Abstract
The settlement of Sahul, the lost continent of Oceania, remains one of the most ancient and debated human migrations. Modern New Guineans inherited a unique genetic diversity tracing back 50,000 years, and yet there is currently no model reconstructing their past population dynamics. We generated 58 new whole-genome sequences from Papua New Guinea, filling geographical gaps in previous sampling, specifically to address alternative scenarios of the initial migration to Sahul and the settlement of New Guinea. Here, we present the first genomic models for the settlement of northeast Sahul considering one or two migrations from Wallacea. Both models fit our data set, reinforcing the idea that ancestral groups to New Guinean and Indigenous Australians split early, potentially during their migration in Wallacea where the northern route could have been favored. The earliest period of human presence in Sahul was an era of interactions and gene flow between related but already differentiated groups, from whom all modern New Guineans, Bismarck islanders, and Indigenous Australians descend. The settlement of New Guinea was probably initiated from its southeast region, where the oldest archaeological sites have been found. This was followed by two migrations into the south and north lowlands that ultimately reached the west and east highlands. We also identify ancient gene flows between populations in New Guinea, Australia, East Indonesia, and the Bismarck Archipelago, emphasizing the fact that the anthropological landscape during the early period of Sahul settlement was highly dynamic rather than the traditional view of extensive isolation.
Keywords: Papuan, human genome, demographic history, Sahul, Oceania
Introduction
New Guineans are descendants of one of the most exceptional and ancient migrations in human history, but the nature of their initial settlement of New Guinea is still veiled in mystery (Allen and O’Connell 2020). When modern human first arrived in Southeast Asia 50–70 thousand years ago (ka) (Westaway et al. 2017), the lower sea level during a colder climate caused two gigantic ancient continents to emerge: Sunda joined many current southeast Asian islands with mainland Asia, whereas Australia and New Guinea formed a single landmass called Sahul. These vast territories were separated by a substantial water crossing, Wallacea, containing a scattering of small islands and creating the major ecological barrier known as the Wallace line (Wallace 1869).
The first anatomically modern humans to cross from Sunda to Sahul numbered probably no more than a few hundred, but migration through the Wallacean islands was nonetheless likely a deliberate effort (Bird et al. 2019; Bradshaw et al. 2019). Several initial settlement paths are possible based on paleogeological reconstructions: a northern route through what today are the Maluku islands (Kealy et al. 2018; Norman et al. 2018), and a southern route through Flores and Timor (Birdsell 1977; Norman et al. 2018). Although not mutually exclusive, for decades the consensus has been that a single migration from Sunda to Sahul created the entire biological diversity of both New Guineans and Indigenous Australians today (Malaspinas et al. 2016; O’Connell et al. 2018). Both populations have unique genomic diversities marked by their high percentage of genetic inheritance coming from Denisovans (3–3.5%) (Reich et al. 2011; Sankararaman et al. 2016; Vernot et al. 2016; Jacobs et al. 2019). However, recent studies now suggest that the arrival of just one human group is unlikely to be sufficient to explain the deep genetic diversity still observed in this region (Allen and O’Connell 2020; Pedro et al. 2020). In part, this argument draws from the uniparental genetic markers found in New Guineans and Indigenous Australians. Although both populations undoubtedly share a common maternal genetic ancestry, marked by the presence of haplogroups M*, N*, and R*, the diversity of their respective mitochondrial DNA (mtDNA) subhaplogroups indicate stark genetic differences (Redd and Stoneking 1999). Most mtDNA subhaplogroups are exclusive to either New Guinea (e.g., P1, P2, P4a, M27, M28, Q*) (Pedro et al. 2020) or Australia (e.g., P8, P4b, M42, S*) (Tobler et al. 2017), in agreement with local evolution of some of these region-specific haplogroups. Coalescence dates between Australian and New Guinean maternal lineages suggest a split at the time of the initial settlement of Sahul (Tobler et al. 2017), or potentially before, as shown by the coalescence dates of P2’P8’P10 (49 ka), P4 (50 ka) (Pedro et al. 2020). Similar patterns are observed for paternal genetic lineages on the Y chromosome (Bergstrom et al. 2016; Nagle et al. 2016). Such different uniparental genetic patterns suggest that either New Guineans and Indigenous Australians separated very early after the initial migration to Sahul and that uniparental lineages segregated accordingly, or that both populations descend from two initially separate migrations (Redd and Stoneking 1999), perhaps at different times as suggested by some archaeological records (Clarkson et al. 2017).
Once in Sahul, modern human dispersals within what is now the island of New Guinea began by at least 49 ka (Summerhayes et al. 2010). Given the paucity of archaeological records, how New Guinea was settled remains poorly resolved. Of particular note, the initial entry point into New Guinea has proven particularly difficult to locate, leading to alternative hypotheses of migration via the New Guinean north coast, through the mountainous highlands, along the southern foothills, or even further south via a vast plain that is now submerged and forms the Arafura Sea (Summerhayes et al. 2017). Archaeological records perhaps favor the Arafura plain pathway, since the oldest known archaeological sites in New Guinea are located in its southeast region. The suggestion is that even earlier sites, marking those first movements, now lie flooded under the Arafura Sea (Summerhayes et al. 2017). Nevertheless, the alternative hypotheses remain feasible. Although the highlands were not hospitable during the Upper Pleistocene, long-term human presence is attested there from at least 25 ka (Summerhayes et al. 2017). Major rivers carved into the New Guinean geology may have formed early natural corridors during initial settlement, allowing human mobility as they still do along the Markham and Ramu Rivers in the northeast, the Fly River in the south and the Sepik River in the north (Pawley 2005). The Sepik route has been supported by close genetic relationships between modern New Guinea highlanders and lowland groups from the Sepik region, but this association may date to a later time period (Bergstrom et al. 2017).
After the initial settlement of New Guinea, independent development of agricultural practices in New Guinea between 10 and 7 ka led to a unique Neolithic revolution (Denham et al. 2003; Golson et al. 2017), driving demographic expansion in the highlands that heavily impacted the genetic and linguistic diversity of these populations (Reesink et al. 2009; Greenhill 2015; Bergstrom et al. 2017). By the time Austronesian settlers arrived around 3 ka (Bellwood 2007; Lipson et al. 2014), New Guineans had already developed a complex network of diverse cultures (Pawley 2005).
Given its extraordinary diversity today, comparable to large continental areas elsewhere, the genetics of New Guinean populations are poorly explored, and scenarios of initial and postsettlement movements are lacking. We have therefore generated 58 high-quality whole-genome sequences (WGS) from Papua New Guinean individuals (supplementary table 1, Supplementary Material online). Coupling these with a data set of 905 genomes from diverse human groups, but particularly individuals from neighboring Indonesia and Australia (supplementary table 2, Supplementary Material online), we reconstruct the key genetic events during the Upper Pleistocene that led to the settlement of New Guinea. Our two primary questions are: was the migration to Sahul conducted by one or several human groups, and what migratory routes did they take? And how did New Guineans subsequently settle the diverse environments of the modern island, and what were the dynamics of this settlement?
Results
Samples and Diversity
Fifty-eight Papua New Guinean participants were included in this study to increase the geographical coverage of New Guinea, especially in the southern regions underrepresented in published whole-genome data sets (supplementary fig. 1 and table 1, Supplementary Material online). Whole-genome sequencing data were obtained at an average depth of 36×, characterized for 41,250,479 SNPs. Uniparental haplogroups were obtained from mitochondrial DNA and Y chromosome data (supplementary table 1, Supplementary Material online). New Guinean mitochondrial DNA diversity was previously described (Pedro et al. 2020), showing mostly haplogroups specific to the region (Q*:38%; P*:29%) as well as the presence of haplogroups frequent in Austronesian populations (B4a1a1:20%). New Guinean Y chromosome diversity is dominated by M1 (39%) and S* (18%) haplogroups frequent in the region, and haplogroups of Austronesian origin (O1:14%) as previously described (Mona et al. 2007; Karafet et al. 2010). Autosomal data were combined with published genomes, notably from Indonesia including the western half of New Guinea Island (Jacobs et al. 2019), northeast Australia (Mallick et al. 2016), and the Bismarck Archipelago (Vernot et al. 2016), and with genotyping data from North Maluku (Kusuma et al. 2017) and imputed genotyping data from Papua New Guinea (Bergstrom et al. 2017), resulting in a data set of 966 individuals (including genomes from Denisovan, Neandertal, and Chimpanzee, supplementary table 2, Supplementary Material online). By increasing the number of New Guinean whole-genome sequencing data as well as the geographic resolution of New Guinean genetic diversity, we created a high-quality data set (e.g., with good phasing; Delaneau et al. 2011 and imputation; Howie et al. 2009), thus allowing us to perform fine-scale demographic analyses.
Characterization of Genetic Structure
We analyzed genetic patterns in individuals from New Guinea, northeast Australia, the Bismarck Archipelago, and Wallacea to identify key regional genetic groups. A coancestry matrix was built by scanning each pair of genomes to detect identity-by-descent (IBD) fragments using fineSTRUCTURE (Lawson et al. 2012) (supplementary fig. 2, Supplementary Material online). Most genomes clustered according to their respective geographical locations, defining 15 regional groups, including seven regional groups from the Wallacea/Sahul region: New Guinean southeast lowlands, New Guinean north and south lowlands, New Guinean east highlands, New Guinean west highlands, northeast Australia, the Bismarck Archipelago, and Wallacea. Of these seven regional groups of interest, we also characterized 58 subgroups based on the fineSTRUCTURE dendrogram (fig. 1A and supplementary table 2, Supplementary Material online). Each of these 58 subgroups defines a “population” based on genetic similarity (Lawson and Falush 2012), a definition that can differ from a linguistic or anthropological perspective but which is more accurate regarding the analyses performed in our work.
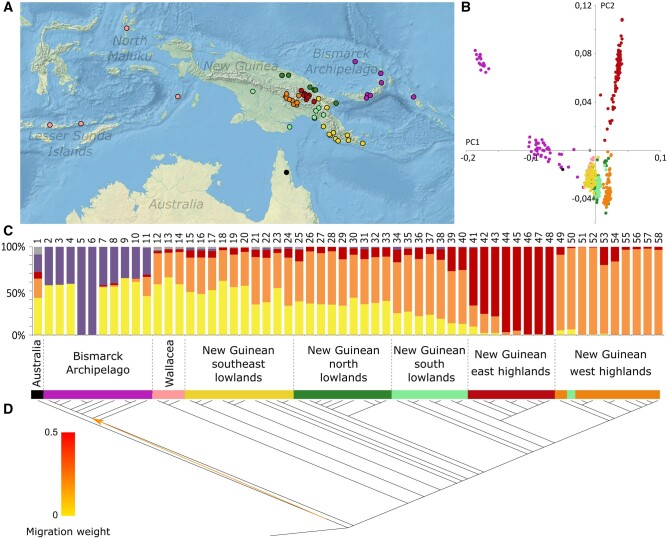
Fig. 1.
Genetic diversity of individuals from New Guinea, Wallacea, the Bismarck Archipelago, and northeast Australia. (A) Geographic locations of all 58 genetic subgroups defined by the fineSTRUCTURE analysis. (B) Principal component analysis representing 26.7% of the total variability of all individual genomes from the 58 subgroups, masked for Asian genetic ancestry. The two first components are represented (PC1 and PC2). (C) ADMIXTURE plot for eight genetic components summarized by genetic subgroups (based on K = 8 from supplementary fig. 7, Supplementary Material online). The gray component combines four components mainly present in African and Eurasian groups. Numbers above bar plots identify genetic subgroups referred in supplementary table 2, Supplementary Material online. (D) TREEMIX plot for all 58 subgroups (based on supplementary fig. 7, Supplementary Material online). All four panels used the same color scheme to identify the 58 genetic subgroups.
As previously described (Bergstrom et al. 2017; Hudjashov et al. 2017), recent gene flow from Asian groups, mostly linked to the Austronesian dispersal ∼3 ka, has substantially impacted regional genetic diversity. Since our study focused on Upper Pleistocene movements, we decided to mask this later Asian genetic ancestry using haplotypic information with PCAdmix (Brisbin et al. 2012). Principal component analysis (PCA) shows that this procedure was efficient; prior to masking the Asian genetic ancestry, all genomes from New Guinea, Wallacea, northeast Australia, and the Bismarck Archipelago were spread along the first principal component toward Asian genomes, whereas after masking, these genomes instead clustered together (supplementary fig. 3, Supplementary Material online). The data set was also analyzed using ADMIXTURE prior and after the masking of Asian tracks (supplementary fig. 4, Supplementary Material online). Asian genetic ancestry is present at an average of 22.7% in New Guinean lowlands, the Bismarck Archipelago, Wallacea, and northeast Australia. It ranges between 0% and 62.1% in individuals from New Guinean lowlands, but absent in New Guinean highlanders, as previously reported (Bergstrom et al. 2017), and between 0% and 49.2% in individuals from the Bismarck Archipelago, between 38.8% and 63.7% in individuals from Wallacea, and between 8.1% and 12.1% in individuals northeast Australia, as previously reported (Mallick et al. 2016; Hudjashov et al. 2017) (supplementary fig. 4, Supplementary Material online). After the masking, the average Asian genetic ancestry is greatly reduced (supplementary fig. 4, Supplementary Material online), which allows us to infer demographic scenarios that are not strongly biased by recent Asian genetic admixture.
We first described the masked data set using a PCA focused on individuals in the seven regional groups of interest (fig. 1B). All genomes from the same geographical regions are grouped together in distinctive clusters. Clusters of New Guinean lowlanders, Wallacean islanders, and northeast Indigenous Australians fall relatively close together. New Guinean highlanders, especially individuals from the New Guinean east highlands, are separated on the second principal component. Individuals from the Bismarck Archipelago form two clusters separated by both main components. This apparent genetic structure coincides with FST genetic distances (supplementary fig. 5, Supplementary Material online). The genetic diversity of the Bismarck Archipelago is particularly differentiated from other New Guineans, Wallaceans, and northeast Indigenous Australians (0.18<FST<0.29; supplementary fig. 5, Supplementary Material online). Within New Guinea, larger genetic distances are found between New Guinean lowlanders and highlanders (0.05<FST<0.18; supplementary fig. 5, Supplementary Material online) than between any other New Guinean subgroups, as previously reported (Bergstrom et al. 2017). Wallaceans are genetically closer to New Guineans (0.04<FST<0.26) than to Bismarck Islanders or northeast Indigenous Australians (0.06<FST<0.318). We further confirm this geographical structure using EEMS (Petkova et al. 2016), which places the biggest genetic barriers between the Bismarck Archipelago and New Guinea, between the New Guinean lowlands and highlands, as previously noted (Bergstrom et al. 2017), and between northeast Australia and New Guinea (supplementary fig. 6, Supplementary Material online). We do not detect any strong genetic barriers in Wallacea.
All of these analyses clearly show that there is strong geographical structure of human genetic diversity in Sahul today. However, surprisingly, it also reveals a relative genetic continuity between New Guinea and Wallacea, suggesting that Wallacea might have represented less of a barrier to humans than other floral and faunal species.
Identification of Gene Flow
We analyzed the genetic diversity of our data set to identify possible connections between each individual in the seven regional groups from New Guinea, northeast Australia, the Bismarck Archipelago, and Wallacea using ADMIXTURE(Alexander et al. 2009). The genetic diversity in our data set is best deconstructed into eight genetic components (supplementary figs. 7–9, Supplementary Material online), four of which are dominated by New Guinean, northeast Indigenous Australian, Bismarck Archipelago, and Wallacean groups (fig. 1C and supplementary fig. 9, Supplementary Material online). This strong genetic structure is confirmed by the genetic ancestries defined in New Guinean east and west highlanders as well as in the Bismarck Archipelago (red, orange, and purple components, respectively). The fourth genetic component (yellow) is more widespread, as it is present in individuals from Wallacea, the New Guinean lowlands, northeast Australia, and the Bismarck Archipelago. It is the main component in Wallacea and New Guinea, and reaches its highest value in the southeast region (31–63%; fig. 2A and supplementary figs. 7 and 9, Supplementary Material online).
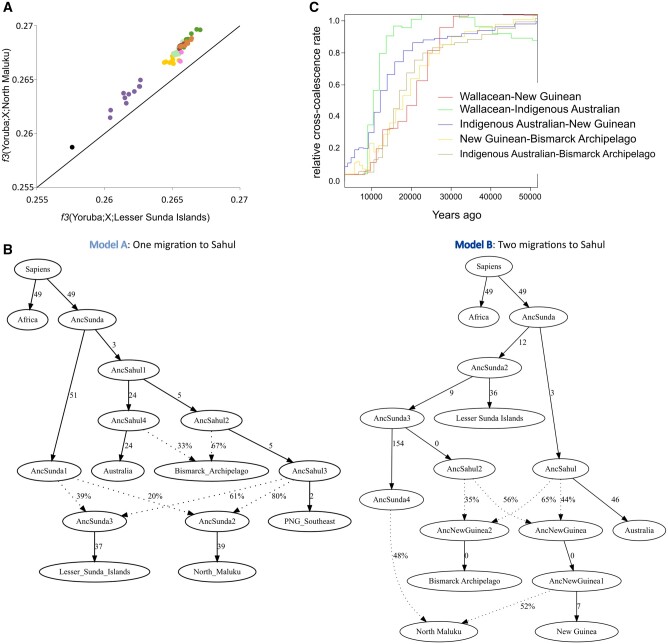
Fig. 2.
Scenario of the settlement of Sahul. (A) f3-outgroup analysis for all Sahul groups compared with populations from the North Maluku Islands (y axis) or Lesser Sunda Islands (x axis). Populations are identified by the same color scheme as in figure 1. (B) Best fitting qpGraph models considering one migration to Sahul (model A; Z score=−2.570) or two migrations to Sahul (model B; Z score = 2.301). One subgroup was used as representant of a region: populations from North Maluku (Wallacea subgroup 23, following fig. 1C and defined in supplementary table 2, Supplementary Material online), Lesser Sunda Islands (Wallacea subgroup 13), New Guinea (New Guinea subgroup 16), the Bismarck Archipelago (Bismarck subgroup 5), northeast Australia (Australia subgroup 1), and Africa (Africa_5). Solid arrows are labeled with branch lengths in 1,000 times drift units. Dotted arrows indicate gene flows whose proportions are indicated by percentages. (C) Relative cross-coalescence rate curves estimated by MSMC2 between populations from Wallacea (Lesser Sunda Islands, Wallacea subgroup 13), New Guinea (New Guinea subgroup 16), the Bismarck Archipelago (Bismarck subgroup 5), and northeast Australia (Australia subgroup 1).
To determine the history of regional mixing, population splits and admixture events were estimated without a priori assumptions with TREEMIX (Pickrell and Pritchard 2012) (fig. 1D and supplementary fig. 10, Supplementary Material online). An optimal model is obtained including five gene flows, one of which is in the Wallacea/Sahul region: from ancestors of northeast Indigenous Australians to a group ancestral to most Bismarck Archipelago islanders (fig. 1D and supplementary fig. 10, Supplementary Material online). This may partly explain the shared genetic ancestry observed in the ADMIXTURE plot (purple, fig. 1C).
The limited number of admixture events is confirmed by f3-statistics performed between all pairs of the 58 subgroups (Patterson et al. 2012). Only seven genetic admixture events are detected (supplementary table 3, Supplementary Material online), four of which show multiples waves of admixture. For all of these gene flows, we estimated the dates of the admixture events using MALDER (Loh et al. 2013). Gene flows are detected between subgroups of New Guinean west highlands, mostly between 9.9 and 11.9 ka (supplementary table 3, Supplementary Material online). Multiple gene flow events between New Guinean east highlanders and New Guinean subgroups from the south lowlands occurred between 25.4 and 30.2 ka and 2.6 and 4.1 ka. During the same two time frames, we also observe gene flows between a New Guinean subgroup from the south lowlands and a Wallacean subgroup from the Lesser Sunda Islands (supplementary table 3, Supplementary Material online). Although limited in number, these gene flows occurred at different time frames spanning several millennia, indicative of a population dynamics model involving multiple periods of isolation and contact.
Modeling the Genetic Scenario of the Migration in Wallacea
Our analyses show that New Guinean groups appear to be more closely related to populations in Wallacea than to groups in northeast Australia and the Bismarck Archipelago (fig. 1D and supplementary fig. 5, Supplementary Material online). Since human populations settled Sahul from Sunda through Wallacea (Allen and O’Connell 2020), we expected that genetic diversity from New Guinea, northeast Australia, and the Bismarck Archipelago would branch from the diversity of Wallacean subgroups. This highest genetic affinity between New Guineans and Wallaceans could be due to ancient gene flow between these populations (supplementary table 3, Supplementary Material online), or to a genetic divergence between northeast Indigenous Australians and Wallaceans prior to the genetic divergence between New Guineans and Wallaceans (fig. 1D). We therefore explored different scenarios that could have led to the observed genetic patterns.
We first sought to identify the most likely route taken by the first settlers of Sahul. We used an f3-outgroup approach to compare the genetic diversity present in New Guinean, northeast Indigenous Australian and Bismarck Archipelago islander subgroups to each Wallacean subgroup, using African Yoruba as an outgroup (supplementary fig. 11, Supplementary Material online). All subgroups from New Guinea, northeast Australia, and the Bismarck Archipelago have f3-statistics values equally distant to all Wallacean subgroups located on the possible southern route to Sahul (Lesser Sunda Islands and Flores Island) (supplementary fig. 11A–C, Supplementary Material online). However, when we included a subgroup located on the northern route (North Maluku islands, Kei Island), we detected a stronger genetic link with all studied subgroups (supplementary fig. 11D–F, Supplementary Material online). Most populations from New Guinea and the Bismarck Archipelago are significantly genetically closer to the population from North Maluku using f4-statistics (f4(Yoruba, X; Lesser Sunda Islands, North Maluku)>0.001, Z score > 3, supplementary fig. 12, Supplementary Material online). We found higher f3-statistics and more significant f4-statistics values for New Guinean subgroups with North Maluku than for subgroups from the Bismarck Archipelago and northeast Australia (fig. 2A and supplementary fig. 12, Supplementary Material online). This suggests that the ancestors of New Guineans, Bismarck Archipelago islanders, and northeast Indigenous Australians are all more closely related to North Maluku islanders than to groups from Lesser Sunda Islands or Flores Island, and that this link is probably more important in New Guineans. This analysis supports two nonexclusive hypotheses: 1) the northern route as the most likely path for the initial settlement of Sahul, 2) secondary gene flows between groups of North Maluku and New Guinea, the Bismarck Archipelago and northeast Australia.
We modeled these hypotheses using qpGraph (Patterson et al. 2012). For completeness, we tested several models that consider migrations from Sunda to Sahul along either the southern route (represented by the Lesser Sunda Islands) or the northern route (represented by the North Maluku Islands) (supplementary fig. 13, Supplementary Material online), as well as different possibilities of admixture between Wallacean and New Guinean groups as suggested by our other analyses (fig. 1C and supplementary fig. 6 and table 3, Supplementary Material online). Since our analyses suggested that Wallaceans share a closer genetic link with New Guineans than northeast Indigenous Australians (figs. 1D and 2A; supplementary fig. 6, Supplementary Material online), we decided to model demographic scenarios with one or two migrations from Sunda to Sahul. Considering one migration from Sunda to Sahul, meaning that both New Guineans and Indigenous Australians share a common ancestor more recently than with Wallaceans, we obtained one model fitting the data (Z =−2.558, fig. 2B and supplementary fig. 13, Supplementary Material online). It describes a common ancestry to both New Guineans and Indigenous Australians, after the split from Wallaceans, followed by major secondary gene flows from New Guinea to Wallacea (between 61% and 79%, fig. 2B and supplementary fig. 13, Supplementary Material online). In this model no preferable route in Wallacea can be distinguished. We then tested models considering two migrations from Sunda to Sahul, in which northeast Indigenous Australians and New Guineans descended separately from groups moving along the northern or southern routes (supplementary fig. 14, Supplementary Material online). We obtained one significant model: two migrations to Sahul, including one via the northern route, an admixture event between the ancestors of New Guineans and northeast Indigenous Australians, and a subsequent gene flow from ancestral groups from New Guinea to North Maluku (52%, Z = 2.337, supplementary fig. 14, Supplementary Material online). In this model, the ancestral group to New Guineans is related to the northern route, via North Maluku, but no route can be proposed for the ancestral Indigenous Australian group. Our qpGraph analyses thus support both models of migrations from Sunda to Sahul, with one or two migrations, followed by high secondary gene flows from New Guinea to Wallacea.
Modeling the Genetic Scenario of the Settlement of Sahul
We refined our models to include a group from the Bismarck Archipelago (represented by New Britain), considering it either to be a sister group to New Guineans or the result of a separate admixture event with ancestral northeast Indigenous Australians (supplementary fig. 15, Supplementary Material online), as suggested by other analyses (fig. 1D). For both models, with one or two migrations to Sahul, we obtained models fitting the data when the Bismarck Archipelago group results from a separate admixture event between ancestral New Guineans and northeast Indigenous Australians (One migration: Z = −2.570, Two migrations: Z = 2.301, fig. 2B and supplementary fig. 15, Supplementary Material online). This corresponds to the gene flow observed between an ancestral Indigenous Australian group and ancestral Bismarck Archipelago group detected by TREEMIX (fig. 1D).
We added further detail to this scenario by reconstructing the effective population sizes of these groups, and estimating divergence dates between all studied groups using MSMC2 (Schiffels and Durbin 2014). Since archaic admixture can be a confounding factor in these analyses (Mallick et al. 2016), we masked both Neandertal and Denisovan genomic tracks using ChromoPainter (Lawson et al. 2012). The archaic genetic ancestry in the newly sequenced genomes ranges between 1.8% and 2.8% of Denisovan ancestry and ranges between 2.3% and 2.5% of Neandertal ancestry, corresponding to previous estimations (Vernot et al. 2016; Jacobs et al. 2019). All estimated effective population size curves show a bottleneck around 60–70 ka, as found for European and Asian populations (supplementary fig. 16, Supplementary Material online). Around 50 ka, estimated effective population sizes for populations from Wallacea and Sahul are between 1,671 and 2,100 individuals, and between 2,540 and 2,999 individuals in Eurasia and 9,506 individuals in Africa. All divergence dates for Wallacea/Sahul groups from Africa, represented by Yoruba genomes, show similar results (74 ± 4.2 ka, supplementary fig. 17, Supplementary Material online). We note here that this date is older than that obtained between Eurasian and African genomes (66.5 ± 3.7 ka), a result previously reported and interpreted as a potential methodological bias or the signal of an even earlier human migration from Africa (Malaspinas et al. 2016; Mallick et al. 2016; Pagani et al. 2016; Bergstrom et al. 2017). We cannot confirm any of these interpretations but the similar dates of cross-coalescence with African genomes and the similar profiles of effective population size curves clearly show that all genomes from Wallacea, New Guinea, northeast Australia, and the Bismarck Archipelago descend from a common ancestor, probably located in Sunda, as represented by our TREEMIX analysis (fig. 1D).
We then estimated divergence dates between each of the studied groups (fig. 2C). Genomes from New Guinean and Wallacean groups diverged between 23.7 and 26.7 ka. Groups from New Guinea and the Bismarck Archipelago show genetic divergence between 17.5 and 19.8 ka. Genomes of New Guineans diverged from the genomes of northeast Indigenous Australians between 13.5 and 15.3 ka. We found that genomes from northeast Indigenous Australians diverged from the genomes of Wallaceans 13.3–15.2 ka, and from genomes of Bismarck Archipelago islanders between 15.2 and 17.3 ka. We caution that these analyses are sensitive to recent admixture events, and the masking of Asian genomic tracks reduces the amount of data to accurately estimate divergence dates. It likely affected results on Wallacean genomes for which the masked Asian genetic data were large (>50%). Moreover, the limited number of Indigenous Australian genomes likely resulted in an insufficient phasing quality for the two available genomes which could strongly affect MSMC2 analyses, as previously noted (Malaspinas et al. 2016). A previous study on Indigenous Australians genomes (Malaspinas et al. 2016) (not available for this study) found that all Indigenous Australians groups diverged from each other around 25 ka, according to MSMC2 analyses, and from New Guinean highlanders around 30–40 ka. Our estimation of the divergence between Indigenous Australians and New Guinean is even more recent, potentially caused by the identified gene flow between groups of northeast Australia and the Bismarck Archipelago (fig. 1D). Although these dates cannot be interpreted as being part of the initial migration to Sahul, they strongly indicate long-term contacts between all of these groups, as seen in our models. The population dynamics between ancestral groups was probably important, over long distances and periods of time, leading to the emergence of modern populations like New Guineans.
Modeling the Genetic Scenario of the Settlement of New Guinea
Our TREEMIX analysis show that New Guinean subgroups branch together. The root of the tree lies in the genetic diversity present today in New Guinean southeast lowlanders (fig. 1D). This result reflects the ADMIXTURE analysis, which shows that the main genetic component in Wallacea is also the main component in southeast New Guinea, reinforcing the hypothesis of an ancient link (yellow component, fig. 1C and supplementary fig. 9, Supplementary Material online). Subsequently, subgroups from the New Guinean north and south lowlands form two different genetic clusters that are genetically drifted from the New Guinean southeast lowlanders (fig. 1D). This suggests that both clusters emerged from two separate migrations, south and north, initiated from the southeast New Guinean region. We modeled the settlement of New Guinea based on our results showing that most of New Guinean genetic diversity could be described by genetic drift with limited occurrences of gene flows (fig. 1D and supplementary table 3, Supplementary Material online). We used qpGraph to determine the best scenario fitting data from the five New Guinean groups (southeast lowlanders, south lowlanders, north lowlanders, east highlanders, and west highlanders) and one African outgroup (Yoruba). First, we modeled the most probable entry point into New Guinea, considering possible migrations from the north lowlands, the south lowlands, and the southeast lowlands, with or without admixture events (supplementary fig. 18, Supplementary Material online). One model is significant (Z score = −0.06); this places southeast lowlands as the most likely entry point but only if there is a secondary admixture event with ancestral New Guinean north lowlanders (supplementary fig. 18, Supplementary Material online). This model corresponds with results from other analyses, which positions the New Guinean southeast group at the root of the New Guinean diversity (fig. 1D).
Highlanders are the last to branch off in our TREEMIX analysis (fig. 1D). Our analyses showed that New Guinean west and east highlanders form two clearly separated clusters (fig. 1A and B; supplementary figs. 2 and 5, Supplementary Material online), and that both New Guinean highlander groups drifted from New Guinean lowlanders located in the southern region (fig. 1D). We first added a New Guinean east highlander group to model dispersal in the highlands from the north, south, or southeast lowlands; using qpGraph (supplementary fig. 19, Supplementary Material online). The model best fitting the data connects New Guinean east highlanders to a branch leading to the New Guinean north lowlanders (Z score =−0.068, supplementary fig. 19, Supplementary Material online). Finally, we added a New Guinean west highlander group, defined as a sister group to New Guinean east highlanders or coming from a second migration from the lowlands (supplementary fig. 20, Supplementary Material online). The best model places New Guinean west highlanders with New Guinean south lowlanders (Z score = 2.759, supplementary fig. 20, Supplementary Material online). This branching describes two separate migrations leading to the genetic diversity observed in New Guinean east and west highlanders, which would explain the stark genetic difference observed in other analyses (fig. 1B and C). However, we note here that it could appear to be in contrast to the TREEMIX analysis which showed the New Guinean east highlanders branched with some PNG South lowlands groups (fig. 1D). In particular these South lowlands groups (PNG South lowlands subgroups 39 and 40, fig. 1D) are geographically located close to North lowlands groups. We computed an f3-outgroup analysis to further explore the connection between PNG highlanders and lowlanders. It confirmed the closer link between PNG West highlanders and PNG South lowlanders but also showed that PNG East highlanders are almost genetically equidistant to both PNG North and South lowlanders, although still more closely related to PNG South highlanders (supplementary fig. 21, Supplementary Material online). Based on our results, we postulate that PNG east highlanders came from a place intermediate to both South and North lowlanders, potentially in the current Morobe province. The qpGraph model also supports a secondary admixture event between New Guinean west highlanders and north lowlanders (fig. 3A and supplementary fig. 20, Supplementary Material online), which could explain the presence of the main New Guinean west highlanders genetic component in many lowlanders (fig. 1C) as well as previous reports on the link between PNG north lowlanders and PNG highlanders (Bergstrom et al. 2017). This final model identifies the southeast lowlands as the entry point into New Guinea, followed by two migrations to the south and north lowlands, which in turn led to the independent settlements of the east and west highlands.
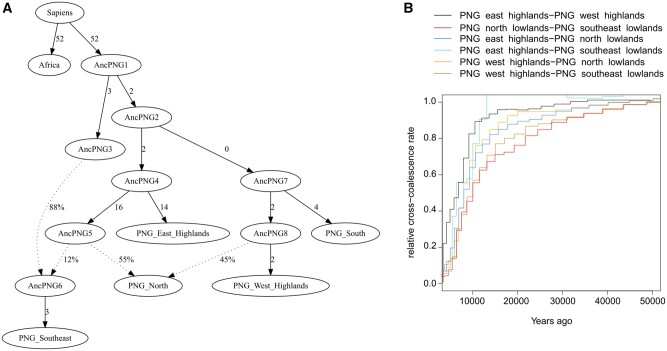
Fig. 3.
Scenario of the settlement of New Guinea. (A) Best fitting qpGraph model for populations from New Guinean southeast lowlands (New Guinea subgroup 16, following fig. 1C and defined in supplementary table 2, Supplementary Material online), north lowlands (New Guinea subgroup 28), south lowlands (New Guinea subgroup 39), east highlands (New Guinea subgroup 45), west highlands (New Guinea subgroup 55), and Africa (Africa_5) (Z score = 2.759). Solid arrows are labeled with branch lengths in 1,000 times drift units. Dotted arrows indicate gene flows whose proportions are indicated by percentages. (B) Relative cross-coalescence rate curves estimated by MSMC2 between populations from New Guinean lowlands and highlands.
Given this scenario, we estimated divergence dates between each New Guinean group of interest using MSMC2 (fig. 3B). Divergence dates between New Guinean lowlanders and New Guinean highlanders ranged between 8.8 and 12.8 ka, which broadly correspond to previous estimations (12–16 ka) (Bergstrom et al. 2017). West and east New Guinean highlander groups diverged between 7.6 and 8.8 ka, similarly to previously reported (6–12 ka) (Bergstrom et al. 2017). These rather recent dates of genetic divergence probably reflect long-term contacts between New Guinean groups following the initial settlement of the island, initiated as early as 49 ka based on archaeological records (Summerhayes et al. 2010). They thus probably illustrate the gene flows between highlander groups, and between highlanders and lowlanders, which are estimated to have occurred at the same time (9.9–11.9 ka, supplementary table 3, Supplementary Material online). Effective population sizes estimated with MSMC2 for each New Guinean group do not show any marked differences (supplementary fig. 16, Supplementary Material online). It was previously interpreted as a bias due to phasing quality (Bergstrom et al. 2017), but it could also mean that contacts between New Guinean highlanders and lowlanders were frequent enough so that populations grew similarly.
Discussion
Our study provides the first models of the settlement of north Sahul and New Guinea based on whole-genome data (fig. 4). Our models support an early split of the ancestral groups of New Guineans from northeast Indigenous Australians, potentially in Wallacea. Among the possible migratory routes, our model brings new evidence for the use of the northern route, via the North Maluku islands. Our genetic data describe a settlement of New Guinea initiated from what is now its southeast lowland via the Arafura Plain, and long-term population contact between ancestral populations of New Guinea, northeast Australia, Wallacea, and the Bismarck Archipelago.
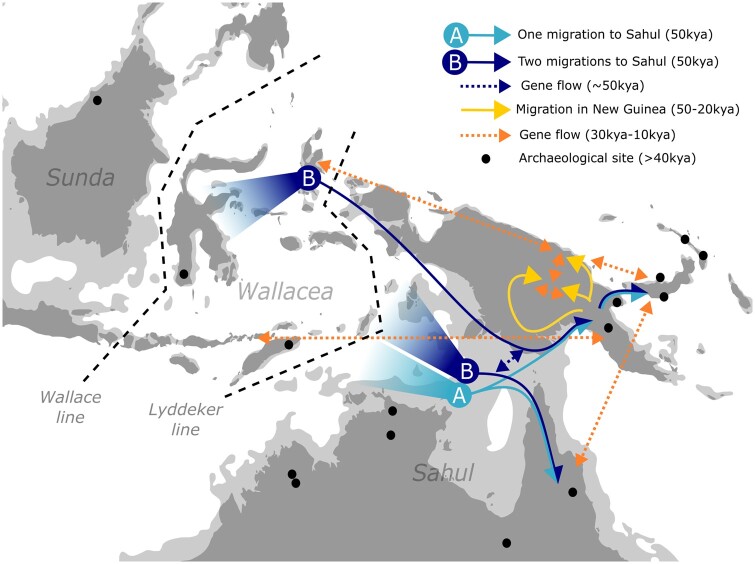
Fig. 4.
Genomic scenario of the human dynamics in north Sahul during the Upper Pleistocene. Light blue arrows represent the settlement of north Sahul following a scenario with one migration to Sahul (model A). The dark blue arrows represent the settlement of north Sahul following a scenario with two migrations to Sahul (model B). The dotted dark blue arrow represents a gene flow following a scenario with two migrations to Sahul (model B). Yellow arrows represent the settlement of New Guinea. The dotted orange arrows represent gene flows after the initial settlement of Sahul. The black dots represent approximate locations of archaeological sites older than 40 ka (O’Connor 2007; Summerhayes et al. 2017). The dashed black lines represent ecological lines in Wallacea. The dark gray areas represent current land area, with light gray areas representing estimated land area around 50,000 years ago based on paleogeological reconstructions (Coller 2009).
Multiple Migrations between Wallacea and Sahul
To reach Sahul, the first human settlers had to cross Wallacea, a region always composed of small islands, large water crossings, and strong ecological barriers (Wallace 1869). These restrictions on movement are reflected in the substantial genetic structuring of current populations (fig. 1 and supplementary fig. 5, Supplementary Material online). But our results also show relative genetic continuity between groups from Wallacea to New Guinea, with stronger genetic barriers between New Guinea and Australia, as well as around the Bismarck Archipelago (supplementary fig. 6, Supplementary Material online), as described in a previous report (Bergstrom et al. 2017). Wallacea was probably less a strong geographical frontier than a place of possible migrations to Sahul (Birdsell 1977; Kealy et al. 2017; Allen and O’Connell 2020). The ancestral population to New Guineans, Bismarck Archipelago Islanders, and northeast Indigenous Australians might have been composed of around 1,300 individuals based on ecological information (Bradshaw et al. 2019) and 1,671–2,100 individuals based on genetic data (supplementary fig. 16, Supplementary Material online).
Our models, like those of others (Malaspinas et al. 2016; Teixeira and Cooper 2019), show that this ancestral population split into different groups early during this migration, upon the arrival in Sahul 50 ka (model A, figs. 3B and 4), or even before in Wallacea, from which multiple migrations could have been conducted within a small time frame (i.e., in less than 900 years; Bradshaw et al. 2019) (model B, fig. 3B and 4). A model with a single migration is still valid given our genetic data but there is growing nongenetic evidence that the initial settlement of Sahul might have been made by multiple migrations (Clarkson et al. 2017; Bird et al. 2019; Bradshaw et al. 2019, 2021; Crabtree et al. 2021). This hypothesis has never been clearly explored using genomic data. Our study offers the first opportunity to test this model, and we found that our genomic data support a model with two migrations to Sahul to explain the current genetic diversity in New Guineans, Bismarck Archipelago Islanders, and Indigenous Australians (model B, fig. 3B and 4). Among the possible routes from Wallacea to Sahul, this model describes the northern route through North Maluku as a preferable path taken by the ancestors of New Guineans (fig. 2A). It is consistent with studies on paleogeological reconstructions of coastlines (fig. 4) (Kealy et al. 2017; Bird et al. 2019) as well as studies using ecological models (Bradshaw et al. 2021). We cannot propose a location for the second route given the limited number of available Indigenous Australian genomes, but it may have been different, and potentially used at different times during the Upper Pleistocene, as was recently described (i.e., possible entry though the northwest Sahul Shelf around 75 ka; Bradshaw et al. 2021). Our genomic model B also agrees with previous studies on uniparental markers that propose stark differences in genetic diversity between New Guineans and northeast Indigenous Australians as a sign of multiple migrations (Redd and Stoneking 1999; Pedro et al. 2020). The genomic model B thus brings together a consensus view that New Guineans and northeast Indigenous Australians share a common ancestor, in Sunda, (Malaspinas et al. 2016; Teixeira and Cooper 2019), therefore integrating recent perspectives on multiple migrations to Sahul (Redd and Stoneking 1999; Allen and O’Connell 2020). Future work on modern and ancient DNA in Wallacea, New Guinea and Australia will surely help to disentangle the two models presented here.
Wallacea was clearly not a major barrier to human migrations (supplementary fig. 6, Supplementary Material online) but it was also not necessarily a simple highway to Sahul (fig. 4). We hypothesize that the multitude of Wallacean islands might have driven genetic differentiation between related groups. These human groups would have progressively migrated east, in a planned effort spanning over a millennium or more (Bradshaw et al. 2019). This time was probably necessary for the populations to grow large enough before migrating further east (Bradshaw et al. 2021), leading to the emergence of genetic differentiation. Several human populations could have crossed Wallacea to land on Sahul, using different routes (Bradshaw et al. 2021).
During this very early period of the settlement of Sahul, it is likely that the demographic successes of these groups were different, as some archaeological sites might indicate (Clarkson et al. 2017). Admixture between ancestral populations, which countered the potential effects of inbreeding and drift in isolated groups, might have been an important factor for the dispersal success of certain human groups in Sahul. A previous study found relatively late divergence dates between New Guinean highlanders and Indigenous Australians (30–40 ka), as well as between Indigenous Australians groups (25 ka) (Malaspinas et al. 2016). It also found signals of postdivergence contacts between ancestral groups of New Guineans and Indigenous Australians in the north of Australia (Malaspinas et al. 2016), which agrees with the late genomic divergence dates that we obtain between southeast New Guinean lowlanders and northeast Indigenous Australians (13.5–15.3 ka, fig. 2C). These dates need to be interpreted with caution, because of technical issues raised above and by others (Malaspinas et al. 2016), but they do show that Sahul, and especially the north region, was probably a place of interactions between human groups with related, but already differentiated, genomes. These interactions probably involved few individuals given the number of region-specific mitochondrial DNA (Pedro et al. 2020) and Y chromosome lineages (Bergstrom et al. 2016) shared between New Guineans and Indigenous Australians. This led to the emergence of the unique genetic diversities passed on to the modern populations of northeast Australia, the Bismarck Archipelago, and New Guinea.
The Complex Settlement of New Guinea
New Guinea was settled rapidly after the initial migrations to Sahul as revealed by archaeological records (Summerhayes et al. 2010). This fast dispersal has made it challenging to determine the route taken to reach this territory. Our study strongly supports an entry point located in what is now the New Guinean southeast region (fig. 4). The genetic distances between current New Guinean southeast lowlanders and Wallaceans are smaller than with any other New Guineans (supplementary fig. 5, Supplementary Material online). The main genetic component in Wallaceans is also mostly present in groups from the New Guinean southeast lowlands (fig. 1C and supplementary fig. 9, Supplementary Material online). Among the different models tested, the only statistically significant scenario places New Guinean southeast lowlanders at the root of New Guinean genetic diversity, even considering secondary gene flows (fig. 3A). This remarkably corresponds to the location of the oldest known archaeological sites, in the Ivané valley, dated at 45–49 ka (Summerhayes et al. 2010). Our genetic model located the, apparently ancient, genetic background to the far east of New Guinea bringing new support for an initial migration via the ancient plain currently under the Arafura Sea (Allen and O’Connell 2020). Recent studies using ecological modeling identified the most likely route to New Guinea in the ancient plain currently under the Arafura Sea along the southeast coast of New Guinea (Bradshaw et al. 2021; Crabtree et al. 2021), converging with our genetic model. Other hypothetical routes through the New Guinean north coast, the highlands or the south foothills could have been taken by other groups of settlers (Summerhayes et al. 2017) but their demographic success, based on genetic data, must have been limited. During the Upper Pleistocene, the Arafura Plain south of modern New Guinea was dominated by savannah and lakes (Summerhayes et al. 2017). It would have been a favorable ecosystem for human dispersal from the west coast to the east coast of Sahul, before reaching what is now the southeast coast of the island of New Guinea (fig. 4).
Settlement within New Guinea was initiated from the lowlands. We found that the genetic diversity of groups from the New Guinean north and south lowlands derived separately from groups in the southeast region (figs. 1D and 3A). During the first stages of the settlement of New Guinea, the lowlands were probably the most hospitable places due to their access to littoral resources (Summerhayes et al. 2017). However, the dry conditions and steep geography might have created locations unfavorable to permanent human settlements, potentially numerous in the highlands and the north lowlands (Terrell 2004). Around 35 ka, these adverse conditions appeared to have significantly reduced human mobility in New Guinea (Allen and O’Connell 2020). This would explain the relative slow population growth we observe at this period for all New Guinean groups, in comparison to European or African groups (supplementary fig. 16, Supplementary Material online). A similar explanation was put forward to interpret the late coalescence dates of many New Guinean maternal lineages, postdating 30 ka (Pedro et al. 2020). This lack of resources could have induced New Guinean populations to explore other locations, such as the highlands (Summerhayes et al. 2017).
Our analyses show that the New Guinean highlands were settled by two separate migratory waves: one migration from the east foothills to the east highlands, and another migration from the south lowlands to the west highlands (figs. 1D, 3A, and 4). These two migrations could have used two major geological corridors defined by the Markham and Ramu Rivers in the east, and the Fly River in the south (Summerhayes et al. 2017). Although we do not find support for the Sepik River as a major migratory route for the initial settlement of the highlands (Bergstrom et al. 2017), all these natural corridors were likely a major path for secondary gene flows between New Guinean highlanders and New Guinean lowlanders, as indicated by our analyses (fig. 3A). Many gene flows appear to have occurred during the early period of the settlement of the highlands, around 25.4–30.2 ka, suggesting continuous contacts between highlanders and lowlanders (fig. 3A and supplementary table 3, Supplementary Material online). This is consistent with archaeological data arguing for long-term, but not permanent, settlements in the highlands around 25 ka (Summerhayes et al. 2017). New Guinean populations probably lived in semipermanent settlements, gathering resources in the highlands and the foothills, but keeping contacts with lowlander groups. These populations would have progressively fixed their settlement in the highlands, reducing contacts with the lowlanders, since no gene flow was detected between 20 and 3 ka, and increasing the genetic differentiation between highlanders and lowlanders (figs. 1 and 3; supplementary fig. 5, Supplementary Material online), as previously reported (Bergstrom et al. 2017).
However, an intense period of population dynamics occurred locally in the New Guinean highlands around 10 ka. We detected several admixture events between west highlander groups at this time (supplementary table 3, Supplementary Material online). This period was marked by the development of agricultural practices and the emergence of a Neolithic culture, especially in the Waghi valley of the west highlands (Denham et al. 2003; Shaw et al. 2020). As previously reported (Bergstrom et al. 2017), this was a major cultural change that likely influenced gene flows in the highlands and the dispersal of cultural traits, such as the Trans-New Guinean linguistic family (Reesink et al. 2009). Lowlanders might not have been significantly influenced by the development of a Neolithic culture in the highlands (Pawley 2005), but their population size was not necessarily much lower than those found in the highlands (supplementary fig. 16, Supplementary Material online) thanks to exchanges of goods and genes between the two regions (supplementary table 3, Supplementary Material online). A second wave of gene flows between highlanders and lowlanders is only detected around 2.6–4.1 ka (supplementary table 3, Supplementary Material online). At the same period, gene flow is also detected between New Guinean lowlanders and Wallaceans (supplementary table 3, Supplementary Material online). These events likely correspond to the influence of the Austronesian culture 3 ka (Lipson et al. 2014), but rather than initiating new contacts between New Guinean groups, the arrival of Austronesians seems to have only reignited New Guinean population dynamics that already existed around 25 ka (supplementary table 3, Supplementary Material online). Genes, material culture, and ideas were already moving within New Guinea (Pawley 2005; Torrence and Swadling 2008). The last stage of the settlement of New Guinea was probably another pulse in a network of population interactions built up over millennia.
New Guinean genomes reveal possible scenarios describing the migrations from Sunda to Sahul and a settlement of New Guinea initiated in the southeast lowlands, followed by two separate dispersals to the highlands (fig. 4). Our models will necessarily be confronted by future genetic and archaeological data, particularly in the western half of New Guinea Island and Australia. Paleogenomics in Oceania is an emerging field and it has already proven to be highly informative about the history of populations with New Guinean genetic ancestry (Skoglund et al. 2016; Choin et al. 2021). Future research will certainly bring key information on the complex population dynamics that existed during the past 50,000 years in north Sahul.
Materials and Methods
Sampling
Fifty-eight individuals from Papua New Guinea participated in the study (supplementary fig. 1 and table 1, Supplementary Material online). These new samples were obtained in Papua New Guinea between 2016 and 2019. The study was approved by the Medical Research Advisory Committee of Papua New Guinea (National Department of Health) under research ethics clearance MRAC 16.21 and by the French Ethics Committees (Committees of Protection of Persons IE-2015-837 (1)). Permission to conduct research in Papua New Guinea was granted by the National Research Institute of Papua New Guinea (permit 99902292358), with full support from the School of Humanities and Social Sciences, University of Papua New Guinea. All samples were collected from healthy unrelated adult donors, all of whom provided written informed consent (supplementary text, Supplementary Material online). DNA was extracted from saliva samples with the Oragene sampling kit according to the manufacturer’s instructions. After a full presentation of the project to a wide audience, a discussion with each individual willing to participate ensured that the project was fully understood. Once participants had signed informed-consent forms, we interviewed them to obtain information on their date and place of birth, their spoken language(s), and similar data related to their genealogy (up to second or third generation) to establish local ancestry. Based on the participants’ genealogy, we chose the samples to be sequenced so as to cover most of Papua New Guinean genomic diversity.
Data Sets
Sequencing libraries were prepared using the TruSeq DNA PCR-Free HT kit. About 150-bp paired-end sequencing was performed on the Illumina HiSeq X5 sequencer. All raw data are available on the European Genome-Phenome data repository (EGA; https://www.ebi.ac.uk/ega/): EGAS00001005393. The 58 new Papuan genomes were combined with raw reads from the Simons Genome Diversity Project (SGDP, n = 292, EBI European Nucleotide Archive: PRJEB9586 and ERP010710) (Mallick et al. 2016); 60 Papuan genomes (EGA: EGAS00001001247 dbGAP: phs001085.v1.p1) (Malaspinas et al. 2016; Vernot et al. 2016); and 161 genomes from Indonesia (EGA: EGAS00001003054) (Jacobs et al. 2019). SNP calling was performed on the combined data set. Reads were aligned to the “decoy” version of the GRCh37 human reference sequence (hs37d5) using BWA MEM (Li 2014). Duplicated reads were removed with picard-tools v.2.12.0 (http://broadinstitute.github.io/picard) and local realignment around indels were performed with GATK v. 3.5 (Poplin et al. 2018). After alignment, the average sequencing depth across the samples was 36×, with a minimum of 31× coverage. Base-calling and per-sample gVCF files were generated using GATK HaplotypeCaller (reads mapping quality ≥20) (Poplin et al. 2018). Multisample VCF files were obtained with CombineGVCFs and GenotypeGVCFs. Genotype calls were subsequently filtered for base depth (≥8× and ≤400×) and genotyping quality (≥30) using BCFtools v. 1.4 (Li 2014). We removed sites within segmental duplications, repeats, and low complexity regions based on available masks (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/genomicSuperDups.txt.gz; http://software.broadinstitute.org/software/genomestrip/node_ReferenceMetadata.html). Only biallelic sites with high-quality variant calls in at least 99% of samples were retained, leading to a whole-genome data set of 571 individuals typed for 41,250,479 SNPs (supplementary table 2, Supplementary Material online). All whole genomes were phased using SHAPEIT v. 2.r79033 (Delaneau et al. 2011) with default settings, except for iterations (50) and states (200) and used the HapMap phase 2 genetic map (International HapMap Consortium et al. 2007) without a reference panel.
In addition to the whole-genome data set, we compiled another data set including genotyping data from Papua New Guinea (Bergstrom et al. 2017). These genotyping data were phased and imputed using all phased New Guinean whole genomes as a reference panel (N = 166) with IMPUTE2 v. 2.3.2 (Howie et al. 2009). Ten Papuan WGS randomly selected were downgraded to the same set of SNPs as the genotyping data to serve as an internal control. All SNPs with an imputation score above 0.4 were kept, leaving 12,168,294 SNPs. The ten downgraded genomes are over 98% consistent with actual genotypes from whole genomes for the same individuals. We also added genotyping data from North Maluku (Kusuma et al. 2017), but we did not impute missing SNPs since no whole genomes are available for this group. Finally, we included data from Neandertal (Prufer et al. 2014), Denisovan (Reich et al. 2010), and Chimpanzee (Mikkelsen et al. 2005). The final data set of 966 genomes was phased with SHAPEIT v. 2.r79033 (Delaneau et al. 2011) without a reference panel with the same settings as above.
To focus on Upper Pleistocene genetic history, recent Asian ancestry was masked in admixed individuals from New Guinea, Wallacea, northeast Australia, and the Bismarck Archipelago using PCAdmix v.1.0 (Brisbin et al. 2012). Since both available Indigenous Australian genomes showed signs of recent European admixture (Mallick et al. 2016), we also masked European genetic ancestry for all individuals. Two parental metapopulations were defined by randomly choosing 100 Asian-European individuals and 100 New Guinean individuals. For the latter group, only unadmixed New Guinean WGSs were selected, as defined by ADMIXTURE v. 1.3 (Alexander et al. 2009). Local ancestry was estimated for all individuals from New Guinea, Wallacea, northeast Australia, and the Bismarck Archipelago showing non-Eurasian genetic ancestry above 20%. Posterior probabilities for New Guinean ancestry above 0.9 and Asian ancestry below 0.1 were criteria to define SNPs with New Guinean ancestry. We applied quality controls using Plink v. 1.932 (Chang et al. 2015) to filter for and exclude: 1) close relatives by using an IBD estimation with an upper threshold of 0.25 (second degree relatives); 2) SNPs that failed the Hardy–Weinberg exact test (P < 10−6) within each group; 3) samples with a call rate <0.99 and displaying missing rates > 0.05 across all samples in each population. For analyses requiring linkage disequilibrium pruning, we used Plink to filter for variants in high linkage disequilibrium (r2 > 0.2), leaving 74,067 SNPs. Some Wallacean groups were pooled together because of the high rate of masked SNPs, according to the fineSTRUCTURE analysis and geography, in order to perform population-based analyses such as TREEMIX, f3 (ex: Wallacea_3-4_6-7 refers to the pool of data from the subgroups Wallacea_1, Wallacea_2, Wallacea_6, Wallacea_7).
Analyses
Principal components analyses were run with smartpca from the EIGENSOFT v.7.2.1 package (Patterson et al. 2006). Populations were genetically defined using fineSTRUCTURE v. 2.07 (Lawson et al. 2012). This package performs model-based Bayesian clustering of genotypes based on the shared IBD fragments between each pair of individuals, without self-copying, calculated with ChromoPainter v. 2.0 (Lawson et al. 2012). Five million MCMC runs were performed. A coancestry heatmap and a dendrogram were inferred to visualize the number of statistically defined clusters that describe the data (supplementary fig. 2, Supplementary Material online). About 15 genetic groups were defined, and subdivided into 106 subgroups, including 58 subgroups from Wallacea and Sahul (supplementary fig. 2 and table 2, Supplementary Material online). Genotyping data from North Maluku were defined as an additional subgroup (Wallacea_11) since the SNP density is much lower than whole-genome sequencing data and no imputation could be performed.
Genetic barriers and corridors were defined using EEMS v. 1 (Petkova et al. 2016). We used approximate geographic coordinates, a genetic dissimilarity matrix between populations and a map of New Guinea and the surrounding areas defining a grid of 700 modeled demes. Depending on their location, several subgroups can be included in one deme, whose size increased accordingly. We ran 5 million MCMC iterations before checking for convergence of the MCMC chain (supplementary fig. 6, Supplementary Material online). Plots were generated in R v. 3.4.3 according to instructions in the EEMS manual. FST genetic distances were obtained with the EIGENSOFT v.7.2.1 package (Patterson et al. 2006).
Genetic ancestries present in the data set were estimated by ADMIXTURE v. 1.3 (Alexander et al. 2009), with default settings, for components K = 2 to K = 15. Ten iterations with randomized seeds were run and compiled with CLUMPAK v. 1 (Kopelman et al. 2015). We used the minimum average cross-validation value to define the most informative K components (here, K = 8), and the major modes defined by CLUMPAK v. 1 were reported. Geographical distributions of the main genetic components found in Wallacea and Sahul were obtained using inverse distance weighting interpolation between data points with QGIS v. 3.16.1 (GDAL-SOFTWARE-SUITE 2013 ; QGIS).
f3-Statistics and f3-outgroup were computed with qp3pop from the ADMIXTOOLS v.6.0 package (Patterson et al. 2012) between all trios of populations. f4-statistics was performed with qp4pop from the ADMIXTOOLS v.6.0 package (Patterson et al. 2012). Admixture dates were estimated with MALDER v. 1.0 for all negatively significant scenarios from the f3-statistics analysis (Loh et al. 2013). All dates mentioned in the manuscript are converted into years based on 30 years per generation. TREEMIX v. 1.12 analysis (Pickrell and Pritchard 2012) was performed by setting blocks to 500 SNPs, accounting for linkage disequilibrium, and migration edges were added sequentially until the model explained 99% of the variance, rooting the tree with Chimpanzee (panTro5).
Scenarios of the settlement of Sahul were tested for the full data set using qpgraph from the ADMIXTOOLS v.6.0 package (Patterson et al. 2012). One subgroup was chosen as representative population for a given regional cluster (as defined in supplementary table 2, Supplementary Material online), based on the number of individuals, the number of SNPs after masking for Asian SNPs and their location. Populations were added one-by-one, testing different scenarios with or without admixture events, following recommendations in (Lipson and Reich 2017). Models with a |Z score|<3 were defined as fitting the data. Over one hundred different models were tested. Only models fitting the data and scenarios based on major hypotheses were represented. Results were visualized with GraphViz v. 2.46.0 (GraphViz; Available from: http://graphviz.org). Population effective sizes and population divergence dates were estimated with MSMC2 v. 2.1.2 (Schiffels and Durbin 2014). Two individuals from each population were randomly chosen. Sample-specific masks were generated from the corresponding BAM files using bamCaller.py from MSMC-Tools packages (Schiffels and Wang 2020). Individual masks for Asian admixture were generated from previously mentioned analyses with PCAdmix v.1.0 (Brisbin et al. 2012). For all analyzed individual, the mask of Asian genetic tracks ranged between 0% and 49% of the total genome. The individual archaic masks were generated with ChromoPainter v.2.0 (Lawson et al. 2012). Two metapopulations were defined: one metapopulation from 100 randomly selected Sub-Saharan Africans; and one metapopulation including Neandertal, Denisovan, and Chimpanzee genomes. Only stretches of more than four SNPs “painted” by the Sub-Saharan metapopulations were kept. A fourth individual mask was added based on human genome mappability hs37d5 (Schiffels and Durbin 2014). MSMC2 was run with a pattern of fixed time segments (1*5 + 40*1 + 5*3 + 3*5) and 25 iterations. Ambiguously phased SNPs were skipped using the –s flag. We used 1.25e-8 as the mutation rate.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Georgia Kaipu (National Research Institute, Papua New Guinea), Christopher Kinipi (University of Papua New Guinea Clinic), Alois Kuaso, and Kenneth Miamba (National Museum and Art Gallery, Papua New Guinea) for their help during the sampling campaigns. We thank Chris Clarkson for his comments on the manuscript. We thank J. Stephen Lansing (Santa Fe Institute) for financially supporting some of the sequencing. We thank Monika Karmin for her help on the analysis of Y chromosome data. We thank Opale Coutant (University of Toulouse) and Adeline Morez (Liverpool University) for their help in data analysis. We acknowledge support from the GenoToul bioinformatics facility of Genopole Toulouse Midi-Pyrénées, France; the LabEx TULIP, France; and the Centre National de Recherche en Génomique Humaine (CNRGH), Evry, France. We especially thank all of our study participants. We acknowledge support from the National Geographic Society (Grant No. HJ-156R-17) and the Leakey Foundation. This research was supported by the European Union through Horizon 2020 research and innovation program under Grant Number 810645 and through the European Regional Development Fund Project Number MOBEC008. This work was supported by the French Ministry of Research grant Agence Nationale de la Recherche (ANR PAPUAEVOL), the French Ministry of Foreign and European Affairs (French Prehistoric Mission in Papua New Guinea), and the French Embassy in Papua New Guinea, as well as a fellowship from the Alexander von Humboldt Foundation.
Author Contributions
N.B., W.P., J.M., M.L., and F.X.R. organized the sampling campaigns. N.B., R.T., J.K., K.S., T.B., M.L., and F.X.R. collected biological samples. C.B., A.B., and J.F.D. generated whole-genome sequences. L.S. and N.B. processed raw whole-genome sequences. N.B., M.M., M.P.C., M.L., and F.X.R. designed the study. N.B. performed the analyses and wrote the paper based on the input from all the other authors. All authors participated in the interpretation of the results and approved the submission.
Data Availability
All raw data are available on the European Genome-Phenome data repository: EGAS00001005393.
References
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J, O’Connell JF.. 2020. A different paradigm for the initial colonisation of Sahul. Archaeol Ocean. 55(1):1–14. [Google Scholar]
- Bellwood P. 2007. Prehistory of the Indo-Malaysian Archipelago. Canberra (Australia: ): ANU E Press. [Google Scholar]
- Bergstrom A, Nagle N, Chen Y, McCarthy S, Pollard MO, Ayub Q, Wilcox S, Wilcox L, van Oorschot RA, McAllister P, et al. 2016. Deep roots for aboriginal Australian Y chromosomes. Curr Biol. 26(6):809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom A, Oppenheimer SJ, Mentzer AJ, Auckland K, Robson K, Attenborough R, Alpers MP, Koki G, Pomat W, Siba P, et al. 2017. A Neolithic expansion, but strong genetic structure, in the independent history of New Guinea. Science 357(6356):1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird MI, Condie SA, O’Connor S, O’Grady D, Reepmeyer C, Ulm S, Zega M, Saltré F, Bradshaw CJA.. 2019. Early human settlement of Sahul was not an accident. Sci Rep. 9(1):8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell JB. 1977. The recalibration of a paradigm for the first peopling of Greater Australia. In: Allen J, Golson J, Jones R, editors. Sunda and Sahul: Prehistoric Studies in Southeast Asia, Melanesia, and Australia. London: Academic Press. p. 113–167. [Google Scholar]
- Bradshaw CJA, Norman K, Ulm S, Williams AN, Clarkson C, Chadœuf J, Lin SC, Jacobs Z, Roberts RG, Bird MI, et al. 2021. Stochastic models support rapid peopling of Late Pleistocene Sahul. Nat Commun. 12(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw CJA, Ulm S, Williams AN, Bird MI, Roberts RG, Jacobs Z, Laviano F, Weyrich LS, Friedrich T, Norman K, et al. 2019. Minimum founding populations for the first peopling of Sahul. Nat Ecol Evol. 3(7):1057–1063. [DOI] [PubMed] [Google Scholar]
- Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J, Reynolds A, Ostrer H, Mezey JG, Bustamante CD.. 2012. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 84(4):343–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ.. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choin J, Mendoza-Revilla J, Arauna LR, Cuadros-Espinoza S, Cassar O, Larena M, Ko AM-S, Harmant C, Laurent R, Verdu P, et al. 2021. Genomic insights into population history and biological adaptation in Oceania. Nature 592(7855):583–587. [DOI] [PubMed] [Google Scholar]
- Clarkson C, Jacobs Z, Marwick B, Fullagar R, Wallis L, Smith M, Roberts RG, Hayes E, Lowe K, Carah X, et al. 2017. Human occupation of northern Australia by 65,000 years ago. Nature 547(7663):306–310. [DOI] [PubMed] [Google Scholar]
- Coller M. 2009. SahulTime: rethinking archaeological representation in the digital age. Archaeologies 5(1):110–123. [Google Scholar]
- Crabtree SA, White DA, Bradshaw CJ, Saltré F, Williams AN, Beaman RJ, Bird MI, Ulm S.. 2021. Landscape rules predict optimal superhighways for the first peopling of Sahul. Nat Hum Behav. 1–11. doi: 10.1038/s41562-021-01106-8. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF.. 2011. A linear complexity phasing method for thousands of genomes. Nat Methods. 9(2):179–181. [DOI] [PubMed] [Google Scholar]
- Denham TP, Haberle SG, Lentfer C, Fullagar R, Field J, Therin M, Porch N, Winsborough B.. 2003. Origins of agriculture at Kuk Swamp in the highlands of New Guinea. Science 301(5630):189–193. [DOI] [PubMed] [Google Scholar]
- Golson J, Denham T, Hughes P, Swadling P, Muke J.. 2017. Ten thousand years of cultivation at Kuk Swamp in the highlands of Papua New Guinea. Terra Australis 46: ANU Press.
- Greenhill SJ. 2015. TransNewGuinea.org: an online database of New Guinea languages. PLoS One 10(10):e0141563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J.. 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudjashov G, Karafet TM, Lawson DJ, Downey S, Savina O, Sudoyo H, Lansing JS, Hammer MF, Cox MP.. 2017. Complex patterns of admixture across the Indonesian archipelago. Mol Biol Evol. 34(10):2439–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, et al. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449(7164):851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GS, Hudjashov G, Saag L, Kusuma P, Darusallam CC, Lawson DJ, Mondal M, Pagani L, Ricaut FX, Stoneking M, Metspalu M, Sudoyo H, et al. 2019. Multiple deeply divergent Denisovan ancestries in Papuans. Cell 177(4):1010–1021.e1032. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Hallmark B, Cox MP, Sudoyo H, Downey S, Lansing JS, Hammer MF.. 2010. Major east–west division underlies Y chromosome stratification across Indonesia. Mol Biol Evol. 27(8):1833–1844. [DOI] [PubMed] [Google Scholar]
- Kealy S, Louys J, O’Connor S.. 2017. Reconstructing palaeogeography and inter-island visibility in the Wallacean archipelago during the likely period of Sahul colonization, 65-45000 years ago. Archaeol Prospect. 24(3):259–272. [Google Scholar]
- Kealy S, Louys J, O’Connor S.. 2018. Least-cost pathway models indicate northern human dispersal from Sunda to Sahul. J Hum Evol. 125:59–70. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I.. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 15(5):1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuma P, Brucato N, Cox MP, Letellier T, Manan A, Nuraini C, Grangé P, Sudoyo H, Ricaut F-X.. 2017. The last sea nomads of the Indonesian archipelago: genomic origins and dispersal. Eur J Hum Genet. 25(8):1004–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, Falush D.. 2012. Population identification using genetic data. Annu Rev Genomics Hum Genet. 13:337–361. [DOI] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D.. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8(1):e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2014. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30(20):2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M, Loh P-R, Patterson N, Moorjani P, Ko Y-C, Stoneking M, Berger B, Reich D.. 2014. Reconstructing Austronesian population history in Island Southeast Asia. Nat Commun. 5(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M, Reich D.. 2017. A working model of the deep relationships of diverse modern human genetic lineages outside of Africa. Mol Biol Evol. 34(4):889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, Berger B.. 2013. Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193(4):1233–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas AS, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergstrom A, Athanasiadis G, Cheng JY, Crawford JE, Heupink TH, et al. 2016. A genomic history of Aboriginal Australia. Nature 538(7624):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538(7624):201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T, Hillier L, Eichler E, Zody M, Jaffe D, Yang S-P, Enard W, Hellmann I, Lindblad-Toh K, Altheide T.. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Mona S, Tommaseo-Ponzetta M, Brauer S, Sudoyo H, Marzuki S, Kayser M.. 2007. Patterns of Y-chromosome diversity intersect with the Trans-New Guinea hypothesis. Mol Biol Evol. 24(11):2546–2555. [DOI] [PubMed] [Google Scholar]
- Nagle N, Ballantyne KN, van Oven M, Tyler-Smith C, Xue Y, Taylor D, Wilcox S, Wilcox L, Turkalov R, van Oorschot RAH, et al. 2016. Antiquity and diversity of aboriginal Australian Y‐chromosomes. Am J Phys Anthropol. 159(3):367–381. [DOI] [PubMed] [Google Scholar]
- Norman K, Inglis J, Clarkson C, Faith JT, Shulmeister J, Harris D.. 2018. An early colonisation pathway into northwest Australia 70-60,000 years ago. Quat Sci Rev. 180:229–239. [Google Scholar]
- O’Connell JF, Allen J, Williams MAJ, Williams AN, Turney CSM, Spooner NA, Kamminga J, Brown G, Cooper A.. 2018. When did Homo sapiens first reach Southeast Asia and Sahul? Proc Natl Acad Sci U S A. 115(34):8482–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor S. 2007. New evidence from East Timor contributes to our understanding of earliest modern human colonisation east of the Sunda Shelf. Antiquity 81(313):523–535. [Google Scholar]
- Pagani L, Lawson DJ, Jagoda E, Morseburg A, Eriksson A, Mitt M, Clemente F, Hudjashov G, DeGiorgio M, Saag L, Wall JD, et al. 2016. Genomic analyses inform on migration events during the peopling of Eurasia. Nature 538(7624):238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2(12):e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson NJ, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawley A. 2005. Papuan pasts: cultural, linguistic and biological histories of Papuan-speaking peoples. Canberra: Pacific Linguistics, Research School of Pacific and Asian Studies, Australian National University. [Google Scholar]
- Pedro N, Brucato N, Fernandes V, Andre M, Saag L, Pomat W, Besse C, Boland A, Deleuze JF, Clarkson C, et al. 2020. Papuan mitochondrial genomes and the settlement of Sahul. J Hum Genet. 65(10):875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova D, Novembre J, Stephens M.. 2016. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 48(1):94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. 2018. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv: 201178. doi:10.1101/201178.
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AJ, Stoneking M.. 1999. Peopling of Sahul: mtDNA variation in aboriginal Australian and Papua New Guinean populations. Am J Hum Genet. 65(3):808–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reesink G, Singer R, Dunn M.. 2009. Explaining the linguistic diversity of Sahul using population models. PLoS Biol. 7(11):e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468(7327):1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Kircher M, Delfin F, Nandineni MR, Pugach I, Ko AMS, Ko YC, Jinam TA, Phipps ME, et al. 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 89(4):516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Patterson N, Reich D.. 2016. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr Biol. 26(9):1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R.. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46(8):919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Wang K.. 2020. MSMC and MSMC2: the multiple sequentially markovian coalescent. In: Dutheil JY, editor. Statistical population genomics. New York: Humana Press. p. 147–166. [DOI] [PubMed] [Google Scholar]
- Shaw B, Field JH, Summerhayes GR, Coxe S, Coster ACF, Ford A, Haro J, Arifeae H, Hull E, Jacobsen G, et al. 2020. Emergence of a Neolithic in highland New Guinea by 5000 to 4000 years ago. Sci Adv. 6(13):eaay4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Posth C, Sirak K, Spriggs M, Valentin F, Bedford S, Clark GR, Reepmeyer C, Petchey F, Fernandes D, et al. 2016. Genomic insights into the peopling of the Southwest Pacific. Nature 538(7626):510–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerhayes GR, Field JH, Shaw B, Gaffney D.. 2017. The archaeology of forest exploitation and change in the tropics during the Pleistocene: the case of Northern Sahul (Pleistocene New Guinea). Quat Int. 448:14–30. [Google Scholar]
- Summerhayes GR, Leavesley M, Fairbairn A, Mandui H, Field J, Ford A, Fullagar R.. 2010. Human adaptation and plant use in highland New Guinea 49,000 to 44,000 years ago. Science 330(6000):78–81. [DOI] [PubMed] [Google Scholar]
- Teixeira JC, Cooper A.. 2019. Using hominin introgression to trace modern human dispersals. Proc Natl Acad Sci U S A. 116(31):15327–15332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell JE. 2004. The ‘sleeping giant’ hypothesis and New Guinea’s place in the prehistory of Greater Near Oceania. World Archaeol. 36(4):601–609. [Google Scholar]
- Tobler R, Rohrlach A, Soubrier J, Bover P, Llamas B, Tuke J, Bean N, Abdullah-Highfold A, Agius S, O’Donoghue A, et al. 2017. Aboriginal mitogenomes reveal 50,000 years of regionalism in Australia. Nature 544(7649):180–184. [DOI] [PubMed] [Google Scholar]
- Torrence R, Swadling P.. 2008. Social networks and the spread of Lapita. Antiquity 82(317):600–616. [Google Scholar]
- Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, Scheinfeldt LB, et al. 2016. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352(6282):235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AR. 1869. The Malay Archipelago. Vol. II. Oxford: Oxford University.
- Westaway KE, Louys J, Awe RD, Morwood MJ, Price GJ, Zhao JX, Aubert M, Joannes-Boyau R, Smith TM, Skinner MM, et al. 2017. An early modern human presence in Sumatra 73,000-63,000 years ago. Nature 548(7667):322–325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available on the European Genome-Phenome data repository: EGAS00001005393.