invasion
- NES
Normalized enrichment scores
- PD-1
Programmed cell death protein 1
- COPD
Chronic obstructive pulmonary disease
- CVD
Cardiovascular diseases
- ICGC
International Cancer Genome Consortium
Authors’ contributions
Yi Fu, and Hailong Wu designed the project. Yi Fu, Xindong Wei and Yuhui Xu performed bioinformatics and statistical analyses. Xindong Wei and Xinjie Ling performed the experiments. Yi Fu, Qiuqin Han, Jiamei Le, Yujie Ma, Yuhui Xu, Ning Liu, Xuan Wang and Ying Tong performed extensive literature search and discussion. Yi Fu, and Hailong Wu drafted the manuscript. Hailong Wu, Xiaoni Kong and Jinyang Gu edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31671453, 31870905 to Hailong Wu, 81772507 to Jinyang Gu), the Scientific Program of Shanghai Municipal Health Commission (SHWJ2019211 to Hailong Wu), Zhejiang Medical and Health Science and Technology Project (No.: WKJ2009-2-036), Construction Project of Shanghai Key Laboratory of Molecular Imaging (18DZ2260400), Shanghai Municipal Education Commission (Class II Plateau Disciplinary Construction Program of Medical Technology of SUMHS, 2018–2020), the Key Program of National Natural Science Foundation of China (Grant No.81830052), the Hundred Teacher Talent Program (B3–0200–20-311008-23 to Yi Fu) and the University-level Scientific Fund (E3–0200–21-201011-42 to Yi Fu) of Shanghai University of Medicine and Health Sciences.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the TCGA-LIHC repository, http://cancergenome.nih.gov/.
Declarations
Ethics approval and consent to participate
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of Shanghai University of Medicine & Health Sciences. Written informed consent was obtained from individual or guardian participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinyang Gu, Email: gjynyd@126.com.
Ying Tong, Email: lilytongy@hotmail.com.
Hailong Wu, Email: wuhl@sumhs.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Shah SA, Cleary SP, Wei AC, Yang I, Taylor BR, Hemming AW, Langer B, Grant DR, Greig PD, Gallinger S. Recurrence after liver resection for hepatocellular carcinoma: risk factors, treatment, and outcomes. Surgery. 2007;141(3):330–339. doi: 10.1016/j.surg.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Sherman M. Recurrence of hepatocellular carcinoma. N Engl J Med. 2008;359(19):2045–2047. doi: 10.1056/NEJMe0807581. [DOI] [PubMed] [Google Scholar]
- 4.Portolani N, Coniglio A, Ghidoni S, Giovanelli M, Benetti A, Tiberio GA, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg. 2006;243(2):229–235. doi: 10.1097/01.sla.0000197706.21803.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P-H, Hsu C-Y, Hsia C-Y, Lee Y-H, Su C-W, Huang Y-H, Lee FY, Lin HC, Huo TI. Prognosis of hepatocellular carcinoma: assessment of eleven staging systems. J Hepatol. 2016;64(3):601–608. doi: 10.1016/j.jhep.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Tang B, Li J, Shi Y, Chen M, Lv X, Meng M, Weng Q, Zhang N, Fan K, Xu M, Ji J. Identification and validation of the angiogenic genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Aging (Albany NY) 2020;12(9):7848–7873. doi: 10.18632/aging.103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68(2):723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 8.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 10.Esserman LJ, Yau C, Thompson CK, van 't Veer LJ, Borowsky AD, Hoadley KA, et al. Use of molecular tools to identify patients with indolent breast cancers with ultralow risk over 2 decades. JAMA Oncol. 2017;3(11):1503–1510. doi: 10.1001/jamaoncol.2017.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buyse M, Loi S, van't Veer L, Viale G, Delorenzi M, Glas AM, d'Assignies MS, Bergh J, Lidereau R, Ellis P, Harris A, Bogaerts J, Therasse P, Floore A, Amakrane M, Piette F, Rutgers E, Sotiriou C, Cardoso F, Piccart MJ, TRANSBIG Consortium Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 12.Mook S, Schmidt MK, Weigelt B, Kreike B, Eekhout I, van de Vijver MJ, Glas AM, Floore A, Rutgers EJT, van ‘t Veer LJ. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann Oncol. 2010;21(4):717–722. doi: 10.1093/annonc/mdp388. [DOI] [PubMed] [Google Scholar]
- 13.Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga J, Bruin S, Kerr D, Kuppen P, van de Velde C, Morreau H, van Velthuysen L, Glas AM, van't Veer LJ, Tollenaar R. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29(1):17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 14.Maak M, Simon I, Nitsche U, Roepman P, Snel M, Glas AM, Schuster T, Keller G, Zeestraten E, Goossens I, Janssen KP, Friess H, Rosenberg R. Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann Surg. 2013;257(6):1053–1058. doi: 10.1097/SLA.0b013e31827c1180. [DOI] [PubMed] [Google Scholar]
- 15.Kopetz S, Tabernero J, Rosenberg R, Jiang Z-Q, Moreno V, Bachleitner-Hofmann T, Lanza G, Stork-Sloots L, Maru D, Simon I, Capellà G, Salazar R. Genomic classifier ColoPrint predicts recurrence in stage II colorectal cancer patients more accurately than clinical factors. Oncologist. 2015;20(2):127–133. doi: 10.1634/theoncologist.2014-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei R, Huang GL, Zhang MY, Li BK, Zhang HZ, Shi M, Chen XQ, Huang L, Zhou QM, Jia WH, Zheng XFS, Yuan YF, Wang HY. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res. 2013;19(17):4780–4791. doi: 10.1158/1078-0432.CCR-12-2728. [DOI] [PubMed] [Google Scholar]
- 17.Nault JC, De Reynies A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145(1):176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Sohn BH, Lee HS, Kim SB, Yoo JE, Park YY, Jeong W, Lee SS, Park ES, Kaseb A, Kim BH, Kim WB, Yeon JE, Byun KS, Chu IS, Kim SS, Wang XW, Thorgeirsson SS, Luk JM, Kang KJ, Heo J, Park YN, Lee JS. Genomic predictors for recurrence patterns of hepatocellular carcinoma: model derivation and validation. PLoS Med. 2014;11(12):e1001770. doi: 10.1371/journal.pmed.1001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y, Lin H, Chen J, Wu Y, Yu S. Identification of prognostic glycolysis-related lncRNA signature in tumor immune microenvironment of hepatocellular carcinoma. Front Mol Biosci. 2021;8:645084. doi: 10.3389/fmolb.2021.645084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng J, Zhou C, Zhou Q, Chen W, Yin Y, Atyah M, Dong Q, Shi Y, Ren N. Development and validation of a metabolic gene-based prognostic signature for hepatocellular carcinoma. J Hepatocell Carcinoma. 2021;8:193–209. doi: 10.2147/JHC.S300633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X, Bi Q, Chen S, Chen X, Li S, Zhong Z, Guo W, Li X, Deng Y, Yang Y. Identification of a five-autophagy-related-lncRNA signature as a novel prognostic biomarker for hepatocellular carcinoma. Front Mol Biosci. 2020;7:611626. doi: 10.3389/fmolb.2020.611626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schönbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y, FANTOM Consortium. RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 23.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu TP, et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol. 2015;8(50):1–14. doi: 10.1186/s13045-015-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, Karni R. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77(5):1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan S-X, Yang F, Yang Y, Tao Q-F, Zhang J, Huang G, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology (Baltimore, Md) 2012;56(6):2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 27.Huang J-L, Cao S-W, Ou Q-S, Yang B, Zheng S-H, Tang J, Chen J, Hu YW, Zheng L, Wang Q. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer. 2018;17(1):93. doi: 10.1186/s12943-018-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model. New York: Springer; 2000. [Google Scholar]
- 31.Pathan M, Keerthikumar S, Ang C-S, Gangoda L, Quek CYJ, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, Stroud DA, Ryan MT, Agbinya JI, Mariadason JM, Burgess AW, Mathivanan S. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 32.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. J Stat Softw. 2011;39(5):1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, Tibshirani RJ. Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Series B Stat Methodol. 2012;74(2):245–266. doi: 10.1111/j.1467-9868.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall R. regplot: Enhanced Regression Nomogram Plot. R package version 1.1 2020 [Available from: https://CRAN.R-project.org/package=regplot.
- 36.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14(1):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer Immunogenomic analyses reveal genotype-Immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, Liu XS. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020;12(1):21. doi: 10.1186/s13073-020-0721-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoshida Y, Brunet JP, Tamayo P, Golub TR, Mesirov JP. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16(4):385–395. doi: 10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Liu J, Chen J, Wang H, Yang L, Chen F, Fan S, Wang J, Shao B, Yin D, Zeng M, Li M, Li J, Su F, Liu Q, Yao H, Su S, Song E. A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat Commun. 2018;9(1):1614. doi: 10.1038/s41467-018-03537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y-Q, Zhang Y, Jiang W, Chen Y-P, Xu S-Y, Liu N, Zhao Y, Li L, Lei Y, Hong XH, Liang YL, Li JY, Zhang LL, Yun JP, Sun Y, Li YQ, Ma J. Development and validation of an immune checkpoint-based signature to predict prognosis in nasopharyngeal carcinoma using computational pathology analysis. J Immunother Cancer. 2019;7(1):298. doi: 10.1186/s40425-019-0752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Zhang L, Xu Y, Wu X, Zhou Y, Mo J. Immune-related long noncoding RNA signature for predicting survival and immune checkpoint blockade in hepatocellular carcinoma. J Cell Physiol. 2020;235(12):9304–9316. doi: 10.1002/jcp.29730. [DOI] [PubMed] [Google Scholar]
- 46.Mao X, Qin X, Li L, Zhou J, Zhou M, Li X, Xu Y, Yuan L, Liu QN, Xing H. A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol Oncol. 2018;149(1):181–187. doi: 10.1016/j.ygyno.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Zhu X, Tian X, Yu C, Shen C, Yan T, Hong J, Wang Z, Fang JY, Chen H. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15(1):60. doi: 10.1186/s12943-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai J, Li B, Zhu Y, Fang X, Zhu M, Wang M, Liu S, Jiang X, Zheng J, Zhang XX, Chen P. Prognostic biomarker identification through integrating the gene signatures of hepatocellular carcinoma properties. EBioMedicine. 2017;19:18–30. doi: 10.1016/j.ebiom.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fluss R, Faraggi D, Reiser B. Estimation of the Youden index and its associated cutoff point. Biom J. 2005;47(4):458–472. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- 50.Cai C, Yang L, Tang Y, Wang H, He Y, Jiang H, Zhou K. Prediction of overall survival in gastric Cancer using a nine-lncRNA. DNA Cell Biol. 2019;38(9):1005–1012. doi: 10.1089/dna.2019.4832. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Chen S-W, Liu L-L, Yang X, Cai S-H, Yun J-P. A model combining TNM stage and tumor size shows utility in predicting recurrence among patients with hepatocellular carcinoma after resection. Cancer Manag Res. 2018;10:3707–3715. doi: 10.2147/CMAR.S175303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. 2013;70(24):4785–4794. doi: 10.1007/s00018-013-1423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, Patterson JW, Slingluff CL., Jr Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012;72(5):1070–1080. doi: 10.1158/0008-5472.CAN-11-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 55.Santoiemma PP, Powell DJ., Jr Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol Ther. 2015;16(6):807–820. doi: 10.1080/15384047.2015.1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shirabe K, Matsumata T, Maeda T, Sadanaga N, Kuwano H, Sugimachi K. A long-term surviving patient with hepatocellular carcinoma including lymphocytes infiltration--a clinicopathological study. Hepatogastroenterology. 1995;42(6):996–1001. [PubMed] [Google Scholar]
- 57.Wada Y, Nakashima O, Kutami R, Yamamoto O, Kojiro M. Clinicopathological study on hepatocellular carcinoma with lymphocytic infiltration. Hepatology. 1998;27(2):407–414. doi: 10.1002/hep.510270214. [DOI] [PubMed] [Google Scholar]
- 58.Shen Y, Peng X, Shen C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics. 2020;112(3):2640–2646. doi: 10.1016/j.ygeno.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 59.Shrestha R, Prithviraj P, Anaka M, Bridle KR, Crawford DHG, Dhungel B, Steel JC, Jayachandran A. Monitoring immune checkpoint regulators as predictive biomarkers in hepatocellular carcinoma. Front Oncol. 2018;8:269. doi: 10.3389/fonc.2018.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roh W, Chen PL, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379):eaah3560. doi: 10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543(7644):199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruan X, Li P, Chen Y, Shi Y, Pirooznia M, Seifuddin F, Suemizu H, Ohnishi Y, Yoneda N, Nishiwaki M, Shepherdson J, Suresh A, Singh K, Ma Y, Jiang CF, Cao H. In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nat Commun. 2020;11(1):45. doi: 10.1038/s41467-019-13688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015. doi: 10.3390/cells8091015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Imamura H, Matsuyama Y, Tanaka E, Ohkubo T, Hasegawa K, Miyagawa S, Sugawara Y, Minagawa M, Takayama T, Kawasaki S, Makuuchi M. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38(2):200–207. doi: 10.1016/S0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 66.Shah SA, Greig PD, Gallinger S, Cattral MS, Dixon E, Kim RD, Taylor BR, Grant DR, Vollmer CM. Factors associated with early recurrence after resection for hepatocellular carcinoma and outcomes. J Am Coll Surg. 2006;202(2):275–283. doi: 10.1016/j.jamcollsurg.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Jung SM, Kim JM, Choi GS, Kwon CHD, Yi NJ, Lee KW, Suh KS, Joh JW. Characteristics of early recurrence after curative liver resection for solitary hepatocellular carcinoma. J Gastrointest Surg. 2019;23(2):304–311. doi: 10.1007/s11605-018-3927-2. [DOI] [PubMed] [Google Scholar]
- 68.Ward M, McEwan C, Mills JD, Janitz M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J Hum Transcr. 2015;1(1):2–9. doi: 10.3109/23324015.2015.1077591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evangelou K, Havaki S, Kotsinas A. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol. 2014;20(29):10212–10216. doi: 10.3748/wjg.v20.i29.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and Cancer. Cancer Discov. 2015;5(10):1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7(11):861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 72.Gavande NS, VanderVere-Carozza PS, Hinshaw HD, Jalal SI, Sears CR, Pawelczak KS, et al. DNA repair targeted therapy: the past or future of cancer treatment? Pharmacol Ther. 2016;160:65–83. doi: 10.1016/j.pharmthera.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pages F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29(8):1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 74.Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, Lim KH, Weber A, Chow P, Chung A, Ooi LLPJ, Toh HC, Heikenwalder M, Ng IOL, Nardin A, Chen Q, Abastado JP, Chew V. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66(2):342–351. doi: 10.1136/gutjnl-2015-310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chew V, Chen J, Lee D, Loh E, Lee J, Lim KH, Weber A, Slankamenac K, Poon RTP, Yang H, Ooi LLPJ, Toh HC, Heikenwalder M, Ng IOL, Nardin A, Abastado JP. Chemokine-driven lymphocyte infiltration: an early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut. 2012;61(3):427–438. doi: 10.1136/gutjnl-2011-300509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset supporting the conclusions of this article is available in the TCGA-LIHC repository, http://cancergenome.nih.gov/.