Abstract
Phosphoinositides are lipids that play a critical role in processes such as cellular signalling, ion channel activity and membrane trafficking. When mutated, several genes that encode proteins that participate in the metabolism of these lipids give rise to neurological or developmental phenotypes. PI4KA is a phosphoinositide kinase that is highly expressed in the brain and is essential for life. Here we used whole exome or genome sequencing to identify 10 unrelated patients harbouring biallelic variants in PI4KA that caused a spectrum of conditions ranging from severe global neurodevelopmental delay with hypomyelination and developmental brain abnormalities to pure spastic paraplegia. Some patients presented immunological deficits or genito-urinary abnormalities. Functional analyses by western blotting and immunofluorescence showed decreased PI4KA levels in the patients’ fibroblasts. Immunofluorescence and targeted lipidomics indicated that PI4KA activity was diminished in fibroblasts and peripheral blood mononuclear cells. In conclusion, we report a novel severe metabolic disorder caused by PI4KA malfunction, highlighting the importance of phosphoinositide signalling in human brain development and the myelin sheath.
Keywords: PI4KA, phosphoinositol, inborn errors of metabolism, hypomyelinating leukodystrophy, spastic paraplegia
Verdura et al. report a novel rare brain metabolic disorder caused by recessive mutations in PI4KA, which encodes an enzyme with a pivotal role in phosphoinositide metabolism at the cell membrane. The description of this syndrome will simplify the identification of undiagnosed cases with similar clinical features.
Introduction
Phosphoinositide lipids at the plasma membrane, which are important determinants of membrane identity, play roles in cell signalling, controlling cell shape and motility and thus also affect the interaction of cells with their environment.1 Phosphoinositide metabolism at the plasma membrane begins with the phosphorylation of phosphatidylinositol (PI) to PI(4)P, which is later further phosphorylated to PI(4,5)P2 and PI(3,4,5)P3. This first step is catalysed by phosphatidylinositol 4 kinase A (PI4KA, MIM *600286) and is regulated by FAM126A and TTC7, which are subunits of the same complex.2,3 Animal models in which PI4KA homologues are inactivated show profound abnormalities: downregulation of Pi4ka expression in zebrafish leads to multiple developmental defects affecting the brain, heart, trunk and most prominently the loss of pectoral fins, while the Pi4ka orthologue knockout is lethal in flies, mice and yeast.4-7
It is intriguing that pathogenic variants in FAM126A, a gene that encodes another partner of the PI4KA/TTC7/FAM126A protein complex, cause hypomyelinating leukodystrophy [hypomyelination and congenital cataracts(HCC); MIM #610532], with the main pathogenic mechanism of which is defective PI(4)P production in oligodendrocytes, a process coregulated by the myelin basic protein (MBP).2,8 Notably, mutations in other genes that encode proteins that are involved in PI metabolism are associated with neurodevelopmental disorders. For instance, variants in the PIK3CA, PIK3R2, AKT3, and FIG4 genes have been linked to the development of polymicrogyria, which can be associated or not with megalencephaly and capillary malformation,5,9–12 and PI4K2A variants have been associated with intellectual disability and epilepsy.13
In this study, we identified 10 patients from unrelated families carrying biallelic variants in PI4KA that caused a spectrum of conditions ranging from severe global neurodevelopmental delay with hypomyelination/delayed myelination and developmental brain abnormalities to pure spastic paraplegia. Analysis of protein structure by 3D modelling and functional studies using western blotting, immunofluorescence and targeted lipidomic studies of the PI4KA pathway in patient cells were performed to confirm the pathogenicity of the identified variants.
Materials and methods
Genetic studies and variant assessment
We identified PI4KA variants in probands by whole-exome or whole-genome sequencing in clinical diagnostic or research settings. Candidate variants were validated by Sanger sequencing and tested for co-segregation in all family members except for Patient 5’s mother (unavailable). The Genome Aggregation Database (gnomAD v.2.1.1; https://gnomad.broadinstitute.org/; accessed 30 November 2020) was used to determine variant frequency in control populations. Variants were annotated with ANNOVAR. High-quality variants with an effect on the coding sequence or splice site regions with a frequency lower than 0.01 were retrieved from public databases (gnomAD and in-house databases). The functional impact of variants was analysed with various prediction tools, including PolyPhen-2, M-CAP, CADD, MutationTaster and LRT pred. Sequence alignment was performed using ClustalOmega (https://www.ebi.ac.uk/tools/msa/clustalo/; accessed 30 November 2020) with sequences extracted from NCBI databases. The PI4KA complex (6bq1 template from the RCSB PDB database)3 was visualized with PyMOL version 2.2.3 (https://www.pymol.org; accessed 30 November 2020).
Patient recruitment
We obtained genotypic and phenotypic information from patients with probable pathogenic PI4KA variants (after in silico criteria), from different hospitals identified with GeneMatcher.14 All patients were analysed by the neurologists and/or clinical geneticists of their respective referral centre, who determined the paraclinical analyses to be performed in each case. We reviewed clinical information related to neurodevelopment, growth parameters, neurological manifestations, behaviour, dysmorphology, and the results of other exams. Hypomyelination was diagnosed when neuroimaging evidenced mildly elevated hyperintensity of most cerebral white matter in T2-weighted images and mild hypointensity, isointensity or mild hyperintensity relative to the cortex in T1-weighted images.15 Blood samples were obtained using standard methods. Written informed consent for genetic testing and publication of the clinical information, including clinical pictures, was obtained from the parents or legal guardians of each patient according to the Declaration of Helsinki.
The research project was approved by the Clinical Research Ethics Committees of IDIBELL Institute (Patients 1 and 9, PR076/14), VHIR Institute [Patient 2, PR(AG)223/2017], CHU Bordeaux (Patient 4, Comité de Protection des Personnes Bordeaux—Outre Mer III), the Broad Institute (Patient 5, Partners IRB Protocol #:2016P001422), the University of Washington (Patient 6, #28853), Città della Salute e della Scienza University Hospital (Patient 7, n. 0060884), the Comitato Etico Brianza (Patient 8; Monza, Italy; PID-GENMET), the IIGM Institute (Patient 10, Comité de Ética de la Investigación con Medicamentos, CEIm) and the Montpellier Local Ethics Committee (Patient 3).
Western blotting
Human fibroblasts were homogenized in RIPA buffer (150 mM NaCl, 1% NonidetTM P40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0), sonicated for 1 min at 4°C, centrifuged for 10 min at 1000g mixed with 4× NuPAGE LDS Sample Buffer (Invitrogen) and heated for 5 min at 100°C. Protein concentrations were quantified using a BCA protein assay kit (Thermo Fisher Scientific). Twenty-five micrograms of each protein sample was subjected to polyacrylamide gel electrophoresis for 30 min at 100 V and for 1 h at 120 V in NuPAGE MOPS SDS Running Buffer (Invitrogen) supplemented with 5 mM sodium bisulfite (Ref. 243973, Sigma-Aldrich). The proteins were transferred to nitrocellulose membranes. After blocking with 5% bovine serum albumin (BSA, Sigma-Aldrich) in 0.05% TBS-Tween (TBS-T) for 1 h at room temperature, the membranes were incubated with primary antibodies for 2 h at room temperature. Following incubation with secondary antibodies for 1 h at room temperature, the proteins were detected with the Chemidoc™ Touch Imaging System (Bio-Rad). The bands were quantified with ImageLab (Bio-Rad). The primary antibodies that were used were anti-PI4KA (12411-1-AP Proteintech) and anti-β-Actin (A2228, Sigma). The secondary antibodies that were used were polyclonal goat anti-mouse (P0447, Dako Cytomation) and polyclonal goat anti-rabbit (P0448, Dako Cytomation). The unmodified full-length blot is shown in Supplementary Fig. 3.
Immunofluorescence
A total of 150 000 cells were seeded on coverslips in wells containing 2 ml of 10% formalin and fixed in for 30 min at room temperature. To permeabilize and block the cells, the coverslips were incubated for 20 min at room temperature in blocking buffer (1% BSA, 0.2% powdered milk, 2% NCS, 0.1 M glycine, and 0.1% TritonTM X-100). The cells were immunostained with primary antibodies for 2 h at room temperature. Following incubation with secondary antibodies for 1 h at room temperature, the slides were mounted using Mowiol®. Confocal images were acquired using a Leica TCS SL laser scanning confocal spectral microscope (Leica Microsystems) and analysed with ImageJ (NIH, USA). The primary antibodies that were used were anti-PI4KA (12411-1-AP, Proteintech) and anti-PI(4)P (Z-P004, Echelon Biosciences Inc.). The secondary antibodies that were used were Alexa Fluor® 555-conjugated goat anti-rabbit IgG (A-21428, Invitrogen) and Alexa Fluor® 488-conjugated goat anti-mouse IgG (A-11001, Invitrogen).
Lipidomics analysis
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood using standard methods. Quantification of phosphoinositide species was performed at ATK-Analytics Innovation and Discovery using 200 μg of TCA-precipitated PBMCs as the starting material. Lipids were extracted and derivatized using TMS-Diazomethane prior to mass spectrometry, as described previously.16,17 PI 17:0 and 20:4 and PI(4)P 17:0 and 20:4 (LM1502, LM1901, Avanti Polar Lipids) were used in parallel as internal standards. The results were analysed using MassLynx software.
Statistical analysis
Statistical significance was assessed using Student’s t-test when two groups were compared, and P < 0.05 was considered significant.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.
Results
Gene discovery and variant assessment
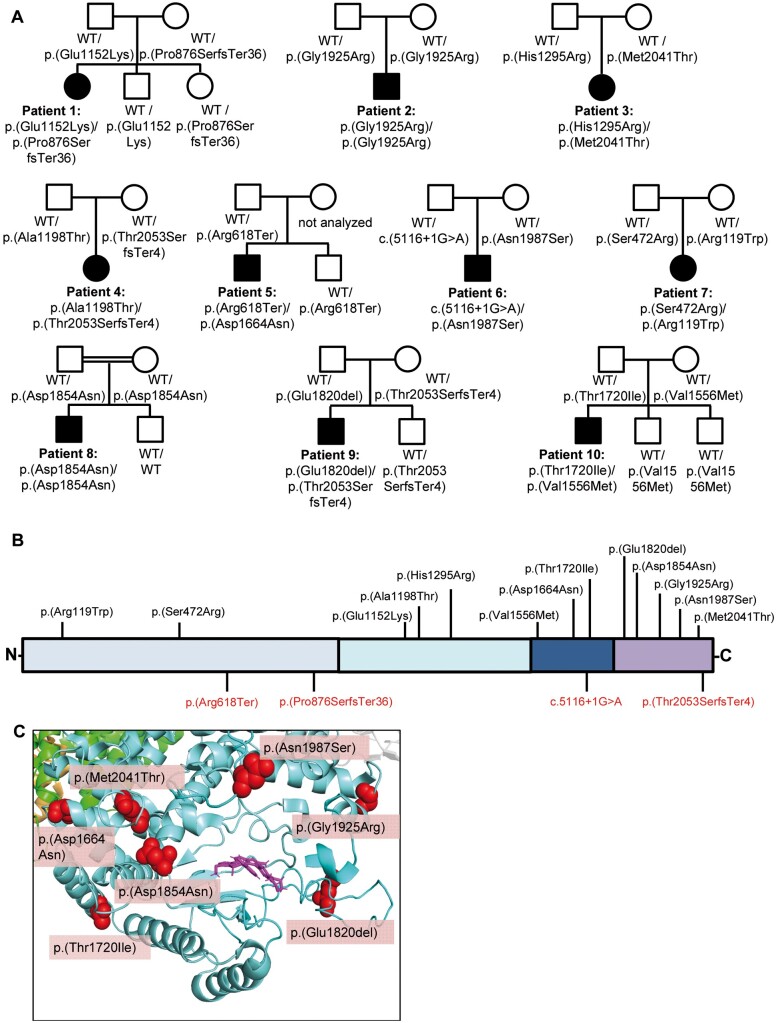
In the framework of a research project aiming to end the diagnostic odyssey in patients with neurogenetic diseases (URD-Cat, Undiagnosed Rare Diseases Consortium of Catalonia), we performed whole-exome sequencing and identified biallelic variants in PI4KA in two patients (Patients 1 and 2) affected with hypomyelinating leukodystrophy. Patient 1 was a compound heterozygote for a missense variant and a loss-of-function variant (p.Glu1152Lys, p.Pro876SerfsTer36), while Patient 2 harboured a homozygous missense variant (p.Gly1925Arg) located close to the PI4KA catalytic site, suggesting possible consanguinity. Through GeneMatcher,14 we recruited eight additional patients from independent families carrying biallelic variants in PI4KA with compatible phenotypes (Figs 1 and 2). Accordingly, the pLI value of PI4KA was 0.00031, and the pRec value was 1, indicating that biallelic deleterious variants in this gene are pathogenic.18 All identified variants were ultra-rare with minor allele frequencies (MAF) <0.00001 in the Genome Aggregation Database (gnomAD) (Supplementary Table 1). Eight patients were compound heterozygous, while two [Patient 2, p.(Gly1925Arg) and Patient 8, p.(Asp1854Asn)] were homozygous. Twelve of the identified variants were missense variants, one was an in-frame deletion of one amino acid, and the remaining four variants were predicted to result in loss-of-function. All missense/in-frame variants affected conserved amino acids and were predicted to be deleterious by various prediction tools (Table 1 and Supplementary Fig. 1). PI4KA contains an α-solenoid domain, a dimerization domain, and a ‘cradle’ domain (which contains three contact surfaces with TTC7, with which PI4KA interacts directly), as well as a catalytic domain at the C-terminus that is responsible for phosphorylation activity and is anchored at the cell membrane.2,3 Interestingly, 7 of 13 conserved missense/in-frame variants were clustered near the active site of PI4KA in the catalytic and ‘cradle’ domains (Fig. 1C and Supplementary Fig. 2). All patients harbouring one loss-of-function variant were compound heterozygotes with a missense or in-frame variant, with no patients harbouring two loss-of-function variants. Patients 4 and 9 shared one of the truncating variants (p.Thr2053SerfsTer4) but displayed discordant phenotypes. Patient 9, who carried an in-frame variant near the catalytic domain of the second allele had a milder presentation, showing spastic paraparesis with an age of onset of 17. Patient 4 harboured a missense variant in the α-solenoid domain and presented with severe hypomyelinating leukodystrophy with onset at 1 year of age.
Figure 1.
PI4KA variant features. (A) Family trees. Square = male; circle = female; filled symbols = affected individuals; open symbols = unaffected carriers; WT = wild-type allele. (B) Structure of PI4KA protein and the mutations identified in this study. Top: Missense/in-frame variants. Bottom: Loss-of-function variants. Light blue = α-solenoid domain; cyan = dimerization domain; dark blue = ‘cradle’ domain; dark purple = catalytic domain. (C) 3D representation of the PI4KA catalytic domain. Blue = PI4KA; green = TTC7B; pink = A1, PI4KA inhibitor occupying the ATP-binding space in the catalytic domain. The red balls represent the location of the missense/in-frame variants found in our cohort. Note the clustering of missense/in-frame variants near the catalytic site of PI4KA.
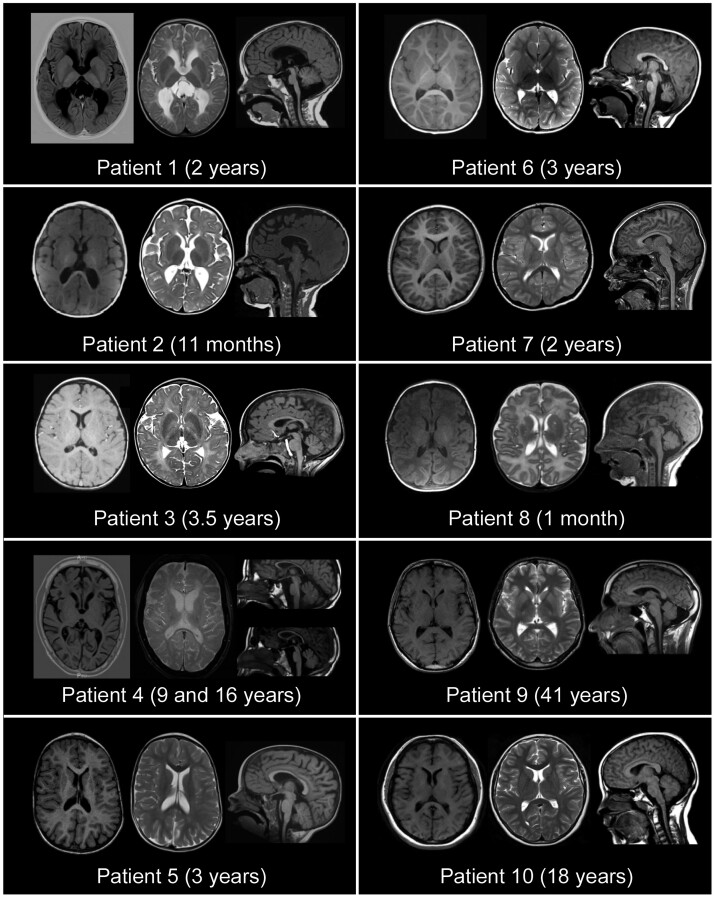
Figure 2.
MRI features of patients harbouring biallelic PI4KA variants. Left: Axial T1-weighted sequences. Middle: Axial T2-weighted sequences. Right: Sagittal T1-weighted sequences. Patients 1 and 2 exhibited diffuse hypomyelination, global white matter atrophy with posterior predominance, a thin corpus callosum and colpocephaly. Patient 1 also presented with brainstem and cerebellar hypoplasia. Patients 3 and 4 had diffuse hypomyelination with a thin corpus callosum and progressive cerebellar atrophy. Patient 4 also had brainstem atrophy. Patients 5–7 exhibited delayed myelination with mild ventriculomegaly. Patients 5 and 6 showed a dysplastic corpus callosum. Patient 6 had brainstem and cerebellar atrophy and Patient 7 exhibited atrophy of the cerebellar inferior lobe. Patient 8 showed bilateral perisylvian polymicrogyria. Patients 9 and 10 showed cervical spinal cord atrophy, and arachnoid cyst of the posterior fossa was observed in Patient 9, the images of whom were otherwise normal.
Table 1.
Main clinical features of the 10 patients with biallelic PI4KA variants
| General information | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Female | Male | Female | Female | Male | Male | Female | Male | Male | Male |
| Ethnicity | Caucasian | Caucasian | Caucasian | Caucasian | Caucasian | Asian/Caucasian | Caucasian | Turkish | Caucasian | Latin American |
| Age of onset | Newborn | Newborn | 6 months | 1 year | Newborn | Newborn | 4 months | Newborn | 17 years | 2 years |
| Age at exam | 4 years | 3 years | 13 years | 19 years | 10 years | 6 years | 11 years | 5 years | 40 years | 18 years |
| Variant | ||||||||||
| cDNA |
c.2624dupC c.3454G>A |
c.5773G>C c.5773G>C |
c.3884A>G c.6122T>C |
c.3592G>A c.6156_6159delGACA |
c.1852C>T c.4990G>A |
c.5116+1G>A c.5960A>G |
c.1414A>C c.355C>T |
c.5560G>A c.5560G>A |
c.5459_5461 delAAG c.6156_6159delGACA |
c.4666G>A c.5159C>T |
| Protein |
p.(Pro876 SerfsTer36) p.(Glu1152Lys) |
p.(Gly1925Arg) p.(Gly1925Arg) |
p.(His1295Arg) p.(Met2041Thr) |
p.(Ala1198Thr) p.(Thr2053 SerfsTer4) |
p.(Arg618Ter) p.(Asp1664Asn) |
p.? p.(Asn1987Ser) |
p.(Ser472Arg) p.(Arg119Trp) |
p.(Asp1854Asn) p.(Asp1854Asn) |
p(.Glu1820del) p.(Thr2053SerfsTer4) |
p.(Val1556Met) p.(Thr1720Ile) |
| Examination | ||||||||||
| Weight (SD) | 11.4 kg (−2.6) | 10.5 kg (−2.4) | 39 kg (−1) | 49 kg (−1) | 26.8 kg (1.23) | 15.8 kg (−2.3) | 40 kg (−0.4) | 14.5 kg (−1.4) | 64 kg (−0.9) | Not available |
| Height (SD) | 92 cm (−3.8) | Not available | 141 cm (−2.7) | 169 cm (+0.8) | 133.6 cm (0.77) | 105.8 cm (−2) | 148 cm (−0.6) | 100 cm (−2) | 171 cm (−1) | Not available |
| HC (SD) | 48.5 cm (−1.4) | 46.2 cm (−3.3) | 52 cm (−2) | Not available | 50 cm (0.5) | 47 cm (−3.8) | 52.3 cm (−1.9) | 47 cm (−3.5) | Not available | 56 cm (−0.3) |
| Motor signs | Spastic tetraparesis | Hypotonia | Spastic paraparesis | Spastic tetraparesis | Spastic paraparesis | Spastic tetraparesis | Spastic paraparesis | Spastic tetraparesis | Spastic paraparesis | Spastic paraparesis |
| Ataxia | + | − | + | + | + | + | + | − | − | − |
| Epilepsy | ++ (IS) | ++ | + | + | − | + | − | ++ | − | − |
| Additional features | Dystonia, choreoathetosis, hyperexcitability, nystagmus | Axonal sensory neuropathy, nystagmus, startle | Irritability (first months), tremor, very smiley, nystagmus | Tremor, nystagmus | Hand flapping when excited | Stereotypic movements, ocular dyspraxia | Dystonia | |||
| Development | ||||||||||
| Gross motor | Severely delayed | Severely delayed | Severy delayed | Delayed | Delayed | Delayed | Delayed | Severely delayed | No delay | Delayed |
| Walking | Not achieved | Not achieved | Not achieved | Delayed | 2.8 years | 4.5 years | 5years | Not achieved | Normal | 15 months |
| Language | None | None | Delayed | Single words | Delayed | None | Delayed | None | Normal | Normal |
| Intellectual disability | Severe | Severe | Moderate | Severe | Moderate | Severe | Severe | Severe | Normal | Mild |
| MRI description | ||||||||||
| Diffuse hypomyelination, WM atrophy, ventriculomegaly, thin CC, brainstem and cerebellar hypoplasia | Diffuse hypomyelination, WM atrophy, ventriculomegaly, thin CC | Diffuse hypomyelination, dysplastic thin CC, cerebellar atrophy with calcifications | Diffuse hypomyelination, thin CC, cerebral, brainstem and cerebellar atrophy | Delayed myelination, dysplastic CC, cerebral atrophy, ventriculomegaly | Delayed myelination, dysplastic CC, brainstem and cerebellar atrophy | Delayed myelination, external hydrocephalus, cerebellar atrophy | Bilateral perisylvian polymicrogyria | Arachnoid cyst of the posterior fossa, cervical spinal cord atrophy | Cervical spinal cord atrophy | |
+ = present; − = absent; ++ = severe; CC = corpus callosum; HC = head circumference; ID = intellectual disability; IS = infantile spasms; WM = white matter.
Clinical features
We studied 10 patients harbouring biallelic, ultrarare and probably deleterious variants after in silico predictions in PI4KA. Table 1 outlines the main clinical features of the patients, and the Supplementary material includes the patients’ clinical summaries and demographics. Weight, length and cranial circumference were all normal at birth, while at the last examination (median age 7 years; 3–42 years), three patients (Patients 1, 2 and 6) had low weight, four (Patients 1, 3, 6 and 8) had short height and four (Patients 2, 3, 6 and 8) had a head circumference less than two standard deviations (SD) below the mean. The median age of disease onset was 2 years (birth to 17 years).
Based on neurological involvement, patients in this cohort fall into two main groups.
Group 1
Patients with developmental encephalopathy with hypomyelinating leukodystrophy/delayed myelination and structural brain anomalies (Patients 1–8). These patients presented global developmental delay in the first 12 months of life; in five of these patients, clinical manifestations started in the neonatal period, mainly seizures and/or severe hypotonia. At the last examination (3–19 years of age), all of the patients presented moderate-to-severe intellectual disability, except for Patient 5, in whom intellectual disability was mild. Four patients were non-verbal, and four had not achieved ambulation, with Patient 3 being non-ambulatory even at the age of 13 years. Axial hypotonia with limb spasticity and pyramidal signs were present in all patients except Patient 2, in whom severe global hypotonia predominated and a neurophysiological study performed at 11 months of age showed axonal sensory neuropathy. Six patients had cerebellar ataxia, and five presented a movement disorder, including tremor in two patients, dystonia in two patients, choreoathetosis in one patient and stereotypies in another patient. There were no formal diagnoses of behavioural disorders, but two patients showed hyperexcitable behaviour. Six patients developed epileptic seizures, which appeared in the neonatal period in four patients and were very frequent initially but became less frequent later. Patient 6 remained without seizures after stopping anti-epileptic treatment at 18 months of age. Seizures were generalized, focal and myoclonic, and in Patient 1, episodes suggestive of infantile spasms were described at 3.5 months of age. In Patients 1–3, seizures usually appeared to be associated with fever or microbial infections and could evolve into status epilepticus. The EEG results of Patients 1 and 2 showed multifocal epileptiform discharges, with paracentral predominance being observed in Patient 1, which improved over time, and slow background activity. In Patient 4, frontotemporal spike-wave discharges were reported, and in Patient 8, bilateral frontal anomalies evolved into marked abnormalities of background activity and frequent diffuse spike and spike-wave discharges; however, in the other three patients, interictal EEG was normal.
Other clinical features included nystagmus in four patients, strabismus in two patients, and myopia and bilateral iris and retinal coloboma in one patient. Sensorineural hearing loss was reported in one patient. Four patients had feeding difficulties in early childhood, and two required nasogastric tube feeding during the first weeks of life. Patients 2, 3, 5 and 8 had immunological problems, including hypogammaglobulinaemia in three patients and lymphopaenia and autoimmune neutropaenia in one patient each. It is worth noting that symptoms of bowel dysfunction (vomiting, diarrhoea, constipation or gastroesophageal reflux disease) were reported in Patients 3, 5, 7 and 8. Finally, Patients 2, 6 and 8 presented abnormalities of the genito-urinary system, such as cryptorchidism, renal cysts and duplication of the collecting system.
Group 2
Patients with predominant spastic paraparesis (Patients 9 and 10). These patients presented a milder phenotype characterized by progressive spastic paraparesis with onset at 2 and 17 years and pes cavus. One showed mild intellectual disability, and the other patient had normal cognition. Patient 9 received a diagnosis of Crohn’s disease with a stenosing-inflammatory pattern and corticosteroid-dependent course at 21 years of age.
Brain MRI findings
The MRI images of all the patients are shown in Fig. 2. Group 1 patients showed a pattern of marked, diffuse supratentorial and infratentorial hypomyelination associated with white matter atrophy and a thin corpus callosum (Patients 1–4), incomplete/delayed myelination (Patients 5–7) and bilateral perisylvian polymicrogyria (Patient 8). The corpus callosum was dysplastic in Patients 3, 5 and 6. In Patients 1, 4 and 6, there was brainstem and cerebellar hypoplasia/atrophy, which remained unchanged up to age 2 in Patient 1. In addition, there was a component of cerebellar atrophy that progressed in successive controls in Patients 3, 4 and 7 (inferior lobe). The severity of myelin involvement correlated with clinical manifestations, which were more severe in Patients 1–3. In contrast, cerebral neuroimaging in the clinically milder patients (Patients 9 and 10) was normal, except for an arachnoid cyst of the posterior fossa in Patient 9, which was considered an unrelated finding. However, in these patients (and in Patients 1–3), cranial MRI revealed cervical spinal cord atrophy.
PI4KA activity in vitro and in vivo
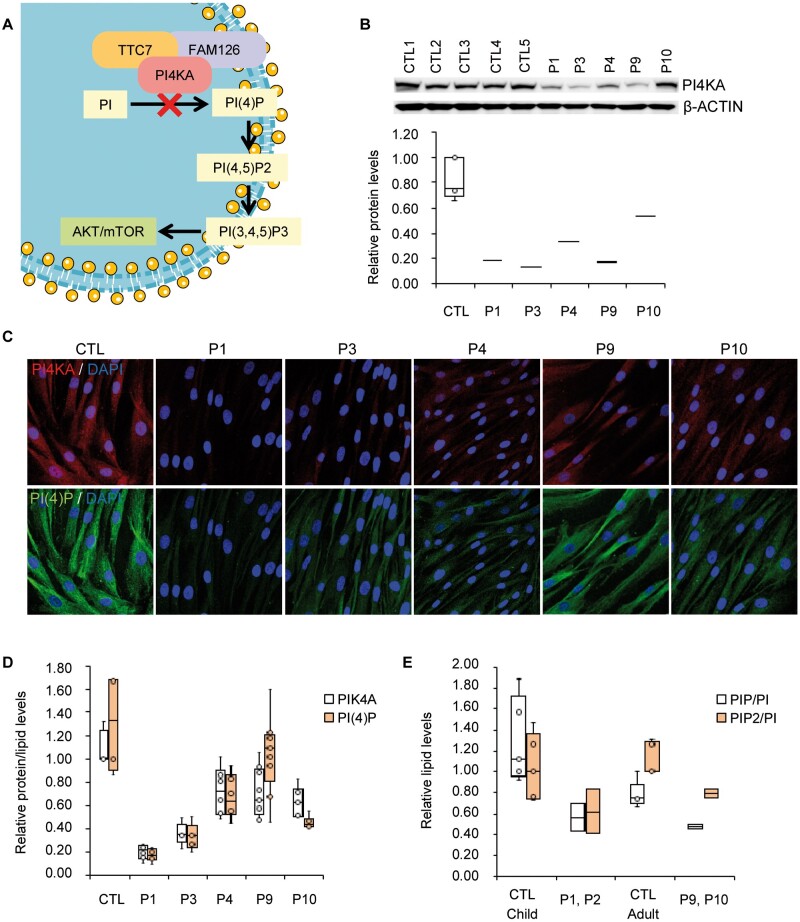
We were able to obtain skin biopsies and/or blood samples from Patients 1,3, 4, 9 and 10, and performed functional analysis of their variants. Western blot analysis showed that the expression of the PI4KA protein was strongly decreased in the fibroblasts of Patients 1, 3, 4, 9 and 10 compared to those of age-matched controls (Fig. 3B). This result was supported by immunofluorescence images (Fig. 3C and D). To evaluate the activity of PI4KA, we used an antibody against the head group of its reaction product, PI(4)P, as previously described.2 Immunofluorescence revealed lower levels of PI(4)P in our patients’ fibroblasts (Fig. 3C and D). Finally, we also performed lipidomics analysis to quantify global phosphoinositide (PI, PIP and PIP2) levels in Patients 1, 2, 9 and 10. All of the patients showed a significantly decreased PIP/PI ratio compared to age-matched control subjects, indicating decreased PI4KA activity in these patients (Fig. 3E).
Figure 3.
PI4KA activity evaluation. (A) Schematic representation of P4KA pathway. (B) Western blot of PI4KA protein and quantification. Patients’ fibroblasts (n = 5) and controls (CTL, n = 5). (C) Immunofluorescence of PI4KA protein and the head group of the lipid PI(4)P and (D) its quantification on a minimum of 100 cells. Patient’s fibroblasts (n = 5) and controls (CTL, n = 4). (E) Targeted lipidomics against phosphatidylinositol (PI), PIP and PIP2, in human PBMCs from control (CTL) children (n = 5), control adults (n = 5); and PI4KA deficient patients (n = 4). Data presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Student’s t-test). Box plot centre line corresponds to the median, lower and upper limits to the first and third quartiles (25th and 75th percentiles), respectively, and whiskers to 1.5× the interquartile range.
Discussion
We describe here a novel neurological syndrome caused by biallelic mutations in the PI4KA gene. The clinical spectrum ranges from a neurodevelopmental disorder of neonatal onset associated with severe hypomyelinating leukodystrophy with pontocerebellar hypoplasia (or even polymicrogyria in one case) to spastic paraparesis beginning in adolescence. This cohort illustrates how mutations in genes causing childhood leukodystrophies at the most severe end of the clinical spectrum, can also give rise to milder presentations, such as juvenile or adult-onset spastic paraplegias. TUBB4A-related leukodystrophy, X-linked adrenoleukodystrophy, Pelizaeus-Merzbacher disease and Alexander disease are paradigmatic for this well-described phenomenon.19–24 Although in most cases recessive mutations in this gene will manifest as hypomyelinating leukodystrophy, we propose the term ‘PI4KA-spectrum’ to describe the different neurological conditions associated with this gene.
Pyramidal tract involvement with increased limb muscle tone predominates in most of the patients in this cohort. Remarkably, Patient 2 shows severe global hypotonia with areflexia and abnormal nerve conduction studies, indicating peripheral nervous system involvement, which is in line with the neuropathy described in a mouse model with Pi4ka inactivation restricted to Schwann cells.25 In comparison with patients with mutations in FAM126A (HCC), another member of the PI4KA/TTC7/FAM126A complex,2 four of our patients had an earlier clinical onset in the neonatal period and were neurologically more severely affected. Furthermore, epilepsy appears to be more common than in HCC patients,26 and immunological or gastrointestinal symptoms have not been reported in patients with HCC. On the other hand, it is striking that none of the patients in the present study developed cataracts, which suggests that their presence may be directly due to malfunction of FAM126A. Peripheral neuropathy is an invariable feature of HCC, whereas it was only observed in one of the patients reported here. Regarding neuroimaging, Patient 4 exhibited a hypomyelinating pattern with a more conspicuous periventricular T2 hyperintensity, similar to the pattern described in patients with FAM126A mutations.26 In contrast, the polymicrogyria and brainstem and cerebellar hypoplasia reported in our cohort have not been described in FAM126A patients.
Further differential diagnosis should be made with other diseases associated with hypomyelination, mainly PLP1 disorders, and specifically disorders that present with cerebellar atrophy or hypoplasia, such as Pol III-related disorders, 18q syndrome, TUBB4A- and RARS-associated hypomyelination and hereditary spastic paraplegia (both pure and complex forms). Disorders that cause pontocerebellar hypoplasia and myelination delay should also be taken into account.27 The combination of severe developmental delay, motor impairment and early-onset seizures is a feature of developmental and epileptic encephalopathies (DEEs), and patients with these diseases often show posterior fossa anomalies, cerebral dysgenesis or delayed myelination. Hence, this group of diseases should also be considered.28 Finally, other disorders associated with neuronal migration and polymicrogyria should be included in the differential diagnosis.29,30
Previously, mutations in PI4KA were reported in three foetuses from a single family who showed bilateral polymicrogyria with hypoplasia/dysplasia of the cerebellum, olivary and dentate nucleus abnormalities, joint contractures and overlapping fingers.31 All three foetuses were compound heterozygous for two loss-of-function variants: a premature stop variant [p.(Arg796Ter)] and a missense variant located in the active site that abrogated kinase activity [p.(Asp1854Asn)]. Therefore, the phenotypes presented by these foetuses may represent the most severe end of the phenotypic spectrum of diseases associated with PI4KA mutations since no abnormalities were detected during pregnancy in our cases. This suggests that our patients may retain higher residual PI4KA activity than previously reported patients.31 In support of this hypothesis, Pi4ka mouse models harbouring two complete loss-of-function alleles are embryonic lethal,5 while pi4ka knockdown zebrafish, which retain some Pi4ka activity, show multiple developmental abnormalities.4 Interestingly, the only patient in our study who shared a variant with the family reported by Pagnamenta et al.31 [p.(Asp1854Asn)] also developed polymicrogyria, suggesting that this variant may be specifically associated with this developmental brain anomaly. Furthermore, the fact that this variant in homozygosity leads to a milder phenotype than the foetuses in Pagnamenta et al.,31 who had the p.Asp1854Asn variant in compound heterozygous state with a stop codon variant, strongly suggests that this missense variant would not cause a complete loss-of-function, and a certain degree of residual PI4KA activity is retained. Despite a clear enrichment of missense variants close to PI4KA’s active site hence supporting the pathogenic role of these variants, we did not find a patient with two loss-of-function alleles, suggesting that the missense variants identified in this cohort may be hypomorphic. Patients 1, 4, 5 and 9 harboured loss-of-function variants (out-of-frame indels or stop variants) that were always in trans with missense variants located far from the active site [p.(Glu1152Lys), p.(Asp1664Asn), p.(Ala1198Thr)] or small in-frame mutations [Patient 9, p.(Glu1820del)], suggesting that the impairment of PI4KA activity resulting from these variants may have been less severe than that caused by other variants. Two patients harbouring the p.(Thr2053SerfsTer4) variant presented different phenotypes, indicating that p.(Ala1198Thr) (Patient 4) might be more deleterious than p.(Glu1820del) (Patient 9); however, phenotype-genotype correlation in a small cohort of patients poses a challenge.
Although the clinical manifestations presented by our cohort of patients were prominently neurological, five patients presented with an immune disorder and/or a history of recurrent infections. On the other hand, one patient suffered from Crohn’s disease, whereas more non-specific manifestations, such as vomiting, diarrhoea, constipation or gastroesophageal reflux disease, were present in four patients. It is tempting to speculate that some of these extraneurological manifestations could be caused by disruption of the interactions between PI4KA and TTC7A, defects that have been associated with immunodeficiency and gastrointestinal manifestations.32
The pathogenic effects of PI4KA mutations remain poorly understood. PI4KA and PI(4)P levels were significantly diminished in the fibroblasts of five patients. PI(4,5)P2, a product of PI(4)P phosphorylation, is a substrate for the PI3K-AKT-mTOR pathway, which is critical for the myelination process.2,8 PIP levels were reduced in the PBMCs of four patients, and specifically PI(4)P was diminished as shown in fibroblasts of five patients, suggesting that their use as biomarkers for diagnosis and prognosis may be indicated. Moreover, and given the important role of PI(4)P in the transport of phosphatidylserine, a phospholipid abundantly found in myelin, to the plasma membrane,33,34PI4KA malfunction may lead to disturbances in the brain cell membrane lipidome, resulting in aberrant myelination. Concordantly, PI4KA inhibition has been shown to significantly reduce phosphatidylserine levels in cultured cells by inhibiting its transport to the plasma membrane, a phenomenon also observed in Lenz-Majewski syndrome, a developmental condition that involves similar abnormalities in PI(4)P/phosphatidylserine metabolism.35 Furthermore, inositol phospholipids are also important in processes such as actin remodelling and oligodendrocyte membrane polarization, which are essential for myelination. This process is similar to the actin polymerization-depolymerization sequence required for correct cell migration during brain development25 and may underlie both the myelination and neurodevelopmental defects we describe (polymicrogyria and brainstem-cerebellar hypoplasia). Interestingly, mice with knockout of Pi4k2a, another kinase involved in PI(4)P formation from PI, develop late-onset features resembling hereditary spastic paraplegia, and the inhibition of PI4K activity disrupts the retrograde axonal transport of neurotrophins.36 The alteration of axonal transport is one of the main processes involved in hereditary spastic paraplegia37 and is also dependent on oligodendrocyte function.38
Given that lipid metabolism defects appear to play an important role in the pathophysiology of PI4KA-associated disorders and other leukodystrophies and hereditary spastic paraplegias, therapeutic avenues for these patients could involve normalization of lipidic disturbances impacting myelination.39,40 Alternatively, upregulation of PI4KA expression or treatment with drugs that specifically treat secondary deficiencies in interacting proteins (TTC7B and FAM126A) may prove helpful in alleviating symptoms in the most severe patients in the future.41,42
In summary, we describe a novel inherited error of metabolism caused by PI4KA malfunction resulting in a broad phenotypic spectrum ranging from severe global neurodevelopmental delay associated with hypomyelinating leukodystrophy and/or brain developmental anomalies such as pontocerebellar hypoplasia/atrophy (or even polymicrogyria) in the most severe forms to spastic paraplegia in milder cases. The presence of immunological deficits, gastrointestinal manifestations or genito-urinary abnormalities may be helpful for the diagnosis of these patients.
Supplementary Material
Acknowledgements
We are grateful to the patients and their families. We also thank Cristina Guilera and Juanjo Martínez (Neurometabolic Disease Laboratory) and Lise Larrieu and Morgane Pointaux (Laboratoire de Génétique Moléculaire) for their excellent technical assistance; Eric Jeziorski for the initial referral of Patient 3; Lucia A. Baselli (Department of Pediatrics, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan), Gaia Kullman (Department of Child Neurology and Psychiatry, San Gerardo Hospital, University of Milan-Bicocca) and Silvia Maitz (Clinical Pediatric Genetic Unit, Pediatric Clinic, Fondazione MBBM, San Gerardo Hospital) for the initial referral of Patient 8; Alexis Traynor-Kaplan for help interpreting the lipidomics results; the University of Washington's School of Pharmacy's Mass Spectrometry Center; and the Cellular Biotechnology Department and Biobank, Hospices Civils de Lyon.
Funding
We thank the CERCA Program/Generalitat de Catalunya for institutional support. This study was supported by grants from the Hesperia Foundation, the Asociación Española contra las Leucodistrofias (ALE-ELA España), the Autonomous Government of Catalonia (SGR 2017SGR1206 and PERIS program URD-Cat SLT002/16/00174) and the Center for Biomedical Research on Rare Diseases (CIBERER) (ACCI19-759 to A.P.). This study was also funded by Fundació La Marató de TV3 (595/C/2020) as well as Instituto de Salud Carlos III (FIS PI20/00758 to C.C.) (co-funded by European Regional Development Fund. ERDF, a way to build Europe). This study was also funded by the Instituto de Salud Carlos III (Rio Hortega, CM18/00145 to V.V.; PFIS, FI18/00141 to L.P.; and Sara Borrell, CD19/00221 to E.V.), co-funded by European Social Fund. ESF investing in your future; the Ministerio de Ciencia e Innovación y Universidades (Juan de la Cierva, FJCI-2016-28811 to E.V.), and the Center for Biomedical Research on Rare Diseases (CIBERER to M.R.).
Sequencing and analysis of Patient 5 were performed by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and were funded by the National Human Genome Research Institute, the National Eye Institute, the National Heart, Lung and Blood Institute grants UM1 HG008900 and R01 HG009141 and the Chan Zuckerberg Initiative to the Rare Genomes Project. This work was in part supported by the association ‘Connaître les Syndromes Cérébelleux’ (CSC). This research received funding specifically appointed to the Department of Medical Sciences from the Italian Ministry for Education, University and Research (Ministero dell’istruzione, dell’università e della ricerca-MIUR) under the programme ‘Dipartimenti di Eccellenza 2018-2022’ Project code D15D18000410001. Whole-exome sequencing was performed as part of the Autism Sequencing Consortium and was supported by the NIMH (MH111661). D.R.A. and A.P. are members of the Undiagnosed Disease Network International (UDNI).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- PI
phosphatidylinositol
References
- 1. Burke JE. Structural basis for regulation of phosphoinositide kinases and their involvement in human disease. Mol Cell. 2018;71(5):653–673. [DOI] [PubMed] [Google Scholar]
- 2. Baskin JM, Wu X, Christiano R, et al. The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat Cell Biol. 2016;18(1):132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lees JA, Zhang Y, Oh MS, et al. Architecture of the human PI4KIIIα lipid kinase complex. Proc Natl Acad Sci U S A. 2017;114(52):13720–13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma H, Blake T, Chitnis A, Liu P, Balla T.. Crucial role of phosphatidylinositol 4-kinase IIIα in development of zebrafish pectoral fin is linked to phosphoinositide 3-kinase and FGF signaling. J Cell Sci. 2009;122(Pt 23):4303–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakatsu F, Baskin JM, Chung J, et al. Ptdins4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 2012;199(6):1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tan J, Oh K, Burgess J, Hipfner DR, Brill JA.. PI4KIIIα is required for cortical integrity and cell polarity during Drosophila oogenesis. J Cell Sci. 2014;127(Pt 5):954–966. [DOI] [PubMed] [Google Scholar]
- 7. Cutler NS, Heitman J, Cardenas ME.. STT4 is an essential phosphatidylinositol 4-kinase that is a target of Wortmannin in Saccharomyces cerevisiae. J Biol Chem. 1997;272(44):27671–27677. [DOI] [PubMed] [Google Scholar]
- 8. Wolf NI, Ffrench-Constant C, van der Knaap MS.. Hypomyelinating leukodystrophies - unravelling myelin biology. Nat Rev Neurol. 2021;17(2):88–103. [DOI] [PubMed] [Google Scholar]
- 9. Nellist M, Schot R, Hoogeveen-Westerveld M, et al. Germline activating AKT3 mutation associated with megalencephaly, polymicrogyria, epilepsy and hypoglycemia. Mol Genet Metab. 2015;114(3):467–473. [DOI] [PubMed] [Google Scholar]
- 10. Baulac S, Lenk GM, Dufresnois B, et al. Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology. 2014;82(12):1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rivière J-B, Mirzaa GM, O'Roak BJ, et al. ; Finding of Rare Disease Genes (FORGE) Canada Consortium. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44(8):934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poduri A, Evrony GD, Cai X, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74(1):41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alkhater RA, Scherer SW, Minassian BA, Walker S.. PI4K2A deficiency in an intellectual disability, epilepsy, myoclonus, akathisia syndrome. Ann Clin Transl Neurol. 2018;5(12):1617–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sobreira N, Schiettecatte F, Valle D, Hamosh A.. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schiffmann R, Van Der Knaap MS.. Invited Article: An MRI-based approach to the diagnosis of white matter disorders. Neurology. 2009;72(8):750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Traynor-Kaplan A, Kruse M, Dickson EJ, et al. Fatty-acyl chain profiles of cellular phosphoinositides. Biochim Biophys Acta. 2017;1862(5):513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De La Cruz L, Traynor-Kaplan A, Vivas O, Hille B, Jensen JB.. Plasma membrane processes are differentially regulated by type I phosphatidylinositol phosphate 5-kinases and RASSF4. J Cell Sci. 2020;133(2):jcs233254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Casasnovas C, Verdura E, Vélez V, et al. A novel mutation in the GFAP gene expands the phenotype of Alexander disease. J Med Genet. 2019;56(12):846–849. [DOI] [PubMed] [Google Scholar]
- 20. Elitt MS, Barbar L, Shick HE, et al. Suppression of proteolipid protein rescues Pelizaeus-Merzbacher disease. Nature. 2020;585(7825):397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Köhler W, Curiel J, Vanderver A.. Adulthood leukodystrophies. Nat Rev Neurol. 2018;14(2):94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van der Knaap MS, Schiffmann R, Mochel F, Wolf NI.. Diagnosis, prognosis, and treatment of leukodystrophies. Lancet Neurol. 2019;18(10):962–972. [DOI] [PubMed] [Google Scholar]
- 23. Kancheva D, Chamova T, Guergueltcheva V, et al. Mosaic dominant TUBB4A mutation in an inbred family with complicated hereditary spastic paraplegia. Mov Disord. 2015;30(6):854–858. [DOI] [PubMed] [Google Scholar]
- 24. Di Bella D, Magri S, Benzoni C, et al. Hypomyelinating leukodystrophies in adults: Clinical and genetic features. Eur J Neurol. 2021;28(3):934–944. [DOI] [PubMed] [Google Scholar]
- 25. Alvarez-Prats A, Bjelobaba I, Aldworth Z, et al. Schwann-cell-specific deletion of phosphatidylinositol 4-kinase alpha causes aberrant myelination. Cell Rep. 2018;23(10):2881–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Biancheri R, Zara F, Rossi A, et al. Hypomyelination and congenital cataract: Broadening the clinical phenotype. Arch Neurol. 2011;68(9):1191–1194. [DOI] [PubMed] [Google Scholar]
- 27. van Dijk T, Baas F, Barth PG, Poll-The BT.. Poll-The BT. What’s new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet J Rare Dis. 2018;13(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheffer IE, Liao J.. Deciphering the concepts behind “Epileptic encephalopathy” and “Developmental and epileptic encephalopathy”. Eur J Paediatr Neurol. 2020;24:11–14. [DOI] [PubMed] [Google Scholar]
- 29. Schiller S, Rosewich H, Grünewald S, Gärtner J.. Inborn errors of metabolism leading to neuronal migration defects. J Inherit Metab Dis. 2020;43(1):145–155. [DOI] [PubMed] [Google Scholar]
- 30. Juric-Sekhar G, Hevner RF.. Malformations of cerebral cortex development: Molecules and mechanisms. Annu Rev Pathol Mech Dis. 2019;14:293–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pagnamenta AT, Howard MF, Wisniewski E, et al. Germline recessive mutations in PI4KA are associated with perisylvian polymicrogyria, cerebellar hypoplasia and arthrogryposis. Hum Mol Genet. 2015;24(13):3732–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jardine S, Dhingani N, Muise AM.. TTC7A: Steward of intestinal health. Cell Mol Gastroenterol Hepatol. 2019;7(3):555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chung J, Torta F, Masai K, et al. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER - Plasma membrane contacts. Science. 2015;349(6246):428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Von Filseck JM, Čopič A, Delfosse V, et al. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349(6246):432–436. [DOI] [PubMed] [Google Scholar]
- 35. Sohn M, Ivanova P, Brown HA, et al. Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER-PM and ER-golgi junctions. Proc Natl Acad Sci U S A. 2016;113(16):4314–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bartlett SE, Reynolds AJ, Weible M, Hendry IA.. Phosphatidylinositol kinase enzymes regulate the retrograde axonal transport of NT-3 and NT-4 in sympathetic and sensory neurons. J Neurosci Res. 2002;68(2):169–175. [DOI] [PubMed] [Google Scholar]
- 37. Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A.. Hereditary spastic paraplegia: Clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol. 2014;261:518–539. [DOI] [PubMed] [Google Scholar]
- 38. Edgar JM, McLaughlin M, Yool D, et al. Oligodendroglial modulation of fast axonal transport in a mouse model of hereditary spastic paraplegia. J Cell Biol. 2004;166(1):121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rickman OJ, Baple EL, Crosby AH.. Lipid metabolic pathways converge in motor neuron degenerative diseases. Brain. 2019;143(4):1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pant DC, Boespflug-Tanguy O, Pujol A, et al. Loss of the sphingolipid desaturase DEGS1 causes hypomyelinating leukodystrophy. J Clin Invest. 2019;129(3):1240–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jardine S, Anderson S, Babcock S, et al. Drug screen identifies leflunomide for treatment of inflammatory bowel disease caused by TTC7A deficiency. Gastroenterol. 2020;158(4):1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wong M-T, Chen SS.. Hepatitis C virus subverts human choline kinase-α to bridge phosphatidylinositol-4-kinase IIIα (PI4KIIIα) and NS5A and upregulates PI4KIIIα activation, thereby promoting the translocation of the ternary complex to the endoplasmic reticulum for viral replication. J Virol. 2017;91(16):e00355-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.