Abstract
In addition to its roles in translocation of preproteins across membranes, Ydj1 facilitates the maturation of Hsp90 substrates, including mammalian steroid receptors, which activate transcription in yeast in a hormone-dependent manner. To better understand Ydj1's function, we have constructed and analyzed an array of Ydj1 mutants in vivo. Both the glucocorticoid receptor and the estrogen receptor exhibited elevated activity in the absence of hormone in all ydj1 mutant strains, indicating a strict requirement for Ydj1 activity in hormonal control. Glucocorticoid receptor containing a mutation in the carboxy-terminal transcriptional activation domain, AF-2, retained elevated basal activity, while mutation of the amino-terminal transactivation domain, AF-1, eliminated the elevated basal activity observed in ydj1 mutant strains. This result indicates that the source of activity is AF-1, which is normally repressed by the carboxy-terminal hormone binding domain in the absence of hormone. Chimeric proteins containing the hormone binding domain of the estrogen or glucocorticoid receptor fused to heterologous activation and DNA binding domains also exhibited elevated activity in the absence of hormone. Thus, Ydj1 mutants appear to increase basal receptor activity by altering the ability of the hormone binding domain of the receptor to repress nearby activation domains. We propose that Ydj1 functions to present steroid receptors to the Hsp90 pathway for folding and hormonal control. In the presence of Ydj1 mutants that fail to bind substrate efficiently, some receptor escapes the Hsp90 pathway, resulting in constitutive activity.
Hsp90, a highly conserved, abundant, and essential cytosolic molecular chaperone, functions in the maturation of a limited set of substrate proteins. Hsp90 was originally found to be involved in the activation of proteins involved in signaling pathways such as steroid hormone receptors and the oncogenic tyrosine kinase v-Src. Recent studies have revealed interaction with the hepatitis B retrovirus, nitric oxide synthase, telomerase, and the mutant form of p53 (7, 23), suggesting that Hsp90 may be involved in the maturation of many diverse substrates.
Hsp90 functions with a number of cochaperones in an apparently dynamic but ordered pathway leading to substrate maturation (48). Much of what is known about the interaction of Hsp90 and cochaperones with substrate proteins comes from studies of steroid hormone receptors (45). Besides promoting receptor activity, Hsp90 and cochaperones prevent steroid receptor activity in the absence of hormone, as receptors bound to Hsp90 cannot bind DNA and are therefore inactive as transcription factors. In vitro studies in reticulocyte lysates have led to the identification of proteins required for steroid receptor activation. Purified receptor is unable to bind hormone, but incubation in reticulocyte lysate results in a form of the receptor bound to Hsp90, p23, and immunophilins of the cyclophilin and FK506-binding protein classes (45). This mature form is capable of binding hormone, at which point chaperones are released, forming transcriptionally active receptor. Other components of the reticulocyte lysate that interact with the receptor in this pathway include Hsp70, Hop, and a member of the Hsp40 family (45). While the specific function of each of these proteins in complex assembly is unclear, Hsp70 and Hsp40 appear to function very early in the pathway.
Steroid receptors have three domains: an amino-terminal activation domain (AF-1), a DNA binding domain, and a carboxy-terminal hormone binding domain (HBD). The HBD contains the hormone binding site, the Hsp90 binding site, and a second activation domain, AF-2. Hormone binding triggers a series of events, including release of Hsp90 and cochaperones, DNA binding, and activation of AF-1 and AF-2. Mutational analysis of the estrogen receptor (ER) and the glucocorticoid receptor (GR) revealed that the two activation domains AF-1 and AF-2 are regulated separately (21, 37). AF-2 is strictly hormone dependent, as hormone binding is necessary to cause the conformation change that exposes the AF-2 site. However, truncation mutants of the ER and the GR that contain the amino-terminal AF-1 but lack the HBD are constitutively active (20, 29, 51). This observation led to the concept that the HBD suppresses AF-1 activity in the absence of hormone. In support of this role, fusion of the HBD of either the ER or the GR to heterologous DNA binding and activation domains conferred both hormonal control and Hsp90 binding (44, 46). The mechanism by which the HBD suppresses AF-1 activity is unknown but is likely a consequence of Hsp90 binding to the HBD.
In vivo studies of Saccharomyces cerevisiae have recapitulated the requirement for Hsp90 and cochaperones in steroid receptor maturation (42). While S. cerevisiae does not contain endogenous steroid receptors, steroid receptors expressed in yeast bind homologs of the proteins mentioned above—Hsp82 (Hsp90), Ssa (Hsp70), Sti1 (Hop), Sba1 (p23), Ydj1 (Hsp40), and Cpr6/7 (Cyp-40)—and activate transcription in a hormone-dependent manner (17, 27). Mutations in the yeast genes encoding Hsc82/Hsp82, Sti1, Ydj1, and Sba1 affect the ability of steroid hormone receptors expressed in yeast to activate reporter genes in response to hormone (4, 17, 42). The maturation of v-Src in S. cerevisiae is also affected by mutations in the Hsp90 pathway. v-Src is toxic to wild-type yeast, due to the aberrant tyrosine phosphorylation of yeast proteins. Expression of v-Src in yeast strains containing mutations in hsp82, ydj1, sti1, or sba1 results in decreased lethality and/or decreased phosphotyrosine activity relative to wild-type yeast (17, 27). Therefore, S. cerevisiae is a useful model for studying the interactions of Hsp90 and cochaperones with substrate proteins.
The focus of this report is the role of Ydj1 in Hsp90 pathways. Ydj1 is a member of the Hsp40 class of molecular chaperones, an important class of chaperones found in virtually every organism and cellular compartment (12, 28). Hsp40s regulate the function of partner Hsp70s by stimulating the ATPase activity of Hsp70. In some cases, Hsp40s also direct partner Hsp70s to interact with substrate polypeptides. Ydj1 is known to stimulate the ATP hydrolysis of the cytosolic Hsp70 Ssa and is capable of binding some denatured substrates (14, 35). Ydj1 is not essential in S. cerevisiae, but disruption of YDJ1 results in slow-growing cells that grow poorly at 30°C and very poorly in liquid media at all temperatures (1, 9). Ydj1 is a class 1 DnaJ homolog (12), sharing the general domain structure of Escherichia coli DnaJ, in which the amino-terminal J domain is followed by a G/F-rich region (constituting the J + G/F region), a zinc finger region, and a less conserved carboxy-terminal domain. The amino-terminal J + G/F region is sufficient to stimulate the ATPase activity of Ssa, while the substrate binding region has been localized to the zinc finger region and/or the carboxy-terminal region (35, 49). Ydj1 also possesses a CAAX box, which specifies the posttranslational addition of a farnesyl group to the carboxy terminus of Ydj1, a modification that facilitates attachment of Ydj1 to endoplasmic reticulum membranes (11).
Previous reports have shown differing roles for Ydj1 in Hsp90 pathways. Supporting a role in activating steroid receptors, Caplan et al. (10) found that the ydj1-151 mutation decreased the ability of the androgen receptor to activate a reporter gene in the presence of hormone. However, Kimura et al. (29) found that the ydj1-G315D mutation greatly increased the activity of the GR and ER in the absence of hormone, suggesting Ydj1 has a role in repressing receptor activity in the absence of hormone. As different Ydj1 mutants and different receptors were used in these studies, the generality of these results and the role of Ydj1 is unresolved. To clarify the role of Ydj1 in Hsp90 pathways in vivo, we have used multiple Ydj1 mutants to examine the role of Ydj1 in the maturation of three different Hsp90 substrates, v-Src, the GR, and the ER.
MATERIALS AND METHODS
Generation of Ydj1 mutants.
With the exception of C406S, which was a gift of Avrom Caplan, Ydj1 mutants were selected from a random PCR-mutagenized library of YDJ1 in one of two genetic screens. The initial screen, which yielded mutants N104, N134, N172, N206, and G315E, was designed to isolate Ydj1 mutants that were capable of growth in the presence of v-Src at 30°C. The majority of mutants isolated in this screen were truncation mutants. Ydj1 mutants F47L, G153R, N274, and N363 were isolated in a screen for temperature-sensitive Ydj1 mutants that produced proteins of 30 kDa or larger.
Construction of ydj1 library.
The 3.6-kb SacI-MunI fragment of YDJ1 was cloned into the SacI-EcoRI sites of the centromeric plasmid pRS316 to create pRS316-YDJ1. A library of PCR-generated ydj1 mutants was generated with Taq polymerase under standard PCR conditions using the primers Ydj1-forward (5′-CTTTTGATAGAACATAA-3′) and Ydj1-reverse (5′-TTGGTACCATTATTACTTTAC-3′). The Ydj1-reverse primer introduces a KpnI site downstream of the stop codon. The PCR products were digested with EcoRI-KpnI and cloned into the same sites of pRS316-YDJ1. The mutagenized 1.4-kb EcoRI-KpnI fragment encompasses 40 bp upstream of the ATG through 100 bp downstream of the stop codon.
(i) Screen 1.
The mutagenized URA3-marked YDJ1 library was transformed into strain JJ160 (ydj1::HIS3) expressing a LEU2-marked multicopy plasmid expressing v-Src under the control of a GAL1 promoter (pLv-src; a gift from F. Boschelli [5]). Transformants were plated onto leucine-uracil-selective medium plates with 2% galactose as the carbon source and grown at 30°C for 5 days. Colonies that appeared were restreaked on selective galactose medium to verify the phenotype. Cell lysates from these strains were then checked for expression of Ydj1, using an antibody raised against full-length Ydj1. Only candidate mutant strains expressing near-wild-type levels of Ydj1 were chosen for further study.
(ii) Screen 2.
To obtain mutants F47L, G153R, N274, and N363, the ydj1 library was subcloned into the LEU2-marked pRS315 vector. Library DNA was transformed into strain JJ160 expressing pRS316-YDJ1. After 2 days, transformants were streaked onto plates containing 5′-fluoro-orotic acid to counterselect for the presence of wild-type YDJ1. Cell lysates from strains exhibiting temperature-sensitive growth were tested for the presence of Ydj1 by immunoblot analysis. Mutant strains producing proteins that migrated between 30 and 44 kDa were selected for further study.
ydj1 mutants were subcloned using internal sites to localize the mutagenized regions and then sequenced. All of the ydj1 mutations are due to a single amino acid mutation. N104, N134, N172, N274, and N363 arise from mutation of a codon to a stop codon. N206 arose from a frameshift mutation that introduced nine amino acids prior to a stop codon. The truncation mutants produce single bands of the expected size, with no full-length protein expressed. All mutants were expressed at similar levels by Western blot analysis (data not shown). Ydj1 mutants were expressed from the LEU2-marked pRS315 vector and/or the LYS2-marked pRS317 vector (47). In most cases, yeast expressing a particular ydj1 mutant displayed uniform colony size. However, slight growth differences between colonies expressing F47L and truncations N104-N206 may be due to variations in plasmid copy number.
v-Src expression.
ydj1 disruption strain JJ160 expressing various Ydj1 mutants was transformed with a 2μm plasmid expressing GAL1–v-src (pBv-src) or the control plasmid (pB656) (5). Yeast cultures were grown overnight at 30°C in raffinose-uracil dropout medium until mid-log phase; 20% galactose was added to a final concentration of 2%. After 6 h, cultures were harvested for growth assays or preparation of cell lysates for immunoblot analysis. Experiments shown were performed using v-Src overexpressed on a multicopy 2μm plasmid, but the same results were obtained if v-Src was expressed from a low-copy-number centromeric plasmid (data not shown).
Plasmids.
These experiments were greatly aided by generous gifts of the following plasmids from Didier Picard, University of Geneva: the wild-type GR (pTCA/GZ), mutant GR (pG/N768) and wild-type ER [pG/ER(G)] expression plasmids and their corresponding reporter plasmids pUCΔ55-26X and pUCΔSS-ERE; the two-hybrid fusion constructs expressing the Gal4 DNA binding domain fused to the ER HBD [pTCA/GAL4(1-93).ER] and the Gal4 activation domain fused to SRC-1 (pGAD424-SRC1); and the chimeric plasmids pHCA/GAL4(1-93).ER.VP16 and pTCA/GAL4(1-93).GR.VP16. The pHCA-TRP/GAL4(1-93).ER.VP16 plasmid was constructed from pHCA/GAL4(1-93).ER.VP16 by moving a 3.0-kb EcoRI/NotI fragment into the pRS314 vector. A plasmid containing the AF-1 mutation Δ108-317 (25) was a gift from the laboratory of Keith Yamamoto, and the YEpCUP1-HSE-M-lacZ plasmid was a gift from Dennis Thiele (50). Plasmid ZF3 was constructed by placing lacZ coding sequences under control of the SSB2 promoter.
β-Galactosidase assays.
β-Galactosidase assays were performed as described elsewhere (39). Briefly, cultures were grown in selective medium overnight, diluted to an optical density at 600 nm (OD600) of 0.05 in fresh selective medium, and grown overnight at 25°C. As described, hormone (final concentration, 10 μM deoxycholate [DOC] or 0.1 μM β-estradiol; Sigma) diluted 1:1,000 from stocks in ethanol or ethanol alone was added prior to the second night of incubation. Cells were harvested at an OD600 of 0.2 to 1.0. β-Galactosidase units were calculated as 103 × OD420 divided by the OD600 × elapsed time (in minutes). Wild-type and ydj1 mutant cells expressing the GR showed similar dose-response curves at DOC concentrations between 0.1 and 10 μM. The results shown are for cells grown overnight at 25°C in the presence of 10 μM DOC or 0.1 μM β-estradiol, which is the minimum saturation level for wild-type cells. Additional experiments with either a 30°C incubation or 2-h hormone induction in the ydj1 mutant strains showed similar results in overall GR or ER activation of a reporter gene relative to wild type.
Yeast strains.
The following yeast strains were used in this study: PJ51-3a (a trp1-1 ura3-1 leu2-3,112 his3-11,15 ade2-1 can1-100 GAL2+ met2-Δ1 lys2-Δ2); JJ160 (a trp-1 ura3-1 leu2-3,112 his3-11,15 ade2-1 can1-100 GAL2+ met2-Δ1 lys2-Δ2 ydj1::HIS3); PJ69-4a (a trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL-HIS3 met2::GAL7-lacZ); JJ257 (α Δtrp1 leu2-3,112 ura3-52 his3-11,15 GAL2 lys1 lys2 ydj1::HIS3); and JJ290 (α trp1 leu2-3,112 ura3-52 his3 gal4Δ gal80Δ GAL2-ADE2 lys2 met2::GAL7-lacZ ydj1::HIS3).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
The v-Src antibody (15) was a generous gift of Frank Boschelli. Polyclonal antiserum was prepared against full-length purified yeast Ydj1 by standard procedures. The antibody against Mge1 has been described previously (38). The GR antibody was obtained from Affinity Bioreagents (catalog no. MA1-510). The anti-phosphotyrosine residue antibody was obtained from Upstate Biologicals (catalog no. 4G10).
Yeast cell lysates were prepared as described by Kimura (29). Briefly, 10 OD600 units of cells were washed once with water and resuspended in cold ethanol containing 1 μM phenylmethylsulfonyl fluoride. After addition of glass beads, cells were vigorously vortexed in a cold room for 2 min. Proteins were precipitated in an ethanol-dry ice bath for 15 min, pelleted, dried, and resuspended in 100 μl of 2× SDS-sample buffer; 5 μl of each sample was subjected to SDS-PAGE on a standard 7.5 or 10% polyacrylamide gel, transferred to nitrocellulose, and probed with antibodies against v-Src, phosphotyrosine residues, or the GR. Chemiluminescence immunoblotting was performed as instructed by the manufacturer (NEN, Boston, Mass.).
RESULTS
The J + G/F region of Ydj1 is sufficient for wild-type growth at 30°C.
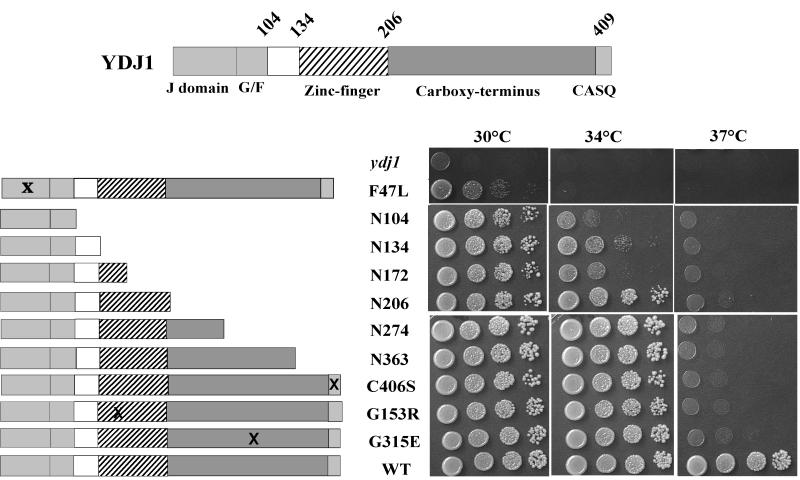
To better understand the in vivo requirements for Ydj1 function, we analyzed an array of Ydj1 mutants in a number of in vivo assays. Figure 1 lists the six carboxy-terminal truncations and three point mutants that we obtained using two separate genetic screens (see Materials and Methods). Also included in this list is the mutation C406S, which inactivates the CAAX box, thus preventing the farnesylation modification of Ydj1 (11). All mutants produce Ydj1 protein levels similar to wild-type cells (data not shown). Plasmids carrying each of these mutants were transformed into a ydj1::HIS3 strain, and the resulting transformants were assayed for growth. Yeast cells containing the J-domain mutation F47L were the only mutant cells that failed to exhibit wild-type growth at 23°C (data not shown) and 30°C (Fig. 1). Yeast expressing even the shortest truncation mutant, N104, which contains only the J + G/F region, grew as well as the wild type at 30°C, the optimal growth temperature for S. cerevisiae. However, none of the mutants were able to grow at 37°C (Fig. 1). The biggest growth difference between the mutants is evident at the intermediate temperature of 34°C. Only truncations containing the entire zinc finger region were able to grow at 34°C, suggesting that this domain has an essential in vivo role at elevated temperatures.
FIG. 1.
Schematic of the domain structure of Ydj1 and the Ydj1 mutants used in the study. Ydj1 mutants were obtained as described in Materials and Methods. The ydj1 disruption strain JJ160 expressing various Ydj1 mutants from low-copy-number plasmids was grown overnight at 25°C and then serially diluted 1:10 prior to plating onto YPD. YPD plates were grown for 2 days at 30, 34, or 37°C.
The carboxy terminus of Ydj1 is required for v-Src activity.
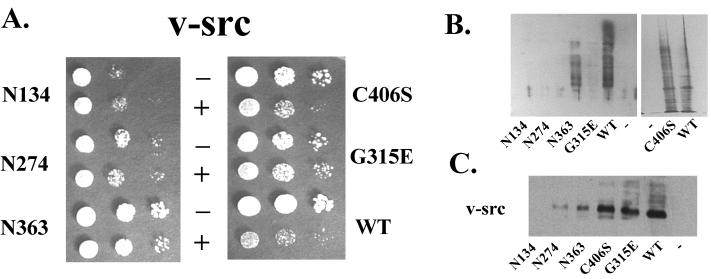
v-Src binds Hsp90 and cochaperones shortly after synthesis and remains bound during transport to the plasma membrane, where it is released from chaperones prior to becoming active as a phosphotyrosine kinase (6). The expression of v-Src is toxic to wild-type yeast but not to yeast containing deletions or mutations of genes involved in the Hsp90 pathway (27). Many of these ydj1 mutants were isolated in a screen for ydj1 mutants that were defective in v-Src processing (see Materials and Methods). We further tested the effect of the ydj1 mutants N134, N274, N363, C406S, and G315E on v-Src maturation. Wild-type and ydj1 mutant yeast strains containing a multicopy plasmid expressing v-Src under the control of a GAL1 promoter or a control vector were assayed for growth on galactose-based media (Fig. 2A). In addition, cell lysates from wild-type and mutant yeast strains were prepared to determine the activity of v-Src using an anti-phosphotyrosine residue antibody (Fig. 2B) and the level of v-Src protein using an antibody against v-Src (Fig. 2C).
FIG. 2.
v-Src phenotype of ydj1 mutant strains. Wild-type (WT) or ydj1 mutant yeast cultures expressing either the GAL1–v-src (pBv-src) multicopy plasmid or the control plasmid (pB656) were grown overnight in selective medium containing raffinose as the carbon source. v-Src expression was induced by addition of 20% galactose to a final concentration of 2%. Cells were harvested 6 h after induction. (A) Yeast cultures were serially diluted 1:10, plated on selective medium containing galactose as the carbon source, and incubated for 2 days at 30°C. (B) Immunoblot of yeast lysates using anti-phosphotyrosine residue antibody 4G10 (Upstate Biologicals). (C) Immunoblot of yeast lysates using anti-v-Src antibody.
Surprisingly, Ydj1 mutants had differing effects on v-Src activity and accumulation. Yeast cells expressing N134 and N274 display neither phosphotyrosine activity nor growth inhibition relative to cells carrying the control vector. This lack of activity was due to the lack of v-Src protein accumulation, as little or no v-Src was detected in cell lysates from these strains. The slow growth of N134 and N274 observed (Fig. 2A) is unrelated to v-Src function; rather, it is a reflection of slower growth of these mutant strains on galactose media (unpublished results).
N363 and C406S exhibited v-Src activity; however, these strains had reduced levels of active v-Src relative to the wild type. We observed growth inhibition in the presence of v-Src and increased levels of phosphotyrosine residues relative to control lysates (Fig. 2). Although some variation in the level of v-Src protein and degree of growth inhibition was observed, v-Src activity was consistently observed in N363 and C406S. These results indicate that the farnesylation signal is not required for v-Src function.
A third pattern of v-Src expression was observed in the presence of the ydj1 mutant G315E. v-Src protein levels were near that of the wild type, but the protein was inactive, resulting in no increased phosphotyrosine activity and no growth inhibition. When expressed in the same yeast cells, the G315E mutation is recessive to wild type and v-Src is active (data not shown), suggesting that the G315E mutation does not prevent further productive interaction with the Hsp90 complex. Therefore, even though cells expressing G315E produce stable v-Src protein, the mutant Ydj1 is unable to facilitate folding of v-Src into the active form. These results indicate that Ydj1 has roles in both the accumulation of v-Src protein and the maturation of active kinase. Interestingly, sequences between 274 and 363 appear to be essential for the stable accumulation of active v-Src protein.
GR activity in ydj1 mutant strains.
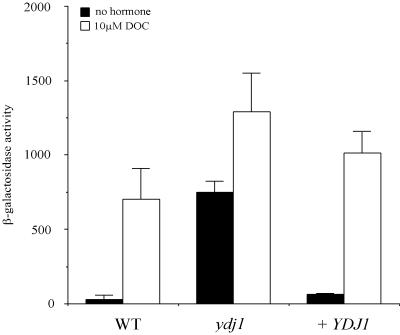
To further investigate whether the carboxy terminus of Ydj1 is required for function in Hsp90 pathways, we assayed the activity of another Hsp90 substrate, the GR, in ydj1 mutant strains. Although not native to S. cerevisiae, steroid receptors expressed in yeast activate transcription of reporter genes in a hormone-dependent manner. The function of the GR was assayed in the ydj1 null strain expressing the rat GR on one plasmid along with a plasmid expressing the lacZ reporter gene under the control of the glucocorticoid response element (GRE). In wild-type strains, very low levels of β-galactosidase activity were produced in the absence of hormone, while hormone addition resulted in an approximate 26-fold induction. In contrast, in a ydj1 null strain, the basal activity of the GR was elevated 28-fold, reaching the activity of wild-type cells in the presence of hormone (Fig. 3). GR activity in the null strain increased further in the presence of hormone, surpassing the level of wild-type cells. Restoring wild-type YDJ1 on a plasmid dramatically reduced the basal levels, resulting in only a twofold increase over cells with a wild-type chromosomal copy of YDJ1.
FIG. 3.
ydj1 null strain exhibits elevated GR activity in the absence of hormone. Yeast cultures were grown in selective medium overnight at 25°C and then diluted into fresh medium with the addition of ethanol or 10 μM DOC. β-Galactosidase assays were performed after overnight (16-h) incubation. Experiments were repeated at least twice. A representative experiment with triplicate samples is shown. Wild-type (WT; PJ51-3A) and ydj1 disruption (JJ160) strains were transformed with plasmids expressing the GR (pTCA/GZ) and corresponding GRE-lacZ reporter plasmid (pUCΔ55-26X). Activity in the wild type, the ydj1 null strain, and a ydj1 null strain with wild-type YDJ1 (+ YDJ1) supplied on a low-copy-number plasmid were determined.
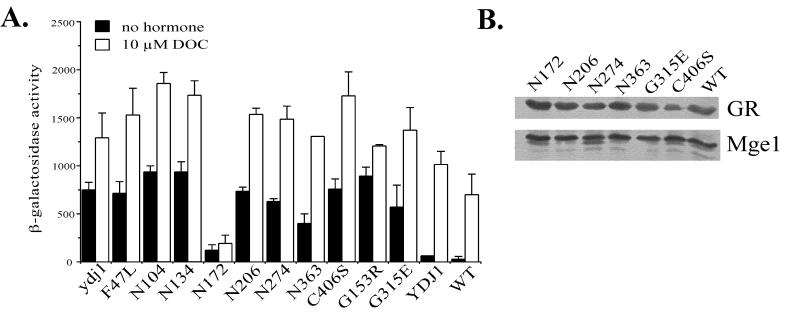
All of the ydj1 mutants were transformed into the ydj1::HIS3 disruption strain and assayed for GR activation of the reporter gene (Fig. 4A). Surprisingly, increased basal activity of the GR was observed in all of the ydj1 mutants. The range of increase in basal activity varied from 5-fold for N172 to 36-fold for N134. The addition of saturating concentrations of hormone (10 μM DOC) resulted in a substantial increase in β-galactosidase production in wild-type and all mutant strains except N172, which expressed only 19% of wild-type activity. Despite the increased activity of many of the Ydj1 mutants compared to the wild type, the levels of GR protein produced in the ydj1 mutant strains was similar to that produced in the wild-type strain (Fig. 4B).
FIG. 4.
GR activity in ydj1 mutant strains. (A) ydj1 disruption strain JJ160 containing GR (pTCA/GZ) and corresponding GRE-lacZ reporter plasmid (pUCΔ55-26X) was transformed with indicated ydj1 mutants expressed on low-copy-number plasmids. ydj1 indicates the null strain, and YDJ1 indicates the presence of wild-type YDJ1 on a plasmid. Wild-type (WT) strain (PJ51-3A) is included for comparison. Assays were performed as described in the legend to Fig. 3 and Materials and Methods. (B) Immunoblot showing level of GR expressed in indicated ydj1 mutant strains. As a control for protein loading, the immunoblot was reprobed with antibodies against the mitochondrial GrpE homolog, Mge1.
Previously it has been shown that the degradation of wild-type β-galactosidase is unaffected by mutation in YDJ1 (31). However, to rule out the possibility that increased activity in ydj1 mutant strains is due to altered degradation of β-galactosidase, we examined the levels of the GRE-lacZ mRNA in ydj1 mutant strains. Expression of GRE-lacZ mRNA in the absence of hormone was observed in lysates from cells expressing F47L, N206, and N363 but not those expressing wild-type YDJ1 (data not shown). These results demonstrate that the level of GRE-lacZ mRNA is directly affected by ydj1 mutation, indicating that in the absence of functional Ydj1, a portion of the GR becomes capable of binding DNA and activating transcription in the absence of hormone.
All of the mutants have increased activity in the absence of hormone, suggesting that the repression of GR activity in the absence of hormone is defective in ydj1 mutant strains. However, one mutant, N172, seems to have a specific defect in hormone-dependent activation of the GR, as it shows little increase in GR activity in response to hormone. We assayed the GR activity in cells expressing both wild-type YDJ1 and N172 and found that the cells behaved like the wild type (data not shown), indicating that N172 does not have a dominant negative effect on receptor activity. All of the other mutations display a hormone-dependent increase in β-galactosidase activity, surpassing the level of activity in wild-type cells. Due to the high basal activity, the fold induction of activity in ydj1 mutant strains appears reduced relative to wild-type cells, but this effect is most likely due to saturation of receptor activity, as similar fold induction levels are observed when the hormone concentration is lowered to 0.1 μM DOC (data not shown).
ER activity in ydj1 mutant strains.
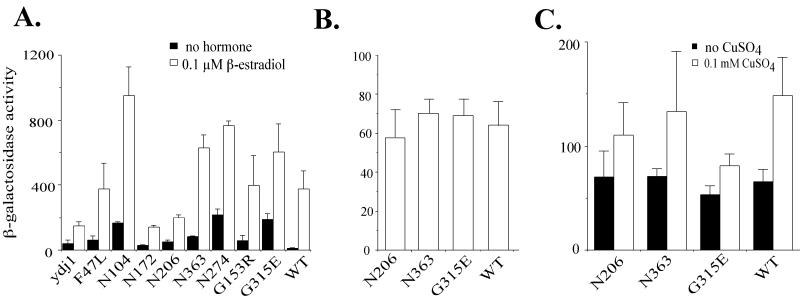
We next examined whether the basal activity of another steroid hormone receptor, the ER, is elevated in ydj1 mutant strains. Like the GR, the ER was expressed from one plasmid while the estrogen response element (ERE) upstream of the lacZ gene was present on another plasmid. The ydj1 null strain and some of the ydj1 mutant strains were examined for ER activation of the lacZ reporter gene. The effects of ydj1 mutations on ER basal activity, shown in Fig. 5A, were similar to those obtained for the GR. The basal activity of the ER in the absence of hormone increased, ranging from 2-fold in the case of N172 to 18-fold for N363. For many mutants, the ER activity in the presence of hormone surpassed that of wild-type cells. However, in the absence of any Ydj1 or in the presence of N172 or N206, the overall activity in the presence of hormone was only 40, 37, or 54% of wild-type levels, respectively, suggesting some ydj1 mutants have decreased hormone-dependent activation of the ER. Thus, elevated basal activity of steroid receptors in the presence of ydj1 mutants is shared between the ER and the GR. These results indicate that a major role of Ydj1 is to facilitate repression of receptor activity in the absence of hormone. However, the ydj1 null and some ydj1 mutants displayed a defect in hormonal activation of the ER but not the GR, suggesting the requirement for Ydj1 in hormonal activation may be receptor dependent (compare Fig. 4A and 5A).
FIG. 5.
ER activity in ydj1 mutant strains. (A) ydj1 disruption strain JJ160 containing plasmids expressing the ER [pG/ER(G)] and the corresponding ERE-lacZ reporter construct (pUCΔSS-ERE) was transformed with indicated ydj1 mutants expressed on low-copy-number plasmids. ydj1 indicates the null strain, and WT indicates the presence of wild-type YDJ1 on a plasmid. Assays were performed as described in the legend to Fig. 3 and Materials and Methods except that 0.1 μM β-estradiol was used instead of 10 μM DOC. (B and C) ydj1 disruption strain containing indicated ydj1 mutants was transformed with a plasmid expressing lacZ under the constitutive promoter SSB2 (B) or the inducible CUP1 promoter containing a mutation in the heat shock element (C). (B) β-Galactosidase assays were performed after overnight incubation at 25°C. (C) β-Galactosidase assays were performed after an overnight incubation in the presence or absence of 0.1 mM CuSO4.
To ensure that the observed effect of ydj1 mutations on steroid receptor activity was not due to a general effect on transcription, we examined the effect of ydj1 mutation on the activity of two additional promoter constructs that drive expression of the lacZ reporter gene. YDJ1 mutations had no significant effect on the activity of the constitutive promoter SSB2 (13) (Fig. 5B) or the copper-inducible promoter CUP1 (50) (Fig. 5C). The finding that β-galactosidase activity arising from these reporter genes is not affected by ydj1 mutations provides further evidence that the dramatic increase in β-galactosidase activity observed for steroid receptors is due to increased transcription and not to altered β-galactosidase degradation in ydj1 mutant strains.
We also examined whether elevated activity of steroid receptors is a common feature of ydj1 mutant S. cerevisiae strains, as we and others have noted strain-dependent differences in the phenotypes of ydj1 mutant strains (reference 42 and unpublished results). The experiments shown in Fig. 1 to 5 were conducted in the W303 strain background (JJ160). We transformed strain JJ257, a ydj1::HIS3 disruption in the S288C background, with plasmids to assay ER and GR activity to confirm that high basal activity of the steroid receptors is observed in this strain background. Strain JJ257 expressing the N363 mutant displayed a 23-fold increase in the basal activity of the ER and a 20-fold increase in the basal activity of the GR (data not shown). These numbers are comparable to results for strain JJ160, in which N363 displayed increases in inductions of 18-fold for the ER and 30-fold for the GR.
These experiments have shown that two separate steroid hormone receptors, the ER and the GR, become active in ydj1 mutant strains in the absence of hormone. This effect is seen in two distinct yeast strains and for all ydj1 mutants tested, indicating it is not a limited phenomenon. Intriguingly, increased basal activity has not been observed in yeast strains containing mutations in other components of the Hsp90 pathway such as Hsc82/Hsp82, Sba1, Sti1, or Cpr7 (4, 16, 17, 42), suggesting that Ydj1 has a unique role in repressing steroid receptor activity. The experiments described in the following sections were designed to attempt to uncover the source of this activity in order to determine the mechanism by which Ydj1 functions in steroid receptor maturation.
ER AF-2 activity is unaffected by mutations in YDJ1.
We examined whether the source of receptor activity in the absence of hormone arose from the amino-terminal AF-1 domain or the AF-2 domain, located in the HBD. First we determined whether the AF-2 domain in ydj1 mutant cells has properties similar to those of hormone-bound receptor. Upon ligand binding and Hsp90 release, the HBD undergoes a large conformational change, exposing AF-2, which is the site of interaction for a number of transcriptional coactivators (19, 24). One such coactivator, SRC-1, was originally isolated in a yeast two-hybrid screen designed to find proteins that interact with the HBD in a hormone-dependent manner (41). We confirmed that we could observe a hormone-dependent two-hybrid interaction between a SRC-1–Gal4 activation domain fusion and a Gal4 DNA binding domain-ER-HBD fusion in a wild-type strain PJ69-4a (data not shown), commonly used in two-hybrid assays (26). A hormone-dependent two-hybrid interaction between a Gal4 DNA binding domain-GR-HBD fusion and the SRC-1 fusion was also observed (data not shown).
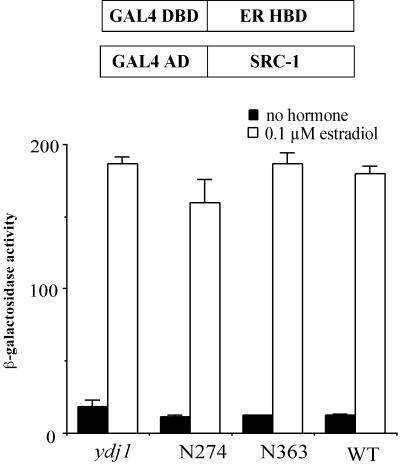
To directly test whether ydj1 mutations result in functional AF-2 domains in the absence or presence of hormone, we examined the two-hybrid interaction between the AF-2 of the ER and SRC-1 in ydj1 mutant strains. To construct a ydj1::HIS3 disruption strain for two-hybrid analysis, we created strain JJ290 by crossing PJ69-4a and the ydj1 disruption strain JJ257, both of which are in the S288C background. As expected, this strain had elevated basal activity of the ER and the GR (data not shown). We monitored the two-hybrid interaction between the Gal4 DNA binding domain-ER-HBD fusion and the Gal4 activation domain–SRC-1 fusion by assaying β-galactosidase levels produced from the GAL7-lacZ reporter gene. In cells expressing wild-type YDJ1, a strong hormone-dependent interaction between the HBD of the ER and SRC-1 was observed (Fig. 6). Almost identical patterns of interaction were observed in the ydj1 null strain or in the presence of N274 or N363. No two-hybrid interaction was observed between the Gal4 DNA binding domain-ER-HBD fusion and the Gal4 activation domain plasmid lacking SRC-1 sequences (data not shown), indicating the interaction requires SRC-1. These results suggest that the basal activity of the ER activity is not due to hormone-independent activation of the AF-2. In addition, the results demonstrate that Ydj1 is not required for the HBD to undergo the hormone-dependent conformational change that exposes AF-2, as wild-type and ydj1 mutant strains displayed similar two-hybrid interactions in the presence of hormone.
FIG. 6.
Two-hybrid interaction between SRC-1 and the ER HBD in ydj1 mutant strains. The Gal4 DNA binding domain (DBD)-ER-HBD fusion plasmid (pTCA-Gal4.ER) and the Gal4 activation domain (AD)–SRC-1 plasmid (424-SRC-1) were transformed into the ydj1 disruption strain JJ290 and strain JJ290 expressing wild-type (WT) YDJ1 or mutant ydj1 from low-copy-number plasmids. β-Galactosidase assays were performed after overnight incubation in the presence or absence of 0.1 μM β-estradiol.
GR containing AF-2 mutation but not AF-1 mutation displays elevated basal activity.
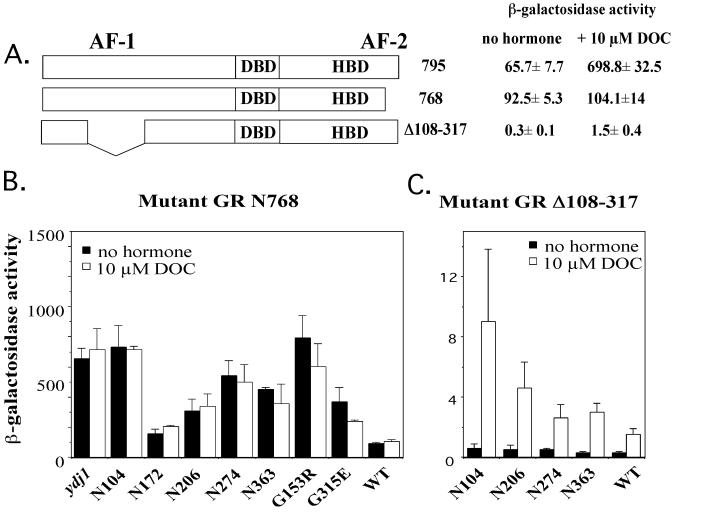
The above results suggest that basal activity of steroid receptors in ydj1 mutant strains does not arise from hormone-independent activation of the AF-2 domain. In addition to AF-2, steroid receptors contain an amino-terminal activation domain, AF-1 (Fig. 7A) (2, 21). To localize the source of receptor activity, we used mutations in the GR that abolish activity of either AF-2 (GR N768) or AF-1 (Δ108-317). In wild-type cells, each of these mutations dramatically reduced GR activity (Fig. 7A), which agrees with earlier findings that the AF-1 and AF-2 domains activate transcription synergistically in yeast (21, 51). First, we tested whether a GR truncation mutant that preserves the Hsp90 binding site but disrupts the AF-2 activation domain had elevated basal activity in ydj1 mutant strains. For this test, we used the previously described GR truncation mutant GR N768 (20), which removes 27 amino acids from the carboxy terminus of the HBD, disrupting helix 11, the core of the AF-2 activating domain (19, 24). In yeast and mammalian cells, GR N768 was unable to activate transcription from a GRE either in the presence or in the absence of hormone (18). GR N768 was transformed into wild-type and ydj1 mutant strains and assayed for GR activity (Fig. 7B). As expected, the GR N768 mutation ablated the hormone response in wild-type cells. In the ydj1 mutant strains, however, the basal activity of GR N768 was as high as in wild-type GR, although no additional increase in response to hormone was observed. These results, combined with those presented in Fig. 6, indicate that increased basal activity is not due to hormone-independent activation of the AF-2 domain.
FIG. 7.
Activity of mutant GR in ydj1 mutant strains. (A) Locations of AF-2 (GR N768) and AF-1 (Δ108-317) mutations in the GR and corresponding activity in wild-type yeast. DBD, DNA binding domain. (B and C) Similar to Fig. 4A except that a plasmid expressing mutant GR N768 (B) or GR Δ108-317 (C) was transformed into the indicated wild-type (WT) or ydj1 mutant strain containing the GRE reporter plasmid.
We next examined the activity of a GR construct containing mutations in the AF-1 domain. We used a mutation that deletes amino acids 108 to 317 of the GR. The Δ108-317 mutation was transformed into wild-type and ydj1 mutant strains and assayed for GR activity (Fig. 7C). As in mammalian cells, this mutation results in very low receptor activity (25) but still displays a hormone-dependent increase in activity. In contrast to the clear increase in basal activity in ydj1 mutant strains expressing the GR N768 mutation, we observed no significant difference in basal receptor activity between wild-type and ydj1 mutant cells expressing GR Δ108-317. Together, the results of these experiments using GR mutants indicate that the receptor activity present in ydj1 mutant cells arises from the AF-1 domain.
Heterologous activation domains fused to the HBD of the ER or GR exhibit elevated basal activity in ydj1 mutant strains.
The AF-1 site is normally repressed in steroid receptors in the absence of hormone. However, deletion of the HBD of either the ER (51) or the GR (20) results in constitutive AF-1 activity, suggesting that the HBD represses the AF-1 site. The HBD of the ER or GR is able to confer hormonal regulation and Hsp90 binding on proteins containing heterologous activation and DNA binding domains (44, 46), indicating that HBDs and Hsp90 can repress a variety of nearby activation domains.
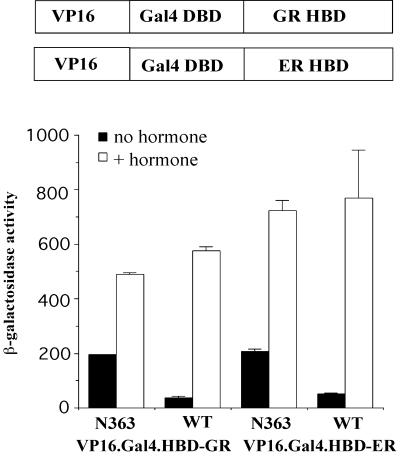
To directly examine whether Ydj1 acts through the HBD to inhibit nearby activation domains, we examined whether chimeric proteins containing heterologous activation and DNA binding domains linked to the HBD of the ER or the GR exhibit elevated activity in ydj1 mutant strains. We used constructs in which the VP16 activation domain was linked to the Gal4 DNA binding domain and the HBD of either the GR (pTCA/Gal4.GR.VP16) or ER [pHCA-TRP/Gal4(1-93).ER.VP16]. We transformed ydj1 disruption strain JJ290 with the above plasmids and measured the activity of the GAL7-lacZ reporter gene. In the presence of wild-type YDJ1, little β-galactosidase activity was observed in the absence of hormone, but addition of DOC or β-estradiol resulted in increased β-galactosidase production. When mutant N363 was expressed, the basal activity of the reporter gene increased fivefold in the case of the GR and fourfold in the case of the ER (Fig. 8). These results demonstrate that the HBD fails to repress heterologous activation domains in ydj1 mutant strains and suggest that Ydj1 acts through the HBD to repress nearby activation domains, including AF-1, in the absence of hormone.
FIG. 8.
Chimeric proteins containing the HBD of the GR or the ER display elevated basal activity in ydj1 mutant strains. ydj1 disruption strain JJ290 expressing either ydj1-N363 or wild-type (WT) YDJ1 was transformed with plasmids expressing chimeric proteins containing the VP16 activation domain and Gal4 DNA binding domain (DBD) fused to the HBD or either the GR [pTCA/GAL4(1-93).GR.VP16] or ER [pHCA-TRP/GAL4(1-93).ER.VP16].
DISCUSSION
To better understand the role of Ydj1 in Hsp90 pathways, we isolated a number of Ydj1 mutants that confer a temperature-sensitive phenotype in the absence of wild-type YDJ1 and analyzed their specific defects in the maturation of diverse Hsp90 substrates.
The J + G/F region confers wild-type growth at 30°C.
Both genetic and biochemical evidence indicates that the J + G/F region of Hsp40s interacts with Hsp70s (12, 28). Our results provide further evidence for the in vivo significance of this portion of Ydj1. The only mutation used in our study that fails to exhibit wild-type growth at 23°C (data not shown) and 30°C is F47L, which contains a mutation in a highly conserved residue of helix 3 in the J domain (28). This result suggests that the interaction between Ydj1 and Hsp70 is critical for both growth and the maturation of Hsp90 substrates.
Expression of N104, which contains the J + G/F region of Ydj1, enables wild-type growth at 30°C, the optimal growth temperature of S. cerevisiae, indicating that this short region is sufficient to carry out some basic important roles. Interestingly, the J + G/F region of the Hsp40, Sis1, is also sufficient to support robust growth at 30°C in a normally inviable sis1 disruption strain (52). Ydj1 and Ssa cooperate in the translocation of preproteins across mitochondrial and endoplasmic reticulum membranes (3). It is likely that disruption of these functions is responsible for the slow-growth phenotype of a ydj1 disruption strain, as N104 relieves the accumulation of the precursor for the mitochondrial protein Hsp60 observed in the ydj1 null strain (data not shown).
Ydj1 and Sis1, which share sequence homology only in the J + G/F region, are both located in the cytosol of S. cerevisiae and are each able to stimulate the ATPase activity of Ssa (36). Previously, it was shown that overexpression of either E. coli DnaJ or SIS1 relieved the 30°C growth defect of a ydj1 null strain (9) and also relieved translocation defects of the ydj1-151 mutant (8). We examined the ability of SIS1 overexpression to rescue the defects in Hsp90 substrate maturation observed in ydj1 mutant strains. Despite relieving the 30°C growth defect, SIS1 overexpression resulted in only a slight decrease in basal activity of the GR in a ydj1 disruption strain (data not shown) and did not affect the ability of ydj1 mutant strains to live in the presence of v-Src (data not shown). It is possible that the inability of SIS1 to rescue the Hsp90 pathway defects of a ydj1 null strain is a consequence of the localization of Sis1 or reduced levels of Sis1 relative to Ydj1 in vivo (52). However, since SIS1 is able to rescue other defects of a ydj1 mutant strain, this result suggests that the carboxy terminus of Ydj1 has a specific role in Hsp90 pathways that cannot be rescued by Sis1.
v-Src maturation requires the carboxy terminus of Ydj1.
Even though yeast expressing N104 grows well at 30°C, the activity of the Hsp90 substrates v-Src, ER, and GR is severely affected at both 25 and 30°C. These results suggest that the carboxy terminus of Ydj1 has specific functions in Hsp90 pathways, but these functions are not essential at 30°C. Within the carboxy terminus of Ydj1 lies the zinc finger region, the neighboring less conserved region, and the farnesylation signal (Fig. 1). The zinc finger region must be present for wild-type growth at the intermediate temperature of 34°C. The farnesylation signal has previously been shown to be required for growth at 37°C (11), and so it is not surprising that all of the truncation mutants fail to grow at 37°C. We constructed pairs of truncation mutations containing wild-type (CASQ) or mutant (SASQ) farnesylation signals after amino acids 209 and 310. The addition of the farnesylation signal did not restore 37°C growth (data not shown). Additionally, the truncation pairs displayed similar v-Src and GR activities (data not shown), indicating that sequences in the carboxy terminus in addition to the farnesylation signal are likely required for growth at elevated temperatures and maturation of substrates.
All but two Ydj1 mutants used in this study resulted in dramatically reduced v-Src activity. Yeast expressing ydj1 mutants N363 and C406S had near-wild-type levels of v-Src activity, indicating that the farnesylation signal is not essential for v-Src activity in vivo. The remaining Ydj1 mutants had differing effects on v-Src activity and accumulation. v-Src protein did not accumulate in ydj1 mutant strains expressing N134 and N274, but it is unknown whether this is due to lower levels of v-Src mRNA or reduced protein stability. In cells expressing G315E, the level of v-Src is unaffected, but the protein is much less active, results similar to those observed for the ydj1-G315D (29) and ydj1-151 mutations (15). Analysis of Hsp90 mutants also revealed affects on v-Src activity as well as stability (40), suggesting multiple roles for both Hsp90 and Ydj1 in v-Src maturation.
Steroid receptors have increased basal activity in ydj1 mutant strains.
The common feature of all ydj1 mutant strains was increased steroid receptor activity in the absence of hormone. This effect on steroid receptors appears to be unique to ydj1 mutations, as no previously described mutations in genes encoding other components of the Hsp90 pathway have affected the basal activity of steroid receptors (4, 16, 17, 42). In addition, it seems unlikely that ydj1 mutations indirectly affect steroid receptor activity, as activation of other signaling pathways does not lead to hormone-independent activation of the GR in yeast (43). Basal receptor activity does not appear to come from unmasked AF-2 domains, as there is no detectable two-hybrid interaction between the transcriptional coactivator SRC-1 and the HBD of the ER in the absence of hormone. Additionally, mutant GR N768 containing an AF-2 mutation retained elevated basal activity in ydj1 mutant strains, whereas GR Δ108-317 containing an AF-1 mutation did not, indicating that the activity arises from the AF-1 domain. Consistent with this interpretation, chimeric proteins containing heterologous activation and DNA binding domains fused to the ER or GR HBD also exhibited constitutive activity in ydj1 mutant strains. These results suggest that increased basal activity in ydj1 mutant strains arises from a defect in the ability of the HBD to repress the amino-terminal AF-1 domain.
In contrast to the dramatic effect on basal activity, no ydj1 mutants except N172 decreased the ability of the GR to activate transcription in a hormone-dependent manner. ER activity appears to have a greater requirement for Ydj1, as the ydj1 null and some ydj1 mutants display significant induction defects. These results suggest that Ydj1 has functions in the activation as well as repression of steroid receptors.
Many ydj1 mutant strains exhibited greater than wild-type activity of the GR and ER in response to hormone. Consistent with this hormone-dependent activation, the ability of the HBD of the ER to undergo a conformational change that exposes the AF-2 was not decreased in the presence of ydj1 mutations. The reason for increased receptor activity is unknown, but does not appear to be due to increased receptor levels in ydj1 mutant cells. Intriguingly, increased hormone-dependent receptor activity was observed in ydj1 mutant cells expressing GR Δ108-317, suggesting a role for the HBD in this activity. Recently, Hsp90 has been found to be involved in the recycling of active DNA-bound receptors back to the inactive form complexed with Hsp90 (33). It is possible that Ydj1 also has a role in receptor recycling, so that loss of Ydj1 would increase the length of time receptor is active.
Model of Ydj1 function in Hsp90 pathways.
The data presented in this report can be integrated into a working model of Ydj1 function in the Hsp90 pathway. According to our model, interaction of Ydj1 with Hsp90 substrates is a critical step resulting in entry of substrates into the Hsp90 folding pathway. In the absence of functional Ydj1, transfer of the receptor to the Hsp90 pathway is not complete, and some of the receptor escapes the regulation imposed by the Hsp90 association, becoming capable of binding DNA and activating transcription. Our results suggest that the interaction of Ydj1 with the HBD of steroid receptors is required for strict hormonal control. When Ydj1 is not functional, the HBD is unable to suppress the activity of AF-1, resulting in active receptor. We have not shown that this interaction is direct, but Ydj1 is capable of binding nonnative polypeptides (14, 35). The ability to bind substrate proteins for presentation to other chaperones would be consistent with the function of DnaJ, which binds ς32 or other substrates before presentation to Hsp70 for subsequent folding (22, 30, 32).
In vertebrate cells, virtually all of the steroid receptors present in cell extracts in the absence of hormone is bound to Hsp90 and cochaperones. Our results suggest the presence of (at least) two distinct populations of steroid receptor complexes in ydj1 mutant cells: one population is in the constitutively active, DNA-bound form, while another population remains bound to Hsp90 until exposure to hormone. Numerous experiments have shown that Hsp90 binding inhibits DNA binding and transcriptional activity, suggesting that active receptor is free of Hsp90 and cochaperones (45). However, since previous studies have shown that continual interaction with Hsp90 is necessary for efficient hormone response in vivo (40), it is likely that there is also a population of receptors in ydj1 mutant cells that remains associated with Hsp90 and cochaperones until hormone binding. Accordingly, Kimura et al. (29) found that GR complexes immunoprecipitated from ydj1 mutant cells exhibiting elevated basal activity contained near-wild-type levels of Hsp90 and cochaperones.
We have found that sequences throughout Ydj1 are required for Hsp90 substrate maturation. Our model predicts that the carboxy terminus of Ydj1 directly binds Hsp90 substrates. While such a direct interaction has not been demonstrated, Lu and Cyr (35) found that a fragment of Ydj1 containing amino acids 173 to 384 is capable of substrate binding. As the carboxy terminus was required for the function of both v-Src and steroid receptors, it appears that substrate binding is required for the function of Ydj1 in Hsp90 pathway, but further studies will be necessary to demonstrate such interactions in vitro and in vivo. It will be particularly informative to extend these studies to examine the effect of ydj1 mutations on the activity of native Hsp90 substrates such as Ste11 (34) and Hap1 (53) to gain a fuller understanding of the roles of Ydj1 in the Hsp90 pathway.
ACKNOWLEDGMENTS
We thank Didier Picard for helpful advice and many of the plasmids used in these studies, Avrom Caplan for the ydj1 mutant C406S, Frank Boschelli for the v-Src antibody and expression plasmids, Dennis Thiele for the YEpCUP1-HSE-M-lacZ plasmid, Keith Yamamoto's laboratory for a plasmid containing the Δ108-317 mutation, and Michael Garabedian for additional plasmids and suggestions. We thank Chris Pfund and David Toft for critical reading of the manuscript and helpful advice.
This work was supported by NSRA 5F32 GM17139 (J.L.J.), and NIH grant 5R01 GM31107 (E.A.C.).
REFERENCES
- 1.Atencio D P, Yaffe M P. MAS5, a yeast homolog of DnaJ involved in mitochondrial protein import. Mol Cell Biol. 1992;12:283–291. doi: 10.1128/mcb.12.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 3.Becker J, Walter W, Yan W, Craig E A. Functional interaction of cytosolic Hsp70 and DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohen S P. Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol Cell Biol. 1998;18:3330–3339. doi: 10.1128/mcb.18.6.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boschelli F, Uptain S M, Lightbody J J. The lethality of p60v-src in Saccharomyces cerevisiae and the activation of p34CDC28 kinase are dependent on the integrity of the SH2 domain. J Cell Sci. 1993;105:519–528. doi: 10.1242/jcs.105.2.519. [DOI] [PubMed] [Google Scholar]
- 6.Brugge J. Interaction of the Rous sarcoma virus protein pp60src with the cellular proteins pp50 and pp90. Curr Top Microbiol Immunol. 1986;123:1–22. doi: 10.1007/978-3-642-70810-7_1. [DOI] [PubMed] [Google Scholar]
- 7.Buchner J. Hsp90 & Co.—a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 8.Caplan A J, Cyr D M, Douglas M G. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 9.Caplan A J, Douglas M G. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caplan A J, Langley E, Wilson E M, Vidal J. Hormone-dependent transactivation by the human androgen receptor is regulated by a dnaJ protein. J Biol Chem. 1995;270:5251–5257. doi: 10.1074/jbc.270.10.5251. [DOI] [PubMed] [Google Scholar]
- 11.Caplan A J, Tsai J, Casey P J, Douglas M G. Farnesylation of YDJ1p is required for function at elevated growth temperatures in S. cerevisiae. J Biol Chem. 1992;267:18890–18895. [PubMed] [Google Scholar]
- 12.Cheetham M E, Caplan A J. Structure, function and evolution of DnaJ: conservation and adaption of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig E A, Jacobsen K. Mutations in cognate gene of Saccharomyces cerevisiae HSP70 result in reduced growth rates at low temperatures. J Biol Chem. 1985;5:3517–3524. doi: 10.1128/mcb.5.12.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cyr D M. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- 15.Dey B, Caplan A J, Boschelli F. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol Biol Cell. 1996;7:91–100. doi: 10.1091/mbc.7.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duina A A, Chang H-C J, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 17.Fang Y, Fliss A E, Rao J, Caplan A J. SBA1 encodes a yeast Hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garabedian M J, Yamamoto K R. Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol Biol Cell. 1992;3:1245–1257. doi: 10.1091/mbc.3.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 20.Godowski P J, Rusconi S, Miesfeld R, Yamamoto K R. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987;325:365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- 21.Gronemeyer H. Transcription activation by estrogen and progesterone receptors. Annu Rev Genet. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- 22.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 23.Holt S E, Aisner D L, Baur J, Tesmer V M, Dy M, Ouellette M, Trager J B, Morin G B, Toft D O, Shay J W, Wright W E, White M A. Functional requirement of p23 and hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 25.Inigues-Lluhi J A, Lou D Y, Yamamoto K R. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 26.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson J L, Craig E A. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 28.Kelley W L. The J-domain family and the recruitment of chaperone power. Trends Biochem. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 29.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 30.Langer T, Lu C, Echols H, Flanagan J, Hayer M K, Hartl F U. Successive action of DnaK, DnaJ, and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 31.Lee D H, Sherman M Y, Goldberg A L. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberek K, Wall D, Georgopoulos C. The DnaJ chaperone catalytically activates the DnaK chaperone to preferentially bind the sigma 32 heat shock transcriptional regulator. Proc Natl Acad Sci USA. 1995;92:6224–6228. doi: 10.1073/pnas.92.14.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, DeFranco D B. Chromatin recycling of glucocorticoid receptors: implications for multiple roles of heat shock protein 90. Mol Endocrinol. 1999;13:355–365. doi: 10.1210/mend.13.3.0258. [DOI] [PubMed] [Google Scholar]
- 34.Louvion J-F, Abbas-Terki T, Picard D. Hsp90 is required for pheromone signaling in yeast. Mol Biol Cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Z, Cyr D M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 36.Lu Z, Cyr D M. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 37.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor family: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao B, Davis J E, Craig E A. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 40.Nathan D F, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 42.Picard D. The role of heat-shock proteins in the regulation of steroid receptor function. In: Freedman L P, editor. Molecular biology of steroid and nuclear hormone receptors. Boston, Mass: Birkhauser; 1998. pp. 1–18. [Google Scholar]
- 43.Picard D, Bunone G, Liu J W, Donze O. Steroid-independent activation of steroid receptors in mammalian and yeast cells and in breast cancer. Biochem Soc Trans. 1997;25:597–602. doi: 10.1042/bst0250597. [DOI] [PubMed] [Google Scholar]
- 44.Picard D, Salser S J, Yamamoto K R. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 45.Pratt W B, Toft D O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocrine Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 46.Scherrer L C, Picard D, Massa E, Harmon J M, Simons S S, Jr, Yamamoto K R, Pratt W B. Evidence that the hormone binding domain of steroid receptors confers hormonal control on chimeric proteins by determining their hormone-regulated binding to heat-shock protein 90. Biochemistry. 1993;32:5381–5386. doi: 10.1021/bi00071a013. [DOI] [PubMed] [Google Scholar]
- 47.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith D F. Dynamics of heat shock protein 90- progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- 49.Szabo A, Korszun R, Hartl F U, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 50.Tamai K T, Liu X, Silar P, Sosinowski T, Thiele D J. Heat shock transcription factor activates yeast methallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White J H, Metzger D, Chambon P. Expression and function of the human estrogen receptor in yeast. Cold Spring Harbor Symp Quant Biol. 1988;53:819–828. doi: 10.1101/sqb.1988.053.01.093. [DOI] [PubMed] [Google Scholar]
- 52.Yan W, Craig E A. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Hach A, Wang C. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol Cell Biol. 1998;18:3819–3828. doi: 10.1128/mcb.18.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]