Abstract
SH3 and OB are the simplest, oldest, and most common protein domains within the translation system. SH3 and OB domains are β-barrels that are structurally similar but are topologically distinct. To transform an OB domain to a SH3 domain, β-strands must be permuted in a multistep and evolutionarily implausible mechanism. Here, we explored relationships between SH3 and OB domains of ribosomal proteins, initiation, and elongation factors using a combined sequence- and structure-based approach. We detect a common core of SH3 and OB domains, as a region of significant structure and sequence similarity. The common core contains four β-strands and a loop, but omits the fifth β-strand, which is variable and is absent from some OB and SH3 domain proteins. The structure of the common core immediately suggests a simple permutation mechanism for interconversion between SH3 and OB domains, which appear to share an ancestor. The OB domain was formed by duplication and adaptation of the SH3 domain core, or vice versa, in a simple and probable transformation. By employing the folding algorithm AlphaFold2, we demonstrated that an ancestral reconstruction of a permuted SH3 sequence folds into an OB structure, and an ancestral reconstruction of a permuted OB sequence folds into a SH3 structure. The tandem SH3 and OB domains in the universal ribosomal protein uL2 share a common ancestor, suggesting that the divergence of these two domains occurred before the last universal common ancestor.
Keywords: diversification of protein domains, ribosome, remote homology, protein structure prediction
Introduction
Proteins commonly perform their specialized functions (binding, catalysis, translocation, or structure) by acquiring specific globular states. A globular protein can contain one or more domains, which are independent units of intramolecular assembly (Chothia and Finkelstein 1990; Chothia and Gerstein 1997; Rose et al. 2006). Domains are also units of evolution; their DNA sequences undergo duplication, deletion, combination with other genes, and adaptation (Bornberg-Bauer et al. 2005). Each domain is characterized by a fold, which is the layout and topology of secondary structural elements (loops, α-helices, and β-strands) (Chothia and Gerstein 1997). From a limited number of folds, evolution has generated a diverse universe of protein function (Chothia and Gerstein 1997; Efimov 1997; Levitt 2009). The hundreds of thousands of known protein structures can be clustered into fewer than a thousand folds (Chothia 1992). Protein structures preserve historical information that can help us understand fold diversification and early steps in protein evolution (Grishin 2001; Lupas et al. 2001; Alva et al. 2015; Nepomnyachiy et al. 2017; Kolodny et al. 2021). Here, we describe the fold evolution of some of the oldest, simplest, and best-preserved proteins in biology.
The deep history of life is recorded within its universal components. The translation system dominates the Universal Gene Set of Life (Harris et al. 2003; Koonin 2003; Charlebois and Doolittle 2004), which is the set of orthologous genes that are universal and are assumed to have been present at or before the last universal common ancestor (LUCA). Among these genes are the components of the ribosome, the oldest RNA/protein assembly in biology (Fox 2010; Lupas and Alva 2017; Bernier et al. 2018). In extant biology, the ribosome catalyzes the translation of all coded proteins from mRNA. In ancestral biology, the ribosome was the nursery of protein folding; the earliest coded proteins were polymerized by the ribosome and for the ribosome.
Within the ribosome, conformations of ribosomal protein (rProtein) segments and their interactions with ribosomal RNA (rRNA) have persisted for nearly 4 Gy (Petrov et al. 2015; Kovacs et al. 2017; Lupas and Alva 2017; Bowman et al. 2020). Intrinsically disordered regions and β-hairpins, deep within the core of the ribosome, are echoes of ancient ancestors of chiral polypeptide (Bowman et al. 2020). Next in the chronology are simple β-domains such as SH3 and OB. More mature rProtein domains show complex combinations of secondary structural elements (Kovacs et al. 2017).
The present study focuses on relationships between the distinct β-barrel folds adopted by SH3 domain and OB domain proteins. Both SH3 and OB domains are partners for nucleic acids and participate in central biological processes including transcription, translation, replication, and maintenance of genome stability, among other functions (Murzin 1993; Achsel et al. 2001; Theobald et al. 2003; Kang et al. 2018).
Here, we resolve the question of common ancestry of SH3 domains and OB domains. Despite high structural similarity, SH3 and OB domains have distinct sequences and β-sheet topologies; β-strands that overlay in the 3D structure have different connectivities in the primary structure. A hypothesis of common ancestry, thus far, is not supported by likelihood or mechanistic precedent for conversion between folds, nor is common ancestry supported at the level of sequence similarity. Conversion of OB topology to SH3 topology requires a multistep noncircular permutation that is complex and improbable (Agrawal and Kishan 2001; Youkharibache et al. 2019). A proposed evolutionary mechanism for converting one of these domains to another involves fragmentation of an ancestral gene at sites that code for just one of the β-strands, 15–30 nucleotides in length, followed by the reinsertion of the fragment at a new position in the same gene (Agrawal and Kishan 2001) (supplementary fig. S1, Supplementary Material online).
SH3 and OB domains are classified as separate architectures and in multiple homology groups in classification systems such as ECOD (Schaeffer et al. 2017), CATH (Sillitoe et al. 2019), and SCOPe (Chandonia et al. 2017) that aim to reflect the interrelatedness between protein domains with similar 3D structures. Thus, current protein classification schemes implicitly assume the independent origins of SH3 and OB domains. However, global similarities in 3D structures of SH3 and OB domains have been interpreted to support a hypothesis of common ancestry (Agrawal and Kishan 2001; Theobald and Wuttke 2005; Youkharibache et al. 2019).
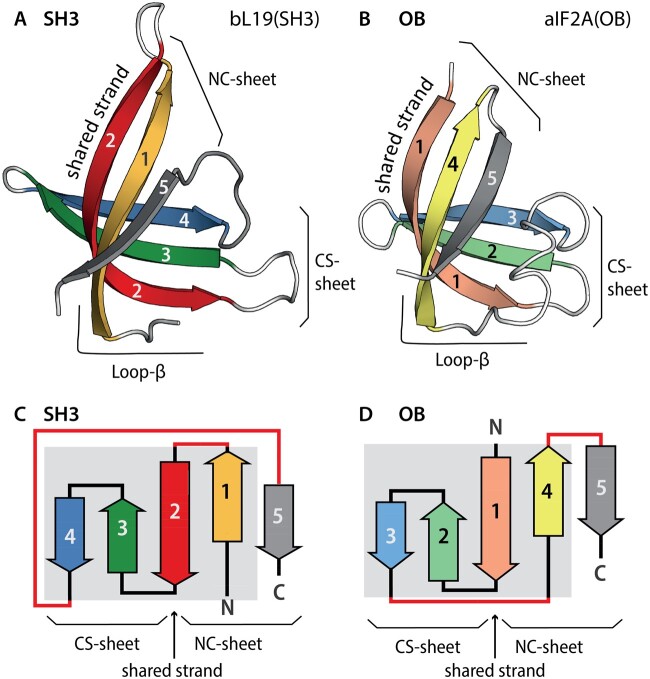
OB and SH3 domains both contain five β-strands (named β1–β5) that form two antiparallel β-sheets (Murzin 1993). One β-sheet contains β-strands that are consecutive in the linear sequence (the consecutive strands sheet, CS-sheet), whereas the other β-sheet contains both the N- and the C-terminal β-strands (the NC-sheet). The CS-sheet is formed by strands β2–β4 in SH3 domains, and by strands β1–β3 in OB domains. In both OB and SH3 domains, the two β-sheets share a β-strand (fig. 1). The shared strand, which links the CS-sheet and the NC-sheet, is β2 in SH3 domains and is β1 in OB domains.
Fig. 1.
The anatomy of SH3 and OB domains. (A) Ribbon and (C) topological representations of the SH3 domain of ribosomal protein bL19 [PDB entry 1VY4, Chain BT]. Strands are colored: β1 yellow; β2 red; β3 green; β4 blue; and β5 gray. (B) Ribbon and (D) topological representations of the OB domain of initiation factor aIF5A [PDB entry 1IZ6, Chain A]. Strands are colored: β1 pink; β2 pale green; β3 light blue; β4 pale yellow; and β5 gray. The shared β-strand, which participates in both β-sheets, is indicated. Differences in strand connectivity are highlighted in red. The common core is indicated by a gray background on the topology diagrams. The consecutive strands sheet (CS-sheet); the N- and the C-terminal β-strands (the NC-sheet).
The goal of our work here is to probe the interrelatedness and ancestral relationship of SH3 and OB domains. We performed analysis of SH3 and OB domains in the context of ancient components of the translation system: rProteins, initiation factors, and elongation factors. We demonstrate that SH3 and OB domains share a common origin, consistent with homologous cores that are related by circular permutation. Using this information, we propose a parsimonious evolutionary scenario that explains the topological differences between these domains. We propose an evolutionary mechanism for interconversion between these folds. In our model, OB domain topology converts to SH3 domain topology, and vice versa, by a conventional single-step circular permutation, not by improbable multistep fragmentation. We demonstrate that a common origin is supported by sequence similarity. Overall, our data suggest that the largest homology groups of SH3 and OB domains originated from a common ancestor and can interconvert.
Results
Anatomies of the SH3 and OB Domains
We based our analysis on structures of SH3 and OB domains within universal, bacteria, or archaea-specific ribosomal proteins, as well as within initiation and elongation factors (table 1). Each of these domain families is highly conserved in sequence and in structure across phylogeny (supplementary figs. S3 and S4, Supplementary Material online). The sequence patterns that we describe for this specific set of domain structures are also present in the ancestral reconstructions of SH3 and OB domains (supplementary fig. S4, Supplementary Material online).
Table 1.
SH3 and OB Domains within Proteins of the Translation System.
| Protein Name | PDB Code: Chain | UniProt Accession | ECOD H-Group Name | Fold Residues | Residues of the Common Corea |
|---|---|---|---|---|---|
| uL2 | 1VY4: BD | P60405 | Nucleic acid-binding proteins (2.1.1.378) | 73–120 | 74–85, 92–110, [111–119] |
| uL2 | 1VY4: BD | P60405 | SH3 (4.1.1.447) | 126–197 | [136–143], 161–169, 172–189 |
| uL24 | 1VY4: BY | Q5SHP9 | SH3 (4.1.1.786) | 1–107 | [7–16], 23–40, 64–71 |
| uS12 | 1VY4: AL | Q5SHN3 | Nucleic acid-binding proteins (2.1.1.370) | 28–100 | 32–40, 54–69, [77–84] |
| uS17 | 1VY4: AQ | P0DOY7 | Nucleic acid-binding proteins (2.1.1.51) | 2–78 | 6–24, 39–46, [51–59] |
| bL19 | 1VY4: BT | P60490 | SH3 (4.1.1.57) | 1–131 | [22–31], 44–53, 59–78 |
| bS1 | 6H4N: y | P0AG67 | Nucleic acid-binding proteins (2.1.1.570) | 276–348 | 280–307, [321–332] |
| aL14 | 4V6U: B5 | Q8U2L5 | SH3 (4.1.1.776) | 3–61 | [4–12], 20–38, 44–50 |
| aL21 | 4V6U: BR | Q8U217 | SH3 (4.1.1.59) | 31–97 | [35–44], 60–92 |
| aS4 | 4V6U: AE | Q8U011 | Nucleic acid-binding proteins (2.1.1.45) | 121–179 | 122–150, [158–167] |
| aS4 | 4V6U: AE | Q8U011 | SH3 (4.1.1.449) | 178–236 | [179–188], 194–204, 212–230 |
| aS28 | 4V6U: AX | Q8U159 | Nucleic acid-binding proteins (2.1.1.20) | 1–71 | 6–17, 25–45, [49–58] |
| bIF1 | 5LMV: W | Q5SHR1 | Nucleic acid-binding proteins (2.1.1.21) | 1–72 | 9–37, [48–57] |
| EF-P | 1UEB: A | Q76G20 | SH3 (4.1.1.27) | 1–63 | [7–15], 17–26, 37–54 |
| EF-P | 1UEB: A | Q76G20 | Nucleic acid-binding proteins (2.1.1.19) | 64–126 | 64–98, [103–115] |
| EF-P | 1UEB: A | Q76G20 | Nucleic acid-binding proteins (2.1.1.22) | 127–184 | 127–136, 149–161, [165–173] |
| aIF1 | 4MNO: A | Q9V138 | Nucleic acid-binding proteins (2.1.1.20) | 25–90 | 25–54, [63–72] |
| aIF2A | 1YZ6: A | Q9V0E4 | Nucleic acid-binding proteins (2.1.1.80) | 9–83 | 13–45, [57–66] |
| aIF5A | 1IZ6: A | O50089 | e1iz6A2 (4.1.1.111) | 2–70 | 11–20, 23–32, [43–59] |
| aIF5A | 1IZ6: A | O50089 | Nucleic acid-binding proteins (2.1.1.21) | 71–135 | 73–104, [113–122] |
The residues of the Lβ element are given in brackets.
Here, domain families are specified by the protein name followed by the domain type in parentheses, as in uL24(SH3) or uS17(OB). Multiple domains of a single type within the same protein are differentiated by superscripts as in bEF-P(OB1) and bEF-P(OB2). Lowercase letters u, b, and a denote universal, bacterial, and archaeal proteins, respectively.
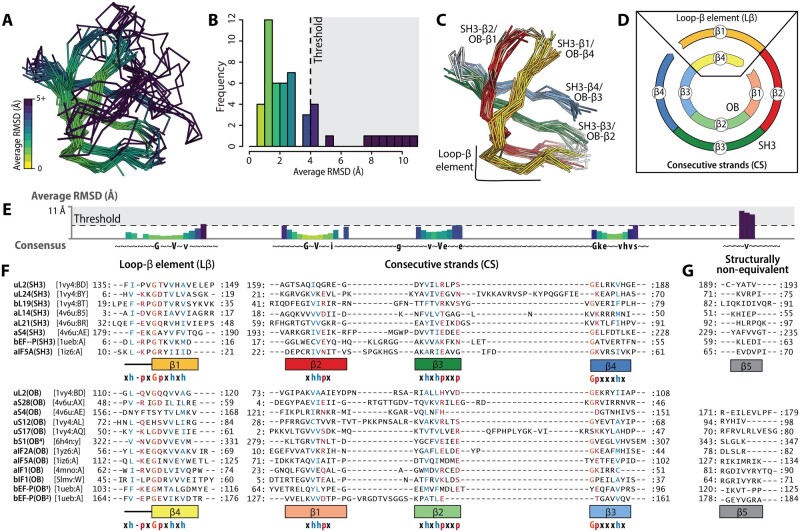
A superimposition of all SH3 and OB domains here shows that for four β-strands, at least three-quarters of amino acid residues of both domain families occupy structurally equivalent positions, with average root mean square deviation (RMSDs) below 4 Å (fig. 2A, B, and E). Common structural elements between SH3 and OB domains (fig. 1) include: 1) the shared strand, 2) the entire CS-sheet, 3) the central strand of the NC-sheet, and 4) a short loop of the NC-sheet. We refer to elements 1 through 4 as the SH3/OB common core and elements 3 and 4 as loop β (Lβ). Lβ in SH3 domains includes strand β1 and its N-terminal loop, and in OB domains includes strand β4 and its N-terminal loop. SH3 elements of the common core are called the SH3 core and OB components of the common core are called the OB core. Strand β5, which displays significantly greater structure variability between and within SH3 and OB domain proteins, is excluded from the SH3/OB common core.
Fig. 2.
The SH3/OB common core. (A) Multiple-structure superimposition of SH3 and OB domains of translation-related proteins colored by the average RMSD between equivalent Cα atoms. (B) Distribution of the average RMSD between equivalent Cα atoms in the multiple-structure superimposition. (C) Multiple-structure superimposition of SH3 and OB domain structures trimmed to the common core. (D) Schematic representation of the circular permutation relationship between the SH3 core and the OB core. The colors in panels (C) and (D) are the same as in figure 1. (E) Average RMSD between equivalent Cα atoms, positions are represented by the HMM consensus sequence. (F) Structure-derived multiple sequence alignment constructed from the segments of SH3 and OB domains that constitute the common core. Columns in which more than 70% of residues have the same physicochemical property are colored: glycine (orange), hydrophobic (blue) and polar (red). (G) Structure-derived multiple sequence alignment for residues in the β5 strands of SH3 and of OB domains, calculated using a maximum average RMSD threshold of 12 Å.
Strand β5 is excluded from the common core because it is structurally variable and nonequivalent across SH3 and OB domain proteins. The average RMSD between strand β5 Cα atoms of SH3 and OB domains is greater than 4 Å (fig. 2B and E). We detected several types of variations of β5 (supplementary fig. S2, Supplementary Material online). First, OB domains of both uL2(OB) and aS28(OB) lack strand β5 altogether, such that these domains have a four-stranded β-barrel that is equivalent to the OB core. Second, in aIF2A(OB) and bS1(OB4), β5 is significantly displaced from its canonical position within the NC-sheet due to the elongation of β4. Third, the edge of the NC-sheet is distorted in bEF-P(SH3), uL2(SH3), and aIF5A(SH3) due to the displacement of β5 by an additional N-terminal β-strand.
Other regions that are variable between SH3 and OB domains were also excluded from the common core. These variable regions include loop insertions and β-strand elongations, which in general contribute only small perturbations to the cores (supplementary fig. S2, Supplementary Material online). The only notable exception is uL24(SH3), for which an insertion (residues 43–64 in 1VY4: BY) distorts the CS-sheet. All other structural variations involve differences in strand conformation of β5 and affect the NC-sheet.
Unlike the transformations between the five-stranded SH3 and OB domains, the SH3 core can be converted to the OB core (and vice versa) by well-established mechanisms. The SH3 core is topologically related to the OB core by a simple circular permutation. The exclusion of strand β5 from the common core is the key to enabling this interconversion. In the SH3 core, the Lβ element is located at the N-terminus of the CS-sheet (Lβ–CS-sheet topology), whereas in the OB core, the Lβ element is located at the C-terminus of the CS-sheet (CS-sheet–Lβ topology) (fig. 2C and D). The sequential order of these elements can be represented in two ways, depending on which topology (SH3 or OB) is used as the linear reference. Throughout this work, we used the SH3 core (Lβ–CS-sheet) as the reference topology, and permuted the OB core (fig. 2F).
Structure Comparison Points to Sequence Similarity of the Common Core
We produced a multiple sequence alignment (MSA) that successfully combines SH3 and OB domain sequences. The superimposed structures of the SH3 and OB domains were used as a guide to produce a MSA of the common core (fig. 2F) in which MSA columns correspond to amino acid residues with equivalent Cα atoms in the structural superimposition. Integration of 3D information indicated that the sequence of either the OB core or the SH3 core must be circularly permuted (fig. 2C and D) to produce a composite MSA of both domain types. To accomplish the circular permutation, residues in the Lβ element of SH3 domains (N-terminus of the SH3 core) were aligned to residues in the Lβ element of OB domains (C-terminus of the OB core), and residues in the CS-sheets of SH3 domains (strands β2–β4) were aligned to residues in the CS-sheets of OB domains (strands β1–β3). The resulting MSA allows the integration of both structural and sequence information and contains the sequences in the topological order of SH3 domains (Lβ–CS-sheet).
Within the common core, superimposed SH3 and OB domains exhibit common 3D localization of amino acids with similar physicochemical properties (fig. 2F). In the structure-derived MSA, with circularly permuted OB domain sequences, the first four blocks of aligned sequences correspond to the four β-strands of the common core. Each block contains columns in which most of the residues possess the same physicochemical property (h, hydrophobic; p, polar; or G, glycine).
The first block has a characteristic pattern of 11-amino acid residues (h–x–h–x2–G–p–x–h–x–h) identified previously (Alva et al. 2009) as the GD-box motif (fig. 2F). The GD-box motif, which can be found either in homologous or in analogous protein structures (Alva et al. 2009), was previously described in OB domains but not in SH3 domains. The second block corresponds to the shared β-strand between the NC-sheets and the CS-sheets. This block exhibits a short pattern (h–h–p) located at the β-bulge of the shared strand, where it switches its connectivity from the NC-sheet to the CS-sheet. The third and fourth blocks of the composite MSA also exhibit a few positions at which the physicochemical properties of amino acids are preserved (fig. 2F).
The combined MSA reveals that strand β5 from the SH3 domain does not align with strand β5 from OB domains. Strand β5 of SH3 domains does not share sequence similarity with strand β5 of OB domains. To generate the MSA of strands β5, the average RMSD threshold was increased up to 12 Å per position (fig. 2E).
Unveiling the Hidden Sequence Similarity within the SH3/OB Core
By performing alignment comparisons, we assessed the statistical significance of circular permutation on sequence similarity within the SH3/OB common core. The similarity between each possible pairing of SH3 and OB domains was probed with HHalign (Steinegger et al. 2019), which incorporates sensitive methods for homology detection. We constructed a MSA of orthologous sequences from archaeal and bacterial species for every domain family in table 1. These MSAs were trimmed to the common core (fig. 2 and table 1). Three types of comparisons were performed. First, MSAs of native OB sequences were compared with MSAs of native SH3 sequences. Second, MSAs of native OB sequences were compared with MSAs of circularly permuted SH3 sequences, with CS-sheet–Lβ arrangements (equivalent to OB topology). Third, MSAs of native SH3 sequences were compared with MSAs of circularly permuted OB sequences with an Lβ–CS-sheet arrangement (equivalent to the SH3 topology). The MSAs were compared by computing the HHalign probability of homology. Additional information about the aligned regions of high-sequence similarity is given in supplementary table S1, Supplementary Material online.
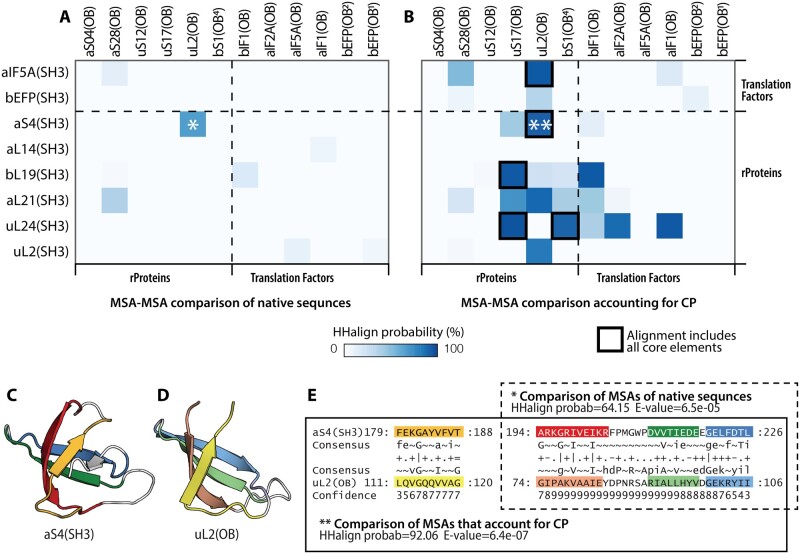
Comparisons of MSAs of native (nonpermuted) sequences yielded little or no sequence similarity between SH3 and OB domains (fig. 3A). The only significant sequence similarity (HHalign probability of 64%) was between aS4(SH3) and uL2(OB). The sequence similarity between these two domains spans all three strands of the CS-sheet (fig. 3).
Fig. 3.
Comparisons of alignments of native sequence and circularly permuted SH3 and OB domains. Heatmap plots showing the HHalign probability values of profile–profile comparisons of the common core of (A) MSAs in native sequences, and (B) MSAs after correction for circular permutation. Ribbon representation of (C) aS4(SH3), and (D) uL2(OB). (E) Comparison of MSAs of aS4(SH3) and uL2(OB) in the native sequence (*) and after correction for circular permutation (**). The colors in panels (C), (D), and (E) are the same as in figure 1. CP, circular permutation.
However, by comparing pairs of alignments with equivalent sequence arrangements (i.e., by correcting for circular permutation), we uncovered 11 SH3/OB pairs with significant sequence similarity (HHalign probability above 50%). In six of these pairs, the sequence similarity was detected across the entire length of the core elements. These six pairs are aS4(SH3) and uL2(OB): 91%; aIF5A(SH3) and uL2(OB): 84%; uL2(SH3) and uL2(OB): 82%; uL24(SH3) and bS1(OB): 90%; uL24(SH3) and uS17(OB): 94%; and aL21(SH3) and uS17(OB): 70% (fig. 3 and supplementary table S1, Supplementary Material online). We note that three of these pairs involve uL2(OB), which is a four-stranded β-barrel. The structure of uL2(OB) (β1, β2, β3, and β4) is essentially equivalent to the OB core. Thus, the full-length uL2(OB) domain (CS-sheet–Lβ) can be transformed to the SH3 core topology (Lβ–CS-sheet) by a simple circular permutation. These data are consistent with a simple mechanism by which the topologies of these two domains may be related.
Discussion
SH3 and OB are ancient protein domains with a wide range of binding specificities that include oligosaccharide, RNA, DNA, peptide, and protein (Youkharibache et al. 2019). The high similarity in 3D structure of SH3 and OB domains has previously suggested common evolutionary origin (Agrawal and Kishan 2001; Theobald and Wuttke 2005; Youkharibache et al. 2019). These domains are structurally very similar (fig. 1); however, direct comparison of SH3 and OB domains has led to the suggestion that the sequential order of their secondary structural elements cannot be interconverted by a single topological transformation (i.e., by a circular permutation, see supplementary fig. S1, Supplementary Material online) (Agrawal and Kishan 2001; Youkharibache et al. 2019). The complex mechanism required for interconversion appeared to invalidate the hypothesis of common evolutionary origin. Furthermore, previous sequence-based analyses failed to detect sequence similarity between SH3 and OB domains.
A Simple Evolutionary Relationship
In the present study, we show that β-strands that overlay in the 3D structure display high-sequence similarity, suggesting a homologous relationship between SH3 and OB domains. Yet, paradoxically, interconversion between five-stranded SH3 and OB domains requires a complex multistep rearrangement of the strands. These complex permutations are evolutionary unlikely. To solve this paradox, we hypothesize that SH3 and OB domains emerged from a common ancestral gene that encoded for a four-stranded precursor of the modern SH3 and OB domains.
The topological differences between canonical SH3 and OB domains can be fully resolved by a model that: 1) excludes strand β5 from the common core, 2) incorporates structural and sequence similarity between the elements of the common core, and 3) explains the transformation between the folds at the gene level. We support our proposal by predicting the structure of the circular permutation of an ancestral reconstruction of the SH3 core of a universal ribosomal protein that results in a five-stranded OB topology, and vice versa.
The Common Core
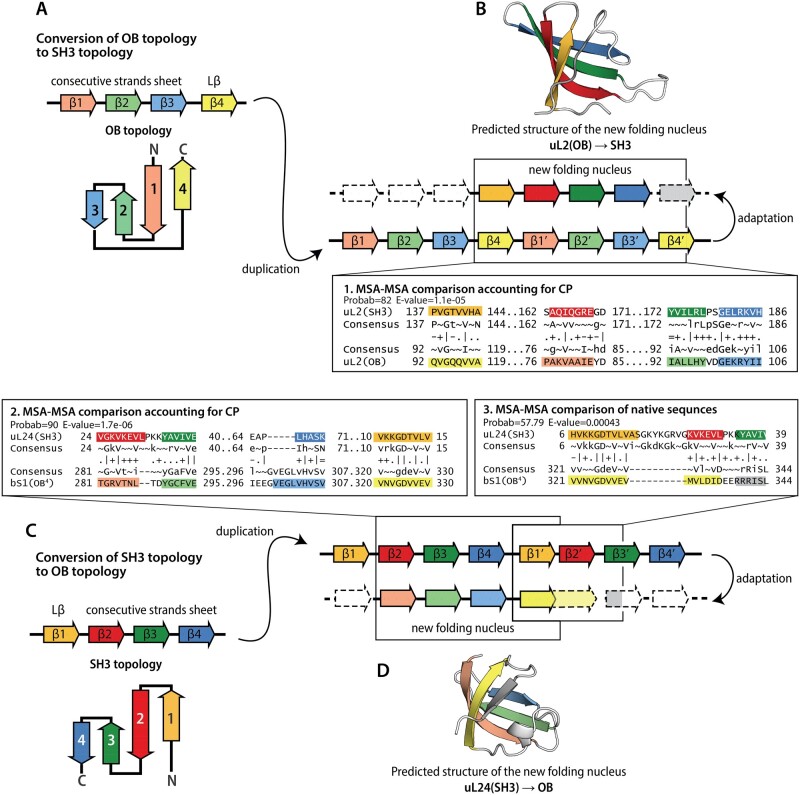
Most translation-related SH3 and OB domains contain five strands. Our structure and sequence analyses revealed a shared common core that includes only the four strands of SH3 and OB domains and excludes β5. We showed that β5 exhibits a far more idiosyncratic 3D structure than the other strands in the folds and is absent from some proteins. Within the common core, the structure of the OB domain forms a Greek-key motif (Lβ–CS-sheet) (fig. 4), whereas SH3 domains form an up and down antiparallel β-sheet (CS-sheet–Lβ) (fig. 4). These two distinct topologies of the common core can be interconverted by a circular permutation. The ancestor of SH3 and OB domains may have been similar to extant uL2(OB) and aS28(OB) domains, which contain only the four strands of the common core, and lack β5.
Fig. 4.
Probable mechanisms of circular permutation between SH3 and OB domains. (A) Schematic representation of the proposed mechanism of the circular permutation of an OB core topology to a SH3 topology. Inset 1: Comparison of the core of SH3 and OB domains in uL2 using the MSA of uL2(SH3) core in native order and the MSA of circularly permuted uL2(OB). (B) Predicted structure of the circular permutation of the core of uL24(SH3). (C) Schematic representation of the proposed mechanism for the circular permutation of a SH3 domain topology that results in an OB domain topology. (D) Predicted structure of the circular permutation of the core of uL2(OB). Comparison between uL24(SH3) and bS1(OB4) using MSAs that account for a circular permutation. Inset 3: Comparison between uL24(SH3) and bS1(OB4) using MSAs in native order. Dotted lines indicate elements that are lost or highly modified.
Domain Duplication
Domain duplication and fusion at the level of genes result in two notable outcomes: 1) tandem repetition of domains, or 2) circular permutation (Grishin 2001; Schmidt-Goenner et al. 2010). There is evidence of duplication and fusion of SH3 and OB domains in early proteins of the translation system. As previously described, OB domains appear in tandem in two translation-related proteins: bS1 and bEF-P. Moreover, tandem arrangements of SH3 and OB domains exist in four translation-related proteins: uL2, aS4, eIF5A, and bEF-P.
Universal ribosomal protein uL2 is a remarkable documentation of early evolutionary history; it contains both a SH3 domain and an OB domain (uL2(SH3) and uL2(OB)) in tandem. Universal ribosomal proteins characteristically have blocks of high-sequence conservation (Vishwanath et al. 2004; Fox 2010) that have been preserved over nearly 3.8 Gy of evolution. The domains in uL2 are highly similar in structure (RMSD of the superimposition: 1.5 Å) and in sequence (HHalign probability: 82%). Altogether, our data point to a shared evolutionary history of the two domains uL2(SH3) and uL2(OB) and suggest that they emerged from a single common ancestor. Because uL2 is arguably one of the oldest ribosomal proteins (Fox 2010), the presence of both domains in its structure also suggests that the divergence between SH3 and OB domains occurred before LUCA and before maturation of the ribosome.
Circular Permutation
The common origin of SH3 and OB domains from a four-stranded ancestor can be explained by both scenarios depicted in figure 4. The two scenarios initiate with different topologies of the ancestral four-stranded unit. One scenario (fig. 4A) initiates with SH3 domain topology (Lβ–CS-sheet); the other scenario (fig. 4C) initiates with OB domain topology (CS-sheet–Lβ). We cannot currently exclude either scenario. In both scenarios an ancestral gene is duplicated to form a tandem repeat in which the C-terminus of the first unit and the N-terminus of the second unit are linked. The close proximity of the N- and C-termini within the tandem repeat (Lβ–CS-sheet–Lβ′–CS-sheet′ or CS-sheet–Lβ–CS-sheet′–Lβ′) appears to favor the emergence of a new folding nucleus (Xia et al. 2016) for a β-barrel fold that differs in topology from the ancestral unit. Once the new folding nucleus is established, adaptation and loss of the terminal structural elements of the tandem repeat (Grishin 2001; Weiner and Bornberg-Bauer 2006; Schmidt-Goenner et al. 2010) would lead to the emergence of a circularly permutated topology (CS-sheet–Lβ′ or Lβ–CS-sheet′).
Adaptations of the Termini
The proposed duplication schemas (fig. 4B) suggest the strand β5 may have emerged as an adaptation of the C-terminus of the tandem repeat. By comparing the MSAs of the full-length domains in the topological sequence order, we identified two instances (uL24(SH3)/bS1(OB) and uL24(SH3)/aIF2a(OB)), in which strand β5 of the OB domain is similar in sequence to strands β2–β3 of the SH3 domain, with HH-align probabilities above 50% (fig. 4 and supplementary table S1, Supplementary Material online). This pattern of sequence similarity fully supports the proposed duplication/adaptation hypothesis. Alternatively, in some SH3 and OB domains, strand β5 may have evolved independently by convergent evolution, thus representing a decoration of the core fold.
Folding of Ancestral Reconstructions
To further test the hypothesis that new folding nuclei could form from duplicated ancestral genes, we predicted structures of the circular permutation of the reconstructed ancestral sequence of the uL2(OB) core, and of the circular permutation of the reconstructed ancestral sequence of the uL24(SH3) core (fig. 4B and D). To generate these circular permutations, we duplicated the sequences of the cores and trimmed the termini of the tandem repeats following the schemas in figure 4A and C. De novo structure prediction using AlphaFold2 (Jumper et al. 2021) of the circular permutation of uL2(OB) produced a four-stranded barrel that is very similar to the SH3 core (supplementary data set 1 and fig. S3, Supplementary Material online). The circular permutation of the uL24(SH3) core produced a five-stranded structure that contains the OB core, with a fifth strand that forms from the duplicated sequence (supplementary data set 2 and fig. S3, Supplementary Material online). Thus, the final folded conformation is not an exact circular permutation but a new fold. We believe that our work here is the first application of AlphaFold2 to recapitulate the emergence of a new fold from an ancestral unit.
Excavating the Most Ancient Records
The translation system has preserved records of the ancient history of protein folding and fold diversification, prior to LUCA. Fold change occurred by circular permutation of bacterial and archaeal versions of ribosomal protein uL33 (Kovacs et al. 2018). The current study provides support for the hypothesis of a common evolutionary origin of SH3 and OB domains, two of the most ancient protein folds. Comparative methods support a mechanism of divergence via gene duplication and adaptation. This process caused circular permutation of core elements and differentiation of noncore elements. The mechanism is independently corroborated here by modeling of permuted ancestral reconstructions using AlphaFold2. The duplication and adaptation mechanism is supported by a strong signal from the SH3/OB common core, and from β5, a noncore element.
Proteins of the translation system consist of a small number of folds. Is this repetitiveness of the fold repertoire an indication of convergent evolution of the earliest protein folds? Our results indicate that the structures of SH3 and OB domains evolved by divergent rather than convergent evolution. We explored deep time relationships between two of the simplest, oldest, and most common beta-barrel folds within proteins of the translation system and identified the minimal patterns, or regularities, within their structures and sequences. The coexistence of the SH3 and OB domains within some of the oldest multidomain proteins of the translation system pointed to a possible mechanism: gene duplication. Our results show that a combination of evolutionary studies with detailed structure and sequence analyses of the oldest known molecular fossils reveals general mechanisms of diversification of folds at the dawn of life.
Materials and Methods
Our analysis includes 11 ribosomal proteins (rProteins) and four translational GTPases that have SH3 and/or OB domains. Orthologous sequences of proteins of the translation system from a Sparse and Efficient Representation of Extant Biology (SEREB) within Bacteria and Archaea (Bernier et al. 2018; Penev et al. 2021) were retrieved from UniProt (UniProt Consortium 2019) and NCBI RefSeq (O’Leary et al. 2016) databases. This set includes ancient and well-characterized orthologous proteins (table 1). Within the set of rProteins and translation-related GTPases analyzed here, SH3 domains belong to the SH3 ECOD H-level, and OB domains belong to the “Nucleic acid-binding proteins” ECOD H-level (Cheng et al. 2014).
Domain Definitions
Coordinate files for rProteins and translation-related factors (bacterial elongation factor P, initiation factors bIF1, aIF1, eIF2a, and eIF5a) were retrieved from the Protein Data Bank (PDB) (Berman et al. 2000). Individual coordinate files were produced for each SH3- or OB domain using the domain-boundary definitions in ECOD (table 1). ECOD is the only hierarchical classification system in which the homology level explicitly allows for variations of the linear connectivity (topology) between elements of secondary structure (Cheng et al. 2014). This means that, in ECOD, sequence rearrangements, such as those produced by circular permutation and by large insertions/deletions, are explicitly taken into consideration in determining homology.
Multiple Sequence Alignment of Orthologous Sequences
The full-length protein sequences were aligned with MAFFT L-INS-I (Katoh and Standley 2013). The resulting MSAs were manually curated based on structural superimposition. The alignments of rProteins are available in ProteoVision, an in house web server for visualization and data mapping of proteins (Penev et al. 2021). The final manually curated MSAs of orthologous proteins were then trimmed to match the domain definitions in ECOD (Schaeffer et al. 2017). The domain boundaries of rProteins were further adjusted to match the Phase definitions in the Accretion Model schema (Kovacs et al. 2017) (table 1). Structural visualizations of protein domains were performed by PyMOL (Schrödinger LLC 2021) and ProteoVision (Penev et al. 2021).
Conserved Core Definition
For the definitions of the conserved core, we first calculated a separate multiple-structure superimpositions for SH3 domains and for OB domains. For these superimpositions, we used MATRAS (Kawabata 2003), which outputs the superimposed coordinates files as well as a structure-derived MSA. The structures of uL24(SH3) and aS4(SH3) failed to be superimposed by MATRAS and were manually adjusted.
Then, SH3/OB pairs were structurally superimposed using CLICK (Nguyen and Madhusudhan 2011) in an all-versus-all fashion. To generate a combined MSA including SH3 sequences and OB sequences, the columns of the structure-derived MSAs of SH3 domains and of OB domains were merged using the output of the CLICK program. To find equivalent columns, we used the list of matched residue pairs produced by CLICK.
Finally, we used the columns of the combined MSA as well as the coordinate files of the superimposed structures to calculate the average RMSD between all pairs of Cα in the same column. We used the distribution of the average RMSD per position in the combined MSA to define the threshold.
Pairwise MSA Comparisons
Three distinct sets of MSAs were produced: Set 1, MSAs matching the full-length domain definitions; Set 2, MSAs that were trimmed to contain only residues within the conserved core definition; and Set 3, circularly permuted MSAs. The sets of alignments that were compared together were: SH3 domains in Set 1 versus OB domains in Set 1; SH3 domains in Set 2 versus OB domains in Set 3; and SH3 domains in set 3 versus OB domains in Set 2. The pairs of MSAs were compared with HHalign with default parameters, only adjusting one parameter to filter columns with more than 60% of gaps (Söding 2005). HHalign produces profiles from the input MSAs. Sequence profiles are position-specific representations of the probability of occurrence of each amino acid. The heatmap of HHalign probabilities includes only the scores of alignments over 25 columns. All the pairs that yielded HHalign probabilities > 50% are summarized in supplementary table S1, Supplementary Material online, including those with less than 25 columns aligned.
Ancestral Sequence and Structure Reconstructions
Ancestral sequence reconstructions were calculated with ANCESCON (Cai et al. 2004) from MSAs matching the full-length domain definitions. Ancestral sequences were reconstructed only for the root (midpoint of the tree) using maximum likelihood rate factor and an alignment-based equilibrium (background) amino acid frequency vector.
To construct the circularly permuted sequences for de novo structure prediction, the ancestral reconstruction of uL24(SH3) was trimmed to the SH3 core and the ancestral reconstruction of uL2(OB) was trimmed to the OB core. These sequences were duplicated in tandem, and their N- and C-terminal ends were removed. De novo structure prediction with single-sequence and no-template was done on the AlphaFold2 Google Colab notebook by Sergey Ovchinnikov, Milot Mirdita, and Martin Steinegger (Ovchinnikov et al. 2021). The sequence constructs for de novo structure prediction are shown in supplementary figure S4, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was funded by the National Aeronautics and Space Administration (Grant No. 80NSSC18K1139) awarded to L.D.W. and A.S.P. C.A.-C. was supported by the NASA Postdoctoral Program, administered by Universities Space Research Association under contract with NASA. The authors thank Dr. Eric Smith for insightful discussions.
Data Availability
The alignments of rProteins are available in ProteoVision at https://proteovision.chemistry.gatech.edu (last accessed August 23, 2021), an in house web server for visualization and data mapping of proteins. All other relevant data are available within this article, the Supplementary Material online, or from the corresponding authors upon request.
References
- Achsel T, Stark H, Lührmann R.. 2001. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc Natl Acad Sci U S A. 98(7):3685–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal V, Kishan RK.. 2001. Functional evolution of two subtly different (similar) folds. BMC Struct Biol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva V, Dunin-Horkawicz S, Habeck M, Coles M, Lupas AN.. 2009. The GD box: a widespread noncontiguous supersecondary structural element. Protein Sci. 18(9):1961–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva V, Söding J, Lupas AN.. 2015. A vocabulary of ancient peptides at the origin of folded proteins. eLife 4:e09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE.. 2000. The Protein Data Bank. Nucleic Acids Res. 28(1):235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier CR, Petrov AS, Kovacs NA, Penev PI, Williams LD.. 2018. Translation: the universal structural core of life. Mol Biol Evol. 35(8):2065–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer E, Beaussart F, Kummerfeld SK, Teichmann SA, Weiner J 3rd. 2005. The evolution of domain arrangements in proteins and interaction networks. Cell Mol Life Sci. 62(4):435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JC, Petrov AS, Frenkel-Pinter M, Penev PI, Williams LD.. 2020. Root of the tree: the significance, evolution, and origins of the ribosome. Chem Rev. 120(11):4848–4878. [DOI] [PubMed] [Google Scholar]
- Cai W, Pei J, Grishin NV.. 2004. Reconstruction of ancestral protein sequences and its applications. BMC Evol Biol. 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia JM, Fox NK, Brenner SE.. 2017. SCOPe: manual curation and artifact removal in the structural classification of proteins – extended database. J Mol Biol. 429(3):348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois RL, Doolittle WF.. 2004. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 14(12):2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Schaeffer RD, Liao Y, Kinch LN, Pei J, Shi S, Kim BH, Grishin NV.. 2014. ECOD: an evolutionary classification of protein domains. PLoS Comput Biol. 10(12):e1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. 1992. Proteins. One thousand families for the molecular biologist. Nature 357(6379):543–544. [DOI] [PubMed] [Google Scholar]
- Chothia C, Finkelstein AV.. 1990. The classification and origins of protein folding patterns. Annu Rev Biochem. 59:1007–1039. [DOI] [PubMed] [Google Scholar]
- Chothia C, Gerstein M.. 1997. Protein evolution. How far can sequences diverge? Nature 385(6617):579–581. [DOI] [PubMed] [Google Scholar]
- Efimov AV. 1997. Structural trees for protein superfamilies. Proteins 28(2):241–260. [DOI] [PubMed] [Google Scholar]
- Fox GE. 2010. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2(9):a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV. 2001. Fold change in evolution of protein structures. J Struct Biol. 134(2–3):167–185. [DOI] [PubMed] [Google Scholar]
- Harris JK, Kelley ST, Spiegelman GB, Pace NR.. 2003. The genetic core of the universal ancestor. Genome Res. 13(3):407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with alphafold. Nature 596(7873):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, Landick R, Darst SA.. 2018. Structural basis for transcript elongation control by Nusg family universal regulators. Cell 173(7):1650–1662.e1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T. 2003. Matras: a program for protein 3D structure comparison. Nucleic Acids Res. 31(13):3367–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodny R, Nepomnyachiy S, Tawfik DS, Ben-Tal N.. 2021. Bridging themes: short protein segments found in different architectures. Mol Biol Evol. 38(6):2191–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 1(2):127–136. [DOI] [PubMed] [Google Scholar]
- Kovacs NA, Penev PI, Venapally A, Petrov AS, Williams LD.. 2018. Circular permutation obscures universality of a ribosomal protein. J Mol Evol. 86(8):581–592. [DOI] [PubMed] [Google Scholar]
- Kovacs NA, Petrov AS, Lanier KA, Williams LD.. 2017. Frozen in time: the history of proteins. Mol Biol Evol. 34(5):1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. 2009. Nature of the protein universe. Proc Natl Acad Sci U S A. 106(27):11079–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Alva V.. 2017. Ribosomal proteins as documents of the transition from unstructured (poly)peptides to folded proteins. J Struct Biol. 198(2):74–81. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Ponting CP, Russell RB.. 2001. On the evolution of protein folds: are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J Struct Biol. 134(2–3):191–203. [DOI] [PubMed] [Google Scholar]
- Murzin AG. 1993. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12(3):861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepomnyachiy S, Ben-Tal N, Kolodny R.. 2017. Complex evolutionary footprints revealed in an analysis of reused protein segments of diverse lengths. Proc Natl Acad Sci U S A. 114(44):11703–11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MN, Madhusudhan MS.. 2011. Biological insights from topology independent comparison of protein 3D structures. Nucleic Acids Res. 39(14):e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, et al. 2016. Reference sequence (Refseq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44(D1):D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov S, Mirdita M, Steinegger M.. 2021. ColabFold - Making protein folding accessible to all. doi: 10.1101/2021.08.15.456425. [DOI] [PMC free article] [PubMed]
- Penev PI, McCann HM, Meade CD, Alvarez-Carreño C, Maddala A, Bernier CR, Chivukula VL, Ahmad M, Gulen B, Sharma A, et al. 2021. Proteovision: web server for advanced visualization of ribosomal proteins. Nucleic Acids Res. 49(W1):W578–W588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, et al. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A. 112(50):15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose GD, Fleming PJ, Banavar JR, Maritan A.. 2006. A backbone-based theory of protein folding. Proc Natl Acad Sci U S A. 103(45):16623–16633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer RD, Liao Y, Cheng H, Grishin NV.. 2017. ECOD: new developments in the evolutionary classification of domains. Nucleic Acids Res. 45(D1):D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Goenner T, Guerler A, Kolbeck B, Knapp EW.. 2010. Circular permuted proteins in the universe of protein folds. Proteins 78(7):1618–1630. [DOI] [PubMed] [Google Scholar]
- Schrödinger LLC. 2021. The Pymol molecular graphics system. Version 2.4.0.
- Sillitoe I, Dawson N, Lewis TE, Das S, Lees JG, Ashford P, Tolulope A, Scholes HM, Senatorov I, Bujan A, et al. 2019. CATH: expanding the horizons of structure-based functional annotations for genome sequences. Nucleic Acids Res. 47(D1):D280–D284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21(7):951–960. [DOI] [PubMed] [Google Scholar]
- Steinegger M, Meier M, Mirdita M, Vöhringer H, Haunsberger SJ, Söding J.. 2019. HH-Suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 20(1):473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS.. 2003. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 32:115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS.. 2005. Divergent evolution within protein superfolds inferred from profile-based phylogenetics. J Mol Biol. 354(3):722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47(D1):D506–D515. [DOI] [PMC free article] [PubMed]
- Vishwanath P, Favaretto P, Hartman H, Mohr SC, Smith TF.. 2004. Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes. Mol Phylogenet Evol. 33(3):615–625. [DOI] [PubMed] [Google Scholar]
- Weiner J, Bornberg-Bauer E.. 2006. Evolution of circular permutations in multidomain proteins. Mol Biol Evol. 23(4):734–743. [DOI] [PubMed] [Google Scholar]
- Xia X, Longo LM, Sutherland MA, Blaber M.. 2016. Evolution of a protein folding nucleus. Protein Sci. 25(7):1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youkharibache P, Veretnik S, Li Q, Stanek KA, Mura C, Bourne PE.. 2019. The small b-barrel domain: a survey-based structural analysis. Structure 27(1):6–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The alignments of rProteins are available in ProteoVision at https://proteovision.chemistry.gatech.edu (last accessed August 23, 2021), an in house web server for visualization and data mapping of proteins. All other relevant data are available within this article, the Supplementary Material online, or from the corresponding authors upon request.