Abstract
Microorganisms striving in extreme environments and exhibiting optimal growth and reproduction at low temperatures, otherwise known as psychrophilic microorganisms, are potential sources of cold-active enzymes. Owing to higher stability and cold activity, these enzymes are gaining enormous attention in numerous industrial bioprocesses. Applications of several cold-active enzymes have been established in the food industry, e.g., β-galactosidase, pectinase, proteases, amylases, xylanases, pullulanases, lipases, and β-mannanases. The enzyme engineering approaches and the accumulating knowledge of protein structure and function have made it possible to improve the catalytic properties of interest and express the candidate enzyme in a heterologous host for a higher level of enzyme production. This review compiles the relevant and recent information on the potential uses of different cold-active enzymes in the food industry.
Keywords: Psychrophiles, Cold-active enzymes, Food industry, Enzyme engineering
Introduction
The term “psychrophiles” is a Greek word coming from psukhros meaning ‘cold’ and philein, ‘loving’. Psychrophiles are defined as microorganisms that can grow and sustain in cold environments, such as deep sea, high elevations and Polar Regions of the planet Earth, and can tolerate the temperature range 0–20 °C; however, the optimal temperature for their growth is 5 °C (Cavicchioli 2016; Salwan and Sharma 2020). The first known and taxonomically described species of psychrophiles are Vibrio marinus and V. psychroerythrus (Morita and Moyer 2001). Nonetheless, several species of bacteria (Bacillus, Bacteroides, Arthrobacter, Clostridium, Pseudomonas, and Methanogenium) and fungi (Pseudogymnoascus, and Geomyces) have also been described to be psychrophilic (Morita and Moyer 2001; Meteyer and Verant 2019). Psychrophiles are well adapted to extreme environments and possess complex metabolic adaptations, such as altered nutrient transport mechanisms, intracellular ice formation and cold denaturation of proteins (Feller and Gerday 2003). Besides, these microorganisms are potentially rich enzymes that can maintain high activity even at low temperatures by reducing the temperature dependence of the reaction (Yayanos 2009; Cavicchioli et al. 2011; Irwin 2020; Rai and Rakshak 2015).
These cold-active or psychrophilic enzymes are advantageous for many industrial applications as they are (i) cost-effective, (ii) able to catalyze reaction without any additional heat aid and (iii) can be inactivated selectively by gentle heat input. Cold-active enzymes are also used in several biotechnology applications to prevent many undesirable reactions and restrict the loss of volatile components (Gerday 2013; Kuddus 2018). Due to these inevitable applications, the role of cold-active enzymes in the coming years is expected to witness generous market growth (Santiago et al. 2016; Al-Ghanayem and Joseph 2020). Numerous possible prospects of these cold-active enzymes have also become evident in food processing applications as the low temperature reduces many adverse reactions and microbial contamination (Kuddus and Ramteke 2012; Javed and Qazi 2016). Production of fruit juices, alcoholic beverages, chocolates, sweeteners, cheese, bakery products and milk products are typical applications of cold-active enzymes.
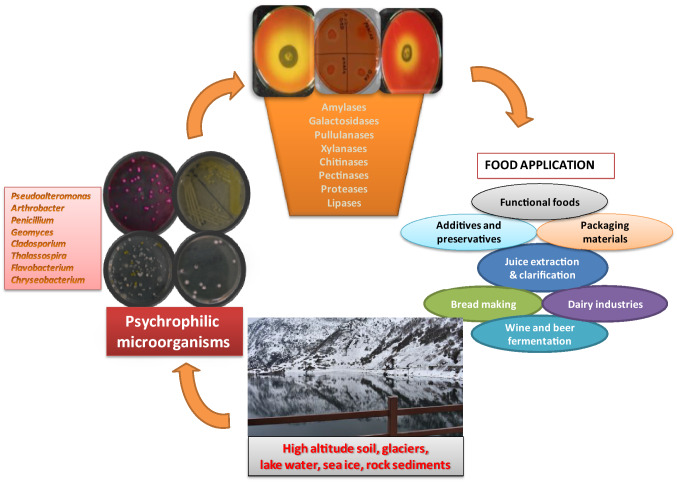
Despite the tremendous importance of cold-active enzymes, finding novel enzymes for their commercial applications in the food industry is a growing challenge (Kuddus 2018). On the other hand, less availability and low stability of these enzymes are significant signs. Furthermore, biotechnologists are more concerned about the specific catalytic activity of these cold-active enzymes, which need to be explored at the industrial level (Kuddus 2018; Hamid and Mohiddin 2018). Therefore, the development and invention of new technologies are needed to boost the quality and production of cold-active enzymes. Moreover, with the advent of approaches, such as protein engineering, rDNA technology and metagenomics, these enzymes can be devised to be used competently for diverse food applications (Thakur et al. 2021a). This review provides insights into applying cold-active enzymes in food processing, emphasizing the recombinant clones and their industrial significance (Fig. 1).
Fig. 1.
Schematic representation showing application of cold active enzymes produced by psychrophilic microorganisms in food processing industry
Psychrophilic microbes as producers of cold-active enzymes
Psychrophiles represent an extensive range of microbial taxa such as bacteria, archaea, algae and yeasts that can thrive in permanently cold habitats, including glaciers, high mountains, ocean depths, shallow subterranean and polar regions. They experience severe physicochemical constraints, including decreased biochemical reaction rates, reduced membrane fluidity, altered transport systems and cold denaturation of proteins (D’Amico et al. 2006; Siddiqui and Cavicchioli 2006). Adaptive features to overcome these constraints have frequently been detected in the genomes of several psychrophiles sequenced so far (Saunders et al. 2003; Riley et al. 2008; Ayala-Del-Rio et al. 2010). Moreover, the production of cold-active enzymes that drive metabolism and the cell cycle in these microorganisms has been considered a characteristic adaptation. The enzymes offer high specific activity even at low temperatures, compensating for the rapid decrease in chemical reaction rates. This adaptive feature is thought to be genetically encoded in the amino acid sequence and reportedly resulted from a long term selection process (Feller and Gerday 2003; Feller 2013).
Psychrophilic microorganisms contain many critical proteins and other metabolites as determinative factors in their adaptation to cold environments. They evolve mechanisms to maintain maximal translation and protein folding under cold conditions (Siddiqui and Cavicchioli 2006; Feller 2013). The protein components liable for the maintenance of RNA for carrying out the translation process, specifically the ribosomal proteins and RNA helicases, appeared to be overexpressed in many cold-adapted microorganisms (Lim et al. 2000; Jung et al. 2010). Furthermore, diminution in the number and strength of non-covalent interactions outside the catalytic cavity offers motional and dynamic strength to the active site amino acids upholding catalytic efficiency of the enzymes in cold temperatures (Feller 2013). More importantly, the chaperones including DnaK, GroEL, ribosome-bound trigger factor (TF) and RNA chaperones, such as cold-shock protein A, which have reportedly played a crucial role in the folding of newly synthesized polypeptides, contribute significantly to the adaptive feature of psychrophilic microorganisms and conformational flexibility of the cold-active enzymes (Strocchi et al. 2006; Sung et al. 2011; Piette et al. 2011). Numerous microorganisms, including bacteria (species of Arthrobacter, Pseudolteromonas, Paracoccus, Pseudomonas, etc.) and fungi (species of Geomyces, Candida, Penicillium, Cladosporium, etc.) residing in the Antarctic and Arctic habitats, and high elevation regions have been investigated for the production of cold-active enzymes, which demonstrate the remarkable potential to be used in various industries (Santiago et al. 2016; Hamid and Mohiddin 2018; Bruno et al. 2019).
Cold-active enzymes in the food industry
Cold-active enzymes inherit flexible structures that presumably recompense for the low kinetic energy present in the cold habitats. Because of this reason, these enzymes often demonstrate a reduction in the activation enthalpy (ΔH) while showing more negative activation entropy (ΔS) as compared to their thermophilic and mesophilic counterparts. That is why the reaction rate of cold-active enzymes tends to decrease very slowly concerning a decrease in temperature around the surrounding. This balancing act of activation parameters is transformed to the high catalytic activity of these enzymes at low temperatures (Siddiqui and Cavicchioli 2006; Cavicchioli et al. 2011). Therefore, the biotechnological potential of cold-active enzymes reflects several factors, including their high activity in low to moderate temperatures, increasing thermolability at high temperatures, and the ability of these enzymes to carry out the reaction in organic solvents (Marx et al. 2007; Margesin and Feller 2010). Cold-active enzymes could also be able to catalyze reactions at temperatures that play down many undesirable and competitive chemical reactions. These low-temperature active enzymes are of particular interest for transforming heat-sensitive substrates (Jeon et al. 2009). Because of these unique features, cold-active enzymes are considered as most germane to the food and feed industry. Importance is being given to evade spoilage, change in flavour and nutritional parameters of the native thermolabile substrates and products. Cold-active enzymes are widely used in food applications, such as meat tenderization, baking, brewing, flavouring, cheese production, and animal feed processing (Tables 1 and 2).
Table 1.
Cold-active enzymes produced by psychrophilic microorganisms and their potential applications in food industry
| Cold-active enzymes | Sample source | Microorganisms | Potential/recommended application | References |
|---|---|---|---|---|
| Xylanase | Soil sample (Antarctica) | Pseudoalteromonas haloplanktis | Xylo-oligosaccharides production | Collins et al. (2002) |
| β-Galactosidase | Sea sediment (Antarctica) | Guehomyces pullulans 17–1 | Dairy industry for production of lactose free products | Song et al. (2010) |
| Soil (Antarctica) | Rahnellainusitata | -Do- | Núñez-Montero et al. (2021) | |
| Chitinase | Soil, seal and penguin feces, and marine sediment (Antarctica) | Pseudomonas sp. | Biocontrol of microbial spoilage of cold-stored foods and against vegetable spoilage pathogenic fungi | Liu et al. (2019) |
| Protease | Deep sea sediment (China) | Pseudoaltermonas sp. SM9913 | Taste enhancement of cold-stored meat | He et al. (2004) |
| Sausage | Penicillium nalgiovense PNA9 | Meat ripening | Papagianni and Sergelidis (2014) | |
| NS | Arsukibacteriumikkense | Production of bioactive dairy products and other functional foods | De Gobba et al. (2014) | |
| East Rathong Glacier, Sikkim | Chryseobacterium polytrichastri | Production of antioxidant peptides | Mukhia et al. (2021) | |
| Sea ice (Antarctica) | Pseudoalteromonas sp. NJ276 | As additives in baking industry, food processing and preservation industries | Wang et al. (2008) | |
| Refrigerator of a meat factory (China) | Serratia sp. WJ39 | Food processing industry | Ji et al. (2014) | |
| Phytase | Deep sea sediment (Antarctica) | Rhodotorula mucilaginosa JMUY14 | Feed industry especially aquaculture feed processing | Yu et al. (2015) |
| Antarctic sample | Cryptococcus laurentii AL27 | Feed industry | Pavlova et al. (2008) | |
| Pectinase | Marine sponge (Antarctica) | Geomyces sp. F09-T3-2 | Production of white wine | Poveda et al. (2018) |
| Fruit orchard soil and spoiled refrigerated fruits and vegetables (Himalaya) | Saccharomyces sp. | Fruit juice clarification | Naga Padma et al. (2011) | |
| Grapes (Argentina) | Bacillus sp. CH15 | Red wine making | Martín and Morata de Ambrosini (2013) | |
| Grape wine and wineries (Argentina) | Aureobasidium pullulans strains | Cold-wine making | Merín and de Ambrosini (2015) |
NS not specified
Table 2.
Cold-active recombinant microbial enzymes and their potential applications in food industry
| Cold-active enzymes | Sample source | Source microorganisms | Cloning/expression host | Expression vector | Potential/recommended applications | References |
|---|---|---|---|---|---|---|
| Xylanase | Sea water (China) | Zunongwangia profunda | Escherichia coli BL21, E. coli DH5α | pGEX-6p-1 | Food industry for processing of sea and saline foods | Liu et al. (2014) |
| NS | Saccharophagusdegradans 2–40 | E. coli DH5α, E. coli BL21 | pET21a | Baking industry | Ko et al. (2016) | |
| NS | Pseudoalteromons haloplanktis TAH3A, Flavobacterium sp. MSY-2 | E. coli BL21 | pET22b-XFH-His6, pET22b-Xyn8-His6 | Baking industry | Dornez et al. (2011) | |
| NS | Glaciecola mesophile | E. coli BL21 | pET22b | Baking industry | Zheng et al. (2011) | |
| NS | Flavobacterium johnsoniae | E. coli JM109, E. coli DH5α, E. coli BL21 | pSCH602, pSCH643 | Food and feed industry | Chen et al. (2013) | |
| β-Galactosidase | Calcium carbonate (Ikka columns, South-West Greenland) | Alkalilactibacillusikkense | E. coli TOP10 | pUC18dLacZ | -Do- | Schmidt and Stougaard (2010) |
| Soil (Antarctica) | Arthrobacter sp. 20B | E. coli TOP10F’ | NS | -Do- | Białkowska et al. (2009) | |
| Soil (Antarctica) | Paracoccus sp. 32d | E. coli LMG 194 | pBAD/Myc-His A | -Do- | Wierzbicka-Woś et al. (2011) | |
| Deep sea water (Mariana Trench) | Alteromonas sp. ML52 | E. coli BL21 | pET-gal | -Do- | Sun et al. (2018) | |
| Alimentary tract (krill Thyssanoessamacrura) (Southern Shetlands) | Pseudoalteromonas sp. 22b | E. coli ER2566 | pETbeta22b | -Do- | Cieśliński et al. (2005) | |
| Frozen soil (China) | Rahnella sp. R3 | E. coli BL21 | pCold I | -Do- | Fan et al. (2015) | |
| α-amylase | Surface sea water (China) | Zunongwangia profunda (MCCC 1A01486) | E. coli DH5α, E. coli BL21 | pGEX-6P-1 | Baking industry | Qin et al. (2014) |
| Deep sea sediment (China) | Bacillus sp. dsh19‑1 | E. coli JM109, E. coli BL21 | pColdI | Starch hydrolysis | Dou et al. (2018) | |
| Sea ice (Antarctica) | Pseudoalteromonas sp. M175 | E. coli BL21 | pET-amy175 | Food industry | Wang et al. (2018a) | |
| Soil (Tehran) | Exiguobacterium sp. SH3 | E. coli DH5α, E. coli BL21 | pEamy | Food industry | Mojallali et al. (2014), Emampour et al. (2015) | |
| Soil (King George Island, Antarctica) | Geomycespannorum | E. coli DH5α, Aspergillus oryzae | pBC-hygro | Food industry | Krishnan et al. (2011), Mao et al. (2015) | |
| NS | Bifidobacterium longum | E. coli MC1061 | NS | In food processing industry for production of slowly digestible starch | Lee et al. (2016) | |
| β-Glucosidase | Konjac field (South Korea) | Paenibacillusxylanilyticus KJ-03 | E. coli JM109, E. coli BL21 | pColdI | Food processing industry | Park et al. (2013) |
| Pullulanase | Soil (Tehran) | Exiguobacterium sp. SH3 | E. coli BL21 | pET-26b (+) | Cold hydrolysis of starch and in sugar syrup production | Mojallali et al. (2014), Rajaei et al. (2015) |
| Soil (China) | PaenibacilluspolymyxaNws-pp2 | E. coli DH5α, E. coli BL21 | pET-28a, pET-32a and pET-42a | Food processing industry | Wei et al. (2015) | |
| Hot-spring (Sikkim, India) | Hot-spring metagenome | E. coli BL21 | pET28a(+) | Production of resistant starch | Thakur et al. (2021b) | |
| Waste water (Europe) | Bacillus methanolicus PB1 | E. coli DH5α, E. coli BL21 | pET-28a | Cold hydrolysis of starch | Heggeset et al. (2012), Zhang et al. (2020) | |
| Chitinase | Sea sediment (China) | Pseudoalteromonas sp. DL-6 | E. coli BL21 | pET28a | Biocontrol of microbial spoilage of cold-stored foods | Wang et al. (2014a; b) |
| Sea ice (Antarctica) | Glaciozyma antarctica PI12 | E. coli JM109, Pichia pastoris | pPICZα A | Biocontrol of microbial spoilage of cold-stored foods | Ramli et al. (2013) | |
| Lipase/esterase | Deep sea water (Indian ocean) | Aeromicrobium sp. SCSIO 25071 | E. coli DH5α, E. coli Rosetta (DE3) | pET28a | As food additives | Su et al. (2016) |
| Marine sediment (Arctic) | Halocynthiibacter arcticus | E. coli BL21 | pET-21a | Production of flavored compounds | Le et al. (2020) | |
| NS | Streptomyces coelicolor A3(2) | E. coli BL21 (DE3) | pET16b | Production of flavored compounds | Brault et al. (2012) | |
| NS | Malassezia globose | E. coli Top10, P. pastoris X-33 | pGAPZαA | Oil modification and food processing industry | Xu et al. (2015) | |
| Car service contaminated soil (Malaysia) | Staphylococcus epidermidis AT2 | E. coli Tuner (DE3)pLacI | pTrcHis2-TOPO | Food processing industry | Kamarudin et al. (2014) | |
| Cold seafloor dwelling Atlantic hagfish stomach (North Norway) | Rhodococcus sp. AW25M09 | E. coli DH5-α, E. coli BL21 (DE3) | pET-22b | Food industry for preserving food flavor and integrity | De Santi et al. (2014) | |
| NS | Neisseria meningitides | E. coli | pQE-30 | Production of organic esters for use in food industry | Yoo et al. (2019) | |
| NS | Lactobacillus acidophilus NCFM | E. coli DH5-α, E. coli BL21 (DE3) | pET-21a | Dairy and food processing industry | Wang et al. (2018b) | |
| NS | Bacillus licheniformis ATCC 14580 | E. coli BL21 | pGEMT-Easy Vector | Food processing industry | Borgi et al. (2014) | |
| Pectinase | Soil (Sub-Antarctic region) | Tetracladium sp. | E. coli, P. pastoris | pPinkα-HC | Production of fruit juice and fermented beverages | Carrasco et al. (2019) |
| Mannanase | Soil (China) | Bacillus subtilis Bs5 | E. coli JM109, E. coli Rosetta-gami (DE3) | pET-32a | Food and feed industry | Huang et al. (2012) |
NS not specified
According to the International Union of Biochemistry and Molecular Biology (IUBMB), enzymes are classified into six major categories based on the type of reactions they catalyze: oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases. Though these enzymatic reactions are vital in food production, food biotechnologists are mainly concerned with oxidoreductases and hydrolases (Chourasia et al. 2020; Raveendran et al. 2018; Bruno et al. 2019). Hydrolases are given preference, considering their significant roles in cheese production, malting and brewing, conversion of starch to glucose and fructose, and human metabolism (Fernandes 2010; Adrio and Demain 2014; Sarmiento et al. 2015). The anticipated food applications of cold-active hydrolytic enzymes are discussed in the subsequence sections.
Hydrolases
Hydrolases constitute a very composite collection of enzymes that catalyze bond cleavages upon reaction with water. The innate role of most hydrolases is digestion that is breaking down nutrients into smaller units suitable for digestion, and because of this, they usually have a broad range of substrate specificity. Examples include proteases (hydrolyze proteins to oligopeptides and amino acids), lipases (hydrolyze triglycerides to glycerol and fatty acids), phosphatases, pectinases, etc.
β-Galactosidases
β-Galactosidases offer an obvious example of glycosidases, with the advantage of being active at low temperatures and cost-effective (Bruno et al. 2019). β-Galactosidases are used to remove lactose from milk and dairy products, where the enzyme hydrolyzes lactose into glucose and galactose. With more than half of the world's population suffering from lactose intolerance and also because of the difficulties in crystallizing lactose, β-galactosidases have drawn the growing attention of food biotechnologists (Dalmaso et al. 2015). Cold-active β-galactosidases (acting at acidic pH) can improve the technical effectiveness of whey by producing syrups rich in glucose and galactose that can be utilized in a variety of food products (Gerday et al. 2000). These enzymes also possess transglycosylation activities to catalyze the hydrolysis of lactose with the instantaneous transfer of the monosaccharides to higher oligosaccharides forming tri and tetrasaccharides (Karasova-Lipovova et al. 2003; Benesova et al. 2005). Such galacto-oligosaccharides have potential uses as prebiotic additives that enhance the growth of Bifidobacteria in the large intestine. Several studies have reported the isolation of cold-active β-galactosidases from psychrophilic bacteria, and many are from the Antarctic and Arctic regions. Karan et al. (2013) and Laye et al. (2017) described the cold-active β-galactosidases from Halorubrum lacusprofundi, a bacterial psychrophile isolated from the hypersaline Deep Lake of Antarctica. Several β-galactosidases, isolated from species of Paracoccus, Arthrobacter and Pseudoalteromonas (P. haloplanktis TAE 79 and LMGP-19143) of the Antarctic region, are functional at low temperatures and have potential application in the milk and dairy industry (Trimbur et al. 1994; Hoyoux et al. 2001; Turkiewicz et al. 2003; Cieslinski et al. 2005; Makowski et al. 2007; Hildebrandt et al. 2009). Cold-active β-galactosidases showing considerable lactose hydrolyzing efficiency were isolated from Enterobacter ludwigii and Alkalilactibacillus ikkense inhabiting the Arctic region (Schmidt and Stougaard 2010; Alikkunju et al. 2016). Besides, filamentous psychrophilic fungi colonizing the Antarctic environments hold the similar potential of producing β-galactosidases as those of bacterial psychrophiles. For instance, β-galactosidases isolated from Tausonia pullulans and Cryptococcus albidus colonizing Antarctic sea sediments reportedly hydrolyze lactose (Koleva et al. 2006; Song et al. 2010; Zhang et al. 2012).
Xylanases
Xylanases that catalyze the endohydrolysis of 1,4-β-d-xylosidic linkages in xylan grasp the enormous potential for extensive use in biomass processing and the food industry (Joshi et al. 2020; Phukon et al. 2020a). They have an essential role in bread-making, since they transform insoluble hemicellulose in the dough to soluble sugars, giving in elastic and soft bread. Few cold-active enzymes, purified from psychrophilic bacteria inhabiting Antarctic habitats, such as Pseudoalteromonas haloplanktis and Flavobacterium sp., have been observed to improve the dough properties along with the bread volume (Collins et al. 2002; Dornez et al. 2011). Few shreds of evidence of filamentous fungi (e.g., Geomyces pannorum and Cladosporium sp.), colonizing extreme habitats such as Polar Regions, producing xylanases have also been described (Del-Cid et al. 2014). The xylanolytic activity of the fungi was reportedly intact at low temperatures with very low stability; however, detailed characterization of xylanases and their physicochemical properties from fungi are truly limited. Therefore, further studies on the psychrophilic xylanases from microorganisms of diverse cold habitats should be driven forward, and efforts must be assumed to facilitate them as model candidates for different food processing applications.
Amylases
Amylases are another kind of glycoside hydrolases that catalyze starch hydrolysis by acting upon α-1,4-glycosidic bonds, a covalent bond joining two α-d-glucose together. They hydrolyze starch to form malotriose, maltose, glucose monomers, and limit dextrins. Amylases are further classified based on the specificity of the reactions catalyzed by them, and are exoamylases (e.g., β-amylases, glucoamylases and α-glucosidases), endoamylases (e.g., α-amylases), debranching amylases (e.g., pullulanases, isoamylases, and dextrinases), and transferases (e.g., 4-α-glucanotransferases, and cyclodextrin glycosyltransferases) (van der Maarel et al. 2002). Amylases have tremendous potential for use in food applications, including wine and beer fermentation and bread and fruit juices. Several cold-active amylases have been purified from psychrophilic microorganisms (Srimathi et al. 2007; Roohi et al. 2013; Ramli et al. 2013; Qin et al. 2014; Rajaei et al. 2015). The most studied cold-active amylases are α-amylases, but only a few have been proposed for possible application in the food industry, including a novel α-amylase isolated from a psychrophilic fungus, Geomyces panorum, that is reported to have latent application for the baking industry (He et al. 2017).
Chitinases
Chitinases can break down chitin by hydrolyzing N-acetyl-β-d-glucosaminide (1→4)-β-linkages randomly into oligo- and monomeric components. Chitinases have been influential in various biotechnological sectors because of their role in the bioconversion of chitinous biomass into several value-added products. Although poorly studied, cold-active chitinases and their high activity at fairly low temperatures can be used in the production of chitooligosaccharides (COS), the oligomers of N-acetyl-d-glucosamine (GlcNAc) and d-glucosamine (GlcN) linked by β-1,4-O-glycoside bond released as a result of chitin degradation (Le and Yang 2019; Rajabi 2019). COS are low weight water-soluble substances having interesting bioactivities and can easily be absorbed by the small intestine. COS can find application in the food industry as a food additive and packaging materials as its antimicrobial and antioxidant properties enable protection from deterioration (Liu et al. 2019; Rajabi 2019). Cold-active chitinases can be used to produce single-cell proteins for use as cheaper and alternative protein dietary supplements to soymeal and fish meal (Patil and Jadhav 2014; Wadhwa and Bakshi 2016). In addition, cold-active chitinases offer antifungal activity, which could be helpful in bio-control of post-harvest fungal pathogens and, therefore, demonstrating the considerable potential for the food industry in safe storage of vegetables and fruits (Castillo et al. 2016; Le and Yang 2019). Reports on cold-active chitinase from psychrophilic microorganisms are not many except a cell-bound cold-active chitobiase, exochitinase isolated from psychrophilic Arthrobacter sp. TAD20 (Lohienne et al. 2001). Despite the tremendous significance, the use of cold-active chitinases has not witnessed any considerable progress in food industries, which could be because of their less availability and specificity. We must prioritize their isolation and exploration from varied sources, including the psychrophilic microorganisms. Furthermore, they can be tailored and expressed in selected hosts to improve their stability and yield towards enhanced applicability in food biotechnology.
Other hydrolases
Pullulanases are critical glycosidase enzymes in starch processing and are primarily used in making maltose and glucose syrups (Thakur et al. 2021a). Pullulanases catalyze the hydrolysis of α-1,6-and α-1,4-linkages in starch, pullulan, amylopectin and other related oligosaccharides (Hii et al. 2012). Many cold-active pullulanases have been identified and characterized and shown their efficiency for other biotechnology applications, such as starch degradation for bio-ethanol production and transformation of biomass-derived starch into resistant starch (Elleuche et al. 2014; Thakur et al. 2021a, b).
Other glycosidases that represent a prospective candidate for the food industry are cellulases. They catalyze total hydrolysis of cellulose into sugars and are used in coffee processing and making wine (Kumar et al. 2018; Jayasekara and Ratnayake 2019). Cellulases, pectinases, and hemicellulase are also used to extract fruit juices and minimize food spoilage (Kuhad et al. 2011). Three cellulolytic activities have been cited from cellulases of natural origin: endoglucanase, β-glucosidase, and β-1,4-cellobiohydrolase. Over the decades, several cold-active cellulases sourced from psychrophilic microorganisms have been purified and characterized (Benesova et al. 2005; Shipkowski and Brenchley 2005; Zeng et al. 2006; Fu et al. 2010); however, their potential applicability in the food industry is scarcely known. Conversely, bioprospecting novel enzymes of these kinds from diverse cold-adapted microbial resources could address the present limitations of cost and energy associated with the use of thermophilic and mesophilic enzymes in various industrial processes.
Proteases
Proteases or peptidases are an interesting class of hydrolyzing enzymes that catalyze the hydrolysis of large proteins into minor peptides. Basing upon their ability to hydrolyze N- or C-terminal peptide bonds and internal peptide bonds, proteases are classified, respectively, into exopeptidases and endopeptidases (Chourasia et al. 2021). Exopeptidases cleaving N-terminal peptide bonds are called aminopeptidases, whereas those hydrolyze C terminal peptide linkage are called carboxypeptidases. Cold-active proteases hold enormous potential due to their several unique features, including optimal activity at low temperature, enabling an easy transformation of thermolabile products (Kuddus and Ramteke 2012; Kuddus 2018). In addition, they have specific applications in processes, such as the fermentation of fish and soya sauce, contributing nil change in nutritional value and flavour. They can also be considered alternatives to rennet, speeding up the ripening process of slow ripening cheeses when used with lipase. Cold-active proteases can also be profitable in taste development and softening of frozen meat products (Joshi and Satyanarayana 2013). Several proteases have been recovered from psychrophilic microorganisms, including bacteria, fungi and algae (Peterson et al. 2013; Vaca et al. 2013; Joshi and Satyanarayana 2013). A cold-active protease isolated from psychrophilic Pseudoalteromonas sp. has been reported to secrete essential amino acids that selectively enhance the taste of frozen meat (He et al. 2004). The psychrophilic serine protease belonging to Chryseobacterium sp. has been proposed for its applicability in improving meat quality because of its tolerance to salt and optimal activity at low temperature (Mageswari et al. 2017).
Cold-active protease obtained from Pseudoalteromonas sp. produced free amino acids from milk protein at 4 °C, implicating its significance in low-temperature food processing (Wang et al. 2008). A metalloprotease isolated from psychrophilic Enterococcus faecalis displayed no side effects while administered orally and can be used in functional foods, increasing their stability and solubility (Yuan et al. 2009). Similarly, cold-adapted proteases from Arsukibacterium ikkense, releasing bioactive peptides from milk caseins, have been accounted for indicating the practical suitability of these enzymes in dairy industry (De Gobba et al. 2014). Besides psychrophilic bacteria, fungi colonizing the extreme habitats have been known to produce proteases active at low to moderate temperatures (Duarte et al. 2018; Lario et al. 2015). One such protease (aspartic protease) purified from psychrotrophic yeast Sporobolomyces roseus was proposed as a potent biocatalyst in the production of soy-sauce and cheese, meat tenderization, and also as bread additive (Bialkowska et al. 2018). In a recent study, cold-active protease from Chryseobacterium polytrichastri isolated from East Rathong Glacier has been reported to produce soybean derived bioactive peptides (Mukhia et al. 2021).
In the present biotechnological era, cold-active proteases can be applied in exciting food bioprocess industries if they can be economically produced at a large scale. There is a need of exploring novel cold-active proteases that can find applications in cheese ripening, recovering heat-sensitive nutrients, such as lipids rich in polyunsaturated fatty acids (Rai et al. 2012, 2017; Hathwar et al. 2011). Cold-active proteases can be applied in the recovery of protein hydrolysates from food processing byproducts that are rich in heat-sensitive lipids and antioxidants (Rai et al. 2013). Furthermore, they can be explored for softening of meat products and winemaking against haze-producing proteins. Due to their tremendous potential in the food industry, novel cold-active enzymes producing isolates from unexplored niches and genomic resources need to be studied. Extensive research is needed on factors affecting the enhanced the production of these cold-active proteases and molecular mechanisms for improvement of the activity to meet their demand at the industrial level.
Esterases and lipases
Esterases, also referred to as carboxyl ester hydrolases, are drawing vast attention for biotechnological applications. They catalyze synthesis and hydrolysis of ester bonds and are classified into lipases (act on lipids) and non-lipolytic esterases (act on water-soluble ester substrates) (Thierry et al. 2017). The curiosity on cold-active lipases is associated with their inherent physiological and structural adaptive mechanisms (Phukon et al. 2020b). Cold-active lipases are becoming a fundamental part of the modern food industry, where they can be used in protein polymerization and gelling in fish, clearing of drains clogged by lipids in food processing, upgrading food texture and modifying flavor, and also in the production of fatty acids and interesterification of fats (Joseph et al. 2008). A cold-active lipase from psychrophilic Pseudomonas fluorescence has been used for the synthesis of flavoring compound butyl caprylate (Tan et al. 1996). Similarly, cold-active lipases from fungal psychrophiles, such as Candida antarctica, C. cylindracea, Hansinuela lanuginose, and Geotrichum candidum, have been applied for the esterification of functionalized phenols giving inlipophilic antioxidants for use in sunflower oil (Buisman et al. 1998; Pandey et al. 1999). Esterases that catalyze simple esters with short-chain fatty acids (e.g., triglycerides) are attracting candidates for use in the cheese ripening process. Several enzymes with cold-adapted esterase activity have been identified from psychrophilic bacteria, such as Pseudoalteromonas spp., and Thalassospira sp., Oleispira sp. (D’Auria et al. 2009; Al-Khudary et al. 2010; Lemak et al. 2012). As a whole, cold-active lipases and esterases offer various advantages as an alternate to the usual biochemical processes in the food industry. However, several challenges have to be surmounted in prioritizing their enhanced utility in diverse biotechnology applications, including the food biotechnology sector, which is discussed further in this review.
Phytases
Phytases are gaining importance in the food and feed industries as they catalyse phosphate removal from phytate, which is considered antinutrient as they bind to divalent minerals (Pable et al. 2019). For decades, they have increasingly been used to enrich absorbable phosphate groups in animal foods, particularly for non-ruminant livestock and fish (Kumar et al. 2012). Numerous studies have reported the isolation and identification of cold-active phytases of psychrophilic microbial origin (Yu et al. 2015; Park and Cho 2011), which can be used in the food manufacturing and processing industry as they mediate phytate degradation increasing the bioavailability of minerals. Because of this property, phytases may also find an application in functional food production (Hamid et al. 2014). Nevertheless, little is known regarding cold-active phytases real-time application within the food industry.
Pectinases
Pectinases or pectinolytic enzymes are an assorted group of enzymes that collectively break down pectin, a significant polysaccharide present in the plant cell wall (Patidar et al. 2018). Pectinolytic enzymes are classified based on the cleavage site (polygalacturonases, lyase/trans-eliminases including pectinlyase, pectate lyase, and pectin esterases). On the pH range, they may be categorized into alkaline and acid active enzymes. They are an integral part of the food industry and contribute more than 40% of the total share of food enzymes (Adapa et al. 2014). Most pectinases used in the food industry are originated from thermophilic or mesophilic microorganisms. Recently, efforts have been growing to modify the thermophilic enzymes for developing cold-active counterparts because of certain unique features, including their high catalytic activity at cold temperature (˂ 25 °C) and easy inactivation upon mild heat treatment (Truong et al. 2001; Pulicherla et al. 2011). Cold catalysis offers less energy input in the enzymatic reaction. Cold-active pectinases can be used in efficient fruit juice extraction, settling the suspended particles in the fermented mash, winemaking, secreting polymeric colour pigments, releasing terphenols for aroma and purification of coffee and tea (Blanco et al. 2009; Jayani et al. 2005; Margesin et al. 2007). Few cold-active pectinases have been identified from the psychrophilic microorganisms (Brigisson et al. 2003); however, experimental evidence supporting the use of native pectinases within the food industry is scarce, except some heterologously expressed pectinolytic enzymes that have excellent potential for juice making and winemaking industries (Pan et al. 2014). Cold-active pectinases can be applied in juice clarification at a lower temperature, useful in the prevention of heat-sensitive phytonutrients. Cold-active pectinases have excellent potential in the food industry, and microorganisms from cold regions should be screened for secretion of pectinases, functional at cold temperature and acidic pH.
Other enzymes
Besides the major class of cold-active enzymes increasingly gaining the attention of the food industry, few other enzymes have little been explored; however, they could hold remarkable potential for use in the food industry. Cold-active tannases have been identified in many psychrophilic bacteria and yeasts. They can be used in several ways as their thermophilic counterparts, including the manufacturing of instant tea, fruit juices, beer, and wines (Kasieczka-Burnecka et al. 2007; Yao et al. 2014). β-Mannanases, possessing optimal activity at low temperature, have been reported from some microbial psychrophiles and can be applied to produce manno-oligosaccharides (Nguyen et al. 2019; Dawood and Ma 2020). β-Mannanases can find application as food and feed additives due to their health improving properties. Psychrophilic microbes are also reported as producers of invertases, an important enzyme used in sucrose hydrolysis that yields an equimolar mixture of glucose and fructose. Cold-active invertases can be used for making confectionery, syrup, infant milk, condensed milk, and beverages (Turkiewicz et al. 2005; Madhusudhan and Raghavarao 2011). There are opportunities for exploring cold-active enzymes from new niches for application in the food processing industries.
Cloning, expression and protein engineering of cold-active enzymes
The elegant specificity of enzymes to catalyze varied sets of reactions makes them essential for biochemical transformation beneficial to humanity. In the beginning, most of the commercial enzymes having industrial applications were produced using native microorganisms. This has restricted the application of enzymes from the native cultivable microorganisms, which are generally produced in low yield (Sarmiento et al. 2015; Santiago et al. 2016). Moreover, the natural enzymes display several complexities at the structural and functional level, limiting their prospective in many biotechnological applications (Huston 2008). Therefore, recombinant expression of these enzymes in heterologous hosts has been preferred as a conventional approach to obtain a high yield for desired enzymes. This has also improved the catalytic efficiency, stereo-selectivity and enzyme stability (Santiago et al. 2016; Duarte et al. 2018). Mesophilic hosts are generally favoured for such recombinant strategies; however, the folding temperature of cold-active enzymes required for their structural and functional integrity may not be compatible with the expressed host (Bjerga et al. 2016; Longwell et al. 2017). To overcome this challenge, incubation temperature is lowered after induction of the desired gene in the host (Feller et al. 1998; Santiago et al. 2016). Besides, the expression of such enzymes from eukaryotic microorganisms such as fungi in bacterial hosts can be cumbersome and substandard, which may be because they lack the ability to secrete extracellular proteins (Duarte et al. 2018). In addition, the post-translational modification mechanism is different from those of prokaryotes, and therefore, eukaryotic microbial hosts must be chosen for heterologous expression of eukaryotic cold-active enzymes (Duarte et al. 2018).
Numerous cold-active enzymes identified from psychrophiles have been successfully expressed in heterologous hosts and hold enormous potential for use in the food industry (Wierzbicka-Wos et al. 2011; Pan et al. 2014). Escherichia coli represents an ideal host for the cold-active enzymes sourced from prokaryotes (Krishna 2002). Quite a few β-galactosidases from cold adaptive bacteria such as Arthrobacter sp. and Paracoccus sp. were cloned and expressed in E. coli, making them exceptional candidates for industrial removal of lactose (Białkowska et al. 2009; Wierzbicka-Wos et al. 2011; Pawlak-Szukalska et al. 2014). Similarly, the gene coding for a cold-active and acidic pectin methyl esterase, isolated from an Antarctic fungus Penicillium chrysogenum PE8F46, was expressed in Pichia pastoris. This enzyme was found to improve the firmness of the pineapple dices, representing its significance in the fruit and vegetable industry (Pan et al. 2014). The DNA segment encoding a novel α‐amylase, identified from the Antarctic psychrotolerant fungi Geomyces pannorum, was overexpressed in P. pastoris (Gao et al. 2016). It resulted in high glucose yielding ability, which can be a potential application in the syrup industry. In the same way, a cold-active aspartic protease from G. pannorum was cloned and expressed in Aspergillus oryzae displaying itself a promising biocatalyst for cheese making (Gao et al. 2018).
Signs of progress in recombinant technology have further revolutionized the calibration, development and cost-effective production of customized enzymes that possess some industrial relevance. Cold-active enzymes isolated from psychrophilic hosts can be tailored to convene the process specifications by introducing mutations in the protein in a controlled manner (Bornscheuer et al. 2012). Enzyme engineering can be achieved either by site-directed mutagenesis or by directed evolution strategy. Site-directed mutagenesis is one of the earliest and widely used enzyme engineering techniques based on known structural features of the desired characteristic, such as protein sequence or crystal structures (Coker and Brenchley 2006; Wang et al. 2014a, b). A combined directed and random mutagenesis approach to alter the cold-active β-galactosidase, identified from an Antarctic Arthrobacter sp., has provided interesting mutations leading to increased lactose hydrolysis at low temperature, suggesting its potential use in the dairy industry (Coker and Brenchley 2006). A cold-active endo-1,5-α-L-arabinanase (pectinase) from Paenibacillus polymyxa was engineered using site-directed mutagenesis, which shifted its optimal activity pH towards acidic conditions, making it a promising candidate for pectin extraction from vegetables and fruits, and juice clarification (Wang et al. 2014a, b).
The most successful strategy in engineering novel cold-active enzymes is the directed evolution method that involves random mutagenesis of a gene translating the enzyme of interest. This is chiefly carried out using polymerase chain reaction (PCR) based methods, followed by screening or selection of the protein variants showing desired features from the resulting library of mutants. Saturation mutagenesis, cassette mutagenesis, error-prone PCR, random-priming recombination, DNA shuffling, and staggered extension process (StEP recombination) are some of the random mutagenesis techniques that have been used so far (Arnold 2001). Alteration in the enzyme properties mainly requires multiple amino acid substitutions simultaneously, producing several protein variants for screening. In addition, present-day high throughput screening methods, including fluorescence-activated cell sorting (Bernath et al. 2004; Becker et al. 2008; Fernandez-Alvaro et al. 2011), permit high-throughput screening and selection of a large number of variants within a short time. Besides, various statistical approaches and bioinformatic methods (e.g., protein structure–activity relationship algorithms) are being used for creating multiple mutations, and at the same time, identifying whether a particular mutation is beneficial or not (Fox et al. 2007). Many cold-active enzymes have been engineered using directed evolution methods, making them relevant for numerous industrial applications (Zhang et al. 2003; Gatti-Lafranconi et al. 2008). Hopefully, these relevant cold-active enzyme variants in the coming days are going to witness a steady growth leading to the manufacturing of value-added nutraceuticals of food and pharmaceutical significance.
Commercial cold-active enzymes used in the food industry
There is a growing demand for processed food products and beverages across the globe, with respect to nutritional excellence as well as favorable taste. This might be because of consumer fondness that shifted their preferences towards healthy food products of high nutritional quality, ultimately promoting the demand for food enzymes. The global market size for food enzymes was estimated at $1944.8 million in the year 2018 and is predicted to achieve $3056.9 million by 2025, with an anticipated compound annual growth rate (CAGR) of ~ 5.5% within the period 2019–2029 (https://www.persistencemarketresearch.com/market-research/food-enzymes-market.asp). North American countries such as United States of America, Canada and Mexico are leading consumers of food enzymes. Moreover, Asia–Pacific region is perched to grow with a substantial market share in the food enzyme market globally (https://www.alliedmarketresearch.com/food-enzyme-market). Commercial biocatalysts have several applications in the food industry as they are applied in cheese production, in bread making, maintaining color and clarity of the wine, and reducing its sulfur content (Aehle 2007; Dewan 2014; Sarmiento et al. 2015). The application of enzymes during food processing increases the nutritional quality, flavour, appearance, and taste (Fernandes and Carvalho 2016; Raveendran et al. 2018). Some enzymes (e.g., lactases, α-amylases, proteases, and lipases) are used as processing aids that act on food components without affecting the nutritional and organoleptic properties of the food. Nevertheless, narrow temperature ranges, pH instability, and side effects, such as allergies associated with a few enzymes, limit food enzymes' market growth.
Cold-active enzymes in food industries are in their infancy and are yet to witness significant growth. The introduction of psychrophilic enzymes in the food processes can be considered as a significant driver that will escalate the growth reconnoitre of food enzymes as they possess unique economic and environmental benefits (Pulicherla et al. 2011; Sarmiento et al. 2015). Therefore, food enzymes are expected to evidence significant adoptions in the coming years, straddling a wide range of materiality and high production efficiency. Nonetheless, very few cold-active enzymes have been developed and commercialized for use in the food market (Table 3). The cold-active xylanase produced by P. haloplanktis, which was reported to be efficient in bread making, is now sold by Puratos (Grand-Bigard, Belgium) (Collins et al. 2002). Selected pectinases that are not considered psychrophilic but display activity at low temperatures are also being used within the food and beverages industries. These enzymes include Novoshape®, Novozymes (pectin methyl esterase from recombinant A. oryzae) (Kitamoto et al. 1999), Pectinase 62L, Biocatalysts (mix of polygalactorunase and pectin lyase from Aspergillus sp.) (Combo et al. 2012) and Lallzyme®, Lallemand (mix of polygalactorunase, pectin esterase and pectin lyase from A. niger) (Zavala-Páramo et al. 2021). Research on potential cold-active enzymes can lead to several commercial biocatalysts in future, having applications in food processing industries.
Table 3.
Selected commercial cold active enzymes used in food industry
| Enzyme name | Brand name | Source microorganism | Enzyme properties | Manufacturing company |
|---|---|---|---|---|
| Xylanase | Premix X-618 | Pseudoalteromonashaloplanktis | Active at temperature range 5–25 °C | Puratos NV, Grand-Bigard, Belgium |
| Pectin methyl esterase | Novoshape® | Aspergillus oryzae | Active at temperature range 10–60 °C | Novozymes Biopharma, United States |
| Mixture of polygalactorunase and pectin lyase | Pectinase 62L | Aspergillus sp. | Active at temperature range 10–60 °C | Biocatalysts Ltd, Wales, United Kingdom |
| Mixture of polygalactorunase, pectin esterase and pectin lyase | Lallzyme® | A. niger | Active at temperature range 5–15 °C | Lallemand, Canada |
Challenges and opportunities
Cold-active enzymes are prospective substitutes to their conventional mesophilic and thermophilic matching parts in many ways. They possess high activity at low and moderate temperatures and carry out processes without any apparent loss in their catalytic efficiency, which consequences savings of consumption energy. On the contrary, at low temperatures, sometimes enzyme activity is compromised due to low substrate solubility and decreased substrate specificity (Collins and Margesin 2019; Mangiagalli and Lotti 2021). There are certain challenges associated with their instability at higher temperatures and at alkaline pH, which might impede the potential use of such cold adaptive enzymes. Furthermore, the low diversity of the psychrophilic microorganisms paired with non-specific isolation techniques to study such microbes is a major limiting factor in discovering novel cold-active enzymes and unfolding their biotechnological potential. Nonetheless, with the growing discovery and invention of up-to-the-minute techniques and instrumentation facilities, it is highly doable that the challenges could be conquered. Engineering the active site or the whole enzyme by introducing changes in the amino acid type and its positioning could be considered a potential strategy in enhancing structural and functional stability at ambient temperatures. Similar strategies could also be applied to increase the enzymatic function in the alkaline or acidic pH.
The metagenomic approaches that have been instrumental in identifying novel genes encoding proteins of high stability and activity in wide ranges of temperature and pH would be a valuable loom to clone and characterize the cold adaptive enzymes even from the uncultivable microbial community. Furthermore, undertaking genetic changes at the cellular level in the microorganisms producing the cold-active enzymes would be more hopeful in revitalizing the quality and quantity of the cold-active enzymes to meet the commercial food market’s expectations.
Conclusion and future directions
Cold-active enzymes reported so far are distinguished by low stability at high temperatures and demonstrated high catalytic efficiency at low temperatures. Besides their easy inactivation, few additional features, including saving energy and reaction time, and stabilizing thermolabile compounds in the reaction mixture, can be considered exceptional alternates to their thermophilic counterparts for use in many industrial applications. However, a minimal number of cold-active enzymes have been used in real food applications. Their low structural stability and cost constraints associated with isolating and purifying these enzymes have been remaining as a significant holdup. Modern molecular and enzyme engineering techniques have drastically predisposed the quality and productivity of enzymes, though enough efforts have to be undertaken in identifying novel cold-active genes that can be further improved to meet the industrial need. Therefore, extensive investigations are on-demand to explore diverse sources of psychrophilic microorganisms to discover unique and novel cold-active enzymes for diverse applications within the biotechnology industries and potential service to humanity.
Acknowledgements
The authors would like to thank Institute of Bioresources and Sustainable Development, Regional Centre, Sikkim and Department of Biotechnology, Govt. of India for the support and encouragement. The manuscript corresponds to manuscript ID IBSD/2020/01/062.
Footnotes
Megha Kumari and Srichandan Padhi have contributed equally.
Contributor Information
Sudhir P. Singh, Email: sudhirsingh@ciab.res.in
Amit Kumar Rai, Email: amitraikvs@gmail.com.
References
- Adapa V, Ramya LN, Pulicherla KK, Sambasiva Rao KRS. Cold-active pectinases: advancing the food industry to the next generation. Appl Biochem Biotechnol. 2014;172:2324–2337. doi: 10.1007/s12010-013-0685-1. [DOI] [PubMed] [Google Scholar]
- Adrio JL, Demain AL. Microbial enzymes: tools for biotechnological processes. Biomolecules. 2014;4:117–139. doi: 10.3390/biom4010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aehle W. Enzymes in industry productions and applications. 3. Weinheim: Wiley-VCH Verlag; 2007. [Google Scholar]
- Al-Ghanayem AA, Joseph B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl Microbiol Biotechnol. 2020;104:2871–2882. doi: 10.1007/s00253-020-10429-x. [DOI] [PubMed] [Google Scholar]
- Alikkunju AP, Sainjan N, Silvester R, Joseph A, Rahiman M, Antony AC, Kumaran RC. Screening and characterization of cold-active beta-galactosidase producing psychrotrophic Enterobacter ludwigii from the sediments of Arctic fjord. Appl Biochem Biotechnol. 2016;180:477–490. doi: 10.1007/s12010-016-2111-y. [DOI] [PubMed] [Google Scholar]
- Al-Khudary R, Venkatachalam R, Katzer M, Elleuche S, Antranikian G. A cold-adapted esterase of a novel marine isolate, Pseudoalteromonas arctica: gene cloning, enzyme purification and characterization. Extremophiles. 2010;14:273–285. doi: 10.1007/s00792-010-0306-7. [DOI] [PubMed] [Google Scholar]
- Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- Ayala-del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW, Bartolo GD, Hauser L, Land M, Bakermans C, Rodrigues D, Klappenbach J, Zarka D, Larimer F, Richardson P, Murray A, Thomashow M, Tiedje JM. The genome sequence of Psychrobacter arcticus, a psychroactive siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol. 2010;76:2304–2312. doi: 10.1128/AEM.02101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Hobenreich H, Vogel A, Knorr J, Wilhelm S, Rosenau F, Jaeger KE, Reetz MT, Kolmar H. Single-cell high-throughput screening to identify enantioselective hydrolytic enzymes. Angew Chem Int Ed Engl. 2008;47:5085–5088. doi: 10.1002/anie.200705236. [DOI] [PubMed] [Google Scholar]
- Benesova E, Markova M, Kralova B. α-glucosidase and β-glucosidase from psychrotrophic strain Arthrobacter sp. C2–2. Czech J Food Sci. 2005;23:116–120. [Google Scholar]
- Bernath K, Hai M, Mastrobattista E, Griths AD, Magdassi S, Tawfik DS. In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Anal Biochem. 2004;325:151–157. doi: 10.1016/j.ab.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Białkowska AM, Cieśliński H, Nowakowska KM, Kur J, Turkiewicz M. A new β-galactosidase with a low temperature optimum isolated from the Antarctic Arthrobacter sp. 20B: gene cloning, purification and characterization. Arch Microbiol. 2009;191:825–835. doi: 10.1007/s00203-009-0509-4. [DOI] [PubMed] [Google Scholar]
- Bialkowska AM, Krysiak J, Florczak T, Szulczewska KM, Wanarska M, Turkiewicz M. The psychrotrophic yeast Sporobolomyces roseus LOCK 1119 as a source of a highly active aspartic protease for the in vitro production of antioxidant peptides. Biotechnol Appl Biochem. 2018;65:726–738. doi: 10.1002/bab.1656. [DOI] [PubMed] [Google Scholar]
- Birgisson HO, Delgado L, García Arroyo R, Kaul H, Mattiasson B. Cold-adapted yeasts as producers of cold-active polygalacturonases. Extremophiles. 2003;7:185–193. doi: 10.1007/s00792-002-0310-7. [DOI] [PubMed] [Google Scholar]
- Bjerga GEK, Lale R, Williamson AK. Engineering low temperature expression systems for heterologous production of cold-adapted enzymes. Bioengineered. 2016;7:33–38. doi: 10.1080/21655979.2015.1128589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco P, Sieiro C, Villa TG. Production of pectic enzymes in yeasts. FEMS Microbiol Lett. 2009;175:1–9. doi: 10.1111/j.1574-6968.1999.tb13595.x. [DOI] [PubMed] [Google Scholar]
- Borgi MA, Khila M, Boudebbouze S, Aghajari N, Szukala F, Pons N, Maguin E, Rhimi M. The attractive recombinant phytase from Bacillus licheniformis: biochemical and molecular characterization. Appl Microbiol Biotechnol. 2014;98:5937–5947. doi: 10.1007/s00253-013-5421-9. [DOI] [PubMed] [Google Scholar]
- Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- Brault G, Shareck F, Hurtubise Y, Lépine F, Doucet N. Isolation and characterization of EstC, a new cold-active esterase from Streptomyces coelicolor A3(2) PLoS ONE. 2012;7:e32041. doi: 10.1371/journal.pone.0032041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno S, Coppola D, di Prisco G, Giordano D, Verde C. Enzymes from marine polar regions and their biotechnological applications. Mar Drugs. 2019;17:544. doi: 10.3390/md17100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisman GJH, van Helteren CTW, Kramer GFH, Veldsink JW, Derksen JTP, Cuperus FP. Enzymatic esterifications of functionalized phenols for the synthesis of lipophilic antioxidants. Biotechnol Lett. 1998;20:131–136. [Google Scholar]
- Carrasco M, Rozas JM, Alcaíno J, Cifuentes V, Baeza M. Pectinase secreted by psychrotolerant fungi: Identification, molecular characterization and heterologous expression of a cold-active polygalacturonase from Tetracladium sp. Microb Cell Fact. 2019;18:45–45. doi: 10.1186/s12934-019-1092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo BM, Dunn MF, Navarro KG, Meléndez FH, Ortiz MH, Guevara SE, Palacios GH. Antifungal performance of extracellular chitinases and culture supernatants of Streptomyces galilaeus CFFSUR-B12 against Mycosphaerella fijiensis Morelet. World J Microbiol Biotechnol. 2016;32:44. doi: 10.1007/s11274-015-1993-0. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R. On the concept of a psychrophiles. ISME J. 2016;10:793–795. doi: 10.1038/ismej.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli R, Charlton T, Ertan H, Omar SM, Siddiqui KS, Williams TJ. Biotechnological uses of enzymes from psychrophiles. Microb Biotechnol. 2011;4:449–460. doi: 10.1111/j.1751-7915.2011.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Kaufman MG, Miazgowicz KL, Bagdasarian M, Walker ED. Molecular characterization of a cold-active recombinant xylanase from flavobacterium johnsoniae and its applicability in xylan hydrolysis. Bioresour Technol. 2013;128:145–155. doi: 10.1016/j.biortech.2012.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia R, Phukon LC, Singh SP, Sahoo D, Rai AK. Role of enzymatic bioprocesses for functional food development. In: Singh SP, Pandey A, Kondo A, Singhania RR, Larroche C, editors. Advances in enzyme catalysis and technology. Elsevier; 2020. pp. 309–334. [Google Scholar]
- Chourasia R, Phukon LC, Abedin M, Sahoo D, Singh SP, Rai AK. Biotechnological approaches for the production of designer cheese with improved functionality. Comp Rev Food Sci Food Saf. 2021;20:960–979. doi: 10.1111/1541-4337.12680. [DOI] [PubMed] [Google Scholar]
- Cieśliński H, Kur J, Białkowska A, Baran I, Makowski K, Turkiewicz M. Cloning, expression, and purification of a recombinant cold-adapted β-galactosidase from antarctic bacterium Pseudoalteromonas sp. 22B. Protein Expr Purif. 2005;39:27–34. doi: 10.1016/j.pep.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Coker JA, Brenchley JE. Protein engineering of a cold-active β- galactosidase from Arthrobacter sp. SB to increase lactose hydrolysis reveals new sites affecting low temperature activity. Extremophiles. 2006;10:515–524. doi: 10.1007/s00792-006-0526-z. [DOI] [PubMed] [Google Scholar]
- Collins T, Margesin R. Psychrophilic lifestyles: Mechanisms of adaptation and biotechnological tools. Appl Microbiol Biotechnol. 2019;103:2857–2871. doi: 10.1007/s00253-019-09659-5. [DOI] [PubMed] [Google Scholar]
- Collins T, Meuwis MA, Stals I, Claeyssens M, Feller G, Gerday C. A novel family 8 xylanase, functional and physicochemical characterization. J Biol Chem. 2002;277:35133–35139. doi: 10.1074/jbc.M204517200. [DOI] [PubMed] [Google Scholar]
- Combo AMM, Aguedo M, Goffin D, Wathelet B, Paquot M. Enzymatic production of pectic oligosaccharides from polygalacturonic acid with commercial pectinase preparations. Food Bioprod Process. 2012;90:588–596. [Google Scholar]
- D’Amico S, Collins T, Marx JC, Feller G, Gerday C. psychrophilic microorganisms: challenges for life. EMBO Rep. 2006;7:385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria S, Aurilia V, Marabotti A, Gonnelli M, Strambini G. Structure and dynamics of cold-adapted enzymes as investigated by phosphorescence spectroscopy and molecular dynamics studies. 2. The case of an esterase from Pseudoalteromonas haloplanktis. J Phys Chem. 2009;113:13171–13178. doi: 10.1021/jp9043286. [DOI] [PubMed] [Google Scholar]
- Dalmaso GZ, Ferreira D, Vermelho AB. Marine extremophiles: a source of hydrolases for biotechnological applications. Mar Drugs. 2015;13:1925–1965. doi: 10.3390/md13041925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood A, Ma K. Applications of microbial β-mannanases. Front Bioeng Biotechnol. 2020;8:598–630. doi: 10.3389/fbioe.2020.598630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gobba C, Tompa G, Otte J. Bioactive peptides from caseins released by cold-active proteolytic enzymes from Arsukibacterium ikkense. Food Chem. 2014;165:205–215. doi: 10.1016/j.foodchem.2014.05.082. [DOI] [PubMed] [Google Scholar]
- De Santi C, Tedesco P, Ambrosino L, Altermark B, Willassen NP, de Pascale D. A new alkaliphilic cold-active esterase from the psychrophilic marine bacterium Rhodococcus sp.: functional and structural studies and biotechnological potential. Appl Biochem Biotechnol. 2014;172:3054–3068. doi: 10.1007/s12010-013-0713-1. [DOI] [PubMed] [Google Scholar]
- Del-Cid A, Ubilla P, Ravanal MC, Medina E, Vaca I, Levican G, Eyzaguirre J, Chavez R. Cold-active xylanase produced by fungi associated with Antarctic marine sponges. Appl Biochem Biotechnol. 2014;172:524–532. doi: 10.1007/s12010-013-0551-1. [DOI] [PubMed] [Google Scholar]
- Dewan SS. Global markets for enzymes in industrial applications. Wellesly: BCC Research; 2014. [Google Scholar]
- Dornez E, Verjans P, Arnaut F, Delcour JA, Courtin CM. Use of psychrophilic xylanases provides insight into the xylanase functionality in bread making. J Agric Food Chem. 2011;59:9553–9562. doi: 10.1021/jf201752g. [DOI] [PubMed] [Google Scholar]
- Dou S, Chi N, Zhou X, Zhang Q, Pang F, Xiu Z. Molecular cloning, expression, and biochemical characterization of a novel cold-active α-amylase from Bacillus sp. dsh19-1. Extremophiles. 2018;22:739–749. doi: 10.1007/s00792-018-1034-7. [DOI] [PubMed] [Google Scholar]
- Duarte AWF, dos Santos JA, Vianna MV, Vieira JMF, Mallagutti VH, Inforsato FJ, Wentzel LCP, Lario LD, Rodrigues A, Pagnocca FC, Junior AP, Sette LD. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit Rev Biotechnol. 2018;38:600–619. doi: 10.1080/07388551.2017.1379468. [DOI] [PubMed] [Google Scholar]
- Elleuche S, Schröder C, Sahm K, Antranikian G. Extremozymes biocatalysts with unique properties from extremophilic microorganisms. Curr Opin Biotechnol. 2014;29:116–123. doi: 10.1016/j.copbio.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Emampour M, Noghabi KA, Zahiri HS. Molecular cloning and biochemical characterization of a novel cold-adapted alpha-amylase with multiple extremozyme characteristics. J Mol Catal B Enzym. 2015;111:79–86. [Google Scholar]
- Fan Y, Hua X, Zhang Y, Feng Y, Shen Q, Dong J, Zhao W, Zhang W, Jin Z, Yang R. Cloning, expression and structural stability of a cold-adapted β-galactosidase from Rahnella sp. R3. Protein Expr Purif. 2015;115:158–164. doi: 10.1016/j.pep.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Feller G. Psychrophilic Enzymes: From folding to function and biotechnology. Scientifica. 2013;2013:512840. doi: 10.1155/2013/512840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller G, Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol. 2003;1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- Feller G, Le Bussy O, Gerday C. Expression of psychrophilic genes in mesophilic hosts: assessments of the folding state of a recombinant alpha-amylase. Appl Environ Microbiol. 1998;64:1163–1165. doi: 10.1128/aem.64.3.1163-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P. Enzymes in food processing: a condensed overview on strategies for better biocatalysts. Enzyme Res. 2010;2010:862537. doi: 10.4061/2010/862537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P, Carvalho F. Enzymes in food processing. In: Dhillon GS, Kaur S, editors. Agro-industrial wastes as feedstock for enzyme production. Elsevier; 2016. pp. 173–199. [Google Scholar]
- Fernandez-Alvaro E, Snajdrova R, Jochens H, Davids T, Bottcher D, Bornscheuer UT. A combination of in vivo selection and cell sorting for the identification of enantioselective biocatalysts. Angew Chem Int Ed Engl. 2011;50:8584–8587. doi: 10.1002/anie.201102360. [DOI] [PubMed] [Google Scholar]
- Fox RJ, Davis SC, Mundor EC, Newman LM, Gavrilovic V, et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol. 2007;25:338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- Fu X, Liu P, Lin L, Hong Y, Huang X, Meng X, Liu Z. A novel endoglucanase (Cel9P) from a marine bacterium Paenibacillus sp. BME-14. Appl Biochem Biotechnol. 2010;160:1627–1636. doi: 10.1007/s12010-009-8648-2. [DOI] [PubMed] [Google Scholar]
- Gao B, Mao Y, Zhang L, He L, Wei D. A novel saccharifying a-amylase of Antarctic psychotolerant fungi Geomyces pannorum: gene, cloning, functional expression, and characterization. Starch. 2016;67:1–9. [Google Scholar]
- Gao B, He L, Wei D, Zhang L. Identification and magnetic immobilization of a pyrophilous aspartic protease from Antarctic psychrophilic fungus. J Food Biochem. 2018;42:12691. [Google Scholar]
- Gatti-Lafranconi P, Caldarazzo SM, Villa A, Alberghina L, Lotti M. Unscrambling thermal stability and temperature adaptation in evolved variants of a cold−active lipase. FEBS Lett. 2008;582:2313–2318. doi: 10.1016/j.febslet.2008.05.037. [DOI] [PubMed] [Google Scholar]
- Gerday C. Psychrophily and catalysis. Biology. 2013;2:719–741. doi: 10.3390/biology2020719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerday C, Aittaleb M, Bentahir M, Chessa JP, Claverie P, et al. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 2000;18:103–107. doi: 10.1016/s0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
- Hamid B, Mohiddin FA. Cold-active enzymes in food Processing. In: Kuddus M, editor. Enzymes in food technology. Singapore: Springer; 2018. pp. 383–400. [Google Scholar]
- Hamid B, Ravinder SR, Deepak C, Singh P, Mohiddin FA, Sahay S, Abidi I. Psychrophilic yeasts and their biotechnological applications—a review. Afr J Biotechnol. 2014;13:2188–2197. [Google Scholar]
- Hathwar SC, Bijinu B, Rai AK, Bhaskar N. Simultaneous recovery of lipids and proteins by enzymatic hydrolysis of fish industry waste using different commercial proteases. Appl Biochem Biotechnol. 2011;164:115–124. doi: 10.1007/s12010-010-9119-5. [DOI] [PubMed] [Google Scholar]
- He HL, Chen XL, Li JW, Zhang YZ, Gao PJ. Taste improvement of refrigerated meat treated with cold-adapted protease. Food Chem. 2004;84:307–311. [Google Scholar]
- He L, Mao Y, Zhang L, Wang H, Alias SA, Gao B, et al. Functional expression of a novel α-amylase from Antarctic psychrotolerant fungus for baking industry and its magnetic immobilization. BMC Biotechnol. 2017;17:22–38. doi: 10.1186/s12896-017-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggeset TM, Krog A, Balzer S, Wentzel A, Ellingsen TE, Brautaset T. Genome sequence of thermotolerant Bacillus methanolicus: features and regulation related to methylotrophy and production of l-lysine and l-glutamate from methanol. Appl Environ Microbiol. 2012;78:5170–5181. doi: 10.1128/AEM.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hii SL, Tan JS, Ling TC, Ariff AB. Pullulanase: role in starch hydrolysis and potential industrial applications. Enzyme Res. 2012;9:213–262. doi: 10.1155/2012/921362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt P, Wanarska M, Kur J. A new cold-adapted beta-d-galactosidase from the Antarctic Arthrobacter sp. 32c—gene cloning, overexpression, purification and properties. BMC Microbiol. 2009;27:151. doi: 10.1186/1471-2180-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyoux A, Jennes I, Dubois P, Genicot S, Dubail F, Francois JM, Baise E, Feller G, Gerday C. Cold-adapted beta-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl Environ Microbiol. 2001;67:1529–1535. doi: 10.1128/AEM.67.4.1529-1535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JL, Bao LX, Zou HY, Che SG, Wang GX. High-level production of a cold-active β-mannanase from Bacillus subtilis Bs5 and its molecular cloning and expression. Mol Genet Microbiol Virol. 2012;27:147–153. [PubMed] [Google Scholar]
- Huston AL. Biotechnological aspects of cold-adapted enzymes. In: Margesin R, Schinner F, Marx JC, Gerday C, editors. Psychrophiles: from biodiversity to biotechnology. Berlin, Heidelberg: Springer; 2008. pp. 347–363. [Google Scholar]
- Irwin JA. Overview of extremophiles and their food and medical applications. In: Salwan R, Sharma V, editors. Physiological and biotechnological aspects of extremophiles. Academic Press; 2020. pp. 65–87. [Google Scholar]
- Javed A, Qazi JI. Psychrophilic microbial enzymes implications in coming biotechnological processes. Am Sci Res J Eng Technol Sci. 2016;23:103–120. [Google Scholar]
- Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: a review. Process Biochem. 2005;40:2931–2944. [Google Scholar]
- Jayasekara S, Ratnayake R. Microbial cellulases: an overview and applications. In: Pascual AR, Eugenio Martín ME, editors. Cellulose. IntechOpen; 2019. pp. 1–21. [Google Scholar]
- Jeon JH, Kim JT, Kang SG, Lee JH, Kim SJ. Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar Biotechnol. 2009;11:307–316. doi: 10.1007/s10126-008-9145-2. [DOI] [PubMed] [Google Scholar]
- Ji XL, Taj MK, Lu XB, Lin LB, Zhang Q, Wei YL. Purification and characterization of an extracellular cold-active protease produced by the psychrotrophic bacterium Serratia sp. WJ39. In: Zhang KL, editor. Applied mechanics and materials. Trans Tech Publications Ltd; 2014. pp. 330–334. [Google Scholar]
- Joseph B, Ramteke PW, Thomas G. Cold-active microbial lipases: some hot issues and recent developments. Biotechnol Adv. 2008;26:457–470. doi: 10.1016/j.biotechadv.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Joshi S, Satyanarayana T. Biotechnology of cold-active proteases. Biology. 2013;2:755–783. doi: 10.3390/biology2020755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Sharma M, Singh SP. Characterization of a novel xylanase from an extreme temperature hot spring metagenome for xylooligosaccharide production. Appl Microbiol Biotechnol. 2020;104:4889–4901. doi: 10.1007/s00253-020-10562-7. [DOI] [PubMed] [Google Scholar]
- Jung YH, Yi JY, Jung HJ, Lee YK, Lee HK, Naicker MC, Uh JH, Jo IS, Jung EJ, Im H. Overexpression of cold shock protein A of Psychromonas arctica KOPRI 22215 confers cold-resistance. Protein J. 2010;29(2):136–142. doi: 10.1007/s10930-010-9233-9. [DOI] [PubMed] [Google Scholar]
- Kamarudin NH, Rahman RN, Ali MS, Leow TC, Basri M, Salleh AB. A new cold-adapted, organic solvent stable lipase from mesophilic Staphylococcus epidermidis AT2. Protein J. 2014;33:296–307. doi: 10.1007/s10930-014-9560-3. [DOI] [PubMed] [Google Scholar]
- Karan R, Capes MD, DasSarma P, DasSarma S. Cloning, overexpression, purification and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeon Halorubrum lacusprofundi. BMC Biotechnol. 2013;13:3. doi: 10.1186/1472-6750-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasova-Lipovova P, Strnad H, Spiwok V, Mala S, Kralova B, Russell NJ. The cloning, purification and characterization of a cold-active β-galactosidase from the psychrotolerant Antarctic bacterium Arthrobacter sp. C2–2. Enzyme Microb Technol. 2003;33:836–844. [Google Scholar]
- Kasieczka-Burnecka M, Kuc K, Kalinowska H, Knap M, Turkiewicz M. Purification and characterization of two cold-adapted extracellular tannin acyl hydrolases from an Antarctic strain Verticillium sp. P9. Appl Microbiol Biotechnol. 2007;77:77–89. doi: 10.1007/s00253-007-1124-4. [DOI] [PubMed] [Google Scholar]
- Kitamoto N, Okada H, Yoshino S, Ohmiya K, Tsukagoshi N. Pectin methylesterase gene (pmeA) from Aspergillus oryzae KBN616: Its sequence analysis and over expression, and characterization of the gene product. Biosci Biotechnol Biochem. 1999;6:120–124. doi: 10.1271/bbb.63.120. [DOI] [PubMed] [Google Scholar]
- Ko JK, Ko H, Kim KH, Choi IG. Characterization of the biochemical properties of recombinant Xyn10C from a marine bacterium, Saccharophagus degradans 2–40. Bioprocess Biosyst Eng. 2016;39:677–684. doi: 10.1007/s00449-016-1548-2. [DOI] [PubMed] [Google Scholar]
- Koleva L, Pishtiyski I, Pavlova K. Purification and properties of extracellular b glucosidase from the Antarctic yeast strain Cryptococcus albidus AL3. Bulg J Agric Sci. 2006;12:713–720. [Google Scholar]
- Krishna SH. Developments and trends in enzyme catalysis in non-conventional media. Biotechnol Adv. 2002;20:239–267. doi: 10.1016/s0734-9750(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Alias SA, Wong CMVL, Pang KL, Convey P. Extracellular hydrolase enzyme production by soil fungi from King George Island, Antarctica. Polar Biol. 2011;34:1535–1542. [Google Scholar]
- Kuddus M. Cold-active enzymes in food biotechnology: an updated mini review. J Appl Biol Biotechnol. 2018;6:58–63. [Google Scholar]
- Kuddus M, Ramteke PW. Recent developments in production and biotechnological applications of cold-active microbial proteases. Crit Rev Microbiol. 2012;38:330–338. doi: 10.3109/1040841X.2012.678477. [DOI] [PubMed] [Google Scholar]
- Kuhad RC, Gupta R, Singh A. Microbial cellulases and their industrial applications. Enzyme Res. 2011;2011:280696. doi: 10.4061/2011/280696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Sinha AK, Makkar HPS, De Boeck G. Phytate and phytase in fish nutrition. J Anim Physiol Anim Nutr. 2012;96(3):335–364. doi: 10.1111/j.1439-0396.2011.01169.x. [DOI] [PubMed] [Google Scholar]
- Kumar B, Bhardwaj N, Alam A, Agrawal K, Prasad H, Verma P. Production, purifcation and characterization of an acid/ alkali and thermo tolerant cellulase from Schizophyllum commune NAIMCC-F-03379 and its application in hydrolysis of lignocellulosic wastes. AMB Expr. 2018;8:173–185. doi: 10.1186/s13568-018-0696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lario LD, Chaud L, Almeida MG, Converti A, Sette LD, Pessoa A., Jr Production, purification and characterization of extracellular acid protease from the marine Antarctic yeast Rhodotorula mucilaginosa L7. Fungal Biol. 2015;119:1129–1136. doi: 10.1016/j.funbio.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Laye VJ, Karan R, Kim JM, Pecher WT, Das-Sarma P, Das-Sarma S. Key amino acid residues conferring enhanced enzyme activity at cold temperatures in an Antarctic polyextremophilic β-galactosidase. PNAS. 2017;114(47):12530–12535. doi: 10.1073/pnas.1711542114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le B, Yang SH. Microbial chitinases: properties, current state and biotechnological applications. World J Microbiol Biotechnol. 2019;35:144. doi: 10.1007/s11274-019-2721-y. [DOI] [PubMed] [Google Scholar]
- Le LTHL, Yoo W, Jeon S, Lee C, Kim KK, Lee JH, Kim TD. Biodiesel and flavor compound production using a novel promiscuous cold-adapted SGNH-type lipase (HaSGNH1) from the psychrophilic bacterium Halocynthiibacter arcticus. Biotechnol Biofuels. 2020;13:1–13. doi: 10.1186/s13068-020-01696-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Jeon HY, Choi HJ, Kim NR, Choung WJ, Koo YS, Ko DS, You S, Shim JH. Characterization and application of BiLA, a psychrophilic α-amylase from Bifidobacterium longum. J Agric Food Chem. 2016;64:2709–2718. doi: 10.1021/acs.jafc.5b05904. [DOI] [PubMed] [Google Scholar]
- Lemak S, Tchigvintsev A, Petit P, Flick R, Singer AU, Brown G, Evdokimova E, Egorova O, Gonzalez CF, Chernikova TN, Yakimov MM, Kube M, Reinhardt R, Golyshin PN, Sevchenko A, Yakunin AF. Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica. Biochem J. 2012;4452:193–203. doi: 10.1042/BJ20112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Thomas T, Cavicchioli R. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon Methanococcoides Burtonii. J Mol Biol. 2000;297(3):553–567. doi: 10.1006/jmbi.2000.3585. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang Z, Zhang X, Shao Z, Liu Z. Cloning, expression and characterization of a novel cold-active and halophilic xylanase from Zunongwangia profunda. Extremophiles. 2014;18:441–450. doi: 10.1007/s00792-014-0629-x. [DOI] [PubMed] [Google Scholar]
- Liu K, Ding H, Yu Y, Chen B. A cold-adapted chitinase-producing bacterium from Antarctica and its potential in biocontrol of plant pathogenic fungi. Mar Drugs. 2019;17:695. doi: 10.3390/md17120695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longwell CK, Labanieh L, Cochran JR. High-throughput screening technologies for enzyme engineering. Curr Opin Biotechnol. 2017;48:196–202. doi: 10.1016/j.copbio.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Lonhienne T, Mavromatis K, Vorgias CE, Buchon L, Gerday C, Bouriotis V. Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: isolation and partial characterization of the enzymes. J Bacteriol. 2001;183:1773–1779. doi: 10.1128/JB.183.5.1773-1779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudhan MC, Raghavarao KSMS. Aqueous two phase extraction of invertase from baker’s yeast: effect of process parameters on partitioning. Process Biochem. 2011;46:2014–2020. [Google Scholar]
- Mageswari A, Subramanian P, Chandrasekaran S, Karthikeyan S, Gothandam KM. Systematic functional analysis and application of a cold-active serine protease from a novel Chryseobacterium sp. Food Chem. 2017;217:18–27. doi: 10.1016/j.foodchem.2016.08.064. [DOI] [PubMed] [Google Scholar]
- Makowski K, Białkowska A, Szczęsna-Antczak M, Kalinowska H, Kur J, Cieśliński H, Turkiewicz M. Immobilized preparation of cold-adapted and halotolerant Antarctic β-galactosidase as a highly stable catalyst in lactose hydrolysis. FEMS Microbiol Ecol. 2007;59:535–542. doi: 10.1111/j.1574-6941.2006.00208.x. [DOI] [PubMed] [Google Scholar]
- Mangiagalli M, Lotti M. Cold-active β-galactosidases: insight into cold adaptation, mechanisms and biotechnological exploitation. Mar Drugs. 2021;19:43. doi: 10.3390/md19010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Yin Y, Zhang L, Alias SA, Gao B, Wei D. Development of a novel Aspergillus uracil deficient expression system and its application in expressing a cold-adapted α-amylase gene from Antarctic fungi Geomyces pannorum. Process Biochem. 2015;50:1581–1590. [Google Scholar]
- Margesin R, Feller G. Biotechnological applications of Psychrophiles. Environ Technol. 2010;31:835–844. doi: 10.1080/09593331003663328. [DOI] [PubMed] [Google Scholar]
- Margesin R, Neuner G, Storey KB. Cold-loving microbes, plants and animals fundamental and applied aspects. Natursch. 2007;94:77–99. doi: 10.1007/s00114-006-0162-6. [DOI] [PubMed] [Google Scholar]
- Martín MC, Morata de Ambrosini VI. Cold-active acid pectinolytic system from psychrotolerant Bacillus: color extraction from red grape skin. Am J Enol Vitic. 2013;64:495–504. [Google Scholar]
- Marx JC, Collins T, Amico SD, Feller G, Gerday C. Cold-adapted enzymes from marine Antarctic microorganisms. Mar Biotech. 2007;9:293–304. doi: 10.1007/s10126-006-6103-8. [DOI] [PubMed] [Google Scholar]
- Merín MG, de Ambrosini VIM. Highly cold-active pectinases under wine-like conditions from non-Saccharomyces yeasts for enzymatic production during winemaking. Lett Appl Microbiol. 2015;60:467–474. doi: 10.1111/lam.12390. [DOI] [PubMed] [Google Scholar]
- Meteyer CU, Verant ML. White-nose syndrome: cutaneous invasive ascomycosis in hibernating bats. In: Eric-Miller R, Lamberski N, Calle P, editors. Fowler's zoo and wild animal medicine current therapy. Elsevier; 2019. pp. 508–513. [Google Scholar]
- Mojallali L, Shahbani Zahiri H, Rajaei S, Akbari Noghabi K, Haghbeen K. A novel ∼34-kDa α-amylase from psychrotroph Exiguobacterium sp. SH3: production, purification, and characterization. Biotechnol Appl Biochem. 2014;61(2):118–125. doi: 10.1002/bab.1140. [DOI] [PubMed] [Google Scholar]
- Morita RY, Moyer CL. Origin of psychrophiles. In: Levin SA, Colwell R, Daily G, Lubchenco J, Mooney HA, Schulze E, Tilman D, editors. Encyclopedia of biodiversity. San Diego: Academic Press; 2001. pp. 917–924. [Google Scholar]
- Mukhia S, Kumar A, Kumar R. Generation of antioxidant peptides from soy protein isolate through psychrotrophic Chryseobacterium sp. derived alkaline broad temperature active protease. LWT Food Sci Technol. 2021;143:111–152. [Google Scholar]
- Naga Padma P, Anuradha K, Reddy G. Pectinolytic yeast isolates for cold-active polygalacturonase production. Innov Food Sci Emerg Technol. 2011;12:178–181. [Google Scholar]
- Nguyen TTH, Myrold DD, Mueller RS. Distributions of extracellular peptidases across prokaryotic genomes reflect phylogeny and habitat. Front Microbiol. 2019;10:413. doi: 10.3389/fmicb.2019.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Montero K, Salazar R, Santos A, Gómez-Espinoza O, Farah S, Troncoso C, Hoffmann C, Melivilu D, Scott F, Barrientos Díaz L. Antarctic Rahnella inusitata: a producer of cold-stable β-galactosidase enzymes. Int J Mol Sci. 2021;22(8):4144. doi: 10.3390/ijms22084144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pable AA, Shah S, Kumar R, et al. Use of Plackett–Burman design for enhanced phytase production by Williopsis saturnus NCIM 3298 for applications in animal feed and ethanol production. 3Biotech. 2019;9:237. doi: 10.1007/s13205-019-1764-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Tu T, Wang L, Luo H, Ma R, Shi P, Meng K, Tao B. A novel low-temperature active pectin methyl esterase from Penicillium chrysogenum F46 with high efficiency in fruit firming. Food Chem. 2014;162:229–234. doi: 10.1016/j.foodchem.2014.04.069. [DOI] [PubMed] [Google Scholar]
- Pandey A, Benjamin S, Soccol CR, Nigam P, Krieger N, Soccol V. The realm of microbial lipases in biotechnology. Biotechnol Appl Biochem. 1999;29:119–131. [PubMed] [Google Scholar]
- Papagianni M, Sergelidis D. Purification and biochemical characterization of a novel alkaline protease produced by Penicillium nalgiovense. Appl Biochem Biotechnol. 2014;172:3926–3938. doi: 10.1007/s12010-014-0824-3. [DOI] [PubMed] [Google Scholar]
- Park I, Cho J. The phytase from antarctic bacterial isolate, Pseudomonas sp. JPK1 as a potential tool for animal agriculture to reduce manure phosphorus excretion. Afr J Agric Res. 2011;6:1398–1406. [Google Scholar]
- Park DJ, Lee YS, Choi YL. Characterization of a cold-active β-glucosidase from Paenibacillus xylanilyticus KJ-03 capable of hydrolyzing isoflavones daidzin and genistin. Protein J. 2013;32:579–584. doi: 10.1007/s10930-013-9520-3. [DOI] [PubMed] [Google Scholar]
- Patidar MK, Nighojkar S, Kumar A, Nighojkar A. Pectinolytic enzymes-solid state fermentation, assay methods and applications in fruit juice industries: a review. 3Biotech. 2018;8:199. doi: 10.1007/s13205-018-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil NS, Jadhav JP. Single cell protein production using Penicillium ochrochloron chitinase and its evaluation in fish meal formulations. J Microb Biochem Technol. 2014;S4:2–4. [Google Scholar]
- Pavlova K, Gargova S, Hristozova T, Tankova Z. Phytase from antarctic yeast strain Cryptococcus laurentii AL27. Folia Microbiol (praha) 2008;53:29–34. doi: 10.1007/s12223-008-0004-3. [DOI] [PubMed] [Google Scholar]
- Pawlak-Szukalska A, Wanarska M, Popinigis AT, Kur J. A novel cold-active β-galactosidase with transglycosylation activity from the Antarctic Arthrobacter sp. 20B: gene cloning, purification and characterization. Process Biochem. 2014;49:2122–2133. [Google Scholar]
- Peterson R, Kautto L, Sluyter SV, Nevalainen H. In search of clarity: Do cold-active proteases from Antarctic fungi provide alternatives to heat-stabilisation with bentonite? Wine Vitic J. 2013;28:20–23. [Google Scholar]
- Phukon LC, Abedin M, Chourasia R, Sahoo D, Parameswaran P, Rai AK. Production of xylanase under submerged fermentation from Bacillus firmus HS11 isolated from Sikkim Himalayan region. Indian J Exp Biol. 2020;58:563–570. [Google Scholar]
- Phukon LC, Chourasia R, Kumari M, Godan TK, Sahoo D, Parameswaran P, Rai AK. Production and characterisation of lipase for application in detergent industry from a novel Pseudomonas helmanticensis HS6. Bioresour Technol. 2020;309:123352. doi: 10.1016/j.biortech.2020.123352. [DOI] [PubMed] [Google Scholar]
- Piette F, Struvay C, Feller G. The protein folding challenge in psychrophiles: facts and current issues. Environ Microbiol. 2011;13(8):1924–1933. doi: 10.1111/j.1462-2920.2011.02436.x. [DOI] [PubMed] [Google Scholar]
- Poveda G, Gil-Durán C, Vaca I, Levicán G, Chávez R. Cold-active pectinolytic activity produced by filamentous fungi associated with Antarctic marine sponges. Biol Res. 2018;51(1):28. doi: 10.1186/s40659-018-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulicherla KK, Ghosh M, Kumar S, Rao KRSS. Psychrozymes: the next generation industrial enzymes. J Mar Sci Res Dev. 2011;1:102–124. [Google Scholar]
- Qin Y, Huang Z, Liu Z. A novel cold-active and salt-tolerant alpha-amylase from marine bacterium Zunongwangia profunda: molecular cloning, heterologous expression and biochemical characterization. Extremophiles. 2014;18:271–281. doi: 10.1007/s00792-013-0614-9. [DOI] [PubMed] [Google Scholar]
- Rai AK, Kumar R. Potential of microbial bio-resources of Sikkim Himalayan region. Envis Himal Bull. 2015;23:99–105. [Google Scholar]
- Rai AK, Swapna HC, Bhaskar N, Baskaran V. Potential of seafood industry byproducts as sources of recoverable lipids: fatty acid composition of meat and non meat component of selected Indian marine fishes. J Food Biochem. 2012;36:441–448. [Google Scholar]
- Rai AK, Bhaskar N, Baskaran V. Bioefficacy of EPA-DHA from lipids recovered from fish processing wastes through biotechnological approaches. Food Chem. 2013;136:80–86. doi: 10.1016/j.foodchem.2012.07.103. [DOI] [PubMed] [Google Scholar]
- Rai AK, Sanjukta S, Jeyaram K. Production of Angiotensin I converting enzyme inhibitory (ACE-I) peptides during milk fermentation and its role in treatment of hypertension. Critical Rev Food Sci Nutr. 2017;57:2789–2800. doi: 10.1080/10408398.2015.1068736. [DOI] [PubMed] [Google Scholar]
- Rajabi M. Chitooligosaccharides in food industry. Open Access J Biomed Sci. 2019;3:130–131. [Google Scholar]
- Rajaei S, Noghabi KA, Sadeghizadeh M, Zahiri HS. Characterization of a pH and detergent-tolerant, cold-adapted type I pullulanase from Exiguobacterium sp. SH3. Extremophiles. 2015;19:1145–1155. doi: 10.1007/s00792-015-0786-6. [DOI] [PubMed] [Google Scholar]
- Ramli AN, Azhar MA, Shamsir MS, Rabu A, Murad AM, Mahadi NM, Illias RM. Sequence and structural investigation of a novel psychrophilic α-amylase from Glaciozyma antarctica PI12 for cold-adaptation analysis. J Mol Model. 2013;19:3369–3383. doi: 10.1007/s00894-013-1861-5. [DOI] [PubMed] [Google Scholar]
- Raveendran S, Parameswaran B, Ummalyma SB, Abraham A, Mathew AK, Madhavan A, Rebello S, Pandey A. Applications of microbial enzymes in food industry. Food Technol Biotechnol. 2018;56:16–30. doi: 10.17113/ftb.56.01.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Staley JT, Danchin A, Antoine D, Ting ZW, Thomas SB, Loren JH, Land ML, Thompson LS. Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genomics. 2008;9:210–218. doi: 10.1186/1471-2164-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roohi R, Kuddus M, Saima S. Cold-active detergent-stable extracellular α-amylase from Bacillus cereus GA6: biochemical characteristics and its perspectives in laundry detergent formulation. J Biochem Technol. 2013;4:636–644. [Google Scholar]
- Salwan R, Sharma V. Physiological and biotechnological aspects of extremophiles. Academic Press; 2020. [Google Scholar]
- Santiago M, Ramírez-Sarmiento CA, Zamora RA, Parra LP. Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front Microbiol. 2016;7:1408. doi: 10.3389/fmicb.2016.01408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento F, Peralta R, Blamey JM. Cold and hot extremozymes: industrial relevance and current trends. Front Bioeng Biotechnol. 2015;3:148–158. doi: 10.3389/fbioe.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders NF, Thomas T, Curmi PM, Mattick JS. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 2003;13:1580–1588. doi: 10.1101/gr.1180903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Stougaard P. Identification, cloning and expression of a cold-active β-galactosidase from a novel Arctic bacterium, Alkalilactibacillus ikkense. Environ Technol. 2010;31:1107–1114. doi: 10.1080/09593331003677872. [DOI] [PubMed] [Google Scholar]
- Shipkowski S, Brenchley JE. Characterization of an unusual cold-active beta-glucosidase belonging to family 3 of the glycoside hydrolases from the psychrophilic isolate Paenibacillus sp. strain C7. Appl Environ Microbiol. 2005;71:4225–4232. doi: 10.1128/AEM.71.8.4225-4232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annu Rev Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- Song C, Chi Z, Li J, Wang X. β-galactosidase production by the psychrotolerant yeast Guehomyces pullulans 17–1 isolated from sea sediment in Antarctica and lactose hydrolysis. Bioprocess Biosyst Eng. 2010;33:1025–1031. doi: 10.1007/s00449-010-0427-5. [DOI] [PubMed] [Google Scholar]
- Srimathi S, Jayaraman G, Feller G, Danielsson B, Narayanan PR. Intrinsic halotolerance of the psychrophilic α-amylase from Pseudoalteromonas haloplanktis. Extremophiles. 2007;11:505–515. doi: 10.1007/s00792-007-0062-5. [DOI] [PubMed] [Google Scholar]
- Strocchi M, Ferrer M, Timmis KN, Golyshin PN. Low temperature-induced systems failure in Escherichia coli: insights from rescue by cold-adapted chaperones. Proteomics. 2006;6(1):193–206. doi: 10.1002/pmic.200500031. [DOI] [PubMed] [Google Scholar]
- Su H, Mai Z, Yang J, Xiao Y, Tian X, Zhang S. Cloning, expression, and characterization of a cold-active and organic solvent-tolerant lipase from Aeromicrobium sp. SCSIO 25071. J Microbiol Biotechnol. 2016;26:1067–1076. doi: 10.4014/jmb.1511.11068. [DOI] [PubMed] [Google Scholar]
- Sun J, Yao C, Wang W, Zhuang Z, Liu J, Dai F, Hao J. Cloning, expression and characterization of a novel cold-adapted β-galactosidase from the deep-sea bacterium Alteromonas sp ML52. Mar Drugs. 2018;16(12):469. doi: 10.3390/md16120469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M, Im H, Lee K. Molecular cloning and chaperone activity of DnaK from cold-adapted bacteria, KOPRI22215. Bull Korean Chem Soc. 2011;32(6):1925–1930. [Google Scholar]
- Tan S, Owusu ARK, Knapp J. Low temperature organic phase biocatalysis using cold adapted lipase from psychrotrophic Pseudomonas P38. Food Chem. 1996;57:415–418. [Google Scholar]
- Thakur M, Sharma N, Rai AK, Singh SP. A novel cold-active type I pullulanase from a hot-spring metagenome for effective debranching and production of resistant starch. Bioresour Technol. 2021;320:124288. doi: 10.1016/j.biortech.2020.124288. [DOI] [PubMed] [Google Scholar]
- Thakur M, Rai AK, Mishra BB, Singh SP. Novel insight into valorization of potato peels biomass into resistant starch III and malto oligosaccharide molecules. Environ Technol Innov. 2021;24:101827. [Google Scholar]
- Thierry A, Collins YF, Abeijón Mukdsi MC, McSweeney PMH, Wilkinson MG, Spinnler HE. Lipolysis and metabolism of fatty acids in cheese. In: Mc Sweeney PLH, editor. Cheese: chemistry, physics and microbiology. Elsevier; 2017. pp. 423–444. [Google Scholar]
- Trimbur DE, Gutshall KR, Prema P, Brenchley JE. Characterization of a psychrotrophic Arthrobacter gene and its cold-active β-galactosidase. Appl Environ Microbiol. 1994;60:4544–4552. doi: 10.1128/aem.60.12.4544-4552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong LV, Tuyen H, Helmke E, Binh LT, Schweder T. Cloning of two pectate lyase genes from the marine Antarctic bacterium Pseudoalteromonas haloplanktis strain ANT/505 and characterization of the enzymes. Extremophiles. 2001;5:35–44. doi: 10.1007/s007920000170. [DOI] [PubMed] [Google Scholar]
- Turkiewicz M, Pazgier M, Kalinowska H, Bielecki S. A cold-adapted extracellular serine proteinase of the yeast Leucosporidium antarcticum. Extremophiles. 2003;7:435–442. doi: 10.1007/s00792-003-0340-9. [DOI] [PubMed] [Google Scholar]
- Turkiewicz M, Pazgier M, Donachie SP, Kalinowska H. Invertase and α-glucosidase production by the endemic Antarctic marine yeast Leucosporidium antarcticum. Pol Polar Res. 2005;26:125–136. [Google Scholar]
- Vaca I, Faudez C, Maza F, Paillavil B, Hernandez V, Acosta F, Levican G, Martinez C, Chavez R. Cultivable psychrotolerant yeasts associated with Antarctic marine sponges. World J Microbiol Biotechnol. 2013;29:183–189. doi: 10.1007/s11274-012-1159-2. [DOI] [PubMed] [Google Scholar]
- van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L. Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol. 2002;94:137–155. doi: 10.1016/s0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- Wadhwa M, Bakshi MPS. Application of waste-derived proteins in the animal feed industry. In: Dhillon GS, editor. Protein byproducts. Academic Press; 2016. pp. 161–192. [Google Scholar]
- Wang QF, Hou YH, Xu Z, Miao JL, Li GY. Purification and properties of an extracellular cold-active protease from the psychrophilic bacterium Pseudoalteromonas sp. NJ276. Biochem Eng J. 2008;38:362–368. [Google Scholar]
- Wang S, Yang Y, Yang R, Zhang J, Chen M, Matsukawa S, Xie J, Wei D. Cloning and characterization of a cold-adapted endo-1,5-a-l-arabinanase from Paenibacillus polymyxa and rational design for acidic applicability. J Agri Food Chem. 2014;62:8460–8469. doi: 10.1021/jf501328n. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao Y, Tan H, Chi N, Zhang Q, Du Y, Yin H. Characterisation of a chitinase from Pseudoalteromonas sp. DL-6, a marine psychrophilic bacterium. Int J Biol Macromol. 2014;70:455–462. doi: 10.1016/j.ijbiomac.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Wang X, Kan G, Ren X, Yu G, Shi C, Xie Q, Wen H, Betenbaugh M. Molecular cloning and characterization of a novel -Amylase from antarctic sea ice bacterium pseudoalteromonas sp M175 and its primary application in detergent. Biomed Res Int. 2018;2018:3258383. doi: 10.1155/2018/3258383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ryu BH, Yoo W, Lee CW, Kim KK, Lee JH, Kim TD. Identification, characterization, immobilization, and mutational analysis of a novel acetylesterase with industrial potential (LaAcE) from Lactobacillus acidophilus. Biochim Biophys Acta - Gen Subj. 2018;1862:197–210. doi: 10.1016/j.bbagen.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Wei W, Ma J, Chen SQ, Cai XH, Wei DZ. A novel cold-adapted type I pullulanase of Paenibacillus polymyxa Nws-pp2: in vivo functional expression and biochemical characterization of glucans hydrolyzates analysis. BMC Biotechnol. 2015;15:96. doi: 10.1186/s12896-015-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicka-Woś A, Cieśliński H, Wanarska M, Kozłowska-Tylingo K, Hildebrandt P, Kur J. A novel cold-active β-d-galactosidase from the Paracoccus sp. 32d—gene cloning, purification and characterization. Microb Cell Fact. 2011;10:1–12. doi: 10.1186/1475-2859-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Lan D, Yang B, Wang Y. Biochemical properties and structure analysis of a DAG-like lipase from Malassezia globosa. Int J Mol Sci. 2015;16:4865–4879. doi: 10.3390/ijms16034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Guo GS, Ren GH, Liu YH. Production, characterization and applications of tannase. J Mol Catal B Enzym. 2014;101:137–147. [Google Scholar]
- Yayanos AA. High-pressure habitats. In: Schaechter M, editor. Encyclopedia of microbiology. 3. Academic Press; 2009. pp. 228–239. [Google Scholar]
- Yoo W, Le LTHL, Lee JH, Kim KK, Kim TD. A novel enantioselective SGNH family esterase (NmSGNH1) from Neisseria meningitides: characterization, mutational analysis, and ester synthesis. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:1438–1448. doi: 10.1016/j.bbalip.2019.07.007. [DOI] [PubMed] [Google Scholar]
- Yu P, Wang XT, Liu JW. Purification and characterization of a novel cold-adapted phytase from Rhodotorula mucilaginosa strain JMUY14 isolated from Antarctic. J Basic Microbiol. 2015;55:1029–1039. doi: 10.1002/jobm.201400865. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Hayashi A, Kitamura Y, Shimada T, Na R, Jin X. Purification and characterization of cold-adapted metalloprotease from deep sea water lactic acid bacteria Enterococcus faecalis TN-9. Int J Biol. 2009;1:12. [Google Scholar]
- Zavala-Páramo MG, Villa-Rivera MG, Lara-Márquez A, López-Romero E, Cano-Camacho H. Applications of fungal pectinases. In: Zaragoza O, Casadevall A, editors. Encyclopedia of mycology. Elsevier; 2021. pp. 316–325. [Google Scholar]
- Zeng R, Xiong P, Wen J. Characterization and gene cloning of a cold-active cellulose from a deep-sea psychrotrophic bacterium Pseudoalteromonas sp. DY3. Extremophiles. 2006;10:79–82. doi: 10.1007/s00792-005-0475-y. [DOI] [PubMed] [Google Scholar]
- Zhang N, Suen WC, Windsor W, Xiao L, Madison V, Zaks A. Improving tolerance of Candida antarctica lipase B towards irreversible thermal inactivation through directed evolution. Protein Eng. 2003;16:599–605. doi: 10.1093/protein/gzg074. [DOI] [PubMed] [Google Scholar]
- Zhang X, Hua M, Song C, Chi Z. Occurrence and diversity of marine yeasts in Antarctica environments. J Ocean Uni China. 2012;11:70–74. [Google Scholar]
- Zhang SY, Guo ZW, Wu XL, Ou XY, Zong MH, Lou WY. Recombinant expression and characterization of a novel cold-adapted type I pullulanase for efficient amylopectin hydrolysis. J Biotechnol. 2020;313:39–47. doi: 10.1016/j.jbiotec.2020.03.007. [DOI] [PubMed] [Google Scholar]
- Zheng H, Guo B, Chen XL, Fan SJ, Zhang YZ. Improvement of the quality of wheat bread by addition of glycoside hydrolase family 10 xylanases. Appl Microbiol Biotechnol. 2011;90:509–515. doi: 10.1007/s00253-011-3088-7. [DOI] [PubMed] [Google Scholar]