Abstract
Severe lung inflammation is common in life-threatening coronavirus disease 2019 (COVID-19). This study tested the hypothesis that polymorphonuclear (PMN, neutrophil) phenotype early in the course of disease progression would predict peak lung disease severity in patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It is increasingly evident that PMN activation contributes to tissue injury resulting from extracellular reactive oxygen species generation, granule exocytosis with release of proteases, neutrophil extracellular trap (NET) formation, and release of cytokines. The current study focuses on PMN activation in response to SARS-CoV-2 infection, specifically, the association between NETs and lung disease. This is a prospective cohort study at an academic medical center with patients enrolled within 4 days of admission at 3 tertiary hospitals: Clements University Hospital, Parkland Memorial Hospital, and Children’s Health in Dallas, TX. Patients were categorized as having minimal or moderate to severe lung disease based on peak respiratory support. Healthy donor controls matched for age, sex, race, and ethnicity were also enrolled. Neutrophils from COVID-19 patients displayed greater IL-8 expression, elastase release, and NET formation as compared with neutrophils from healthy donors. Importantly, neutrophils from COVID-19 patients had enhanced NET formation in the absence of any additional stimulus, not seen in PMN from healthy donors. Moreover, PMA-elicited NET formation by circulating PMN correlated with severity of lung disease. We speculate that neutrophil immuno-phenotyping can be used to predict lung disease severity in COVID-19 patients.
SUPPLEMENTARY INFORMATION
The online version contains supplementary material available at 10.1007/s10753-021-01585-x.
KEY WORDS: elastase, SARS-CoV-2, SIRS, IL-8, immunophenotyping.
INTRODUCTION
The COVID-19 pandemic has placed unparalleled strain on healthcare facilities and providers and has resulted in millions of deaths worldwide [1]. Patients with SARS-CoV-2 sepsis experience symptoms ranging from asymptomatic infection to multi-organ dysfunction syndrome and death. As reported with SARS-CoV and MERS-CoV, a cytokine storm has been linked with disease severity, and acute lung injury (ALI) accounts for a significant proportion of morbidity and mortality [2–4]. A better understanding of the innate immune response is crucial to the clinical management of COVID-19 and may have relevance in other viral respiratory infections.
In the SARS-CoV outbreak of 2002–2003, polymorphonuclear leukocyte (PMN, neutrophil) count was an independent predictor of adverse outcome [5], and PMN to lymphocyte ratio has been correlated with severity of disease in COVID-19 patients [6, 7]. It is increasingly evident that PMN play a critical role in the pathophysiology of many diseases ranging from infection to cancer [8], but relatively little is known about the role of neutrophils in viral illnesses. Despite an unequivocal role in host defense, PMN activation contributes to tissue injury resulting from granule exocytosis with release of proteases, neutrophil extracellular trap (NET) formation, and release of cytokines [9–12]. In addition to direct damage to tissues, NETs are a source of autoantigens and immunostimulatory proteins that serve as damage-associated molecular patterns that may amplify the host response [13].
Our lab has a longstanding interest in the drivers of inflammation and the resolution phase of inflammation. Our current focus is on the role of neutrophils in the systemic inflammatory response syndrome and the molecular mechanisms eliciting lung injury. There is a gap in knowledge regarding neutrophil responses in viral infections and the contribution of neutrophil activation to organ injury. Several studies have demonstrated the presence of NET fragments or cell-free DNA in the plasma of COVID-19 patients [14–17], and post-mortem analysis reveals the presence of NETs in lung tissue of COVID-19 patients [17–19]. An elevation in cytokines, including IL-8, has also been observed in the plasma of COVID-19 patients [20], but the cellular source is not clear. Neutrophil elastase has been implicated in the pathogenesis of lung disease, and elastase inhibitors have been used in clinical trials with mixed results [9, 21–24]. Treatment options for COVID-19 are limited and primarily consist of supportive therapy and suppression of the systemic inflammatory response. These approaches, along with some use of anti-viral medications, are based on the knowledge and management of other severe viral respiratory infections and empirical evidence gained over the course of the current pandemic. These therapies and aggressive supportive care have resulted in a slight improvement in case fatality rates, but significant morbidity and mortality remain. Monoclonal antibody therapies have been authorized in certain circumstances as a prophylactic after known exposure and in the early stages of infection [25]. Remdesivir, the only authorized antiviral treatment, was shown to shorten the time to recovery in hospitalized adults with evidence of lower respiratory infection [26]. However, a recent Cochrane review concluded that remdesivir probably had minimal impact on mortality, considering all causes, in adults requiring hospitalization with SARS-CoV-2 infection [27]. Therapies that target the neutrophil response have been proposed [28, 29], and some have progressed to clinical trials [30]. A better understanding of the role of neutrophils and the underlying molecular mechanisms driving inflammatory complications and organ injury is needed to more precisely identify potential therapeutic targets and inform the design of clinical trials.
Based on the early reports of neutrophilia and NET formation in the lungs of severely ill COVID-19 patients, we sought to investigate the neutrophil phenotype in patients hospitalized with SARS-CoV-2. Our primary aim was to determine whether neutrophil phenotype early in the course of disease could be used to predict peak lung disease severity. We hypothesized that the NET formation potential of circulating PMN would correlate with peak lung disease. The ability to stratify risk and predict which patients may require intensive care and mechanical ventilation would allow for a more personalized approach to clinical care. Moreover, predictive capacity could have a significant impact on resource utilization, particularly intensive care unit bed assignment, at a time when this resource is limited. A secondary aim was to pinpoint molecular targets for future study as potential therapeutic targets. This study focuses on PMN activation in response to SARS-CoV-2 infection, specifically the association between elastase release and NET formation potential and severity of lung disease. We found that freshly isolated neutrophils from COVID-19 patients produced IL-8 and released elastase and NETs in the absence of exogenous stimulation. In addition, neutrophils from COVID-19 patients had enhanced elastase release upon exogenous stimulation. Importantly, PMNs from COVID-19 patients displayed greater NET formation potential than PMN from healthy donors, and moreover, NET formation potential correlated with the severity of lung disease.
MATERIALS AND METHODS
Patients and Data Collection
This was a prospective cohort study with 2 cohorts: (1) hospitalized patients with SARS-CoV-2 infection and (2) healthy donors matched for age, sex, race, and ethnicity. COVID-19 patients at Clements University Hospital, Parkland Memorial Hospital, and Children’s Health were screened for enrollment by the UT Southwestern SARS-CoV-2 Biorepository (COVID-19 BR), and 32 patients were enrolled for our study between May 2020 and January 2021. Approval was obtained from the Institutional Review Board at UT Southwestern. Samples were collected and processed within 2 h by the COVID-19 BR. Eight patients were excluded in the final analysis due to the timing of the blood draw. Of the 24 patients included in our study, neutrophil elastase release and NET formation were measured in 20 patients due to staff availability. These assays are performed immediately upon neutrophil isolation. Neutrophil IL-8 production was measured in 21 patients due to sample availability. H3CIT-elastase fragments were assessed in all 24 patients.
Peak respiratory support during hospital stay was used to categorize patients by severity of lung disease. Based on guidelines from the World Health Organization, minimal lung disease (n = 14) is defined as room air to 6 LPM on the nasal cannula. Moderate to severe lung disease (n = 10) is 6 LPM on the high flow nasal cannula up to 50 LPM (n = 7), BiPAP (n = 1), or mechanical ventilation (n = 2) [31, 32]. Due to our limited sample size, we combined the moderate and severe groups for analysis. Clinical data were collected from study entry until discharge and included demographic data, BMI, white blood cell (WBC) count, PMN percentage, C-reactive protein, and respiratory support; dates of symptom onset, admission, and blood draw; length of stay; pre-existing conditions; and measures of organ dysfunction. There were no significant differences in age, sex, BMI, race, ethnicity, length of stay from symptom onset, or time of sample acquisition in the COVID-19 cohort based on lung disease severity. The average BMI was 27.9, slightly above the reference range (18.5–24.9). Peak PMN percentage (74.0 ± 4% for minimal lung disease vs 84.4 ± 15% for mod/severe lung disease) and C-reactive protein (47.7 mg/L for minimal lung disease vs 111.9 mg/L for mod/severe lung disease) were elevated in COVID-19 patients and correlated with lung disease severity, consistent with published reports [3, 31]. The reference ranges for PMN percentage and C-reactive protein are 40–60% and less than 10 mg/L, respectively.
Materials
HBSS and Dulbecco’s PBS (DPBS) were purchased from Mediatech (Manassas, VA, USA), dextran from Pharmacosmos (Holbaek, Denmark), Ficoll-Paque from GE Healthcare (Piscataway, NJ, USA), and human serum albumin from CSL Behring (King of Prussia, PA, USA). Abs included IL-8 (clone 8CH) from Invitrogen (Waltham, MA, USA), citrullinated histone H3 (R2 + R8 + R17) (H3CIT) from Cayman Chemical (Ann Arbor, MI, USA), and biotinylated elastase from R&D Systems (Minneapolis, MN, USA). SYTOX™ Green was purchased from Thermo Fisher Scientific, MeOSuc-Ala-Ala-Pro-Val-7-amino-4-methylcoumarin from MP Biomedicals (Solon, OH, USA), and PMA and fMLF from Sigma (St. Louis, MO, USA). Additional reagents were obtained from Fisher Scientific (Pittsburgh, PA, USA). All buffers and reagents were strictly endotoxin free.
Human PMN Purification
PMNs were isolated from acid citrate dextrose anticoagulated whole blood according to standard techniques as previously described and approved by the Institutional Review Board at UT Southwestern and in collaboration with the COVID-19 BR. [33, 34].
Elastase Activity
Freshly isolated PMNs were diluted in PICM-G buffer (10 mM sodium phosphate buffer with 2.7 mM KCl, 138 mM NaCl, 0.6 mM CaCl2, 1.0 mM MgCl2, and 0.1% glucose) containing MeOSuc-Ala-Ala-Pro-Val-7-amino-4-methylcoumarin (20 μM) and no agonist, 1 nM PMA, or 3 nM PMA before addition to a 96-well microplate. Duplicate wells (1 × 105 PMN/well) were read in the CLARIOstar from BMG Labtech (Cary, NC, USA) at an excitation/emission of 360 ± 20/450 ± 30 nm, with readings every 5 min. Based on the kinetics of elastase release and before substrate consumption, data are expressed as relative fluorescence units (RFUs) at 90 min.
NET Formation
Sytox Green:
Freshly isolated PMNs were diluted in HBSS containing 0.1% glucose and 1% human serum albumin containing 5 µM Sytox Green and no agonist, 1 µM fMLF or 3 nM PMA before addition to a 96-well microplate. Duplicate wells (1 × 105 PMN/well) were read in the CLARIOstar at an excitation/emission of 491 ± 15/535 ± 20nm with readings every 5 min. Endpoint readings are expressed as RFUs.
H3CIT-elastase complexes:
Plasma was collected and stored at –80 °C by the COVID-19 BR. ELISA was performed, as previously described [35], with H3CIT and biotinylated elastase as the capture and detection antibodies, respectively. Results are expressed as OD.
IL-8 Intracellular Flow
PMN pellets were collected and stored at –80 °C by the COVID-19 BR. Pellets were thawed and processed according to the two-step protocol for intracellular proteins as previously described [36]. Twenty thousand events were acquired on a FACSCalibur from BD Biosciences (San Jose, CA, USA) and analyzed using FlowJo version 9.9.6 from Tree Star (Ashland, OR, USA). The neutrophil population was identified by forward versus side scatter.
Statistical Analysis
Data for each individual are presented as well as the mean ± standard error of the mean for the cohort. Statistical analysis was performed using GraphPad Prism 9 for Windows from GraphPad Software (La Jolla, CA, USA). Comparisons between cohorts were performed using one-way ANOVA with multiple comparison or Student’s t test. Results were considered statistically significant with a p value less than 0.05, and relative p values are noted in the figure legends and text.
RESULTS
Elevated WBC Count in COVID-19 Patients with Moderate to Severe Lung Disease
Our primary hypothesis centered on the analysis of PMN phenotype early in the course of COVID-19 as a predictor for the development and severity of lung disease. Thus, we divided the COVID-19 cohort into two groups based on peak respiratory support during hospital stay: no/minimal vs mod/severe lung disease. The baseline characteristics of each cohort are displayed in Table 1. COVID-19 patients and healthy donors did not differ in regard to age, sex, race, or ethnicity. Peak WBC count was higher in patients who developed moderate to severe lung disease (14.36 ± 1.61 × 109/L) versus patients who developed no/minimal lung disease (8.80 ± 0.86 × 109/L, p < 0.01) consistent with a recently published prospective study in Wuhan that reported significantly higher WBC count in COVID-19 patients with severe-critically severe versus mild-moderate disease [37]. The reference range for (WBC) count is 4.5–11.0 × 109/L.
Table 1.
Characteristics of Study Cohorts
| Characteristics | COVID-19 (n = 24) | Lung disease severity | Healthy (n = 23) | p valuea | |
|---|---|---|---|---|---|
| Minimal (n = 14) | Mod/Severe (n = 10) | ||||
| Age (years) | 41.7 (28.0–62.3) | 39.7 (27.1–64.5) | 44.8 (30.5–59.3) | 41 (30.0–48.5) | 0.799 |
| Female | 10 (42%) | 6 (43%) | 4 (40%) | 12 (52%) | 0.382 |
| BMI (kg/m2) | 27.9 (24.9–36.4) | 27.9 (24.3–35.8) | 27.9 (25.4–44.0) | 0.989 | |
| Ethnicity | 0.778 | ||||
| Hispanic | 8 (33%) | 5 (36%) | 3 (30%) | 5 (22%) | |
| Non-Hispanic | 16 (67%) | 9 (64%) | 7 (70%) | 18 (78%) | |
| Race | 0.584 | ||||
| White | 16 (67%) | 10 (71%) | 6 (60%) | 16 (70%) | |
| Black | 3 (13%) | 1 (7%) | 2 (20%) | 2 (8%) | |
| Asian | 1 (4%) | 1 (7%) | 0 (0%) | 5 (21%) | |
| Other | 2 (8%) | 1 (7%) | 1 (10%) | 0 (0%) | |
| Unknown | 2 (8%) | 1 (7%) | 1 (10%) | 0 (0%) | |
| Length of stay from symptom onset (days) | 12 (9–15) | 12 (9–13.2) | 14 (8–15) | 0.967 | |
| Day of sample acquisition | 3 (2–3) | 3 (2–3) | 2.5 (1–3) | 0.403 | |
Categorical values are presented as n (%) and continuous values are presented as median (interquartile range).
BMI body mass index.
ap values calculated using one-way ANOVA and chi-squared analysis as appropriate.
PMN From COVID-19 Patients Produce IL-8
COVID-19 patients are reported to have increased serum IL-8 levels that correlate with disease severity, but the cellular source(s) have not been definitively identified [20, 38, 39]. Neutrophils are a source of IL-8 production, and neutrophil IL-8 production and release may result in amplification of systemic inflammation. We measured intracellular IL-8 levels in freshly isolated PMN from COVID-19 patients and compared this with levels in healthy donors. PMN from COVID-19 patients had significantly more intracellular IL-8 than PMN from healthy donors (Fig. 1). Interestingly, PMN from COVID-19 patients who went on to develop mod/severe lung disease tended to produce less IL-8 than COVID-19 patients with minimal lung disease. This may represent the timing of sampling, early in the course of disease, but suggests that PMN IL-8 would not serve to predict future disease severity.
Fig. 1.
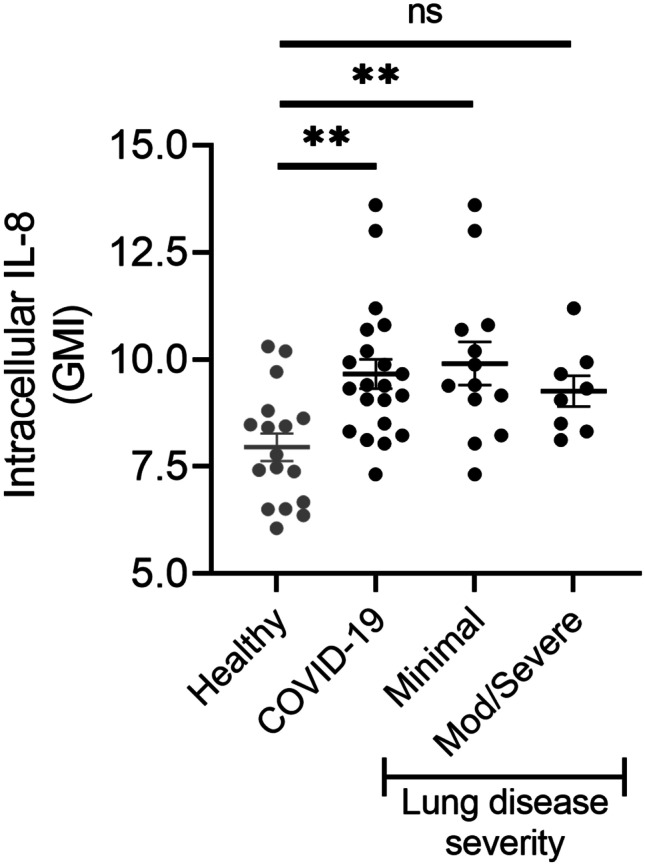
COVID-19 patients have increased neutrophil IL-8 production. Flow cytometry was used to determine neutrophil IL-8 protein levels in healthy donors (n = 17) and COVID-19 patients (n = 21). Data are presented as the geometric mean intensity (GMI) demonstrating increased intracellular IL-8 production in freshly isolated neutrophils from COVID-19 patients compared to healthy donors. There is no difference in IL-8 expression in patients who developed moderate to severe lung disease versus those with minimal lung disease. **p < 0.01.
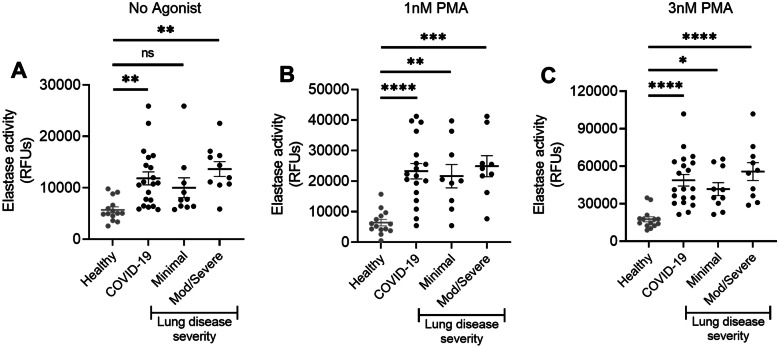
PMN From COVID-19 Patients Release Elastase at Baseline and Upon Stimulation with PMA
Neutrophil azurophilic granules contain proteases, including elastase, and release of these granule contents is tightly regulated. Neutrophil elastase has been implicated in the pathogenesis of lung disease in numerous disease settings [9]. Given the purported role of elastase in lung injury, and the correlation between high PMN number and disease severity in COVID-19 patients, we sought to assess elastase release by PMN from COVID-19 patients with and without exogenous stimulation. Importantly, in previous studies [35, 40], we have noted that PMNs from healthy donors have minimal elastase release in the absence of stimulation with agonists. Freshly isolated PMN from COVID-19 patients released significantly increased amounts of elastase as compared to healthy donors without exogenous stimulation (Fig. 2A). A trend toward greater elastase release was noted in patients with mod/severe lung disease. Further study with a larger patient population is merited to determine if elastase release correlates with lung disease severity. Exogenous stimulation with PMA does not elicit elastase release by PMN from healthy donors [35]. Interestingly, PMA elicits elastase release from COVID-19 patient neutrophils in a concentration-dependent manner suggesting a primed phenotype (Fig. 2B, C). Our findings support further investigation of neutrophil elastase as a potential therapeutic target.
Fig. 2.
Increased neutrophil elastase release in COVID-19 patients. A Freshly isolated neutrophils from COVID-19 patients release more elastase at baseline (no exogenous stimulation) than neutrophils from healthy donors. B Neutrophils from COVID-19 patients release more elastase than neutrophils from healthy donors when stimulated with 1 nM PMA or 3 nM PMA (C). n = 14 healthy donors, n = 10 COVID-19 patients with minimal lung disease, n = 10 COVID-19 patients who developed mod/severe lung disease. One patient in each group did not receive stimulation with 1 nM PMA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Circulating H3CIT-Elastase Complexes in COVID-19 Patients
Circulating cell-free DNA and myeloperoxidase-DNA complexes have been used as surrogates for NET formation and found to be elevated in plasma from COVID-19 patients [14, 15]. Interestingly, we found no change in circulating H3CIT-elastase complexes in COVID-19 patients vs healthy donors (Supplemental Fig. 1). Our H3CIT-elastase ELISA is specific for NET fragments that are produced following the citrullination of H3. NET formation that does not require the citrullination of H3 has been reported [41] and could explain the lack of detection in our ELISA. It is also possible that NET fragments are sequestered in damaged organs of patients, and that NET fragments present in the circulation do not correlate with the level of lung disease. In addition, we sampled plasma at a single time point which may not have been optimal for detection of circulating NETs. Our samples were collected within 4 days of admission and processed within 2 h. Many studies of circulating NET components rely on clinical waste samples that have been collected throughout the hospital stay and stored for hours or days [14–16].
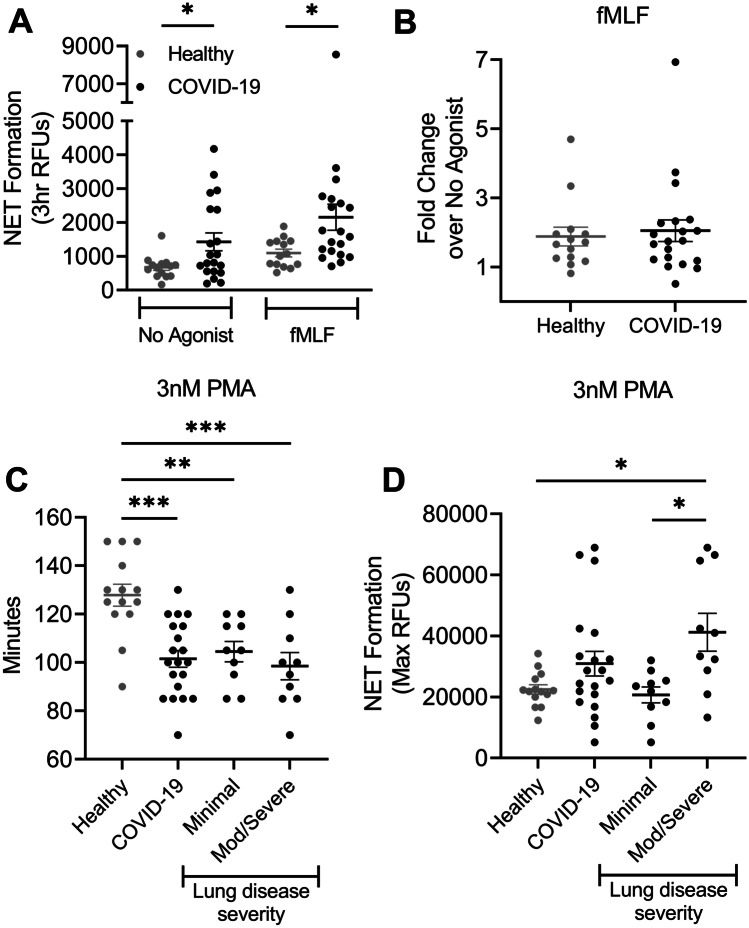
NET Formation Potential Correlates With Lung Disease Severity
NETs have been implicated in the pathogenesis of numerous disease processes including ALI [8, 10–12, 42]. Based on our expertise in neutrophil biology and interest in understanding the role of neutrophils in ALI, we sought to determine whether the NET formation potential of circulating PMN could predict future severity of lung disease in patients infected with SARS-CoV-2. Unexpectedly, we found that PMNs from COVID-19 patients generate NETs without exogenous stimulus (Fig. 3A) that is not significantly increased by stimulation with fMLF (Fig. 3A, B). Interestingly, there was a much greater degree of interindividual variability in basal NET generation among PMNs from COVID-19 patients, suggesting that a subset of patients display a primed phenotype. This phenotype, however, was not enhanced by the addition of fMLF. Based on the increased IL-8 production, elastase release, and NET formation in unstimulated PMN from COVID-19 patients, we sought to assess NET formation potential by measuring NET formation in response to PMA, a well-characterized agonist for NET formation. Interestingly, when stimulated with PMA, PMN from COVID-19 patients began generating NETs much more quickly than healthy donors (Fig. 3C). Whereas PMN from healthy donors began generating NETs on average 127.7 ± 4.9 min following stimulation with 3 nM PMA, PMN from COVID-19 patients began generating NETs on average 101.5 ± 3.5 min following stimulation again suggestive of a primed neutrophil phenotype in COVID-19 patients. In addition to a significant difference in the kinetics of NET formation, PMN from COVID-19 patients who would later develop mod/severe lung disease generated an overall increased abundance of NETs than either healthy donors or COVID-19 patients with minimal lung disease (Fig. 3D).
Fig. 3.
Rapid onset of NET formation in PMN from COVID-19 patients and correlation with lung disease severity. A Increased basal NET formation in freshly isolated neutrophils from COVID-19 patients without exogenous stimulus. Increased NET generation in neutrophils from COVID-19 patients upon stimulation with 1 μM fMLF compared to healthy donors. B Using paired samples from the same patient/donor, no significant fold change in NET formation upon stimulation with 1 µM fMLF. C NET formation occurs more rapidly in neutrophils from COVID-19 patients stimulated with 3 nM PMA compared to healthy controls. D Neutrophils from COVID-19 patients who developed mod/severe lung disease exhibit greater NET production in response to 3 nM PMA compared to COVID-19 patients with minimal lung disease or healthy controls. n = 14 healthy donors, n = 10 COVID-19 patients with minimal lung disease, n = 10 COVID-19 patients who developed mod/severe lung disease. *p < .05; **p < 0.01; ***p < 0.0001.
Discussion
The essential antibacterial role of neutrophils is unequivocal. Much less is known about the role of neutrophils in viral infections. Neutrophils exhibit antiviral properties including the capture of viral particles by NETs [43] that may be beneficial for the host immune response, but neutrophils are also implicated in amplification of inflammation and tissue destruction in influenza and respiratory syncytial virus infections, among other pulmonary diseases [10–12, 42]. Several therapeutic strategies targeting neutrophilic inflammation have been proposed. It is important to determine the pathophysiologic role of neutrophils in viral lung disease to better define therapeutic targets. Due to their critical role in host defense, previous attempts at therapeutically targeting neutrophils have had mixed results, suggesting that more mechanistic understanding will be required to identify narrow targets. Moreover the kinetics of neutrophil activation in COVID-19 infection and other processes may have critical relevance to time therapeutic interventions. In this study, we examined whether neutrophil phenotype can predict lung disease severity in hospitalized patients with COVID-19 and sought to identify potential therapeutic targets for further study.
We present 3 novel findings from our prospective study of freshly isolated neutrophils from COVID-19 patients early in the course of disease progression: (1) NET formation potential correlates with the severity of lung disease, (2) neutrophils from patients infected with SARS-CoV-2 release elastase and generate NETs with or without exogenous stimulation, and (3) IL-8 production is higher in PMN isolated from COVID-19 patients as compared with healthy donors. Our findings add to the current body of literature regarding the role of neutrophils in the pathophysiology of COVID-19 with a focus on ex vivo elastase release and NET formation in freshly isolated neutrophils from COVID-19 patients early in disease progression.
Notwithstanding a small sample size, we observed significant differences in neutrophil phenotype in patients infected with SARS-CoV-2 compared with healthy donors. Our finding that intracellular neutrophil IL-8 levels do not correlate with lung disease severity suggests that another cell type or types may be the source of high levels of circulating IL-8 measured in the plasma of COVID-19 patients. It is likely that neutrophil IL-8 levels change over the course of disease progression and that the early time point selected for our study is prior to the peak expression. A longitudinal study of plasma, neutrophil, and monocyte IL-8 levels is warranted and beyond the scope of this pilot study. Circulating IL-10 levels are significantly increased in COVID-19 patients [44], and IL-10 can inhibit LPS-induced IL-8 production in human neutrophils [45]. The trend toward lower neutrophil IL-8 in COVID-19 patients with moderate to severe lung disease could be the result of inhibition by IL-10 and represents an important avenue of future study.
There is mounting evidence suggesting a role for neutrophil elastase and NETs in the pathogenesis of lung injury [9–12, 42]. Moreover, both elastase and other NET components are currently under investigation as therapeutic targets [17, 21–24, 30]. However, we are just beginning to recognize the breadth of unique agonists that elicit NET formation and the disparate molecular mechanisms driving both regulated elastase release and NET formation [35, 41, 46]. A recent proteomic analysis of NETs found stimulus-dependent differences in protein composition and post-translational modifications that may impact biologic activity [46]. The partnering of ex vivo analysis of patient neutrophils with basic mechanistic studies allows for more precise identification of potential therapeutic targets with greater chance of success. In this study, we sought to determine whether basal elastase release and NET formation in freshly isolated neutrophils from COVID-19 patients correlated with lung injury. Moreover, we wanted to test the ability of these cells to respond to an exogenous stimulus to determine if neutrophils from COVID-19 patients demonstrated a primed phenotype with greater potential for elastase release and NET formation than neutrophils from healthy donors. We chose to study both fMLF and PMA as the exogenous stimuli for this pilot study based on the relatively small interindividual variability in response among healthy donor neutrophils [35]. Although we observed a significant increase in neutrophil elastase release in COVID-19 patients compared to healthy donors, it did not correlate with lung disease severity. A larger cohort is needed to further investigate this finding. Elastase inhibitors are currently used to treat cystic fibrosis and are in clinical trials for the treatment of numerous other inflammatory conditions including COVID-19 associated ALI [23, 24, 47].
Our data strongly suggest a primed neutrophil phenotype in COVID-19 patients with significant clinical implications as primed circulating neutrophils enter organs with the potential for over exuberant elastase release and NET formation. Neutrophil priming, a state of preactivation, is a well-characterized phenotype [48, 49]. In our study, neutrophils from COVID-19 patients stimulated with PMA demonstrated concentration-dependent elastase release and greater potential for NET formation. Interestingly, NET formation in response to fMLF was only modestly increased in neutrophils from COVID-19 patients. Neutrophil formyl-peptide receptor 1 (FPR1) recognizes a large number of ligands including fMLF and endogenous N-formylated peptides from damaged cells [50]. Upon engagement, FPR1 becomes resistant to subsequent stimulation [51]. Differential cell surface expression has also been demonstrated as a regulatory mechanism of FPR1 [52], providing one possible explanation for our findings.
In contrast to previous studies, we did not observe an increase in circulating NET fragments in COVID-19 patients. Our assay is specific for H3CIT-elastase fragments and is less sensitive than measures of cell-free DNA. Further investigation is needed to determine the source(s) and clinical implications of circulating DNA fragments in COVID-19 patients to better target them therapeutically. Broad targeting of DNA fragments with DNase has demonstrated promising results [53, 54] in pilot studies, but there is some evidence that the timing and route of treatment may be critical for positive outcomes [55].
Although our study is limited by the size of the cohorts resulting in the inability to distinguish between moderate and severe lung disease, it adds to the growing body of literature regarding the role of neutrophils in the pathogenesis of COVID-19 lung disease. The neutrophil isolation protocol used for our study resulted in the loss of low-density neutrophils (LDNs) which may result in an underestimate of NET formation in our samples, as LDNs are more likely to generate NETs and have been associated with COVID-19 severity [18, 19, 56]. Several studies have demonstrated NET formation in COVID-19 patients, but the underlying mechanisms driving neutrophil activation and NET formation are unknown. Among the many possible candidates, histidine-rich glycoprotein (HRG) merits further investigation as both a biomarker and potential therapeutic target. HRG serves numerous functions in the maintenance of immune homeostasis [57]. In a murine sepsis model, HRG is significantly reduced, and HRG supplementation improves survival [58]. A recent serum proteomics study of patients with COVID-19 found that among the proteins tested, HRG was the best single predictor of mortality. HRG levels in blood samples collected within the first 4 days of hospitalization were significantly lower in non-survivors than survivors [59]. The role of HRG and other plasma factors in the regulation of neutrophil activity during sepsis represents an intriguing area of future investigation.
In conclusion, infection with SARS-CoV-2 initiates a wide range of symptom severity and COVID-19 progresses over the course of days to weeks. Regardless of known comorbidities and risk factors, it is difficult to predict which patients will experience severe inflammatory complications including organ dysfunction and death. Assessment of neutrophil phenotype and function early in the course of disease progression may provide a tool for risk stratification and the opportunity to alter therapeutic strategies.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Supplemental Fig. 1. Plasma H3CIT-elastase complexes. There was no difference in plasma H3CIT-elastase complexes in healthy donors (n = 16) vs COVID-19 patients (n = 24). (PDF 224 KB)
ACKNOWLEDGEMENTS
We would like to thank Drs. Dwight Towler, Benjamin Greenberg, and David Russell for their leadership in establishing the UT Southwestern SARS-CoV-2 Biorepository. We would also like to thank staff members of the biorepository, Christine Pybus, Jana Hadas, and George Adams, and the Neurology Translational Research Center, Manuel Huichapa, Phoebe Pittman, Martha Mann, and Morgan McCreary, for enrolling patients, collecting samples, isolating plasma and neutrophils, and providing the clinical data for our study.
AUTHOR CONTRIBUTION
The study was designed by Jessica Moreland and Jessica Hook. Nedha Kinnare, Jessica Hook, and Parth Patel performed the research and analyzed the data. Jessica Moreland, Nedha Kinnare, and Jessica Hook wrote the manuscript. Nancy Monson directed the biorepository and participated in the final editing of the manuscript.
AVAILABILITY OF DATA AND MATERIAL
All data supporting the conclusions are presented in the manuscript. Additional information will be made available by the corresponding author upon reasonable request.
CODE AVAILABILITY
Not applicable.
DECLARATIONS
Ethics Approval
Study approval was obtained from the Institutional Review Board at UT Southwestern, and this study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments.
Consent to Participate
Informed consent was obtained from all individual participants or legal guardians in the study.
Consent for Publication
Informed consent was obtained from all individual participants or legal guardians to publish non-identifiable data.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.WHO, World Health Organization. 2021. Coronavirus Disease (COVID-19) Situation Reports. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---17-august-2021. Accessed Aug 2021.
- 2.Hu B, Huang S, Yin L. The cytokine storm and COVID-19. Journal of Medical Virology. 2021;93(1):250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, et al. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. Journal of Leukocyte Biology. 2020;108(1):17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alosaimi B, et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. 2020;126:154895. doi: 10.1016/j.cyto.2019.154895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam CW, Chan MH, Wong CK. Severe acute respiratory syndrome: Clinical and laboratory manifestations. Clinical Biochemist Reviews. 2004;25(2):121–132. [PMC free article] [PubMed] [Google Scholar]
- 6.Ma A, et al. Neutrophil-to-lymphocyte ratio as a predictive biomarker for moderate-severe ARDS in severe COVID-19 patients. Critical Care. 2020;24(1):288. doi: 10.1186/s13054-020-03007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, et al. Predictive values of neutrophil-to-lymphocyte ratio on disease severity and mortality in COVID-19 patients: A systematic review and meta-analysis. Critical Care. 2020;24(1):647. doi: 10.1186/s13054-020-03374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayadas TN, Cullere X, Lowell CA. The multifaceted functions of neutrophils. Annual Review of Pathology: Mechanisms of Disease. 2014;9:181–218. doi: 10.1146/annurev-pathol-020712-164023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamakuma S, et al. Relationship between neutrophil elastase and acute lung injury in humans. Pulmonary Pharmacology & Therapeutics. 2004;17(5):271–279. doi: 10.1016/j.pupt.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Narasaraju T, et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. American Journal of Pathology. 2011;179(1):199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porto BN, Stein RT. Neutrophil Extracellular Traps in Pulmonary Diseases: Too Much of a Good Thing? Frontiers in Immunology. 2016;7:311. doi: 10.3389/fimmu.2016.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortjens B, van Woensel JB, Bem RA. Neutrophil Extracellular Traps in Respiratory Disease: Guided anti-microbial traps or toxic webs? Paediatric Respiratory Reviews. 2017;21:54–61. doi: 10.1016/j.prrv.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Denning NL, et al. DAMPs and NETs in Sepsis. Frontiers in Immunology. 2019;10:2536. doi: 10.3389/fimmu.2019.02536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo, Y., et al. 2020. Neutrophil extracellular traps in COVID-19. JCI Insight 5(11). 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed]
- 15.Zuo, Y., et al. 2020. Neutrophil extracellular traps (NETs) as markers of disease severity in COVID-19. medRxiv. 10.1101/2020.04.09.20059626.
- 16.Zuo, Y., et al. 2020. Neutrophil extracellular traps and thrombosis in COVID-19. medRxiv. 10.1101/2020.04.30.20086736.
- 17.Barnes, B.J., et al. 2020 Targeting potential drivers of COVID-19: Neutrophil extracellular traps. Journal of Experimental Medicine 217(6). 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed]
- 18.Leppkes M, et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. eBioMedicine. 2020;58:102925. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obermayer A, et al. Neutrophil Extracellular Traps in Fatal COVID-19-Associated Lung Injury. Disease Markers. 2021;2021:5566826. doi: 10.1155/2021/5566826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masso-Silva JA, et al. Increased peripheral blood neutrophil activation phenotypes and NETosis in critically ill COVID-19 patients: A case series and review of the literature. Clinical Infectious Diseases. 2021 doi: 10.1093/cid/ciab437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeiher BG, et al. Neutrophil elastase inhibition in acute lung injury: Results of the STRIVE study. Critical Care Medicine. 2004;32(8):1695–1702. doi: 10.1097/01.ccm.0000133332.48386.85. [DOI] [PubMed] [Google Scholar]
- 22.Iwata K, et al. Effect of neutrophil elastase inhibitor (sivelestat sodium) in the treatment of acute lung injury (ALI) and acute respiratory distress syndrome (ARDS): A systematic review and meta-analysis. Internal Medicine. 2010;49(22):2423–2432. doi: 10.2169/internalmedicine.49.4010. [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi S, et al. Usefulness of a selective neutrophil elastase inhibitor, sivelestat, in acute lung injury patients with sepsis. Drug Des Devel Ther. 2013;7:305–316. doi: 10.2147/DDDT.S42004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohira S, et al. Effect of the neutrophil elastase inhibitor sivelestat on perioperative inflammatory response after pediatric heart surgery with cardiopulmonary bypass: A prospective randomized study. Artificial Organs. 2013;37(12):1027–1033. doi: 10.1111/aor.12103. [DOI] [PubMed] [Google Scholar]
- 25.Nathan R, et al. A Narrative Review of the Clinical Practicalities of Bamlanivimab and Etesevimab Antibody Therapies for SARS-CoV-2. Infectious Disease and Therapy. 2021 doi: 10.1007/s40121-021-00515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beigel JH, et al. Remdesivir for the Treatment of Covid-19 - Final Report. New England Journal of Medicine. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansems, K., et al. 2021. Remdesivir for the treatment of COVID-19. Cochrane Database of Systematic Reviews 8:CD014962. 10.1002/14651858.CD014962. [DOI] [PMC free article] [PubMed]
- 28.Hazeldine J, Lord JM. Neutrophils and COVID-19: Active Participants and Rational Therapeutic Targets. Frontiers in Immunology. 2021;12:680134. doi: 10.3389/fimmu.2021.680134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reusch N, et al. Neutrophils in COVID-19. Frontiers in Immunology. 2021;12:652470. doi: 10.3389/fimmu.2021.652470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang CC, et al. Targeting Neutrophils to Treat Acute Respiratory Distress Syndrome in Coronavirus Disease. Frontiers in Pharmacology. 2020;11:572009. doi: 10.3389/fphar.2020.572009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. The Journal of Clinical Investigation. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO. 2020. WHO R&D Blueprint novel Coronavirus COVID-19 Therapeutic Trial Synopsis.
- 33.Nauseef WM. Isolation of human neutrophils from venous blood. Methods in Molecular Biology. 2007;412:15–20. doi: 10.1007/978-1-59745-467-4_2. [DOI] [PubMed] [Google Scholar]
- 34.Kuhns, D.B., et al. 2015. Isolation and Functional Analysis of Human Neutrophils. Current Protocols in Immunology 111:7231–72316. 10.1002/0471142735.im0723s111. [DOI] [PMC free article] [PubMed]
- 35.Hook JS, et al. Lipoproteins from Staphylococcus aureus Drive Neutrophil Extracellular Trap Formation in a TLR2/1- and PAD-Dependent Manner. The Journal of Immunology. 2021;207(3):966–973. doi: 10.4049/jimmunol.2100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hook JS, et al. Mycobacterium tuberculosis Lipoarabinomannan Activates Human Neutrophils via a TLR2/1 Mechanism Distinct from Pam3CSK4. The Journal of Immunology. 2020;204(3):671–681. doi: 10.4049/jimmunol.1900919. [DOI] [PubMed] [Google Scholar]
- 37.Luo J, et al. The selection of indicators from initial blood routine test results to improve the accuracy of early prediction of COVID-19 severity. PLoS ONE. 2021;16(6):e0253329. doi: 10.1371/journal.pone.0253329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Valle DM, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nature Medicine. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma A, et al. High Levels of Circulating IL-8 and Soluble IL-2R Are Associated With Prolonged Illness in Patients With Severe COVID-19. Frontiers in Immunology. 2021;12:626235. doi: 10.3389/fimmu.2021.626235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potera RM, et al. Neutrophil azurophilic granule exocytosis is primed by TNF-alpha and partially regulated by NADPH oxidase. Innate Immunity. 2016;22(8):635–646. doi: 10.1177/1753425916668980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeltz S, et al. To NET or not to NET:Current opinions and state of the science regarding the formation of neutrophil extracellular traps. Cell Death and Differentiation. 2019;26(3):395–408. doi: 10.1038/s41418-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortjens B, et al. Neutrophil extracellular traps cause airway obstruction during respiratory syncytial virus disease. The Journal of Pathology. 2016;238(3):401–411. doi: 10.1002/path.4660. [DOI] [PubMed] [Google Scholar]
- 43.Galani IE, Andreakos E. Neutrophils in viral infections: Current concepts and caveats. Journal of Leukocyte Biology. 2015;98(4):557–564. doi: 10.1189/jlb.4VMR1114-555R. [DOI] [PubMed] [Google Scholar]
- 44.Han H, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9(1):1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P, et al. IL-10 inhibits transcription of cytokine genes in human peripheral blood mononuclear cells. The Journal of Immunology. 1994;153(2):811–816. [PubMed] [Google Scholar]
- 46.Petretto A, et al. Neutrophil extracellular traps (NET) induced by different stimuli: A comparative proteomic analysis. PLoS ONE. 2019;14(7):e0218946. doi: 10.1371/journal.pone.0218946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahebnasagh A, et al. Neutrophil elastase inhibitor (sivelestat) may be a promising therapeutic option for management of acute lung injury/acute respiratory distress syndrome or disseminated intravascular coagulation in COVID-19. Journal of Clinical Pharmacy and Therapeutics. 2020;45(6):1515–1519. doi: 10.1111/jcpt.13251. [DOI] [PubMed] [Google Scholar]
- 48.Vogt KL, et al. Priming and de-priming of neutrophil responses in vitro and in vivo. European Journal of Clinical Investigation. 2018;48(Suppl 2):e12967. doi: 10.1111/eci.12967. [DOI] [PubMed] [Google Scholar]
- 49.Miralda I, Uriarte SM, McLeish KR. Multiple Phenotypic Changes Define Neutrophil Priming. Frontiers in Cellular and Infection Microbiology. 2017;7:217. doi: 10.3389/fcimb.2017.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss E, Kretschmer D. Formyl-Peptide Receptors in Infection, Inflammation, and Cancer. Trends in Immunology. 2018;39(10):815–829. doi: 10.1016/j.it.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Didsbury JR, et al. Receptor class desensitization of leukocyte chemoattractant receptors. Proc Natl Acad Sci U S A. 1991;88(24):11564–11568. doi: 10.1073/pnas.88.24.11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gemperle C, et al. Regulation of the formyl peptide receptor 1 (FPR1) gene in primary human macrophages. PLoS ONE. 2012;7(11):e50195. doi: 10.1371/journal.pone.0050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee YY, et al. Long-acting nanoparticulate DNase-1 for effective suppression of SARS-CoV-2-mediated neutrophil activities and cytokine storm. Biomaterials. 2021;267:120389. doi: 10.1016/j.biomaterials.2020.120389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fisher J, et al. Proteome Profiling of Recombinant DNase Therapy in Reducing NETs and Aiding Recovery in COVID-19 Patients. Molecular and Cellular Proteomics. 2021;20:100113. doi: 10.1016/j.mcpro.2021.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mai SH, et al. Delayed but not Early Treatment with DNase Reduces Organ Damage and Improves Outcome in a Murine Model of Sepsis. Shock. 2015;44(2):166–172. doi: 10.1097/SHK.0000000000000396. [DOI] [PubMed] [Google Scholar]
- 56.Carissimo G, et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nature Communications. 2020;11(1):5243. doi: 10.1038/s41467-020-19080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poon IK, et al. Histidine-rich glycoprotein: The Swiss Army knife of mammalian plasma. Blood. 2011;117(7):2093–2101. doi: 10.1182/blood-2010-09-303842. [DOI] [PubMed] [Google Scholar]
- 58.Wake H, et al. Histidine-Rich Glycoprotein Prevents Septic Lethality through Regulation of Immunothrombosis and Inflammation. eBioMedicine. 2016;9:180–194. doi: 10.1016/j.ebiom.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vollmy, F., et al. 2021. A serum proteome signature to predict mortality in severe COVID-19 patients. Life Science Alliance 4(9). 10.26508/lsa.202101099. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Supplemental Fig. 1. Plasma H3CIT-elastase complexes. There was no difference in plasma H3CIT-elastase complexes in healthy donors (n = 16) vs COVID-19 patients (n = 24). (PDF 224 KB)
Data Availability Statement
All data supporting the conclusions are presented in the manuscript. Additional information will be made available by the corresponding author upon reasonable request.
Not applicable.