Abstract
Background
Kuruma shrimp, a major commercial shrimp species in the world, has two cryptic or sibling species, Marsupenaeus japonicus and Marsupenaeus pulchricaudatus. Codon usage analysis would contribute to our understanding of the genetic and evolutionary characteristics of the two Marsupenaeus species. In this study, we analyzed codon usage and related indices using coding sequences (CDSs) from RNA-seq data.
Results
Using CodonW 1.4.2 software, we performed the codon bias analysis of transcriptomes obtained from hepatopancreas tissues, which indicated weak codon bias. Almost all parameters had similar correlations for both species. The gene expression level (FPKM) was negatively correlated with A/T3s. We determined 12 and 14 optimal codons for M. japonicus and M. pulchricaudatus, respectively, and all optimal codons have a C/G-ending. The two Marsupenaeus species had different usage frequencies of codon pairs, which contributed to further analysis of transcriptional differences between them. Orthologous genes that underwent positive selection (ω > 1) had a higher correlation coefficient than that of experienced purifying selection (ω < 1). Parity Rule 2 (PR2) and effective number of codons (ENc) plot analysis showed that the codon usage patterns of both species were influenced by both mutations and selection. Moreover, the average observed ENc value was lower than the expected value for both species, suggesting that factors other than GC may play roles in these phenomena. The results of multispecies clustering based on codon preference were consistent with traditional classification.
Conclusions
This study provides a relatively comprehensive understanding of the correlations among codon usage bias, gene expression, and selection pressures of CDSs for M. japonicus and M. pulchricaudatus. The genetic evolution was driven by mutations and selection pressure. Moreover, the results point out new insights into the specificities and evolutionary characteristics of the two Marsupenaeus species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-08106-y.
Keywords: Codon usage pattern, Marsupenaeus japonicus, Marsupenaeus pulchricaudatus, Orthologous genes, Phylogenetics
Background
The codon is the basic information unit for translation of messenger RNA (mRNA), and 62 codons encode 20 different amino acids [1–3]. For different genes or genomes, the selection of synonymous codons is nonrandom, which is called codon usage bias (CUB) [4, 5]. Codon preference is specific to the organism and may be influenced by GC content, gene expression level, and gene length [6–8]. In addition, codon usage patterns may affect the biological functions of mRNA biosynthesis, translation elongation rate, protein folding, and other downstream expressions [7, 9–12]. It is now thought that CUB is mainly affected by selection and mutational pressure [13–17]. Vicario et al. inferred that selection has acted on codon usage in the genus Drosophila, at least often enough to leave a footprint of selection in modern genomes [18]. Correspondence analysis proved that both selection and mutation pressure affect the codon usage pattern in Bungarus species [19]. Translational selection shapes codon and amino acid usage in three Pancrustacean arthropods [20]. In general, the pattern of codon usage is similar among closely related species but differs significantly among distantly related organisms [3, 18, 21–23]. Based on relative synonymous codon usage (RSCU) values, 27 species were clustered into two primary groups, which was consistent with the evolutionary status of these species [24]. According to these mentioned studies, codon usage showed evolutionary conservation and could be used for taxonomic differentiation.
The majority of past researches has studied the codon preference of species with genome-wide information [25–27]. Recent rapid development of next-generation sequencing has provided large amounts of genomic and transcriptome data. Machado et al., detected and quantified strong selection on synonymous sites of Drosophila melanogaster by using deep genomic population sequencing [28]. Utilizing Ribo-seq and RNA-seq approaches, Chu et al., studied how codon usage bias could impact the translation patterns of Arabidopsis thaliana [29]. Guan et al., analyzed codon usage of Hirudinaria manillensis RNA-seq data and found that genetic evolution was driven by mutation pressure and selection [30]. Based on the transcriptional sequence, Yi et al., found that the expression-linked patterns of codon usage revealed that higher expression was associated with higher GC3 and lower effective number of codons (ENC) [24]. Additional studies of codon usage bias based on transcriptome data include Bombyx mori [31], Taenia multiceps [32], and Megalobrama amblycephala [33].
The kuruma shrimp (Marsupenaeus japonicus) includes two cryptic species, distributed allopatrically but co-occurring in the northern South China Sea [34]. Previous studies showed obvious genetic differentiation between both shrimp species [35, 36]. Transcriptome analyses for these Marsupenaeus species evidenced a large number of putative orthologs, and the divergence time between M. japonicus and M. pulchricaudatus was approximately 0.26–0.69 Mya according to the peak of synonymous rates [37]. In Arachis duranensis and Arachis ipaënsis, Song et al., found the complex correlation among gene expression, codon usage bias, and substitution rate orthologs [38]. Orthologous genes typically perform equivalent functions across different species, which are closely related to gene expression [39]. However, the relationship between differentially expressed genes and codon usage patterns is still unknown in Marsupenaeus species.
This study performed codon usage bias analysis based on transcriptomes from M. japonicus and M. pulchricaudatus using CodonW software. We systematically compared the codon usage patterns of the two Marsupenaeus species and evaluated the comprehensive effects of various factors, including GC content, gene expression levels and gene length. The results provide new insights into the genetic divergence and the phylogenetic relationships of these two Marsupenaeus species.
Results
Nucleotide composition and PR2-plot analysis
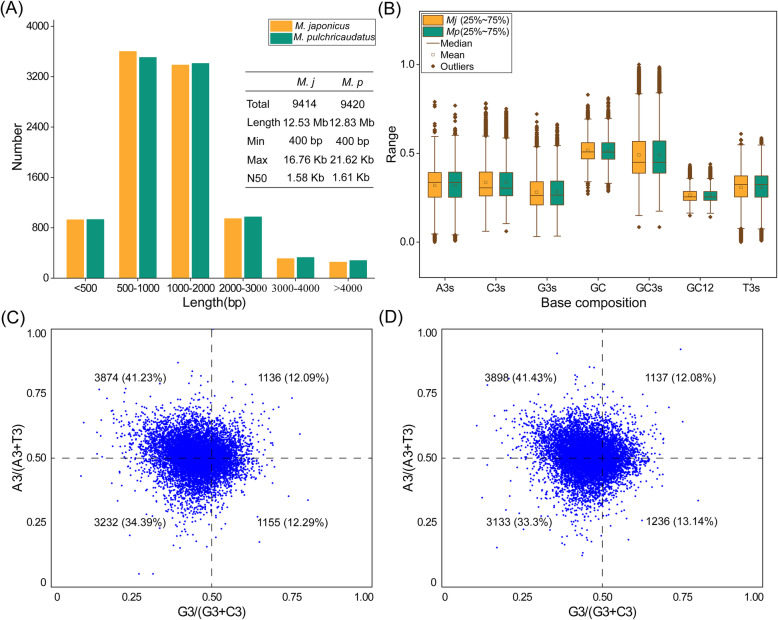
A total of 9414 and 9420 unigenes with lengths larger than 400 bp were screened from M. japonicus and M. pulchricaudatus libraries, respectively (Fig. 1a). The length distribution of the two groups was similar. In the M. japonicus, the mean contents of A and T nucleotides were 31.89% (SD = 10.45%) and 30.8% (SD = 9.04%), respectively, and the mean contents of C and G nucleotides were 33.63% (SD = 10.71%) and 28.03% (SD = 9.21%), respectively. In the M. pulchricaudatus, the average contents of A and T nucleotides were 31.86% (SD = 10.55%) and 30.8% (SD = 9.18%) respectively, and the average contents of C and G nucleotides were 33.56% (SD = 10.73%) and 28.27% (SD = 9.29%), respectively (Fig. 1b). The average contents of GC were 51.61 and 51.54% for M. japonicus and M. pulchricaudatus, respectively. The mean contents of GC3s were 49.1 and 49.17% for M. japonicus and M. pulchricaudatus, respectively, which were significantly higher than that of GC12. For M. japonicus and M. pulchricaudatus, the median of GC biases [G3/(G3 + C3)] were 0.4563 and 0.4582, and the median of AT biases [A3/(A3 + T3)] were 0.5047 and 0.5051, respectively (Fig. 1c, d). Parity Rule 2 (PR2) plot analysis showed that purines (A and G) were used more frequently than pyrimidines (C and T) in the two Marsupenaeus species. The unbalanced use of the third base suggested that mutation pressure and selection contribute to codon usage bias.
Fig. 1.
Length distribution of unigenes (a), nucleotide composition (b), and PR2 plot analysis for M. japonicus (c) and M. pulchricaudatus (d)
Correlation analysis of codon usage parameters
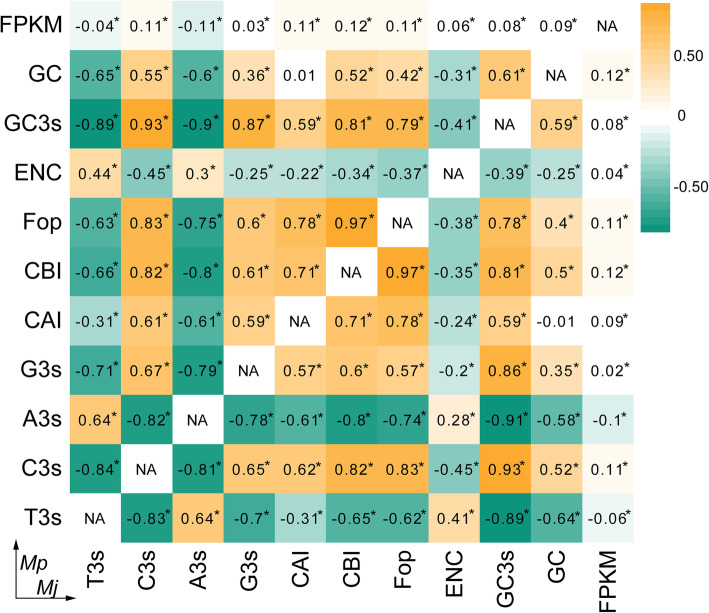
All parameters had similar correlations between M. japonicus and M. pulchricaudatus (Fig. 2). The results indicated that FPKM was negatively correlated with T3s and A3s (p < 0.05) and positively correlated with other parameters (p < 0.05) in M. japonicus and M. pulchricaudatus. There was a significant (p < 0.05) positive correlation among T3s, A3s, and ENc values. These three values were negatively (p < 0.05) correlated with other parameters. Correlation analysis indicated that the third base content of synonymous codons significantly affects gene expression and codon usage bias. The significant correlation (p < 0.05) between GC3 and GC content indicated that the nucleotide contents play an important role in codon usage bias. The first and second base contents were often determined by selection and the third base content was affected by mutation pressure [40, 41].
Fig. 2.
Correlation analysis of codon usage parameters. Significant difference at p < 0.05; ** significant difference at p < 0.01
The average ENc values were 52.1 and 52.22 for M. japonicus and M. pulchricaudatus, respectively. The number of genes with ENc values equal to 61was 268 (2.85%) and 249 (2.64%) for M. japonicus and M. pulchricaudatus, which indicates that all synonymous codons have the same probability. The number of genes with ENc values less than 35 was 187 (1.99%) for M. japonicus and 133 (1.41%) for M. pulchricaudatus, while the minimum values were 23.6 and 27.46, for these species, respectively. The S1|c21076_g1 unigene sequence had the lowest ENc, with 23.6 for M. japonicus. The gene was annotated as nesprins-1 (nuclear envelope spectrin repeat 1), a new member of the nuclear membrane protein family. The S2|c17052_g1 sequence had the lowest ENc with 27.46 for M. pulchricaudatus, which was annotated as the vrille (vri) gene.
A value of 35 was the standard for codon bias [42, 43]. To explore the effect of GC3s on codon usage bias, we performed ENc plot analysis. In Fig. S1A and S1B, most genes were aggregated close to the expected curve, which showed that codon usage bias was mainly affected by mutation pressure. We found lower ENc values in M. japonicus than in M. pulchricaudatus. Meanwhile, we estimated the difference between the expected and the observed ENc values and calculated the (ENcexp - ENcobs)/ENcexp (Fig. S1c, d). The frequency distribution of unigenes with values within 0–0.1 was highest, which showed that most ENc values from GC3s were larger than the observed ENc values. For M. japonicus, the average observed and expected ENc values were 52.1 and 56.67, respectively, and for M. pulchricaudatus, these values were 52.2 and 56.59, respectively. Moreover, there was a significant positive correlation between GC3s and CAI values (Fig. S1e, f).
Gene ontology (GO) annotation based on GC3s
To further understand the influence of GC3s on gene function, we performed GO annotation for the CDSs with low, mid, and high GC3, including 1000, 1001, and 1005 genes in M. japonicus and 1005, 1001, and 1002 genes in M. pulchricaudatus. The gene ontology terms presented similar functional categories for both shrimp species (Fig. S2). The biological process categories, including 13 subtypes and most corresponding genes, were involved in cellular processes, metabolic processes, single-organism processes, and biological regulation. Thirteen subtypes were annotated with cellular component, and the highest gene number was observed in the “cell part” and “cell” categories. In the molecular function category, the “binding” was the highest category.
Correlation analysis between codon usage parameters and the substitution rate
A total of 5036 pairs of single-copy orthologous genes were previously identified between the M. japonicus and M. pulchricaudatus libraries [37]. Among these orthologs, the Ka/Ks values of 2491 pairs were calculated, showing mean values equal to 0.002, 0.019, and 0.175 for Ka, Ks, and Ka/Ks (ω), respectively. There were 49 pairs of orthologous genes with a ω value greater than 1 (positive selection) and 2225 pairs with a ω value less than 1 (purifying selection).
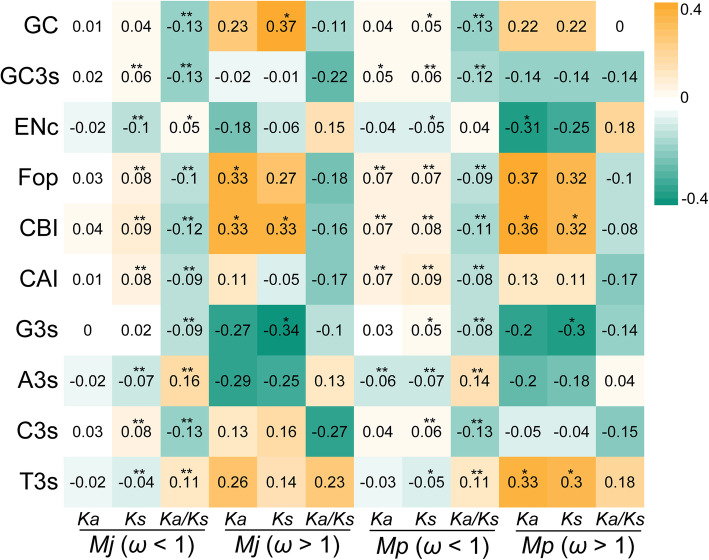
Overall, orthologous genes that underwent positive selection (ω > 1) had a higher correlation coefficient than those that experienced purifying selection (ω < 1), which could be because more genes with ω < 1 lead to large differences. Almost all parameters had different significance levels with Ka, Ks, or Ka/Ks (Fig. 3). In M. japonicus, the Ka/Ks of genes with ω less than 1 was positively correlated with ENc, A3s, and T3s (p < 0.01) but negatively correlated with other parameters (p < 0.01). There was no significant correlation between any parameters and the Ka/Ks of genes with ω greater than 1. However, GC content and CBI value were positively correlated with Ks, and G3s was negatively correlated with Ks. In addition, Fop and CBI values were positively correlated with Ka. In M. pulchricaudatus, the Ka/Ks of genes with ω less than 1 were positively correlated with A3s and T3s but negatively correlated with other parameters. Similar to M. japonicus, there was no significant correlation between all parameters and the Ka/Ks of genes with ω greater than 1. However, CBI and T3s values were positively correlated with Ka and Ks.
Fig. 3.
Correlation analysis between Ka and Ks values and codon preference. *Significant difference at p < 0.05; ** significant difference at p < 0.01
Correspondence analysis (COA)
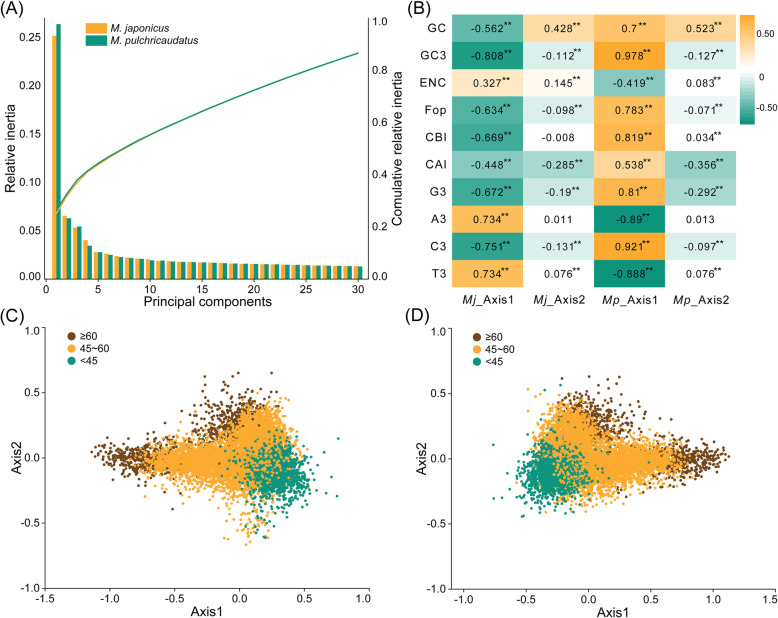
Based on the RSCU values, correspondence analysis was used to investigate the factors related to codon usage patterns and to reflect the variation trend in codon usage. The results indicated that the first five axes accounted for 43.8 and 44.3% of the amino-acid variation for M. japonicus and M. pulchricaudatus, respectively (Fig. 4a). In M. japonicus, Axis 1 and Axis 2 explained 25.16 and 6.54% of the variance, respectively. In M. pulchricaudatus, Axis 1 and Axis 2 explained 26.38 and 6.29% of the variance, respectively. In M. japonicus, the relationships were highly significantly positive between Axis 1 and A3, T3, and ENc (p < 0.01), and others were significantly negatively correlated (p < 0.01) (Fig. 4b). In M. pulchricaudatus, the relationships were highly significantly negative between Axis 1 and A3, T3, and ENc (p < 0.01) (Fig. 4b).
Fig. 4.
Correspondence analysis (a), correlation analysis (b) and GC content effect codon preference of M. japonicus (c) and M. pulchricaudatus (d). Different colors represent different GC contents, green represents GC% < 45%, yellow represents GC between 45and 60, and brown represents GC% > 60%
To identify the effect of GC content on codon bias, GC contents of genes were color-coded on the plot, which uses Axis 1 as the abscissa and Axis 2 as the ordinate (Fig. 4c for M. japonicus and Fig. 4d for M. pulchricaudatus). Overall, the distribution of GC content was the opposite along Axis 1. In M. japonicus, the larger the value of Axis 1, the smaller the GC content. The negative correlation (− 0.562 with p-value < 0.01) between Axis 1 and GC content is presented in the Fig. 4b. Instead, the larger the value of Axis 1, the larger the GC content of M. pulchricaudatus, and the positive correlation was 0.7 (p < 0.01).
Determination of optimal codons
There were 32 codons with the RSCU values > 1 in M. japonicas and M. pulchricaudatus, which indicated that these codons were preferred by the two species (Table S1). Except for Trp and Met, the codons of Ala, Arg, Gly, Pro, Ser, and Thr had a higher bias. In addition, the codons with the RSCU value > 1 mainly ended with C and A. Based on ENc values, we obtained the RSCU datasets of high and low expression genes and calculated the △RSCU value (Table S2). We determined 12 and 14 optimal codons for M. japonicus and M. pulchricaudatus, respectively (Table 1). In M. japonicus, 9 optimal codons were C-ending, and 3 optimal codons were G-ending. In M. pulchricaudatus species, 9 optimal codons were C-ending, and 5 optimal codons were G-ending. Most optimal codons were the same in the two Marsupenaeus species, except ACC (Thr), CCG (Pro), GCG (Ala), and GGC (Gly).
Table 1.
The optimal codons based on high and low levels of expression. AA: amino acids
| M. japonicus | M. pulchricaudatus | ||||||
|---|---|---|---|---|---|---|---|
| AA | Codon | RSCU-H | RSCU-L | △RSCU | RSCU-H | RSCU-L | △RSCU |
| Val | GUC | 1.557 | 0.995 | 0.563 | 1.454 | 0.984 | 0.470 |
| Ser | UCG | 1.005 | 0.682 | 0.324 | 1.202 | 0.717 | 0.486 |
| Pro | CCC | 1.552 | 0.906 | 0.646 | 1.764 | 0.920 | 0.844 |
| CCG | 0.896 | 0.614 | 0.282 | 1.047 | 0.606 | 0.441 | |
| Thr | ACC | 1.756 | 0.959 | 0.797 | 1.627 | 1.001 | 0.626 |
| ACG | 1.170 | 0.698 | 0.472 | 1.356 | 0.720 | 0.636 | |
| Ala | GCG | 0.836 | 0.564 | 0.271 | 1.002 | 0.537 | 0.464 |
| Tyr | UAC | 1.602 | 0.944 | 0.658 | 1.609 | 0.943 | 0.666 |
| His | CAC | 1.551 | 0.986 | 0.565 | 1.541 | 0.998 | 0.544 |
| Asn | AAC | 1.605 | 0.966 | 0.639 | 1.590 | 0.998 | 0.591 |
| Asp | GAC | 1.470 | 0.991 | 0.479 | 1.545 | 0.986 | 0.559 |
| Glu | GAG | 1.329 | 0.924 | 0.405 | 1.465 | 0.917 | 0.548 |
| Cys | UGC | 1.407 | 0.929 | 0.478 | 1.469 | 0.980 | 0.488 |
| Arg | CGC | 1.832 | 0.835 | 0.997 | 2.120 | 0.830 | 1.289 |
| Gly | GGC | 1.819 | 1.001 | 0.818 | 2.108 | 0.978 | 1.130 |
Codon pairs in two Marsupenaeus species
A synonymous codon that encodes two amino acids is called a duplex codon or codon pairs and is more commonly used than a single codon. The two Marsupenaeus species had different use frequencies of codon pairs (Table 2), such as GlyAla (GGAGCU vs GGAGCA), GlnArg (CAGAGA vs CAAAGA), and GluAsn (GAGAAC vs GAAAAU). In M. japonicus, the high-frequency codon pair of ArgArg was AGAAGA, while the optimal codon of Arg was CGC (Fig. S3). The high-frequency codon pair of AspAsp was GAUGAU, while the optimal codon of Asp was GAC. The high-frequency codon pair of GluGlu was GAAGAA, while the optimal codon of Glu was GAG. There were other inconsistencies, including GlyGly (GGAGGA) and Gly (GGC), HisHis (CAUCAU) and His (CAC), ProPro (CCACCA) and Pro (CCC), SerSer (AGCAGC) and Ser (UCG), ThrThr (ACAACA) and Thr (ACC/ACG), and ValVal (GUGGUG) and Val (GUC) (Fig. S3). In M. pulchricaudatus, the high-frequency codon pair of HisHis was CACCAC, which differentiates it from that of M. japonicus. The high-frequency codon pair of ProPro was CCACCA, while the optimal codons of Pro were CCC and CCG. The high-frequency codon pair of AlaAla was GCAGCA, while the optimal codon of Ala was GCG (Fig. S4). Codon pair utilization biases play an important role in protein synthesis by interacting with tRNA isoacceptors [44]. Codon pair analysis enables us to obtain a clear picture of the codon usage bias during transcription and translation.
Table 2.
The different duplex codons of two Marsupenaeus species
| Codons | M. japonicus | M. pulchricaudatus | Codons | M. japonicus | M. pulchricaudatus |
|---|---|---|---|---|---|
| Arg_Pro | AGGCCA | AGACCA | Phe_Thr | TTCACA | TTCACC |
| Asn_Ile | AACATT | AACATC | Pro_His | CCTCAT | CCTCAC |
| Asn_Leu | AACCTC | AACCTG | Pro_Lys | CCCAAG | CCAAAG |
| Asp_Pro | GATCCA | GACCCA | Pro_Ser | CCTTCA | CCATCA |
| Asp_Val | GATGTG | GATGTT | Pro_Val | CCAGTG | CCTGTG |
| Cys_Met | TGTATG | TGCATG | Ser_Gln | TCACAG | AGCCAG |
| Gln_Arg | CAGAGA | CAAAGA | Ser_Met | TCCATG | TCAATG |
| Gln_Ile | CAGATC | CAGATT | Thr_Leu | ACCCTC | ACTTTG |
| Glu_Asn | GAGAAC | GAAAAT | Trp_His | TGGCAC | TGGCAT |
| Gly_Ala | GGAGCT | GGAGCA | Trp_Tyr | TGGTAC | TGGTAT |
| Gly_Val | GGAGTG | GGTGTT | Tyr_Ala | TATGCT | TATGCA |
| His_His | CATCAT | CACCAC | Tyr_Pro | TACCCA | TATCCA |
| Leu_Leu | CTGCTG | CTCCTC | Val_Cys | GTGTGC | GTGTGT |
| Leu_Met | CTGATG | TTGATG | Val_Gly | GTGGGC | GTTGGA |
| Lys_Glu | AAAGAA | AAGGAA | Val_Ser | GTGTCA | GTCAGC |
| Phe_Ala | TTTGCA | TTTGCT |
Multispecies clustering analysis
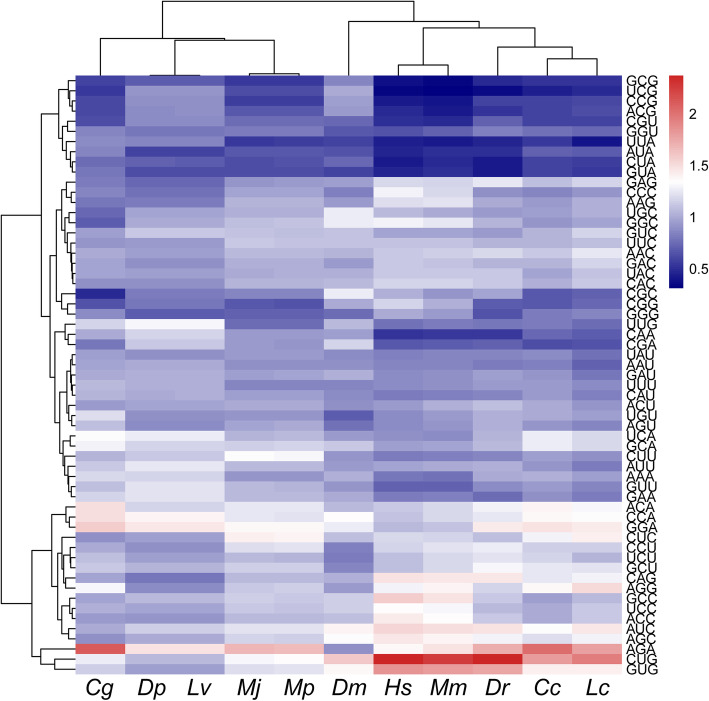
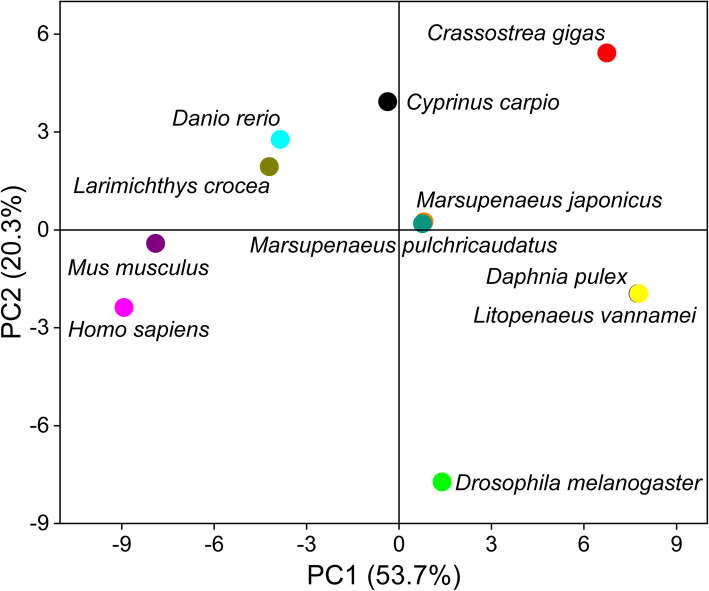
Based on the RSCU values of 59 codons (except Met, Trp, Taa, Tag, and Tga), the heat map (Fig. 5) showed that two Marsupenaeus species were clustered with Daphnia pulex and Litopenaeus vannamei and then Crassostrea gigas. The Larimichthys crocea, Cyprinus carpio, and Danio rerio were classified into the same cluster. Homo sapiens and Mus musculus were clustered into one group. Interestingly, Drosophila melanogaster and mammals were grouped at first, and then Arthropoda and Crassostrea gigas joined in them. This may be mainly because D. melanogaster has a stronger codon preference than other arthropods. Similar to the clustering results, the PCA showed that the two Marsupenaeus species overlapped almost completely, and the relationship between C. gigas and arthropods was not as strong as indicated by the results of heat map clustering (Fig. 6). These clustering results were consistent with traditional species classification.
Fig. 5.
Clustering analysis based on RSCU values. Cg = Crassostrea gigas, Dp = Daphnia pulex, Lv = Litopenaeus vannamei, Mj = Marsupenaeus japonicus, Mp = Marsupenaeus pulchricaudatus, Dm = Drosophila melanogaster, Hs = Homo sapiens, Mu = Mus musculus, Dr. = Danio rerio, Cc = Cyprinus carpio, Lc = Larimichthys crocea
Fig. 6.
Principal component analysis (PCA) of RSCU values
Discussion
Given the significant biological effects of different codon patterns, identifying these patterns in a given gene or genome is important to understand the molecular mechanisms of gene expression and to uncover the effects of long-term evolution on the genome [15, 45, 46]. Moreover, identifying these patterns is helpful for the phylogenetic analysis of species and to improve the expression of a target gene by optimizing codons [23, 47–49].
In this study, we analyzed the codon preferences of transcripts of two Marsupenaeus species, which were consistent overall. There was no significant difference in the content of AT and GC of the third base. The first and second base contents of a codon are usually affected by selection, while the third base content is affected by mutation pressure [40, 41]. The gene expression level (FPKM) was significantly negatively correlated with A/T3s. This result indicated that the third codon base significantly affects codon preference and gene expression level. Many studies have shown that codon usage bias correlates with gene expression levels, and codon usage patterns of highly expressed genes affect proteome-wide translation efficiency [12, 50, 51]. Whittle et al. found that translational selection shapes codon and amino acid usage in three Pancrustacean arthropods [20]. In Parasteatode tepidariorum, highly expressed genes favored amino acids with low or intermediate size/complexity (S/C) scores (glycine and alanine) and disfavored those with large S/C scores (such as cysteine) [50]. Further studies must consider correlation analysis between codon usage, amino acid frequency and expression levels.
The mean effective codon numbers (ENc) of the two cryptic species were 52.1 and 52.2, respectively, indicating the weak codon preference of both species. The S1|c21076_g1 sequence had the lowest ENc, with 23.6 for M. japonicus, and was annotated as nesprins-1, which is involved in the formation of the gamete cytoskeleton at different developmental stages [52]. Our previous study showed that there was significant variation in spermatheca traits, including the ratio of spermatheca length and width to body length. It remains to be further verified whether the dynamic expression level of this gene is different in the same developmental period of both species. The S2|c17052_g1 sequence, with the lowest ENc for M. pulchricaudatus, was annotated as the vrille (vri) gene, which encodes a core transcriptional repressor required for circadian behavior in Drosophila [53]. The two Marsupenaeus species have distinct geographical features with significant environmental differences, including temperature, sunlight and ocean currents. Long-term selection effects of different environments may affect the biorhythm, which still needs to be further tested. The FPKM of the S1|c21076_g1 and the S2|c17052_g1 sequences indicated low and high expression levels, respectively. Genes using the codons that are recognized by more abundant tRNA molecules may be translated more efficiently and with fewer mistakes than genes that use less frequent codons [54, 55]. Nelson et al. found that the high frequency of AGA/AGG codons present in the HCcAg and HUIFNa2 genes could be one of the factors limiting its expression in Escherichia coli [47]. In future studies, we will consider measuring the tRNA gene copy numbers and performing the correlation analysis with gene expression levels.
The codon preference of different species is generally influenced by mutation and selection pressure [56, 57]. The PR2 analysis showed that the usage frequencies of the four bases were not equal in the two Marsupenaeus species, suggesting that mutation pressure and selection contribute to codon usage patterns [58]. The ENc-GC3 plot reflects underlying factors governing CUB, which is based on the assumption that only GC content determines variations [59]. When the codons are affected only by GC compositional constraints, the gene lies on or very close to the curve. In the present study, the average observed ENc value was lower than the expected value for both species, suggesting that factors other than GC may act. Hiroshi Akashi et al. conducted seminal studies using population genetic approaches to corroborate the major codon preference model in Drosophila, which showed that selection does indeed affect the silent sites of proteins [60]. Based on 75 orthologous gene pairs from Drosophila, McVean and Nielsen estimated parameters of both mutation and selection, and the results showed considerable variation in the strength of selection between different Drosophila species [61].
Overall, the correlation analysis between the Ka/Ks value and codon preference parameters of orthologous genes in the two cryptic species was consistent. Nielsen et al. used a more complex mutation model to simultaneously estimate mutation rates, dN/dS, and the results supported the major codon preference model, and the notch gene of Drosophila melanogaster showed evidence of selection on synonymous sites [62]. In the group with ω < 1, the Ka/Ks of variety I was significantly positively correlated with ENc. In the group with ω > 1, Ka and Ks were significantly positively correlated with T3s, and the Fop and GC contents of variety I were significantly positively correlated with Ka and Ks, respectively. In Arachis duranensis and Arachis ipaënsis orthologs, highly expressed genes were subjected to stronger selective pressure than genes with low expression levels based on the negative correlation between selection constrain and both gene expression [38]. The positively selected orthologous genes related to the immune process mainly comprised single Von Willebrand factor, type C (VWC) domain protein, legumain, ras-related C3 botulinum, caspase, protein kinases, profilin family protein [37]. These genes were mainly annotated with the GO terms biological process (innate immune response, response to abiotic or biotic stimulus). The main reason for selection of codon bias may be that the increased use of major codons leads to more efficient and more accurate translation. However, some genes have been found to be under selection in the opposite direction, and the exact relative contribution of selection remains unclear [63]. The results of the correspondence analysis showed that the codon preference parameters of the two cryptic species had an opposite correlation with axis 1, which has been considered the most important evaluation index, and here showed a highly significant correlation with C3 and GC3. The gene expression level was significantly positively correlated with GC content. GC content is likely to be determined mostly by genome-wide processes rather than by selective forces acting specifically on coding regions, being the most significant parameter explaining codon bias differences between different organisms [64]. The results from Camiolo et al., indicated that gene sequences with higher GC content showed a higher expression level and better codon preference [65]. More efficient transcription and translation by the use of optimal synonymous codons increases the fitness of the organism [66, 67].
In this study, RSCU and ENc values were combined to determine 12 optimal codons in variety I, among which 9 ended in C and 3 ended in G, and 14 optimal codons in variety II, among which 9 ended in C and 5 ended in G. These results showed that Marsupenaeus species are genetically more likely to end in C/G, which was similar to the codon usage characteristic of carp (Cyprinus carpio), zebrafish (Danio rerio), Acanthopagrus schlegelii and Pagrus major [68, 69]. This may be because the evolution of M. japonicus is mainly mutated from AT to CG. Based on RNA-seq data, Whittle et al. found that three Pancrustacean arthropods have different optimal codons in highly expressed genes, and the majority of optimal codons from Parhyale hawaiensis were GC3 codons [20]. In Parasteatoda tepidariorum, highly expressed genes exhibited preferential usage of T3 codons, suggestive of selection [50]. Al-Saif et al., showed that reducing the proportion of UU or UA could enhance the resistance to mRNA attenuation, thus increasing protein expression [70]. In recent decades, the roles of codon usage bias in fine-tuning transcription, post-transcriptional processing, mRNA stability, translation initiation, elongation, and peptide folding have been revealed. The expression of functional proteins in heterologous hosts is a cornerstone of modern biotechnology, and the existence of slightly different codes in different organisms is a very significant barrier to heterologous expression [49]. The peptide LBDv (lipopolysaccharide binding domain) was synthesized based on the modified sequence of LBD (named LBD2) from FcALF2 and exhibited an apparently enhanced antimicrobial activity [71]. There were 31 different double codon pairs between the two Marsupenaeus species, and the optimization of the codon pair could improve the efficiency of protein translation compared with the single optimal codon [72–74]. The genetic distance of species is closely related to the codon preference difference, which can be used for species classification [33]. Based on the RSCU values of mitochondrial genomes among shrimp, the multidimensional scaling (MDS) plot showed that, for the most part, members of each infraorder clustered together and were largely distinct from the samples from the other infraorders [75]. The results of multispecies heat map analysis and clustering based on RSCU values are consistent with traditional species classification, which supported our previous results based on genotyping-by sequencing (GBS) and single copy nuclear genes (SCNGs) [35]. The results indicated that the size of interspecies codon preference differences can reflect the proximity of species, which is also verified in other species [21, 74, 76, 77].
Conclusions
In conclusion, we systematically compared the codon usage patterns of two Marsupenaeus species and evaluated the comprehensive effects of various factors. The codon usage patterns of both species were affected by mutations and selection. This study provides a relatively comprehensive understanding of the correlations among codon usage bias, gene expression, and selection pressure of CDS from M. japonicus and M. pulchricaudatus. Moreover, the results point out new insights into the specificities and evolutionary characteristics of these two cryptic species. However, the effect of codon usage bias on gene expression and the biological implications of different optimal codons in both species need further exploration.
Methods
Data collection and filtering
cDNA libraries were constructed from hepatopancreas of ten healthy M. japonicus (weight: 12.67 ± 3.22 g) and ten healthy M. pulchricaudatus (weight: 11.36 ± 4.2 g) from Huilai (Guangdong, China), and then sequenced for transcriptome assembly and functional annotation, as previously reported [37]. Raw Illumina sequences are accessible from NCBI Sequence Read Archive (SRA) (https://trace.ncbi.nlm.nih.gov/Traces/sra/) under accession SRR7786082 (Marsupenaeus pulchricaudatus) and SRR7786083 (Marsupenaeus japonicus). A total of 14,126 and 13,695 unigenes with CDS regions were identified from the M. japonicus and M. pulchricaudatus libraries, respectively. Orthologous groups were screened using OrthoMCL with default settings [78]. Gene expression levels as fragments per kilobase million [79] were estimated by RSEM software [80]. Coding sequences of the other nine species (Table S3) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). All CDSs were filtered using the OmicShare online platform (http://www.omicshare.com/tools), and those sequences with lengths less than 400 bp or unknown bases were eliminated.
Codon usage indices analysis
The GC1, GC2, and GC3 contents were calculated using Perl GitHub, and GC12 was the average value of GC1 and GC2. Using the CodonW 1.4.2 software (http://codonw.sourceforge.net), we performed codon bias analysis. The calculation indices included GC content, nucleotide composition at the 3rd codon position (A3s, T3s, G3s, and C3s), effective number of codons (ENc), the codon adaptation index (CAI), codon bias index (CBI), frequency of optional codon (Fop), and relative synonymous codon usage (RSCU), and so on. The parity rule 2 (PR2) plot analysis was based on the third codon position, using A3/(A3 + T3) as the ordinate and G3/(G3 + C3) as the abscissa. The PR2 plot can be used to estimate the impact of selection and mutation pressure on codon usage bias [81].
ENc-plot and GO annotation
The effective number of codons (ENc), with a value between 20 and 61, is a key parameter to interpret codon bias. The value 20 indicates that only one synonymous codon is chosen, 61 represents no usage bias, and all synonymous codons have the same probability. The lower the value for a coding sequence, the stronger the codon usage bias [42, 82]. In general, a gene possesses strong codon usage bias when the ENc value is lower than 35 [43, 83]. The ENc plot was drawn by Origin 2020 (OriginLab Corporation, USA), which uses the ENc value as the ordinate and GC3s as the abscissa. The expected ENc values were calculated based on the equation: Enc (exp) = 2 + GC3s + 29/[GC3s 2 + (1- GC3s)2] [84]. The codon adaptation index (CAI) is an important index for estimating synonymous codon usage bias and gene expression levels, and a higher CAI value signifies the stronger codon usage bias [85–87]. Gene ontology annotation was performed using Blast2GO v2.5 (E-value <1e− 6) [88]. GO classifications were compared among different groups of GC3s (High, Mid, and Low) using the OmicShare online platform.
Correlation analysis
The codon usage patterns were often shaped by many factors, such as GC content, expression level, tRNA abundance, protein structure, and hydrophilicity [89, 90]. We performed a correlation analysis between codon bias parameters and expression level (FPKM). Using the PAML toolkit [91], we calculated the nonsynonymous substitution ratio (Ka) and synonymous substitution ratio (Ks). The Ka/Ks (ω) can be used to determine whether there is selective pressure on protein-encoding genes [92, 93]. Values of ω > 1 suggest that the gene evolved under positive selection, whereas ω close to zero indicates that the gene is under heavy selection pressure [92, 94].
Correspondence analysis (COA)
To further investigate the factors related to the codon usage pattern, correspondence analysis was conducted by CodonW based on the RSCU values. The COA was used to compare the usage patterns of 59 codons (except Met, Trp, Taa, Tag, and Tga) and reflect the variation trend in codon usage. COA creates a series of orthogonal axes, which were used to estimate the main source of variation. Using SPSS v22 (https://www.ibm.com/support/pages/spss-statistics-220-available-download), the relative coefficient between ten codon bias parameters and Axis1 and Axis2 was calculated.
Relative synonymous codon usage and optimal codons
According to Sharp et al. [95], the relative synonymous codon usage (RSCU) is an index to measure the codon usage preference. The higher the RSCU value, the stronger the preference. Based on the calculated ENc values, 10% of the genes with extremely high and low ENc values were regarded as the high and low RSCU datasets [96]. The optimal codons were confirmed based on the △RSCU value and chi-square test [66, 83, 97].
Clustering and principal component analysis
The protein-coding sequences of nine species (Table S3) were downloaded from the ensemble database (http://asia.ensembl.org/index.html) and NCBI (https://www.ncbi.nlm.nih.gov/), and codon usage preference was analyzed using CodonW. The heatmap was generated based on RSCU values using the OmicShare online platform. Based on the RSCU of 59 codons, principal component analysis (PCA) was performed using Origin 2020 (OriginLab Corporation, Northampton, MA, USA).
Supplementary Information
Additional file 1: Fig. S1. ENc plot, ENc frequency and GC3s-CAI for M. japonicus (a, c, e) and M. pulchricaudatus (b, d, f).
Additional file 2: Fig. S2. Gene ontology (GO) annotation.
Additional file 3: Fig. S3. The codon pairs of M. japonicus.
Additional file 4: Fig. S4. The codon pairs of M. pulchricaudatus.
Additional file 5: Table S1. The relative synonymous codon usage (RSCU) of synonymous codons. Table S2. The RSCU datasets of high and low expression genes. Table S3. Genome information of nine species.
Acknowledgements
The authors are grateful to Dongshan Swire Marine Station (Xiamen University) for providing the laboratory space.
Abbreviations
- NCBI
National Center for Biotechnology Information
- GO
Gene Ontology
- CDS
the coding sequences
- SRA
Sequence Read Archive
- ENc
the effective number of codons
- CAI
the codon adaptation index
- CBI
codon bias index
- Fop
frequency of optional codon
- RSCU
relative synonymous codon usage
- COA
correspondence analysis
- PR2
parity rule 2
- FPKM
Fragments Per Kilobase per Million
- PCA
principal component analysis
- Ka
non-synonymous substitution ratio
- Ks
synonymous substitution ratio
Authors’ contributions
PPW and YM conceived and designed the experiments, YM, YQS, and JW obtained funds for the study. PPW, YQS, and JW performed bioinformatics analysis. PPW and YM drafted the manuscript. YM, YQS, and JW participated in the manuscript revision. All authors read and approved the final manuscript.
Funding
This study was supported by the Project of China Agriculture Research System (Grant No. CARS-48), the Science and Technology Plan Project of Ningbo (2019B10011), the Natural Science Foundation of Jiangsu Province (No. BK20210924), the Open Research Fund of Jiangsu Key Laboratory of Marine Biotechnology (HS2020001), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All the necessary information needed to support the results of this paper are included within the article. All of the RNA sequencing data used in the study are available through the NCBI SRA database (https://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession numbers SRR7786082 (Marsupenaeus pulchricaudatus) and SRR7786083 (Marsupenaeus japonicus). The protein-coding sequences of other species (Table S3) were downloaded from the ensemble database (http://asia.ensembl.org/index.html) and NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers: PRJNA629593; PRJEB14656; PRJNA508983; PRJNA164; PRJNA168; PRJNA169; PRJNA13922; PRJNA352247; PRJNA354443.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Duret L. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 2002;12(6):640–649. doi: 10.1016/S0959-437X(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 2.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biro JC. Studies on the origin and evolution of codon bias. Biomolecules. 2008:0807–3901.
- 4.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 5.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12(1):32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komar AA. The yin and Yang of codon usage. Hum Mol Genet. 2016;25(R2):R77–R85. doi: 10.1093/hmg/ddw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C-H, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell. 2015;59(5):744–754. doi: 10.1016/j.molcel.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galtier N, Roux C, Rousselle M, Romiguier J, Figuet E, Glémin S, Bierne N, Duret L. Codon usage bias in animals: disentangling the effects of natural selection, effective population size, and GC-biased gene conversion. Mol Biol Evol. 2018;35(5):1092–1103. doi: 10.1093/molbev/msy015. [DOI] [PubMed] [Google Scholar]
- 9.Eyre-Walker AC. An analysis of codon usage in mammals: selection or mutation bias? J Mol Evol. 1991;33(5):442–449. doi: 10.1007/BF02103136. [DOI] [PubMed] [Google Scholar]
- 10.Powell JR, Dion K. Effects of codon usage on gene expression: empirical studies on Drosophila. J Mol Evol. 2015;80(3–4):219–226. doi: 10.1007/s00239-015-9675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zalucki YM, Power PM, Jennings MP. Selection for efficient translation initiation biases codon usage at second amino acid position in secretory proteins. Nucleic Acids Res. 2007;35(17):5748–5754. doi: 10.1093/nar/gkm577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frumkin I, Lajoie MJ, Gregg CJ, Hornung G, Church GM, Pilpel Y. Codon usage of highly expressed genes affects proteome-wide translation efficiency. Proc Natl Acad Sci. 2018;115(21):E4940–E4949. doi: 10.1073/pnas.1719375115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1997;25(1):244–245. doi: 10.1093/nar/25.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp PM, Li W-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1–2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 15.Yannai A, Katz S, Hershberg R. The codon usage of lowly expressed genes is subject to natural selection. Genome biology and evolution. 2018;10(5):1237–1246. doi: 10.1093/gbe/evy084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archetti M. Codon usage bias and mutation constraints reduce the level of errorminimization of the genetic code. J Mol Evol. 2004;59(2):258–266. doi: 10.1007/s00239-004-2620-0. [DOI] [PubMed] [Google Scholar]
- 17.Dhindsa RS, Copeland BR, Mustoe AM, Goldstein DB. Natural selection shapes codon usage in the human genome. Am J Hum Genet. 2020;107(1):83–95. doi: 10.1016/j.ajhg.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7(1):1–17. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakraborty S, Nag D, Mazumder TH, Uddin A. Codon usage pattern and prediction of gene expression level in Bungarus species. Gene. 2017;604:48–60. doi: 10.1016/j.gene.2016.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Whittle CA, Extavour CG. Codon and amino acid usage are shaped by selection across divergent model organisms of the Pancrustacea. G3: genes. Genomes, Genetics. 2015;5(11):2307–2321. doi: 10.1534/g3.115.021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Cataluña A, Salas-Massó N, Diéguez AL, Balboa S, Lema A, Romalde JL, Figueras MJ. Revisiting the taxonomy of the genus Arcobacter: getting order from the chaos. Front Microbiol. 2018;9:2077. doi: 10.3389/fmicb.2018.02077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athey J, Alexaki A, Osipova E, Rostovtsev A, Santana-Quintero LV, Katneni U, Simonyan V, Kimchi-Sarfaty C. A new and updated resource for codon usage tables. BMC bioinformatics. 2017;18(1):1–10. doi: 10.1186/s12859-017-1793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller JB, Whiting MF, Kauwe JS, Ridge PG. How Codon Usage Bias Affects Our Ability to Recover the Tree of Life. 2019(2019100086 (doi: 10.20944/preprints201910.0086.v1)).
- 24.Yi S, Li Y, Wang W. Selection shapes the patterns of codon usage in three closely related species of genus Misgurnus. Genomics. 2018;110(2):134–142. doi: 10.1016/j.ygeno.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42(1):287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay P, Basak S, Ghosh TC. Differential selective constraints shaping codon usage pattern of housekeeping and tissue-specific homologous genes of rice and Arabidopsis. DNA Res. 2008;15(6):347–356. doi: 10.1093/dnares/dsn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trotta E. Selection on codon bias in yeast: a transcriptional hypothesis. Nucleic Acids Res. 2013;41(20):9382–9395. doi: 10.1093/nar/gkt740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado HE, Lawrie DS, Petrov DA. Pervasive strong selection at the level of codon usage Bias in Drosophila melanogaster. Genetics. 2020;214(2):511–528. doi: 10.1534/genetics.119.302542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu D, Wei L. Characterizing the heat response of Arabidopsis thaliana from the perspective of codon usage bias and translational regulation. J Plant Physiol. 2019;240:153012. doi: 10.1016/j.jplph.2019.153012. [DOI] [PubMed] [Google Scholar]
- 30.Guan D-L, Ma L-B, Khan MS, Zhang X-X, Xu S-Q, Xie J-Y. Analysis of codon usage patterns in Hirudinaria manillensis reveals a preference for GC-ending codons caused by dominant selection constraints. BMC Genomics. 2018;19(1):542. doi: 10.1186/s12864-018-4937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia X, Liu S, Zheng H, Li B, Qi Q, Wei L, Zhao T, He J, Sun J. Non-uniqueness of factors constraint on the codon usage in Bombyx mori. BMC Genomics. 2015;16(1):356. doi: 10.1186/s12864-015-1596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, Xu J, Chen L, Wang Y, Gu X, Peng X, Yang G. Analysis of transcriptome data reveals multifactor constraint on codon usage in Taenia multiceps. BMC Genomics. 2017;18(1):308. doi: 10.1186/s12864-017-3704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan X, Yi S, Guo X, Wang W. A comprehensive analysis of codon usage patterns in blunt snout bream (Megalobrama amblycephala) based on RNA-Seq data. Int J Mol Sci. 2015;16(6):11996–12013. doi: 10.3390/ijms160611996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsoi KH, Ma KY, Wu TH, Fennessy ST, Chu KH, Chan TY. Verification of the cryptic species Penaeus pulchricaudatus in the commercially important kuruma shrimp P. japonicus (Decapoda : Penaeidae) using molecular taxonomy. Invertebr Syst. 2014;28(5):476–490. doi: 10.1071/IS14001. [DOI] [Google Scholar]
- 35.Wang P, Chen B, Zheng J, Cheng W, Zhang H, Wang J, et al. Fine-Scale Population Genetic Structure and Parapatric Cryptic Species of Kuruma Shrimp (Marsupenaeus japonicus), Along the Northwestern Pacific Coast of China. Frontiers in Genetics. 2020;11(118). [DOI] [PMC free article] [PubMed]
- 36.Tsoi KH, Chan TY, Chu KH. Molecular population structure of the kuruma shrimp Penaeus japonicus species complex in western Pacific. Mar Biol. 2007;150(6):1345–1364. doi: 10.1007/s00227-006-0426-x. [DOI] [Google Scholar]
- 37.Wang P, Xing C, Wang J, Su Y, Mao Y. Evolutionary adaptation analysis of immune defense and hypoxia tolerance in two closely related Marsupenaeus species based on comparative transcriptomics. Fish & shellfish immunology. 2019;92:861–870. doi: 10.1016/j.fsi.2019.06.055. [DOI] [PubMed] [Google Scholar]
- 38.Song H, Gao H, Liu J, Tian P, Nan Z. Comprehensive analysis of correlations among codon usage bias, gene expression, and substitution rate in Arachis duranensis and Arachis ipaënsis orthologs. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-13981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24(11):539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Hu C, Chen J, Ye L, Chen R, Zhang L, Xue X. Codon usage bias in human cytomegalovirus and its biological implication. Gene. 2014;545(1):5–14. doi: 10.1016/j.gene.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 41.RoyChoudhury S, Mukherjee D. A detailed comparative analysis on the overall codon usage pattern in herpesviruses. Virus Res. 2010;148(1–2):31–43. doi: 10.1016/j.virusres.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 42.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87(1):23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 43.Comeron JM, Aguadé M. An evaluation of measures of synonymous codon usage bias. J Mol Evol. 1998;47(3):268–274. doi: 10.1007/PL00006384. [DOI] [PubMed] [Google Scholar]
- 44.Irwin B, Heck JD, Hatfield GW. Codon pair utilization biases influence translational elongation step times. J Biol Chem. 1995;270(39):22801–22806. doi: 10.1074/jbc.270.39.22801. [DOI] [PubMed] [Google Scholar]
- 45.Quax TE, Claassens NJ, Söll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell. 2015;59(2):149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul P, Malakar AK, Chakraborty S. Codon usage and amino acid usage influence genes expression level. Genetica. 2018;146(1):53–63. doi: 10.1007/s10709-017-9996-4. [DOI] [PubMed] [Google Scholar]
- 47.Acosta-Rivero N, Sánchez JC, Morales J. Improvement of human interferon HUIFNα2 and HCV core protein expression levels in Escherichia coli but not of HUIFNα8 by using the tRNAAGA/AGG. Biochem Biophys Res Commun. 2002;296(5):1303–1309. doi: 10.1016/S0006-291X(02)02056-9. [DOI] [PubMed] [Google Scholar]
- 48.Mauro VP, Chappell SA: Considerations in the use of codon optimization for recombinant protein expression. In: volume Recombinant Protein Expression in Mammalian Cells, chapter 7. Springer; 2018: 275–288, Considerations in the Use of Codon Optimization for Recombinant Protein Expression, DOI: 10.1007/978-1-4939-8730-6_18. [DOI] [PubMed]
- 49.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Whittle CA, Extavour CG. Expression-linked patterns of codon usage, amino acid frequency, and protein length in the basally branching arthropod Parasteatoda tepidariorum. Genome biology and evolution. 2016;8(9):2722–2736. doi: 10.1093/gbe/evw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williford A, Demuth JP. Gene expression levels are correlated with synonymous codon usage, amino acid composition, and gene architecture in the red flour beetle, Tribolium castaneum. Mol Biol Evol. 2012;29(12):3755–3766. doi: 10.1093/molbev/mss184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114(24):4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 53.Gunawardhana KL, Rivas GB, Caster C, Hardin PE. Crosstalk between vrille transcripts, proteins, and regulatory elements controlling circadian rhythms and development in Drosophila. Iscience. 2021;24(1):101893. doi: 10.1016/j.isci.2020.101893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Victor MP, Acharya D, Chakraborty S, Ghosh TC. The combined influence of codon composition and tRNA copy number regulates translational efficiency by influencing synonymous nucleotide substitution. Gene. 2020;745:144640. doi: 10.1016/j.gene.2020.144640. [DOI] [PubMed] [Google Scholar]
- 55.Whittle CA, Kulkarni A, Chung N, Extavour CG. Adaptation of codon and amino acid use for translational functions in highly expressed cricket genes. BMC Genomics. 2021;22(1):1–21. doi: 10.1186/s12864-021-07411-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Huang Y, Du X, Chen Z, Zeng X, Chen Y, et al. Patterns of synonymous codon usage bias in the model grass Brachypodium distachyon. Genet Mol Res. 2012;11(4):4695–4706. doi: 10.4238/2012.October.17.3. [DOI] [PubMed] [Google Scholar]
- 57.Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol Biol Evol. 2009;27(3):650–660. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- 58.Yang X, Luo X, Cai X. Analysis of codon usage pattern in Taenia saginata based on a transcriptome dataset. Parasit Vectors. 2014;7(1):1–11. doi: 10.1186/s13071-014-0527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nair RR, Nandhini MB, Monalisha E, Murugan K, Sethuraman T, Nagarajan S, et al. Synonymous codon usage in chloroplast genome of Coffea arabica. Bioinformation. 2012;8(22):1096–1104. doi: 10.6026/97320630081096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akashi H, Kliman RM, Eyre-Walker A. Mutation pressure, natural selection, and the evolution of base composition in Drosophila. Mutation and Evolution. 1998:49–60. 10.1007/978-94-011-5210-5_5. [PubMed]
- 61.McVean GA, Vieira J. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics. 2001;157(1):245–257. doi: 10.1093/genetics/157.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen R, Bauer DuMont VL, Hubisz MJ, Aquadro CF. Maximum likelihood estimation of ancestral codon usage bias parameters in Drosophila. Mol Biol Evol. 2007;24(1):228–235. doi: 10.1093/molbev/msl146. [DOI] [PubMed] [Google Scholar]
- 63.Singh ND, Bauer DuMont VL, Hubisz MJ, Nielsen R, Aquadro CF. Patterns of mutation and selection at synonymous sites in Drosophila. Mol Biol Evol. 2007;24(12):2687–2697. doi: 10.1093/molbev/msm196. [DOI] [PubMed] [Google Scholar]
- 64.Knight RD, Freeland SJ, Landweber LF. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2001;2(4):1–13. doi: 10.1186/gb-2001-2-4-research0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camiolo S, Melito S, Porceddu A. New insights into the interplay between codon bias determinants in plants. DNA Res. 2015;22(6):461–470. doi: 10.1093/dnares/dsv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci. 1999;96(8):4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akashi H. Inferring weak selection from patterns of polymorphism and divergence at" silent" sites in Drosophila DNA. Genetics. 1995;139(2):1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao G, Zhang Z, Zhang Z, Chen S, Zhu F, Jia C, et al. The analysis of the microsatellite sequences and codon bias of the coding sequence in acanthopagrus schlegelii, pagrus major and their hybrid progenies. Oceanology and Limnology. 2019;50(5):1108–1115. [Google Scholar]
- 69.Zhang X, Shun X. The sysnonymous codon bias of carp (Cyprinus carpio L.) and Zebrafish (Danio rerio L.) Chinese Journal Fisheries. 2010;23(4):23–29. [Google Scholar]
- 70.Al-Saif M, Khabar KS. UU/UA dinucleotide frequency reduction in coding regions results in increased mRNA stability and protein expression. Mol Ther. 2012;20(5):954–959. doi: 10.1038/mt.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang H, Li S, Li F, Xiang J. Structure and bioactivity of a modified peptide derived from the LPS-binding domain of an anti-lipopolysaccharide factor (ALF) of shrimp. Marine drugs. 2016;14(5):96. doi: 10.3390/md14050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shao Z-Q, Zhang Y-M, Feng X-Y, Wang B, Chen J-Q. Synonymous codon ordering: a subtle but prevalent strategy of bacteria to improve translational efficiency. PLoS One. 2012;7(3):e33547. doi: 10.1371/journal.pone.0033547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qian W, Yang J-R, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8(3):e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng C, C-j X, Wang Y, W-l L, X-r Y, Li X, et al. Codon usage patterns in Chinese bayberry (Myrica rubra) based on RNA-Seq data. BMC genomics. 2013;14(1):732. doi: 10.1186/1471-2164-14-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan MH, Gan HM, Lee YP, Poore GC, Austin CM. Digging deeper: new gene order rearrangements and distinct patterns of codons usage in mitochondrial genomes among shrimps from the Axiidea, Gebiidea and Caridea (Crustacea: Decapoda) PeerJ. 2017;5:e2982. doi: 10.7717/peerj.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu Y, Peng C, Chen Z, Chang X, Qiu J, Lin Z, et al. Analysis of codon usage Bias in Clausena lansium Transcriptome. Molecular Plant Breeding. 2018;16(18):5904–5913. [Google Scholar]
- 77.Chen Z, Hu F, Wang X, Fan H, Zhang Z. Analysis of codon usage bias of Ananas comosus with genome sequencing data. Journal of Fruit Science. 2017;34(08):946–955. [Google Scholar]
- 78.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sueoka N, Kawanishi Y. DNA G+ C content of the third codon position and codon usage biases of human genes. Gene. 2000;261(1):53–62. doi: 10.1016/S0378-1119(00)00480-7. [DOI] [PubMed] [Google Scholar]
- 82.Fuglsang A. The ‘effective number of codons’ revisited. Biochem Biophys Res Commun. 2004;317(3):957–964. doi: 10.1016/j.bbrc.2004.03.138. [DOI] [PubMed] [Google Scholar]
- 83.Jiang Y, Deng F, Wang H, Hu Z. An extensive analysis on the global codon usage pattern of baculoviruses. Arch Virol. 2008;153(12):2273–2282. doi: 10.1007/s00705-008-0260-1. [DOI] [PubMed] [Google Scholar]
- 84.Hartl DL, Moriyama EN, Sawyer SA. Selection intensity for codon bias. Genetics. 1994;138(1):227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee S, Weon S, Lee S, Kang C. Relative codon adaptation index, a sensitive measure of codon usage bias. Evol Bioinforma. 2010;6:47–55. doi: 10.4137/ebo.s4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496(7443):57–63. doi: 10.1038/nature12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng H, Zhang W, Zhang L, Zhang Z, Li J, Lu G, Zhu Y, Wang Y, Huang Y, Liu J, Kang H, Chen J, Wang L, Chen A, Yu S, Gao Z, Jin L, Gu W, Wang Z, Zhao L, Shi B, Wen H, Lin R, Jones MK, Brejova B, Vinar T, Zhao G, McManus DP, Chen Z, Zhou Y, Wang S. The genome of the hydatid tapeworm Echinococcus granulosus. Nat Genet. 2013;45(10):1168–1175. doi: 10.1038/ng.2757. [DOI] [PubMed] [Google Scholar]
- 88.Götz S, Garcíagómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shah P, Gilchrist MA. Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet. 2010;6(9):e1001128. doi: 10.1371/journal.pgen.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu C, Cai X, Chen Q, Zhou H, Cai Y, Ben A. Factors affecting synonymous codon usage bias in chloroplast genome of oncidium Gower Ramsey. Evol Bioinforma. 2011;7:271–278. doi: 10.4137/EBO.S8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 92.Nekrutenko A, Makova KD, Li W-H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 2002;12(1):198–202. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends in genetics: TIG 2002; 18(9):486–486, 487, DOI: 10.1016/S0168-9525(02)02722-1. [DOI] [PubMed]
- 94.Zhang Z, Li J, Zhao X-Q, Wang J, Wong GK-S, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics, proteomics & bioinformatics. 2006;4(4):259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sharp PM, Li W-H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bellgard M, Schibeci D, Trifonov E, Gojobori T. Early detection of G+ C differences in bacterial species inferred from the comparative analysis of the two completely sequenced helicobacter pylori strains. J Mol Evol. 2001;53(4–5):465–468. doi: 10.1007/s002390010236. [DOI] [PubMed] [Google Scholar]
- 97.Liu Q. Analysis of codon usage pattern in the radioresistant bacterium Deinococcus radiodurans. Biosystems. 2006;85(2):99–106. doi: 10.1016/j.biosystems.2005.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. ENc plot, ENc frequency and GC3s-CAI for M. japonicus (a, c, e) and M. pulchricaudatus (b, d, f).
Additional file 2: Fig. S2. Gene ontology (GO) annotation.
Additional file 3: Fig. S3. The codon pairs of M. japonicus.
Additional file 4: Fig. S4. The codon pairs of M. pulchricaudatus.
Additional file 5: Table S1. The relative synonymous codon usage (RSCU) of synonymous codons. Table S2. The RSCU datasets of high and low expression genes. Table S3. Genome information of nine species.
Data Availability Statement
All the necessary information needed to support the results of this paper are included within the article. All of the RNA sequencing data used in the study are available through the NCBI SRA database (https://trace.ncbi.nlm.nih.gov/Traces/sra/) under the accession numbers SRR7786082 (Marsupenaeus pulchricaudatus) and SRR7786083 (Marsupenaeus japonicus). The protein-coding sequences of other species (Table S3) were downloaded from the ensemble database (http://asia.ensembl.org/index.html) and NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers: PRJNA629593; PRJEB14656; PRJNA508983; PRJNA164; PRJNA168; PRJNA169; PRJNA13922; PRJNA352247; PRJNA354443.