Abstract
How early stages of speciation in free-spawning marine invertebrates proceed is poorly understood. The Western Pacific abalones, Haliotis discus, H. madaka, and H. gigantea, occur in sympatry with shared breeding season and are capable of producing viable F1 hybrids in spite of being ecologically differentiated. Population genomic analyses revealed that although the three species are genetically distinct, there is evidence for historical and ongoing gene flow among these species. Evidence from demographic modeling suggests that reproductive isolation among the three species started to build in allopatry and has proceeded with gene flow, possibly driven by ecological selection. We identified 27 differentiation islands between the closely related H. discus and H. madaka characterized by high FST and dA, but not high dXY values, as well as high genetic diversity in one H. madaka population. These genomic signatures suggest differentiation driven by recent ecological divergent selection in presence of gene flow outside of the genomic islands of differentiation. The differentiation islands showed low polymorphism in H. gigantea, and both high FST, dXY, and dA values between H. discus and H. gigantea, as well as between H. madaka and H. gigantea. Collectively, the Western Pacific abalones appear to occupy the early stages speciation continuum, and the differentiation islands associated with ecological divergence among the abalones do not appear to have acted as barrier loci to gene flow in the younger divergences but appear to do so in older divergences.
Keywords: marine speciation, ecological speciation, fertilization protein, GRAS-Di, introgression
Introduction
Long-distance gene flow in the ocean without apparent physical barriers to it is expected to constrain population differentiation and speciation (Palumbi 1992). In particular, speciation in broadcast spawning marine invertebrates is expected to be challenging due to extensive gene flow (Bierne et al. 2003). However, these expectations are contradicted by the high diversity of marine invertebrates in the oceans. Hence, the question how speciation proceeds in the oceans in the absence of geographic isolation remains an important topic in marine biology (Bierne et al. 2003; Palumbi 2009; Puebla 2009).
The process of speciation can be described as a continuum of divergence leading to reproductive isolation, referred to as the “speciation continuum” (Nosil 2012; Seehausen et al. 2014). The later stages of divergence along the speciation continuum are characterized by the evolution of strong barriers to gene flow between species. An example demonstrating such barriers in broadcast spawning marine invertebrates is the coevolution of interacting fertilization proteins, Lysin in sperm and VERL in egg, in the North American abalones (Clark et al. 2009). Many nonsynonymous substitutions among abalone species in the Lysin- and VERL-coding genes have been suggested to prevent interspecies fertilization and cause reproductive isolation in the absence of physical barriers to gene flow. This coevolution was hypothesized to be caused by sexual conflict resulting from polyspermy (Clark et al. 2009); that is, sperm evolves to increase fertilization efficiency, followed by the evolution of eggs to decrease fertilization efficiency, thereby reducing the fertilization of an egg with multiple sperms (i.e., polyspermy) which halts embryonic development. Previous theoretical studies have suggested a model where sexual conflict and ensuing coevolution of interacting fertilization proteins also drive reproductive isolation in the early stages of speciation in broadcast spawning marine invertebrates (Van Doorn et al. 2001). In this model, intraspecific competition for fertilization increases variance in male gamete protein repertoire initiating speciation (Van Doorn et al. 2001). Therefore, interacting fertilization proteins may be responsible for the initial reproductive isolation that eventually led to speciation of the North American abalones.
Ecological factors, such as habitat or temporal isolation, are suggested to be fundamental mechanisms of speciation under situations characterized by the absence of physical barriers to gene flow (Nosil 2012). Barriers to gene flow evolving between populations as a result of ecologically based divergent selection have become known as “ecological speciation” (Nosil 2012). Although this speciation process is expected to be common in the early stages of speciation in marine environments (Lessios 2007; Bester-van der Merwe et al. 2012; Puritz et al. 2012; Rhode et al. 2013; Momigliano et al. 2017), examples of ecological speciation in the ocean are still rare. According to the genomic view of ecological speciation, gene flow is restricted at barrier loci (i.e., loci underlying ecological isolation) clustered in a few genomic regions, but continued in the outside of these regions (speciation with gene flow), leading to the emergence of peaks of genetic differentiation surrounding the barrier loci (i.e., heterogeneous genomic differentiation; Wu 2001; Feder et al. 2012). Also, the speciation with gene flow model must further be distinguished between models where there is no initial period of allopatry or reduced gene flow (i.e., sympatric speciation) and those where gene flow occurs after a substantial period of independent evolution between diverging taxa (i.e., secondary contact; Cruickshank and Hahn 2014). However, empirical genomic data demonstrating such heterogenous differentiation and demographic processes in the early stages of marine speciation are scarce (Palumbi 2009; Pogson 2016; Momigliano et al. 2017). Hence, investigation of genome-wide patterns of genetic differentiation at multiple stages of the speciation continuum in the ocean has been called forth (Puebla 2009; Miglietta et al. 2011).
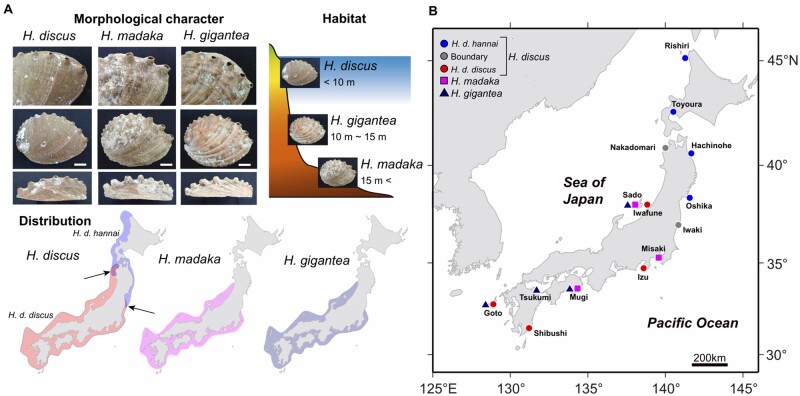
Here, we focus on the speciation continuum in three species of closely related Western Pacific abalones: Haliotis discus, H. madaka, and H. gigantea (fig. 1A; Ino 1952). Of these, H. discus is distributed widely along the coasts of the Japanese archipelago and the Korean Peninsula, and it is classified into two subspecies due to slight morphological differences in dorsal surface of the shell (Ino 1952). Haliotis discus hannai is distributed throughout northern Japanese and Korean coasts, whereas H. discus discus has more southern distribution along Japanese and Korean coasts (Ino 1952; Hara and Sekino 2005; Nam et al. 2021; fig. 1A). However, their geographic distribution ranges have not been accurately determined in Japan (fig. 1A). On the other hand, H. madaka and H. gigantea tend to be endemic to the Japanese archipelago and occur in sympatry with the subspecies H. discus discus in the southern Japanese coast (fig. 1A) sharing the same breeding season (Ino 1952). Previous genetic studies using allozyme (Hara and Fujio 1992), mitochondrial DNA (An et al. 2005), microsatellite DNA (Sekino and Hara 2007a), and the Lysin gene (Lee and Vacquier 1995) variability have shown that the three species are closely related but differ in the degree of divergence from each other; H. gigantea is genetically distinct from H. discus and H. madaka. The three species are also morphologically distinct and occupy different depth zones (fig. 1A; Ino 1952; Itami et al. 1978; Sakai and Shimamoto 1978), but can produce viable F1, F2, and backcross generation offspring in experimental settings (Koike et al. 1988; Ahmed et al. 2008), suggesting that prezygotic and postzygotic isolation is still incomplete. Hence, it appears that the Western Pacific abalones in the Japanese archipelago may be in the early stages of speciation triggered by ecological factors such as the differences in habitat selection, providing an opportunity to investigate the evolutionary processes and genomic architecture along the speciation continuum in broadcast spawning marine invertebrates.
Fig. 1.
(A) Morphological and ecological features, as well as geographical distributions of the Western Pacific abalones in Japan. The scale bars in the (A) corresponds to 2 cm. Arrows in the distribution map of Haliotis discus show boundary zones between the two subspecies, H. discus hannai and H. discus discus. (B) Sampling locations of the Western Pacific abalones used in this study.
To address how far speciation has proceeded along the speciation continuum, we examined the patterns of genomic divergence in the Western Pacific abalones using nuclear single nucleotide polymorphism (SNP) loci obtained with a cost-effective nontargeting polymerase chain reaction (PCR)-based sequencing method, genotyping by random amplicon sequencing, direct (GRAS-Di; Enoki and Takeuchi 2018; Hosoya et al. 2019) and whole-genome sequencing, as well as with mitochondrial genome sequencing. Based on these genomic data, we examined the patterns and degree of genomic divergence among the three abalone species, with focus on identifying genomic regions showing evidence for divergent selection. We also performed demographic modeling to infer divergence times and patterns of gene flow among the three species based on analyses of the site-frequency spectrum (SFS).
Results
Close Phylogenetic Relationships among the Western Pacific Abalones
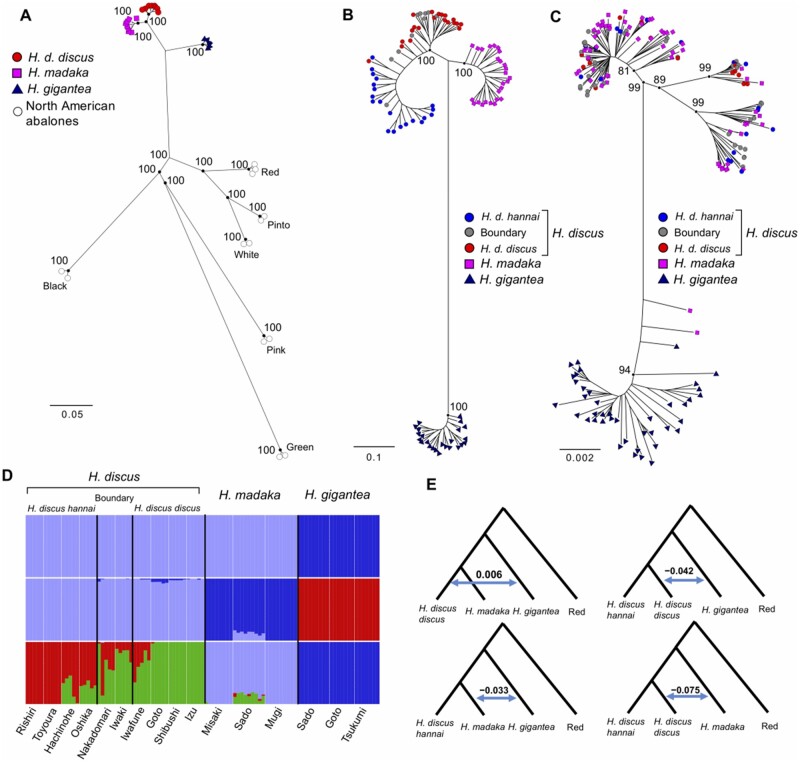
The RAxML tree (Stamatakis 2014) based on 46,366 SNP loci common to the North American and the Western Pacific abalones derived from whole-genome sequencing data suggested that the genetic distances among the Western Pacific abalones are very small in comparison to those among six North American abalones (fig. 2A). This result suggests that speciation of the Western Pacific abalones has occurred more recently than that of the North American abalones which are thought to be reproductively isolated (Clark et al. 2009). Similarly, the RAxML tree of the Western Pacific abalones based on 18,109 SNP loci derived from GRAS-Di demonstrated small but clear genetic differentiation among H. discus, H. madaka, and H. gigantea (fig. 2B). Given that the three species are caught in sympatry by local fishermen in our study sites (fig. 1B), the result suggests that the species are undoubtedly reproductively isolated.
Fig. 2.
(A) ML tree of the Western Pacific and the North American abalones (Red: Haliotis rufescens, Pinto: H. kamtschatkana, White: H. sorenseni. Black: H. cracherodii, Pink: H. corrugate, Green: H. fulgens) based on whole-genome sequencing data (46,366 SNP loci). (B) ML tree of the Western Pacific abalones based on GRAS-Di data (18,109 SNP loci). (C) Neighbor-joining tree based on the mitochondrial genomes of the Western Pacific abalones. The values in the nodes in (A)–(C) are bootstrap values. (D) Individual admixture proportions (q-values) among the Western Pacific abalones estimated by ADMIXTURE. (E) D-statistics of the Western Pacific abalones (z-score [D-statistic/standard error] > 3). The Red abalone (H. rufescens) was used as an outgroup in calculation of D-statistics.
However, the phylogenetic tree based on mitochondrial genome data was not consistent with the phylogenetic tree based on nuclear genomic data; whereas all H. gigantea individuals fell into one of the four clades, the other three clades were shared among H. discus, and H. madaka populations (fig. 2C). Moreover, analysis of molecular variance (AMOVA) based on mitochondrial genome data did not show genetic differentiation between H. discus and H. madaka (AMOVA, P > 0.05). These results may be attributed to the recent establishment of reproductive isolation between H. discus and H. madaka and/or ongoing hybridization between these species. Two mitochondrial haplotypes detected in two H. madaka individuals were close to the H. gigantea clade and located at basal positions (fig. 2C), suggesting that this haplotype sharing is not attributable to recent hybridization.
Coevolution of Lysin- and VERL- Genes Is Not Associated with Reproductive Isolation
Interspecies nonsynonymous substitutions in the Lysin and VERL genes are involved in the reproductive isolation among the North American abalones (Clark et al. 2009). Therefore, interspecies nonsynonymous substitutions in the two genes may be also associated with reproductive isolation among the Western Pacific abalones. However, such nonsynonymous substitutions within the Western Pacific abalones have been reported only in the Lysin gene of H. gigantea (Lee and Vacquier 1995); intraspecific polymorphisms in Lysin genes of the Western Pacific abalones and substitutions in VERL genes of the Western Pacific abalones have not been investigated yet. Our gene annotation suggested that both of Lysin (HDSC01187: 6,572–33,666) and VERL (HDSC09152: 4,839–9,463) genes were included in the H. discus hannai reference genome (Nam et al. 2017). Hence, we next examined whether nonsynonymous substitutions occur in the Lysin and VERL genes of H. madaka and H. gigantea against the H. discus hannai reference genome (Nam et al. 2017) using whole-genome sequencing data.
Consistent with the results of the previous study (Lee and Vacquier 1995), there were one synonymous and four nonsynonymous substitutions in the first exon of the Lysin gene, for which positive selection was suggested (Clark et al. 2009), in H. gigantea (supplementary table 1, Supplementary Material online). In contrast, we found no nonsynonymous substitution between H. discus and H. madaka in the Lysin gene (supplementary table 1, Supplementary Material online). In all species, intraspecific polymorphism was absent, suggesting selective sweep in this gene as suggested in the North American abalones (Clark et al. 2009). VERL comprised 22 protein repeats, and the first two repeats (repeat 1 and 2) are the most different from other repeats. Among these repeats, VERL repeat 2 is considered to play a role in species-specific sperm selection (Galindo et al. 2003). However, we found nonsynonymous substitution in the coding region of VERL repeats 1 and 2 neither in H. madaka nor in H. gigantea, suggesting that the coevolution of Lysin and VERL genes has not occurred in all Western Pacific abalones. Therefore, this finding raises the possibility that some other factors rather than the coevolution of fertilization proteins, such as ecological factor(s), contribute to reproductive isolation in the Western Pacific abalones.
Footprints of Recent and Historical Hybridization
Given the close phylogenetic relationships and shared mitogenome haplotypes among H. discus, H. madaka, and H. gigantea (figs. 2A–C), hybridization may have occurred among the three abalone species. To get insight into this possibility, we set to evaluate the degree of gene flow among the three abalone species. In clustering analyses using ADMIXTURE (Alexander et al. 2009) with GRAS-Di data, the cross-validation error reached a plateau at around K = 4 (supplementary fig. 1, Supplementary Material online). When assuming K = 4, H. discus was assigned into two genetic clusters corresponding to H. discus hannai and H. discus discus, and these two genetic clusters admixed in putative boundary zones (fig. 2D). Haliotis madaka and H. gigantea were also assigned into different genetic clusters (fig. 2D), suggesting limited gene flow among three abalone species.
However, the H. madaka population in Sado showed clear signatures of genomic introgression from H. discus discus in the ADMIXTURE plot (fig. 2D). For each of the putative hybrid individuals of H. madaka population in Sado, we genotyped 76 SNP loci in 46 scaffolds (parts of these SNP loci are located closely to each other in same scaffold) that were fixed for different alleles in each of H. discus and H. madaka. Consistent with the ADMIXTURE results, the genotype data based on the 76 SNP loci showed clear signatures of allele sharing between H. discus and the Sado population of H. madaka (supplementary fig. 2, Supplementary Material online). One can expect that first-generation (F1) hybrids would be consistently heterozygous at nearly all of the 76 SNP loci. However, because all of the putative hybrids failed to conform to this expectation, they could be second-generation (F2) hybrids, backcrosses, or later-generation hybrids. NewHybrids analysis (Anderson and Thompson 2002) based on 76 SNP loci assigned four putative hybrids to be backcrosses to H. madaka, and five putative hybrids to be pure H. madaka with 100% posterior probability (supplementary table 2, Supplementary Material online). Given that H. discus discus and H. madaka are co-distributed in Sado, it can be inferred that H. discus discus and pure (or nearly pure) H. madaka have hybridized and even backcrossed to H. madaka there recently. It is notable that number of inter-scaffold SNP pairs showed high r2 values (>0.5). This suggests that they are either in close physical proximity of each other, or strong positive selection due to possible functional associations maintains this high linkage disequilibrium (LD) (supplementary table 3, Supplementary Material online). Although ADMIXTURE analyses did not find any evidence of hybridization except in H. madaka in Sado, ABBA–BABA analyses based on whole-genome sequencing data showed signals of hybridization among H. discus discus, H. madaka, and H. gigantea, which overlap in their geographic distributions (z-score [D-statistic/standard error] > 3; fig. 2E). Altogether, these findings suggest that the strength of reproductive isolation between H. discus and H. madaka varies regionally and that the Western Pacific abalones have hybridized historically.
Demographic Modeling Supported Speciation-with-Gene Flow Model
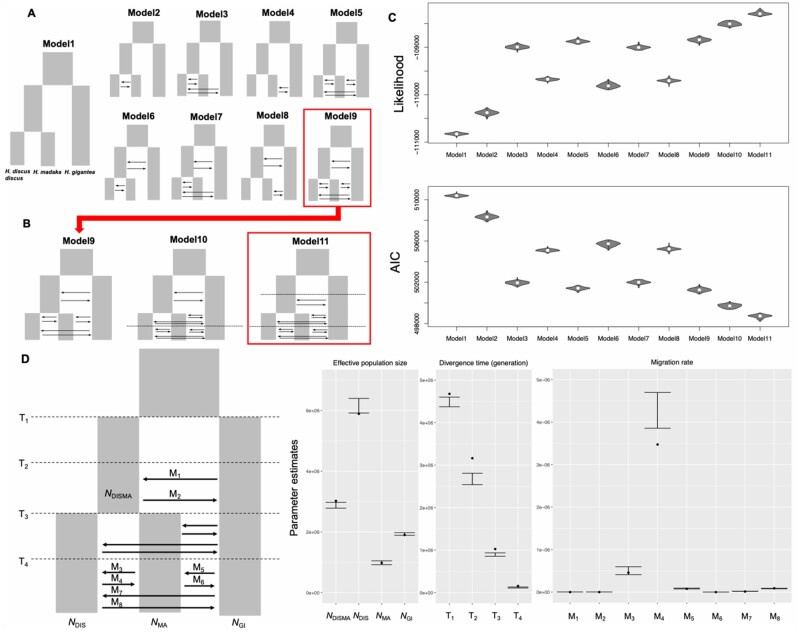
The aforementioned genomic analyses implied that the Western Pacific abalones have speciated in the face of gene flow. To test this hypothesis, we performed demographic modeling based on the SFS using fastsimcoal2 (Excoffier et al. 2013). In this analysis, we used GRAS-Di data from sympatric H. discus discus, H. madaka (except for the Sado population with recent hybridization), and H. gigantea populations. First, we compared nine demographic models (fig. 3A); one model assumed no gene flow during the speciation process (Model 1), whereas the other eight models (Models 2, 3, 4, 5, 6, 7, 8, and 9) assumed gene flow in different pairs of species or an ancestor. To simplify the models, we assumed constant population size for each species. The highest log-likelihood and lowest Akaike information criterion (AIC) were obtained for Model 9 (fig. 3A and C). Model 9 assumed speciation with gene flow for both the initial divergence of H. gigantea and subsequent divergence between H. discus discus and H. madaka, and gene flow between H. discus discus and H. gigantea, and between H. madaka and H. gigantea, which is consistent with the result of ABBA–BABA analyses (fig. 2E).
Fig. 3.
SFS-based coalescent analyses for modeling Western Pacific abalone speciation. (A) Nine demographic models tested. (B) Three demographic models with and without secondary contact based on Model 9 that supported by first-round demographic modeling. (C) Distributions of log10 likelihood and AIC from 100 expected SFS each approximated using 1 million coalescent simulations under the parameters that maximize the likelihood for each model. (D) Parameter estimates of effective population sizes, the times of divergences, migration rates, and their associated 95% CIs derived from nonparametric block bootstraps. NDISMA, effective population size (Ne) of a common ancestor of Haliotis discus and H. madaka; NDIS, Ne of H. discus; NMA, Ne of H. madaka; NGI, Ne of H. gigantea; T1 and T2, times of divergences (in generations); M1–M8, migration rates.
For speciation with gene flow model, we must further distinguish between models where there is no initial period of allopatry or reduced gene flow (i.e., sympatric speciation) and those where gene flow occurs after a substantial period of independent evolution between diverging taxa (i.e., secondary contact; Cruickshank and Hahn 2014). Hence, we compared the Model 9 with its derivate model that assumes allopatric phase or reduced gene flow in the divergence event between H. discus discus and H. madaka (Model 10), and higher log-likelihood and lower AIC were obtained for Model 10 (fig. 3B and C). Next, we compared Model 10 with its derivate model that assumes allopatric phase or reduced gene flow between H. gigantea and an ancestor of H. discus discus and H. madaka (Model 11), and the higher log-likelihood and lower AIC were obtained for Model 11 assuming secondary contact between H. gigantea and an ancestor of H. discus discus and H. madaka (fig. 3B and C). Consequently, demographic modeling suggested that both speciation events in the Western Pacific abalones started with allopatric or reduced gene flow phase followed by secondary contact.
The parameter estimates based on Model 11 show an interesting pattern of gene flow; the rate of gene flow from H. discus discus to H. madaka was higher than that from H. madaka to H. discus discus (fig. 3D). This pattern is consistent with the results of the clustering analyses using ADMIXTURE, which suggested genomic introgression from H. discus discus to H. madaka abalones in Sado (fig. 2D). Although the H. madaka population in Sado was not included in this demographic modeling, this suggests that directional genomic introgression may have occurred there too.
Potential Genomic Regions Resistant to Gene Flow
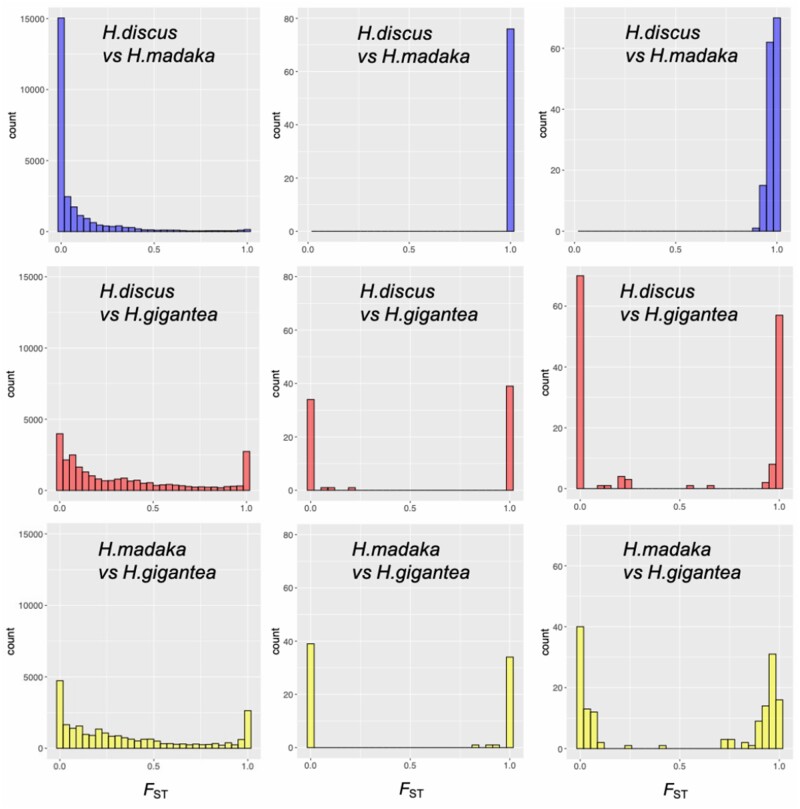
Given the close relationship (median FST = 0.007; fig. 4) as well as historical and ongoing hybridization between H. discus and H. madaka, highly divergent genomic regions between H. discus and H. madaka are candidate targets of divergent selection, that is, “barrier loci” (Feder et al. 2012). The above mentioned 76 SNP loci that were fixed for different alleles in H. discus and H. madaka (except for Sado population) are such candidates (fig. 4). To identify such genomic regions further, we performed SNP-based genome scans based on GRAS-Di data using pcadapt (Luu et al. 2017), Fdist (Beaumont and Nichols 1996), BayeScan (Foll and Gaggiotti 2008), and FLK (Bonhomme et al. 2010) using all H. discus and H. madaka individuals. As a result, we found 148 outlier SNP loci (16 fixed SNP loci were also included) that were detected by all four methods (fig. 4). Similarly to loci fixed to different alleles between H. discus and H. madaka, some of these outlier SNP loci also showed high inter-scaffold r2 values (>0.5) in H. madaka in Sado (supplementary table 4, Supplementary Material online).
Fig. 4.
Histograms of site-FST values of all SNPs (left), 76 fixed SNPs (middle), and 148 common outlier SNPs (right).
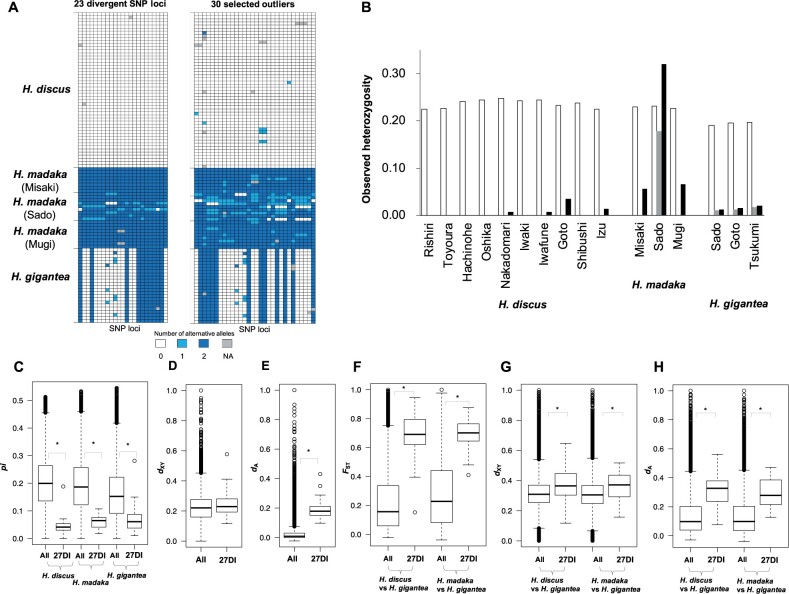
We next investigated whether the 76 fixed and the 148 outlier SNP loci were located in divergent genomic regions identified with the aid of 5.9 million SNP loci obtained from the whole-genome sequencing data. We did this by searching divergent genomic regions (in 20-kb windows) showing up in the top 1% of the FST distribution between H. discus and H. madaka. This search yielded 23 fixed and 30 outlier SNP loci overlapping divergent genomic regions in 27 different scaffolds (fig. 5A;table 1; henceforth referred to as “23 divergent SNP loci,” “30 selected outliers,” and “27 differentiation islands” [Burri 2017]). The number of these overlaps were confirmed to be significantly higher than expected on the basis of random sampling of SNP loci (max. 6 overlapping loci for 148 SNP loci and max. 4 overlapping loci for 76 SNP loci; P < 0.001). Observed heterozygosity in the 30 selected outliers, as well as that in the 23 divergent SNP loci, was lower than that of other loci in all populations with the exception of H. madaka population in Sado where genomic introgression from H. discus was detected (fig. 5B). Nucleotide diversity (pi) in 27 differentiation islands was also lower than other genomic regions in all abalones, suggesting that these islands are under divergent selection (fig. 5C).
Fig. 5.
(A) Genotypes in 23 divergent and 30 selected outlier SNP loci showing the genetic divergence between Haliotis discus and H. madaka abalones. NA shows loci that were not gentyped. (B) Observed heterozygosity in each of the three Western Pacific abalone populations across all SNP loci (white), 23 divergent SNP loci (gray), and 30 selected outlier (black). (C) Boxplot of nucleotide diversity (pi) in H. discus, H. madaka, and H. gigantea in all and 27 differentiation islands (27DI). (D) Boxplot of absolute sequence divergence (dXY) between H. discus and H. madaka in all and 27DI. (E) Boxplot of net sequence divergence (dA) between H. discus and H. madaka in all and 27DI. (F) Boxplot of sliding-window FST values between H. discus and H. gigantea, and between H. discus and H. gigantea in all and 27DI. (G) Boxplot of dXY between H. discus and H. gigantea, and between H. discus and H. gigantea in all and 27DI. (H) Boxplot of dA between H. discus and H. gigantea, and between H. discus and H. gigantea in all and 27DI. Asterisks show significant differences (Wilcoxon rank-sum test; P < 0.05).
Table 1.
Twenty-Seven Divergent Genomic Regions between Closely Related Haliotis discus and H. madaka.
| Scaffold | Divergent Loci | Selected Outliers | top 1% FST Window(HD vs. HM) | d XY(HD vs. HM) | d A(HD vs. HM) | fd(HD–HG) | f d(HM–HG) | pi(HD) | pi(HM) | pi (HG) | F ST(HD vs. HG) | F ST(HM vs. HG) | d XY(HD vs. HG) | d XY(HM vs. HG) | d A(HD vs. HG) | d A(HM vs. HG) | Linkage group | Annotated gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HDSC00060 | 639957 | 639786, 639957 | 620,001–640,000 | 0.169 | 0.138* | 0.011 | 0.0295† | 0.0312† | 0.0786 | 0.793* | 0.810* | 0.487* | 0.487* | 0.433* | 0.432* | LG17 | ||
| HDSC00194 | 596133 | 580,001–600,000 | 0.273 | 0.299** | 0.134 | 0.0712† | 0.0707 | 0.0323† | 0.635* | 0.694* | 0.351 | 0.294 | 0.299 | 0.243 | LG16 | SSTR2 | ||
| HDSC00283 | 87998, 88035 | 80,001–100,000 | 0.235 | 0.454** | 0.027 | 0.0274† | 0.0655† | 0.0506 | 0.834* | 0.766* | 0.493* | 0.439 | 0.454* | 0.381* | LG01 | |||
| HDSC00375 | 244254 | 240,001–260,000 | 0.336* | 0.265** | 0.130 | 0.0709† | 0.0701 | 0.0869 | 0.632* | 0.628* | 0.432 | 0.336 | 0.353* | 0.257 | SPR | |||
| HDSC00833 | 448991, 448996, 449000 | 440,001–460,000 | 0.246 | 0.192* | 0.041 | 0.0115† | 0.0613† | 0.0390† | 0.796* | 0.640* | 0.218 | 0.231 | 0.192 | 0.181 | LG07 | NCL | ||
| HDSC00857 | 128388, 128389, 128528 | 120,001–140,000 | 0.208 | 0.163* | 0.004 | 0.0355† | 0.0551† | 0.0610 | 0.693* | 0.650* | 0.306 | 0.269 | 0.258 | 0.211 | CNOT11 | |||
| HDSC00971 | 73261, 73267, 73295, 73302 | 60,001–80,000 | 0.288 | 0.213** | 0.279* | 0.0686† | 0.0818 | 0.0100† | 0.567* | 0.717* | 0.238 | 0.300 | 0.199 | 0.254 | LG13 | |||
| HDSC01372 | 307426 | 300,001–320,000 | 0.251 | 0.134* | 0.035 | 0.0413† | 0.0552† | 0.0512 | 0.687* | 0.699* | 0.311 | 0.298 | 0.334* | 0.337* | LG01 | GATA3 | ||
| HDSC01400 | 162134, 162148, 162000 | 162134, 162148, 162185 | 160,001–180,000 | 0.180 | 0.203** | 0.014 | 0.0500† | 0.0410† | 0.0696 | 0.689* | 0.763* | 0.394 | 0.392 | 0.265 | 0.244 | SLC6A14 | ||
| HDSC01500 | 127929 | 120,001–140,000 | 0.300 | 0.251** | 0.045 | 0.0390† | 0.0586† | 0.1017 | 0.610* | 0.580 | 0.301 | 0.290 | 0.231 | 0.209 | LG11 | |||
| HDSC01503 | 62447, 62466 | 60,001–80,000 | 0.348* | 0.288** | 0.107 | 0.0509† | 0.0686† | 0.2813 | 0.395 | 0.411 | 0.354 | 0.394 | 0.187 | 0.219 | ||||
| HDSC01562 | 42647, 42668 | 40,001–60,000 | 0.411* | 0.350** | 0.298* | 0.0575† | 0.0649† | 0.1066 | 0.529* | 0.558 | 0.295 | 0.292 | 0.213 | 0.206 | ||||
| HDSC01641 | 284248, 284249 | 280,001–300,000 | 0.115 | 0.342** | 0.009 | 0.0195† | 0.0175† | 0.0249† | 0.857** | 0.872** | 0.365 | 0.299 | 0.342* | 0.278 | LG04 | |||
| HDSC02293 | 286533 | 280,001–300,000 | 0.229 | 0.179* | 0.006 | 0.0713† | 0.0283† | 0.0303† | 0.657* | 0.875** | 0.378 | 0.432 | 0.327* | 0.403* | BECN1 | |||
| HDSC02343 | 93665 | 80,001–100,000 | 0.151 | 0.349** | 0.019 | 0.0314† | 0.0437† | 0.0307† | 0.802* | 0.817* | 0.380 | 0.371 | 0.349* | 0.334* | ||||
| HDSC02435 | 46386 | 40,001–60,000 | 0.222 | 0.405** | 0.037 | 0.0457† | 0.0701 | 0.0644 | 0.746* | 0.745* | 0.460* | 0.457* | 0.405* | 0.390* | ||||
| HDSC02598 | 98883, 98953, 98963 | 80,001–100,000 | 0.231 | 0.166* | 0.062 | 0.0473† | 0.0826 | 0.0901 | 0.676* | 0.663* | 0.399 | 0.419 | 0.330* | 0.332* | ||||
| HDSC03044 | 88188 | 80,001–100,000 | 0.224 | 0.477** | 0.023 | 0.0308† | 0.0300† | 0.0619 | 0.820* | 0.846* | 0.524* | 0.517* | 0.477* | 0.471* | ||||
| HDSC03119 | 57856 | 40,001–60,000 | 0.163 | 0.336** | 0.0518† | 0.0264† | 0.0558 | 0.698* | 0.828* | 0.390 | 0.412 | 0.336* | 0.370* | |||||
| HDSC03434 | 5718 | 1–20,000 | 0.240 | 0.276** | 0.035 | 0.0366† | 0.0846 | 0.0710 | 0.691* | 0.480 | 0.329 | 0.221 | 0.276 | 0.143 | ||||
| HDSC03507 | 14619 | 1–20,000 | 0.202 | 0.157* | 0.029 | 0.0558† | 0.0341† | 0.1123 | 0.747* | 0.745* | 0.646* | 0.462* | 0.562* | 0.389* | ||||
| HDSC03534 | 35699 | 20,001–40,000 | 0.183 | 0.250** | 0.017 | 0.0087† | 0.0701 | 0.0365† | 0.857** | 0.655* | 0.272 | 0.261 | 0.250 | 0.208 | SCRN3 | |||
| HDSC04345 | 116238 | 100,001–120,000 | 0.218 | 0.159* | 0.037 | 0.0321† | 0.0857 | 0.0869 | 0.771* | 0.704* | 0.476* | 0.491* | 0.416* | 0.404* | ||||
| HDSC04393 | 10800, 11024, 11028 | 1–20,000 | 0.580** | 0.431** | 0.682** | 0.1880 | 0.1043 | 0.0395† | 0.152 | 0.702* | 0.190 | 0.427 | 0.076 | 0.355* | LG02 | DPEP1 | ||
| HDSC04994 | 205736 | 200,001–220,000 | 0.152 | 0.119* | n.e | 0.022 | 0.0247† | 0.0412† | 0.0202† | 0.590* | 0.666* | 0.117 | 0.157 | 0.094 | 0.127 | LG11 | DHODH, ZNF121 | |
| HDSC06394 | 27870, 27872 | 20,001–40,000 | 0.225 | 0.242** | 0.006 | 0.0437† | 0.0959 | 0.1501 | 0.563* | 0.489 | 0.340 | 0.347 | 0.243 | 0.224 | CFAP221 | |||
| HDSC06434 | 33688, 33720, 33772 | 33688 | 20,001–40,000 | 0.289 | 0.235** | n.e | 0.0000† | 0.1073 | 0.0417† | 0.945** | 0.733* | 0.479* | 0.503* | 0.458* | 0.428* | PSMD10 |
Note.—HD, Halitis discus; HM, Haliotis madaka; HG, Haliotis gigantea; pi, nucleotide diversity; dXY, absolute sequence divergence; dA, net sequence divergence. The fd value of HDSC06434 was not estimated due to low nucleotide diversity **Above 99 percentile; *above 90 percentile; †below 10 percentile.
If the 27 differentiation islands have been involved in reducing gene flow between sympatric H. discus and H. madaka since the early stages of divergence, they are expected to show both high FST and high absolute sequence divergence (dXY: Nei and Li 1979) as pointed out by Cruickshank and Hahn (2014). This is because divergence is expected to be initiated in the regions with reduced gene flow, resulting in both high FST and elevated nucleotide divergence. To test this, we estimated dXY for the 27 differentiation islands. Unexpectedly, the values of dXY of the identified differentiation islands were not significantly higher than the genomic background (Wilcoxon rank-sum test; P > 0.05; fig. 5D;table 1). On the other hand, 4 of the 27 differentiation islands (scaffolds HDSC00375, HDSC01503, HDSC01562, and HDSC04393) had dXY values above the 90th percentile of the genome-wide distribution (table 1). Because introgression from H. gigantea to either H. discus or H. madaka can also elevate both FST and dXY (Jones et al. 2018; Oziolor et al. 2019), the possibility of introgression in 27 differentiation islands was evaluated based on sliding-window-based fd-statistics, a modified version of ABBA–BABA statistics (Martin et al. 2015). Of the four differentiation islands with both high FST and high dXY, differentiation islands within the HDSC01562 and HDSC04393 scaffolds showed high fd values above 90th percentile, suggesting that they might originate from introgression (table 1). Consequently, only two of the differentiation islands (in scaffolds HDSC00375 and HDSC01503) exhibited features of high FST and high dXY expected from barrier loci allowing speciation with gene flow.
Although dXY between H. discus and H. madaka in the 27 differentiation islands was not high, the values of dA (Nei and Li 1979) were significantly higher than the genomic background (Wilcoxon rank-sum test; P < 0.05; fig. 5E;table 1). dA value is obtained by subtracting pi of the two lineages from dXY, and this measure captures pairwise differences that arose since the lineage split (Burri 2017). Therefore, the high dA value suggests that although nucleotide divergence between H. madaka and H. discus due to divergent selection has certainly been accumulating in the differentiation islands, relatively high within-species pi have elevated absolute dXY in their genomic backgrounds (fig. 5C) and lead to the low dXY in the differentiation islands.
Several previous studies demonstrate that high FST is seldom associated with high dXY (Cruickshank and Hahn 2014). Such pattern of differentiation can be caused by recurrent background selection before and after speciation, but not by reduced gene flow (Begun et al. 2007; Cruickshank and Hahn 2014; Burri 2017; Ravinet et al. 2017). If background selection has contributed to high FST and unaffected dXY in differentiation islands, comparisons between H. discus and H. gigantea, and between H. madaka and H. gigantea should also show high FST and unaffected dXY because background selection is not expected to be a lineage-specific (Burri 2017). However, contrary to this expectation, these FST and dXY values exceeded significantly the genomic background levels as well as dA values (Wilcoxon rank-sum test; P < 0.05; fig. 5F–H; table 1). In addition, pi in each of 27 differentiation islands was low in not all species (table 1). These results suggest that background selection cannot explain unaffected dXY between H. discus and H. madaka and that most of the differentiation islands have reduced gene flow between H. gigantea and H. discus or H. madaka, with older divergences. On the other hand, one differentiation island (in scaffold HDSC04994) showed low pi in all three species and unaffected dXY in all species pairs, and thus, this island may have been generated by background selection. In addition to background selection, local adaptation following or unrelated to speciation has also been suggested to result in high FST and unaffected dXY values (Cruickshank and Hahn 2014; Ravinet et al. 2017). However, given high FST and high dXY between H. discus and H. gigantea, and between H. madaka and H. gigantea, it is unlikely that the 27 differentiation islands are involved in local adaptation of H. madaka and H. discus.
The 27 differentiation islands included 13 annotated genes (table 1). Among these genes, the 26S proteasome non-ATPase regulatory subunit 10 (PSMD10) that was included in the differentiation island of HDSC 06434 has been reported to change oxidative stress-related pathways in the muscle of the white leg shrimp Litopenaeus vannamei in response to increased salinity (Wang et al. 2015). Also, the Ankyrin gene, the motif of which is included in PSMD10, is a candidate gene of local adaptation in Red abalone (De Wit and Palumbi 2013). Therefore, this gene might be related to the adaptive differentiation also in Western Pacific abalones. Indeed, SnpEff (Cingolani et al. 2012) analyses revealed the presence of divergent nonsynonymous substitutions among the three abalones in PSMD10 genes. Each species has different combinations of these substitutions, that is, haplotypes, which may be associated with the expression of different phenotypes among the three abalone species (supplementary fig. 3, Supplementary Material online). Other genomic regions also include functionally interesting genes (table 1). For example, the sex peptide receptor (SPR) that included in the differentiation island of HDSC00375 scaffold has been studied in the fruit fly D. melanogaster, and it has an important function in determining reproductive behavior (Yapici et al. 2008). Nowland et al. (2019) suggested that this gene may play a role in adaptive differences between the tropical black-lip rock oyster populations. Many genes of solute carrier family (SLC) differentially expressed in hyposalinity- and temperature-stressed oysters (Ertl et al. 2019), and solute carrier family 6 member 14 (SLC6A14) was included in one differentiation island of the HDSC01400 scaffold.
Extension of Haliotis discus hannai Reference Genome Based on Linkage Information
The genome assembly of H. discus hannai used in this study (Nam et al. 2017) is fragmented (scaffold N50 = 211,346 bp). The reason for this is that Haliotis species have the largest sequenced genomes among gastropods (∼1.9 Gb in H. discus hannai), making it impossible to infer genome-wide distribution of the detected differentiation islands. More contiguous genome assemblies of H. rufescens (scaffold N50 = 1,895,871 bp) and H. rubra (scaffold N50 = 1,227,833 bp) have been obtained using the Chicago method and Nanopore sequencing (Gan et al. 2019; Masonbrink et al. 2019). Nevertheless, chromosome-level genome assemblies of Haliotis have not been available. To overcome this limitation, we increased the contiguity of H. discus hannai genome using linkage information from a full-sib family bred in captivity. This family has been used for linkage map construction with microsatellite DNA markers (Sekino and Hara 2007b). The parents and 96 full-sibs were genotyped for 175,768 SNP loci across the genome using the GRAS-Di method, and these SNP loci were used for scaffolding with SELDLA, which extend scaffolds based on linkage maps generated with low-depth sequencing data (Yoshitake et al. 2018). We determined locations of 4,345 scaffolds and orientations of 1,149 scaffolds out of 80,105 scaffolds, representing 54.3% and 24.7% of the H. discus hannai genome sequences, respectively. This extension led to 21-fold increase in N50 (scaffold N50 = 4,499,196 bp). The extended scaffolds were further anchored onto 18 linkage groups (LGs; Sekino and Hara 2007b) and assembled using the linkage maps with 167 microsatellite DNA markers (Sekino and Hara 2007b) with ALLMAPS (Tang et al. 2015). High collinearity (Pearson correlation coefficient > 0.9) of the locations of microsatellite DNA markers was indicated in 12 LGs, but not in six LGs (LG04, 11, 12, 14, 15, and 16; supplementary fig. 4, Supplementary Material online). Moreover, dot plots comparing the 18 LGs with 26 H. rufescens and 8 H. rubra scaffolds larger than 5 Mb indicated successful alignment of LG01, LG05, and LG11 (supplementary figs. 5 and 6, Supplementary Material online). Although there are still limitations in the quality of the extended genome, we were able to map 10 of the 27 differentiation islands into LG01, LG02, LG04, LG07, LG11, LG13, LG16, and LG17 of the extended H. discus hannai genome (table 1). Furthermore, we observed high interchromosomal LD among divergent SNP loci/selected outliers in H. madaka in Sado (supplementary fig. 7, Supplementary Material online), which can be suggestive of positive selection maintaining functional associations among distant loci.
Discussion
Speciation with Gene Flow Model Fits the Western Pacific Abalones
Our population genomic analyses revealed that the sympatric Western Pacific abalones are genetically distinct even though they are capable of hybridization with high fertility (Koike et al. 1988; Ahmed et al. 2008). We found no nonsynonymous substitutions in the Lysin and VERL genes between the most closely related H. discus and H. madaka (supplementary table 1, Supplementary Material online). For H. gigantea that diverged from a common ancestor of H. discus and H. madaka, several nonsynonymous substitutions were identified in the Lysin gene but not in the VERL gene. These findings suggest that the coevolution of fertilization proteins is not the main factor for the nearly complete reproductive isolation of the Western Pacific abalones in the wild. On the other hand, each of the three abalone species occupies different depth zones (fig. 1A). A previous study suggested that fertilization rates of abalones decrease when the physical distance between males and females is about 2 m or more during spawning (Babcock and Keesing 1999). Also, although geographical distributions of H. discus and H. madaka overlap, Sakai and Shimamoto (1978) reported slight differences in the microgeographical distribution of these species. Therefore, it seems likely that reproductive isolation of the Western Pacific abalones is caused by ecological factors, such as habitat separation.
Although the three species are genetically distinct and reciprocally monophyletic, ABBA–BABA analyses suggested historical hybridization among all three species. This inference was backed up by results of demographic modeling, suggesting that the speciation of these taxa could have occurred in the face of gene flow (cf., Nosil 2012). Given the extensive sharing of mitogenome haplotypes between H. discus discus and H. madaka, as well as clear signs of introgression from H. discus discus to H. madaka in one H. madaka population shown by clustering analyses and demographic modeling, the reproductive isolation between the sympatric H. discus discus and H. madaka is still developing in the face of spatially varying interspecies gene flow. Also, lack of shared mitogenome haplotypes and admixture in the clustering analyses suggest that recent hybridization has not occurred, or has rarely occurred, between H. gigantea and H. discus discus/H. madaka, but demographic modelling results indicated that hybridization between these species has occurred during their speciation processes, suggesting speciation-with-gene flow in the Western Pacific abalones. Our demographic modeling also suggested secondary contact in both speciation events, suggesting that initial allopatric or reduced gene flow phase is important for speciation in the free-spawning marine invertebrates. This implies that marine invertebrate speciation consists of allopatric and sympatric phases, which conforms to traditional ideas in ecological speciation (Rundle and Nosil 2005).
Past phylogenetic analyses suggested close genetic relationships between the Western Pacific and North American abalones (Lee and Vacquier 1995; Streit et al. 2006), and North American abalones are considered to have more derived morphological characters than the Western Pacific abalones (Geiger and Groves 1999). Based on this, the Western Pacific abalones are considered to have diverged from the amphi-Pacific abalones, which radiated in California during glacial cycles (Geiger and Groves 1999). This hypothesis is in line with the result of our phylogenomic analyses. Although it is difficult to estimate the divergence times among the Western Pacific abalones because information on the mutation rates of marine invertebrates is limited, our demographic modeling using the human mutation rate (2.5 × 10−8; Nachman and Crowell 2000) suggests that H. gigantea diverged 14 million years ago (Ma) and H. discus and H. madaka diverged 3 Ma assuming generation time of 3 years (H. discus in the wild reach sexual maturity as 3- or 4-year-olds; Yu et al. 2018). Fossil records of H. discus and H. gigantea in Pliocene have been reported (Geiger and Groves 1999). It is notable that 14 Ma is the time when the Sea of Japan opened by the rotation of the Japanese archipelago (Chinzei 1986) and that 3 Ma corresponds to the time after which the Sea of Japan was repeatedly closed (Tada 1994). These paleogeographic isolation events of the Sea of Japan have led to several allopatric divergences in coastal species (Kojima et al. 1997, 2004; Akihito et al. 2008; Kokita and Nohara 2011; Hirase et al. 2012; Hirase and Ikeda 2014). Therefore, speciation of Western Pacific abalones in Japanese archipelago might have started before the Pleistocene due to these complex paleogeographic events (Kitamura et al. 2001; Kitamura and Kimoto 2006), and interspecific gene flow has continued until now.
It is noteworthy that relatively more nonsynonymous substitutions than synonymous substitutions in the Lysin gene were detected only in H. gigantea that diverged first. This means that the evolution of Lysin and VERL begins after long-term ecological separation of H. gigantea, and which is consistent with the assumption that allopatry is the essence of coevolution of gamete recognition (Palumbi 2009). This coevolution was hypothesized to be induced by sexual conflict resulting from polyspermy (Clark et al. 2009), and polyspermy is more likely to occur in high population density. One common ecological situation in which population densities might increase is when a species invades a new habitat (Palumbi 2009). Therefore, we can speculate that long-term ecological separation and increase of population density of H. gigantea led to sperm competition and evolution of Lysin gene with selective sweep as suggested by Clark et al. (2009). Alternatively, given that continued gene flow between H. gigantea and other two species, reinforcement driven by hybrid disadvantage may also be involved in the Lysin evolution (Palumbi 2009). In any case, the absence of any mutations in VERL suggests that the evolution of fertilization genes after ecological separation is ongoing in H. gigantea.
Differentiation Islands of the Western Pacific Abalones
Under a secondary contact scenario fitting to the Western Pacific abalones, genomic differentiation first builds up over time due to genetic drift during a period of geographical isolation. After secondary contact, gene flow between populations gradually erodes differentiation outside barrier loci and regions with increased background selection (Cruickshank and Hahn 2014; Burri 2017; Ravinet et al. 2017). Whether differentiation becomes erased in most genomic regions or only in a few depends on the level of gene flow and the duration of allopatric isolation. The low-level differentiation between H. discus and H. madaka (fig. 4) appears to fit to the former scenario with the 27 differentiation islands representing possible barrier loci. Previous studies suggest that loci causing speciation with gene flow are likely to be located in relatively few linked clusters within the genome (Feder et al. 2012; Cruickshank and Hahn 2014). Extension of the reference genome of H. discus hannai based on linkage information made it possible to investigate the genomic locations of the 27 differentiation islands, and to discover that the candidate genomic regions were located at least in eight different LGs. Therefore, our data suggest that genomic regions responsible for abalone speciation are distributed in multiple chromosomes, even if the species are at early stages of speciation continuum.
A key prediction of speciation with gene flow model is that causal loci of speciation should have both high relative divergence (FST) and absolute divergence (dXY; Cruickshank and Hahn 2014). However, dXY values of most of the differentiation islands between most closely related H. discus and H. madaka were not high in comparison to other regions. Background selection has been identified as a mechanism generating such pattern of differentiation (Cruickshank and Hahn 2014; Burri 2017). However, background selection is not likely to explain this differentiation pattern because high FST and low dXY were specific for the pair of most closely related H. discus and H. madaka. Contrary to the dXY, the dA between H. discus and H. madaka was relatively high in the differentiation islands. The likely explanation for relatively low dXY in the differentiation islands is recent ecological selection, which reduces pi in H. discus and H. madaka, whereas continued gene flow in the genomic background can likely explain the relatively high pi for these two species. These findings are consistent with a simulation study of Cruickshank and Hahn (2014) showing that there is a difference in the power of FST and dXY in identifying barrier loci contributing to recent divergences in the presence of high gene flow. On the other hand, the differentiation islands showed high dXY as well as FST and dA values between H. discus and H. gigantea, as well as between H. madaka and H. gigantea, suggesting that the low background pi in H. gigantea is due to long-term isolation (cf., fig. 5C).
The H. madaka population in Sado with enhanced genomic introgression from H. discus showed high observed heterozygosity in 23 divergence and 30 outlier loci within the 27 differentiation islands. In particular, genomic introgression in the differentiation islands around 23 divergence loci seems robust because these loci were fixed for different alleles in each of H. discus and H. madaka. Simultaneously, this population showed high interchromosomal LD, which supports the hypothesis that these genomic regions are associated with reductions in hybrid survival and reproductive isolation (Bradbury et al. 2014). These findings suggest that the differentiation islands are indeed associated with the ecological divergence between H. discus and H. madaka, but have not worked as stable barrier loci between the two species (cf., Han et al. 2017); temporal and/or spatial weakening of the ecologically based divergent selection has facilitated gene flow and prevented accumulation of fixed differences. Available evidence suggests that fluctuating selective pressures are a widespread phenomenon (e.g., Nosil 2012). All this said, the observed high FST, dXY, and dA characterizing differentiation between H. gigantea and H. discus or H. madaka suggest that the differentiation islands are likely to work as barrier loci on longer time scales.
In conclusion, our findings suggest that the Western Pacific abalones, H. discus, H. madaka, and H gigantea, are going through speciation-with-gene flow under ecological separation, which was initiated in allopatry. Speciation in the abalones has proceeded to different stages in different species pairs: Barriers to gene flow are incomplete among the younger divergence of H. discus and H. madaka but fairly complete in the case of older divergences of H. gigantea and other two species in which evolution of fertilization proteins has taken place. Hence, the Western Pacific abalones provide insight into the genomic processes taking place in the early phase of speciation continuum before coevolution of interacting fertilization proteins has started.
Materials and Methods
Samples
From 2000 to 2008, we collected abalone individuals from 15 sampling locations along the coastline of Japan (fig. 1B;supplementary table 5, Supplementary Material online). These samples were provided by local fishermen, and species identification was conducted by us. Hatiois discus is classified into two subspecies, H. discus hannai and H. discus discus on the basis of sampling locations (Ino 1952; see fig. 1A for their geographic distributions). This is because these two subspecies show only slight morphological difference in dorsal surface of the shell (Ino 1952), and hence, are difficult to distinguish based on morphological features. Therefore, we assigned H. discus populations into H. discus hannai, H. discus discus, and into boundary populations based on the geographic location of the sampling sites (Ino 1952). Tissue samples of the Red abalones (H. rufescens) from California were used as outgroups. For all species, DNA was extracted from foot muscles using the phenol/chloroform method with TNES-urea buffer (Asahida et al. 1996) or Gentra Puregene Tissue Kit (QIAGEN) and stored at −30 °C before analyses.
Mitogenome Sequencing and Analyses
Primer sets for PCR used for mitochondrial DNA sequencing were designed for 15 genes, including COI, COII, COIII, ND1, ND2, ND3, ND4, ND4L, ND5, ND6, ATP6, ATP8, Cyt b, 12SrRNA, and 16SrRNA (supplementary table 6, Supplementary Material online). PCR mixture (26 μl) contained 50 ng of template DNA, 2.5 μl of 10× Blend Taq-Plus- Buffer, 0.25 µl of Blend Taq-Plus- polymerase (0.65 units) (Toyobo), 2.5 µl of dNTPs Mixture (2 mM), and 1 μl of each primer (50 µM). Thermal cycling in PCR consisted of initial denaturation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min. The PCR products were purified with AMPure XP Kit (Beckman Coulter) and were bidirectionally sequenced with the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) in combination with an ABI 3730 capillary sequencer (Thermo Fisher Scientific) using the forward and reverse primers used for PCR.
Sequences were aligned using ClustalW in the MEGA 5 with default parameters. DNAcollapser of FaBox (Villesen 2007) was used to assign individuals to respective haplotypes. Obtained haplotype sequences were deposited to the EMBL/DDBJ/GenBank database. The length of the concatenated sequences (13 protein and 2 rRNA-coding genes) was trimmed to 7,837 base pairs due to the poor quality of sequences in some individuals. Neighbor-joining tree (Saitou and Nei 1987) was constructed in MEGA 5 (Tamura et al. 2011) using the Jukes–Cantor distance measure (Jukes and Cantor 1969), with an option of 95%-cutoff partial deletion. The statistical credibility of the resulting tree was evaluated with 1,000 bootstrap replications. We note that the Jukes–Cantor distance performs well in estimating phylogenetic relationships among closely related lineages (Nei and Kumar 2000; Hirase et al. 2016). Genetic differentiation among species was assessed by AMOVA (Excoffier et al. 1992) using Arlequin ver. 3.5 (Excoffier et al. 2005).
GRAS-Di Sequencing
The GRAS‐Di method, which was developed by Toyota Motor Corp. (Enoki and Takeuchi 2018), was used for genome-wide DNA sequencing. The patent for GRAS‐Di is currently available under the license agreement or contract‐based analysis services. This method generates a sequence library consisting of amplicons created by two rounds of PCR using random primers and sequence adapter sequences. The final PCR product is purified using either columns or magnetic beads without size selection, and then sequenced on an Illumina platform. This simple method allowed us to obtain the genotypes at several thousands of loci across the genome. Details of this method are given in Hosoya et al. (2019). All library constructions and sequencing were carried out at Eurofins Japan. The GRAS-Di reads were filtered by Trimmomatic ver. 0.38 (-phred33 ILLUMINACLIP TruSeq3-SE.fa:2:40:15 LEADING:10 TRAILING:10 SLIDINGWINDOW:4:20 MINLEN:32) to remove Illumina adapters and low-quality regions. The filtered reads were aligned to the H. discus hannai genome sequence (Nam et al. 2017) using the BWA-MEM algorithm in BWA ver. 0.7.8 (Li and Durbin 2009) with default settings, followed by local realignment around indels by GATK ver. 3.7 (McKenna et al. 2010) with default settings. After these alignment procedures, multimapped reads (indicated by “XA” and “SA” tags in their BAM records) were removed. Mapping statistics are shown in supplementary table 7, Supplementary Material online. SNPs were called using SAMtools ver. 1.9 mpileup and bcftools and filtered using VCFtools ver. 0.1.12 (--minQ 20 --remove-indels --maf 0.05 –max-alleles 2 --min-meanDP 15 –minDP 10) (Danecek et al. 2011). The MAF filtering option was omitted when we generated a vcf file for demographic modeling. To remove potential genotyping errors (e.g., due to allele dropout), SNP loci were removed if the proportion of missing genotypes was >10% at least in one of five genetic groups (H. discus hannai, boundary populations of H. discus hannai and H. discus discus, H. discus discus, H. madaka, and H. gigantea) that were pruned using pop_missing_filter.sh of dDocent ver. 2.2.7 package (Puritz et al. 2014). Finally, SNP loci that showed departure from the Hardy–Weinberg equilibrium (P < 0.001) in all genetic groups were removed using the script filter_hwe_by_pop.pl of the dDocent package. (Of note, no SNP locus was filtered with this criterion.)
Whole-Genome Sequencing
Genome sequencing was performed for the four abalone species (supplementary table 8, Supplementary Material online) using Illumina HiSeq X Ten with the 250-bp paired-end protocol. All library construction and sequencing were carried out at BGI, China. The paired-end reads were filtered by Trimmomatic to remove the adapter, Illumina-specific sequences and low-quality regions (-phred33 ILLUMINACLIP TruSeq3-SE.fa:2:40:15 LEADING:10 TRAILING:10 SLIDINGWINDOW:4:20 MINLEN:32). The filtered paired-end reads were aligned to the H. discus hannai genome sequence (Nam et al. 2017) using the BWA-MEM algorithm in BWA with default settings, followed by local realignment around indels by GATK ver. 3.7 with default settings. Multimapped reads were removed in the same manner as described above. Mapping statistics are shown in supplementary table 8, Supplementary Material online. SNPs were called using SAMtools mpileup (-Q 30) and bcftools, and filtered using VCFtools (--minQ 20 --remove-indels --maf 0.05 –max-alleles 2 –minDP 6 -max-missing-count 1).
Gene Prediction and Annotation of H. discus hannai genome
We downloaded RNA sequencing data for H. discus hannai individuals from the NCBI Sequence Read Archive (SRA, Leinonen et al. 2011): SRR1910554, SRR1943392, SRR1944101, SRR1944137, SRR1985199, SRR1985200, SRR1985201, SRR1987402, SRR1987403, SRR1987404, SRR2057965, SRR2057966, SRR2057969, SRR2058004, SRR2058005, SRR2058006, SRR2059857, SRR2059863, SRR2059864, SRR2059865, SRR5082552, SRR5082554, SRR5082555, SRR5083065, SRR5083066, SRR5083067, SRR6459616, SRR6459617, SRR6459618, SRR6459619, SRR7956997, SRR7956998, SRR7956999, SRR7957000, SRR7957001, SRR7957002, SRR7967467, SRR7967468, and SRR7967469. All RNA sequencing reads were aligned to the H. discus hannai genome sequence as single-end reads using HISAT2 ver. 2.1.0 (Kim et al. 2019). Stringtie ver. 1.3.5 (Pertea et al. 2015) was used to predict transcript structures based on the aligned reads. Third, we identified candidate coding regions within transcript sequences using TransDecoder ver. 5.5.0 (https://github.com/TransDecoder/TransDecoder, last accessed July 14, 2021). These analyses were performed with default set tings. An annotation file is available from Dryad (https://doi.org/10.5061/dryad.gf1vhhmq5).
Gene functions were annotated using BlastP searches (E-value cut-off: e-10) against the nr database of NCBI.
Phylogenomic Analyses of the Western Pacific and North American Abalones
To infer genetic relationships among the Western Pacific and North American abalones, we constructed a phylogenomic tree using our whole-genome sequencing data of the Western Pacific abalones and North American abalones. Whole-genome sequencing data of North American abalones, except for H. rufescens, were downloaded from the NCBI SRA (SRR7958743−SRR7958752; Masonbrink et al. 2019). The paired-end reads were filtered and aligned to the H. discus hannai genome sequence in the same manner as described above. SNPs were called using SAMtools mpileup (-Q 30) and bcftools and filtered using VCFtools (--minDP 10 --remove-indels --max-missing-count 0). A maximum likelihood (ML) tree was constructed using RAxML ver. 8.2.12 (Stamatakis 2014) with 1,000 bootstrap replicates using the GTRGAMMA model of nucleotide substitution with a correction for ascertainment bias (-m ASC_GTRGAMMA --asc-corr=stamatakis -f a -# 1000) (Leaché et al. 2015). Invariant sites were removed using ascbias.py (https://github.com/btmartin721/raxml_ascbias, last accessed July 14, 2021).
Population Genomics Analyses Based on GRAS-Di Data
Genetic diversity (observed heterozygosity) of each abalone species and population was estimated using GenoDive ver. 2.0b27 (Meirmans and Van Tienderen et al. 2004). LD between SNP loci was evaluated based on r2 values calculated using PLINK ver. 1.07 (Purcell et al. 2007). Site-FST values were calculated using VCFtools and those in monomorphic sites and under zero were converted to zero. To infer the genetic relationships among the Western Pacific abalones, we constructed an ML tree using RAxML as mentioned above. A clustering analysis was performed using ADMIXTURE (Alexander et al. 2009) assuming K from 1 to 10 with default parameters . To alleviate the effect of LD on the clustering analysis, we retained one SNP locus every 5,000 bp using “–thin 5,000” command in vcftools. Cross-validation scores were used to determine the most appropriate K (Alexander and Lange 2011). We analyzed genotypes of the Sado population of H. madaka using NewHybrids ver. 1.1. This analysis was performed using the genotypes of H. discus and H. madaka (except for the Sado population) as parental genotypes with 10,000 burnin iterations, and 50,000 iterations.
We used four outlier tests to search for signatures of selection between H. discus and H. madaka: BayeScan (Foll and Gaggiotti 2008), pcadapt (Luu et al. 2017), FLK (Bonhomme et al. 2010), and the coalescent method of Beaumont and Nichols (1996) as implemented in Arlequin (referred to as the Fdist method). For BayeScan analyses, we set prior odds to 10 to minimize chances of false positives and ran 5,000 pilot runs, followed by 100,000 iterations (5,000 samples, a thinning interval of 10, and a burn-in of 50,000) (FDR < 0.05). For pcadapt analyses, SNP loci that significantly contributed to the first principal component separating H. discus and H. madaka were retrieved (FDR < 0.01). For the Fdist analyses, we used the island model with 100 simulated demes and 20,000 coalescent simulations (P < 0.05). FLK analyses were performed with default settings (P < 0.05). Outliers identified consistently in all tests were considered as putative loci under selection. To annotate and predict the effects of SNPs, we used SnpEff (Cingolani et al. 2012) based on a custom database using a gff3 file of H. discus hannai genome.
Evolutionary history of Western Pacific abalone speciation process was investigated with ML-based demographic inference based on SFS using the program fastsimcoal2. The folded three-population multidimensional observed SFS from GRAS-Di SNP data was obtained with easySFS.py (https://github.com/isaacovercast/easySFS, last accessed July 14, 2021). As sample sizes varied among populations, we narrowed down to 30 chromosomes per population (i.e., 15 diploid individuals) to project the SFS. Five models of divergence were tested (fig. 3A), and for all them, priors were drawn from a uniform log distribution. Because there are no estimates of nuclear mutation rates for abalones, we applied a rate of 2.5 × 10−8 in humans (Nachman and Crowell 2000). For each model, we performed 100 independent runs of 100,000 coalescent simulations to obtain ML estimates of model parameters. Model selection was performed using the run with the highest likelihood for each of the models. Following Meier et al. (2017), AIC was used for model selection and assessing the likelihood distribution for each model by calculating the likelihoods of 100 SFS derived from 1,000,000 coalescent simulations. We derived 95% confidence intervals for parameter estimates from the models using nonparametric site bootstrapping where we resampled SNP loci with replacement. We created 100 bootstrapped SFSs and then performed ten independent runs of likelihood estimation on them. Estimates from each of the best runs were then used to derive the 95% confidence intervals around all parameter estimates from the focal model. Details of each model are available from Dryad (https://doi.org/10.5061/dryad.gf1vhhmq5).
Population Genomics Analyses Based on Whole-Genome Sequencing Data
Genome-wide genetic diversity and differentiation between abalones were investigated by calculating pi, FST, and dXY values for nonoverlapping 20-kb sliding windows using a python script of Martin et al. (2013) (https://github.com/simonhmartin/genomics_general, last accessed July 14, 2021). dA was calculated as dXY − (piX + piY)/2. To test for gene flow among abalones, we performed the ABBA–BABA test using popstats (D statistics were calculated as (BABA − ABBA)/(BABA + ABBA); Skoglund et al. 2015). A z-score with an absolute value of 3 or more was considered to be evidence of significant gene flow (Reich et al. 2009; Green et al. 2010). A test for interspecies gene flow based on fd-statistics, a modified version of ABBA–BABA statistics, in nonoverlapping 20-kb sliding windows was also performed using the python script of Martin et al. (2015).
Elongation of H. discus hannai scaffolds
To increase the contiguity of H. discus hannai genome (Nam et al. 2017), we used linkage information from one captive bred full-sib family of this species (family “F”) used earlier for microsatellite DNA-based linkage map construction (Sekino and Hara 2007b). The parents and their 96 full-sib offspring were genotyped for SNPs obtained with the GRAS-Di method; the obtained SNP loci were scaffolded with SELDLA ver. 2.0.9 (Yoshitake et al. 2018). The minimum depth in SNP filtering (DP) was set to 5, and the extended scaffolds were anchored into 18 LGs (36 diploid chromosomes) containing 167 microsatellite DNA markers (Sekino and Hara 2007b) with ALLMAPS (Tang et al. 2015). The genomic position of each microsatellite marker in the SELDLA-extended H. discus hannai reference genome was determined by the BlastN search. Dot plots used to explore homology were generated using minimap2 (Li 2018). Minimap2 results were converted to tab-delimited text using a portable pipeline software (https://github.com/c2997108/OpenPortablePipeline, last accessed July 14, 2021), and regions with continuous alignment of at least 1,000 bp were retained.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are indebted to T. Kobayashi, who provided us the parental and offspring samples of a full-sib family used for GRAS-Di analysis. We thank M. Ravinet and K. Yoshitake for helpful advice about genetic analyses. We are grateful to members of the Kikuchi Laboratory for helpful comments on this research. This work was supported by the Japan Society for the Promotion of Science (KAKENHI 21580240, 17K19280). Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Author Contributions
S.H., M.S., and K.K. designed the study. M.S. and M.H. provided tissue and DNA samples. S.H., M.N., M.I., and M.S. performed wet laboratory experiments and collected data. S.H. and Y.Y. performed genome analyses. S.H., J.M., and K.K. wrote the manuscript with the help of Y.Y., M.S., M.I., and M.H. All authors approved the final version of the manuscript.
Data Availability
All mitochondrial sequence data are available from DDBJ under accession numbers: LC613253–LC616030; the DDBJ accession numbers of the data of the whole-genome sequencing (DRR276780–DRR276816), GRAS-Di sequencing (DRR276817–DRR277014). Other data for analysis are available on Dryad (https://doi.org/10.5061/dryad.gf1vhhmq5).
References
- Ahmed F, Koike Y, Strüssmann CA, Yamasaki I, Yokota M, Watanabe S.. 2008. Genetic characterization and gonad development of artificially produced interspecific hybrids of the abalones, Haliotis discus discus Reeve, Haliotis gigantea Gmelin and Haliotis madaka Habe. Aquac Res. 39(5):532–541. [Google Scholar]
- Akihito, Fumihito A, Ikeda Y, Aizawa M, Makino T, Umehara Y, Kai Y, Nishimoto Y, Hasegawa M, Nakabo T, et al. 2008. Evolution of Pacific Ocean and the Sea of Japan populations of the gobiid species, Pterogobius elapoides and Pterogobius zonoleucus, based on molecular and morphological analyses. Gene 427(1–2):7–18. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Lange K.. 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H-S, Jee Y-J, Min K-S, Kim B-L, Han S-J.. 2005. Phylogenetic analysis of six species of Pacific abalone (Haliotidae) based on DNA sequences of 16s rRNA and cytochrome c oxidase subunit I mitochondrial genes. Mar Biotechnol (NY). 7(4):373–380. [DOI] [PubMed] [Google Scholar]
- Anderson E, Thompson E.. 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160(3):1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahida T, Kobayashi T, Saitoh K, Nakayama I.. 1996. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish Sci. 62(5):727–730. [Google Scholar]
- Babcock R, Keesing J.. 1999. Fertilization biology of the abalone Haliotis laevigata: laboratory and field studies. Can J Fish Aquat Sci. 56(9):1668–1678. [Google Scholar]
- Beaumont MA, Nichols RA.. 1996. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B Biol Sci. 263(1377):1619–1626. [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh Y-P, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, et al. 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5(11):e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester-van der Merwe AE, D'Amato ME, Swart BL, Roodt-Wilding R.. 2012. Molecular phylogeny of South African abalone, its origin and evolution as revealed by two genes. Mar Biol Res. 8(8):727–736. [Google Scholar]
- Bierne N, Bonhomme F, David P.. 2003. Habitat preference and the marine-speciation paradox. Proc R Soc Lond B Biol B. 270(1522):1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme M, Chevalet C, Servin B, Boitard S, Abdallah J, Blott S, SanCristobal M.. 2010. Detecting selection in population trees: the Lewontin and Krakauer test extended. Genetics 186(1):241–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury IR, Bowman S, Borza T, Snelgrove PV, Hutchings JA, Berg PR, Rodríguez-Ezpeleta N, Lighten J, Ruzzante DE, Taggart C. 2014. Long distance linkage disequilibrium and limited hybridization suggest cryptic speciation in Atlantic cod. PLoS One 9:e106380. [DOI] [PMC free article] [PubMed]
- Burri R. 2017. Interpreting differentiation landscapes in the light of long‐term linked selection. Evol Lett. 1(3):118–131. [Google Scholar]
- Chinzei K. 1986. Opening of the Japan Sea and marine biogeography during the Miocene. J Geomagn Geoelec. 38(5):487–494. [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ.. 2009. Coevolution of interacting fertilization proteins. PLoS Genet. 5(7):e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW.. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 23(13):3133–3157. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit P, Palumbi SR.. 2013. Transcriptome‐wide polymorphisms of red abalone (Haliotis rufescens) reveal patterns of gene flow and local adaptation. Mol Ecol. 22(11):2884. [DOI] [PubMed] [Google Scholar]
- Enoki, H., Takeuchi, Y, editors. 2018. New genotyping technology, GRAS-Di, using next generation sequencer. Proceedings of the Plant and Animal Genome Conference XXVI. San Diego, CA. Available from: https://pag.confex.com/pag/xxvi/meetingapp.cgi/Paper/29067.
- Ertl NG, O'Connor WA, Elizur A.. 2019. Molecular effects of a variable environment on Sydney rock oysters, Saccostrea glomerata: thermal and low salinity stress, and their synergistic effect. Mar Genomics. 43:19–32. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M.. 2013. Robust demographic inference from genomic and SNP data. PLoS Genet. 9(10):e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S.. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 1:117693430500100–117693430500150. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM.. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131(2):479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Egan SP, Nosil P.. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28(7):342–350. [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O.. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180(2):977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo BE, Vacquier VD, Swanson WJ.. 2003. Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci U S A. 100(8):4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan HM, Tan MH, Austin CM, Sherman C, Wong YT, Strugnell J, Gervis M, McPherson L, Miller A.. 2019. Best foot forward: nanopore long reads, hybrid meta-assembly and haplotig purging optimises the first genome assembly for the Southern Hemisphere blacklip abalone (Haliotis rubra). Front Genet. 10:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger DL, Groves LT.. 1999. Review of fossil abalone (Gastropoda: Vetigastropoda: Haliotidae) with comparison to recent species. J Paleontol. 73(5):872–885. [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Lamichhaney S, Grant BR, Grant PR, Andersson L, Webster MT.. 2017. Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin's finches. Genome Res. 27(6):1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Fujio Y.. 1992. Geographic distribution of isozyme genes in natural abalone. Bull Tohoku Natl Fish Res Inst. 54:115–124. [Google Scholar]
- Hara M, Sekino M.. 2005. Genetic difference between Ezo-awabi Haliotis discus hannai and Kuro-awabi H. discus discus populations: microsatellite-based population analysis in Japanese abalone. Fish Sci. 71(4):754–766. [Google Scholar]
- Hirase S, Ikeda M.. 2014. Divergence of mitochondrial DNA lineage of the rocky intertidal goby Chaenogobius gulosus around the Japanese Archipelago: reference to multiple Pleistocene isolation events in the Sea of Japan. Mar Biol. 161(3):565–574. [Google Scholar]
- Hirase S, Ikeda M, Kanno M, Kijima A.. 2012. Phylogeography of the intertidal goby Chaenogobius annularis associated with paleoenvironmental changes around the Japanese Archipelago. Mar Ecol Prog Ser. 450:167–179. [Google Scholar]
- Hirase S, Takeshima H, Nishida M, Iwasaki W.. 2016. Parallel mitogenome sequencing alleviates random rooting effect in phylogeography. Genome Biol Evol. 8(4):1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya S, Hirase S, Kikuchi K, Nanjo K, Nakamura Y, Kohno H, Sano M.. 2019. Random PCR‐based genotyping by sequencing technology GRAS‐Di (genotyping by random amplicon sequencing, direct) reveals genetic structure of mangrove fishes. Mol Ecol Resour. 19(5):1153–1163. [DOI] [PubMed] [Google Scholar]
- Ino T. 1952. Biological study on the propagation of Japanese abalone (genus Haliotis). Bull Tokai Reg Fish Res Lab. 5:1–102. [Google Scholar]
- Itami K, Takeda R, Sakai T, Shimamoto N.. 1978. Distribution and growth of the young abalone, Nordotis gigantea (GMELIN), from south coast of Awaji Island in Hyogo Prefecture. Bull Hyogo Pref Fish Exp Stn. 18:15–28. [Google Scholar]
- Jones MR, Mills LS, Alves PC, Callahan CM, Alves JM, Lafferty DJ, Jiggins FM, Jensen JD, Melo-Ferreira J, Good JM.. 2018. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360(6395):1355–1358. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR.. 1969. Evolution of protein molecules. In: Munro RE, editor. Mammalian protein metabolism. New York: Academic Press, ; pp. 21–132. [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Kimoto K.. 2006. History of the inflow of the warm Tsushima Current into the Sea of Japan between 3.5 and 0.8 Ma. Palaeogeogr Palaeoclimatol Palaeoecol. 236(3–4):355–366. [Google Scholar]
- Kitamura A, Takano O, Takata H, Omote H.. 2001. Late Pliocene-early Pleistocene paleoceanographic evolution of the Sea of Japan. Palaeogeogr Palaeoclimatol Palaeoecol. 172(1–2):81–98. [Google Scholar]
- Koike Y, Sun Z-X, Takashima F.. 1988. On the feeding and growth of juvenile hybrid abalones. Aquac Sci. 36(3):231–235. [Google Scholar]
- Kojima S, Hayashi I, Kim D, Iijima A, Furota T.. 2004. Phylogeography of an intertidal direct-developing gastropod Batillaria cumingi around the Japanese Islands. Mar Ecol Prog Ser. 276:161–172. [Google Scholar]
- Kojima S, Segawa R, Hayashi I.. 1997. Genetic differentiation among populations of the Japanese turban shell Turbo (Batillus) cornutus corresponding to warm currents. Mar Ecol Prog Ser. 150:149–155. [Google Scholar]
- Kokita T, Nohara K.. 2011. Phylogeography and historical demography of the anadromous fish Leucopsarion petersii in relation to geological history and oceanography around the Japanese Archipelago. Mol Ecol. 20(1):143–164. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Banbury BL, Felsenstein J, De Oca AN-M, Stamatakis A.. 2015. Short tree, long tree, right tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies. Syst Biol. 64(6):1032–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-H, Vacquier V.. 1995. Evolution and systematics in Haliotidae (Mollusca: Gastropoda): inferences from DNA sequences of sperm lysin. Mar Biol. 124(2):267–278. [Google Scholar]
- Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration. 2011. The sequence read archive. Nucleic Acids Res. 39(Database issue):D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios HA. 2007. Reproductive isolation between species of sea urchins. Bull Mar Sci. 81(2):191–208. [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu K, Bazin E, Blum MG.. 2017. pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol Ecol Resour. 17(1):67–77. [DOI] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD.. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol Biol Evol. 32(1):244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masonbrink RE, Purcell CM, Boles SE, Whitehead A, Hyde JR, Seetharam AS, Severin AJ.. 2019. An annotated genome for Haliotis rufescens (red abalone) and resequenced green, pink, pinto, black, and white abalone species. Genome Biol Evol. 11(2):431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JI, Sousa VC, Marques DA, Selz OM, Wagner CE, Excoffier L, Seehausen O.. 2017. Demographic modelling with whole‐genome data reveals parallel origin of similar Pundamilia cichlid species after hybridization. Mol Ecol. 26(1):123–141. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Van Tienderen PH.. 2004. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 4(4):792–794. [Google Scholar]
- Miglietta MP, Faucci A, Santini F.. 2011. Speciation in the sea: overview of the symposium and discussion of future directions. Integr Comp Biol. 51(3):449–455. [DOI] [PubMed] [Google Scholar]
- Momigliano P, Jokinen H, Fraimout A, Florin A-B, Norkko A, Merilä J.. 2017. Extraordinarily rapid speciation in a marine fish. Proc Natl Acad Sci U S A. 114(23):6074–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL.. 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156(1):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam B-H, Kim H, Seol D, Kim H, Noh ES, Kim EM, Noh JK, Kim Y-O, Park JY, Kwak W.. 2021. Genotyping-by-Sequencing of the regional Pacific abalone (Haliotis discus) genomes reveals population structures and patterns of gene flow. PLoS One 16(4):e0247815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam B-H, Kwak W, Kim Y-O, Kim D-G, Kong HJ, Kim W-J, Kang J-H, Park JY, An CM, Moon J-Y, et al. 2017. Genome sequence of pacific abalone (Haliotis discus hannai): the first draft genome in family Haliotidae. Gigascience 6(5):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S.. 2000. Molecular evolution and phylogenetics. Oxford: Oxford University Press. [Google Scholar]
- Nei M, Li W-H.. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 76(10):5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P. 2012. Ecological speciation. Oxford: Oxford University Press. [Google Scholar]
- Nowland SJ, Silva CN, Southgate PC, Strugnell JM.. 2019. Mitochondrial and nuclear genetic analyses of the tropical black-lip rock oyster (Saccostrea echinata) reveals population subdivision and informs sustainable aquaculture development. BMC Genomics 20(1):711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oziolor EM, Reid NM, Yair S, Lee KM, VerPloeg SG, Bruns PC, Shaw JR, Whitehead A, Matson CW.. 2019. Adaptive introgression enables evolutionary rescue from extreme environmental pollution. Science 364(6439):455–457. [DOI] [PubMed] [Google Scholar]
- Palumbi S. 2009. Speciation and the evolution of gamete recognition genes: pattern and process. Heredity (Edinb) 102(1):66–76. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. 1992. Marine speciation on a small planet. Trends Ecol Evol. 7(4):114–118. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson GH. 2016. Studying the genetic basis of speciation in high gene flow marine invertebrates. Curr Zool. 62(6):643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla O. 2009. Ecological speciation in marine vs. freshwater fishes. J Fish Biol. 75(5):960–996. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puritz JB, Hollenbeck CM, Gold JR.. 2014. dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ. 2:e431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puritz JB, Keever CC, Addison JA, Byrne M, Hart MW, Grosberg RK, Toonen RJ.. 2012. Extraordinarily rapid life-history divergence between Cryptasterina sea star species. Proc R Soc Lond B Biol Sci. 279(1744):3914–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravinet M, Faria R, Butlin R, Galindo J, Bierne N, Rafajlović M, Noor M, Mehlig B, Westram A.. 2017. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J Evol Biol. 30(8):1450–1477. [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 461(7263):489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode C, Vervalle J, Bester-van der Merwe AE, Roodt-Wilding R.. 2013. Detection of molecular signatures of selection at microsatellite loci in the South African abalone (Haliotis midae) using a population genomic approach. Mar Genomics. 10:27–36. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Nosil P.. 2005. Ecological speciation. Ecol Lett. 8(3):336–352. [Google Scholar]
- Saitou N, Nei M.. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425. [DOI] [PubMed] [Google Scholar]
- Sakai T, Shimamoto N.. 1978. Distribution of abalones from the coastal waters of Awaji Island in Hyogo Prefecture. Bull Hyogo Pref Fish Exp Stn. 18:11–14. [Google Scholar]
- Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, Peichel CL, Saetre G-P, Bank C, Brännström A, et al. 2014. Genomics and the origin of species. Nat Rev Genet. 15(3):176–192. [DOI] [PubMed] [Google Scholar]
- Sekino M, Hara M.. 2007a. Individual assignment tests proved genetic boundaries in a species complex of Pacific abalone (genus Haliotis). Conserv Genet. 8(4):823–841. [Google Scholar]
- Sekino M, Hara M.. 2007b. Linkage maps for the Pacific abalone (genus Haliotis) based on microsatellite DNA markers. Genetics 175(2):945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Mallick S, Bortolini MC, Chennagiri N, Hünemeier T, Petzl-Erler ML, Salzano FM, Patterson N, Reich D.. 2015. Genetic evidence for two founding populations of the Americas. Nature 525(7567):104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed]
- Streit K, Geiger DL, Lieb B.. 2006. Molecular phylogeny and the geographic origin of Haliotidae traced by haemocyanin sequences. J Molluscan Stud. 72(1):105–110. [Google Scholar]
- Tada R. 1994. Paleoceanographic evolution of the Japan Sea. Palaeogeogr Palaeoclimatol Palaeoecol. 108(3–4):487–508. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Zhang X, Miao C, Zhang J, Ming R, Schnable JC, Schnable PS, Lyons E, Lu J.. 2015. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 16(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorn GS, Luttikhuizen PC, Weissing FJ.. 2001. Sexual selection at the protein level drives the extraordinary divergence of sex–related genes during sympatric speciation. Proc R Soc Lond B Biol B. 268(1481):2155–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P. 2007. FaBox: an online toolbox for fasta sequences. Mol Ecol Notes. 7(6):965–968. [Google Scholar]
- Wang X, Wang S, Li C, Chen K, Qin JG, Chen L, Li E.. 2015. Molecular pathway and gene responses of the pacific white shrimp Litopenaeus vannamei to acute low salinity stress. J Shellfish Res. 34(3):1037–1048. [Google Scholar]
- Wu CI. 2001. The genic view of the process of speciation. J Evol Biol. 14(6):851–865. [Google Scholar]
- Yapici N, Kim Y-J, Ribeiro C, Dickson BJ.. 2008. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451(7174):33–37. [DOI] [PubMed] [Google Scholar]
- Yoshitake K, Igarashi Y, Mizukoshi M, Kinoshita S, Mitsuyama S, Suzuki Y, Saito K, Watabe S, Asakawa S.. 2018. Artificially designed hybrids facilitate efficient generation of high-resolution linkage maps. Sci Rep. 8(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Xu D, Ye H, Yue H, Ooka S, Kondo H, Yazawa R, Takeuchi Y.. 2018. Gonadal transcriptome analysis of pacific abalone Haliotis discus discus: identification of genes involved in germ cell development. Mar Biotechnol (NY). 20(4):467–480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mitochondrial sequence data are available from DDBJ under accession numbers: LC613253–LC616030; the DDBJ accession numbers of the data of the whole-genome sequencing (DRR276780–DRR276816), GRAS-Di sequencing (DRR276817–DRR277014). Other data for analysis are available on Dryad (https://doi.org/10.5061/dryad.gf1vhhmq5).