Abstract
Alzheimer disease (AD) is a neurodegenerative disease characterized by a cognitive decline leading to dementia. The most impactful genetic risk factor is apolipoprotein E (APOE). APOE-ε4 significantly increases AD risk, APOE-ε3 is the most common gene variant, and APOE-ε2 protects against AD. However, the underlying mechanisms of APOE-ε4 on AD risk remains unclear, with APOE-ε4 impacting many pathways. We investigated how the APOE isoforms associated with the neuroinflammatory state of the brain with and without AD pathology. Frozen brain tissue from the superior and middle temporal gyrus was analyzed from APOE-ε3/3 (n = 9) or APOE-ε4/4 (n = 10) participants with AD pathology and APOE-ε3/3 (n = 9) participants without AD pathology. We determined transcript levels of 757 inflammatory related genes using the NanoString Human Neuroinflammation Panel. We found significant pathways impaired in APOE-ε4/4-AD individuals compared to APOE-ε3/3-AD. Of interest, expression of genes related to microglial activation (SALL1), motility (FSCN1), epigenetics (DNMT1), and others showed altered expression. Additionally, we performed immunohistochemistry of P2RY12 to confirm reduced microglial activation. Our results suggest APOE-ε3 responds to AD pathology while potentially having a harmful long-term inflammatory response, while APOE-ε4 shows a weakened response to pathology. Overall, APOE isoforms appear to modulate the brain immune response to AD-type pathology.
Keywords: Alzheimer disease, Apolipoprotein E, Microglia, NanoString, Neuroinflammation, Neuropathology
INTRODUCTION
Alzheimer disease (AD) is the fifth leading cause of death for those older than 65. Currently, AD affects over 6 million Americans and that number is expected to more than double by 2050 if no treatment to delay or prevent onset is found (1, 2). Hallmark pathologies that distinguish AD from other dementia-inducing diseases include Aβ plaques and neurofibrillary tangles (3–5). These lesions are both typically present in brain before cognitive tests can register impairment (6, 7). Of the 2 pathologies, cognitive deficits are most closely associated with tangle pathology (7–10). While the majority of AD cases are considered sporadic, there are genetic risk factors that can increase an individual’s risk of developing AD (11).
The AD risk gene that confers the largest known public health impact is an isoform of apolipoprotein E (APOE). APOE has 3 common isoforms: APOE-ε2, APOE-ε3, and APOE-ε4. The APOE-ε4 allele confers an increased risk of AD and has an allele frequency of 14% in the general population but is present in between 40% and 60% of AD cases (12–15). The primary role of APOE in the central nervous system is to transport cholesterol and lipids to provide neurons and other cells with the energy supply needed to function properly (16–19). In the presence of APOE-ε4, there is a decrease in this energy supply and overall decrease in support APOE can give to the neurons (20, 21). Outside of the primary role of APOE in the brain, APOE-ε4 has many secondary affects that are thought to impact AD pathology and progression. Clearance of intracellular debris and toxic proteins through autophagy has also been shown to be impacted by APOE-ε4 (22, 23). With AD pathology, APOE-ε4 has been shown to increase the amount of Aβ plaques (24–29), possibly due to decreased Aβ clearance from the brain, as well as increase tau tangles (30). APOE genotypes have been shown to impact brain development patterns as early as infancy (31) and APOE-ε4 has been shown to have differential impacts on regional pathology in AD (32, 33).
Inflammation has been associated with AD pathology for decades, and research suggests that this inflammation could be a double-edged sword in AD, exacerbating or ameliorating pathology and disease based on a specific type of response (34, 35). Studies suggest that microglia become activated in AD, releasing cytotoxic factors which may facilitate phagocytosis of Aβ, removing the inflammatory stimuli, eventually leading to induction of anti-inflammatory factors (36, 37). However, this cascade has also been shown to cause detrimental long-term consequences if the stimulus is not removed. Additionally, in AD, studies have suggested that alterations in inflammatory status vary over the disease course and the presence of multiple inflammatory profiles simultaneously (38, 39). In a histological study, a reduced level of IBA-1 has been seen suggesting that with AD pathology, the microglia are unable to provide neuronal support leading to neurotoxic environments (40). In another histological study, researchers found a higher level of microglia in AD brains, specifically in female patients suggesting sex differences in inflammation (41). Furthermore, with a growing understanding of the complexity of inflammation, studies have suggested that inflammation could exacerbate Aβ production, Aβ deposition, and tau propagation (42, 43). It is clear that, functionally, inflammation in AD is multifaceted and can have both beneficial and detrimental consequences depending on when and where the inflammation occurs.
The role of APOE in AD-related inflammation has been examined in vitro and in vivo. Studies using mouse models have suggested that APOE-ε4 induces a proinflammatory state in microglia in the presence of AD pathology and lipopolysaccharides (44–48). Further, a study in human autopsy tissue found that individuals with APOE-ε4 have a pro-inflammatory state of inflammation in their brains regardless of the level of pathology (49, 50). In a human study investigating levels of microglia using the frontal and temporal cortex, researchers found an increased level of microglia in APOE-ε4 brain suggesting APOE plays a role with microglia in the presence of AD pathology (51). Interestingly, recent studies have found impairments in other inflammatory cascades. For instance, reduction in microglial activation upon repeated traumatic brain injury in a humanized APOE-ε4 mouse model compared to APOE-ε3 suggesting an impaired inflammatory response (52). Another study found APOE-ε4 directly impairs autophagy which has been implicated in inflammation (22). With the complexity of the role of APOE-ε4, the present study aims to explore these findings further and expand upon these previous findings.
In the current study, we investigated neuroinflammatory processes, and other pathways related to neuroinflammation, using human brain tissue from the superior and middle temporal gyrus (SMTG) from APOE-ε3 or APOE-ε4 participants with and without AD pathology to characterize the inflammatory processes occurring. Our findings suggest that with APOE-ε3, there is a change in inflammatory profile based on the level of AD pathology in the brain. However, with AD pathology in APOE-e4 patients, there is an inflammatory profile similar to control brains suggesting an impaired inflammatory response to pathology.
MATERIALS AND METHODS
Sample Collection
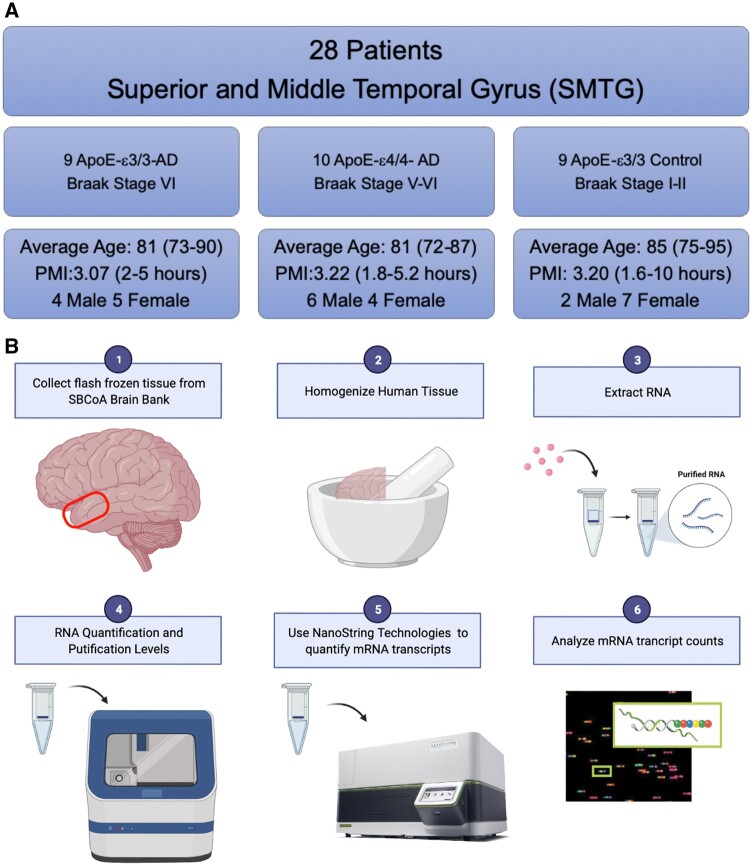
SMTG samples were obtained through the University of Kentucky Alzheimer’s Disease Research Center (UK-ADRC) at the Sanders-Brown Center on Aging (SBCoA). The UK-ADRC recruitment details have been previously described as well as the pathological assessments that were performed on the tissue at the University of Kentucky (53–55). Sample breakdown can be seen in Table 1. Frozen samples of the SMTG were obtained and are briefly summarized in Figure 1A. A piece of SMTG was pulverized and, therefore, gray and white matter were not separated out for this study. All samples were homozygous for their APOE isoform; therefore, they will be described as follows for this manuscript: APOE-ε3/3 with AD pathology: APOE-ε3-AD; APOE-ε4/4 with AD pathology: APOE-ε4-AD, APOE-ε3/3 control: APOE-ε3-Control.
TABLE 1.
AD Subject Characteristics
| Alzheimer Disease |
|||||
|---|---|---|---|---|---|
| Combined | APOE-ε3 Control | APOE-ε3-AD | APOE-ε4-AD | p Value | |
| Number of participants | 28 | 9 | 9 | 10 | |
| Age at death, years, mean (SD) | 82.1 (5.4) | 85.2 (5.4) | 80.8 (5.1) | 80.5 (4.8) | 0.583 |
| Male, N (%) | 12 (42.8) | 2 (22.0) | 4 (44.4) | 6 (60.0) | 0.381 |
| PMI, mean (SD) | 3.2 (1.6) | 3.2 (2.6) | 3.1 (1.0) | 3.2 (1.0) | 0.395 |
| Cancer, N (%) | 13 (46) | 6 (66) | 4 (50.0) | 3 (42.9) | 0.240 |
| Average RIN | 8.7 | 8.92 | 8.51 | 8.76 | |
| Atherosclerosis, N (%) | 0.011 | ||||
| 0 | 2 (7.2) | 1 (11.0) | 0 (0.0) | 1 (10.0) | |
| 1 | 6 (21.4) | 3 (33.3) | 0 (0.0) | 3 (30.0) | |
| 2 | 5 (17.9) | 3 (33.3) | 0 (0.0) | 2 (20.0) | |
| 3 | 11 (39.3) | 2 (22.2) | 7 (77.8) | 2 (20.0) | |
| 4 | 4 (14.3) | 0 (0.0) | 2 (22.2) | 2 (20.0) | |
| Arteriosclerosis severity, N (%) | 0.017 | ||||
| None | 4 (14.2) | 4 (44.0) | 0 (0.0) | 0 (0.0) | |
| Mild | 20 (71.4) | 5 (56.0) | 6 (66.7) | 9 (90.0) | |
| Moderate | 3 (10.7) | 0 (0.0) | 3 (33.3) | 0 (0.0) | |
| Unknown | 1 (3.6) | 0 (0.0) | 0 (0.0) | 1 (10.0) | |
| Total infarcts, N (%) | 0.098 | ||||
| 0 | 17 (60.7) | 5 (55.6) | 4 (44.4) | 8 (80.0) | |
| 1 | 6 (21.4) | 3 (33.3) | 3 (33.3) | 0 (0.0) | |
| 2 | 3 (10.7) | 1 (11.1) | 2 (22.2) | 0 (0.0) | |
| >2 | 2 (7.2) | 0 (0.0) | 0 (0.0) | 2 (20.0) | |
| Neuritic plaques, N (%) | <0.001 | ||||
| None | 6 (21.4) | 6 (66.7) | 0 (0.0) | 0 (0.0) | |
| Sparse | 3 (10.6) | 3 (33.3) | 0 (0.0) | 0 (0.0) | |
| Moderate | 3 (12.5) | 0 (0.0) | 1 (11.1) | 2 (20.0) | |
| Severe | 16 (66.7) | 0 (0.0) | 8 (88.9) | 8 (80.0) | |
| Braak stage, N (%) | <0.001 | ||||
| I | 6 (21.4) | 6 (66.7) | 0 (0.0) | 0 (0.0) | |
| II | 3 (12.5) | 3 (33.3) | 0 (0.0) | 0 (0.0) | |
| V | 3 (12.5) | 0 (0.0) | 0 (0.0) | 3 (30.0) | |
| VI | 16 (66.7) | 0 (0.0) | 9 (100.0) | 7 (70.0) | |
Detailed breakdown of AD subject characteristics. Combined shows both AD and control subjects. One-way ANOVA was used for all variables that were reported using means and SDs, Kruskal-Wallis tests were used for atherosclerosis, total infarcts, and Braak stage, and chi-square tests were used on the remaining values. Bolded values indicate those that reached statistical significance.
FIGURE 1.
Participants breakdown/study design. (A) Basic depiction of participant samples selected with ApoE status, Braak stage, postmortem interval (PMI), and gender differences. (B) Graphical depiction of the study design highlighting the region of interest, superior and middle temporal gyrus (SMTG), and important components of NanoString Analysis. Figure generated with BioRender.com.
RNA Isolation
Frozen SMTG was pulverized using a mortar and pestle on dry ice with liquid nitrogen. Brain powder was stored at −80°C. RNA was extracted from approximately 40 mg of frozen pulverized tissue using the E.Z.N.A total RNA kit II (Omega-Bio-Tek, Norcross, GA) according to manufacturer’s instructions. RNA was assessed with Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) at the University of Kentucky Genomics Core Laboratory. RIN values are shown in Table 1; samples with RIN values lower than 7 were removed from analysis. The Genomics Core ran 20 ng of RNA per sample on the NanoString Technologies nCounter Human Neuroinflammatory Panel (Nanostring Technologies, Seattle, WA). These genes were preselected from NanoString to allow for a relatively unbiased panel. All cases were assigned study identifiers allowing for blinded data analysis.
Statistical Analysis—NanoString Data
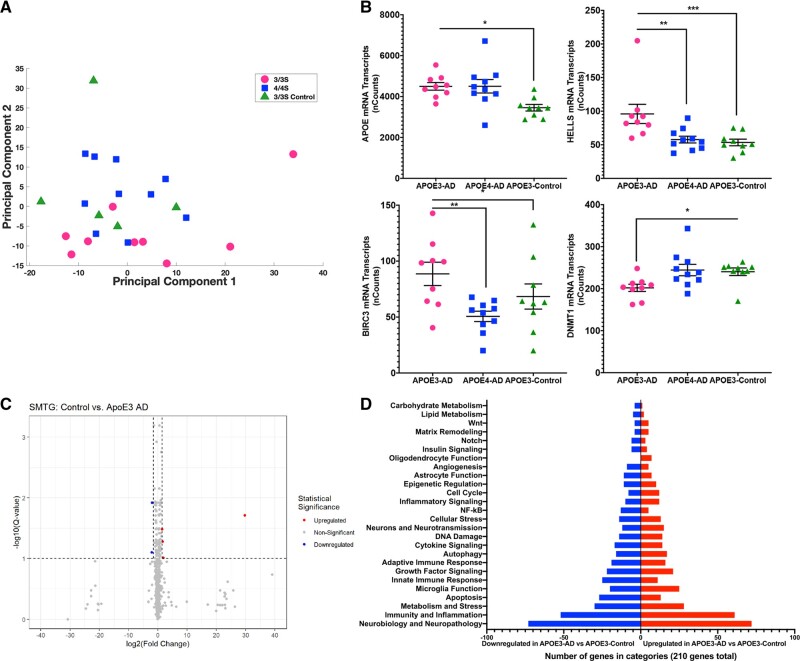
NanoString data analysis was performed using 2 software packages, nCounter and the NanoStringDiff package in R (56). Using the raw NanoString data, NanoStringDiff analyses were used to model the unnormalized NanoString reads using a negative binomial generalized linear model which incorporates normalization into its parameter estimation (57). Since many genes were tested simultaneously, a false discovery rate (FDR) of 10% was used to determine statistical significance. To allow for this study to mimic the general population, outliers were not removed from the data analysis. Figure 1B is a graphical representation of these methods. Additionally, a principal component analysis (PCA) was run to identify variation between groups. This was performed by standardizing across genes using a z score on the normalized nCounter data (Fig. 2A).
FIGURE 2.
AD impact on neuroinflammation. (A) Principal component analysis was performed using normalized NanoString counts for each participant that were then z scored (n = 28). Principal component 2 has 2 directions that are distinguishing the clusters: the positive direction represents microglial function, and the negative direction represents neurodegeneration. (B) Normalized NanoString nSolver counts for 4 of the top significant genes of interest. Significance markers: * p = 0.05–0.01, **p = 0.01–0.001, ***p = 0.001–0.0001. (C) Volcano plot showing all 757 genes with colored dots representing statistically significant genes if their p value was less than 0.05 and their log2 (fold change) was either less than −1.5 or greater than 1.5. (D) Of the 210 significant genes (p < 0.05), number of genes up- or downregulated categorized based on NanoString annotations.
Quantitative Real-Time RT-PCR and Statistical Analysis
A subset of genes on the NanoString panel was additionally analyzed using qPCR to verify trends observed (Table 2). RNA previously used for NanoString Analysis was quantified using the Biospec-Nano spectrophotometer (Shimadzu, Kyoto, Japan). About 100 ng of RNA was reverse transcribed to cDNA using the cDNA High-Capacity kit (Life Technologies, Carlsbad, CA) according to manufacturer’s instructions. Real-time PCR was performed using a 96-well plate. In each well, 1 µL of the appropriate gene probe was added to 10 µL of the Fast-TaqMan reagent along with 0.5 µL of cDNA and 6.5 µL of RNase-free water. TaqMan Gene Expression probes used are provided in Table 2 (Life Technologies). Real-time PCR was performed using the ViiA 7 Real-Time PCR system (Applied Biosystems, Grand Island, NY). All genes were normalized to 18S rRNA and the fold change was determined using the −ΔΔCt method (58). Statistical analysis was done using a Student t-test. Outliers were removed using the Grubbs test for a single outlier (59). Results from qPCR can be seen in Supplementary Data Figures S1 and S2.
TABLE 2.
Genes Used for Real-Time PCR
| Gene | PMID | Probe ID |
|---|---|---|
| APOE | NP_000032.1 | Hs00171168_m1 |
| ARC | NP_056008.1 | Hs01045540_g1 |
| C6 | NP_001108603.2 | Hs01110040_m1 |
| CLSTN1 | NP_001009566.1 | Hs00208929_m1 |
| CX3CL1 | NP_002987.1 | Hs00171086_m1 |
| FSCN1 | NP_003079.1 | Hs00602051_mH |
| GRN | NP_002078.1 | Hs00173570_m1 |
| IL-1b | NP_000567.1 | Hs01555410_m1 |
| IL1RN | NP_000568.1 | Hs00893626_m1 |
| ITGAM | NP_001139280.1 | Hs00167304_m1 |
| KDM4A | NP_055478.2 | Hs04285637_m1 |
| LINGO1 | NP_116197.4 | Hs01081148_m1 |
| LTBR | NP_002333.1 | Hs01101194_m1 |
| MCM2 | NP_004517.2 | Hs01091564_m1 |
| NRGN | NP_001119653.1 | Hs00183469_m1 |
| P2RY12 | NP_073625.1 | Hs1881698_s1 |
| PIK3R1 | NP_852664.1 | Hs00933163_m1 |
| PINK1 | NP_115785.1 | Hs00260868_m1 |
| PSEN2 | NP_000438.2 | Hs01577197_m1 |
| SALL1 | NP_001121364.1 | Hs04285637_m1 |
| STX18 | NP_058626.1 | Hs01099207_m1 |
| TGFb1 | NP_000651.3 | Hs00998133_m1 |
| TMEM88B | NP_001140157.1 | Hs04188764_m1 |
| TNFa | NP_000585.2 | Hs00174128_m1 |
| TREM2 | NP_061838.1 | Hs00219132_m1 |
String Pathway Analysis
All pathway analysis was performed through string-db.org (60). In brief, the top 60 significant genes names were input into the multiple proteins search option. The pathway image displayed was used to visualize clusters of genes related to specific functions. The number of genes was capped at 60 to allow for visualization.
Immunohistochemistry
Human SMTG brain sections were stained immunohistochemically for P2RY12 (Purinergic Receptor P2Y12). Formalin-fixed paraffin-embedded tissue was cut into 8 µm-thick sections. Immunohistochemistry (IHC) staining was performed using anti-P2RY12 (rabbit polyclonal, 1:1000 IHC, Lot No. D118480) (Sigma-Aldrich, St. Louis, MO). A biotinylated goat antirabbit IgG (1:200; Lot No. ZG0818) secondary antibody (Vector Laboratories, Burlington, CA) was used with avidin-biotin substrate (ABC solution, Vector Laboratories). Color development was preformed using DAB (DAKO). Stained sections were dehydrated, and cover slipped in DPX (Electron Microscopy Sciences, Hatfield, PA). All slides were scanned using Zeiss Axio Scan Slide Scanner (Carl Zeiss Microscopy, White Plains, NY) and analyzed using the Halo image analysis system (Indica Labs, Albuquerque, NM).
RESULTS
AD Participant Characterization
All participants were selected from the UK-ADRC at the Sanders-Brown Center on Aging (SBCoA). The AD cohort for this study was defined as Braak stage V-VI, Thal Aβ phase 4 or higher, and consensus diagnosis of AD in APOE-ε3 and APOE-ε4 individuals. In the AD cases selected, there was minimal cerebrovascular pathology, and no detectable pathologies for other neurodegenerative diseases in the areas observed with pathological assessments (egTDP-43, α-synuclein proteinopathies). Sample size of APOE-ε4 participants for inclusion in the study was limited by there being few cases that lacked cerebrovascular comorbidities. Matching age, sex, and postmortem interval across groups also limited our APOE-ε3 AD participant size. The non-AD participant cohort was defined as a Braak stage I–II with a consensus clinical diagnosis of normal before death. More information about the cohort is presented in Table 1 and Figure 1A. Due to the prevalence of AD pathology in APOE-ε4 carriers, there was only one APOE-ε4 case that met our non-AD control criteria in the entire UK-ADRC autopsy cohort. For this reason, our control group is only APOE-ε3 participants. All samples were homozygous for their APOE isoform; therefore, they will be described as follows for this study: APOE-ε3/3 with AD pathology: APOE-ε3-AD; APOE-ε4/4 with AD pathology: APOE-ε4-AD, APOE-ε3/3 without AD pathology: APOE-ε3-Control.
Neuroinflammatory Profiles Separate Groups Based on Disease State and APOE Isoform
To characterize the differences between the disease state and APOE isoform, verification that all groups were different was essential. A PCA was run to visualize the variation between the groups. The normalized NanoString counts were used from the SMTG. Figure 2A shows the analysis of the disease states and APOE isoforms. The second dimension of the PCA (PCA2) shows variation between the groups and allows for separation between both APOE isoforms and disease states. Separation in PCA2 is driven by genes involved in microglial function, with higher expression resulting in increased PCA2 levels, as seen in APOE-ε4-AD patients. Additionally, decreased levels of PCA2 were driven by increased expression of genes associated with multiple aspects of neurodegeneration, as seen in APOE-ε3-AD patients.
APOE Isoforms Alter the Neuroinflammatory Profile With or Without AD Pathology
A large number of genes showed significant differential expression between APOE-ε3-Control and APOE-ε3-AD cases. We found 210 genes (p < 0.05) (27% of total genes analyzed) were significantly different at an alpha level of 0.05. After controlling for multiple comparisons, we found 167 genes (FDR q value <0.1) (22% of total genes analyzed) showed significant differential expression. Overall, the data show a significant difference due to AD pathology with APOE-ε3. Figure 2B shows 4 genes that are associated with pathways connected with AD in APOE-ε3/3 carriers that have also been shown to be implicated in AD disease progression. The volcano plot in Figure 2C shows gene changes between APOE-ε3-AD relative to APOE-ε3-Control and Figure 2D highlights the gene categories altered. Overall, this shows significant cellular responses to AD pathology in the APOE-ε3 genotype. We further investigated if these changes were also seen comparing APOE-ε4-AD to APOE-ε3-Control. Of the 757 genes assessed, 101 genes (13.3% of total genes analyzed) were significant with a p value <0.05, and 5 remained significant after correction for multiple comparisons (FDR q value <0.1). The pathways and specific genes altered between the groups show changes in microglial function and immune response. These findings are striking, showing the significant similarities between APOE-ε4-AD with APOE-ε3-Control. However, these changes are not related to the typical proinflammatory phenotype associated with APOE-ε4 (Supplementary DataFig. S2).
APOE Isoforms Influence the Inflammatory Profile in the Presence of AD Pathology
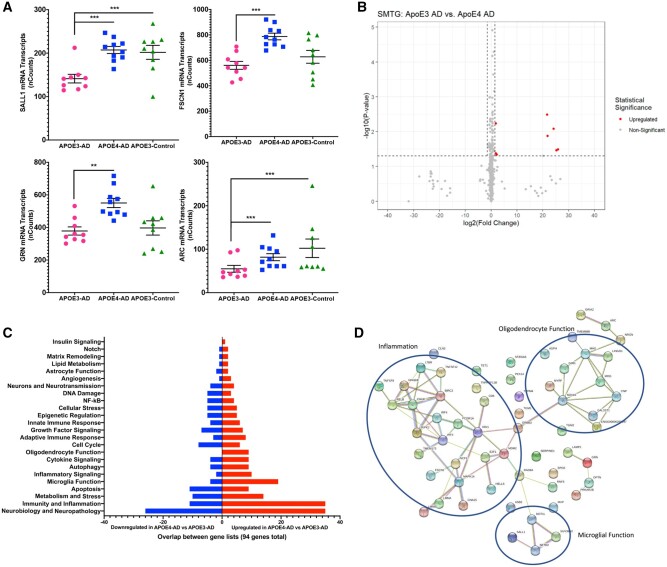
To see if the same pathways affected in AD are impacted by APOE isoforms, APOE-ε3-AD was compared with APOE-ε4-AD. A large number of genes were differentially expressed between APOE isoforms: 94 total genes (12.4% of total genes analyzed) were significantly different (p < 0.05) with 10 genes (1.3% of total genes analyzed) significant after correction for multiple comparisons (FDR q value <0.1). The majority of the differentially expressed genes are involved with microglial function, epigenetic regulation, and oligodendrocyte function (Fig. 3A–D). These data show alterations between the 2 APOE genotypes with AD; however, these changes parallel those seen between APOE-ε3-AD and APOE-ε3-Control. Of the 94 genes that were differentially expressed between the APOE isoforms, 33% of these genes overlap with the genes altered between APOE-ε3-AD and APOE-ε3-Control.
FIGURE 3.
ApoE isoform impact on neuroinflammation with AD. (A) Normalized NanoString nSolver counts for 4 of the top significant genes of interest. Significance markers: *p = 0.05–0.01, **p = 0.01–0.001, ***p = 0.001–0.0001 (all *** have passed FDR). (B) Volcano plot showing all 757 genes with colored dots representing statistically significant genes if their p value was less than 0.05 and their log2 (fold change) was either less than −1.5 or greater than 1.5. (C) Of the significant genes (p < 0.05), number of genes up- or downregulated categorized based on NanoString annotations. (D) string-db.org pathway analysis of the top 60 significant genes.
Association Between APOE Isoforms and Microglial Morphology
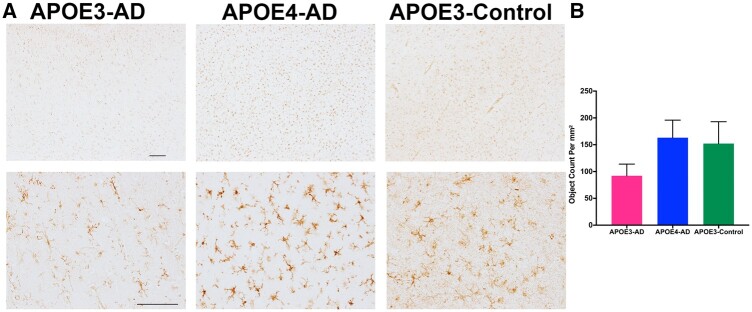
To confirm the changes seen in the transcriptional analysis, a limited histological analysis was performed on a select group of samples used in RNA analysis (4 samples/group) using the SMTG. Due to the small sample size for histology, overall trends in the expression level were examined. Tissue was stained with P2RY12, a homeostatic microglial marker, to determine the activation levels of microglia in the SMTG region. The data show a trend with APOE−ε4-AD and APOE- ε3-Controls having higher levels of P2RY12 and APOE- ε3-AD having lower expression levels of P2RY12 (Fig. 4), suggesting activation of microglia in APOE- ε3-AD patients but not in APOE−ε4-AD patients.
FIGURE 4.
Association between APOE isoforms and microglial morphology. (A) Immunohistochemistry staining of P2RY12 was scanned on the Axio Slide scanner and representative images from the regions were selected from Halo. Top row is 4× with a scale bar of 200 mM and bottom row is 20× with a scale bar of 100 mM. (B) Halo analysis was used to determine the quantification of microglia stained with P2RY12 using object analysis. PRISM 8 was used to generate graphical representation of data measuring object count per mm2.
DISCUSSION
APOE-ε4 is the most impactful known sporadic AD risk factor that has been implicated in many biological pathways including inflammation (61); however, the impact of APOE genotype on inflammatory profiles in human AD brain has not been fully explored. In the current study, we identified significant differences in several key inflammatory pathways when comparing APOE-ε3 and APOE-ε4 participants with and without AD pathology. The inflammatory pathways in APOE-ε4-AD participants appeared to be impaired and generally resembled a control brain with no AD pathology. This is in contrast to APOE-ε3-AD, which showed significant changes in inflammatory pathways compared to APOE-ε3-Control, suggesting that APOE-ε3 brains have an inflammatory response to AD pathology. We also investigated genes that were differentially expressed between APOE-ε3-AD and APOE-ε4-AD and found genes altered in key pathways such as microglial function, epigenetic regulation, neuronal support, cellular motility, oligodendrocyte function, and many other pathways. While this is not a complete list of pathways that our study investigated, these are key pathways that have been supported by animal and cellular research.
Since we are studying the impact of APOE isoforms, it was important to determine the expression levels of APOE. We found a significant increase in APOE-ε3 mRNA expression levels with AD and, while not statistically significant, a similar trend in APOE-ε4 compared to control participants. APOE gene expression was similar in APOE-ε3-AD and APOE-ε4-AD brains (Fig. 2B) (62). This indicates there is an increase in mRNA levels of APOE in the presence of AD pathology regardless of APOE isoform. While studies have shown a decrease in APOE-ε4 protein compared to APOE-ε3 in AD, we have only quantified transcript levels and have not confirmed with protein levels in this study (45, 63, 64).
APOE- ε4 microglia are known to produce a proinflammatory phenotype upon activation and in AD and the present study was able to confirm the previous literature. However, outside of this response, there are additional inflammatory cascades necessary for phagocytosis and an overall response to AD pathology. Because the gene expression panel we used investigated inflammation, we identified significantly altered genes related specifically to microglia and their role in inflammation. Of interest, spalt like transcription factor 1 (SALL1) is a transcriptional regulator of microglial function and is important in maintaining microglia in a homeostatic and probing state (65). Upon activation, SALL1 expression is downregulated to allow for phagocytosis, proliferation (65), activation (66), and in the presence of neurodegeneration, conversion from resting microglia to a disease-associated microglia (DAM) (67) or the neurodegenerative microglia (MGnD) (68) phenotype. Studies suggest that the DAM/MGnD phenotype is beneficial in the early and middle stages of disease to phagocytize and cluster microglia around plaques, yet this can lead to an increase in neurotoxicity in late stages if they remain in this activation state (69). In the present study, in the presence of AD pathology with APOE-ε3 genotype, SALL1 was downregulated, which would facilitate the switch to an activation state, thus permitting microglia to respond to AD pathology. However, in the comparison between APOE-ε4-AD and APOE-ε3-Control participants, the same change was not seen and the expression of SALL1 between the 2 groups was not significantly changed. This suggests that APOE-ε4 microglia may be unable to switch from a homeostatic state to an activated state necessary to clear the AD pathology, perhaps explaining the increase in pathology seen in APOE-ε4-AD participants (Fig. 3A).
The suggestion that APOE- ε4 microglia cannot respond to AD pathology can further be seen in our immunohistochemical staining for P2RY12. P2RY12 has been classified as a homeostatic microglial marker that becomes downregulated upon microglial activation (68). In the presence of AD pathology, it is predicted that P2RY12 expression levels would decline to allow for a response to the accumulating pathology (70). This was seen in our APOE-ε3-AD samples while our APOE-ε3-Control samples still had an increased expression, as expected with lack of pathology. However, APOE-ε4-AD samples had similar levels to that of APOE-ε3-Control, which suggests that APOE-ε4 microglia cannot respond to AD pathology (Fig. 4). Further studies on SALL1 and P2RY12 are required to determine how APOE-ε4 impacts the DAM/MGnD pathways and overall microglial activation states. Additionally, understanding when the DAM/MGnD pathway and activation is beneficial versus neurotoxic will be necessary in future studies targeting inflammation.
Studies have consistently shown the impact AD pathology has on neuronal connectivity, especially through reduced long-term potentiation (LTP), a major molecular mechanism in memory formation. Our data show that the activity-regulated cytoskeleton-associated protein (ARC) which is an intermediate early gene specifically involved with LTP, is reduced in the APOE-ε3-AD participants relative to APOE-ε3-Control and APOE-ε4-AD tissue (Fig. 3A). ARC impacts recycling of glutamatergic receptors necessary for making synaptic connections and long-term memories and upon overactivation of ARC, can increase Aβ production (71). While previous labs have shown that there is an increase in ARC expression in AD, our data suggest a decrease in the APOE-ε3-AD brain. This suggests there is a potential negative role of AD pathology in the context of LTP with ARC. A caveat for this finding is that depending on the agonal state prior to autopsy, there could be a confounding element impacting expression levels of ARC.
Another transcript that was significantly altered in our study is fascin actin-bundling protein 1 (FSCN1), which is involved in white matter and general motility. FSCN1 is present on all cell types in the brain and is responsible for cytoskeleton remodeling which can allow for an increase in motility. Studies have implicated FSCN1 in white matter dysfunction and in AD as a whole due to the impact on motility. One study specifically highlighted that FSCN1 was downregulated in the white matter of AD participants (72). Our findings show a significant decrease in FSCN1 expression in the presence of AD pathology with APOE-ε3 compared to APOE-ε4-AD (Fig. 3A). FSCN1 is important in motility, and some studies have surprisingly showed an increase in motility of microglia in APOE-ε4 (73), which aligns with the results we have seen where FSCN1 is unchanged in APOE-ε4-AD.
Epigenetics has been an area of research in many diseases, especially cancer research, for many years and is a growing area of study in the neurodegenerative field (74). DNA methyltransferase 1 (DNMT1) is a gene responsible for maintaining methylation patterns after DNA replication. Methylation patterns are necessary to maintain proper transcription. Our results show that DNMT1 expression was decreased in APOE-ε3-AD participants compared to APOE-ε3-Control (Fig. 2B). With the decrease seen in the AD brain, this could lead to altered gene regulation. Studies have shown that in the presence of AD pathology, methylation patterns have been altered suggesting an important role of epigenetics in AD. Another gene of interest from the present study is helicase, lymphoid specific (HELLS), which is responsible for normal development and survival in addition to DNA methylation (75, 76). The present study shows an elevated level of expression in APOE-ε3-AD participants compared to APOE-ε4-AD participants and APOE-ε3-Control (Fig. 2B). While the results of DNMT1 and HELLS are opposite, it suggests a potential implication of DNA methylation alterations between APOE isoforms and AD status. Further investigations into the epigenetic link in AD, especially in the context of DNMT1 and HELLS, are needed to identify a possible connection.
Due to the nature of the study, there are limitations that need to be considered. Biochemical studies of material obtained at autopsy provide inherently cross-sectional insights. As mentioned previously, the human brain tissue samples evaluated comprised a mixture of gray and white matter in the SMTG region, which could skew some results because different samples will have differing proportions of gray or white matter. Additionally, due to the small sample sizes, covariates in our participant selection could not be parsed out. And finally, because we included APOE-ε4/4 individuals, there were relatively many included cases with very severe AD pathology.
It has long been known that APOE-ε4 affects many aspects of AD as well as brain health as a whole. In this study, we have identified areas in which APOE isoforms influence inflammatory pathways in AD. Using both RNA and histology data, our results suggest that APOE-ε4 patients have a reduced inflammatory response to AD pathology. It has previously been found that APOE-ε4 individuals respond differently to anti-Aβ immunotherapies, with increased rates of vasogenic edema, microhemorrhage, and other adverse events (77–79), possibly due to this altered inflammatory response. By understanding the specific differences between APOE-ε3 and APOE-ε4 individuals, especially in terms of inflammatory responses to AD pathology, we can begin to determine insights relevant to future therapies. In addition, these data show that when stratifying and grouping participants, APOE status must be considered carefully as they clearly respond to AD pathology differently.
Supplementary Material
ACKNOWLEDGMENTS
We are sincerely grateful for the research volunteers and clinical colleagues at the University of Kentucky Alzheimer’s Disease Research Center.
Research reported in this article was funded by grants P30-AG028383 (UK-ADRC), 1RF1AG057754-01 (DMW), 1T32AG057461-01 (CMK), and 1F31AG069372-01 (CMK) from the National Institute on Aging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare that they have no competing interests.
Supplementary Data can be found at academic.oup.com/jnen.
REFERENCES
- 1.Center for Disease Control, Centers for Training E, Assistance T. A Public Health Approach to Alzheimer’s and Other Dementias – A Public Health Crisis. Atlanta, GA: Alzheimer’s Association, Center for Disease Control, Emory Centers for Training and Technical Assistance 2019
- 2.Alzheimer’s Association. 2021 Alzheimer’s Disease Facts and Figures. Chicago, IL: Alzheimer’s Association 2021
- 3. Maurer K, Volk S, Gerbaldo H, et al. Alzheimer’s disease. Lancet 1997;349:1546–9 [DOI] [PubMed] [Google Scholar]
- 4. Thal DR, Walter J, Saido TC, et al. Neuropathology and biochemistry of Abeta and its aggregates in Alzheimer’s disease. Acta Neuropathol 2015;129:167–82 [DOI] [PubMed] [Google Scholar]
- 5. Braak H, Braak E.. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging 1995;16:271–8; discussion 278–84 [DOI] [PubMed] [Google Scholar]
- 6. Jack CR, Knopman DS, Jagust WJ, et al. Tracking pathophysiological processes in Alzheimer’s disease: An updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013;12:207–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson PT, Braak H, Markesbery WR.. Neuropathology and cognitive impairment in Alzheimer disease: A complex but coherent relationship. J Neuropathol Exp Neurol 2009;68:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nasreddine ZS, Phillips NA, Bedirian V, et al. The Montreal Cognitive Assessment, MoCA: A brief screening tool for mild cognitive impairment. J Am Geriatr Soc 2005;53:695–9 [DOI] [PubMed] [Google Scholar]
- 9. Bergeron D, Flynn K, Verret L, et al. Multicenter validation of an MMSE-MoCA conversion table. J Am Geriatr Soc 2017;65:1067–72 [DOI] [PubMed] [Google Scholar]
- 10. Folstein MF, Folstein SE, McHugh PR.. Mini-mental state. J Psychiatr Res 1975;12:189–98 [DOI] [PubMed] [Google Scholar]
- 11. Kunkle BW, Grenier-Boley B, Sims R, et al. ; Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES). Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 2019;51:414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993;261:921–3 [DOI] [PubMed] [Google Scholar]
- 13. Saunders AMP, Strittmatter WJMD, Schmechel DMD, et al. Association of apolipoprotein E allele E4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 1993;43:1467–72 [DOI] [PubMed] [Google Scholar]
- 14. Strittmatter WJ, Weisgraber KH, Huang DY, et al. Binding of human apolipoprotein E to synthetic amyloid beta peptide: Isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A 1993;90:8098–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pastor P, Roe CM, Villegas A, et al. Apolipoprotein Eε4 modifies Alzheimer’s disease onset in an E280A PS1 kindred. Ann Neurol 2003;54:163–9 [DOI] [PubMed] [Google Scholar]
- 16. Boehm-Cagan A, Bar R, Liraz O, et al. ABCA1 agonist reverses the ApoE4-driven cognitive and brain pathologies. J Alzheimers Dis 2016;54:1219–33 [DOI] [PubMed] [Google Scholar]
- 17. Boehm-Cagan A, Bar R, Harats D, et al. Differential effects of apoE4 and activation of ABCA1 on brain and plasma lipoproteins. PLoS One 2016;11:e0166195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rawat V, Wang S, Sima J, et al. ApoE4 alters ABCA1 membrane trafficking in astrocytes. J Neurosci 2019;39:9611–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rebeck GW. The role of APOE on lipid homeostasis and inflammation in normal brains. J Lipid Res 2017;58:1493–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu CC, Liu CC, Kanekiyo T, et al. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat Rev Neurol 2013;9:106–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minagawa H, Gong JS, Jung CG, et al. Mechanism underlying apolipoprotein E (ApoE) isoform-dependent lipid efflux from neural cells in culture. J Neurosci Res 2009;87:2498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parcon PA, Balasubramaniam M, Ayyadevara S, et al. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimers Dement 2018;14:230–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cortes CJ, la Spada AR.. TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: Molecular mechanisms, cellular processes, and emerging therapeutic opportunities. Neurobiol Dis 2019;122:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vitek MP, Snell J, Dawson H, et al. Modulation of nitric oxide production in human macrophages by apolipoprotein-E and amyloid-beta peptide. Biochem Biophys Res Commun 1997;240:391–4 [DOI] [PubMed] [Google Scholar]
- 25. Verghese PB, Castellano JM, Garai K, et al. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci U S A 2013;110:E1807–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ulrich JD, Ulland TK, Mahan TE, et al. ApoE facilitates the microglial response to amyloid plaque pathology. J Exp Med 2018;215:1047–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stephen TL, Cacciottolo M, Balu D, et al. APOE genotype and sex affect microglial interactions with plaques in Alzheimer’s disease mice. Acta Neuropathol Commun 2019;7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagy ZS, Esiri MM, Jobst KA, et al. Influence of the apolipoprotein E genotype on amyloid deposition and neurofibrillary tangle formation in Alzheimer’s disease. Neuroscience 1995;69:757–61 [DOI] [PubMed] [Google Scholar]
- 29. Beffert U, Cohn JS, Petit-Turcotte C, et al. Apolipoprotein E and β-amyloid levels in the hippocampus and frontal cortex of Alzheimer’s disease subjects are disease-related and apolipoprotein E genotype dependent. Brain Res 1999;843:87–94 [DOI] [PubMed] [Google Scholar]
- 30. Shi Y, Yamada K, Liddelow SA, et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 2017;549:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dean DC, Jerskey BA, Chen K, et al. Brain differences in infants at differential genetic risk for late-onset alzheimer disease: A cross-sectional imaging study. JAMA Neurol 2014;71:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sabbagh MN, Malek-Ahmadi M, Dugger BN, et al. The influence of Apolipoprotein E genotype on regional pathology in Alzheimer’s disease. BMC Neurol 2013;13:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murray ME, Graff-Radford NR, Ross OA, et al. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: A retrospective study. Lancet Neurol 2011;10:785–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wyss-Coray T, Rogers J.. Inflammation in Alzheimer disease-A brief review of the basic science and clinical literature. Cold Spring Harb Perspect Med 2012;2:a006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wyss-Coray T, Mucke L.. Review inflammation in neurodegenerative disease-a double-edged sword. Neuron 2002;35:419–32 [DOI] [PubMed] [Google Scholar]
- 36. Combs CK, Karlo JC, Kao S-C, et al. β-amyloid stimulation of microglia and monocytes results in TNFα-dependent expression of inducible nitric oxide synthase and neuronal apoptosis. J Neurosci 2001;21:1179–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hickman SE, Allison EK, el Khoury J.. Microglial dysfunction and defective β-amyloid clearance pathways in aging alzheimer’s disease mice. J Neurosci 2008;28:8354–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sudduth TL, Schmitt FA, Nelson PT, Wilcock DM.. Neuroinflammatory phenotype in early Alzheimer’s disease. Neurobiol Aging 2013;34:1051–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cullen NC, Mälarstig AN, Stomrud E, et al. Accelerated inflammatory aging in Alzheimer’s disease and its relation to amyloid, tau, and cognition. Sci Rep 2021;11:1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Minett T, Classey J, Matthews FE, et al. Microglial immunophenotype in dementia with Alzheimer’s pathology. J Neuroinflammation 2016;13:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Overmyer M, Helisalmi S, Soininen H, et al. Reactive microglia in aging and dementia: An immunohistochemical study of postmortem human brain tissue. Acta Neuropathol 1999;97:383–92 [DOI] [PubMed] [Google Scholar]
- 42. Hur JY, Frost GR, Wu X, et al. The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer’s disease. Nature 2020;586:735–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clayton K, Delpech JC, Herron S, et al. Plaque associated microglia hyper-secrete extracellular vesicles and accelerate tau propagation in a humanized APP mouse model. Mol Neurodegen 2021;16:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lynch JR, Tang W, Wang H, et al. APOE genotype and an ApoE-mimetic peptide modify the systemic and central nervous system inflammatory response. J Biol Chem 2003;278:48529–33 [DOI] [PubMed] [Google Scholar]
- 45. Lanfranco MF, Sepulveda J, Kopetsky G, et al. Expression and secretion of apoE isoforms in astrocytes and microglia during inflammation. Glia 2021;69:1478–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu Y, Nwabuisi-Heath E, Dumanis SB, et al. APOE genotype alters glial activation and loss of synaptic markers in mice. Glia 2012;60:559–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Colton C, Brown C, Cook D, et al. APOE and the regulation of microglial nitric oxide production: A link between genetic risk and oxidative stress. Neurobiol Aging 2002;23:777–85 [DOI] [PubMed] [Google Scholar]
- 48. Vitek MP, Brown CM, Colton CA.. APOE genotype-specific differences in the innate immune response. Neurobiol Aging 2009;30:1350–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tzioras M, Davies C, Newman A, et al. Invited review: APOE at the interface of inflammation, neurodegeneration and pathological protein spread in Alzheimer’s disease. Neuropathol Appl Neurobiol 2019;45:327–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Friedberg JS, Aytan N, Cherry JD, et al. Associations between brain inflammatory profiles and human neuropathology are altered based on apolipoprotein E ε4 genotype. Sci Reports 2020;10:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Egensperger R, Kösel S, von Eitzen U, et al. Microglial activation in Alzheimer disease: Association with APOE genotype. Brain Pathol 1998;8:439–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Muza P, Bachmeier C, Mouzon B, et al. APOE genotype specific effects on the early neurodegenerative sequelae following chronic repeated mild traumatic brain injury. Neuroscience 2019;404:297–313 [DOI] [PubMed] [Google Scholar]
- 53. Schmitt FA, Nelson PT, Abner E, et al. University of Kentucky Sanders-Brown healthy brain aging volunteers: Donor characteristics, procedures and neuropathology. Curr Alzheimer Res 2012;9:724–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmitt FA, Wetherby MM, Wekstein DR, et al. Brain donation in normal aging: Procedures, motivations, and donor characteristics from the Biologically Resilient Adults in Neurological Studies (BRAiNS) Project. Gerontologist 2001;41:716–22 [DOI] [PubMed] [Google Scholar]
- 55. Nelson PT, Jicha GA, Schmitt FA, et al. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: Neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol 2007;66:1136–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2020 [Google Scholar]
- 57. Wang H, Horbinski C, Wu H, et al. NanoStringDiff: A novel statistical method for differential expression analysis based on NanoString nCounter data. Nucleic Acids Res 2016;44:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 59. Burns MJ, Nixon GJ, Foy CA, et al. Standardisation of data from real-time quantitative PCR methods – Evaluation of outliers and comparison of calibration curves. BMC Biotechnol 2005;5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kloske CM, Wilcock DM.. The important interface between Apolipoprotein E and neuroinflammation in Alzheimer’s disease. Front Immunol 2020;11:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Talwar P, Sinha J, Grover S, et al. Meta-analysis of apolipoprotein e levels in the cerebrospinal fluid of patients with Alzheimer’s disease. J Neurol Sci 2016;360:179–87 [DOI] [PubMed] [Google Scholar]
- 63. Bekris LM, Galloway NM, Montine TJ, et al. APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am J Med Genet B Neuropsychiatr Genet 2010;153B:409–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Riddell DR, Zhou H, Atchison K, et al. Impact of Apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci 2008;28:11445–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Buttgereit A, Lelios I, Yu X, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol 2016;17:1397–406 [DOI] [PubMed] [Google Scholar]
- 66. Xiang X, Werner G, Bohrmann B, et al. TREM2 deficiency reduces the efficacy of immunotherapeutic amyloid clearance. EMBO Mol Med 2016;8:992–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deczkowska A, Keren-Shaul H, Weiner A, et al. Disease-associated microglia: A universal immune sensor of neurodegeneration. Cell 2018;173:1073–81 [DOI] [PubMed] [Google Scholar]
- 68. Krasemann S, Madore C, Cialic R, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 2017;47:566–81.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shi Y, Holtzman DM.. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol 2018;18:759–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walker DG, Tang TM, Mendsaikhan A, et al. Patterns of expression of purinergic receptor P2RY12, a putative marker for non-activated microglia, in aged and Alzheimer’s disease brains. Int J Mol Sci 2020;21:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kerrigan TL, Randall AD.. A new player in the “synaptopathy” of Alzheimer’s disease – Arc/Arg 3.1. Front Neurol 2013;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Castaño EM, Maarouf CL, Wu T, et al. Alzheimer disease periventricular white matter lesions exhibit specific proteomic profile alterations. Neurochem Int 2013;62:145–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Muth C, Hartmann A, Sepulveda-Falla D, et al. Phagocytosis of apoptotic cells is specifically upregulated in apoe4 expressing microglia in vitro. Front Cell Neurosci 2019;13:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qazi TJ, Quan Z, Mir A, Qing H.. Epigenetics in Alzheimer’s disease: Perspective of DNA methylation. Mol Neurobiol 2018;55:1026–44 [DOI] [PubMed] [Google Scholar]
- 75. Mathys H, Adaikkan C, Ransohoff RM, et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep 2017;21:366–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Han Y, Ren J, Lee E, et al. Lsh/HELLS regulates self-renewal/proliferation of neural stem/progenitor cells. Sci Rep 2017;7:1–1428127051 [Google Scholar]
- 77. Sevigny J, Chiao P, Bussière T, et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 2016;537:50–6 [DOI] [PubMed] [Google Scholar]
- 78. Sperling R, Salloway S, Brooks DJ, et al. Amyloid-related imaging abnormalities in patients with Alzheimer’s disease treated with bapineuzumab: A retrospective analysis. Lancet Neurol 2012;11:241–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Salloway S, Sperling R, Gilman S, et al. ; Bapineuzumab 201 Clinical Trial Investigators. A phase 2 multiple ascending dose trial of bapineuzumab in mild to moderate Alzheimer disease. Neurology 2009;73:2061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.