Abstract
The Nrf2 transcription factor governs the expression of hundreds genes involved in cell defense against oxidative stress, the hallmark of numerous diseases such as neurodegenerative, cardiovascular, some viral pathologies, diabetes and others. The main route for Nrf2 activity regulation is via interactions with the Keap1 protein. Under the normoxia the Keap1 binds the Nrf2 and targets it to the proteasomal degradation, while the Keap1 is regenerated. Upon oxidative stress the interactions between Nrf2 and Keap1 are interrupted and the Nrf2 activates the transcription of the protective genes. Currently, the Nrf2 system activation is considered as a powerful cytoprotective strategy for treatment of different pathologies, which pathogenesis relies on oxidative stress including viral diseases of pivotal importance such as COVID-19. The implementation of this strategy is accomplished mainly through the inactivation of the Keap1 “guardian” function. Two approaches are now developing: the Keap1 modification via electrophilic agents, which leads to the Nrf2 release, and direct interruption of the Nrf2:Keap1 protein-protein interactions (PPI). Because of theirs chemical structure, the Nrf2 electrophilic inducers could non-specifically interact with others cellular proteins leading to undesired effects. Whereas the non-electrophilic inhibitors of the Nrf2:Keap1 PPI could be more specific, thereby widening the therapeutic window.
Keywords: Oxidative stress, Nrf2, Keap1, ROS, Influenza virus, SARS-CoV-2
Graphical abstract
1. Introduction
Oxidative stress is one of the key factors affecting our cells due to the reactive oxygen species (ROS) and free radicals [1], constantly produced from internal sources (cell metabolism) as well as external exposure (UV light, ionizing radiation, ozone, toxicants such as cigarette smoke, heavy metals, insecticides and pesticides, viruses and bacteria, altering cellular balance). The mitochondria are the main source of the endogenous ROS, generating them during cellular respiration. Some cytosolic enzymes such as cytochrome P450, lipoxygenases, and NADPH oxidase also generate ROS as well as peroxisomal oxidases [2]. ROS participates in normal cellular and organismal processes, acting as redox signaling molecules [3], [4] with the ability to tight control of subcellular localization and rapid changes in oxidants levels. When ROS generation exceeds their catabolism, oxidation of the intracellular proteins, lipids, DNA increases, leads to the oxidative damage of their functions, formation of toxic molecules, and possible transformation of the normal cellular processes to the pathological ones [5]. Besides ROS our cells are exposed to other toxicants such as environmental pollutants, bacterial and viral toxins. To resist to the wide spectrum of oxidative and chemical omnipresent stresses our cells have developed an adaptive defense system, consisting of dozens cytoprotective genes, which products provide adaptation to different stressors. These genes are components of different systems, such as xenobiotic detoxication, glutathione- and thioredoxin-based antioxidant metabolic pathways, drug-resistance protein transporters, proteasomal degradation, autophagy, iron and lipid metabolism, NADPH regeneration, required as a cofactor for some of above mentioned systems [6].
The majority of these cytoprotective genes contains antioxidant-responsive element, ARE, cis-acting sequence in the 5′-upstream regulatory region, through which modulation of transcription occurs via binding of basic domain and leucine zipper (bZIP) transcription factors (TFs), the most prominent of which is Nrf2 (NF-E2 p45-related factor 2) [6]. The existence of the common cellular system, responsible for chemoprotection had been predicted a decade before such system was discovered. In the 1988 Talalay et al. paper [7] it had been proposed that the cell ability to express set of chemoprotective enzymes can be induced by a variety of different small molecules but with a similar chemical property. This implies existence of a cellular system, regulating chemoprotection, and that the system should have key element(s), which could be activated during stress. In 1994 Nrf2 was isolated [8], now referred to as a master regulator of cellular antioxidant defense [9]. A four years later the cellular protein Keap1 (Kelch-like ECH-associated protein 1) was identified as a suppressor of Nrf2 activity [10], and subsequently have been demonstrated as a sensor [11], which thiol group of cysteine residues react with inducers molecules. Conventional wisdom suggests that Keap1 is a redox/electrophile sensitive negative regulator of Nrf2/ARE signaling pathway, which, in turn, mediates the expression of hundred genes, involved in the cytoprotective systems. After the Nrf2/ARE signaling discovery the field has quickly exploded and still is an area of ongoing research and focus of interest of numerous articles. As several thorough reviews can be found in [12], [13], [14], [15] we only mention some key points and we focus primarily on the recent data about the role of Nrf2 in emerging viral diseases and ways to modulate Nrf2 activity more specific through competitive regulation.
2. Nrf2
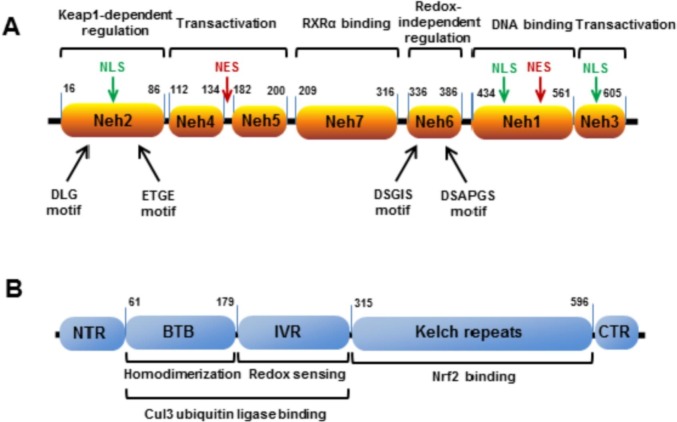
From the evolutionary standpoint the Nrf2 system is a defense mechanism against oxidants and xenobiotics, widely conserved across a broad spectrum of organisms [16], with first Nrf2 orthologues appeared, according to some estimates, in fungi more than one billion years ago [17]. The biology of Nrf2:Keap1 interaction is relatively well elucidated. Nrf2 is a 66 kDa protein, consisting of 7 conserved functional regions (Fig. 1a), named Nrf2-ECH homology domains (Neh) [6], [18], [19]. Neh1 domain contains a basic leucine zipper motif, and is responsible for heterodimerization with its transcriptional partner Maf TF and for DNA binding. Neh2 domain interacts with Keap1 through two motifs: high affinity (Kd ~ 5 nM) ETGE and low affinity (Kd ~ 1 μM) DLG [20], [21], [22]. Both motifs are indispensable for Keap1 regulation [23]. A recent study has shown that Keap1 uses several mechanisms to activate Nrf2 in response to a wide range of environmental stresses [24]. A model for the molecular mechanisms leading to Nrf2 activation is the Hinge-Latch model, where the DLGex-binding motif of Nrf2 dissociates from Keap1 as a latch, while the ETGE motif remains attached to Keap1 as a hinge. The DLG latch dissociation is triggered by inhibitors of Keap1-Nrf2 interaction and occurs during stress-inducible protein p62-mediated Nrf2 activation, but not by electrophilic Nrf2 inducers [24]. Also Neh2 domain contains lysine residues, which are substrates for ubiquitination and participate in Keap1-dependent Nrf2 proteasomal degradation [25]. Neh3, Neh4 and Neh5 are transactivator domains, interacting with intracellular co-activator molecules. Deletion of Neh3 abolishes the Nrf2 ability to activate ARE-mediated gene expression, keeping intact dimerization and DNA-binding capability as well as subcellular localization [26]. Neh4 and Neh5 act synergistically and cooperatively recruit co-activator molecule CBP [27], which acetylates Nrf2, hence enhancing it activity [28]. Neh6 allows Keap1-independent regulation, triggered by GSK3β phosphorylation of DSGIS and DSAPGS motifs, creating a phosphodegron for Nrf2 ubiquitination by Cul1-β-TrCP complex [29]. Neh7 domain also regulates Nrf2 activity, binding retinoic acid receptor α, a nuclear receptor, which represses Nrf2 activity [30].
Fig. 1.
Protein domains structure of Nrf2 (A) and Keap1 (B). Nrf2 and Keap1 domain boundaries are given according to papers by Canning et al., 2015 [31] and Jung et al., 2018 [32].
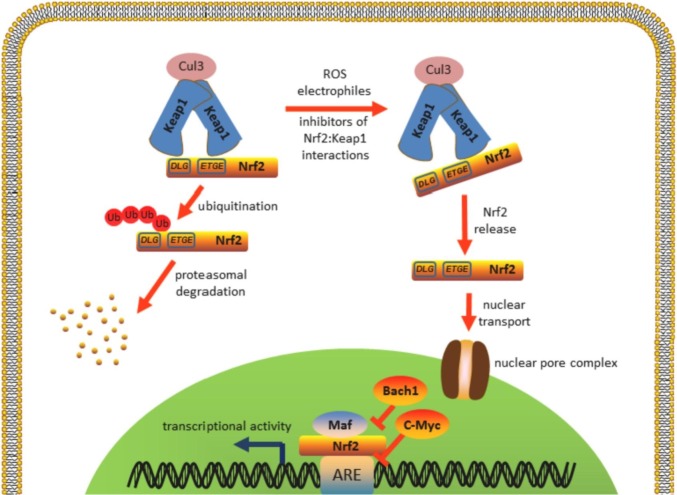
Under unstressed conditions, Nrf2 is localized in the cytoplasm (Fig. 2 ), and fraction of cytoplasmic Nrf2 is tethered to the outer mitochondrial membrane through mitochondrial serine/threonine-protein phosphatase PGAM5, forming a ternary complex with the Keap1 and the Nrf2 [33]. Nrf2 nuclear localization is regulated through the import/export balance. Nrf2 contains three nuclear localization sequences (in the Neh1, Neh2, and Neh3 domains [34]) and two nuclear export sequences (NES) (NESzip [35] in the bZIP region of Neh1 domain and NESTA [36] in the Neh5 transactivation domain). NESTA is a redox-sensitive signal: oxidation of cysteine 183 in its structure promotes Nrf2 nuclear accumulation, possibly, via preventing access of the nuclear exportin CRM1 to the NESTA [36]. Nuclear retention of Nrf2 is enhanced after heterodimerization with Maf in the nucleus perhaps due to the NESzip masking [37]. Under unstressed condition Bach1 TF competes with Nrf2 for binding Maf, repressing transcription of ARE-regulated genes [38], [39], wherein Nrf2 activates Bach1 expression [40]. Increase of intracellular heme level inhibits Bach1 negative regulation of Nrf2, allowing Nrf2 to activate target genes, including heme oxygenase-1 (HO-1), the key enzymes, regulating heme catabolism [38]. Another Nrf2 negative regulator is c-Myc, competing for ARE and increasing Nrf2 degradation [41]. Additional cellular mechanisms for Nrf2 control, including epigenetics, transcriptional, post-transcriptional regulations are reviewed in detail elsewhere [42], [43], [44], [45], [18], [6].
Fig. 2.
The Nrf2 signaling pathway.
3. Keap1
Human Keap1 is 69 kDa protein containing 27 cysteine residues, most of them are accessible for redox oxidation or electrophiles conjugation. The Keap1 cysteine thiols reactivity depends on concentration and varies for different inducers (so-called “cysteine code”), providing a basis for fine-tuning and differentiating response pattern for different stressors [46], [47]. In addition to the N- and C-end domains, three major functional Keap1 domains (Fig. 1b) have been described: BTB, homodimerization domain; IVR domain with most reactive cysteines, acting as sensors; Kelch repeat domain, binding Nrf2. BTB and IVR also participate in binding Cul3 ubiquitin ligase, carrying out Nrf2 ubiquitination. To date, the mechanism of Keap1-dependent regulation is considered as following. Two Keap1 molecules bind high affinity ETGE and low affinity DLG sites of Nrf2, resulting in a stoichiometric ratio 2:1. After homodimerization Keap1 bridges Nrf2 to the Cul3/RING-box protein complex, which ubiquitinates Nrf2 lysine residues, located in the Neh2 domain. The ubiquitinated Nrf2 is extracted from the complex with Keap1-Cul3 by the p97 protein [48] and targeted for proteasomal degradation whereas the Keap1 is regenerated [49]. Thus, under basal unstressed conditions the Keap1 binds Nrf2, sequestering it from translocation to the nucleus. During stress, oxidants or electrophilic molecules modify Keap1 cysteine residues, which lead to the conformational changes and to the switching off Keap1-dependent negative Nrf2 regulation [50], [51]. The concentration of Keap1 in the nucleus is several times lower than in the cytoplasm and this results in a low level of Nrf2 basal activity [52]. Nuclear Keap1 import is Nrf2-independent and is mediated through importin α7 [53]. Similar to Nrf2, Keap1 contains NES, ensuring mechanism to terminate Nrf2 activity through its export to cytoplasm after recovery of cellular redox homeostasis [54], [55]. Another probable mechanism to turn off Nrf2 activation is the Nrf2 ubiquitination and degradation inside the nucleus via nuclear translocation of Keap1/Cul3 complex [56], [57]. Additional mechanism of nuclear Nrf2 repression was discovered during investigation of Hutchinson-Gilford progeria syndrome (HGPS), premature aging syndrome, when Nrf2 is switched off due to the trapping to the nuclear periphery by mutated lamin A (progerin) protein, resulting in chronic oxidative stress and aging effects [58]. The reactivation of the Nrf2 pathway in HGPS patient cells reversed oxidative stress and cellular HGPS effects [59], [60]. During aging Nrf2 activity declines, whereas level of its inhibitors increases [61], [62]. Lewis et al. found a positive correlation between constitutive Nrf2 signaling activity and maximum lifespan potential, studying long-lived naked mole rats and other rodent species [63]. Remarkably, the elevated Nrf2 activity significantly correlated with level of Nrf2 negative regulators Keap1 and β-TrCP, but not with the expression of the Nrf2 itself. Taken together, these data suggest that if free radical theory of aging holds, Nrf2 could be a key regulator of this process.
The absolute quantity of Nrf2, Keap1, and Cul3 were determined in the paper of Iso et al. using quantitative immunoblot analyses of five murine cell lines in basal and induced states [52]. Surprisingly, the Nrf2 and Keap1 tend to be very abundant cellular proteins with the total level, ranging in a basal state from 50,000 to 300,000 Keap1 molecules per cell and from 49,000 to 190,000 Nrf2 molecules per cell [52] exceeding the amount of many other TFs [64]. In the Raw264.7 cells upon electrophilic stimuli the Keap1 and Cul3 level does not change, whereas the Nrf2 level increased fourfold in the cytoplasm/organelles and tenfold in the nucleus, reaching concentrations of 0.6 μM and 2.7 μM, respectively [52]. Tight control of the Nrf2 via different mechanisms results in a rapid turnover of the Nrf2: in a basal state the Nrf2 half-life in different cells is 7–18 min [65], [66], [67], [68] and different inducers prolong its half-life by several times [65], [68]. The Keap1 half-live is 12.7 h and, unlike Nrf2, it is shortened after reacting with stressors [69]. Besides Nrf2, Keap1 also regulates Bcl-2 [70] and IKKβ [71], promoting theirs degradation.
Keap1 have been reported to interact with more than a dozen cellular proteins with a motif, resembling the Nrf2 ETGE [72], [73]. Furthermore, recent databases analysis identified 40 possible Keap1-interacting cellular proteins with a similar motif [74]. These interactions are considered as a non-canonical Nrf2 activation mechanism, since other cellular partners compete with Nrf2 for the Keap1 and increase in their concentration could activate the Nrf2 via direct displacement it from the complex with the Keap1 [75], [72]. The IKKβ, intracellular activator of NF-κB, contains both ETGE and DLG motifs, resembling Nrf2, and the ETGE is essential for Keap1 interaction [71]. The p62, stress-inducible protein, captures the Keap1 with 349DPSTGEL355 motif and is the well-studied example of such cellular proteins, disrupting Nrf2:Keap1 interactions. The p62, positively regulated by Nrf2, participates in autophagy, shuttling proteins for autophagic degradation [75]. Phosphorylation of the S351 in the p62 interacting motif by mTORC1 significantly (30-fold) improve affinity to the Keap1, but still several times weaker than Nrf2 ETGE motif [76]. Besides Nrf2 displacement p62 binding triggers Keap1 autophagic degradation, resulting in a more sustained Keap1 inactivation [77]. Table 1 listed several cellular proteins with a motif, resembling ETGE, shown to compete for the Keap1. Unlike aforementioned examples, p21, cyclin dependent kinase inhibitor, binds to Nrf2, both DLG and ETGE motifs, through 154KRR156 sequence, but compete with the Keap1 only for a weaker DLG site [78]. p21 has many functions, including those involved in the cell cycle regulation, DNA repair, apoptosis [79], [80]. DNA damage activates p21, which blocks cell cycle progression and simultaneously induces cellular protective response through the interplay with the Nrf2 pathway.
Table 1.
Cellular proteins, competing with Nrf2 for the Keap1.
| Protein | Interacting motif | Description |
|---|---|---|
| p62 | 349DPSTGEL355 | autophagy protein, shuttling target proteins, including Keap1, for degradation [76], [77], [81]. |
| prothymosin α | 42EENGE46 | anti-apoptotic protein, involved in cell proliferation [56], [82]. |
| DPP3 | 480ETGE483 | zinc-dependent metallopeptidase that hydrolyses dipeptides at the N-terminal site and contributes to the protein turnover [83], [84]. |
| WTX | 286SPETGE291 | tumor suppressor mediates degradation of β-catenin, thereby downregulating WNT/β-catenin signaling pathway [85]. |
| PALB2 | 91ETGE94 | a major BRCA2 binding partner, controlling its nuclear localization, DNA repair and checkpoint function [86]. |
| iASPP | 239DLT241 | a binding partner and transactivity inhibitor of NF-kB and p53 [87]. |
| CDK20 | 25ETGE28 | а protein kinase, promoting cell proliferation and radiochemoresistance [88]. |
| gankyrin | 21ELKE24 and 201ENKE204 | oncoprotein, facilitating degradation of p53 and Rb [89]. |
| HBXIP | 110GLNLG114 | oncoprotein, transactivator of several oncogenes, regulator of cellular apoptosis and division [90]. |
| MCM3 | 387ETGE390 | a subunit of the replicative DNA helicase [91]. |
| IKKβ | 36ETGE39 | part of the IKK complex, which activates NF-κB after pro-inflammatory stimuli [71]. |
| FAM129B | 708DLG710 and 718ETGE721 | antioxidative protein [92] |
| RMP | 215EELERQE221 and 246EEKE249 | oncogene whose product acts as an inhibitor of PP1γ (protein serine/threonine phosphatase gamma) [93] |
The network of Nrf2-regulated genes consists of hundreds of genes [94], [95], some modulated directly and others indirectly/secondarily through Nrf2-regulated TFs, such as Notch1, MafG, C/EBPβ, retinoid X receptor α, arylhydrocarbon receptor [95], [96], [97], [98], [99]. Also, regulation of such wide battery of genes partially could be realized through crosstalk with other signaling pathways such as Notch1 [99], heat shock proteins [100], Nf-kB [101], [71], p53 [102], PI3K-AKT, and mTOR [15]. Genome scale analysis using chromatin immunoprecipitation with parallel sequencing revealed 645 and 654 genes, which are direct targets for basal Nrf2 activity and induced Nrf2 activation, correspondingly, wherein only 244 genes are intersected [98].
4. Clinical significance
Most of the Nrf2 regulated genes are associated with cell defense pathways, such as protection against oxidative stress. Oxidative stress is a common feature of many diseases, such as neurodegenerative (Alzheimer's disease (AD), Parkinson's disease (PD), amyotrophic lateral sclerosis (ALS), multiple sclerosis (MS), frontotemporal dementia, Friedrich ataxia, Huntigton disease [5], [103]), cardiovascular [104], [105], inflammation diseases (atherosclerosis [106], rheumatoid arthritis [107], inflammatory bowel disease), airway diseases (asthma, chronic obstructive pulmonary disease (COPD) [108]) and a number of viral infections [109], [110], [111]. Since the excessive generation of ROS leads to the deregulation of the cellular redox balance during these pathologies, stimulation of the Nrf2 signaling could be beneficial offering a therapeutic strategy. Multiple lines of evidence from the literature support this approach potential for different indications. A meta-analysis of 9 PD and 7 AD microarray datasets revealed several dozen common downregulated ARE-contained genes [112]. Variations in the Nrf2 gene are associated with the alteration of PD risk and the age of its onset [113], [114], [115], affect AD progression [116], alter the risk of ALS [117] and ulcerative colitis [118]. Neuroprotective effect of Nrf2 activation has been shown in several studies with the mouse models of PD [119], [120], [121], [122]. In AD neurons Nrf2 localizes predominantly in inactive cytoplasmic pool, in contrast to normal hippocampus neurons with mainly nuclear localization of Nrf2 [123]. Intracranial delivery of Nrf2-expressing lentivirus into hippocampus resulted in significant improvement of spatial learning in murine AD model, comparing with the control vector [124]. In other study inhibition of GSK3β, Nrf2 repressor, increased Nrf2 nuclear level, its transcriptional activity, decreased oxidative stress with improved learning and memory [125]. During COPD, caused by air pollutants, including cigarette smoke, Nrf2 pathway activity is decreased and the disease severity is negatively correlated with the Nrf2 expression [126], [127]. Nrf2 activation increases pulmonary bacterial clearance by alveolar macrophages and improves protection against opportunistic bacterial infections [128]. Tecfidera (BG-12, dimethyl fumarate [129]) is FDA-approved first line treatment for relapsing MS with the mechanism, widely accepted, involving Nrf2 activation (possibly, through blocking its degradation [112]). Treatment with the Nrf2 activators also has been shown encouraging results in mouse model of ALS [130]. Additional information regarding the Nrf2 role in the neurodegenerative processes are given in recent reviews [103], [131], [132], [133], [134], [135].
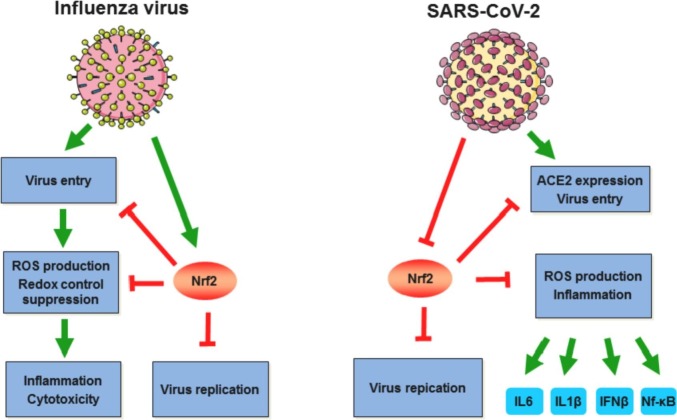
Oxidative stress contributes to the pathogenesis of wide range of viral diseases and expression of different Nrf2-regulated genes was shown to be altered during viral infections. The Nrf2 role in viral diseases could be of opposite sign, depending on virus type, cell type or stage of infection: different viral proteins during replication promote oxidative stress, which should be controlled by the virus in order to secure optimal replication conditions [136], [137], [138]. As Nrf2 exerts multiple effects, most of the studied viruses (for example, influenza virus) induce it to facilitate their replication through protection against virus-induced cytopathic effects by increasing antioxidant genes expression (for example, influenza virus (Fig. 3 ) [139]). For influenza virus it was observed on a single cell level that Nrf2-regulated genes expression was associated with a higher viral transcript expression [140]. Other example of Nrf2 activating virus is the Dengue virus, which activates Nrf2 via nuclear translocation and after endoplasmic reticular stress this signaling lead to the Nrf2-mediated TNF-α production which, in turn, contributes to the severity of illness in patients with DENV infection [141]. Recently, Ferrari et al. demonstrated that during first 24 h of the DENV infection Nrf2 is activated with increased expression of antioxidant genes, restricted IFN type I response, decreased ROS production and inflammation [142], thereby limiting antiviral response. Later (24 h–48 h after infection) viral protease NS2B3 cleaves Nrf2, resulting in ROS accumulation and increased inflammation, creating conditions favoring viral replication [142]. The contrary the replication strategy of respiratory syncytial virus is to promote Nrf2 proteasomal degradation, thereby suppressing its antioxidant activity [143], [144]. Nrf2-deficient mice had significantly higher RSV infection severity comparing with wild type mice and pretreatment with Nrf2 inductor limits virus replication and inflammation in mice with functional Nrf2 [145]. In a similar fashion Nrf2 activation inhibited replication and virus-induced cytotoxicity of several rotavirus strains [146]. Likewise, the Nrf2 agonists impaired herpes simplex virus 1 replication, whereas infected cells with high Nrf2 activity demonstrated lower level of the HSV-1 replication [147]. For some viral infections data are conflicting and mixed. Bender and Hildt in their recent review summarized reports about the Nrf2 role in viral hepatitis infections [148]. Authors surmised that the contradictory results regarding HCV and Nrf2 could be explained by different cellular stress response induction in acute and chronic HCV viral models (high ROS production and depletion of GSH in acute phase and lower ROS level and increased GSH in chronic infection) [148]. A flood of paper over past year regarding SARS-CoV-2 infection demonstrates a major role of oxidative stress and excessive inflammation response in COVID-19 pathogenesis [149], [111], [150]. Besides oxidative stress Nrf2 also modulates inflammation through downregulating proinflammatory cytokines IL-6 and IL-1β expression, suppressing type I IFN response and acting as an antagonist of NF-kB signaling [151], [152], [153]. It is noteworthy that, Il-6 blockade has been considered as a treatment against COVID-19 and in some conditions Il-6 antagonists deployment may be beneficial [154]. Olagnier et al. have demonstrated that NRF2 pathway is suppressed in lung biopsies obtained from COVID-19 patients, whereas Nrf2 activators such as 4-octyl-itaconate and dimethyl fumarate trigger cellular antiviral response, resulting in SARS-CoV-2 replication inhibition [155]. Moreover, Nrf2 activity suppression was shown to upregulated ACE2, SARS-CoV-2 receptor, while Nrf2 activation reduce ACE2 level [156], which could led to a lower virus internalization (Fig. 3). Moreover, ACE2 is an enzyme, which cleaves angiotensin II (Ang II), a vasoconstrictor, to angiotensin 1–7, a vasodilator. Ang II produces ROS by stimulating membrane-bound NADPH oxidase [157], [158]. The degradation of Ang II by ACE2 reduces oxidative stress as it inhibits NADPH oxidase, and therefore, Ang II-induced ROS production. If ACE2 is bound to the S protein, the level of Ang II will increase, leading to an increased presence of superoxide species and subsequent cell damage, inevitably creating a cycle of oxidative stress, and ultimately, increasing the risk of severe course of COVID-19 [150]. These findings are consistent with previous report, describing antiviral activity of heme oxygenase 1, a well-known target of the Nrf2 induction, against a panel of viruses including HIV, HCV, HBV, influenza virus, Ebola virus and others [159]. At the same time, the activation of NRF2 pathway by ROS is nonlinear. Low doses of an oxidizing agent such as ozone can have a therapeutic effect through the activation of NRF2 in viral diseases. In particular, ozone therapy is considered as a promising method of treatment for COVID-19 [160]. These efforts, investigating Nrf2 role, set the foundation for further studies that can establish the effects of Nrf2 pathway modulation in different viral diseases.
Fig. 3.
Nrf2 and its role in influenza and SARS-CoV-2 infections.
This figure was created using images from Servier Medical Art by Servier under a Creative Commons Attribution 3.0 Unported License.
The dark side of the Nrf2, the other facet of this system, is its role in cancer promotion, where Nrf2 is usually up-regulated, providing to the malignant cells survival and growth benefits [161], [162], [32], [45]. Whereas short-term Nrf2 activation counteracts oxidative stress, which could lead to clinical benefits and even suppress (trough ROS and chemical carcinogens neutralization) cancer initiation, especially in its earliest stages, long-term activation could promote the development of carcinogenesis and help cancer cells to evade chemo- and radiotherapy [163], [164], [165]. The constitutive Nrf2 pathway activation is a feature of different cancers and often is associated with the poor prognosis. The meta-analysis of the Nrf2 prognostic value in solid tumor patients showed that the high Nrf2 expression level had negative impact on survival [166]. The Nrf2 up-regulation could be realized through oncogenes (KRAS, BRAF, MYC [167]), via mutations in Nrf2 Neh2 domain [168] or via Keap1 [169], [170], including epigenetic Keap1 silencing [171], Keap1 inactivation through oncometabolites [172], stress, and hormone signaling, accumulation of Keap1:Nrf2 disrupting cellular proteins [32]. The dual role of Nrf2 in cancer suggests that for therapy activation or inhibition strategy for Nrf2 should be chosen depending on the context [161]. For malignancies inhibition of the Nrf2 pathway could be beneficial. On the other hand, taking into account that the Nrf2 activity in many degenerative processes usually plays a protective role, it seems straightforward to augment it through additional activators.
5. Electrophilic Nrf2 inducers
To date, two main approaches have been developed for Nrf2 pathway activation: electrophilic activators and inhibitors of Nrf2:Keap1 interactions. Several electrophilic activators are in advanced clinical stages with one molecule (Tecfidera) approved by FDA [173]. The mechanism of their action is to imitate the natural stress-induced Nrf2 activation through forming covalent adducts with the sulfhydryl groups of the Keap1 cysteines [174]. Magesh et al. have reviewed a dozen of chemical agents classes, inducing Nrf2 activity: Michael acceptors (2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid, CDDO, and dimethyl fumarate), oxidizable diphenols and quinones (tert-butylhydroquinone, tBHQ), isothiocyanates and sulfoxythiocarbamates (sulphoraphane), dithiolethiones and diallyl sulfides (oltipraz), vicinal dimercaptans (lipoic acid), trivalent arsenicals, selenium-based compounds, polyenes (lycopene), hydroperoxides (tert-butylhydroperoxide), heavy metals and metal complexes [175]. Reactivity of the Keap1 different cysteines toward different inducers varies [176] with some residues (Cys151, Cys273, Cys288) have been shown to be critical for Keap1 function [47], [174]. The apparent mechanism of stress-induced Nrf2 induction is the Keap1 conformation changes, followed with the Nrf2 release from the complex with it [51], [50]. The most studied Nrf2-induced compounds are dimethyl fumarate (Tecfidera), sulforaphane, naturally occurring isothiocyanate isolated from cruciferous vegetables, such as broccoli and cauliflower, and CDDO-Me, also originated from the natural product, isolated from pharmaceutically active plants [177]. The Nrf2-inducing activity of all three agents is highly dependent on the Keap1 Cys151 modification. Despite the exact mechanism of the dimethyl fumarate remains unknown, its Nrf2-inducing activity has been demonstrated in vitro [178] as well as in vivo [179]. Structural studies of the CDDO-Me revealed formation of the adduct with the Keap1 Cys151, inhibiting binding to the Cul3. Blocking the Keap1:Cul3 interactions has been proposed also as a possible sulforaphane-mediated mechanism. The importance of Nrf2 pathway activation as an anti-viral approach has been reinforced by several electrophilic compounds. Thus, dimethyl fumarate and sulforaphane have been shown to inhibit HIV replication in macrophages [180], [181]. Besides HIV, sulforaphane-mediated antiviral activity has been demonstrated against influenza virus [182], respiratory syncytial virus [145], hepatitis C virus [183], and herpes virus [184], [147].
The Achilles heel of electrophilic agents is their non-discriminative concentration-dependent [161] reactivity toward a variety of cellular nucleophilic molecules, laying the foundation for off-target effects. Some Nrf2-inducing electrophilic molecules have been shown to react with other cellular targets [185], [186], raising questions about the selectivity of such agents to Nrf2 signaling, complicated mechanism of their action and possible influence on other pathways. Even dimethyl fumarate and sulforaphane, two of the most popular Nrf2 inductors, have possible NRf2-independent effects [187], [188], [189], [190]. Furthermore dimethyl fumarate demonstrated equal therapeutic effects on multiple sclerosis murine model in Nrf2−/− and WT mice, raising concerns about mechanism of its action [188]. For these reasons it is recommended to minimize unspecific reactive molecules in drug development process, despite there is not a clear link between toxicity and drug-protein adducts formation [188]. Also, similarity of Nrf2 inducers with pan assay interference compounds (PAINS, molecules, showing false bioactivity in many assays [191]) was noticed by several authors [192], [193], [194]. For instance, one of the most insidious PAINS, curcumin [191], is often recognized as a potent Nrf2 activator [195], [196]. Besides specificity issue, efficiency of such electrophilic induction is challenged due to the unfavorable cytoplasmic milieu. Thus, enzymes of cellular xenobiotic metabolism system conjugate reactive electrophilic molecules with abundant polar compounds with subsequent excretion out of cells [197], [198], [199]. Thereby being able to affect whole thiol proteome, provoking Nrf2-independent effects, electrophilic inducers have narrow therapeutic window.
For this reason researchers are making progress toward more specific Nrf2 inducers with decreased toxicity, through development of new possible Nrf2 activators or chemically “tuning” previously studied inducers [200]. Thus, Copple et al. compared the Nrf2 activating potency of sulforaphane, DMF, and some structurally related molecules [201]. Using cell line with stable expression of luciferase ARE reporter they found that for several electrophilic inducers increase in Nrf2 activating ability is associated with much smaller increase in toxicity and better in vitro therapeutic index than for well-known clinically studied Nrf2 inducers. Other recent work also demonstrated that medicinal chemistry offer an opportunity to modify previously described Nrf2 activators to less cytotoxic molecules with preserving Nrf2 activating activity [202]. Authors from another study pointed toward pro-electrophilic Nrf2 inducers, which become activated only by strong oxidation in injured cells [203]. Such mechanism accounts for preserving GSH level in uninjured cells, whereas electrophile inducers, for instance DMF, deplete GSH in a concentration-dependent manner through non-specific reaction with its thiol group, restraining normal cellular protection ability [204]. More specific derivatives such as monoethyl fumarate (MEF), reactive only to the sole Cys151 on KEAP1 instead of DMF could be more safe Nrf2 activators [204].
Another way for Nrf2 activation is concurrent inhibition of its interaction with Keap1. It is important to note that such an approach enables more specific regulatory impact. Furthermore, taking into account the side effects of the constant activation of Nrf2 and a long time during which the pathogenesis of neurodegenerative diseases takes place [205], creation of drugs for cell- or tissue-specific activation of this pathway is of high interest.
6. Direct inhibitors of Nrf2:Keap1 interactions
Alternative widely used approach for Nrf2 signaling activation is non-electrophilic activators of the Nrf2 system, based on direct interruption of Nrf2:Keap1 interactions. The Nrf2:Keap1 PPI are well described for the complexes Keap1 Kelch domain with the ETGE and DLG motifs from the Nrf2. The Keap1 Kelch domain constitutes a six-bladed β-propeller structure, while the Neh2 domain is intrinsically disordered [206] . Protein data bank (PDB) discloses several dozen of the Keap1 structures, in bound and unbound states [207], [22], [169], [208]. For the ETGE and DLG motifs the interface surfaces are approximately 529 Å2 and 820 Å2 [209] correspondingly. The relatively small interacting areas make possible the development of direct inhibitors or the Nrf2:Keap1 PPI, both peptides and small molecules.
A number of peptide ligands were screened, capable to displace the endogenous Nrf2 from the complex with the Keap1. The minimal length of the ETGE motif peptides was estimated by several groups by truncating the Nrf2 Neh2 domain. Hu's group investigated by surface plasmon resonance method a dozen of ETGE peptides from 7mer to 16mer [210]. 7mer (H-EETGEFL-OH) and 8mer (H-DEETGEFL-OH) peptides had affinity to Keap1 much greater than 1 μM. The 9mer peptide Nrf2 (H-LDEETGEFL-OH) was more active with a Kd of 352 nM. One order increase in potency was demonstrated with 10mer peptide (H-QLDEETGEFL-OH), whereas further consistent elongation of Nrf2 peptides till 16mer didn't improve affinity. Fluorescence polarization (FP) assay confirmed the similar ranking of the same peptides with a significant jump in activity for the peptides greater than 10mer [211], while 16mer Nrf2 peptide was significantly better than shorter variants. Well's group with the aid of FP assay also investigated a series of the DLG and ETGE peptides [212], [213]. Peptides of various lengths were examined and the Kd of 7mer, 9mer and 10mer Nrf2 peptides were 96 nM, 54 nM and 51 nM, correspondingly [212]. In a further study authors increased lipophilicity of several 7mer Nrf2 peptides through including stearoyl group to the N-terminus [213]. Such modification significantly increased affinity with the most potent molecule (St-DPETGEL-OH) was almost 4 times more effective, than Neh2 domain of the Nrf2 [213].
Other non-proteinogenic chemical peptide modifications such as N-terminal acetylation improve binding of some peptide variants to the Keap1, possibly through decreasing unfavoured electric charges of N-terminal free amino group [210]. Insertion of terminal cysteine residues for further head-to-tail cyclization strengthened binding to the Keap1 [214]. The tighter variant (Ac-c[CLDPETGEYLC]-OH) demonstrated 2.8 nM affinity to Keap1 and 9.4 nM IC50 in FP activity assay. Recent paper from the same laboratory evaluated potential of the strong linear Keap1 binder (Ac-LDPETGEYL-OH, Kd = 42 nM [214]) as moiety to recruit Keap1 in vivo in order to downregulate cellular target protein, Tau [215]. The artificial peptide consisted of three functional parts: Keap1 and Tau recognizing modules as well as cell penetration sequence. The results confirmed the viability of such approach for the Keap1-mediated and proteasome-dependent intracellular target degradation. Representative Nrf2:Keap1 peptide inhibitors with evaluated Kd less than 100 nM are listed in the Table 2 .
Table 2.
Some promising Nrf2:Keap1 peptide inhibitors.
| Compound | Affinity to the Keap1 | Link |
|---|---|---|
| H-QLDEETGEFL-OH | 27 nM | [210] |
| FITC-AFFAQLQLDEETGEFL-OH | 29 nM | [211] |
| FITC-β-DEETGEF-OH | 96 nM | [212] |
| FITC-β-LDEETGEFL-OH | 54 nM | [212] |
| FAM-LDEETGEFLP-OH | 51 nM | [212] |
| St-DPETGEL-OH | 22 nM | [213] |
| Ac-c[CLDPETGEYLC]-OH | 3 nM | [214] |
| Ac-LDPETGEYL-OH | 42 nM | [214] |
| monobody R1 | 300 pM | [216] |
| GQLDPETGEFL | 87 nM | [217] |
| c[GQLDPETGEFL] | 18 nM | [217] |
The intensive search for potential peptide inhibitors of NRF2-Keap1 interaction inhibitors continues [218], [219]. Many linear and cyclic peptides show promising results in vitro. However, neither cyclization nor conjugation with cell penetrating peptides (CPP) could generate cell-active peptides in many cases [218]. Thus, the development of methods for the effective intracellular delivery of antibodies, their fragments, or antibody-like molecules is extremely important. Various strategies for intracellular targeting of antibodies via protein-transduction domains or their mimics, liposomes, polymer vesicles, and viral envelopes are now under the scrutiny of developers [220].
An appealing strategy for design the Keap1 binders with better than wild type Nrf2 affinity is the use of antibody-like alternative protein scaffold. To date, the sole example of this strategy is the article, published by Guntas et al., who engineered artificial protein with a subnanomolar affinity to the Keap1 Kelch-domain [216]. As a scaffold authors used monobody, 10th type lll domain of human fibronectin, which forms three unstructured flexible loops, similar to complementarity-determining regions of the traditional antibodies, and is well tolerated to the mutagenesis [221], [222], [223]. The best binder clone R1 had the RDEETGEFHWP in one of the loops and demonstrated affinity toward the Keap1 15-time tighter (Kd = 300 pM) than full-size Nrf2. Meanwhile, despite reported superb affinity, cellular effects after monobody R1 gene transfection were quite moderate [216].
Despite promising in vitro activity in a low nanomolar concentrations cellular effect of NRF-derived peptides were exhibited in a micromolar range, implying quite poor cell entry [213]. Peptide-inhibitors ineffective access to the cytoplasmic Keap1 necessitated efforts for delivery approaches development. The most popular strategy for peptides delivery is the use of CPP, cell penetrating peptides [224], [225]. Steel et al. [226] generated a series of Nrf2-peptides (10mer, 14mer and 16mer), fused to the TAT-peptide, derived from the HIV-1 Trans-Activator of Transcription protein. The most potent inducer of HO-1 expression (Nrf2 activation target) was Nrf2 14mer peptide (LQLDEETGEFLPIQ), conjugated with TAT. HO-1 expression was up-regulated in a time- and dose-dependent manner and reached maximum after 6 h and 12 h, measured as mRNA and protein, respectively [226]. Similar approach was described in Zhao et al. article, who also synthesized TAT-Nrf2 peptide [227]. Unlike to previous studies, they used shorter Nrf2-peptide (DEETGE) and designed it for Nrf2 activation in injured brain. Intracerebroventricular infusion of TAT-DEETGE into mouse with model traumatic brain injury had no positive effects on Nrf2 regulation [227]. Modified peptide with calpain cleavage site between TAT and DEETGE (TAT-CAL-DEETGE) increased expression of Nrf2-target genes in brain of injured mice, comparing with healthy controls [227]. In a further study TAT-CAL-DEETGE peptide was tested as a potential neuroprotectant against global cerebral ischemia [228]. On a cellular level it decreased Keap1:Nrf2 interactions in the cytoplasm and enhanced nuclear Nrf2 translocation. Both intracerebroventricular pretreatment as well as peripheral post-treatment administration of this peptide exerted neuroprotection and preserved cognitive functions in rats after ischemia inducement [228].
Small molecules approach is an alternative to peptide-inhibitors due to the better stability and oral bioavailability. Different screening techniques have identified dozen of small molecules inhibitors of Nrf2:Keap1 interactions with several compounds exerted activity in a nanomolar range [209], [229], [230]. Mimicking the natural Nrf2:Keap1 interactions Jiang et al. [231] have discovered a compound with high affinity to the Keap1 (Kd – 3.6 nM) and 28.6 nM IC50 in the FP assay. This compound demonstrated dose-dependent cellular activity in Nrf2 induction and increased expression of Nrf2-regulated genes. Further modification of this molecule enhanced two-fold cellular activity (IC50 was determined to be 14.4 nM [232]), induced Nrf2 downstream genes and exhibited anti-inflammatory effects in mice. Davies et al. [233] developed other potential small molecule inhibitor with high affinity to the Keap1 (Kd = 1.3 nM) and demonstrated its ability to activate Nrf2 in both cell-based assay as well as in vivo in rat respiratory disease model.
These results demonstrated the potential of small molecules and Nrf2-derived peptides to inhibit Nrf2:Keap1 interactions and thereby to activate Nrf2/ARE pathway. Measured in vitro high affinity of such inhibitors could be translated in a clinical relevant activity in case of taking into account huge intracellular Keap1 concentration and delivery issues.
7. Room for competitive Nrf2 regulation by drugs
Modulation of PPI depends on relative quantities and interaction strength of interacting proteins (including intracellular endogenous competitors), which determines the inhibitor amount inside cells for effective inhibition.
Thereby efficient PPI inhibitors delivery inside cells is the critical issue. For small molecules it's relatively straightforward to alter their structure for transmembrane diffusion, however it could decrease affinity to the target [234]. Peptides delivery to the cytosol or nucleus requires use of the vectors, the most popular of which is CPPs, a group of short peptides with high proportion of basic amino acids [235], [236], [237]. CPP translocation to the cytoplasm occurs in a receptor-independent pathway, mechanism of which is still questionable [238], [239]. CPP delivered various cargos inside cells [237], [236], but typically required high concentration for their activity [226], [240], therefore without an active transport the intracellular concentration of delivered cargos is less than their environment. Also, the penetration inherent non-specificity greatly hampers in vivo use of CPP technology, restricting it mostly to the in vitro/ex vivo applications. Thus, there is a desperate need for the development of technologies, able [241] to deliver Nrf2-derived peptides for Nrf2:Keap1 interactions efficient inhibition in target cells.
Nrf2, one of the stimuli-sensitive TFs, sits at the hub of the complex network, regulating cell defense pathways, including antioxidant protection. The involvement of oxidative stress in pathogenesis of numerous pathologies presents short-term Nrf2 activation in target cells as a viable cytoprotective therapeutic strategy for the treatment of some neurodegenerative, cardiovascular, inflammation diseases and possibly some viral diseases [242], [243]. Several small molecules Nrf2 inducers are in clinical trials with one compound approved as a first-line oral therapy for MS. To circumvent the drawbacks of unspecific cysteine-modifying Nrf2 activators direct Nrf2:Keap1 PPI inhibitors have been developed. However, up to 1 μmol the Keap1 cytoplasmic concentration [52] set a high bar for this approach, stimulating search for threshold estimation of minimal concentration of disrupting compound, capable to outcompete endogenous Nrf2 for binding to the Keap1. The rise of antibody-like scaffolds provides tremendous opportunities for the development of small high-affinity PPI inhibitors for virtually any intracellular protein [244]. The concern of high intracellular concentration of targets like Keap1 could be alleviated by the deployment of targeted protein degradation technologies [245], which are shown to achieve substoichiometric target inhibition [246], [247]. A significant challenge is the creation of multi-functional systems that can provide specific in vivo delivery of effective Nrf2 activators into cells that require the protection minimally affecting other organs and tissues. A possible solution to this problem could be new drugs based on the development of polypeptide modular nanotransporters [248] or multifunctional nanoparticles [249].
This work was supported by the Russian Science Foundation Grants 17-14-01304 and 21-14-00130.
CRediT authorship contribution statement
Alexey V. Ulasov: Writing – original draft. Andrey A. Rosenkranz: Writing – review & editing. Georgii P. Georgiev: Funding acquisition. Alexander S. Sobolev: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Halliwell B. Free radicals and other reactive species in disease. eLS. 2015 doi: 10.1002/9780470015902.a0002269.pub3. [DOI] [Google Scholar]
- 2.Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 3.Finkel T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 4.D'Autreaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell. Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 5.Barnham K.J., Masters C.L., Bush A.I. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 6.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Talalay P., Long M.J., Prochaska H.J. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moi P., Chan K., Asunis I., Cao A., Kan Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T., Yamamoto M. Molecular basis of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2015;88:93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell. Biol. 2020;40 doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baird L., Yamamoto M. The Keap1-Nrf2 pathway: From mechanism to medical applications. 2020. pp. 125–147. [DOI] [Google Scholar]
- 14.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., Dinkova-Kostova A.T. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 15.Dodson M., Vega M.R., Cholanians A.B., Schmidlin C.J., Chapman E., Zhang D.D. Modulating NRF2 in disease: timing is everything. Annu. Rev. Pharmacol. Toxicol. 2019;59:555–575. doi: 10.1146/annurev-pharmtox-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuse Y., Kobayashi M. Conservation of the Keap1-Nrf2 system: an evolutionary journey through stressful space and time. Molecules. 2017;22 doi: 10.3390/molecules22030436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gacesa R., Dunlap W.C., Barlow D.J., Laskowski R.A., Long P.F. Rising levels of atmospheric oxygen and evolution of Nrf2. Sci. Rep. 2016;6 doi: 10.1038/srep27740. (27740-doi:10.1038/srep27740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 2017 doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenkov N.K., Kozhin P.M., Chechushkov A.V., Martinovich G.G., Kandalintseva N.V., Menshchikova E.B. Mazes of Nrf2 regulation. Biochemistry (Mosc) 2017;82:556–564. doi: 10.1134/S0006297917050030. [DOI] [PubMed] [Google Scholar]
- 20.Eggler A.L., Liu G., Pezzuto J.M., Breemen R.B., Mesecar A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukutomi T., Takagi K., Mizushima T., Ohuchi N., Yamamoto M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol. 2014;34:832–846. doi: 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo S.C., Li X., Henzl M.T., Beamer L.J., Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a "tethering" mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 24.Horie Y., Suzuki T., Inoue J., Iso T., Wells G., Moore T.W., Mizushima T., Dinkova-Kostova A.T., Kasai T., Kamei T., Koshiba S., Yamamoto M. Molecular basis for the disruption of Keap1-Nrf2 interaction via Hinge & Latch mechanism. Commun. Biol. 2021;4:576. doi: 10.1038/s42003-021-02100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nioi P., Nguyen T., Sherratt P.J., Pickett C.B. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell. Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 28.Sun Z., Chin Y.E., Zhang D.D. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell. Biol. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/beta-TrCP. Free Radic. Biol. Med. 2015;88:147–157. doi: 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Wang H., Liu K., Geng M., Gao P., Wu X., Hai Y., Li Y., Li Y., Luo L., Hayes J.D., Wang X.J., Tang X. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013;73:3097–3108. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 31.Canning P., Sorrell F.J., Bullock A.N. Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med. 2015;88:101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung B.J., Yoo H.S., Shin S., Park Y.J., Jeon S.M. Dysregulation of NRF2 in cancer: from molecular mechanisms to therapeutic opportunities. Biomol. Ther. (Seoul.) 2018;26:57–68. doi: 10.4062/biomolther.2017.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo S.C., Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theodore M., Kawai Y., Yang J., Kleshchenko Y., Reddy S.P., Villalta F., Arinze I.J. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J. Biol. Chem. 2008;283:8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., Jain M.R., Chen C., Yue X., Hebbar V., Zhou R., Kong A.N. Nrf2 possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J. Biol. Chem. 2005;280:28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 36.Li W., Yu S.W., Kong A.N. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J. Biol. Chem. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 37.Li W., Yu S., Liu T., Kim J.H., Blank V., Li H., Kong A.N. Heterodimerization with small maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim. Biophys. Acta. 2008;1783:1847–1856. doi: 10.1016/j.bbamcr.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davudian S., Mansoori B., Shajari N., Mohammadi A., Baradaran B. BACH1, the master regulator gene: a novel candidate target for cancer therapy. Gene. 2016;588:30–37. doi: 10.1016/j.gene.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 39.Dhakshinamoorthy S., Jain A.K., Bloom D.A., Jaiswal A.K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 40.Jyrkkanen H.K., Kuosmanen S., Heinaniemi M., Laitinen H., Kansanen E., Mella-Aho E., Leinonen H., Yla-Herttuala S., Levonen A.L. Novel insights into the regulation of antioxidant-response-element-mediated gene expression by electrophiles: induction of the transcriptional repressor BACH1 by Nrf2. Biochem. J. 2011;440:167–174. doi: 10.1042/BJ20110526. [DOI] [PubMed] [Google Scholar]
- 41.Levy S., Forman H.J. C-myc is a Nrf2-interacting protein that negatively regulates phase II genes through their electrophile responsive elements. IUBMBLife. 2010;62:237–246. doi: 10.1002/iub.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Y., Yu S., Zhang C., Kong A.N. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic. Biol. Med. 2015;88:337–349. doi: 10.1016/j.freeradbiomed.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayes J.D., Chowdhry S., Dinkova-Kostova A.T., Sutherland C. Dual regulation of transcription factor Nrf2 by Keap1 and by the combined actions of beta-TrCP and GSK-3. Biochem. Soc. Trans. 2015;43:611–620. doi: 10.1042/BST20150011. [DOI] [PubMed] [Google Scholar]
- 44.Krajka-Kuzniak V., Paluszczak J., Baer-Dubowska W. The Nrf2-ARE signaling pathway: an update on its regulation and possible role in cancer prevention and treatment. Pharmacol. Rep. 2017;69:393–402. doi: 10.1016/j.pharep.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Rojo D.L., Chapman V.E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto T., Suzuki T., Kobayashi A., Wakabayashi J., Maher J., Motohashi H., Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tao S., Liu P., Luo G., Vega R.M., Chen H., Wu T., Tllotson J., Chapman E., Zhang D.D. p97 negatively regulates NRF2 by extracting ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex. Mol. Cell. Biol. 2017;37 doi: 10.1128/MCB.00660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dinkova-Kostova A.T., Abramov A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015;88:179–188. doi: 10.1016/j.freeradbiomed.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baird L., Lleres D., Swift S., Dinkova-Kostova A.T. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baird L., Swift S., Lleres D., Dinkova-Kostova A.T. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol. Adv. 2014;32:1133–1144. doi: 10.1016/j.biotechadv.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iso T., Suzuki T., Baird L., Yamamoto M. Absolute amounts and status of the Nrf2-Keap1-Cul3 complex within cells. Mol. Cell. Biol. 2016;36:3100–3112. doi: 10.1128/MCB.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Z., Wu T., Zhao F., Lau A., Birch C.M., Zhang D.D. KPNA6 (Importin {alpha}7)-mediated nuclear import of Keap1 represses the Nrf2-dependent antioxidant response. Mol. Cell. Biol. 2011;31:1800–1811. doi: 10.1128/MCB.05036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Z., Zhang S., Chan J.Y., Zhang D.D. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velichkova M., Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol. Cell. Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karapetian R.N., Evstafieva A.G., Abaeva I.S., Chichkova N.V., Filonov G.S., Rubtsov Y.P., Sukhacheva E.A., Melnikov S.V., Schneider U., Wanker E.E., Vartapetian A.B. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol. Cell. Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf 2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gordon L.B., Rothman F.G., Lopez-Otin C., Misteli T. Progeria: a paradigm for translational medicine. Cell. 2014;156:400–407. doi: 10.1016/j.cell.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabriel D., Roedl D., Gordon L.B., Djabali K. Sulforaphane enhances progerin clearance in Hutchinson-Gilford progeria fibroblasts. Aging Cell. 2015;14:78–91. doi: 10.1111/acel.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kubben N., Zhang W., Wang L., Voss T.C., Yang J., Qu J., Liu G.H., Misteli T. Repression of the antioxidant NRF2 pathway in premature aging. Cell. 2016;165:1361–1374. doi: 10.1016/j.cell.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silva-Palacios A., Ostolga-Chavarria M., Zazueta C., Konigsberg M. Nrf2: molecular and epigenetic regulation during aging. Ageing Res. Rev. 2018;47:31–40. doi: 10.1016/j.arr.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Yu C., Xiao J.H. The Keap1-Nrf2 system: a mediator between oxidative stress and aging. Oxidative Med. Cell. Longev. 2021 doi: 10.1155/2021/6635460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis K.N., Wason E., Edrey Y.H., Kristan D.M., Nevo E., Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3722–3727. doi: 10.1073/pnas.1417566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biggin M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 65.Itoh K., Wakabayashi N., Katoh Y., Ishii T., O'Connor T., Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 66.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. (1128/MCB.24.16.7130-7139.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen T., Sherratt P.J., Huang H.C., Yang C.S., Pickett C.B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 68.Stewart D., Killeen E., Naquin R., Alam S., Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 69.Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Niture S.K., Jaiswal A.K. INrf2 (Keap1) targets Bcl-2 degradation and controls cellular apoptosis. Cell Death Differ. 2011;18:439–451. doi: 10.1038/cdd.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Kim J.E., You D.J., Lee C., Ahn C., Seong J.Y., Hwang J.I. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell. Signal. 2010;22:1645–1654. doi: 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 72.Silva-Islas C.A., Maldonado P.D. Canonical and non-canonical mechanisms of Nrf2 activation. Pharmacol. Res. 2018;134:92–99. doi: 10.1016/j.phrs.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 73.Kopacz A., Kloska D., Forman H.J., Jozkowicz A., Grochot-Przeczek A. Beyond repression of Nrf2: an update on Keap1. Free Radic. Biol. Med. 2020;157:63–74. doi: 10.1016/j.freeradbiomed.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karttunen M., Choy W.Y., Cino E.A. Prediction of binding energy of Keap1 interaction motifs in the Nrf2 antioxidant pathway and design of potential high-affinity peptides. J. Phys. Chem. B. 2018;122:5851–5859. doi: 10.1021/acs.jpcb.8b03295. [DOI] [PubMed] [Google Scholar]
- 75.Dodson M., Zhang D.D. Non-canonical activation of NRF2: new insights and its relevance to disease. Curr. Pathobiol. Rep. 2017;5:171–176. doi: 10.1007/s40139-017-0131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., Hoshii T., Hirao A., Takagi K., Mizushima T., Motohashi H., Lee M.S., Yoshimori T., Tanaka K., Yamamoto M., Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 78.Chen W., Sun Z., Wang X.J., Jiang T., Huang Z., Fang D., Zhang D.D. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El-Deiry W.S. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016;76:5189–5191. doi: 10.1158/0008-5472.CAN-16-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karimian A., Ahmadi Y., Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 81.Lau A., Wang X.J., Zhao F., Villeneuve N.F., Wu T., Jiang T., Sun Z., White E., Zhang D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Padmanabhan B., Nakamura Y., Yokoyama S. Structural analysis of the complex of Keap1 with a prothymosin alpha peptide. Acta Crystallogr.Sect. F. Struct. Biol. Cryst. Commun. 2008;64:233–238. doi: 10.1107/S1744309108004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hast B.E., Goldfarb D., Mulvaney K.M., Hast M.A., Siesser P.F., Yan F., Hayes D.N., Major M.B. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 2013;73:2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu K., Alcivar A.L., Ma J., Foo T.K., Zywea S., Mahdi A., Huo Y., Kensler T.W., Gatza M.L., Xia B. NRF2 induction supporting breast cancer cell survival is enabled by oxidative stress-induced DPP3-KEAP1 interaction. Cancer Res. 2017;77:2881–2892. doi: 10.1158/0008-5472.CAN-16-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Camp N.D., James R.G., Dawson D.W., Yan F., Davison J.M., Houck S.A., Tang X., Zheng N., Major M.B., Moon R.T. Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. J. Biol. Chem. 2012;287:6539–6550. doi: 10.1074/jbc.M111.316471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma J., Cai H., Wu T., Sobhian B., Huo Y., Alcivar A., Mehta M., Cheung K.L., Ganesan S., Kong A.N., Zhang D.D., Xia B. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol. Cell. Biol. 2012;32:1506–1517. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ge W., Zhao K., Wang X., Li H., Yu M., He M., Xue X., Zhu Y., Zhang C., Cheng Y., Jiang S., Hu Y. iASPP is an antioxidative factor and drives cancer growth and drug resistance by competing with Nrf2 for Keap1 binding. Cancer Cell. 2017;32:561–573. doi: 10.1016/j.ccell.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 88.Wang Q., Ma J., Lu Y., Zhang S., Huang J., Chen J., Bei J.X., Yang K., Wu G., Huang K., Chen J., Xu S. CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells. Oncogene. 2017;36:5321–5330. doi: 10.1038/onc.2017.161. [DOI] [PubMed] [Google Scholar]
- 89.Yang C., Tan Y.X., Yang G.Z., Zhang J., Pan Y.F., Liu C., Fu J., Chen Y., Ding Z.W., Dong L.W., Wang H.Y. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J. Exp. Med. 2016;213:859–875. doi: 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou X.L., Zhu C.Y., Wu Z.G., Guo X., Zou W. The oncoprotein HBXIP competitively binds KEAP1 to activate NRF2 and enhance breast cancer cell growth and metastasis. Oncogene. 2019;38:4028–4046. doi: 10.1038/s41388-019-0698-5. [DOI] [PubMed] [Google Scholar]
- 91.Mulvaney K.M., Matson J.P., Siesser P.F., Tamir T.Y., Goldfarb D., Jacobs T.M., Cloer E.W., Harrison J.S., Vaziri C., Cook J.G., Major M.B. Identification and characterization of MCM3 as a kelch-like ECH-associated protein 1 (KEAP1) substrate. J. Biol. Chem. 2016;291:23719–23733. doi: 10.1074/jbc.M116.729418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng K.C., Lin R.J., Cheng J.Y., Wang S.H., Yu J.C., Wu J.C., Liang Y.J., Hsu H.M., Yu J., Yu A.L. FAM129B, an antioxidative protein, reduces chemosensitivity by competing with Nrf2 for Keap1 binding. EBioMedicine. 2019;45:25–38. doi: 10.1016/j.ebiom.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wan Z.H., Jiang T.Y., Shi Y.Y., Pan Y.F., Lin Y.K., Ma Y.H., Yang C., Feng X.F., Huang L.F., Kong X.N., Ding Z.W., Tan Y.X., Dong L.W., Wang H.Y. RPB5-mediating protein promotes cholangiocarcinoma tumorigenesis and drug resistance by competing with NRF2 for KEAP1 binding. Hepatology. 2020;71:2005–2022. doi: 10.1002/hep.30962. [DOI] [PubMed] [Google Scholar]
- 94.Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K., Engel J.D., Yamamoto M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S.R., Bell D.A. Identification of novel NRF2-regulated genes by ChIP-seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hou Y., Xue P., Bai Y., Liu D., Woods C.G., Yarborough K., Fu J., Zhang Q., Sun G., Collins S., Chan J.Y., Yamamoto M., Andersen M.E., Pi J. Nuclear factor erythroid-derived factor 2-related factor 2 regulates transcription of CCAAT/enhancer-binding protein beta during adipogenesis. Free Radic. Biol. Med. 2012;52:462–472. doi: 10.1016/j.freeradbiomed.2011.10.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shin S., Wakabayashi N., Misra V., Biswal S., Lee G.H., Agoston E.S., Yamamoto M., Kensler T.W. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol. Cell. Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T.W., Wasserman W.W., Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wakabayashi N., Shin S., Slocum S.L., Agoston E.S., Wakabayashi J., Kwak M.K., Misra V., Biswal S., Yamamoto M., Kensler T.W. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci. Signal. 2010;3 doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niture S.K., Jaiswal A.K. Hsp90 interaction with INrf2(Keap1) mediates stress-induced Nrf2 activation. J. Biol. Chem. 2010;285:36865–36875. doi: 10.1074/jbc.M110.175802. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Lee D.F., Kuo H.P., Liu M., Chou C.K., Xia W., Du Y., Shen J., Chen C.T., Huo L., Hsu M.C., Li C.W., Ding Q., Liao T.L., Lai C.C., Lin A.C., Chang Y.H., Tsai S.F., Li L.Y., Hung M.C. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol. Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.You A., Nam C.W., Wakabayashi N., Yamamoto M., Kensler T.W., Kwak M.K. Transcription factor Nrf2 maintains the basal expression of Mdm2: an implication of the regulation of p53 signaling by Nrf2. Arch. Biochem. Biophys. 2011;507:356–364. doi: 10.1016/j.abb.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 103.Johnson D.A., Johnson J.A. Nrf2–a therapeutic target for the treatment of neurodegenerative diseases. Free Radic. Biol. Med. 2015;88:253–267. doi: 10.1016/j.freeradbiomed.2015.07.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith R.E., Tran K., Smith C.C., McDonald M., Shejwalkar P., Hara K. The role of the Nrf2/ARE antioxidant system in preventing cardiovascular diseases. Diseases. 2016;4 doi: 10.3390/diseases4040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alfieri A., Srivastava S., Siow R.C., Modo M., Fraser P.A., Mann G.E. Targeting the Nrf2-Keap1 antioxidant defence pathway for neurovascular protection in stroke. J. Physiol. 2011;589:4125–4136. doi: 10.1113/jphysiol.2011.210294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Q.M., Maltagliati A.J. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genomics. 2018;50:77–97. doi: 10.1152/physiolgenomics.00041.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ferrandiz M.L., Nacher-Juan J., Alcaraz M.J. Nrf2 as a therapeutic target for rheumatic diseases. Biochem. Pharmacol. 2018;152:338–346. doi: 10.1016/j.bcp.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 108.Cuadrado A., Manda G., Hassan A., Alcaraz M.J., Barbas C., Daiber A., Ghezzi P., Leon R., Lopez M.G., Oliva B., Pajares M., Rojo A.I., Robledinos-Anton N., Valverde A.M., Guney E., Schmidt H.H.H.W. Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacol. Rev. 2018;70:348–383. doi: 10.1124/pr.117.014753. [DOI] [PubMed] [Google Scholar]
- 109.Paracha U.Z., Fatima K., Alqahtani M., Chaudhary A., Abuzenadah A., Damanhouri G., Qadri I. Oxidative stress and hepatitis C virus. Virol. J. 2013;10:251. doi: 10.1186/1743-422X-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ivanov A.V., Valuev-Elliston V.T., Ivanova O.N., Kochetkov S.N., Starodubova E.S., Bartosch B., Isaguliants M.G. Oxidative stress during HIV infection: mechanisms and consequences. Oxid. Med. Cell Longev. 2016 doi: 10.1155/2016/8910396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Delgado-Roche L., Mesta F. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) infection. Arch. Med. Res. 2020;51:384–387. doi: 10.1016/j.arcmed.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Q., Li W.X., Dai S.X., Guo Y.C., Han F.F., Zheng J.J., Li G.H., Huang J.F. Meta-analysis of Parkinson's disease and Alzheimer's disease revealed commonly impaired pathways and dysregulation of NRF2-dependent genes. J. Alzheimers Dis. 2017;56:1525–1539. doi: 10.3233/JAD-161032. [DOI] [PubMed] [Google Scholar]
- 113.von Otter M., Landgren S., Nilsson S., Celojevic D., Bergstrom P., Hakansson A., Nissbrandt H., Drozdzik M., Bialecka M., Kurzawski M., Blennow K., Nilsson M., Hammarsten O., Zetterberg H. Association of Nrf2-encoding NFE2L2 haplotypes with Parkinson's disease. BMC Med. Genet. 2010;11:36. doi: 10.1186/1471-2350-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gui Y., Zhang L., Lv W., Zhang W., Zhao J., Hu X. NFE2L2 variations reduce antioxidant response in patients with parkinson disease. Oncotarget. 2016;7:10756–10764. doi: 10.18632/oncotarget.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.von Otter M., Bergstrom P., Quattrone A., De Marco E.V., Annesi G., Soderkvist P., Wettinger S.B., Drozdzik M., Bialecka M., Nissbrandt H., Klein C., Nilsson M., Hammarsten O., Nilsson S., Zetterberg H. Genetic associations of Nrf2-encoding NFE2L2 variants with Parkinson's disease - a multicenter study. BMC Med. Genet. 2014;15:131. doi: 10.1186/s12881-014-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.von Otter M., Landgren S., Nilsson S., Zetterberg M., Celojevic D., Bergstrom P., Minthon L., Bogdanovic N., Andreasen N., Gustafson D.R., Skoog I., Wallin A., Tasa G., Blennow K., Nilsson M., Hammarsten O., Zetterberg H. Nrf2-encoding NFE2L2 haplotypes influence disease progression but not risk in Alzheimer's disease and age-related cataract. Mech. Ageing Dev. 2010;131:105–110. doi: 10.1016/j.mad.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 117.Bergstrom P., Von O.M., Nilsson S., Nilsson A.C., Nilsson M., Andersen P.M., Hammarsten O., Zetterberg H. Association of NFE2L2 and KEAP1 haplotypes with amyotrophic lateral sclerosis. Amyotroph. Lateral. Scler. Frontotemporal. Degener. 2014;15:130–137. doi: 10.3109/21678421.2013.839708. [DOI] [PubMed] [Google Scholar]
- 118.Arisawa T., Tahara T., Shibata T., Nagasaka M., Nakamura M., Kamiya Y., Fujita H., Yoshioka D., Okubo M., Sakata M., Wang F.Y., Hirata I., Nakano H. Nrf2 gene promoter polymorphism is associated with ulcerative colitis in a japanese population. Hepato-Gastroenterology. 2008;55:394–397. [PubMed] [Google Scholar]
- 119.Burton N.C., Kensler T.W., Guilarte T.R. In vivo modulation of the parkinsonian phenotype by Nrf2. Neurotoxicology. 2006;27:1094–1100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 120.Chen P.C., Vargas M.R., Pani A.K., Smeyne R.J., Johnson D.A., Kan Y.W., Johnson J.A. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's disease: critical role for the astrocyte. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jazwa A., Rojo A.I., Innamorato N.G., Hesse M., Fernandez-Ruiz J., Cuadrado A. Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid. Redox.Signal. 2011;14:2347–2360. doi: 10.1089/ars.2010.3731. [DOI] [PubMed] [Google Scholar]
- 122.Williamson T.P., Johnson D.A., Johnson J.A. Activation of the Nrf2-ARE pathway by siRNA knockdown of Keap1 reduces oxidative stress and provides partial protection from MPTP-mediated neurotoxicity. Neurotoxicology. 2012;33:272–279. doi: 10.1016/j.neuro.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ramsey C.P., Glass C.A., Montgomery M.B., Lindl K.A., Ritson G.P., Chia L.A., Hamilton R.L., Chu C.T., Jordan-Sciutto K.L. Expression of Nrf2 in neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 2007;66:75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kanninen K., Heikkinen R., Malm T., Rolova T., Kuhmonen S., Leinonen H., Yla-Herttuala S., Tanila H., Levonen A.L., Koistinaho M., Koistinaho J. Intrahippocampal injection of a lentiviral vector expressing Nrf2 improves spatial learning in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16505–16510. doi: 10.1073/pnas.0908397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Farr S.A., Ripley J.L., Sultana R., Zhang Z., Niehoff M.L., Platt T.L., Murphy M.P., Morley J.E., Kumar V., Butterfield D.A. Antisense oligonucleotide against GSK-3beta in brain of SAMP8 mice improves learning and memory and decreases oxidative stress: involvement of transcription factor Nrf2 and implications for alzheimer disease. Free Radic. Biol. Med. 2014;67:387–395. doi: 10.1016/j.freeradbiomed.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goven D., Boutten A., Lecon-Malas V., Marchal-Somme J., Amara N., Crestani B., Fournier M., Leseche G., Soler P., Boczkowski J., Bonay M. Altered Nrf2/Keap1-Bach1 equilibrium in pulmonary emphysema. Thorax. 2008;63:916–924. doi: 10.1136/thx.2007.091181. [DOI] [PubMed] [Google Scholar]
- 127.Suzuki M., Betsuyaku T., Ito Y., Nagai K., Nasuhara Y., Kaga K., Kondo S., Nishimura M. Down-regulated NF-E2-related factor 2 in pulmonary macrophages of aged smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2008;39:673–682. doi: 10.1165/rcmb.2007-0424OC. [DOI] [PubMed] [Google Scholar]
- 128.128. Harvey C.J., Thimmulappa R.K., Sethi S., Kong X., Yarmus L., Brown R.H., Feller-Kopman D., Wise R., Biswal S. 2011;3 doi: 10.1126/scitranslmed.3002042. [DOI] [PMC free article] [PubMed] [Google Scholar]; 128. Harvey C.J., Thimmulappa R.K., Sethi S., Kong X., Yarmus L., Brown R.H., Feller-Kopman D., Wise R., Biswal S. 2011;3 doi: 10.1126/scitranslmed.3002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu M.C., Ji J.A., Jiang Z.Y., You Q.D. The Keap1-Nrf2-ARE pathway as a potential preventive and therapeutic target: an update. Med. Res. Rev. 2016;36:924–963. doi: 10.1002/med.21396. [DOI] [PubMed] [Google Scholar]
- 130.Neymotin A., Calingasan N.Y., Wille E., Naseri N., Petri S., Damiano M., Liby K.T., Risingsong R., Sporn M., Beal M.F., Kiaei M. Neuroprotective effect of Nrf2/ARE activators, CDDO ethylamide and CDDO trifluoroethylamide, in a mouse model of amyotrophic lateral sclerosis. Free Radic. Biol. Med. 2011;51:88–96. doi: 10.1016/j.freeradbiomed.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Todorovic M., Mellick G.D. 2016. Nrf2 and Parkinson's Disease. [DOI] [PubMed] [Google Scholar]
- 132.Buendia I., Michalska P., Navarro E., Gameiro I., Egea J., Leon R. Nrf2-ARE pathway: an emerging target against oxidative stress and neuroinflammation in neurodegenerative diseases. Pharmacol. Ther. 2016;157:84–104. doi: 10.1016/j.pharmthera.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 133.Dinkova-Kostova A.T., Kostov R.V., Kazantsev A.G. The role of Nrf2 signaling in counteracting neurodegenerative diseases. FEBS J. 2018;285:3576–3590. doi: 10.1111/febs.14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Joshi G., Johnson J.A. The Nrf2-ARE pathway: a valuable therapeutic target for the treatment of neurodegenerative diseases. Recent Pat. CNS Drug. Discov. 2012;7:218–229. doi: 10.2174/157488912803252023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Niu Y., Zhang J., Dong M. Nrf2 as a potential target for Parkinson's disease therapy. J. Mol. Med. (Berl) 2021;99:917–931. doi: 10.1007/s00109-021-02071-5. [DOI] [PubMed] [Google Scholar]
- 136.Lee C. Therapeutic modulation of virus-induced oxidative stress via the Nrf2-dependent antioxidative pathway. Oxidative Med. Cell. Longev. 2018;2018:6208067. doi: 10.1155/2018/6208067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ramezani A., Nahad M.P., Faghihloo E. The role of Nrf2 transcription factor in viral infection. J. Cell. Biochem. 2018;119:6366–6382. doi: 10.1002/jcb.26897. [DOI] [PubMed] [Google Scholar]
- 138.Herengt A., Thyrsted J., Holm C.K. NRF2 in viral infection. Antioxidants. (Basel) 2021;10:1491. doi: 10.3390/antiox10091491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kosmider B., Messier E.M., Janssen W.J., Nahreini P., Wang J., Hartshorn K.L., Mason R.J. Nrf2 protects human alveolar epithelial cells against injury induced by influenza A virus. Respir. Res. 2012;13:43. doi: 10.1186/1465-9921-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Russell A.B., Trapnell C., Bloom J.D. Extreme heterogeneity of influenza virus infection in single cells. elife. 2018;7 doi: 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cheng Y.L., Lin Y.S., Chen C.L., Tsai T.T., Tsai C.C., Wu Y.W., Ou Y.D., Chu Y.Y., Wang J.M., Yu C.Y., Lin C.F. Activation of Nrf2 by the dengue virus causes an increase in CLEC5A, which enhances TNF-alpha production by mononuclear phagocytes. Sci Rep. 2016;6:32000. doi: 10.1038/srep32000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ferrari M., Zevini A., Palermo E., Muscolini M., Alexandridi M., Etna M.P., Coccia E.M., Fernandez-Sesma A., Coyne C., Zhang D.D., Marques E.T.A., Olagnier D., Hiscott J. Dengue Virus Targets Nrf2 for NS2B3-Mediated Degradation Leading to Enhanced Oxidative Stress and Viral Replication. J. Virol. 2020;94 doi: 10.1128/JVI.01551-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Komaravelli N., Tian B., Ivanciuc T., Mautemps N., Brasier A.R., Garofalo R.P., Casola A. Respiratory syncytial virus infection down-regulates antioxidant enzyme expression by triggering deacetylation-proteasomal degradation of Nrf2. Free Radic. Biol. Med. 2015;88:391–403. doi: 10.1016/j.freeradbiomed.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Komaravelli N., Ansar M., Garofalo R.P., Casola A. Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein - ring finger protein 4 dependent pathway. Free Radic. Biol. Med. 2017;113:494–504. doi: 10.1016/j.freeradbiomed.2017.10.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cho H.Y., Imani F., Miller-DeGraff L., Walters D., Melendi G.A., Yamamoto M., Polack F.P., Kleeberger S.R. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am. J. Respir. Crit. Care Med. 2009;179:138–150. doi: 10.1164/rccm.200804-535OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Patra U., Mukhopadhyay U., Sarkar R., Mukherjee A., Chawla-Sarkar M. RA-839, a selective agonist of Nrf2/ARE pathway, exerts potent anti-rotaviral efficacy in vitro. Antivir. Res. 2019;161:53–62. doi: 10.1016/j.antiviral.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 147.Wyler E., Franke V., Menegatti J., Kocks C., Boltengagen A., Praktiknjo S., Walch-Ruckheim B., Bosse J., Rajewsky N., Grasser F., Akalin A., Landthaler M. Single-cell RNA-sequencing of herpes simplex virus 1-infected cells connects NRF2 activation to an antiviral program. Nat. Commun. 2019;10:4878. doi: 10.1038/s41467-019-12894-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bender D., Hildt E. Effect of hepatitis viruses on the Nrf2/Keap1-signaling pathway and its impact on viral replication and pathogenesis. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20184659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cecchini R., Cecchini A.L. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.110102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Suhail S., Zajac J., Fossum C., Lowater H., McCracken C., Severson N., Laatsch B., Narkiewicz-Jodko A., Johnson B., Liebau J., Bhattacharyya S., Hati S. Role of oxidative stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) infection: a review. Protein J. 2020;39:644–656. doi: 10.1007/s10930-020-09935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Townsend B.E., Johnson R.W. Sulforaphane induces Nrf2 target genes and attenuates inflammatory gene expression in microglia from brain of young adult and aged mice. Exp. Gerontol. 2016;73:42–48. doi: 10.1016/j.exger.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Olagnier D., Lababidi R.R., Hadj S.B., Sze A., Liu Y., Naidu S.D., Ferrari M., Jiang Y., Chiang C., Beljanski V., Goulet M.L., Knatko E.V., Dinkova-Kostova A.T., Hiscott J., Lin R. Activation of Nrf2 signaling augments vesicular stomatitis virus oncolysis via autophagy-driven suppression of antiviral immunity. Mol. Ther. 2017;25:1900–1916. doi: 10.1016/j.ymthe.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jones S.A., Hunter C.A. Is IL-6 a key cytokine target for therapy in COVID-19? Nat. Rev. Immunol. 2021;21:337–339. doi: 10.1038/s41577-021-00553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Olagnier D., Farahani E., Thyrsted J., Blay-Cadanet J., Herengt A., Idorn M., Hait A., Hernaez B., Knudsen A., Iversen M.B., Schilling M., Jorgensen S.E., Thomsen M., Reinert L.S., Lappe M., Hoang H.D., Gilchrist V.H., Hansen A.L., Ottosen R., Nielsen C.G., Moller C., van der Horst D., Peri S., Balachandran S., Huang J., Jakobsen M., Svenningsen E.B., Poulsen T.B., Bartsch L., Thielke A.L., Luo Y., Alain T., Rehwinkel J., Alcami A., Hiscott J., Mogensen T.H., Paludan S.R., Holm C.K. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11:4938. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao S., Ghosh A., Lo C.S., Chenier I., Scholey J.W., Filep J.G., Ingelfinger J.R., Zhang S.L., Chan J.S.D. Nrf2 deficiency upregulates intrarenal angiotensin-converting Enzyme-2 and angiotensin 1–7 receptor expression and attenuates hypertension and nephropathy in diabetic mice. Endocrinology. 2018;159:836–852. doi: 10.1210/en.2017-00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Petry A., Weitnauer M., Gorlach A. Antioxid. RedoxSignal. 2010;13:467–487. doi: 10.1089/ars.2009.3026. [DOI] [PubMed] [Google Scholar]
- 158.Griendling K.K., Minieri C.A., Ollerenshaw J.D., Alexander R.W. Angiotensin II stimulates NADH and NADPH oxidase activity in cultured vascular smooth muscle cells. Circ. Res. 1994;74:1141–1148. doi: 10.1161/01.res.74.6.1141. [DOI] [PubMed] [Google Scholar]
- 159.Espinoza J.A., Gonzalez P.A., Kalergis A.M. Modulation of antiviral immunity by heme Oxygenase-1. Am. J. Pathol. 2017;187:487–493. doi: 10.1016/j.ajpath.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 160.Martinez-Sanchez G., Schwartz A., Donna V.D. Potential cytoprotective activity of ozone therapy in SARS-CoV-2/COVID-19. Antioxidants (Basel) 2020;9 doi: 10.3390/antiox9050389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sporn M.B., Liby K.T. NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Taguchi K., Yamamoto M. The KEAP1-NRF2 system in cancer. Front. Oncol. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kensler T.W., Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hayes J.D., McMahon M., Chowdhry S., Dinkova-Kostova A.T. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox.Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 165.Hu R., Saw C.L., Yu R., Kong A.N. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid. Redox.Signal. 2010;13:1679–1698. doi: 10.1089/ars.2010.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang L., Zhang C., Qin L., Xu J., Li X., Wang W., Kong L., Zhou T., Li X. The prognostic value of NRF2 in solid tumor patients: a meta-analysis. Oncotarget. 2018;9:1257–1265. doi: 10.18632/oncotarget.19838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., Scrimieri F., Winter J.M., Hruban R.H., Iacobuzio-Donahue C., Kern S.E., Blair I.A., Tuveson D.A. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim Y.R., Oh J.E., Kim M.S., Kang M.R., Park S.W., Han J.Y., Eom H.S., Yoo N.J., Lee S.H. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 2010;220:446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 169.Padmanabhan B., Tong K.I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M.I., Kobayashi A., Yokoyama S., Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 170.Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S., Hoque M.O., Herman J.G., Baylin S.B., Sidransky D., Gabrielson E., Brock M.V., Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang R., An J., Ji F., Jiao H., Sun H., Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 172.Adam J., Hatipoglu E., O'Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G.W., Wolhuter K., Stevens M., Fischer R., Carmeliet P., Maxwell P.H., Pugh C.W., Frizzell N., Soga T., Kessler B.M., El-Bahrawy M., Ratcliffe P.J., Pollard P.J. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sheridan C. Second oral MS drug wins FDA nod. Nat. Biotechnol. 2013;31:373. doi: 10.1038/nbt0513-373a. [DOI] [PubMed] [Google Scholar]
- 174.Hur W., Gray N.S. Small molecule modulators of antioxidant response pathway. Curr. Opin. Chem. Biol. 2011;15:162–173. doi: 10.1016/j.cbpa.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 175.Magesh S., Chen Y., Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012;32:687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.McMahon M., Lamont D.J., Beattie K.A., Hayes J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wilson A.J., Kerns J.K., Callahan J.F., Moody C.J. Keap calm, and carry on covalently. J. Med. Chem. 2013;56:7463–7476. doi: 10.1021/jm400224q. [DOI] [PubMed] [Google Scholar]
- 178.Albrecht P., Bouchachia I., Goebels N., Henke N., Hofstetter H.H., Issberner A., Kovacs Z., Lewerenz J., Lisak D., Maher P., Mausberg A.K., Quasthoff K., Zimmermann C., Hartung H.P., Methner A. Effects of dimethyl fumarate on neuroprotection and immunomodulation. J. Neuroinflammation. 2012;9:163. doi: 10.1186/1742-2094-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Venci J.V., Gandhi M.A. Dimethyl fumarate (Tecfidera): a new oral agent for multiple sclerosis. Ann. Pharmacother. 2013;47:1697–1702. doi: 10.1177/1060028013509232. [DOI] [PubMed] [Google Scholar]
- 180.Cross S.A., Cook D.R., Chi A.W., Vance P.J., Kolson L.L., Wong B.J., Jordan-Sciutto K.L., Kolson D.L. Dimethyl fumarate, an immune modulator and inducer of the antioxidant response, suppresses HIV replication and macrophage-mediated neurotoxicity: a novel candidate for HIV neuroprotection. J. Immunol. 2011;187:5015–5025. doi: 10.4049/jimmunol.1101868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Furuya A.K., Sharifi H.J., Jellinger R.M., Cristofano P., Shi B., Noronha C.M. Sulforaphane inhibits HIV infection of macrophages through Nrf2. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kesic M.J., Simmons S.O., Bauer R., Jaspers I. Nrf2 expression modifies influenza a entry and replication in nasal epithelial cells. Free Radic. Biol. Med. 2011;51:444–453. doi: 10.1016/j.freeradbiomed.2011.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yu J.S., Chen W.C., Tseng C.K., Lin C.K., Hsu Y.C., Chen Y.H., Lee J.C. Sulforaphane suppresses hepatitis C virus replication by up-regulating heme Oxygenase-1 expression through PI3K/Nrf2 pathway. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schachtele S.J., Hu S., Lokensgard J.R. Modulation of experimental herpes encephalitis-associated neurotoxicity through sulforaphane treatment. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yore M.M., Kettenbach A.N., Sporn M.B., Gerber S.A., Liby K.T. Proteomic analysis shows synthetic oleanane triterpenoid binds to mTOR. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Blewett M.M., Xie J., Zaro B.W., Backus K.M., Altman A., Teijaro J.R., Cravatt B.F. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci. Signal. 2016;9:rs10. doi: 10.1126/scisignal.aaf7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hosseini A., Masjedi A., Baradaran B., Hojjat-Farsangi M., Ghalamfarsa G., Anvari E., Jadidi-Niaragh F. Dimethyl fumarate: regulatory effects on the immune system in the treatment of multiple sclerosis. J. Cell. Physiol. 2019;234:9943–9955. doi: 10.1002/jcp.27930. [DOI] [PubMed] [Google Scholar]
- 188.Schulze-Topphoff U., Varrin-Doyer M., Pekarek K., Spencer C.M., Shetty A., Sagan S.A., Cree B.A., Sobel R.A., Wipke B.T., Steinman L., Scannevin R.H., Zamvil S.S. Dimethyl fumarate treatment induces adaptive and innate immune modulation independent of Nrf2. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4777–4782. doi: 10.1073/pnas.1603907113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hahm E.R., Singh S.V. Sulforaphane inhibits constitutive and interleukin-6-induced activation of signal transducer and activator of transcription 3 in prostate cancer cells. Cancer Prev. Res. (Phila.) 2010;3:484–494. doi: 10.1158/1940-6207.CAPR-09-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Heiss E., Herhaus C., Klimo K., Bartsch H., Gerhauser C. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J. Biol. Chem. 2001;276:32008–32015. doi: 10.1074/jbc.M104794200. [DOI] [PubMed] [Google Scholar]
- 191.Baell J., Walters M.A. Chemistry: chemical con artists foil drug discovery. Nature. 2014;513:481–483. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 192.Richardson B.G., Jain A.D., Speltz T.E., Moore T.W. Non-electrophilic modulators of the canonical Keap1/Nrf2 pathway. Bioorg. Med. Chem. Lett. 2015;25:2261–2268. doi: 10.1016/j.bmcl.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dahlin J.L., Walters M.A. The essential roles of chemistry in high-throughput screening triage. FutureMed. Chem. 2014;6:1265–1290. doi: 10.4155/fmc.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhuang C., Wu Z., Xing C., Miao Z. Small molecules inhibiting Keap1-Nrf2 protein-protein interactions: a novel approach to activate Nrf2 function. Med. Chem. Comm. 2017;8:286–294. doi: 10.1039/c6md00500d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Balogun E., Hoque M., Gong P., Killeen E., Green C.J., Foresti R., Alam J., Motterlini R. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem. J. 2003;371:887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Soetikno V., Sari F.R., Lakshmanan A.P., Arumugam S., Harima M., Suzuki K., Kawachi H., Watanabe K. Curcumin alleviates oxidative stress, inflammation, and renal fibrosis in remnant kidney through the Nrf2-keap1 pathway. Mol. Nutr. Food Res. 2013;57:1649–1659. doi: 10.1002/mnfr.201200540. [DOI] [PubMed] [Google Scholar]
- 197.Coles B.F., Kadlubar F.F. Detoxification of electrophilic compounds by glutathione S-transferase catalysis: determinants of individual response to chemical carcinogens and chemotherapeutic drugs? Biofactors. 2003;17:115–130. doi: 10.1002/biof.5520170112. [DOI] [PubMed] [Google Scholar]
- 198.Pompella A., Visvikis A., Paolicchi A., De T., Casini V.A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003;66:1499–1503. doi: 10.1016/s0006-2952(03)00504-5. [DOI] [PubMed] [Google Scholar]
- 199.Kosower N.S., Kosower E.M. The glutathione status of cells. Int. Rev. Cytol. 1978;54:109–160. doi: 10.1016/s0074-7696(08)60166-7. [DOI] [PubMed] [Google Scholar]
- 200.Satoh T., McKercher S.R., Lipton S.A. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2013;65:645–657. doi: 10.1016/j.freeradbiomed.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Copple I.M., Shelton L.M., Walsh J., Kratschmar D.V., Lister A., Odermatt A., Goldring C.E., Dinkova-Kostova A.T., Honda T., Park B.K. Chemical tuning enhances both potency toward nrf2 and in vitro therapeutic index of triterpenoids. Toxicol. Sci. 2014;140:462–469. doi: 10.1093/toxsci/kfu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jo J., Ibrahim L., Iaconelli J., Kwak J., Kumar M., Jung Y., Lairson L.L., Chatterjee A.K., Schultz P.G., Bollong M.J., Yun H. Discovery and SAR studies of 3-amino-4-(phenylsulfonyl)tetrahydrothiophene 1,1-dioxides as non-electrophilic antioxidant response element (ARE) activators. Bioorg. Chem. 2021;108:104614. doi: 10.1016/j.bioorg.2020.104614. [DOI] [PubMed] [Google Scholar]
- 203.Lipton S.A., Rezaie T., Nutter A., Lopez K.M., Parker J., Kosaka K., Satoh T., McKercher S.R., Masliah E., Nakanishi N. Therapeutic advantage of pro-electrophilic drugs to activate the Nrf2/ARE pathway in Alzheimer's disease models. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.389. (e2499-doi:10.1038/cddis.2016.389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Satoh T., Lipton S. Recent advances in understanding NRF2 as a druggable target: development of pro-electrophilic and non-covalent NRF2 activators to overcome systemic side effects of electrophilic drugs like dimethyl fumarate. F1000Res. 2017;6:2138. doi: 10.12688/f1000research.12111.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bains J.S., Shaw C.A. Neurodegenerative disorders in humans: the role of glutathione in oxidative stress-mediated neuronal death. Brain. Res. BrainRes. Rev. 1997;25:335–358. doi: 10.1016/s0165-0173(97)00045-3. [DOI] [PubMed] [Google Scholar]
- 206.Tong K.I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Beamer L.J., Li X., Bottoms C.A., Hannink M. Conserved solvent and side-chain interactions in the 1.35 angstrom structure of the kelch domain of Keap1. Acta Crystallogr.D Biol. Crystallogr. 2005;61:1335–1342. doi: 10.1107/S0907444905022626. [DOI] [PubMed] [Google Scholar]
- 208.Tong K.I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S., Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jiang Z.Y., Lu M.C., You Q.D. Discovery and development of kelch-like ECH-associated protein 1. Nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein-protein interaction inhibitors: achievements, challenges, and future directions. J. Med. Chem. 2016;59:10837–10858. doi: 10.1021/acs.jmedchem.6b00586. [DOI] [PubMed] [Google Scholar]
- 210.Chen Y., Inoyama D., Kong A.N., Beamer L.J., Hu L. Kinetic analyses of Keap1-Nrf2 interaction and determination of the minimal Nrf2 peptide sequence required for Keap1 binding using surface plasmon resonance. Chem. Biol. Drug Des. 2011;78:1014–1021. doi: 10.1111/j.1747-0285.2011.01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Inoyama D., Chen Y., Huang X., Beamer L.J., Kong A.N., Hu L. Optimization of fluorescently labeled Nrf2 peptide probes and the development of a fluorescence polarization assay for the discovery of inhibitors of Keap1-Nrf2 interaction. J. Biomol. Screen. 2012;17:435–447. doi: 10.1177/1087057111430124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hancock R., Bertrand H.C., Tsujita T., Naz S., El-Bakry A., Laoruchupong J., Hayes J.D., Wells G. Peptide inhibitors of the Keap1-Nrf2 protein-protein interaction. Free Radic. Biol. Med. 2012;52:444–451. doi: 10.1016/j.freeradbiomed.2011.10.486. [DOI] [PubMed] [Google Scholar]
- 213.Hancock R., Schaap M., Pfister H., Wells G. Peptide inhibitors of the Keap1-Nrf2 protein-protein interaction with improved binding and cellular activity. Org. Biomol. Chem. 2013;11:3553–3557. doi: 10.1039/c3ob40249e. [DOI] [PubMed] [Google Scholar]
- 214.Lu M.C., Chen Z.Y., Wang Y.L., Jiang Y.L., Yuan Z.W., You Q.D., Jiang Z.Y. Binding thermodynamics and kinetics guided optimization of potent Keap1-Nrf2 peptide inhibitors. RSC Adv. 2015;5:85983–85987. doi: 10.1039/c5ra16262a. [DOI] [Google Scholar]
- 215.Lu M., Liu T., Jiao Q., Ji J., Tao M., Liu Y., You Q., Jiang Z. Discovery of a Keap1-dependent peptide PROTAC to knockdown tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 2018;146:251–259. doi: 10.1016/j.ejmech.2018.01.063. [DOI] [PubMed] [Google Scholar]
- 216.Guntas G., Lewis S.M., Mulvaney K.M., Cloer E.W., Tripathy A., Lane T.R., Major M.B., Kuhlman B. Engineering a genetically encoded competitive inhibitor of the KEAP1-NRF2 interaction via structure-based design and phage display. Protein Eng. Des. Sel. 2016;29:1–9. doi: 10.1093/protein/gzv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lu M.C., Jiao Q., Liu T., Tan S.J., Zhou H.S., You Q.D., Jiang Z.Y. Discovery of a head-to-tail cyclic peptide as the Keap1-Nrf2 protein-protein interaction inhibitor with high cell potency. Eur. J. Med. Chem. 2018;143:1578–1589. doi: 10.1016/j.ejmech.2017.10.052. [DOI] [PubMed] [Google Scholar]
- 218.Colarusso S., De S.D., Frattarelli T., Andreini M., Cerretani M., Missineo A., Moretti D., Tambone S., Kempf G., Augustin M., Steinbacher S., Munoz-Sanjuan I., Park L., Summa V., Tomei L., Bresciani A., Dominguez C., Toledo-Sherman L., Bianchi E. Optimization of linear and cyclic peptide inhibitors of KEAP1-NRF2 protein-protein interaction. Bioorg. Med. Chem. 2020;28:115738. doi: 10.1016/j.bmc.2020.115738. [DOI] [PubMed] [Google Scholar]
- 219.Mou Y., Wen S., Li Y.X., Gao X.X., Zhang X., Jiang Z.Y. Recent progress in Keap1-Nrf2 protein-protein interaction inhibitors. Eur. J. Med. Chem. 2020;202 doi: 10.1016/j.ejmech.2020.112532. [DOI] [PubMed] [Google Scholar]
- 220.Slastnikova T.A., Ulasov A.V., Rosenkranz A.A., Sobolev A.S. Targeted intracellular delivery of antibodies: the state of the art. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.01208. (1208-doi:10.3389/fphar.2018.01208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Koide A., Koide S. Monobodies: antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol. Biol. 2007;352:95–109. doi: 10.1385/1-59745-187-8:95. [DOI] [PubMed] [Google Scholar]
- 222.Koide A., Wojcik J., Gilbreth R.N., Hoey R.J., Koide S. Teaching an old scaffold new tricks: monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 2012;415:393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gilbreth R.N., Koide S. Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr. Opin. Struct. Biol. 2012;22:413–420. doi: 10.1016/j.sbi.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lindgren M., Hallbrink M., Prochiantz A., Langel U. Cell-penetrating peptides. Trends Pharmacol. Sci. 2000;21:99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 225.Heitz F., Morris M.C., Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Steel R., Cowan J., Payerne E., O'Connell M.A., Searcey M. Anti-inflammatory effect of a cell-penetrating peptide targeting the Nrf2/Keap1 interaction. ACS Med. Chem. Lett. 2012;3:407–410. doi: 10.1021/ml300041g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhao J., Redell J.B., Moore A.N., Dash P.K. A novel strategy to activate cytoprotective genes in the injured brain. Biochem. Biophys. Res. Commun. 2011;407:501–506. doi: 10.1016/j.bbrc.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tu J., Zhang X., Zhu Y., Dai Y., Li N., Yang F., Zhang Q., Brann D.W., Wang R. Cell-permeable peptide targeting the Nrf2-Keap1 interaction: a potential novel therapy for global cerebral ischemia. J. Neurosci. 2015;35:14727–14739. doi: 10.1523/JNEUROSCI.1304-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pallesen J.S., Tran K.T., Bach A. Non-covalent small-molecule kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 (Keap1-Nrf2) inhibitors and their potential for targeting central nervous system diseases. J. Med. Chem. 2018 doi: 10.1021/acs.jmedchem.8b00358. [DOI] [PubMed] [Google Scholar]
- 230.Schmoll D., Engel C.K., Glombik H. The Keap1-Nrf2 protein-protein interaction: a suitable target for small molecules. Drug Discov. Today Technol. 2017;24:11–17. doi: 10.1016/j.ddtec.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 231.Jiang Z.Y., Lu M.C., Xu L.L., Yang T.T., Xi M.Y., Xu X.L., Guo X.K., Zhang X.J., You Q.D., Sun H.P. Discovery of potent Keap1-Nrf2 protein-protein interaction inhibitor based on molecular binding determinants analysis. J. Med. Chem. 2014;57:2736–2745. doi: 10.1021/jm5000529. [DOI] [PubMed] [Google Scholar]
- 232.Jiang Z.Y., Xu L.L., Lu M.C., Chen Z.Y., Yuan Z.W., Xu X.L., Guo X.K., Zhang X.J., Sun H.P., You Q.D. Structure-activity and structure-property relationship and exploratory in vivo evaluation of the nanomolar Keap1-Nrf2 protein-protein interaction inhibitor. J. Med. Chem. 2015;58:6410–6421. doi: 10.1021/acs.jmedchem.5b00185. [DOI] [PubMed] [Google Scholar]
- 233.Davies T.G., Wixted W.E., Coyle J.E., Griffiths-Jones C., Hearn K., McMenamin R., Norton D., Rich S.J., Richardson C., Saxty G., Willems H.M., Woolford A.J., Cottom J.E., Kou J.P., Yonchuk J.G., Feldser H.G., Sanchez Y., Foley J.P., Bolognese B.J., Logan G., Podolin P.L., Yan H., Callahan J.F., Heightman T.D., Kerns J.K. Monoacidic inhibitors of the kelch-like ECH-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein-protein interaction with high cell potency identified by fragment-based discovery. J. Med. Chem. 2016;59:3991–4006. doi: 10.1021/acs.jmedchem.6b00228. [DOI] [PubMed] [Google Scholar]
- 234.Matsson P., Doak B.C., Over B., Kihlberg J. Cell permeability beyond the rule of 5. Adv. Drug Deliv. Rev. 2016;101:42–61. doi: 10.1016/j.addr.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 235.Liu H., Zeng F., Zhang M., Huang F., Wang J., Guo J., Liu C., Wang H. Emerging landscape of cell penetrating peptide in reprogramming and gene editing. J. Control. Release. 2016;226:124–137. doi: 10.1016/j.jconrel.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 236.Koren E., Torchilin V.P. Cell-penetrating peptides: breaking through to the other side. Trends Mol. Med. 2012;18:385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 237.Bechara C., Sagan S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013;587:1693–1702. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 238.Copolovici D.M., Langel K., Eriste E., Langel U. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 239.Richard J.P., Melikov K., Vives E., Ramos C., Verbeure B., Gait M.J., Chernomordik L.V., Lebleu B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 240.Takenobu T., Tomizawa K., Matsushita M., Li S.T., Moriwaki A., Lu Y.F., Matsui H. Development of p53 protein transduction therapy using membrane-permeable peptides and the application to oral cancer cells. Mol. Cancer Ther. 2002;1:1043–1049. [PubMed] [Google Scholar]
- 241.Ulasov A.V., Rosenkranz A.A., Sobolev A.S. Transcription factors: time to deliver. J. Control. Release. 2018;269:24–35. doi: 10.1016/j.jconrel.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 242.Zinovkin R.A., Grebenchikov O.A. Transcription factor Nrf2 as a potential therapeutic target for prevention of cytokine storm in COVID-19 patients. Biochemistry (Mosc) 2020:833–837. doi: 10.1134/S0006297920070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Cuadrado A., Pajares M., Benito C., Jimenez-Villegas J., Escoll M., Fernandez-Gines R., Garcia Yague A.J., Lastra D., Manda G., Rojo A.I., Dinkova-Kostova A.T. Can activation of NRF2 be a strategy against COVID-19? Trends Pharmacol. Sci. 2020;41:598–610. doi: 10.1016/j.tips.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gebauer M., Skerra A. Engineered protein scaffolds as next-generation therapeutics. Annu. Rev. Pharmacol. Toxicol. 2020;60:391–415. doi: 10.1146/annurev-pharmtox-010818-021118. [DOI] [PubMed] [Google Scholar]
- 245.Wu T., Yoon H., Xiong Y., Dixon-Clarke S.E., Nowak R.P., Fischer E.S. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol. 2020;27:605–614. doi: 10.1038/s41594-020-0438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Daniels D.L., Riching K.M., Urh M. Monitoring and deciphering protein degradation pathways inside cells. Drug Discov. Today Technol. 2019;31:61–68. doi: 10.1016/j.ddtec.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 247.Bondeson D.P., Mares A., Smith I.E., Ko E., Campos S., Miah A.H., Mulholland K.E., Routly N., Buckley D.L., Gustafson J.L., Zinn N., Grandi P., Shimamura S., Bergamini G., Faelth-Savitski M., Bantscheff M., Cox C., Gordon D.A., Willard R.R., Flanagan J.J., Casillas L.N., Votta B.J., Besten W., Famm K., Kruidenier L., Carter P.S., Harling J.D., Churcher I., Crews C.M. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Slastnikova T.A., Rosenkranz A.A., Gulak P.V., Schiffelers R.M., Lupanova T.N., Khramtsov Y.V., Zalutsky M.R., Sobolev A.S. Modular nanotransporters: a multipurpose in vivo working platform for targeted drug delivery. Int. J. Nanomedicine. 2012;7:467–482. doi: 10.2147/IJN.S28249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Torchilin V.P. Multifunctional, stimuli-sensitive nanoparticulate systems for drug delivery. Nat. Rev. Drug Discov. 2014;13:813–827. doi: 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]