Abstract
Background
Brain tumor surgery must balance the benefit of maximal resection against the risk of inflicting severe damage. The impact of increased resection is diagnosis-specific. However, the precise diagnosis is typically uncertain at surgery due to limitations of imaging and intraoperative histomorphological methods. Novel and accurate strategies for brain tumor classification are necessary to support personalized intraoperative neurosurgical treatment decisions. Here, we describe a fast and cost-efficient workflow for intraoperative classification of brain tumors based on DNA methylation profiles generated by low coverage nanopore sequencing and machine learning algorithms.
Methods
We evaluated 6 independent cohorts containing 105 patients, including 50 pediatric and 55 adult patients. Ultra-low coverage whole-genome sequencing was performed on nanopore flow cells. Data were analyzed using copy number variation and ad hoc random forest classifier for the genome-wide methylation-based classification of the tumor.
Results
Concordant classification was obtained between nanopore DNA methylation analysis and a full neuropathological evaluation in 93 of 105 (89%) cases. The analysis demonstrated correct diagnosis in 6/6 cases where frozen section evaluation was inconclusive. Results could be returned to the operating room at a median of 97 min (range 91-161 min). Precise classification of the tumor entity and subtype would have supported modification of the surgical strategy in 12 out of 20 patients evaluated intraoperatively.
Conclusion
Intraoperative nanopore sequencing combined with machine learning diagnostics was robust, sensitive, and rapid. This strategy allowed DNA methylation-based classification of the tumor to be returned to the surgeon within a timeframe that supports intraoperative decision making.
Keywords: brain tumor, DNA methylation, extent of resection, intraoperative diagnostics, nanopore
Key Points.
Nanopore DNA methylation classification of brain tumors is robust and sensitive.
The method is effective over a diverse spectrum of both adult and pediatric tumors.
It can be done fast enough to impact intraoperative neurosurgical strategy and patient outcome.
Importance of the Study.
The recently established DNA methylation classification of brain tumors refines prognosis and can direct more detailed individualized treatment of patients. The currently used pipeline for this classification usually takes weeks to return results. We aimed to establish a workflow that could be used intraoperatively, as a precise diagnosis is typically uncertain at surgery. Nanopore sequencing combined with a machine learning algorithm generated robust, sensitive, and rapid classification of the tumor entity. The method is well-functioning over a broad range of tumors without pre-analytic presumption of tumor entity. This workflow could be performed intraoperatively, yielding diagnostics as fast as 91 min from tumor sampling. The improved intraoperative diagnostics could support personalized surgical strategy choices in a high fraction of patients with preoperative uncertainty of tumor entity. The study demonstrates a feasible workflow that merges precision molecular diagnostics with modern neurosurgical strategies for individualized treatment of patients with brain tumors.
Brain tumors are the most common cause of cancer-related death in children and young adults.1 The 5-year survival rate for malignant central nervous system (CNS) tumors for all ages is approximately 20%, but the expected survival differs primarily depending on the tumor entity.1 The first line of treatment is most often surgical resection of the tumor. Tumor resection reduces tumor burden, often alleviates symptoms and can in some cases be curative. Resection also allows for diagnostic sampling which guides further treatment.2,3 The extent of tumor resection is balanced against the risk of inflicting neurological damage and other postoperative complications (onco-functional balance).4 The overall goal is often stated as “maximal safe resection,” but this concept is vague. What could be considered an acceptable risk is dependent on the gain of a complete vs subtotal resection? In tumors close to eloquent brain the risk of resection can increase dramatically when the resection is pushed from near complete to a complete resection.5 Choosing the optimal neurosurgical strategy depends on a precise diagnosis as the prognosis is highly variable among tumor entities and even among subgroups.6 For example, the 5-year survival of WNT-activated medulloblastoma patients is 95% and a small residual tumor after surgery has no impact on survival. In contrast, group 3 medulloblastoma patients have a 5-year survival rate of 50%, and gross total resection (GTR) likely improves survival.3 Also, the expected available time for patients to recover and rehabilitate after surgery should influence the surgical risk. Patients with IDH-mutated gliomas will have time to recover postoperatively, and aggressive GTR has been demonstrated beneficial.7 For other entities, such as IDH-wildtype glioma with features of glioblastoma, the short expected survival allows for less surgical risk to be accepted, as the patient will have less time to recover from even reversible neurological deficits.8 Preoperative imaging modalities cannot reliably distinguish between these glial tumor entities.
Currently, surgical decision making relies on preoperative imaging combined with intraoperative histological evaluation of hematoxylin- and eosin-stained frozen sections to give a preliminary diagnosis. This process can be time-consuming and error-prone due to inter-observer variability in the histopathological diagnosis of many CNS tumors.9 Tumor localization, growth pattern, low cellularity, and small biopsy size may further complicate intraoperative histological classification.10 Diagnostic biopsies of CNS tumors before resection are rarely performed, especially in children, as they postpone treatment and lead to additional surgical interventions. However, there is still an urgent need for accurate classification of CNS tumors to guide surgical strategy and postoperative therapy. Liquid biopsies,11 intraoperative Raman spectroscopy,12 mass spectrometry,13 or targeted qPCR10 have been proposed as methods to provide pre- or intraoperative diagnosis of CNS tumors. However, these methods largely provide hypothesis-driven tests and do not discriminate wider ranges of tumor entities. Definitive preoperative differentiation between subtypes of medulloblastomas or between an IDH-mutated oligodendroglioma and a molecular glioblastoma is not possible with widely with available preoperative imaging or any other analysis.
DNA methylation-based classification of CNS tumors can improve diagnostic precision. This can revise the WHO grading of a tumor and refine patient management.6 It has been shown to impact treatment of children by providing additional molecular subtyping and amending final diagnosis.14 The basis of this classification is the DNA methylation pattern of cancer cells which is unique for each entity.15 Machine learning algorithms can be trained to identify these patterns and use them to discriminate the type of tumor entity as well as the subgroups, which are now included in the WHO tumor grading system.16 This method is highly robust and reproducible even for small samples and poor-quality material but current implementations have a turnaround time of days to weeks.6
Nanopore sequencing can be used to detect base modifications such as 5-methylcytosine (5mC) in native DNA without the need for bisulfite conversion.17 This provides an ideal platform for rapid generation of tumor methylation profiles. We have previously demonstrated the potential of nanopore DNA methylation analysis (NDMA) for multimodal and rapid molecular diagnostics in brain cancer.18 This was used for subclassification of the tumor entities into over 80 subgroups according to the Heidelberg cohort.6 Here, we demonstrate how low-pass whole-genome nanopore sequencing can be used to generate DNA methylation profiles of a tumor biopsy that is sufficient for accurate classification in less than 2 h.
Methods
Patient Samples and Ethical Approval
A total of 107 tissue biopsies and the corresponding clinical data were obtained from patients treated at Oslo University Hospital (OUH), Norway and from the “Barnebiobank-barnekreft” (REK 2016/943) and “Biobank for nevrokirurgisk forskning” (REK 2016/1791) general biobanks after signed, written, informed consent was obtained from patients, or for pediatric patients from their parents. The study was approved by the Regional Committee for Medical Research Ethics (REK 2018/2465 and 2019/173). Of the 107 samples, 44 frozen tissue samples were obtained from the biobanks, 39 frozen tissue sections were from the Section of Neuropathology (OUH), and 24 fresh biopsies were sampled at the time of surgery. Two non-CNS tumors (a metastatic sarcoma and a teratoma) were analyzed but not included in the study. The tumors were assessed according to standard diagnostic protocols and classified according to the WHO 2016 Classification of Central Nervous System Tumors. Diagnostically challenging cases were evaluated by a second opinion at the German Brain Tumor Reference Center in Bonn, Germany. In one intraoperative case, the frozen section was not evaluated. The samples were grouped into 6 non-overlapping cohorts for different analytic strategies: (1) A validation of archival material form patients before 2018 (n = 12), (2) a prospective case series of patients operated at OUH from 2018 to 2020 (n = 36), (3) a historic collection of diagnostically challenging embryonal tumors operated 2005-2012 (n = 21), (4) a selected cohort of double-blinded cases operated 2018-2019 (n = 10), (5) a selection of inconclusive frozen sections operated 2019-2020 (n = 6), and (6) intraoperative cases operated 2019-2020 (n = 20), (Supplementary Table 1; Supplementary Figure 1).
Sample Processing and DNA Extraction
DNA was extracted using spin columns (QIAamp DNA Micro Kit, Qiagen, NL) according to the manufacturer’s protocol with ~15 mg of tumor tissue. Quantification of eluted DNA was performed on a Qubit 4.0 fluorometer using a dsDNA BR Assay (Thermo Fisher, USA) and assessed for purity using the NanoDrop 260/280 ratio (NanoDrop, Thermo Fisher).
For intraoperative NDMA analysis, the tumor biopsy was immediately placed in lysis buffer (Qiagen) at the operating room, allowing digestion to begin during transport to the laboratory.10 Upon arrival, the biopsy lysate was homogenized for 30 s (TissueLyser, Qiagen) and digested for 10 min at 56°C and 5 min at 70°C. This reduced the total sample handling and DNA extraction time to 30 min. The DNA quantity and quality were still above the required standards.
Nanopore Whole-Genome Sequencing for NDMA
To allow the pooling of samples and reduce the risk of cross-contamination, all libraries were barcoded using a Rapid Barcoding Kit (RBK004, Oxford Nanopore Technologies, UK). Briefly, a fragmentation mix containing unique barcodes for each patient was added to 200 to 400 ng of genomic DNA. A PCR step of 30°C for 1 min, then 80°C for 1 min followed. The barcoded libraries were then pooled and cleaned with AMPure XP beads (Beckman Coulter, USA). A rapid adapter was added to the libraries before loading them onto a flow cell. Whole-genome sequencing was performed for a minimum of 3 h on a MinION Mk 1B device using an R9.4.1 Flow Cell (FLO-MIN106D, Oxford Nanopore Technologies). Each flow cell was reused up to 4 times after washing with the Flow Cell Wash Kit (EXP-WSH003, Oxford Nanopore Technologies). When multiplexing retrospective samples, up to 10 libraries were pooled and sequenced for up to 24 h.
Nanopore Data Analysis Pipeline
A MinIT (Oxford Nanopore Technologies), a small computing device with an advanced GPU, SSD storage capacity, and accelerated base-calling function, was used for base-calling the raw sequencing reads in real time. Equipped with the manufacturer software MinKNOW (v20.06.17), the MinIT can obtain raw data (FAST5) in real time, separate barcoded reads and filter low-quality reads while simultaneously base-calling using the built-in proprietary software guppy (v4.0.11, GPU based, fast mode, Oxford Nanopore Technologies). FAST5 and FASTQ files were analyzed using an adapted snakemake19 v5.4.0 workflow from the nanoDx pipeline20 that enabled analysis on a local laptop computer. Within this pipeline, reads were aligned to the hg19 human reference genome (minimap2 v2.15),21 and copy number profiles were generated. The methylation status of the genome-wide CpG sites (5mC) was called (nanopolish v0.11.0)17 and binarized (cutoff beta value >0.6) together with 5mC signals for overlapping sites probed by the Illumina BeadChip 450K array. For each sample, an ad hoc random forest classifier (R/ranger package v0.10.1)22 was trained using the Heidelberg reference cohort of brain tumor methylation profiles, and a final report containing sequencing statistics and classification results was generated. The nanoDx pipeline was initiated when at least 3 FAST5 file packages had been generated containing a mean of 13 321 reads per sample and a mean of 6165 CpG sites per sample (Supplementary Table 3). This was achieved after 20-60 min of sequencing depending on active flow-cell pores. Absolute copy number estimation (ACE, v1.6.0)23 was implemented to assess tumor purity, and the returned estimations were evaluated manually. For all the 105 patients, mean read length was 2803 base pairs (range; 522-6959), mean number of CpG sites covered were 30 694 CpGs (range 3396-100 000), mean coverage was 0.12× (range 0.01-0.74), and the mean random forest out-of-bag error rate was 5.81% (range 4.8-7.9).
Methylation Array Processing
The 450k array was used to obtain genome-wide DNA methylation profiles for tumor samples, according to the manufacturer’s instructions (Illumina). DNA methylation data were generated at the Institute for Neuropathology, Charite Universitaets Medizin (Berlin, Germany) using >400 ng of DNA was used as input material. The tumor classification was performed according to the established protocol.6
Code and Data Availability
The current NDMA classification and analysis pipeline are publicly available at https://gitlab.com/pesk/nanoDx (v2.0.X). The source code to reproduce all analyses and figures in this manuscript is available at https://gitlab.com/kuscheluis/nanoINTRAOP. Raw sequencing data (fast5 files) from all samples used in this study were deposited at the European Nucleotide Archive (accession PRJEB48142).
Results
Patient and Tumor Characteristics
The study includes 105 patients with a median age of 26 years (range 0-84). 50 patients were pediatric (≤20 years) and 55 were adults. There was a selection of cases where diagnostic subclassification could be challenging and where such subclassification could be advantageous intraoperatively (Supplementary Table 1). Ninety-seven samples were from first surgery, while eight samples were from the second or later surgery. Of these, four had received radiotherapy to the lesion and two of these also received temozolomide chemotherapy. Final histopathological classification WHO grades of these were 18 grade I, 13 grade II, 16 grade III, and 57 grade IV tumors (Table 1).
Table 1.
Clinical Characteristics of the Patient Cohort
| Clinical Characteristics | Adults (n = 55) | Pediatric (n = 50) | Overall (n = 105) |
|---|---|---|---|
| Gender | |||
| Male | 34 (62%) | 30 (60%) | 64 (61%) |
| Female | 21 (38%) | 20 (40%) | 41 (39%) |
| Age at surgery | |||
| Range | 24-84 | 0-19 | 0-84 |
| Mean | 50.2 | 8.5 | 30.4 |
| Median | 48 | 7.5 | 26 |
| Tumor location | |||
| Frontal | 28 (51%) | 8 (16%) | 36 (34%) |
| Midline | 1 (2%) | 6 (12%) | 7 (7%) |
| Parietal | 5 (9%) | 5 (5%) | |
| Temporal | 13 (24%) | 3 (6%) | 16 (15%) |
| Posterior fossa | 3 (6%) | 31 (62%) | 34 (32%) |
| Ventricular | 1 (2%) | 1 (1%) | |
| Extra-axial | 2 (4%) | 2 (3%) | |
| Occipital | 1 (2%) | 1 (1%) | |
| Spinal | 2 (4%) | 2 (2%) | |
| Sella | 1 (2%) | 1 (1%) | |
| WHO grade | |||
| I | 6 (11%) | 12 (24%) | 18 (17%) |
| II | 11 (20%) | 3 (6%) | 14 (13%) |
| III | 13 (24%) | 2 (4%) | 15 (14%) |
| IV | 24 (44%) | 33 (66%) | 57 (54%) |
| NA | 1 (2%) | 1 (1%) | |
| Biopsy type | |||
| Fresh | 14 (25%) | 10 (20%) | 24 (23%) |
| Frozen | 41 (75%) | 40 (80%) | 81 (77%) |
NDMA Robustness
In total, we performed NDMA on 105 individual patient samples using public reference data for more than 80 CNS tumor entities and compared methylation-based classification to the definite WHO integrated diagnosis obtained by full neuropathological workup. NDMA was concordant with final neuropathological diagnosis in 93 of the 105 cases (89%) (Figure 1; Supplementary Table 1). Most adult cases were classified as gliomas (20 glioblastomas, 14 oligodendrogliomas, and 10 astrocytomas) by NDMA (Figure 1A). Among the pediatric cases in the study (Figure 1B), medulloblastomas were the most common tumor type (23 cases) followed by pilocytic astrocytomas (10 cases).
Figure 1.
Nanopore DNA methylation analysis (NDMA) gives a robust classification of brain tumors. Final neuropathological diagnosis (left) compared to NDMA diagnosis (center) and concordance of methods (right). “A” represents results from the 55 CNS tumors from adult cases reported in the study while “B” represents results from the 50 pediatric cases reported. In summary, NDMA was concordant with final neuropathological diagnosis in 93 of the 105 cases. Abbreviations: A, astrocytoma; ATRT, atypical teratoid rhabdoid tumor; CNS, central nervous system; CPH, craniopharyngioma; DMG, diffuse midline glioma; EPN, ependymoma; GBM, glioblastoma; MB, medulloblastoma; MNG, meningioma; O, oligodendroglioma; PLEX, plexus papilloma; PXA, pleomorphic xanthoastrocytoma; SCHW, schwannoma; SHH, sonic hedgehog.
To initially evaluate the robustness of NDMA we analyzed 79 samples from 4 independent cohorts and compared these results to the final neuropathological diagnosis (Supplementary Table 1; Supplementary Figure 1). NDMA afforded classification of all cases, including cases not subclassified by standard diagnostic evaluation. Overall, this demonstrated a robust pipeline for NDMA with low coverage sequencing and methylation profiling.
Out of the total 105 samples, discordant results between NDMA and neuropathological evaluation were observed in 12 cases (11%). These made up 4 of the 55 adult cases (7.3%), and 8 of the 50 pediatric cases (16%). Discordance was primarily observed in diagnostically challenging cases (8 cases) and 1 case with low tumor cell content (Supplementary Table 2). Clear misclassification by NDMA was observed for 3/105 samples (2.8%). Of note, 5/12 (42%) discordant cases were from recurring tumors despite only eight recurring tumors being found in the complete data (7.6%). All cases that produced results that were discordant between NDMA and standard neuropathology were further analyzed by Illumina Infinium HumanMethylation450 Bead Chip (450k) array (Table 2). The classification results of the Illumina sequencing matched NDMA in 3 cases and neuropathology in another 3 cases. A methylation classification could not be made in 4 cases due to DNA quality or quantity insufficiencies while a new tumor subtype was concluded for the remaining 2 cases (Supplementary Table 2).
Table 2.
Overview of Discordant Cases
| Patient ID | Final Pathology Diagnosis | NDMA Diagnosis | Illumina Methylation Classification | Calibrated Score (Illumina) |
|---|---|---|---|---|
| NDMA_5 | Recurrent ganglioglioma | Low-grade glioma, DNET | No match with calibrated score >= 0.3 | – |
| NDMA_88 | Paraganglioma | Meningioma | Meningioma | 0.98 |
| NDMA_28 | High-grade glioma | Atypical teratoid/rhabdoid tumor, SHH | Atypical teratoid/rhabdoid tumor, subclass SHH | 0.87 |
| NDMA_29 | High-grade glioma | Medulloblastoma, subclass group 3. | Medulloblastoma, subclass group 3 | 0.78 |
| NDMA_51 | Ganglioglioma | Posterior fossa pilocytic astrocytoma | Pilocytic astrocytoma (MCF) | 0.43 |
| Low-grade glioma, subclass hemispheric pilocytic astrocytoma, and ganglioglioma | 0.37 | |||
| NDMA_57 | Diffuse astrocytoma, IDHwt (pediatric) | Posterior fossa pilocytic astrocytoma | N/A—due to lack of material | – |
| NDMA_70 | Ganglioglioma | (Anaplastic) pleomorphic xanthoastrocytoma | Control tissue, reactive tumor microenvironment | 0.46 |
| Pilocytic astrocytoma (MCF) | 0.45 | |||
| Low-grade glioma, subclass hemispheric pilocytic astrocytoma, and ganglioglioma | 0.44 | |||
| NDMA_76 | Astrocytoma, IDHwt | Diffuse midline glioma, H3 K27M mutant | Diffuse leptomeningeal glioneuronal tumor | 0.97 |
| NDMA_77 | Recurrent pleomorphic xanthoastrocytoma | Glioblastoma, subclass RTK II | (Anaplastic) pleomorphic xanthoastrocytoma | 0.88 |
| NDMA_84 | Recurrent GBM with granular cell component | Posterior fossa pilocytic astrocytoma | No match with calibrated score >= 0.3 | – |
| NDMA_96 | Recurrent pilocytic astrocytoma | Diffuse midline glioma, H3 K27M mutant | Diffuse leptomeningeal glioneuronal tumor | 0.99 |
| NDMA_105 | Recurrent astroblastoma | Glioblastoma, subclass RTK II | No match with calibrated score >= 0.3 | – |
Abbreviations: DNET, dysembryoplastic neuroepithelial tumor; MCF, methylation class family; NDMA, nanopore DNA methylation analysis; RTK II, receptor tyrosine kinase II; SHH, sonic hedgehog.
Final neuropathology diagnosis compared to the results of NDMA classification and full Illumina methylation classification.
NDMA Sensitivity
To evaluate the sensitivity of NDMA, we performed NDMA on 6 samples with non-informative intraoperative frozen section evaluations. NDMA was performed on the same tissue block used for intraoperative frozen section histology. NDMA classification was consistent with the final neuropathological diagnosis in 6/6 cases (Supplementary Table 3). This suggests that NDMA can accurately classify tumors even when intraoperative frozen section histology is difficult.
Intraoperative NDMA Feasibility
We further optimized the NDMA workflow to obtain rapid classification results within a surgically relevant time period, typically below 120 min. Benchmarking of rapid NDMA was established on biopsies from 20 independent surgical procedures. The median transport time from the operating room to the laboratory was 6 min (range 5-24 min); the median time for DNA isolation, purification, and library preparation was 41 min (22-61 min); real-time computational analysis was initiated after 20-60 min of sequencing, and the median runtime of each bioinformatics analysis cycle was 28 min (17-60 min) (Figure 2A). Reports could be returned to the operating room as fast as 91 min from the time when the sample was obtained. Fifteen of the 20 surgeries were still ongoing when the results were ready. A median of 4140 (696-10 803) CpG methylation sites were detected after 30 min of sequencing (Figure 2B, left) and ad hoc random forest classifiers with an average out-of-bag error rate of 7.6% were generated (Figure 2B, right). The error rate was reduced to 3.5% when tumors were classified on the level of clinically relevant methylation class families (MCF) rather than all subclasses (Figure 2C). Using the MCF classifier and a minimum of 3500 CpG sites as cutoff for reproducible classification20 yielded correct classification in 20/20 patients (Supplementary Table 4). This cutoff was reached after a median of 30 min of sequencing (Supplementary Table 4). In summary, NDMA can provide accurate classification of CNS tumors within a timeframe relevant for intraoperative decision making.
Figure 2.
Nanopore DNA methylation analysis (NDMA) enables the detailed and rapid classification of brain tumors. (A) The NDMA workflow timeline demonstrated rapid feedback for the ongoing surgery. Time range stated at each step. (B) Dot plots demonstrating the relationship between sequencing time and total CpG sites detected (left panel) and between ad hoc random forest out-of-bag (OOB) error rate and sequencing time using the subgroup classifier (right panel). Dotted line demarks 3500 CpG cutoff for analysis; black line demarks polynomial regression with 95% CI. (C) Comparison of random forest OOB error using the methylation class family (MCF) classifier or full subclassification after 30 min of nanopore sequencing. Color indicates the number of CpG sites.
NDMA’s Impact on Surgical Strategy
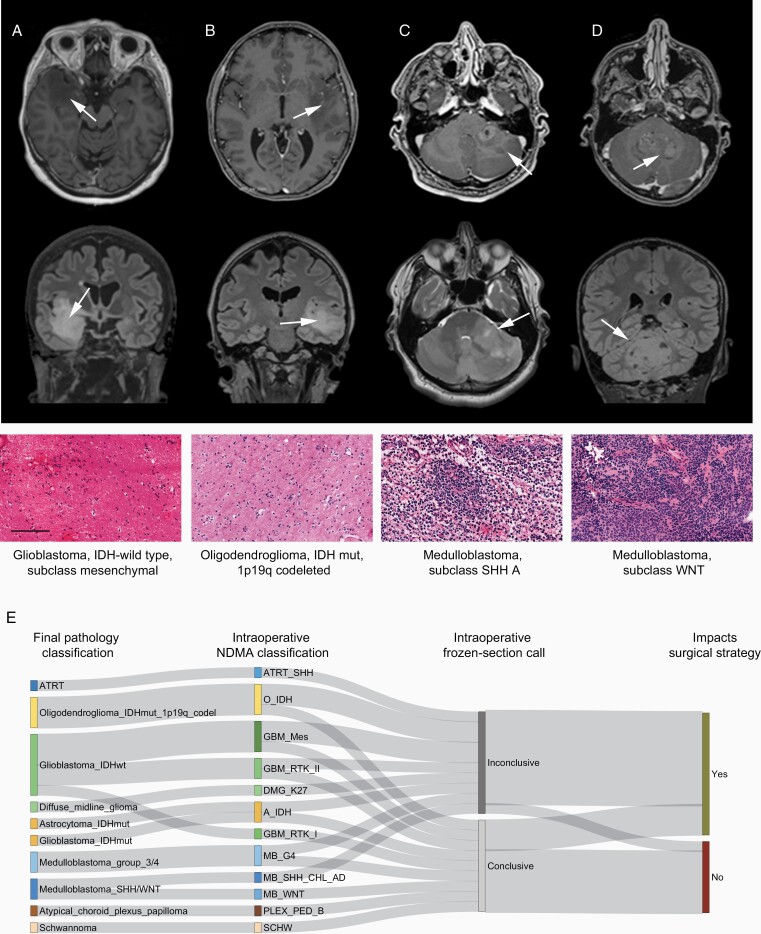
In this intraoperative cohort, surgery was stopped for 2 out of 20 patients (Figure 3A and B) due to uncertain tumor classification and for 1 patient (Figure 3C) due to classification as a possible lymphoma based on frozen section evaluation; NDMA correctly classified all 3 samples and the results would have warranted further resection. Conversely, another patient (Figure 3D) was returned to surgery after intraoperative imaging for resection of a small tumor remnant, while NDMA correctly classified the tumor as a WNT-activated subgroup medulloblastoma, for which further surgery was unnecessary. Precise classification of the tumor entity and subtype would have supported modification of the surgical strategy in 12 out of 20 patients (Figure 3E; Supplementary Table 4). In the complete set of analyses, all instances of NDMA discordance with the final neuropathological diagnosis would not have negatively impacted the surgical strategy if the result was combined with preoperative imaging and previous medical history (Supplementary Table 2). In summary, NDMA can accurately discriminate between tumor entities intraoperatively and guide surgical procedures when preoperative imaging and frozen section evaluation are ambiguous.
Figure 3.
Intraoperative nanopore DNA methylation analysis (NDMA) impacts neurosurgical strategy. Four cases are shown to demonstrate where an improved intraoperative diagnosis would have impacted the intraoperative surgical strategy. Preoperative MRI imaging (top row T1 contrast-enhanced, second row T2-fluid-attenuated inversion recovery, arrows demark tumors) in (A) and (B) demonstrates temporal lobe tumors with diffuse infiltration in the surrounding tissue and spots of contrast enhancement. Frozen section H&E staining (third row) demonstrates no definite tumor characteristics. This led to termination of the resection, resulting in a larger postoperative residual tumor. The NDMA diagnosis (bottom) supports further resection. Importantly, differentiating IDH-wildtype glioma showing molecular features of a glioblastoma from IDH-mutant and 1p/19q-codeleted oligodendroglioma allows for modulation of the intraoperative risk. In another case, ambiguous preoperative imaging (C) combined with frozen section evaluation suggested a possible lymphoma, resulting in cessation of surgery. The final diagnosis was in line with the NDMA findings and confirmed a medulloblastoma, SHH subtype, resulting in reoperation. (D) Preoperative imaging identifies a likely medulloblastoma, and an early postoperative MRI demonstrates a tumor remnant. The NDMA identification of a WNT-subtype medulloblastoma negated the need for further resection. Scale bar 100 µm. (E) Sankey plot demonstrating the final neuropathological diagnosis (left), the intraoperative NDMA analysis, the result from intraoperative frozen section pathology, and whether improved intraoperative diagnostic classification would have impacted surgical decision making. Abbreviations: A, astrocytoma; ATRT, atypical teratoid rhabdoid tumor; CPH, craniopharyngioma; DMG, diffuse midline glioma; EPN, ependymoma; GBM, glioblastoma; MB, medulloblastoma; MNG, meningioma; O, oligodendroglioma; PLEX, plexus papilloma; PXA, pleomorphic xanthoastrocytoma; SCHW, schwannoma; SHH, sonic hedgehog.
Discussion
In this study, we used NDMA to classify 105 CNS tumors. The overall concordance between NDMA classification and standard neuropathological workup was over 88%. Furthermore, we showed that NDMA can be performed in an intraoperative setting, and in doing so it can produce accurate classification that outperforms frozen section analysis.
The use of nanopore technology as a research and possibly clinical tool is gaining momentum. The technique is relatively cost-efficient and easy to set up.18 Wongsurawat and colleagues have shown that nanopore can be used to assess IDH mutation status and MGMT methylation level simultaneously in fresh brain tumor biopsies and cell lines of diffuse gliomas.24 Their nanopore Cas9-targeted sequencing pipeline was able to generate results within 36 h of sequencing, that compared favorably with standard clinical workup. The technology has also been used in diagnosing and monitoring treatment response in pediatric patients with high-grade gliomas. Using cell-free tumor DNA from cerebral spinal fluid, Bruzek and colleagues have recently demonstrated that 0.1 femtomole DNA and 12 h of nanopore sequencing was sensitive and specific enough to detect H3.3A and H3C2 mutations. These results were comparable to standard next-generation sequencing.25
As in previous studies on DNA methylation classification of CNS tumors,6,26 we found some discordance between a full neuropathological workup and NDMA. Most of these cases were difficult-to-diagnose tumors, especially among the pediatric subpopulation which often represent complex cases.14 Some discordant cases had low tumor cell content, but further optimization of the machine learning algorithm, especially with respect to the skewness of the training set, may decrease such misclassifications. Of note, none of these NDMA misclassifications would have misled the surgical decision making when evaluated in the context of preoperative imaging and previous clinical history.
Limitations
An efficient workflow is imperative in intraoperative diagnostics. The current pipeline is not quick enough to impact shorter surgeries. The impact of a specific diagnose is however likely to be highest in longer, complex surgeries, where tumor is situated in deep and eloquent areas. In such cases, where intraoperative monitoring and imaging also are used, the current pipeline is fast enough to return data to ongoing surgeries. Faster DNA isolation techniques, more computational resources and improved flow-cell quality will further shorten turn-around time. Six intraoperative frozen section biopsies with non-informative results during surgery were analyzed as part of this study. NDMA classification of all 6 cases was concordant with the final neuropathological evaluation, even in samples with low estimated tumor cell content. Although these results are promising, further evaluation of NDMA sensitivity with regard to tumor cell content and tumor classification is warranted.
The current study is limited by its restriction to tumor entities represented in the reference dataset, which currently does not contain non-CNS malignancies.27
Conclusion
In conclusion, we demonstrate the accuracy, sensitivity, feasibility, and impact of ultra-low coverage nanopore whole-genome sequencing for intraoperative neuropathological classification. Even within our moderately sized cohort, we identified a substantial number of patients where improved intraoperative diagnostic accuracy would have impacted surgical decision making in real time. Importantly, methylation-based classification is a diagnostic approach generalizable far beyond neuro-oncology, and ongoing efforts to include a wider range of malignancies hold promise for pan-cancer classification.6,28,29
Supplementary Material
Acknowledgments
We thank the Norwegian National Childhood Tumor Biobank (Norsk National Barnekreftbiobanken, funded by Barnekreftforeningen #210002) for providing frozen tissue samples of pediatric tumors.
Funding
L.D. and E.O.V.M. were supported by a grant from the Childhood Cancer Society of Norway (Barnekreftforeningen #19006), J.P. was supported by a grant from the Childhood Cancer Society of Norway (Barnekreftforeningen #19008), and S.H. was supported by a grant from the Regional Health authorities (HSØ #2017073). P.E. is a participant in the BIH-Charité Clinical Scientist Program funded by the Charité-Universitätsmedizin Berlin and the Berlin Institute of Health.
Conflict of interest statement. None of the authors have competing interests related to the presented method or data.
Authorship Statement. L.D., S.H., P.E., and E.O.V.M. conceived and designed the study. B.D.T., I.A.L., C.J.S., and E.O.V.M. were responsible for clinical care of patients and collected all the biomaterials. L.D. and S.H. performed the laboratory testing. D.C. was responsible for methylation array testing and analysis. P.N., H.L., and J.P. provided neuropathological evaluations and diagnoses. L.P.K., S.H., and C.J.S. processed data. P.E. and E.O.V.M. supervised all aspects of the study. L.D., S.H., P.E., and E.O.V.M. wrote the first draft of the manuscript. All authors contributed to final data interpretation and critical revision of the manuscript and approved the final version of the manuscript.
Unpublished Paper Cited
Kuschel et al., submitted, preprint available at https://www.medrxiv.org/content/10.1101/2021.03.06.21252627v1
References
- 1. Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol. 2020;22(Supplement_1):IV1–IV96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weller M, van den Bent M, Preusser M, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 2021;18(3):170–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thompson EM, Hielscher T, Bouffet E, et al. Prognostic value of medulloblastoma extent of resection after accounting for molecular subgroup: a retrospective integrated clinical and molecular analysis. Lancet Oncol. 2016;17(4):484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duffau H, Mandonnet E. The “onco-functional balance” in surgery for diffuse low-grade glioma: integrating the extent of resection with quality of life. Acta Neurochir (Wien). 2013;155(6):951–957. [DOI] [PubMed] [Google Scholar]
- 5. Yong RL, Lonser RR. Surgery for glioblastoma multiforme: striking a balance. World Neurosurg. 2011;76(6):528–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jakola AS, Myrmel KS, Kloster R, et al. Comparison of a strategy favoring early surgical resection vs a strategy favoring watchful waiting in low-grade gliomas. JAMA. 2012;308(18):1881–1888. [DOI] [PubMed] [Google Scholar]
- 8. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136(5):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol. 2010;120(3):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shankar GM, Francis JM, Rinne ML, et al. Rapid intraoperative molecular characterization of glioma. JAMA Oncol. 2015;1(5):662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nassiri F, Chakravarthy A, Feng S, et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat Med. 2020;26(7):1044–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hollon TC, Pandian B, Adapa AR, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26(1):52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu H, Xia YK, Li CJ, et al. Rapid diagnosis of IDH1-mutated gliomas by 2-HG detection with gas chromatography mass spectrometry. Lab Invest. 2019;99(4):588–598. [DOI] [PubMed] [Google Scholar]
- 14. Pickles JC, Fairchild AR, Stone TJ, et al. DNA methylation-based profiling for paediatric CNS tumour diagnosis and treatment: a population-based study. Lancet Child Adolesc Health. 2020;4(2):121–130. [DOI] [PubMed] [Google Scholar]
- 15. Fernandez AF, Assenov Y, Martin-Subero JI, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22(2):407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 17. Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14(4):407–410. [DOI] [PubMed] [Google Scholar]
- 18. Euskirchen P, Bielle F, Labreche K, et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017;134(5):691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Köster J, Rahmann S. Snakemake – a scalable bioinformatics workflow engine. Bioinformatics. 2012;28(19):2520–2522. [DOI] [PubMed] [Google Scholar]
- 20. Kuschel LP, Hench J, Frank S, et al. Robust methylation-based classification of brain tumors using nanopore sequencing. medRxiv. doi: 10.1101/2021.03.06.21252627, March 8, 2021, preprint: not peer reviewed. [DOI] [PubMed] [Google Scholar]
- 21. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wright MN, Ziegler A. Ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw. 2017;77(1):1–17. [Google Scholar]
- 23. Poell JB, Mendeville M, Sie D, Brink A, Brakenhoff RH, Ylstra B. ACE: absolute copy number estimation from low-coverage whole-genome sequencing data. Bioinformatics. 2019;35(16):2847–2849. [DOI] [PubMed] [Google Scholar]
- 24. Wongsurawat T, Jenjaroenpun P, De Loose A, et al. A novel Cas9-targeted long-read assay for simultaneous detection of IDH1/2 mutations and clinically relevant MGMT methylation in fresh biopsies of diffuse glioma. Acta Neuropathol Commun. 2020;8(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bruzek AK, Ravi K, Muruganand A, et al. Electronic DNA analysis of CSF cell-free tumor DNA to quantify multi-gene molecular response in pediatric high-grade glioma. Clin Cancer Res. 2020;26(23):6266–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jaunmuktane Z, Capper D, Jones DTW, et al. Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre. Acta Neuropathol Commun. 2019;7(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Capper D, Engel NW, Stichel D, et al. DNA methylation-based reclassification of olfactory neuroblastoma. Acta Neuropathol. 2018;136(2):255–271. [DOI] [PubMed] [Google Scholar]
- 28. Koelsche C, Hartmann W, Schrimpf D, et al. Array-based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol. 2018;31(8):1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moran S, Martínez-Cardús A, Sayols S, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17(10):1386–1395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The current NDMA classification and analysis pipeline are publicly available at https://gitlab.com/pesk/nanoDx (v2.0.X). The source code to reproduce all analyses and figures in this manuscript is available at https://gitlab.com/kuscheluis/nanoINTRAOP. Raw sequencing data (fast5 files) from all samples used in this study were deposited at the European Nucleotide Archive (accession PRJEB48142).