Abstract
Pieris rapae and Phyllotreta nemorum are Brassicaceae specialists, but do not feed on Iberis amara spp. that contain cucurbitacins. The cucurbitacins are highly oxygenated triterpenoid, occurring widespread in cucurbitaceous species and in a few other plant families. Using de novo assembled transcriptomics from I. amara, gene co-expression analysis and comparative genomics, we unraveled the evolutionary origin of the insect deterrent cucurbitacins in I. amara. Phylogenetic analysis of five oxidosqualene cyclases and heterologous expression allowed us to identify the first committed enzyme in cucurbitacin biosynthesis in I. amara, cucurbitadienol synthase (IaCPQ). In addition, two species-specific cytochrome P450s (CYP708A16 and CYP708A15) were identified that catalyze the unique C16 and C22 hydroxylation of the cucurbitadienol backbone, enzymatic steps that have not been reported before. Furthermore, the draft genome assembly of I. amara showed that the IaCPQ was localized to the same scaffold together with CYP708A15 but spanning over 100 kb, this contrasts with the highly organized cucurbitacin gene cluster in the cucurbits. These results reveal that cucurbitacin biosynthesis has evolved convergently via different biosynthetic routes in different families rather than through divergence from an ancestral pathway. This study thus provides new insight into the mechanism of recurrent evolution and diversification of a plant defensive chemical.
Keywords: evolution, cytochrome P450, biosynthesis, triterpenoid, cucurbitacin
Introduction
It is well known that plants produce many specialized metabolites, yet we know little of how they evolved and how new ecological functions are gained. Due to coevolution, plants can become susceptible to specialized insect species, whereas some plant species evolve new chemical defense compounds to overcome this problem. Insect species such as Pieris rapae and Phyllotreta nemorum are serious agricultural pests of crops in the Brassicaceae family. They have overcome the toxicity of the well-known glucosinolates that function as oviposition stimulants and feeding stimulants for them. Iberis spp., however, that also belong to the Brassicaceae, are free from these herbivores because they contain alternative feeding inhibitors, cucurbitacins (Nielsen et al. 1977; Sachdev-Gupta et al. 1993). Interestingly, in cucurbitacous species the cucurbitacins act as feeding stimulants for specialized herbivores such as spotted cucumber beetles (Diabrotica undecimpunctata howardi) (Martin et al. 2002). Species in the genus Iberis, may therefore be used as a model to investigate the evolutionary reoccurrence of the insect deterrent cucurbitacin.
Cucurbitacins are structurally diverse tetracyclic triterpenes that are well known for their bitter taste and strong toxicity (Chen et al. 2005). Cucurbitacins are particularly well known to occur in the Cucurbitaceae family. At least 100 species belonging to 30 genera of the Cucurbitaceae family have been reported to contain cucurbitacins (Raemisch and Turpin 1984). Accordingly, studies of cucurbitacin biosynthesis have been predominantly carried out within members of the Cucurbitaceae family (Shang et al. 2014; Zhou et al. 2016). Besides in cucurbitacous families, cucurbitacins have been reported in several other taxonomically distant families (Chen et al. 2005; Gry et al. 2006), mainly in angiosperms but also in the monocot species Phormium tenax (Kupchan et al. 1978). Cucurbitacins are also found outside of the plant kingdom occurring in mushrooms (Fujimoto et al. 1987; Jian-Wen et al. 2002) and marine mollusks (Chen et al. 2005) (fig. 1A). The origin of cucurbitacin biosynthesis in these species remains unknown. Likewise, it remains enigmatic how the entire cucurbitacin pathway in these species may have evolved.
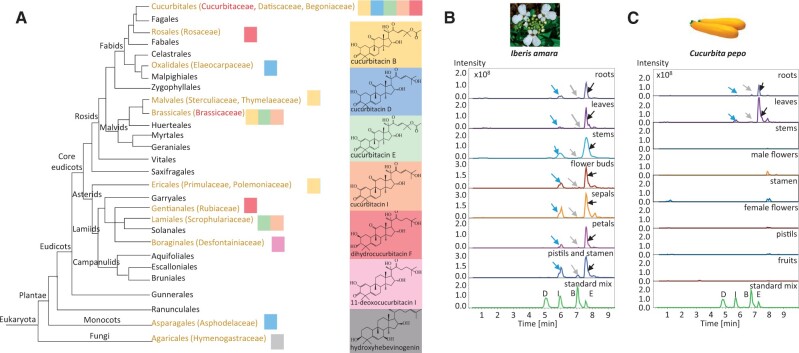
Fig. 1.
Distribution of cucurbitacins across families in the Eukaryota and their accumulation pattern in a brassicaceous and a cucurbitaceous species. (A) Phylogenetic relationships in Eukaryota indicating multiple origins of cucurbitacin biosynthesis. The picture was adapted according to Stevens, P. F. (2001 onwards). Angiosperm Phylogeny Website. Version July 14, 2017 (http://www.mobot.org/MOBOT/research/APweb/). Note, only the major cucurbitacins in each order are shown color-coded. (B) LC-ESI-MS extracted ion chromatograms (EIC) of extracts from roots, leaves, stems, flower buds, sepals, petals, pistils, and stamen of 8-week-old Iberis amara plants. (C) LC-ESI-MS extracted ion chromatograms (EIC) of extracts from Cucurbita pepo roots, leaves, stems, male flowers, stamen, female flowers, pistils, and fruits. The extracted ions at m/z 539, 537, 581, and 579 are representative of sodium adducts of cucurbitacin D, I, B, E, respectively. Peaks indicated by blue, gray, and black arrows were verified by comparing the corresponding mass spectra to those of authentic cucurbitacin I, B, and E standards, respectively.
To address this knowledge gap, we investigated cucurbitacin biosynthesis in the brassicaceous species I. amara, a species that accumulates cucurbitacin E and I (Nielsen et al. 1977) and is taxonomically distant from the Cucurbitaceae. Using de novo assembled transcriptomics from I. amara, gene co-expression analysis and comparative genomics, phylogenetic analysis and heterologous reconstitution of biosynthetic pathways, we identified the first committed enzyme, an oxidosqualene cyclase (OSC) that catalyzes the formation of cucurbitadienol, as well as two novel cytochromes P450 (P450s) catalyzing C16β- and C22-hydroxylation, in the cucurbitacin biosynthetic pathway of I. amara. We demonstrate that these three key enzymes in the cucurbitacin biosynthetic pathway have been independently recruited in brassicaceous and cucurbitaceous species and show that the biosynthetic routes are different, thus implying that the cucurbitacin pathway has evolved more than once in the angiosperm kingdom.
Results
Cucurbitacin E, I, and B Accumulate Ubiquitously in I. amara
Cucurbitacin (Cuc) E and I have previously been isolated from the aerial part of I. amara and identified based on co-migration on thin layer chromatography (Nielsen et al. 1977). To evaluate more precisely where cucurbitacins accumulate in I. amara plant, methanol extracts from different organs of 1-week-old, 4-week-old, and 8-week-old plants were profiled by LC-ESI-MS/MS. CucE and CucI and a trace amount of CucB were detected in both roots and aerial parts, implicating that Cucs accumulate constitutively (supplementary fig. 1, Supplementary Material online and fig. 1B). In contrast to I. amara, Cuc accumulation in the cucurbitaceous species Cucurbita pepo (fig. 1C) and Citrullus lanatus (supplementary fig. 2, Supplementary Material online) displayed an organ-specific manner. CucE and trace amounts of CucI and CucB were only present in roots and leaves, whereas absent (or in trace amounts) in the stem, fruit, and flower parts. Some cucurbitaceous species have been reported to contain CucD, but we could not detect CucD C. pepo and Ci. lanatus.
Identification of I. amara Cucurbitadienol Synthase
To elucidate the cucurbitacin biosynthesis pathway in I. amara, transcriptomes of 4-week-old leaves and roots were generated, assembled, and analyzed, as both organs accumulate cucurbitacins (supplementary fig. 1B, Supplementary Material online). After de novo transcriptome assembly, a total of 43,548 and 60,975 contigs were obtained for leaves and roots, respectively (supplementary table 1, Supplementary Material online). As cucurbitacins are triterpenes, we anticipate that the first step of cucurbitacin biosynthesis in I. amara is catalyzed by an OSC. Five full-length OSC sequences were retrieved from the assembled transcriptome using 13 Arabidopsis thaliana OSC cDNA sequences as queries in BlastN searches. Of the five full-length OSCs identified in the transcriptome, only two, denoted I. amara cucurbitadienol synthase (IaCPQ) and I. amara cycloartenol synthase (IaCAS) (see below), respectively, were highly expressed in both roots and aerial organs (supplementary fig. 3, Supplementary Material online), and accordingly selected for cloning and further characterization as their expression profile followed the accumulation pattern of cucurbitacins.
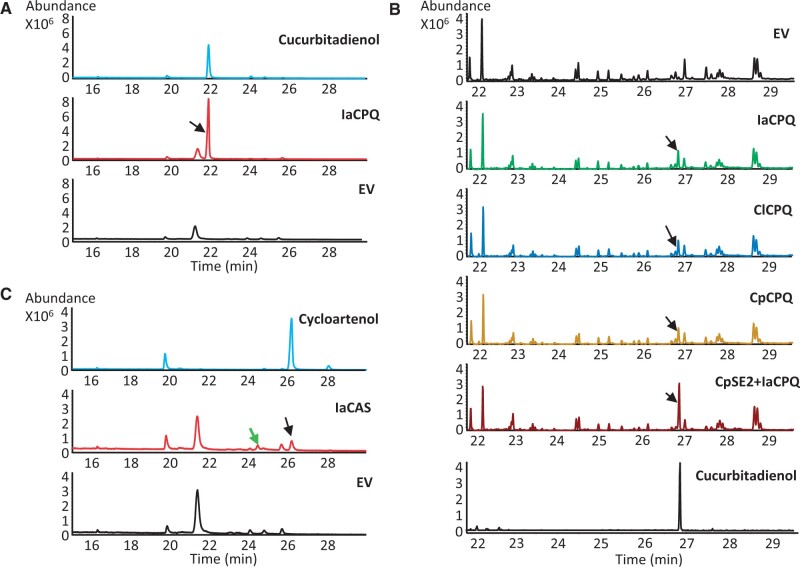
Both IaCPQ and IaCAS were expressed in the Saccharomyces cerevisiae strain PA14 (Calegario et al. 2016) for functional characterization. When IaCPQ was expressed, extracts of the yeast spent medium revealed a single peak eluting at 21.9 min by GC-MS analysis (fig. 2A), but not in the empty vector control culture. The compound eluting at 21.9 min was purified from a 1.6-l yeast culture and its structure was determined by NMR analysis as 4,9-cyclo-9,10-secocholesta-5,24-diene (cucurbitadienol) (supplementary fig. 4A and B and NMR data, compound 3, Supplementary Material online). As different host environments could affect the enzyme activity, the function of IaCPQ was compared with that of the previously characterized C. pepo CPQ (CpCPQ) (Dong et al. 2018) and a CPQ candidate cloned from Ci. lanatus cDNA (ClCPQ), by transient expression in Nicotiana benthamiana leaves. GC-MS analysis of ethyl acetate extracts of N. benthamiana leaves infiltrated with IaCPQ, CpCPQ, or ClCPQ showed a single new peak at retention time 26.9 min that co-eluted with the prepared cucurbitadienol standard (fig. 2B). Some CPQs may also use 2,3; 22,23-dioxidosqualene as substrate. An example of this is the CPQ from the cucurbitaceous species, Siraitia grosvenorii, that catalyze the cyclization of 2,3; 22,23-dioxidosqualene to 24,25-epoxycucurbitadienol in addition to the cyclization of 2,3-oxidosqualene to cucurbitadienol (Itkin et al. 2016). To determine if IaCPQ similarly can use both 2,3-oxidosqualene and 2,3; 22,23-dioxidosqualene as substrates, the previously identified C. pepo squalene epoxidase 2 (CpSE2), previously shown to utilize both 2,3-oxidosqualene and 2,3; 22,23-dioxidosqualene as substrates (Dong et al. 2018), was co-agroinfiltrated with IaCPQ into N. benthamiana. No peak corresponding to 24,25-epoxycucurbitadienol could be detected, but an increased cucurbitadienol peak was observed as compared with the empty vector control (fig. 2B), which demonstrates that 2,3-oxidosqualene is the substrate for IaCPQ and that only one cyclized product, cucurbitadienol, is produced.
Fig. 2.
Identification and characterization of Iberis amara cucurbitadienol synthase. (A) Total ion current (TIC) GC-MS chromatograms of extracts from an empty vector (EV) control yeast strain (black) and a strain expressing IaCPQ (red), and of purified cucurbitadienol as a standard (blue). The peak indicated with a black arrow at 21.9 min corresponds to cucurbitadienol. (B) TIC GC-MS chromatograms of extracts from Nicotiana benthamiana leaves agro-infiltrated with an EV control, IaCPQ, ClCPQ, CpCPQ, and CpSE2 with IaCPQ, and the chromatogram of the cucurbitadienol standard. Black arrows indicate the cucurbitadienol peak. (C) TIC GC-MS chromatograms of extracts from an EV control yeast strain (black) and a strain expressing IaCAS (red), and the chromatogram of a cycloartenol standard (blue). The peak indicated with a black arrow eluting at 26.2 min corresponds to cycloartenol. The second peak indicated with a green arrow eluting at 24.5 min likely corresponds to 31-norcycloartenol. Note, the GC-MS analysis program for yeast samples differs slightly from the one used for GC-MS analysis for plant extracts, giving rise to differences in retention times.
In addition, GC-MS analysis of the spent medium of yeast strain PA14 expressing IaCAS revealed two new chromatographic peaks that eluted at 24.5 and 26.2 min, respectively (fig. 2C), as compared with the yeast strain PA14 expressing an empty vector control. The latter peak has the same retention time and EI-MS fragmentation pattern as an authentic cycloartenol standard (fig. 2C and supplementary fig. 4C and D, Supplementary Material online), indicating IaCAS encodes a cycloartenol synthase. The second peak at 24.5 min in the IaCAS chromatogram has a parent mass 14 Da lower than cycloartenol (supplementary fig. 4E, Supplementary Material online) and thus likely corresponds to 31-norcycloartenol, a demethylation product of cycloartenol previously shown to accumulate in yeast expressing a functional cycloartenol synthase (Gas-Pascual et al. 2014).
Iberis amara Cucurbitadienol Synthase Evolved Independently from I. amara Cycloartenol Synthase
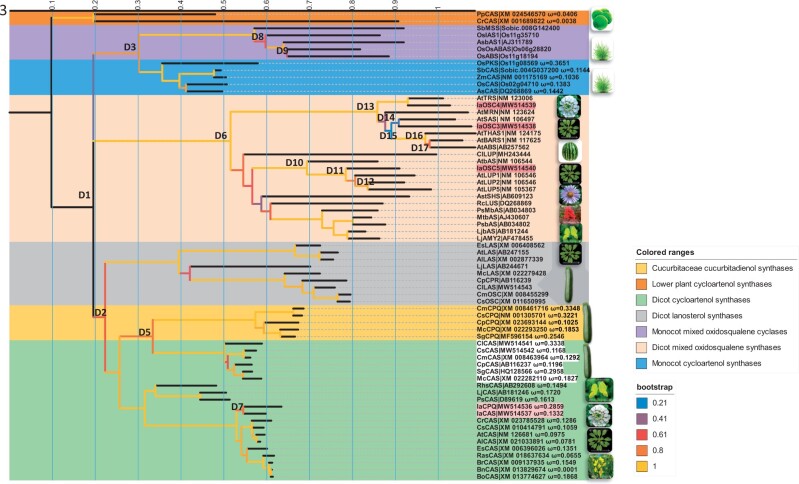
To determine the evolutionary origin of IaCPQ, a phylogenetic analysis was performed. About 67 full-length OSC-cDNA sequences were selected, which based on the literature code for OSCs generating the main known triterpenoid scaffolds. The coding sequences were aligned based on codons. Seven distinct triterpenoid scaffold clades could be identified, of which structures configured through protosteryl cations (CASs, CPQs, and lanosterol synthases) and dammarenyl cations (β-amyrin synthases, lupeol synthases, and multifunctional OSCs) (Thimmappa et al. 2014) were clearly separated (fig. 3). There appears to be a duplication event (D1) before the split of monocot and dicot plants which resulted in three ancestor genes, which gave rise to monocot CASs group (fig. 3, blue colored background) and other OSCs (fig. 3, purple colored background), dicot protosteryl cation group (fig. 3, green, yellow, and gray colored background), and dicot dammarenyl cation groups (light orange colored background). This is corresponding to the study of Xue et al. (2012). The second duplication event D2 gave rise to lanosterol group and a CAS ancestor to both cucurbitaceous species and brassicaceous species. Interestingly, based on the relative branch length the D5 duplication separating cucurbitaceous CPQ and CAS happened before the divergence of the cucurbitaceous species, whereas IaCPQ duplicated (D7) from IaCAS (fig. 3) after the Iberis spp. diverged from other brassicaceous species. This might explain why only Iberis spp. produce cucurbitacin in Brassicaceae, whereas cucurbitacins are widely spread over the cucurbitaceous species. In agreement with a relatively more recent evolutionary event of IaCPQ, the amino acid identity between IaCPQ and IaCAS is 86%, whereas CPQ and CAS from the Cucurbitaceae family are relatively more diverged and display around 70% identical (supplementary fig. 5, Supplementary Material online).
Fig. 3.
Iberis amara cucurbitadienol synthase evolved from I. amara cycloartenol synthase. The evolutionary history of OSCs was inferred by using the maximum likelihood method to build a rooted tree. About 67 OSCs nucleotide sequences were selected so they broadly represent OSCs from species with known product profiles. The coding sequences were aligned based on codons. Branches of CASs and CPQs were labeled with ω values. Branches were color coded corresponding to their bootstrap value. Predicted gene duplications were identified by searching for all branching points in the topology with at least one species that is present in both subtrees of the branching point. D1–D17 indicate duplicate events. The scale bar represents the number of substitution sites corresponding to the branch length. Evolutionary analyses were conducted in MEGA X. Note, some species pictures on the tree represent the cucurbitaceous species, brassicaceous specious, fabaceous species, Arabidopsis species, monocot species, lower plants. Only pictures for I. amara, Citrullus lanatus, Ricinus communi, Aster tataricus represent single species. Gene names from I. amara were coded with light red background. CAS names from cucurbitacecous species were colored with white background. Lj, Lotus japonicus; Ps, Pisum sativum; As, Avena strigose; Rc, Ricinus communi; Ast, Aster tataricus; Rhs, Rhizophora stylosa; Mt, Medicago truncatula; Sg, Siraitia grosvenorii; Mc, Momordica charantia; Cp, Cucurbita pepo; Cl, Citrullus lanatus; Cs, Cucumis sativus; Cm, Cucumis melo; Cr, Capsella rubella; Rs, Raphanus sativus; At, Arabidopsis thaliana; Al, Arabidopsis lyrate; Br, Brassica rapa; Bo, Brassica oleracea; Es, Eutrema salsugineum; Cs, Camelina sativa; Ia, I. amara; Os, Oryza sativa; Sb, Sorghum bicolor; Zm, Zea mays; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii. IAS1, isoarborinol synthase 1, ABAS, mixed α- and β-amyrin synthase; ABS, achilleol B synthase; MSS, mixed simiarenol synthase; PKS, parkeol synthase; ABS, arabidiol synthase; SAS, seco-amyrin synthase; TRS, Tirucalladienol synthase; OSC, oxidosqualene synthase; PEN1, pentacyclic triterpene synthase 1; PEN6, pentacyclic triterpene synthase 6; PEN3, pentacyclic triterpene synthase 3; THAS1, thalianol synthase 1; BARS1, baruol synthase 1; MRN1, marneral synthase 1; LUP, Lupeol synthase; CPR, putative oxidosqualene cyclase; CPX, cycloartenol synthase; CPQ, cucurbitadienol synthase; CAS, cycloartenol synthase; LAS, lanosterol synthase; AMY2, mixed β-amyrin synthase; MbAS, multifunctional β-amyrin synthase; bAS, β-amyrin synthase.
To further determine the relative selection pressures on the CPQs, the nonsynonymous to synonymous substitution rate ratios (ω=dN/dS) were calculated by phylogenetic analysis by maximum likelihood (PALM) analysis (fig. 3). The free-ratio model was significantly better than both the one-ratio model and the two-ratio model. For the two-ratio model, the IaCPQ branch was chosen as a foreground branch (P < 0.01). This supports the hypothesis of variable selective constraints across the phylogeny (details of the log-likelihood are in supplementary table 2, Supplementary Material online). Both IaCPQ and IaCAS are under purifying selection (ω < 1), however as would be expected, IaCPQ has a higher ω value (0.2859) than the conserved IaCAS (ω= 0.1332), indicating a relaxed selection constraint for IaCPQ as compared with IaCAS. In agreement with a difference in selection pressure, the longer branch length for IaCPQ as compared with IaCAS possibly reflects a relatively higher evolutionary rate, supporting that IaCPQ has been neofunctionalized from a duplicated IaCAS. Since the Iberis spp. has undergone the whole genome duplication event (Yang et al. 2020), indicate this might be the origin of IaCPQ.
Iberis amara Cucurbitadienol Synthase Is a Monotopic Membrane Protein Expressed in All Organs
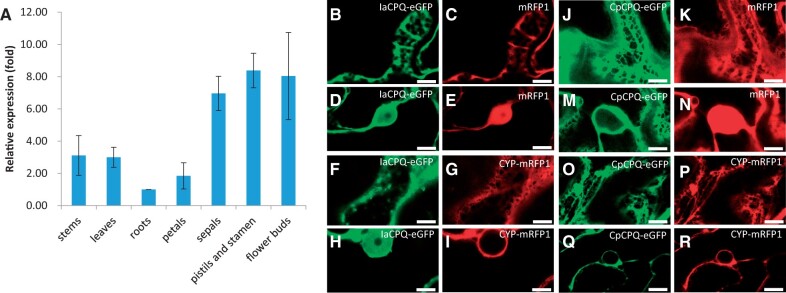
Cucurbitacins accumulate in all the examined organs of I. amara (fig. 1 and supplementary fig. 1, Supplementary Material online). To determine whether cucurbitacins are also biosynthesized in all examined organs, we analyzed relative IaCPQ expression levels in leaf, root, stem, and flower organs by quantitative real-time PCR (qPCR). IaCPQ was expressed in all the tested organs, with expression being lowest in roots, 2- to 3-fold higher in stems, leaves, and petals, and highest in flower buds, pistils and stamen, and sepals (7- to 8-fold higher compared with roots) (fig. 4A). This agrees with the relative accumulation of cucurbitacins in these organs (fig. 1B). In comparison, the functional ClCPQ homolog in Ci. lanatus was highest expressed in leaves and roots (supplementary fig. 6A, Supplementary Material online), and also here expression levels are also in accordance with the organ-specific accumulation of cucurbitacins in this species (supplementary fig. 2, Supplementary Material online).
Fig. 4.
Iberis amara cucurbitadienol synthase is expressed in all plant organs tested and is a monotopic membrane protein. (A) Relative expression of IaCPQ in different I. amara organs quantified by qPCR. (B–R) Representative confocal images of the epidermal cell space ([B], [C], [F], [G], [J], [K], [O], [P]) and the nucleus ([D], [E], [H], [I], [M], [N], [Q], [R]) of Nicotiana benthamiana leaves expressing C-terminally tagged proteins. mRFP1 and CYP98A1 (P450) are used as controls for cytosolic and ER localization, respectively. Soluble mRFP1 is found in the nucleus and fills in gaps between plant cell compartments. ER-localized proteins (CpCPQ and CYP98A1) are restrained to nuclear membranes and a well-defined ER membrane network. IaCPQ is observed on the nuclear membrane as a monotopic membrane protein, however, excess protein appears to permeate the nucleus. Similar images were obtained with N-terminal fluorescent fusion constructs for IaCPQ and CpCPQ (supplementary fig. 6B, Supplementary Material online). Scale bars=10 µm.
To investigate whether cucurbitadienol synthases from the Brassicaceae and the Cucurbitaceae have the same subcellular localization, the enhanced green fluorescent protein (eGFP) was fused to either the N- or C-termini of both IaCPQ and CpCPQ, and transiently expressed in N. benthamiana leaves. Confocal imaging of the IaCPQ fused to eGFP at either N- or C-terminal suggests that IaCPQ is a monotopic membrane protein that adheres to the plasma membrane and around the cortical ER (fig. 4B and F; supplementary fig. 6B, Supplementary Material online). The P450 CYP98A1 N-terminally fused to mRFP was included as a positive control for an ER-localized enzyme (fig. 4G and I) (Laursen et al. 2016). In addition, green fluorescent signal was observed in the nucleus (fig. 4D and H; supplementary fig. 6B, Supplementary Material online), likely originating from overexpression of tagged IaCPQ resulting in saturation of membranes and subsequent accumulation in the nuclei; nevertheless this signal was much less intense than what was observed for the soluble mRFP positive control (fig. 4C and E; supplementary fig. 6B, Supplementary Material online). The observed soluble fraction of green fluorescent signal is not due to cleavage of the eGFP tag, which is confirmed by Western blot (data not shown). Compared with IaCPQ, localization of CpCPQ fused to eGFP at either the N- or C-terminal was more restricted to a characteristic ER membrane network (fig. 4J and O; supplementary fig. 6B, Supplementary Material online) and the nuclear membrane (fig. 4M and Q; supplementary fig. 6B, Supplementary Material online), as evidenced from the same localization to the ER-localized control (CYP98A1-mRFP) (fig. 4P and R; supplementary fig. 6B, Supplementary Material online) but different localization as the soluble mRFP (fig. 4K and N). Similarly, ClCPQ was also shown to be localized to the ER membrane network (supplementary fig. 6C, Supplementary Material online).
In line with the subcellular localization, homology modeling of IaCPQ, ClCPQ, and CpCPQ using the crystalized Homo sapiens lanosterol synthase (HsLAS, PDB ID: 1W6J) as a template (Thoma et al. 2004), and membrane interaction analysis using the Positioning of Proteins in Membrane (PPM) server predicted that these proteins have membrane-inserting hydrophobic segments (supplementary fig. 7A, Supplementary Material online). Interestingly, all three CPQs were predicted as monotopic membrane localized proteins, like HsLAS. The membrane inserting residues were identified (supplementary fig. 7A, Supplementary Material online) and the residues that interact with lipids were highlighted (supplementary fig. 7B, Supplementary Material online).
CYP708A16 Hydroxylates Cucurbitadienol to 16β-Hydroxycucurbitadienol
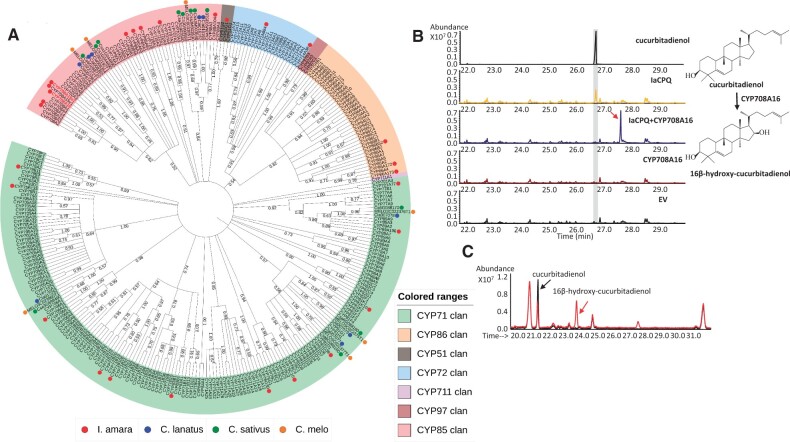
The subsequent steps in the cucurbitacin biosynthetic pathway are likely oxidations carried out by P450s. Attempts to find candidates based on homology searches within the I. amara transcriptome data set using known Ci. lanatus P450s involved in cucurbitacin biosynthesis (Zhou et al. 2016) as baits were not successful, indicating that the P450-mediated oxidations are not evolutionary conserved between I. amara and Ci. lanatus. Therefore, we mined our I. amara transcriptome using 245 A. thaliana P450s as query sequences (Paquette et al. 2009) and retrieved 25 full-length and around 200 partial I. amara P450 genes. We expected that P450 transcripts involved in the cucurbitacin pathway would be relatively highly represented in our transcriptome data set. Phylogenetic analysis of these full-length genes showed that 12 of them grouped with the P450 families 51, 708, 71, 72, 81, 85, 88, 89, 90, and 93, families known to harbor members that can oxidize triterpenes (fig. 5A) (Ghosh 2017). Thus, these 12 P450s were agroinfiltrated separately or in combinations with IaCPQ in N. benthamiana leaves to see which one of them could catalyze the first oxidation step(s). Of the 12 tested P450s, only CYP708A16 showed activity on cucurbitadienol (fig. 5B). When CYP708A16 and IaCPQ were co-agroinfiltrated into N. benthamiana leaves, the cucurbitadienol peak disappeared, and a new peak at RT of 27.7 min was detected by GC-MS. This peak was absent in the leaves agro-infiltrated with only IaCPQ, CYP708A16, or empty vector alone (fig. 5B). The estimated molecular mass of the new peak fit with a hydroxylated cucurbitadienol (supplementary fig. 8, Supplementary Material online). This result was confirmed by the co-expression of CYP708A16 with a CPQ from Momordica charantia (Cucurbitaceae) in yeast (fig. 5C). To determine the chemical structure, the metabolite corresponding to the new peak was purified from N. benthamiana leaves by preparative HPLC and the purified compound was subjected to 1- and 2D NMR experiments. The NMR analysis determined that the new metabolite is 16β-hydroxy-4,9-cyclo-9,10-secocholesta-5,24-diene (16β-hydroxycucurbitadienol) (Supplementary NMR data, compound 2, Supplementary Material online). None of the reported P450 genes from cucurbitaceous species were shown to hydroxylate at the C16 position of cucurbitadienol (Zhou et al. 2016). Thus, CYP708A16 is, to our knowledge, the first enzyme in a cucurbitacin biosynthetic pathway hydroxylating cucurbitadienol at the C16 position.
Fig. 5.
CYP708A16 belongs to the CYP708 family and catalyzes the conversion of cucurbitadienol to 16β-hydroxy-cucurbitadienol. (A) Maximum likelihood phylogenetic tree of Iberis amara and selected Arabidopsis thaliana P450 amino acid sequences. Bootstrap values in % are shown at the branch points. The tree was constructed with MEGA X and depicted using the online tool iTOL v5. (https://itol.embl.de/). The assembled 25 full-length I. amara P450s (MW514544, MW514545, MW514546, MW514547, MW514552, MW514553, MW514554, MW514555, MW514557, MW514558, MW514559, MW514560, MW514561, MW514562, MW514563, MW514564, MW514565, MW514566, MW514567, MW514568, MW514569, MW514570, MW514571, MW514572) from the Miseq data and additional full-length CYP708As (MW514548, MW514549, MW514550, MW514551, MW514556) assembled from the Hiseq data are labeled with red dots. Cucurbitacin biosynthetic related P450 genes from Citrullus lanatus (blue dot), Cucumis sativus (green dot), Cucumis melo (orange dot) (Zhou et al. 2016), and selected P450 sequences from A. thaliana are also included in the tree. (B) GC-MS chromatograms of the cucurbitadienol standard and extracts from Nicotiana benthamiana leaves agro-infiltrated with an EV control and vectors expressing IaCPQ, CYP708A16, and IaCPQ together with CYP708A16. Peaks in the gray box indicate cucurbitadienol. The red arrow indicates the unique peak in the extract of the IaCPQ and CYP708A16 co-expressing leaves. (C) TIC GC-MS of extracts from yeast strains expressing McCPQ, and McCPQ with CYP708A16. Cucurbitadienol is indicated with a black arrow. The peak of 16β-hydroxy-cucurbitadienol is indicated with a red arrow. Note, the GC-MS analysis program for yeast samples differs slightly from the one used for GC-MS analysis for plant extracts, giving rise to the observed differences in retention times.
IaCYP708A15v2 Catalyzes 22-Hydroxylation in the Cucurbitacin Biosynthesis
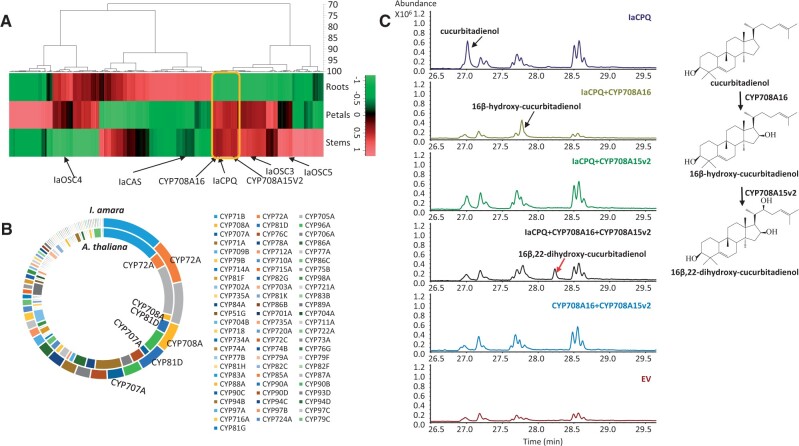
Since only one P450 gene (CYP706A16) was found to be involved in cucurbitacin biosynthesis among the 25 genes we cloned from our Iberis transcriptome of 4-week-old roots and leaves, we opted for a new strategy to obtain additional candidate genes. Cucurbitacin biosynthetic genes in cucurbitaceous species have been shown to be co-expressed (Zhou et al. 2016). Thus, we expected other candidate genes involved in the cucurbitacin biosynthesis in I. amara would show a similar expression pattern as IaCPQ and CYP708A16. Accordingly, to search for additional P450 candidates, co-expression analysis was performed on a new I. amara transcriptome data set (HiSeq 4000) generated from stems, roots, and petals of 8-week-old I. amara plants. The transcriptome was de novo assembled by Trinity, which resulted in 167,939 contigs (supplementary table 3, Supplementary Material online). A BlastN analysis identified 287 full-length or nearly full-length P450 contigs. The P450 candidates were together with the above five identified OSCs subjected to hierarchical cluster analysis (fig. 6A). In agreement with a role in cucurbitacin biosynthesis, CYP708A16 identified above clustered together with IaCPQ but not with any of the other four OSCs. Thus, P450s that co-expressed with IaCPQ and CYP708A16 were selected as additional candidates for cucurbitacin biosynthesis (supplementary table 4, Supplementary Material online).
Fig. 6.
IaCYP708A15v2 catalyzes 22-hydroxylation in the cucurbitacin biosynthesis. (A) Hierarchical cluster heat map of expression patterns of the 287 P450 contigs and five oxidosqualene cyclases expressed in the roots, petals, and stems of Iberis amara. Putative biosynthetic P450 genes involved in cucurbitacin biosynthesis are marked by a yellow box. (B) Doughnut chart was drawn based on the exact number of genes from each P450 subfamily of I. amara (outer doughnut) and Arabidopsis thaliana (inner doughnut). Each subfamily was color coded. In particular, the CYP72A, CYP708A, CYP81D, and CYP707A subfamilies were expanded in I. amara as compared with A. thaliana. (C) GC-MS chromatograms of extracts from Nicotiana benthamiana leaves agro-infiltrated with EV control and vectors expressing IaCPQ, IaCPQ+CYP708A16, IaCPQ+CYP708A15v2, IaCPQ+CYP708A16+CYP708A15v2, and CYP708A16+CYP708A15v2. Cucurbitadienol and 16β-hydroxy-cucurbitadienol are indicated with black arrows. The new peak that appeared only in the extract from IaCPQ+CYP708A16+CYP708A15v2 expressing leaves is indicated by a red arrow.
To further narrow down candidates for further analysis, we compared the number of contigs per P450 family identified in I. amara to the known families in A. thaliana (fig. 6B and supplementary table 5, Supplementary Material online). Four P450 subfamilies: CYP72A, CYP708A, CYP707A, and CYP81D, appeared expanded in I. amara compared with A. thaliana (fig. 6B and supplementary table 5, Supplementary Material online). Since there is no cucurbitacin biosynthesis in A. thaliana, we hypothesized that gene members within these expanded families could be responsible for the newly evolved cucurbitacin pathway. In accordance, the highly expressed CYP708A15v2 (supplementary table 4, Supplementary Material online) was selected as a promising candidate for catalyzing the second oxidative step in cucurbitacin biosynthesis. When CYP708A15v2 was co-agroinfiltrated with IaCPQ and CYP708A16 in N. benthamiana (fig. 6C) the cucurbitadienol and the 16β-hydroxycucurbitadienol peak disappeared, and a new peak was detected by GC-MS (fig. 6C). The new peak was purified by preparative HPLC, and NMR experiments identified the peak as 16β,22-dihydroxy-4,9-cyclo-9,10-secocholesta-5,24-diene (16β,22-dihydroxy-cucurbitadienol, supplementary NMR data, compound 1, Supplementary Material online). When CYP708A15v2 was co-infiltrated with IaCPQ into N. benthamiana leaves, our GC-MS analysis did not indicate a hydroxylation product of cucurbitadienol (fig. 6C). However, in the LC-MS analysis, two small new peaks were observed in the leaves expressing both CYP708A15v2 and IaCPQ. These peaks are likely further oxidized 22-hydroxycucurbitadienol products (supplementary fig. 9, Supplementary Material online).
IaCPQ and CYP708A15v2 Are Localized to the Same Scaffold in the Draft Genome of I. amara
In the Ci. lanatus and Cucumis sativus genomes, the cucurbitacin pathways are organized in gene clusters, where the cucurbitadienol synthase and other genes in the pathway are colocated (Zhou et al. 2016). However, in the C. pepo (zucchini) genome the pathway is organized slightly differently, only one P450 belonging to the CYP81 family, (Cp4.1LG12g10100), based on the homology of Ci. lanatus gene which catalyzes the C2-hydroxylation of 11-carbonyl-20β-hydroxy cucurbitadienol, is colocated with the cucurbitadienol synthase (Cp4.1LG12g10070) (supplementary fig. 10A and C, Supplementary Material online). Thus, we were interested in investigating how the cucurbitacin pathway is organized in the I. amara genome. For this purpose, a draft genome assembly of I. amara was generated, which include 50,001 scaffolds with an average size of 12,535 bp. The estimated genome size is ∼799 MB. The IaCPQ was localized to scaffold 711 together with CYP708A15v2 catalyzing the C22-hydroxylation, however, the two genes were separated by at least 15 genes spanning over 100 kb. Furthermore, CYP708A16 catalyzing the 16β-hydroxylation was localized to scaffold 16365 (supplementary fig. 10C, Supplementary Material online).
Discussion
Iberis amara in Brassicaceae has previously been used as a model species to understand the evolution of monosymmetric petals (Busch et al. 2014), in this paper, we report how I. amara evolved to produce cucurbitacins, a new chemical defense in the Brassicaceae for insect herbivores like Pieris rapae and Phyllotreta nemorum, which otherwise feed on many brassicaceous species (Nielsen et al. 1977; Sachdev-Gupta et al. 1993). We elucidated the first three committed steps of the cucurbitacin biosynthesis in I. amara, and showed that the cucurbitacin pathway in this species has evolved independently to the corresponding pathway in cucurbitaceous species. Thus, cucurbitacin biosynthesis is an example of convergent evolution and represents an evolutionary innovation, where the same substrates are catalyzed by different cytochrome P450 families. This is in contrast to some of the other types of convergent evolutionary events in specialized metabolite biosynthesis, such as geraniol biosynthesis in valerian and rose where unrelated enzymes utilized the same substrate (Dong et al. 2013; Magnard et al. 2015), and methyl anthranilate biosynthesis in maize and grape where different substrates, as well as unrelated enzymes, were recruited (Wang and Luca 2005; Köllner et al. 2010). Interestingly, a somewhat similar situation is observed in momilactone biosynthesis. The two independently evolved momilactone biosynthetic gene clusters from rice (Oryza sativa) and bryophyte Calohypnum plumiforme contain exactly the same enzymatic activities but have been recruited from different P450 families (Mao et al. 2020; Zhang and Peters 2020).
Cucurbitacins are found in 13 plant families that are spread over three major clades of core eudicots: Malvids (Brassicaceae, Sterculiaceae, Thymelaeaceae), Fabids (Cucurbitaceae, Datiscaceae, Begoniaceae, Rosaceae, Elaeocarpaceae), Lamiids (Desfontainiaceae, Scrophulariaceae, Rubiaceae, Primulaceae, Polemoniaceae) (fig. 1A). To determine whether this fragmented distribution is a result of convergent evolution or a divergent ancestral core eudicot pathway that was selectively lost in most plant families, we compared the first committed step of cucurbitacin biosynthesis of I. amara from the Brassicaceae (Malvids clade) and of plant species from the Cucurbitaceae (Fabids clade). The first committed step in cucurbitacin biosynthesis in brassicaceous species as well as in cucurbitaceous species is the cyclization of 2,3-oxidosqualene to cucurbitadienol, catalyzed by cucurbitadienol synthases (fig. 2). Our phylogenetic analysis shows that cucurbitadienol synthase of both I. amara and cucurbitaceous species evolved independently from cycloartenol synthases in sterol metabolism (fig. 3). IaCPQ has a relatively shorter branch length for the branchpoint with IaCAS than cucurbitaceous CPQs has to cucurbitaceous CASs, indicative of that the pathway in cucurbitaceous plants is older than in I. amara (fig. 3). Furthermore, compared with IaCAS, IaCPQ seems to be under a more relaxed purifying selection, with an ω value of 0.3071 for IaCPQ as compared with the IaCAS branch (ω= 0.1426). Several studies have shown that neo- and subfunctionalization of duplicated genes may be facilitated by relaxation of selective constraints (Lynch and Conery 2000; Wertheim et al. 2015), thus, supporting our finding that IaCPQ is derived from the conserved CAS enzymes and accordingly is under relaxed selection pressure. Taken together, our results show that IaCPQ was neofuctionalized from a duplicated I. amara CAS and not inherited from a common ancestor shared with the cucurbitaceous species.
To elucidate how the subsequent steps in the cucurbitacin biosynthetic pathway were recruited in I. amara, we identified the two subsequent P450-catalyzed steps. The first oxidation step, C16-hydroxylation, is catalyzed by CYP708A16 in I. amara (fig. 5). A corresponding C16-hydroxylation has not been reported for other cucurbitacin producing plants, despite most cucurbitacins are hydroxylated at C16 (fig. 1A). To our surprise, CYP708A16, when expressed in tobacco leaves, produces 16β-hydroxycucurbitadienol, despite the general conception in the literature that the C16-hydroxylation configuration in cucurbitaceous plants is α and not β. Accordingly, we isolated cucurbitacins from I. amara seeds and performed NMR experiments, which showed a C16α-hydroxylation. NMR experiments of cucurbitacin E and I standards derived from cucurbitaceous plants confirmed that the C16 position has an α configuration (supplementary NMR data and figs. 16 and 18, Supplementary Material online). Considering that CYP708A16 completely converted cucurbitadienol to 16β-hydroxycucurbitadienol, is an indication that CYP708A16 has cucurbitadienol as in planta substrate and that our data are not an artifact. This raises the question of why we detect 16β-hydroxycucurbitadienol and not 16α-hydroxycucurbitadienol as would be expected from the C16α configuration of the end product cucurbitacin E and I. A theoretical possibility is that when CYP708A16 is expressed in I. amara under native conditions 16α-hydroxycucurbitadienol is produced. A more likely possibility is that additional enzymes in I. amara epimerize the C16-hydroxy moiety (supplementary fig. 11, Supplementary Material online); similar as has been observed in thalianin biosynthesis in A. thaliana (Huang et al. 2019). In that paper, Huang et al. (2019) showed that 3β,7β-thaliandiol, and 3β,7β-dihydroxy-16-keto-thalian-15-yl acetate (T7) are intermediates of thalianin biosynthesis. However, in the root extract, they did not detect compounds with a 3β-hydroxyl group. Instead, they found 3α,7β-dihydroxy-16-keto-thalian-15-yl acetate and a thalianin analog with a 3α-hydroxyl group. In addition, they found two promiscuous oxidoreductases (THAR1 and THAR2) capable of epimerizing of C3 hydroxy moiety of T7. In brief, THAR1 converted the C3β-hydroxyl of T7 into the corresponding C3-ketone, and subsequently THAR2 reduced the C3-ketone into the 3α-alcohol. The same orientation of the 16-hydroxyl moiety of cucurbitacins evolved in two lineages independently suggests that the α-orientation must be important for an in planta or plant defense functions.
An interesting fact of this study is that the two characterized I. amara P450s both belong to the CYP708A subfamily (fig. 5A). CYP708A is a Brassicaceae-specific P450 subfamily (Nelson and Werck-Reichhart 2011), which includes CYP708A2 known to hydroxylate the triterpene thalianol in A. thaliana (Field and Osbourn 2008). This clearly demonstrates that CYP708A15v2 and CYP708A16 are evolutionary unrelated to the corresponding P450 mediated steps in Cucurbitaceae but rather to thalianol biosynthesis (fig. 5). Based on our RNAseq analysis, there are at least 17 CYP708A subfamily members in I. amara, thus in I. amara this subfamily is expanded as compared with the four CYP708As in A. thaliana (fig. 6B and supplementary table 5, Supplementary Material online), six in Brassica rapa, and three in Brassica oleracea (Yu et al. 2017). The evolution and expansion of the CYP708A subfamily in I. amara could reflect the emergence of the cucurbitacin biosynthetic pathway in the Brassicaceae, and thus we speculate that some of the additional CYP708As we identified may be involved in additional oxidative steps in the cucurbitacin biosynthesis or represent putative pseudogenes.
In contrast to I. amara, the first two hydroxylation steps in the cucurbitaceous species are the 11-carbonylation step catalyzed by a CYP87 in Ci. lanatus (Zhou et al. 2016), and the 19-hydroxylation step is catalyzed by a CYP88 in Cucumis sativus (Shang et al. 2014) (supplementary fig. 10C, Supplementary Material online). Although both I. amara and Ci. lanatus produce cucurbitacin E, the biosynthetic routes appear to proceed differently in each lineage. Nevertheless, the brassicaceous CYP708 and cucurbitaceous CYP87 and CYP88 families acting on the cucurbitadienol scaffold have been recruited from the ancient CYP85 clan, which is the P450 clan primarily responsible for sterol metabolism in plants (Nelson and Werck-Reichhart 2011); which lies in agreement with the evolution of cucurbitadienol synthase from a sterol metabolism predisposition in both the Brassicaceae and the Cucurbitaceae. Taken together, our results support that the cucurbitacin biosynthetic genes in I. amara evolved independently and via a different biosynthetic route as in the cucurbitaceous species, but from the same predisposition of sterol biosynthesis.
Gene clusters containing biosynthetic-pathway and/or co-regulated genes are common in prokaryotic genomes. An increasing number of gene clusters have also been found in eukaryotes, particularly in fungal genomes, and more recently also in a number of plant species (Boycheva et al. 2014). Most of such clustered genes are involved in the biosynthesis of specialized metabolites (Boycheva et al. 2014). This is also the case for the cucurbitacin biosynthetic gene cluster in melon, cucumber, and watermelon (Zhou et al. 2016) (supplementary fig. 10, Supplementary Material online). By comparative genomic analysis, we revealed an additional cucurbitaceous species, Lagenaria siceraria (bottle gourd), which diverged from watermelon around 10.4–14.6 Ma (Wu et al. 2017), that has a similar biosynthetic gene cluster (supplementary fig. 10C, Supplementary Material online). Bottle gourd together with melon, cucumber, and watermelon, all descend from a single tribe (Benincaseae) in the Cucurbitaceae (Purseglove 1976; Walters et al. 1991; Ghebretinsae et al. 2007; Sebastian et al. 2010). However, similar gene clusters do not occur in all cucurbitaceous species. In supplementary figure 10A and C, Supplementary Material online, we show that only one of the biosynthetic genes, that is, CYP81, is clustered together with CPQ in C. pepo, C. maxima, and C. moschata (Cucurbiteae tribe). This suggests that the cucurbitacin cluster in the Cucurbitaceace either originated after the split between the Benincaseae and Cucurbitae tribes or was not present in the basal cucurbitaceous ancestor, alternatively, that it was disassembled in the genome of the Cucurbita genus. In brassicaceous species CYP708As have been reported to contribute to triterpenoid scaffold diversification and have been found recurrently to cluster with OSCs (Liu et al. 2020). The case of cucurbitacin biosynthesis in I. amara appears to do not follow the general pattern of triterpenoid diversification in Brassicaceae. De novo assembly of the I. amara genome showed that CYP708A15v2 and IaCPQ are in the same scaffold (150 kb) separated by 100 kb that contains at least 15 additional genes (supplementary fig. 10C, Supplementary Material online). This raises the question of whether the cluster of CYP708As and IaCPQ was disassembled or is in the process of being assembled in I. amara. Interestingly, Zhang and Peters in a commentary paper for the discovery of biosynthetic gene cluster for momilactone formation mentioned that momilactone biosynthetic gene clusters from both the Poaceae and bryophyte are not self-sufficient, meaning that other biosynthetic genes are located outside of the cluster. This fits our discovery for cucurbitacin biosynthetic gene clusters in cucurbitaceous species and might also be the case for the I. amara. Further investigation of cucurbitacin biosynthetic gene organization in genome of other cucurbitaceous tribes besides Benincaseae, Cucurbitae and in I. amara may eventually lead to a better understanding of the mechanism of gene clustering of specialized metabolites such as cucurbitacins.
In this paper, we show that cucurbitacin biosynthesis has evolved independently in two lineages. Interestingly, the pathways are catalyzed by different P450 families and the sequence of the enzymatic steps differs, yet the same products are achieved. This raises the research questions as to whether the convergent evolution of cucurbitacins serves similar ecological and physiological roles in both lineages. Further investigation on this question will provide deeper insight into the convergent evolution of not only cucurbitacins but chemical defense compounds in general in different plant lineages.
Materials and Methods
Metabolite Analysis of Different Organs by LC-MS/MS
For analysis of cucurbitacins in various organ extracts of I. amara and C. pepo, aliquots of 100 mg of frozen, powdered material were extracted with 0.3 ml of 100% MeOH in 1.5-ml Eppendorf vials. After a brief vortex and 10 min incubation at room temperature, the extracts were centrifuged for 10 min at 13,000 × g and filtered through 0.45-µm filters (SRP4, Sartorius, Germany). Liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) was carried out using an Agilent 1100 Series LC (Agilent Technologies, Santa Clara, CA) coupled to a Bruker HCT-Ultra ion trap mass spectrometer (Bruker Daltonics, Billerica, MA). An Agilent Zorbax SB-C18 column (1.8 µm, 2.1 × 50 mm i.d., Agilent Technologies, Santa Clara, CA) maintained at 35 °C was used for separation at a flow rate of 0.2 ml min1, using the following linear elution gradient of mobile phase A (water with 0.1% (v/v) HCOOH) and B (acetonitrile with 0.1% (v/v) HCOOH): 0 min, 30% B; 1.0 min, 30% B, 9.0 min, 98% B; 12.4 min, 98% B. ESI-MS was run in positive mode, and MS and MS2 spectra were acquired in the range 100–1,200 Da.
Extraction of Ci. lanatus var. citroides were performed as described above, and extracts were analyzed by ultrahigh-performance liquid chromatography quadrupole time-of-flight tandem mass spectrometry (UHPLC-ESI-QTOF-MS/MS) using a Dionex UltiMate 3000 RS UHPLC (Thermo Fisher Scientific, Waltham, MA) hyphenated with a Bruker compact QTOF mass spectrometer (Bruker Daltonics, Billerica, CA) operated in positive ionization mode. Samples were separated on a Kinetix XB-C18 column (2.1 × 100 mm i.d., 1.7 µm particle size; Phenomenex, Torrance, CA) using the same mobile phases, MS settings, and injection volumes as previously described (Dong et al. 2018). Data were analyzed using DataAnalysis ver. 4.3 (Bruker Daltonics, Billerica, CA). Authentic standards of cucurbitacins B, D, E, and I were purchased from Extrasynthese (Genay, France) and Merck (Darmstadt, Germany).
Cloning and Sequence Analysis of OSCs
Total RNA was isolated from C. pepo and Ci. lanatus var. citroides 7-day-old seedlings and from I. amara 1-month-old leaves and roots. The C. pepo cucurbitadienol synthase (CpCPQ) was retrieved from GenBank: AB116238 (Shibuya et al. 2004). A putative Ci. lanatus cucurbitadienol synthase (ClCPQ,) was identified from the Cucurbit Genomics Database (http://cucurbitgenomics.org/) using BlastP searches with CpCPQ as a query. OSCs (IaCAS, IaCPQ, IaOSC3, IaOSC4, and IaOSC5) were identified by searching the I. amara Miseq database using 13 published A. thaliana oxidosqualene cyclases (Morlacchi et al. 2009; Xue et al. 2012). Primers with USER compatible sites (https://international.neb.com/applications/cloning-and-synthetic-biology/user-cloning) for cloning full-length OSCs are listed in supplementary table 6, Supplementary Material online. Amplification was done by polymerase chain reaction (PCR) in a Labcycler (SensoQuest GmbH, Göttingen, Germany) using homemade Phusion X7 polymerase. The following program was applied: 98 °C for 1 min; 30 cycles of 98 °C for 20 s, 55 °C for 30 s, 72 °C for 1 min and 30 s; 72 °C for 7 min. Gel purified amplicons were ligated with the USER modified binary vector pEAQ: HT-DEST by USER Enzyme (New England Biolabs, Ipswich, MA) directly, and transformed into E. coli competent cells using the heat shock method. Three clones for each construct were sent for sequencing (http://www.macrogen.com/). Putative OSC sequences were blasted against the NCBI ENTREZ database (NCBI BLAST) and Cucurbit Genomics Database (http://cucurbitgenomics.org/).
A bootstrapped maximum likelihood phylogenetic tree, based on a codon-based multiple alignment of OSC sequences, was generated in MEGA X (Kumar et al. 2018). The optimal model was first selected based on the maximum likelihood method using the model selection function in MEGA X. The evolutionary history was inferred by using the maximum likelihood method and the general time reversible model (Nei and Kumar 2000). The tree with the highest log likelihood (−76,579.33) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter=1.2854]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 14.40% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. About 1,000 bootstrap replicates were carried out. This analysis involved 67 nucleotide sequences. Nonsynonymous to synonymous substitution rate ratios (ω = (dN/dS)) were calculated for codon-based nucleotide alignments with the program codeml from the PAML package, version PAML X1.3.1 (Xu and Yang 2013). The one-ratio model was tested with the settings model M0, and branch models were tested with the free-ratio model (1:b) and two-ratio model (2). Models were tested against each other with likelihood ratio tests (Yang 1998).
Expression of OSCs in S. cerevisiae
For expression in S. cerevisiae, the full-length coding sequences of the I. amara OSCs were amplified with the primers listed in supplementary table 6, Supplementary Material online and Gateway recombined into the donor vector pDONR207. Sequence-verified entry clones were Gateway recombined into the high-copy number yeast destination vector pESC-URA-tHMG1-DEST (Fiallos-Jurado et al. 2016) for expression in the BY4742-derived sterol-engineered yeast strain PA14 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, trp1Δ0, Perg7::PMET3-ERG7) (Calegario et al. 2016). For transformation into strain PA14, the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Gietz and Woods 2002) was used and transformed cells were selected on SD medium supplemented with a dropout mix without uracil (Takara Bio, Mountain View, CA). The resulting yeast cells were cultivated in the presence of MβCD as described (Moses et al. 2014). For GC-MS analysis, 1 ml of the spent medium was extracted thrice with 0.5 volumes of hexane after which the organic extracts were pooled, evaporated to dryness, and trimethylsilylated with 10 µl of pyridine and 50 µl of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Merck, Darmstadt, Germany). GC-MS analysis was carried out using a GC model 6890 and MS model 5973 (Agilent Technologies, Santa Clara, CA) as previously described (Moses et al. 2014).
Purification of Cucurbitadienol from Yeast
Yeast strain PA14 expressing IaCPQ was cultivated for five days in a total volume of 1.6 l in the presence of methyl-β-cyclodextrin (MβCD) as described (Moses et al. 2014) and the resulting yeast culture was extracted thrice with 200 ml of hexane. The obtained organic phases were pooled and evaporated to dryness. Cucurbitadienol was purified from the residue by column chromatography using 20 ml of silica gel as stationary phase and 10% (v/v) ethyl acetate in hexane as mobile phase. Using thin-layer chromatography, fractions containing cucurbitadienol were identified, after which they were pooled and the solvent evaporated. This yielded ∼ 8.5 mg of cucurbitadienol, of which the structure was confirmed by NMR analysis (supplementary data 1, Supplementary Material online).
Transient Expression of OSCs and P450s in N. benthamiana
Agrobacterium tumefaciens infiltration (agro-infiltration) was performed as previously described (Dong et al. 2013). All the OSCs and P450s in the binary vector pEAQ: HT-DEST were individually electroporated into Agrobacterium tumefaciens. Agrobacterium tumefaciens harboring constructs with the OSCs alone or combined with the P450s were infiltrated into leaves of 5-week-old N. benthamiana plants. Five days after agro-infiltration the infiltrated leaves were harvested for metabolites analysis. For isolation of 16β-hydroxycucurbitadienol and 16β,22-dihydroxycucurbitadienol for NMR analysis, 100 leaves on 50 plants were infiltrated for each construct.
Subcellular Localization
Preparation of the plasmids pCAMBIA1300-UeGFP or pCAMBIA1300-UmRFP for in-frame N-and C-terminal fusions of enhanced green fluorescent protein (eGFP) and monomeric red fluorescent protein (mRFP), respectively, were described in (Laursen et al. 2016). Construction of the plasmids for N- and C-terminal eGFP and mRFP fusion with the full-length coding sequences of IaCPQ, CpCPQ, and ClCPQ was done by amplification of the coding sequences with USER cloning primers without stop codon (supplementary table 6, Supplementary Material online) and insertion of the amplicons in pCAMBIA1300-UeGFP or pCAMBIA1300-UmRFP vectors by the single insert USER cloning technique (Geu-Flores et al. 2007). The CYP98A1-mRFP construct was used as a control (Laursen et al. 2016). For in planta transient expression, Confocal Laser Scanning Microscopy (CLSM) was used essentially as previously described (Laursen et al. 2016). Briefly, the cauliflower tobacco mosaic virus 35S promoter-driven genes were introduced into N. benthamiana leaves by agroinfiltration for transient expression. At 4-day postagroinfiltration, leaf discs were excised, mounted in water, and observed by CLSM. Cell imaging was performed using an SP5x laser scanning confocal microscope equipped with a DM6000 microscope (Leica, Wetzlar, Germany). Images were recorded using a 63× water immersion objective lens. Excitation/emission wavelengths were 488/500–550 nm for eGFP and 561/570–650 nm for mRFP constructs. The images were sequentially acquired and processed using the LAS Advanced Fluorescence version 2.7.3.9723 software (Leica, Wetzlar, Germany).
Homology Modeling and Membrane Interactions Analysis
The homology models of OSCs were built using YASARA Structure (Krieger et al. 2009), version 19.12.14. Lanosterol synthase (LAS, PDB ID: 1W6J) (Thoma et al. 2004) was used as a template to build the models. The percentage identity of LAS and CPQ sequences is ∼ 50%. The models with the highest Z score were selected and subjected to membrane interaction analysis using Positioning of Proteins in Membrane server tools (http://opm.phar.umich.edu) (Lomize et al. 2012).
GC-MS Analysis of Triterpenes Produced by Agroinfiltrated N. benthamiana Leaves
Agroinfiltrated N. benthamiana leaves were extracted with ethyl acetate and analyzed using GC–MS. Aliquots of 200 mg of frozen, powdered material were extracted with 1 ml ethyl acetate. Extracts were shortly vortexed, incubated at 37 °C for 1 h, centrifuged for 10 min at 4,000 × g, and 60 µl of supernatant was transferred to a 200 µl glass vial insert and evaporated to dryness using MaxiVac Beta system (Labogene, Alleroed, Denmark) at 40 °C for 1 h. The pellet was derivatized with 40 µl of trimethylsilyl cyanide (TMSCN) (Khakimov et al. 2013). All steps involving sample derivatization and injection were automated using a MultiPurpose Sampler (MPS) (Gerstel, Mülheim an der Ruhr, Germany). After reagent addition, the sample was transferred into the agitator of the MPS and incubated at 40 °C for 40 min with an agitation speed of 750. Immediately after derivatization, 1 µl of the derivatized sample was injected in a splitless mode. The spilt/splitless injector port was operated at 320 °C. The septum purge flow and purge flow to split vent at 2.1 min after injection were set to 3 and 15 ml , respectively. The GC-MS consisted of an Agilent 7890A GC and an Agilent 5975C series MSD (Agilent Technologies, Santa Clara, CA). GC separation was performed on an Agilent HP-5MS column (30 m × 250 µm × 0.25 µm) by using hydrogen carrier gas at a constant flow rate of 1.2 ml . The GC oven temperature program, mass spectra range, and MS detector setting were performed as described (Dong et al. 2018). The GC-MS analysis program differs slightly from the one used for GC-MS analysis for yeast extracts, giving rise to the observed differences in retention times. The mass spectrometer was tuned according to the manufacturer’s recommendation by using perfluorotributylamine (PFTBA). The MPS and GC-MS were controlled using Maestro software (Gerstel, Mülheim an der Ruhr, Germany).
Quantitative Real-Time PCR Analysis
Total RNA was isolated from selected organs from I. amara and Ci. lanatus when they were just fruiting. qPCR assays were performed with SYBR Green supermixes (Bio-Rad, Hercules, CA) on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Two-step amplification conditions were used: 30 s at 95 °C, 40 cycles of 10 s at 95 °C and 30 s at 58 °C. After qPCR, a melt curve was generated by heating the samples from 55 to 95 °C with a 0.5 °C elevated gradient. Primers targeting the transcript of I. amara and Ci. lanatus cucurbitadienol synthase and the housekeeping genes (actin) were designed (supplementary table 6, Supplementary Material online). The method was used to calculate the relative fold change in gene expression. All qPCR experiments were performed with three technical and three biological replicates for I. amara and five biological replicates for Ci. lanatus, and the mean ± SE of biological replications was used for generating the bar chart.
De Novo Transcriptome Assembly
Transcriptome analysis was done by two types of sequencing: Miseq was aiming at obtaining more full-length genes whereas Hiseq was aiming for the generation of large amounts of reads for gene expression analysis. For Miseq, total RNA was isolated from I. amara 4-week-old leaves and roots. For Hiseq, total RNA was isolated from I. amara 8-week-old stems, roots, and petals. The polyadenylated mRNA was purified using poly T oligo attached magnetic beads. For both sequencing technologies, the library preparation is the same and followed the instruction of the standard TruSeq RNA Sample Preparation Kit. The runs were set up as paired-end (2 × 300 bp) sequencing on a MiSeq sequencer and paired-end (2×100 bp) sequencing on a HiSeq 4000 sequencer (Illumina). Sequencing was carried out at Macrogen (South Korea). The raw reads were quality checked by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then the adapters were trimmed off using the Trimmomatic tool (http://www.usadellab.org/cms/?page=trimmomatic). Eventually, trinity (http://sihua.ivyunion.org/trinity.htm) was used for data assembly. RNA-Seq by expectation-maximization (RSEM) method was used for quantifying transcript abundances from RNA-Seq data (http://deweylab.github.io/RSEM/), and fragments per kilobase million for each transcript. For gene annotation, the six-frame conceptual translation products of the nucleotide query sequence (both strands) were compared against the NCBI protein sequence database (UniProt Swiss-Prot) by using the BlastX program. A hierarchical cluster heat map of expression patterns of the cytochrome P450 genes was made with GeneMathsXT 2.12.
De Novo Genome Assembly
High-quality genomic DNA was isolated from 1-month-old I. amara leaves using the DNeasy Plant Maxi Kit (QIAGEN). Nextera mate-pair (8 kb insert) and 20-kb SMRTbell libraries were generated from 8 and 20 µg DNA by following the instruction of Nextera mate-pair Library Construction Kit (Illumina) and SMRTbell Express Template Prep Kit (Pacific Biosciences), respectively. The corresponding libraries were set up to run as paired-end (2 × 150 bp) sequencing on a HiSeq 4000 sequencer or to run on a Pacbio RS sequencing platform at Macrogen (South Korea). De novo assembly was first performed using the data generated from the Pacbio RS sequencing platform by using Platanus assembler (v. 1.2.4) (Kajitani et al. 2014). For trimming, contig generation, scaffolding, and gap closing, well established Platanus modules with default options were used. After assembly from PacBio data, in total 61,701 scaffolds were generated with an N50 of 17,338 bp. About 23,340 gaps were found in the assembled sequences. To close the gaps and generate longer scaffolds, PBJelly (v. 15.8.24) was used for gap closing and re-scaffolding by using mate-pair reads. This step reduced the number of scaffolds to 50,001 and an N50 of 30,211 bp. For running modules of setup, support, extraction, assembly, and output modules of PBJelly, default settings were used. BLASR in the PBJelly package was used to map the PacBio long reads against contigs and the following parameter were used: minMatch 8, minPctIdentity 70, bestn 1, nCandidates 20, maxScore 500, and noSplitSubreads. The locations of the putative genes/proteins in the scaffolds were predicted using the tool Maker (v2.31.8) and their functions were annotated using Protein BLAST+ (v2.4.0) searching against UniProt Swiss-Prot (20150214).
Extraction and Isolation of 16β-Hydroxy-Cucurbitadienol and 16β,22-Dihydroxy-Cucurbitadienol
For isolation of 16β-hydroxy-cucurbitadienol and 16β,22-dihydroxy-cucurbitadienol, 100 agroinfiltrated N. benthamiana leaves co-expressing IaCPQ with CYP708A16 or combined with CYP708A15v2, were separately ground in liquid nitrogen and extracted. In general, 2 g of the ground leaf powder was extracted with 50 ml of ethyl acetate essentially as previously described (Khakimov et al. 2013). Extracts were shortly vortexed, incubated at 37 °C for 1 h, centrifuged for 20 min at 4,000 × g, and all the supernatant was transferred to a new vial and evaporated under vacuum. The pellet was suspended in 4 ml methanol. About 90 µl of the methanol suspensions from both extracts were separated using a Dionex UltiMate 3000 semipreparative high-performance liquid chromatography (Thermo Fisher Scientific, Waltham, MA) equipped with a Supelco C18 column (150 × 4.0 mm i.d., 5 µm particle size), an ultraviolet-visible detector, and an automated fraction collector. The mobile phases were water (A) and acetonitrile (B). The gradient program was: 0–1 min, isocratic 50% B; 1–15 min, linear gradient 50–100% B; 15–25 min isocratic 100% B; 25–26 min with a flow rate of 1.5 ml . Fractions for 16β-hydroxy-cucurbitadienol and 16β, 22-dihydroxy-cucurbitadienol were collected at intervals of 20.95–21.45 min and 17.17–17.55 min, and for both compounds, about 1 mg was collected. Purified 16β-hydroxy cucurbitadienol and 16β,22-dihydroxy cucurbitadienol were reanalyzed using GC-MS as described above to confirm the purity of the compound.
Isolation of Cucurbitacins from I. amara
To isolate cucurbitacins from the Iberis genus, I. amara seeds were used. In short, two grams of I. amara seeds were ground in liquid nitrogen and extracted with 2 ml of methanol by incubating in an ultrasonicator for 15 min at room temperature (24 °C). This was repeated independently twelve times and the extracts were pooled. About 90 µl injections of cucurbitacins E and I were separated using the abovementioned semipreparative HPLC and the following linear elution gradient of water (A) and acetonitrile (B) maintained at 35 °C and with a flow rate of 1.5 ml : 0 min, 10% B, 3.0 min, 10% B, 15.5 min, 50% B, 33 min, 70% B; 42.5 min, 100% B. Fractions for cucurbitacin I and cucurbitacin E were collected at intervals of 16.47–16.62 min and 19.51–19.7 min, respectively, according to the retention times of cucurbitacin I and E reference standards. The purity of fractions was confirmed by LC-MS/MS as mentioned above.
Structure Elucidation of Isolated Compounds
Nuclear Magnetic Resonance (NMR) spectroscopy-based structure elucidation of 16β,22-hydroxy-cucurbitadienol, 16β-hydroxy cucurbitadienol, cucurbitadienol, cucurbitacin E, and I are described in detail in supplementary NMR data, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement 613692-TriForC and the Research Foundation Flanders for a postdoctoral fellowship to J.P. A.A. was supported by the grant from the Independent Research Fund Denmark (Grant No. 7017-00275B). S. B. was supported by The Novo Nordisk Foundation, Distinguished Investigator grant (Grant No. NNF20OC0060298). We are grateful to Dr David Nelson for naming the P450s.
Author Contributions
L.D. designed and performed most of the experiments, analyzed the data and drafted the paper. J.P. and K.M. designed, performed the yeast experiments, and assisted in writing the manuscript. A.A. and J.-E.B. assisted in the design, performance of subcellular localization experiments and writing. B.K. provided the GC-MS analysis, performed NMR analysis and NMR data interpretation, and assisted in writing the manuscripts. M.S.M. assisted in NMR data interpretation. C.E.O. performed LC-MS analysis. D.S. assisted in NMR data interpretation and writing the manuscript. M.R. performed the homology modeling of OSC proteins. A.G. supervised the yeast experiments and helped in editing the manuscript. S.B. supervised the project and assisted in writing the manuscript.
Data Availability
The data underlying this article are available in the article and its Supplementary Material online. The genome and transcriptome data underlying this article are available in European Nucleotide Archive at https://www.ebi.ac.uk/ena/browser/view/PRJEB45578, and can be accessed with accession number PRJEB45578. Genome assembly data and annotation data can be accessed at (https://zenodo.org/record/4943411#.YMoLlUyxVPZ) with doi: 10.5281/zenodo.4943411.
References
- Boycheva S, Daviet L, Wolfender J-L, Fitzpatrick TB.. 2014. The rise of operon-like gene clusters in plants. Trends Plant Sci. 19(7):447–459. [DOI] [PubMed] [Google Scholar]
- Busch A, Horn S, Zachgo S.. 2014. Differential transcriptome analysis reveals insight into monosymmetric corolla development of the crucifer Iberis amara. BMC Plant Biol. 14(1):285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calegario G, Pollier J, Arendt P, de Oliveira LS, Thompson C, Soares AR, Pereira RC, Goossens A, Thompson FL.. 2016. Cloning and functional characterization of cycloartenol synthase from the red seaweed Laurencia dendroidea. PLoS One 11(11):e0165954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Chiu MH, Nie RL, Cordell GA, Qiu SX.. 2005. Cucurbitacins and cucurbitane glycosides: structures and biological activities. Nat Prod Rep. 22(3):386–399. [DOI] [PubMed] [Google Scholar]
- Dong L, Miettinen K, Goedbloed M, Verstappen FW, Voster A, Jongsma MA, Memelink J, van der Krol S, Bouwmeester HJ.. 2013. Characterization of two geraniol synthases from Valeriana officinalis and Lippia dulcis: similar activity but difference in subcellular localization. Metab Eng. 20:198–211. [DOI] [PubMed] [Google Scholar]
- Dong L, Pollier J, Bassard J-E, Ntallas G, Almeida A, Lazaridi E, Khakimov B, Arendt P, de Oliveira LS, Lota F, et al. 2018. Co-expression of squalene epoxidases with triterpene cyclases boosts production of triterpenoids in plants and yeast. Metab Eng. 49:1–12. [DOI] [PubMed] [Google Scholar]
- Fiallos-Jurado J, Pollier J, Moses T, Arendt P, Barriga-Medina N, Morillo E, Arahana V, de Lourdes Torres M, Goossens A, Leon-Reyes A.. 2016. Saponin determination, expression analysis and functional characterization of saponin biosynthetic genes in Chenopodium quinoa leaves. Plant Sci. 250:188–197. [DOI] [PubMed] [Google Scholar]
- Field B, Osbourn AE.. 2008. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science 320(5875):543–547. [DOI] [PubMed] [Google Scholar]
- Fujimoto H, Hagiwara H, SuzukI K, Yamazaki M.. 1987. New toxic metabolites from a mushroom, Hebeloma vinosophyllum. II. isolation and structures of hebevinosides VI, VII, VIII, IX, X, and XI. Chem Pharm Bull (Tokyo). 35(6):2254–2260. [DOI] [PubMed] [Google Scholar]
- Gas-Pascual E, Berna A, Bach TJ, Schaller H.. 2014. Plant oxidosqualene metabolism: cycloartenol synthase–dependent sterol biosynthesis in Nicotiana benthamiana. PLoS One 9(10):e109156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA.. 2007. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 35(7):e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebretinsae AG, Thulin M, Barber JC.. 2007. Relationships of cucumbers and melons unraveled: molecular phylogenetics of Cucumis and related genera (Benincaseae, Cucurbitaceae). Am J Bot. 94(7):1256–1266. [DOI] [PubMed] [Google Scholar]
- Ghosh S. 2017. Triterpene structural diversification by plant cytochrome P450 enzymes. Front Plant Sci. 8:1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA.. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. In: Christine Guthrie GRF, editor. Methods enzymol. Academic Press: Elsevier. p. 87–96. [DOI] [PubMed] [Google Scholar]
- Gry J, Søborg I, Andersson H.. 2006. Cucurbitacins in plant food. Copenhagen (Denmark: ): Nordic Council of Ministers. [Google Scholar]
- Huang AC, Jiang T, Liu Y-X, Bai Y-C, Reed J, Qu B, Goossens A, Nützmann H-W, Bai Y, Osbourn A.. 2019. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364(6440):eaau6389. [DOI] [PubMed] [Google Scholar]
- Itkin M, Davidovich-Rikanati R, Cohen S, Portnoy V, Doron-Faigenboim A, Oren E, Freilich S, Tzuri G, Baranes N, Shen S, et al. 2016. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc Natl Acad Sci U S A. 113(47):E7619–E7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian-Wen T, Ze-Jun D, Zhi-Hui D, Ji-Kai L.. 2002. Lepidolide, a novel seco-ring-A cucurbitane triterpenoid from Russula lepida (Basidiomycetes). Z Naturforsch C J Biosci. 57(11–12):963–965. [DOI] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakimov B, Motawia MS, Bak S, Engelsen SB.. 2013. The use of trimethylsilyl cyanide derivatization for robust and broad-spectrum high-throughput gas chromatography–mass spectrometry based metabolomics. Anal Bioanal Chem. 405(28):9193–9205. [DOI] [PubMed] [Google Scholar]
- Köllner TG, Lenk C, Zhao N, Seidl-Adams I, Gershenzon J, Chen F, Degenhardt J.. 2010. Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using S-adenosyl-L-methionine. Plant Physiol. 110–158360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K.. 2009. Improving physical realism, stereochemistry, and side‐chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77(S9):114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupchan SM, Meshulam H, Sneden AT.. 1978. New cucurbitacins from Phormium tenax and Marah oreganus. Phytochemistry 17(4):767–769. [Google Scholar]
- Laursen T, Borch J, Knudsen C, Bavishi K, Torta F, Martens HJ, Silvestro D, Hatzakis NS, Wenk MR, Dafforn TR, et al. 2016. Characterization of a dynamic metabolon producing the defense compound dhurrin in Sorghum. Science 354(6314):890–893. [DOI] [PubMed] [Google Scholar]
- Liu Z, Suarez Duran HG, Harnvanichvech Y, Stephenson MJ, Schranz ME, Nelson D, Medema MH, Osbourn A.. 2020. Drivers of metabolic diversification: how dynamic genomic neighbourhoods generate new biosynthetic pathways in the Brassicaceae. New Phytol. 227(4):1109–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL.. 2012. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40(Database issue):D370–D376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155. [DOI] [PubMed] [Google Scholar]
- Magnard J-L, Roccia A, Caissard J-C, Vergne P, Sun P, Hecquet R, Dubois A, Hibrand-Saint Oyant L, Jullien F, Nicolè F, et al. 2015. Biosynthesis of monoterpene scent compounds in roses. Science 349(6243):81–83. [DOI] [PubMed] [Google Scholar]
- Mao L, Kawaide H, Higuchi T, Chen M, Miyamoto K, Hirata Y, Kimura H, Miyazaki S, Teruya M, Fujiwara K, et al. 2020. Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants. Proc Natl Acad Sci U S A. 117(22):12472–12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PA, Blackburnsmall M, Schroder RF, Matsuo K, Li BW.. 2002. Stabilization of cucurbitacin E-glycoside, a feeding stimulant for diabroticite beetles, extracted from bitter Hawkesbury watermelon. J Insect Sci. 2: 19. [PMC free article] [PubMed] [Google Scholar]
- Morlacchi P, Wilson WK, Xiong Q, Bhaduri A, Sttivend D, Kolesnikova MD, Matsuda SP.. 2009. Product profile of PEN3: the last unexamined oxidosqualene cyclase in Arabidopsis thaliana. Org Lett. 11(12):2627–2630. [DOI] [PubMed] [Google Scholar]
- Moses T, Pollier J, Almagro L, Buyst D, Van Montagu M, Pedreño MA, Martins JC, Thevelein JM, Goossens A.. 2014. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc Natl Acad Sci U S A. 111(4):1634–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S.. 2000. Molecular evolution and phylogenetics. Oxford, England: Oxford University Press. [Database] [Google Scholar]
- Nelson D, Werck‐Reichhart D.. 2011. A P450‐centric view of plant evolution. Plant J. 66(1):194–211. [DOI] [PubMed] [Google Scholar]
- Nielsen JK, Larsen LM, Søorensen H.. 1977. Cucurbitacin E and I in Iberis amara: feeding inhibitors for Phyllotreta nemorum. Phytochemistry 16(10):1519–1522. [Google Scholar]
- Paquette SM, Jensen K, Bak S.. 2009. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http://www.P450.kvl.dk). Phytochemistry 70(17–18):1940–1947. [DOI] [PubMed] [Google Scholar]
- Purseglove J. 1976. The origins and migrations of crops in tropical Africa. Orig Afr Plant Domest. 291–310. [Google Scholar]
- Raemisch DR, Turpin F.. 1984. Field tests for an adult western corn rootworm aggregation pheromone associated with the phagostimulatory characteristic of bitter Cucurbita spp. J Agric Entomol. 1:339–344. [Google Scholar]
- Sachdev-Gupta K, Radke CD, Renwick JAA.. 1993. Antifeedant activity of cucurbitacins from Iberis amara against larvae of Pieris rapae. Phytochemistry 33(6):1385–1388. [Google Scholar]
- Sebastian P, Schaefer H, Telford IR, Renner SS.. 2010. Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci U S A. 107(32):14269–14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Ma Y, Zhou Y, Zhang H, Duan L, Chen H, Zeng J, Zhou Q, Wang S, Gu W, et al. 2014. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346(6213):1084–1088. [DOI] [PubMed] [Google Scholar]
- Shibuya M, Adachi S, Ebizuka Y.. 2004. Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron 60(33):6995–7003. [Google Scholar]
- Thimmappa R, Geisler K, Louveau T, O’Maille P, Osbourn A.. 2014. Triterpene biosynthesis in plants. Annu Rev Plant Biol. 65(1):225–257. [DOI] [PubMed] [Google Scholar]
- Thoma R, Schulz-Gasch T, D’Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A.. 2004. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 432(7013):118–122. [DOI] [PubMed] [Google Scholar]
- Walters TW, Decker-Walters DS, Posluszny U, Kevan P.. 1991. Determination and interpretation of comigrating allozymes among genera of the Benincaseae (Cucurbitaceae). Syst Bot. 16(1):30–40. [Google Scholar]
- Wang J, Luca VD.. 2005. The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’methylanthranilate. Plant J. 44(4):606–619. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Shamimuzzaman M, Sun H, Salse J, Sui X, Wilder A, Wu Z, Levi A, Xu Y, Ling K-S, et al. 2017. The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring‐spot virus resistance locus. Plant J. 92(5):963–975. [DOI] [PubMed] [Google Scholar]
- Xu B, Yang Z.. 2013. PAMLX: a graphical user interface for PAML. Mol Biol Evol. 30(12):2723–2724. [DOI] [PubMed] [Google Scholar]
- Xue Z, Duan L, Liu D, Guo J, Ge S, Dicks J, ÓMáille P, Osbourn A, Qi X.. 2012. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 193(4):1022–1038. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bi H, Yang W, Li T, Jiang J, Zhang L, Liu J, Hu Q.. 2020. The genome sequence of alpine Megacarpaea delavayi identifies species-specific whole-genome duplication. Front Genet. 11:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 15(5):568–573. [DOI] [PubMed] [Google Scholar]
- Yu J, Tehrim S, Wang L, Dossa K, Zhang X, Ke T, Liao B.. 2017. Evolutionary history and functional divergence of the cytochrome P450 gene superfamily between Arabidopsis thaliana and Brassica species uncover effects of whole genome and tandem duplications. BMC Genomics 18(1):733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peters RJ.. 2020. Why are momilactones always associated with biosynthetic gene clusters in plants? Proc Natl Acad Sci U S A. 117(25):13867–13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ma Y, Zeng J, Duan L, Xue X, Wang H, Lin T, Liu Z, Zeng K, Zhong Y, et al. 2016. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat Plants. 2:16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and its Supplementary Material online. The genome and transcriptome data underlying this article are available in European Nucleotide Archive at https://www.ebi.ac.uk/ena/browser/view/PRJEB45578, and can be accessed with accession number PRJEB45578. Genome assembly data and annotation data can be accessed at (https://zenodo.org/record/4943411#.YMoLlUyxVPZ) with doi: 10.5281/zenodo.4943411.