Abstract
Spectral editing in in vivo 1H-MRS provides an effective means to measure low-concentration metabolite signals that cannot be reliably measured by conventional MRS techniques due to signal overlap, e.g., γ-aminobutyric acid (GABA), glutathione (GSH) and D-2-hydroxyglutarate (2HG). Spectral editing strategies utilize known J-coupling relationships within the metabolite of interest to discriminate their resonances from overlying signals. This consensus recommendation paper provides a brief overview of commonly used homo-nuclear editing techniques and considerations for data acquisition, processing, and quantification. Also, we have listed the experts’ recommendations for minimum requirements to achieve adequate spectral editing and reliable quantification. These include selecting the right editing sequence, dealing with frequency drift, handling unwanted co-edited resonances, spectral fitting of edited spectra, setting up multi-center clinical trials, and recommending sequence parameters to be reported in publications.
Keywords: Spectral editing, J-difference editing, multiple quantum filtering, GABA, glutathione (GSH), consensus recommendations
Introduction
Proton magnetic resonance spectroscopy (1H-MRS) provides a non-invasive way to investigate in vivo metabolite concentrations in health and disease. However, in vivo 1H MRS signals of metabolites with low concentration, including γ-aminobutyric acid (GABA), glutathione (GSH) and D-2-hydroxyglutarate (2HG), are often obscured by signals from more concentrated metabolites. This makes accurate detection and quantification challenging. One way to obtain information about such signals is to remove the strong overlapping resonances via spectral editing. The majority of editing strategies for separating overlapped metabolite resonances are based on utilizing the J-coupling (or scalar coupling) and chemical shift as a means of discriminating between coupled and non-coupled spins. Editing pulses are utilized to select a narrowband region of the spectrum within which the target spins resonate, making those techniques spin-system-specific. One class of editing techniques is J-difference editing which involves the subtraction of two separate acquisitions, one with an editing pulse and the other without. Editing pulses are used to modulate the evolution of J-coupling, such that subtraction maintains the target signal while removing the contribution of other metabolites. Another class is multiple quantum coherence filtering (MQF), which is a single-shot technique that achieves spectral simplification by removing unwanted signals through coherence selection. This consensus is mainly focused on such homo-nuclear editing techniques, with a brief mentioning of other alternatives such as long echo modulation, echo time (TE) averaging, polarization transfer, and multidimensional J-resolved and correlation methods.
While both J-difference editing and MQF can disambiguate spectral quantification of strongly overlapped J-coupled resonances, the last decade has seen a significant trend towards J-difference-based editing techniques such as MEGA editing1,2 as these are straightforward to implement, more sensitive, and enable quantification of other metabolites using non-edited spectra. They have been integrated into a multitude of different spatial MRS localization (i.e., volume selection) techniques (PRESS,1 STEAM,1 semi-LASER,3 LASER,4 and SPECIAL5) and MR spectroscopic imaging (MRSI) sequences.4,6 Spectral editing typically focuses on the selective editing of a single target metabolite; however, recently several methods demonstrated simultaneous editing of several J-coupled target metabolites.7–9 Reducing the inevitable co-editing of other unwanted resonances has also been the target of research efforts, including (but not limited to) macromolecules (MM) in GABA editing.10,11 J-difference editing relies on the subtraction of two spectra and highly frequency-selective inversion of a target resonance, thereby making it susceptible to artifacts related to subject motion and scanner instabilities. There are ongoing efforts to improve MRS acquisition and post-processing approaches to mitigate motion and instability-related artifacts.4,12–16
Spectral editing is highly relevant at clinical magnetic field strengths (≤ 3 T), which presents appreciable overlapping resonances of J-coupled metabolites. Recent developments towards MR systems with ultra-high magnetic field strengths (≥ 7 T) provide improved spectral separation of metabolites, particularly for J-coupled spin systems.17 The use of ultra-high field MR systems may relax the need for spectral editing that can come with a significant loss in SNR and additional sources of variability in exchange for chemical selectivity. Nevertheless, the availability of ultra-high field MR systems is currently limited. Higher fields present other challenges, e.g., B0 shimming due to increased susceptibility effects and achieving sufficient RF power due to the specific absorption rate (SAR) limit and hardware limitations. Thus, advanced MRS editing techniques are the preferred choice for unambiguous detection of several important J-coupled metabolites (e.g., GABA, GSH and 2HG) for the vast majority of the scientific community using clinical MR systems. The purpose of this experts’ consensus recommendation paper is to summarize editing-specific technical considerations and to provide minimum requirements for data acquisition, processing and quantification.
1. J-difference editing
As the performance of spectral editing depends strongly on the experimental conditions during data acquisition, several minimum requirements can be formulated for successful data acquisition of a spectral editing study. The quality of MRS measures depends on the homogeneity of the static magnetic field (B0) and spectral editing is no exception. Inadequate B0 homogeneity can lead to a decreased spectral resolution, loss of detection sensitivity, and poor water suppression. The requirement for water suppression may be less stringent in spectral editing compared with non-edited MRS due to spectral subtraction in some metabolites (e.g., GABA and 2HG). However, editing of some metabolites (e.g., Asc, GSH) can be influenced by poor water suppression because water signals are within the bandwidth of the editing pulse. The detection error of a metabolite, e.g., expressed by Cramér-Rao lower bounds (CRLB), increases with increasing spectral overlap and spectral linewidth when the B0 homogeneity decreases.
The editing efficiency of spectral editing is a critical factor in determining its usability. In the consensus recommendation paper on terminology and concepts for MRS,18 editing efficiency is defined as a ratio of the integral of absolute values under the edited resonances (with relaxation-weighting removed) to the same measure of those resonances in a spectrum acquired with zero echo time. The editing efficiency defined in this manner has theoretical (i.e., spin physics) and practical (i.e., measurement imperfections) contributions. Theoretically, the editing efficiency for the J-difference editing of lactate (an AX3 spin-system) and GABA (an AX2 spin-system) is 100% and 50%, respectively. The theoretical GABA editing efficiency is lower because only 50% of the GABA multiplet modulates with the echo-time TE. In addition to the theoretical contributions, which include editing sequence type (J-difference, MQF) and TE, there is a wide range of factors that contribute to the practical or experimental editing efficiency. These factors include, but are not limited to, chemical shift displacement error (CSDE), RF amplitude B1+ miscalibration and inhomogeneity, frequency drift, and subject motion. This consensus recommendation paper covers these effects and discusses means to minimize their contributions.
For localized in vivo MRS, the maximum available radiofrequency (RF) amplitude dictates the maximum available bandwidth of RF pulses, which determines the CSDE. This CSDE leads to the selection of different voxel locations for different moieties, which results in variations in scalar coupling evolution because spins experience a different number of refocusing pulses leading to a change in editing efficiency.19 As the CSDE increases with the B0 strength, this effect becomes unacceptably large for MEGA-PRESS at B0 above 3 T, if no dedicated local transmit RF coil is available to achieve sufficient RF amplitude and bandwidth. To overcome this problem, adiabatic RF pulses that can achieve a higher bandwidth for the same maximum RF amplitude have been increasingly adopted in editing sequences, e.g., LASER- or semi-LASER-based MEGA editing at 3 T4,20 and 7 T.3,10,21,22
One of the most important experimental aspects of J-difference editing methods is the temporal stability of the subject and MR systems. Any instability during the acquisition of the two sub-spectra can result in a subtraction artifact that could lead to an under- or over-estimation of the edited signal of interest. Besides advanced methods like B0 field locks23 and prospective motion and/or frequency correction,24 a common and recommended approach for single voxel MRS techniques25 is to deal with repetition-to-repetition variations by acquiring and storing each transient separately. This approach allows phase- and frequency-correction of each transient and the removal of spectra that are beyond correction. However, note that this approach cannot address potentially inaccurate spectral editing outcomes via a reduced editing efficiency or increased contribution of spectral co-editing of other resonances in the presence of frequency drift and/or subject motion.
RF power calibration
Incorrect calibration of the transmit RF magnetic field (B1+) across the localized volume leads to signal loss and potential artifacts, along with compromised localization performance and editing efficiency.26 This type of imperfection is always present when dealing with inhomogeneous B1+ distributions across the volume of interest as encountered at high B0 fields and surface coil transmission.
Recommendations regarding RF pulses and power calibration are:
Perform transmit RF power calibration on the localized volume for both single voxel MRS and MR spectroscopic imaging (MRSI).
The average B1+ amplitude and the editing pulse efficiency need to be estimated empirically by applying the editing pulses to the water resonance with water suppression turned off. The water intensity ratio between the scans with the on- and off-resonance editing pulses offers information on the B1+ amplitude.
The effects of incorrect B1+ amplitudes need to be accounted for through simulations using experimentally measured B1+ values and editing sequence details, e.g., sequence timing, real RF pulses, and localization gradients.
RF pulse durations should not vary from subject to subject to achieve the desired flip angle. Even though some of the vendor-supplied sequences use this feature, this will lead to variation in CSDE, co-editing, and editing efficiency from subject to subject. The users should verify that only the RF power is adjusted during B1+ calibration of the refocusing pulses used for localization while the pulse duration is fixed for the duration of the cohort study.
In circumstances where RF power is limited and pulse length is increased substantially to accommodate the coil load, the magnitude of the CSDE and the influence on the study outcomes need to be considered.
Co-editing of unwanted resonances
Unwanted signals from other metabolites frequently co-edit with the desired resonances of the metabolite of interest. Typical examples include co-editing of macromolecules (MM) in GABA editing, co-editing of N-acetylaspartate (NAA) in GSH editing, and BHB co-editing in lactate editing. Table 1 provides an overview of commonly observed editing/co-editing partners. The effect and importance of the co-edited signals depend on whether the co-edited signals overlap with the target signals (Table 1). For example, co-editing of glutamate/glutamine and 2HG in GABA editing has no consequences for the accuracy of GABA editing as the co-edited glutamate/glutamine and 2HG signals are well separated from the GABA signal of interest at ~3 ppm. This can be even useful for editing multiple non-overlapping metabolite peaks with a single editing pulse when the resonances of the edited peaks have very similar chemical shifts for all co-edited metabolites. Multiplexed editing can use co-editing to measure multiple overlapping metabolites as described below.7 In contrast, co-edited NAA+NAAG in GSH editing can lead to baseline distortion caused by a more intense NAA+NAAG signal, which in turn can affect the quantification of GSH. On the other hand, the co-editing of MM in GABA editing directly affects the GABA measurements because the edited GABA and MM signals at ~3 ppm overlap completely and are indistinguishable. Co-editing of metabolite signals needs to be investigated on in vitro phantoms containing the metabolites of interest and measured at an appropriate pH (7.0 – 7.4), RF coil load, and temperature (37 °C). The experimentally measured co-editing profile can also be confirmed with quantum-mechanical simulations and incorporated into simulated metabolite basis sets for subsequent spectral fitting and quantification.
Table 1.
Common editing target metabolites and associated co-edited metabolites.
| Target metabolite(s) | Co-edited metabolite(s) | Ref | |||
|---|---|---|---|---|---|
| Target name (chemical group) | Target chemical shift (ppm) | Edit chemical group | Edit chemical shift (ppm) | ||
| Asc (H6/H6′) | 3.72 – 3.74 | H5 | 4.00 | BHB, lactate, threonine | 8,66,92 |
| aspartate (H3/H3′) | 2.67 – 2.80 | H2 | 3.89 | 106 | |
| BHB (H4) | 1.19 | H3 | 4.13 | lactate, threonine | 20 |
| cystathionine (H4/H4′) | 2.72 | H3/H3′ | 2.13–2.19 | NAA, NAAG, 2HG, GABA, homocarnosine, MM, glutamate, glutamine | 126 |
| GABA (H4) | 3.01 | H3 | 1.89 | MM, glutamate, glutamine, homocarnosine, NAA, NAAG | 11,63 |
| glucose (bH2) | 3.23 | bH1 | 4.63 | GSH, NAAG | 109,110 |
| GSH (cysteine H3/H3) | 2.93 – 2.98 | H2 | 4.56 | NAA | 42,80,90 |
| 2HG (H2) | 4.01 | H3/H3′ | 1.82 – 1.98 | GABA, homocarnosine, MM, cystathionine, glutamate, glutamine, cysteine, NAA, NAAG | 96,97,127 |
| lactate (H3) | 1.31 | H2 | 4.10 | BHB, threonine | 103,128 |
| NAAG (aspartate H3/H3′) | 2.52 – 2.72 | H2 | 4.61 | NAA | 7,104 |
| threonine (H4) | 1.32 | H3 | 4.25 | BHB, lactate | 37,102 |
| serine | 3.94–3.98 | H | 3.83 | NAA, glutamate, myo-inositol | 112 |
Note: Spectral editing is recommended for metabolites listed above at 3 T or lower. In general, both J-difference and MQF editing methods are equally applicable. While the need for spectral editing is less at ultra-high magnetic fields (7 T or above) due to an increased spectral separation, editing is still recommended for some metabolites (e.g., BHB) to improve the reliability of detection. Spectral editing could also help to establish the presence of previously unobserved metabolites in vivo.113
2HG = D-2-hydroxyglutarate; Asc = ascorbate; Asp = aspartate; BHB = β-hydroxybutyrate; GABA = g-aminobutyric acid; GSH = glutathione; MM = macromolecules; NAA = N-acetylaspartate; NAAG = N-acetylaspartylglutamate
The in vitro evaluation of MM co-editing is more challenging since no single protein is a perfect approximation of the in vivo MM profile. In some cases, the co-editing can be evaluated on an amino acid that approximates a part of the MM profile. For example, lysine can be used to simulate co-edited MM in GABA editing at 3 ppm. However, in general, the in vitro approximations are not accurate enough to quantify the amount of co-editing in vivo. MM co-editing can be minimized by using editing pulses with a narrow frequency profile and/or employing a symmetric editing approach.11 Both of these methods make the measurement more sensitive to frequency drift and subject motion, thus are not recommended when real-time frequency updates are not utilized. In addition, symmetric editing reduces the SNR of GABA because the OFF pulse partially inverts the coupled spins, especially at < 7 T. Alternatively, inversion recovery can be used for MM nulling. However, most of the MM reducing methods decrease the editing efficiency of the GABA signals. Hence, the common approach in clinical studies has been to accept the MM contamination to the edited GABA signals and label the total GABA and MM signals as GABA+. While this is a practical solution, it should always be remembered that any changes in GABA+ could be caused by changes in GABA as well as the confounding MM signals. In addition, any changes in B0 caused by motion or frequency drift will influence the proportion of MM and GABA that contribute to GABA+. For a standardized editing method (e.g., MEGA-PRESS) at a given B0 strength, it is recommended to establish the MM contribution and the influence of frequency drift. The MM contribution can be determined in a separate study by utilizing differences in T1 relaxation through inversion recovery or performing symmetric editing.11
Recommendations regarding co-editing during spectral editing are:
Identify co-editing partners for the editing target resonances using known chemical shifts27,28 (as listed in Table 1 for various metabolites), or in vitro phantoms with appropriate pH, temperature, and RF coil load.
In the case of no or partial spectral overlap, the co-edited metabolites can be quantified through spectral fitting using appropriately constructed basis sets.
In case of complete or nearly complete spectral overlap (e.g., GABA, MM, and homocarnosine), additional experiments are recommended to estimate the co-editing contribution.
Co-editing of MM is more complicated because no well-defined small molecule approximation exist that allow an in vitro investigation. It is recommended that the contribution of MM to the edited signals is established in a separate dedicated study and reported in subsequent studies. However, when the contribution of MM to the edited signals is not available, a practical recommendation is to specify the MM contribution in quantification of the target edited signals (e.g., GABA + MM = GABA+).
Frequency drift due to subject motion/system instability
Uncorrected hardware-related system frequency drift during data acquisition adversely affects edited signals by changing the contribution of co-edited signals and editing efficiency. In addition, phase or frequency variations due to subject motion or system instability can lead to subtraction errors in J-difference editing. Overall, spectral editing requires greater stability of frequency, phase, and the subject than conventional MRS. Navigator scans that provide real-time frequency information could be utilized to update the system frequency before the acquisition of each transient or each data block. Further details can be found in the consensus recommendation paper on motion and frequency correction in this special issue.16
When real-time frequency correction is not available, the following recommendations are proposed:
The bandwidth of the editing pulses needs to be sufficiently narrow as to minimize (unwanted) co-editing, but sufficiently broad to provide some immunity to frequency drift. Spectral editing data should be discarded (or frequency drift should be accounted for by quantum-mechanical simulations when feasible for fitting29) when the frequency drift is more than 25% of the editing pulse bandwidth (i.e., full width at half maximum).
Acquire spectral editing data in acquisition blocks of 2 to 5 min to monitor frequency drift from system instability or subject motion with interleaved scanner frequency adjustments between acquisition blocks. The duration of the block should be short enough to keep drift at or below 5 Hz.
When possible, perform spectral editing measurements before high gradient duty-cycle MRI sessions to minimize temporal frequency drift. A high gradient duty-cycle MRI session can lead to significant frequency drift (up to 5 Hz/min), which can last for 30 min or longer.30,31 In cases when spectral editing is performed during strong frequency drift, use real-time frequency correction. The detailed recommendations on this topic are described in the consensus recommendation paper on motion and frequency correction for MRS.16
Spectral editing data within an acquisition block of several minutes need to be stored as single-transients, thus enabling the possibility of post-acquisition frequency (and phase) alignment. Although post-acquisition correction of frequency and phase can reduce the subtraction artifacts, it cannot recover the signal loss caused by motion and/or frequency drift during data acquisition.
Eddy currents with short time constants during signal acquisition are invariably present in all MRS scans and the effect can be recognized by asymmetrical line shapes of all resonances in spectra. The standard approach using the phase profile in a reference water signal is recommended to correct the lineshape distortion in the metabolite signals as described in the consensus recommendation paper on data analysis and quantification.32
In cases of MR systems with strong B0 eddy currents, it is recommended to establish the effects of eddy currents on the editing performance in phantoms because the effect of eddy currents on the editing yield caused by frequency shift during the editing pulse cannot be corrected by the standard approach.32
Data processing
J-difference editing data stored as single transients should undergo correction of frequency and phase shifts to minimize line broadening and subtraction artifacts.14,25,33 When the sensitivity of single-transient signals is not sufficient for an accurate frequency and phase estimation, post-acquisition signal averaging is recommended to achieve a sufficient SNR. The water signal needs to be acquired with the same sequence for eddy current correction and to establish the phase and amplitude factors necessary for optimally combining multi-channel receive signals. During the frequency- and phase-alignment procedure, data quality assurance and data rejection can be performed. Excessive and abrupt frequency and phase shifts, broadened lines, and a large unsuppressed water signal are all indications of subject motion and such transients should be excluded from signal summation. Following summation of the sub-spectra, a small frequency and/or phase correction between the two sub-spectra may be required before subtraction. The difference spectrum should contain the target and co-edited signals with their expected relative phase relations. Subtraction of singlet signals (e.g., total creatine = creatine + phosphocreatine, total choline, NAA) should be complete without requiring an amplitude adjustment between the two sub-spectra. After all data processing corrections, if the amplitude of singlet subtraction artifacts in the difference spectrum is still greater than 10% of the target edited signals, the data should be excluded as they are likely affected by subject motion and/or system instability. The data with significant and variable signal intensity at around 1.5 ppm may require a careful examination of the data quality as it is indicative of lipid contamination from outside of the voxel or subject motion during scans.
Recommendations regarding data processing in J-difference editing are summarized below:
Correct frequency and phase shifts of single transients to reduce subtraction artifacts.
Exclude severely corrupted transients due to subject motion before signal averaging.
Exclude spectra with subtraction artifacts > 10% of the target edited signals singlet.
If significant lipid signals are present at around 1.5 ppm, examine the causes (e.g., subject motion, frequency drift, voxel locations, insufficient signal crushing, and CSDE) to determine the inclusion/exclusion of edited spectra.
Data quantification
Once edited spectra meet the recommended data quality criteria in consideration of singlet subtraction artifacts, overwhelming lipid/water signals, frequency drift, and subject motion,16 the data can undergo quantification processes. Spectral fitting using measured or simulated basis sets is recommended to quantify edited spectra in J-difference editing. Standard fitting algorithms (e.g., LCModel or jMRUI) designed for non-edited spectra need to be adapted for the edited spectra or specifically designed algorithms (e.g., GANNET for GABA34) could be used.
The generation of appropriate basis sets using simulations rests on accurate chemical shifts and scalar couplings for all metabolites, and proper matching of sequence timings, RF pulses, and B0 gradients between the simulation and experiment. A large number of publications are available detailing and refining chemical shifts and scalar couplings for metabolites.27,28,35,36 While the values for common metabolites have steadily converged, uncertainty remains for less commonly detected metabolites. When spectral patterns of experiment and simulation do not properly match, it is recommended to measure and characterize the metabolite of interest on a well-constructed phantom with B0 and B1+ distributions similar to the in vivo condition. Especially temperature and pH of the phantoms need to be carefully matched to in vivo conditions as temperature and pH can influence the chemical shifts of metabolites. The sequence parameters in experiments, including RF pulse power and editing frequency setting, also need to match those in simulations. For simulations of 3D-localized MRS pulse sequences, it is generally not acceptable to replace the sequence with ideal, hard RF pulses without B0 gradients. With the RF bandwidth typically used in vivo, the spectral pattern of J-coupled resonances can vary significantly or may lead to a complete cancelation of the signal. Therefore, it is recommended to simulate the MRS pulse sequence including RF pulses and gradients as closely matched as possible to the actual experiments.
As spectral editing is typically performed at medium to long TE, T2 relaxation effects are significant in most edited spectra, necessitating T2 relaxation correction for quantification. Edited signals (e.g., GABA) are typically quantified using an internal reference signal (e.g., total creatine) for which the concentration is assumed. T1 and T2 relaxation times and their B0 dependence are fairly well characterized for total choline, total creatine and NAA,37,38 while those of the edited signals are often unknown with few exceptions.39–46 The simplest option to address the T1 and T2 relaxation effect is to assume equal T1 and T2 values for edited and reference signals. However, it is suggested that an effort is made to establish T1 and T2 values for the metabolites of interest. The measurement of T2 for J-coupled metabolites (edited metabolites) is difficult since their signal intensity decreases and their spectral pattern changes with TE, as evidenced by a large range of reported T2 values for these metabolites. The use of unsuppressed water signals as a concentration reference is recommended when changes in reference metabolites are expected in pathologic conditions as described in the consensus recommendation paper on data analysis and quantification.32
Recommendations regarding data quantification are summarized below:
The use of measured or simulated basis sets is recommended for the spectral fitting of edited spectra in J-difference editing to provide more reliable quantification.
For simulations of basis sets, proper matching of sequence timing, RF pulses and B0 gradients between simulation and actual experiment is required.
The measurements of T1 and T2 values for the metabolites of interest could help accurate quantification with the relaxation correction. When the relaxation correction is performed, the source of T1 and T2 values or assumptions made for those values need to be reported.
Minimum system requirements for spectral editing
In addition to the technical considerations and complications described above, successful spectral editing requires other considerations related to the MR system, including good B0 homogeneity and achievable RF amplitudes.
Static magnetic field (B0) homogeneity
Optimizing the B0 homogeneity through shimming is important for any MRS application, including spectral editing. The general considerations regarding B0 shimming are discussed in the consensus recommendation paper on B0 shimming for MRS.47 Achieving B0 homogeneity through shimming improves the quantification accuracy as it reduces the linewidths and spectral overlaps. B0 inhomogeneity can become an important factor when spectral editing is combined with MRSI. While the local B0 homogeneity of individual voxels may be sufficient to provide spectra with adequate quality, global B0 inhomogeneity across voxels will lead to frequency offsets and spatial variations in the editing efficiency when narrowband selective editing pulses are used. Thus, spatial variations of the editing efficiency need to be corrected using the information from singlet signal frequency shifts within the MRSI42 or MRI-based B0 maps, and modeling of edited spectra using simulated basis sets with the appropriate frequency offset.
RF amplitude
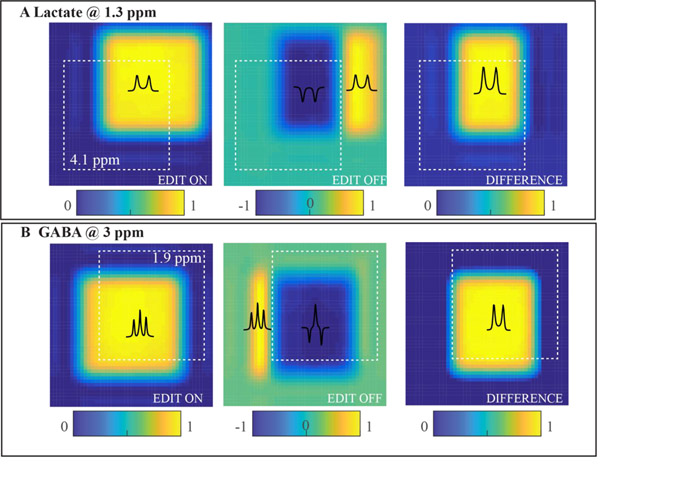
The maximum achievable RF amplitude is a key parameter in determining the performance of spatial localization and editing efficiency. For built-in body transmit RF coils at 3 T, the maximum RF amplitude, B1+(max), is typically limited to 15 – 30 μT for 1H MRS. The bandwidth of a conventional 180° slice selective refocusing pulse would then be limited to ~1 kHz, leading to significant CSDE even at 3 T. The presence of CSDE for scalar-coupled spins, i.e., coupling partners, leads to four distinct spatial compartments within the volume of interest in which coupled spins experiences zero, one or two refocusing localization pulses, which modify the spin evolution.4,19,20 Figure 1 demonstrates the effect of limited RF bandwidth of the localization pulses on the performance of MEGA-PRESS for lactate (Fig. 1A) and GABA (Fig. 1B). During EDIT ON scan (with editing pulses selecting the coupled spins), target spins (lactate at 1.3 ppm and GABA at 3 ppm) are in-phase in all compartments. Therefore, the editing pulses are effectively suppressing the influence of CSDE on the spin evolution. However, during the EDIT OFF scan, target spins in all compartments are not all in-phase, leading to a loss of signal intensity in the edited spectra. The effect of CSDE during spectral editing is a loss of editing efficiency that is a complicated function of various parameters including bandwidth and profile of refocusing pulses, TE, scalar coupling constants, resonance patterns, and chemical shifts. For example, the effect of misregistration/signal loss is increased with the increased separation between coupled spins (compare 345 Hz vs 135 Hz for lactate and GABA at 3 T) (Fig. 1). The most efficient way to minimize this effect and thus maximize the editing efficiency is using high-bandwidth RF pulses. Two possible approaches include the use of dedicated head-only transmit RF coils that provide higher B1+(max) and/or high bandwidth adiabatic RF pulses as in (semi-)LASER sequences.
Figure 1. Effect of CSDE on editing efficiency for PRESS editing of lactate (TE = 144 ms) and GABA (TE = 70 ms).
(A) Numerical simulations of the 2D signal distribution of 1.3 ppm lactate resonance (a coupled spin at 4.1 ppm 2D distribution is depicted by the white dashed box). The 2D plane represents two spatial dimensions selected by two 180° localization RF pulses (1 kHz refocusing bandwidth). While severe spatial misregistration between two coupled spins is evident during the EDIT ON scan, the overall signal intensity is relatively uniform. This is due to the editing RF pulses refocusing voxel compartments that did not experience the effect of the 180° localization pulses. However, for the EDIT OFF case, one can observe distinct compartments where the spins of interest exhibit phases opposite that of the theoretical non-localized dual spin-echo experiment.124 (B) Numerical simulations of the 2D signal distribution of 3.0 ppm GABA resonance (coupled spins at 1.9 ppm 2D distribution is depicted by the white dashed box). The same dynamics are observed for GABA editing, albeit to the less extent due to smaller frequency difference between GABA coupled spins compared to lactate. Image intensity represents the integral of doublets of lactate or two outer peaks of GABA signals and color bars indicate respective intensity ranges. The significant reduction of the signal intensity in the final DIFFERENCE spectra (e.g., ~50% for lactate, ~20% for GABA) due to CSDE is observed at 3T.
2. Multiple quantum filtering (MQF) editing
Multiple quantum coherence filtering (MQF) is a single-shot editing method based on the selection of target coherences in J-coupled spin systems using MQ gradient filters and frequency selective RF pulses. This MQF has its origin in the first observation of multiple quantum transitions in 195648 and the subsequent introduction of pulsed field gradients to select the desired coherence pathway in multiple quantum transitions in 1980.49 Practical implementation of MQF in vivo was achieved by incorporating double quantum coherence filtering gradients50 and three-dimensional localization.51
In MQF, 3D localization can be effectively incorporated into the pulses used for basic MQ generation and refocusing using slice selection gradients. Various slice selection schemes have been incorporated into MQF, including STEAM,52 PRESS,51,53–55 and hybrid of STEAM and PRESS.56,57 The use of 90° pulses for slice selection is preferable to refocusing pulses because of higher achievable bandwidth of 90° pulses than 180° refocusing pulses in minimizing signal loss associated with CSDE.
MQF sequences typically use a selective MQ conversion pulse to increase the editing yield.58 A double-band selective refocusing pulse is often used during the MQ preparation period to enhance editing yields by inhibiting coherence leakage to non-selected coherences and to suppress other non-selected J-coupled resonances.59 Due to the single-shot nature of MQF, the selectivity of the doubly selective pulse could be set higher than that of J-difference editing to reduce co-editing of unwanted J-coupled signals. By combining the selectivity of the doubly selective refocusing pulse and the selective MQ conversion pulse, MQF provides better suppression of interfering J-coupled resonances than J-difference editing.60
Because MQF removes all singlet signals including water, total creatine, and NAA, there is no discernable frequency or concentration reference within the MQF spectra. To overcome this lack of reference signals, simultaneous acquisition of singlet signals can be added to MQF within the same TR using dual echo approaches.56,61,62 Simultaneously measured singlet signals such as water or creatine could serve as a frequency, phase, and concentration reference for the edited signals and a reference for coil combination. With the advantage of the single-shot nature of MQF, it can readily be converted to MRSI to measure GABA,61,63 GSH42, lactate58,64,65 and ascorbate.66 Inaccurate RF calibration and frequency drift can lead to signal loss in MQF like any other MRS measurements. However, frequency drift and subject motion do not cause subtraction artifacts in MQF due to its single-shot nature, unlike J-difference editing methods.
Most of the recommendations for J-difference editing applies to MQF editing including items a, d and e in the RF power calibration section, items a and b in the co-editing of unwanted resonances section, items a, b and c in the frequency drift due to subject motion/system instability section and all items in the minimum requirements for spectral editing section.
Recommendations for MQF editing are summarized below:
The use of a doubly selective refocusing pulse is recommended to enhance editing yields and to suppress non-selected J-coupled resonances.
Simultaneous acquisition of singlet signals (e.g., total creatine or water) within the same TR is recommended to provide a reference for frequency, phase, concentration, and coil combination.
3. Alternative techniques to J-difference or MQF editing
J-difference and MQF editing are arguably the most used methods to selectively detect metabolite signals that exhibit spectral overlap. Other spectral editing methods including editing through bond polarization transfer such as Hartman-Hahn transfer for GABA editing 67 and total correlation spectroscopy (TOCSY) for glucose editing68 have been demonstrated in the human brain, which provides the theoretical editing efficiency of up to 100%. Where spectral editing is not available, detection of a few metabolites with low concentration and other overlapping resonances could be achieved using standard MRS methods. In cases of partial spectral overlap (e.g., 2HG and glutamate), the TE of a standard PRESS method69 can be optimized to enhance signals of the metabolite of interest (e.g., 2HG) while simultaneously reducing the overlap with neighboring metabolites (e.g., glutamate) through a peak narrowing effect due to lineshape modulation by scalar coupling evolution 70. While it is very difficult to obtain absolute separation with this strategy, it can be combined with spectral fitting algorithms to estimate the metabolite of interest. However, reliable quantification requires very good B0 homogeneity with narrow linewidths (≤ 0.05 ppm). Note that selecting an optimal TE has little value when the overlapping metabolite has no scalar coupling and needs to be removed either by subtraction or MQF.
For some metabolites, the B0 strength can have a dramatic effect on signal patterns and overlaps. For example, glutamate and glutamine are strongly overlapping below 3 T, but readily separated at 7 T and above (see Fig. 5 in Reference17). To improve the linewidth and reduce the spectral overlapping, homonuclear decoupling techniques such as multiple TE averaging can be used to simplify the multiplet splitting and provide singlet-like lines for most metabolites. 71 However, the number of TEs necessary to achieve decoupling is relatively large (> 10), and any measurement instability will interfere with the narrowing effect, making this method prone to artifacts and incomplete signal separation. Alternatively, 2D J-resolved and correlation spectroscopy could be used to separate metabolite signals,72–76 including the overlap between GABA and macromolecules. However, this class of methods have the challenge of long acquisition times due to encoding of the second spectral dimension and low SNR of the 2D cross-peaks compared to MEGA or MQF editing. On the other hand, it should be noted that while most metabolites, including glutamate, glutamine and lactate, are overlapping with a range of MM resonances in 1D spectra, the cross-peaks of metabolites and MM are separated in 2D spectra. Further comparison between 1D editing and 2D spectroscopic methods has been described with more details in a recent review paper,77 and here we focus on the 1D editing methods. A method to reduce the contribution of MM in 1D spectra is based on the selective nulling of short T1 MM signals with an inversion recovery while simultaneously retaining part (~60%) of long T1 metabolite signals.10 While MM nulling does not improve the separation of glutamate and glutamine directly, it provides spectral simplification by removing the MM baseline, which in turn will translate into an improved and more reliable spectral fit at a cost of reduced SNR. Alternatively, a reliable spectral fit can be achieved by independent measurement of the MM spectrum and including it in the basis set.
4. Metabolite-specific considerations
GABA
GABA, the major inhibitory neurotransmitter, is the most common target for spectral editing due to its complete overlap with total creatine singlet signals that are an order of magnitude larger than those of the GABA multiplet. GABA editing was first described in 1993,78 but reached broad usage with the development of BASING-2 and MEGA-based1 editing methods. In addition to the overlapping signal of total creatine, the potential co-editing of MM has been recognized as a complication to selective GABA detection and the contribution of MM to the GABA signals has been investigated using various techniques, including T1-based signal nulling79,80 and symmetric editing.11 At 3 T or lower, it has been reported that MM contributions comprise over 50% of GABA signals in MEGA-based editing methods.78,81 While editing methods such as doubly selective MQF and Hartman-Hahn transfer have achieved a significant reduction of MM contributions even at 3 T,56,60,67,82 co-editing of MM remains the biggest challenge for spectral editing of GABA. A pragmatic solution has been to consider the combination of edited GABA and MM (typically labeled GABA+) with the understanding that any changes in GABA+ signals may be due to GABA and/or MM. Field drift and/or subject motion can also change the relative GABA and MM contributions to the edited signal.16 Homocarnosine, which is a combination of GABA and histidine, is another metabolite known to contribute to GABA+ signals, but signals of the GABA moiety of homocarnosine are not easily distinguishable from GABA.83
Glutathione (GSH) and ascorbate (Asc)
The reduced form of glutathione (GSH) and ascorbate (Asc, commonly known as vitamin C) are the two most highly concentrated and important nonenzymatic antioxidants in the human body. In its reduced form, GSH protects cells from oxidative stress that plays a critical role in various brain disorders and is transformed into its oxidized form (GSSG) when scavenging free radicals.84 Consequently, increased GSSG/GSH ratios and total glutathione (GSH + GSSG) have been shown to reflect oxidative stress using biochemical assays ex vivo.85 The concentration of GSSG is known to be two orders of magnitude lower than that of GSH,86 thus it is beyond the detection limit of in vivo MRS.84 Therefore, GSH has been an important research target in aging87 and various neurological disorders 62,88 where oxidative stress has been implicated. While GSH can be measured directly via short TE 1H MRS at 7 T and above,80 the resonances of GSH overlap considerably with the resonances of more abundant metabolites at lower B0 strength, leading to poor reproducibility.89 Thus, in vivo detection has mostly been achieved via spectral editing.53,55,62,80,90 The GSH resonance at 2.95 ppm can be well detected in the difference spectrum when spectral editing pulses are applied at the GSH resonance at 4.56 ppm without major quantification problems caused by the co-editing of other metabolites. The co-editing of aspartyl resonances of NAA is unavoidable but does not significantly affect the quantification of GSH.
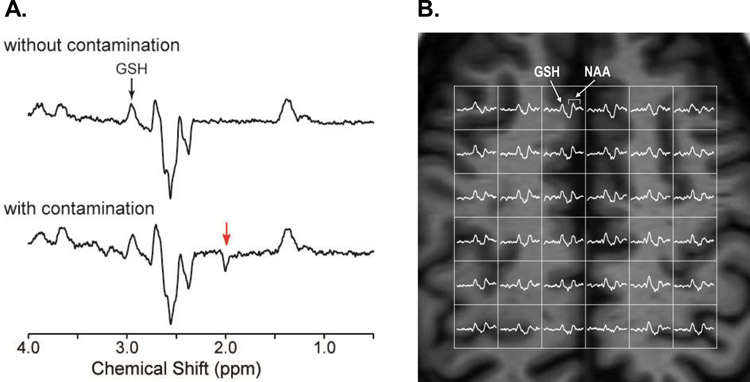
Ascorbate has been mostly measured via spectral editing,66,91,92 while direct detection is feasible at 7 T92 and above with the use of short TE MRS. Ascorbate and GSH can be co-edited using a double spectral editing approach.8 Figure 2 shows typical examples of GSH detection in the human brain using MEGA-PRESS (Fig. 2A) and MQF (Fig. 2B) editing methods.
Figure 2.
(A) Representative single voxel MR spectra of GSH from the motor cortex (3.5 × 2.5 × 2.3 cm3) of a subject with ALS using MEGA-PRESS (TE = 68 ms, TR = 2 s, 512 averages, editing pulse at 4.56 ppm for edit on and at 7.5 ppm for edit off) at 3 T. The bottom spectrum is shown without processing. Slight frequency drifts over time lead to small subtraction artifacts as can be seen for NAA (red arrow). The top spectrum shows the same spectrum as below after the individual transients are frequency aligned leading to a clean, flat baseline. Both spectra are shown with 1 Hz exponential line broadening. (B) GSH MRSI measured from the fronto-parietal region of the human brain using the doubly selective MQF editing sequence (TE = 115 ms, TR = 1.5 s, matrix size = 8 × 8, nominal voxel size = 1.25 × 1.25 × 3.0 cm3, field of view = 20 cm, and scan time = 16 min) at 3 T. All spectra were processed with 2 Hz exponential line broadening and are shown in the range from 3.6 to 2.2 ppm (adapted from Reference125).
D-2-Hydroxyglutarate (2HG)
Overproduction of the oncometabolite 2HG is the metabolic hallmark of isocitrate dehydrogenase (IDH) mutations in cancer.93 IDH mutations are highly frequent in several cancers, such as glioma, acute myeloid leukemia, chondrosarcoma, and cholangiocarcinoma. In these cancers, IDH mutations are early genetic events and the accumulation of 2HG plays a major role in modulating metabolism, epigenome, microenvironment, and immunity to further drive tumor formation. Due to its unique biology and high tumor specificity 2HG can be used as a biomarker to probe multiple mechanisms in mutant IDH cancers. 2HG measured by 1H MRS has emerged as an important clinical application, and it was shown to be highly valuable for diagnosing and monitoring glioma,94 or assessing treatment response and pharmacodynamics of targeted mutant IDH1 inhibitors.95
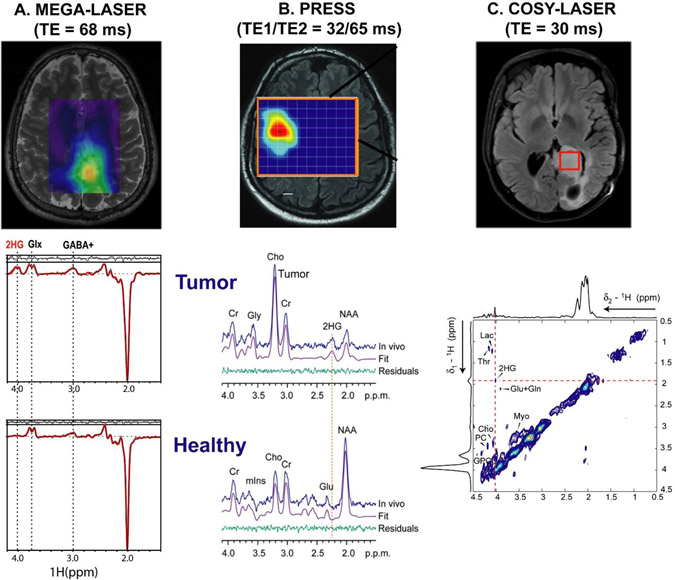
2HG is a strongly coupled spin system of five protons96,97 with complicated multiplet peak patterns in three chemical shift regions at 4.02 ppm (H2), 2.25 ppm (H4,H4’), and 1.9 ppm (H3,H3’). 2HG signals are overlapped by nearby signals of major metabolites such as glutamine and glutamate (2.1–2.3 ppm), total creatine (3 ppm) and myo-inositol (4 ppm), and NAA (2 ppm).27 Several editing methods have been used to resolve the spectral overlap between 2HG and other metabolites: 1) long TE schemes with dual,96 triple70 or quadruple refocusing,98 2) J-difference editing,96,97 and 3) 2D correlation spectroscopy (COSY).97 Long TE schemes create a particular phase modulation for the 2HG signal at 2.25 ppm to maximize the separation and difference from neighboring glutamate and glutamine peak lineshapes. The longer TE schemes96 have the advantage of a simpler implementation and shorter acquisition times, but the background of normal brain metabolites is not removed and can still contaminate the fitting of the 2HG peak pattern, especially in situations with suboptimal shimming (linewidth > 0.08 ppm). J-difference editing removes unwanted background signals and selectively detects the 2HG signal at 4.02 ppm. However, this method requires longer acquisition times and is sensitive to measurement instability that may produce subtraction artifacts, requiring more advanced implementation for robust performance.99 Figure 3 shows a summary of 2HG detection strategies, including J-difference editing (Fig. 3A), optimal echo-time selection (Fig. 3B), and 2D COSY (Fig. 3C) methods.
Figure 3.
Optimized in vivo MRS detection of 2HG from mutant-IDH glioma patients using (A) J-difference editing 3D MEGA-LASER sequence (TR/TE = 1600/68 ms, matrix 10 × 10 × 10, FOV=200 × 200 × 200 mm3, and acquisition time = 9.5 min; reproduced with permission from Reference99); (B) long echo 2D PRESS (TR/TE = 1300/97 ms, matrix 16 × 16, FOV = 160 × 160 mm2, slice 15 mm, and acquisition time = 10 min; reproduced with permission from Reference96); and (C) 2D COSY-LASER (TR/TE = 1500/30 ms, voxel 3.0 × 3.0 × 3.0 cm3, 64 t1 increments, acquisition time = 11.5 min; reproduced with permission from Reference97) at 3 T.
Lactate, threonine, and β-hydroxybutyrate (BHB)
Lactate, the anaerobic metabolic product, was an earlier target of spectral editing due to its simple spin system, which is weakly-coupled even at 1.5 T.100 The concentration of lactate is fairly low in normal brain tissue at ~0.5 to 1mM,101 but it can reach several mM in pathologies such as tumors or stroke.2 In the normal brain, lactate can be observed at 1.31 ppm in well-localized, short TE MR spectra,17. However, the lactate signals at 1.31 ppm are almost completely overlapping with the threonine signals at 1.32 ppm. Thus, the observed signals at 1.31 ppm are likely from both metabolites. Threonine editing has been shown at 3 T, 4 T, and 9.4 T.37,102 In pathological tissues, most extracranial tissues and in the absence of high-quality spatial localization, the lactate signal is quickly overwhelmed by intense signals from lipids. In these cases, lactate has been detected through J-difference editing2,102 or MQF editing.103 In applications with the presence of lipid signals in the brain (e.g., brain tumors), lactate detection with J-difference editing is desirable as it has 100% theoretical editing efficiency, when neglecting T2 losses. However, in the presence of large lipid signals or small subject/system instabilities, lactate detection via MQF editing is recommended over J-difference editing.103 A common co-editing partner of lactate, and a valid editing target in its own right, is β-hydroxybutyrate (BHB).20 BHB, together with acetone and acetoacetate, is a ketone body that can function as an alternative substrate to glucose. Under normal (fed) conditions, BHB levels in the blood are low, but can rise to several mM after 72 hours of fasting. The methine proton chemical shifts of BHB at 4.13 ppm and lactate at 4.10 ppm, together with the same scalar coupling, make the editing efficiency of BHB and lactate identical.
Other less frequently edited metabolites (aspartate, cystathionine, glucose, NAAG, serine, taurine)
N-acetylaspartylglutamate (NAAG), a glutamatergic modulator, can be detected via spectral editing at 3 T7,104 or directly detected at 7 T and above.105 Aspartate106 and taurine107 have been measured using spectral editing. Recently, direct detection of taurine has been demonstrated at 7 T.17,108 Glucose has been detected using various editing methods.68,109,110 At 4 T and higher, glucose can be detected directly via the αH1 resonance at 5.22 ppm.111 Serine, another important amino acid implicated in cancer and psychosis, was shown at 7 T with a combination of triple refocusing and difference techniques that edits the 3CH2 group of serine signals at 3.94 and 3.98 ppm and subtracts the overlapping total creatine signal at 3.92 ppm.112 Recently, cystathionine has been detected via spectral editing in brain tumors and has been suggested as a non-invasive biomarker for 1p/19q-codeleted gliomas.113
Simultaneous editing of multiple metabolites
Conventional J-difference editing (e.g., MEGA-PRESS) prioritizes the measurement of a single low-concentration metabolite of interest over the assessment of a broad neurochemical profile. This precludes the interrogation of a large number of editable metabolites. However, simultaneous editing allows more than one metabolite (e.g., GSH/Asc editing8 and GSH/Lac editing114), while its application is restricted to spin systems with resolved editing targets and detected resonances using double editing with MEGA (DEW-MEGA) PRESS. Spectral Hadamard-encoding techniques address this problem by segregating overlapping edited resonances into separate difference spectra. This multiplexed approach allows more than one editing experiment to be performed in a single acquisition. Hadamard-encoding and reconstruction of MEGA-edited spectroscopy (HERMES) is a four-step editing scheme that simultaneously resolves signals from two or three J-coupled spin systems in a single experiment without affecting the SNR.7 HERMES has been implemented for NAA/NAAG, 7 GABA+/GSH,21,115 MM-suppressed GABA/GSH,116 and NAA/NAAG/aspartate,106 and has been extended to MRSI for GABA+/GSH.117
DEW-MEGA, HERMES, and other multi-target editing approaches share common traits in that they are more SNR- and time-efficient than performing multiple consecutive editing experiments, but also the likelihood for subtraction artifacts or co-editing of unwanted metabolites increases with the complexity of the editing scheme. The SNR efficiency can be significantly influenced by the editing efficiency of each of the target metabolite at a chosen echo time of the experiment. This effect could be minimal in some cases, e.g., for GSH and GABA, but could be substantial for other metabolites. Nevertheless, the use of HERMES for GSH/GABA+ in a multi-site study118 and a multi-vendor study119 show promising results.
5. Technical implementation of editing in multi-site clinical studies
As with any multi-center clinical trial using an MR-based outcome measure, standardization of data acquisition is an important consideration. In addition to the usual considerations for MRS sequence parameters (e.g., standardization of TR, TE, voxel size, localization technique, scan time, sampling rate, and the number of acquired points) and RF power calibration, it is important to standardize the properties of the editing pulses, e.g., pulse frequencies, shapes, and bandwidths. Particularly for GABA editing, the degree of MM co-editing due to off-resonance inversion of the 1.7 ppm MM resonance will depend on the bandwidth of the editing pulses as well as the frequency profile. Efforts to standardize acquisition parameters without full sequence standardization have been moderately successful,120–122 with 28% of the total variance in a 24-site, 272-subject dataset being explained by vendor and site effects.
More recently, efforts have been made to standardize the full sequence timing (e.g., RF pulse shapes, durations, amplitudes, and timings) for MEGA-PRESS on four different commercial MR platforms,119 which resulted in substantial reductions in vendor sequence discrepancies. Aside from the data acquisition and system requirements discussed earlier, the most impactful difference in sequences lies in editing pulse timing, which is sub-optimal in common implementations on some platforms, i.e., editing pulses are not constrained to be TE/2 apart which gives most efficient editing. The universal sequence represents the current best-practice for standardized multi-vendor studies. When MQF editing is used for multi-site clinical studies, full harmonization of editing sequences with identical RF pulses (e.g., pulse shapes, frequencies, and bandwidths), sequence timing, and gradients are necessary, considering the complex influence of these factors on the edited signals.
Currently, spectral editing is mostly performed as a single voxel MRS technique. Thus, technical consideration pertinent to the single-voxel techniques is important, including standardized anatomical landmarks for voxel location. Automated voxel placement based on co-registration to ‘model’ anatomical images has been recently implemented123 and can reduce variability between subjects and operators within the site as well as between sites. However, since there is approximately a 10% variation between subjects in brain size, and MRS voxel size is usually fixed regardless of head size, there is a practical lower limit on the matching anatomical positions between subjects. Reproducibility of voxel placement can be improved by systematic training of operators and providing templates and anatomical landmark-based rules for voxel placement.
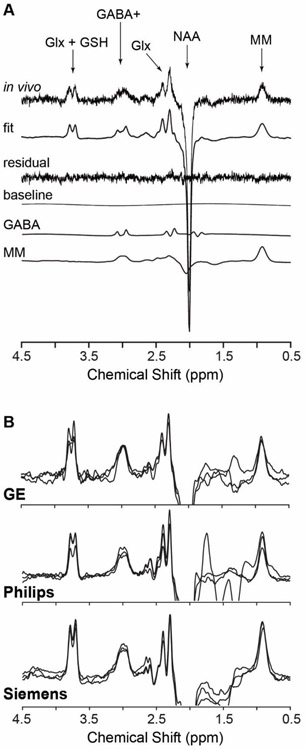
Figure 4 shows typical results of GABA detection in a research setting (Fig. 4A) and as part of a multi-site, multi-vendor clinical trial (Fig. 4B). Contributions of MM to GABA+ signals can be clearly identified in (A). While both acquisitions display prominent GABA+ signals, the spectra in (B) show significant and variable lipid-related signals in the 1–2 ppm range. While this does not necessarily negatively affect the GABA detection and quantification, the presence of a non-negligible baseline distortion with spurious signals should be a warning sign. Further investigation into the spatial position of the voxel, subject motion, frequency drift, or outlier spectra may be warranted to establish the reliability of the edited GABA+ signals.
Figure 4.
(A) A representative GABA spectrum from the human occipital lobe (4.0 × 2.3 × 3.0 cm3) using MEGA-PRESS (TR/TE = 3000/68 ms, 256 averages) at 3 T. Sub-spectra show traces of best fit, residuals, baseline, GABA and MM. The MM spectrum is an average metabolite-nulled GABA-edited spectrum from 13 subjects. Both metabolite and metabolite-nulled spectra were acquired with editing pulse at 1.9 ppm with resolved averages to monitor motion and frequency drifts and resetting the frequency at 64 scan blocks. (B) GABA+ spectra from the medial parietal region randomly selected from the Big GABA dataset show typical data quality for GE (B, top), Philips (B, middle), Siemens (B, bottom) using MEGA-PRESS (TR/TE = 2000/68 ms, voxel size = 3.0 × 3.0 × 3.0 cm3, 320 averages). See details in Reference121.
6. Overall consensus recommendation statements:
Sequence selection for J-difference editing: High-bandwidth refocusing pulses for spatial localization are recommended for optimal editing efficiency and sensitivity. At 3 T and below, PRESS-based methods can be used for GABA editing. At higher B0 with limited B1, (semi-)LASER-based editing methods are recommended as CSDE is greatly reduced by the higher bandwidth of adiabatic refocusing pulses.
RF power calibration: Transmit RF power calibration should be performed on the localized volume. Maintain RF pulse durations identical regardless of the RF coil load to prevent any variations in CSDE, co-editing, and editing efficiency.
Co-editing: Co-editing is always present and researchers need to be aware of it. Co-editing of metabolite signals should be quantitatively investigated through in vitro phantom studies or quantum-mechanical simulations. It is recommended to minimize MM contributions using strategies, e.g., T1 nulling and symmetric editing, and to characterize MM contributions and incorporated them into the spectral fitting algorithm. In a clinical setting when GABA+ is reported without further investigation of MM contributions, interpretation of findings should consider MM contributions as part of GABA+ and potential changes of MM contributions by frequency drift and subject motion associated B0 changes as limitations of the study.
Frequency drift: Frequency and phase instabilities are the rule, not the exception. Ideally, frequency drift and B0 changes associated with subject motion need to be monitored and corrected in real time with navigators during data acquisition. When vendor-supplied navigators are used, rigorous testing is advised because not all implementations perform as intended. When an effective frequency update is not available, data need to be stored as single-transients and occasionally interleaved with approximately 2–5 min, depending on the frequency drift, with a global frequency adjustment. Post-acquisition correction of frequency and phase can be used when the frequency drift is less than ~25% of the editing pulse bandwidth, to reduce the subtraction artifacts while it cannot recover the signal loss. When possible, spectral editing data should be acquired before high-gradient-duty-cycle methods such as functional or diffusion-weighted MRI to minimize system frequency drifts.
Exclusion of spectra: Any corrupted transients due to subject motion and edited spectra with substantial subtraction artifacts (> 10% of the target edited signals) need to be excluded from further data processing.
Data quantification: Quantification of edited spectra can be performed similarly to that of non-edited spectra. However, especially for J-coupled spins, it is crucial that the quantum-mechanical simulations used to generate metabolite basis sets include all details of the pulse sequence, including RF pulse profiles.
Editing in clinical studies: Multi-site studies applying edited MRS should use the same acquisition and data processing methods for all sites, to the greatest degree possible. If sequences with standardized pulse shapes and timings are not available across multiple vendors, an identical bandwidth of the editing pulse is recommended at a minimum.
Publication guidelines: In publications, the bandwidth of spatially-selective refocusing pulses for estimating the chemical shift displacement and the bandwidth of the editing pulse should be included. Data acquisition and processing details such as real-time or post-acquisition frequency correction methods need to be included as well as the amount of frequency drift. It is also recommended to discuss the origin and the approximate amounts of co-edited signals. Scaling factors for relaxation (e.g., T1 and T2) and other corrections need to be reported if used for quantification.
Acknowledgements:
The authors are grateful to Mark Mikkelsen for his assistance in generating Fig. 4.
List abbreviations
- NAA
N-acetylaspartate
- NAAG
N-acetylaspartylglutamate
- GABA
γ-aminobutyric acid
- Asc
ascorbate
- CSDE
chemical shift displacement error
- COSY
correlation spectroscopy
- CRLB
Cramér-Rao lower bounds
- DEW-MEGA
double editing with MEGA
- TE
echo time
- GSH
glutathione
- HERMES
Hadamard-encoding and reconstruction of MEGA-edited spectroscopy
- BHB
β-hydroxybutyrate
- 2HG
D-2-hydroxyglutarate
- IDH
isocitrate dehydrogenase
- MM
macromolecules
- MRSI
MR spectroscopic imaging
- MQF
multiple quantum coherence filtering
- GSSG
oxidized form of glutathione
- 1H-MRS
proton magnetic resonance spectroscopy
- RF
radiofrequency
- SAR
specific absorption rate
- B0
static magnetic field
- TOCSY
total correlation spectroscopy
- B1+
transmit RF magnetic field
Contributor Information
In-Young Choi, Department of Neurology, Hoglund Biomedical Imaging Center, University of Kansas Medical Center, Kansas City, KS, USA.
Ovidiu C. Andronesi, Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
Peter Barker, Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore MD, USA, F.M. Kirby Center for Functional MRI, Kennedy Krieger Institute, Baltimore, MD, USA.
Wolfgang Bogner, High-field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Austria.
Richard Edden, Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore MD, USA, F.M. Kirby Center for Functional MRI, Kennedy Krieger Institute, Baltimore, MD, USA.
Lana G. Kaiser, Henry H. Wheeler, Jr. Brain Imaging Center, University of California, Berkeley, CA, USA
Phil Lee, Department of Radiology, Hoglund Biomedical Imaging Center, University of Kansas Medical Center, Kansas City, KS, USA.
Małgorzata Marjańska, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, USA.
Melissa Terpstra, Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, USA.
Robin A. de Graaf, Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, USA
References:
- 1.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. October 1998;11(6):266–272. [DOI] [PubMed] [Google Scholar]
- 2.Star-Lack J, Spielman D, Adalsteinsson E, Kurhanewicz J, Terris DJ, Vigneron DB. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson. August 1998;133(2):243–254. [DOI] [PubMed] [Google Scholar]
- 3.Andreychenko A, Boer VO, Arteaga de Castro CS, Luijten PR, Klomp DW. Efficient spectral editing at 7 T: GABA detection with MEGA-sLASER. Magn Reson Med. October 2012;68(4):1018–1025. [DOI] [PubMed] [Google Scholar]
- 4.Bogner W, Gagoski B, Hess AT, et al. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. December 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Near J, Simpson R, Cowen P, Jezzard P. Efficient gamma-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. December 2011;24(10):1277–1285. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H, Edden RA, Ouwerkerk R, Barker PB. High resolution spectroscopic imaging of GABA at 3 Tesla. Magn Reson Med. March 2011;65(3):603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan KL, Puts NA, Schar M, Barker PB, Edden RA. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med. July 2016;76(1):11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terpstra M, Marjanska M, Henry PG, Tkac I, Gruetter R. Detection of an antioxidant profile in the human brain in vivo via double editing with MEGA-PRESS. Magn Reson Med. December 2006;56(6):1192–1199. [DOI] [PubMed] [Google Scholar]
- 9.Oeltzschner G, Saleh MG, Rimbault D, et al. Advanced Hadamard-encoded editing of seven low-concentration brain metabolites: Principles of HERCULES. Neuroimage. January 15 2019;185:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moser P, Hingerl L, Strasser B, et al. Whole-slice mapping of GABA and GABA(+) at 7T via adiabatic MEGA-editing, real-time instability correction, and concentric circle readout. Neuroimage. January 1 2019;184:475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magnet Reson Med. March 2001;45(3):517–520. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya PK, Lowe MJ, Phillips MD. Spectral quality control in motion-corrupted single-voxel J-difference editing scans: an interleaved navigator approach. Magn Reson Med. October 2007;58(4):808–812. [DOI] [PubMed] [Google Scholar]
- 13.Evans CJ, Puts NA, Robson SE, et al. Subtraction artifacts and frequency (mis-)alignment in J-difference GABA editing. J Magn Reson Imaging. October 2013;38(4):970–975. [DOI] [PubMed] [Google Scholar]
- 14.Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn Reson Med. January 2015;73(1):44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saleh MG, Alhamud A, Near J, van der Kouwe AJ, Meintjes EM. Volumetric navigated MEGA-SPECIAL for real-time motion and shim corrected GABA editing. NMR Biomed. March 2016;29(3):248–255. [DOI] [PubMed] [Google Scholar]
- 16.Andronesi OC, Bhattacharyya PK, Bogner W, et al. Motion Correction Methods for Magnetic Resonance Spectroscopy: Experts’ Consensus Recommendations. NMR in Biomedicine. 2020; 10.1002/nbm.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tkac I, Andersen P, Adriany G, Merkle H, Ugurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at 7 T. Magn Reson Med. September 2001;46(3):451–456. [DOI] [PubMed] [Google Scholar]
- 18.Kreis R, Boer V, Choi I-Y, et al. Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: Background and experts’ consensus recommendations. NMR Biomed. 2020:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser LG, Young K, Matson GB. Elimination of spatial interference in PRESS-localized editing spectroscopy. Magn Reson Med. October 2007;58(4):813–818. [DOI] [PubMed] [Google Scholar]
- 20.Dacko M, Lange T. Improved detection of lactate and beta-hydroxybutyrate using MEGA-sLASER at 3 T. NMR Biomed. July 2019;32(7):e4100. [DOI] [PubMed] [Google Scholar]
- 21.Saleh MG, Mikkelsen M, Oeltzschner G, et al. Simultaneous editing of GABA and glutathione at 7T using semi-LASER localization. Magn Reson Med. August 2018;80(2):474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnusson PO, Boer VO, Marsman A, Paulson OB, Hanson LG, Petersen ET. Gamma-aminobutyric acid edited echo-planar spectroscopic imaging (EPSI) with MEGA-sLASER at 7T. Magn Reson Med. February 2019;81(2):773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry PG, van de Moortele PF, Giacomini E, Nauerth A, Bloch G. Field-frequency locked in vivo proton MRS on a whole-body spectrometer. Magn Reson Med. October 1999;42(4):636–642. [DOI] [PubMed] [Google Scholar]
- 24.Hess AT, Tisdall MD, Andronesi OC, Meintjes EM, van der Kouwe AJ. Real-time motion and B0 corrected single voxel spectroscopy using volumetric navigators. Magn Reson Med. August 2011;66(2):314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waddell KW, Avison MJ, Joers JM, Gore JC. A practical guide to robust detection of GABA in human brain by J-difference spectroscopy at 3 T using a standard volume coil. Magn Reson Imaging. September 2007;25(7):1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deelchand DK, Marjanska M, Henry PG, Terpstra M. MEGA-PRESS of GABA+: Influences of acquisition parameters. NMR Biomed. October 28 2019:e4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. May 2000;13(3):129–153. [DOI] [PubMed] [Google Scholar]
- 28.Govind V, Young K, Maudsley AA. Corrigendum: proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V, Young K, Maudsley AA, NMR Biomed. 2000; 13: 129–153. NMR Biomed. July 2015;28(7):923–924. [DOI] [PubMed] [Google Scholar]
- 29.van der Veen JW, Marenco S, Berman KF, Shen J. Retrospective correction of frequency drift in spectral editing: The GABA editing example. NMR Biomed. August 2017;30(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris AD, Glaubitz B, Near J, et al. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. October 2014;72(4):941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee CY, Choi IY, Lee P. Prospective frequency correction using outer volume suppression-localized navigator for MR spectroscopy and spectroscopic imaging. Magn Reson Med. December 2018;80(6):2366–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Near J, Harris AD, Juchem C, et al. Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: experts’ consensus recommendations. NMR Biomed. February 21 2020:e4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA-editing at 7 Tesla. Magn Reson Med. May 2002;47(5):1009–1012. [DOI] [PubMed] [Google Scholar]
- 34.Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging. December 2014;40(6):1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4 T. NMR Biomed. January 2008;21(1):22–32. [DOI] [PubMed] [Google Scholar]
- 36.Kaiser LG, Marjanska M, Matson GB, et al. 1H MRS detection of glycine residue of reduced glutathione in vivo. J Magn Reson. February 2010;202(2):259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marjanska M, Henry PG, Ugurbil K, Gruetter R. Editing through multiple bonds: threonine detection. Magn Reson Med. February 2008;59(2):245–251. [DOI] [PubMed] [Google Scholar]
- 38.Deelchand DK, Van de Moortele PF, Adriany G, et al. In vivo 1H NMR spectroscopy of the human brain at 9.4 T: initial results. J Magn Reson. September 2010;206(1):74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emir UE, Deelchand D, Henry PG, Terpstra M. Noninvasive quantification of T2 and concentrations of ascorbate and glutathione in the human brain from the same double-edited spectra. NMR Biomed. April 2011;24(3):263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marjanska M, Auerbach EJ, Valabregue R, Van de Moortele PF, Adriany G, Garwood M. Localized 1H NMR spectroscopy in different regions of human brain in vivo at 7 T: T2 relaxation times and concentrations of cerebral metabolites. NMR Biomed. February 2012;25(2):332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edden RA, Intrapiromkul J, Zhu H, Cheng Y, Barker PB. Measuring T2 in vivo with J-difference editing: application to GABA at 3 Tesla. J Magn Reson Imaging. January 2012;35(1):229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi IY, Lee P. Doubly selective multiple quantum chemical shift imaging and T1 relaxation time measurement of glutathione (GSH) in the human brain in vivo. NMR Biomed. January 2013;26(1):28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreychenko A, Klomp DW, de Graaf RA, Luijten PR, Boer VO. In vivo GABA T2 determination with J-refocused echo time extension at 7 T. NMR Biomed. November 2013;26(11):1596–1601. [DOI] [PubMed] [Google Scholar]
- 44.Madan A, Ganji SK, An Z, et al. Proton T2 measurement and quantification of lactate in brain tumors by MRS at 3 Tesla in vivo. Magn Reson Med. June 2015;73(6):2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanberg KM, Prinsen H, Coman D, de Graaf RA, Juchem C. Quantification of glutathione transverse relaxation time T2 using echo time extension with variable refocusing selectivity and symmetry in the human brain at 7 Tesla. J Magn Reson. May 2018;290:1–11. [DOI] [PubMed] [Google Scholar]
- 46.Murali-Manohar S, Borbath T, Wright AM, Soher B, Mekle R, Henning A. T2 relaxation times of macromolecules and metabolites in the human brain at 9.4 T. Magn Reson Med. August 2020;84(2):542–558. [DOI] [PubMed] [Google Scholar]
- 47.Juchem C, Cudalbu C, de Graaf RA, et al. B0 Shimming for In Vivo MR Spectroscopy: Experts’ consensus recommendations. NMR Biomed. 2020:in press. [DOI] [PubMed] [Google Scholar]
- 48.Anderson W Nuclear resonance saturation effects and multiple-quantum transitions. Phys Rev. 1956(104):850. [Google Scholar]
- 49.Bax A, De Jong PG, Mehlkopf AF, Smidt J. Separation of the different orders of NMR multiple-quantum transitions by the use of pulsed field gradients. Chem Phy Lett. 1980;69(3):567–570. [Google Scholar]
- 50.Nosel W, Trimble LA, Shen JF, Allen PS. On the use of double-quantum coherence from an AX3 system (protons in lactate) for spectral editing. Magn Reson Med. September 1989;11(3):398–404. [DOI] [PubMed] [Google Scholar]
- 51.Keltner JR, Wald LL, Frederick BD, Renshaw PF. In vivo detection of GABA in human brain using a localized double-quantum filter technique. Magn Reson Med. 1997;37(3):366–371. [DOI] [PubMed] [Google Scholar]
- 52.Sotak CH. A volume-localized, two-dimensional NMR method for the determination of lactate using zero-quantum coherence created in a stimulated echo pulse sequence. Magn Reson Med. July 1988;7(3):364–370. [DOI] [PubMed] [Google Scholar]
- 53.Trabesinger AH, Weber OM, Duc CO, Boesiger P. Detection of glutathione in the human brain in vivo by means of double quantum coherence filtering. Magn Reson Med. August 1999;42(2):283–289. [DOI] [PubMed] [Google Scholar]
- 54.McLean MA, Busza AL, Wald LL, Simister RJ, Barker GJ, Williams SR. In vivo GABA+ measurement at 1.5T using a PRESS-localized double quantum filter. Magn Reson Med. August 2002;48(2):233–241. [DOI] [PubMed] [Google Scholar]
- 55.Zhao T, Heberlein K, Jonas C, Jones DP, Hu X. New double quantum coherence filter for localized detection of glutathione in vivo. Magn Reson Med. March 2006;55(3):676–680. [DOI] [PubMed] [Google Scholar]
- 56.Choi I-Y, Lee S-P, Merkle H, Shen J. Single-shot two-echo technique for simultaneous measurement of GABA and creatine in the human brain in vivo. Magn Reson Med. June 2004;51(6):1115–1121. [DOI] [PubMed] [Google Scholar]
- 57.Choi C, Coupland NJ, Hanstock CC, et al. Brain gamma-aminobutyric acid measurement by proton double-quantum filtering with selective J rewinding. Magn Reson Med. August 2005;54(2):272–279. [DOI] [PubMed] [Google Scholar]
- 58.He Q, Shungu DC, van Zijl PC, Bhujwalla ZM, Glickson JD. Single-scan in vivo lactate editing with complete lipid and water suppression by selective multiple-quantum-coherence transfer (Sel-MQC) with application to tumors. J Magn Reson B. March 1995;106(3):203–211. [DOI] [PubMed] [Google Scholar]
- 59.Shen J, Rothman DL, Brown P. In vivo GABA editing using a novel doubly selective multiple quantum filter. Magn Reson Med. March 2002;47(3):447–454. [DOI] [PubMed] [Google Scholar]
- 60.Choi IY, Lee SP, Shen J. In vivo single-shot three-dimensionally localized multiple quantum spectroscopy of GABA in the human brain with improved spectral selectivity. J Magn Reson. January 2005;172(1):9–16. [DOI] [PubMed] [Google Scholar]
- 61.Choi I-Y, Lee S-P, Merkle H, Shen J. In vivo detection of gray and white matter differences in GABA concentration in the human brain. NeuroImage. October 15 2006;33(1):85–93. [DOI] [PubMed] [Google Scholar]
- 62.Choi IY, Lee SP, Denney DR, Lynch SG. Lower levels of glutathione in the brains of secondary progressive multiple sclerosis patients measured by 1H magnetic resonance chemical shift imaging at 3 T. Mult Scler. March 2011;17(3):289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen J, Shungu DC, Rothman DL. In vivo chemical shift imaging of gamma-aminobutyric acid in the human brain. Magn Reson Med. January 1999;41(1):35–42. [DOI] [PubMed] [Google Scholar]
- 64.Hurd RE, Freeman DM. Metabolite specific proton magnetic resonance imaging. Proc Natl Acad Sci U S A. June 1989;86(12):4402–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lei H, Peeling J. Multiple-voxel double-quantum lactate-edited spectroscopy using two-dimensional longitudinal Hadamard encoding. Magn Reson Med. July 1999;42(1):19–23. [DOI] [PubMed] [Google Scholar]
- 66.Choi I-Y, Lee S-P. Two-echo multiple quantum chemical shift imaging of ascorbic acid (vitamin C) in the human brain in vivo at 3 T. Paper presented at: Proc Intl Soc Mag Reson Med 162008; Toronto. [Google Scholar]
- 67.Choi I-Y, Lee S-P, Shen J. Selective homonuclear Hartmann-Hahn transfer method for in vivo spectral editing in the human brain. Magn Reson Med. March 2005;53(3):503–510. [DOI] [PubMed] [Google Scholar]
- 68.Marjanska M, Henry PG, Bolan PJ, et al. Uncovering hidden in vivo resonances using editing based on localized TOCSY. Magn Reson Med. April 2005;53(4):783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganji SK, An Z, Tiwari V, et al. In vivo detection of 2-hydroxyglutarate in brain tumors by optimized point-resolved spectroscopy (PRESS) at 7T. Magn Reson Med. March 2017;77(3):936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.An Z, Ganji SK, Tiwari V, et al. Detection of 2-hydroxyglutarate in brain tumors by triple-refocusing MR spectroscopy at 3T in vivo. Magn Reson Med. July 2017;78(1):40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. March 2004;51(3):435–440. [DOI] [PubMed] [Google Scholar]
- 72.Aue W, Karhan J, Ernst R. Homonuclear broad band decoupling and two‐dimensional J‐resolved NMR spectroscopy. The Journal of Chemical Physics. 1976;64(10):4226–4227. [Google Scholar]
- 73.Dreher W, Leibfritz D. On the use of two-dimensional-J NMR measurements for in vivo proton MRS: measurement of homonuclear decoupled spectra without the need for short echo times. Magn Reson Med. September 1995;34(3):331–337. [DOI] [PubMed] [Google Scholar]
- 74.Schulte RF, Lange T, Beck J, Meier D, Boesiger P. Improved two-dimensional J-resolved spectroscopy. NMR Biomed. April 2006;19(2):264–270. [DOI] [PubMed] [Google Scholar]
- 75.Velan SS, Lemieux SK, Raylman RR, et al. Detection of cerebral metabolites by single-voxel-based PRESS and COSY techniques at 3T. J. Magn. Reson. Imaging. August 2007;26(2):405–409. [DOI] [PubMed] [Google Scholar]
- 76.Andronesi OC, Ramadan S, Mountford CE, Sorensen AG. Low-power adiabatic sequences for in-vivo localized two-dimensional chemical shift correlated MR spectroscopy. Magnet Reson Med. 2010;64(6):1542–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogner W, Hangel G, Esmaeili M, Andronesi OC. 1D-spectral editing and 2D multispectral in vivo(1)H-MRS and (1)H-MRSI - Methods and applications. Anal Biochem. July 15 2017;529:48–64. [DOI] [PubMed] [Google Scholar]
- 78.Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of g-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. June 15 1993;90(12):5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shungu DC, Mao X, Gonzales R, et al. Brain gamma-aminobutyric acid (GABA) detection in vivo with the J-editing (1) H MRS technique: a comprehensive methodological evaluation of sensitivity enhancement, macromolecule contamination and test-retest reliability. NMR Biomed. July 2016;29(7):932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prinsen H, de Graaf RA, Mason GF, Pelletier D, Juchem C. Reproducibility measurement of glutathione, GABA, and glutamate: Towards in vivo neurochemical profiling of multiple sclerosis with MR spectroscopy at 7T. J Magn Reson Imaging. January 2017;45(1):187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harris AD, Puts NA, Barker PB, Edden RA. Spectral-editing measurements of GABA in the human brain with and without macromolecule suppression. Magn Reson Med. December 2015;74(6):1523–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi C, Bhardwaj PP, Kalra S, et al. Measurement of GABA and contaminants in gray and white matter in human brain in vivo. Magn Reson Med. July 2007;58(1):27–33. [DOI] [PubMed] [Google Scholar]
- 83.Petroff OA, Mattson RH, Behar KL, Hyder F, Rothman DL. Vigabatrin increases human brain homocarnosine and improves seizure control. Ann Neurol. December 1998;44(6):948–952. [DOI] [PubMed] [Google Scholar]
- 84.Rae CD, Williams SR. Glutathione in the human brain: Review of its roles and measurement by magnetic resonance spectroscopy. Anal Biochem. July 15 2017;529:127–143. [DOI] [PubMed] [Google Scholar]
- 85.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52(1):711–760. [DOI] [PubMed] [Google Scholar]
- 86.Satoh T, Yoshioka Y. Contribution of reduced and oxidized glutathione to signals detected by magnetic resonance spectroscopy as indicators of local brain redox state. Neurosci Res. May 2006;55(1):34–39. [DOI] [PubMed] [Google Scholar]
- 87.Fletcher RH, Fletcher SW. Glutathione and ageing: ideas and evidence. Lancet. November 19 1994;344(8934):1379–1380. [DOI] [PubMed] [Google Scholar]
- 88.Weiduschat N, Mao X, Hupf J, et al. Motor cortex glutathione deficit in ALS measured in vivo with the J-editing technique. Neurosci Lett. June 6 2014;570:102–107. [DOI] [PubMed] [Google Scholar]
- 89.Terpstra M, Cheong I, Lyu T, et al. Test-retest reproducibility of neurochemical profiles with short-echo, single-voxel MR spectroscopy at 3T and 7T. Magn Reson Med. October 2016;76(4):1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Terpstra M, Henry PG, Gruetter R. Measurement of reduced glutathione (GSH) in human brain using LCModel analysis of difference-edited spectra. Magn Reson Med. July 2003;50(1):19–23. [DOI] [PubMed] [Google Scholar]
- 91.Terpstra M, Gruetter R. 1H NMR detection of vitamin C in human brain in vivo. Magn Reson Med. February 2004;51(2):225–229. [DOI] [PubMed] [Google Scholar]
- 92.Terpstra M, Ugurbil K, Tkac I. Noninvasive quantification of human brain ascorbate concentration using 1H NMR spectroscopy at 7 T. NMR Biomed. April 2010;23(3):227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. December 10 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Choi C, Raisanen JM, Ganji SK, et al. Prospective Longitudinal Analysis of 2-Hydroxyglutarate Magnetic Resonance Spectroscopy Identifies Broad Clinical Utility for the Management of Patients With IDH-Mutant Glioma. J Clin Oncol. November 20 2016;34(33):4030–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Andronesi OC, Arrillaga-Romany IC, Ly KI, et al. Pharmacodynamics of mutant-IDH1 inhibitors in glioma patients probed by in vivo 3D MRS imaging of 2-hydroxyglutarate. Nat Commun. April 16 2018;9(1):1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choi C, Ganji SK, DeBerardinis RJ, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. January 26 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Andronesi OC, Kim GS, Gerstner E, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. January 11 2012;4(116):116ra114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Emir UE, Larkin SJ, de Pennington N, et al. Noninvasive Quantification of 2-Hydroxyglutarate in Human Gliomas with IDH1 and IDH2 Mutations. Cancer Res. January 1 2016;76(1):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Andronesi OC, Loebel F, Bogner W, et al. Treatment Response Assessment in IDH-Mutant Glioma Patients by Noninvasive 3D Functional Spectroscopic Mapping of 2-Hydroxyglutarate. Clin Cancer Res. April 1 2016;22(7):1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sotak CH, Freeman DM, Hurd RE. The unequivocal determination of in vivo lactic acid using two-dimensional double-quantum coherence-transfer spectroscopy. J Magn Reson. January 01, 1988 1988;78:355–361. [Google Scholar]
- 101.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. June 2009;61(6):1279–1285. [DOI] [PubMed] [Google Scholar]
- 102.Choi C, Coupland NJ, Kalra S, Bhardwaj PP, Malykhin N, Allen PS. Proton spectral editing for discrimination of lactate and threonine 1.31 ppm resonances in human brain in vivo. Magn Reson Med. September 2006;56(3):660–665. [DOI] [PubMed] [Google Scholar]
- 103.Payne GS, Harris LM, Cairns GS, et al. Validating a robust double-quantum-filtered 1H MRS lactate measurement method in high-grade brain tumours. NMR Biomed. October 2016;29(10):1420–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Edden RA, Pomper MG, Barker PB. In vivo differentiation of N-acetyl aspartyl glutamate from N-acetyl aspartate at 3 Tesla. Magn Reson Med. June 2007;57(6):977–982. [DOI] [PubMed] [Google Scholar]
- 105.Gruber S, Heckova E, Strasser B, et al. Mapping an Extended Neurochemical Profile at 3 and 7 T Using Accelerated High-Resolution Proton Magnetic Resonance Spectroscopic Imaging. Invest Radiol. October 2017;52(10):631–639. [DOI] [PubMed] [Google Scholar]
- 106.Chan KL, Saleh MG, Oeltzschner G, Barker PB, Edden RAE. Simultaneous measurement of Aspartate, NAA, and NAAG using HERMES spectral editing at 3 Tesla. Neuroimage. July 15 2017;155:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hardy DL, Norwood TJ. Spectral editing technique for the in vitro and in vivo detection of taurine. J Magn Reson. July 1998;133(1):70–78. [DOI] [PubMed] [Google Scholar]
- 108.Bogner W, Gruber S, Trattnig S, Chmelik M. High-resolution mapping of human brain metabolites by free induction decay (1)H MRSI at 7 T. NMR Biomed. June 2012;25(6):873–882. [DOI] [PubMed] [Google Scholar]
- 109.de Graaf RA, Dijkhuizen RM, Biessels GJ, Braun KP, Nicolay K. In vivo glucose detection by homonuclear spectral editing. Magn Reson Med. May 2000;43(5):621–626. [DOI] [PubMed] [Google Scholar]
- 110.Kaiser LG, Hirokazu K, Fukunaga M, G BM. Detection of glucose in the human brain with 1H MRS at 7 Tesla. Magn Reson Med. December 2016;76(6):1653–1660. [DOI] [PubMed] [Google Scholar]
- 111.Gruetter R, Garwood M, Ugurbil K, Seaquist ER. Observation of resolved glucose signals in 1H NMR spectra of the human brain at 4 Tesla. Magn Reson Med. July 1996;36(1):1–6. [DOI] [PubMed] [Google Scholar]
- 112.Choi C, Dimitrov I, Douglas D, et al. In vivo detection of serine in the human brain by proton magnetic resonance spectroscopy (1H-MRS) at 7 Tesla. Magn Reson Med. October 2009;62(4):1042–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Branzoli F, Deelchand DK, Sanson M, Lehericy S, Marjanska M. In vivo (1) H MRS detection of cystathionine in human brain tumors. Magn Reson Med. October 2019;82(4):1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chan KL, Snoussi K, Edden RAE, Barker PB. Simultaneous detection of glutathione and lactate using spectral editing at 3 T. NMR Biomed. December 2017;30(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saleh MG, Oeltzschner G, Chan KL, et al. Simultaneous edited MRS of GABA and glutathione. Neuroimage. November 15 2016;142:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Oeltzschner G, Chan KL, Saleh MG, Mikkelsen M, Puts NA, Edden RAE. Hadamard editing of glutathione and macromolecule-suppressed GABA. NMR Biomed. January 2018;31(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chan KL, Oeltzschner G, Saleh MG, Edden RAE, Barker PB. Simultaneous editing of GABA and GSH with Hadamard-encoded MR spectroscopic imaging. Magn Reson Med. July 2019;82(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mikkelsen M, Saleh MG, Near J, et al. Frequency and phase correction for multiplexed edited MRS of GABA and glutathione. Magn Reson Med. July 2018;80(1):21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saleh MG, Rimbault D, Mikkelsen M, et al. Multi-vendor standardized sequence for edited magnetic resonance spectroscopy. Neuroimage. April 1 2019;189:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Harris AD, Puts NAJ, Wijtenburg SA, et al. Normalizing data from GABA-edited MEGA-PRESS implementations at 3 Tesla. Magn Reson Imaging. October 2017;42:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mikkelsen M, Barker PB, Bhattacharyya PK, et al. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage. October 1 2017;159:32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mikkelsen M, Rimbault DL, Barker PB, et al. Big GABA II: Water-referenced edited MR spectroscopy at 25 research sites. Neuroimage. May 1 2019;191:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park YW, Deelchand DK, Joers JM, et al. AutoVOI: real-time automatic prescription of volume-of-interest for single voxel spectroscopy. Magn Reson Med. November 2018;80(5):1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yablonskiy DA, Neil JJ, Raichle ME, Ackerman JJ. Homonuclear J coupling effects in volume localized NMR spectroscopy: pitfalls and solutions. Magn Reson Med. February 1998;39(2):169–178. [DOI] [PubMed] [Google Scholar]
- 125.Choi IY, Lee P, Denney DR, et al. Dairy intake is associated with brain glutathione concentration in older adults. The American journal of clinical nutrition. February 2015;101(2):287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Branzoli F, Pontoizeau C, Tchara L, et al. Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy. Neuro Oncol. June 10 2019;21(6):765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Branzoli F, Di Stefano AL, Capelle L, et al. Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro Oncol. June 18 2018;20(7):907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arteaga de Castro CS, Boer VO, Andreychenko A, et al. Improved efficiency on editing MRS of lactate and gamma-aminobutyric acid by inclusion of frequency offset corrected inversion pulses at high fields. NMR Biomed. October 2013;26(10):1213–1219. [DOI] [PubMed] [Google Scholar]