Abstract
Adaptor protein complex 4-associated hereditary spastic paraplegia is caused by biallelic loss-of-function variants in AP4B1, AP4M1, AP4E1 or AP4S1, which constitute the four subunits of this obligate complex. While the diagnosis of adaptor protein complex 4-associated hereditary spastic paraplegia relies on molecular testing, the interpretation of novel missense variants remains challenging. Here, we address this diagnostic gap by using patient-derived fibroblasts to establish a functional assay that measures the subcellular localization of ATG9A, a transmembrane protein that is sorted by adaptor protein complex 4. Using automated high-throughput microscopy, we determine the ratio of the ATG9A fluorescence in the trans-Golgi-network versus cytoplasm and ascertain that this metric meets standards for screening assays (Z′-factor robust >0.3, strictly standardized mean difference >3). The ‘ATG9A ratio’ is increased in fibroblasts of 18 well-characterized adaptor protein complex 4-associated hereditary spastic paraplegia patients [mean: 1.54 ± 0.13 versus 1.21 ± 0.05 (standard deviation) in controls] and receiver-operating characteristic analysis demonstrates robust diagnostic power (area under the curve: 0.85, 95% confidence interval: 0.849–0.852). Using fibroblasts from two individuals with atypical clinical features and novel biallelic missense variants of unknown significance in AP4B1, we show that our assay can reliably detect adaptor protein complex 4 function. Our findings establish the ‘ATG9A ratio’ as a diagnostic marker of adaptor protein complex 4-associated hereditary spastic paraplegia.
Keywords: hereditary spastic paraplegia, adaptor protein complex 4, functional assay, high-throughput imaging, biomarker
Ebrahimi-Fakhari et al. establish the intracellular ATG9A distribution as a diagnostic marker for AP-4-associated hereditary spastic paraplegia using a novel high-throughput assay in patient-derived fibroblasts. This approach enables the functional validation of novel variants in AP4B1, AP4M1, AP4S1 and AP4E1 with robust diagnostic power and opens opportunities for biomarker discovery.
Graphical Abstract
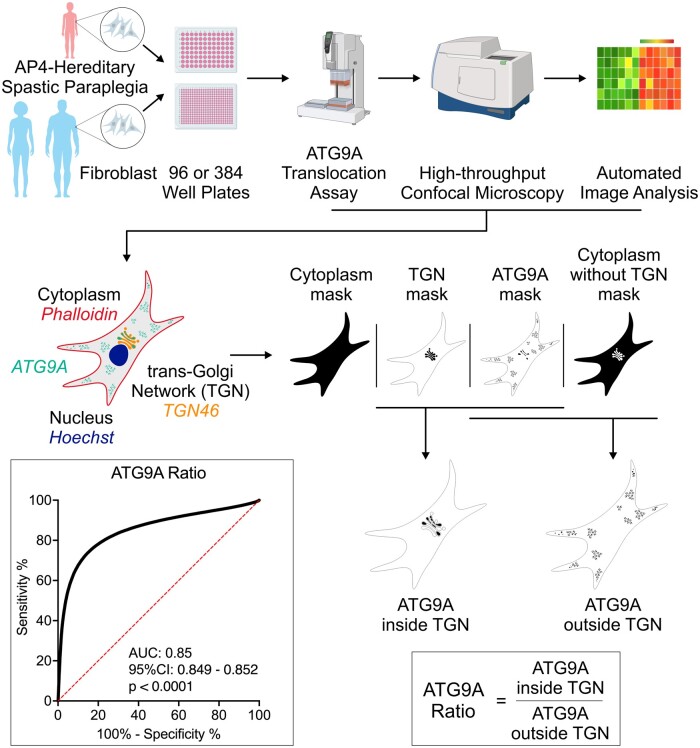
Graphical Abstract.
Introduction
The hereditary spastic paraplegias (HSPs) are a heterogeneous group of over 80 different neurogenetic disorders that share the clinical feature of spasticity due to corticospinal tract dysfunction.1 Childhood-onset forms of HSP often pose a diagnostic challenge given their non-specific clinical presentation and slow progression in many cases. The increasing availability of next-generation sequencing is beginning to close this diagnostic gap, yet, at the same time, has broadened the phenotypic spectrum associated with HSP genes. As for other rare disorders, the interpretation of genetic variants remains a significant challenge, particularly for missense variants associated with novel or atypical phenotypes. In this setting, functional assays in patient-derived cells can aid the interpretation of variants and help establish a clinical diagnosis. This is supported by the current American College of Medical Genetics and Genomics (ACMG) recommendations for variant classification (criterion PS3), which integrate results from ‘well-established’ functional assays.2
Adaptor protein complex 4 (AP-4)-associated hereditary spastic paraplegia (AP-4-HSP) encompasses four forms of childhood-onset HSP, caused by biallelic variants in the subunits of AP-4.3,4 This includes AP4B1-associated SPG47 (OMIM #614066), AP4M1-associated SPG50 (OMIM #612936), AP4E1-associated SPG51 (OMIM #613744) and AP4S1-associated SPG52 (OMIM #614067). AP-4 is a member of the adaptor protein complex family of coated vesicle adaptors, which functions in the generation of intracellular transport vesicles carrying transmembrane cargo proteins (recently reviewed in Sanger et al.5). Owing to the obligate nature of the heterotetrameric AP-4 complex,6–8 loss of AP-4 function is common to all forms of AP-4-HSP. Recent work has shown that AP-4 deficiency leads to prominent changes in the subcellular localization of the AP-4 cargo protein ATG9A, including in patient-derived cells.9–11 ATG9A is the only conserved transmembrane autophagy-related protein and in mammalian cells cycles between the trans-Golgi network (TGN) and ATG9A vesicles, which associate with endosomes and autophagosome formation sites.12,13 ATG9A is a lipid scramblase important for autophagic vesicle formation14–17 and is essential for axonal function, as demonstrated in CNS-specific Atg9a knockout mice.18
Informed by this emerging understanding of AP-4 biology, we here set out to validate a high-throughput ATG9A translocation assay in patient-derived fibroblasts as a diagnostic marker of AP-4-HSP. Using an automated microscopy platform that allows us to measure the subcellular localization of ATG9A in thousands of cells in a single run, we show that the ATG9A fluorescence signal in the TGN is robustly increased in well-characterized AP-4-HSP patients. Our findings establish the ‘ATG9A ratio’ as a robust indicator of AP-4 function that can provide a functional validation of novel variants in AP-4 subunit genes.
Materials and methods
Patients and clinical information
This study was approved by the Institutional Review Board at Boston Children’s Hospital (IRB-P00033016 and IRB-P00016119). Patients with AP-4-HSP were recruited from the AP-4-HSP International Registry (ClinicalTrials.gov Identifier: NCT04712812). All probands underwent clinical exome sequencing showing biallelic variants in AP4B1, AP4M1 or AP4S1. Clinical information was collected systematically using the AP-4-HSP Natural History Study Questionnaire.3 Severity of motor symptoms and corticospinal tract dysfunction were assessed using the Spastic Paraplegia Rating Scale19 and a four-stage mobility score.3 Clinical information of 13 patients has been reported previously.3
Variant interpretation and scoring
Variants were standardized to the following transcripts: AP4B1 (NM_001253852.3, GRCh37/hg19), AP4M1 (NM_004722.4, GRCh37/hg19), AP4E1 (NM_007347.5, GRCh37/hg19) and AP4S1 (NM_007077.4, GRCh37/hg19). Variants were classified following ACMG guidelines2 using VarSome20 and InterVar.21 For data shown in Figs 1 and 2 and Supplementary Fig. 1, all reported variants from the AP-4-HSP International Registry were annotated as described previously,22 using the most recent version of all tools and scores.23–28
Figure 1.
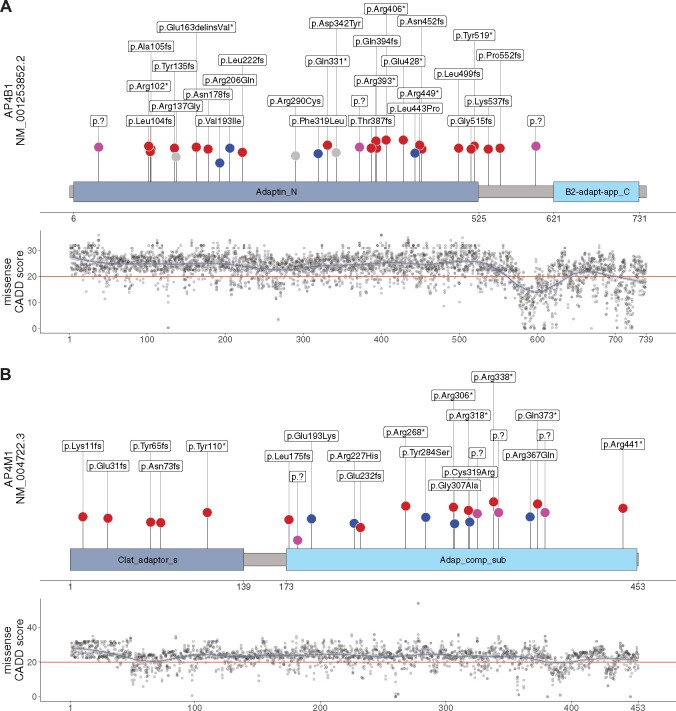
AP4B1 and AP4M1 subunit structures with variant distribution and CADD scores for all possible missense variants. (A and B) Schematic representation of AP4B1 (based on UniProt Q9Y6B7) and AP4M1 (based on UniProt O00189) proteins with their respective aligned units identified by pfam. Blue dots represent reported missense variants, splice variants are depicted in pink and truncating variants in red. Novel variants are highlighted in grey. Length of vertical lines corresponds to the CADD score for each variant. Also shown are the CADD PHRED (version 1.6) scores across the protein secondary structure for all possible missense variants in AP4B1 and AP4M1 respectively. The red horizontal line marks the recommended cut-off of 20. Similar analyses for M-CAP and REVEL scores are found in Supplementary Fig. 1. Data are summarized in Supplementary File 1.
Figure 2.
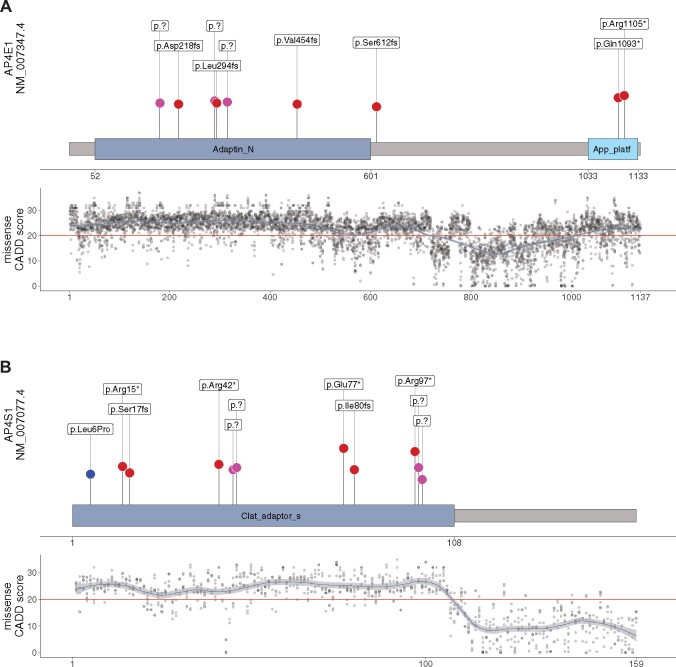
AP4E1 and AP4S1 subunit structures with variant distribution and CADD scores for all possible missense variants. (A and B) Schematic representation of AP4E1 (based on UniProt Q9UPM8) and AP4S1 (based on UniProt Q9Y587-2) proteins with their respective aligned units identified by pfam. Blue dots represent reported missense variants, splice variants are depicted in pink and truncating variants in red. Length of vertical lines corresponds to the CADD score for each variant. Also, shown are the CADD PHRED (version 1.6) scores across the protein secondary structure for all possible missense variants in AP4E1 and AP4S1, respectively. The red horizontal line marks the recommended cut-off of 20. Similar analyses for M-CAP and REVEL scores can be found in Supplementary Fig. 1. Data are summarized in Supplementary File 1.
Analysis of variant spectrum and missense variant distribution in AP-4 protein domain structures
All variants from individuals included in the AP-4-HSP International Registry were mapped to the secondary protein structures of the AP-4 subunits derived from UniProt (Supplementary File 1). Next, the available domain information from UniProt29 and pfam30 were compared with the homology model of the AP-4 complex reported recently.31 Domains predicted by pfam showed a greater overlap and were thus chosen for further analyses. For AP4S1, alignment was transferred from isoform 1 and isoform 2 of Q9Y6B7 using InterPro.32 Constraint scores of protein regions were analysed using CADD PHRED,27 M-CAP and REVEL scores (Supplementary File 1).
Reagents and antibodies
The following reagents were used: Bovine serum albumin (AmericanBIO, #9048–46-8), saponin (Sigma, #47036-50G-F), Molecular Probes Hoechst 33258 (#H3569) and Alexa Fluor 647-labelled phalloidin (Thermo Fisher Scientific, #A22287). The following primary antibodies were used: Anti-ATG9A at 1:400 (Abcam # ab108338), anti-TGN46 at 1:800 (Bio-Rad #AHP500G), anti-AP4E1 and anti-AP4B1 (both produced in house6). Fluorescently labelled secondary antibodies used in this study were all purchased from Thermo Fisher Scientific and used at 1:2000 (#A11008, #A11016, #A21245). Horseradish peroxidase-conjugated secondary antibodies were purchased from Sigma-Aldrich and used at 1:5000 dilution. Lentivirus to express human AP4B1 under a PGK promoter was generated as described previously.11
Fibroblast cell culture
Fibroblast lines were established from routine skin punch biopsies.11 Primary human skin fibroblasts were cultured and maintained as described previously.33 Briefly, cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) high glucose (Gibco, #11960044) supplemented with 20% foetal bovine serum (FBS) (Gibco, #10082147), penicillin 100 U/ml and streptomycin 100 µg/ml (Gibco, #15140122). Cells were kept in culture for up to 8 passages and routinely tested for the presence of mycoplasma contamination. For high-throughput imaging shown in Figs 3–5, fibroblasts were seeded onto 96-well plates (Greiner Bio-One, #655090) at a density of 3 × 103 per well or onto 384-well plates (Greiner Bio-One, #781090) at a density of 2 × 103 per well using the Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific, #11388-558).
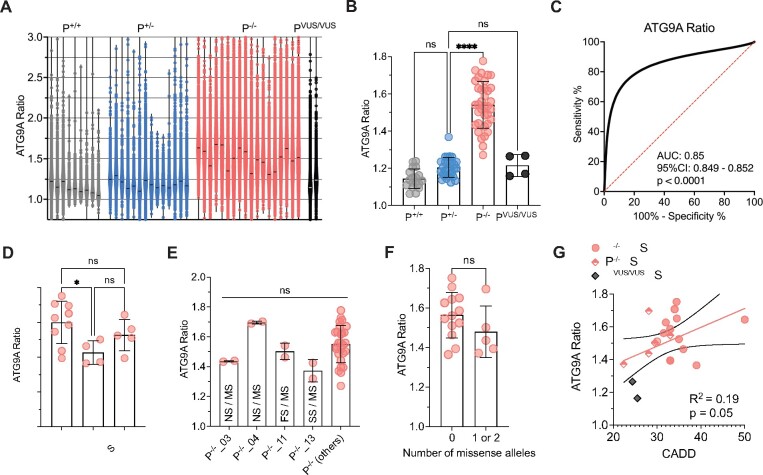
Figure 3.
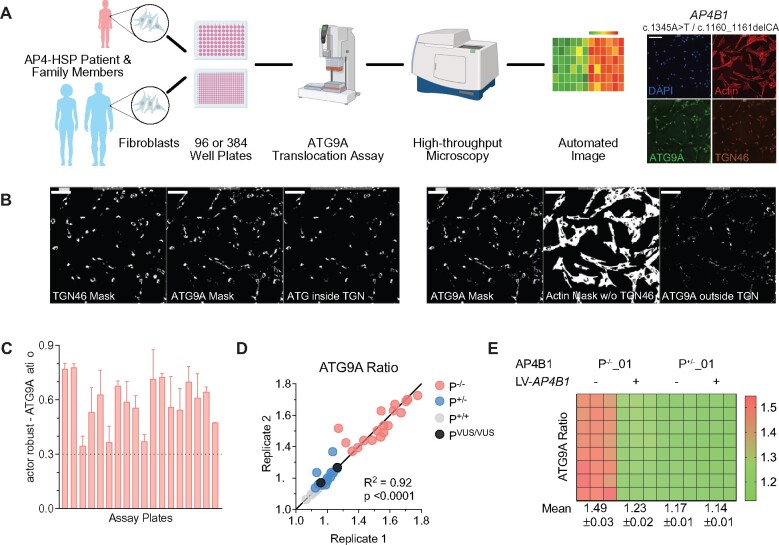
Development of a high-throughput ATG9A translocation assay to determine AP-4 function. (A) Schematic overview of the high-throughput assay and workflow. Fibroblasts are derived by a routine skin punch biopsy, cultured and plated in 96- or 384-well plates. Cells are stained for DAPI (nuclear marker), actin (cytoplasmic marker), TGN46 (trans-Golgi network marker) and ATG9A. Plates are then subjected to confocal microscopy using a high-content imager and analysed using an automated image analysis pipeline. This figure was, in part, created with BioRender.com. (B) Automated image analysis to determine the ATG9A ratio on the level of individual cells. First cells are identified and outlined based on the presence of a DAPI-positive nucleus inside a phalloidin (actin marker)-positive area. Next, different masks are generated: an actin mask to outline the cell, a TGN46 mask to outline the area of the trans-Golgi network, and an ATG9A mask based on intracellular ATG9A fluorescence. ATG9A fluorescence intensity is then measured inside the TGN mask as well as inside the actin-staining positive cytoplasm outside the trans-Golgi network (a mask generated by subtracting the TGN46 mask from the actin mask). The ATG9A ratio is calculated for each cell by dividing the ATG9A fluorescence in both compartments. (C) Z′-factor robust scores for the ATG9A ratio show a robust separation of positive (P−/−) and negative controls (P+/−) and meet a predefined threshold of 0.3 in all assay plates. (D) Replicate plot showing the distribution of ATG9A ratio levels for each of two assay plates (biological replicates) in fibroblasts from patients with AP-4-HSP and biallelic loss-of-function variants (P−/−, Table 1), asymptomatic, heterozygous controls (P+/−), healthy unrelated controls (P+/+, Supplementary Table 2), and two individuals with novel biallelic missense variants in AP4B1 (novel variants, Table 1, Supplementary Table 1). Correlation analysis of 36 pairs shows a Pearson correlation coefficient of 0.95 with a P-value of <0.0001. (E) Heatmap of the ATG9A ratio in a 96-well plate with fibroblasts from patient P−/−_01 with biallelic truncating variants in AP4B1 and a heterozygous control (P+/−_01) in the absence and presence of lentivirus to re-express the missing AP4B1 subunit. Re-expression over 24 h restores the ATG9A ratio close to that of controls indicating that the ATG9A assay can detect dynamic changes in AP-4 function.
Figure 4.
Validation of the ‘ATG9A ratio’ as an indicator of AP-4 function and a diagnostic marker for AP-4-HSP. (A) Violin plots showing the distribution of data points for each fibroblast line with a mean of 6765 ± 4536 (SD) cells from two assay plates for each cell line. Each data point corresponds to the ATG9A ratio of a single fibroblast. Horizontal lines indicate the median (334 data points are outside the axis limits). (B) Grouped analysis of the mean ATG9A ratio of all assay plates demonstrates a significant increase of the ATG9A ratio in AP-4-HSP patients with biallelic loss-of-function variants (P−/−) compared to heterozygous carriers (P+/−) (Kruskal–Wallis test with Dunn’s multiple comparisons test, P < 0.0001). ATG9A ratios of the two individuals with novel biallelic missense variants in AP4B1 (PVUS/VUS) and that of healthy controls (P+/+) are not significantly different from heterozygous controls. Each data point represents the mean ATG9A ratio of each assay plate. (C) Receiver-operating characteristic (ROC) curve for the pooled ATG9A ratio data of all AP-4-HSP patients with biallelic loss-of-function variants (P−/−, n = 160 229 cells) versus heterozygous carriers (P+/−, n = 86 691 cells) shows an area under curve (AUC) of 0.85 with a 95% confidence interval between 0.8488 and 0.8519 and a P-value of <0.0001. (D) Grouped analysis of the mean ATG9A ratios of AP-4-HSP patients with biallelic loss-of-function variants (P−/−) separated by their AP-4-HSP subtype shows that patients with AP4M1-associated SPG50 have lower mean ATG9A ratios compared to AP4B1-associated SPG47 (Kruskal–Wallis test with Dunn’s multiple comparisons test, P < 0.05). Each data point represents the mean ATG9A ratio of each assay plate. (E) Mean ATG9A ratios of three individuals with missense variants previously classified as variant of unclear significance by ACMG criteria, in trans with pathogenic variants, are not different compared to patients with biallelic variants classified as pathogenic or likely pathogenic. Similarly, the mean ATG9A ratio of fibroblasts from an individual with a novel missense variant in trans with a novel splice-site variant (P−/−_13), both previously classified as of uncertain significance, is not significantly different. This confirms the detrimental impact of these variants on AP-4 function (Kruskal-Wallis test with Dunn’s multiple comparisons test, P > 0.05). Each data point represents the mean ATG9A ratio of each assay plate. FS, frameshift variant; NS, nonsense variant; MS, missense variant; SS, splice site variant. (F) Analysis of the mean ATG9A ratio from AP-4-HSP patients with biallelic loss-of-function variants grouped by the presence or absence of alleles with missense variants. While there is a trend towards lower ATG9A ratios in patients with 1 or 2 missense variants, this did not reach statistical significance (Mann–Whitney test, P = 0.23). Each data point represents the mean ATG9A ratio of each assay plate. (G) ATG9A ratio levels in correlation to CADD scores in individuals with biallelic loss-of-function variants with no missense variant (red circles represent the mean of the CADD scores from both alleles), biallelic loss-of-function variants with a missense variant on one allele (diamonds half-filled with red represent CADD score of the missense variant) and individuals with novel biallelic missense variants (PVUS/VUS_01 and PVUS/VUS_02, grey diamonds represent missense variant with the lowest CADD score). There is a trend towards a correlation that does not however reach statistical significance (Pearson correlation coefficient: r = 0.19, P = 0.05). Each data point represents the mean ATG9A ratio of each assay plate. *P < 0.05; ****P < 0.0001.
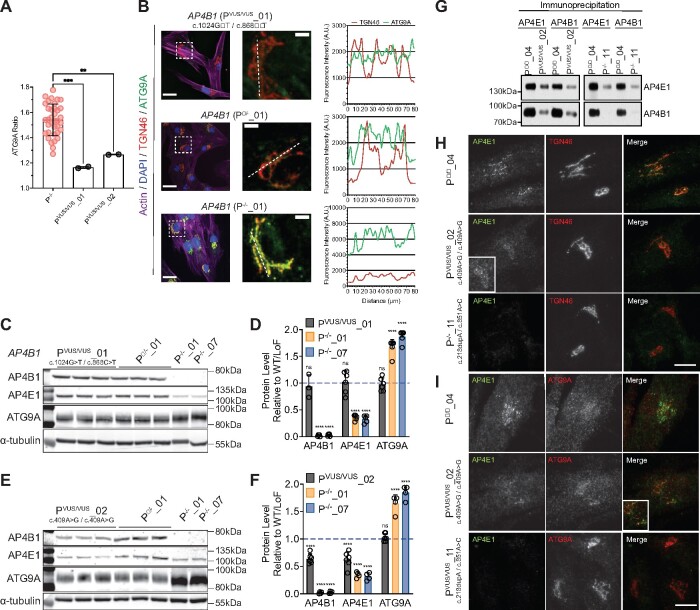
Figure 5.
The ATG9A ratio can provide a functional assessment of novel missense variants. (A) Mean ATG9A ratio of all assay plates in AP-4-HSP patients with biallelic loss-of-function variants (P−/−) and two patients with novel biallelic missense variants (PVUS/VUS_01 and PVUS/VUS_02) (one-way ANOVA with Dunnett’s post hoc test, P−/− versus PVUS/VUS_01, P = 0.0003; P−/− versus PVUS/VUS_02, P = 0.0037). (B) Fibroblasts from an individual with novel biallelic missense variants in AP4B1 (PVUS/VUS_01), an asymptomatic individual with a heterozygous variant in AP4B1 (P+/−_01) and an AP-4-HSP patient with biallelic loss-of-function variants in AP4B1 (P−/−_01) stained with phalloidin (actin marker) and antibodies against TGN46 (trans-Golgi network marker) and ATG9A. Right panel shows magnification of inserts. Dotted line indicates from which line the plots for the TGN46 and corresponding ATG9A signal are generated. Fibroblasts from PVUS/VUS_01 show no elevation of ATG9A fluorescence while the signal is increased in P−/−_01. (C and D) Western blot of whole-cell lysates from fibroblasts of PVUS/VUS_01 compared to P+/−_01 and two AP-4-HSP patients with biallelic loss-of-function variants in AP4B1 (P−/−_01 and P−/−_07). Fibroblasts from PVUS/VUS_01 show levels of AP4B1, AP4E1 and ATG9A that are not different compared to P+/−_01, whereas levels of AP4B1 are robustly reduced and levels of ATG9A elevated in P−/−_01 and P−/−_07 (one-way ANOVA with Dunnett’s post hoc test, P < 0.0001). Bar graph shows the mean and standard deviation of 4–6 (PVUS/VUS_01) and 5 (P−/−_01 and P−/−_07) samples. (E and F) Western blot of whole-cell lysates from fibroblasts of PVUS/VUS_02 compared to P+/−_01 and two AP-4-HSP patients with biallelic loss-of-function variants in AP4B1 (P−/−_01 and P−/−_07). Fibroblasts from PVUS/VUS_02 show a reduction of AP4B1 and AP4E1, indicative of lower AP-4 complex formation. There is, however, no significant change in ATG9A compared to P+/−_01 (one-way ANOVA with Dunnett’s post hoc test, P < 0.0001). Bar graph shows the mean and standard deviation of 6 (PVUS/VUS_02) and 4 (P−/−_01 and P−/−_07) samples. (G) Co-immunoprecipitation of AP4E1 and AP4B1 demonstrates reduced binding of both subunits in lysates from PVUS/VUS_02 compared to a control (P+/+_04). For comparison, the same experiment with lysates from a patient with biallelic loss-of-function variants in AP4M1 is shown (P−/−_11), demonstrating near complete absence of AP-4 complex assembly. (H and I) Fibroblasts from an individual with novel biallelic missense variants in AP4B1 (PVUS/VUS_02), an asymptomatic control (P+/+_04) and an AP-4-HSP patient with biallelic loss-of-function variants in AP4M1 (P−/−_11) stained with antibodies against AP4E1 and TGN46 showing reduced levels of AP4E1 associated with the TGN in PVUS/VUS_02. No significant change in ATG9A localization is found. **P < 0.01; ***P < 0.001; ****P < 0.0001.
AP4B1 knockout SH-SY5Y cells
AP4B1 knockout SH-SY5Y cells (AP4B1KO) were a mixed population of CRISPR/Cas9-edited cells, as previously described.10 SH-SY5Y cells were maintained in DMEM/F12 (Gibco, Cat# 11320033) supplemented with 10% heat-inactivated FBS (Gibco, Cat# 10438026), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C under 5% CO2. SH-SY5Y cells were differentiated into a neuron-like state by culture in the presence of retinoic acid as described previously.34 Cells were seeded at 25% confluency in standard culture medium, and the following day the medium was replaced with neurobasal medium with B27 (Thermo Fisher Scientific, #11320082, #17504-044) containing 10 μM all-trans-retinoic acid (MedChemExpress, #HY-14649). After 48 h, cells were split 1:4 and cultured for 3 further days in the presence of retinoic acid (5 days total).
Nucleofection
AP4B1 plasmid DNA constructs were purchased from Twist Bioscience (Supplementary File 2). Plasmid to express pmaxGFP™ was purchased from Lonza (Lonza, #VCA-1003). Nucleofection of AP4B1KO SH-SY5Y cells with full-length wild type AP4B1 or AP4B1 carrying variants found in PVUS/VUS_01 and PVUS/VUS_02 was carried out using the Cell Line Nucleofector Kit V (Lonza, #VCA-1003) and Amaxa Nucleofector 2b device (Lonza, #AAB-1001). Per 2 × 106 cells, 2 μg of AP4B1 plasmid and 2 μg of GFP plasmid were used. After nucleofection cells were resuspended in differentiation medium with retinoic acid and seeded onto 96-well plates (Greiner Bio-One, Cat# 655090) at a density of 14 × 103 per well. After 48 h, cells were fixed using 4% paraformaldehyde (PFA) and stained and imaged as described below.
Immunocytochemistry
Automation of each step in the immunocytochemistry protocol was maximized by using automated pipettes and reagent dispensers (Thermo Fisher Scientific Multidrop Combi Reagent Dispenser, Integra VIAFLO 96/384 liquid handler, Integra VOYAGER pipette). After 24 h in a humidity-controlled incubator, plates were fixed using 3% PFA, permeabilized with 0.1% saponin in phosphate-buffered saline (PBS) and blocked in PBS containing 1% bovine serum albumin (BSA) and 0.01% saponin (blocking solution). Primary antibody (diluted in blocking solution) was added for 1 h at room temperature. Plates were washed three times in blocking solution and then fluorochrome-conjugated secondary antibody, Hoechst 33258 and Phalloidin were added for 30 min at room temperature. Plates were then washed three times with PBS. For experiments shown in Fig. 5H and I, cells were grown onto 13 mm glass coverslips and fixed in 3% formaldehyde in PBS or, for AP4E1 labelling, ice-cold methanol. Formaldehyde fixed cells were permeabilized and blocked in PBS with 0.5% BSA and 0.01% saponin. Methanol fixed cells were blocked in 0.5% BSA. Primary antibody (diluted in blocking solution) was added for 60 min at room temperature. Coverslips were washed three times in PBS and then fluorochrome-conjugated secondary antibody was added in blocking solution for 40 min at room temperature. Coverslips were then washed three times in PBS. Widefield and confocal images were captured on a Zeiss LSM 710 confocal microscope on an inverted AxioImagerZ1 using a Zeiss Plan Apochromat 63× oil immersion objective (NA 1.4) and ZEN Black software version 2.3. Images were always processed in the same way with the same exposure times and the same manipulations to optimize brightness and contrast. For experiments shown in Fig. 6, plates with differentiated SH-SY5Y cells were fixed in 4% PFA at 48 h after nucleofection, permeabilized with 0.1% saponin in PBS and blocked in 1% BSA/0.01% saponin in PBS. Primary antibody diluted in blocking solution was added for 1 h at room temperature. Plates were washed three times in blocking solution and fluorochrome-conjugated secondary antibodies and Hoechst 33258 were added for 30 min at room temperature.
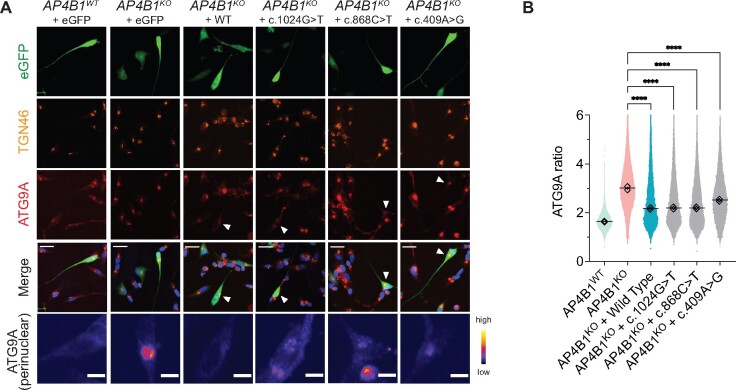
Figure 6.
Expression of novel AP4B1 missense variants (c.1024G>T, c.868C.T, c.409A>G) in AP4B1KO SH-SY5Y cells reduces the ATG9A ratio. (A) Differentiated AP4B1KO SH-SY5Y cells transfected with plasmids to express full-length wild-type or mutant AP4B1 alleles found in PVUS/VUS_01 (c.1024G>T and c.868C.T) and PVUS/VUS_02 (c.409A>G). Differentiated AP4B1WT and AP4B1KO SH-SY5Y cells transfected with eGFP only serve as controls. Cells are stained with antibodies against TGN46 (trans-Golgi network marker) and ATG9A. Transfected cells are detected based on the eGFP signal. White arrowheads indicate transfected cells. The bottom row shows a magnified and pseudocoloured image (colour code indicates grey-scale value) of the transfected cell. (B) After 48 h, expression of full-length wild-type AP4B1, AP4B1c.409A>G, AP4B1c.868C>T, AP4B1c.1024G>T in AP4B1KO SH-SY5Y cells reduced the ATG9A ratio, indicating the restoration of AP-4 function (one-way ANOVA with Dunnett’s post hoc test, P < 0.0001). Each data point represents the ATG9A ratio of a single cell [mean number of cells per group: 27 834 ± 4401 (SD), each diamond represents the mean of each assay plate (n = 2)], horizontal lines indicate the mean from two assay plates. A total of 615 data points are outside the axis limits. ****P < 0.0001.
High-content imaging and automated image analysis
High-throughput confocal imaging was performed on an ImageXpress Micro Confocal Screening System (Molecular Devices) using an experimental pipeline modified from the pipeline described in Behne et al.11 For experiments shown in Figs 3–5A and B, images were acquired using a 20× S Plan Fluor objective (NA 0.45 μM, WD 8.2–6.9 mm). Per well, 9 fields were acquired in a 3 × 3 format (96-well plates) or 4 fields in a 2 × 2 format (384-well plates). Each experiment was performed in biological duplicate on two separate plates with each plate carrying loss-of-function (LoF) and matched wild-type (WT)/LoF cells. Analysis was performed using a customized image analysis pipeline in MetaXpress (Molecular Devices): Briefly, cells were identified based on the presence of DAPI signal inside a phalloidin-positive cell body. Sequential masks were generated for (1) the TGN by outlining the area covered by TGN marker TGN46 (TGN46+ area) and (2) for the cell area outside the TGN (actin+ area minus TGN46+ area). ATG9A fluorescence intensity was measured in both compartments in each cell and the ‘ATG9A’ ratio was calculated by dividing the ATG9A fluorescence intensity in the TGN by the ATG9A fluorescence intensity in the remaining cell body. For experiments shown in Fig. 6, images were acquired using a 40× S Plan Fluor objective (NA 0.60 µm, WB 3.6–2.8 mm). Per well, 64 fields were acquired in an 8×8 format. Automated image analysis was performed as described above using the eGFP signal, instead of phalloidin, to outline the cytoplasm. Z′-factor robust values {Z′ robust = 1–3*[mad(a)+mad(b)]/abs[median(a)-median(b)]} and strictly standardized mean difference (SSMD)35 were calculated for each plate and only plates that met the predefined quality metrics of a Z′-factor robust ≥ 0.3 and SSMD ≥ 3 were included in subsequent analyses.
Western blot
Western blotting was done as previously described.36 Western blots shown in Fig. 5G were developed using chemiluminescence detection of HRP-conjugated secondary antibody using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) and X-ray film.
Immunoprecipitation
For immunoprecipitations of AP4E1 and AP4B1, equal starting material (similar passage number and growth rate) of fibroblasts from P+/+_4 (unaffected control), PVUS/VUS_02 (patient with novel biallelic missense variants in AP4B1) and P−/−_11 (patient with biallelic loss-of-function variants in AP4M1) were lysed in PBS with 0.1% Triton X-100 and cleared by centrifugation at 21 000×g for 20 min. Antibodies against AP4E1 and AP4B1 (both produced in house6) were incubated with the lysates for 90 min at 4°C with rotation, then Protein A sepharose beads (GE Healthcare, #17-0780-01) were added, and samples were incubated for a further 45 min. Beads were washed four times in PBS with 0.1% Triton X-100, once in PBS, and the immunoprecipitates were recovered in NuPAGE LDS Sample Buffer at 75°C for 10 min to prepare for Western blot analysis. All steps were performed on ice, unless otherwise noted.
Statistical analysis
Statistical analysis was performed in GraphPad Prism version 9.0. Normality of the data was assessed with the Shapiro–Wilk test and QQ plots. Figures 3C, 4B, D–F, 5A, D and F show the mean with standard deviation. Figure 4A shows violin plots of all data points with horizontal lines indicating the median (334 data points were outside the axis limits). Data shown in Fig. 4F were compared using the Mann–Whitney test. For experiments shown in Fig. 4B, D and E groups were compared using Kruskal–Wallis test with posthoc Dunn’s test for multiple comparisons. Data shown in Figs 5A, D, F and 6B were compared using one-way ANOVA with Dunnett’s posthoc test after a normal distribution was established. The Receiver Operator Characteristic (ROC) curve shown in Fig. 4C was computed by pooling all values for patients (P−/−_1 − P−/−_18) and heterozygous controls (P+/−_1 − P+/−_17). For Figs 3D and 4G, a simple linear regression analysis was performed and the Pearson correlation coefficient was calculated. Statistical significance was determined at P < 0.05 for all analyses.
Data availability
Data are available from the corresponding authors upon reasonable request. Fibroblast lines generated in this study are available with a material transfer agreement.
Results
Clinical and molecular characterization
We established fibroblast lines from 20 individuals with biallelic variants in subunits of the AP-4 complex (Table 1, Supplementary Table 1), as well as 14 fibroblast lines from parents or siblings (confirmed asymptomatic, heterozygous carriers) and 10 healthy, unrelated control individuals (Supplementary Table 2). Demographic, clinical and genetic information are provided in Table 1. Clinical information of 13 patients has been reported previously.3 In the current cohort, 18/20 of referred individuals showed ≥6 of the 9 core clinical and radiographic features of AP-4-HSP (according to Ebrahimi-Fakhari et al.,3 a list can be found in Supplementary Table 3). The majority of patients presented with developmental delay and subsequent intellectual disability (19/20), progressive spasticity (19/20), microcephaly (17/20) and epilepsy (14/20). Most individuals ambulated with assistance or used a wheelchair as indicated by 4-Stage Mobility Scores of III (8/20) and IV (7/20). The mean Spastic Paraplegia Rating Scale (SPRS) score was 24.4 ± 10.8 (SD) indicating a moderate degree of corticospinal tract dysfunction and associated complications. Common neuroimaging features consisted of a thin corpus callosum (17/20), abnormal signal in the periventricular white matter (18/20) and ex-vacuo ventriculomegaly (15/20).
Table 1.
Clinical and molecular characterization of the study cohort
| Patient ID | AP4-HSP subunit | Allele 1 | Allele 2 |
CADD PHRED
score |
ACMG classification a | Sex | Age | SPRS | 4-FMS | AP-4-HSP core features | ATG9A ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P−/−_01 | AP4B1 | c.1345A>T (p.Arg449Ter) (Nonsense) | c.1160_1161delCA (p.Thr387ArgfsTer30) (Frameshift) | 36/33 | Pathogenic/pathogenic | M | 3 y 11 m | 18 | 2 | 9/9 | 1.65 |
| P−/−_02 | AP4B1 | c.530_531insA (p.Asn178GlufsTer20) (Frameshift) | c.114-2A>C p.? (Splice site) | 32/34 | Pathogenic/pathogenic | F | 5 y 3 m | 15 | 3 | 7/9 | 1.59 |
| P−/−_03 | AP4B1 | c.1216C>T (p.Arg406Ter) (Nonsense) | c.1328T>C (p.Leu443Pro) (Missense) | 41/28.1 | Pathogenic/uncertain significance | F | 4 y 11 m | 35 | 4 | 8/9 | 1.44 |
| P−/−_04 | AP4B1 | c.1216C>T (p.Arg406Ter) (Nonsense) | c.1328T>C (p.Leu443Pro) (Missense) | 41/28.1 | Pathogenic/uncertain significance | F | 4 y 10 m | 22 | 3 | 8/9 | 1.70 |
| P−/−_05 | AP4B1 | c.114-2A>C p.? (Splice site) | c.114-2A>C p.? (Splice site) | 34/34 | Pathogenic/pathogenic | F | 7 y 5 m | 32 | 3 | 8/9 | 1.71 |
| P−/−_06 | AP4B1 | c.617G>A (p.Arg206Gln) (Missense) | c.617G>A (p.Arg206Gln) (Missense) | 33/33 | Likely pathogenic/likely pathogenic | M | 4 y 8 m | 22 | 3 | 8/9 | 1.39 |
| P−/−_07 | AP4B1 | c.1160_1161delCA (p.Thr387ArgfsTer30) (Frameshift) | c.1160_1161delCA (p.Thr387ArgfsTer30) (Frameshift) | 33/33 | Pathogenic/pathogenic | M | 8 y 10 m | 41 | 4 | 9/9 | 1.55 |
| P−/−_08 | AP4B1 | c.664delC (p.Leu222CysfsTer31) (Frameshift) | c.1177C>T (p.Arg393Ter) (Nonsense) | 28.9/40 | Pathogenic/pathogenic | M | 1 y 9 m | NA | 4 | 8/9 | 1.75 |
| P−/−_09 | AP4B1 | c.1608_1609insCA (p.Lys537GlnfsTer18) (Frameshift) | c.1608_1609insCA (p.Lys537GlnfsTer18) (Frameshift) | 32/32 | Pathogenic/pathogenic | M | 7 y 2 m | 22 | 2 | 8/9 | 1.63 |
| P−/−_10 | AP4M1 | c.916C>T (p.Arg306Ter) (Nonsense) | c.916C>T (p.Arg306Ter) (Nonsense) | 39/39 | Pathogenic/pathogenic | F | 31 y 11m | NA | 4 | 8/9 | 1.36 |
| P−/−_11 | AP4M1 | c.218dupA (p.Asn73LysfsTer43) (Frameshift) | c.851A>C (p.Tyr284Ser) (Missense) | 24.5/29.5 | Pathogenic/Uncertain Significance | M | 9 y 1 m | 41 | 4 | 9/9 | 1.50 |
| P−/−_12 | AP4M1 | c.916C>T (p.Arg306Ter) (Nonsense) | c.694dupG (p.Glu232GlyfsTer21) (Frameshift) | 39/33 | Pathogenic/pathogenic | M | 18 m | 29 | 4 | 9/9 | 1.46 |
| P−/−_13 | AP4M1 | c.1025 + 2dupT p.? (Splice site) | c.205A>C (p.Thr69Pro) (Missense) | 34/22.4 | Uncertain significance/uncertain significance | M | 5 y 10 m | 0 | 2 | 6/9 | 1.37 |
| P−/−_14 | AP4S1 | c.138 + 3_138 + 6delAAGT p.? (Splice site) | c.138 + 3_138 + 6delAAGT p.? (Splice site) | 33/33 | Pathogenic/pathogenic | F | 4 y 11 m | NA | 3 | 7/9 | 1.39 |
| P−/−_15 | AP4S1 | c.294 + 1G>T p.? (Splice site) | c.294 + 1G>T p.? (Splice site) | 35/35 | Pathogenic/pathogenic | M | 3 y 6 m | 19 | 3 | 9/9 | 1.52 |
| P−/−_16 | AP4S1 | c.289C>T (p.Arg97Ter) (Nonsense) | c.289C>T (p.Arg97Ter) (Nonsense) | 50/50 | Pathogenic/pathogenic | F | 4 y 2 m | 22 | 2 | 9/9 | 1.64 |
| P−/−_17 | AP4S1 | c.49dupT (p.Ser17PhefsTer2) (Frameshift) | c.49dupT (p.Ser17PhefsTer2) (Frameshift) | 30/30 | Pathogenic/pathogenic | F | 12 y 9 m | NA | 3 | 8/9 | 1.51 |
| P−/−_18 | AP4S1 | c.49dupT (p.Ser17PhefsTer2) (Frameshift) | c.238_239insG (p.Ile80SerfsTer3) (Frameshift) | 30/33 | Pathogenic/pathogenic | F | 17 y 1 m | 24 | 3 | 8/9 | 1.56 |
| PVUS/VUS_01 | AP4B1 | c.1024G>T (p.Asp342Tyr) (Missense) | c.868C>T (p.Arg290Cys) (Missense) | 28.6/25.5 | Uncertain significance/uncertain significance | F | 3 y 0 m | NA | 4 | 3/9 | 1.16 |
| PVUS/VUS_02 | AP4B1 | c.409A>G (p.Arg137Gly) (Missense) | c.409A>G (p.Arg137Gly) (Missense) | 24.4/24.4 | Uncertain significance/uncertain significance | F | 41 y 3 m | NA | 2 | 1/8 | 1.27 |
F, female; M, male; m, months; NA, not available; SPRS, Spastic Paraplegia Rating Scale; y, years. Reference sequences: AP4B1: NM_001253852.3; AP4M1: NM_004722.4; AP4E1: NM_007347.5; AP4S1: NM_007077.4.
Classification of variants based on ACMG criteria using VarSome (Kopanos et al., 2019). CADD PHRED scores were computed using version 1.6 (https://cadd.gs.washington.edu/score).27
AP4B1 was most commonly mutated in our cohort. Variants covered a wide spectrum including frameshift/nonsense (22/40 alleles), missense (10/40 alleles) and splice site variants (8/40 alleles), in homozygous or compound heterozygous states (Table 1). Eight alleles carried variants of uncertain significance by ACMG criteria,2 classified in VarSome,20 including one individual with a compound heterozygous splice site and missense variant in AP4S1 (P−/−_13), and two individuals with a compound heterozygous or homozygous missense variant in AP4B1, respectively (PVUS/VUS_01 and PVUS/VUS_02). CADD scores for all variants were above the recommended cut-off for significance of 20.
Individuals PVUS/VUS_01 and PVUS/VUS_02 presented with atypical clinical features and novel variants in AP4B1 (Table 1, Supplementary Table 1), and were thus referred for a functional interpretation of their variants.
Briefly, PVUS/VUS_01 is a 3-year-old white female, born to non-consanguineous parents of mixed-European heritage, who presented in the neonatal period with refractory epilepsy with myoclonic seizures, followed by severe global development delay and hypotonia. Brain MR imaging was normal. Exome sequencing showed compound-heterozygous missense variants in AP4B1 (NM_001253852.3), classified as a variant of unclear significance (VUS) and a likely benign variant, respectively. The c.1024G>T (p.Asp342Tyr) variant showed an allele frequency of 0.0000119 in gnomAD exomes,37 with a homozygous allele count of 0. In silico analysis revealed 9 pathogenic predictions from BayesDel_addAF, DANN, EIGEN, FATHMM-MKL, LIST-S2, M-CAP, MutationAssessor, MutationTaster and SIFT, versus 4 benign predictions from DEOGEN2, MVP, PrimateAI and REVEL, annotated using dbNSFP.38 The second variant, c.868C>T (p.Arg290Cys), had an allele frequency of 0.00000398 in gnomAD exomes, with a homozygous allele count of 0. There were 7 benign predictions from BayesDel_addAF, DEOGEN2, EIGEN, MVP, MutationAssessor, PrimateAI and REVEL versus 6 pathogenic predictions from DANN, FATHMM-MKL, LIST-S2, M-CAP, MutationTaster and SIFT. CADD PHED scores (version 1.6)27 were 28.6 and 25.5, respectively (Table 1).
PVUS/VUS_02 is a 41-year-old white woman of Belgian ancestry, with no history of parental consanguinity, who presented with adult-onset mild spastic paraplegia, mild intellectual disability, ataxia and dysarthria. There was no history of seizures or microcephaly. She continues to ambulate with a spastic gait and is independent with most activities of daily living. Brain MR imaging showed mild cerebellar atrophy but a normally formed corpus callosum, lateral ventricles and periventricular white matter. Exome sequencing showed a homozygous missense variant in AP4B1 (NM_001253852.3), classified as a variant of uncertain significance. The variant, c.409A>G (p.Arg137Gly), was absent from gnomAD exomes and genomes and was predicted to be pathogenic based on 11 pathogenic predictions from BayesDel_addAF, DANN, DEOGEN2, EIGEN, FATHMM-MKL, LIST-S2, M-CAP, MutationAssessor, MutationTaster, REVEL and SIFT, versus 2 benign predictions from MVP and PrimateAI. The CADD score was 24.4.
AP-4 subunit structures with variant distribution and computational scores
The distribution of variants in the AP-4 subunit genes of individuals included in the AP-4-HSP International Registry and Natural History Study (ClinicalTrials.gov Identifier: NCT04712812) is shown in Figs 1 and 2. The CADD score of each variant is indicated and a model of CADD scores is provided for all possible missense variants across the protein secondary structures of AP4B1, AP4M1, AP4E1 and AP4S1 (similar analyses for M-CAP and REVEL scores can be found in Supplementary Fig. 1, data are summarized in Supplementary File 1). All types of variants observed in patients were predominately located in the known functional domains. This observation was particularly evident for missense variants (blue dots) in AP4B1 and AP4M1, which were exclusively located in either the Adaptin N terminal region (in the case of AP4B1, Fig. 1A) or the Adaptor complexes medium subunit domain (in the case of AP4M1, Fig. 1B). Novel missense variants present in PVUS/VUS_01 and PVUS/VUS_02 localized to the Adaptin N terminal region of AP4B1. In AP4B1, a few truncating or splice variants were located in the region between the functional domains (Fig. 1A). However, the C-terminal B2-adapt-app_C domain of AP4B1 is notable, since no disease-associated variants have been identified here to date. In AP4B1, AP4E1 and AP4S1, higher CADD scores in functional domains indicated greater restrain for missense variation (Figs 1A, 2A and B). AP4M1 deviates from this pattern since no lowering of CADD scores was found for missense variants that fall between the functional domains (Fig. 1B). The observed higher conservation of this shortest linker region of all AP-4 proteins possibly indicates that this region is important to precise inter-domain spacing and positioning.
Validation of the ‘ATG9A ratio’ as an indicator of AP-4 function and a diagnostic marker for AP-4-HSP
The AP-4 complex mediates transport of transmembrane proteins from the TGN to the periphery of the cell.5 Three independent groups identified the core autophagy protein ATG9A as a major cargo of AP-4.9,10,39 In a small cohort of AP-4-HSP patients, using conventional immunocytochemistry, we previously showed that ATG9A accumulates in the TGN.11
Building on this finding, we developed an automated, high-throughput imaging platform that allows us to measure the intracellular distribution of ATG9A in thousands of patient-derived fibroblasts in a single run (Fig. 3A and B). Specifically, we determined the ratio of ATG9A fluorescence signal in the area of the TGN in relation to the signal in the remainder of the cell (Fig. 3B). Using this ‘ATG9A ratio’, we established that our assay has favourable metrics for cell-based high-throughput assays (Z′-factor robust > 0.3, SSMD > 3; Fig. 3C and Supplementary Fig. 2) and reproducibility between replicates (Fig. 3D). Confirming that our assay provides a surrogate of AP-4 function and is sensitive to its restoration, we found that re-expression of the missing AP4B1 subunit in fibroblasts with biallelic truncating variants in AP4B1 restored the ATG9A ratio to a level close to controls (Fig. 3E).
We found that fibroblast lines from patients with typical clinical features of AP-4-HSP (≥6 core clinical features), most (16/18) with at least one loss-of-function variant (frameshift, nonsense or essential splice site), showed a significant increase in the ATG9A ratio compared to heterozygous controls and healthy unrelated controls (Fig. 4A and B, Table 2). This increase was ∼1.3-fold compared to heterozygous controls from the same assay plate (Table 2). Figure 4A shows violin plots demonstrating the distribution of the ATG9A ratio on the level of individual cells with a mean of 6765 ± 4536 (SD) cells per cell line. Figure 4B shows a grouped analysis summarized by assay plate with two biological replicates per cell line. ROC analysis showed that the ATG9A ratio can separate patients with biallelic loss-of-function variants from heterozygous controls with robust diagnostic power (area under the curve 0.85, 95% confidence interval 0.849–0.852) (Fig. 4C). Individuals with AP4M1-associated SPG50 showed a lower mean ATG9A ratio compared to individuals with AP4B1-associated SPG47 (Fig. 4D).
Table 2.
ATG9A ratio can discriminate AP-4-HSP patients from controls
| Typical AP-4-HSP patients (n = 18) | Heterozygous carriers (n = 14) | |
|---|---|---|
| ATG9A ratio | 1.54 ± 0.13 (mean, SD), 95% CI of mean: 1.50–1.58 | 1.21 ± 0.05 (mean, SD), 95% CI of mean: 1.18–1.23 |
| 1.54 (median), 95% CI of median: 1.49–1.62 | 1.20 (median), 95% CI of median: 1.17–1.24 |
Fibroblasts were derived from three patients with compound heterozygous missense variants where one allele was classified as of uncertain significance and the other as pathogenic (P−/−_3, P−/−_4, P−/−_11) showed an ATG9A ratio that was not significantly different from that of patients with two variants classified as pathogenic (Fig. 4E). This finding supports the pathogenicity of the variants. Similarly, in one individual with a novel missense variant in trans with a novel splice-site variant (P−/−_13), both previously classified as of uncertain significance, we found an ATG9A ratio that confirms the detrimental impact of these variants on AP-4 function (Fig. 4E). In a grouped analysis, patients with one or more missense alleles tended to have lower ATG9A ratios than patients with two nonsense, frameshift, or splice site variants, though this difference did not reach statistical significance (Fig. 4F). Exploring correlations with in silico prediction tools, we found that there was a trend, but no significant correlation, between the CADD scores of each patient’s variants and their corresponding ATG9A ratio levels (Fig. 4G).
The ATG9A ratio can provide a functional assessment of novel missense variants
Having established the ATG9A ratio in a well-characterized cohort of patients with AP-4-HSP, we next explored if our assay could be used to functionally characterize novel variants in AP-4 subunit genes. We found that fibroblasts from two individuals with biallelic missense variants in AP4B1 and atypical phenotypes (PVUS/VUS_01 and PVUS/VUS_02, Table 1) showed no elevation in the ATG9A ratio (Fig. 5A and B), suggesting that the variants may not disrupt AP-4 function.
In PVUS/VUS_01, the protein levels of AP4B1 and AP4E1, which can be used as a surrogate for AP-4 stability,7,11 were not different compared to controls (Fig. 5C and D). In addition, the level of ATG9A was unaffected, whereas ATG9A levels were increased in cells from patients with bona fide AP-4-HSP (Fig. 5C and D). Since these proteins are known to change with the loss of AP-4 function, including in AP-4 knockout cells, AP-4 knockout mice and cells from AP-4-HSP patients,9–11,40,41 the absence of change argues against a functionally-relevant impact of the novel missense variants in PVUS/VUS_01.
In PVUS/VUS_02, as in PVUS/VUS_01, the ATG9A ratio was significantly lower than that of patients with typical AP-4-HSP and biallelic loss-of-function variants (Fig. 5A). However, looking at protein levels in whole-cell lysates, we found a reduction of AP4B1 and AP4E1, by about 30% compared to heterozygous controls, although no change in ATG9A protein levels (Fig. 5E and F). To further investigate the effect of the variant on AP-4, we used immunocytochemistry experiments (Fig. 5H and I). Immunostaining revealed a reduced presence of AP4E1 at the TGN, suggestive of reduced levels of AP-4 complex associated with membranes (Fig. 5H). In contrast, there was no significant impact on ATG9A at the TGN (Fig. 5I). Taken together, these findings argue that the homozygous c.409A>G (p.Arg137Gly) missense variant leads to reduced AP-4 complex formation. Supporting this hypothesis, immunoprecipitation experiments showed reduced binding of AP4E1 and AP4B1 (Fig. 5G). Importantly, however, there was residual complex formation, whereas no apparent complex formation was seen in cells with biallelic loss of function variants (Fig. 5G and I).
Collectively, findings in PVUS/VUS_02 indicate that while this homozygous missense variant leads to lower AP-4 complex formation, the level of AP-4 complex expression appears to be sufficient for efficient sorting of ATG9A at the TGN in fibroblasts.
To further characterize the functional impact of each variant present in PVUS/VUS_01 and PVUS/VUS_02, we expressed each mutant allele in human AP4B1 knockout SH-SY5Y cells,10 a neuronal model of AP-4 deficiency. AP4B1 knockout SH-SY5Y cells show an elevated ATG9A ratio, indicating accumulation of ATG9A in the TGN area (Fig. 6A and B). Using our high-throughput imaging platform, we established that this assay can reliably detect differences between negative (AP4B1 knockout) and positive (AP4B1 wild type) controls (mean: Z′-factor robust: 0.92, SSMD: 30.9). We found that re-expression of each mutant allele (c.1024G>T, c.868C>T, c.409A>G) over 48 h led to a significant reduction in the ATG9A ratio, providing evidence that they, at least in part, restore AP-4 function in a neuronal model of AP-4 deficiency.
In summary, based on data in well-characterized AP-4-HSP patients and controls, we establish that an elevated ATG9A ratio in patient-derived fibroblasts indicates clinically relevant AP-4 dysfunction with a high degree of sensitivity and specificity, providing a diagnostic functional assay for cases with atypical features and variants of uncertain significance.
Discussion
In the absence of a biochemical biomarker in blood, urine or CSF, a functional assay in patient-derived fibroblasts can aid the interpretation of novel gene variants and help establish a clinical diagnosis. Having a functional assay is of particular importance to rare disorders with a wide or non-specific clinical spectrum. Cell-based functional assays similar to ours exist for a small number of disorders, mainly inborn errors of metabolism, where the function of an enzyme or accumulation of a metabolite can be assayed. Such disorders include other forms of HSP, for example, SPG5 where oxysterol levels can be measured42 and SPG26 where GM2 synthase activity can be approximated,43 and lysosomal storage diseases, for example, the use of filipin staining used in the diagnosis of Niemann–Pick disease type C.44
With just over 200 individuals identified to date,3 AP-4-HSP is an ultra-rare, yet paradigmatic, form of childhood-onset HSP and a genetic mimic of cerebral palsy. The diagnosis relies on molecular testing, and there is often a significant diagnostic delay.45 To aid the diagnostic process and to provide a tool for gene variant interpretation, we developed a diagnostic functional assay that measures a known and well-established downstream target of AP-4.
ATG9A is a transmembrane autophagy-related protein and cycles between the TGN and ATG9A vesicles. There is increasing evidence that ATG9A acts as a lipid scramblase that enables the expansion of nascent autophagosome membranes.15–17 In neurons, autophagosomes form in the distal axon46,47 and are subject to active transport.36,48–50 Thus, transport of ATG9A to distal axons may act as a cue to compartmentalize autophagosome biogenesis.39,41,51,52 From recent work,9-11,39–41,53 and overlapping phenotypes of AP-439,41,54 and Atg9a18 knockout mice, the following working model for AP-4 deficiency emerges: (i) AP-4 is required for sorting of ATG9A from the TGN; (ii) loss-of-function variants in any of the four AP-4 subunits lead to a loss of AP-4 assembly and function; (iii) ATG9A accumulates in the TGN leading to a reduction of axonal delivery of ATG9A; and (iv) lack of ATG9A at the distal axon impairs autophagosome biogenesis and axonal function, leading to the length-dependent axonal degeneration. While loss of AP-4 function clinically leads to a neurological disorder, the mislocalization of ATG9A can be assayed in multiple cell types and tissues, including in fibroblasts, as shown here.
Our assay employs automated high-throughput microscopy of ATG9A in patient-derived fibroblasts and can reliably detect reduced AP-4 function and thus AP-4-associated HSP. The assay can be performed following a standard diagnostic skin biopsy and using standard laboratory resources. Following fibroblast derivation, results can be obtained within days. Based on the data from 18 patients with genetically and clinically confirmed AP-4-HSP, we define a value between 1.49–1.62 (representing the 95% CI of the median in this cohort) as strongly indicative of clinically significant AP-4 dysfunction. ROC analysis shows an AUC of 0.85 indicating robust separation of patients and controls, with an ATG9A ratio of >1.37 showing a sensitivity of 69% and a specificity of 89% for detecting AP-4-HSP. Relying on a ratio rather than an absolute fluorescence value and by deriving data from thousands of cells in each run, our assay performs robustly and reliably. Controls are included on each assay plate and suitable positive controls may include fibroblast lines from patients with biallelic early frameshift variants and established loss of function, whereas negative controls can be fibroblasts derived from a family member with a heterozygous variant or unrelated wild type controls.
In cases where the pathogenicity of variants is unclear and/or the phenotype deviates from core clinical features present in the majority of AP-4-HSP patients, our platform can provide an approximation of AP-4 function. As demonstrated in two atypical cases with novel missense variants, this can help substantiate or disprove a clinical diagnosis and, thus, can aid the diagnostic workup, with implications for counselling and treatment. Findings in PVUS/VUS_02 illustrate that biallelic missense variants may lower the probability of AP-4 complex formation, yet this might not lead to a lack of trafficking of AP-4 cargo. There is likely a threshold of residual expression that allows sufficient AP-4 complex to form and cargo to be transported. This notion is supported by additional re-expression experiments in AP4B1KO SH-SY5Y cells, a human neuronal model for AP-4 deficiency. We found that expression of the c.409A>G allele reduced the ATG9A ratio 48 h after transfection, yet to a lower degree compared to expression of the full-length wild-type protein or the c.1024G>T and c.868C>T alleles found in PVUS/VUS_01. Thus, it remains possible that the c.409A>G variant conveys a partial reduction in AP-4 function possibly by reducing the amount of AP-4 assembly. From knockdown experiments of AP-1, however, we estimate that expression of adaptor protein complexes would have to be reduced substantially, i.e. >90%, to lead to a significant impact on downstream targets.55,56
We recognize that a subtle impairment of ATG9A trafficking in neurons, particularly neuronal populations known to be vulnerable to changes in autophagy57,58 and involved in AP-4-HSP,59 might not be reflected in fibroblasts or other cell culture-based experiments. We also acknowledge that our assay relies on ATG9A distribution only and does not take into account other cargo proteins that are potentially important in neurons.10,60 Thus, an interpretation of results from our fibroblast-based assay should integrate detailed clinical information, neuroimaging, molecular testing and in silico predictions, including those illustrated in Figs 1 and 2 and Supplementary Fig. 1.
The overlap of CADD PHRED scores of missense variants across AP4B1, AP4E1 and AP4S1 with the annotated domains underscores their relevance for function and variant interpretation (i.e. ACMG criterion PM1), but as shown for variants in individuals PVUS/VUS_01 and PVUS/VUS_02 this has limitations. In a large cohort, our functional assay used in combination with variant location and in silico prediction scores may provide an opportunity to further refine the impact of missense variants, aid the interpretation and classification of variants and enable further exploration of differences between subunits and functional domains in combination with structural biology approaches.31 This could be done posthoc focusing on missense variants identified in individuals through molecular testing or prospectively using a massive parallel screen of all possible missense variants, as has been recently shown for a cancer predisposition gene.61
For the future development of biomarkers, it will be important to establish if changes in ATG9A can be detected in other specimens including in blood, saliva or cerebrospinal fluid, which would allow for the investigation of longitudinal changes with disease progression or the impact of interventions.
In conclusion, we here establish a high-throughput ATG9A translocation assay in patient-derived fibroblasts as a diagnostic test for AP-4-HSP. Our approach enables the functional validation of novel variants in AP4B1, AP4M1, AP4S1 and AP4E1 and can thus provide additional support for a clinical and molecular diagnosis of this debilitating childhood neurological disorder.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
The authors thank the patients and their families for participating in this study.
Funding
J.E.A. is supported by the Deutsche Forschungsgemeinschaft (German Research Foundation, 270949263/GRK2162), the German National Academic Foundation and the Max Weber-Program of the State of Bavaria. B.P. received support by the Deutsche Forschungsgemeinschaft (German Research Foundation, PO2366/2-1). B.B. received a scholarship from the German National Academic Foundation and the Carl Duisberg Program. M.Z. received a scholarship from the German National Academic Foundation and the German National Exchange Service (Deutscher Akademischer Austauschdienst). J.H. received support from the Wellcome Trust (086598 and 214272 to M.S. Robinson, and 100140 to C.I.M.R.). D.E.F. received support from the CureAP4 Foundation, the CureSPG50 Foundation, the Spastic Paraplegia Foundation, the Manton Center for Orphan Disease Research and the National Institute of Health/National Institute of Neurological Disorders and Stroke (2R25NS070682 and 1K08NS123552-01). A.K.D. received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 896725. A.S. was funded by the Deutsche Forschungsgemeinschaft (German Research Foundation, SA 4171/1-1). The Rosamund Stone Zander Translational Neuroscience Center at Boston Children’s Hospital is supported by the Massachusetts Life Sciences Center and the J.P. Fletcher Foundation. The BCH Intellectual and the Developmental Disabilities Research Center Clinical/Translational Core is supported by the National Institutes of Health (BCH IDDRC, 1U54HD090255).
Competing interests
D.E.F. received an honorarium for speaking at the Movement Disorder International Congress in 2019, royalties from Cambridge University Press as Editor of ‘Movement Disorders and Inherited Metabolic Disorders’, a consulting fee from Alcimed Inc. M.S. and D.E.F. received research funding through a joint research agreement between Boston Children’s Hospital and Astellas Pharmaceuticals Inc. and Mitobridge Inc. M.S. reports grant support from Novartis, Roche, Biogen, Astellas, Aeovian, Bridgebio, Aucta and Quadrant Biosciences. M.S. has served on Scientific Advisory Boards for Novartis, Roche, Celgene, Regenxbio, Alkermes and Takeda.
Glossary
- ACMG =
American College of Medical Genetics and Genomics
- AP-4 =
adaptor protein complex 4
- AP-4-HSP =
AP-4 associated hereditary spastic paraplegia
- AUC =
area under curve
- BSA =
bovine serum albumin
- DMEM =
Dulbecco’s Modified Eagle Medium
- FBS =
foetal bovine serum
- HSP =
hereditary spastic paraplegia
- LoF =
loss of function
- PFA =
paraformaldehyde
- ROC =
receiver operating characteristic
- SPRS =
Spastic Paraplegia Rating Scale
- SD =
standard deviation
- SSMD =
strictly standardized mean difference
- TGN =
trans-Golgi network
- VUS =
variant of unclear significance
- WT =
wild type
References
- 1. Shribman S, Reid E, Crosby AH, Houlden H, Warner TT.. Hereditary spastic paraplegia: From diagnosis to emerging therapeutic approaches. Lancet Neurol. 2019;18(12):1136–1146. [DOI] [PubMed] [Google Scholar]
- 2. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ebrahimi-Fakhari D, Teinert J, Behne R, et al. Defining the clinical, molecular and imaging spectrum of adaptor protein complex 4-associated hereditary spastic paraplegia. Brain. 2020;143(10):2929–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ebrahimi-Fakhari D, Behne R, Davies AK, Hirst J, et al. AP-4-associated hereditary spastic paraplegia. In: Adam MP, Ardinger HH, Pagon RA, eds. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 2018, 1993–2021. PMID: 30543385. [PubMed] [Google Scholar]
- 5. Sanger A, Hirst J, Davies AK, Robinson MS.. Adaptor protein complexes and disease at a glance. J Cell Sci. 2019;132(20):jcs222992. [DOI] [PubMed] [Google Scholar]
- 6. Hirst J, Bright NA, Rous B, Robinson MS.. Characterization of a fourth adaptor-related protein complex. Mol Biol Cell. 1999;10(8):2787–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hirst J, Irving C, Borner GH.. Adaptor protein complexes AP-4 and AP-5: New players in endosomal trafficking and progressive spastic paraplegia. Traffic. 2013;14(2):153–164. [DOI] [PubMed] [Google Scholar]
- 8. Dell'Angelica EC, Mullins C, Bonifacino JS.. AP-4, a novel protein complex related to clathrin adaptors. J Biol Chem. 1999;274(11):7278–7285. [DOI] [PubMed] [Google Scholar]
- 9. Mattera R, Park SY, De Pace R, Guardia CM, Bonifacino JS.. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc Natl Acad Sci U S A. 2017;114(50):E10697–E10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davies AK, Itzhak DN, Edgar JR, et al. AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat Commun. 2018;9(1):3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Behne R, Teinert J, Wimmer M, et al. Adaptor protein complex 4 deficiency: A paradigm of childhood-onset hereditary spastic paraplegia caused by defective protein trafficking. Hum Mol Genet. 2020;29(2):320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orsi A, Razi M, Dooley HC, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23(10):1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Young AR, Chan EY, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119(Pt 18):3888–3900. [DOI] [PubMed] [Google Scholar]
- 14. Guardia CM, Tan XF, Lian T, et al. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep. 2020;31(13):107837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maeda S, Yamamoto H, Kinch LN, et al. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol. 2020;27(12):1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matoba K, Kotani T, Tsutsumi A, et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol. 2020;27(12):1185–1193. [DOI] [PubMed] [Google Scholar]
- 17. Orii M, Tsuji T, Ogasawara Y, Fujimoto T.. Transmembrane phospholipid translocation mediated by Atg9 is involved in autophagosome formation. J Cell Biol. 2021;220(3):e202009194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamaguchi J, Suzuki C, Nanao T, et al. Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy. 2018;14(5):764–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schule R, Holland-Letz T, Klimpe S, et al. The Spastic Paraplegia Rating Scale (SPRS): A reliable and valid measure of disease severity. Neurology. 2006;67(3):430–434. [DOI] [PubMed] [Google Scholar]
- 20. Kopanos C, Tsiolkas V, Kouris A, et al. VarSome: The human genomic variant search engine. Bioinformatics. 2019;35(11):1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Q, Wang K.. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100(2):267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Popp B, Ekici AB, Thiel CT, et al. Exome Pool-Seq in neurodevelopmental disorders. Eur J Hum Genet. 2017;25(12):1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cingolani P, Patel VM, Coon M, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cingolani P, Platts A, Wang Le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu X, Jian X, Boerwinkle E.. dbNSFP v2.0: A database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34(9):E2393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jian X, Boerwinkle E, Liu X.. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42(22):13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M.. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiong HY, Alipanahi B, Lee LJ, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347(6218):1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. UniProt C., UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gadbery JE, Abraham A, Needle CD, et al. Integrating structural and evolutionary data to interpret variation and pathogenicity in adapter protein complex 4. Protein Sci. 2020;29(6):1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hunter S, Apweiler R, Attwood TK, et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009;37(Database issue):D211–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ebrahimi-Fakhari D, Wahlster L, Bartz F, et al. Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-pick type C1 disease cells. Hum Mol Genet. 2016;25(16):3588–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kovalevich J, Langford D.. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol. 2013;1078:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bray MA, Carpenter A.. Advanced assay development guidelines for image-based high content screening and analysis. 2004. ed. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2017. vol. https://www.ncbi.nlm.nih.gov/books/NBK53196/. [PubMed]
- 36. Ebrahimi-Fakhari D, Saffari A, Wahlster L, et al. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep. 2016;17(4):1053–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu X, Li C, Mou C, Dong Y, Tu Y.. dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ivankovic D, Drew J, Lesept F, et al. Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy. 2020;16(3):391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. D'Amore A, Tessa A, Naef V, et al. Loss of ap4s1 in zebrafish leads to neurodevelopmental defects resembling spastic paraplegia 52. Ann Clin Transl Neurol. 2020;7(4):584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. De Pace R, Skirzewski M, Damme M, et al. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLOS Genetics. 2018;14(4):e1007363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schols L, Rattay TW, Martus P, et al. Hereditary spastic paraplegia type 5: Natural history, biomarkers and a randomized controlled trial. Brain. 2017;140(12):3112–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harlalka GV, Lehman A, Chioza B, et al. Mutations in B4GALNT1 (GM2 synthase) underlie a new disorder of ganglioside biosynthesis. Brain. 2013;136(Pt 12):3618–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tangemo C, Weber D, Theiss S, Mengel E, Runz H.. Niemann-Pick Type C disease: Characterizing lipid levels in patients with variant lysosomal cholesterol storage. J Lipid Res. 2011;52(4):813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ebrahimi-Fakhari D, Cheng C, Dies K, et al. CureSPG47. Clinical and genetic characterization of AP4B1-associated SPG47. Am J Med Genet A. 2018;176(2):311–318. [DOI] [PubMed] [Google Scholar]
- 46. Bunge MB. Fine structure of nerve fibers and growth cones of isolated sympathetic neurons in culture. J Cell Biol. 1973;56(3):713–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hollenbeck PJ. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol. 1993;121(2):305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maday S, Holzbaur EL.. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014;30(1):71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maday S, Wallace KE, Holzbaur EL.. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196(4):407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL.. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206(5):655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stavoe AK, Hill SE, Hall DH, Colon-Ramos DA.. KIF1A/UNC-104 transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev Cell. 2016;38(2):171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamura H, Shibata M, Koike M, Sasaki M, Uchiyama Y.. Atg9A protein, an autophagy-related membrane protein, is localized in the neurons of mouse brains. J Histochem Cytochem. 2010;58(5):443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mattera R, Williamson CD, Ren X, Bonifacino JS.. The FTS-Hook-FHIP (FHF) complex interacts with AP-4 to mediate perinuclear distribution of AP-4 and its cargo ATG9A. Mol Biol Cell. 2020;31(9):963–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matsuda S, Miura E, Matsuda K, et al. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57(5):730–745. [DOI] [PubMed] [Google Scholar]
- 55. Hirst J, Borner GH, Antrobus R, et al. Distinct and overlapping roles for AP-1 and GGAs revealed by the "knocksideways" system. Curr Biol. 2012;22(18):1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinson MS, Sahlender DA, Foster SD.. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell. 2010;18(2):324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ebrahimi-Fakhari D, Saffari A, Wahlster L, et al. Congenital disorders of autophagy: An emerging novel class of inborn errors of neuro-metabolism. Brain. Feb. 2016;139(Pt 2):317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Teinert J, Behne R, Wimmer M, Ebrahimi-Fakhari D.. Novel insights into the clinical and molecular spectrum of congenital disorders of autophagy. J Inherit Metab Dis. 2020;43(1):51–62. [DOI] [PubMed] [Google Scholar]
- 59. Verkerk AJ, Schot R, Dumee B, et al. Mutation in the AP4M1 gene provides a model for neuroaxonal injury in cerebral palsy. Am J Hum Genet. 2009;85(1):40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Davies AK, Ziegler M, Jumo H, Saber WA, Ebrahimi-Fakhari D, Borner GHH. AP-4 mediates vesicular transport of the 2-AG endocannabinoid producing enzyme DAGLB. BiorXiv, doi: 10.1101/2020.10.25.353995, 1 January 2020, preprint: not peer reviewed. [DOI]
- 61. Jia X, Burugula BB, Chen V, et al. Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk. Am J Hum Genet. 2021;108(1):163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding authors upon reasonable request. Fibroblast lines generated in this study are available with a material transfer agreement.