Abstract
A sensitive PCR assay that detects the thermophilic campylobacters C. jejuni, C. coli, C. lari, and C. upsaliensis is reported. Furthermore, by digestion of the PCR products with two restriction enzymes, species differentiation was demonstrated. Thus, the present method has the potential to be used for both detection and identification of thermophilic Campylobacter species.
The rate of Campylobacter infections has been increasing, with the number of cases often exceeding those of Salmonella and Shigella (14, 16). The clinically most important campylobacters are the members of the thermophilic group, C. jejuni, C. coli, C. lari, and C. upsaliensis, with C. jejuni responsible for the majority of human cases (1). The true incidences, however, might be underestimated, since most cultivation methods are designed for C. jejuni alone and therefore may not be optimal for the other species (3). Furthermore, in many clinical laboratories, typing is only done to the genus level, since species determination is cumbersome and sometimes gives incorrect or intermediate results.
Thus, there is a great need for simple methods for detection and reliable differentiation of the thermophilic Campylobacter species. Indeed, a number of genetically based detection and typing methods have been developed for Campylobacter species in food, clinical, and environmental samples (4–6, 9, 12, 13, 17, 22, 23). However, the majority of these methods suffer from drawbacks like the need for multiple PCRs or restriction enzymes or the requirement of a hybridization step with species-specific probes after the PCR.
This study describes a sensitive and simple method for both the detection and differentiation of thermophilic campylobacters. Detection is achieved by the amplification of 491 bp of a highly polymorphic part of the 23S rRNA gene, and further species differentiation is accomplished by digestion of the PCR product by two restriction enzymes, AluI and Tsp509I, resulting in specific restriction fragments for each species.
The bacterial strains used in this study are listed in Table 1. The bacteria were cultured aerobically or in a microaerophilic atmosphere on blood agar at 37 or 42°C.
TABLE 1.
Bacterial strains used in this study
| Species | No. of strains tested | No. of PCR-positive strains | Species according to PCR typing | Sourcea |
|---|---|---|---|---|
| C. jejuni | ||||
| Human isolates | 66 | 66 | 65 C. jejuni, 1 C. colib | UU, RSÖ, Telelab |
| Veterinary isolates | 7 | 7 | C. jejuni | NVI |
| SLV-167 | 1 | 1 | C. jejuni | NFA |
| SLV-267 | 1 | 1 | C. jejuni | NFA |
| SLV-264 | 1 | 1 | C. jejuni | NFA |
| CCUG 11284 | 1 | 1 | C. jejuni | CCUG |
| C. coli | ||||
| Human isolate | 1 | 1 | C. coli | Telelab |
| Veterinary isolates | 14 | 14 | 13 C. coli, 1 C. jejunib | NVI |
| SLV-271 | 1 | 1 | C. coli | NFA |
| CCUG 11283 | 1 | 1 | C. coli | CCUG |
| C. lari | ||||
| Veterinary isolates | 3 | 3 | C. lari | NVI |
| CCUG 23947 | 1 | 1 | C. lari | CCUG |
| From seagulls | 2 | 2 | C. lari | NVI |
| 4219/92 | 1 | 1 | C. lari | NIPH |
| 1557/93 | 1 | 1 | C. lari | NIPH |
| 1089/94 | 1 | 1 | C. lari | NIPH |
| 2138/94 | 1 | 1 | C. lari | NIPH |
| C. upsaliensis | ||||
| Human isolate | 1 | 1 | C. upsaliensis | Telelab |
| Veterinary isolates | 6 | 6 | C. upsaliensis | NVI |
| From dogs | 5 | 5 | C. upsaliensis | NVI |
| CCUG 14913 | 1 | 1 | C. upsaliensis | CCUG |
| 14913 | 1 | 1 | C. upsaliensis | NIPH |
| C. hyointestinalis | 2 | 0 | NVI | |
| C. fetus subsp. fetus | 2 | 0 | NVI | |
| C. fetus subsp. venerealis | 1 | 0 | NVI | |
| C. mucosalis | 2 | 1c | NVI | |
| Helicobacter pylori | 8 | 0 | UU | |
| Salmonella enteritidis SLV-397 | 1 | 0 | NFA | |
| Salmonella typhimurium SLV-248 | 1 | 0 | NFA | |
| Salmonella dublin SLV-242 | 1 | 0 | NVI | |
| Salmonella bovismorbificans SLV-443 | 1 | 0 | SDIIC | |
| Shigella flexneri 820/92 | 1 | 0 | SDIIC | |
| Shigella boydii 915/94 | 1 | 0 | SDIIC | |
| Shigella sonnei 637/94 | 1 | 0 | SDIIC | |
| Escherichia coli | ||||
| SLV-165 | 1 | 0 | NFA | |
| O157:H7 81186 | 1 | 0 | SDIIC | |
| O157:H7 ATCC 43889 | 1 | 0 | SDIIC | |
| O157:H7 ATCC 43890 | 1 | 0 | SDIIC | |
| Yersinia enterocolitica SLV-408 | 1 | 0 | NFA | |
| Vibrio cholerae CCUG 34649 | 1 | 0 | CCUG | |
| Vibrio parahaemolyticus CCUG 19113 | 1 | 0 | CCUG | |
| Vibrio vulnificus CCUG 13448 | 1 | 0 | CCUG | |
| Staphylococcus aureus SLV-350 | 1 | 0 | NFA | |
| Staphylococcus saprophyticus SLV-013 | 1 | 0 | NFA | |
| Enterobacter cloacae SLV-164 | 1 | 0 | NFA | |
| Enterococcus durans SLV-078 | 1 | 0 | NFA | |
| Bacillus cereus SLV-160 | 1 | 0 | NFA | |
| Klebsiella oxytoca SLV-089 | 1 | 0 | NFA | |
| Clostridium perfringens SLV-442 | 1 | 0 | NFA |
NFA, National Food Administration, Uppsala, Sweden; SDIIC, Swedish Institute for Infectious Disease Control, Stockholm, Sweden; NVI, National Veterinary Institute, Uppsala, Sweden; Telelab, A/S Telelab, Skien, Norway; UU, University of Uppsala, Uppsala, Sweden; RSÖ, Örebro Medical Center Hospital, Örebro, Sweden; CCUG, Culture Collection of the University of Göteborg, Göteborg, Sweden; NIPH, National Institute of Public Health, Oslo, Norway.
The disagreements between the biochemical and genetic typing are discussed in the text.
One strain of C. mucosalis gave a faint band of the correct size when using 1.5 mM MgCl2. This band disappeared when the MgCl2 concentration was lowered to 0.5 mM.
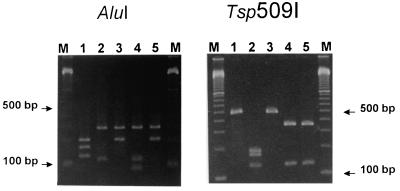
Chromosomal DNA was prepared either by simply breaking the bacterial cells with heat or by the method of Pitcher et al. (15). The PCR mixtures contained a 0.25 μM concentration of each oligonucleotide (Scandinavian Gene Synthesis, Köping, Sweden), 50 mM KCl, 10 mM Tris-HCl (pH 8.3 at room temperature), 1.5 mM MgCl2, a 0.1 mM concentration of each deoxynucleotide, and 0.02 U of AmpliTaq Gold (PE Applied Biosystems, Branchburg, N.J.) per μl. The temperature cycling was performed on a PTC-200 DNA engine (MJ Research, Watertown, Mass.) at 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min for 45 cycles, followed by a final extension at 72°C for 5 min. The temperature protocol was preceded by a 12-min incubation at 94°C in order to activate the AmpliTaq Gold polymerase. The PCR products were visualized by 3% agarose gel electrophoresis. To differentiate between species, 10-μl samples were removed from PCR-positive mixtures and digested in two separate reaction tubes for 1 h with either 0.5 U of AluI at 37°C or 0.5 U of Tsp509I at 65°C. PCR products of type strains with known banding patterns were digested and analyzed in parallel with unknown samples in order to ensure cleavage activity. Banding patterns, easy to interpret, were reproducibly obtained and visualized by gel electrophoresis (Fig. 1).
FIG. 1.
Restriction fragments generated by digestion with AluI and Tsp509I of PCR products of the thermophilic campylobacters. Lanes: 1, C. jejuni CCUG 11284; 2, C. lari CCUG 23947; 3, C. coli CCUG 11283; 4, C. upsaliensis C434 (from a dog); 5, C. upsaliensis CCUG 14913. Lanes M contain a 100-bp ladder.
The most variable part of the 23S rRNA gene, located between helices 43 and 69 (7), of different Campylobacter species has been extensively sequenced and used as a target for species-specific PCR methods (4, 8). One primer pair, THERM1 and THERM2, has been shown to specifically detect the thermophilic Campylobacter species (4). In the present study, THERM1 was used together with a new primer, THERM4 (5′-CTTCGCTAATGCTAACCC-3′). THERM4 was constructed to perfectly match the sequences of the thermophilic species by using published DNA sequences (Table 2). The primer pair THERM1 and THERM4 demonstrated high specificity, giving a PCR product of the correct size for all strains of the thermophilic group, but not for other Campylobacter species or other bacteria (Table 1). One isolate of Campylobacter mucosalis, though, gave a faint band of the correct size. This band, however, disappeared when the MgCl2 concentration was lowered to 0.5 mM. Nevertheless, the higher MgCl2 concentration was used throughout the study, since amplification of the C. mucosalis isolate resulted in a barely visible band on the gel and the restriction fragments of this PCR product generated unique banding patterns clearly distinguishable from those of the target species.
TABLE 2.
23S rRNA gene sequences accessible in the GenBank sequence database used for primer design and for choosing discriminatory restriction enzymes
| Strain | Accession no. | Source or reference |
|---|---|---|
| C. jejuni | ||
| LMG 6444 | X67767 | 21 |
| LMG 6629 | X67765 | 21 |
| ATCC 33250 | X66616 | 20 |
| LMG 7790 | X67766 | 21 |
| ATCC 43431 | Z29326 | 11 |
| C. coli | ||
| LMG 7535 | X67770 | 21 |
| LMG 6440 | X67764 | 21 |
| VC167 | U09611 | 19 |
| C. lari | ||
| LMG 8846 | X67769 | 21 |
| NCTC 11352 | Y11764 | Direct submission |
| C. hyoilei NCTC 33450 | Y11765 | Direct submission |
| C. upsaliensis | ||
| LMG 8854 | X67774 | 21 |
| LMG 7533 | X67763 | 21 |
| C. sputorum subsp. sputorum LMG 7795 | X67758 | 21 |
| C. sputorum subsp. fecalis LMG 6617 | X67758 | 21 |
| C. sputorum subsp. bubulus LMG 6447 | X67772 | 21 |
| C. mucosalis | ||
| LMG 6448T | X80636 | 2 |
| LMG 7794 | X67760 | 21 |
| C. fetus subsp. venerealis LMG 6570 | X67773 | 21 |
| C. hyointestinalis LMG 2817 | X67761 | 21 |
| C. fetus LMG 6442 | X67762 | 21 |
| C. concisus LMG 7788 | X67759 | 21 |
The PCR assay detected as few as one to five cells or chromosomes of C. jejuni SLV-428. The number of chromosomes was calculated according to the chromosomal size estimated by Karlyshev et al. (10). The detection of such a low number of cells is probably enhanced by the occurrence of three copies of the 23S rRNA genes on the chromosome (18).
Restriction enzymes specifically recognizing polymorphic areas within the amplified segment were identified with the WWWtacg version 2.38 software (Harry Mangalam, University of California, Irvine). Digestion with AluI gave unique combinations of fragments for C. jejuni and C. lari, respectively (Fig. 1). Isolates of C. upsaliensis showed either of two AluI banding patterns, one of which was the same as that for C. coli (Fig. 1). The two different AluI patterns of C. upsaliensis strains were expected, since both sequence variants have been published (Table 2). Differentiation between C. coli and C. upsaliensis was achieved by using a second enzyme, Tsp509I, which resulted in discriminatory banding patterns (Fig. 1). There was a discrepancy between the expected identity and the result of the PCR assay in two cases of 118 thermophilic campylobacters tested (Table 1). One human isolate previously identified as C. jejuni was typed as C. coli according to the restriction banding patterns. This isolate was positive for hippurate hydrolysis and was accordingly PCR positive for the hippuricase gene only present in this species (12). Thus, the PCR assay failed to classify this isolate correctly. The second isolate, a veterinary strain previously typed as C. coli, was according to the PCR assay C. jejuni. The PCR typing result was supported by a positive amplification of the hippuricase gene. However, the DNA fragment generated was about 300 bp shorter than expected (12). Nevertheless, all other C. coli isolates analyzed were negative for the hippuricase gene PCR, indicating that this particular isolate in fact is a C. jejuni strain with a truncated hippuricase gene, thus explaining the negative hippurate hydrolysis phenotype characteristic of strains of C. coli.
The PCR method presented in this study identifies with a single PCR low numbers of thermophilic campylobacters (1 to 5 CFU). In addition, by two simple restriction enzyme digestions performed directly in the PCR mixture, it differentiates between species within this group of bacteria. Thus, the method has the potential to be used for detection and identification of thermophilic campylobacters in complex samples, such as foods in which low numbers are present. This method also has the possibility of complementing or replacing phenotypic methods for identifying thermophilic Campylobacter species.
Acknowledgments
We thank Christina Normark for excellent technical assistance and Bjørn-Erik Kristiansen, Telemark Biomedical Center, Skien, Norway; Per Olcén, Central Hospital of Örebro, Sweden; Ameera Gibreel, Department of Pharmaceutical Biosciences, University of Uppsala, Uppsala, Sweden; Anders Gunnarsson, National Veterinary Institute, Uppsala, Sweden; and Astrid Wage and Traute Vardund, National Institute of Public Health, Oslo, Norway, for supplying bacteria.
REFERENCES
- 1.Allos B M, Blaser M J. Campylobacter jejuni and the expanding spectrum of related infections. Clin Infect Dis. 1995;20:1092–1099. doi: 10.1093/clinids/20.5.1092. [DOI] [PubMed] [Google Scholar]
- 2.Bastyns K, Chapelli S, Vandamme P, Goossens H, de Wachter R. Species-specific detection of campylobacteria important in veterinary medicine by PCR amplification of 23S rRNA fragments. Syst Appl Microbiol. 1994;17:563–568. [Google Scholar]
- 3.Corry J E, Post D E, Colin P, Laisney M J. Culture media for the isolation of campylobacters. Int J Food Microbiol. 1995;26:43–76. doi: 10.1016/0168-1605(95)00044-k. [DOI] [PubMed] [Google Scholar]
- 4.Eyers M, Chapelle S, van Camp G, Goossens H, De Wachter R. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J Clin Microbiol. 1993;31:3340–3343. doi: 10.1128/jcm.31.12.3340-3343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giesendorf B A J, Quint W G V, Henkens M H C, Stegeman H, Huf F A, Niesters H G M. Rapid and sensitive detection of Campylobacter spp. in chicken products by using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3804–3808. doi: 10.1128/aem.58.12.3804-3808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez I, Grant K A, Richardson P T, Park S F, Collins M D. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J Clin Microbiol. 1997;35:759–763. doi: 10.1128/jcm.35.3.759-763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopfl P, Ludwig W, Schleifer K H, Larsen N. The 23S ribosomal RNA higher-order structure of Pseudomonas cepacia and other prokaryotes. Eur J Biochem. 1989;185:355–364. doi: 10.1111/j.1432-1033.1989.tb15123.x. [DOI] [PubMed] [Google Scholar]
- 8.Hurtado A, Owen R J. A molecular scheme based on 23S rRNA gene polymorphisms for rapid identification of Campylobacter and Arcobacter species. J Clin Microbiol. 1997;35:2401–2404. doi: 10.1128/jcm.35.9.2401-2404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson C J, Fox A J, Jones D M. A novel polymerase chain reaction assay for the detection and speciation of thermophilic Campylobacter spp. J Appl Bacteriol. 1996;81:467–473. doi: 10.1111/j.1365-2672.1996.tb03534.x. [DOI] [PubMed] [Google Scholar]
- 10.Karlyshev A V, Henderson J, Ketley J M, Wren B W. An improved physical and genetic map of Campylobacter jejuni NCTC 11168 (ua580) Microbiology. 1998;144:503–508. doi: 10.1099/00221287-144-2-503. [DOI] [PubMed] [Google Scholar]
- 11.Kim N W, Gutell R R, Chan V L. Complete sequences and organization of the rRNA operon from Campylobacter jejuni TGH9011 (ATCC43431) Gene. 1995;164:101–106. doi: 10.1016/0378-1119(95)00471-h. [DOI] [PubMed] [Google Scholar]
- 12.Linton D, Lawson A J, Owen R J, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metherell L A, Logan J M J, Stanley J. PCR-enzyme-linked immunosorbent assay for detection and identification of Campylobacter species: application to isolates and stool samples. J Clin Microbiol. 1999;37:433–435. doi: 10.1128/jcm.37.2.433-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philips A C. Incidence, epidemiology and prevention of foodborne Campylobacter species. Trends Food Sci Technol. 1995;6:83–86. [Google Scholar]
- 15.Pitcher D G, Saunders N A, Owen R J. Raid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 16.Skirrow M B. Campylobacter. Lancet. 1990;336:921–923. doi: 10.1016/0140-6736(90)92282-m. [DOI] [PubMed] [Google Scholar]
- 17.Stucki U, Frey J, Nicolet J, Burnens A P. Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J Clin Microbiol. 1995;33:855–859. doi: 10.1128/jcm.33.4.855-859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor D E, Eaton M, Yan W, Chang N. Genome maps of Campylobacter jejuni and Campylobacter coli. J Bacteriol. 1992;174:2332–2337. doi: 10.1128/jb.174.7.2332-2337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trust T J, Logan S M, Gustafson C E, Romaniuk P J, Kim N W, Chan V L, Ragan M A, Guerry P, Gutell R R. Phylogenetic and molecular characterization of a 23S rRNA gene positions the genus Campylobacter in the epsilon subdivision of the Proteobacteria and shows that the presence of transcribed spacers is common in Campylobacter spp. J Bacteriol. 1994;176:4597–4609. doi: 10.1128/jb.176.15.4597-4609.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Camp G, Chapelle S, de Wachter R. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA genes with conserved primer sequences. Curr Microbiol. 1993;27:147–151. doi: 10.1007/BF01576012. [DOI] [PubMed] [Google Scholar]
- 21.van Camp G, Van de Peer Y, Neefs J M, Vandamme P, de Wachter R. Presence of internal transcribed spacers in the 16S and 23S ribosomal RNA genes of Campylobacter. Syst Appl Microbiol. 1993;16:361–368. [Google Scholar]
- 22.van Doorn L J, Giesendorf B A, Bax R, van der Zeijst B A, Vandamme P, Quint W G. Molecular discrimination between Campylobacter jejuni, Campylobacter coli, Campylobacter lari and Campylobacter upsaliensis by polymerase chain reaction based on a novel putative GTPase gene. Mol Cell Probes. 1997;11:177–185. doi: 10.1006/mcpr.1997.0100. [DOI] [PubMed] [Google Scholar]
- 23.Wegmüller B, Lüthy J, Candrian U. Direct polymerase chain reaction detection of Campylobacter jejuni and Campylobacter coli in raw milk and dairy products. Appl Environ Microbiol. 1993;59:2161–2165. doi: 10.1128/aem.59.7.2161-2165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]