Abstract
Recently, long noncoding RNAs (lncRNAs) have attracted great attention from researchers. LncRNAs are non-protein-coding RNAs of more than 200 nucleotides in length. Multiple studies have been published on the relationship between lncRNA expression and the progression of human diseases. LncRNA small nucleolar RNA host gene 4 (SNHG4), a member of the lncRNA SNHG family, is abnormally expressed in a variety of human diseases, including gastric cancer, renal cell carcinoma, glioblastoma, neuroblastoma, prostate cancer, colorectal cancer, osteosarcoma, cervical cancer, liver cancer, lung cancer, non-small-cell lung cancer, neonatal pneumonia, diabetic retinopathy, neuropathic pain, acute cerebral infarction, acute myeloid leukaemia, and endometriosis. In this paper, the structure of SNHG4 is first introduced, and then studies in humans, animal models and cells are summarized to highlight the expression and function of SNHG4 in the above diseases. In addition, the specific mechanism of SNHG4 as a competing endogenous RNA (ceRNA) is discussed. The findings indicate that SNHG4 can be used as a biomarker for disease prognosis evaluation and as a potential target for disease diagnosis and treatment.
Keywords: LncRNA SNHG4, ceRNA, Human diseases, Function, Molecular mechanism
Introduction
RNA is mainly divided into coding RNA and noncoding RNA (ncRNA). According to transcriptome sequencing data, 70–90% of the human genome is involved in transcription; only 2% of the transcripts encode proteins, while the majority are non-protein-coding RNAs [1, 2]. ncRNAs principally include ribosomal RNA (rRNA), long noncoding RNA (lncRNA), transfer RNA (tRNA), microRNA (miRNA), small nuclear RNA (snRNA), circular RNA (circRNA), small nucleolar RNA (snoRNA), and piwi-interacting RNA (piRNA) [3]. Among them, lncRNAs, accounting for approximately 80% of ncRNAs [4], are the most studied. LncRNAs are ncRNAs that are more than 200 nucleotides in length and do not encode proteins [5, 6]. They have been reported to participate in many pathophysiological processes, including gene expression, protein activity, cell proliferation, apoptosis, and inflammation [7, 8]. Multiple studies have been published on the association between the expression of lncRNAs and the progression of human diseases.
LncRNA small nucleolar RNA host gene 4 (SNHG4) is a member of the SNHG family. SNHGs are the host genes of snoRNAs present in the nucleus and cytoplasm [9]. To date, it has been reported that the SNHG family has 22 members, from SNHG1 to SNHG22 [10]. They play significant roles in human cancers and other diseases. Xu et al. [11] pointed out that SNHG3 is a novel oncogenic lncRNA, which is aberrantly expressed in osteosarcoma, hepatocellular carcinoma (HCC), lung cancer, etc. Upregulation of SNHG3 contributes to biological functions, including tumour cell proliferation, migration, and invasion. Thin [12], Huang [13], and Xiao [14] have published reviews on SNHG1. They mainly summarized the relationship between SNHG1 and cancer and revealed that SNHG1 may act as a useful biomarker for the diagnosis, prognosis and treatment of human cancer. In addition, SNHG5, SNHG7, SNHG12, and SNHG16 have all been reported to promote the progression of cancers [15–18].
In this paper, we mainly summarized the research progress regarding SNHG4 in most tumours and some non-tumour diseases. Although an increasing number of studies on SNHG4 and human diseases have been published, they still do not cover all diseases. For example, there is no research report on SNHG4 in pancreatic cancer, breast cancer, etc. at present. In addition, there is a clear gap between existing research and clinical practice. In the future, more large-scale and multifaceted studies are needed to further verify the role of SNHG4 in various diseases. SNHG4 mainly plays a carcinogenic role in tumours. Reducing the expression of SNHG4 can inhibit the proliferation of tumour cells and is expected to become a potential target for cancer treatment. In some non-neoplastic diseases, in addition to affecting proliferation, SNHG4 is closely related to the immune response and can play dual pro-inflammatory and anti-inflammatory roles [20, 22]. In cerebral ischaemia–reperfusion injury, the SNHG4/miR-449c-5p/STAT6 axis participates in and inhibits the inflammatory process. In contrast, SNHG4 can also increase the levels of pro-inflammatory factors (IL-6, IL-12, and TNF-α) and promote neuroinflammation. In addition, Horikawa et al. [23] disclosed a transcript containing an intron sequence of SNHG4, which is expressed in podocytes. Podocytes play an indispensable role in the kidney [24]. Overall, we summarize the function of SNHG4 from many aspects, including human studies and in vivo and in vitro studies. Some mechanisms by which SNHG4 acts as a competing endogenous RNA (ceRNA) are also discussed, and it is finally speculated that SNHG4 may be used as a target for disease treatment, diagnosis and prognosis evaluation.
LncRNA SNHG4, a member of the lncRNA SNHG family
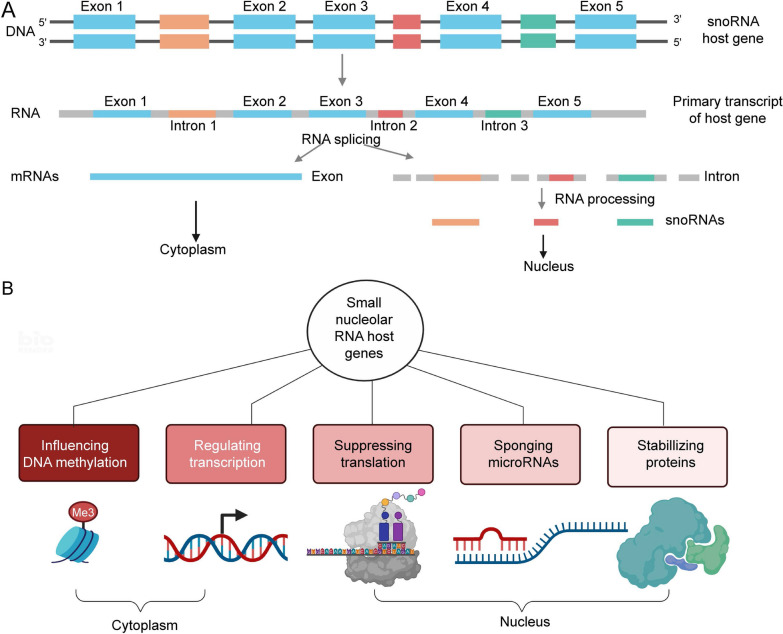
SNHGs are the host genes of snoRNAs (small nucleolar RNAs), including exons and introns [10, 25]. The introns are mainly processed into snoRNAs, while the exons are reassembled and play roles in the cytoplasm [10, 26]. Zimta et al. [27] first summarized the five main molecular mechanisms of SNHGs. SNHGs can influence DNA methylation, regulate transcription, repress translation, act as ceRNAs and prevent protein ubiquitination. LncRNA SNHG4, which is located at 5q31.2 [28], is a member of the SNHG family. SNHG4 also consists of exons and introns [23] (Fig. 1). In 2014, Chaudhry [29] first found that SNHG4 expression was increased in irradiated TK6 cells but was downregulated in bystander cells versus control cells. SNHG4 has been reported to be abnormally expressed in many diseases. It is downregulated in patients with neonatal pneumonia (NP) [30], diabetic retinopathy (DR) [19], acute cerebral infarction (ACI) [20], and acute myeloid leukaemia (AML) [21] and is upregulated in gastric cancer (GC) [31], renal cell carcinoma (RCC) [32], glioblastoma (GBM) [33], neuroblastoma [34], colorectal cancer (CRC) [35], etc. The aberrant expression of SNHG4 has been proven to be closely related to the genesis, occurrence and progression of various diseases.
Fig. 1.
A The structure of the SNHG family members and the synthetic pathway of snoRNAs. B SNHGs are present in the cytoplasm and nucleus. They have five main types of molecular mechanisms of action
SNHG4 in human studies
GC
GC, with its high incidence and mortality, has become the third leading cause of cancer-associated death worldwide [36, 37]. Due to the lack of specific symptoms in the early stage, most patients with GC are in the advanced stage at the time of diagnosis, and the treatment effect and prognosis are poor [38–40]. Thus, there is an urgent need to find new biomarkers for early diagnosis to achieve the best treatment response. Recently, lncRNAs have been constantly reported to play important roles during development and disease, including cancer [41–43]. Wang et al. [44] reported that SNHG4 was upregulated in GC tissues compared to normal adjacent tissue. The high expression level of SNHG4 was related to a lower survival rate. In addition, SNHG4 expression was noticeably associated with tumour-node-metastasis (TNM) stage (P < 0.001), lymph node metastasis (P = 0.003), and tumour invasion (P = 0.004). In summary, SNHG4 may act as an oncogenic lncRNA in GC.
RCC
RCC, a renal-epithelial cancer, accounts for almost 90% of renal tumours [45, 46]. It is the second most common malignant cancer of the urinary system [47] and has a high distant metastasis rate. Currently, the most ideal treatment is standard nephrectomy; however, almost 25% of clear cell renal cell carcinoma (ccRCC) patients are prone to relapse after surgery [48, 49]. Therefore, it is significant to further explore the mechanisms of RCC and develop more effective target therapeutic drugs. The Cancer Genome Atlas (TCGA) data show that SNHG4 is upregulated in many cancers, including RCC [32], and Wu and colleagues [32] confirmed its high expression in RCC. Moreover, they further explored the correlation between SNHG4 and the clinical characteristics and prognosis of RCC patients. The results showed that high SNHG4 expression was closely related to relapse status, poor tumour grade, advanced T stage, lymph node invasion, and distant metastasis. Kaplan–Meier survival curve and multivariate Cox survival analysis both showed that high expression of SNHG4 predicted a poor prognosis. It was an independent prognostic factor for low survival rate and tumour relapse in RCC patients.
GBM
Glioma is one of the most common types of primary intracranial carcinoma, and GBM is one of the gliomas [50]. GBM is highly malignant and aggressive, with a median survival of approximately one year [51, 52]. Despite therapies with surgery, radiotherapy, and chemotherapy, patient prognosis has not substantially improved [53]. It was reported that aberrant gene expression is vital in the development of GBM [54]. LncRNAs are a group of long-chain RNAs that do not encode proteins but can regulate gene expression. Lu [55] pointed out that LINC00511 facilitates temozolomide resistance in GBM cells by binding with miR-126-5p. In addition, studies have revealed that lncRNA HULC stimulates the epithelial-mesenchymal transition (EMT) process in human glioblastoma [56]. Wang and his team found [33] that SNHG4 is also upregulated in GBM. High expression of SNHG4 represents a worse prognosis.
PCa
Prostate cancer (PCa) is one of the most universal malignant tumours [57, 58] and the fifth most common cause of cancer-related death in men [59]. Notably, approximately 15% of PCa cases are fatal, indicating that patients have a high risk of death. Despite the drastically improved PCa survival rate, patients at the metastatic stage still have a poor prognosis [60]. Thus, there is an urgent need to understand the molecular mechanism of PCa for the development of novel biomarkers that can be used for diagnosis and treatment [61]. Recently, it has been reported that lncRNAs can participate in the regulation of gene expression and changes in biological behaviour during the carcinogenesis of prostate cells [62]. Wang et al. [63] found that SNHG4 expression was higher in PCa tissues than in paracancerous tissues. High levels of SNHG4 decreased overall survival (OS) and were distinctly linked to lymph node metastasis (P = 0.029) and the tumour stage (P = 0.014) of PCa patients. SNHG4 is expected to become an effective biomarker for the diagnosis, treatment and prognosis of PCa.
Osteosarcoma
Osteosarcoma is the most recognized primary malignant bone tumour and is mainly seen in adolescents [64, 65]; it is the second leading cause of cancer-related mortality among children and adolescents [66]. Due to the strong metastatic and highly invasive characteristics of osteosarcoma, the prognosis of patients is generally poor, and the five-year survival rate is only approximately 50% [67–69]. Thus, finding new molecular biomarkers for the diagnosis, treatment, and evaluation of metastasis is of great importance. Researchers used TCGA RNA-seq data to evaluate the expression of SNHG4 in human osteosarcoma samples. The results showed that SNHG4 expression was considerably increased in osteosarcoma tissues compared with adjacent non-tumour tissues. High SNHG4 expression was positively linked to tumour size (P = 0.020). Patients with high SNHG4 expression had a higher tumour recurrence and a lower survival rate [70]. Huang et al. [71] confirmed the above findings. In addition, they pointed out that a high level of SNHG4 is closely related to advanced pathological stage (P = 0.036) and distant metastasis (P = 0.039) in osteosarcoma patients.
Liver cancer
Liver cancer is one of the most common cancers of the digestive system [72] and the second leading cause of cancer-related deaths worldwide [73–75]. In almost all patients, liver cancer is caused by chronic liver disease [76]. HCC is the most prominent histological form of liver cancer [76]. The economic and medical burden caused by liver cancer worldwide is very high [77]. ncRNAs are involved in the gene regulation of liver cancer. Researchers [78, 79] have discussed the role of SNHG4 in liver cancer. SNHG4 is upregulated in liver cancer. High SNHG4 expression was found to be correlated with histologic grade (P = 0.001), histological type (P = 0.01), stage (P = 0.01), survival status (P = 0.013), and T classification (P = 0.004). Compared with patients with low SNHG4 expression, those with high SNHG4 expression had poorer OS and relapse-free survival. Multivariate analysis further identified SNHG4 as an independent prognostic factor of poor survival in liver cancer.
Other diseases
In ACI [20], NP [30], DR [19], AML [21], neuroblastoma [33], CRC [35], cervical cancer (CC) [80], non-small-cell lung cancer (NSCLC) [28], and endometriosis [81], researchers explored the expression of SNHG4 in patients with these diseases. However, they did not seem to have studied the relationship between SNHG4 and the clinical features of these diseases, nor have they explored the relationship between SNHG4 and disease prognosis. Overall, SNHG4 is abnormally expressed in a variety of diseases, suggesting that SNHG4 may be involved in their occurrence and development (Table 1). Further research on specific molecular mechanisms is expected to provide new ideas for the diagnosis and treatment of these diseases.
Table 1.
The expression of SNHG4 and its clinical significance in a variety of diseases
| Disease type | Number of clinical samples | Expression level | Clinicopathologic features | HR | P value | Prognostic implication of SNHG4 overexpression | Property | PMID |
|---|---|---|---|---|---|---|---|---|
| Gastric cancer (GC) | 53 tissue samples from GC patients | Upregulation | Lymph node metastasis, tumour invasion, TNM stage | 1.54 | P < 0.001 | Poor | Oncogene | 33236157 |
| Renal cell carcinoma (RCC) | 99 pairs of RCC tissues and adjacent normal tissues | Upregulation | Advanced T stage, node invasion, distant metastasis status, poor tumour grade, relapse status | 1.50 | P = 0.007 | Poor | Oncogene | 33088220 |
| Glioblastoma (GBM) | 62 pairs of GBM tissues and adjacent normal tissues | Upregulation | – | 3.32 | – | Poor | Oncogene | 32427712 |
| Neuroblastoma | 30 pairs of neuroblastoma tissues and adjacent normal tissues | Upregulation | – | – | P = 0.018 | Poor | Oncogene | 32614236 |
| Prostate cancer (PCa) | 113 pairs of PC tissues and adjacent normal tissues | Upregulation | Tumour stage, lymph node metastasis | – | P = 0.005 | Poor | Oncogene | 31608997 |
| Colorectal cancer (CRC) | 12 pairs of CRC tissues and adjacent normal tissues | Upregulation | – | – | – | – | Oncogene | 33744866 |
| Osteosarcoma | 24 pairs of osteosarcoma tissues and adjacent normal tissues | Upregulation | Distant metastasis, lager tumour size, advanced pathological stage | – | P = 0.047 | Poor | Oncogene | 32537941 |
| 136 cases of osteosarcoma patients and 40 adjacent normal tissue | Upregulation | Tumour size | – | P = 0.001 | Poor | Oncogene | 30152090 | |
| Cervical cancer (CC) | 27 pairs of CC tissues and adjacent normal tissues | Upregulation | – | – | – | – | Oncogene | 31590627 |
| Liver cancer | 371 liver cancer tissues and 50 normal liver tissues | Upregulation | Histological type, histologic grade, stage, T classification, survival status | – | P < 0.001 | Poor | Oncogene | 31967298 |
| Hepatocellular carcinoma (HCC) | 49 pairs of HCC tissues and adjacent normal tissues | Upregulation | – | – | P < 0.001 | Poor | Oncogene | 30537372 |
| Non-small-cell lung cancer (NSCLC) | 50 pairs of NSCLC tissues and adjacent normal tissues | Upregulation | – | – | – | Poor | Oncogene | 33816782 |
| Neonatal pneumonia (NP) | 15 peripheral venous blood from NP patients and healthy controls | Downregulation | – | – | – | – | – | 34450538 |
| Diabetic retinopathy (DR) | 60 tissue samples from DR patients, diabetic patients without complication and healthy controls | Downregulation | – | – | – | – | – | 32454787 |
| Acute cerebral infarction (ACI) | 30 tissue samples from ACI patients and healthy controls | Downregulation | – | – | – | – | – | 32982175 |
| Acute myeloid leukaemia (AML) | 60 tissue samples from AML patients and healthy controls | Downregulation | – | – | – | – | – | 32319862 |
| Endometriosis | 25 EC samples and EU samples | Upregulation | – | – | – | – | – | 32525017 |
EC, ectopic endometrium; EU, eutopic endometrium
SNHG4 in in vivo studies
Some researchers have used xenotransplantation mouse models to explore the functional role of SNHG4 (Table 2). Cheng et al. [31] revealed that SNHG4 promotes GC cell growth in vivo. They injected HGC-27 and MKN-45 cells transfected with shNC and shSNHG4 subcutaneously into nude mice. Then, they measured the tumour size and weight of the two groups. Compared with the control, the tumours of the shSNHG4 group were significantly smaller and lighter. In addition, in an in vivo study of neuroblastoma, researchers found that SNHG4 promotes tumour cell growth, invasion, and migration, especially lung metastasis, and induces apoptosis [34]. Similarly, SNHG4 knockdown could inhibit RCC [32], CRC [35], osteosarcoma [71], CC [80], lung cancer [82], and NSCLC [28] tumour growth. In addition, Liu and colleagues [81] pointed out that silencing SNHG4 inhibited the growth of endometrial tissue outside the uterine cavity in vivo. In summary, the results of in vivo studies showed that SNHG4 not only plays a tumorigenic role in some cancers but also promotes the growth of intimal tissues in non-tumour diseases such as endometriosis. The development of drugs targeting SNHG4 has certain clinical value in the future.
Table 2.
Function and mechanism of SNHG4 in different diseases
| Disease type | Cell lines | Expression level | Effect in vitro | Effect in vivo | Genes/proteins/pathways affected | PMID |
|---|---|---|---|---|---|---|
| Gastric cancer (GC) | GES-1, AGS, HGC-27, NCI-N87, MKN-45, 293T | Upregulation | Proliferation ↑, migration ↑, invasion ↑, cell cycle arrest ↓ | Tumour growth ↑ | miR-204-5p, RRM2 | 33567852 |
| GES-1, SNU719, AGS, HGC-27 | Upregulation | Proliferation ↑, migration ↑, invasion ↑, EMT ↑, apoptosis ↓ | – | miR-204-5p | 33236157 | |
| Renal cell carcinoma (RCC) | Caki-1, Caki-2, ACHN, 786-O, 769-P, HK-2 | Upregulation | Proliferation ↑, migration ↑, invasion ↑, apoptosis ↓ | Tumour growth ↑ | miR-204-5p, RUNX2 | 33088220 |
| Glioblastoma (GBM) | U-251 | Upregulation | Proliferation ↑ | – | miR-138, c-Met axis | 32427712 |
| Neuroblastoma | SH-SY5Y, CHP-212, SK-N-FI, and IMR-32, 293T | Upregulation | Proliferation ↑, migration ↑, invasion ↑, EMT ↑, apoptosis ↓ | Tumour growth ↑ lung metastasis ↑ | miR-377-3p | 32614236 |
| Prostate cancer (PCa) | PC-3, LNCaP, DU145, 22RV1, WPMY-1 | Upregulation | Proliferation ↑, migration ↑, invasion ↑, apoptosis ↓ | – | miR‐377, ZIC5 | 31608997 |
| Colorectal cancer (CRC) | FHC, HCT8, LoVo, HCT116, SW620, HT29 | Upregulation | Proliferation ↑, migration ↑, invasion ↑, cell cycle arrest ↓ | Tumour growth ↑ | miR-590-3p, CDK1 | 33744866 |
| Osteosarcoma | hFOB1.19, HOS, MG63, Saos2, SJSA1, U2OS | Upregulation | Proliferation ↑, migration ↑, invasion ↑, apoptosis ↓ | Tumour growth ↑ | miR-377-3p | 32537941 |
| MG-63, HOS, 143B, SW-1353, Saos2, U2OS | Upregulation | Proliferation ↑, colony formation ↑ | – | miR-224- 3p | 30152090 | |
| Cervical cancer (CC) | NCEC, c4-1, Caski, HeLa, SiHa | Upregulation | Proliferation ↑, apoptosis ↓ | Tumour growth ↑ | miR-148a-3p, c-Met | 31590627 |
| Lung cancer | NCI-H2170, NCI-H520, SK-MES-1, NCI-H1975, NCI-H1437, SPC-A-1 | Upregulation | Proliferation ↑, migration ↑, invasion ↑, EMT ↑, cell cycle arrest ↓ | Tumour growth ↑ | miR-98-5p | 31220419 |
| Non-small-cell lung cancer (NSCLC) | H1299, H1650, H1975, SPCA1, 16HBE | Upregulation | Proliferation ↑, migration ↑, invasion ↑, apoptosis ↓ | Tumour growth ↑ | miR-let-7e, KDM3A, p21 | 33816782 |
| Neonatal pneumonia (NP) | WI-38 | Downregulation | Proliferation ↑, migration ↑, SOD concentration ↑, Inflammation ↓, apoptosis ↓ | – | METTL3, NF-κB pathway | 34450538 |
| Diabetic retinopathy (DR) | ARPE-19 | Downregulation | Apoptosis ↓ | – | miR-200b, Oxr1 | 33822671 |
| Neuropathic pain | PC12 | Upregulation | Inflammation ↑ | – | miR-423-5p | 32454787 |
| Acute cerebral infarction (ACI) | HAPI, HEK293 | Downregulation | Inflammation ↓ | – | miR-449c-5p, STAT6 | 32982175 |
| Acute myeloid leukaemia (AML) | Kasumi-6 | Downregulation | Proliferation ↓ | – | miR-10a, PTEN | 32319862 |
| Endometriosis | HESCs | Upregulation | Proliferation ↑ | Endometrial tissue growth ↑ | miR-148a-3p, c-Met | 32525017 |
SNHG4 in cell lines studies
In addition to discussing SNHG4 expression and related clinicopathologic features for the diseases above, we summarized the potential biological functions and specific molecular mechanisms of SNHG4, which have been described in detail in in vivo studies. Next, we will mainly introduce the in vitro cell function verification of SNHG4. A large number of studies have revealed that SNHG4 mainly promotes proliferation, invasion, migration, apoptosis inhibition and EMT [31, 35, 70, 81]. In addition, Zhang and Pan et al. [20, 22] found that SNHG4 has dual pro-inflammatory and anti-inflammatory functions (Table 2).
Proliferation
Cell proliferation is one of the most fundamental attributes of organisms [83]. Almost all living beings sustain life via cell proliferation [83]. This process also plays a vital role in the initiation and development of cancers [84, 85]. Researchers usually use Cell Counting Kit-8 (CCK-8), MTT, colony formation, and flow cytometry assays to assess the level of cell proliferation. Cyclin D1, Ki67, CDK1, CDK4, CDK6, and PCNA are all proliferation-related proteins, and their levels indirectly represent the level of cell proliferation. Cyclin D1 is a G1 checkpoint protein [86, 87]. Ki67, a well-known proliferation marker, can be used to evaluate cancer progression [88, 89]. CDK1, CDK4, and CDK6 are cyclin-dependent kinases that phosphorylate the corresponding proteins to drive the cell cycle process and thus affect cell proliferation [86, 90]. Proliferating cell nuclear antigen (PCNA) is also a cell proliferation-related marker [91, 92]. Cheng et al. [31] pointed out that, compared with the shNC group, the expression levels of cyclin D1, CDK6, and Ki67 were decreased in the shSNHG4 group. SNHG4 knockdown reduced cell viability and promoted cell cycle arrest, thus inhibiting GC cell proliferation. In CRC, researchers demonstrated an SNHG4/miR-590-3p/CDK1 axis that influenced the cell cycle and ultimately regulated cell proliferation [35]. SNHG4 binds to miR-590-3p to inactivate it, thereby alleviating the inhibitory effect of miR-590-3p on the downstream target gene CDK1. Finally, SNHG4 promotes the proliferation of CRC cells by promoting the transition of the cell cycle from the G2 phase to the M phase. In lung cancer, Tang and colleagues found that SNHG4 promotes cell growth by increasing the protein levels of Ki67, CDK4 and CDK6 [82]. SNHG4 was also found to promote the proliferation and migration of ectopic endometrium by regulating c-Met mediated by miR-148a-3p [81]. In addition, SNHG4 can function as a ceRNA, sponge miR-138 to upregulate c-Met [33], sponge miR-204-5p to upregulate RUNX2 [32], and then promote glioblastoma cell and RCC cell proliferation. Although SNHG4 plays a proliferation-promoting role in most diseases, it is downregulated in AML and inhibits proliferation by regulating the miR-10a/PTEN axis [21]. SNHG4 sponges miR-10a to upregulate PTEN, which acts as a tumour suppressor and can inhibit cancer cell proliferation.
In summary, the effect of SNHG4 on proliferation depends on the type of disease, although in most cases, it promotes proliferation. Future research can further explore the mechanisms and pathways by which SNHG4 affects proliferation.
Migration, invasion and metastasis
Cell migration and invasion are two crucial initial processes of cancer cell metastasis [93, 94]. Cell migration is necessary for the normal growth and development of organisms. However, abnormal cell migration may lead to serious consequences such as tumour formation and metastasis [95]. Cell invasion refers to cell dissemination into adjacent organ tissues, thus inducing further development and distant metastasis of cancer cells [96]. Cell metastasis is the main cause of the widespread transformation of cancer cells to multiple organs [97]. All of these factors ultimately trigger cancer-related death. It has been reported that SNHG4 promotes migration, invasion, and metastasis in a variety of cancers. Cheng et al. [31] found that two metastasis markers, MMP2 and MMP9, were reduced after the knockdown of SNHG4, indicating that SNHG4 knockdown efficaciously decreases the metastatic capacity of GC cells. In addition, high expression of SNHG4 was associated with lung metastasis in neuroblastoma [34], lymph node metastasis in PCa [63], and distant metastasis in osteosarcoma [71].
EMT refers to the ability of epithelial cells to transform into mesenchymal cells during development [98, 99]. During this process, cancer cells lose cell-to-cell adhesion, gain the ability to migrate and invade, and finally trigger cancer cell metastasis [100, 101]. During EMT, E-cadherin expression levels are increased, while N-cadherin and Snail expression levels are decreased [102, 103]. In GC [44], neuroblastoma [34], and lung cancer [82], it has been reported that SNHG4 promotes EMT by upregulating E-cadherin and downregulating N-cadherin, thereby exerting a tumorigenic effect. Overall, SNHG4 plays an important role in invasion and migration.
Apoptosis
Apoptosis, which is a type of programmed cell death (PCD), is a basic process of multicellular biological development [104, 105]. It can effectively remove damaged cells and maintain a stable internal environment. However, abnormal apoptosis can also trigger a series of diseases [106, 107]. Neurodegenerative diseases, anaemia, and transplant rejection are common causes of excessive apoptosis. Insufficient apoptosis can cause cancer and autoimmune diseases. Apoptosis is strictly controlled by multiple genes, including the Bcl-2 family, caspase family, some oncogenes, and the tumour suppressor gene P53 [108, 109]. As mentioned above, SNHG4 plays a carcinogenic role in most tumour diseases, and it can promote cancer cell proliferation and migration and inhibit apoptosis. SNHG4 silencing increases the expression of pro-apoptotic proteins such as Bax and caspase-3, thereby promoting neuroblastoma cell apoptosis [34]. Wu and colleagues also confirmed that overexpression of SNHG4 could inhibit RCC cell apoptosis [32]. Overexpression of SNHG4 resulted in an increase in Bcl-2 protein expression and a decrease in caspase-3, -8, and -9 activities in 769-P and ACHN cell lines, which proved that SNHG4 significantly inhibited the apoptosis of RCC cells. In another study on diabetic retinopathy, researchers found that SNHG4 upregulates the expression of Oxr1 by sponging miR-200b and thus inhibits ARPE-19 cell apoptosis [19]. In summary, SNHG4 acts to inhibit apoptosis in most diseases. Downregulating the SNHG4 level is expected to alleviate disease progression by promoting apoptosis.
Inflammation
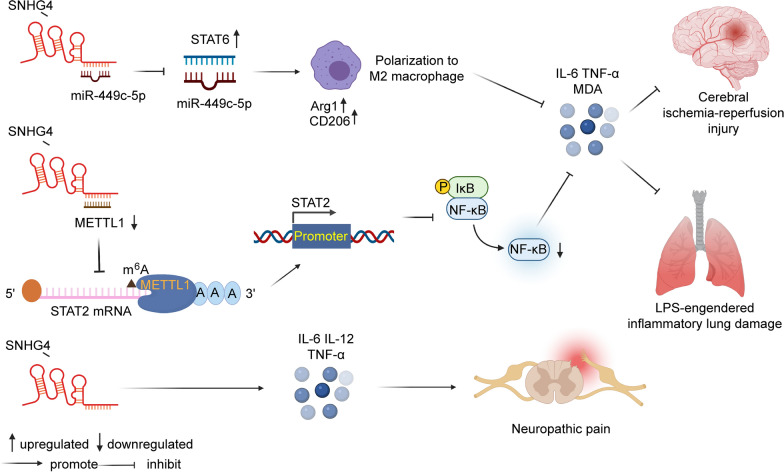
Inflammation is the basic physiological response of the body to external stimuli; it is usually coordinated by immune cells to restore homeostasis [110]. In ACI, however, hyperactivated microglia produce massive amounts of inflammatory cytokines, causing inflammatory storms and eventually aggravating disease progression [111]. Zhang et al. [20] reported that SNHG4 can repress inflammation through the miR-449c-5p/STAT6 axis in microglia during cerebral ischaemia–reperfusion injury. The STAT signalling pathway is closely associated with immune regulation, and STAT6 is a cytoplasmic transcription factor that can be activated by IL-4 and IL-13. Their work revealed that the upregulation of SNHG4 inhibited the expression of miR-449c-5p and activated the STAT6 signalling pathway, thus inhibiting the M1 polarization of microglia and reducing inflammation. In NP, researchers found that SNHG4 inhibited the production of IL-6, TNF-α, and malondialdehyde (MDA), and inhibited the expression of NF-κB pathway proteins. Mechanistically, SNHG4 bound to and downregulated the expression of METLL3. METTL3 interference reduced the m6A level of STAT2 mRNA and promoted the STAT2 translation level. Finally, SNHG4 reduced the inflammatory lung injury caused by lipopolysaccharide (LPS) [30]. In another study on neuropathic pain, SNHG4 played the opposite role. During neuropathic pain progression, SNHG4 increases the levels of proinflammatory factors (IL-6, IL-12, and TNF-α) and promotes neuroinflammation [22]. In summary, SNHG4 can play a proinflammatory role and an anti-inflammatory role (Fig. 2), and the specific mechanism deserves further investigation.
Fig. 2.
The dual role of SNHG4 on inflammation. In NP and ACI, SNHG4 had the effect of inhibiting inflammation. In neuropathic pain, SNHG4 promoted the expression of inflammation
SNHG4 involved in ceRNA regulation
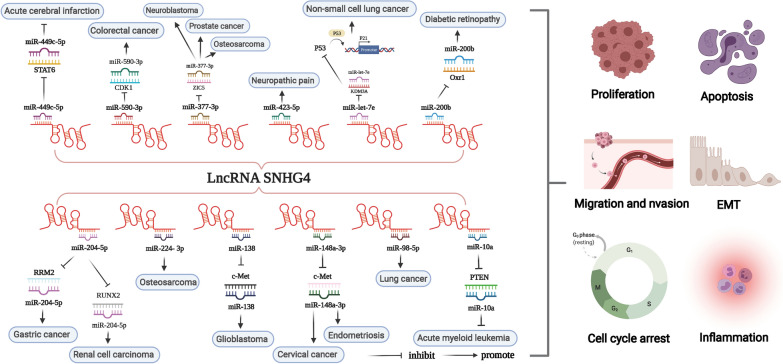
In 2011, Salmena and colleagues first proposed a hypothesis about ceRNA: it forms a large-scale regulatory network that plays a role in both physiological and pathological conditions [112]. Theoretically, ceRNAs include all RNAs that contain microRNA response elements (MREs) and can recognize and bind miRNAs. At present, the most common ceRNAs are lncRNAs, circRNAs and pseudogene RNAs [113–115]. It is common knowledge that miRNAs mainly inhibit the expression of target genes by binding to mRNAs. ceRNAs can competitively bind to and inactivate miRNAs through MREs, thereby affecting the mRNA level of target genes [116–118]. Multiple studies have shown that lncRNAs, as ceRNAs, play an important role in malignant tumours and other diseases. This paper mainly summarized the lncRNA-miRNA-mRNA regulatory network formed by SNHG4 acting as a ceRNA (Table 2) (Fig. 3).
Fig. 3.
Function and molecular mechanisms of lncRNA SNHG4 in a variety of diseases. MiRNAs can cause gene silencing by binding to mRNAs, while ceRNAs can regulate gene expression by competitively binding to miRNAs. SNHG4 can serve as a ceRNA and sponge miRNA, thereby affecting the expression of target genes
Conclusions and perspectives
In recent years, there have been a variety of studies on lncRNAs. Researchers have focused on exploring the relationship between specific lncRNAs and diseases, especially cancer, as a breakthrough to provide new ideas for disease diagnosis and treatment [119]. Although lncRNAs do not encode proteins, they can regulate the expression of protein-coding genes in a variety of ways, most commonly through the ceRNA mechanism [120, 121]. LncRNAs can act as molecular sponges of miRNAs to negatively regulate their expression, resulting in the inhibition of miRNA targets, thus participating in a variety of important biological processes, such as embryonic development, stem cell maintenance, cell proliferation, differentiation, tumorigenesis, and cancer progression [122, 123].
SNHG4, a newly discovered lncRNA and one of the members of the SNHG family, has attracted great attention from researchers. First, researchers explored its expression in diseases and found that SNHG4 is highly expressed in most diseases and low in DR, ACI, and AML. They found that high expression of SNHG4 is closely associated with the clinicopathologic features and prognosis of some cancers. SNHG4 may therefore be used as a prognostic biomarker for these diseases. Next, they conducted a series of in vitro and in vivo studies to investigate the biological behaviour of SNHG4 in diseases. Experiments have proven that SNHG4 can promote cell proliferation, invasion, migration, and EMT and inhibit apoptosis. Targeted inhibition of SNHG4 seems to be beneficial for inhibiting the survival and development of tumour cells, thereby achieving therapeutic effects. Therefore, SNHG4 is expected to become a potential therapeutic target for various diseases. Finally, they revealed the molecular mechanism of SNHG4 in the pathogenesis of diseases. SNHG4 mainly acts as a sponge for miRNA. For example, SNHG4 sponges miR-204-5p and then upregulates RRM2 expression to exert a tumorigenic effect in GC. Similarly, the miR-148a-3p/c-Met axis and miR-let-7e/KDM3A/p21 axis play a role in the occurrence and development of CC and NSCLC, respectively.
Although there have been many types of research on SNHG4 and cancer, they are mainly in the basic research stage and more clinical application research needs to be carried out in the future. In addition, there are few reports of SNHG4 in non-neoplastic diseases; therefore, future research may be tilted towards non-neoplastic diseases. The development of lncRNA detection technology is very important for the diagnosis and treatment of diseases. The latest advances in CRISPR/Cas9 gene knockout, knock-in, and point mutation technologies may help us better understand the biological effects of lncRNAs to develop and apply clinically targeted therapeutic drugs. Currently, there is no approved or tested drug-related to SNHG4, however, as summarized in Fig. 3, SNHG4 regulates many miRNAs, thereby affecting the mRNA level of downstream target genes and promoting the expression of target genes. Targeted inhibition of SNHG4 or downstream genes such as STAT6, PTEN, RRM2, etc. may play an important role in inhibiting proliferation, invasion, migration, etc., and alleviate disease progression. In conclusion, SNHG4 is a novel target for disease diagnosis, treatment and prognosis evaluation, although the specific targeting mechanism and its true clinical application deserve further investigation.
Acknowledgements
Not applicable.
Abbreviations
- lncRNAs
Long non-coding RNAs
- SNHG4
Small nucleolar RNA host gene 4
- ceRNA
Competing endogenous RNA
- ncRNA
Non-coding RNA
- rRNA
Ribosomal RNA
- tRNA
Transfer RNA
- miRNA
MicroRNA
- snRNA
Small nuclear RNA
- circRNA
Circular RNA
- snoRNA
Small nucleolar RNA
- piRNA
Piwi-interacting RNA
- NP
Neonatal pneumonia
- DR
Diabetic retinopathy
- ACI
Acute cerebral infarction
- AML
Acute myeloid leukaemia
- GC
Gastric cancer
- RCC
Renal cell carcinoma
- GBM
Glioblastoma
- CRC
Colorectal cancer
- TNM
Tumour-node-metastasis
- ccRCC
Clear cell renal cell carcinoma
- TCGA
The Cancer Genome Atlas
- PCa
Prostate cancer
- OS
Overall survival
- HCC
Hepatocellular carcinoma
- CC
Cervical cancer
- NSCLC
Non-small-cell lung cancer
- CCK-8
Cell counting kit-8
- PCNA
Proliferating cell nuclear antigen
- EMT
Epithelial-mesenchymal transition
- PCD
Programmed cell death
- MREs
MicroRNA response elements
- MDA
Malondialdehyde
- LPS
Lipopolysaccharide
Authors’ contributions
QFC and XYG contributed equally to this study. QFC and XYG designed the study. QFC drafted the manuscript. XYG and QXZ drew the mechanism diagrams. ZXG, DDS and JW made relevant edits to the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by grants awarded by the National Science and Technology Major Project of China (No. 2018ZX10302206), Science and Technology Major Projects of Zhejiang Province (No. 2018C04016), and the Science and Technology Major Projects of Ningbo (No. 2016C51008).
Availability of data and materials
All data in our study are available upon request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qingfei Chu, Xinyu Gu contributed equally to this work
References
- 1.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Wei DM, Jiang MT, Lin P, Yang H, Dang YW, Yu Q, Liao DY, Luo DZ, Chen G. Potential ceRNA networks involved in autophagy suppression of pancreatic cancer caused by chloroquine diphosphate: a study based on differentially-expressed circRNAs, lncRNAs, miRNAs and mRNAs. Int J Oncol. 2019;54(2):600–626. doi: 10.3892/ijo.2018.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4(11):e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SH, Lim KH, Yang S, Joo JY. Long non-coding RNAs in brain tumors: roles and potential as therapeutic targets. J Hematol Oncol. 2021;14(1):77. doi: 10.1186/s13045-021-01088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cen X, Huang XQ, Sun WT, Liu Q, Liu J. Long noncoding RNAs: a new regulatory code in osteoarthritis. Am J Transl Res. 2017;9(11):4747–4755. [PMC free article] [PubMed] [Google Scholar]
- 8.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12(2):84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 10.Qin Y, Sun W, Wang Z, Dong W, He L, Zhang T, Zhang H. Long non-coding small nucleolar RNA host genes (SNHGs) in endocrine-related cancers. Onco Targets Ther. 2020;13:7699–7717. doi: 10.2147/OTT.S267140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu B, Mei J, Ji W, Bian Z, Jiao J, Sun J, Shao J. LncRNA SNHG3, a potential oncogene in human cancers. Cancer Cell Int. 2020;20(1):536. doi: 10.1186/s12935-020-01608-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thin KZ, Tu JC, Raveendran S. Long non-coding SNHG1 in cancer. Clin Chim Acta. 2019;494:38–47. doi: 10.1016/j.cca.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Huang L, Jiang X, Wang Z, Zhong X, Tai S, Cui Y. Small nucleolar RNA host gene 1: a new biomarker and therapeutic target for cancers. Pathol Res Pract. 2018;214(9):1247–1252. doi: 10.1016/j.prp.2018.07.033. [DOI] [PubMed] [Google Scholar]
- 14.Xiao B, Huang Z, Zhou R, Zhang J, Yu B. The prognostic value of expression of the long noncoding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) in patients with solid malignant tumors: a systematic review and meta-analysis. Med Sci Monit. 2018;24:5462–5472. doi: 10.12659/MSM.911687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YH, Hu YQ, Wang SC, Li Y, Chen DM. LncRNA SNHG5: a new budding star in human cancers. Gene. 2020;749:144724. doi: 10.1016/j.gene.2020.144724. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Tian B, Tang J, Wu J, Wang H, Wu Z, Li X, Yang D, Zhang B, Xiao Y, et al. SNHG7: a novel vital oncogenic lncRNA in human cancers. Biomed Pharmacother. 2020;124:109921. doi: 10.1016/j.biopha.2020.109921. [DOI] [PubMed] [Google Scholar]
- 17.Tamang S, Acharya V, Roy D, Sharma R, Aryaa A, Sharma U, Khandelwal A, Prakash H, Vasquez KM, Jain A. SNHG12: an LncRNA as a potential therapeutic target and biomarker for human cancer. Front Oncol. 2019;9:901. doi: 10.3389/fonc.2019.00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Y, Xiao T, Ou W, Wu Z, Wu J, Tang J, Tian B, Zhou Y, Su M, Wang W. LncRNA SNHG16 as a potential biomarker and therapeutic target in human cancers. Biomark Res. 2020;8:41. doi: 10.1186/s40364-020-00221-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Qin M, Li J, Cui S. LncRNA SNHG4 sponges miR-200b to inhibit cell apoptosis in diabetic retinopathy. Arch Physiol Biochem. 2021 doi: 10.1080/13813455.2021.1900873. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Sun WC, Liang ZD, Yin XR, Ji ZR, Chen XH, Wei MJ, Pei L. LncRNA SNHG4 attenuates inflammatory responses by sponging miR-449c-5p and up-regulating STAT6 in microglial during cerebral ischemia-reperfusion injury. Drug Des Devel Ther. 2020;14:3683–3695. doi: 10.2147/DDDT.S245445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Z, Wang W. LncRNA SNHG4 regulates miR-10a/PTEN to inhibit the proliferation of acute myeloid leukemia cells. Hematology. 2020;25(1):160–164. doi: 10.1080/16078454.2020.1754636. [DOI] [PubMed] [Google Scholar]
- 22.Pan X, Shen C, Huang Y, Wang L, Xia Z. Loss of SNHG4 attenuated spinal nerve ligation-triggered neuropathic pain through sponging miR-423-5p. Mediators Inflamm. 2020;2020:2094948. doi: 10.1155/2020/2094948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horikawa A, Yoneda T, Yaoita E, Yamaguchi K, Shigenobu S, Kuramochi M, Yamate J, Inui T, Ishibashi O. A novel splicing variant of small nucleolar RNA host gene 4 is a podocyte-selective non-coding RNA upregulated in response to puromycin aminonucleoside-induced podocyte injury. J Biochem. 2019;165(5):447–454. doi: 10.1093/jb/mvy118. [DOI] [PubMed] [Google Scholar]
- 24.Brinkkoetter PT, Ising C, Benzing T. The role of the podocyte in albumin filtration. Nat Rev Nephrol. 2013;9(6):328–336. doi: 10.1038/nrneph.2013.78. [DOI] [PubMed] [Google Scholar]
- 25.Gong J, Li Y, Liu CJ, Xiang Y, Li C, Ye Y, Zhang Z, Hawke DH, Park PK, Diao L, et al. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. 2017;21(7):1968–1981. doi: 10.1016/j.celrep.2017.10.070. [DOI] [PubMed] [Google Scholar]
- 26.Yang H, Jiang Z, Wang S, Zhao Y, Song X, Xiao Y, Yang S. Long non-coding small nucleolar RNA host genes in digestive cancers. Cancer Med. 2019;8(18):7693–7704. doi: 10.1002/cam4.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimta AA, Tigu AB, Braicu C, Stefan C, Ionescu C, Berindan-Neagoe I. An emerging class of long non-coding RNA with oncogenic role arises from the snoRNA host genes. Front Oncol. 2020;10:389. doi: 10.3389/fonc.2020.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F, Quan Q. The long non-coding RNA SNHG4/microRNA-let-7e/KDM3A/p21 pathway is involved in the development of non-small cell lung cancer. Mol Ther Oncolytics. 2021;20:634–645. doi: 10.1016/j.omto.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhry MA. Small nucleolar RNA host genes and long non-coding RNA responses in directly irradiated and bystander cells. Cancer Biother Radiopharm. 2014;29(3):135–141. doi: 10.1089/cbr.2013.1574. [DOI] [PubMed] [Google Scholar]
- 30.Li SX, Yan W, Liu JP, Zhao YJ, Chen L. Long noncoding RNA SNHG4 remits lipopolysaccharide-engendered inflammatory lung damage by inhibiting METTL3—mediated m(6)A level of STAT2 mRNA. Mol Immunol. 2021;139:10–22. doi: 10.1016/j.molimm.2021.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Cheng XB, Zhang T, Zhu HJ, Ma N, Sun XD, Wang SH, Jiang Y. Knockdown of lncRNA SNHG4 suppresses gastric cancer cell proliferation and metastasis by targeting miR-204-5p. Neoplasma. 2021;68(3):546–556. doi: 10.4149/neo_2021_200914N981. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Liu T, Sun L, Zhang S, Dong G. Long noncoding RNA SNHG4 promotes renal cell carcinoma tumorigenesis and invasion by acting as ceRNA to sponge miR-204-5p and upregulate RUNX2. Cancer Cell Int. 2020;20:514. doi: 10.1186/s12935-020-01606-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Tian W, Wu L, Wei Z, Li W, Xu Y, Li Y. LncRNA SNHG4 regulates miR-138/c-Met axis to promote the proliferation of glioblastoma cells. NeuroReport. 2020;31(9):657–662. doi: 10.1097/WNR.0000000000001469. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Guo JF, Zhang ML, Li AM. LncRNA SNHG4 promotes neuroblastoma proliferation, migration, and invasion by sponging miR-377-3p. Neoplasma. 2020;67(5):1054–1062. doi: 10.4149/neo_2020_191023N1081. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Z, Tan F, Pei Q, Li C, Zhou Y, Li Y, Pei H. lncRNA SNHG4 modulates colorectal cancer cell cycle and cell proliferation through regulating miR-590-3p/CDK1 axis. Aging. 2021;13(7):9838–9858. doi: 10.18632/aging.202737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du M, Zheng R, Ma G, Chu H, Lu J, Li S, Xin J, Tong N, Zhang G, Wang W, et al. Remote modulation of lncRNA GCLET by risk variant at 16p13 underlying genetic susceptibility to gastric cancer. Sci Adv. 2020;6(21):eaay5525. doi: 10.1126/sciadv.aay5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seidlitz T, Koo BK, Stange DE. Gastric organoids-an in vitro model system for the study of gastric development and road to personalized medicine. Cell Death Differ. 2021;28(1):68–83. doi: 10.1038/s41418-020-00662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Fu M, Liu J, Chong W, Fang Z, Du F, Liu Y, Shang L, Li L. The role and application of small extracellular vesicles in gastric cancer. Mol Cancer. 2021;20(1):71. doi: 10.1186/s12943-021-01365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Xu Q, Wang W, Sun S. MIR100HG: a credible prognostic biomarker and an oncogenic lncRNA in gastric cancer. 2019. Biosci Rep. 10.1042/BSR20190171. [DOI] [PMC free article] [PubMed]
- 40.Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ, Cheng XD. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer. 2020;19(1):96. doi: 10.1186/s12943-020-01219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K, Huang J, Ni J, Song D, Ding M, Wang J, Huang X, Li W. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle. 2017;16(6):578–587. doi: 10.1080/15384101.2017.1288324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5(22):10976–10996. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou C, Duan S. The role of long non-coding RNA NNT-AS1 in neoplastic disease. Cancers. 2020 doi: 10.3390/cancers12113086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Zhu W, Qiu J, Chen F. lncRNA SNHG4 promotes cell proliferation, migration, invasion and the epithelial-mesenchymal transition process via sponging miR-204-5p in gastric cancer. Mol Med Rep. 2021 doi: 10.3892/mmr.2020.11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choueiri TK, Vaishampayan U, Rosenberg JE, Logan TF, Harzstark AL, Bukowski RM, Rini BI, Srinivas S, Stein MN, Adams LM, et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol. 2013;31(2):181–186. doi: 10.1200/JCO.2012.43.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh D. Current updates and future perspectives on the management of renal cell carcinoma. Life Sci. 2021;264:118632. doi: 10.1016/j.lfs.2020.118632. [DOI] [PubMed] [Google Scholar]
- 47.Guo R, Zou B, Liang Y, Bian J, Xu J, Zhou Q, Zhang C, Chen T, Yang M, Wang H, et al. LncRNA RCAT1 promotes tumor progression and metastasis via miR-214-5p/E2F2 axis in renal cell carcinoma. Cell Death Dis. 2021;12(7):689. doi: 10.1038/s41419-021-03955-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin C, Shi L, Li K, Liu W, Qiu Y, Zhao Y, Zhao B, Li Z, Li Y, Zhu Q. Mechanism of tumor-derived extracellular vesicles in regulating renal cell carcinoma progression by the delivery of MALAT1. Oncol Rep. 2021 doi: 10.3892/or.2021.8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H, Li W, Gu W, Yan Y, Yao X, Zheng J. MALAT1 accelerates the development and progression of renal cell carcinoma by decreasing the expression of miR-203 and promoting the expression of BIRC5. Cell Prolif. 2019;52(5):e12640. doi: 10.1111/cpr.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159(7):1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu F, Chen Y, Zhao C, Wang H, He D, Xu L, Wang J, He X, Deng Y, Lu EE, et al. Olig2-dependent reciprocal shift in PDGF and EGF receptor signaling regulates tumor phenotype and mitotic growth in malignant glioma. Cancer Cell. 2016;29(5):669–683. doi: 10.1016/j.ccell.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen PY, Touat M, Alexander BM, Mellinghoff IK, Ramkissoon S, McCluskey CS, Pelton K, Haidar S, Basu SS, Gaffey SC, et al. Buparlisib in patients with recurrent glioblastoma harboring phosphatidylinositol 3-kinase pathway activation: an open-label, multicenter, multi-arm, phase II trial. J Clin Oncol. 2019;37(9):741–750. doi: 10.1200/JCO.18.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith AA, Huang YT, Eliot M, Houseman EA, Marsit CJ, Wiencke JK, Kelsey KT. A novel approach to the discovery of survival biomarkers in glioblastoma using a joint analysis of DNA methylation and gene expression. Epigenetics. 2014;9(6):873–883. doi: 10.4161/epi.28571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Y, Tian M, Liu J, Wang K. LINC00511 facilitates Temozolomide resistance of glioblastoma cells via sponging miR-126-5p and activating Wnt/β-catenin signaling. J Biochem Mol Toxicol. 2021 doi: 10.1002/jbt.22848. [DOI] [PubMed] [Google Scholar]
- 56.Yin T, Wu J, Hu Y, Zhang M, He J. Long non-coding RNA HULC stimulates the epithelial-mesenchymal transition process and vasculogenic mimicry in human glioblastoma. Cancer Med. 2021;10(15):5270–5282. doi: 10.1002/cam4.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drozdz-Afelt JM, Koim-Puchowska B, Klosowski G, Kaminski P. Polymorphism of glutathione S-transferase in the population of Polish patients with carcinoma of the prostate. Environ Sci Pollut Res Int. 2020;27(16):19375–19382. doi: 10.1007/s11356-020-08435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia L, Bouamar H, Gu X, Zeballos C, Qin T, Wang B, Zhou Y, Wang Y, Yang J, Zhu H, et al. Gli2 mediates the development of castration-resistant prostate cancer. Int J Oncol. 2020;57(1):100–112. doi: 10.3892/ijo.2020.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S, Sugawara Y, Chen S, Beelman RB, Tsuduki T, Tomata Y, Matsuyama S, Tsuji I. Mushroom consumption and incident risk of prostate cancer in Japan: a pooled analysis of the Miyagi Cohort Study and the Ohsaki Cohort Study. Int J Cancer. 2020;146(10):2712–2720. doi: 10.1002/ijc.32591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Li J, Ma Y, Xu C, Wang Y, He Y. MicroRNA miR-145-5p inhibits phospholipase D 5 (PLD5) to downregulate cell proliferation and metastasis to mitigate prostate cancer. Bioengineered. 2021;12(1):3240–3251. doi: 10.1080/21655979.2021.1945361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Piano M, Manuelli V, Zadra G, Otte J, Edqvist PD, Pontén F, Nowinski S, Niaouris A, Grigoriadis A, Loda M, et al. Lipogenic signalling modulates prostate cancer cell adhesion and migration via modification of Rho GTPases. Oncogene. 2020;39(18):3666–3679. doi: 10.1038/s41388-020-1243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cai X, Dai Y, Gao P, Ren G, Cheng D, Wang B, Wang Y, Yu J, Du Y, Wang X, et al. LncRNA CCAT1 promotes prostate cancer cells proliferation, migration, and invasion through regulation of miR-490-3p/FRAT1 axis. Aging. 2021;13(14):18527–18544. doi: 10.18632/aging.203300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang ZY, Duan Y, Wang P. SP1-mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR-377 to facilitate prostate cancer progression through regulation of ZIC5. J Cell Physiol. 2020;235(4):3916–3927. doi: 10.1002/jcp.29285. [DOI] [PubMed] [Google Scholar]
- 64.Mirabello L, Koster R, Moriarity BS, Spector LG, Meltzer PS, Gary J, Machiela MJ, Pankratz N, Panagiotou OA, Largaespada D, et al. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Discov. 2015;5(9):920–931. doi: 10.1158/2159-8290.CD-15-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S, Li H, Chen S, Wang Z, Yao Y, Chen T, Ye Z, Lin P. Andrographolide induces apoptosis in human osteosarcoma cells via the ROS/JNK pathway. Int J Oncol. 2020;56(6):1417–1428. doi: 10.3892/ijo.2020.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Durfee RA, Mohammed M, Luu HH. Review of osteosarcoma and current management. Rheumatol Ther. 2016;3(2):221–243. doi: 10.1007/s40744-016-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Y, Xu J, Pan X, Zhang Y, Weng Y, Zhou D, He S. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020;11(4):278. doi: 10.1038/s41419-020-2485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.ESMO/European Sarcoma Network Working Group Bone sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25 Suppl 3:iii113–123. doi: 10.1093/annonc/mdu256. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Wang F, Wang L, Zhang Q. MiR-363 suppresses cell migration, invasion, and epithelial-mesenchymal transition of osteosarcoma by binding to NOB1. World J Surg Oncol. 2020;18(1):83. doi: 10.1186/s12957-020-01859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu R, Feng F, Yu X, Liu Z, Lao L. LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma. Cell Prolif. 2018;51(6):e12515. doi: 10.1111/cpr.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang YF, Lu L, Shen HL, Lu XX. LncRNA SNHG4 promotes osteosarcoma proliferation and migration by sponging miR-377-3p. Mol Genet Genomic Med. 2020;8(8):e1349. doi: 10.1002/mgg3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 73.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132(5):1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 74.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Supp(0):S2–6. doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cornella H, Alsinet C, Sayols S, Zhang Z, Hao K, Cabellos L, Hoshida Y, Villanueva A, Thung S, Ward SC, et al. Unique genomic profile of fibrolamellar hepatocellular carcinoma. Gastroenterology. 2015;148(4):806–18.e810. doi: 10.1053/j.gastro.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suk KT, Mederacke I, Gwak GY, Cho SW, Adeyemi A, Friedman R, Schwabe RF. Opposite roles of cannabinoid receptors 1 and 2 in hepatocarcinogenesis. Gut. 2016;65(10):1721–1732. doi: 10.1136/gutjnl-2015-310212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu Q, Yang H, Cheng P, Han Q. Bioinformatic analysis of the prognostic value of the lncRNAs encoding snoRNAs in hepatocellular carcinoma. BioFactors. 2019;45(2):244–252. doi: 10.1002/biof.1478. [DOI] [PubMed] [Google Scholar]
- 79.Jiao Y, Li Y, Jia B, Chen Q, Pan G, Hua F, Liu Y. The prognostic value of lncRNA SNHG4 and its potential mechanism in liver cancer. 2020. Biosci Rep. 10.1042/BSR20190729. [DOI] [PMC free article] [PubMed]
- 80.Li H, Hong J, Wijayakulathilaka W. Long non-coding RNA SNHG4 promotes cervical cancer progression through regulating c-Met via targeting miR-148a-3p. Cell Cycle. 2019;18(23):3313–3324. doi: 10.1080/15384101.2019.1674071. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Liu Y, Huang X, Lu D, Feng Y, Xu R, Li X, Yin C, Xue B, Zhao H, Wang S, et al. LncRNA SNHG4 promotes the increased growth of endometrial tissue outside the uterine cavity via regulating c-Met mediated by miR-148a-3p. Mol Cell Endocrinol. 2020;514:110887. doi: 10.1016/j.mce.2020.110887. [DOI] [PubMed] [Google Scholar]
- 82.Tang Y, Wu L, Zhao M, Zhao G, Mao S, Wang L, Liu S, Wang X. LncRNA SNHG4 promotes the proliferation, migration, invasiveness, and epithelial-mesenchymal transition of lung cancer cells by regulating miR-98-5p. Biochem Cell Biol. 2019;97(6):767–776. doi: 10.1139/bcb-2019-0065. [DOI] [PubMed] [Google Scholar]
- 83.López-Sáez JF, de la Torre C, Pincheira J, Giménez-Martín G. Cell proliferation and cancer. Histol Histopathol. 1998;13(4):1197–1214. doi: 10.14670/HH-13.1197. [DOI] [PubMed] [Google Scholar]
- 84.Morgan EL, Patterson MR, Ryder EL, Lee SY, Wasson CW, Harper KL, Li Y, Griffin S, Blair GE, Whitehouse A, et al. MicroRNA-18a targeting of the STK4/MST1 tumour suppressor is necessary for transformation in HPV positive cervical cancer. PLoS Pathog. 2020;16(6):e1008624. doi: 10.1371/journal.ppat.1008624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ersvær E, Kildal W, Vlatkovic L, Cyll K, Pradhan M, Kleppe A, Hveem TS, Askautrud HA, Novelli M, Wæhre H, et al. Prognostic value of mitotic checkpoint protein BUB3, cyclin B1, and pituitary tumor-transforming 1 expression in prostate cancer. Mod Pathol. 2020;33(5):905–915. doi: 10.1038/s41379-019-0418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med. 2016;94(12):1313–1326. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9(1):13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 88.Yang C, Zhang J, Ding M, Xu K, Li L, Mao L, Zheng J. Ki67 targeted strategies for cancer therapy. Clin Transl Oncol. 2018;20(5):570–575. doi: 10.1007/s12094-017-1774-3. [DOI] [PubMed] [Google Scholar]
- 89.Ishibashi N, Maebayashi T, Aizawa T, Sakaguchi M, Nishimaki H, Masuda S. Correlation between the Ki-67 proliferation index and response to radiation therapy in small cell lung cancer. Radiat Oncol. 2017;12(1):16. doi: 10.1186/s13014-016-0744-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14(3):2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tu B, Du L, Fan QM, Tang Z, Tang TT. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012;325(1):80–88. doi: 10.1016/j.canlet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 92.Mori H, Sugie S, Yoshimi N, Hara A, Tanaka T. Control of cell proliferation in cancer prevention. Mutat Res. 1999;428(1–2):291–298. doi: 10.1016/S1383-5742(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 93.Tochhawng L, Deng S, Pervaiz S, Yap CT. Redox regulation of cancer cell migration and invasion. Mitochondrion. 2013;13(3):246–253. doi: 10.1016/j.mito.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 94.Lu Y, Sha H, Sun X, Zhang Y, Wu Y, Zhang J, Zhang H, Wu J, Feng J. CRNDE: an oncogenic long non-coding RNA in cancers. Cancer Cell Int. 2020;20:162. doi: 10.1186/s12935-020-01246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mak M, Spill F, Kamm RD, Zaman MH. Single-cell migration in complex microenvironments: mechanics and signaling dynamics. J Biomech Eng. 2016;138(2):021004. doi: 10.1115/1.4032188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krakhmal NV, Zavyalova MV, Denisov EV, Vtorushin SV, Perelmuter VM. Cancer invasion: patterns and mechanisms. Acta Naturae. 2015;7(2):17–28. doi: 10.32607/20758251-2015-7-2-17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K, Willbanks A, Sarkar S. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Singh M, Yelle N, Venugopal C, Singh SK. EMT: mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 100.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 101.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27(20):2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 103.Gao J, Liu R, Feng D, Huang W, Huo M, Zhang J, Leng S, Yang Y, Yang T, Yin X, et al. Snail/PRMT5/NuRD complex contributes to DNA hypermethylation in cervical cancer by TET1 inhibition. Cell Death Differ. 2021 doi: 10.1038/s41418-021-00786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Call JA, Eckhardt SG, Camidge DR. Targeted manipulation of apoptosis in cancer treatment. Lancet Oncol. 2008;9(10):1002–1011. doi: 10.1016/S1470-2045(08)70209-2. [DOI] [PubMed] [Google Scholar]
- 105.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147(4):742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12(7):439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 107.Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D’Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging. 2016;8(4):603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20(3):175–193. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carrà G, Lingua MF, Maffeo B, Taulli R, Morotti A. P53 vs NF-κB: the role of nuclear factor-kappa B in the regulation of p53 activity and vice versa. Cell Mol Life Sci. 2020;77(22):4449–4458. doi: 10.1007/s00018-020-03524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lavy M, Gauttier V, Poirier N, Barillé-Nion S, Blanquart C. Specialized pro-resolving mediators mitigate cancer-related inflammation: role of tumor-associated macrophages and therapeutic opportunities. Front Immunol. 2021;12:702785. doi: 10.3389/fimmu.2021.702785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lucas SM, Rothwell NJ, Gibson RM. The role of inflammation in CNS injury and disease. Br J Pharmacol. 2006;147 Suppl 1(Suppl 1):S232–240. doi: 10.1038/sj.bjp.0706400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7(12):13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan Y, Shi Q, Yuan X, Xue C, Shen S, He Y. DANCR: an emerging therapeutic target for cancer. Am J Transl Res. 2020;12(7):4031–4042. [PMC free article] [PubMed] [Google Scholar]
- 116.Su K, Wang N, Shao Q, Liu H, Zhao B, Ma S. The role of a ceRNA regulatory network based on lncRNA MALAT1 site in cancer progression. Biomed Pharmacother. 2021;137:111389. doi: 10.1016/j.biopha.2021.111389. [DOI] [PubMed] [Google Scholar]
- 117.Chen Y, Lin Y, Bai Y, Cheng D, Bi Z. A long noncoding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network identifies eight lncRNA biomarkers in patients with osteoarthritis of the knee. Med Sci Monit. 2019;25:2058–2065. doi: 10.12659/MSM.915555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang C, Zheng J, Liu X, Xue Y, He Q, Dong Y, Wang D, Li Z, Liu L, Ma J, et al. Role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glioma angiogenesis via Staufen1-mediated mRNA decay. Mol Ther Nucleic Acids. 2020;20:866–878. doi: 10.1016/j.omtn.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qu C, Dai C, Guo Y, Qin R, Liu J. Long non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role in the regulatory mechanism of ovarian cancer. 2020. Biosci Rep. 10.1042/BSR20200800. [DOI] [PMC free article] [PubMed]
- 121.Xu Z, Wu Z, Xu J, Zhang J, Yu B. Identification of hub driving genes and regulators of lung adenocarcinoma based on the gene co-expression network. 2020. Biosci Rep. 10.1042/BSR20200295. [DOI] [PMC free article] [PubMed]
- 122.Wang FY, Kang CS, Wang-Gou SY, Huang CH, Feng CY, Li XJ. EGFL7 is an intercellular EGFR signal messenger that plays an oncogenic role in glioma. Cancer Lett. 2017;384:9–18. doi: 10.1016/j.canlet.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 123.Zhao H, Ahirwar DK, Oghumu S, Wilkie T, Powell CA, Nasser MW, Satoskar AR, Li DY, Ganju RK. Endothelial Robo4 suppresses breast cancer growth and metastasis through regulation of tumor angiogenesis. Mol Oncol. 2016;10(2):272–281. doi: 10.1016/j.molonc.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data in our study are available upon request.