Abstract
The current evolutionary biology theory primarily involves genetic alterations and random DNA sequence mutations to generate the phenotypic variation required for Darwinian natural selection to act. This neo-Darwinian evolution is termed the Modern Evolution Synthesis and has been the primary paradigm for nearly 100 years. Although environmental factors have a role in neo-Darwinian natural selection, Modern Evolution Synthesis does not consider environment to impact the basic molecular processes involved in evolution. An Extended Evolutionary Synthesis has recently developed that extends the modern synthesis to consider non-genetic processes. Over the past few decades, environmental epigenetics research has been demonstrated to regulate genetic processes and directly generate phenotypic variation independent of genetic sequence alterations. Therefore, the environment can on a molecular level through non-genetic (i.e. epigenetic) mechanisms directly influence phenotypic variation, genetic variation, inheritance and adaptation. This direct action of the environment to alter phenotype that is heritable is a neo-Lamarckian concept that can facilitate neo-Darwinian (i.e. Modern Synthesis) evolution. The integration of genetics, epigenetics, Darwinian theory, Lamarckian concepts, environment, and epigenetic inheritance provides a paradigm shift in evolution theory. The role of environmental-induced epigenetic transgenerational inheritance in evolution is presented to describe a more unified theory of evolutionary biology.
Keywords: review, evolution, epigenetic, transgenerational, inheritance, genetics, adaptation, phenotype, Darwin, Lamarck, molecular
Current Evolution Paradigm
Charles Darwin’s theory of evolution through natural selection provides the basis for our current concepts of evolutionary biology [1]. Adaptive evolution occurs with four biological processes: (i) variation within a population; (ii) variation is heritable; (iii) competition occurs between offspring for limited resources; and (iv) the survival and reproduction of the offspring are not random, but are associated with heritable variation [1, 2]. These concepts of evolution through natural selection developed in the late 1800s and were then advanced in the early 1900s with the rediscovery of Mendelian genetics and the identification of DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) as the molecular mechanism involved in phenotypic variation and inheritance. Adaptation of the classic Darwinian theory with molecular genetics led to the concept of the “Modern Evolutionary Synthesis” proposed in the mid-1900s by Huxley [3]. Subsequently, the role of genetics in phenotypic variation, inheritance and adaptation was established in large part by the field of population genetics. This neo-Darwinian (i.e. Modern Evolution Synthesis) theory of evolution has developed over the past century and is the current paradigm in evolutionary biology [2, 4, 5].
The advances in molecular genetics, genome-wide DNA sequence mutation analysis and understanding of genetic variation are inadequate in considering the complexities of phenotypic variation observed and in rapid evolutionary events for the current evolutionary biology theory (i.e. Modern Synthesis). This is in large part due to the low frequency of associated genetic mutations [4–10]. Phenotypic mutation rates and genetic mutation rates are dramatically different [10]. Understanding the origins of genetic variation and environmental pressure induced evolutionary phenomena are difficult to explain with the Modern Evolution Synthesis theory [4, 11]. Over the past 50 years, molecular technology has been used to investigate evolutionary biology, but many examples of finding no correlated genetic mutations or a low frequency of DNA sequence mutations suggest that additional mechanisms are also involved. Phenotypic plasticity is a very good example of how physiological change can facilitate adaptation, but most phenotypic plasticity has not been related to genetic DNA sequence alterations [2, 4, 5, 11, 12]. Darwin proposed a critical role for environmental impacts to mediate natural selection, but genetic changes alone cannot explain these phenomena well. This has led to the debate that a reevaluation of the current evolution paradigm is needed [2, 4, 5, 11, 12].
Recently an Extended Evolutionary Synthesis (EES) has been developed to expand the Modern Synthesis concepts and include non-classical genetic concepts [13]. This EES incorporates non-genetic processes and non-genetic inheritance and expands on classic genetic considerations, but does not fully develop the molecular processes or elements that integrate with the genetic mechanism [13]. This demonstrates an appreciation that the Modern Synthesis, in considering our current molecular biology, falls short of effectively explaining all mechanisms of evolution. Other aspects of EES not well developed include a lack of detail on how environment can directly impact developmental and biological processes independent of classic genetics. The current review discusses the role of environmental epigenetics and epigenetic transgenerational inheritance as a major molecular component to integrate classic evolution concepts of Lamarck, Darwin and Modern Synthesis and the more recent EES to develop a more unified theory.
Environment and Evolution
Environment has a critical role in Darwinian Evolution [1] as one of the primary factors to facilitate natural selection processes. These environmental factors act on the survival and reproductive fitness of individuals having different phenotypes. The current paradigm in evolutionary biology is that DNA sequence alterations promote this phenotypic variation that responds to the environmental pressure through natural selection [4, 11]. In addition to evolutionary biology, a large number of biological phenomena suggest major impacts of the environment. Ecological parameters such as chemical exposures, temperature, and limited nutrition all impact an individual’s physiology and phenotypes, but do not have the ability to alter DNA sequence. Identical twins have essentially the same genetics, but generally develop discordant disease as they age [1, 2, 4–6, 14–25]. Only a low frequency (generally 1% or less) of individuals that have a specific disease have a correlated genetic mutation, and the dramatic increase in disease frequency in the population cannot be explained with genetics alone [26]. Many phenomena such as regional disease frequency or the fact that environmental toxicant exposures can promote disease, but do not generally have the ability to alter DNA sequence, cannot be easily explained with genetic mutations alone [27]. Therefore, many biological phenomena do not follow normal Mendelian genetic rules and are difficult to explain with classic genetic processes or mechanisms alone [17].
One of the first evolutionary biology theories proposed in 1802 by Jean Baptiste Lamarck suggested that environment promotes the phenotypic alterations associated with evolution [20, 21]. This Lamarckian concept was that environment directly promotes phenotypic variation, which becomes heritable for subsequent generations. This is distinct from the role of environmental factors providing a selection pressure in Darwin’s natural selection theory. This Lamarckian concept was interpreted as conflicting with Darwin’s natural selection evolutionary theory, so was discounted and not considered in the Modern Synthesis (i.e. neo-Darwinian) evolution theory [6]. As will be discussed, a molecular mechanism that could promote direct alterations in phenotype generationally, independent of DNA sequence, and alter genome activity would support this Lamarckian concept [11].
Environment and Phenotypic Variation
A number of evolutionary biology observations suggest that the rates of molecular and morphological evolution are largely decoupled [18]. One of the first observations that environment directly promotes phenotypic variation was the report of Daphnia magna to respond to the presence of predators in the environment [19]. This morphological phenotype induced by the environment was termed the Baldwin effect and later thought to be due to genetics and considered a neo-Darwinian phenomenon [28]. However, this phenomenon does not follow normal Mendelian genetics and is a good example of environmentally induced phenotypic variation. In the early 1900s Paul Kammerer demonstrated in the midwife toad an environmentally induced parent-of-origin non-genetic acquired reproductive trait with arid or aquatic environments [23]. One of the more significant series of studies was pioneered by Conrad Waddington in the 1940s and 1950s [24]. Several generations of Drosophila (i.e. fruit fly) following heat shock exposure promoted a wing structure change that was transmitted for sixteen generations, so was inherited. This adaptive wing shape became “canalized” in the population. This non-genetic phenomenon that became inherited was referred to as “epigenetics” [24]. Although the specific molecular aspects of the epigenetic process were not known or proposed, epigenetics was, by definition, a non-genetic process that did not follow normal Mendelian genetic rules. As will be discussed, the molecular mechanisms have now been characterized and provide a new science to help explain this and other non-genetic (i.e. independent of DNA sequence) processes and inheritance.
The initial genetic terminology used to describe phenotypic variation effects such as those observed by Baldwin, Kammerer, and Waddington was genetic accommodation [22]. These non-genetic heritable changes occur in response to novel environmental pressure. Although these early observations were critical phenomena, interest waned in favor of strictly genetic inheritance of traits in the absence of any known non-genetic mechanisms. When the Modern Synthesis (i.e. neo-Darwinian) theory was formalized, Ernst Mayr described these non-genetic (soft) forms of inheritance as “gradual change of genetic material itself, either by use or disuse, or by some internal presence of tendencies, or through a direct effect of the environment” [25]. Therefore, environmental impacts on phenotypic variation and non-genetic inheritance were strictly left out of the Modern Evolution Synthesis without a specific mechanism to be considered [2]. In contrast, environmental impacts on genetic processes such as horizontal gene transfer in bacteria in adaptation to extreme environments [29], generational maternal effects in both plants and animals [30, 31], and generational prion protein transmission [32, 33] are processes acceptable for inclusion in the Modern Synthesis and EES theories. These processes are examples of multigenerational direct exposure (i.e. intergenerational) phenomena. Therefore, the current Modern Evolution Synthesis involves a genetic determinism focus that cannot explain the environmental impacts on phenotypic variation or non-genetic forms of inheritance. The “curse of complexity” in adaptive evolution of complex traits has been suggested to be resolved by the non-genetic molecular mechanisms of epigenetics [34]. To address these issues, an EES does consider non-genetic components [13], but does not effectively consider a direct environmental impact on phenotypic variation, adaptation, and evolution.
Epigenetics
As discussed, the term epigenetics was coined by Conrad Waddington in the 1940s [24]. Studies in embryology and development were known as “epigenesis,” which was a concept from Aristotle’s time. The integration of epigenesis and genetics provided the origins for the term epigenetics [24, 35]. As discussed, Waddington’s experiments with drosophila demonstrated that a heat induced wing structure developed and was inherited for multiple generations, which was termed “epigenetics.” The definition of epigenetics has changed with greater understanding of the molecular mechanisms. The initial definition of Waddington focused on gene–environment interactions but had no molecular insights [24, 36, 37]. As our molecular understanding has developed, the definition has evolved [27], the current definition of epigenetics is “molecular factors and processes around DNA that regulate genome activity independent of DNA sequence, and are mitotically stable” [38]. The genome activity not only involves gene expression, but also genome stability components such as silencing repeat regions and transposable elements to maintain genome integrity [38]. The mitotic stability of the epigenome is critical to maintain cell specificity and differentiation following cell proliferation [38]. Therefore, as a cell undergoes mitosis, the DNA sequence is replicated, as well as the epigenome is replicated to allow cells and tissues to maintain the normal state of differentiation and development acquired [38]. The history of epigenetics is summarized in Table 1 and involved the initial definition of the term in the 1940s, discovery of DNA methylation in the 1970s by Holliday and Pugh, and Riggs [39, 40], demonstration of the role of DNA methylation in X-inactivation in the late 1980s [41], role in imprinted genes for allelic gene expression in the 1990s [42], role of histone modifications in the 1990s [43], role of non-coding RNA (ncRNA) in 2000s [44], epigenome mapping in 2005 [45], and the identification of epigenetic transgenerational inheritance in 2005 [27] (Table 1).
Table 1:
History of epigenetics
| 1940s | Conrad Waddington defined epigenetics as environment–gene interactions that induce developmental phenotypes |
| 1975 | Holliday and Pugh, and Riggs identify DNA methylation |
| 1988 | X-chromosome inactivation and DNA methylation |
| 1990s | Imprinted genes, allelic expression, and DNA methylation |
| 1995 | Histone modifications and chromatin structure |
| 2000s | Non-coding RNAs |
| 2005 | Epigenome mapping |
| 2005 | Epigenetic transgenerational inheritance |
The currently known molecular epigenetic factors include DNA methylation, histone modifications, changes to chromatin structure, expression of non-coding RNA, and RNA methylation [46] (Fig. 1). All these epigenetic factors can directly regulate gene expression independent of DNA sequence. The first factor identified was DNA methylation that occurs at a cytosine residue adjacent to a guanine residue (CpG) sites to form 5-methylcytosine [47]. Although other DNA modifications exist, such as 5-hydroxymethyctyosine or methyl adenine, they are far less frequent and their potential roles in mechanisms of non-genetic adaptation have not been identified. Histone modifications can also act as an epigenetic factor to regulate gene expression independent of DNA sequence (Fig. 1). The chemical modification of histone proteins with methylation or acetylation can modify the gene expression of the associated DNA [48–50]. These histone modifications also effect chromatin structure, impact regulatory protein (e.g. transcription factor) binding, and promote heterochromatin or euchromatin regions of the genome. Histone variants can also alter chromatin structure and gene regulation [51]. In the male germline histone retention is impacted by histone modifications and alters early embryonic development [52]. Non-coding RNAs are another critical epigenetic factor that regulates gene expression independent of DNA sequence (Fig. 1). A number of different classes of ncRNA exist that are not translated into protein, but can regulate gene expression by binding to DNA or proteins involved in gene expression [53, 54]. The small and large ncRNA act as epigenetic regulatory factors to alter gene expression [55]. RNA methylation at N6-methyl adenine is also an epigenetic factor that can regulate the ncRNA secondary structure to influence protein or DNA binding, and regulates gene expression [56, 57] (Fig. 1). The different epigenetic factors do not only act independently, but integrate with each other to provide a level of epigenetic complexity to accommodate the needs of cellular development and differentiation. For example, ncRNA can facilitate DNA methylation processes [58] (Fig. 1). The DNA methylation can modify histone modifications and chromatin structure [59] (Fig. 1). Therefore, the complexity of the epigenome with its various epigenetic factors can accommodate the requirements for the cellular, organ and phenotypic variation observed. The integration of all these epigenetic processes has been shown to be critical for epigenetic inheritance in response to environmental factors [60]. These epigenetic molecular factors and processes provide the ability for environmental factors to alter gene expression independent of DNA sequence. Therefore, gene expression and genome activity require a precursor epigenetic process to occur, which allows classic genetic processes to function.
Figure 1:
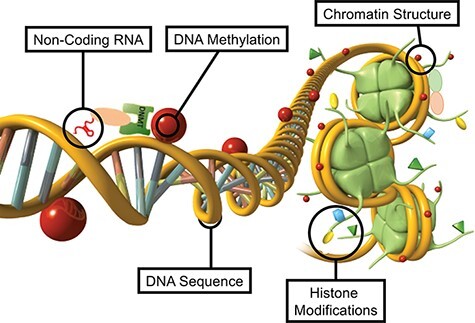
Representation of the primary epigenetic factors and processes schematic of non-coding RNA, DNA methylation, chromatin structure, histone modifications, and DNA structure presented. Modified from Nilsson et al. [46]
Environmental epigenetics is the primary molecular mechanism in any organism that is used to promote physiological and phenotypic alterations [2, 14–16, 34, 38, 47–57, 61–71]. Factors such as nutrition, temperature, light, toxicants, exposures, stress, or trauma [27, 46] directly alter epigenetics to promote the cellular response and environmental phenotypic variation. The actions of environmental factors early in development can permanently program the cellular molecular function, which then impacts later life disease or phenotypes [27, 46]. Since cellular identity and function is determined by the epigenetics which regulates the transcriptome, environmental epigenetics is the molecular factor that essentially controls cellular phenotypic variation. Mary Jane West-Eberhard proposed that environmental pressures result in selection of novel phenotypic traits, which results in genetic alterations, and ultimately speciation [68]. This theory has been coined “genes as followers” and was suggested to be a genetic phenomenon. However, environmental epigenetics has now been suggested to provide the molecular mechanism involved, not genetics [2, 14–16, 34, 38, 47–57, 61–71]. Therefore, the current epigenetic science developed over the past decades indicates environmental epigenetics is the primary molecular mechanism promoting phenotypic variation that is associated with adaptation processes [11]. A large number of studies and literature supports a direct role for non-genetic processes (i.e. epimutations) in phenotypic variation (Table 2). The role of epigenetics in regulating this phenotypic variation to impact evolution is the content of this literature.
Table 2:
Non-genetic (epigenetic) association with phenotypic variation references
| Reference title | ||
|---|---|---|
| Recherches sur l’organisation des corps vivans | Lamarck J. 1802 | [21] |
| A New Factor in Evolution | Baldwin J. 1896 | [19] |
| Organisers and Genes | Waddington CH. 1940 | [24] |
| An epigenetic mutation responsible for natural variation in floral symmetry | Cubas P. 1999 | [77] |
| Developmental plasticity and evolution | West-Eberhard MJ. 2003 | [68] |
| The Baldwin effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity | Crispo E. 2007 | [22] |
| Transgenerational epigenetic imprints on mate preference | Crews D. 2007 | [82] |
| Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior | Skinner MK. 2008 | [93] |
| Identical but not the same: the value of discordant monozygotic twins in genetic research | Zwijnenburg PJ. 2010 | [15] |
| Progressive, Transgenerational Changes in Offspring Phenotype and Epigenotype following Nutritional Transition | Burdge GC. 2011 | [84] |
| Exploring the correlations between sequence evolution rate and phenotypic divergence across the Mammalian tree provides insights into adaptive evolution | Janecka J. 2012 | [18] |
| Adaptive evolution: evaluating empirical support for theoretical predictions | Olson-Manning CF. 2012 | [5] |
| Environmental heterogeneity and phenotypic divergence: can heritable epigenetic variation aid speciation? | Flatscher R. 2012 | [79] |
| Epigenetic variation, inheritance, and selection in plant populations | Hirsch S. 2012 | [97] |
| Epigenetic variation: origin and transgenerational inheritance | Becker C. 2012 | [100] |
| General-purpose genotype or how epigenetics extend the flexibility of a genotype | Massicotte R. 2012 | [119] |
| Epigenetics and the evolution of Darwin’s finches | Skinner MK. 2014 | [122] |
| How stable “should” epigenetic modifications be? Insights from adaptive plasticity and bet hedging | Herman JJ. 2014 | [91] |
| Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations | Jiang C. 2014 | [72] |
| Genetic and epigenetic analysis of monozygotic twins discordant for testicular cancer | Kratz CP. 2014 | [14] |
| Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences | Vogt G. 2015 | [89] |
| Environmental Epigenetics and a Unified Theory of the Molecular Aspects of Evolution: A Neo-Lamarckian Concept that Facilitates Neo-Darwinian Evolution | Skinner MK. 2015 | [11] |
| Landscape of natural epigenetic variation in humans | Chatterjee A. 2015 Heyn H. 2013 |
[140, 141] |
| Facing environmental predictability with different sources of epigenetic variation | Leung C. 2016 | [120] |
| Epigenetic programming alterations in alligators from environmentally contaminated lakes | Guillette LJ, Jr, 2016 | [125] |
| Epigenetics in natural animal populations | Hu J. 2017 | [92] |
| Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants | Lamke J. 2017 | [132] |
| Epigenetic variation between urban and rural populations of Darwin’s finches | McNew SM. 2017 | [123] |
| Natural epigenetic variation within and among six subspecies of the house sparrow, Passer domesticus | Riyahi S. 2017 | [115] |
| Epigenetics and adaptive phenotypic variation between habitats in an asexual snail | Thorson JLM. 2017 | [117] |
| Facilitation of environmental adaptation and evolution by epigenetic phenotype variation: insights from clonal, invasive, polyploid, and domesticated animals | Vogt G. 2017 | [102] |
| An Epigenetic Perspective on the Midwife Toad Experiments of Paul Kammerer (1880–1926) | Vargas AO. 2017 | [23] |
| Epigenetic and genetic variation among three separate introductions of the house sparrow (Passer domesticus) into Australia | Sheldon EL. 2018 | [116] |
| Contribution of epigenetic variation to adaptation in Arabidopsis | Schmid MW. 2018 | [108] |
| Regional epigenetic variation in asexual snail populations among urban and rural lakes | Thorson JLM. 2019 | [118] |
| Sources of epigenetic variation and their applications in natural populations | Angers B. 2020 | [90] |
| Understanding natural epigenetic variation | Richards CL. 2010 | [99] |
| Rapid Epigenetic Adaptation in Animals and Its Role in Invasiveness | Carneiro VC. 2020 | [114] |
| Epigenetic regulation in plant abiotic stress responses | Chang YN. 2020 | [133] |
| Epimutations Define a Fast-Ticking Molecular Clock in Plants | Yao N. 2021 | [73] |
| Epigenetic variation in animal populations: Sources, extent, phenotypic implications, and ecological and evolutionary relevance | Vogt G. 2021 | [70] |
| Epigenetic inheritance of DNA methylation changes in fish living in hydrogen sulfide-rich springs | Kelley JL. 2021 | [121] |
| Differential DNA Methylation in Somatic and Sperm cells of Hatchery versus Wild (Natural-Origin) Steelhead Trout Populations | Nilsson E. 2021 | [124] |
The current neo-Darwinian (i.e. Modern Synthesis) suggests that genetic variation is essential for evolution and drives phenotypic variation and adaptation. Interestingly, genetic mutations most often require an epigenetic precursor [11]. This includes the most predominant point mutation of C to T conversion being promoted by DNA methylation, or DNA methylation regulation of copy number variation, or histone modifications and DNA methylation altering translocation sites, or DNA methylation regulating transposable element activity [11]. The environmental induction of epigenetic transgenerational inheritance has been shown to increase the frequency of point mutations and copy number mutations generationally [11, 71]. Therefore, environmental epigenetics and epigenetic inheritance promote genetic variation that may facilitate adaptive phenotypic variation [71]. The frequency of epigenetic alterations is five orders of magnitude higher than genetic mutation frequency [72, 73]. As will be discussed, the ability of environmental epigenetics to be a primary driver of phenotypic and genetic variation needs to be integrated into a unified evolutionary biology theory.
Epigenetic Transgenerational Inheritance and Evolution
Environmentally induced epigenetic transgenerational inheritance was first described on a molecular level with a toxicant exposure in a rat model [74] and later with a stress exposure in a mouse model [75]. This is a non-genetic form of inheritance mediated through epigenetic alterations in the germline (sperm or egg) that transmit altered phenotypes to subsequent generations [46, 76]. The phenomenon has been extensively demonstrated in hundreds of laboratories in all organisms investigated from plants to humans. In plants, worms, and flies transgenerational inheritance has been transmitted hundreds of generations [46, 77]. Environmental exposures include industrial toxicants, nutrition, smoking, alcohol, and stress or trauma. The direct environmental exposure of an individual impacts the exposed individual and the germline within that individual that will generate the next generation, so this is referred to as a multigenerational exposure (i.e. intergenerational phenomenon) (Fig. 2). The first transgenerational generation not involving direct exposure is the grand-offspring F2 generation. For a gestating female, the F0 generation mother, F1 generation fetus, and germline within the fetus that will generate the F2 generation grand-offspring are all directly exposed, such that the first transgenerational generation is the F3 generation great grand-offspring (Fig. 2). The repeated demonstration of epigenetic transgenerational inheritance of altered phenotypes suggests that this molecular mechanism plays a significant role in ecology and medicine and should be integrated into evolutionary biology theory [4, 6, 11, 12, 16, 77–92]. Environmentally induced epigenetic transgenerational inheritance has also been observed in a number of field populations responding to natural selection [11, 88, 89, 92]. A number of these studies are summarized below and have been reviewed in regard to a role for epigenetics in evolution [1, 2, 4, 7, 8, 12, 16, 29, 33, 46, 74, 76, 77, 82, 93–101].
Figure 2:
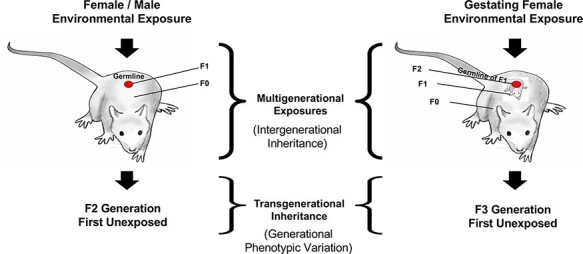
Environmentally induced transgenerational epigenetic inheritance: schematic of environmental exposure and affected generations for both gestating female and adult male or female. The multigenerational direct exposures are indicated in contrast to the transgenerational generation without direct exposure. Modified from Nilsson et al. [46]
This non-genetic form of inheritance is induced early in development to reprogram the epigenetics of the sperm or egg to allow transmission to the next generation. The next generation will have its embryonic stem cell epigenome and transcriptome altered, which will impact all the somatic cells derived from the stem cells epigenetics and transcriptomes [46]. Those cells sensitive to the epigenetic alterations will have an increased susceptibility to develop disease later in life, such that the generational physiology and phenotype of the individual will be modified. A large number of different transgenerational pathologies develop, and the toxicant exposures result in generational toxicology [46]. Since the impacts are transgenerationally inherited, they have the potential to impact evolution [11]. The origins of the transgenerational germline epigenetic alterations have been shown to be throughout gametogenesis from the primordial germ cells to the mature gametes [52]. Therefore, like genetic changes, epigenetic changes can have an important role in short-term microevolution [6] and contribute to macroevolutionary (i.e. at or above the level of species) processes, such as speciation and adaptive radiation [2, 4, 12, 79, 80] (Table 3).
Table 3:
Non-genetic (epigenetic) association with evolution references
| Reference title | ||
|---|---|---|
| The significance of responses of the genome to challenge | McClintock B. 1984 | [17] |
| Maternal Effects as Adaptations | Mousseau TA, 1998 | [31] |
| Epigenetic transgenerational actions of endocrine disruptors and male fertility | Anway MD, 2005 | [74] |
| Environmental epigenomics and disease susceptibility | Jirtle RL, 2007 | [76] |
| Chromatin structure and the inheritance of epigenetic information | Margueron R, 2010 | [51] |
| Epigenetic transgenerational actions of environmental factors in disease etiology | Skinner MK, 2010 | [27] |
| A unified approach to the evolutionary consequences of genetic and nongenetic inheritance | Day T, 2011 | [6] |
| Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans | Rechavi O, 2011 | [110] |
| Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans | Greer EL, 2011 | [111] |
| Transgenerational epigenetic inheritance in plants | Hauser MT, 2011 | [131] |
| Remembering the prolonged cold of winter | Song J, 2013 | [86] |
| Nongenetic inheritance and the evolution of costly female preference | Bonduriansky R, 2013 | [129] |
| Transgenerational developmental programming | Aiken CE, 2014 | [101] |
| Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice | Gapp K, 2014 | [75] |
| Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance | Yan W. 2014 | [44] |
| Does evolutionary theory need a rethink? | Laland K. 2014 | [4] |
| Transgenerational epigenetic inheritance in mammals: how good is the evidence? | van Otterdijk SD, 2016 | [135] |
| The evolutionary implications of epigenetic inheritance | Jablonka E. 2017 | [2] |
| Transgenerational epigenetics: Integrating soma to germline communication with gametic inheritance | Sharma A. 2017 | [66] |
| Principles of Transgenerational Small RNA Inheritance in Caenorhabditis elegans | Rechavi O, 2017 | [113] |
| Lamarck rises from his grave: parental environment-induced epigenetic inheritance in model organisms and humans | Wang Y, 2017 | [137] |
| Functions and mechanisms of epigenetic inheritance in animals | Skvortsova K, 2018 | [134] |
| Plant epigenetic mechanisms: role in abiotic stress and their generational heritability | Sudan J, Raina M, Singh R. 2018 | [103] |
| Quantitative epigenetics and evolution | Banta JA, 2018 | [69] |
| Environmentally Induced Epigenetic Transgenerational Inheritance of Disease | Nilsson E, 2018 | [46] |
| Protein-Based Inheritance: Epigenetics beyond the Chromosome | Harvey ZH, 2018 | [32] |
| Histone Methylation and Memory of Environmental Stress | Fabrizio P, 2019 | [112] |
| Maternal transmission of the epigenetic “memory of winter cold” in Arabidopsis | Luo X, 2020 | [109] |
| Molecular mechanisms of epigenetic inheritance: Possible evolutionary implications | Sarkies P. 2020 | [88] |
Examples of epigenetics and epigenetic transgenerational inheritance impacts on evolution (e.g. natural population) are provided below for natural populations and laboratory models (Table 4). Empirical tests of the potential role of environmental epigenetic mechanisms in environmental adaptation and evolution have been described [102]. A role for environmentally induced epigenetic variation and inheritance in plants has been observed [77, 97–100]. The high level of developmental plasticity in changing environments is proposed to be due to environmental epigenetics facilitating adaptation and evolution in plants [103, 104]. Plant models of reproduction lack sequestered germ cells [105], so have adaptive epigenetics and phenotypes. Specific examples include Taraxacum officinale [106, 107] and Arabidopsis [108, 109]. The Caenorhabditis elegans is a model worm with high epigenetic variation and epigenetic transgenerational inheritance [110–112]. Epigenetic inheritance of histone modifications and ncRNA can alter adaptive responses in C. elegans [113]. The house sparrow (Passer domesticus) demonstrates high levels of epigenetic variation facilitating rapid phenotypic change and adaptive evolution [102, 114]. The invasive house sparrow exhibits phenotypic and epigenetic variation in subpopulations in the Middle East [115] and Australia [116]. Clonal lineages of animals that are not reliant on genetic variation also have been used to investigate environmentally induced epigenetic variation. The asexual snail Potamopyrgus antipodarum is a widespread invasive species in North America. Alteration of environmental conditions (i.e. water flow) was found to associate with adaptive phenotypic variation that correlated with epigenetic alterations [117, 118]. Chrosomus eos-neogaeus is a hybrid clonal fish that inhabits lakes and intermittent stream environments that has epigenetic variation in the divergent environments [119, 120]. Another fish example is Poecilia, a fish that survives in fresh water and sulfur environments, having distinct generational epigenetic changes in the distinct environments [121]. Combined observations support a role for environmentally induced epigenetic variation and inheritance to promote adaptive phenotypic variation.
Table 4:
Examples of epigenetic transgenerational inheritance impacts on evolution
| Organism | Reference | |
|---|---|---|
| Plant | (Taraxacum officinale, Arabidopsis) | [77, 97–100, 103–109] |
| Worm | (Caenorhabditis elegans) | [110–113] |
| Bird | (House sparrow, Passer domesticus) | [102, 114–116] |
| Asexual snail | (Potamopyrgus antipodarum) | [117, 118] |
| Hybrid clonal fish | (Chrosomus eos-neogaeus) | [119, 120] |
| Fish | (Poeciliamexicana) | [121] |
| Bird | (Darwin finch, Geospiza fortis) | [122, 123] |
| Fish | (Steelhead trout, Oncorhynchus mykiss) | [124] |
| Alligator | (Alligatormississippiensis) | [125] |
| Mouse | (Mus musculus) | [46, 75] |
| Rat | (Rattus norvegicus) | [46, 74, 82, 93] |
Adaptive radiations also provide additional examples of epigenetically mediated evolutionary change. One of the first examples of this used five species of Darwin’s finches in the Galapagos islands to assess genetic and epigenetic relatedness [122]. The epigenetic variation observed statistically correlated with the phylogenic relatedness of the different species, in contrast with the genetic variation, which did not correlate, and was at a higher frequency than the genetic alterations [122]. Observations were extended in the Darwin’s finches with a comparison of epigenetic variation between two Darwin finch species having distinct urban and rural populations with phenotypic variation [123]. Finches in the urban environment with altered nutrition were found to have epigenetic variation, but no genetic variation [123]. Another example of human-mediated alterations in phenotypic variation and epigenetic variation used the steelhead trout (Oncorhynchus mykiss) [124]. The hatchery and wild populations of steelhead trout have significant phenotypic variation in growth, maturation rates, and subsequent fitness and survival. Epigenetic differences between the fish populations were dramatic in somatic and germ cells, with minimal genetic alterations, between the hatchery and wild populations [124]. A similar observation was made with the American alligator (Alligator mississippiensis) in Florida USA, where animals in a pristine uncontaminated environment had dramatic epigenetic alterations when compared to alligators in contaminated environments, associated with corresponding reproductive pathologies and phenotypic variation [125]. Observations demonstrate that invaders, founder populations, clonal lineages, and adaptive radiations involve environmental epigenetics and epigenetic inheritance for adaptive phenotypic variation and evolution [102] (Table 4).
Observations in a growing number of mammalian species also support a role for environmentally induced epigenetic transgenerational inheritance in adaptation and evolution [126] (Table 4). The first studies to investigate the role of environmental epigenetics and epigenetic transgenerational inheritance used ancestral toxicant or stress exposure types of experimental design [74, 75]. Darwin proposed that one of the critical determinants of evolution was sexual selection [1]. A study was designed to investigate the environmental impacts of a toxicant exposure on the epigenetic transgenerational inheritance of altered mate preference associated with sexual selection [82]. An F0 generation female gestating rat was exposed to the agricultural fungicide vinclozolin transiently during gonadal sex determination and then subsequent F3 generation animals (great grand-offspring) were obtained to assess mate preference behavior alterations and epigenetic alterations. A significant mate preference alteration was identified along with epigenetic alterations in the germline [82]. Transgenerational brain transcriptome changes associated with the mate preference behaviors were observed [93]. Therefore, environmentally induced epigenetic transgenerational inheritance of mate preference, known to be critical for evolution, was observed [27, 82, 93]. A number of reviews have suggested a role for epigenetics in both microevolution and macroevolution [6–8, 16, 34, 79, 80, 127–130], Table 3.
The current Modern Synthesis and EES theories support the role of genetics as being the primary molecular factor involved in adaptation and evolution. Genetic variation was thought to be the driver for phenotypic variation in the Modern Synthesis, while the EES suggests epigenetics can facilitate phenotypic variation as well. The above examples demonstrate that epigenetics and epigenetic inheritance are stable and could independently impact evolution alongside genetics. The environmentally induced epigenetic transgenerational inheritance of phenotypic variation needs to be considered an equally important molecular mechanism. The promotion of adaptive or maladaptive traits through epigenetic variation is orders of magnitudes more frequent than genetic induced adaptive or maladaptive traits. The distinction is that environment can readily promote the epigenetic variation that becomes inherited to facilitate the natural selection and evolutionary process. Since epigenetic alterations also facilitate formation of genetic mutations and genetic variation, indirectly environmental epigenetics can facilitate and drive genetic variation. These advances in our understanding of molecular biology, physiology and inheritance need to be incorporated into a more unified evolutionary theory.
Integration of Epigenetic Transgenerational Inheritance and Unified Theory of Evolution
The advances in epigenetics over the last three decades have demonstrated that epigenetics is equally important as genetics in the regulation of gene expression, phenotypic variation, and adaptation (Table 2). Epigenetics is the precursor for all genomic activity (i.e. gene expression) and genetic stability (e.g. transposable element mobilization or copy number variation). Therefore, all biological processes from evolution to disease etiology will require the incorporation of epigenetics and genetics. The evidence for a functional role of environmental epigenetics and epigenetic transgenerational inheritance in the phenotypic variation and adaptation in plants [131–133] and animals [46, 134–136] is now compelling. This new science demonstrates a role for the environment to directly promote phenotypic variation that is heritable. This provides significant support for the previously discarded ideas of “soft inheritance” (i.e. epigenetic inheritance) from the late 1800s and early 1900s [11]. This neo-Lamarckian concept provides a redemption for some of the ideas of Jean Baptiste Lamarck [21], who first described the inheritance of acquired characteristics [11, 137, 138] (Fig. 3). Since the neo-Darwinian theory (i.e. Modern Synthesis) did not include direct environmental impacts to promote phenotypic (i.e. non-genetic) variation, the integration of epigenetics, environmental epigenetics and epigenetic transgenerational inheritance need to be integrated in a Unified Theory of Evolution (Table 4 and Fig. 3). The EES supports this concept and promotes a role for non-genetic phenomenon such as epigenetics and epigenetic inheritance [13]. This is also supported by computational modeling of evolution [6, 139]. However, the direct actions of the environment to promote heritable phenotypic variation independent of DNA sequence mutations and genetics are not a major component of the EES. Therefore, there is a consensus that the current concepts need to be integrated and incorporated into our understanding of evolutionary biology [11, 13] (Table 3).
Figure 3:
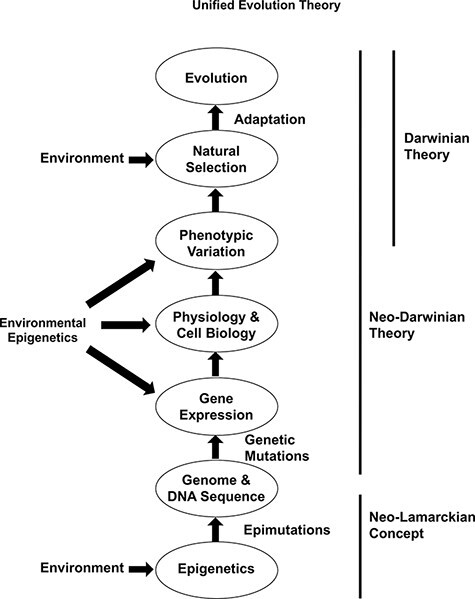
Schematic of the Unified Theory of Evolution. No dominance is suggested by the appearance of specific circles (e.g. epimutations versus genetics) such that all are equally important components. Modified from Skinner [11]
The integration of epigenetic transgenerational inheritance expands the role of epigenetics beyond the short-term impacts of gene expression to also promote phenotypic variation within a population permanently. Therefore, environmentally induced epigenetic transgenerational inheritance has macroevolution impacts equally as important as genetics and genetic mutations [11] (Table 3). Due to the molecular frequency of epigenetic variation being several orders of magnitude higher than genetic variation [73, 105], and being environmentally responsive, the impacts of epigenetic transgenerational inheritance and epigenetic variation on the evolutionary and adaptive trajectory of species are supported [2, 11]. Previous studies have documented epigenetic variation at the level of cellular epigenetics for tissue function and disease [140], and the level of the organism to correlate with phenotypic variation [141]. As with genetics, epigenetics is anticipated to produce adaptive and maladaptive phenotypes and effects. The increased frequency of epigenetic variation is anticipated to provide a spectrum of adaptive and maladaptive phenotypes. An example is the thrifty phenotype induced by caloric restriction during fetal development inducing epigenetic metabolism effects that allow the offspring to survive on fewer calories, but on a normal diet, develop obesity [142]. Therefore, both adaptive and maladaptive phenotypes are expected in an evolutionary setting. The postulates of natural selection are supported by the evidence of epigenetic transgenerational inheritance and phenotypic change. Therefore, the integration of epigenetics and genetics provides a more efficient molecular mechanism and theory than the current Modern Synthesis (i.e. neo-Darwinian) theory alone [11] (Fig. 3). In addition, this is supported by and expands the concepts of the EES [13]. The Unified Theory of environmental epigenetics facilitating (i) the neo-Lamarckian concept of environment directly impacting phenotype that is heritable and (ii) subsequently altering genetic variation and phenotypic variation through the neo-Darwinian theory, (iii) for adaptation and natural selection as per classic Darwinian theory, is proposed as the Unified Theory of Evolution (Fig. 3 and Table 5).
Table 5:
Evolution theory components
| Neo-Lamarckian Concept: |
| Environment directly alters phenotype generationally |
| Darwinian Evolution Theory: |
| Natural selection acts on phenotypic variation for adaptation |
| Neo-Darwinian Evolution Theory: |
| Genetic mutations promote phenotypic variation on which natural selection acts |
| Environmental Epigenetics: |
| Environmental epigenetic alterations promote phenotypic variation and facilitate genetic mutations to influence adaptation and natural selection |
| Unified Evolution Theory: |
| Environmental epigenetics and genetic mutations both promote heritable phenotypic variation on which natural selection acts |
Paradigm Shifts in Science
The vast majority of biological theories today are primarily based on “genetic determinism” in which genetic mutations are the basis for all phenomena from disease etiology to evolution. This genetic determinism paradigm has been in place for the past century and became ingrained in all aspects of the biological sciences. Following the development of modern genomics and sequencing of the human and other species genomes, the past 20 years has demonstrated that most biological phenomena cannot be explained with genetics alone. The crisis developing is that the frequency of genetic mutations is rare, such that correlations with function, pathology, and disease or phenotypes are not common. In the 1970s, Thomas Kuhn described the concept of paradigm shifts in science to help explain the historic development of the sciences and provide insights into the shifts required in rapidly developing science technology and theory [143]. The concept was that when the current paradigm cannot explain phenomena observed, a crisis develops that promotes new science development that allows a new scientific paradigm to develop. Due to the vested interest in the current paradigms, this generally takes a generation of scientists to develop [143]. The developments over the past several decades in the molecular epigenetic area provides a new science that can help explain the deficiencies and problems with genetic determinism. The integration of epigenetics and genetics provides a more complete and efficient molecular explanation of the processes in biology. We now know environmental epigenetics can dramatically influence genetic variation through the promotion of genetic mutations and alterations. Independent of genetics, environmental epigenetics can also promote phenotypic variation, since the epigenetic processes precede and directly regulate the transcriptomes required for the phenotypes observed. The role of epigenetic transgenerational inheritance as a form of non-genetic inheritance allows the transmission of phenotypes for generations. The integration of genetics and epigenetics provides a novel and more efficient molecular model of evolution, not considered within the Modern Synthesis or EES theories (Fig. 3 and Table 4). This does not detract or minimize the essential aspects of Darwinian theory nor advances of the Modern Synthesis, but simply provides new science not previously considered in evolution theory [11]. This Kuhnian paradigm shift in evolutionary biology theory is the integration of the classic established concepts with the new science to create a new paradigm of a more Unified Theory presented (Fig. 3). This is not restricted to evolutionary biology and is now needed in all areas of biology from disease etiology theory, cell and developmental biology theory, and environmental sciences.
Summary
The support for environmental epigenetics and epigenetic transgenerational inheritance in regulation of phenotypic variation and genetic variation to impact both microevolution and macroevolution is now compelling [6, 8, 34, 79–81, 127, 128] (Tables 2 and 3). This information has been reviewed and the need to integrate epigenetics and genetics in a unified theory of evolution discussed [6, 8, 16, 34, 79–81, 127, 128]. The integration and Unified Evolution Theory proposed (Fig. 3) has a number of parameters to consider: (i) environmental epigenetics provides a molecular mechanism for Lamarck’s concept that environment can directly alter phenotype in a heritable manner; (ii) environmental exposures at critical developmental windows promote the epigenetic transgenerational inheritance of germline (e.g. sperm and egg) epimutations that alter phenotypic variation generationally; (iii) direct environmental exposures of developing somatic tissue can alter somatic epigenomes and phenotype in the individual exposed, but this will not be heritable and the phenotypes will often be distinct from transgenerational phenotypes; (iv) phenotypic variation is derived from a combination of integrated genetic and epigenetic processes on which natural selection acts; and (v) environment has a critical role in natural selection, as well as in the induction of heritable adaptive phenotypic variation (i.e. epigenetic transgenerational inheritance). Therefore, the environment has a more direct role, independent of genetics, in driving phenotypic variation, adaptation, and evolutionary biology.
As shown in Fig. 3 and Table 5, the integration of epigenetics and genetics contribute to a Unified Theory of Evolution that explains environmental impacts, phenotypic variation, genetic variation, and adaptation that natural selection acts on. All classic and previously established elements of evolution theory are included, and this unified theory brings in the new advanced science of environmental epigenetics. This shift is a new paradigm in evolution, as defined by classic Kuhn paradigm shifts in science [143]. This does not detract from the critical aspects of Darwinian theory, Modern Synthesis, or EES, but simply integrates the new advanced science of epigenetics and epigenetic transgenerational inheritance. The current review expands this proposed concept [11] and provides a significant amount of supporting literature (Tables 2 and 3) and experimental models to support the role of environmentally induced epigenetic transgenerational inheritance in evolution.
Acknowledgements
We thank the critical insights and assistance of Dr Jennifer L.M. Thorson, as well as Drs Millissia Ben Maamar and Daniel Beck for critically reviewing the manuscript. We acknowledge Ms Amanda Quilty for editing and Ms Heather Johnson for assistance in preparation of the manuscript.
Contributor Information
Michael K Skinner, Center for Reproductive Biology, School of Biological Sciences, Washington State University, Pullman, WA 99164-4236, USA.
Eric E Nilsson, Center for Reproductive Biology, School of Biological Sciences, Washington State University, Pullman, WA 99164-4236, USA.
Funding
This study was supported by the John Templeton Foundation (50183 and 61174) (https://templeton.org/) grants to M.K.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
None declared
Conflict of interest statement
References
- 1. Darwin C. On the Origin of Species. London: John Murray, 1859, 488. [Google Scholar]
- 2. Jablonka E. The evolutionary implications of epigenetic inheritance. Interface Focus 2017;7:20160135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huxley J. Evolution: The Modern Synthesis. London: George Allen & Unwin Ltd, 1942, 645. [Google Scholar]
- 4. Laland K, Uller T, Feldman M. et al. Does evolutionary theory need a rethink? Nature 2014;514:161–4. [DOI] [PubMed] [Google Scholar]
- 5. Olson-Manning CF, Wagner MR, Mitchell-Olds T. Adaptive evolution: evaluating empirical support for theoretical predictions. Nat Rev Genet 2012;13:867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Day T, Bonduriansky R. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am Nat 2011;178:E18–36. [DOI] [PubMed] [Google Scholar]
- 7. Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol 2009;84:131–76. [DOI] [PubMed] [Google Scholar]
- 8. Kuzawa CW, Thayer ZM. Timescales of human adaptation: the role of epigenetic processes. Epigenomics 2011;3:221–34. [DOI] [PubMed] [Google Scholar]
- 9. Nei M, Nozawa M. Roles of mutation and selection in speciation: from Hugo de Vries to the modern genomic era. Genome Biol Evol 2011;3:812–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burger R, Willensdorfer M, Nowak MA. Why are phenotypic mutation rates much higher than genotypic mutation rates? Genetics 2006;172:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skinner MK. Environmental epigenetics and a unified theory of the molecular aspects of evolution: a neo-Lamarckian concept that facilitates neo-Darwinian evolution. Genome Biol Evol 2015;7:1296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pigliucci M. Do we need an extended evolutionary synthesis? Evolution 2007;61:2743–9. [DOI] [PubMed] [Google Scholar]
- 13. Laland KN, Uller T, Feldman MW. et al. The extended evolutionary synthesis: its structure, assumptions and predictions. Proc Biol Sci Royal Soc 2015;282:20151019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kratz CP, Edelman DC, Wang Y. et al. Genetic and epigenetic analysis of monozygotic twins discordant for testicular cancer. Int J Mol Epidemiol Genet 2014;5:135–9. [PMC free article] [PubMed] [Google Scholar]
- 15. Zwijnenburg PJ, Meijers-Heijboer H, Boomsma DI. Identical but not the same: the value of discordant monozygotic twins in genetic research. Am J Med Genet B Neuropsychiatr Genet 2010;153B:1134–49. [DOI] [PubMed] [Google Scholar]
- 16. Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol 2014;398:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McClintock B. The significance of responses of the genome to challenge. Science 1984;226:792–801. [DOI] [PubMed] [Google Scholar]
- 18. Janecka J, Chowdhary B, Murphy W. Exploring the correlations between sequence evolution rate and phenotypic divergence across the Mammalian tree provides insights into adaptive evolution. J Biosci 2012;37:897–909. [DOI] [PubMed] [Google Scholar]
- 19. Baldwin J. A new factor in evolution. Am Nat 1896;30:441–51. [Google Scholar]
- 20. Calabi L. On Darwin’s ‘metaphysical notebooks’. I: teleology and the project of a theory. Riv Biol 2001;94:123–59. [PubMed] [Google Scholar]
- 21. Lamarck J. Recherches sur l’organisation des corps vivans. Paris: Chez L’auteur, Maillard, 1802. [Google Scholar]
- 22. Crispo E. The Baldwin effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution 2007;61:2469–79. [DOI] [PubMed] [Google Scholar]
- 23. Vargas AO, Krabichler Q, Guerrero-Bosagna C. An epigenetic perspective on the midwife toad experiments of Paul Kammerer (1880–1926). J Exp Zool B Mol Dev Evol 2017;328:179–92. [DOI] [PubMed] [Google Scholar]
- 24. Waddington CH. Organisers and Genes. Cambridge: Cambridge University Press, 1940. [Google Scholar]
- 25. Mayr E. Prologue: some thoughts on the history of the evolutionary synthesis. In: Mayr E, Provine WB (eds.), The Evolutionary Synthesis: Perspectives on the Unification of Biology. Cambridge, MA: Harvard University Press, 1980, 1–48. [Google Scholar]
- 26. Schork NJ, Murray SS, Frazer KA. et al. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev 2009;19:212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab 2010;21:214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paenke I, Sendhoff B, Kawecki TJ. Influence of plasticity and learning on evolution under directional selection. Am Nat 2007;170:E47–58. [DOI] [PubMed] [Google Scholar]
- 29. Husnik F, McCutcheon JP. Functional horizontal gene transfer from bacteria to eukaryotes. Nat Rev Microbiol 2018;16:67–79. [DOI] [PubMed] [Google Scholar]
- 30. Falconer DS. Introduction to Quantitative Genetics. Suffolk, Great Britain: Benjamin-Cummings Publishing Company, 1996, 464. [Google Scholar]
- 31. Mousseau TA, Fox CW (eds). Maternal Effects as Adaptations. Oxford, United Kingdom: Oxford University Press, 1998, 400. [Google Scholar]
- 32. Harvey ZH, Chen Y, Jarosz DF. Protein-based inheritance: epigenetics beyond the chromosome. Mol Cell 2018;69:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harvey ZH, Chakravarty AK, Futia RA. et al. A prion epigenetic switch establishes an active chromatin state. Cell 2020;180:928–40 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Badyaev AV. Epigenetic resolution of the ‘curse of complexity’ in adaptive evolution of complex traits. J Physiol 2014;592:2251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci 2002;981:61–81. [PubMed] [Google Scholar]
- 36. Waddington CH. Principles of Embryology. London: George Allen & Unwin Ltd., 1956. [Google Scholar]
- 37. Waddington CH. The genetic assimilation of the bithorax phenotype. Evolution 1956;10:1–13. [Google Scholar]
- 38. Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 2011;6:838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science 1975;187:226–32. [PubMed] [Google Scholar]
- 40. Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 1975;14:9–25. [DOI] [PubMed] [Google Scholar]
- 41. Dossin F, Heard E. The molecular and nuclear dynamics of X-chromosome inactivation. Cold Spring Harb Perspect Biol 2021:a040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. SanMiguel JM, Bartolomei MS. DNA methylation dynamics of genomic imprinting in mouse development. Biol Reprod 2018;99:252–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kan RL, Chen J, Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet 2021;S0168–9525:00170–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan W. Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Mol Cell Endocrinol 2014;398:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Argelaguet R, Clark SJ, Mohammed H. et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 2019;576:487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nilsson E, Sadler-Riggleman I, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet 2018;4:1–13, dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singer J, Roberts-Ems J, Riggs AD. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science 1979;203:1019–21. [DOI] [PubMed] [Google Scholar]
- 48. Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta 2014;1839:627–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bartova E, Krejci J, Harnicarova A. et al. Histone modifications and nuclear architecture: a review. J Histochem Cytochem 2008;56:711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taylor BC, Young NL. Combinations of histone post-translational modifications. Biochem J 2021;478:511–32. [DOI] [PubMed] [Google Scholar]
- 51. Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 2010;11:285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ben Maamar M, Nilsson EE, Skinner MK. Epigenetic transgenerational inheritance, gametogenesis and germline development . Biol Reprod 2021;105:570–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wei JW, Huang K, Yang C. et al. Non-coding RNAs as regulators in epigenetics. Oncol Rep 2017;37:3–9. [DOI] [PubMed] [Google Scholar]
- 54. Huang B, Jiang C, Zhang R. Epigenetics: the language of the cell? Epigenomics 2014;6:73–88. [DOI] [PubMed] [Google Scholar]
- 55. Kornfeld JW, Bruning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet 2014;5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 2015;29:1343–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fu Y, Dominissini D, Rechavi G. et al. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014;15:293–306. [DOI] [PubMed] [Google Scholar]
- 58. Urquiaga MCO, Thiebaut F, Hemerly AS. et al. From trash to luxury: the potential role of plant LncRNA in DNA methylation during abiotic stress. Front Plant Sci 2020;11:603246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lobo J, Henrique R, Jeronimo C. The role of DNA/histone modifying enzymes and chromatin remodeling complexes in testicular germ cell tumors. Cancers (Basel) 2018;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beck D, Ben Maamar M, Skinner MK. Integration of sperm ncRNA-directed DNA methylation and DNA methylation-directed histone retention in epigenetic transgenerational inheritance. Epigenetics Chromatin 2021;14:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tan Q, Christiansen L, von Bornemann Hjelmborg J. et al. Twin methodology in epigenetic studies. J Exp Biol 2015;218:134–9. [DOI] [PubMed] [Google Scholar]
- 62. Waddington CH. Epigenetics and evolution. Symp Soc Exp Biol 1953;7:186–99. [Google Scholar]
- 63. Jablonka E. Genes as followers in evolution – a post-synthesis synthesis? Biol Philos 2006;21:143–54. [Google Scholar]
- 64. Quina AS, Buschbeck M, Di Croce L. Chromatin structure and epigenetics. Biochem Pharmacol 2006;72:1563–9. [DOI] [PubMed] [Google Scholar]
- 65. Sibbritt T, Patel HR, Preiss T. Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA 2013;4:397–422. [DOI] [PubMed] [Google Scholar]
- 66. Sharma A. Transgenerational epigenetics: integrating soma to germline communication with gametic inheritance. Mech Ageing Dev 2017;163:15–22. [DOI] [PubMed] [Google Scholar]
- 67. Skinner MK, Guerrero-Bosagna C, Haque MM. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 2015;10:762–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. West-Eberhard MJ. Developmental Plasticity and Evolution. New York: Oxford University Press, 2003, 816. [Google Scholar]
- 69. Banta JA, Richards CL. Quantitative epigenetics and evolution. Heredity 2018;121:210–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vogt G. Epigenetic variation in animal populations: sources, extent, phenotypic implications, and ecological and evolutionary relevance. J Biosci 2021;46:24. [PubMed] [Google Scholar]
- 71. McCarrey JR, Lehle JD, Raju SS. et al. A novel aspect of epigenetic transgenerational inheritance promoting genome instability. PLoS One 2016;11:1–15, e0168038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jiang C, Mithani A, Belfield EJ. et al. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res 2014;24:1821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yao N, Schmitz RJ, Johannes F. Epimutations define a fast-ticking molecular clock in plants. Trends Genet 2021;37:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Anway MD, Cupp AS, Uzumcu M. et al. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005;308:1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gapp K, Jawaid A, Sarkies P. et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 2014;17:667–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 1999;401:157–61. [DOI] [PubMed] [Google Scholar]
- 78. Skinner MK. Environmental stress and epigenetic transgenerational inheritance. BMC Med 2014;12:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Flatscher R, Frajman B, Schonswetter P. et al. Environmental heterogeneity and phenotypic divergence: can heritable epigenetic variation aid speciation? Genet Res Int 2012;2012:698421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rebollo R, Horard B, Hubert B. et al. Jumping genes and epigenetics: towards new species. Gene 2010;454:1–7. [DOI] [PubMed] [Google Scholar]
- 81. Jablonka E, Lamb MJ. Evolution in Four Dimensions, revised edn. Cambridge, MA: MIT Press, 2014. [Google Scholar]
- 82. Crews D, Gore AC, Hsu TS. et al. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci U S A 2007;104:5942–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Seisenberger S, Peat JR, Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr Opin Cell Biol 2013;25:281–8. [DOI] [PubMed] [Google Scholar]
- 84. Burdge GC, Hoile SP, Uller T. et al. Progressive, transgenerational changes in offspring phenotype and epigenotype following nutritional transition. PLoS One 2011;6:e28282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pembrey ME, Bygren LO, Kaati G. et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 2006;14:159–66. [DOI] [PubMed] [Google Scholar]
- 86. Song J, Irwin J, Dean C. Remembering the prolonged cold of winter. Curr Biol 2013;23:R807–11. [DOI] [PubMed] [Google Scholar]
- 87. Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol 2008;25:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sarkies P. Molecular mechanisms of epigenetic inheritance: possible evolutionary implications. Semin Cell Dev Biol 2020;97:106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vogt G. Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences. J Biosci 2015;40:159–204. [DOI] [PubMed] [Google Scholar]
- 90. Angers B, Perez M, Menicucci T. et al. Sources of epigenetic variation and their applications in natural populations. Evol Appl 2020;13:1262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Herman JJ, Spencer HG, Donohue K. et al. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 2014;68:632–43. [DOI] [PubMed] [Google Scholar]
- 92. Hu J, Barrett RDH. Epigenetics in natural animal populations. J Evol Biol 2017;30:1612–32. [DOI] [PubMed] [Google Scholar]
- 93. Skinner MK, Anway SMI, Gore AC. et al. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoSOne 2008;3:1–11, e3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Darwin C. The Varriation of Animals and Plants under Domestication. London: John Murray, 1868. [Google Scholar]
- 95. Jenkins F. The origins of species. North Br Rev 1867;46:277–318. [Google Scholar]
- 96. Guerrero-Bosagna C, Settles M, Lucker B. et al. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 2010;5:1–17, e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hirsch S, Baumberger R, Grossniklaus U. Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harb Symp Quant Biol 2012;77:97–104. [DOI] [PubMed] [Google Scholar]
- 98. Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecol Lett 2008;11:106–15. [DOI] [PubMed] [Google Scholar]
- 99. Richards CL, Bossdorf O, Verhoeven KJ. Understanding natural epigenetic variation. New Phytol 2010;187:562–4. [DOI] [PubMed] [Google Scholar]
- 100. Becker C, Weigel D. Epigenetic variation: origin and transgenerational inheritance. Curr Opin Plant Biol 2012;15:562–7. [DOI] [PubMed] [Google Scholar]
- 101. Aiken CE, Ozanne SE. Transgenerational developmental programming. Hum Reprod Update 2014;20:63–75. [DOI] [PubMed] [Google Scholar]
- 102. Vogt G. Facilitation of environmental adaptation and evolution by epigenetic phenotype variation: insights from clonal, invasive, polyploid, and domesticated animals. Environ Epigenet 2017;3:dvx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sudan J, Raina M, Singh R. Plant epigenetic mechanisms: role in abiotic stress and their generational heritability. 3 Biotech 2018;8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Miryeganeh M, Saze H. Epigenetic inheritance and plant evolution. Popul Ecol 2019;62:17–27. [Google Scholar]
- 105. Quadrana L, Colot V. Plant transgenerational epigenetics. Annu Rev Genet 2016;50:467–91. [DOI] [PubMed] [Google Scholar]
- 106. Wilschut RA, Oplaat C, Snoek LB. et al. Natural epigenetic variation contributes to heritable flowering divergence in a widespread asexual dandelion lineage. Mol Ecol 2016;25:1759–68. [DOI] [PubMed] [Google Scholar]
- 107. Ferreira de Carvalho J, Oplaat C, Pappas N. et al. Heritable gene expression differences between apomictic clone members in Taraxacum officinale: insights into early stages of evolutionary divergence in asexual plants. BMC Genomics 2016;17:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schmid MW, Heichinger C, Coman Schmid D. et al. Contribution of epigenetic variation to adaptation in Arabidopsis. Nat Commun 2018;9:4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Luo X, Ou Y, Li R. et al. Maternal transmission of the epigenetic ‘memory of winter cold’ in Arabidopsis. Nat Plants 2020;6:1211–8. [DOI] [PubMed] [Google Scholar]
- 110. Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 2011;147:1248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Greer EL, Maures TJ, Ucar D. et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011;479:365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Fabrizio P, Garvis S, Palladino F. Histone methylation and memory of environmental stress. Cells 2019;8:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Rechavi O, Lev I. Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr Biol 2017;27:R720–30. [DOI] [PubMed] [Google Scholar]
- 114. Carneiro VC, Lyko F. Rapid epigenetic adaptation in animals and its role in invasiveness. Integr Comp Biol 2020;60:267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Riyahi S, Vilatersana R, Schrey AW. et al. Natural epigenetic variation within and among six subspecies of the house sparrow, Passer domesticus. J Exp Biol 2017;220:4016–23. [DOI] [PubMed] [Google Scholar]
- 116. Sheldon EL, Schrey A, Andrew SC. et al. Epigenetic and genetic variation among three separate introductions of the house sparrow (Passer domesticus) into Australia. R Soc Open Sci 2018;5:172185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Thorson JLM, Smithson M, Beck D. et al. Epigenetics and adaptive phenotypic variation between habitats in an asexual snail. Sci Rep 2017;7:14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Thorson JLM, Smithson M, Sadler-Riggleman I. et al. Regional epigenetic variation in asexual snail populations among urban and rural lakes. Environ Epigenet 2019;5:dvz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Massicotte R, Angers B. General-purpose genotype or how epigenetics extend the flexibility of a genotype. Genet Res Int 2012;2012:317175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Leung C, Breton S, Angers B. Facing environmental predictability with different sources of epigenetic variation. Ecol Evol 2016;6:5234–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kelley JL, Tobler M, Beck D. et al. Epigenetic inheritance of DNA methylation changes in fish living in hydrogen sulfide-rich springs. Proc Natl Acad Sci U S A 2021;118:e2014929118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Skinner MK, Guerrero-Bosagna C, Haque MM. et al. Epigenetics and the evolution of Darwin’s finches. Genome Biol Evol 2014;6:1972–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. McNew SM, Beck D, Sadler-Riggleman I. et al. Epigenetic variation between urban and rural populations of Darwin’s finches. BMC Evol Biol 2017;17:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Nilsson E, Sadler-Riggleman I, Beck D. et al. Differential DNA methylation in somatic and sperm cells of hatchery versus wild (natural-origin) steelhead trout populations. Environ Epigenet 2021;7:1–17, dvab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Guillette LJ Jr., Parrott BB, Nilsson E. et al. Epigenetic programming alterations in alligators from environmentally contaminated lakes. Gen Comp Endocrinol 2016;238:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Legoff L, D’Cruz SC, Tevosian S. et al. Transgenerational inheritance of environmentally induced epigenetic alterations during mammalian development. Cells 2019;8:1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jaeger J, Monk N. Bioattractors: dynamical systems theory and the evolution of regulatory processes. J Physiol 2014;592:2267–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Klironomos FD, Berg J, Collins S. How epigenetic mutations can affect genetic evolution: model and mechanism. BioEssays 2013;35:571–8. [DOI] [PubMed] [Google Scholar]
- 129. Bonduriansky R, Day T. Nongenetic inheritance and the evolution of costly female preference. J Evol Biol 2013;26:76–87. [DOI] [PubMed] [Google Scholar]
- 130. Zeh JA, Zeh DW. Maternal inheritance, epigenetics and the evolution of polyandry. Genetica 2008;134:45–54. [DOI] [PubMed] [Google Scholar]
- 131. Hauser MT, Aufsatz W, Jonak C. et al. Transgenerational epigenetic inheritance in plants. Biochim Biophys Acta 2011;1809:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lamke J, Baurle I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol 2017;18:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chang YN, Zhu C, Jiang J. et al. Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol 2020;62:563–80. [DOI] [PubMed] [Google Scholar]
- 134. Skvortsova K, Iovino N, Bogdanovic O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol 2018;19:774–90. [DOI] [PubMed] [Google Scholar]
- 135. van Otterdijk SD, Michels KB. Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J 2016;30:2457–65. [DOI] [PubMed] [Google Scholar]
- 136. Xu Q, Xie W. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol 2018;28:237–53. [DOI] [PubMed] [Google Scholar]
- 137. Wang Y, Liu H, Sun Z. Lamarck rises from his grave: parental environment-induced epigenetic inheritance in model organisms and humans. Biol Rev Camb Philos Soc 2017;92:2084–111. [DOI] [PubMed] [Google Scholar]
- 138. Nilsson EE, Ben Maamar M, Skinner MK. Environmentally induced epigenetic transgenerational inheritance and the Weismann barrier: the dawn of neo-Lamarckian theory. J Dev Biol 2020;8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bonduriansky R, Crean AJ, Day T. The implications of nongenetic inheritance for evolution in changing environments. Evol Appl 2012;5:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chatterjee A, Stockwell PA, Rodger EJ. et al. Genome-wide DNA methylation map of human neutrophils reveals widespread inter-individual epigenetic variation. Sci Rep 2015;5:17328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Heyn H, Moran S, Hernando-Herraez I. et al. DNA methylation contributes to natural human variation. Genome Res 2013;23:1363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Demetriou CA, van Veldhoven K, Relton C. et al. Biological embedding of early-life exposures and disease risk in humans: a role for DNA methylation. Eur J Clin Invest 2015;45:303–32. [DOI] [PubMed] [Google Scholar]
- 143. Kuhn TS. The Structure of Scientific Revolutions. Chicago, IL, USA: University of Chicago Press, 1962. [Google Scholar]


