Abstract
Acinetobacter baumannii is a multidrug resistant pathogen that causes numerous infections associated with high mortality rates. Exposure to human body fluids, such as human pleural fluid (HPF) and human serum, modulates gene expression in A. baumannii, leading to changes in its pathogenic behavior. Diverse degrees of effects at the transcriptional level were observed in susceptible and carbapenem-resistant strains. The transcriptional analysis of AB5075, a hyper-virulent and extensively drug-resistant strain showed changes in genes associated with quorum sensing, quorum quenching, fatty acids metabolism, and high-efficient iron uptake systems. In addition, the distinctive role of human serum albumin (HSA) as a critical component of HPF was evidenced. In the present work, we used model strain to analyze more deeply into the contribution of HSA in triggering A. baumannii’s response. By qRT-PCR analysis, changes in the expression level of genes associated with quorum sensing, biofilm formation, and phenylacetic acid pathway were observed. Phenotypic approaches confirmed the transcriptional response. HSA, a predominant component of HPF, can modulate the expression and behavior of genes not only in a hyper-virulent and extensively drug-resistant A. baumannii model, but also in other strains with a different degree of susceptibility and pathogenicity.
Keywords: Acinetobacter baumannii, human serum albumin, human pleural fluid, quorum sensing, biofilm formation
Introduction
Acinetobacter baumannii accounts for high mortality rates in hospitalized patients. Capable of persisting under desiccation, nutrient starvation, and exposure to high concentrations of antimicrobial agents, A. baumannii prevails in diverse clinical settings and within the human host. Human pleural fluid (HPF) was recently shown to alter the phenotypic behavior and expression of genes in A. baumannii (1). The effect was observed in an antibiotic-susceptible model strain A. baumannii A118, and in a hypervirulent and multi-drug resistant representative strain A. baumannii AB5075 (2). In the AB5075 strain, the expression of genes associated with quorum sensing, quorum quenching, fatty acid metabolism, and high-efficient iron uptake systems was affected by HPF; moreover, the distinctive role of human serum albumin (HSA) as a central component of HPF was found to be involved in AB5075’s response (2). To further understand the specific contribution of HSA as an important component of HPF in triggering A. baumannii’s response, we studied its effects on the antibiotic-susceptible model A118 strain.
Materials and Methods
Bacterial Strains
Acinetobacter baumannii strain A118 was used as bacterial models. This strain has been shown to be susceptible to a variety of antibiotics (3, 4).
HSA Depletion
To obtain HSA-depleted HPF (dHPF), 1 mL of HPF was placed into a 3 kDa Amicon™ Ultra Centrifugal Filter (Millipore, Temecula, CA, USA) and the solution was centrifuged at 20,000 g for 10 min, as previously described (2). SDS-PAGE was carried out to confirm HSA depletion as previously stated (2).
N-Acyl Homoserine Lactone (AHL) Detection
Agrobacterium tumefaciens NT1 (pZLR4) was used to detect the presence of AHLs in A. baumannii cultures as previously described (5, 6). The A. tumefaciens NT1 (pZLR4) AHL biosensor, which contains a plasmid-localized traG-lacZ fusion (pZLR4) responds to AHLs of chain lengths ranging from C6 to C12 (7). Briefly, 500 μL of the homogenate were loaded in a central well of 0.7% LB agar plates supplemented with 40 μg of 5-bromo-3- indolyl-b-D-galactopyranoside (X-Gal) per mL and 250 μL (OD = 2.5) of overnight cultures of A. tumefaciens biosensor. The presence of AHL was determined by the development of the blue color. As a positive control, 100 μL of N-Decanoyl-DL-homoserine lactone (C10-AHL) 12.5 mg/mL was utilized. Quantification of 5,5’-dibromo-4,4’-dichloro-indigo production in different conditions was determined by measuring the intensity of each complete plate and subtracting the intensity measured in the negative control, using ImageJ software (NIH). The values were normalized to the positive control, which received the arbitrary value of 100. Experiments were performed in triplicate. Significance differences were determined by ANOVA followed by Tukey’s multiple comparison test (p < 0.05), using GraphPad Prism (GraphPad Software, San Diego, CA, USA).
Biofilm Assays
Biofilms assays were performed as previously described (2, 8). A118 cells were cultured in LB or LB supplemented with 4% HPF, 4% dHPF, or 4% dHPF + 0.2% HSA with agitation for 18 h at 37 °C. Following, the optical density at 600 nm (OD600) of each culture was adjusted to 0.9–1.1, vortexed, and diluted 1:100 in LB broth before being plated in technical triplicate in a 96-well polystyrene microtiter plate and being incubated at 37 °C for 24 h without agitation. The following day, the OD600 (ODG) was measured using a microplate reader (SpectraMax M3 microplate/ cuvette reader with SoftMax Pro v6 software) to determine the total biomass. Wells were emptied with a vacuum pipette, washed three times with 1X phosphate-buffered saline (PBS), and stained with 1% crystal violet (CV) for 15 m. Excess CV was removed by washing three more with 1X PBS and the biofilm-associated with the CV was solubilized in ethanol acetate (80:20) for 30 m. The OD580 (ODB) was measured using a microplate reader and results were reported as the ratio of biofilm to total biomass (ODB/ODG). Experiments were performed in triplicate, statistical analysis (Mann-Whitney test) was performed using GraphPad Prism (GraphPad Software, San Diego, CA, USA), and a p < 0.05 was considered significant.
RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
RNA extracted and DNase-treated from A. baumannii strain A118 grown in LB and LB supplemented with 4% HPF, 4% dHPF, or 4% dHPF + 0.2% HSA, was used to synthesized cDNA using the manufacturer protocol provided within the iScriptTM Reverse Transcription Supermix for qPCR (Bio-Rad, Hercules, CA, USA). The cDNA concentrations were adjusted to a concentration of 50 ng/μL. qPCR was conducted using the iQTM SYBR®Green Supermix through the manufacturer’s instructions. At least three biological replicates of cDNA were used and were run in quadruplets. All samples were then run on the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The transcript levels of each sample were normalized to the rpoB transcript levels for each cDNA sample. The relative quantification of gene expression was performed using the comparative threshold method 2-ΔΔCt. The ratios obtained after normalization were expressed as folds of change compared with cDNA samples isolated from bacteria cultures on LB. Significance differences were determined by ANOVA followed by Tukey’s multiple comparison test (p < 0.05), using GraphPad Prism (GraphPad Software, San Diego, CA, USA).
Results and Discussion
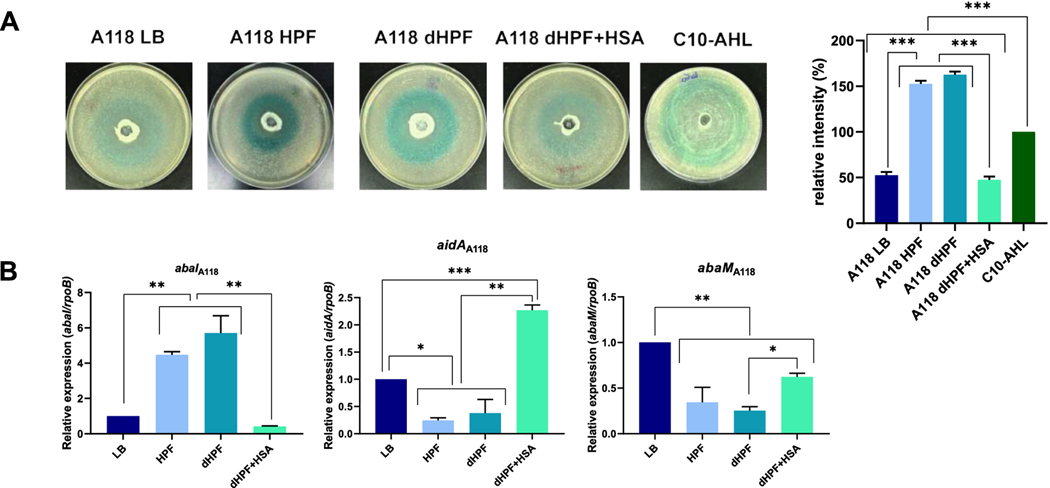
Quorum sensing was identified as one of the mechanisms modified in A. baumannii AB5075 when HSA was present in the growth medium. To further confirm this response in an unrelated A118 strain, the impact of HSA on quorum sensing, assessed via N-acyl-homoserine lactone (AHL) production, was determined using Agrobacterium tumefaciens-based solid plate assays (6). Figure 1A shows that the supernatant recovered from the A118 strain cultured in LB broth produced a blue halo, indicating the presence of AHLs. In addition, when cultured in the presence of HPF or dHPF (HPF depleted of HSA), A. baumannii A118 produced darker blue halos indicative of higher amounts of AHLs. On the other hand, when cultured in the presence of dHPF supplemented with HSA, strain A118 produced a low-intensity blue halo (Fig. 1A). HSA induced degradation or inhibition of synthesis of large chain AHLs in A. baumannii A118, an effect that was also observed with the AB5075 strain (2).
Figure 1. Phenotypic and genetic analysis of quorum sensing coding genes.
A) N-Acyl Homoserine Lactone (AHL) detection in A. baumannii A118 supernatants of bacteria cultured in LB or LB supplemented with HPF, dHPF or dHPF + HSA using Agrobacterium tumefaciens NT1 pZLR4 biosensor strain. The presence of AHLs was revealed by the development of a blue color. Quantification of 5,5’-dibromo-4,4’-dichloro-indigo was estimated as the percentage relative to the C10-AHL standard, measured with ImageJ (NIH). B) qRT-PCR of A118 strain of the abaI, aidA and abaM genes associated with quorum sensing expressed in LB or LB supplemented with HPF, dHPF, or dHPF + HSA. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used. LB was used as the reference condition. The mean ± SD is informed. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: p < 0.05; two asterisks: p < 0.01 and three asterisks: p < 0.001.
To evaluate this response at the transcriptional level, we carried out qRT-PCR assays using total RNA extracted from A. baumannii A118 cells cultured in LB or LB supplemented with HPF, HSA-depleted HPF (dHPF), or dHPF supplemented with HSA (dHPF + HSA) (Figure 1 B). The results of these experiments revealed that the level of expression of abaI, which codes for AHLs synthase (9), was higher in cultures containing HPF and dHPF (4.46- and 5.7-fold increase, respectively), while in presence of dHPF + HSA the expression of abaI was decreased (0.417-fold) (Fig. 1 B). The qRT-PCR results further demonstrated that the expression levels of aidA, which is an A. baumannii lactonase responsible for AHL degradation (10), were 0.24- and 0.37- fold decreased when grown under HPF and dHPF conditions, respectively. On the other hand, A. baumannii A118 cultured in the dHPF + HSA condition produced a 2.86-fold increase in expression of aidA. The abaM gene, which encodes a negative regulator of quorum sensing, AbaM (9), was down-regulated in all culture conditions respect LB (0.34-fold, 0.25-fold and 0.62-fold decrease under HPF, dHPF and dHPF + HSA treatments, respectively) (Fig. 1B).
These results suggest that HSA induces changes in the expression of genes associated with quorum sensing, leading to a lower synthesis and/or increased degradation of AHLs. The intense blue color observed with the addition of HPF may be the consequence of reactive oxygen species (ROS) present in this fluid that induces higher production of AHLs.
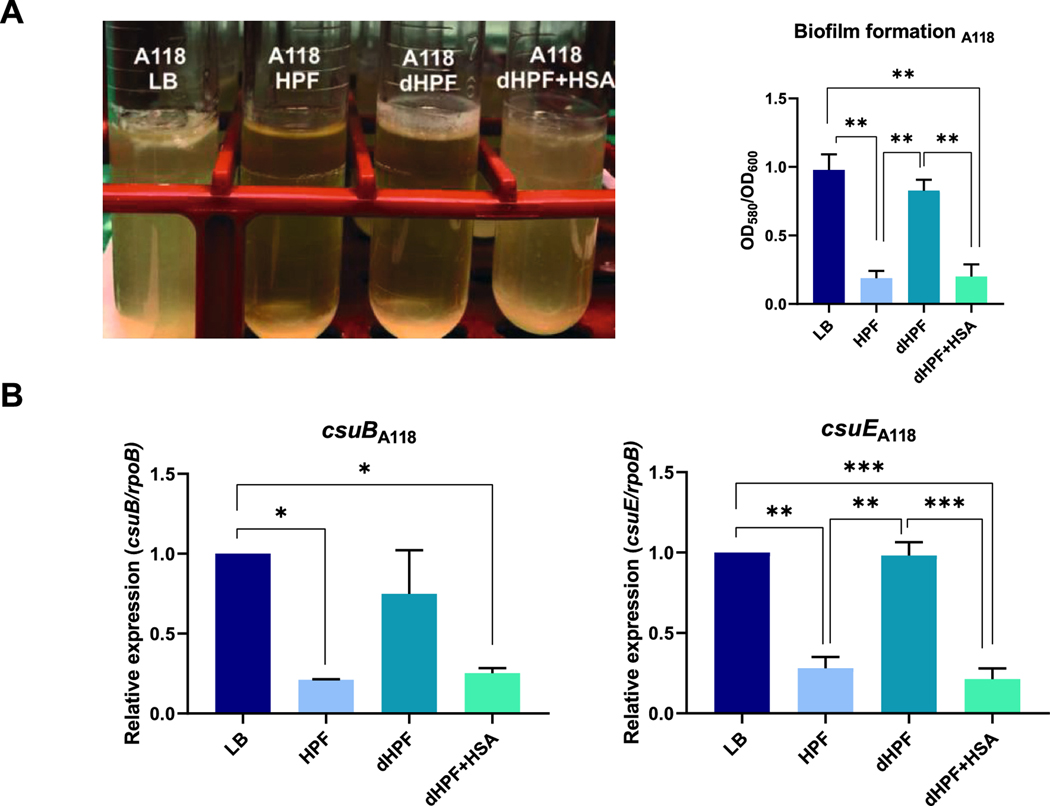
Consistent with the presumed role of HSA in biofilm formation, discernible differences occurred when A. baumannii A118 was cultured in a medium with different supplements (Fig. 2A). The presence of HSA, i.e., medium supplemented with HPF or dHPF + HSA, was associated with a less developed biofilm formation, while under LB or dHPF conditions a robust biofilm was observed (Fig. 2A). Also, two components of the csu operon, csuB and csuE, were downregulated in cells cultured in the presence of HPF or dHPF + HSA (Fig. 2B). This operon plays an important role in biofilm formation on abiotic surfaces in A. baumannii (11).
Figure 2. Phenotypic and genetic analysis of biofilm formation coding genes.
A) Biofilm assays performed with the A118 strain grown in LB or LB supplemented with HPF, dHPF, or dHPF + HSA. B) qRT-PCR of the csuB and csuE biofilm formation-related genes in cells cultured in LB or LB supplemented with HPF, dHPF or dHPF + HSA. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used. LB was used as the reference condition. The mean ± SD is informed. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: p < 0.05; two asterisks: p < 0.01 and three asterisks: p < 0.001.
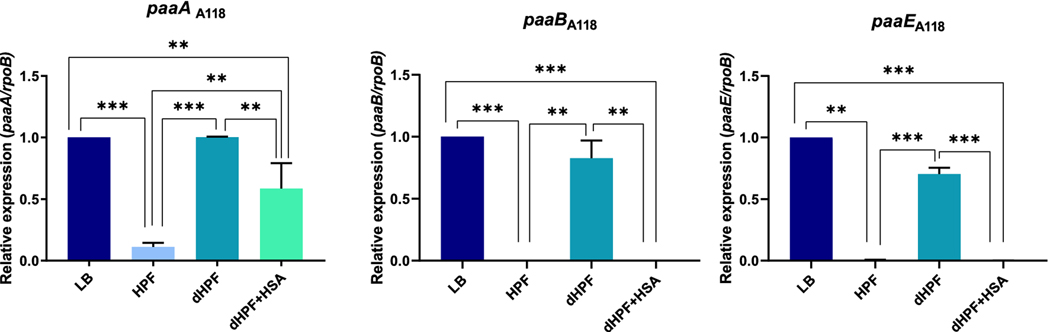
Lastly, as the phenylacetic acid pathway (PA) contributes to host immune evasion and virulence, we analyzed the expression of representative genes of this pathway, paaA, paaB and paaE (12). We observed decreases in the expression levels of all paa genes analyzed in cells grown in media supplemented with HPF or dHPF + HSA (Fig.3).
Figure 3. Genetic analysis of phenylacetic acid pathway related coding genes.
qRT-PCR of paaA, paaB and paaE, genes related with the phenylacetic acid pathway, in A118 cells grown in LB or LB supplemented with HPF, dHPF or dHPF + HSA. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used. LB was used as the reference condition. The mean ± SD is informed. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: p < 0.05; two asterisks: p < 0.01 and three asterisks: p < 0.001.
Conclusion
The results described herein support the role of HSA as one of the HPF components that affect the expression of genes related to quorum sensing, biofilm formation, and phenylacetic acid production, all features associated with A. baumannii pathobiology. A similar HSA modulatory response was previously observed with the AB5075 strain (2). Taken together, the results obtained with A. baumannii A118 and AB5075 suggest a general response of A. baumannii when grown in the presence of HSA. This data showed that A. baumannii’s ability to control its transcriptional and phenotypical response could be one of the factors contributing to A. baumannii’s adaptability to overcome stressful conditions within the host. Future work will be performed using a panel of different strains to observe if the modulatory response is ubiquitous, as well as, to identify the mechanism involved in trigerring A. baumannii’s response.
Acknowledgments
Funding: The authors’ work was supported by NIH SC3GM125556 to MSR, R01AI100560 to RAB, R01AI063517, R01AI072219 to RAB, and 2R15 AI047115 to MET. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to RAB and 1I01BX002872 to KMP-W from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to RAB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. MRT and TS are recipient of a postdoctoral fellowship from CONICET.
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
Publisher's Disclaimer: This AM is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect post-acceptance improvements, or any corrections. Use of this AM is subject to the publisher’s embargo period and AM terms of use. See here for Springer Nature’s terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
REFERENCES
- 1.Rodman Nyah MJ, Fung Sammie, Nakanouchi Jun, Myers Amber L., Harris Caitlin M., Dang Emily, Fernandez Jennifer S., Liu Christine, Mendoza Anthony M., Jimenez Veronica, Nikolaidis Nikolas, Brennan Catherine A., Bonomo Robert A., Sieira Rodrigo, Ramirez Maria Soledad. Human Pleural Fluid Elicits Pyruvate and Phenylalanine Metabolism in Acinetobacter baumannii to Enhance Cytotoxicity and Immune Evasion. Frontiers in Microbiology. 2019;10:1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pimentel C, Le C, Tuttobene MR, Subils T, Martinez J, Sieira R, et al. Human Pleural Fluid and Human Serum Albumin Modulate the Behavior of a Hypervirulent and Multidrug-Resistant (MDR) Acinetobacter baumannii Representative Strain. Pathogens. 2021;10(4):471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, et al. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol. 2010;48(4):1488–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Traglia GM, Chua K, Centron D, Tolmasky ME, Ramirez MS. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol Evol. 2014;6(9):2235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez J, Razo-Gutierrez C, Le C, Courville R, Pimentel C, Liu C, et al. Cerebrospinal fluid (CSF) augments metabolism and virulence expression factors in Acinetobacter baumannii. Sci Rep. 2021;11(1):4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulk Tierney AR, Rather PN. Methods for Detecting N-Acyl Homoserine Lactone Production in Acinetobacter baumannii. Methods Mol Biol. 2019;1946:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant Microbe Interact. 1998;11(11):1119–29. [DOI] [PubMed] [Google Scholar]
- 8.Quinn B, Rodman N, Jara E, Fernandez JS, Martinez J, Traglia GM, et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci Rep. 2018;8(1):14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Martin M, Dubern JF, Alexander MR, Williams P. AbaM Regulates Quorum Sensing, Biofilm Formation and Virulence in Acinetobacter baumannii. J Bacteriol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez M, Mayer C, Fernandez-Garcia L, Blasco L, Muras A, Ruiz FM, et al. Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS One. 2017;12(3):e0174454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154(Pt 11):3398–409. [DOI] [PubMed] [Google Scholar]
- 12.Bhuiyan MS, Ellett F, Murray GL, Kostoulias X, Cerqueira GM, Schulze KE, et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci U S A. 2016;113(34):9599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]