ABSTRACT
Infectious endocarditis (IE) is an uncommon disease with significant morbidity and mortality. The pathogenesis of IE has historically been described as a cascade of host-specific events beginning with endothelial damage and thrombus formation and followed by bacterial colonization of the nascent thrombus. Enterococcus faecalis is a Gram-positive commensal bacterial member of the gastrointestinal tract microbiota in most terrestrial animals and a leading cause of opportunistic biofilm-associated infections, including endocarditis. Here, we provide evidence that E. faecalis can colonize the endocardial surface without pre-existing damage and in the absence of thrombus formation in a rabbit endovascular infection model. Using previously described light and scanning electron microscopy techniques, we show that inoculation of a well-characterized E. faecalis lab strain in the marginal ear vein of New Zealand White rabbits resulted in rapid colonization of the endocardium throughout the heart within 4 days of administration. Unexpectedly, ultrastructural imaging revealed that the microcolonies were firmly attached directly to the endocardium in areas without morphological evidence of gross tissue damage. Further, the attached bacterial aggregates were not associated with significant cellular components of coagulation or host extracellular matrix damage repair (i.e. platelets). These results suggest that the canonical model of mechanical surface damage as a prerequisite for bacterial attachment to host sub-endothelial components is not required. Furthermore, these findings are consistent with a model of initial establishment of stable, endocarditis-associated E. faecalis biofilm microcolonies that may provide a reservoir for the eventual valvular infection characteristic of clinical endocarditis. The similarities between the E. faecalis colonization and biofilm morphologies seen in this rabbit endovascular infection model and our previously published murine gastrointestinal colonization model indicate that biofilm production and common host cell attachment factors are conserved in disparate mammalian hosts under both commensal and pathogenic contexts.
Keywords: Enterococcus faecalis, biofilms, endocarditis, animal models, microbiome, electron microscopy
Enterococcus faecalis forms microcolonies in vivo on undamaged rabbit endovasculature.
INTRODUCTION
Enterococcus faecalis is a Gram-positive commensal bacterial resident of the mammalian gastrointestinal (GI) tract and an opportunistic pathogen that causes persistent infections often associated with antimicrobial resistance (Ch'ng et al. 2019). The formation of biofilms—surface-associated bacteria enveloped by a bacterially-derived extracellular matrix (ECM)—occurs during E. faecalis colonization of the GI tract in a murine model (Barnes et al. 2017) and is a major virulence factor in the pathogenesis of enterococcal catheter-associated urinary tract infection and endocarditis (Singh, Nallapareddy and Murray 2007; Thurlow et al. 2010; Frank et al. 2013; Sillanpää et al. 2013; Kafil and Mobarez 2015; Madsen et al. 2017). We have previously reported that the E. faecalis biofilm microcolony morphology in an in vivo germ-free mouse model of GI colonization recapitulated the structural and ultrastructural forms observed in our reported in vitro and ex vivo models (Barnes et al. 2017). The goal of this work was to further investigate E. faecalis in vivo biofilm morphologies by extending our imaging studies to a rabbit model of endocarditis, which is a clinically relevant animal model of enterococcal infection.
Here, we define infectious endocarditis (IE) as an infection of the endothelium lining the heart and associated vasculature as proposed by Veltrop, Beekhuizen and Thompson (1999). The formation of a vegetation—a mass composed of bacteria, fibrin and platelets, at a site of endocardial damage, most often a damaged heart valve surface—is a classic clinical hallmark of IE. From a human disease standpoint, IE is uncommon, but affected patients are burdened with significant morbidity and mortality (Holland et al. 2016) and recently reported one year mortality rates are over 30% (Toyoda et al. 2017).
Microbiologically, staphylococci, streptococci and enterococci are the top three etiologic agents of IE (Liesman et al. 2017; Dahl et al. 2019); E. faecalis is reported to be responsible for ∼10% of all infective endocarditis cases (Murdoch et al. 2009; Chirouze et al. 2013; Baddour et al. 2015; Fernández-Hidalgo, Escolà-Vergé and Pericàs 2020). Notably, the etiology of infective endocarditis has been reported to be shifting towards an increased prevalence of enterococci as the causative agent in several recent regional reports (Fernández-Hidalgo, Escolà-Vergé and Pericàs 2020).
While the most prominent symptoms are focused in the heart, endocarditis is considered a multi-system disease with complications caused by vegetation embolization and hematogenous spread of the pathogenic microbe (Holland et al. 2016; Rezar et al. 2020). Disease onset can be acute, with rapid progression after primary bacteremia with valvular damage and vegetation formation common within a few days, or insidious, with patient presentations often evolving slowly over the course of weeks to months; this latter presentation is classically, though imprecisely, categorized as sub-acute endocarditis (McDonald 2009). While the broad clinical manifestations of endocarditis are largely a function of cardiac valve damage, endocarditis itself is simply microbial colonization of any of the endothelial surfaces.
The standard model of IE describes an ordered, stepwise progression leading to the pathophysiological state: (1) disruption or damage of the endothelial surface, (2) recruitment of host clotting factors and platelets, (3) colonization of the clot by bacterial adhesion and (4) bacterial microcolony and biofilm development on and within the nascent vegetation (Holland et al. 2016; Liesenborghs et al. 2020). Given this conceptual framework, most animal models of endocarditis use catheter-based systems in which the endothelium is mechanically disrupted to expose the sub-endothelial layer and stimulate a pro-thrombotic environment prior to bacterial inoculation (Freedman, Arnold and Valone 1974; Liesenborghs et al. 2020). Here, we present data demonstrating that, in a rabbit model of IE, colonization of the non-valvular cardiac endovascular surface by E. faecalis occurs even in the absence of endothelial disruption and without an apparent contribution by host cellular immune factors or platelets. These findings support a model in which enterococcal colonization can occur on the undamaged endothelium and may provide insight into the delay in development of clinical symptomatology in these naturally-occurring chronic infections.
MATERIALS AND METHODS
Reagents
Fixative, buffers and electron microscopy supplies and reagents were obtained from Electron Microscopy Sciences (Hatfield, PA), Alcian Blue 8GX was procured from Sigma-Aldrich (St. Louis, MO) and fluorescent labels (BODIPY-FL : vancomycin conjugate) were from ThermoFisher Scientific (Waltham, MA Todd-Hewitt Broth (THB), Brain Heart Infusion (BHI) Broth and Difco Granulated Agar (Becton, Dickinson and Company, Sparks, MD) were used for bacterial cultivation.
Bacterial growth conditions, animal infection inoculum preparation and strains
All bacterial strains were stored as glycerol stocks at −80°C. Bacterial cultures for rabbit infections were prepared in 20 mL THB in 125 mL baffled Erlenmeyer flasks inoculated either with an aliquot from the frozen glycerol stock or with three isolated colonies obtained by first streaking an aliquot from the frozen glycerol stock on BHI agar. Cultures were incubated overnight without agitation at 37°C in 5–7% CO2. Cells were pelleted and re-suspended in potassium phosphate-buffered saline (KPBS) to a final target concentration of 1 × 108 CFU/mL; actual concentrations were verified by plating serial dilutions of the inoculum on BHI agar. All BHI agar plates were incubated at 37°C in ambient air.
Derivatives of E. faecalis strain OG1RF (Dunny, Brown and Clewell 1978) were used throughout this work. Strains OG1RF(pCF10; Dunny, Funk and Adsit 1981), OG1RF(pCF10-GFP; previously referred to as pCF10-LC1; Cook et al. 2011), OG1RF CFP+ (Barnes et al. 2017) and OG1RF(pCF10ΔprgABUC; plasmid previously referred to as pCF10ΔprgA-C; Bhatty et al. 2015) have been previously described. Note that the inclusion of the CFP gene was incidental to the results reported here due to elevated autofluorescence in the host tissue. Except where indicated, the OG1RF strain in images shown herein was passaged one time through the rabbit model of endocarditis, as follows: aortic valve homogenate from a previously-infected rabbit was diluted 1:100 in THB and incubated overnight at 37°C in 5–7% CO2, from which an aliquot was mixed with an equal volume of 80% sterile glycerol and stored at −80°C. The animal-passaged strain was originally generated to address variation in aortic valve colonization observed in our standard infective endocarditis model in unrelated studies that were being conducted at the same time as the infections described herein. Use of the animal-passaged strain was the standard operating procedure for all rabbit endovascular experiments in our laboratory.
The animal-passaged strain and two other frozen stocks of OG1RF from our laboratory's historic repository were analyzed by whole genome sequencing (WGS). Strains were streaked on BHI agar from glycerol stocks stored −80°C; agar plates were then incubated at 37°C overnight. Single colonies of each strain were inoculated into 1 mL BHI, incubated at 37°C overnight, pelleted and stored at −20°C until extraction for genomic DNA with the Qiagen DNeasy Blood and Tissue kit (Germantown, MD). gDNA was eluted in Qiagen Buffer AE. Nextera XT library creation and Illumina HiSeq 2500 high-output mode paired end 125 bp were performed at the University of Minnesota Genomics Center (San Diego, CA). Illumina sequence reads of all three strains have been deposited at the NCBI Sequence Read Archive (BioProject Accession PRJNA723360). Sequence alignment and SNP identification were performed in Geneious Prime v.2020.2.4. A total of eight SNPs were identified in the animal-passaged OG1RF strain relative to the OG1RF reference sequence used (NCBI Reference Sequence: NC_017316.1). The same SNPs were also found in the genomes of the non-animal passaged OG1RF stocks, indicating that the animal-passaged OG1RF strain did not accrue any mutations as a result of the single in vivo passaging event.
Rabbit endovascular infection model
All animal experiments were approved by the University of Minnesota Institutional Animal Care and Use Committee (IACUC; protocol number 1211A24283). New Zealand White rabbits (1.8–2.3 kg) were obtained from a regional breeder (Bakkom Rabbitry, Viroqua, WI) or a national supplier of specific pathogen-free (SPF) animals (Charles River Laboratories, Wilmington, MA). Rabbits were housed singly with pellets, water and hay cubes available ad libitum; animals were allowed to acclimate for at least three days before use.
An established rabbit model of infective endocarditis was used as the basis for the endovascular infection model (Chuang et al. 2009). Animals from Bakkom Rabbitry or Charles River Laboratories underwent one or more steps, as indicated in the text, of the following surgical procedure to produce aortic valve damage prior to the induction of bacteremia.
Sedation
Rabbits were anesthetized with subcutaneous injection of 0.8 mL ketamine (100 mg/mL) and 0.4 mL xylazine (100 mg/mL). Sedated animals undergoing the surgical procedure were placed in a supine position, fur was removed from the neck region with clippers and the site was cleaned with betadine solution.
Anesthesia
Surgical plane anesthesia was maintained for the duration of the surgical procedure by subcutaneously boosting each animal with the sedation doses of ketamine and xylazine at regular intervals (generally 60 min).
Surgery
Once surgical plane anesthesia was verified in an animal, an ∼2 cm region of the left carotid artery was isolated and ligated distally with 3–0 silk (Ethicon, Somerville, NJ). Blood flow from the caudal direction was temporarily manually occluded, then the artery was nicked with a scalpel.
Cardiac catheterization
A length of clamped polyethylene catheter (OD 1.27 mm; BD Intramedic, Franklin Lakes, NJ) was advanced into the carotid artery until resistance was felt and the tubing pulsed, indicating placement adjacent to or across the aortic valve.
Mechanical ablation of the aortic valve
Two 3–0 silk ligatures were placed around the artery to anchor the tubing to induce mechanical damage to the aortic valve. Tubing was retracted after 2 h, the ligatures around the artery were tightened, and the skin was closed with simple interrupted sutures.
Bacteremia
Infection was initiated in post-procedure rabbits by injecting 2 mL of prepared E. faecalis into the marginal vein of either ear. The total bacterial load inoculated ranged from 4.6×107 to 2.2×108 CFU for animals from which images shown herein originated.
Post-surgical animals received 0.1 mL of buprenorphine (0.3 mg/mL) by subcutaneous injection shortly after surgery and twice a day thereafter for 3 days. Rabbits were euthanized by intravenous overdose of Beuthanasia-D Special at 4 days post-infection, except for rabbits from which hearts were imaged by optical microscopy, which were euthanized at 3 days post-infection. Hearts were removed upon necropsy for processing as described in separate sections below.
Control animals that underwent full anesthesia included (a) rabbits that underwent procedures through the Surgery step, were kept anesthetized but were not catheterized for the 2 h dwell time and were sham-infected with 2 mL sterile KPBS, and (b) rabbits that were fully anesthetized for the full duration of the catheterization procedure carried out in other animals in parallel, but which did not undergo any procedures and were injected with E. faecalis as described above. In addition, three rabbits that only underwent the Sedation step above and were not infected were used as unadulterated controls; two of these rabbits were euthanized, as described above, within 1 h of sedation and the third rabbit died under sedation due to unknown causes.
Two additional rabbits from Bakkom Rabbitry were directly made bacteremic as described above. These animals were not sedated or anesthetized and did not undergo any surgical procedures, but did receive a single subcutaneous injection of 0.1 mL of buprenorphine (0.3 mg/mL) on the day of infection. Rabbits were euthanized and necropsied as described above at 14 days post-infection.
Electron microscopy
Cardiac tissue samples were processed as previously described with minor modifications (Barnes et al. 2017), using a cationic-dye-based ECM stabilization technique originally pioneered by Erlandsen (Erlandsen et al. 2004). Briefly, explanted tissue was placed in ice cold KPBS immediately after necropsy (within 20 min of euthanasia) and kept cold until dissection (typically within 3 h). After dissection, samples were rinsed thoroughly in KPBS, then underwent primary fixation in a 2% paraformaldehyde, 2% glutaraldehyde solution with 0.15% Alcian Blue 8GX in a 150 mM sodium cacodylate buffer (Na-Cac) for 22 h at room temperature. After washing in Na-Cac (× 3), samples went through secondary fixation in partially-reduced 1% osmium tetroxide for 60 min, were washed again (× 3), then chemically dehydrated in a graded ethanol series (25–100%). Chemically dried tissue went through physical dehydration using a CO2-based critical point dryer (Tousimis, Inc., Rockville, MD). Fully dried samples were mounted on aluminum stubs with conductive carbon tape and coated in an argon plasma coater with either platinum (1–2 nm; VCR Inc., Burlingame, CA) or iridium (3–5 nm; ACE 600, Leica Microsystems, Inc., Buffalo Grove, IL).
Low-voltage scanning electron microscopy (LV-SEM) was done on a Hitachi SU-8230 using the mixed low-angle backscatter and secondary electron (SE) detector mode at 0.8 kV acceleration voltage (typical) using SE suppression as necessary to reduce excessive surface charging throughout except for supplemental images in Figure S1 (Supporting Information) where a Hitachi S-4700 (3 kV, mixed upper and lower SE detectors) was used. A liquid nitrogen-cooled decontaminator was used for all sample imaging and all images were collected and stored as lossless TIF data files.
Optical microscopy
Explanted cardiac tissue from rabbits infected with strain OG1RF(pCF10-GFP), which carries GFP on a large, low-copy number, conjugative plasmid, was rapidly cooled to 4°C and the aortic root was dissected and examined for GFP signal (see below). Due to high levels of interfering autofluorescence from the cardiac endothelium, the sample was immediately labeled with sub-inhibitory levels (0.5 µg/mL) of BODIPY-FL : vancomycin, a conjugated fluorescent probe, for 20 min at room temperature, rinsed in cold KPBS and the unfixed, hydrated tissue was immediately imaged using a confocal dissecting microscope. Fluorescent micrographs were obtained using an AZ100M motorized microscope (Nikon Instruments, Melville, NY) with an AZ-PlanApo 4× objective (NA: 0.4) and dual cameras; gross tissue images were taken with a DS-Ri1 color camera (Nikon). Immunofluorescent (IF) images were taken as a z-stack (n = 70) using Elements software and collected as ND2 files (v. 3.2, Nikon), the Nikon GFP filter set (Ex: 488 nm; Em: 520 nm) and a cooled CCD camera (CoolSNAP ES2, Photometrics, Tucson, AZ).
Samples processed for routine histology (H&E) and tissue Gram staining (Hucker–Twort, Newcomer Supply, Middleton, WI) were prepared by the Comparative Histology Shared Resource of the Masonic Cancer Center at the University of Minnesota. Histology imaging was done on an Axioplan.M1 using an AxioCam MRc5 (Carl Zeiss Microscopy, White Plains, NY) with either a 20× (NA: 0.75) or 100× (NA:1.3) objective through Zen (v 2.1; Carl Zeiss).
Image processing
All micrographs shown are representative and are presented with minimal corrections (histogram stretch, global contrast/brightness adjustments, etc.). IF micrographs were deconvolved using Huygens Pro (v. 19.0.4, SVI, Hilversum, NL); data are shown as a maximum intensity projection (MIP) of the full stack. Resulting MIPs were minimally processed for brightness and contrast using the FIJI package (Schindelin et al. 2012) of ImageJ (v. 1.52p; Schneider, Rasband and Eliceiri 2012) and cropped to illustrate the relevant features. Histologic images are the best focus plane nearest the most representative IF region. All adjustments were made in accordance with the Ethics and Digital Imaging guidelines of the Microscopy Society of America (Mackenzie et al. 2006).
Detection of enterococcal DNA in cardiac formalin fixed, paraffin embedded sections by quantitative PCR (qPCR)
Cardiac tissue sections from formalin fixed, paraffin embedded (FFPE) blocks from six rabbits were processed for total DNA isolation using the Quick-DNA/RNA FFPE Miniprep Kit (Zymo Research, Irvine, CA) in accordance with the instructions; two ∼5 µm thick sections were used per extraction. DNA samples were amplified using a standard iTaq Universal SYBR Green Supermix protocol on a CFX96 qPCR instrument (Bio-Rad, Hercules, CA) to verify the presence of bacterial DNA in rabbits inoculated with E. faecalis; negative controls (cardiac tissue from two uninfected rabbits) and standard qPCR controls were included. Paired experiments using both a universal 16S rRNA primer set (Forward (926F): AAACTCAAAKGAATTGACGG, Reverse (1062R): CTCACRRCACGAGCTGAC) and an E. faecalis specific 16S set (FaecalF: CGCTTCTTTCCTCCCGAGT; FaecalR: GCCATGCGGCATAAACTG) were conducted (Ryu et al. 2013; Yang et al. 2015).
RESULTS
Enterococcus faecalis rapidly colonizes undamaged endovascular surfaces in a rabbit model of aortic valve endocarditis
We previously used an established rabbit model of aortic valve IE to identify biofilm-associated virulence factors in the core genome of E. faecalis strain OG1RF (Frank et al. 2012, 2013). In this model, E. faecalis OG1RF reliably colonizes the aortic valve following ablation of the valve by endovascular catheter-based mechanical impingement to initiate vegetation formation (Fig. 1). We first used immunofluorescent microscopy to verify that the E. faecalis cells were metabolically active. The unfixed aortic valve and root from a heart harvested 3 days post-infection from a catheterized rabbit infected with E. faecalis that expressed GFP on the low-copy conjugative plasmid pCF10 was examined for fluorescence using a confocal dissecting microscope. However, no bacterial signal was detected above the background autofluorescence from the cardiac endothelium. Thus, immediately following tissue dissection, the entire tissue region was briefly labeled with a sub-inhibitory level of BODIPY-FL: vancomycin (∼20 min). Only bacterial cells capable of active vancomycin uptake and incorporation into the growing peptidoglycan structures of the cell well (i.e. only metabolically active Gram-positive cells) concentrate the dye sufficiently for detection. Following staining and rinsing in cold KPBS, the unfixed, hydrated tissue was immediately imaged. The unstained, brightfield image (Fig. 2A) shows the opened aortic root, with asterisks denoting the location of the now-removed aortic valve leaflets. The green fluorescent emission channel (Fig. 2B) shows both some residual tissue autofluorescence and multiple brighter, punctate regions suggestive of metabolically active enterococci. The boxed area shows one region of elevated signal; the higher magnification in the inset shows more detail. In the overlay of the fluorescent and brightfield images (Fig. 2C), it is notable that, while there are several areas with the punctate labeling pattern, there is apparently more labeling around the entrance to one of the cardiac arteries. Note that, as expected, histologic identification of small bacteria is difficult even when using a tissue Gram stain (Fig. 2D); microcolonies are essentially invisible on routine H&E-stained sections (Fig. 2D, inset).
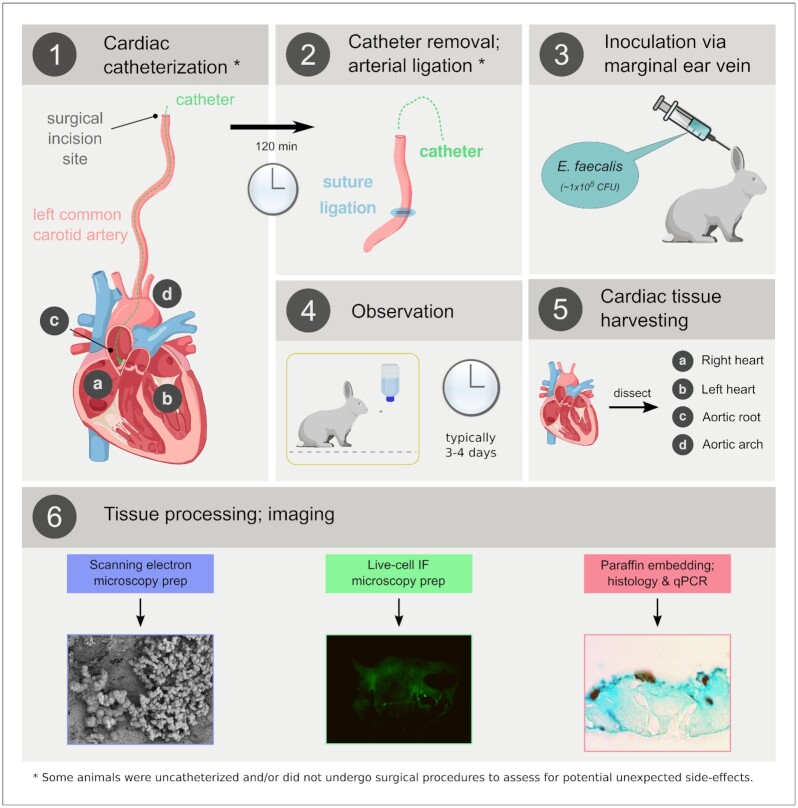
Figure 1.
Generalized established rabbit model of infectious endocarditis. (1) Anesthetized rabbits underwent surgery to expose the left common carotid artery. A catheter was inserted into the left carotid, advanced until it touched the aortic valve and was left in place for 120 min to physically ablate the valve surface. (2) Post-catheter removal, the artery was ligated and the surgical incision was irrigated and closed. (3) After closure of the surgical incision, a bolus of E. faecalis was injected into the marginal ear vein. (4) After 3–4 days of observation (see text), the animals were euthanized and the cardiac tissue removed (5) for processing (6) via a variety of downstream modalities. IF, immunofluorescence.
Figure 2.
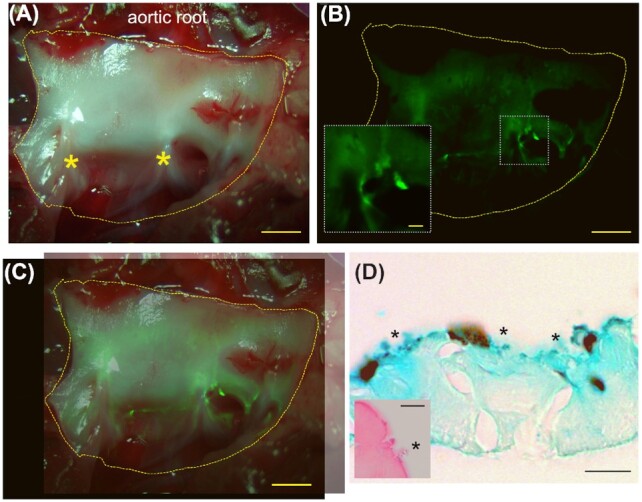
Gross immunofluorescent and histological labeling of E. faecalis microcolonies on the leporine cardiac endothelial surface. (A) Brightfield image of fresh, unfixed explanted rabbit cardiac tissue, including the endothelium adjacent to the aortic valve leaflets (bottom; asterisks denote where leaflets have been removed from in this image) and the aortic root (top). The region shown was labeled with BODIPY-FL conjugated to vancomycin for 20 min at a sub-inhibitory level for E. faecalis. Scale bar = 1 mm. (B) Green fluorescence channel of the valve imaged in (A). Deconvolution of the resulting image stack was able to remove the majority of the bulk tissue autofluorescence signal, highlighting the localized areas of bright BODIPY labeling indicative of live bacterial cells. Scale bar = 1 mm. Inset: Magnified section. Scale bar = 200 µm. (C) Overlay of the histological (A) and IF (B) images (the alignment is imperfect due to different camera frame sizes). (D) Tissue Gram stain (Hucker–Twort) of matched FFPE sections showing small, blue-staining objects immediately adjacent to the endocardial surface, consistent with Gram-positive cocci (asterisks). Scale bar = 10 µm. While microcolonies can occasionally be identified on H&E stained section (inset; asterisk), staining is typically quite poor. Scale bar of inset = 5 µm
Once characterized at the light microscopy level, we proceeded to image in vivo E. faecalis biofilms on the cardiac endothelial surface using SEM (Fig. 3A–D). LV-SEM revealed the presence of microcolonies composed of chains of diplococci juxtaposed with ECM widespread across the surface of the aortic root (Fig. 3C) in the hearts of endocarditis-model rabbits four days after valve ablation and subsequent induction of enterococcal bacteremia. Similar microcolonies were also visualized on the aortic arch (Fig. 3D), an area of the endovasculature that the catheter used for valve ablation was passed through during insertion. Notably, microcolonies were routinely located on endothelial surfaces without evidence of mechanical damage (Fig. 3C and D). This unanticipated observation raised the question of whether E. faecalis microcolonies might be found elsewhere in the heart, particularly in areas remote from where even accidental impingement by the catheter could have occurred. Indeed, microcolonies were found on intact endothelial surfaces in the right and left sides of the heart (Fig. 3A and B). While an animal-passaged OG1RF strain was used throughout these experiments, a non-passaged OG1RF strain showed equivalent formation of microcolonies with similar morphology and attachment (Figure S1, Supporting Information); as noted in Materials and Methods, the passaged strain accumulated no coding mutations as a result of the passaging as shown by WGS. Together, these results indicate that E. faecalis can colonize undamaged endothelial surfaces throughout the heart within 4 days in a rabbit model of aortic valve endocarditis.
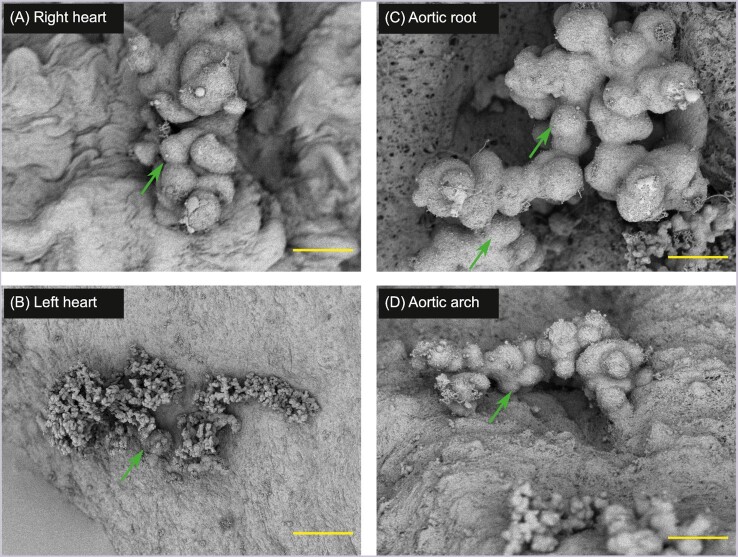
Figure 3.
Enterococcus faecalis colonization and microcolony development on cardiac endothelium throughout the rabbit heart and aorta 4 days after classical catheter ablation of the aortic valve and induction of bacteremia. Representative scanning electron micrographs from three different rabbits (panels A and B are from the same animal) that underwent catheter placement at the aortic valve for 120 min with subsequent induction of bacteremia are shown. Images are shown at a range of magnifications to demonstrate the large number of microcolonies that develop within 4 days, as well as the morphology of the microcolonies themselves. Note that microcolony formation readily occurs in areas distant from the site of mechanical damage, in regions without visible endothelial damage, platelet aggregation, or apparent recruitment of significant host coagulation factors. Additionally, no components of the host cellular immune response (i.e. neutrophils, macrophages) are observed. Green arrows highlight selected regions. Scale bars: A (25 µm); B (90 µm); C (500 µm) and D (25 µm).
Mechanical ablation of the aortic valve as performed in the standard rabbit model of endocarditis is not necessary for E. faecalis microcolony formation on undamaged endothelial surfaces throughout the heart
To further investigate the possibility that non-obvious mechanical damage may have contributed to the E. faecalis colonization observed in Fig. 3, we repeated the experiments in matched animals that were not catheterized and thus had no direct mechanical ablation (Fig. 4). These rabbits were fully anesthetized and either (condition 1) underwent the same surgical procedure up to, and including, nicking of the carotid artery as rabbits from Fig. 3 (Fig. 4C) or (condition 2) did not undergo any surgical procedures after being anesthetized (Fig. 4A, B and D). No bacteria were recovered from homogenized aortic valve leaflets removed from rabbits that underwent condition 1 (n = 2, including the rabbit from which the sample in Fig. 4C was collected) when an undiluted aliquot of the homogenate was plated, suggesting that catheter ablation of the aortic valve is required for E. faecalis OG1RF to colonize the valve at levels above the limit of detection of our CFU plating assay. Whole hearts from condition 2 rabbits were processed for imaging studies, so no CFU plating data were obtained. Regardless, microcolonies were again observed by LV-SEM throughout the right (Fig. 4A) and left (Fig. 4B) sides of the heart, as well as in the ascending aorta through the aortic arch (Fig. 4C) and into the descending aorta (Fig. 4D). Notably, no significant morphological differences relating to E. faecalis attachment, microcolony formation, or biofilm development compared to the catheterized condition in Fig. 3 were identified. Findings from SPF animals were indistinguishable from standard rabbits, and no microcolonies were found in uninfected controls (data not shown).
Figure 4.
Enterococcus faecalis attachment and microcolony development also occur in matched control animals that were anesthetized but not catheterized. In uncatheterized rabbits that otherwise underwent the same full duration of anesthesia and either no surgical procedures (panels A, B and D) or surgical procedures including terminal ligation and nicking of the carotid artery (panel C), enterococcal microcolony development throughout the endocardium proper and the adjacent aortic arch/root was indistinguishable from those seen in Fig. 3. Images are representative from three different rabbits (panels A and B are from the same animal). Note that the adjacent smooth round ‘holes'' seen in panel B and the crack or tear in the lower left portion of panel D are common processing artifacts (typically caused by uneven shrinkage) caused by variations in the chemical and physical dehydration stages. Green arrows highlight selected regions. Scale bars: A (2 µm); B (8 µm); C (200 µm) and D (3 µm)
Enterococcus faecalis microcolonies form and persist on cardiac endothelial surfaces in rabbits following induction of bacteremia without surgical intervention
To rule out the possibility that a process secondary to the sedation, anesthesia, or surgical procedures themselves might be manifesting microscopically as increased endothelial dysfunction sufficient to increase E. faecalis colonization even without gross disruption, we inoculated matched, unsedated rabbits via the marginal ear vein with the same strain at the same concentration as in the experiments described above. Thus, the only procedural manipulation performed on these rabbits was induction of enterococcal bacteremia itself. In order to also assess microcolony longevity, the rabbits were euthanized at 14 days post-infection. Again, the formation of microcolonies by E. faecalis throughout the cardiac endothelium was indistinguishable (Fig. 5) from the processes occurring in the rabbits that received full (Fig. 3) or minimal (Fig. 4) interventions.
Figure 5.
Enterococcus faecalis microcolonies persist on cardiac endothelial surfaces in rabbits given bacteremia without sedation. Biofilm microcolonies that were strikingly similar to those seen in Figs 3 and 4 were observed in rabbits followed out to 14 days post-inoculation. Rabbits showed no external signs of infection. Images are representative from a single rabbit. Green arrows highlight selected regions. Scale bars: A (5 µm); B (25 µm); C (3 µm) and D (5 µm)
Enterococcus faecalis microcolonies are present despite damage, presence of platelets, or innate immune system components
The standard model of bacterial colonization of the endothelial surface posits that prior damage to the endothelium leads to exposure of sub-endothelial factors, recruitment of activated coagulation cascade components (fibrin, fibronectin, etc.) and aggregation of platelets forming a nascent thrombus (Holland et al. 2016; Liesenborghs et al. 2020). This model proposes that it is only after this structure is formed that bacteria in the bloodstream can bind. Curiously, platelets and immune cells were consistently conspicuously absent in all LV-SEM images where microcolonies were observed (Figs 3–5, Figure S2A, Supporting Information and data not shown). Platelets are rarely found in or even near E. faecalis microcolonies (Figure S2B, Supporting Information); definitive neutrophils are so rarely associated that we have no good examples to show. Importantly, our SEM preparation methods are not simply destroying the more sensitive eukaryotic hematologic cells: fragile red blood cells are well-preserved in some fields of view (Figure S2, Supporting Information2).
Molecular detection of E. faecalis DNA in rabbit cardiac tissue sections
To provide an orthogonal method demonstrating the presence of enterococci associated with the cardiac endothelial surface, qPCR was used to detect the presence of E. faecalis-specific 16S rRNA gene sequences on FFPE sections from six of the rabbits imaged here. Molecular analysis of FFPE cardiac tissue from six individual rabbits using two independent primer sets (a universal 16S and an E. faecalis-specific set) run in triplicate demonstrated the presence of E. faecalis 16S DNA in 6/6 samples; 2/2 matched negative controls were appropriately negative, and all standard controls were as expected (Figure S3, Supporting Information).
DISCUSSION
The clinical disease state of endocarditis is focused on the functional changes secondary to bacterial damage to the cardiac valves: colonization of the valve and, typically, formation of a vegetation commonly leads to valvular insufficiency, a decrease in effective cardiac function and, eventually, the predictable patient symptoms, which vary by the causative microbe. Acute infective endocarditis is typically caused by staphylococci and presents as a rapidly-evolving, syndromic febrile disease that can be preceded or accompanied by sepsis. Conversely, chronic (sub-acute) IE is a more insidious disease most often caused by oral streptococci and enterococci that is characterized by weeks-to-months of general malaise, low-grade fevers and other nonspecific findings (McDonald 2009).
Regardless of the causative agent, the classical model of IE has presumed that the bacteria are late-comers to the process: abnormalities in the endothelial surface—congenital structural abnormalities (e.g. bicuspid aorta), mechanical damage to the endothelial surface (e.g. endothelial scarring from rheumatic fever, abrasive foreign material in IV drug use), artificial valves, etc.—lead to activation of the coagulation cascade and recruitment of platelets and other thrombogenic factors (Veltrop, Beekhuizen and Thompson 1999; McDonald 2009; Holland et al. 2016). After the fibrin clot structures form, patients with bacteremia are at risk of colonization of these clots, formation of a vegetation and eventually endocarditis. The data presented here suggest that this model is incomplete for E. faecalis colonization of the endovascular surface.
Notably, while much of the clinical literature published over the past 50 years has insisted on the requirement for pre-existing endothelial damage (Keynan and Rubinstein 2013), a detailed reading of the historic literature both supports animal models in which no endothelial damage is required (Jones 1969) and indicates that the use of animal models using mechanical ablation increases vegetation rates and decreases the number of experimental animals needed for a given investigation (Durack, Beeson and Petersdorf 1973; Durack and Beeson 1972a and b). Given these results, it can be inferred that the routine use of catheter ablation prior to bacterial inoculation in animal models of endocarditis is, at least in part, primarily a function of experimental convenience and efficiency. Indeed, while preexisting endothelial damage or structural cardiac abnormalities in human patients likely increase the rate of bacterial colonization and eventual endocarditis, endothelial damage is not itself a prerequisite for bacterial attachment to the endothelium (Tunkel and Scheld 1992). Even in cases where prior endothelial damage is not required, however, the process is still widely reported to require an active, host-derived thrombus to provide a nidus for bacterial colonization (McDonald 2009). Here we demonstrate that E. faecalis endocardial surface colonization in a rabbit endovascular infection model shows similar patterns of microcolony distribution and morphologies without an apparent requirement for prior endothelial damage and lacking many of the cellular host response factors (i.e. platelets and thrombus formation) previously reported in multiple classical IE model systems.
Using previously described correlative light and SEM—including cationic dye-based ECM stabilization to prevent biofilm loss during processing and high-intensity imaging—we show in this work that one-time inoculation of a well-characterized plasmid-free E. faecalis lab strain (OG1RF) in the marginal ear vein of New Zealand White rabbits resulted in rapid colonization of the endocardium throughout the heart and through the aortic arch within four days of administration (Figs 3 and 4), contrasting sharply with the primary cardiac valve colonization location more commonly reported in similar endocarditis animal models (Keynan and Rubinstein 2013). In addition to the SEM micrographs showing structures strongly consistent with the E. faecalis biofilm microcolony architecture phenotype that we have previously reported in several in vitro and in vivo model systems (Kristich et al. 2008; Barnes et al. 2012, 2017; Frank et al. 2013; Dale et al. 2015, 2017; Korir, Dale and Dunny 2019), the uptake of fluorescently-tagged vancomycin into unfixed cardiac tissue explants (a marker for active peptidoglycan synthesis; Fig. 2A–C) and the presence of small blue cocci on tissue Gram stains (Fig. 2D) all directly support a clear bacterial origin for these structures. Molecular detection via qPCR was also used as an orthogonal detection method: enterococcal DNA was detected in cardiac tissue from infected animals but not in uninfected controls.
The propensity for E. faecalis colonization of the endothelial surface independent of mechanical damage from direct catheter-induced aortic valve abrasion, surgical manipulation and even sedation/anesthesia, suggests that E. faecalis is able to directly colonize the undamaged endothelial surface in the otherwise healthy rabbits used in this model system (Fig. 6A). This hypothesis was also supported from an ultrastructural perspective (Figs 3–5 and Figure S2, Supporting Information): the microcolonies appeared to firmly attach directly to the endocardium without microscopic evidence of adjacent tissue damage. Further, the attached bacterial aggregates were not associated with significant cellular components of coagulation or host ECM damage repair. Thus, the results reported here suggest that neither the standard model of mechanical surface damage as a prerequisite for bacterial attachment to host sub-endothelial components (Fig. 6B, top), nor the involvement of a host cellular innate immune response associated with the nascent biofilm, may be absolute. Given these findings, we propose a second, simpler model through which E. faecalis can directly colonize endovascular epithelium (Fig. 6B, bottom): colonization can occur on native, undisturbed endothelium without active involvement of the cellular components of the host coagulation or innate immune system.
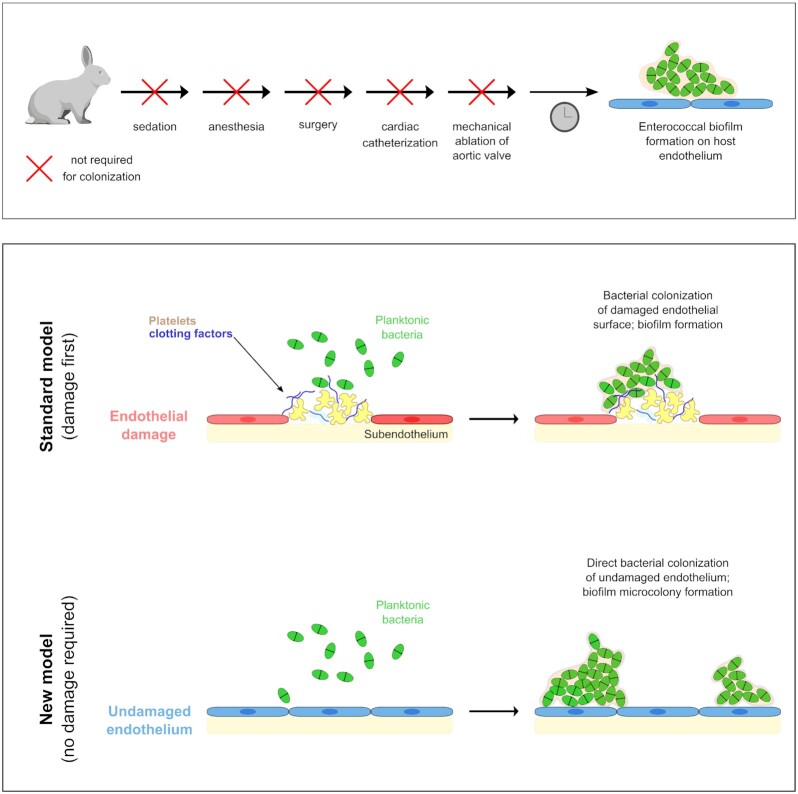
Figure 6.
Summary of experiment variations and proposed model of E. faecalis colonization of undamaged endovasculature. Disruption of the endothelial surface and exposure of the sub-endothelium are not required for E. faecalis surface colonization and microcolony development. In addition, sequential removal of possible inflammatory interventions caused by surgical treatment or even sedation/anesthesia was also not necessary, as peripheral inoculation of E. faecalis was still sufficient to cause endothelial colonization. Bacterial colonization of the cardiac valvular surface—and by extension, vascular endothelial tissue—has classically been described as requiring an initial surface insult/damage to the endothelium (top): this model suggested that colonization occurred at the interface of the clot in a rabbit endocarditis model system. Here, we propose a model in which E. faecalis is able to directly colonize the intact endothelium (bottom) without requiring host coagulation cascade factors, platelets or access to the sub-endothelial layer.
The microcolonies formed on the endothelium appear essentially identical to the E. faecalis microcolonies seen in a wide range of our previously published work (summarized in Fig. 7). This conservation across diverse surfaces, conditions and media types suggests that E. faecalis surface colonization is a fundamental part of its lifestyle. Since endothelium can be viewed as a specialized form of epithelium (Dyer and Patterson 2010), the similarities between the E. faecalis colonization and biofilm morphologies seen in our recently published murine GI model (Barnes et al. 2017) and those in this rabbit endocardial system suggest that biofilm production and common host cell attachment factors are conserved in disparate mammalian hosts under both commensal and pathogenic pressures; we are currently investigating this question via a variety of molecular expression techniques. Whether the factors that are involved in effective colonization and expansion on undamaged surfaces are similar to the well-characterized attachment factors involved in adherence to host factors exposed during damage to endothelial and epithelial surfaces (Rozdzinski et al. 2001) also remains an open question. In addition, soluble coagulation factors (such as fibrinogen/fibrin) may play an important role in this attachment process—an issue which itself deserves further study. Soluble fibrin/fibrinogen itself (or via interactions with fibronectin) may be involved in modification of the enterococcal cell surface prior to endothelial or valvular attachment, as it is in Staphylococcus aureus (Tong et al. 2015). Finally, the lack of a pronounced cellular host immune response suggests that E. faecalis is able to efficiently evade the humoral immune response for extended periods of time. While in vitro evidence for this evasion exists (Park et al. 2007), further characterization in animal models is needed. All of these areas of investigation would benefit from a genome-wide interrogation of critical determinants of endothelial attachment and innate immune evasion.
Figure 7.
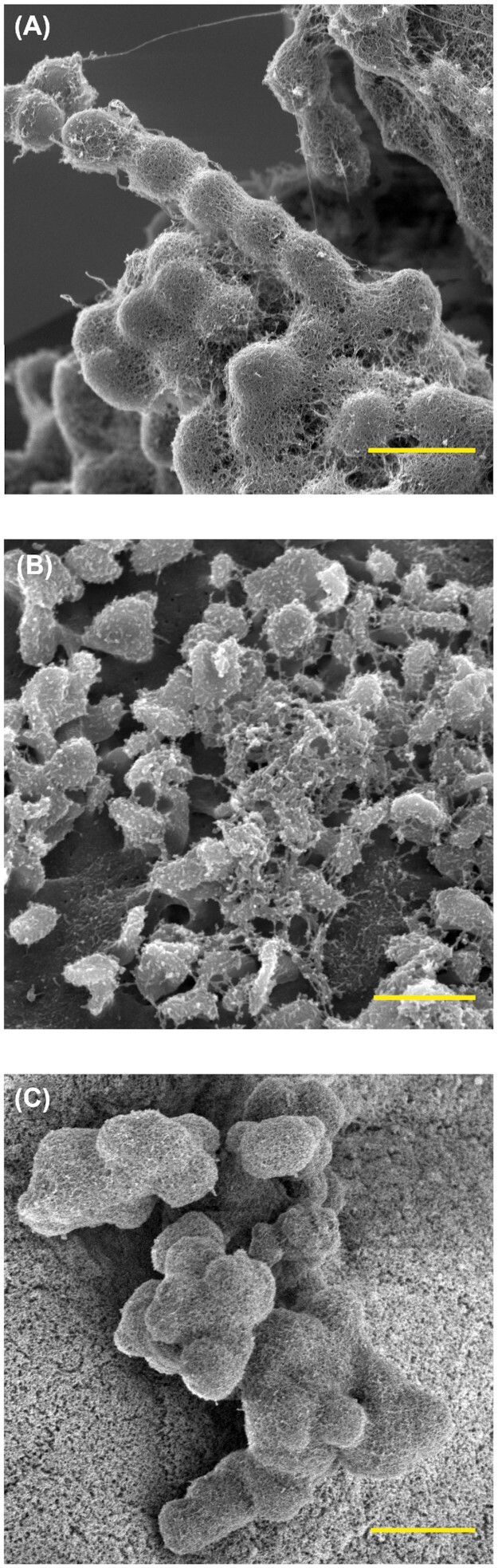
Conservation of E. faecalis microcolony morphology by SEM under a wide range of experimental conditions, from in vitro growth on fluoropolymer surfaces to in vivo model systems. (A) An in vitro E. faecalis biofilm formation on Aclar fluoropolymer membranes (8 h post-inoculation). Scale bar = 1 µm (B)Ex vivo E. faecalis biofilm formation on porcine cardiac valves (4 h). Scale bar = 2 µm (C)In vivo colonization of the germ-free murine GI tract at 96 h. Scale bar = 5 µm
One of the vagaries of enterococcal endocarditis remains the question of primary infection source: where do the enterococci come from? While the cardiac symptoms of E. faecalis endocarditis are largely driven by direct damage of the valve tissue leading to severely decreased cardiac output, the source of the bacteria involved has been presumed to be from direct bacteremic colonization. Translocation of enterococci—primarily, but not exclusively, E. faecalis—across the GI epithelium and into other organs and the lymphatic system has been previously reported both in clinical case reviews (Ceci et al. 2015; Stanley et al. 2016; Khan, Siddiqui and Saif 2018) and in a wide range of animal model systems under a number of stress conditions (Wells, Jechorek and Erlandsen 1990; Mason et al. 2011; Knoop et al. 2016), suggesting that enterococci may have effective (but poorly understood) invasion mechanisms consistent with eventual vascular colonization. Consistent with this, recent work by Brown et al.(2021) has shown that peritoneal inoculation of E. faecalis in a mouse model system can lead to cardiac microlesions, though the specific mechanism may be different there. In addition, work by Thurlow et al. (2010) further supports that non-valvular colonization is a significant factor since cardiac tissue homogenates still showed grossly elevated bacterial loads even after removal of the infected valve.
Here, we present data on the colonization of non-valvular surfaces, which are areas that would have likely been considered outside the region of interest in many previous studies. The data in this study demonstrating that E. faecalis is able to attach to undamaged endothelium without eliciting an apparent immediate cellular immune response by the host suggests the possibility of cryptic, long-term infections that may be relevant in human endocarditis or other disease states. While our current data do not demonstrate that the observed colonization of undamaged endothelium can proceed to the formation of distinct vegetations, our findings are consistent with a model of initial establishment of stable, endothelial-associated E. faecalis biofilm microcolonies that may provide a reservoir for eventual valvular infection.
FUNDING
Parts of this work were carried out in the Characterization Facility, University of Minnesota, which receives partial support from the National Science Foundation (NSF) through the MRSEC (award number DMR-2011401) and the NNCI (award number ECCS-2025124) programs. Fluorescent microscopy was done using equipment at the University of Minnesota University Imaging Centers (UIC; SCR_020997). Tissue sectioning and routine histological staining were done at the Comparative Pathology Shared Resource (CPSR) of the Masonic Cancer Center at the University of Minnesota. This work was supported by the National Institutes of Health, grants GM049530, GM118079, AI120601, AI058134 and AI105454 to GMD. A training fellowship to JLD was provided by T90 DE022732 from the National Institute of Dental and Craniofacial Research. AMTB received support via NIH training grants HL007741 and AI055433 for portions of this work. KLF is supported by NIH/NIAID award AI141961 and Uniformed Services University award R0733973.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Anne-Marie Leuck, M.D., for useful discussions and assistance with endocarditis experiments.
These results were presented in part at Lab Medicine 2017—the 52nd annual meeting of the Academy of Clinical Laboratory Physicians and Scientists; an abstract of those findings can be found as Barnes A, Frank K, Dale J, Manias D, Nilson J and Dunny G. 2018. Enterococcus faecalis colonizes and forms persistent biofilm microcolonies on the undamaged endothelial surface in a rabbit endovascular infection model. Am J Clin Pathol 149: S164–S165.
The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of any section of the National Institutes of Health, the Department of Defense, the Uniformed Services University of the Health Sciences or any other agency of the U.S. Government. The funding agencies had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Contributor Information
Aaron M T Barnes, Department of Microbiology and Immunology, University of Minnesota School of Medicine, Minneapolis, MN 55455, USA; Department of Laboratory Medicine and Pathology, University of Minnesota School of Medicine, Minneapolis, MN 55455, USA.
Kristi L Frank, Department of Microbiology and Immunology, University of Minnesota School of Medicine, Minneapolis, MN 55455, USA; Department of Microbiology and Immunology, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
Jennifer L Dale, Department of Microbiology and Immunology, University of Minnesota School of Medicine, Minneapolis, MN 55455, USA.
Dawn A Manias, Department of Microbiology and Immunology, University of Minnesota School of Medicine, Minneapolis, MN 55455, USA.
Jennifer L Powers, Department of Microbiology and Immunology, University of Minnesota School of Medicine, Minneapolis, MN 55455, USA.
Gary M Dunny, Department of Microbiology and Immunology, University of Minnesota School of Medicine, Minneapolis, MN 55455, USA.
Conflicts of interests
None declared.
REFERENCES
- Baddour LM, Wilson WR, Bayer ASet al. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals from the American Heart Association. Circulation. 2015;132:1435–86. [DOI] [PubMed] [Google Scholar]
- Barnes AMT, Ballering KS, Leibman RS. et al. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio. 2012;3:e00193–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AMT, Dale JL, Chen Y. et al. Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence. 2017;8:282–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatty M, Cruz MR, Frank KL. et al. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol. 2015;95:660–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AO, Singh KV, Cruz MRet al. Cardiac microlesions form during severe bacteremic Enterococcus faecalis infection. J Infect Dis. 2021;223:508–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci M, Delpech G, Sparo Met al. Clinical and microbiological features of bacteremia caused by Enterococcus faecalis. J Infect Dev Count. 2015;9:1195–203. [DOI] [PubMed] [Google Scholar]
- Ch'ng J-H, Chong KKL, Lam LNet al. Biofilm-associated infection by enterococci. Nat Rev Microbiol. 2019;17:82–94. [DOI] [PubMed] [Google Scholar]
- Chirouze C, Athan E, Alla Fet al. Enterococcal endocarditis in the beginning of the 21st century: analysis from the International Collaboration on Endocarditis-Prospective Cohort Study. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2013;19:1140–7. [DOI] [PubMed] [Google Scholar]
- Chuang ON, Schlievert PM, Wells CLet al. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun. 2009;77:539–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook L, Chatterjee A, Barnes Aet al. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol. 2011;81:1499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl A, Iversen K, Tonder Net al. Prevalence of infective endocarditis in Enterococcus faecalis bacteremia. J Am Coll Cardiol. 2019;74:193–201. [DOI] [PubMed] [Google Scholar]
- Dale JL, Cagnazzo J, Phan CQet al. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob Agents Chemother. 2015;59:4094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JL, Nilson JL, Barnes AMTet al. Restructuring of Enterococcus faecalis biofilm architecture in response to antibiotic-induced stress. npj Biofilms and Microbiomes. 2017;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G, Funk C, Adsit J. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid. 1981;6:270–8. [DOI] [PubMed] [Google Scholar]
- Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci. 1978;75:3479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack DT, Beeson PB, Petersdorf RG. Experimental bacterial endocarditis. 3. Production and progress of the disease in rabbits. Br J Exp Pathol. 1973;54:142–51. [PMC free article] [PubMed] [Google Scholar]
- Durack DT, Beeson PB. Experimental bacterial endocarditis. I. Colonization of a sterile vegetation. Br J Exp Pathol. 1972a;53:44–9. [PMC free article] [PubMed] [Google Scholar]
- Durack DT, Beeson PB. Experimental bacterial endocarditis. II. Survival of a bacteria in endocardial vegetations. Br J Exp Pathol. 1972b;53:50–3. [PMC free article] [PubMed] [Google Scholar]
- Dyer L, Patterson C. Development of the endothelium: an emphasis on heterogeneity. Semin Thromb Hemost. 2010;36:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsen SL, Kristich CJ, Dunny GMet al. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J Histochem Cytochem. 2004;52:1427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Hidalgo N, Escolà-Vergé L, Pericàs JM. Enterococcus faecalis endocarditis: what's next?. Fut Microbiol. 2020;15:349–64. [DOI] [PubMed] [Google Scholar]
- Frank KL, Barnes AMT, Grindle SMet al. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect Immun. 2012;80:539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KL, Guiton PS, Barnes AMTet al. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis. Infect Immun. 2013;81:1696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman LR, Arnold S, Valone J. Experimental Endocarditis. Ann N Y Acad Sci. 1974;236:456–65. [DOI] [PubMed] [Google Scholar]
- Holland TL, Baddour LM, Bayer ASet al. Infective endocarditis. Nat Rev Dis Primers. 2016;2:16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JET. The experimental production of streptococcal endocarditis in the pig. J Pathol. 1969;99:307–18. [DOI] [PubMed] [Google Scholar]
- Kafil HS, Mobarez AM. Spread of enterococcal surface protein in antibiotic resistant Enterococcus faecium and Enterococcus faecalis isolates from urinary tract infections. Open Microbiol J. 2015;9:14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keynan Y, Rubinstein E. Pathophysiology of infective endocarditis. Curr Infect Dis Rep. 2013;15:342–6. [DOI] [PubMed] [Google Scholar]
- Khan Z, Siddiqui N, Saif MW. Enterococcus faecalis infective endocarditis and colorectal carcinoma: case of new association gaining ground. Gastroenterol Res. 2018;11:238–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop KA, McDonald KG, Kulkarni DHet al. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016;65:1100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korir ML, Dale JL, Dunny GM. Role of epaQ, a previously uncharacterized Enterococcus faecalis gene, in biofilm development and antimicrobial resistance. J Bacteriol. 2019;201:e00078–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich CJ, Nguyen VT, Le Tet al. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl Environ Microbiol. 2008;74:3377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesenborghs L, Meyers S, Vanassche Tet al. Coagulation: at the heart of infective endocarditis. J Thromb Haemost. 2020;18:995–1008. [DOI] [PubMed] [Google Scholar]
- Liesman RM, Pritt BS, Maleszewski JJet al. Laboratory diagnosis of infective endocarditis. J Clin Microbiol. 2017;55:2599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Burke MG, Carvalho Tet al. Ethics and digital imaging. Microsc Tod. 2006;14:40–1. [Google Scholar]
- Madsen KT, Skov MN, Gill Set al. Virulence factors associated with Enterococcus faecalis infective endocarditis: a mini-review. Open Microbiol J. 2017;11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason KL, Stepien TA, Blum JEet al. From commensal to pathogen: translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta. mBio. 2011;2:e00065–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JR. Acute infective endocarditis. Infect Dis Clin North Am. 2009;23:643–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch DR, Corey GR, Hoen Bet al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the international collaboration on endocarditis-prospective cohort study. Arch Intern Med. 2009;169:463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Kim KM, Lee JHet al. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immun. 2007;75:1861–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezar R, Lichtenauer M, Haar Met al. Infective endocarditis - a review of current therapy and future challenges. Hell J Cardiol HJC Hell Kardiologike Epitheorese. 2020. DOI: 10.1016/j.hjc.2020.10.007. [DOI] [PubMed] [Google Scholar]
- Rozdzinski E, Marre R, Susa Met al. Aggregation substance-mediated adherence of Enterococcus faecalis to immobilized extracellular matrix proteins. Microb Pathog. 2001;30:211–20. [DOI] [PubMed] [Google Scholar]
- Ryu H, Henson M, Elk Met al. Development of quantitative PCR assays targeting the 16s rRNA genes of Enterococcus spp. and their application to the identification of Enterococcus species in environmental samples. Appl Environ Microbiol. 2013;79:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise Eet al. FIJI: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää J, Chang C, Singh KVet al. Contribution of individual Ebp pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLoS ONE. 2013;8:e68813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis. 2007;195:1671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Mason LJ, Mackin KEet al. Translocation and dissemination of commensal bacteria in post-stroke infection. Nat Med. 2016;22:1277–84. [DOI] [PubMed] [Google Scholar]
- Thurlow LR, Thomas VC, Narayanan Set al. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun. 2010;78:4936–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SYC, Davis JS, Eichenberger E. et al. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda N, Chikwe J, Itagaki Set al. Trends in infective endocarditis in California and New York state, 1998-2013. JAMA. 2017;317:1652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunkel A, Scheld W. Experimental models of endocarditis. In: Kaye D (ed). Infective Endocarditis. 2nd edn, New York, NY: Raven Press, 1992, 37–56. [Google Scholar]
- Veltrop MH, Beekhuizen H, Thompson J. Bacterial species- and strain-dependent induction of tissue factor in human vascular endothelial cells. Infect Immun. 1999;67:6130–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CL, Jechorek RP, Erlandsen SL. Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J Infect Dis. 1990;162:82–90. [DOI] [PubMed] [Google Scholar]
- Yang Y-W, Chen M-K, Yang B-Yet al. Use of 16S rRNA gene-targeted group-specific primers for Real-Time PCR analysis of predominant bacteria in mouse feces. Appl Environ Microbiol. 2015;81:6749–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.