Abstract
Throughout its distribution across Eurasia, domestic pig (Sus scrofa) populations have acquired differences through natural and artificial selection, and have often interbred. We resequenced 80 Eurasian pigs from nine different Asian and European breeds; we identify 42,288 reliable SNPs on the Y chromosome in a panel of 103 males, among which 96.1% are newly detected. Based on these new data, we elucidate the evolutionary history of pigs through the lens of the Y chromosome. We identify two highly divergent haplogroups: one present only in Asia and one fixed in Europe but present in some Asian populations. Analyzing the European haplotypes present in Asian populations, we find evidence of three independent waves of introgression from Europe to Asia in last 200 years, agreeing well with the literature and historical records. The diverse European lineages were brought in China by humans and left significant imprints not only on the autosomes but also on the Y chromosome of geographically and genetically distinct Chinese pig breeds. We also find a general excess of European ancestry on Y chromosomes relative to autosomes in Chinese pigs, an observation that cannot be explained solely by sex-biased migration and genetic drift. The European Y haplotype is associated with leaner meat production, and we hypothesize that the European Y chromosome increased in frequency in Chinese populations due to artificial selection. We find evidence of Y chromosomal gene flow between Sumatran wild boar and Chinese pigs. Our results demonstrate how human-mediated admixture and selection shaped the distribution of modern swine Y chromosomes.
Keywords: Y chromosome, admixture, selection, pig, genome sequencing
Introduction
The pig (Sus scrofa) provides humans an important source of animal protein and serves as an important biomedical model for human disease (Lunney 2007). Pigs were domesticated in at least two locations (Anatolia and China) approximately 10k years ago (Larson et al. 2005), and gradually formed a variety of breeds in Europe and Asia (Kijas and Andersson 2001; Wang et al. 2011; Ottoni et al. 2013). There is evidence of long-term gene flow between domestic pigs and wild boars during and after the domestication of pigs across Eurasia (Giuffra et al. 2000; Kijas and Andersson 2001; Frantz et al. 2015; Yang et al. 2017). Eurasia has the largest diversity of pig breeds, about one-third of which can be found in a variety of environments in China (Wang et al. 2011).
Studies on uniparental genetic markers have further revealed interesting details of pig evolutionary history. Analyses of mtDNA revealed two highly divergent lineages, one of which is present exclusively in Europe, whereas the other is present in both Europe and Asia (Kijas and Andersson 2001; Ramirez et al. 2009; Bosse, Megens, Madsen, et al. 2014), interpreted as sex-biased gene flow involving female pigs from Asia to Europe. In contrast, Ramirez et al. (2009), Cliffe et al. (2010), and Guirao-Rico et al. (2018) investigated porcine Y chromosome variation and inferred sex-biased gene flow in the opposite direction, a paternal migration event from Europe to China.
There are limited recapitulative historical records of pigs moving from Europe to China. At least since the 1840s, modern breeds such as Berkshire, Hampshire, local Russian pigs, Duroc, Large White, and Landrace were introduced into China (Xu 2004). But many questions remain open regarding this apparent male-biased migration from Europe to China: When did it happen exactly? Is there archaic migration from Europe to China? What is the origin of the European Y chromosome in Chinese pigs? Which populations in China received this migration pulse? And why is the European Y chromosome preserved in Chinese pigs? To answer these questions, we have resequenced 80 whole genomes of 9 various pig populations and taken advantage of genomic resources recently made available for this species, particularly on the Y chromosome.
Currently, there are 13 de novo assembled pig genomes (Fang et al. 2012; Groenen et al. 2012; Li et al. 2013, 2017; Vamathevan et al. 2013) publicly available. Furthermore, an improved assembly and gene annotation of the pig X chromosome and a draft assembly of the pig Y chromosome (VEGA62) were generated by sequencing BAC and fosmid clones from the Duroc breed and incorporating information from optical mapping and fiber FISH (Skinner et al. 2016). The improved porcine sex chromosomes were included in Build 11.1 of the S. scrofa reference genome with comprehensive gene annotation and variant information. These improved assemblies allowed us to generate a comprehensive analysis of the evolutionary history of pigs from the perspective of sex chromosomes, especially from the unique genetic perspective of patrilineal inheritance.
In this study, we analyzed 205 high-quality pig whole-genome sequence data from diverse populations, including three outgroups: Phacochoerus africanus, Sus verrucosus, and Sus celebensis. We confirm previous findings regarding the demography of Eurasian pigs based on autosomal and mtDNA variation and present new findings regarding the patrilineal evolutionary history of pigs. With a large new Y chromosomal SNP data set, we identify two highly divergent swine Y haplogroups: one present only in Asia and one fixed in Europe but present in some Asian breeds. Our results also provide evidence of three independent waves of introgression from Europe to Asia in the last 200 years. Finally, we identify Y chromosomal gene flow between Sumatran wild boar and Chinese pigs.
Results
Overview of Whole-Genome Sequencing and SNP Calling
We combined previously sequenced genomic data from 59 Chinese indigenous pigs representing the breeds of highest local importance (Ai et al. 2015), 24 other Chinese pigs (Zhu et al. 2017), and publicly available data for 39 pigs and 3 outgroups; in addition to that, we add 80 newly sequenced genomes of European and Chinese pigs, altogether making a high-quality data set of 205 individuals from 27 populations (fig. 1A and supplementary table S1, Supplementary Material online).
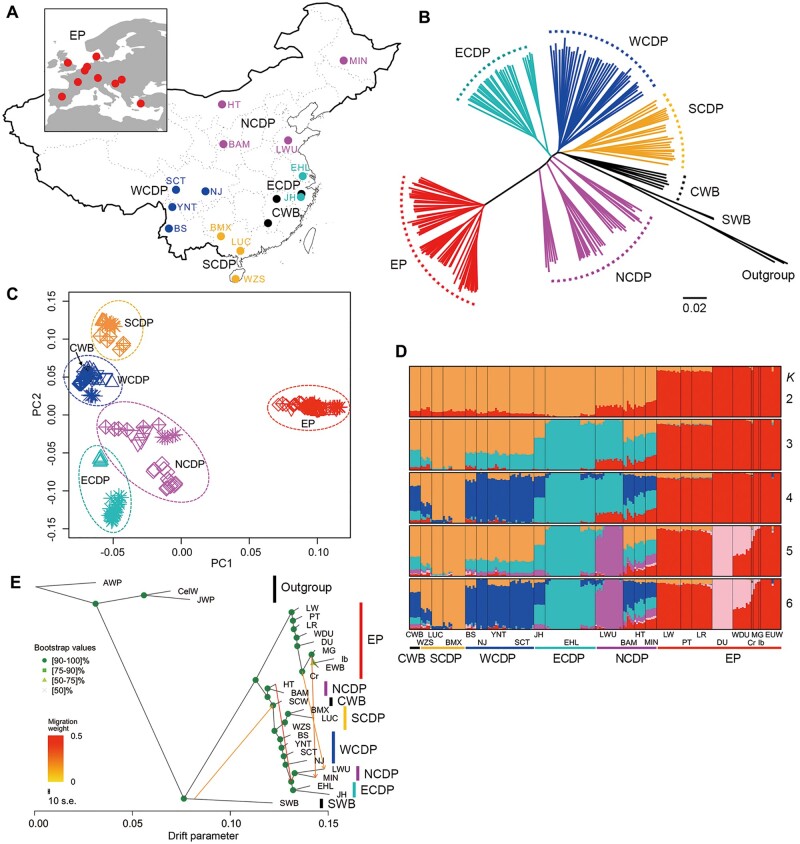
Fig. 1.
Demographic history of Eurasian pigs inferred using autosomal SNP data. (A) Geographical locations of Eurasian pigs analyzed in this study. EP, European pigs including Creole (Cr), Duroc (DU), Iberian (Ib), Large White (LW), Mangalica (MG), Landrace (LR), Pietrain (PT), and wild boars (EUW); ECDP, East Chinese domestic pigs including Erhualian and Jinhua; WCDP, West Chinese domestic pigs including Baoshan (BS), Neijiang (NJ), and Tibetan pigs in Yunnan (YNT) and Sichuan provinces (SCT); SCDP, South Chinese domestic pigs including Bamaxiang (BMX), Luchuan (LUC), and Wuzhishan (WZS); NCDP, North Chinese domestic pigs including Bamei (BAM), Hetao (HT), Laiwu (LWU), and Min (MIN); CWB, Chinese wild boars (CWB). (B) Neighbor joining phylogenic tree of all sequenced pigs. SWB, Sumatran wild boars. Sus celebensis (Celebes wild boar), Sus verrucosus (Java warty pig), and P. africanus (African warthog) were used as outgroups. (C) Principal component (PC) analysis plots based on the first two PCs. (D) ADMIXTURE analysis with K = 2–6. The optimal K value with the lowest cross-validation (CV) error was 6 in ADMIXTURE analyses (supplementary fig. S8, Supplementary Material online). Colors in each column represent ancestry proportion. (E) Relationships among Eurasian pigs inferred using TreeMix. Full names of the pig breeds are detailed in supplementary table S1, Supplementary Material online.
All data were mapped to Build 11.1 of the S. scrofa reference genome using BWA (Li and Durbin 2009). A total of 37,542,852 SNPs were identified in the 205 genomes (supplementary table S2, Supplementary Material online) using Platypus (Rimmer et al. 2014). This SNP data set included 36,332,442 autosomal SNPs, 1,167,598 SNPs on the X chromosome, 42,288 SNPs on the Y chromosome, and 524 SNPs on the mitochondrial DNA. The SNP calls were validated by comparing to a SNP array data set generated using the porcine 60K BeadChip genotyping array (Illumina), by dual resequencing of six pigs (picking six individuals to sequence for twice), and by comparisons to Build 150 of the S. scrofa dbSNP data set from the NCBI database (supplementary note S1 and figs. S1 and S2, Supplementary Material online). Using the latter comparison, we found that 81.3% (29,554,672) of the SNPs on autosomes were already present in dbSNP, and 18.7% (6,777,770) were novel (supplementary fig. S2 and table S3, Supplementary Material online). And the quality of new Y assembly (Skinner et al. 2016) was enough for our subsequent analysis (supplementary note S2 and figs. S3–S7, Supplementary Material online). On the X and Y chromosomes, 20.0% (233,093), 96.1% (40,646) were novel, respectively (supplementary table S3, Supplementary Material online). These novel SNPs, especially the 96.1% Y chromosomal SNPs, considerably expand the catalog of porcine genetic variants.
Demographic History Inferred by Autosomal Data
We constructed a neighbor-joining (NJ) tree for the above 205 animals using 36,332,442 autosomal SNPs. All individuals from the same population clustered together, and European pigs formed a clade clearly separated from Chinese pigs in the NJ tree (fig. 1B). We also conducted principal component (PC) (Price et al. 2006), ADMIXTURE (Alexander et al. 2009) and TreeMix (Pickrell and Pritchard 2012) analyses to further assess the population structure. Chinese wild boars clustered into one group (fig. 1B) and consisted of four major ancestry components (fig. 1D), which correspond roughly to their geographical distributions: South China, West China, East China, and North China (fig. 1B–D). European pigs formed an independent group that was genetically distinct from Chinese pigs in both the ADMIXTURE results (fig. 1D) and the PC plot (fig. 1C). The genomes of the three European commercial pig breeds (LW, PT, and LR) contain approximately uniform proportions of South Chinese ancestry (fig. 1D, orange component), which is consistent with the history of importation of pigs from South China to Europe during the Industrial Revolution (Phillips and Hsu 1944; McLaren 1990; White 2011). Moreover, East Chinese pig ancestry was detected in the genomes of several European commercial pigs as well (LW, PT, and LR; fig. 1D, k = 3 and k = 4), which is in line with records that East Chinese pigs, such as Meishan pigs, were imported to improve reproductive traits of European and US local pigs (McLaren 1990). Significant signals of gene flow from both South and East Chinese pig breeds to European pig breeds were also detected using D statistics (supplementary table S4, Supplementary Material online). Notably, European ancestry was also detected in Chinese pigs in different geographic regions, especially in North Chinese pigs including Min pigs (MIN), Hetao pigs (HT), and Bamei pigs (BAM).
According to the TreeMix results, deep divergences were inferred not only between Sumatran and Eurasian pigs, but also between European and Chinese pigs (fig. 1E). Among Chinese pigs, partial North Chinese (HT and BAM) pigs showed a closer relationship with European pigs, and all other Chinese pig populations showed signals of admixture with Sumatran wild boars (fig. 1E).
When we added four migration edges, 99.4% of variation in the data were explained by TreeMix and OptM suggested optimal results (supplementary fig. S9, Supplementary Material online). Allowing four migration events (m = 4), we infer an admixture event from the Sumatran wild boar (SWB) contributing 16.1% of genetic ancestry to the ancestor of the Chinese wild boar and all Chinese pigs except for the partial North Chinese pigs (HT and BAM; fig. 1E). Admixture from Sumatran wild boar to Chinese pigs was also supported by D statistics (supplementary table S4, Supplementary Material online). One admixture edge is inferred from HT to the common ancestors of partial Northern (LWU and MIN) and Eastern Chinese populations (fig. 1E). We also detected migration events from ancestor of European Mangalica (MG) pigs to MIN and from ancestor of Creole (Cr) pigs to LWU, which exhibited gene flow from European pigs to North Chinese pigs. All of these migration events were also validated by D statistics, which suggested gene flow between Erhualian (EHL) pig and North Chinese pigs, such as HT and MIN, as well as between HT and other North Chinese pigs detected in TreeMix (supplementary table S4, Supplementary Material online). This is possibly a consequence of complicated and frequent historical human migrations among Northern, Central, and Eastern China (Ge 1997).
Together, our analyses of autosomal data support previous conclusions: 1) Chinese and European pigs represent two genetically divergent ancestral populations; 2) Sumatran wild boars are largely different from Eurasian pigs; and 3) Southern and Eastern Chinese pigs contributed to the development of modern European breeds (Giuffra et al. 2000; Larson et al. 2005; White 2011; Groenen et al. 2012; Frantz et al. 2013). Additionally, we describe the evidence of gene-flow from European to North Chinese pigs.
Two Highly Divergent Haplogroups in the Y Chromosomes of Eurasian Pigs
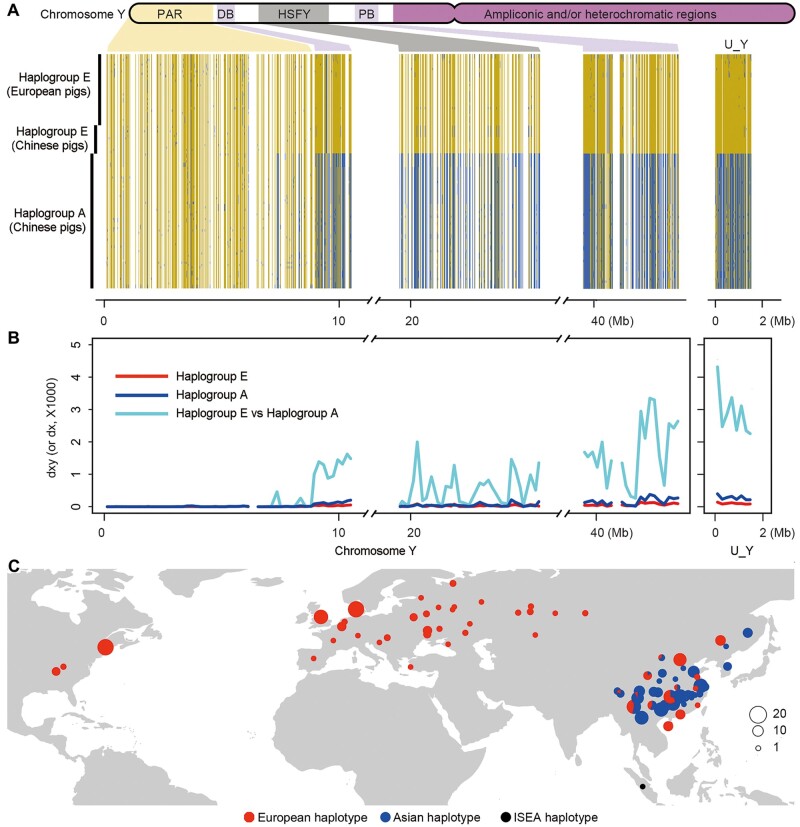
We then examined the Y chromosome in 101 Eurasian male pigs (European pigs, n = 31; Chinese pigs, n = 70). To our surprise, we found a large region consisting of at least 13.3 Mb on chromosome Y segregated in two highly divergent haplogroups (fig. 2A). We hereafter refer to this region as MSY (the male-specific region of the Y chromosome), which only comprises the distal, HSFY (heat shock transcription factor, Y chromosome) and proximal regions on the Y chromosome and ranges from 8.9 to 43.5 Mb (including unmapped repetitive regions from 10.6 to 19.5 Mb and from 25.5 to 39.5 Mb) on Build 11.1 version of the Y chromosome, plus 1.6 Mb unmapped Y-linked contigs (we discarded the most amplicon regions, which is of high identity sustained by frequent gene conversion, to reduce bias). The rest parts on Y chromosome are the homologous region with X chromosome (PAR) and highly repetitive regions, which are excluded from MSY. Pairwise nucleotide diversity (fig. 2B), a Bayesian phylogenetic tree (fig. 3A), and a median joining haplotype network (supplementary fig. S10, Supplementary Material online) all illustrate the large divergence in the MSY region between these two haplotype groups. For example, in the haplotype network (supplementary fig. S10, Supplementary Material online), we infer 12,455 mutations separating the two haplogroups.
Fig. 2.
Demographic history of Eurasian pigs based on Y chromosome data. (A) The haplotype pattern on the Y chromosome shared in Eurasian pigs. The haplotypes were reconstructed for each individual using all qualified SNPs on the Y chromosome. Alleles that are identical or different from the ones on the VEGA62 reference genome are indicated by orange or blue, respectively. PAR, the homologous region with X chromosome on Y chromosome; HSFY, heat shock transcription factor on Y chromosome; PB, proximal block. (B) The plots of dx and dxy (the number of pairwise differences per site) statistics in a window size of 200 kb on the Y chromosome. These statistics were calculated for pigs with the European or Asian haplotypes. U_Y indicates the unmapped Y-linked contig. (C) The geographical distribution of the European (red) and Asian (blue) haplotypes within the proximal and distal regions of the Y chromosome (the MSY region) in Eurasian pigs. European and Asian haplotypes were phased using six tag SNPs on the porcine 60k Chip (Illumina) within this region. ISEA haplotype (black), the haplotype found in a Sumatran wild boar.
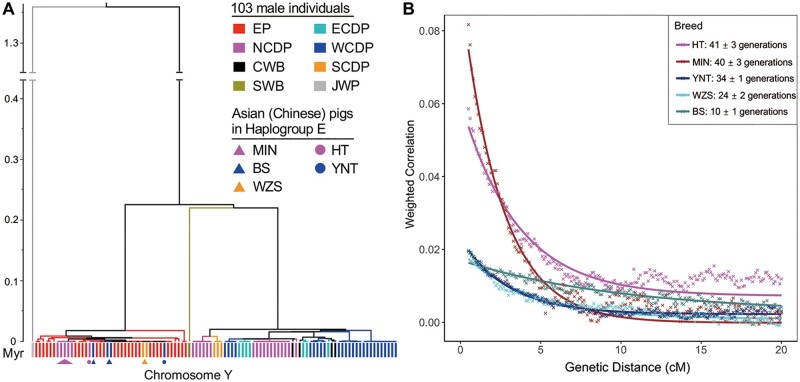
Fig. 3.
Admixture date estimation using phylogenetic trees of Y chromosome of 103 male pigs and weighted LD decay in autosomal regions. (A) Phylogenetic relationships among the 103 male Eurasian pigs constructed using the MSY sequence of the Build 11.1 Y chromosome. Inferred divergence time is shown on the Y-axis of the Bayesian tree. The abbreviations EP, ECDP, NCDP, WCDP, CWB, SCDP, and SWB are described in figure 1. Sus verrucosus (Java warty pig) was set as the outgroup. (B) The five populations are plotted to show their LD decay in autosomal regions (represented by a weighted correlation coefficient) with genetic distance. JH and EUW were used as reference populations for all test breeds. Full names of abbreviations for the pig breeds are detailed in supplementary table S1, Supplementary Material online.
Previously, we observed a similar pattern, with three highly divergent haplotypes present in the large region from 49 to 101 Mb (Build 10.2) on the X chromosome in pigs (Ai et al. 2015). However, the geographical distribution of the Y chromosome haplogroups is different from those on the X chromosome (supplementary fig. S11, Supplementary Material online). For the Y chromosome, one haplogroup is present exclusively in pigs across China and we denote it as the Asian haplogroup (also referred to as Haplogroup A), whereas the other haplogroup is present in European pigs and some Chinese indigenous pigs from North, South, and West China, and we refer to it as the European haplogroup (Haplogroup E) due to its more likely European origin. This distribution of haplogroups is consistent with the one found by Guirao-Rico et al. (2018), who sequenced a more limited amount of genetic markers. Nucleotide diversity of the MSY region was significantly lower compared with autosomes; and moreover, nucleotide diversity within haplogroup E was lower than that within haplogroup A (supplementary note S4, note S5, table S5, and fig. S12, Supplementary Material online).
We also analyzed matrilineal inheritance of mtDNA and found two highly divergent haplotypes with geographical distribution pattern opposite to the Y chromosomes, that is, one haplogroup exclusively in Europe, whereas the other one was present in both European and Chinese pigs (supplementary figs. S13 and S14, Supplementary Material online). This result supports previous findings on pigs’ mtDNA, which are also supported by historic evidence of pig migrations from China to Europe (Ramirez et al. 2009; Bosse, Megens, Madsen, et al. 2014).
European Y Chromosomes May Be under Selection in Certain Chinese Pigs
We had two primary hypotheses to explain the substantial frequency of European-associated Y chromosome haplogroups and the absence of European mtDNA in Chinese pigs: 1) sex-biased migration, with an excess of males moving from Europe to China, or 2) selection favored the European Y haplogroups in China. These hypotheses are not mutually exclusive.
We tested if hypothesis 1, male-biased hybridization, was sufficient to explain the observed frequencies of Haplogroup E in the Chinese pigs given the observed autosomal hybridization rates. Specifically, we fixed the overall hybridization rate based on the autosomal ancestry proportions by population. Then, we calculated the range of possible proportions of males in the European component of admixture following Goldberg et al. (2014), and used a binomial test to test if the frequency of European Y chromosomes is higher than expected under this range of scenarios (see Materials and Methods). Hetao (HT) and Min (MIN) populations were considered a single population because these two populations obtained similar European ancestry proportions in the same wave of gene flow as described below. For Hetao and Min (HT&MIN), Wuzhishan (WZS), and Baoshan (BS) populations, the observed European Y chromosome frequency is higher than expected even if all European contributions were from males (P values ranging from 0 to 0.022, 0 to 0.026, 0 to 0.0034, respectively; see supplementary table S6, Supplementary Material online). Thus, we reject the hypothesis that the excess of European Y chromosomes relative to autosomes is only due to sex-biased hybridization in HT, MIN, WZS, and BS.
Because the combined HT&MIN population has a higher level of European ancestry, we have more confidence in their estimated admixture proportions on the X chromosome. This allowed us to estimate the proportion of males among the individuals that contributed European ancestry to those populations, instead of using a range of values as described above. Following equation 14 from Goldberg and Rosenberg (2015), for mean X-chromosomal ancestry, X, we have X = 2/3f + 1/3 m, where f and m are the female and male European contributions to hybridization, respectively. In the autosomes, the mean European ancestry (A) is given by A = (f + m)/2 (Goldberg et al. 2014). Inputting ADMIXTURE-estimated values for the autosomal (A) and X-chromosomal (X) European ancestry, we solve for m = 0.32 and f = 0.08. Among a total of 12 individuals in the HT&MIN populations, we observed 0 individuals with European mtDNA, which is expected in 37% of scenarios given the estimated male and female contributions to hybridization. For the Y chromosomes, 6 out of 7 Y chromosomes were European, which is higher than expected (P = 0.0055, binomial test).
For WZS, BS, and YNT populations, the European-related ancestry is higher on the autosomes than the X chromosome, consistent with the direction of sex biased for HT&MIN populations. However, solving for male and female contributions using the same equation results in a migration rate outside the possible range of [0,1], which means the model of a single admixture event is violated. That is, the sex bias may occur over multiple generations of migration and even a full male-biased European migration would not be sufficient to produce the observed results. Additionally, the sample sizes per population are small, so small quantitative differences in mean ancestry should be interpreted with caution.
We also performed simulation analysis to further testing male-biased hybridization (hypothesis 1) could explain the observed frequencies of Haplogroup E in certain Chinese pigs alone. This analysis used synonymous sites on autosomes to establish a demographic model of population size, divergence and migration via diffusion approximations for demographic inference (∂a∂i) (Gutenkunst et al. 2009) (supplementary fig. S15 and table S7, Supplementary Material online). And then we simulated sequences of Y chromosome based on the above model to estimate the frequency of European Y haplotype in the target Chinese pig population under the scenario of sex-biased migration, in which all recent migrants to Chinese population are European male pigs. Then we examined if the frequency of European Y haplotype in the target Chinese pig population in the simulated can mimic that was seen in the observed data. If the simulated data under the neutral model cannot do so, we can argue that it must be selection.
In 1,000 independent simulations of MIN&HT Y haplotypes (supplementary table S8, Supplementary Material online), we found a total of 38 simulations in which frequency of European Y haplotypes were consistent with the observed frequency, that is, 6 of the 7 simulated Y sequences of MIN&HT were from European pigs. The probability of occurrence is 0.038. And the situation that the seven simulated Y sequences of MIN&HT are all from Europe only occurred twice. Only 3 of the 1000 simulated Y haplotypes of BS are consistent with the observed frequency, that is, all the 3 Y haplotypes in BS are from European pigs. The corresponding occurrence probability in BS is 0.003. In WZS, 209 simulations matched the actual situation, where a total of 2 Y haplotypes of WZS were from Europe. There are 89 simulations of YNT, which are consistent with the actual situation, in which only one of the 12 Y sequences is from European pigs. These results indicate that the frequency of European Y haplotypes in HT& MIN and BS cannot be explained by the neutral model.
These results suggest sex-biased hybridization alone cannot explain the high frequency of European Y chromosomes in certain Asian pig populations, and other factors, such as selection, could have affected the frequency of European Y chromosomes in those populations.
Origin of the European Y Haplogroup in Chinese Pigs
As mentioned previously, haplogroups A and E form two divergent clusters in the median joining haplotype network (supplementary fig. S10, Supplementary Material online) largely reflecting Asian and European origins. Most Chinese indigenous pig breeds belonging to haplogroup E cluster within clades of modern European commercial pigs and have relatively large genetic distances to European wild boars. A special case is the MIN pigs which clusters with the European Mangalica (MG) pig, a traditional domesticated breed from Hungary, which is highly divergent from all other European breeds (395 mutations separate these haplotypes from the closest European breed, supplementary fig. S10, Supplementary Material online). These results suggest that European Y chromosomes in Chinese pigs more likely originated from European domestic pigs than from wild boars.
We further investigated the geographical distribution of the two haplogroups in a larger panel of 426 male pigs from 82 diverse populations across Eurasia and America using six tag SNPs representing the MSY region (supplementary table S9, Supplementary Material online). Again, the Asian Y haplotypes were observed only in Eastern Asian pigs including Chinese pigs, Korean, and Russian Primorsky wild boars; the European Y haplotypes were found in European and Chinese pigs, as well as in American pigs, which originated from European pigs (fig. 2C).
At Least Two of European Y Chromosome Introgression in Chinese Pigs
We estimated the divergence time of the haplotypes on the MSY region of the Y chromosome using a strict molecular clock implemented in BEAST (Drummond et al. 2012) under the GTR + Γ model (more models in supplementary note S6 and table S10, Supplementary Material online) referred to in previous work (Wu et al. 2007; Wei et al. 2013). We separately used the divergence time between S. verrucosus and S. scrofa, 4.2 Ma (Frantz et al. 2013) and 1.36 Ma (Zhang et al. 2021), as softbound priors. The most recent coalescence time between MSY haplotypes of European pigs and Chinese pigs in haplogroup E gives an upper bound of the admixture time between these groups (i.e., the latest admixture time must be equal to or more recent than the coalescence time ignoring biases due to the “winner’s curse” [Bazerman and Samuelson 1983]). Thus, we estimate the time to the most recent common ancestors (TMRCA) between each Chinese sequence belonging to haplogroup E and their closest European MSY haplotype to calculate the bounds for the time of admixture between Europe and China (fig. 3A and table 1). We also applied the ROLLOFF method (Moorjani et al. 2011) on autosomal regions, which utilizes the decrease of admixture linkage disequilibrium (LD) on autosomes with genetic distance in admixed population to infer date of admixture from autosomal SNPs (fig. 3B).
Table 1.
Estimates of the TMRCA of Phylogenetic Nodes of Particular Interest in the MSY Region.
| Node | Time Estimate of TMRCA (years) | 95% Highest Posterior Density (HPD) Interval |
|---|---|---|
| Hetao pig and Large White | 180 | 113–252 |
| Min pig and SwallowBelly Mangalica | 151 | 83–208 |
| Yunnan Tibetan pig and Large White | 92 | 50–136 |
| Wuzhishan pig and Large White | 85 | 38–132 |
| Baoshan pig and White Duroc | 22 | 2–46 |
Note.—For each group, the most recent TMRCA among all pairs is shown as this value provides an upper bound for the latest admixture time.
Intriguingly, we found the admixture time inferred by BEAST when divergence time between S. verrucosus and S. scrofa was fixed at 1.36 Ma coincided well with the estimation by ROLLOFF method. We found the TMRCA of North Chinese pigs and European pigs to be older than the TMRCA for West and South Chinese pigs and European pigs. Specifically, the oldest TMRCA was estimated to be 180 years between HT pigs (a North Chinese pig breed) and European pigs (table 1), whereas we found TMRCAs with European pigs of 151, 92 and 85 years between MIN pigs (another North Chinese pig breed), YNT (West Chinese), and WZS (South Chinese), respectively. BS (West Chinese) had a much more recent TMRCA of 22 years with White Duroc pigs. Accordingly, we could clearly divide curves of admixed LD inferred by ROLLOFF into three categories: North Chinese pigs (HT and MIN pig) had the fastest decline, corresponding to the admixture time at 41 ± 3 and 40 ± 3 generations ago (123 ± 9 and 120 ± 9 years ago when we set the generation time as 3 years), respectively; the decay rate and amplitude of BS pig were the lowest and its admixture time with European domesticated pigs was 10 ± 1 generations ago (30 ± 3 years ago); the decay rate of YNT and WZS pigs were similar and intermediate between the decay rates of North Chinese pigs and BS pig, and their admixture time with European domesticated pigs are 34 ± 1 and 24 ± 2 generations ago (102 ± 3 and 72 ± 6 years ago; fig. 3B), respectively. These results may suggest that the patrilineal introgression of European ancestry in Chinese pigs occurred in three different time periods in different parts of China: 1) initial gene flow into North Chinese pigs, 2) a possible second gene-flow event into South and West Chinese pigs, and 3) very recent gene-flow into West Chinese pigs.
All the above introgression is well in line with at least three waves of introgression into Chinese pig breeds from Europe since 1840s summarized by Yang et al. (2017) based on historical documents. Furthermore, we clarified which donor of introgression was and where introgression occurred in this study, and there was no archaic introgression from Europe to Asia. It was reported that the first wave of introgression may have occurred around the 1840 s when pig breeds including breeds from Russia, Large White, Duroc, and Tamworth pig were brought in China. Herein, we detected the first wave of gene flow occurred in North Chinese pigs between 120 and 180 years ago (based on the TMRCA and ROLLOFF method; the same applies hereafter), and the introgression donors consisted of ancestors of Large White and SwallowBelly Mangalica pigs (table 1 and supplementary fig. S10, Supplementary Material online). The second introgression we identified in South and West Chinese pigs occurred between 72 and 102 years ago, in accordance with the second pig introduction since the 1910s recorded in historical documents. This wave of gene flow mainly came from ancestors of Large White (table 1 and supplementary fig. S10, Supplementary Material online). The last wave of introgression based on historical records had occurred since the 1980s. Accordingly, we detected an introgression into BS pigs from White Duroc using MSY information ∼30 years ago.
When we fixed the divergence time between S. verrucosus and S. scrofa at 4.2 Ma, the divergence time European pigs in Haplogroup E and Chinese pigs in Haplogroup A was estimated to be 1.25 My (supplementary table S11, Supplementary Material online), which fits well with the previous estimate of divergence time of 1.2 Ma based on autosomal data (Frantz et al. 2013). However, the admixture could farthest reach 1,000 years ago in the North China group, and the most recent introgression from Europe happened 300 years ago in BS breed (supplementary table S11, Supplementary Material online). Obviously, compared with the estimation using 4.2 Ma as the divergence time between S. verrucosus and S. scrofa, both evidence including the results of ROLLOFF and the historical documents unanimously supported the time estimation when fixing the divergence time between S. verrucosus and S. scrofa at 1.36 Ma. Thus, we preferred the time estimation when fixing the divergence time between S. verrucosus and S. scrofa at 1.36 Ma. In reverse, all of these also could be treated as evidence to support the pig population history new estimated using the de novo mutation rate (Zhang et al. 2021).
MSY Haplogroups Are Associated with Fatness Traits
The previously undiscovered Y chromosome gene flow events into Chinese pigs, with a potential role for selection amplifying the signal of introgression, raises the question: on what phenotypes could selection have acted? To date, phenotype–genotype association studies on the Y chromosome of pigs have not been possible due to the lack of a high-quality assembly of the Y chromosome and the fact that few experimental pig populations contained both divergent haplogroups of the Y chromosome. The pig Y chromosome is now well assembled, and to overcome the second challenge, we resorted to a heterogeneous pig population constructed by crossing eight Chinese and European pig breeds using a disc rotation breeding system, including Erhualian, Bamaxiang, Laiwu, Tibetan, Landrace, Large white, Pietran, and white Duroc pigs (supplementary note S7, Supplementary Material online). There was no artificial selection for any traits during the breeding process. We measured more than 200 traits in a total of 836 sixth-generation mosaic offsprings of this population including 448 females and 388 males. All the 836 piglets were sequenced and the average genome depth was approximately 7.8× (Ji et al. 2018; Zhang et al. 2021). Among the 388 males, there were 150 individuals with haplogroup A and 238 individuals with haplogroup E.
Correlation analysis was performed between MSY haplotypes and various traits via two methods: direct Student’s t-test and t-test adjusted with top 10 PCs of genetic structure as covariates. We detected significant differences in six different traits related to fatness between Asian (Chinese) and European Y haplogroups, including four traits of backfat thickness at hip (HBFT), first rib (LRBFT), last rib (FRBFT) and shoulder (SBFT), and two other measurements of leaf and veil fat weight (LFW and VFW) with direct student’s t-test P values of 6.8 × 10−8, 5.5 × 10−6, 1.9 × 10−6, 8.1 × 10−4, 1.7 × 10−4, and 1.5 × 10−3, respectively. The t-test with ten PCs as covariates also supported these results with P values for the same traits ranging from 6.5 × 10−7 to 3.4 × 10−2 (table 2 andsupplementary fig. S16, Supplementary Material online). And the phenotypic variance explained by MSY haplotypes for the six traits, including HBFT, LRBFT, FRBFT, SBFT, LFW, and VFW, is 6.9%, 5.5%, 4.6%, 2.5%, 3.2% and 1.9%, respectively. These results suggest that the European Y haplogroup is associated with reduced fat deposition when compared with Asian Y haplogroup (table 2).
Table 2.
Comparison of Fatness Traits between the Male Pigs with Chinese Chromosome Y and with European Chromosome Y.
| Trait | ChrY-A |
ChrY-E |
P Value (t-test) | P Value (10 PCs as covariates) | Phenotypic Variance Explained by ChrY (%) | ||
|---|---|---|---|---|---|---|---|
| N | Mean ± SD | N | Mean ± SD | ||||
| Backfat thickness at the hip (HBFT, cm) | 150 | 3.48 ± 1.02 | 238 | 2.90 ± 1.03 | 6.83 × 10-8 | 6.51 × 10-7 | 6.9 |
| Backfat thickness at the last rib (LRBFT, cm) | 150 | 2.87 ± 0.78 | 238 | 2.49 ± 0.77 | 1.89 × 10-6 | 1.31 × 10-4 | 5.5 |
| Backfat thickness at the first rib (FRBFT, cm) | 150 | 3.70 ± 0.86 | 238 | 3.30 ± 0.86 | 5.54 × 10-6 | 3.35 × 10-3 | 4.9 |
| Backfat thickness at the shoulder (SBFT, cm) | 150 | 4.20 ± 0.91 | 238 | 3.89 ± 0.96 | 8.05 × 10-4 | 2.51 × 10-2 | 2.5 |
| Leaf fat weight (LFW, kg) | 150 | 2.34 ± 0.79 | 237 | 2.04 ± 0.79 | 1.65 × 10-4 | 8.08 × 10-5 | 3.2 |
| Veil fat weight (VFW, kg) | 149 | 0.87 ± 0.21 | 237 | 0.80 ± 0.23 | 1.52 × 10-3 | 3.39 × 10-2 | 1.9 |
ChrY-A denotes Asian Y haplogroup; ChrY-E denotes European Y haplogroup.
Gene Flow Resulting in Similar Y Chromosomes in Sumatran Wild Boar and Chinese Pigs
Frantz et al. (2013) detected conflicting phylogenetic signals between mtDNA and autosomal chromosomes when comparing Sumatran wild boars (SWB) to Eurasian pigs and Javan Warty Pig (S. verrucosus), a pattern also detected in this study (supplementary fig. S13, Supplementary Material online). Furthermore, we also observe a much more recent divergence between Sumatran wild boars and Chinese pigs in the MSY region than in autosomes (TMRCA results and supplementary fig. S13, Supplementary Material online). In both the autosomes and MSY, S. verrucosus is an outgroup for all S. scrofa (including SWB and Eurasian pigs). In the MSY, SWB clusters within Haplogroup A of Eurasian pigs, and there is a deep split separating this cluster from Haplogroup E. In the autosomes, the SWB is an outgroup for Eurasian pigs, with a deep divergence separating them. Finally, in the mtDNA, S. verrucosus and SWB clustered together, with a deep divergence from Eurasian pigs (supplementary figs. S13 and S14, Supplementary Material online).
We propose two models to explain this inconsistent phenomenon, model A: the Sumatran wild boar exchanged Y chromosomes with Chinese male individuals and received significant contributions from S. verrucosus females (fig. 4A); model B: Sumatran wild boar exchanged Y chromosomes with Chinese pigs and contributed mtDNA to S. verrucosus (fig. 4B). Both models can explain the characteristics of phylogenetic signals on the autosomes, Y chromosome, and mtDNA of Sumatran wild boars (supplementary fig. S13, Supplementary Material online).
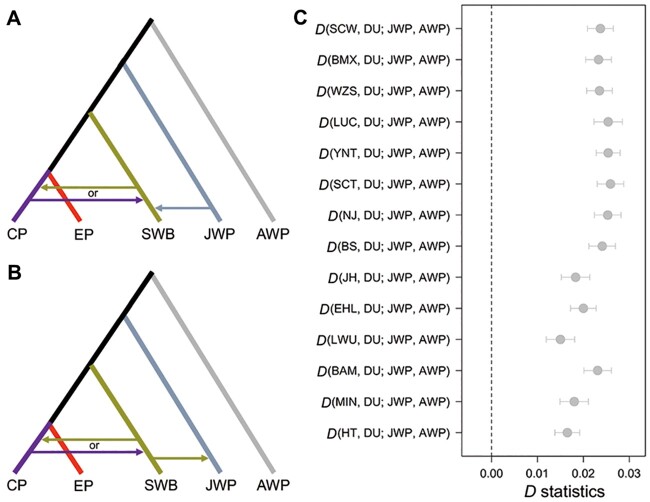
Fig. 4.
Patterson’s D statistic to test for two models explaining the phylogenetic discordance among autosomes, Y chromosome, and mtDNA related to Sumatran wild boar. (A) Model A: Sumatran wild boars (SWB) exchanged Y chromosomes with Chinese male individuals and received significant contributions from Sus verrucosus (JWP). (B) Model B: Sumatra wild pigs (SWB) exchanged Y chromosomes with Chinese pigs (CP) and then contributed mtDNA to S. verrucosus (JWP). (C) D statistic of combination of European pigs (EP) represented by DU and all Chinese pigs involved in this study. See supplementary table S4, Supplementary Material online, for more results of combination of other EP and CP. Positive values indicate an excess of alleles shared with CP and JWP. AWP here denotes African common warthog (Phacochoerus africanus).
We used D statistics in the form of D(CP, EP; JWP, AWP) to test the two models, where Chinese pigs (CP) and European pigs (EP) are treated as ingroups (H1 and H2, respectively, in the notation of Green et al. [2010]), S. verrucosus (JWP) as H3 and African warthog (AWP) as Outgroup (see Materials and Methods). In the case of model A, because Sumatran wild boars exchanged part of the genetic material with CP first, and then passed genetic material to JWP (the divergence time of mtDNA of JWP and SWB is only 20k years ago, far less than that of their MSY region at 0.216 Ma; supplementary table S12 and fig. S13, Supplementary Material online), CP and JWP genetic similarity should be the same as that of EP and JWP. If model B is true, JWP would be more related to CP when compared with EP. We tested different combinations of pig breeds in CP and EP (supplementary table S4, Supplementary Material online). Our results showed that D values of all combinations with Chinese pigs applied in this analysis were significantly greater than zero (fig. 4C and supplementary table S4, Supplementary Material online), in agreement with model B. Our analysis of gene flows detected with TreeMix supports a model with gene flow from SWB to CP (fig. 1E). On the other hand, the TMRCA estimates in the MSY region suggest the Y chromosome of the Sumatran wild boar was possibly derived from Chinese pigs, because when we fixed the divergence time of JWP and S. scrofa at 1.36 (Zhang et al. 2021) or 4.2 Ma (Frantz et al. 2013; Groenen 2016), the TMRCA of 0.225 or 1.25 Ma of haplogroup E and haplogroup A containing Sumatran wild boar was close to the new autosomal estimated (Zhang et al. 2021) or the previously reported (Frantz et al. 2013) divergence time of European pigs and Asian pigs of 0.219 or 1.2 Ma, and smaller than the divergence time of Sumatran wild boar and Eurasian pigs (0.275 or 1.6–2.4 Ma, suggested by Zhang et al. [2021] or Frantz et al. [2013]). Although these models are difficult to distinguish based on the single Sumatran wild boar genome available, we can at least exclude a model in which SWB are formed by admixture between predominantly male S. scrofa and predominantly female S. verrucosus. Moreover, whereas we cannot exclude additional gene flow from JWP to SWB, our results suggest that the predominant pattern is gene flow from SWB to JWP.
Discussion
It is well-recognized that European and Asian pigs are geographically and genetically distinct groups (Groenen et al. 2012; Frantz et al. 2013). However, many studies have recently identified gene flow between these groups (McLaren 1990; White 2011; Groenen et al. 2012; Bosse, Megens, Frantz, et al. 2014; Yang et al. 2017; Guirao-Rico et al. 2018). Here, we illuminate the complex demographic history of Eurasian pigs through the lens of Y chromosomes.
We identify two long and highly divergent Y chromosome haplogroups in the MSY region, which we refer to as haplogroup “A” (the most prevalent in Asia and absent in Europe) and haplogroup “E” (which is fixed in Europe, and present in some Chinese populations). We show evidence of gene flow between Asian pigs within haplogroup A and Sumatran wild boars, but cannot further determine the specific direction of gene flow between them. Furthermore, we describe a complex history of haplogroup E, shaped by three different waves of introgression from European to Chinese pigs, possibly followed by selection increasing its frequency in China.
We find an enrichment of European ancestry on Y chromosomes relative to autosomes in certain Chinese pigs (HT&MIN, BS), more so than would be expected under neutrality. This enrichment could be explained by segregation distortion (i.e., the European Y chromosome is more often passed on to gametes than the Asian Y), natural selection (i.e., European Y chromosome positively affects fertility or survival) or artificial selection (i.e., European Y chromosome affects traits of interest to breeders). Although we cannot exclude any of these possibilities, the last hypothesis seems likely since we find that the European Y chromosome is associated with leaner meat production, using a study of 836 F6 mosaic pigs crossbred by 8 diverse breeds and measured for more than 200 traits.
We note that all domestic Chinese pig breeds used in this study were collected from nucleus herds raised in national conservation farms. These herds are currently geographically isolated populations. The presumed lack of recent gene flow between these populations is consistent with the hypothesis that introgression of European ancestry into Chinese pigs in different locations and at different time points is better explained by multiple independent waves of gene flow. In addition, the results showed that the MSY haplotypes of most Chinese domestic pigs in haplogroup E were closer to European domesticated pigs’ MSY haplotypes than to that of European wild boars (supplementary fig. S10, Supplementary Material online), which indicates that the gene flow was driven by humans rather than by migration of natural populations of wild boars.
Actually, we detected the gene flow happened less than 200 years and in the form of three independent waves based on a new estimated pig population history (Zhang et al. 2021). This genomic evidence perfectly conformed to the three waves of the artificial introduction of European pig breeds into China described in historical documents. The most recent two waves of introgression of Y chromosomes from Europe to China (ranging from 22 to 102 years ago based on the TMRCA and ROLLOFF method) affected mostly South and West Chinese pigs from the YNT, WZS, and BS population. This event is consistent with the known importation of improved European breeds to China during the early twentieth century, due to their desirable performance of high lean pork production. These breeds, including Large White, Berkshire, Duroc, and Landrace, were introduced as terminal sires to crossbreed with female Chinese indigenous breeds such as Kele and Licha, leaving hybridization signals in the genomes of these breeds (Ai et al. 2014; Yang et al. 2017). Accordingly, the MSY haplotypes of YNT and WZS pigs located at the root of the group of European Large White pigs, whereas the MSY haplotypes of BS pigs cluster within the group of European White Duroc pigs, reflecting a more recent admixture (supplementary fig. S10, Supplementary Material online).
Interestingly, a century earlier, during the Industrial Revolution and early nineteenth century, migration happened in the opposite direction: Asian pigs mainly from South China were introduced into Britain to improve local breeds (supplementary note S3 and fig. S17, Supplementary Material online), resulting in mosaic modern genomes of British-derived commercial breeds (such as Large White, Landrace, and Duroc) that possess up to ∼30% Asian admixture (McLaren 1990; Bosse, Megens, Frantz, et al. 2014). We also identify genetic signatures of this event in the autosomes and in the mtDNA, but not in the Y chromosomes. This observation could be explained by female-biased admixture from Asia to Europe. Another nonmutually exclusive explanation is that Chinese MSY haplotypes were selected against by European breeders. Because the pig breeding strategy of Europeans is to cultivate mostly lean meat type pigs, such as Duroc, Large White, and Landrace, even if Chinese MSY haplotypes were once admixed in European pigs, which had not been documented, they would be eliminated through artificial selection.
The oldest wave of gene flow (ranging from 120 to 180 years ago based on the TMRCA and ROLLOFF method) brought haplogroup E to Northern Chinese populations (HT and MIN). Both a Bayesian phylogenetic tree (fig. 3A) and a median joining haplotype network (supplementary fig. S10, Supplementary Material online), show that the MSY haplotypes of HT pigs were very distant from that of MIN pigs. The MSY haplotype of HT pigs in haplogroup E clusters with MSY haplotypes of modern European commercial pigs, whereas the MSY haplotypes of MIN pigs are closer to that of SwallowBelly mangalica (MG) pigs, a Hungarian native breed which is famous for its thick and woolly coat similar to that of a sheep, and is deeply divergent from the modern European commercial pig breeds (Zsolnai et al. 2006). The MG to MIN migration is also supported by TreeMix analyses (fig. 1E). Zhang et al. (2018) found that MIN pigs were in intermediate genetic distance between Asian pig breeds and the European pig breeds, and they speculated it resulted from gene flow from Soviet-Largewhite pigs, which were bred in the Soviet Union by introducing Largewhite (LW) pigs from Britain within the past 100 years. In contrast, we show that MIN pigs actually received earlier gene flow from a European native pig breed (related to the ancestral breeds from which Mangalica was derived), ∼120 to 151 years ago (fig. 3B and table 1). These results illustrate that the first migration involved more diverse donor breeds, including at least one indigenous (into MIN) and one breed related to modern commercial breeds (into HT). Historical documents also indicate more diverse pig breeds including Berkshire, Large White, Duroc, pigs from Russia, and Tamworth were brought into China by Japanese and German around the 1840s than the subsequent two introductions (Xu 2004; Yang et al. 2017).
In summary, we conclude that most Chinese pigs carry Y chromosomes similar to the one in Sumatran wild boars. Furthermore, three waves of gene flow from Europe into China contributed to the genetic diversity of Chinese pigs and had affected the Y chromosome more strongly than the rest of the genome, possibly due to artificial selection for leaner meat.
Materials and Methods
Samples and Genome Sequencing
We sequenced the genomes of 80 Chinese and European pigs. A total of 83 Chinese pigs were sequenced in previous studies of our group (Ai et al. 2015; Zhu et al. 2017). Of these animals, Chinese pigs were from 16 geographically diverse breeds, European pigs were from 4 commercial breeds (supplementary table S1, Supplementary Material online). The genome sequencing was conducted as described previously (Ai et al. 2015). Detailed information for samples and genome sequencing is shown in the supplementary methods, Supplementary Material online.
SNP Calling
We downloaded the genome sequence data for 42 pigs. These data were integrated into the sequence data obtained in this study, resulting in a 205-sample high-quality data set (supplementary tables S1 and S2, Supplementary Material online). The cleaned reads of all individuals were aligned to the S. scrofa reference genome (build 11.1) using BWA (Li and Durbin 2009). The mapped reads were subsequently processed by sorting, duplicate marking, indel realignment, and base quality recalibrating using Picard (http://picard.sourceforge.net, last accessed January 5, 2021) and GATK (McKenna et al. 2010). A two-round procedure of SNP calling was performed using Platypus (Rimmer et al. 2014) (Supplementary table S13, Supplementary Material online). Additional information is provided in the supplementary methods, Supplementary Material online.
Population Genetic Analysis Using Autosomal Data
Qualified autosomal SNPs were used to calculate genetic distance among all individuals using Plink as described previously (Ai et al. 2013). A NJ tree was then constructed for all individuals using Neighbor in PHYLIP v3.69 (Felsenstein 2005) and visualized with the FigTree software (http://beast.bio.ed.ac.uk/, last accessed January 5, 2021, FigTree). Population genetic structure was inferred using the maximum-likelihood approach implemented in ADMIXTURE v1.20 (Alexander et al. 2009). PC analysis was conducted using Smartpca in EIGENSOFT v6.0 (Price et al. 2006). TreeMix (Pickrell and Pritchard 2012) was used to infer patterns of historical splits and mixture among Eurasian pig populations in the context of Suidae. To estimate the optimal number of migration edges for each value of K, we used the R package OptM (Fitak 2021) with the linear method. BITE (Milanesi et al. 2017) was used to do 100 bootstrap replicates, and to visualize the consensus trees with bootstrap values and migration edges when doing TreeMix analyses. We further used qpDstat from AdmixTools (Patterson et al. 2012) to calculate D statistics in the form of D(H1, H2; H3, Outgroup) with default parameters to show if population H3 is symmetrically related to H1 and H2 or shares an excess of alleles with either of the two, with standard errors computed with a block jackknife. To estimate admixture dates, we used ROLLOFF v625 (Moorjani et al. 2011), which uses the decay of admixture LD to estimate the time of gene flow. JH and EUW were used as two reference populations when running ROLLOFF. The details of the analyses are described in the supplementary methods, Supplementary Material online.
Evolutionary History Analysis Using Y Chromosome Data
The BWA software (Li and Durbin 2009) was employed to align filtered cleaned reads from all male individuals to the Y chromosome reference sequence (VEGA62 version; Skinner et al. 2016). For the SNPs-calling procedure on the Y chromosome and on a 1.6-Mb unmapped Y-linked contig, some additional quality control was conducted. To reduce alignment error, we excluded the reads tagged the “SA: Z” (other canonical alignments in a chimeric alignment) and “XA: Z” (alternative hits) flags, keeping only the reads that uniquely aligned to the Y chromosome. Then, the two-round SNP calling was performed using Platypus (Rimmer et al. 2014), as it was for autosomal SNPs. In total, 81,057 SNPs were obtained on the Y chromosome assembly and on the Y-linked contig of 103 male individuals. Notably, 12,670 SNPs on Y chromosome and on the Y-linked contig, which were same as the SNPs called by the reads of female individuals misaligned to Y chromosome reference sequence, were excluded due to the possible bias. Moreover, for SNPs on the Y chromosome, we replaced the heterozygous SNPs with missing data on the Y chromosome and then filter under the criterion MAF > 0.009 and SNP call rates > 80%. After applying these filters, a total of 42,288 high-quality SNPs on the Y chromosome and the Y-linked contig were detected in the 102 male individuals (excepting Sumatran wild boar) for subsequent haplotype construction and other analyses. FASTA-formatted sequence files were used to construct phylogenetic tree via BEAST (Drummond et al. 2012) under the GTR + Γ model of evolution, referred to process of human Y-chromosomal phylogeny construction (Wei et al. 2013). Split times and 95% highest posterior density intervals in the tree were estimated using BEAST with 10,000,000 MCMC samples. Due to the dissimilarity of the population history of pigs estimated via genomic analyses applying a directly estimated mutation rate (Zhang et al. 2021) from the previous study (Frantz et al. 2013), we used two divergence time estimates of S. verrucosus (JVWP) and S. scrofa (CB11-2), 4.2 My (Frantz et al. 2013) and 1.36 My (Zhang et al. 2021), as softbound priors, respectively. We assessed the convergence of MCMC samples using TRACER (Rambaut et al. 2018) (effective sample size [ESS] > 100). Due to the incompleteness of the Y chromosome in the sample of Sumatran wild boar (ERR173178), we carried out two rounds of the above calculations using BEAST. First, we excluded Sumatra wild boars and used the sequences containing 40,586 SNPs on the MSY region of the remaining 102 male pigs to construct a phylogenetic tree. Second, we used the sequences containing 3,967 SNPs on the MSY region of Sumatran wild boars extracted from the other 102 male pigs and used those determine the position and divergence time of Sumatran wild boars in the phylogenetic tree. Phylogenetic analysis of mtDNA was also performed with BEAST using mtDNA sequences of the 103 male individuals. TRACER was also used to assess the convergence of MCMC samples. A median joining haplotype network (Bandelt et al. 1999) was constructed in POPART (Leigh and Bryant 2015) using MSY haplotypes of 101 S. scrofa individuals and 1 S. verrucosus individual to infer relationships between MSY haplotypes of Asian pigs and European pigs. The detailed analyses are described in the supplementary methods, Supplementary Material online.
Pairwise nucleotide differences per site within (dx) and between (dxy) populations were calculated as described previously (Ai et al. 2015). Segregating sites, theta, and Pi values were calculated for autosomes and the MSY region using VariScan (Hutter et al. 2006). To investigate the global distribution of the haplotypes within MSY, we used six tag SNPs (CAHM0000184, chrY : 953841; CAHM0000174, chrY : 1124654; CAHM0000177, chrY : 1125090; CAHM0000179, chrY : 1125745; CAHM0000180, chrY : 1125941; CAHM0000190, chrY : 1190760) from the Illunima PorcineSNP60 Beadchip based on Build 10.2 representing this region in 426 Eurasian pigs from 82 geographically diverse populations.
Next, we tested whether sex-biased hybridization alone could explain the observed frequency of European MSY haplotypes in Chinese pigs. One way to consider sex-biased hybridization is through comparisons of the X chromosome and the autosomes. Using ADMIXTURE to infer X-chromosomal ancestry, we see higher European ancestry on the autosomes than X chromosome in Chinese populations with mixed ancestry, consistent with male-biased contributions from Europe. However, low ADMIXTURE estimates are more prone to error, especially on the X chromosome, and comparison between small quantities may lead to spurious inferences. Therefore, we instead consider the full range of possible sex bias consistent with overall hybridization levels. That is, we fix the contribution from Europe to the Asian gene pool equal to the autosomal ADMIXTURE estimates, A (A is the sum of the two European-related ancestries from K = 6 values, pink and red components). This analysis was based on the ancestry assignment when K = 6 because inferred ancestry best matched population assignment. Following (Goldberg et al. 2014) (equations 1 and 34), we have A = (m + f)/2, where m is the male contributions from Europe and f is the female contributions from Europe. If European contributions are completely male biased, then f = 0 and m = 2A. If all European contributions are from females, then m = 0. The range of m provides an expectation for the range of European Y chromosome frequencies in the Asian pig populations. Therefore, using the inferred A for each population, we have the range of expected Y chromosome frequencies in the population in [0, 2A]. We can then test if the observed number of Y chromosomes in each Chinese population is higher than expected given the range of European male contributions using a binomial test.
We also applied the simulation method to examine whether selection is involved in resulting in a substantial frequency of European-associated Y chromosome haplogroups in certain Chinese pigs. Although some synonymous sites are expected to evolve under purifying selection (Lawrie et al. 2013), they are generally assumed to be under weak selection and nearly neutral (Yang and Nielsen 2008). Here, we used SNPs at 4DTv sites for demographic inference to reduce the impact of natural selection as reported previously (Zhang et al. 2017; Marburger et al. 2019).
We used easySFS (https://github.com/isaacovercast/easySFS) to estimate the site frequency spectrum (sfs), which was the input file for the demographic analysis using GADMA (Noskova et al. 2020). The GADMA approximation software tool was used to compare the expected allele frequency and the observed allele frequency spectrum over the parameter value space by computing a composite-likelihood score for the best plausible evolutionary scenarios. The method of ordinary differential equations was applied using diffusion approximations for demographic inference ∂a∂i software (Gutenkunst et al. 2009) implemented in GADMA. Time for generation we used is 3.0 years; number of repeats was set to 15; the remaining parameters were set as default. Heuristic searches by means of the genetic algorithm implemented by GADMA was used to find the best-fit parameter values in order to automatically infer the best demographic model from the joint sfs data.
We hypotheses that sex-biased migration (with an excess of males moving from Europe to China) can explain the substantial frequency of European-associated Y chromosome haplotypes in certain Chinese pigs (HT&MIN, WZS, BS, YNT). Here, we simulated the sequences of Chinese Y chromosome under the best neutral model from GADMA using coalescent simulations implemented in ms (Hudson 2002), assuming all the recent migrants to Chinese pig population were European male. The effective population size for Y chromosome (Ny) was calculated as:
Therefore, the migration rates (M) here, except when it is male-only migration, is:
where Mauto is the migration rate derived from autosomal data. When it is male-only migration (all migrants are male), the migration rate (M′) is:
We independently repeated 1,000-times simulations to calculate how many repeats where the frequency of European Y haplotypes in the target Chinese pig population in the neutrally simulated can mimic that was seen in the observed data. If the occurrence rate is not greater than 0.05, we considered it to be a small probability event, which cannot occur, and we can defeat the above hypotheses and believe that selection must be involved.
Together, we find support for male-biased migration from Europe, but this demographic history is not sufficient to explain the high levels of Haplogroup E Y chromosomes in Chinese pigs.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was financially supported by the National Swine Industry and Technology System of China (nycytx-009), Innovative Research Team in University (IRT1136), and the National Natural Science Foundation of China (31672383).
Author Contributions
L.H. organized and coordinated the research. L.H. and H.A. designed the study. H.A., M.Z., R.N., A.G., and W.L. performed the bioinformatics, population genetics, and evolutionary history analyses. B.Y., J.M., and Z.Z. performed sample collection. H.A., L.H., M.Z., and R.N. analyzed the results. H.A., M.Z., and D.B. wrote the draft manuscript. L.H. and R.N. revised the paper.
Data Availability
The raw sequence reads from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA751703.
References
- Ai H, Fang X, Yang B, Huang Z, Chen H, Mao L, Zhang F, Zhang L, Cui L, He W, et al. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 47(3):217–225. [DOI] [PubMed] [Google Scholar]
- Ai H, Huang L, Ren J.. 2013. Genetic diversity, linkage disequilibrium and selection signatures in Chinese and Western pigs revealed by genome-wide SNP markers. PLoS One. 8(2):e56001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H, Yang B, Li J, Xie X, Chen H, Ren J.. 2014. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics. 15:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Bazerman MH, Samuelson WF.. 1983. I won the auction but don't want the prize. J Conflict Resol. 27(4):618–634. [Google Scholar]
- Bosse M, Megens H-J, Frantz LA, Madsen O, Larson G, Paudel Y, Duijvesteijn N, Harlizius B, Hagemeijer Y, Crooijmans RP.. 2014. Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat Commun. 5(1):4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse M, Megens HJ, Madsen O, Frantz LA, Paudel Y, Crooijmans RP, Groenen MA.. 2014. Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations. Mol Ecol. 23(16):4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe KM, Day AE, Bagga M, Siggens K, Quilter CR, Lowden S, Finlayson HA, Palgrave CJ, Li N, Huang L, et al. 2010. Analysis of the non-recombining Y chromosome defines polymorphisms in domestic pig breeds: ancestral bases identified by comparative sequencing. Anim Genet. 41(6):619–629. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29(8):1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Mou Y, Huang Z, Li Y, Han L, Zhang Y, Feng Y, Chen Y, Jiang X, Zhao W, et al. 2012. The sequence and analysis of a Chinese pig genome. Gigascience 1(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 2005. Department of Genome Sciences PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author, University of Washington, Seattle.
- Fitak R. Forthcoming 2021. optM: an R package to optimize the number of migration edges using threshold models. Available from: https://cran.rproject.org/web/packages/OptM/index.html.
- Frantz LAF, Schraiber JG, Madsen O, Megens H-J, Bosse M, Paudel Y, Semiadi G, Meijaard E, Li N, Crooijmans RPMA, et al. 2013. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol. 14(9):R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz LA, Schraiber JG, Madsen O, Megens HJ, Cagan A, Bosse M, Paudel Y, Crooijmans RP, Larson G, Groenen MA.. 2015. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet. 47(10):1141–1148. [DOI] [PubMed] [Google Scholar]
- Ge J. 1997. History of Chinese immigration. Fujian: Fujian People’s Publishing House. [Google Scholar]
- Giuffra E, Kijas JM, Amarger V, Carlborg O, Jeon JT, Andersson L.. 2000. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154(4):1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A, Rosenberg NA.. 2015. Beyond 2/3 and 1/3: the complex signatures of sex-biased admixture on the X chromosome. Genetics 201(1):263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A, Verdu P, Rosenberg NA.. 2014. Autosomal admixture levels are informative about sex bias in admixed populations. Genetics 198(3):1209–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MA. 2016. A decade of pig genome sequencing: a window on pig domestication and evolution. Genet Sel Evol. 48(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens H-J, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491(7424):393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao-Rico S, Ramirez O, Ojeda A, Amills M, Ramos-Onsins SE.. 2018. Porcine Y-chromosome variation is consistent with the occurrence of paternal gene flow from non-Asian to Asian populations. Heredity 120(1):63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5(10):e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18(2):337–338. [DOI] [PubMed] [Google Scholar]
- Hutter S, Vilella AJ, Rozas J.. 2006. Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinformatics. 7(1):409–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Zhou L, Huang Y, Zheng M, Liu X, Zhang Y, Huang C, Peng S, Zeng Q, Zhong L, et al. 2018. A whole-genome sequence based association study on pork eating quality traits and cooking loss in a specially designed heterogeneous F6 pig population. Meat Sci. 146:160–167. [DOI] [PubMed] [Google Scholar]
- Kijas J, Andersson L.. 2001. A phylogenetic study of the origin of the domestic pig estimated from the near-complete mtDNA genome. J Mol Evol. 52(3):302–308. [DOI] [PubMed] [Google Scholar]
- Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E, et al. 2005. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307(5715):1618–1621. [DOI] [PubMed] [Google Scholar]
- Lawrie DS, Messer PW, Hershberg R, Petrov DA.. 2013. Strong purifying selection at synonymous sites in D. melanogaster. PLoS Genet. 9(5):e1003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D.. 2015. popart: full‐feature software for haplotype network construction. Methods Ecol Evol. 6(9):1110–1116. [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen L, Tian S, Lin Y, Tang Q, Zhou X, Li D, Yeung CK, Che T, Jin L, et al. 2017. Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies. Genome Res. 27(5):865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, Wang T, Yeung CKL, Chen L, Ma J, et al. 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 45(12):1431–1438. [DOI] [PubMed] [Google Scholar]
- Lunney JK. 2007. Advances in swine biomedical model genomics. Int J Bio Sci. 3(3):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marburger S, Monnahan P, Seear PJ, Martin SH, Koch J, Paajanen P, Bohutínská M, Higgins JD, Schmickl R, Yant L.. 2019. Interspecific introgression mediates adaptation to whole genome duplication. Nat Commun. 10(1):5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren DG. 1990. The potential of Chinese swine breeds to improve pork production efficiency in the US. Urbana. 51(1):61801. [Google Scholar]
- Milanesi M, Capomaccio S, Vajana E, Bomba L, Garcia JF, AjmoneMarsan P, Colli L.. 2017. BITE: an R package for biodiversity analyses. bioRxiv. doi:10.1101/181610.
- Moorjani P, Patterson N, Hirschhorn JN, Keinan A, Hao L, Atzmon G, Burns E, Ostrer H, Price AL, Reich D.. 2011. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 7(4):e1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskova E, Ulyantsev V, Koepfli K-P, O’Brien SJ, Dobrynin P.. 2020. GADMA: genetic algorithm for inferring demographic history of multiple populations from allele frequency spectrum data. GigaScience 9(3):giaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoni C, Flink LG, Evin A, Georg C, De Cupere B, Van Neer W, Bartosiewicz L, Linderholm A, Barnett R, Peters J, et al. 2013. Pig domestication and human-mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics. Mol Biol Evol. 30(4):824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RW, Hsu TY.. 1944. Chinese swine and their performance: compared with modern and crosses between Chinese and modern breeds. J Hered. 35(12):365–379. [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38(8):904–909. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez O, Ojeda A, Tomas A, Gallardo D, Huang LS, Folch JM, Clop A, Sanchez A, Badaoui B, Hanotte O, et al. 2009. Integrating Y-chromosome, mitochondrial, and autosomal data to analyze the origin of pig breeds. Mol Biol Evol. 26(9):2061–2072. [DOI] [PubMed] [Google Scholar]
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SR, Wilkie AO, McVean G, Lunter G, Consortium WGS, 2014. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 46(8):912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner BM, Sargent CA, Churcher C, Hunt T, Herrero J, Loveland JE, Dunn M, Louzada S, Fu B, Chow W, et al. 2016. The pig X and Y Chromosomes: structure, sequence, and evolution. Genome Res. 26(1):130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamathevan JJ, Hall MD, Hasan S, Woollard PM, Xu M, Yang Y, Li X, Wang X, Kenny S, Brown JR, et al. 2013. Minipig and beagle animal model genomes aid species selection in pharmaceutical discovery and development. Toxicol Appl Pharmacol. 270(2):149–157. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang A, Wang L, Li K, Yang G, He R, Qian L, Xu N, Huang R, Peng Z, et al. 2011. Animal genetic resources in China: pigs. Beijing (China: ): China Agricultural Press. In Chinese. [Google Scholar]
- Wei W, Ayub Q, Chen Y, McCarthy S, Hou Y, Carbone I, Xue Y, Tyler-Smith C.. 2013. A calibrated human Y-chromosomal phylogeny based on resequencing. Genome Res. 23(2):388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. 2011. From globalized pig breeds to capitalist pigs: a study in animal cultures and evolutionary history. Environ Hist. 16(1):94–120. [Google Scholar]
- Wu G-S, Yao Y-G, Qu K-X, Ding Z-L, Li H, Palanichamy MG, Duan Z-Y, Li N, Chen Y-S, Zhang Y-P.. 2007. Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biol. 8(11):R245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. 2004. Introduction and domestication of European breeds of pig in modern China. Anc Mod Agric. 154–162. [Google Scholar]
- Yang B, Cui L, Perez-Enciso M, Traspov A, Crooijmans RPMA, Zinovieva N, Schook LB, Archibald A, Gatphayak K, Knorr C, et al. 2017. Genome-wide SNP data unveils the globalization of domesticated pigs. Genet Sel Evol. 49(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 2008. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol Biol Evol. 25(3):568–579. [DOI] [PubMed] [Google Scholar]
- Zhang D, He X, Wang W, Wang L, Liu D.. 2018. Determination and analysis of the whole genome sequence of Min pig. J Northeast Agric Univ. 49(11):9–17. [Google Scholar]
- Zhang L, Su W, Tao R, Zhang W, Chen J, Wu P, Yan C, Jia Y, Larkin RM, Lavelle D, et al. 2017. RNA sequencing provides insights into the evolution of lettuce and the regulation of flavonoid biosynthesis. Nat Commun. 8(1):2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yang Q, Ai H, Huang L.. 2021. Revisiting the evolutionary history of pigs via de novo mutation rate estimation by deep genome sequencing on a three-generation pedigree. bioRxiv 2021.2003.2029.437103. [DOI] [PMC free article] [PubMed]
- Zhang Y, Sun Y, Wu Z, Xiong X, Zhang J, Ma J, Xiao S, Huang L, Yang B.. 2021. Subcutaneous and intramuscular fat transcriptomes show large differences in network organization and associations with adipose traits in pigs. Sci China Life Sci. 1:1–15. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li W, Yang B, Zhang Z, Ai H, Ren J, Huang L.. 2017. Signatures of selection and interspecies introgression in the genome of Chinese domestic pigs. Genome Biol Evol. 9(10):2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsolnai A, Radnóczy L, Fésüs L, Anton I.. 2006. Do Mangalica pigs of different colours really belong to different breeds? Arch Anim Breed. 49(5):477–483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence reads from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA751703.