Abstract
Transforming growth factor β (TGF-β)-activated kinase 1 (TAK1) is a member of the MAPKKK superfamily and has been characterized as a component of the TGF-β/bone morphogenetic protein signaling pathway. TAK1 function has been extensively studied in cultured cells, but its in vivo function is not fully understood. In this study, we isolated a Drosophila homolog of TAK1 (dTAK1) which contains an extensively conserved NH2-terminal kinase domain and a partially conserved COOH-terminal domain. To learn about possible endogenous roles of TAK1 during animal development, we generated transgenic flies which express dTAK1 or the mouse TAK1 (mTAK1) gene in the fly visual system. Ectopic activation of TAK1 signaling leads to a small eye phenotype, and genetic analysis reveals that this phenotype is a result of ectopically induced apoptosis. Genetic and biochemical analyses also indicate that the c-Jun amino-terminal kinase (JNK) signaling pathway is specifically activated by TAK1 signaling. Expression of a dominant negative form of dTAK during embryonic development resulted in various embryonic cuticle defects including dorsal open phenotypes. Our results strongly suggest that in Drosophila melanogaster, TAK1 functions as a MAPKKK in the JNK signaling pathway and participates in such diverse roles as control of cell shape and regulation of apoptosis.
During the development of multicellular organisms, cells receive various extracellular stimuli. These signals are propagated from the cell surface to the nucleus via specific sets of intracellular signaling molecules. Mitogen-activated protein kinase (MAPK) is one of the intracellular signaling molecules commonly activated by various stimuli and plays a crucial role in cell proliferation, differentiation, and regulation of early development (41, 53, 65). MAPK is activated by a sequential cascade of protein kinases; MAPK is activated by dual phosphorylation catalyzed by MAPK kinase (MAPKK), which is itself phosphorylated and activated by a MAPKK kinase (MAPKKK) (41, 47). At least three MAPK modules, the MAPK/extracellular signal-regulated kinase (ERK) pathway, the c-Jun amino-terminal kinase (JNK)/stress-activated protein kinase (SAPK) pathway, and the p38 MAPK pathway, participate in distinct (but sometimes partially overlapping) functions in various biological processes (9, 11, 67).
Transforming growth factor β(TGF-β)-activated kinase 1 (TAK1) is a member of the MAPKKK superfamily (71). The function of TAK1 has been extensively studied in transient transfection assays using cultured cells (44, 52, 54–56, 71). These studies have revealed that TAK1 can function in a signal transduction pathway that is triggered by the TGF-β superfamily of ligands. Stimulation of cells with TGF-β or bone morphogenetic protein 4 (BMP4) activates TAK1 activity in cultured cells and, in the case of TGF-β, leads to induction of plasminogen activator inhibitor 1 (PAI-1), a TGF-β-responsive gene (71). In addition, it has been shown that TAK1 has a role in a TGF-β-independent signaling pathway. Ceramide is thought to be a second messenger molecule which has been implicated in a variety of biological processes (28, 38). Ceramide stimulates the kinase activity of TAK1, and ceramide-induced JNK/SAPK activation can be blocked by expression of a dominant negative form of TAK1 (56). In addition, biochemical studies have revealed that MAPKK4 and MAPKK3/MAPKK6 are substrates of TAK1 (44, 56, 66), indicating that TAK1 can activate the JNK pathway and/or the p38 MAPK pathway in cultured cells.
Despite these extensive biochemical studies of TAK1 in cultured cells, the in vivo function of TAK1 is not fully understood. Overexpression of Xenopus TAK1 (xTAK1) and an upstream activator called TAB1 (TAK1 binding protein 1) can induce ventral mesoderm and inhibit neural differentiation. Overexpression epistasis experiments using activated and dominant negative forms of TAK1 and BMP or activin receptors place the activity of TAK1 downstream of the BMP2/4 receptors. Furthermore, TAK1 has been shown to act downstream or in parallel to Smad1 and Smad4 (71). In addition to its role in BMP signaling, ectopic expression of xTAK1 in early Xenopus embryos also induced apoptosis. This finding suggests that TAK1 may mediate a number of different processes (54).
To further address the in vivo function of TAK1, we took a transgenic approach in the Drosophila model system. There are several advantages to using Drosophila melanogaster as a functional assay system. First, transgenic flies obtained using P-element-mediated germ line transformation (58) can be used for genetic interaction studies to screen for downstream or upstream signaling components (6, 63). Second, expression of exogenous genes can be temporally and spatially controlled using either tissue-specific promoters or the GAL4 upstream activation sequence (UAS) system (5, 6, 16, 45, 64). Third, basic MAPK cascades are conserved between vertebrates and Drosophila. For example, the Drosophila MAPK/ERK cascade consists of D-raf, Dsor1, and Rolled, which correspond to Raf, MAPK/ERK kinase (MEK), and ERK, respectively, in vertebrates (15). The Drosophila JNK pathway consists of Basket (Bsk) and Hemipterous (Hep), corresponding to JNK and MKK7, respectively, in vertebrates (21, 26, 35, 49, 51, 57). In addition, two Drosophila p38 (D-p38) homologs and a Drosophila MKK3 have recently been cloned (1, 25, 26, 61).
In this report, we describe the effects of overexpressing mouse TAK1 (mTAK1) and a newly discovered Drosophila homolog (dTAK1) in developing eyes and embryos. The results indicate that TAK1 can specifically activate the JNK pathway in vivo and mediate both cell shape and apoptotic responses in Drosophila.
MATERIALS AND METHODS
Identification of a Drosophila TAK1-like sequence.
A 1.9-kb SalI-SpeI fragment containing most of the mTAK1 open reading frame (71) was random prime labeled with 32P and used to probe a Drosophila genomic library (Stratagene) under low-stringency conditions (46). Several positive plaques were picked and purified. A 2.0-kb genomic SalI/EcoRI fragment which showed the strongest hybridization to mTAK1 was subcloned into pBluescript KS+ and partially sequenced. This sequence confirmed that the 2.0-kb fragment contained an open reading frame with similarity to mTAK1. The 2.0-kb genomic fragment was labeled and used to probe a directional Drosophila lambda GT22a ovarian cDNA library (59). Positive plaques were purified and grown up as phage, and DNA inserts were excised as SalI/NotI fragments. Two cDNAs of 2.2 and 3.4 kb were subcloned into pBluescript KS+. The 3.4-kb subcloned fragment was sequenced by primer walking using a U.S. Biochemicals cycle sequencing kit. A search of the Drosophila expressed sequence tag (EST) database identified three dTAK1 EST clones, GM05307, GM05309, and GM09711 (Berkeley Drosophila Genome Project).
Plasmid construction and generation of transgenic flies.
Ectopic expression of the mTAK1 and human TAB1 (hTAB1) constructs were achieved using GAL4 /UAS system (6) and an eye-specific expression vector, pGMR (31). The full-length mTAK1 cDNA was cloned as a EcoRI-XhoI fragment from pEF-mTAK1 (71) into EcoRI-XhoI-digested pUAST (6) and as an EcoRI-KpnI fragment into EcoRI-KpnI-digested pGMR (31). The mTAK1ΔN cDNA, which encodes an activated form of mTAK1, a truncation lacking the NH2-terminal 22 amino acids, was cloned as an EcoRI-XbaI fragment from pEF-mTAK1ΔN (71) into EcoRI-XbaI-digested pUAST and as an EcoRI-synthesized blunt end of a XbaI cleavage site fragment into EcoRI-StuI-digested pGMR. The mTAK1-K63W cDNA, which encodes a dominant negative form of mTAK1 (54, 71) in which lysine 63 was replaced by tryptophan, was cloned as an EcoRI-DraI fragment from pBS-mTAK1-K63W (71) into EcoRI-StuI-digested pGMR. The hTAB1 cDNA was cloned as an EcoRI-SmaI fragment from pBS-hTAB1 (55) into EcoRI-HpaI-digested pGMR. dTAK1 was also subcloned into pUAST. The full-length dTAK1 cDNA was cloned as an EcoRI-NotI fragment from pBS-dTAK1 into EcoRI-NotI-digested pUAST. The dTAK1-K46R cDNA, which encodes a dominant negative form of dTAK1 in which lysine 46 was replaced by arginine, was cloned as an EcoRI-NotI fragment from pBS-dTAK1-K46R into EcoRI-NotI-digested pUAST. Flies bearing transgenes were generated by P-element-mediated germ line transformation (58). At least five independent transformant lines were obtained for each transgenic construct. Two copies of the transgenes (pGMR-mTAK1-K63W and UAS-dTAK1-K46R) were required to reveal the dominant negative phenotype. Most of the flies expressing dTAK1, GMR-GAL4; UAS-dTAK1 died at early pupal stages at 25°C, presumably due to leaky GAL4 expression. To circumvent the lethality, we cultured these flies at 18°C.
Fly strains.
Drosophila cultures and crosses were carried out by standard procedures at 25°C. To test for modification of the TAK1 phenotype, each mutant was crossed with three different pGMR-mTAK1ΔN lines which show similar but different extents of the small eye phenotypes. Interaction with the TAK1 phenotype was scored as positive only if a similar effect was observed with all three pGMR-mTAK1ΔN lines. The following mutant strains were used for the genetic interaction assay: tldE4, tld15, tldB4, dppd6, dppd12, scwE2, Mad12, Med4, tkv5, tkv7, tkv8, sax1, put135, put10460, shn1, shn04738, omb3198, N55e11, DlRF, argos257, spi1, sev14, EgfrE3, flbf6, Sos34Ea-7, SosJC2, Gap1B2, Ras1e2F, phl1, Dsor1Su1, Dsor1r2, Dsor1LH110, rlsem, hep1, hepr75, bsk1, bsk2, and flp147E strains. We also tested three transgenic strains, pGMR-p35, pGMR-DIAP1, and pGMR-DIAP2 strains (30, 31). Df(3L)H99 was described previously (23). We also preformed the genetic interaction assay using GMR-GAL4/UAS-dTAK1 and obtained results similar to that seen for pGMR-mTAK1ΔN lines (only the data for GMR-GAL4/UAS-dTAK1 crosses are shown in the figures). Targeted expression of UAS-driven transgenes (6) was induced using the GAL4 lines Dll-GAL4, elav-GAL4, hs-GAL4, en-GAL4, GMR-GAL4, and pnr-Gal4. hepr75 is a lethal allele of the hep mutants; however, some escapers of the hemizygous males survive during the pupal stage. The pupal eyes of the hepr75 males were subjected to cobalt sulfide staining to detect the loss-of-JNK signaling phenotype.
Histology.
Flies were prepared for scanning electron micrographs as described by Kimmel et al. (36). Adult eyes were prepared for tangential histological sections. Adult heads were fixed at 4°C overnight in a mixture of 2.5% glutaraldehyde and 3.7% paraformaldehyde in 0.1 M phosphate buffer (pH 7.0). After a buffer wash, the heads were dehydrated through a graded acetone series. After dehydration, the heads were transferred into 60% LR white resin (Polysciences) in acetone at 4°C for 1.5 h. Then the heads were replaced with pure resin for 8 h and polymerized at 55°C overnight. Embedded heads were sectioned on an ultramicrotome and viewed under phase-contrast optics. Cobalt sulfide staining of pupal retinas was done as described by Wolff and Ready (70). Staging was carried out by aging white prepupae at 25°C. Acridine orange staining was performed by the method of Wolff and Ready (70). For 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining, embryos were collected and stained for β-galactosidase activity according to standard protocols (2). In situ hybridization to whole-mount eye discs was carried out as described previously (62). Riboprobes of rpr, hid, and dpp were made by using a DIG RNA labeling kit (Boehringer Mannheim) and hybridized at 55°C. The probes were detected with a monoclonal antibody against digoxigenin coupled to alkaline phosphatase and BM purple as a substrate (Boehringer Mannheim). Cuticle preparation was performed according to a standard protocol (2) except that embryos were not fixed before mounting.
Phosphorylation analysis.
Anti-JNK1 antibody (Santa Cruz), anti-phospho-JNK (p-JNK) antibody (Promega), anti-D-p38b antibody (a gift from T. Adachi-Yamada [1]), and anti-phospho-p38 (p-p38) antibody (Santa Cruz) were used. Flies of genotypes UAS-dTAK1; hs-GAL4 and hs-GAL4 (as a wild-type control) were subjected to heat shock (twice at 37°C for 30 min with a 30-min interval). Flies at the third larval stage were collected (just before or 5 h after the heat shock) and homogenized. Their extracts were immunoblotted with one of the antibodies described above after separation in a sodium dodecyl sulfate-polyacrylamide gel.
Nucleotide sequence accession number.
The GenBank accession number for the dTAK1 cDNA reported here is AF199466.
RESULTS
Ectopic expression of mTAK1 affects eye formation.
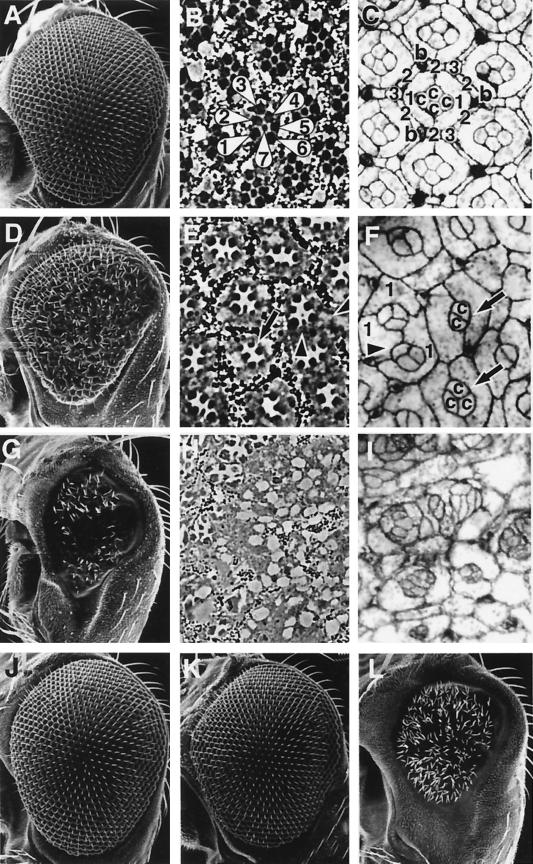
To study the in vivo function of TAK1, we generated transgenic flies carrying the wild-type mouse TAK1 (UAS-mTAK1) or a truncated, constitutively active form of mTAK1 (UAS-mTAK1ΔN). Ectopic expression of the wild-type mTAK1 showed various defects in different adult tissues. For example, UAS-mTAK1/Dll-GAL4 flies were lacking the distal parts of legs and antenna, whereas UAS-mTAK1/elav-GAL4 flies showed a rough eye phenotype (data not shown). Ectopic expression of the activated form of mTAK1 (mTAK1Δ1N) resulted in lethality in early developmental stages. To circumvent the early lethality, we ectopically expressed mTAK1 and mTAK1ΔN in the adult eye using an eye-specific expression vector, pGMR. pGMR contains multimerized Glass-binding sites and promotes gene expression in all cells in and posterior to the morphogenetic furrow in the larval eye disc (16, 45). Eye-specific expression of the TAK1 transgenes induced specific defects in the visual system. pGMR-mTAK1 transgenic flies showed relatively weak defects in the compound eyes. The phenotype varied among transgenic strains: 6 out of 11 transgenic strains revealed a rough eye phenotype and a reduction in size of the compound eye. Figure 1D shows the eye phenotype of a medium-strength pGMR-mTAK1 line. The eye size is reduced to about 70% of the wild-type eye size. Many ommatidia, typically located in the medial region, were fused to each other (Fig. 1D). Electron micrographs of ommatidial cross sections of this mutant revealed that some ommatidia were missing photoreceptors, while others showed disrupted rows of pigment granules (Fig. 1E). In the pupal eye disc, a wild-type ommatidium contains four cone cells, two primary pigment cells, which are surrounded by six secondary pigment cells, three tertiary pigment cells, and three sensory bristles (Fig. 1C). In ommatidia from mTAK1 overexpression lines, only two to three cone cells were observed. The number of primary pigment cells was relatively normal, but occasionally one of the primary pigment cells was also missing (Fig. 1F). Secondary and tertiary pigment cells seemed to be heavily affected by mTAK1 expression, as these cells were rarely observed in the mutant retina (Fig. 1F).
FIG. 1.
Phenotypes induced by ectopic TAK1 signaling. Scanning electron micrographs of the compound eye (A, D, G, J, K, and L), tangential histological sections (B, E, and H), and pupal eyes (at 40 h after puparium formation) stained with cobalt sulfide (C, F, and I) are shown. (A to C) Canton-S. (A) The wild-type eye is composed of a regular array of about 800 ommatidia. (B) Each ommatidium contains six outer photoreceptor cells and an inner photoreceptor cell, R7 (R1 to R7 are indicated with numbers). (C) Cobalt sulfide staining shows the apical profile of cells in the epithelium. Each ommatidium contains four cone cells (indicated with “c”) and two primary pigment cells (“1”), surrounded by six secondary pigment cells (“2”), three tertiary pigment cells (“3”), and three interommatidial bristles (“b”). (D to F) pGMR-mTAK overexpression. (E) Most of the ommatidia contain only five to six photoreceptors (arrow), and visual pigments are also disrupted at many positions (arrowheads). (F) In the pupal eye, most of the ommatidia contain only two or three cone cells (arrows) and occasionally are also missing primary pigment cells (arrowhead). Numerous numbers of interommatidial cells are also missing in this mutant. (G to I) pGMR-mTAK1ΔN overexpression. (G) Expression of mTAK1ΔN in the developing eye results in a severe decrease in size of the compound eyes. (H and I) Ommatidial structures are totally disrupted and hard to discriminate in the adult head section and pupal eye disc. (J) pGMR-hTAB1. Expression of hTAB1 alone in the eye has no effect on eye development. (K) Weak pGMR-mTAK1 overexpression phenotype. (L) Coexpression of hTAB1 and mTAK1 (weak line as shown in panel K) causes a phenotype as severe as pGMR-mTAK1ΔN overexpression, as shown in panel G. All pictures are shown with anterior to the left and dorsal up.
mTAK1ΔN was also expressed using the pGMR vector. In contrast to the pGMR-mTAK1 transgenic line, all pGMR-mTAK1ΔN transgenic files (eight independent transgenic lines) displayed severe eye phenotypes. In the weakest line (Fig. 1G), eye size was less than 30% of the wild type and ommatidial units were rarely identified. In addition, photoreceptor cells and pigment granules were difficult to recognize (Fig. 1H). In the mutant pupal eye disc, cone cells, primary pigment cells, and accessory cells existed in a disorganized array (Fig. 1I). The results suggest that the strength of signaling activity of mTAK1ΔN is stronger than that of intact mTAK1 in Drosophila. Shibuya et al. have also reported that TAB1 activates TAK1 signaling activity in cultured cells and Xenopus (54, 55). If TAK1 is activated by TAB1 in Drosophila, the pGMR-mTAK1 phenotype should be enhanced by the coexpression of TAB1. Expression of hTAB1 alone did not lead to any defect in the eye (Fig. 1J). However, when hTAB1 was coexpressed with mTAK1, the reduced eye phenotype was dramatically enhanced (Fig. 1L), consistent with the observations from the cultured cell and Xenopus (54, 55).
Isolation of dTAK1.
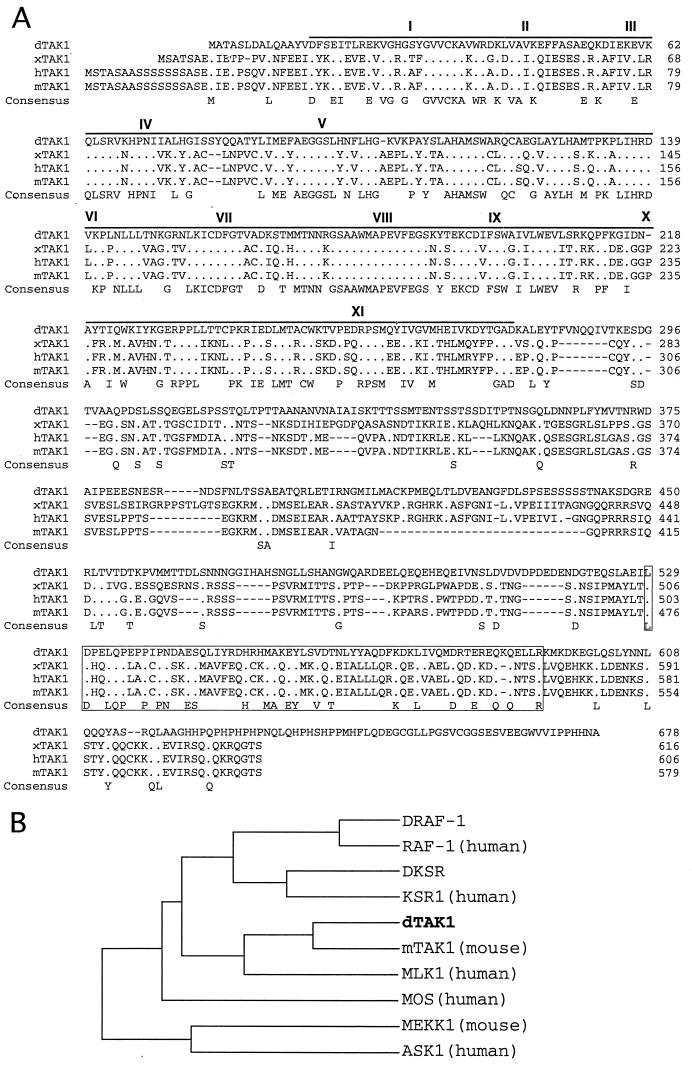
The results of the ectopic mTAK1 and hTAB1 expression indirectly suggest that a TAK1 homolog is likely to exist in Drosophila. To examine this issue further, we searched for a Drosophila homolog of TAK1, using the mTAK1 cDNA as a probe on a Drosophila genomic library under low-stringency conditions. A 2.0-kb genomic fragment was obtained and confirmed by sequencing to be related to vertebrate TAK1. A Drosophila ovarian cDNA library was then screened using the 2.0-kb genomic fragment as a probe. Two cDNAs of 2.2 and 3.4 kb were isolated and sequenced. The 3.4-kb clone contained a full-length cDNA encoding a 678-amino-acid protein which we refer to as dTAK1 (Fig. 2A). The overall protein appeared similar to the vertebrate TAK1s, containing an NH2-terminal protein kinase domain as well as a long COOH-terminal domain (Fig. 2A). The kinase domain showed 56% identity and 73% similarity with the amino acid sequences of mTAK1 (Fig. 2A). Phylogenetic analysis of the catalytic domain of dTAK1 with those of other Drosophila and vertebrate MAPKKK proteins indicated that its closest relatives are the vertebrate TAK1s (Fig. 2B). In the COOH-terminal region, dTAK1 is less well conserved with its vertebrate homologs. However, there is a stretch of 65 amino acids that is relatively well conserved between Drosophila and vertebrates (37% identity and 57% similarity to mTAK1 [Fig. 2A]). Interestingly, this region is almost completely missing in one of the alternative spliced forms of the human TAK1 (hTAK1c) (52). Shibuya et al. have reported that TAB2 binds directly to the COOH-terminal region of mTAK1 (55). The functional significance the TAB2-TAK1 interaction is not known; however, it seems likely that the COOH-terminal region of TAK1, perhaps through this conserved 65-amino-acid stretch, may help regulate TAK1 kinase activity and/or provide signaling specificity through physical interactions with other signaling molecule(s).
FIG. 2.
Primary structure of dTAK1. (A) The dTAK1 primary sequence is compared with that of xTAK1, one of the three alternative splicing forms of hTAK1 (hTAK1b), and mTAK1. The protein sequences are presented in single-letter code. Gaps (−) were introduced to optimize the alignment. Identical residues are indicated with periods. The protein kinase domain sequence is shown by overline, and sequences corresponding to conserved kinase subdomains I to XI (27) are indicated by roman numerals. The 65-residue stretch of amino acids in the COOH-terminal domain that is conserved between TAK1s is boxed. (B) Relationship between catalytic domains of members of the vertebrate and Drosophila MAPKKK group, presented as a dendrogram created using the Gene Works program (version 2.0; IntelliGenetics). The figure presents the analysis of the human MAPKKKs RAF-1, KSR1, MLK1, MOS, and ASK1 (3, 14, 34, 63, 68), the mouse MAPKKKs TAK1 and MEKK1 (40, 71), and the Drosophila MAPKKKs DRAF-1, DKSR (48, 63), and dTAK1.
dTAK1 behaves similarly to mTAK1 and facilitates apoptosis in the eyes.
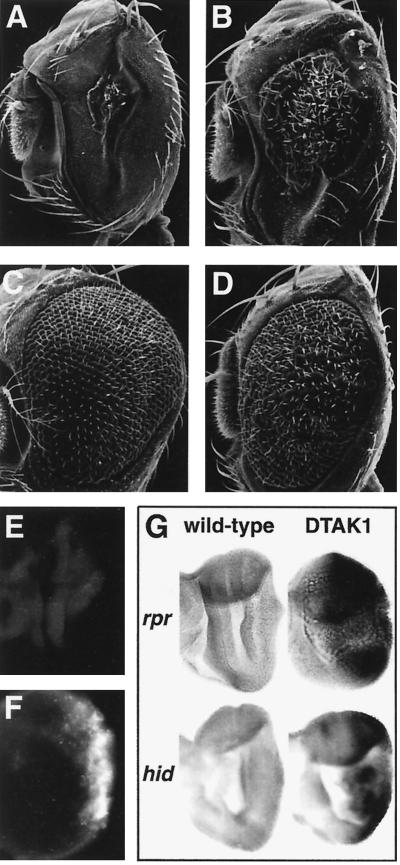
To test whether dTAK1 is a true functional ortholog of mTAK1, we overexpressed dTAK1 under the control of the GMR-GAL4 driver. A small eye phenotype similar to that produced by pGMR-mTAK1ΔN (activated form) was obtained (Fig. 3A and B). The difference in the strength of dTAK1 enzymatic activity compared to the nonactivated mTAK1 might be explained by the difference in the NH2-terminal structures of these proteins. mTAK1 contains a serine-rich NH2-terminal domain which has been shown to down regulate mTAK1 kinase activity (71). Interestingly, dTAK1 lacks this domain, suggesting that dTAK1 may have a higher basal level of activity in the absence of an activator protein(s), such as TAB1, than does the vertebrate gene product (54, 55).
FIG. 3.
Ectopic TAK1 signaling induces apoptosis in the developing eye. (A) GMR-GAL4/UAS-dTAK1(strong); (B) GMR-GAL4/UAS-dTAK1(weak); (C) GMR-GAL4/UAS-dTAK1(weak); pGMR-p35; (D) GMR-GAL4/UAS-dTAK1(weak); Df(3L)H99/+. The deficiency of Df(3L)H99 uncovers three proapoptotic genes, rpr, hid, and grim. The reduced eye phenotype of GMR-GAL4/UAS-dTAK1(weak) (B) is rescued either by coexpression of p35 (C) or by a heterozygous mutant background which removes the proapoptotic genes (D). (E and F) Acridine orange staining of eye discs at late third-instar larval stage of Canton-S (E) and GMR-GAL4/UAS-dTAK1 (F) flies. Acridine orange-positive cells are rare in the wild-type eye disc (E) but abundant in dTAK1 overexpression eye discs predominantly posterior to the morphogenetic furrow (F). (G) Eye discs from wild-type flies (left two discs) or GMR-GAL4/UAS-dTAK1 flies (right) were labeled with an rpr antisense riboprobe (top) or a hid antisense riboprobe (bottom). rpr and hid expression is not evident in the wild-type eye disc. dTAK1 expression induces rpr and hid most significantly in regions posterior to the morphogenetic furrow.
Next, we further analyzed the cause of the small eye phenotype induced by ectopic TAK1 signaling. Acridine orange staining, which identifies dying cells (70), was used to examine the extent of cell death in larval eye discs of dTAK1-overexpressing flies. This study revealed that dying cells increased dramatically in regions posterior to the morphogenetic furrow (Fig. 3F), suggesting that cell death occurred rapidly after dTAK1 induction. There are several possibilities that might account for the induction of cell death in the eye disc in response to ectopic TAK1 signaling. One possibility is that TAK1 signaling might directly activate a cell death signaling pathway leading to apoptosis. Alternatively, overpression of TAK1 leads to necrosis as the result of abnormal cell fate specification. To distinguish between these possibilities, we examined whether the phenotype of flies expressing dTAK1 could be suppressed by specific apoptosis inhibitors. We found that the reduced eye phenotype of dTAK1 overexpression was rescued by coexpression of the apoptotic inhibitor protein p35 (31) (Fig. 3C). We also found that the Drosophila inhibitor of apoptosis proteins 1 and 2 (DIAP1 and DIAP2) (30) effectively rescued the phenotype (data not shown). Moreover, when we examined the effects of introducing the deficiency H99, which uncovers the proapoptotic genes reaper (rpr), head involution defective (hid), and grim (10, 23, 69), into a background expressing dTAK1, the eye was restored to normal size (Fig. 3D). Figure 3G shows that the expression of rpr and hid is dramatically induced in regions posterior to the morphogenetic furrow in the eye discs of GMR-GAL4/UAS-dTAK overexpression lines compared to the wild type. Since overexpression of hid in the eye disc does not induce rpr and vice versa (data not shown), it is likely that rpr and hid are independently induced by the TAK1 signal. These data indicate that the cell death induced by dTAK1 is apoptotic in nature and that its induction is dependent on endogenous proapoptotic gene activity.
We also examined the effect of ectopic TAK1 expression on cell fate determination of the photoreceptor cells. Sequential recruitment of the photoreceptor cells was disrupted in dTAK1-overexpressing discs (Fig. 4C and D). Early photoreceptor cell (R8 and occasionally R2 and R5) induction appeared to occur normally. However, later photoreceptor induction was delayed or disrupted, and photoreceptor clustering showed an abnormal and diffuse pattern even in the presence of p35 (Fig. 4C to F). This result indicates that ectopic TAK1 signaling may also directly or indirectly affect certain aspects of cell fate specification. Maybe this abnormal photoreceptor induction affects later cell development which includes cone cell and pigment cells. However, we found that proper numbers of the cone cells and primary pigment cells were induced in the pupal eye discs from pGMR-mTAK1ΔN; pGMR-p35 (data not shown). We conclude that TAK1 induces cell death after proper cell fate determination in at least some of the ommatidial cells such as R8. However, in other cells, abnormal cell fate determination by the ectopic TAK1 signaling may also contribute to induction of cell death in the TAK1-expressing eye discs.
FIG. 4.
Effect of dTAK1 expression in photoreceptor cell induction. Late third-instar eye discs were doubly labeled with anti-Elav antibody (A, C, E) and for rhomboid lacZ (X81) expression. Anti-Elav antibody stains all of the photoreceptor cells (R1 to R8). X81 expresses lacZ strongly in R8 and relatively weakly in R2 and R5 (19). (A and B) Wild type; (C and D) GMR-GAL4/UAS-dTAK1; (E and F) GMR-GAL4/UAS-dTAK1; pGMR-p35. R8 induction occurs normal in GMR-GAL4/UAS-dTAK1 (D) and GMR-GAL4/UAS-dTAK1; pGMR-p35 (F) eye discs. R2 and R5 are occasionally induced in the GMR-GAL4/UAS-dTAK1; pGMR-p35 disc (indicated with arrows in panel F). Photoreceptor cell markers are disordered and diffuse in more posterior regions (C to F). This result indicates that dTAK1 overexpression altered or delayed specification of the photoreceptor cells, particularly for R3, R4, R1, R6, and R7. All images are presented with anterior to the left.
We repeated all of the experiments described above using mTAK1ΔN overexpression strains and obtained similar results. On this basis, we conclude that dTAK1 is a functional homolog of mTAK1.
TAK1 specifically activates the Hep-Bsk cascade.
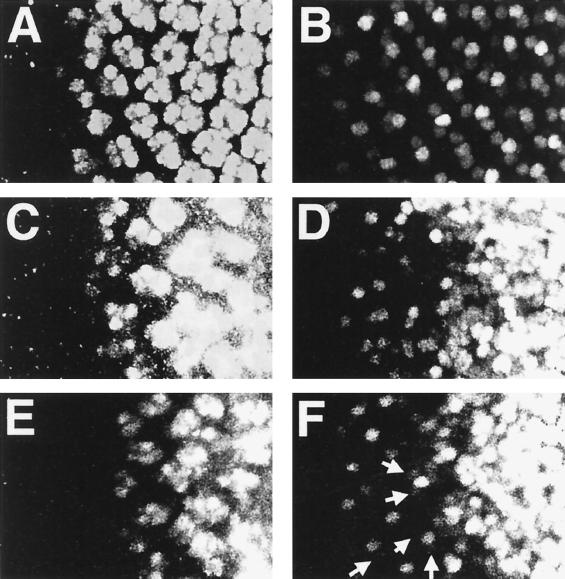
To address which signaling pathway(s) might be activated by TAK1 in vivo, we tested for genetic interactions between mutants in various signaling pathways and a TAK1-overexpressing line (either pGMR-mTAK1ΔN or GMR-GAL4/UAS-dTAK1). Included in this screen were mutations that disrupt BMP signaling, the Raf-MAPK pathway, and the JNK pathway (see Materials and Methods). Among the various mutants tested, only alleles of hep and bsk were found to interact strongly with lines overexpressing TAK1. In hemizygous hep or heterozygous bsk mutant backgrounds, the reduced size of compound eyes of GMR-GAL4/UAS-dTAK1 flies was rescued to that of wild-type flies (Fig. 5A to C). For comparison, Fig. 5D illustrates an example of noninteraction between Dsor1, a member of the Rl/MAPK pathway, and GMR-GAL4/UAS-dTAK1. Overexpression of hep in the eye resulted in a similar small eye phenotype, and this phenotype was rescued by the presence of p35 (Fig. 5E and F), confirming that ectopically activated JNK signaling induces cell death in the Drosophila eye.
FIG. 5.
Genetic interaction of pGMR-mTAK1ΔN. (A to F) Scanning electron micrographs of the compound eye. (A) GMR-GAL4/UAS-dTAK1(weak); (B) hep1/Y; GMR-GAL4/UAS-dTAK1(weak); (C) GMR-GAL4/UAS-dTAK1(weak); bsk2/+; (D) Dsor1LH110/Y; GMR-GAL4/UAS-dTAK1(weak) (E) GMR-GAL4/+; UAS-hep/+; (F) GMR-GAL4/+; UAS-hep/pGMR-p35. Reduced eye phenotype of GMR-GAL4/UAS-dTAK1 (A) is suppressed by one copy reduction of the hep or bsk gene (B and C). In contrast to this, a mutant which is involved in MAPK/ERK cascade, Dsor1, does not show any genetic interaction to this phenotype (D). Ectopic expression of hep also results in the small eye phenotype (E). This phenotype is suppressed by the presence of p35 (F), indicating that ectopic activation of the JNK signal induced apoptosis in the developing eye. GMR-GAL4/UAS-hep flies lost most of the interommatidial bristles (E). Bristle phenotype is not rescued by p35 expression (F), suggesting that apoptosis is not a direct cause of this phenotype. Anterior is to the left, and dorsal is up.
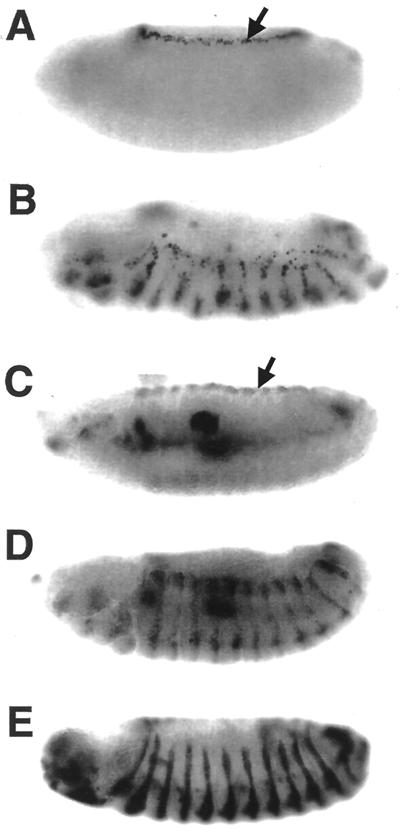
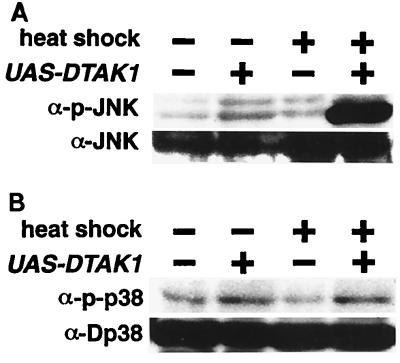
The Hep-Bsk signaling pathway has been shown to regulate the process of dorsal closure during the embryonic development (21, 49, 51, 57). Two genes, puckered (puc) and decapentaplegic (dpp), are downstream targets of Hep-Bsk signaling (21, 22, 42, 49) and are induced in the leading-edge cells in the embryonic epidermis during dorsal closure (Fig. 6A and C). To further examine whether TAK1 signaling is mediated by Hep and Bsk, we misexpressed dTAK1 using the en-GAL4 driver, which promotes GAL4 expression in the posterior compartment of the embryonic ectoderm (22) (Fig. 6E). Ectopic puc and dpp expression was observed in UAS-dTAK1/en-GAL4 embryos in a striped pattern (Fig. 6B and D). This result indicates that exogenous mTAK1 activates the Hep-Bsk pathway in the embryo as well as during imaginal disc development. We also tested whether dTAK1 expression leads to Bsk protein phosphorylation. dTAK1 was transiently expressed in the third-instar larva by expressing UAS-dTAK1 and hs-GAL4. Strong Bsk phosphorylation was observed only in the flies carrying UAS-dTAK1 after heat shock (Fig. 7A). Phosphorylation of D-p38 was not affected by dTAK1 expression (Fig. 7B). We conclude that TAK1 can activate the Hep-Bsk MAPK cascade in vivo.
FIG. 6.
Ectopic induction of puc and dpp by dTAK1 demonstrated by X-Gal staining (A and B) and in situ hybridization for dpp antisense probe (C and D) of stage 14 embryos. (A) puc-lacZ/+; (B) en-GAL4/UAS-dTAK1; puc-lacZ/+; (C) wild type; (D) en-GALY/UAS-dTAK1. puc and dpp expression in the leading-edge cells is indicated by arrows (A and C, respectively). Ectopic expression of dTAK1, controlled by en-GAL4, ectopically induces both puc and dpp in the embryonic ectoderm with a striped pattern (B and D). (E) en-GAL4/UAS-lacZ. en-GAL4 expression pattern is shown. Anterior is to the left, and dorsal is up.
FIG. 7.
In vivo phosphorylation of Bsk and D-p38 by ectopic dTAK1 expression. Third-instar larva carrying only hs-GAL4 or both UAS-dTAK1 and hs-GAL4 were collected with or without heat shock treatment. Extracts prepared from these larva were immunoblotted with either anti-p-JNK or anti-JNK1 antibody (A) and with anti-p-D-p38 or anti-p38 (B). Bsk phosphorylation is increased dramatically only in UAS-dTAK1-carrying animals upon heat shock. However, phosphorylation of D-p38 is not induced by the dTAK1 overexpression.
Expression of the dominant negative dTAK1 in the embryo resulted in several cuticle defects including a dorsal open phenotype.
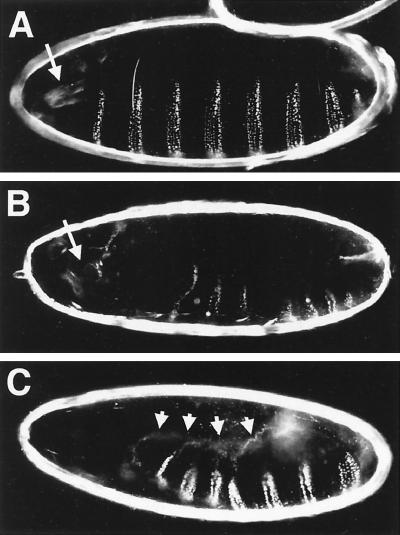
A common feature exhibited by loss-of-function mutations in genes of the JNK signaling pathway is a failure of proper dorsal closure due to disruptions in the movement and the ability of cells to change shape in the leading edge of the lateral epidermis (21, 22, 33, 37, 49–51, 57). Some of these mutants also exhibit defects in head structure and/or problems with germ band retraction (29, 60). To test whether dTAK1 also controls dorsal closure signals during normal development, we overexpressed a kinase-dead form (dTAK1-K46R) in the embryonic epidermis, using a pnr-GAL4 driver. Various types of cuticle defects were observed. The most typical phenotype was a head structure defect in which mouth hooks were missing (36%, n = 591) (Fig. 8B). A certain fraction of these defective embryos also exhibited an anterior open phenotype similar to that exhibited by hep and bsk mutants. In the most extreme cases, almost the entire dorsal cuticle failed to close (Fig. 8C). Occasionally, we also observed embryos with a U-shaped phenotype presumably due to insufficient germ band retraction (data not shown). These phenotypes are consistent with the idea that dTAK1 participates in the JNK signaling pathway.
FIG. 8.
Lateral view of the cuticle phenotypes of dominant negative dTAK1-expressing embryos of the following genotypes: (A) +/pannierMD237GAL4, as a wild-type control; (B and C) UAS-dTAK1-K46R/+; UAS-dTAK1-K46R/pannierMD237GAL4. (A) Wild-type cuticle illustrating the regular spacing of the denticle belt on the ventral side and complete closure of the epidermis on the dorsal side. (B and C) Expression of dTAK1-K46R (two copies of transgene) during embryonic development by means of pnr-GAL4 (32) causes various defects. Defects in the anterior structure, typically loss of the mouth hooks (normal position of the mouth hooks is indicated with arrows in panels A and B), are seen in 37% of embryos (n = 591). Embryos of this type are frequently exhibit a small whole in the anterior and dorsal side of the cuticle (B). In the most extreme cases, the embryo is completely open dorsally (6%, n = 591) (C). Arrows indicate the edge of the dorsal hole of the cuticle.
Expression of dominant negative forms of mTAK1 and dTAK1 in the eye suggest a role of JNK signaling in controlling pigment and bristle cell shape and position.
Little is known about the role of the JNK signaling pathway in eye development. Clones of partial loss-of-function bsk alleles showed no apparent defect in ommatidial development (49); however, mutations in disheveled, which appears to play a dual role in Wnt and JNK signaling pathways, show planar polarity defects in the eye (4). We wished to examine whether loss of TAK1 activity might affect ommatidial development. To explore this issue, we examined the outcome of expressing dominant negative forms of mTAK1 and dTAK1 in the eye disc, using the Glass promoter.
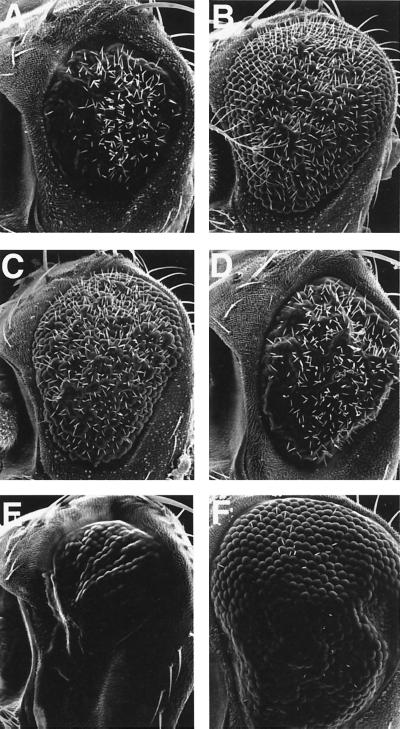
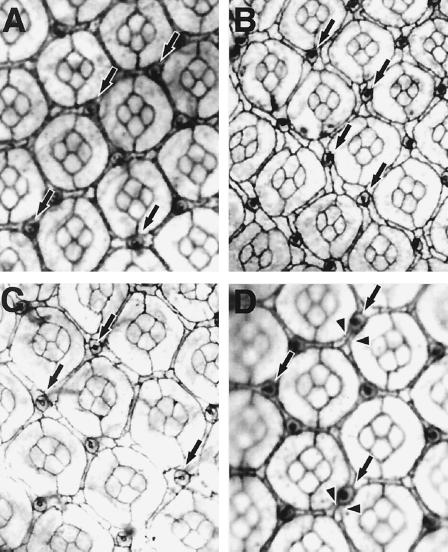
Flies expressing mTAK1-K63W or dTAK1-K46R showed a rough eye phenotype (data not shown), and this phenotype was sensitive to gene dosage. Flies expressing one copy of mTAK1-K63W had an almost wild-type compound eye, at least in the outer structure, while flies carrying two copies of the transgene displayed a rough eye phenotype (data not shown). We analyzed these putative loss-of-function phenotypes at a cellular level. In an apical profile of the pupal eye disc of pGMR-mTAK1-K63W-expressing lines (one copy), we noticed that bristles were mislocalized in the mutant pupal eye disc (Fig. 9A). In the wild-type fly, each ommatidium is surrounded by three bristles (Fig. 1C). These bristles are usually located at the anterior end of each horizontal face of the interommatidial space. In the pGMR-mTAK1-K63W disc, more than 30% of the bristles were located on the posterior end (or sometimes in the middle) of the horizontal face (Fig. 9A). Similar results were seen with overexpression of pGMR-dTAK1-K63R (Fig. 9C). We also observed abnormalities in the secondary and tertiary pigment cells. These two types of pigment cells lie between ommatidia and are distinguishable based on positions and cell shapes. Secondary pigment cells lie between two ommatidia and have rectangular shapes. Tertiary pigment cells are shared among three ommatidia at a vertex and have hexagonal cell shapes (Fig. 1C). In the dominant negative overexpression discs (Fig. 9B and C), it is obvious that the cell shapes of the secondary and tertiary pigment cells are heterogeneous. For instance, normally the secondary pigment cells which lie on the horizontal face are thicker than those lying on the slanting face (Fig. 1C). The hexagonal shape of the tertiary pigment cells is also distorted in the dominant negative overexpressing discs (Fig. 9B and C). We also examined the apical cell profile of hep hemizygous mutant eye discs and found that they too showed mislocation of bristle cells and alterations in positions of pigment cells (Fig. 9D). These observations suggest that the endogenous TAK1 signaling, likely acting through the Hep-Bsk pathway, plays an important role in regulating correct cell shape changes and/or cell movement in the secondary and tertiary pigment cells, as well as in the bristle cells.
FIG. 9.
Phenotypes induced by the expression of dominant negative TAK1. Pupal eyes of the following genotypes at 40 h after puparium formation were stained with cobalt sulfide. (A) pGMR-mTAK1-K63W (one copy). Expression of mTAK1-K63W, a dominant negative form of mTAK1, at a lower level results in defective positioning of the interommatidial bristle (indicated with arrows, compared to the wild-type shown in Fig. 1C). (B) pGMR-mTAK1-K63W (two copies). A higher level of mTAK1-K63W expression totally disrupts the ommatidial array. The cell shapes of secondary and tertiary pigment cells are irregular, and it is hard to discriminate these two cell types by morphology. (C) GMR-GAL4/UAS-dTAK1-K46R; UAS-dTAK1-K46R/+. A similar phenotype is observed in a fly expressing dTAK1-K46R (two copies), a kinase-inactive form of dTAK1. (D) hepr75/Y. The hep mutant disc also shows bristle mislocation (indicated with arrows) and abnormal pigment cell shapes (arrowheads). All images show the phenotype of the center region of the pupal eye discs (even in the wild-type discs, bristle mislocation is occasionally observed in the anterior edge region). Anterior is to the left.
Interestingly, despite the abnormal morphology of the secondary and tertiary pigment cells in the pupal eye disc of pGMR-mTAK1-K63W and an allele of the mutant, hepr75, the total number of secondary and tertiary pigment cells did not change (Fig. 9). Since a wave of apoptosis is known to control pigment cell number (8, 70), this result implies that either endogenous programmed cell death in the visual system is independent of TAK1-JNK signaling or the expression level of dominant negative TAK1 is insufficient for inhibiting endogenous apoptosis.
DISCUSSION
TAK1 activates the JNK signaling pathway in Drosophila.
There are many known members of MAPKKK family, including MEKs, germinal center kinase, mixed-lineage kinases (MLKs), tumor progression locus 2, apoptosis signal-regulating kinase 1 (ASK1) and TAK1. These kinases have been shown to activate JNK and/or p38 MAPK pathways in cell culture experiments, but whether they contribute to the function of these pathways in vivo is unknown (18, 34). In this study we have shown that TAK1 can activate the JNK signaling cascade in Drosophila. This conclusion is based on four lines of evidence. First, ectopic expression of TAK1 induces expression of dpp and puc, known downstream targets of JNK signaling in embryonic ectodermal cells (Fig. 6B and D). Second, ectopic TAK1 expression resulted in the phosphorylation of the Bsk protein (Fig. 7A). Third, heterozygosity for hep and bsk mutants is able to dominantly suppress the dTAK1 overexpression phenotype (Fig. 5B and C). Fourth, dominant negative forms of TAK1 exhibit cuticular phenotypes similar to loss-of-function phenotypes of bsk and hep alleles (Fig. 8B and C). Thus, TAK1 is likely to serve as the missing MAPKKK in the JNK signaling pathway functioning downstream of the misshapen products, a Drosophila MAPKKKK, and upstream of Hep.
In addition to its role in JNK signaling, previous evidence from cell culture experiments has implicated TAK1 in p38 MAPK signaling pathways (44, 56, 66, 71). However, we found no evidence for the involvement of TAK1 in p38 activation in Drosophila. Specifically, the reduced eye phenotype of pGMR-mTAK1ΔN overexpression was not rescued by heterozygosity for a deficiency which uncovers one of the D-p38 loci (D-p38b) (data not shown). Likewise, expression of a dominant negative form of D-p38b, where Thr-183 of the MAPKK target site was replaced with Ala, did not suppress the pGMR-mTAK1ΔN overexpression phenotype (data not shown); furthermore, and most compelling, D-p38 phosphorylation was not observed in the flies expressing dTAK1 (Fig. 7B). Thus, our data suggest that TAK1 may specifically activate JNK signaling and not p38 MAPK signaling in Drosophila; however, this conclusion will need to be tested further once mutations in D-p38 and dTAK1 become available.
Endogenous function of the TAK1 signaling in Drosophila.
Two different biological processes are known to be controlled by the JNK signaling pathway in Drosophila. One is the movement of leading-edge cells during the process of embryonic dorsal closure (21, 22, 33, 37, 49–51, 57), and the other is in planar polarity determination of adult tissues (4, 60). In neither case, however, is the role or identity of the putative MAPKKK molecule(s) that might be involved in these processes known. In this study, gain-of-function experiments revealed that TAK1 could activate the JNK signaling cascade. Loss-of-function experiments, using dominant negative dTAK1 and mTAK1, also revealed that TAK1 is likely to be required for the proper cell movement and/or shape changes in the embryo and visual system. Indeed, the dorsal open phenotype and head structure defects observed in the dominant negative dTAK1-expressing embryos are highly reminiscent of the phenotype produced by JNK pathway loss-of-function mutants (21, 22, 33, 37, 49–51, 57). Impaired control of cell shape was also noted in TAK1 gain-of-function phenotypes obtained by overexpression of dTAK1 in the presence of p35 (Fig. 4F) and also with dominant negative forms of TAK1 (Fig. 9B and C). The function of JNK signaling in cell movement in the visual system has been studied only for ommatidial planar polarity determination (4, 60). Although we did not observe defects in planar polarity, we do see abnormalities in the positioning and shape of interommatidial cells in both dominant negative TAK1-expressing lines and hep mutants (Fig. 9). This result suggests that the endogenous JNK signaling also participates in the positioning and shape of the interommatidial cells. Since a true loss-of-function hep allele also shows this phenotype, albeit more weakly, it is not likely that the phenotype caused by overexpression of dominant negative TAK1 is due to a novel neomorphic property of this protein. The weaker interommatidial phenotype of hemizygous hepr75 flies compared to that of pGMR-mTAK1-K63W (two copies) might indicate that there is some genetic redundancy for this class of kinase in Drosophila. Likewise, the lack of observable planar polarity defects in these animals may indicate either a nonrequirement for a TAK1 kinase in this process or a genetic redundancy for a TAK1-type kinase, or that this particular phenotype is less sensitive to TAK1 loss-of-function and so is not observable under the conditions used in this study. Only when clones of true TAK1 loss-of-function alleles are available will we be able to fully address this issue.
TAK1 function in TGF-β/Dpp signaling.
Previous reports have shown that TAK1 is activated by TGF-β/BMP stimuli and that a kinase-negative form of TAK1 prevents TGF-β/BMP signaling in mammalian cells and in Xenopus (54, 71). Surprisingly, we did not observe any genetic interactions between pGMR-mTAK1ΔN overexpression lines and mutations in the Dpp signaling pathway. We also found that ectopic TAK1 signaling in the wing disc was unable to induce optomotor blind, a known downstream target of Dpp signaling (24) and, furthermore, that ectopic vein formation induced by a constitutively active Dpp receptor could not be suppressed by overexpression of dTAK1-K46R expression (Y. Takatsu et al., unpublished data). These results may indicate that Dpp signaling is not regulated by TAK1 in the visual system and wing or, alternatively, that the effects of Dpp-induced TAK1 signaling are mild compared to the Mad/Medea pathway of Dpp signaling and so were not observable with the genetic tests presently at hand. Once again, the availability of TAK1 mutants will help clarify this issue.
Apoptosis triggered by the TAK1-JNK signaling pathway.
We have shown that the ectopic activation of the TAK1 signaling pathway induces apoptosis in Drosophila. A role for TAK1 in mediating apoptosis has been previously suggested from overexpression studies in cell culture (56). For example, in one study, Shirakabe et al. showed that ceramide, a second messenger molecule that induces various cellular responses such as cell cycle arrest, differentiation, and apoptosis (28, 38, 56), activates TAK1 and that TAK1-K63W blocks ceramide-induced apoptosis (56). The role of the JNK pathway in stress-induced apoptosis in vertebrates has been extensively studied. Various environmental stresses, such as UV irradiation, exposure to toxic agents, osmotic stresses, and heat shock, all trigger JNK pathway activation and result in apoptosis in certain cells and tissues (12, 20, 39, 43, 57). We believe that the TAK1-induced apoptosis that we report here is also likely to be mediated through the JNK pathway. Consistent with this view, we have observed that ectopic hep expression using GMR-GAL4 also results in a reduced eye phenotype (Fig. 5E). Since we have shown that the constitutively active TAK1 phenotype is effectively suppressed by a reduction in gene dosage of the proapoptotic genes rpr, hid, and grim or by coexpression of apoptosis inhibitors such as p35 or DIAPs (Fig. 2C) which block the enzymatic activities of caspases (7, 13), we believe that TAK1-JNK function is positioned upstream of the proapoptotic genes and the caspase cascade. Interestingly, ectopic TAK1 expression in Xenopus embryos also immediately induces cell death, yet no vertebrate homologs of the Drosophila proapoptotic genes have been identified. Recently, however, Evans et al. reported that overexpression of the Drosophila rpr gene can induce apoptosis in Xenopus eggs (17). This may indicate that proapoptotic genes exist in vertebrates and that they could be targets of TAK1-JNK signaling. Further genetic analysis using TAK1 transgenic flies should provide a unique opportunity to identify molecules that help interface JNK activation with the basic apoptotic machinery.
ACKNOWLEDGMENTS
Y. Takatsu and M. Nakamura contributed equally to this work.
We are grateful to N. Nakamura and K. Nakagawa for experimental help. We also thank A. Kreuz and T. Tanimura for comments on the manuscript; T. Adachi-Yamada and K. Sawamoto for fly stocks and technical advice; S. Goto for the UAS-hep construct; H. Steller for rpr and hid cDNAs; N. Patel for Elav antibody; T. Adachi-Yamada for anti-p-D-p38b antibody; and K. Basler, D. Brunner, S. Cohen, M. Freeman, K. Ito, E. Hafen, S. Hayashi, Y. Hiromi, K. Kimura, Y. Nishida, N. Perrimon, G. M. Rubin, L. Raftery, H. Okano, G. Pflugfelder, and the Bloomington Stock Center for fly stocks.
This work was supported by grants-in-aids for scientific research from the Ministry of Education, Science, and Culture of Japan and the Research for the Future program of the Japan Society for the Promotion of Science. M.S. was supported by PHS grant GM47462 to M.B.O. M.C.D. is a Research Associate of the Howard Hughes Medical Institute. M.B.O. is an Associate Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adachi-Yamada T, Nakamura M, Irie K, Tomoyasu Y, Sano Y, Mori E, Goto S, Ueno N, Nishida Y, Matsumoto K. p38 mitogen-activated protein kinase can be involved in transforming growth factor β superfamily signal transduction in Drosophila wing morphogenesis. Mol Cell Biol. 1999;19:2322–2329. doi: 10.1128/mcb.19.3.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M. Drosophila: a laboratory manual. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 3.Bonner T I, Oppermann H, Seeburg P, Kerby S B, Gunnell M A, Young A C, Rapp U R. The complete coding sequence of the human raf oncogene and the corresponding structure of the c-raf-1 gene. Nucleic Acids Res. 1986;14:1009–1015. doi: 10.1093/nar/14.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutros M, Paricio N, Strutt D I, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- 5.Bowtell D D, Kimmel B E, Simon M A, Rubin G M. Regulation of the complex pattern of sevenless expression in the developing Drosophila eye. Proc Natl Acad Sci USA. 1989;86:6245–6249. doi: 10.1073/pnas.86.16.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 7.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 8.Cagan R L, Ready D F. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- 9.Cano E, Mahadevan L C. Parallel signal processing among mammalian MAPKs. Trends Biochem Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Nordstrom W, Gish B, Abrams J M. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 11.Davis R J. MAPKs: new JNK expands the group. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 12.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 13.Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 14.Dorow D S, Devereux L, Dietzsch E, De Kretser T. Identification of a new family of human epithelial protein kinases containing two leucine/isoleucine-zipper domains. Eur J Biochem. 1993;213:701–710. doi: 10.1111/j.1432-1033.1993.tb17810.x. [DOI] [PubMed] [Google Scholar]
- 15.Downward J. KSR: a novel player in the RAS pathway. Cell. 1995;83:831–834. doi: 10.1016/0092-8674(95)90198-1. [DOI] [PubMed] [Google Scholar]
- 16.Ellis M C, O'Neill E M, Rubin G M. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- 17.Evans E K, Kuwana T, Strum S L, Smith J J, Newmeyer D D, Kornbluth S. Reaper-induced apoptosis in a vertebrate system. EMBO J. 1997;16:7372–7381. doi: 10.1093/emboj/16.24.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. MEKKs, GCKs, MLKs, PAKs, TAKs, and tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 19.Freeman M, Kimmel B E, Rubin G M. Identifying targets of the rough homeobox gene of Drosophila: evidence that rhomboid functions in eye development. Development. 1992;116:335–346. doi: 10.1242/dev.116.2.335. [DOI] [PubMed] [Google Scholar]
- 20.Galcheva-Gargova Z, Derijard B, Wu I H, Davis R J. An osmosensing signal transduction pathway in mammalian cells. Science. 1994;265:806–808. doi: 10.1126/science.8047888. [DOI] [PubMed] [Google Scholar]
- 21.Glise B, Bourbon H, Noselli S. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell. 1995;83:451–461. doi: 10.1016/0092-8674(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 22.Glise B, Noselli S. Coupling of Jun amino-terminal kinase and Decapentaplegic signaling pathways in Drosophila morphogenesis. Genes Dev. 1997;11:1738–1747. doi: 10.1101/gad.11.13.1738. [DOI] [PubMed] [Google Scholar]
- 23.Grether M E, Abrams J M, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 24.Grimm S, Pflugfelder G O. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- 25.Han S J, Choi K Y, Brey P T, Lee W J. Molecular cloning and characterization of a Drosophila p38 mitogen-activated protein kinase. J Biol Chem. 1998;273:369–374. doi: 10.1074/jbc.273.1.369. [DOI] [PubMed] [Google Scholar]
- 26.Han Z S, Enslen H, Hu X, Meng X, Wu I-H, Barrett T, Davis R J, Ip Y T. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol Cell Biol. 1998;18:3527–3539. doi: 10.1128/mcb.18.6.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 28.Hannun Y A, Obeid L M. Ceramide: an intracellular signal for apoptosis. Trends Biochem Sci. 1995;20:73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 29.Harden N, Loh H Y, Chia W, Lim L. A dominant inhibitory version of the small GTP-binding protein Rac disrupts cytoskeletal structures and inhibits developmental cell shape changes in Drosophila. Development. 1995;121:903–914. doi: 10.1242/dev.121.3.903. [DOI] [PubMed] [Google Scholar]
- 30.Hay B A, Wassarman D A, Rubin G M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 31.Hay B A, Wolff T, Rubin G M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2127. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 32.Heitzler P, Haenlin M, Ramain P, Calleja M, Simpson P. A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics. 1996;143:1271–1286. doi: 10.1093/genetics/143.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou X S, Goldstein E S, Perrimon N. Drosophila Jun relays the Jun amino-terminal kinase signal transduction pathway to the Decapentaplegic signal transduction pathway in regulating epithelial cell sheet movement. Genes Dev. 1997;11:1728–1737. doi: 10.1101/gad.11.13.1728. [DOI] [PubMed] [Google Scholar]
- 34.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 35.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 36.Kimmel B E, Heberlein U, Rubin G M. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- 37.Kockel L, Zeitlinger J, Staszewski L M, Mlodzik M, Bohmann D. Jun in Drosophila development: redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes Dev. 1997;11:1748–1758. doi: 10.1101/gad.11.13.1748. [DOI] [PubMed] [Google Scholar]
- 38.Kolesnick R, Golde D W. The sphingomyelin pathway in tumor necrosis factor and interleukin-1 signaling. Cell. 1994;77:325–328. doi: 10.1016/0092-8674(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 39.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 40.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 41.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky A M, Martinez-Arias A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriguchi T, Kuroyanagi N, Yamaguchi K, Gotoh Y, Irie K, Kano T, Shirakabe K, Muro Y, Shibuya H, Matsumoto K, Nishida E, Hagiwara M. A novel kinase cascade mediated by mitogen-activated protein kinase kinase 6 and MKK3. J Biol Chem. 1996;271:13675–13679. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 45.Moses K, Rubin G M. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen T, Jamal J, Shimell M J, Arora K, O'Connor M B. Characterization of tolloid-related-1: a BMP-1-like product that is required during larval and pupal stages of Drosophila development. Dev Biol. 1994;166:569–586. doi: 10.1006/dbio.1994.1338. [DOI] [PubMed] [Google Scholar]
- 47.Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18:128–131. doi: 10.1016/0968-0004(93)90019-j. [DOI] [PubMed] [Google Scholar]
- 48.Nishida Y, Hata M, Ayaki T, Ryo H, Yamagata M, Shimizu K, Nishizuka Y. Proliferation of both somatic and germ cells is affected in the Drosophila mutants of raf proto-oncogene. EMBO J. 1988;7:775–781. doi: 10.1002/j.1460-2075.1988.tb02875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noselli S. JNK signaling and morphogenesis in Drosophila. Trends Genet. 1998;14:33–38. doi: 10.1016/S0168-9525(97)01320-6. [DOI] [PubMed] [Google Scholar]
- 50.Riesgo-Escovar J R, Hafen E. Drosophila Jun kinase regulates expression of decapentaplegic via the ETS-domain protein Aop and the AP-1 transcription factor DJun during dorsal closure. Genes Dev. 1997;11:1717–1727. doi: 10.1101/gad.11.13.1717. [DOI] [PubMed] [Google Scholar]
- 51.Riesgo-Escovar J R, Jenni M, Fritz A, Hafen E. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 1996;10:2759–2768. doi: 10.1101/gad.10.21.2759. [DOI] [PubMed] [Google Scholar]
- 52.Sakurai H, Shigemori N, Hasegawa K, Sugita T. TGF-β-activated kinase 1 stimulates NF-kappa B activation by an NF-kappa B-inducing kinase-independent mechanism. Biochem Biophys Res Commun. 1998;243:545–549. doi: 10.1006/bbrc.1998.8124. [DOI] [PubMed] [Google Scholar]
- 53.Seger R, Krebs E G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 54.Shibuya H, Iwata H, Masuyama N, Gotoh Y, Yamaguchi K, Irie K, Matsumoto K, Nishida E, Ueno N. Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 1998;17:1019–1028. doi: 10.1093/emboj/17.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science. 1996;272:1179–1182. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 56.Shirakabe K, Yamaguchi K, Shibuya H, Irie K, Matsuda S, Moriguchi T, Gotoh Y, Matsumoto K, Nishida E. TAK1 mediates the ceramide signaling to stress-activated protein kinase/c-Jun N-terminal kinase. J Biol Chem. 1997;272:8141–8144. doi: 10.1074/jbc.272.13.8141. [DOI] [PubMed] [Google Scholar]
- 57.Sluss H K, Han Z, Barrett T, Davis R J, Ip Y T. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 58.Spradling A C, Rubin G M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 59.Stroumbakis N D, Li Z, Tolias P P. RNA- and single-stranded DNA-binding (SSB) proteins expressed during Drosophila melanogaster oogenesis: a homolog of bacterial and eukaryotic mitochondrial SSBs. Gene. 1994;143:171–177. doi: 10.1016/0378-1119(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 60.Strutt D I, Weber U, Mlodzik M. The role of RhoA in tissue polarity and Frizzled signalling. Nature. 1997;387:292–295. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- 61.Suzanne M, Irie K, Glise B, Agnes F, Mori E, Matsumoto K, Noselli S. The Drosophila p38 MAPK pathway is required during oogenesis for egg asymmetric development. Genes Dev. 1999;13:1464–1474. doi: 10.1101/gad.13.11.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 63.Therrien M, Chang H C, Solomon N M, Karim F D, Wassarman D A, Rubin G M. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 64.Tomlinson A, Bowtell D D, Hafen E, Rubin G M. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51:143–150. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- 65.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 66.Wang W, Zhou G, Hu M, Yao Z, Tan T H. Activation of the hematopoietic progenitor kinase-1 (HPK1)-dependent, stress-activated c-Jun N-terminal kinase (JNK) pathway by transforming growth factor β (TGF-β)-activated kinase (TAK1), a kinase mediator of TGF-β signal transduction. J Biol Chem. 1997;272:22771–22775. doi: 10.1074/jbc.272.36.22771. [DOI] [PubMed] [Google Scholar]
- 67.Waskiewicz A J, Cooper J A. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 68.Watson R, Oskarsson M, Vande Woude G F. Human DNA sequence homologous to the transforming gene (mos) of Moloney murine sarcoma virus. Proc Natl Acad Sci USA. 1982;79:4078–4082. doi: 10.1073/pnas.79.13.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.White K, Grether M E, Abrams J M, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 70.Wolff T, Ready D F. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- 71.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]