Abstract
Although rapid evolution of pericentromeric satellite DNA repeats is theorized to promote hybrid incompatibility (HI) (Yunis and Yasmineh 1971; Henikoff et al. 2001; Ferree and Barbash 2009; Sawamura 2012; Jagannathan and Yamashita 2017), how divergent repeats affect hybrid cells remains poorly understood. Recently, we demonstrated that sequence-specific DNA-binding proteins cluster satellite DNA from multiple chromosomes into “chromocenters,” thereby bundling chromosomes to maintain the entire genome in a single nucleus (Jagannathan et al. 2018, 2019). Here, we show that ineffective clustering of divergent satellite DNA in the cells of Drosophila hybrids results in chromocenter disruption, associated micronuclei formation, and tissue atrophy. We further demonstrate that previously identified HI factors trigger chromocenter disruption and micronuclei in hybrids, linking their function to a conserved cellular process. Together, we propose a unifying framework that explains how the widely observed satellite DNA divergence between closely related species can cause reproductive isolation.
Keywords: satellite DNA, chromocenter, hybrid incompatibility, speciation
Drosophila Hybrids Exhibit Chromocenter Disruption and Micronuclei Formation
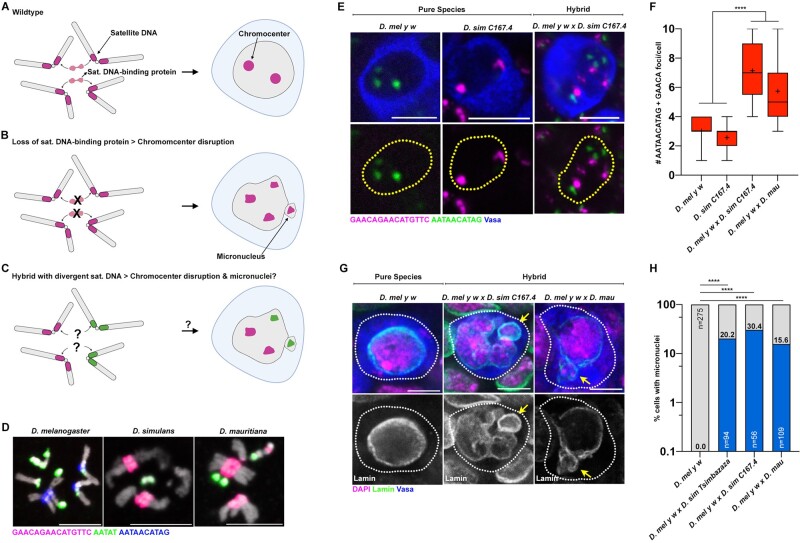
The majority of repetitive satellite DNA are present at heterochromatic regions adjacent to the centromere on eukaryotic chromosomes. Our previous work showed the importance of these pericentromeric satellite DNA in encapsulating the full complement of chromosomes into a single nucleus (Jagannathan et al. 2018, 2019). Sequence-specific DNA-binding proteins cluster their cognate pericentromeric satellite DNA repeats to create physical links between heterologous chromosomes, forming a cytological structure known as chromocenter (fig. 1A). Depletion of satellite DNA-binding proteins led to the detachment of heterologous chromosomes from one another (chromocenter disruption), their subsequent loss from the primary nucleus (micronuclei) and cell death (fig. 1B). Together, we demonstrated how identical satellite DNA repeats on multiple chromosomes are important for chromocenter formation and maintenance of the entire genome in the nucleus. This prompted us to hypothesize that the highly divergent satellite DNA repeats between closely related species may fail to form chromocenters properly, ultimately resulting in hybrid incompatibility (fig. 1C). Drosophila melanogaster diverged ∼2–3 Ma from the Drosophila simulans species complex (e.g., D. simulans and D. mauritiana) (Lachaise et al. 2004) but these species still maintain near complete synteny (Sturtevant and Novitski 1941; Drosophila 12 Genomes Consortium et al. 2007; Chakraborty et al. 2021). However, the satellite DNA content of D. melanogaster is markedly different from that of the D. simulans species complex (fig. 1D) (Lohe and Brutlag 1987; Jagannathan et al. 2017). This satellite DNA divergence takes the form of changes in satellite DNA abundance, changes in chromosomal location of identical satellite DNA repeats and the presence/absence of novel repeat sequences (Jagannathan et al. 2017). Most strikingly, the (AATAACATAG)n satellite DNA whose clustering into chromocenters in D. melanogaster is required for viability (Jagannathan et al. 2019), is completely absent in the D. simulans species complex (fig. 1D). Rather, these species contain an unrelated repeat (GAACAGAACATGTTC)n at the corresponding autosomal locations (fig. 1D) (Lohe and Brutlag 1987; Jagannathan et al. 2017). In addition, the (AATAT)n satellite DNA repeat, whose clustering is important for D. melanogaster fertility, exhibits differential abundance and chromosomal locations between these species (fig. 1D) (Lohe and Brutlag 1987; Jagannathan et al. 2017).
Fig. 1.
Chromocenter disruption and micronuclei in the larval germ cells of D. melanogaster–D. simulans/D. mauritiana hybrids. (A–C) A model of chromocenter formation (A) and function (B) in pure species and proposed dysfunction (C) in hybrids. (D) FISH against the (AATAACATAG)n satellite (blue), the (AATAT)n satellite (green), and the (GAACAGAACATGTTC)n satellite (magenta) on larval neuroblast mitotic chromosomes from the indicated species and costained with DAPI (gray). (E) FISH against the (AATAACATAG)n satellite (green) and the (GAACAGAACATGTTC)n satellite (magenta) in the larval female germ cells from the indicated species and hybrids and costained with Vasa (blue). (F) Box-and-whisker plot of the total number of (AATAACATAG)n and (GAACAGAACATGTTC)n chromocenter foci per larval germ cell from D. mel y w females (n = 38), D. sim C167.4 females (n = 28), D. mel y w×C167.4 hybrid females (n = 21), and D. mel y w×mau hybrid females (n = 42). n indicates the number of germ cells analyzed, **** represents P < 0.0001 based on Tukey’s multiple comparisons test from an ordinary one-way ANOVA and crosshairs mark the mean. (G) IF against Lamin (green) and Vasa (blue) in the larval female germ cells from the indicated species and hybrids and costained with DAPI (magenta). Arrows indicate micronuclei. (H) Quantification of micronuclei-containing cells in the larval female germ cells from the indicated species and hybrids. The percentage of micronuclei-containing cells is indicated above the respective columns. n indicates the number of germ cells analyzed, **** represents P < 0.0001 from Fisher’s exact test. All scale bars are 5 µm, yellow dashed lines demarcate nuclear boundary, and white dashed lines indicate cell boundary.
The cross between D. melanogaster females and D. simulans/D. mauritiana males yields lethal hybrid males and sterile hybrid females (Sturtevant 1920) (supplementary fig. S1A, Supplementary Material online). Although previous reports have indicated that adult hybrid ovaries are nearly entirely devoid of germ cells (Hollocher et al. 2000; Matute et al. 2014) (supplementary fig. S1B, Supplementary Material online), we found that larval L3 ovaries in hybrids raised at 18 °C contained a few surviving Vasa+ germ cells (supplementary fig. S1C and D, Supplementary Material online). We first examined chromocenter formation in female hybrid germ cells using DNA FISH probes against the (AATAT)n repeat (present in both species), the D. melanogaster-specific (AATAACATAG)n repeat, and the D. simulans species complex-specific (GAACAGAACATGTTC)n repeat.
In the larval ovarian germ cells of pure species, both the D. melanogaster-specific (AATAACATAG)n satellite DNA and the D. simulans species complex-specific (GAACAGAACATGTTC)n satellite DNA, were clustered into approximately two to three foci (fig. 1E and F). In contrast, the larval germ cells of female D. melanogaster–D. simulans and D. melanogaster–D. mauritiana hybrids exhibited increased number of foci, indicating a striking declustering of these satellite DNA repeats (fig. 1E and F; supplementary fig. S2A, Supplementary Material online). We also observed that hybrid germ cells exhibited declustering of (AATAT)n satellite DNA in comparison to germ cells from pure species (supplementary fig. S2B, Supplementary Material online).
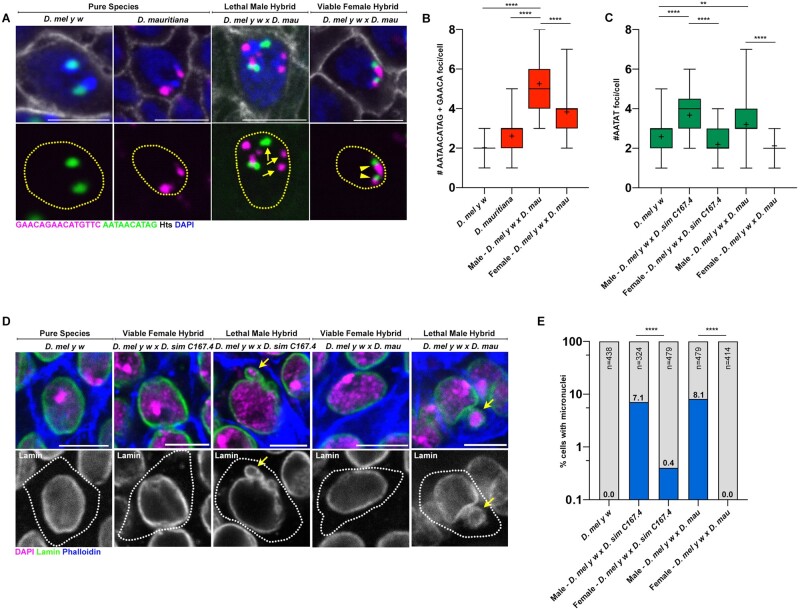
Male hybrids arising from the above cross (supplementary fig. S1A, Supplementary Material online) die during larval development due to the atrophy of the imaginal discs, which are tissues that are fated to form critical adult structures (Sanchez and Dübendorfer 1983; Orr et al. 1997) (supplementary table S1, Supplementary Material online). Similar to female hybrid germ cells, we observed defective chromocenter formation in hybrid male imaginal discs in comparison to the pure species controls as indicated by the increased number of satellite DNA foci (fig. 2A–C, arrows). It is suggested that karyotype and gene dosage differences between male and female hybrids (discussed in the next section) lead to female hybrids that are almost completely viable, especially at lower temperatures (Barbash et al. 2000). Interestingly, chromocenter disruption was not observed in the imaginal discs of viable female hybrids: D. melanogaster-specific (AATAACATAG)n and D. simulans-specific (GAACAGAACATGTTC)n satellite DNA were typically associated with each other, revealing intact chromocenter formation, even though these two satellite DNA repeats never coexist in the cells of pure species (fig. 2A–C, arrowheads).
Fig. 2.
Chromocenter disruption and micronuclei in the larval imaginal discs of lethal male D. melanogaster–D. simulans/D. mauritiana hybrids. (A) FISH against the (AATAACATAG)n satellite (green) and the (GAACAGAACATGTTC)n satellite (magenta) on larval imaginal discs from the indicated species and hybrids and costained with DAPI (blue) and Hts (gray). (B) Box-and-whisker plot of total number of (AATAACATAG)n and (GAACAGAACATGTTC)n foci per cell from D. mel y w (n = 46), D. mau (n = 64), and male (n = 43) and female (n = 60) D. mel y w×D. mau hybrids. n indicates the number of imaginal disc cells analyzed, **** represents P < 0.0001 based on Tukey’s multiple comparisons test from an ordinary one-way ANOVA and crosshairs mark the mean. (C) Box-and-whisker plot of total number of (AATAT)n foci per cell from D. mel y w (n = 48), male (n = 37) and female (n = 51) D. mel y w×D. sim C167.4 hybrids, and male (n = 39) and female (n = 41) D. mel y w×D. mau hybrids. n indicates the number of imaginal disc cells analyzed, ** represents P = 0.0049, and **** represents P < 0.0001 based on Tukey’s multiple comparisons test from an ordinary one-way ANOVA and crosshairs mark the mean. (D) IF against Lamin (green) in larval imaginal discs from the indicated species and hybrids and costained with DAPI (magenta) and phalloidin (blue). (E) Quantification of micronuclei-containing cells in the larval imaginal discs from the indicated species and hybrids. The percentage of micronuclei-containing cells is indicated above the respective columns. n indicates the number of imaginal disc cells analyzed and **** represents P < 0.0001 from Fisher’s exact test. All scale bars are 5 µm, yellow dashed lines demarcate nuclear boundary, and white dashed lines indicate cell boundary.
Consistent with our previous reports that chromocenter disruption leads to micronuclei formation in D. melanogaster, we observed micronuclei in both the hybrid female germ cells (fig. 1G and H) as well as hybrid male imaginal discs (fig. 2D and E). In contrast, micronuclei were not observed in the intact imaginal discs of viable female hybrids (fig. 2D and E). Taken together, our data suggest that cells from atrophied tissues in hybrid animals exhibit defective chromocenter formation and micronuclei. Moreover, these cytological defects are highly correlated with cellular lethality in hybrid animals.
Removal of Hybrid Incompatibility Genes Rescues Chromocenter Formation
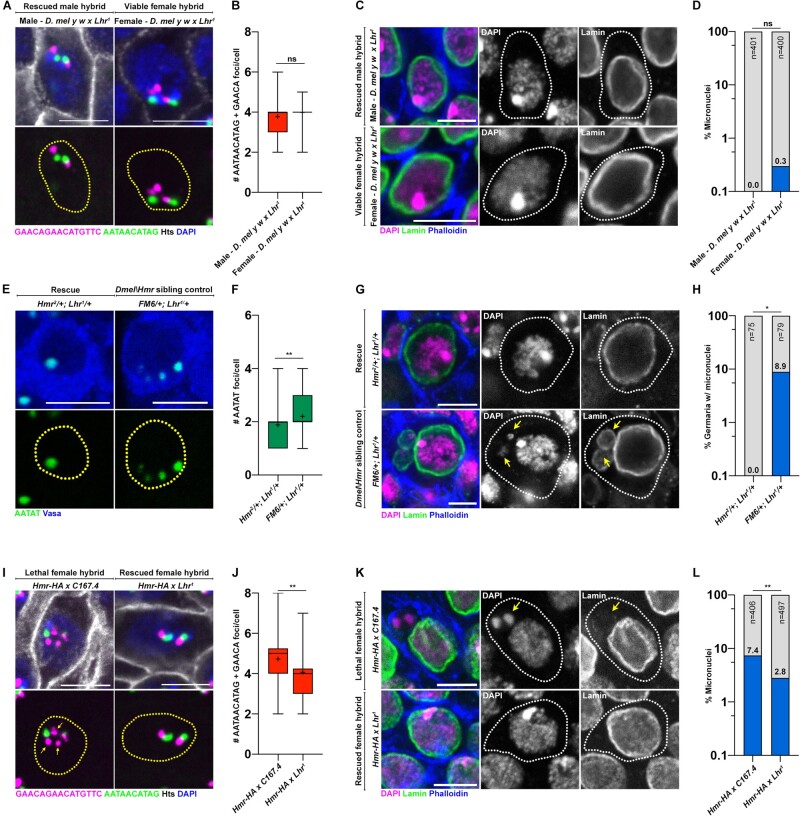
Prior work has identified D. melanogaster Hmr (Hutter and Ashburner 1987; Barbash et al. 2003), D. simulans Lhr (Watanabe 1979), and D. simulans Gfzf (Phadnis et al. 2015) as genes causing lethality in D. melanogaster–D. simulans hybrids. Lethality is primarily observed in male hybrids due to the presence of the Hmr HI gene on the D. melanogaster X chromosome; male hybrids are hemizygous for Hmrmel (Hmrmel/Y) whereas female hybrids are heterozygous for Hmr (Hmrmel/Hmrsim). As such, the D. simulans Hmr ortholog (which is not a HI factor) or the D. simulans X chromosome more generally is thought to protect female hybrids from lethality through an uncharacterized mechanism (Barbash et al. 2000). In lethal male hybrids, the three HI genes (Hmrmel, Lhrsim, and Gfzfsim) are considered to act dominantly and the removal of even one HI gene restores the viability of male hybrids (Barbash et al. 2000; Barbash 2010; Phadnis et al. 2015). Interestingly, Hmr and Lhr also localize to repetitive DNA and chromocenters in the pure species context and have been demonstrated to interact with one another (Brideau et al. 2006; Satyaki et al. 2014; Blum et al. 2017; Kochanova et al. 2020). Strikingly, we observed significant rescue of chromocenter disruption (fig. 3A and B compare with fig. 2A and B) and micronuclei formation (fig. 3C and D compare with fig. 2D and E) in the imaginal discs of hybrid males, whose viability was restored by mutating D. simulans Lhr (supplementary table S1, Supplementary Material online), suggesting that Lhrsim inhibits chromocenter formation in inviable hybrids. We next used a model of hybrid sterility rescue, where crossing D. melanogaster In(1)AB, Hmr2/FM6 females to D. simulans Lhr1 males restores ovary development in female hybrids (Barbash and Ashburner 2003) (supplementary fig. S3A–C, Supplementary Material online). We found that early germ cells of rescued female hybrids (In(1)AB, Hmr2/+; Lhr1/+) exhibited intact chromocenters (fig. 3E and F; supplementary fig. S3D and E, Supplementary Material online) and did not form micronuclei (fig. 3G and H). These results suggested that the incompatibility caused by the HI genes (Hmr and Lhr) is strongly correlated with chromocenter disruption. In contrast, germ cells of nonrescued sibling controls (FM6/+; Lhr1/+) containing Hmr (supplementary fig. S3A–C, Supplementary Material online) exhibited increased declustering of the (AATAT)n satellite DNA (fig. 3E and F; supplementary fig. S3D and E, Supplementary Material online). Consistent with previous results, chromocenter disruption in the early germ cells of nonrescued hybrid ovaries was accompanied by micronuclei formation (fig. 3G and H).
Fig. 3.
HI factors regulate chromocenter disruption and micronuclei in sterile and lethal hybrids. (A) FISH against the (AATAACATAG)n satellite (green) and the (GAACAGAACATGTTC)n satellite (magenta) on larval imaginal discs from the indicated hybrids and costained with DAPI (blue) and Hts (gray). (B) Box-and-whisker plot of total number of (AATAACATAG)n and (GAACAGAACATGTTC)n foci per cell from male (n = 63) and female (n = 63) D. mel y w×Lhr1 hybrids. n indicates the number of imaginal disc cells analyzed, ns represents P = 0.16 from a Student’s t-test and crosshairs mark the mean. (C) IF against Lamin (green) in larval imaginal discs from the indicated hybrids and costained with DAPI (magenta) and phalloidin (blue). (D) Quantification of micronuclei-containing cells in the larval imaginal discs from male and female D. mel y w×Lhr1 hybrids. The percentage of micronuclei-containing cells is indicated above the respective columns. n indicates the number of imaginal disc cells analyzed, ns represents P > 0.9999 from Fisher’s exact test. (E) FISH against the (AATAT)n satellite (green) in the early germ cells of the indicated 0- to 2-day-old adult female hybrids. (F) Box-and-whisker plot of total number of (AATAT)n foci per cell from In(1)AB, Hmr2/+; Lhr1/+ (n = 76) and FM6/+; Lhr1/+ germ cells (n = 73). n indicates the number of germ cells analyzed, ** represents P = 0.0074 from a Student’s t-test and crosshairs mark the mean. (G) IF against Lamin (green) in early germ cells from the indicated adult 0- to 2-day-old female hybrids and costained with DAPI (magenta) and phalloidin (blue). Arrows point to micronuclei. (H) Quantification of micronuclei-containing cells in the early germ cells from the indicated adult 0- to 2-day-old female hybrids. The percentage of micronuclei-containing cells is indicated above the respective columns. n indicates the number of germ cells analyzed, * represents P = 0.013 from Fisher’s exact test. (I) FISH against the (AATAACATAG)n satellite (green) and the (GAACAGAACATGTTC)n satellite (magenta) on larval imaginal discs from the indicated hybrids and costained with DAPI (blue) and Hts (gray). Arrows point to declustered satellite DNA. (J) Box-and-whisker plot of total number of (AATAACATAG)n and (GAACAGAACATGTTC)n foci per cell from Hmr-HA×C167.4 (n = 58) and Hmr-HA×Lhr1 (n = 62) female hybrids raised at 29 °C. n indicates the number of imaginal disc cells analyzed, ** represents P = 0.0022 from a Student’s t-test and crosshairs mark the mean. (K) IF against Lamin (green) in larval imaginal discs from the indicated female hybrids raised at 29 °C and costained with DAPI (magenta) and phalloidin (blue). Arrows point to micronuclei. (L) Quantification of micronuclei-containing cells in larval imaginal discs from the indicated female hybrids. The percentage of micronuclei-containing cells is indicated above the respective columns. n indicates the number of imaginal disc cells analyzed, ** represents P = 0.0017 from Fisher’s exact test. All scale bars are 5 µm, yellow dashed lines demarcate nuclear boundary, and white dashed lines indicate cell boundary.
Finally, we found that female hybrids containing an extra copy of D. melanogaster Hmr raised at 29 °C, which are known to be inviable (Barbash et al. 2000) (supplementary table S2, Supplementary Material online), exhibited chromocenter disruption (fig. 3I and J) and micronuclei formation in the imaginal discs (fig. 3K and L). Furthermore, these defects were rescued in female hybrids with the D. simulans Lhr1 mutation (fig. 3I–L and supplementary table S2, Supplementary Material online). Taken together, our data suggest that HI genes trigger chromocenter disruption and micronuclei formation while causing hybrid incompatibility.
The D1 Satellite DNA-Binding Protein Is Functionally Diverged between Species
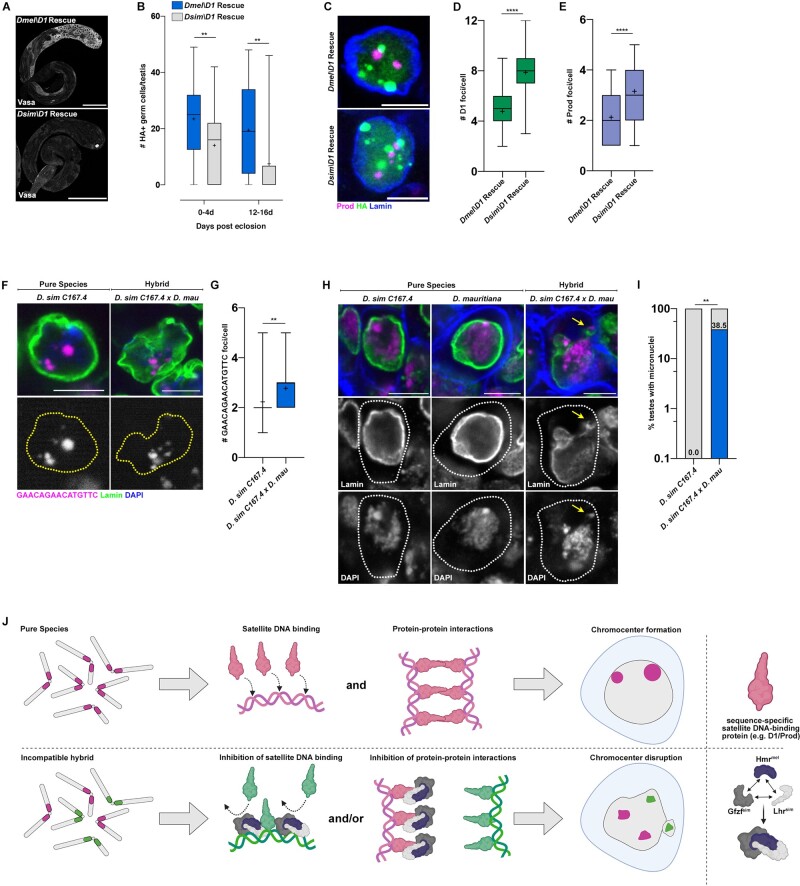
The above data imply that the inability of chromosomes from two species to form chromocenters may cause cellular defects such as micronuclei, leading to hybrid incompatibility. This raised the question as to whether chromocenter forming proteins such as D1 and Prod, which in D. melanogaster bind and cluster the (AATAT)n and (AATAACATAG)n respectively (Jagannathan et al. 2018; Jagannathan et al. 2019), might have functionally diverged. Interestingly, an evolutionary analysis on D. melanogaster chromatin-associated proteins identified that the D1 gene (but surprisingly not Prod) exhibits signs of positive selection (Parey and Crombach 2019). Strikingly, we found that the expression of D. simulans D1 (D1sim) under the control of the nos-gal4 early germ cell driver failed to rescue the germ cell depletion phenotype of the D. melanogaster D1 mutant to the same extent as D. melanogaster D1 (D1mel) (fig. 4A and B; supplementary fig. S4A and B, Supplementary Material online). Moreover, chromocenter formation was clearly impaired in the germ cells of D. melanogaster D1 mutant expressing D1sim, in comparison to germ cells expressing D1mel as determined by immunostaining for the D1 and Prod satellite DNA-binding proteins (fig. 4C–E). Although we did not observe micronuclei in any of the D1sim expressing germ cells, these results reveal a functional divergence between D1sim and D1mel impacting germ cell viability, despite both proteins binding the (AATAT)n satellite DNA (supplementary fig. S4C, Supplementary Material online). Interestingly, we observed that the Prod protein colocalized with clustered (GAACAGAACATGTTC)n satellite DNA in D. simulans (supplementary fig. S4D, Supplementary Material online), even though this repeat is highly diverged from the (AATAACATAG)n satellite DNA that is bound by Prod in D. melanogaster. Surprisingly, D. simulans Prod (Prodsim) was able to fully rescue the loss of viability caused by mutation of D. melanogaster Prod (supplementary fig. S4E and F, Supplementary Material online) and ectopic expression of Prodsim in D. melanogaster spermatocytes resulted in chromatin threads connecting heterologous chromosomes, similar to that of Prodmel (Jagannathan et al. 2019) (supplementary fig. S4G, Supplementary Material online), suggesting that Prod from D. simulans can function in the D. melanogaster background.
Fig. 4.
Functional divergence of the D1 chromocenter forming protein from D. melanogaster and D. simulans. (A) D1 mutant (D1LL03310) testes expressing HA-tagged UASt-D1mel and UASt-D1sim under the control of nos-gal4 were stained for Vasa (gray). Scale bars are 25 µm. (B) Quantification of HA+ germ cells in testes from 0- to 4-day-old and 12- to 16-day-old males of the following genotypes, nos>D1mel; D1LL03310 (n = 29, 0–4 days) and (n = 31, 12–16 days) and nos>D1sim; D1LL03310 (n = 31, 0–4 days) and (n = 32, 12–16 days). n indicates the number of testes analyzed, ** represents P = 0.006 (0–4 days) and P = 0.001 (12–16 days) from a Student’s t-test. (C) IF against Prod (magenta), HA (green), and Lamin (blue) in spermatogonial cells of the indicated genotypes. (D) Box-and-whisker plot of HA foci/cell in D1 mutant (D1LL03310) spermatogonia expressing D1mel-HA (n = 57) or D1sim-HA (n = 60). n indicates the number of germ cells analyzed, **** represents P < 0.0001 from Student’s t-test and crosshairs mark the mean. (E) Box-and-whisker plot of Prod foci/cell in D1 mutant (D1LL03310) spermatogonia expressing D1mel-HA (n = 57) or D1sim-HA (n = 60). n indicates the number of germ cells analyzed, **** represents P < 0.0001 from Student’s t-test and crosshairs mark the mean. (F) FISH against the (GAACAGAACATGTTC)n satellite (magenta) in spermatogonial cells from the indicated 0- to 3-day-old males and costained with DAPI (blue) and Lamin (green). (G) Box-and-whisker plot of large autosomal (GAACAGAACATGTTC)n foci per spermatogonial cell from D. sim C167.4 (n = 39) and D. sim C167.4×D. mau w+ (n = 31). n indicates the number of germ cells analyzed, ** represents P = 0.0067 from a Student’s t-test and crosshairs mark the mean. (H) IF against Lamin (green) in spermatogonial cells from the indicated 0- to 3-day-old males and costained with DAPI (magenta) and phalloidin (blue). Arrows point to micronuclei. (I) Quantification of testes containing at least one cell with a micronucleus from 0- to 4-day-old D. sim C167.4 pure species males (n = 18) and 0- to 4-day-old D. sim C167.4×D. mau hybrid males (n = 26). The percentage of micronuclei-containing testes is indicated above the respective columns. n indicates the number of testes analyzed, ** represents P = 0.0027 from Fisher’s exact test. All scale bars (except panel A) are 5 µm and yellow dashed lines demarcate nuclear boundary. (J) A speculative model of how hybrid incompatibility factors may inhibit chromocenter formation by species-specific satellite DNA-binding proteins in cells of affected hybrid tissues.
We next tested whether D. simulans D1/Prod could complement their D. melanogaster counterparts in hybrids, where the genomes of both species are brought together in the same nucleus. To do so, we separately crossed D. melanogaster strains carrying loss-of-function alleles of Dmel\D1 (D1LL03310) and Dmel\Prod (prodk08810) to D. simulans. Consistent with previous observations that the D. melanogaster autosomes do not have any major effect hybrid lethality loci (Cuykendall et al. 2014), we did not observe a significant rescue of hybrid male lethality in comparison to the control (supplementary tables S3 and S4, Supplementary Material online). A previous study demonstrated that Prodsim was capable of complementing Prodmel function in viable female hybrids and rescued male hybrids (Itoh et al. 1999). We were able to recapitulate these findings by crossing heterozygous prodk08810 females to the D. simulans C167.4 and Lhr1 strains and we observed that hybrids containing only Prodsim are at least as viable as their siblings containing both Prodmel and Prodsim (supplementary fig. S5A and B and table S3, Supplementary Material online). Similarly, we observed that viable female hybrids and rescued male hybrids containing only D1sim are equally as viable as their siblings containing both D1mel and D1sim (supplementary fig. S5C and D and table S4, Supplementary Material online). These data suggest that D1sim and Prodsim are capable of complementing their D. melanogaster counterparts, specifically in the somatic cells of viable female hybrids and rescued male hybrids. Consistently, protein sequence alignment revealed 92.1% identity between the D1 orthologs and 97.4% identity between the Prod orthologs (supplementary fig. S6A and B, Supplementary Material online). However, our data have also shown that chromocenter formation is significantly disrupted in the atrophied imaginal discs of male hybrids and the sterile gonads of female hybrids in a HI factor-dependent manner. Therefore, in these tissues, we propose that the repeat-associated HI factors may exploit differences in the underlying sequence of species-specific D1 and Prod to impede chromocenter formation.
Chromocenter Disruption Is a Common Phenotype among Drosophila Hybrids
Due to their recent divergence ∼250,000 years ago, the satellite DNA content of D. simulans and D. mauritiana are more similar to each other in comparison to D. melanogaster (Jagannathan et al. 2017). However, a few satellite DNA repeats remain distinct between these species (e.g., the D. simulans contains Y-specific (AATAAAC)n and (AAGAGAG)n repeats, which are lacking in D. mauritiana), whereas other satellite DNA show different abundances and locations on the chromosomes. For example, the (GAACAGAACATGTTC)n satellite DNA is restricted to chromosomes 2 and 3 of D. simulans while being present on chromosomes X, Y, 2, and 3 of D. mauritiana (Jagannathan et al. 2017). Crosses between D. simulans females and D. mauritiana males result in fertile female progeny but sterile male progeny (supplementary fig. S7A, Supplementary Material online), with the testes of sterile males reported to exhibit loss of premeiotic germ cells (Lachaise et al. 1986; Kulathinal and Singh 1998). We confirmed that the testes of male hybrids exhibited dramatic early germ cell loss with age (supplementary fig. S7B, Supplementary Material online). Moreover, we observed chromocenter disruption of the (GAACAGAACATGTTC)n satellite DNA (fig. 4F and G) and micronuclei formation (fig. 4H and I), occurring specifically in the male germ cells of hybrid testes. In contrast, germ cell content and tissue morphology were intact in the fertile female hybrids between D. simulans and D. mauritiana (supplementary fig. S7C, Supplementary Material online). Therefore, chromocenter disruption due to differences in satellite DNA composition, even between very recently diverged species, may drive micronuclei formation and cell death and promote hybrid incompatibility.
The divergence of satellite DNA repeats between species has been long postulated to mediate hybrid incompatibility (HI) and reproductive isolation. However, poor conservation of these noncoding repeats across species is also one of the main reasons why satellite DNA is typically considered to be “junk DNA,” making it difficult to speculate on the “incompatibility of (useless) junk.” Our recent work identified a conceptual and mechanistic framework to understand how satellite DNA functions within species: pericentromeric satellite DNA association between heterologous chromosomes (chromocenters) bundles the entire set of chromosomes within the nucleus (Jagannathan et al. 2018; Jagannathan et al. 2019). In this study, we demonstrate that this framework can explain how divergent satellite DNA repeats impede the viability and fertility of hybrids. Indeed, we provide the first proof-of-concept evidence that cells from hybrid animals containing distinct satellite DNA repeats exhibit phenotypes unique to chromocenter disruption. Our study lays the foundation for understanding hybrid incompatibility at a cellular level in Drosophila as well as other eukaryotes.
What is the potential mechanism behind chromocenter disruption in affected hybrid tissues? We have previously shown that binding of satellite DNA repeats by sequence-specific binding proteins forms DNA-protein modules, which interact with each other to form chromocenters. Our data in this study indicate that HI factors (Hmrmel, Lhrsim, and potentially Gfzfsim) are upstream of this process and directly impede chromocenter formation in hybrids. Previous work from other groups have shown that Hmr and Lhr can localize to chromocenters (Brideau et al. 2006; Satyaki et al. 2014; Kochanova et al. 2020) and exhibit increased expression levels in lethal male hybrids (Thomae et al. 2013). Moreover, Hmr and Lhr colocalize/form complexes with Gfzf in both pure species and hybrids (Cooper et al. 2019; Lukacs et al. 2021). We therefore speculate that a complex of HI factors in hybrids could inhibit either the protein–DNA or protein–protein interactions that contribute to chromocenter formation (fig. 4J). Interestingly, recent work has demonstrated that Hmr and Lhr facilitate the detachment of pericentromeric heterochromatin from sister chromatids during anaphase in D. melanogaster (Blum et al. 2017). A tempting possibility is that Hmr/Lhr may perform this function aberrantly in hybrids and detach pericentromeric heterochromatin from heterologous chromosomes during interphase, thus triggering chromocenter disruption.
Finally, we postulate that it is the rapid divergence of satellite DNA repeats that necessitates coevolution of proteins involved in the formation and regulation of chromocenters, with each species continually fine-tuning a “chromocenter strategy” for its own unique repeat content. Encapsulating the genomes of two organisms in a single cell (hybrids) could trigger a clash of strategies that results in chromocenter disruption and loss of cellular and organismal viability. We are therefore struck by the potential for satellite DNA turnover within species to alter evolutionary trajectories of chromocenter-associated proteins, thereby planting the seed for subsequent reproductive isolation and speciation.
Materials and Methods
Fly Husbandry and Strains
All fly stocks were raised on standard Bloomington medium at 25 °C unless otherwise indicated. D. melanogaster y w was used as a wild type stock. The following fly stocks were obtained from the Drosophila species stock center: D. simulans w501 (DSSC#14021-0251.195), D. mauritiana w1 (DSSC#14021-0241.60), D. mauritiana w+ wild type (DSSC#14021-0241.150), D. simulans Lhr1 (DSSC# 14021-0251.023). D1LL03310 (DGRC140754), FRT42D prodk08810 (DGRC111248), and D. simulans C167.4 (DGRC107850) were obtained from the Kyoto stock center. prodU (BDSC42686) was obtained from the Bloomington Drosophila stock center. Hmr-HA (Satyaki et al. 2014) and In(1)AB, Hmr2/FM6 (Barbash and Ashburner 2003) were gifts from Daniel Barbash, whereas the D. simulans Tsimbazaza strain (Hollocher et al. 2000) was a gift from Patricia Wittkopp. nos-gal4 (Van Doren et al. 1998) and bam-gal4 (Chen and McKearin 2003) and pUASt-GFP-Prodmel (Jagannathan et al. 2019) have been previously described.
Transgene Construction
For construction of pUASt-GFP-Prodsim, a codon optimized Prodsim ORF was subcloned into the NotI and KpnI sites of pUASt-EGFP-attB (Salzmann et al. 2013) resulting in pUASt-GFP-Prodsim. For construction of p400-GFP-Prodsim, a 400 bp promoter upstream of the Prod start site from D. melanogaster was PCR amplified using the following primer pair, GATCAAGCTTCTGTTGTTATGCATATCGTTC and GATCGAATTCCCGGGTATCCTTGCTC and subcloned into the HindIII and EcoRI sites on pUASt-GFP-Prodsim, replacing the UAS sequence. Transgenic flies were generated for both plasmids using PhiC31 integrase-mediated transgenesis into the attP40 site (BestGene). The following plasmids, pUASt-D1mel-HA and pUASt-D1sim-HA were obtained from Dan Barbash (Ferree and Barbash 2009) and transgenic flies were generated using PhiC31 integrase-mediated transgenesis into the attP2 site (BestGene).
Immunofluorescence Staining and Microscopy
For Drosophila tissues, immunofluorescence staining was performed as described previously (Cheng et al. 2008). Briefly, tissues were dissected in PBS, transferred to 4% formaldehyde in PBS, and fixed for 30 min. Tissues were then washed in PBS-T (PBS containing 0.1% Triton-X) for at least 60 min, followed by incubation with primary antibody in 3% bovine serum albumin (BSA) in PBS-T at 4 °C overnight. Samples were washed for 60 min (three 20-min washes) in PBS-T, incubated with secondary antibody in 3% BSA in PBS-T at 4 °C overnight, washed as above, and mounted in VECTASHIELD with DAPI (Vector Labs). The following primary antibodies were used: rabbit anti-Vasa (1:200; d-26; Santa Cruz Biotechnology), rat anti-Vasa (Developmental Studies Hybridoma Bank), mouse anti-LaminDm0 (ADL84.12, 1:200, Developmental Studies Hybridoma Bank), mouse anti-Hts (1B1, Developmental Studies Hybridoma Bank), rat anti-HA (Sigma, 3F10), Phalloidin-Alexa488 (Abcam, ab176753), Phalloidin-Alexa546 (ThermoFisher, a22283, 1:200), rabbit anti-Prod (gift from Tibor Torok, 1:5000), and guinea pig anti-D1 (generated using the synthetic peptide DGENDANDGYVSDNYNDSESVAA [Covance]). Fluorescent images were taken using a Leica TCS SP8 confocal microscope with 63× oil-immersion objectives (NA = 1.4). Brightfield images were acquired using a Keyence microscope. Images were processed using Adobe Photoshop software.
DNA Fluorescence In Situ Hybridization
Whole mount Drosophila tissues were prepared as described above, and optional immunofluorescence staining protocol was carried out first. Subsequently, samples were postfixed with 4% formaldehyde for 10 min and washed in PBS-T for 30 min. Fixed samples were incubated with 2 mg/ml RNase A solution at 37 °C for 10 min, then washed with PBS-T + 1 mM EDTA. FISH using heat denaturation was carried out as follows: samples were washed in 2xSSC-T (2xSSC containing 0.1% Tween-20) with increasing formamide concentrations (20%, 40%, and 50%) for 15 min each followed by a final 30-min wash in 50% formamide. Hybridization buffer (50% formamide, 10% dextran sulfate, 2× SSC, 1 mM EDTA, 1 μM probe) was added to washed samples. Samples were denatured at 91 °C for 2 min, then incubated overnight at RT. For hybrid tissues, FISH using acid denaturation was used instead of heat denaturation and carried out as follows: samples were washed in 2xSSC-T, DNA was denatured using a 30-min incubation with 2 N HCl at RT followed by three rinses in ice-cold PBS-T. Hybridization buffer (60% formamide, 10% dextran sulfate, 2× SSC, 1 mM EDTA, 0.5–1 μM probe) was added to washed samples and incubated overnight at RT. For mitotic chromosome spreads, larval 3rd instar brains were squashed according to previously described methods (Larracuente and Ferree 2015). Briefly, tissue was dissected into 0.5% sodium citrate for 5–10 min and fixed in 45% acetic acid/2.2% formaldehyde for 4–5 min. Fixed tissues were firmly squashed with a cover slip and slides were submerged in liquid nitrogen until bubbling ceased. Coverslips were then removed with a razor blade and slides were dehydrated in 100% ethanol for at least 5 min. After drying, hybridization mix (50% formamide, 2× SSC, 10% dextran sulfate, 100 ng of each probe) was applied directly to the slide, samples were heat denatured at 95 °C for 5 min and allowed to hybridize overnight at room temperature. Following hybridization, slides were washed three times for 15 min in 0.2× SSC and mounted with VECTASHIELD with DAPI (Vector Labs). The following probes were used for Drosophila in situ hybridization: (AATAT)6, (AATAACATAG)3 and (GAACAGAACATGTTCGAACAGAACATGTTCGAACA) and have been previously described (Jagannathan et al. 2017).
Rescue Experiments
The pUASt-D1mel-HA and pUASt-D1sim-HA transgenes were each recombined with the D1LL03310 mutant allele to generate two rescue strains (pUASt-D1mel-HA, D1LL03310, and pUASt-D1sim-HA, D1LL03310). These rescue strains were crossed to flies containing the nos-gal4 germ cell driver and the D1 mutant allele (nos-gal4::VP16; D1LL03310) at room temperature as follows,
D1mel rescue: nos-gal4::VP16/CyO; D1LL03310/TM2×pUASt-D1mel-HA, D1LL03310/TM6B
D1sim rescue: nos-gal4::VP16/CyO; D1LL03310/TM2×pUASt-D1sim-HA, D1LL03310/TM6B
We collected 0- to 4-day-old and 12- to 16-day-old D1 mutant flies expressing D1mel and D1sim (nos>UAS-D1mel; D1LL03310/D1LL03310 and nos>UAS-D1sim; D1LL03310/D1LL03310) and dissected testes to score the number of HA+ germ cells and assess the extent of rescue. The Prodsim rescue allele was generated by recombining the p400-GFP-Prodsim transgene with prodk08810. The Prodsim rescue allele and the FRT42D prodk08810 allele as a control were each crossed to prodU in vials at 25 °C. The percent of transheterozygous prod mutant flies was quantified in each replicate.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dan Barbash, Trisha Wittkopp, Tibor Torok, the Bloomington Drosophila Stock Center, the Kyoto Stock Center, the Drosophila Species Stock Center, and the Developmental Studies Hybridoma Bank for reagents and resources. We thank members of the Yamashita lab and Jagannathan lab for discussion and comments on the manuscript. This work was supported by the Howard Hughes Medical Institute (Y.M.Y.) and an American Heart Association Postdoctoral Fellowship (M.J., 17POST33660746). M.J. and Y.M.Y. conceived the project, interpreted the data, and wrote the manuscript. Both authors contributed to conducting experiments and analyzing data.
References
- Barbash DA. 2010. Ninety years of Drosophila melanogaster hybrids. Genetics 186(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA, Ashburner M.. 2003. A novel system of fertility rescue in Drosophila hybrids reveals a link between hybrid lethality and female sterility. Genetics 163(1):217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA, Roote J, Ashburner M.. 2000. The Drosophila melanogaster hybrid male rescue gene causes inviability in male and female species hybrids. Genetics 154(4):1747–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA, Siino DF, Tarone AM, Roote J.. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci U S A. 100(9):5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JA, Bonaccorsi S, Marzullo M, Palumbo V, Yamashita YM, Barbash DA, Gatti M.. 2017. The hybrid incompatibility genes Lhr and Hmr are required for sister chromatid detachment during anaphase but not for centromere function. Genetics 207(4):1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA.. 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314(5803):1292–1295. [DOI] [PubMed] [Google Scholar]
- Chakraborty M, Chang C-H, Khost DE, Vedanayagam J, Adrion JR, Liao Y, Montooth KL, Meiklejohn CD, Larracuente AM, Emerson JJ.. 2021. Evolution of genome structure in the Drosophila simulans species complex. Genome Res. 31:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin DM.. 2003. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130(6):1159–1170. [DOI] [PubMed] [Google Scholar]
- Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM.. 2008. Centrosome misorientation reduces stem cell division during ageing. Nature 456(7222):599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JC, Lukacs A, Reich S, Schauer T, Imhof A, Phadnis N.. 2019. Altered localization of hybrid incompatibility proteins in Drosophila. Mol Biol Evol. 36(8):1783–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuykendall TN, Satyaki P, Ji S, Clay DM, Edelman NB, Kimchy A, Li L-H, Nuzzo EA, Parekh N, Park S, et al. 2014. A screen for F1 hybrid male rescue reveals no major-effect hybrid lethality loci in the Drosophila melanogaster autosomal genome. G3 (Bethesda) 4(12):2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium, Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450(7167):203–218. [DOI] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA.. 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7(10):e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. [DOI] [PubMed] [Google Scholar]
- Hollocher H, Agopian K, Waterbury J, O’Neill RW, Davis AW.. 2000. Characterization of defects in adult germline development and oogenesis of sterile and rescued female hybrids in crosses between Drosophila simulans and Drosophila melanogaster. J Exp Zool. 288(3):205–218. [DOI] [PubMed] [Google Scholar]
- Hutter P, Ashburner M.. 1987. Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibling species. Nature 327(6120):331–333. [DOI] [PubMed] [Google Scholar]
- Itoh M, Yu S, Watanabe TK, Yamamoto MT.. 1999. Structural and genetic studies of the proliferation disrupter genes of Drosophila simulans and D. melanogaster. Genetica 106(3):223–229. [DOI] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM.. 2018. A conserved function for pericentromeric satellite DNA. Elife 7:e34122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM.. 2019. The modular mechanism of chromocenter formation in Drosophila. Elife. 8:e43938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Warsinger-Pepe N, Watase GJ, Yamashita YM.. 2017. Comparative analysis of satellite DNA in the Drosophila melanogaster species complex. G3 (Bethesda) 7(2):693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Yamashita YM.. 2017. Function of junk: pericentromeric satellite DNA in chromosome maintenance. Cold Spring Harb Symp Quant Biol. 82:319–327. [DOI] [PubMed] [Google Scholar]
- Kochanova NY, Schauer T, Mathias GP, Lukacs A, Schmidt A, Flatley A, Schepers A, Thomae AW, Imhof A.. 2020. A multi-layered structure of the interphase chromocenter revealed by proximity-based biotinylation. Nucleic Acids Res. 48(8):4161–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal R, Singh RS.. 1998. Cytological characterization of premeiotic versus postmeiotic defects producing hybrid male sterility among sibling species of the Drosophila melanogaster complex. Evolution 52(4):1067–1079. [DOI] [PubMed] [Google Scholar]
- Lachaise D, Capy P, Cariou M-L, Joly D, Lemeunier F, David JR.. 2004. Nine relatives from one African ancestor: population biology and evolution of the Drosophila melanogaster subgroup species. In: Singh R, Uyenoyama M, editors. The Evolution of Population Biology. Cambridge: Cambridge University Pressp. p. 315–344. [Google Scholar]
- Lachaise D, David JR, Lemeunier F, Tsacas L, Ashburner M.. 1986. The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans, and D. melanogaster from the afrotropical region. Evolution 40(2):262–271. [DOI] [PubMed] [Google Scholar]
- Larracuente AM, Ferree PM.. 2015. Simple method for fluorescence DNA in situ hybridization to squashed chromosomes. J Vis Exp. 95:e52288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Brutlag DL.. 1987. Identical satellite DNA sequences in sibling species of Drosophila. J Mol Biol. 194(2):161–170. [DOI] [PubMed] [Google Scholar]
- Lukacs A, Thomae AW, Krueger P, Schauer T, Venkatasubramani AV, Kochanova NY, Aftab W, Choudhury R, Forne I, Imhof A.. 2021. The integrity of the speciation core complex is necessary for centromeric binding and reproductive isolation in Drosophila. bioRxiv. 429932. [DOI] [PMC free article] [PubMed]
- Matute DR, Gavin-Smyth J, Liu G.. 2014. Variable post-zygotic isolation in Drosophila melanogaster/D. simulans hybrids. J Evol Biol. 27(8):1691–1705. [DOI] [PubMed] [Google Scholar]
- Orr HA, Madden LD, Coyne JA, Goodwin R, Hawley RS.. 1997. The developmental genetics of hybrid inviability: a mitotic defect in Drosophila hybrids. Genetics 145(4):1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parey E, Crombach A.. 2019. Evolution of the Drosophila melanogaster chromatin landscape and its associated proteins. Genome Biol Evol. 11(3):660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Baker EP, Cooper JC, Frizzell KA, Hsieh E, de la Cruz AFA, Shendure J, Kitzman JO, Malik HS.. 2015. An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science 350(6267):1552–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann V, Inaba M, Cheng J, Yamashita YM.. 2013. Lineage tracing quantification reveals symmetric stem cell division in Drosophila male germline stem cells. Cell Mol Bioeng. 6(4):441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez L, Dübendorfer A.. 1983. Development of imaginal discs from lethal hybrids between Drosophila melanogaster and Drosophila mauritiana. Wilhelm Rouxs Arch Dev Biol. 192:48–50. [DOI] [PubMed] [Google Scholar]
- Satyaki PR, Cuykendall TN, Wei KH, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA.. 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 10(3):e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K. 2012. Chromatin evolution and molecular drive in speciation. Int J Evol Biol. 2012:301894. [PMC][10.1155/2012/301894] [22191063] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. 1920. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics 5(5):488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, Novitski E.. 1941. The homologies of the chromosome elements in the genus Drosophila. Genetics 26(5):517–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomae AW, Schade GOM, Padeken J, Borath M, Vetter I, Kremmer E, Heun P, Imhof A.. 2013. A pair of centromeric proteins mediates reproductive isolation in Drosophila species. Dev Cell. 27(4):412–424. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R.. 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 8(4):243–246. [DOI] [PubMed] [Google Scholar]
- Watanabe TK. 1979. A gene that rescues the lethal hybrids between Drosophila melanogaster and D. simulans. Jpn J Genet. 54(5):325–331. [Google Scholar]
- Yunis JJ, Yasmineh WG.. 1971. Heterochromatin, satellite DNA, and cell function. Structural DNA of eucaryotes may support and protect genes and aid in speciation. Science 174(4015):1200–1209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.