Abstract
True sea snakes (Hydrophiini) are among the last and most successful clades of vertebrates that show secondary marine adaptation, exhibiting diverse phenotypic traits and lethal venom systems. To better understand their evolution, we generated the first chromosome-level genomes of two representative Hydrophiini snakes, Hydrophis cyanocinctus and H. curtus. Through comparative genomics we identified a great expansion of the underwater olfaction-related V2R gene family, consisting of more than 1,000 copies in both snakes. A series of chromosome rearrangements and genomic structural variations were recognized, including large inversions longer than 30 megabase (Mb) on sex chromosomes which potentially affect key functional genes associated with differentiated phenotypes between the two species. By integrating multiomics we found a significant loss of the major weapon for elapid predation, three-finger toxin genes, which displayed a dosage effect in H. curtus. These genetic changes may imply mechanisms that drove the divergent evolution of adaptive traits including prey preferences between the two closely related snakes. Our reference-quality sea snake genomes also enrich the repositories for addressing important issues on the evolution of marine tetrapods, and provide a resource for discovering marine-derived biological products.
Keywords: true sea snakes, V2R genes, chromosomal rearrangements, genomic structural variation, three-finger toxin, divergent evolution
Introduction
Snakes are a good model for studying extreme adaptations and venom evolution. Scientists have focused much on the genetic and evolutionary characterization of snakes in terrestrial niches (Castoe et al. 2013; Vonk et al. 2013; Schield et al. 2019; Suryamohan et al. 2020) but little on those in marine habitats. Hydrophiini, the true sea snakes or fully aquatic sea snakes, is one of the most advanced and youngest clades within Serpentes (Sanders et al. 2013). Although over 60 species of true sea snakes have been discovered to date (Uetz et al. 2020), comprising the largest proportion (nearly 87%) of marine reptiles, little is known about the evolutionary trajectories and genetic mechanisms underlying the adaptive radiation and divergent evolution that occurred during their transition from land back to sea. Although a few recent studies have discussed evolutionary issues regarding the marine adaptation of sea snakes from a genomic perspective (Kishida et al. 2019; Peng et al. 2020), high-quality chromosome-scale genomes are still needed to achieve more comprehensive and accurate understandings of the molecular mechanisms driving the evolution of diversified traits within the Hydrophiini lineage.
In addition to anatomical and physiological evolution, the venom system inherited from terrestrial elapids also underwent evolutionary alterations and developments essential for sea snakes to occupy neritic zones. Previous research has characterized the venom composition or expression profiles of marine elapids (Durban et al. 2018; Peng et al. 2020), but the genomic architecture of venom gene families and relationships between the evolution of sea snake venom toxin repository and their feeding strategies remain poorly understood. Moreover, as extremely venomous but medically important animals applied in antivenom production and exploration of therapeutic proteins or peptides (Zheng et al. 2016), sea snakes are faced with overharvesting and populations being damaged (Van Cao et al. 2014). Alternative approaches of directly exploiting snake venom call for massive sequence information from multiomic big data with high-continuity and completeness.
To help settle these issues, we incorporated multiple state-of-the-art sequencing and scaffolding protocols and generated the first chromosome-scale genome assemblies of two Hydrophiinae snakes, Hydrophis cyanocinctus (annulated sea snake) and H. curtus (spine-bellied sea snake, formerly known as Lapemis curtus). Belonging to the largest genus in Hydrophiinae, these two snakes are widespread in shallow-water throughout the tropical and subtropical Indo-Pacific and are among the most commonly captured sea snakes around Southeast Asia (Van Cao et al. 2014). Despite their close affinity in taxonomy, there exist noticeable differences in morphologies, physiologies, and diets. On the basis of reference-quality genomes, we conducted chromosome-wide comparative genomics to lift the veil on the molecular mechanisms shaping varied phenotypes within Hydrophiini. Transcriptomics and proteomics were further integrated with genomic analyses to facilitate comprehensive understanding of species-specific genomic organization and evolution of key venom genes as well as their linkage to divergent traits and adaptation patterns in true sea snakes.
Results and Discussion
Genome Assembly and Annotation
We combined long-read, short-read, and proximity-ligation-based sequencing technologies together with advanced assembly algorithms to obtain superior genome assemblies (supplementary note 2, Supplementary Material online). The results yielded two haplotype assemblies named HCya_v2 (contig N50, 18.99 Mb; scaffold N50, 264.25 Mb) for H. cyanocinctus and HCur_v2 (contig N50, 9.7 Mb; scaffold N50, 266.23 Mb) for H. curtus. 94.4% and 97% of the total sequence was anchored onto 18 and 17 chromosome-level scaffolds in HCya_v2 and HCur_v2, respectively (table 1 and fig. 1a). The Hi-C (chromosome conformation capture) contact maps recognized seven macrochromosomes in both assemblies (supplementary fig. S2, Supplementary Material online). Comparatively, our genomes had better continuity with higher N50 values and shorter gap openings (0.01% in HCya_v2 and 0.02% in HCur_v2) than previously published snake genomes. The same held true for other reported genomes of representative reptiles (supplementary table S6, Supplementary Material online). The contig N50 value of HCya_v2 was 9-fold (18.99 vs. 2.11 Mb) and 62.8-fold (18.99 vs. 0.3 Mb) greater than that of the tiger rattlesnake (Crotalus tigris) and Indian cobra (Naja naja) genome respectively, which presented the two best-quality snake genome assemblies prior to our publication (Suryamohan et al. 2020; Margres et al. 2021). For H. curtus, our assembly HCur_v2 appeared far more continuous than the recently reported one (Peng et al. 2020), with a 53- and 197.7-fold increase in contig N50 and scaffold N50 values, respectively, and a total sequence length closer to the estimated genome size of H. curtus. Using the 3,950 tetrapod genes in the benchmarking universal single-copy orthologs (BUSCO) (Simao et al. 2015) set, we achieved a 90.1% and 89.8% genome completeness score for HCya_v2 and HCur_v2, respectively (table 1 and supplementary table S7, Supplementary Material online). To assess the assembling quality of repeat elements, we ran LTR Assembly Index (LAI) (Ou et al. 2018) evaluation and found that both HCya_v2 and HCur_v2 indexes were near 20, indicating that they had reached the reference genome level and approached the golden genome level (table 1 and supplementary fig. S3, Supplementary Material online). Additionally, the consensus quality values (QVs), which represent Phred-scaled probability of base error in the assembly (Rhie et al. 2020), were 33.735 for HCya_v2 and 33.845 for HCur_v2, indicating a relatively high assembly accuracy for both genomes. These quality control metrics of our genomes were all comparable to those of the C. tigris genome (table 1).
Table 1.
Comparison of Recently Reported High-Quality Snake Genomes.
| Species | Hydrophis cyanocinctus (Annulated Sea Snake) | H. curtus (Spine-Bellied Sea Snake) | H. curtus (Spine-Bellied Sea Snake) | Naja naja (Indian Cobra) | Crotalus tigris a (Tiger Rattlesnake) |
|---|---|---|---|---|---|
| Assembly name | HCya_v2 | HCur_v2 | — | Nana_v5 | — |
| Sequencing technologies | CLR+HiC+PE | CLR+HiC+PE | PE+MP+CLR | CLR+ONT+PE+Chicago+OM+HiC | CLR+PE |
| Estimated genome size (Gb) | 2.02 | 2.03 | 1.91–1.98 | 1.48–1.77 | 1.6 |
| Assembly size (Gb) | 1.98 | 1.96 | 1.63 | 1.77 | 1.59 |
| Gap length (Mb, % of assembly) | 0.29 (0.01) | 0.36 (0.02) | 25.52 (1.57) | 109.67 (6.20) | — |
| Assembly level | Chromosome | Chromosome | Scaffold | Chromosome | Chromosome |
| Contig number | 2,399 | 2,109 | 15,263 | 13,805 | 4,228 |
| Contig N50 (Mb) | 18.99 | 9.70 | 0.18 | 0.30 | 2.11 |
| Scaffold number | 1,163 | 711 | 3,139 | 1,897 | 380 |
| Scaffold N50 (Mb) | 264.25 | 266.23 | 1.35 | 224.09 | 207.72 |
| Chromosome-anchoring percentage (%) | 94.38 | 97.02 | — | 94.30 | — |
| BUSCO completeness (%)b | 90.1 (89.2) | 89.8 (88.8) | 86.6 (86.1) | 88.6 (87.8) | 91.6 (89.9) |
| LAI | 19.56 | 19.56 | 18.96 | 5.55 | — |
| QV scorec | 33.735 | 33.845 | 24.045 | 28.904 | 38.808 |
| Reference | This study | This study | Peng et al. (2020) | Suryamohan et al. (2020) | Margres et al. (2021) |
CLR, PacBio continuous long reads; ONT, Oxford Nanopore; PE, paired-end; MP, mate-pair; OM, optical map.
The C. tigris genome deposited in NCBI (ASM1654583v1) was its initial assembly at contig level and the final chromosome-level assembly had not been available, so the gap information could not be retrieved. The LAI evaluation of the C. tigris genome was performed but no valid LAI value was obtained.
The values in brackets represent the percentages of complete and single-copy orthologs.
QV scores are Phred-scaled where QV=−10log10E for a probability of error (E) at each base in the assembly. A higher QV indicates a higher base-level accuracy of the assembly (Rhie et al. 2020).
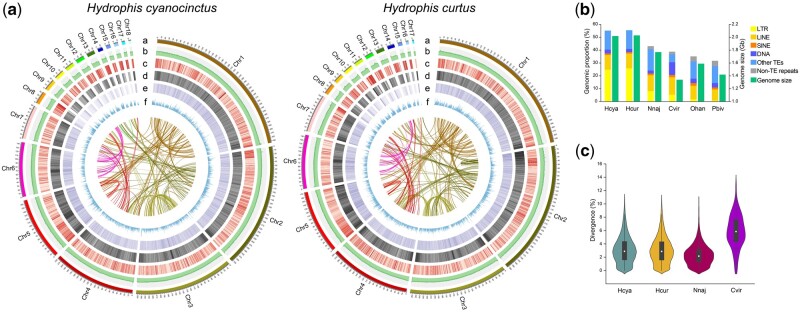
Fig. 1.
Genomic features of Hydrophis cyanocinctus and H. curtus. (a) Circos view of the genome assemblies (HCya_v2 and HCur_v2) of H. cyanocinctus and H. curtus. Tracks depict circular representation of haplotype chromosomes in megabases (Mb, a), GC percentage (b), the density of protein-coding genes, TEs, LTR elements (c–e), and gene expression profile of venom gland (f). Lines inside the circle indicate within-genome links between syntenic paralogs. (b) Bar plot comparing the genomic fractions of different classes of repeat elements and estimated genome size among six snakes. The repeat contents in the genomes of Naja naja, Ophiophagus hannah, Crotalus viridis, and Python bivittatus were retrieved from their publications (Castoe et al. 2013; Schield et al. 2019; Suryamohan et al. 2020). The assembly size of C. viridis genome (UTA_CroVir_3.0) was used as there had been no determined or estimated genome size reported for this species. (c) Violin plot showing the frequency distribution of sequence divergence between LTR pairs in the genomes of four snakes. Species name abbreviations: Hcya, H. cyanocinctus; Hcur, H. curtus; Nnaj, N. naja; Cvir, C. viridis; Ohan, O. hannah; Pbiv, P. bivittatus.
Genome annotation was performed via customized hybrid methods (supplementary note 4, Supplementary Material online). Ultimately, we annotated 23,898 and 23,192 protein-coding genes, as well as 43,062 and 34,061 transcripts as a consequence of alternative splicing in the H. cyanocinctus and H. curtus genomes, respectively. We found 21,487 (90%) and 22,022 (95%) genes on assigned chromosomes of H. cyanocinctus and H. curtus, respectively. We also predicted noncoding RNA (ncRNA) regions in our genomes (supplementary table S13, Supplementary Material online). Overall, we obtained the best reference-quality sea snake genomes with highly continuous and complete assemblies and annotations, which are comparable to other available genomes of nonavian reptiles including common wall lizard, red-eared slider turtle, and saltwater crocodile (supplementary table S6, Supplementary Material online).
Genome Size and Repeat Content
The genome sizes of H. cyanocinctus and H. curtus were estimated at ∼2.02 and ∼2.03 Gb, respectively (table 1 and supplementary note 2.1, Supplementary Material online), which are relatively higher than the majority of those of snakes in the Animal Genome Size Database (Pasquesi et al. 2018; Gregory 2020). Genome annotation results revealed that the larger genome sizes can be attributed to an increase in repetitive sequences primarily comprised transposable elements (TEs) making up 55.20% and 55.59% of the H. cyanocinctus and H. curtus genomes, respectively (fig. 1a and b; supplementary tables S8 and S9, Supplementary Material online). Long terminal repeats (LTRs) appear predominant with significantly higher genomic proportions (24.80% in H. cyanocinctus and 25.86% in H. curtus) in the two sea snake genomes compared with other published non-Hydrophiini snake genomes. Interestingly, both sea snakes displayed extremely identical distribution patterns of LTR divergence levels (fig. 1c). The estimated time points of LTR insertion are also similar, starting from ∼15 Ma with a median value of 3.5 and 3.1 Ma for H. cyanocinctus and H. curtus, respectively (supplementary fig. S4, Supplementary Material online). Moreover, the Gypsy family showed notably earlier insertion than the Copia family. This striking expansion of LTR retrotransposons is likely to have had a bearing on the environmental adaptation and lineage radiation of true sea snakes (see below). Noteworthy is the distance between the data of our study and a previous report on H. curtus (Peng et al. 2020), largely resulting from the distinct qualities of genome assembly particularly in assembly size and LTR completeness (table 1).
The Phylogenetic Status of Hydrophiini
It is pretty essential to understand the evolutionary history and genetic relationship of true sea snakes within Serpentes (snakes); however, previous phylogenetic research on sea snakes were mainly based on morphological and molecular evidence from a few nuclear or mitochondrial gene markers (Sanders et al. 2008; Strickland et al. 2016), resulting in unstable phylogeny and ambiguous divergence times. Leveraging the high integrity of our assemblies, we revisited this issue by phylogenomics with sequenced and annotated genomes of H. cyanocinctus, H. curtus, 14 other species from snakes, lizards, and the outgroup chicken. We constructed a squamate species tree under maximum likelihood (ML) with nearly 100% bootstrapping support at all nodes (fig. 2a and supplementary note 5.2, Supplementary Material online). This phylogenetic tree indicated a sister-group relationship between the Hydrophiini clade (H. cyanocinctus+H. curtus) and Notechis scutatus (mainland tiger snake), with Pseudonaja textilis (eastern brown snake) together clustering into the Hydrophiinae group. Moreover, the subfamily Elapinae represented by Ophiophagus hannah (king cobra) was found paraphyletic in relation to Hydrophiinae. These local topologies strongly support the origin of true sea snakes from Australo-Melanesian terrestrial elapids, coinciding well with previous analyses (Sanders et al. 2008, 2010, 2013; Strickland et al. 2016).
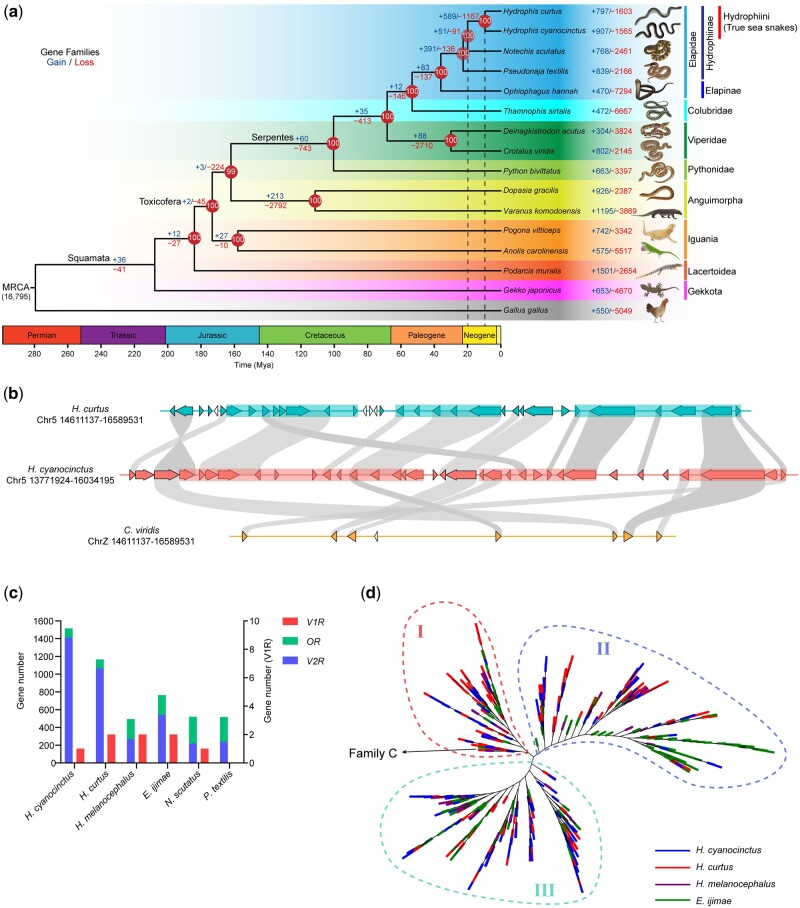
Fig. 2.
Evolutionary trajectories of true sea snakes and associated gene families. (a) A time-calibrated phylogram of 15 representatives in squamata using Gallus gallus (Chicken) as an outgroup. Gains (in blue) and losses (red) of gene families are denoted along nodes and branches. Numbers in circles at nodes refer to the bootstrap support values. The estimated divergence dates of terrestrial Hydrophiinae–Hydrophiini and Hydrophis cyanocinctus–H. curtus are indicated by dashed lines. MRCA, most recent common ancestor. Ma, million years ago. (b) The largest consecutive cluster of vomeronasal type-2 receptor (V2R) genes in H. cyanocinctus comprises 35 tandemly arrayed copies on Chromosome 5. The homologous region in the genome of H. curtus contains 30 V2R copies, whereas only seven copies were found in that of Crotalus viridis. Non-V2R genes are represented by uncolored arrows. Syntenic orthologs between species are linked by gray bands. The translucent rectangles indicate regions of V2R genes that may arise from segmental duplications. (c) Gene counts of V2R, V1R, and OR identified in the genomes of six selected snakes. The values for H. melanocephalus and Emydocephalus ijimae were retrieved from the publication (Kishida et al. 2019). (d) An unrooted phylogenetic tree of V2Rs in H. cyanocinctus, H. curtus, H. melanocephalus, and E. ijimae. The tree was constructed using the extracellular domains of V2Rs with two TAS1R proteins (XP_008122812.1, XP_003228982.3) from Anolis carolinensis as outgroups. The clade ancestral to Group I contains four V2Rs from the four species that are classified into Family C according to (Yang et al. 2005).
We employed Bayesian relaxed molecular clocks calibrated with fossil-based age constraints to estimate the evolutionary timescale throughout the tree. Our analysis indicated that true sea snakes diverged from their terrestrial Hydrophiinae relatives at least 19.9 Ma (early Miocene) and that the split between H. cyanocinctus and H. curtus dates back to 9.5 Ma (fig. 2a and supplementary fig. S6, Supplementary Material online). Given that they demonstrated fairly identical timescales of LTR explosion dating from ∼15 Ma and peaking at 3.1–3.5 Ma (supplementary fig. S4, Supplementary Material online), the fast speciation and clade radiation in Hydrophiini, typically in Hydrophis, were supposed to occur much later than their initial invasion to marine habitats, as previously concluded (Sanders et al. 2008, 2010, 2013). As TEs plays an important role in adaptive responses to environmental changes (Casacuberta and Gonzalez 2013; Schrader and Schmitz 2019), the recent coincident expansion of LTRs implies that Hydrophis snakes may have experienced marine geological events such as sea level fluctuations that drove repeated isolation of populations and acceleration of speciation during the late Pliocene to early Pleistocene (Lukoschek and Keogh 2006; Lee et al. 2016; Ukuwela et al. 2016).
Evolution of Genes in Marine Adaptation
Although inhabiting mainly in shallow sea water, true sea snakes have been recorded to dive to the mesopelagic zone with depth >200 m (Crowe‐Riddell et al. 2019). They are able to perform oxygen exchange through their skins and prevent decompression sickness following deep dives (Seymour 1974; Seymour and Webster 1975; Palci et al. 2019). To explore critical genetic changes in true sea snakes that arose alongside colonization of the oceans, we performed comparative genomic analysis to screen for expansions and contractions of gene families across squamate reptiles based upon our well-supported species phylogeny. We found 589 expanded and 1,167 contracted gene families at the branch of Hydrophiini (fig. 2a). Functional enrichment analysis of gene ontology (GO) terms and KEGG pathways showed that these genes are involved in biological processes and functions mostly including G protein-coupled receptor (GPCR) activity, ion transmembrane transport, sensory perception of sound and light, immune response, and energetic metabolism (supplementary fig. S7a and b, Supplementary Material online). We also identified in Hydrophiini 136 single-copy orthologous genes under positive selection, which were enriched into function terms represented by DNA mismatch repair, alternative mRNA splicing, respiratory chain and oxygen sensing (supplementary fig. S7c, Supplementary Material online). The rapid evolution of these genes and gene families is possibly responsible for the formation of various adaptive morphological and physiological traits of Hydrophiini snakes in marine environments including sublingual salt glands, cutaneous breathing, and resistance to bend.
We focused on the loss and gain of chemosensory GPCR genes in true sea snakes, which were the most remarkable shifts. The vomeronasal organ (VNO) accessory to the main olfactory organ in most tetrapods detects chemical signals for intraspecific communication, foraging, and predator avoidance. Accompanied by olfactory receptors (ORs), terrestrial snakes possess a well-developed vomeronasal system with predominant vomeronasal type-2 receptors (V2Rs) to execute the sense of smell (Schwenk 1995; Brykczynska et al. 2013). In fully aquatic sea snakes, the main olfactory system tends to be degenerated with OR genes rarely expressed (Kishida et al. 2019), but they retain VNOs with tongue-flicking behavior to detect water-soluble chemicals and precisely locate prey (Thewissen and Nummela 2008; Kutsuma et al. 2018). Our genome annotations of the two sea snakes saw contracted OR genes (104 copies in H. cyanocinctus and 101 copies in H. curtus) but gigantic repertoires of V2R genes (1,411 copies in H. cyanocinctus and 1,065 copies in H. curtus), which to our knowledge are the biggest V2R groups in reptilian genomes (Brykczynska et al. 2013; Kishida et al. 2019; Lind et al. 2019). Only one and two V1R genes were found in H. cyanocinctus and H. curtus, respectively, in line with the ancestral state in other snakes. Our analysis of paralogs revealed that most V2R genes are clustered at Chromosome 2 and Chromosome 5 in the two genomes, which expanded mainly through tandem duplication and segmental duplication (supplementary table S21, Supplementary Material online). In our assemblies, the largest clustering region (less than ten consecutive non-V2R genes) contains 77 V2R genes on Chromosome 5 of H. curtus, and the largest continuous cluster consists of 35 tandem copies of V2R on Chromosome 5 of H. cyanocinctus (fig. 2b and supplementary fig. S9, Supplementary Material online).
It is noteworthy that this expansion of V2R repositories was not well manifested in the genomes of another two true sea snakes, H. melanocephalus and Emydocephalus ijimae reported by Kishida et al. (fig. 2c). To deeper explore the molecular evolution of V2R family with the Hydrophiini clade, we conducted phylogenetic analysis of the V2Rs identified in these four sea snakes using the extracellular domains. The ML tree (fig. 2d and supplementary fig. S10a, Supplementary Material online) saw overall clustering of three major groups. The clades in each group basically contained all four species selected, which indicated that the expansion of V2Rs might have occurred in the common ancestor of the four snakes, and more broadly was supposed to had arisen in ancestral species of the Hydrophiini clade. Considering the possibly fewer copies of V2R genes in H. melanocephalus and E. ijimae, the V2R repositories might have undergone varying degrees of expanding in different genus of Hydrophiini or even different species within Hydrophis. The explosive expansion of V2R genes specially observed in H. cyanocinctus and H. curtus could potentially be related to their unique smelling capabilities, which however, needs to be further investigated with more genetic and phenotypic data of this clade and other related species. Furtherly, we construct another V2R tree of the four sea snakes using their seven-transmembrane (7TM) domains (supplementary fig. S10b, Supplementary Material online). Interestingly, we found the two trees differed in their topologies. Two main clustering groups could be generally recognized in the second tree of V2R-7TM, and the branches from E. ijimae mostly gathered in more ancestral clades of Group I. This difference was considered to reveal the divergent evolving patterns of the extracellular and transmembrane domains of V2Rs, of which the latter are relatively more conserved within species, whereas the former exhibit more diversity in line with their role of directly binding and recognizing various pheromone ligands (Yang et al. 2005).
Genomic Synteny and Chromosomal Evolution
We performed genome-wide synteny analysis to demonstrate chromosomal variations during the evolution of venomous Colubroidea snakes. In gross appearance, there exist conserved synteny and collinearity between the genomes of the two marine elapids, the terrestrial elapid N. naja and the viperid C. viridis (prairie rattlesnake). We identified the fifth chromosome of H. cyanocinctus and H. curtus as their Z Chromosome by interchromosomal homologies in conjunction with Z-linked gene markers (Matsubara et al. 2006) mapped to the sea snake chromosomes (fig. 3a and b; supplementary fig. S11 and table S24, Supplementary Material online). Multiple chromosomal rearrangements were observed indicating possible fusion/fission events. Typically, Chromosomes 7 and 14 of H. cyanocinctus are syntenic to Chromosome 7 of H. curtus, and Chromosome 4 of the two sea snakes share synteny with Chromosomes 5 and 6 of C. viridis. The microchromosomal rearrangements appear more complex between the two sea snakes and between elapids and prairie rattlesnake. Particularly, Chromosome 11 of H. curtus shows syntenic regions with Chromosomes 8, 11, and 15 of H. cyanocinctus, whereas Chromosome 15 of C. viridis was mapped to Chromosomes 16, 17, and part of Chromosomes 8, 13, and 14 of H. cyanocinctus. Within Elapidae, notable is the separation of Hydrophis Chromosome 6 into N. naja Chromosomes 5 and 6, as well as the rearrangement of H. cyanocinctus Chromosomes 4 and 7 into N. naja Chromosomes 4, 7, and 10, corresponding to the gained chromosome number in N. naja (Suryamohan et al. 2020) (supplementary fig. S11, Supplementary Material online). These synteny patterns align with previous findings suggesting that colubroids exhibit great karyotype variability and diversification of chromosomal organization, which is especially marked in Elapidae (Olmo 2005), and may play an important role in species differentiation of elapids including Hydrophiini snakes through chromosomal rearrangements (Navarro and Barton 2003; Olmo 2005).
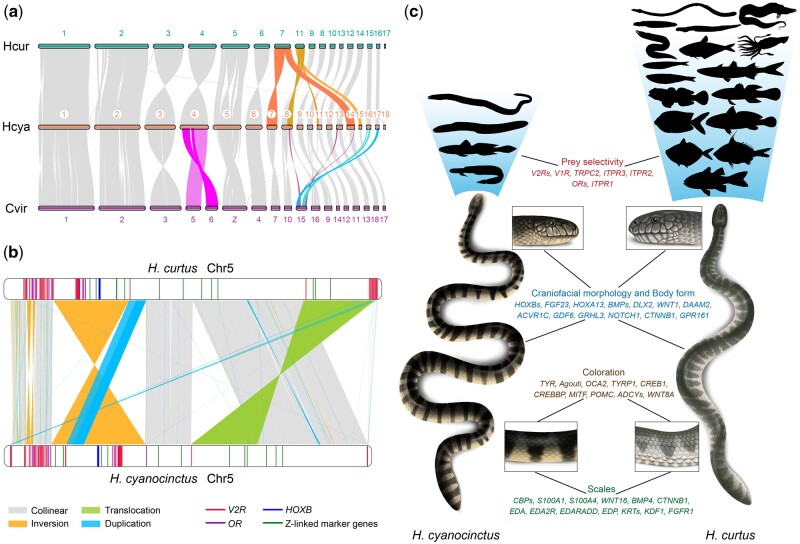
Fig. 3.
Global and local genomic structural differences promote divergent evolution in sea snakes. (a) Chromosomal synteny between Hydrophis cyanocinctus, H. curtus, and Crotalus viridis. Syntenic blocks are linked by shaded bands. Potential chromosome fissions/fusions are highlighted with different colors. (b) Local rearrangements and structural varaitions (SVs) between Chromosome 5 of H. cyanocinctus and H. curtus. Key functional genes are denoted with colored sticks in the chromosome containers. The largest inversion INV16903 and translocation INVTR19226 together cover 90 V2R genes and seven ChrZ-linked markers (NOSIP, MYST2, GH1, TUBG1, GAD2, WAC, and KLF6) in H. curtus. (c) Schematic of distinct phenotypic traits of the two sea snakes and associated genes interfered by candidate SVs. The silhouettes represent varied body shapes and taxonomy of prey as listed in supplementary table S25, Supplementary Material online.
Genomic Structural Variations
Despite their relatively recent evolutionary origin, true sea snakes are considerably diverse in morphological, physiological, and ecological phenotypes, which are well embodied between H. cyanocinctus and H. curtus (supplementary note 6.1, Supplementary Material online). To elucidate the potential mechanisms underlying their differentiated phenotypic traits, we screened the two genomes for local rearrangements and interspecific structural variants (SVs). Defining HCur_v2 as the reference genome, we identified 28,423 SVs ≥500 bp in the HCya_v2 assembly. The set comprises 3,707 insertions, 4,035 deletions, 243 inversions, 4,371 translocations, 8,366 duplications, and other complicated variations (supplementary note 6.2 and table S26, Supplementary Material online).
We explored large variations highlighted in balanced SVs (inversions and translocations). 28 balanced SVs with lengths over 1 Mb were noticed, of which five exceeding 10 Mb are the longest SVs in our analysis (supplementary table S31, Supplementary Material online). Interestingly, the Z Chromosome (Chr5) bore more intrachromosomal rearrangements (43.9% of its length in H. curtus) with the largest inversion INV16903 spanning 35.6 Mb and inverted translocation INVTR19226 spanning 33 Mb (fig. 3b and supplementary fig. S13, Supplementary Material online), which were verified by PacBio long reads (supplementary fig. S14, Supplementary Material online). The two rearranged regions in H. curtus contain 694 and 336 genes respectively, with seven of 12 Z-linked marker genes included. This is discordant with the previous view that there are no large-scale structural differences or gene order changes on Z chromosomes among squamate reptiles (Matsubara et al. 2006; Vicoso et al. 2013), and may reveal the ongoing evolution of heteromorphic (Z/W) sex chromosomes and linked genes in Hydrophiini snakes driven by suppressed recombination (Hughes et al. 2010; Kozak et al. 2017). Notably, we found dozens of V2R genes (39, 51, 24, respectively) harbored within the top three longest SVs (fig. 3b and supplementary table S32, Supplementary Material online). This is probably correlated with the bulkiness of V2R family in the genomes, but it is more likely that these functional genes participating in mating and predation in snakes may have accumulated more mutations or undergone stronger selection in large inverted regions than in collinear regions, leading to phenotypic differences and ecological adaptive divergence between populations, as well as reproductive isolation and subsequent speciation (Noor et al. 2001; Wellenreuther and Bernatchez 2018). Moreover, for V2R/V1R genes, we identified 52 balanced SVs with breakpoints disrupting the genic regions and 39 copy number variations (CNVs) hitting the exons (supplementary tables S33 and S34, Supplementary Material online). We also found two CNVs (INS17118 and CPG26290) affecting ITPR3 and TRPC2, which are essential for calcium ion transmembrane transport in vomeronasal signal transduction (Brykczynska et al. 2013; Silva and Antunes 2017). As the vomeronasal receptors of snakes play a significant part in prey selection (Halpern and Frumin 1979; Burghardt 1993; Saviola et al. 2013), these SVs associated with vomeronasal genes can to some extent reveal the genetic changes that might contribute to the diet divergence, where H. cyanocinctus prefers eels and eel-like elongated (anguilliform) fish and H. curtus—a representative generalist—takes a wide range of benthic, demersal, and pelagic fish from different depths (fig. 3c and supplementary table S25, Supplementary Material online).
We further identified scores of candidate SVs associated with other phenotypic variations between the two sea snakes (supplementary note 6.3, Supplementary Material online). Interestingly but not surprisingly, we found the CNVs among them mostly falling within or across LTRs (supplementary fig. S20, Supplementary Material online). This in some sense reveals the potential functions of TEs in facilitating structural genomic variations (Kazazian 2004; Bohne et al. 2008) and regulating gene expression by altering cis-regulatory elements (Saenko et al. 2015; Woronik et al. 2019). Although they need further experimental validation, these SVs will be a valuable resource in studying the genetic mechanisms underlying differentiated phenotypes and speciation of sea snakes (fig. 3c).
Divergent Evolution of the Venom System
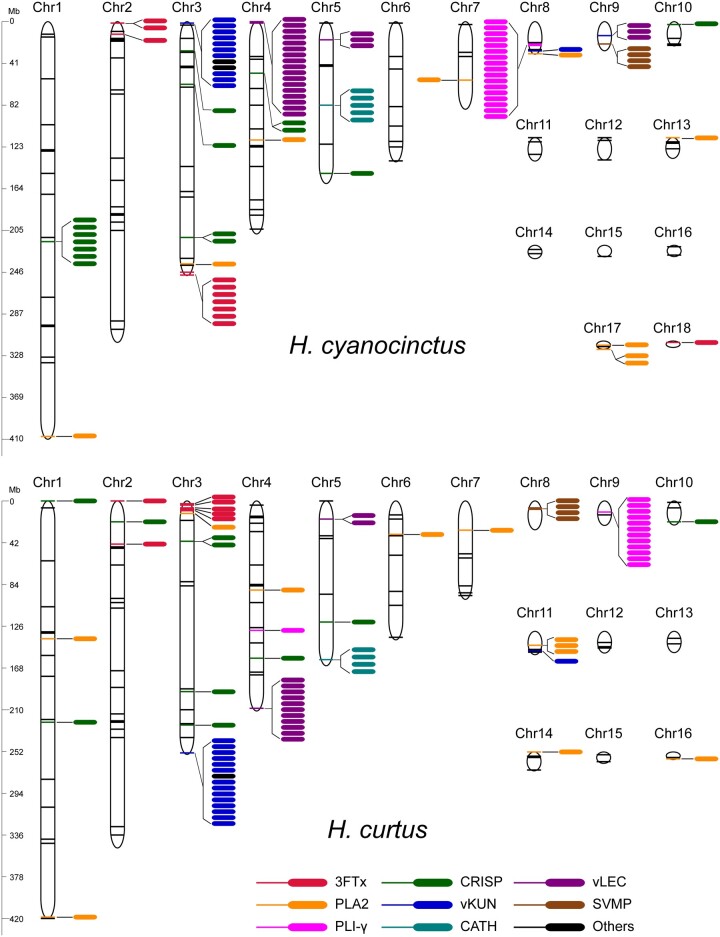
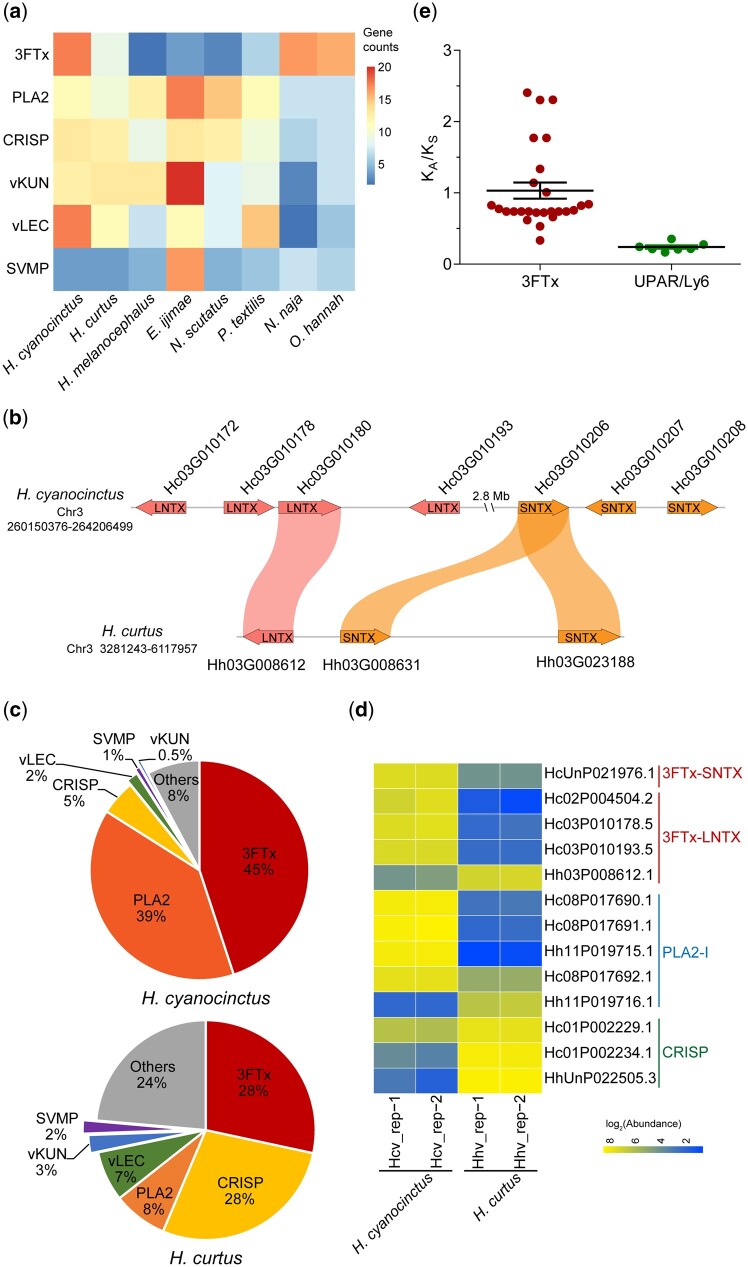
We comprehensively annotated venom-related genes and pinpointed their genomic locations and organization in the two sea snake genomes through a multistep approach (supplementary note 7.1, Supplementary Material online). We identified 118 and 102 venom genes from 32 families in the genome of H. cyanocinctus and H. curtus, respectively, of which there are 60 and 49 venom toxin genes for each species (table 2). Meanwhile, hundreds of nonvenom paralogs were also annotated (supplementary tables S36 and S37, Supplementary Material online). These venom-relevant genes are dispersed throughout the chromosomes but concentrated on macrochromosomes, especially the first four, which is consistent with the N. naja genome but in contrast to the C. viridis genome, where the venom genes are mostly assigned onto microchromosomes (Schield et al. 2019; Suryamohan et al. 2020) (fig. 4 and supplementary table S38, Supplementary Material online). Representative shared venom gene families in the two sea snakes are three-finger toxin (3FTx), phospholipase A2 (PLA2), cysteine-rich secretory protein (CRISP), venom Kunitz (vKUN), venom lectin (vLEC), and snake venom metalloproteinase (SVMP), which have also been found in many other terrestrial and marine elapids (Vonk et al. 2013; Durban et al. 2018; Suryamohan et al. 2020). Comparative statistics showed that most venom gene families have similar sizes across the two genomes with vKUN (15 copies in H. curtus) and vLEC (20 copies in H. cyanocinctus) as the largest two families, demonstrating tandem or proximal duplication mainly on Chromosomes 3 and 4, respectively. The most striking difference of venom gene copy number comes from the 3FTx family, which has 20 copies in H. cyanocinctus but ten in H. curtus. Of note, the genes of 3FTx, PLA2, and CRISP exhibit poor microsynteny relative to those of vKUN, vLEC, and SVMP between the two species (fig. 5b and supplementary fig. S21, Supplementary Material online). We further hunted for six major venom-related gene families and compared their copy numbers in nine Elapidae snakes (fig. 5a). In general, the families of PLA2, CRISP, vKUN, and vLEC may have expanded in the common ancestor of Hydrophiinae snakes, whereas the 3FTx family appeared to have undergone independent contraction in different lineages of Hydrophiinae, demonstrated herein by H. curtus, E. ijimae, and their terrestrial relatives (Notechis scutatus and P. textilis). Another noteworthy point is that the family size of SVMP is extremely high in E. ijimae (19 copies), which is likely to be linked to its specialized diet of fish eggs.
Table 2.
Major Venom Gene Families in the Genomes of Hydrophis cyanocinctus and H. curtus.
| Familya |
H. cyanocinctus
|
H. curtus
|
||||
|---|---|---|---|---|---|---|
| Total Paralogs | Venom Genes | Venom Toxin Genes | Total Paralogs | Venom Genes | Venom Toxin Genes | |
| 3FTx | 20 | 20 | 20 | 10 | 10 | 10 |
| PLA2 | 13 | 4 | 4 | 11 | 4 | 3 |
| CRISP | 15 | 7 | 7 | 14 | 6 | 6 |
| vKUN | 14 | 14 | 2 | 15 | 15 | |
| vLEC | 20 | 20 | 2 | 13 | 13 | |
| SVMP | 4 | 4 | 4 | 4 | 4 | 4 |
| SVSP | 3 | 3 | 2 | 3 | 3 | 2 |
| LAAO | 3 | 3 | 3 | 3 | 3 | 3 |
| VF | 3 | 3 | 3 | 4 | 4 | 4 |
| NGF-β | 5 | 2 | 2 | 5 | 2 | 2 |
| PDGF/VEGF | 11 | 2 | 1 | 11 | 2 | 1 |
| NP | 5 | 1 | 1 | 4 | 1 | 1 |
| AChE/CES | 21 | 3 | 3 | 19 | 2 | 2 |
| CATH | 5 | 5 | 4 | 3 | ||
| Waprin | 5 | 5 | 2 | 2 | ||
| LIP | 21 | 3 | 18 | 2 | ||
| CST | 6 | 3 | 6 | 4 | ||
| 5′-NT | 2 | 2 | 3 | 3 | ||
| PDE | 7 | 1 | 1 | 7 | 1 | 1 |
| PLB | 3 | 1 | 1 | 3 | 1 | 1 |
| Ficolin | 2 | 1 | 1 | 6 | 5 | 5 |
| HYAL | 6 | 1 | 6 | 1 | ||
| MCO | 5 | 1 | 5 | 1 | 1 | |
| TCTP | 2 | 2 | 1 | 1 | ||
| QPCT | 2 | 1 | 2 | 1 | ||
| CAL | 1 | 1 | 1 | 1 | ||
| CREC | 1 | 1 | 1 | 1 | ||
| DPP-IV | 7 | 1 | 5 | 1 | ||
| Others | 3 | 3 | 5 | 3 | ||
| Total | 118 | 60 | 102 | 49 | ||
Full names of the family abbreviations are available in supplementary table S35, Supplementary Material online.
Fig. 4.
Genomic locations of major venom-related genes in Hydrophis cyanocinctus and H. curtus. Colored sticks indicate the locations of venom-related genes along chromosomes, and colored ellipses denote gene copies in clusters. The venom-related genes on unassigned scaffolds are not shown (supplementary tables S36 and S37, Supplementary Material online). 3FTx, three-finger toxin; PLA2, phospholipase A2; CRISP, cysteine-rich secretory protein; vKUN, venom Kunitz-type protease inhibitor; vLEC, venom lectin; SVMP, snake venom metalloproteinase; PLI-γ, gamma-type phospholipase A2 inhibitor; CATH, cathelicidin.
Fig. 5.
Divergent evolution of venom gene families observed in multiomics. (a) Matrix of copy numbers of six venom-related gene families identified in the genomes of eight Elapidae species. (b) Microsynteny of 3FTx genes on Chromosome 3 of Hydrophis cyanocinctus and H. curtus. (c) Pie charts displaying the percentage composition of venom-related transcripts in the venom gland mRNA expression profile of H. cyanocinctus and H. curtus. Transcript abundance levels were measured from three individuals per species using the algorithm of FPKM (fragments per kilobase of gene per million mapped reads). (d) Heat map showing the differential expression of venom proteins of 3FTx, PLA2, and CRISP family. Each row refers to a homologous protein group detected in the pooled venom proteome by mass spectrometry. (e) Comparison of KA/KS values of 3FTx genes and their nontoxin paralogs (UPAR/Ly6 domain-containing genes) in H. cyanocinctus. Horizontal lines among the dots represent the means±SEM. 3FTx, three-finger toxin; SNTX, short neurotoxin; LNTX, long neurotoxin; PLA2-I, type-I phospholipase A2; CRISP, cysteine-rich secretory protein; vKUN, venom Kunitz-type protease inhibitor; vLEC, venom lectin; SVMP, snake venom metalloproteinase.
To compare the functional expression of 3FTx genes, we carried out sequencing and analysis of the venom gland transcriptome and venom proteome of the two sea snakes. As expected, the transcripts of 3FTx, PLA2, CRISP, vLEC, vKUN, and SVMP occupy the major proportion (92.2% in H. cyanocinctus and 76.4% in H. curtus) of expression abundance of venom-related genes. Interestingly, the mRNA composition ratio of the three chief venom families differs obviously between the two snakes. Although 3FTx transcripts constitute the highest portion (45%) in venom-relevant gene expression in H. cyanocinctus, their contribution in H. curtus dips to 28%, just in line with that of CRISP (fig. 5c). The relative proportion of PLA2 versus CRISP is concomitantly inversed in H. cyanocinctus (39% vs. 5%) and H. curtus (8% vs. 28%). Surprisingly, this distinction was remarkably amplified on the protein level. Comparative quantitation of the pooled venom proteins displayed a marked upregulation of four 3FTx groups by 9.8∼66.2-fold whereas only one long-chain (3FTx-LNTX) group less expressed in H. cyanocinctus (fig. 5d and supplementary table S44, Supplementary Material online). Such is also the case with PLA2. On the contrary, the expression of CRISP was downregulated in H. cyanocinctus relative to H. curtus by 2.8∼60.1-fold. Therefore, the gene dosage effect of the 3FTx family is consistent on the mRNA and protein level, indicating partial degeneration and inactivation of 3FTx genes in H. curtus. Nonetheless, the regulating tendencies appeared more complicated than expected in the venom gland transcriptome and venom proteome, which requires more investigation of the regulatory networks and underlying mechanisms of the diversity in venom composition.
For a deeper understanding of the evolution of 3FTx family in sea snakes, we evaluated the evolutionary rates of major venom families in H. cyanocinctus and H. curtus by calculating the numbers of synonymous (KS) and nonsynonymous (KA) nucleotide substitutions per site for each pair of venom genes and nonvenom paralogs. For the 3FTx-like, PLA2, CRISP, LEC, and MP families in H. cyanocinctus, the KA/KS substitution ratios of venom gene pairs present significantly higher than those of nonvenom gene pairs (fig. 5e and supplementary fig. S22, Supplementary Material online). Molecular phylogeny of the 3FTx family across Elapidae reflects the classification of long neurotoxin, short neurotoxin, and nonconventional subfamilies, which is locally in conformity with the species phylogeny within Elapidae. The 3FTx proteins of H. cyanocinctus and H. curtus gather in clades mostly close to those of Hydrophiinae snakes (supplementary fig. S23, Supplementary Material online).
Conclusions
Snakes are an ideal model for studying early vertebrate genome structure, molecular basis linking diverse phenotypes with extreme environments, venom evolution, and the role of TEs in adaptation. Hydrophis is the primary lineage in true sea snakes with a spectacularly rapid radiation of around 48 species and exhibits a significantly greater speciation rate than most genera in Elapidae (Sanders et al. 2010; Lee et al. 2016), making it a suitable model for evolutionary research on secondary marine adaption and associated extreme traits. We generated chromosome-scale genome assemblies of two true sea snakes, representative species demonstrating both convergent evolution within Hydrophiini and divergent evolution within Hydrophis in terms of phenotypic and venom-related traits. Using bioinformatic methodologies of comparative genomics, we uncovered extensive genetic innovations in true sea snakes including LTR expansion, a boom in the V2R family, and gain, loss, and directional selection of genes that were likely to propel novel adaptive traits emerging during the land-to-sea transition. The expansion of the V2R repositories is striking in the two Hydrophis snakes and might have occurred in the common ancestor of Hydrophiini. We built a highly reliable phylogeny of squamata with stable topology supporting the sister-group relationship of these Hydrophis snakes with Australasian terrestrial elapids, whose divergence dated to the early Miocene. The timeline of LTR insertion supports the claim that invasion of the oceans may not have by itself triggered the rapid diversification of Hydrophis, which instead was more likely driven by recent geological and climatic changes that led to repeated vicariance in Plio-Pleistocene, as well as other ecological factors such as diet specialization (Lukoschek and Keogh 2006; Sanders et al. 2010; Lee et al. 2016; Ukuwela et al. 2016).
We focused on genomic variations between the two species that could contribute to divergent evolution in true sea snakes. Interchromosomal fusions/fissions occurred frequently among Elapidae and between Hydrophis snakes, giving rise to variable karyotypes and chromosomal organization. Large intrachromosomal rearrangements including inversions and inverted translocations tend to fall onto the Z Chromosome and cover key gene clusters like V2Rs and HOXBs, which are linked to crucial phenotypic divergence regarding prey selection and body development. We identified a series of SVs in genic and intergenic regions affecting genes potentially associated with differentiated phenotypic traits of craniofacial shape, body form, skin color, scales, and sense of smell. We also found that the expanded TEs might be greatly involved in inducing SVs in regulatory regions, leading to a shift in gene expression. Finally, we compared the size and genomic distribution of venom gene families and detected a loss of ancestral 3FTx genes in H. curtus. The gene dosage effects were observed in venom gland transcriptome and magnified in venom proteome. Given that 3FTx is the principal toxin family in the venom of elapids and acts as the most lethal weapon against the neuromuscular transmission of prey (Phui Yee et al. 2004; Barber et al. 2013), the partial loss-of-function of 3FTx and accompanying regulation of the venom composition of major toxin cocktail in H. curtus is considered to be closely bound up with different prey preferences between the two sea snakes. Collectively, these interspecies genetic variations manifested on a multiomic level can in some degree explain the molecular basis giving impetus to divergent phenotypic evolution in true sea snakes and more specifically, within Hydrophis. Further investigation is needed to validate the candidate SVs associated with phenotype differences and clarify the deeper regulatory mechanisms of gene expression by TEs.
We anticipate that these high-quality sea snake genomes will provide a major resource for addressing high-profile issues in the evolutionary biology of snakes and for comparative studies on marine tetrapods that show convergent extreme adaptation from land back to sea. Furthermore, our sea snake multiomic data can promote antivenom development and help establish and enrich databases used in high-throughput discovery of marine-derived bioactive molecules.
Materials and Methods
A full description of the methods can be found in the supplementary notes, Supplementary Material online.
Animals and Samples
A total of four annulated sea snakes (H. cyanocinctus, QH_1, QH_2, QH_3, QH_4) and four spine-bellied sea snakes (H. curtus, PK_1, PK_2, PK_3, PK_4) were used in this study, which were captured in the South China Sea adjoining Sanya, China. Venom was extracted from the snakes 2–3 days prior to sacrificing. Genomic DNA was extracted from the muscle tissue of two male individuals (QH_4 and PK_4) using the QIAamp DNA mini kit (Qiagen, Hilden, Germany). Total RNA was prepared from the muscle tissue of QH_4 and PK_4 and venom gland tissue of all specimens using the Ambion kits (Ambion-1561).
Genome and Transcriptome Sequencing
For genome survey, short-insert (350 bp) paired-end sequencing libraries were constructed from genomic DNA of the two species using the TruSeq DNA LT Sample Prep kit (Illumina, San Diego, CA) and sequenced on the Illumina HiSeq X Ten platform. This generated 265.28 and 219.18 Gb raw data for H. cyanocinctus and H. curtus, respectively, which were then filtered by Trimmomatic v0.33. The genome sizes were estimated by k-mer distribution analysis (k = 17) with Jellyfish v2.2.3. For long-read single-molecule real-time (SMRT) sequencing (Chaisson et al. 2015), SMRTbell libraries with an average insert size of 20 kb were prepared from genomic DNA using PacBio SMRTbell Express Template Prep Kit v1.0 (Pacific Biosciences, Menlo Park, CA) as per the manufacturer’s instructions. SMRT sequencing was performed in 10–11 SMRT cells for each species on the PacBio Sequel system, producing 146 Gb (72×) and 138 Gb (68×) subreads for H. cyanocinctus and H. curtus, respectively. The Hi-C (chromosome conformation capture) (Mascher et al. 2017) sequencing library of each species was generated from the same snake individual (QH_4 and PK_4) subjected to short- and long-read sequencing. The resulting 150-bp paired-end raw reads were processed by fastp v0.20.0 and we obtained 230.7 and 229.9 Gb clean Hi-C data for H. cyanocinctus and H. curtus, respectively.
For short-read RNA sequencing (RNA-seq), the transcriptomic libraries were constructed from venom glands of three snake individuals for each species (QH_1, QH_2, QH_3, and PK_1, PK_2, PK_3) using TruSeq Stranded mRNA LT Sample Prep Kit following the standard Illumina RNA-seq library preparation pipeline. The libraries were sequenced using Illumina HiSeq X Ten sequencer, yielding 37.27 and 37.1 Gb of 150 bp paired-end raw reads in total for H. cyanocinctus and H. curtus, respectively, which were processed with Trimmomatic to obtain clean data. Thereafter the clean reads were de novo assembled into transcripts using Trinity v2.8.2. For full-length transcript sequencing, Iso-Seq (isoform sequencing) SMRTbell libraries for each species were generated using the pooled RNA sample from muscle and venom gland of individual QH_4 and PK_4, respectively. The libraries were constructed according to the “Procedure & Checklist—Iso-Seq Template Preparation for Sequel Systems.” Two SMRT cells were sequenced on the Pacbio Sequel platform yielding 27.7 and 22 Gb subreads for H. cyanocinctus and H. curtus, respectively. Raw subreads were first processed via IsoSeq v3 to get high-quality consensus isoform sequences. We then run LoRDEC v0.7 for base-correction using Illumina RNA-seq data from the same species.
Genome Assembly and Evaluation
The genomic Pacbio long reads were initially assembled into contigs in Falcon and Falcon-Unzip (Chin et al. 2016) with a coverage cutoff of 35×. Sequence contamination from prokaryotes and plants were removed through a BlastN search. Allelic contigs in highly heterozygous genomic regions were identified and processed with Purge Haplotigs. We adopted Arrow v2.1 to give the first round of polishing with the raw long reads. We then used Hi-C data to produce long-range scaffolds anchored onto chromosomes. The Pacbio-derived draft assemblies and clean Hi-C data were used as input for Juicer v1.5.7 to create high-resolution genomic contact maps. The Hi-C read pairs were mapped to the draft assembly contigs using BWA-MEM and the alignments were imported into 3D-DNA v170123 (Dudchenko et al. 2017), in which the input contigs were ordered, oriented and grouped into chromosome-length scaffolds based on the Hi-C interaction links. After misjoin correction, the exported scaffolding data and interaction matrices were loaded into Juicebox v1.11.08 to perform manual inspection. The output was introduced back into 3D-DNA and Ns were added between adjoining contigs. The final chromosome-length assemblies were worked out by gap filling and several rounds of base error correction with Pacbio and Illumina reads. We identified 18 and 17 chromosomes in HCya_v2 and HCur_v2, respectively, of which we could clearly distinguish seven macrochromosomes in the Hi-C heatmaps of both species (supplementary fig. S2, Supplementary Material online). For H. cyanocinctus, the chromosome number and boundary are identical with the karyotype reported previously (seven macro- and 11 microchromosomes) (Zheng and Hong 1983). The assembly quality of our genomes and recently reported six snake genomes (C. viridis, C. tigris, N. naja, E. ijimae, H. melanocephalus, and H. curtus) were evaluated using BUSCO v3.1.0 based on 3,950 tetrapod vertebrate orthologous genes in the tetrapoda_odb9 data set. The QV scores were assessed by Merqury (Rhie et al. 2020).
Repeat Element Analysis
For repeat annotation, we first applied the EDTA (Extensive De Novo TE Annotator) (Ou et al. 2019) v1.7.0 pipeline and other programs including RepeatMasker v4.0.9 (Tarailo-Graovac and Chen 2009) and RepeatModeler v1.0.11 to de novo identify each repeat class and produce an integrated repeat library. Moreover, we used RepeatMasker to implement homology-based annotation with the EDTA reference repeat library and combined with structural annotation from the raw step. The predicted repeat sequences matching to protein domains were weeded out by BlastX search against the chordata proteins in Swiss-Prot database, resulting in a highly reliable and nonredundant library of repetitive elements. To verify the annotation accuracy, the genomes were hard-masked by replacing the potential repeat sequences with N bases, and afterwards we assessed the BUSCO completeness and mapping rates of RNA-seq reads.
We invoked LTR_retriever (Ou and Jiang 2018) to run the LAI program to evaluate and compare the assembly quality of repeat elements utilizing long terminal repeat (LTR) retrotransposons. The LTR insertion times (T) were estimated employing LTR_retriever which calculated according to the equation T = d/2 μ with μ as the mutation rate and d as the sequence divergence between LTR pairs.
Gene Annotation
We implemented the MAKER (Cantarel et al. 2008) and BRAKER (Hoff et al. 2016) pipeline to predict protein-coding genes using different types of genetic evidence. For transcript evidence, the RNA-seq clean reads were mapped onto the reference genomes and assembled in a genome-guided way through Trinity and StringTie v1.3.3b. Afterwards, PASA was applied to merge the two genome-guided transcript assets and the de novo transcriptome assembly (see above) into a comprehensive assembled transcriptome. For homologous protein evidence, we downloaded protein sequences of closely related species from public databases (supplementary table S12, Supplementary Material online). The integrated transcripts and protein homologs were aligned to the repeat-soft-masked genomes using BLAST and Exonerate to run the first round of MAKER annotation. After quality evaluation by BUSCO analysis, the predicted genes with annotation edit distance (AED) <0.1 were selected to train the gene predictor SNAP. Meanwhile, the RNA-seq and Iso-Seq data were mapped to the repeat-hard-masked genomes using HISAT2 and minimap2, respectively. Based on the resulting alignments together with the homologous proteins, BRAKER performed iterative gene prediction to train and refine gene models invoking Augustus and GeneMark-ES. The second round of MAKER annotation was ab initio gene prediction using all the previous output. The detailed annotation results are listed in supplementary table S13, Supplementary Material online.
For gene functional annotation, we ran DIAMOND (Buchfink et al. 2015) to conduct homolog search of the annotated protein sequences against the NR, Swiss-Prot, TrEMBL, KEGG, KOG, and eggNOG database. The functional prediction of a protein came from the best hit with the highest sequence identity. We also applied InterProscan v5.36-75.0 to annotate the protein domain and motif features mapped in InterPro and Pfam database. The resulting InterPro hits were used to retrieve the associated GO terms (Ashburner et al. 2000).
For annotation of noncoding RNAs, the repeat-hard-masked genomes were queried against the Rfam (Griffiths-Jones et al. 2005) database using BlastN. For “noncoding” identification of long noncoding RNAs (lncRNAs), the predicted lncRNAs were further processed by sequence alignments with the reference transcripts and then going through the pipeline of CPC, CNCI, PLEK, Pfam to screen, and exclude those with protein-coding potential. The transfer RNAs and ribosomal RNAs were annotated by tRNAscan-SE and RNAmmer, respectively.
Ortholog Calling
Annotated protein sets of the 14 related species were downloaded (seven snakes, six lizards, and the outgroup Gallus gallus, supplementary table S12, Supplementary Material online) and the longest protein isoform was selected for each gene locus. We followed the OrthoFinder (Emms and Kelly 2015) pipeline invoking DIAMOND and OrthoMCL to call orthogroups based on sequence identity. Consequently, 16,795 orthologous gene families were clustered, containing 18,795 genes of H. cyanocinctus and 18,484 genes of H. curtus, and 679 single-copy orthologous genes of the 16 species were identified (supplementary tables S15 and S16, Supplementary Material online).
Species Phylogeny
Protein sequences of each single-copy orthologous group from the 16 selected species were aligned using MAFFT and the alignments were then concatenated into a super-gene alignment (494,664 aligned amino acid positions). This supermatrix was used to infer a maximum-likelihood (ML) species phylogenetic tree by RAxML (Stamatakis 2014) and IQ-TREE (Nguyen et al. 2015), respectively, which displayed identical topology structures (supplementary fig. S5, Supplementary Material online). We estimated the divergence times between clades through penalized likelihood calculation implemented by the MCMCtree program in PAML (Yang 2007), using a Bayesian relaxed-molecular clock model calibrated with fossil time constraints (supplementary table S17, Supplementary Material online). The convergence and sufficient sampling were checked by Tracer.
Gene Family Evolution
We applied CAFE (De Bie et al. 2006) to identify the gene families that underwent expansion or contraction across the species phylogeny. Functional enrichment of these evolved genes was then carried out using the GO and KEGG pathway database based on hypergeometric distribution with the genes of H. cyanocinctus or H. curtus assigned as a reference set.
To characterize the duplication events of paralogs, the longest protein isoforms of all annotated genes in each sea snake genome were brought to self-to-self sequence alignments using BlastP with an E-value cutoff of 10−10. The BLAST output together with the gene annotations were imported into MCScanX (Wang et al. 2012) to analyze the gene duplication types depending on their sequence conservation and genomic distribution.
To explore the family evolution of V2Rs within Hydrophiini, the extracellular domain sequences of V2Rs in H. melanocephalus and E. ijimae were retrieved from their publication (Kishida et al. 2019). The two genomes were searched for gene regions containing 7TM domains of V2Rs through TBlastN using the 7TM sequences of V2Rs from H. cyanocinctus and H. curtus as query. The ML phylogenetic trees were constructed by IQ-TREE with the TAS1R (taste receptor type 1) proteins of Anolis carolinensis as outgroups.
Selection Test
We explored changes in the ratio of the nonsynonymous substitution rate (KA) to the synonymous substitutions rate (KS) using the codeml program from PAML with the Hydrophiini clade designated as foreground branch. Branch-site models were implemented and P values were then calculated assuming a χ2 distribution to determine whether a gene was under positive selection according to the likelihood values and differences between degrees of freedom.
Intergenomic Synteny
We first performed reciprocal alignments of the longest transcripts of all annotated genes of selected genomes using LAST to call orthologous gene pairs. MCScanX was then employed to identify intergenomic syntenic blocks using the LAST hits and genome annotation information. For the N. naja genome, as its gene annotations are not publicly available, we alternatively utilized all the gene locations (start and end positions) from the supplementary data of the publication (Suryamohan et al. 2020). The interspecies genome synteny was plotted using the JCVI utility libraries. The results show good synteny between Chr5 of our sea snakes with ChrZ of C. viridis and N. naja (fig. 3a and supplementary figs. S11 and S12, Supplementary Material online). Besides, we mapped the Z-linked marker genes conserved in snakes (Matsubara et al. 2006) to the sea snake genomes to recognize and confirm the assignment of Z Chromosomes (supplementary table S24, Supplementary Material online).
Identification of Genomic Structural Variations
Interspecies structural variations (SVs) between the genomes of H. cyanocinctus and H. curtus were identified using SyRI v1.2 (Goel et al. 2019). First, we ran Nucmer to perform a whole-genome alignment with the HCur_v2 assembly designated as the reference genome. Since there exist interchromosomal fissions/fusions between the two snakes, we manually coalesced the parted chromosomes (Chr7 + Chr14, Chr8 + Chr15 in H. cyanocinctus, and Chr9 + Chr11 in H. curtus) before alignment. The alignments were used as input for SyRI, which initially identified collinear and structurally rearranged (noncollinear) regions between homologous chromosomes. Inversions, translocations, and duplications were then sequentially annotated within rearranged regions. Afterwards, SyRI identified local structural and sequence variations by comparing the overlaps and gaps between consecutive alignments in annotation blocks. For insertions and deletions, we reserved those ≥500 bp in size for further study. The output of SyRI was plotted with the script plotsr (supplementary fig. S13, Supplementary Material online). We separately analyzed the balanced SVs (inversions and translocations) and unbalanced CNVs (copy number variations, herein including insertions, deletions, copy gains, copy losses, and tandem repeats), and recognized the genes that could be affected by or adjacent to the balanced SVs and CNVs respectively, defining the flanking range of 5 kb upstream and downstream from a coding gene as a genic region. The large SVs on Chr5 were verified by aligning the PacBio long reads to assembled genomes in minimap2 and visualizing the alignments in IGV.
Venom-Related Gene Identification
We first collected sequences of known venom-related protein families from public databases (supplementary table S35, Supplementary Material online) and manually reviewed sequences from the Animal toxin annotation project (Jungo et al. 2012) to construct a combined reference library. All annotated proteins of H. cyanocinctus and H. curtus were queried against the combined library using BlastP v2.8.1+ with an E-value cutoff of 10−5 and identity cutoff of 70%. For each candidate venom-related protein, we performed a reciprocal BlastP search against the NR and UniProt database, and a BlastN search against the NT database using the corresponding transcript sequence as query to confirm the hit. Next, we manually curated a venom-related gene according to the annotations of the closest homologous protein hit in Swiss-Prot. If the “tissue specificity” annotation contains “venom,” that gene was hence recognized as a venom gene. The venom genes were further identified as toxin genes if the “molecular function” keywords of the closest homologs are termed as “toxin” or “toxin activity.” We screened the genomes of six other elapids for the 3FTx, PLA2, CRISP, vKUN, vLEC, and SVMP families following the similar method. The local microsynteny of the six venom gene families were analyzed and plotted by JCVI.
Gene Expression Quantification
The RNA-seq clean reads from the venom glands of three snake individuals of each species (QH_1, QH_2, QH_3, and PK_1, PK_2, PK_3) were aligned to the reference genomes using HISAT2. The read counts mapped to each gene in each sample were then calculated with htseq-count v0.9.1. Gene expression abundance was measured by the algorithm of fragments per kilobase of gene per million mapped reads (FPKM) using Cufflinksv2.2.1 (Trapnell et al. 2010).
Venom Proteomes
To compare venom protein expression between H. cyanocinctus and H. curtus, we conducted tandem mass tag (TMT)-based quantitative proteomic analysis (Dayon et al. 2008) of pooled venom from eight individuals of the two species. Detailed experimental procedures of protein digestion, peptide labeling, liquid chromatography separation, and mass spectrometry (MS) are provided in supplementary note 7.3, Supplementary Material online. We used Proteome Discoverer v2.2 (Thermo) to search the MS raw data against the merged database of all the annotated proteins of the two snakes. A global false discovery rate (FDR) for protein identification was set to ≤0.01. The protein groups considered for relative quantification (credible proteins) were picked out following the criteria of Score Sequest HT >0 and unique peptides ≥1. Based on the relative abundance quantitation, we carried out t-test to determine differentially venom-expressed protein groups between the two snakes, according to the criterion of P value <0.05 and fold change ≥2 or ≤0.5.
Molecular Evolution and Phylogeny
For the evaluation of the diversification rates of major venom-related gene families, we ran self-to-self BlastN (E-value ≤10−5) within each sea snake using the longest transcripts annotated to obtain paralog pairs of venom genes and of nonvenom genes, respectively. ParaAT v2.0 was then used to perform pairwise alignment of these gene pairs. The output was imported into KaKs_Calculator v1.2 (Zhang et al. 2006) to compute the nonsynonymous and synonymous substitution rates (KA and KS) for each gene pair with the models of Yang–Nielsen (YN), modified YN (MYN), and Nei–Gojobori (NG). Significant differences between venom and nonvenom gene families were checked by Mann–Whitney test.
We collected the 3FTx protein sequences of 14 elapids and the outgroup C. viridis from their annotated genomes, transcriptomes, and Swiss-Prot. A ML tree of the 3FTx family in Elapidae was constructed with RAxML and visualized using iTOL.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 42076094, 81773627, and 81274162), the National Key Research and Development Program of China (Grant No. 2018YFC0310904), the National Major Scientific and Technological Special Projects for “Significant New Drugs Innovation and Development” (Grant No. 2019ZX09301119), and the Shanghai Science and Technology Innovation Action Plan (Grant No. 21S11902400). The authors would like to thank Xulei Fan, Yincong Gu, and Dr Dong An (OE Biotech, Inc., Shanghai, China) for their technical assistance with the bioinformatics analysis.
Data Availability
PacBio and Illumina sequencing data for genome and transcriptome have been deposited to the NCBI Sequence Read Archive (SRA) as Bioproject PRJNA573877, PRJNA616080, PRJNA608244, and PRJNA622900. The genome assembly sequences of Hydrophis cyanocinctus and H. curtus have been deposited at DDBJ/ENA/GenBank (under accession numbers: JAAZTL000000000 and JABAHG000000000, respectively). All other data are available from the corresponding author upon reasonable request.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CM, Isbister GK, Hodgson WC.. 2013. Alpha neurotoxins. Toxicon 66:47–58. [DOI] [PubMed] [Google Scholar]
- Bohne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN.. 2008. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 16(1):203–215. [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Tzika AC, Rodriguez I, Milinkovitch MC.. 2013. Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles. Genome Biol Evol. 5(2):389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Burghardt GM. 1993. The comparative imperative: genetics and ontogeny of chemoreceptive prey responses in natricine snakes. Brain Behav Evol. 41(3–5):138–146. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M.. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18(1):188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Gonzalez J.. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol. 22(6):1503–1517. [DOI] [PubMed] [Google Scholar]
- Castoe TA, de Koning AP, Hall KT, Card DC, Schield DR, Fujita MK, Ruggiero RP, Degner JF, Daza JM, Gu W, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 110(51):20645–20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, et al. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517(7536):608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13(12):1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe‐Riddell JM, D’Anastasi BR, Nankivell JH, Rasmussen AR, Sanders KL.. 2019. First records of sea snakes (Elapidae: Hydrophiinae) diving to the mesopelagic zone (>200 m). Austral Ecol. 44(4):752–754. [Google Scholar]
- Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC.. 2008. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 80(8):2921–2931. [DOI] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW.. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22(10):1269–1271. [DOI] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356(6333):92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J, Sasa M, Calvete JJ.. 2018. Venom gland transcriptomics and microRNA profiling of juvenile and adult yellow-bellied sea snake, Hydrophis platurus, from Playa del Coco (Guanacaste, Costa Rica). Toxicon 153:96–105. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel M, Sun H, Jiao WB, Schneeberger K.. 2019. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20(1):277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2020. Animal Genome Size Database. Available from: http://www.genomesize.com. Accessed December 15, 2020.
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A.. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33(Database issue):D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern M, Frumin N.. 1979. Roles of the vomeronasal and olfactory systems in prey attack and feeding in adult garter snakes. Physiol Behav. 22(6):1183–1189. [DOI] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M.. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32(5):767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, et al. 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463(7280):536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungo F, Bougueleret L, Xenarios I, Poux S.. 2012. The UniProtKB/Swiss-Prot Tox-Prot program: a central hub of integrated venom protein data. Toxicon 60(4):551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr. 2004. Mobile elements: drivers of genome evolution. Science 303(5664):1626–1632. [DOI] [PubMed] [Google Scholar]
- Kishida T, Go Y, Tatsumoto S, Tatsumi K, Kuraku S, Toda M.. 2019. Loss of olfaction in sea snakes provides new perspectives on the aquatic adaptation of amniotes. Proc Biol Sci. 286(1910): 20191828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak GM, Wadsworth CB, Kahne SC, Bogdanowicz SM, Harrison RG, Coates BS, Dopman EB.. 2017. A combination of sexual and ecological divergence contributes to rearrangement spread during initial stages of speciation. Mol Ecol. 26(8):2331–2347. [DOI] [PubMed] [Google Scholar]
- Kutsuma R, Sasai T, Kishida T.. 2018. How snakes find prey underwater: sea snakes use visual and chemical cues for foraging. Zoolog Sci. 35(6):483–486. [DOI] [PubMed] [Google Scholar]
- Lee MS, Sanders KL, King B, Palci A.. 2016. Diversification rates and phenotypic evolution in venomous snakes (Elapidae). R Soc Open Sci. 3(1):150277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind AL, Lai YYY, Mostovoy Y, Holloway AK, Iannucci A, Mak ACY, Fondi M, Orlandini V, Eckalbar WL, Milan M, et al. 2019. Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. Nat Ecol Evol. 3(8):1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoschek V, Keogh JS.. 2006. Molecular phylogeny of sea snakes reveals a rapidly diverged adaptive radiation. Biol J Linn Soc. 89(3):523–539. [Google Scholar]
- Margres MJ, Rautsaw RM, Strickland JL, Mason AJ, Schramer TD, Hofmann EP, Stiers E, Ellsworth SA, Nystrom GS, Hogan MP, et al. 2021. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proc Natl Acad Sci U S A. 118(4):e2014634118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544(7651):427–433. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y.. 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A. 103(48):18190–18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH.. 2003. Chromosomal speciation and molecular divergence–accelerated evolution in rearranged chromosomes. Science 300(5617):321–324. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J.. 2001. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 98(21):12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo E. 2005. Rate of chromosome changes and speciation in reptiles. Genetica 125(2–3):185–203. [DOI] [PubMed] [Google Scholar]
- Ou S, Chen J, Jiang N.. 2018. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46(21):e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Jiang N.. 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176(2):1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palci A, Seymour RS, Van Nguyen C, Hutchinson MN, Lee MS, Sanders KL.. 2019. Novel vascular plexus in the head of a sea snake (Elapidae, Hydrophiinae) revealed by high-resolution computed tomography and histology. R Soc Open Sci. 6(9):191099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquesi GIM, Adams RH, Card DC, Schield DR, Corbin AB, Perry BW, Reyes-Velasco J, Ruggiero RP, Vandewege MW, Shortt JA, et al. 2018. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat Commun. 9(1):2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Ren JL, Deng C, Jiang D, Wang J, Qu J, Chang J, Yan C, Jiang K, Murphy RW, et al. 2020. The genome of Shaw’s sea snake (Hydrophis curtus) reveals secondary adaptation to its marine environment. Mol Biol Evol. 37:1744–1760. [DOI] [PubMed] [Google Scholar]
- Phui Yee JS, Nanling G, Afifiyan F, Donghui M, Siew Lay P, Armugam A, Jeyaseelan K.. 2004. Snake postsynaptic neurotoxins: gene structure, phylogeny and applications in research and therapy. Biochimie 86(2):137–149. [DOI] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM.. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21(1):245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenko SV, Lamichhaney S, Martinez Barrio A, Rafati N, Andersson L, Milinkovitch MC.. 2015. Amelanism in the corn snake is associated with the insertion of an LTR-retrotransposon in the OCA2 gene. Sci Rep. 5:17118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders KL, Lee MS, Leys R, Foster R, Keogh JS.. 2008. Molecular phylogeny and divergence dates for Australasian elapids and sea snakes (Hydrophiinae): evidence from seven genes for rapid evolutionary radiations. J Evol Biol. 21(3):682–695. [DOI] [PubMed] [Google Scholar]
- Sanders KL, Lee MS, Mumpuni Bertozzi T, Rasmussen AR.. 2013. Multilocus phylogeny and recent rapid radiation of the viviparous sea snakes (Elapidae: Hydrophiinae). Mol Phylogenet Evol. 66:575–591. [DOI] [PubMed] [Google Scholar]
- Sanders KL, Mumpuni, Lee MS.. 2010. Uncoupling ecological innovation and speciation in sea snakes (Elapidae, Hydrophiinae, Hydrophiini). J Evol Biol. 23:2685–2693. [DOI] [PubMed] [Google Scholar]
- Saviola AJ, Chiszar D, Busch C, Mackessy SP.. 2013. Molecular basis for prey relocation in viperid snakes. BMC Biol. 11:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schield DR, Card DC, Hales NR, Perry BW, Pasquesi GM, Blackmon H, Adams RH, Corbin AB, Smith CF, Ramesh B, et al. 2019. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 29(4):590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Schmitz J.. 2019. The impact of transposable elements in adaptive evolution. Mol Ecol. 28(6):1537–1549. [DOI] [PubMed] [Google Scholar]
- Schwenk K. 1995. Of tongues and noses: chemoreception in lizards and snakes. Trends Ecol Evol. 10(1):7–12. [DOI] [PubMed] [Google Scholar]
- Seymour RS. 1974. How sea snakes may avoid the bends. Nature 250(5466):489–490. [DOI] [PubMed] [Google Scholar]
- Seymour RS, Webster MED.. 1975. Gas transport and blood acid-base balance in diving sea snakes. J Exp Zool. 191(2):169–181. [DOI] [PubMed] [Google Scholar]
- Silva L, Antunes A.. 2017. Vomeronasal receptors in vertebrates and the evolution of pheromone detection. Annu Rev Anim Biosci. 5:353–370. [DOI] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland JL, Carter S, Kraus F, Parkinson CL.. 2016. Snake evolution in Melanesia: origin of the Hydrophiinae (Serpentes, Elapidae), and the evolutionary history of the enigmatic New Guinean elapid Toxicocalamus. Zool J Linn Soc. 178(3):663–678. [Google Scholar]
- Suryamohan K, Krishnankutty SP, Guillory J, Jevit M, Schroder MS, Wu M, Kuriakose B, Mathew OK, Perumal RC, Koludarov I, et al. 2020. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat Genet. 52(1):106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N.. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 25(1):Unit 4 10. [DOI] [PubMed] [Google Scholar]
- Thewissen JGM, Nummela S.. 2008. Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates. Oakland (CA: ): University of California Press. [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Freed P, Hošek J. 2020. The Reptile Database. Available from: http://www.reptile-database.org. Accessed December 15, 2020.
- Ukuwela KDB, Lee MSY, Rasmussen AR, de Silva A, Mumpuni Fry BG, Ghezellou P, Rezaie-Atagholipour M, Sanders KL.. 2016. Evaluating the drivers of Indo-Pacific biodiversity: speciation and dispersal of sea snakes (Elapidae: Hydrophiinae). J Biogeogr. 43:243–255. [Google Scholar]
- Van Cao N, Thien Tao N, Moore A, Montoya A, Redsted Rasmussen A, Broad K, Voris HK, Takacs Z.. 2014. Sea snake harvest in the gulf of Thailand. Conserv Biol. 28(6):1677–1687. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D.. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11(8):e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJ, Kerkkamp HM, Vos RA, Guerreiro I, Calvete JJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 110(51):20651–20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L.. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol. 33(6):427–440. [DOI] [PubMed] [Google Scholar]
- Woronik A, Tunstrom K, Perry MW, Neethiraj R, Stefanescu C, Celorio-Mancera MP, Brattstrom O, Hill J, Lehmann P, Kakela R, et al. 2019. A transposable element insertion is associated with an alternative life history strategy. Nat Commun. 10(1):5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shi P, Zhang YP, Zhang J.. 2005. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics 86(3):306–315. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J.. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4(4):259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hong Y.. 1983. On the karyotypes of Hydrophis cyanocinctus Daudin. Sichuan J Zool. 2:7–9. [Google Scholar]
- Zheng Z, Jiang H, Huang Y, Wang J, Qiu L, Hu Z, Ma X, Lu Y.. 2016. Screening of an anti-inflammatory peptide from Hydrophis cyanocinctus and analysis of its activities and mechanism in DSS-induced acute colitis. Sci Rep. 6:25672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PacBio and Illumina sequencing data for genome and transcriptome have been deposited to the NCBI Sequence Read Archive (SRA) as Bioproject PRJNA573877, PRJNA616080, PRJNA608244, and PRJNA622900. The genome assembly sequences of Hydrophis cyanocinctus and H. curtus have been deposited at DDBJ/ENA/GenBank (under accession numbers: JAAZTL000000000 and JABAHG000000000, respectively). All other data are available from the corresponding author upon reasonable request.