ABSTRACT
Thermoacidophilic archaea belonging to the order Sulfolobales thrive in extreme biotopes, such as sulfuric hot springs and ore deposits. These microorganisms have been model systems for understanding life in extreme environments, as well as for probing the evolution of both molecular genetic processes and central metabolic pathways. Thermoacidophiles, such as the Sulfolobales, use typical microbial responses to persist in hot acid (e.g. motility, stress response, biofilm formation), albeit with some unusual twists. They also exhibit unique physiological features, including iron and sulfur chemolithoautotrophy, that differentiate them from much of the microbial world. Although first discovered >50 years ago, it was not until recently that genome sequence data and facile genetic tools have been developed for species in the Sulfolobales. These advances have not only opened up ways to further probe novel features of these microbes but also paved the way for their potential biotechnological applications. Discussed here are the nuances of the thermoacidophilic lifestyle of the Sulfolobales, including their evolutionary placement, cell biology, survival strategies, genetic tools, metabolic processes and physiological attributes together with how these characteristics make thermoacidophiles ideal platforms for specialized industrial processes.
Keywords: Archaea, Thermoacidophiles, Sulfolobales
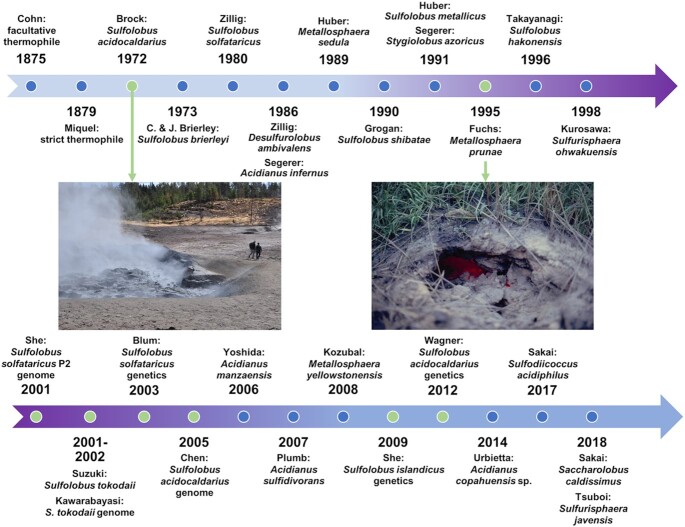
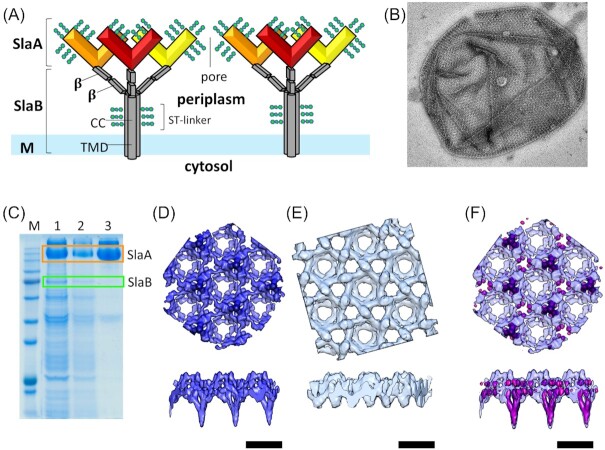
The natural habitat and biological features of the Sulfolobales.
INTRODUCTION
Thermoacidophiles are microorganisms that have developed mechanisms to successfully persist in unusually hot, acidic environments, with optimal conditions of pH ≤4 and temperature ≥55°C. In fact, thermoacidophiles have been isolated from some of the most inhospitable environments on earth, such as acidic hot springs and volcanic solfataras. In 1972, Thomas Brock isolated the thermoacidophile Sulfolobus acidocaldarius from a sulfur hot spring in Yellowstone National Park (left image, Fig. 1) and designated the genus Sulfolobus (Brock et al. 1972). The natural habitat of this microbe, a member of the Crenarchaeota, was Locomotive Spring, an extremely hot acidic environment with a pH of 2.4 and temperature of 83°C. Likewise, in 1980, Wolfram Zillig described Sulfolobus solfataricus (renamed Saccharolobus solfataricus) isolated from a volcanic hot spring in Italy (Zillig et al. 1980) and Desulfurolobus ambivalens (renamed Acidianus ambivalens) from a solfatara in Iceland in 1986 (Zillig et al. 1986). Zillig also discovered the first Japanese isolate belonging to this group in 1990—Sulfolobus shibatae (renamed Saccharolobus shibatae) (Grogan, Palm and Zillig 1990). Beyond these discoveries, Zillig also isolated the first thermoacidophile virus (Martin et al. 1984) (see the section 'Viruses and CRISPR systems of thermoacidophiles') and was the first to describe the eukaryotic-like archaeal RNA polymerase from S. acidocaldarius (Zillig, Stetter and Janekovic 1979) (see the section 'Genetic mechanisms'). In 1986, Karl Stetter established the genus Acidianus with the isolation of Acidianus infernus from a solfatara crater in Italy, which consequently led to the renaming of Sulfolobus brierleyi as Acidianus brierleyi (Segerer et al. 1986). Stetter also established the genus Metallosphaera with the isolation of Metallosphaera sedula in 1989 from a solfataric field in Italy (Huber et al. 1989).
Figure 1.
Timeline of thermoacidophile isolations and major events. Timeline contains the organism's name at the time of the associated event. The following are the current classifications: Sulfolobus brierleyi (f. Acidianus brierleyi), Saccharolobus solfataricus (f. Sulfolobus solfataricus), Acidianus ambivalens (f. Desulfurolobus ambivalens), Saccharolobus shibatae (f. Sulfolobus shibatae) Sulfuracidifex metallicus (f. Sulfolobus metallicus), Metallosphaera hakonensis (f. Sulfolobus hakonensis), Sulfurisphaera tokodaii (f. Sulfolobus tokodaii) and Saccharolobus islandicus (f. Sulfolobus islandicus).
Thermoacidophiles not only thrive in thermal acidic biotopes but also encounter other biologically deleterious conditions, such as oxidative stress caused by high levels of metals in mining environments. For instance, Metallosphaera prunae was isolated from a uranium mine in Germany (Fuchs et al. 1995) and uses an interesting stress response mechanism to withstand high levels of soluble uranium (see the section 'Extreme thermoacidophily and stress response'). Figure 1 (right) shows the features of the isolation site of M. prunae. In addition to Sa. shibatae, several other thermoacidophiles have been isolated from hot springs in Japan, such as Sulfurisphaera ohwakuensis in 1988 (Kurosawa et al. 1998), Sulfolobus hakonensis (renamed Metallosphaera hakonensis) in 1996 (Takayanagi et al. 1996) and Sulfolobus tokodaii (renamed Sulfurisphaera tokodaii) in 2002 (Suzuki et al. 2002), to name a few. It has become clear that thermoacidophiles are globally distributed in hot, acidic features; for example, recent isolates have come from the Copahue volcanic region in Argentina—Acidianus copahuensis in 2014 (Urbieta et al. 2014), and Indonesian hot springs—Sulfurisphaera javensis in 2018 (Tsuboi et al. 2018). Recently, Saccharolobus caldissimus was isolated from an acidic Japanese hot spring, establishing the Saccharolobus genus which, as mentioned above, led to the renaming of both Sulfolobus solfataricus and Sulfolobus shibatae to Saccharolobus solfataricus and Saccharolobus shibatae, respectively (Sakai and Kurosawa 2017). Figure 1 illustrates the timeline of these thermoacidophile isolations. Many thermoacidophiles have leveraged the chemistry of metal and sulfur deposits for bioenergetic benefit through chemolithotrophy (see the section 'Metabolism'). As such, chemolithotrophic metabolism in hot acid can be exploited for biomining applications (see the section 'Potential and current uses of thermoacidophiles in biotechnological applications').
The study of thermoacidophiles was originally restricted to observational microbiology focused on phenotypic characteristics, such as cell morphology and growth physiology. However, following the sequencing of the Sa. solfataricus genome in 2001 (She et al. 2001), several other Sulfolobales genomes were reported, including S. acidocaldarius in 2005 (Chen et al. 2005) (Fig. 1). Genome sequences opened up prospects for transcriptomics (Auernik et al. 2008; Ortmann et al. 2008; Koerdt et al. 2011; Kozubal et al. 2011; Maezato et al. 2012; Ulas et al. 2012; Kouril et al. 2013b; Wolf et al. 2016), proteomics (Ellen et al. 2009; Koerdt et al. 2011), metabolomics and systems biology (Ulas et al. 2012; Kouril et al. 2013b; Wolf et al. 2016), and metagenomics (Inskeep et al. 2013; Campbell et al. 2017) with these archaea, offering further insights into life in hot acid. The development of genetic systems for thermoacidophiles was challenging, given their unique characteristics and practical considerations related to their growth conditions. In 2003, soon after the availability of its genomic sequence, Paul Blum generated the first Sulfolobales mutant in Sa. solfataricus based on lactose autotrophy (Worthington et al. 2003). Later in 2009, a genetic system was developed for Sulfolobus islandicus (renamed Saccharolobus islandicus), based on uracil auxotrophy and the ability to generate uracil through pyrEF as a selectable marker (She et al. 2009). Similarly, in 2012, Wagner et al. developed a genetic system based on a uracil auxotrophic parent strain and 5-FOA toxicity that is widely used today (Wagner et al. 2012) (see the section 'Potential and current uses of thermoacidophiles in biotechnological applications'). Genetic engineering capabilities for thermoacidophiles have expanded over the past decade that have not only supported fundamental microbiological studies but also fueled prospects for biotechnological processes.
While there are moderately thermoacidophilic bacteria (Norris et al. 1996; Goto et al. 2002; Johnson, Okibe and Roberto 2003), most thermoacidophiles are archaea. However, life in thermal, acidic environments is not limited to the order Sulfolobales. There are thermoacidophilic Euryarchaeota belonging to the order Thermoplasmatales. Species in the genus Picrophilus, such as Picrophilus oshimae and Picrophilus torridus from solfataras in Japan (Schleper et al. 1995), have an optimum growth temperature of 60°C and a pH optimum near 0. Thermoplasma acidophilum, isolated from a coal refuse pile, grows optimally at 59°C and pH of 1–2 (Darland et al. 1970). Here, the focus will be on thermoacidophiles from the order Sulfolobales and an examination of what is currently known about their diversity, growth physiology, cell biology and biotechnological prospects.
THE DIVERSITY OF THERMOACIDOPHILIC LIFE
Thermoacidophilic biotopes are ubiquitously distributed across both terrestrial and marine environments, closely associated with volcanic outflows or calderas resulting from tectonic activity. In terrestrial realms, these environments are often isolated features, presenting as steam-saturated/superheated discharges (fumaroles) in the form of geysers, solfatara and pools, and on occasion mixing with soils to form mineral-heavy mud pots. In marine environments, these vents are distinguished by their rapid mixing with dramatically cooler, saline waters (Kelley, Baross and Delaney 2002), resulting in sharp gradients of temperature, pH, oxygen and solutes, and abrupt dislocated niches (Reysenbach et al. 2000). In both environments, water chemistry is shaped by transformation of sulfur species from highly reduced metal sulfides and hydrogen sulfide to highly oxidized sulfate, with concomitant production of protons (i.e. acid) (Nordstrom, McCleskey and Ball 2009). Despite the incredibly exogenic nature of reduced inorganic species, their abiotic transformation at elevated temperatures and low pH is minimal (Chen and Morris 1972), pointing to the importance of sulfur biooxidizers in constructing and occupying this extremophilic niche (Odling-Smee, Laland and Feldman 1996).
While sulfur oxidation is a potential bioenergetic source in these environments, strategies to handle thermal stress, acid stress, high levels of aqueous heavy metals and minimal organic carbon availability must be employed (see the section 'Extreme thermoacidophily and stress response'). These biotopes are dominated by archaeal chemolithoautotrophs (Inskeep et al. 2013; Ward et al. 2017) that have been intrinsically tailored by evolution to inhabit and thrive in these highly selective niches (Valentine 2007; Colman et al. 2018). In contrast, many bacterial and eukaryotic organisms in thermal, acidic biotopes are limited to acid- or temperature-tolerant microorganisms, as opposed to obligate/sustained thermoacidophily (pH < 3.5; T > 65°C).
Diversity of eukaryotic and bacterial thermoacidophiles
Previous efforts have identified the limitations of organisms at the cusp of thermoacidophily. Specifically, in eukaryotes, it appears that the boundary stems from a temperature limitation. As far back as the 1970s, exhaustive sampling and culturing have demonstrated the inability to cultivate eukaryotes (specifically, fungi and algae) from geothermal features in excess of 60°C, despite growth at slightly lower temperatures (Tansey and Brock 1972). Further work showed that algae are limited to ∼60°C (Boyd et al. 2012), and protists to below 70°C (Brown and Wolfe 2006). For unicellular organisms that inhabit more thermophilic locales, hydrogen sulfide levels can be inhibitory. For many more complex organisms that depend upon gaseous compounds for cellular processes, growth is limited by the solubility of many gases (oxygen, carbon dioxide, etc.), which diminishes with rising temperatures (Rothschild and Mancinelli 2001).
As is the case with the Eukarya, there are few lineages of Bacteria that are thermoacidophilic. Some bacteria grow at extreme temperatures, in excess of 70°C (e.g. the genera Thermotoga, Caldicellulosiruptor, Aquifex), albeit at neutralophilic conditions (Counts et al. 2017). Conversely, there are also a number of acidophilic bacteria, primarily from the genera Leptospirillum and Acidithiobacillus, that are also autotrophic and are found in acidic features with low organic carbon concentrations. But these bacteria grow optimally at temperatures far below anything considered thermophilic (i.e. 28–45°C); however, Leptospirillum thermoferrooxidans grows at temperatures up to 50°C (Kondrat'eva et al. 2012; Dopson 2016). As temperatures increase, bacteria from the thermotolerant and acidotolerant genera Sulfobacillus,Alicyclobacillus and Hydrogenobaculum are most common (see Fig. 2).
Figure 2.
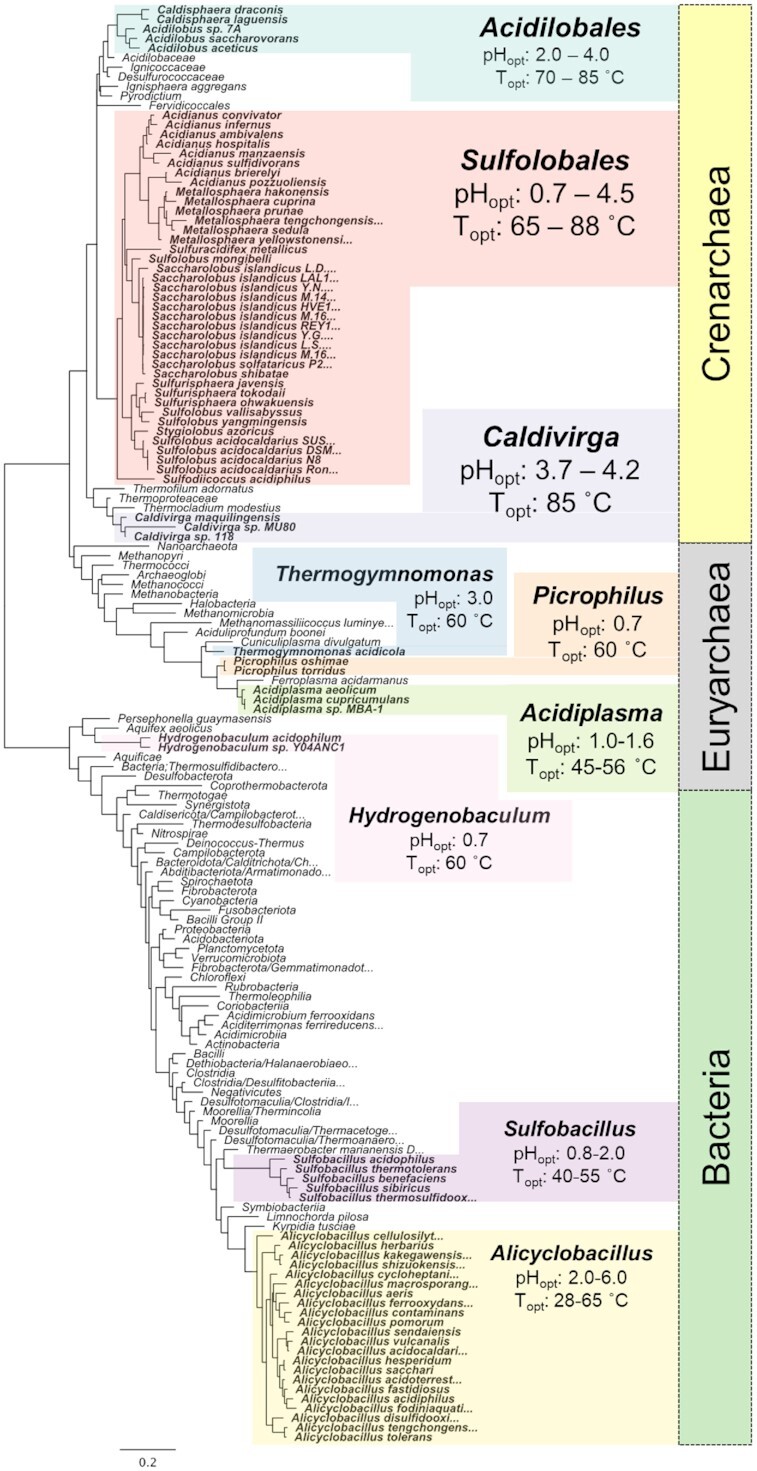
16S phylogeny tree of thermoacidophilic organisms.
Archaeal thermoacidophilic diversity
In contrast to the other domains of life, the Archaea predominate in extremely thermoacidophilic features. While culture-independent techniques are rapidly expanding the number of recognized species in acidic hydrothermal environments [e.g. candidate phyla Geoarchaeota (Kozubal et al. 2013) and Marsarchaeota (Jay et al. 2018)], most of the isolated thermoacidophiles to date originate from the archaeal phyla Crenarchaeota and Euryarchaeota. In both phyla, almost all isolated and named species are native to terrestrial hot acid environments. Currently, the only thermoacidophile with a validly published name from a marine environment is the deep-sea Euryarchaeon Aciduliprofundum boonei, an anaerobic heterotroph growing optimally at 70°C and pH 4.2–4.8, that utilizes sulfur and iron as electron acceptors (Reysenbach et al. 2006).
Other thermoacidophilic Euryarchaeota originate from the order Thermoplasmatales, which includes the thermotolerant Acidiplasma, as well as the moderately thermophilic Picrophilus and Thermogymnomonas. The genus Acidiplasma contains a few moderately thermophilic acidophiles (pH optimum 1–2, Topt 45–55°C), including the cell wall-lacking Acidiplasma aeolicum (Golyshina et al. 2009). While these organisms grow chemoorganotrophically, they also oxidize iron; for example, Acidiplasma cupricumulans (f. Ferroplasma cupricumulans) originates from a copper mine heap (Hawkes et al. 2006), and the recently sequenced Acidiplasmasp. strain MBA-1 originates from a pyrite–arsenopyrite gold-bearing concentrate bioleaching reactor (Bulaev, Kanygina and Manolov 2017). In contrast, the thermophilic genus Picrophilus contains two members, Picrophilus oshimae and Picrophilus torridus, which are aerobic, heterotrophic organisms from solfatara in Hokkaido, Japan, exhibiting remarkable acid tolerance (pH optima < 1.0), with optimal growth near 60°C (Schleper et al. 1996). While most organisms maintain a near circumneutral intracellular pH, P. oshimae actually maintains an intracellular pH of ∼4.6, making it a reservoir for acid-stable cytoplasmic proteins (van de Vossenberg et al. 1998). Additionally, sequencing revealed that Picrophilusspp. have some of the smallest genomes (around 1.5 Mb) isolated from free-living organisms (Futterer et al. 2004). Finally, in addition to the well-studied obligately aerobic heterotrophs from the genus Thermoplasma, Thermoplasma acidophilum and Thermoplasma volcanium (Segerer and Stetter 1988), there is a cell wall-less species, Thermogymnomonas acidicola, that grows near 60°C, but at slightly higher pH (around 3.0 optimally) (Itoh, Yoshikawa and Takashina 2007).
While the temperature optima of the euryarchaeal thermoacidophiles is limited to around 60°C, the thermoacidophiles from the crenarchaeal phylum all grow at temperatures ranging from 65°C to 88°C. These organisms are composed of three major clades, spanning three orders: Acidilobales,Sulfolobales and Thermoproteales. While the Sulfolobales are a well-studied archaeal lineage (over 30 named species, across 7 genera and >20 distinct genomes), the other two lineages, Acidilobales (containing Caldisphaera and Acidilobus) and Thermoproteales (only the Caldivirga are thermoacidophiles), contain just a few named, characterized strains. From the order Acidilobales, there are just two genera, Acidilobus and Caldisphaera, belonging to families derived from the same names. Both groups consist of anaerobic heterotrophs, growing optimally at pH ranging from 2.5 to 4.0 (mostly moderate acidophiles) and temperatures around 70–75°C for the Caldisphaera and slightly elevated temperatures of 50–80°C for the Acidilobusspp. (Prokofeva et al. 2000; Itoh et al. 2003; Boyd et al. 2007; Prokofeva et al. 2009). Meanwhile, the genus Caldivirga is represented by a single member, Caldivirga maquilingensis, isolated from the Philippines, which is capable of anaerobic (and microaerophilic) growth on heterotrophic substrates at moderate pH (optimum 3.7–4.2) and extremely thermophilic conditions (85°C) (Itoh et al. 1999).
The Sulfolobales
As mentioned previously, one of the first archaeal lineages discovered was the Sulfolobales, named for their presence and perceived usage of sulfur by Thomas Brock from his excursions to Yellowstone in the 1960s (Brock et al. 1972). Over the course of the following decades, a number of intriguing microorganisms emerged from terrestrial hot springs throughout the world, representing the seven named genera today from the order: Acidianus,Metallosphaera,Saccharolobus (f. Sulfolobus),Stygiolobus,Sulfodiicoccus,Sulfolobus,Sulfuracidifex (f. Sulfolobus) and Sulfurisphaera (f. Sulfolobus) (Counts, Willard and Kelly 2020). These include organisms with a broad array of physiological traits, ranging from extreme to moderate acidophily (0.7–4.5), thermophily (65–88°C), obligate and facultative aerobes, obligate anaerobes, metal oxidizers, sulfur reducers/oxidizers, chemoheterotrophs and chemolithoautotrophs.
Species in the thermoacidophilic genus Acidianus grow anaerobically, reducing sulfur in its various forms, or aerobically, oxidizing sulfur (Segerer et al. 1986). The genus contains the most acidophilic Sulfolobales member to date: Acidianus sulfidivorans (pHopt ∼ 0.7), and the most thermophilic member: Acidianus infernus (Topt ≈ 88°C) (Segerer et al. 1986; Plumb et al. 2007). The order also contains several members with metal biooxidation capabilities (Huber et al. 1989; Huber and Stetter 1991). Acidianus ambivalens (f. Desulfurolobus ambivalens) has long served as a model for the study of sulfur biotransformation within the Sulfolobales (Laska, Lottspeich and Kletzin 2003; Müller et al. 2004; Brito et al. 2009; Protze et al. 2011).
The genus Metallosphaera was named for the perceived ability of its members to biooxidize iron (and by proxy release other metals from ores, e.g. copper) (Huber et al. 1989). The type species, Metallosphaera sedula, along with the recently isolated Metallosphaera yellowstonensis, serve as model systems for metal biooxidation by extremely thermoacidophilic archaea (Auernik et al. 2008; Kozubal et al. 2011). Further, M. sedula has also been examined for autotrophy catalyzed by the 3-hydroxypropionate/4-hydroxybutyrate cycle (Berg et al. 2010a), which has shown promise for metabolic engineering of biosynthetic pathways (Hawkins et al. 2013; Keller et al. 2013; Lian et al. 2016; Straub et al. 2018).
In contrast to the other genera of the Sulfolobales, the genus Saccharolobus appears to depend less on lithotrophic pathways and more, as its name suggests, on sugar catabolism (Sakai and Kurosawa 2018). Members of the genus Saccharolobus are mostly aerobic and are among the most thermophilic (Topt ≥ 80°C) and least acidophilic organisms in the order (pHopt ≥ 3.0) (Zillig et al. 1980; Grogan 1989; Sakai and Kurosawa 2018). Their original taxonomical placement in Sulfolobus was changed following growing genomics information pointing to evolutionary divergence (Sakai and Kurosawa 2018). In fact, a number of unnamed and informally named species, e.g. ‘Sulfolobus islandicus’, appear to be much more closely related to other members of the genus Saccharolobus, which is fitting given that they use pentoses, hexoses, and di-, tri- and polysaccharides (Grogan 1989).
The main representative of the genus Sulfolobus: S. acidocaldarius, has a much narrower range of carbohydrate utilization. S. acidocaldarius grows best at 75°C and pH 3.0, using only amino acids, sucrose, dextrin and starch (Grogan 1989). This archaeon was originally named for its perceived capability to oxidize sulfur in the sulfur-rich pools of Yellowstone National Park (Brock et al. 1972). Despite these early reports, sulfur biooxidation capacity in strains that are currently available from culture collections is limited. However, recent studies showed that sulfur oxidation can be restored in S. acidocaldarius DSM 639 by inserting genes encoding sulfur oxygenase reductase (SOR) and thiosulfate:quinone oxidoreductase (TQO) (Zeldes et al. 2019), perhaps reflecting an evolutionary connection to this process. S. acidocaldarius has emerged as a tractable genetic platform to understand the physiological features of the Sulfolobales, such as pili structure controlling motility (Albers and Jarrell 2015), UV-stress response (Wagner et al. 2012), biofilm formation (van Wolferen et al. 2020), and cellular division (Pulschen et al. 2020). In addition to S. acidocaldarius, Saccharolobus solfataricus and ‘Sulfolobus islandicus’ are currently the only Sulfolobales with tractable genetic systems (Straub et al. 2018).
The remaining genera are represented by only a few named species, but vary dramatically in some of their observed traits. For example, the genus Stygiolobus contains a single member, Stygiolobus azoricus (Topt 80°C and pH 2.5–3.0), and is the only obligate anaerobe from the order to date, capable of sulfur reduction in the presence of hydrogen (Segerer et al. 1991). The genus Sulfurisphaera contains three species: Sulfurisphaera javaensis,Sulfurisphaera tokodaii and Sulfurisphaera ohwakuensis (the genus type species), all of which are extremely thermophilic (optima 80–85°C), but vary with respect to acidophily (optima 2.0–4.0) (Tsuboi et al. 2018). Sulfurisphaera species are facultative anaerobes and oxidize sulfur and iron to varying extents, and grow on complex organic substrates (Kurosawa et al. 1998; Tsuboi et al. 2018). In contrast, the two current members of the genus Sulfuracidifex: Sulfuracidifex (f. Sulfolobus) metallicus and Sulfuracidifex tepidarius, are less thermophilic acidophiles (temperature optimum: 65°C; pH optima: 2.0–3.5) and obligately aerobic chemolithoautotrophs, capable of mixotrophic growth in the presence of reduced sulfur compounds (Huber and Stetter 1991; Itoh et al. 2020). Sulfuracidifex metallicus has served as a model system for metal biooxidation studies (Bathe and Norris 2007). The genus Sulfodiicoccus is another single-member genus (type species Sulfodiicoccus acidiphilus), growing optimally at 65–70°C and pH 3.0–3.5. This archaeon is different from other Sulfolobales in that it is not only unable to oxidize elemental sulfur, but is possibly inhibited by it (Sakai and Kurosawa 2017). Furthermore, S. acidiphilus also lacks key components for carbon dioxide fixation by the 3-hydroxypropionate/4-hydroxybutyrate cycle and apparently does not grow autotrophically (Sakai and Kurosawa 2017, 2019). See Table 1 for a listing of thermoacidophilic microorganisms.
Table 1.
Thermoacidophile organisms.
| Kingdom | Phylum/division | Genus/species | T opt (°C) | pHopt | Isolation site (locale, country) | Reference |
|---|---|---|---|---|---|---|
| Eukarya | Rhodophyta | Galdieria sulphuraria (Merola) | 45 | 2–3 | Solfatara (Pozzuoli, Campania, Italy) | (Merola et al. 1982) |
| Bacteria | Proteobacteria | Acidithiobacillus (A. caldus) | 25–45 (45) | 2.0–4.0 (2.0–2.5) | Coal spoil enrichment (Belfast, Northern Ireland, United Kingdom) | (Dopson 2016) |
| Nitrospirae | Leptospirillum(L. ferriphilum) | 30–43 (30–37) | 1.4–3.0 (1.4–1.8) | Bioleaching Tank (South Africa) | (Coram and Rawlings 2002) | |
| Firmicutes | Sulfobacillus thermosulfidooxidans | 50–55 | 1.7–2.4 | Copper-zinc-pyrite ore (Nikolaev Mine, East Kazakhstan, Kazakhstan) | (Bogdanova et al. 2006) | |
| Sulfobacillus sibericus | 55 | 2.0–2.5 | Nezhdaninskoe ore deposit (East Siberia, Republic of Sakha, Russian Federation) | (Melamud et al. 2003) | ||
| Alicyclobacillus(A. acidocaldarius) | 35–65 (60–65) | 1.5–4.5 (3.0–4.0) | Hot spring (Yellowstone NP, Wyoming, USA) | (Darland and Brock 1971; Karavaiko et al.2005) | ||
| Aquificae | Hydrogenobaculum acidophilum | 65 | 3.0–4.0 | Solfatara (Tsumagoi, Gunma, Japan) | (Shima and Suzuki 1993) | |
| Archaea | Candidate Geoarchaeota | Uncultured | 60–78 | 3.5 | Norris Geyser Basin (Yellowstone, Wyoming, USA) | (Kozubal et al. 2013) |
| Candidate Marsarchaeota | Uncultured | 50–80 | 3.0–3.5 | Thermal springs (Yellowstone, Wyoming, USA) | (Jay et al. 2018) | |
| Euryarchaea | Aciduliprofundum boonei | 70 | 4.2–4.8 | Deep sea vents (Mariner, Lau Basin, near Tonga) | (Reysenbach et al. 2006) | |
| Acidiplasma cupricumulans | 53.6 | 1.0–1.2 | Mineral bioleaching heap (Undisclosed, Myanmar) | (Hawkes et al. 2006) | ||
| Acidiplasma aeolicum | 45 | 1.4–1.6 | Hydrothermal pool (Vulcano Island, Messina, Italy) | (Golyshina et al. 2009) | ||
| Picrophilus (P. torridus/P. oshimae) | 60 | 0.7 | Solfatara (Hokkaido, Japan) | (Schleper et al. 1996) | ||
| Thermoplasma volcanium | 60 | 2.0 | Solfatara (Vulcano Island, Messina, Italy) | (Segerer and Stetter 1988) | ||
| Thermoplasma acidophilum | 59 | 1.0–2.0 | Coal refuse pile (Friar Tuck Mine, Indiana, USA) | (Darland et al. 1970) | ||
| Thermogymnomomonas acidicola | 60 | 3.0 | Solfatara (Ohwaku-dani, Hakone, Japan) | (Itoh, Yoshikawa and Takashina 2007) | ||
| Crenarchaea(non-Sulfolobales) | Acidilobus aceticus | 85 | 3.9 | Thermal spring (Moutnovski, Kamchatka, Russia) | (Prokofeva et al. 2000) | |
| Acidilobus saccharovorans | 80–85 | 3.5–4.0 | Thermal spring (Uzon Caldera, Kamchatka, Russia) | (Prokofeva et al. 2009) | ||
| Caldisphaera laguensis | 70–75 | 3.5–4.0 | Hot spring (Mt Maquiling, Laguna, Philippines) | (Itoh et al. 2003) | ||
| Caldivirga maquilingensis | 85 | 3.7–4.2 | Hot spring (Mt Maquiling, Laguna, Philippines) | (Itoh et al. 1999) | ||
| Archaea | Crenarchaea(Sulfolobales) | Acidianus ambivalens | 81 | 2.5 | Solfatara (Leihnukur, Iceland) | (Zillig et al. 1986) |
| Acidianus brierleyi | 70 | 1.5–2.0 | Thermal spring drainage (Yellowstone, Wyoming, USA) | (Brierley and Brierley 1973; Segerer et al. 1986) | ||
| Acidianus infernus | 90 | 2.0 | Mud pot (Naples, Campania, Italy) | (Segerer et al. 1986) | ||
| Acidianus sulfidivorans | 74 | 0.8–1.4 | Solfatara (Lihir Island, Papua New Guinea) | (Plumb et al. 2007) | ||
| Metallosphaera cuprina | 65 | 3.5 | Thermal spring (Tengchong, Yunnan, China) | (Liu et al. 2011) | ||
| Metallosphaera hakonensis | 70 | 3.0 | Thermal spring (Ohwaku-dani, Hakone, Japan) | (Takayanagi et al. 1996) | ||
| Metallosphaera prunae | 75 | 2.5 | Uranium slag heap (Ronneburg, Hesse, Germany) | (Fuchs et al. 1995) | ||
| Metallosphaera sedula | 75 | 2.5 | Thermal pool (Naples, Campania, Italy) | (Huber et al. 1989) | ||
| Saccharolobus caldissimus | 85 | 3.0 | Thermal spring (Ohwaku-dani, Hakone, Japan) | (Sakai and Kurosawa 2018) | ||
| Saccharolobus shibatae | 81 | 3.0 | Mud pot (Kyushu, Japan) | (Grogan et al. 1990) | ||
| Saccharolobus solfataricus | 87 | 4.5 | Thermal spring (Agnano, Campania, Italy) | (Zillig et al. 1980) | ||
| Stygiolobus azoricus | 80 | 2.5–3.0 | Solfatara (São Miguel Island, Azores, Portugal) | (Segerer et al. 1991) | ||
| Sulfodiicoccus acidiphilus | 65–70 | 3.0–3.5 | Solfatara (Ohwaku-dani, Hakone, Japan) | (Sakai and Kurosawa 2017) | ||
| Sulfolobus acidocaldarius | 70–75 | 2.0–3.0 | Thermal spring (Yellowstone, Wyoming, USA) | (Brock et al. 1972) | ||
| Sulfuracidifex tepidarius | 65 | 3.5 | Solfatara (Ohwaku-dani, Hakone, Japan) | (Itoh et al. 2020) | ||
| Sulfuracidifex metallicus | 65 | 2.0–3.0 | Solfatara (Krafla, Iceland) | (Itoh et al.2020) | ||
| Sulfurisphaera ohwakuensis | 84 | 2.0 | Thermal spring (Ohwaku, Hakone, Japan) | (Kurosawa et al. 1998) | ||
| Sulfurisphaera tokodaii | 80 | 2.5–3.0 | Hot spring (Beppu, Kyushu, Japan) | (Suzuki et al. 2002) | ||
| Sulfurisphaera javensis | 80–85 | 2.5–4.0 | Thermal spring (Java, Indonesia) | (Tsuboi et al. 2018) |
VIRUSES AND CRISPR SYSTEMS OF THERMOACIDOPHILES
Thermoacidophiles share their natural habitat with viruses (Munson-Mcgee, Snyder and Young 2018). A recent survey of the viral communities in thermal hot springs in Yellowstone National Park showed that >60% of cells were infected by viruses and that the majority even contained two or more virus types at the same time (Munson-Mcgee et al.2018). Consequently, viruses represent an important evolutionary pressure in these archaeal dominated environments. The ongoing arms race between viruses and their hosts has led to the development of anti-viral defense strategies and mechanisms from viruses to circumvent them (Borges, Davidson and Bondy-Denomy 2017; Hwang and Maxwell 2019; Hampton, Watson and Fineran 2020). However, the fact that in many cases cells carry multiple virus types suggests that viruses can also have beneficial relationships with their microbial hosts. Viruses shape microbial populations, are a major driver of microbial evolution and impact host ecology. An excellent example is the virus–host mutualism by which chronically virus infected-Sulfolobus cells kill the virus-resistant cells in the population (DeWerff et al. 2020).
Viruses of thermoacidophiles
In comparison with known bacterial and eukaryotic viruses, only a modest number of archaeal viruses have been isolated to date (Prangishvili et al. 2017). However, thermoacidophiles, especially members of the Sulfolobales, have proven to be a very rich source of archaeal viruses (Prangishvili, Stedman and Zillig 2001; Prangishvili et al. 2017; Munson-Mcgee, Snyder and Young 2018). These viruses are characterized by a large genetic and morphological diversity, including many unique shapes that are not found in viruses infecting bacteria and eukaryotes (Pina et al. 2011; Prangishvili et al. 2017). The evolutionary origin of archaeal viruses is not clear, but the high diversity might have originated during the early stages of evolution of cellular life, maintained in Archaea, and lost in bacterial and eukaryotic lineages (Prangishvili, Forterre and Garrett 2006; Prangishvili 2015; Prangishvili et al. 2017). All isolated viruses from Sulfolobales have DNA genomes, and the majority of their gene products have unknown functions (Prangishvili et al. 2017). Metagenomic analysis indicated the presence of viruses with RNA genomes in high-temperature acidic hot springs (Bolduc et al. 2012). However, viral particles were not isolated, and the exact host remains unknown (Bolduc et al. 2012; Stedman, Kosmicki and Diemer 2013).
Members of at least eight different viral families infect thermoacidophilic archaea: bottle-shaped Ampullaviridae (Haring et al. 2005), tailed Bicaudaviridae (Häring et al. 2005), spindle-shaped Fuselloviridae (Schleper, Kubo and Zillig 1992), droplet-shaped Guttaviridae (Arnold, Ziese and Zillig 2000), filamentous Lipotrixviridae (Bettstetter et al. 2003), polyhedral Portogloboviridae (Liu et al. 2017), rod-shaped Rudiviridae (Prangishvili et al. 1999) and the icosahedral Turriviridae (Rice et al. 2004) (Fig. 3). The diversity of morphotypes encountered among viruses infecting thermoacidophilic Crenarchaea is in stark contrast to that found for euryarchaeal or bacterial viruses, which are dominated by head–tail morphologies (Pietilä et al. 2014; Prangishvili et al. 2017). Interestingly, recently available cryo-EM (cryogenic electron microscopy) structures have shown that several viruses infecting members of the Sulfolobales package their dsDNA genome in A-form (DiMaio et al. 2015; Wang et al. 2019b). A-form DNA was at first thought to be an artifact and have no biological significance, but the widespread usage of A-form DNA by archaeal viruses suggests that this packaging helps to protect the viral genomes against adverse conditions in thermal hot springs (Wang et al.2019b).
Figure 3.
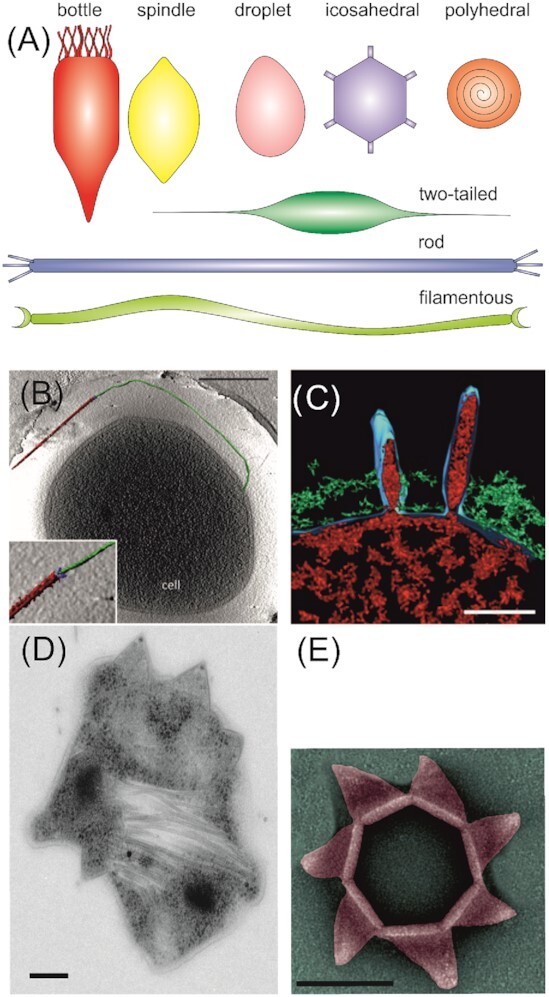
Thermoacidophilic archaeal viruses and their infection mechanisms. (A) Schematic representation of virion morphologies of viruses infecting thermoacidophilic archaea as described in the text. (B) Segmented tomographic volume of an SIRV2 virion (red) attached to a surface filament of Sa. islandicus (green) with help of the three terminal virion fibers (blue). Inset depicts a magnification of the interaction between the tail fibers and the surface structure. Scale bar, 500 nm. (C) Volume segmentations of electron microscopy tomograms showing Sulfolobus spindle-shaped virus 1 maturation and release by budding. Scale bar, 50 nm. (D) Transmission electron micrograph of a thin section of a SIRV2-infected Sa. islandicus cell displaying several pyramidal egress structures. Scale bar, 100 nm. (E)Transmission electron micrographs of an isolated pyramidal egress structure in open conformation isolated after SIRV2 infection of Sa. islandicus. Scale bar, 100 nm. Adapted from Bize et al. (2009); Quax et al. (2011); Quemin et al. (2013, 2016).
Among the thermoacidophiles, Sa. islandicus and Sa. solfataricus are model systems for the study of virus–host interactions in Crenarchaea (Pina et al. 2011; Prangishvili, Koonin and Krupovic 2013; Dellas et al. 2014). The valuable, but still limited, knowledge on infection strategies of crenarchaeal viruses mainly derives from viruses infecting these organisms. For initial attachment and entry into the cell, the various surface appendages with which Sulfolobales are covered (see the section 'Cell cycle and modes of growth') are important for multiple viruses. Sulfolobus turreted icosahedral virus (STIV1) binds with its turrets to thin filaments of unknown identity on the surface of Sa. solfataricus (Hartman et al. 2019). Saccharolobus islandicus rod-shaped virus (SIRV) particles use the three tail fibers that are present at the distal parts of the virion for attachment to adhesive type IV pili on the its surface (Quemin et al. 2013; Deng et al. 2014; Rowland et al. 2020). Like SIRV, the Sulfolobus spindle-shaped virus SSV requires pili for infection, but the particles do not directly attach to the pili, and the role of pili in viral entry is unresolved (Rowland et al. 2020). Primary attachment to filamentous surface structures is a common strategy of bacterial viruses, which can increase the chances of successful infection (Poranen, Daugelavičius and Bamford 2002; Quemin and Quax 2015). The mechanisms by which viruses move along archaeal filaments to the cell surface are unknown and are likely different from those of bacterial viruses, since the archaeal surface filaments have different structural organization (see the section 'Cell cycle and modes of growth') (Quemin and Quax 2015; Chaudhury, Quax and Albers 2018).
Once virions have attached, their genomes can enter the cell. Some viruses, such as SSV, can integrate their genome into that of the host (Muskhelishvili, Palm and Zillig 1993; Serre et al. 2002; Clore and Stedman 2007). Circularization and replication of the integrated SSV genome is induced by UV light (Schleper, Kubo and Zillig 1992; Fröls et al. 2008). SSV has been employed to develop a genetic manipulation system for Sa. solfataricus (see the section 'Potential and current uses of thermoacidophiles in biotechnological applications'). Several other viruses do not integrate, instead replicating directly after entry. The replication mechanism of only a few thermoacidophile model viruses has been studied. For example, replication of the dsDNA genome of Acidianus Filamentous Virus 1 (AFV1) relies on recombination events for initiation and termination and has a terminally bound protein (Pina et al. 2014). Replication of the linear dsDNA genome of SIRV requires a virus-encoded dimeric Rep protein that initiates replication by making single stranded nicks and the virus-encoded Holliday junction resolvase (Hjr) to resolve viral genome concatemers (Blum et al. 2001; Peng et al. 2001; Oke et al. 2010, 2011). SIRV Hjr interacts with proliferating cell nuclear antigen (PCNA), a key replication protein in archaea (Gardner et al. 2014). Interestingly, SIRV forms a distinct replication focus in the cell to which viral and host replication proteins are specifically recruited (Martínez-Alvarez, Deng and Peng 2017).
After genome replication, virions are formed in the cytoplasm. Virion maturation can occur (i) before, (ii) during or (iii) after release. (i) Several lytic viruses infecting Sulfolobales, such as STIV and SIRV, were shown to mature in the cytoplasm and employ an unusual lysis mechanism that relies on the formation of 7-fold symmetric pyramidal egress structures, of which a dozen form during viral infection on the host cell surface (Bize et al. 2009; Brumfield et al. 2009; Prangishvili and Quax 2011; Quax et al. 2011; Quax and Daum 2017) (Fig. 3). These ∼150-nm structures consist of one viral protein and protrude through the protective S-layer (see the section 'Cell cycle and modes of growth') (Fu et al. 2010; Quax et al. 2010; Snyder et al. 2011; Daum et al. 2014). They open outward at the end of the infection cycle to allow for the release of virions (Bize et al. 2009; Brumfield et al. 2009; Fu et al. 2010; Daum et al. 2014). (ii) SSV matures upon egress, as the virions are released via budding and are covered in a lipid layer during this process (Quemin et al. 2016). In fact, this is the first case of budding observed for a prokaryotic virus. Budding viruses allow for a continuous release of virions and the cells remain alive throughout the infection cycle (Schleper, Kubo and Zillig 1992; Quemin et al. 2016). The ESCRT-III system could play a role in the budding of archaeal viruses (Fig. 3) (Liu et al. 2017). (iii) Acidianus two-tailed virus (ATV) and Sulfolobus monocaudavirus (SMV1) are exceptional viruses for which virion maturation (the lengthening of the tails) happens outside and independent of the host cell, after viral release (Häring et al. 2005; Prangishvili et al. 2006; Scheele et al. 2011; Uldahl et al. 2016). These tails consist of helically arranged globular subunits that develop from the two pointed ends of the virion when it is outside the host cell (Prangishvili et al. 2006). High temperatures are required for this morphological transformation. In summary, viruses of thermoacidophiles are unique because of their diverse morphologies and the model virus–host systems of the Sulfolobales have provided important insights into the infection strategies of crenarchaeal viruses in general.
CRISPR-Cas mediated viral defense in thermoacidophiles
The omnipresence of viruses in archaeal habitats has resulted in the development of several anti-viral defense strategies, of which CRISPR-Cas is without doubt the best known. CRISPR (clustered regularly interspaced short palindromic repeats) systems in Sulfolobales have been studied since the ‘early days’ of CRISPR research (Vestergaard et al. 2008; Han and Krauss 2009; Held and Whitaker 2009; Lillestøl et al. 2009; Garrett et al. 2011; Zhang and White 2013), just after these systems were suggested to play a role in defense against viruses in bacteria and archaea (Bolotin et al. 2005; Mojica et al. 2005; Pourcel, Salvignol and Vergnaud 2005; Makarova et al. 2006). CRISPR arrays consist of a series of ∼30 bp genomic repeats, which are interspaced by unique sequences that can match foreign genetic elements (van der Oost, Jackson and Wiedenheft 2014). CRISPR-associated proteins (Cas) are usually encoded in the proximity of the CRISPR array. Upon a viral infection, new spacers, exactly matching the genome of the infecting virus, are integrated between two repeats. The arrays are then processed by Cas proteins, and the spacer is used as a guide to specifically target and interfere with the matching sequences in the viral genome (Barrangou et al. 2007; Barrangou and Horvath 2017; Jackson et al. 2017). Thus, CRISPR-Cas provides specific and inheritable immunity (Barrangou et al. 2007; Brouns et al. 2008). CRISPR arrays can also be used as a map to track previous encounters with viruses and to indicate viral host range (Bautista et al. 2017; Munson-Mcgee et al.2018; Pauly et al. 2019).
CRISPR-Cas systems are present in ∼40% of bacteria, most archaea (85%), and almost all extreme thermophiles (97%) (Makarova et al. 2019). Based on the Cas proteins, the CRISPR systems have been divided into several different groups, and this division keeps evolving as new systems are being discovered (Makarova et al. 2019). Crenarchaea, such as the Sulfolobales, usually harbor multiple CRISPR systems in their genome, and generally have longer CRISPR arrays than bacteria (Zhang and White 2013). Crenarchaeal genomes are substantially enriched for type III systems of class 1 (Zhu et al. 2018; Makarova et al. 2019), which rely on transcription-dependent (specific RNA binding and cleavage) and subsequent (non-specific) DNA degradation (Deng et al. 2013; Goldberg et al. 2014; Zhu et al. 2018). Type III systems typically possess a Cas10 protein with a Palm polymerase domain that can cyclize ATP to generate cyclic oligoadenylate (cOA) to act as a second messenger (Kazlauskiene et al. 2017; Niewoehner et al. 2017; Rouillon et al. 2018). Formation of cOA leads to signal amplification that activates other defense mechanisms, including host and viral DNA degradation that results in immunity or cell dormancy (Rouillon et al. 2018; Rostøl and Marraffini 2019). In Saccharolobus, cyclic tetra-adenylate (cA4) can be degraded by host-encoded ring-nucleases to reset the signal (Athukoralage et al. 2018). Interestingly, some archaeal viruses encode a potent ring nuclease that acts as an anti-CRISPR (Arc) (Athukoralage et al. 2020a,b). More Arcs have been identified in archaeal viruses, such as those that bind and inhibit type III-B or I-D CRISPR systems (He et al. 2018; Bhoobalan-Chitty et al. 2019). Different viral families probably use different strategies to evade CRISPR-Cas immunity, as natural populations of Sulfolobus have developed CRISPR-Cas immunity with a different structure and diversity in response to SIRV and SSV infections (Pauly et al. 2019).
Despite the viral strategies to evade CRISPR-Cas mediated defense, this seems an effective immune system in extreme thermophiles (Topt ≥ 70°C), illustrated by the presence of CRISPR-Cas in nearly all of their genomes. It remains to be seen why CRISPR-Cas systems are so ubiquitous, specifically in extremely thermophilic archaea. One possible explanation relates to the lower mutation rate of viruses in extreme environments that, combined with the lower population sizes of extreme thermophiles compared with mesophiles, gives extremely thermophilic viruses limited possibility to escape immunity (Weinberger et al. 2012; Prangishvili et al. 2017).
Besides CRISPR-Cas, Crenarchaea are specifically enriched in toxin–antitoxin (TA) systems, which play a role in abortive infection in bacteria (Koonin, Makarova and Wolf 2017). Viral infection of different Sulfolobales induces expression of TA systems (Ortmann et al. 2008; Quax et al. 2013; León-Sobrino, Kot and Garrett 2016). Furthermore, several thermoacidophiles encode an Argonaute protein, which has been implicated in defense against foreign genetic elements (Makarova et al. 2009; Swarts et al. 2014a,b; Willkomm et al. 2017). However, it needs to be verified experimentally if archaeal TA or Argonaute systems provide immunity to viral infection. Certainly, thermoacidophiles encode novel viral defense mechanisms that are awaiting discovery (Doron et al. 2018).
GENETIC MECHANISMS
Effective packaging and organization of genomic DNA into the confined space of the nucleus or nucleoid, while at the same time enabling a dynamic and reliable genome replication and gene expression, is essential for every living organism. The underlying molecular mechanisms (replication, transcription and translation) are central to life. The study of genetic mechanisms in archaea lags behind those focused on bacteria and eukaryotes. However, it is clear that archaeal information processing machineries are related to their eukaryotic counterparts, that gene regulation processes are bacteria-like and that chromosome organization has both eukaryote-like and bacteria-like features (Peeters et al. 2015; Blombach et al. 2019; Lemmens et al. 2019a; Greci and Bell 2020).
Thermoacidophilic Crenarchaea belonging to the Sulfolobales order have served as an archaeal model system to study chromosome organization, DNA replication, transcription and translation processes. Although many insights can be extended further to the entire archaeal domain, there are also unique, lineage-specific aspects. For example, while most archaea harbor eukaryote-like histones involved in chromosome structuring, these are completely absent in thermoacidophilic archaea, namely in all Crenarchaeota and in Thermoplasma acidophilum (Peeters et al. 2015; Hocher et al. 2019).
Chromosome packaging and structuring
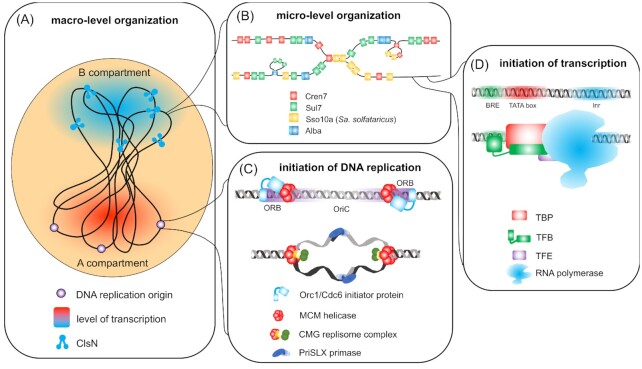
Thermoacidophilic archaea typically have a single circular, relatively small chromosome with a size between 1.5 and 3 Mbp (Chen and Morris 1972; She et al. 2001). This chromosome is packaged into a condensed and organized chromatin structure by the action of different types of chromatin proteins (Fig. 4). There is a large evolutionary divergence in chromatin proteins present in archaea, including the Sulfolobales; while histone orthologs are absent in this lineage, an interplay exists between a variety of nucleoid-associated proteins (NAPs) with different levels of conservation (Peeters et al. 2015). These proteins are small (between 7 and 10 kDa), basic and highly abundant in the cell, constituting up to 5% of soluble cellular protein (Mai et al. 1998). They harbor DNA-binding motifs that are also found in specific transcription regulators (see below the section 'Regulatory transcription factors in Sulfolobales'), such as the winged helix-turn-helix (wHTH) motif, and bind DNA with low or no sequence specificity. In case of low sequence specificity, these NAPs typically prefer GC-rich sequences (Kalichuk et al. 2016; Hocher et al. 2019). The Sulfolobales harbor two paralogs of the archaea-universal NAP Alba, which was initially assumed to be an important chromatin structuring protein (Bell et al. 2002), with the extent of heterodimerization between the two paralogs determining the architectural effects (Laurens et al. 2012). This hypothesis has recently been revisited as Alba was shown to be a general nucleic acid-binding protein interacting with RNA as well (Guo et al. 2014). Besides Alba, the NAP Cren7 is highly conserved in not only the Sulfolobales but also all Crenarchaeota; Cren7 is a versatile architectural protein, bending and also bridging DNA, thereby forming highly condensed chromatin filaments (Guo et al. 2008; Zhang et al. 2019c, 2020). Sul7d, analogous to Cren7, is a monomeric protein, which is capable of bending DNA; it has been found in many Sulfolobales genera: Sulfolobus and Saccharolobus, Acidianus, Metallosphaera, Stygiolobus and Sulfurisphaera (Kalichuk et al. 2016). In addition, there are species-specific NAPs, such as the Sso10a parologs in S. solfataricus, that are dimeric proteins capable of bending DNA and either bridging it or forming filaments (Driessen et al. 2016). Finally, the euryarchaeal T. acidophilum, which also lacks histones, harbors a NAP that is homologous to the bacterial HU family (Hocher et al. 2019).
Figure 4.
Main principles in genome organization and genetic information processing in the Sulfolobales. Conceptual schemes representing the major elements and principles of macro-level organization of the genomic DNA (adapted from Takemata, Samson and Bell 2019) (A), micro-level organization of the genomic DNA (partially adapted from Peeters et al. 2015) (B), initiation of replication (C) and initiation of transcription (D).
The heterogeneity in the NAP protein machinery responsible for packaging DNA in the Sulfolobales (Fig. 4B)—when considering a single species—is hypothesized to accommodate differential local chromatin structuring when expression levels or post-translational modifications (PTMs) of the individual NAPs are altered. This in turn might affect transcriptional expression in a polygenic manner (Peeters et al. 2015). Indeed, PTMs have been observed for the NAPs Alba, Cren7, Sso7d and Sul7d (Vorontsov et al. 2016). Early studies postulated that acetylation of the Lys16 residue of Alba constitutes a global gene regulation mechanism similar to eukaryotic histone modification (Bell et al. 2002). This has later been refuted and determined to be an N-terminal acetylation event instead that does not affect the nucleic acid-binding capacity of Alba and is possibly involved in protein turnover regulation (Ma et al. 2016; Cao et al. 2018a). Besides acetylation, methylation occurs widely on these NAPs (Niu et al. 2013; Vorontsov et al. 2016) and might be linked to thermostabilization of the chromatin, as well as to epigenetic mechanisms of gene regulation. The existence of such epigenetic mechanisms was recently demonstrated for a strain of Sa. solfataricus that was evolved in an adaptive laboratory evolution experiment and displayed a superacid-resistant phenotype. This evolved strain harbored no genomic changes with respect to the original strain (Payne et al. 2018; Johnson et al. 2019). Instead, the acid resistance appeared to be mediated by a different methylation status of the NAPs Cren7 and Sso7d. Other than NAP methylation, methylation of the genomic DNA itself might also be responsible for epigenetic mechanisms of gene regulation. Recently, the DNA methylome of S. acidocaldarius was mapped and shown to consist of base methylations that are more than just a part of restriction modification defense systems (Couturier and Lindås 2018). More specifically, N6-methyl-adenine methylations were found and hypothesized to be involved in the regulation of the cell cycle or other biological functions (Couturier and Lindås 2018).
Besides NAP-mediated local structuring, the genome is also organized in domains at a higher level. This higher order chromosome organization has recently been elucidated for S. acidocaldarius and Sa. islandicus using the Hi-C methodology, a combination of chromosome conformation capture (3C) and high-throughput sequencing (Takemata, Samson and Bell 2019). Similar to what has been observed in metazoan eukaryotes and very different from what is observed in bacteria, the Sulfolobales genome is organized in two distinct sub-Mbp compartments, each characterized by a different average level of transcription (Takemata, Samson and Bell 2019) (Fig. 4A). While the A compartment, which harbors genes mainly involved in core metabolic processes such as protein biogenesis, is transcriptionally active, the B compartment appears to be in a more silent transcriptional state and harbors genes that function in diverse metabolic pathways and physiological processes. For example, gene expression of the B compartment is typically induced in response to environmental stress conditions, such as the archaellum motility apparatus and a fatty acid metabolism gene cluster. In addition, the B compartment is enriched in mobile genetic elements, such as CRISPR-Cas clusters. Although it is still unclear which proteins are responsible for the active structuring of the chromosome into the two compartments, a major role has been described for the novel chromatin structuring protein coalescin (ClsN) (Takemata, Samson and Bell 2019). While the Sulfolobales do not harbor a homolog of the typical condensin complex belonging to the Structural Maintenance of Chromosome (SMC) family, conserved in bacteria, eukaryotes and most other archaea (Kamada and Barillà 2018), they possess the SMC-like ClsN instead, which is significantly smaller than the SMC subunits of condensin and possibly has a zinc hook domain instead of a hinge (Takemata, Samson and Bell 2019). There is an inverse correlation between the presence of ClsN and the transcriptional machinery, with ClsN being mainly associated with transcriptionally less active genes in the B compartment. It is hypothesized that the protein assists in the higher level compartmentalization by mediating intra- and interdomain interactions (Takemata, Samson and Bell 2019).
DNA replication
One of the most important transactions undergone by genomic DNA is its replication as part of the cell division process. It is striking that, similar to eukaryotes, several archaeal lineages are characterized by the chromosome harboring multiple replication origins, genomic sites at which replication is initiated. In contrast, bacteria only have a single origin. Sulfolobales are characterized by three replication origins (OriCs 1, 2 and 3), each of which accommodate a single replication initiation event during the cell cycle (Lundgren et al. 2004; Robinson et al. 2004; Robinson and Bell 2007; Duggin, McCallum and Bell 2008). Mutagenesis analysis indicated that, while none of the individual OriCs is essential, at least one is required (Samson et al. 2013). The observation that all three OriCs are located in the transcriptionally active chromosome compartment A (Fig. 4A) might suggest that DNA replication processes are involved in higher order chromatin structuring. However, this appears not to be the case as OriC mutant strains do not display any differences in their chromatin structure (Takemata, Samson and Bell 2019).
The archaeal replication machinery resembles the eukaryotic machinery, as exemplified by the well-described machinery of Sa. solfataricus (Fig. 4C) (Dionne et al. 2003b; Greci and Bell 2020). Archaeal initiator proteins, responsible for OriC recognition and assembly of the replisome, are related to the eukaryotic Orc1 and Cdc6 replication initiation proteins, which are characterized by an N-terminal AAA+ fold and a C-terminal wHTH domain (Cunningham Dueber et al. 2007). Sulfolobales encode three Orc1/Cdc6-like paralogs, with Orc1–1 specifically recognizing the origin recognition boxes (ORBs) in OriC1 (Samson, Abeyrathne and Bell 2016). Upon binding ATP, two inversely bound Orc1–1 proteins recruit two MCM homohexamers, which are the 3′-to-5′ helicases (Samson, Abeyrathne and Bell 2016; Meagher, Epling and Enemark 2019). Also, analogous to the eukaryotic system, the MCM helicase associates with additional replication proteins, forming the so-called CMG (Cdc45-MCM-GINS) replisome core (Fig. 4C). In S. acidocaldarius and Sa. islandicus, MCM recruits a Cdc45 ortholog and two GINS-like proteins Gins23 and Gins15, each protein having a homodimeric composition in the complex (Xu et al. 2016). In contrast, in T. acidophilum a homotetrameric GINS protein is part of the CMG complex (Ogino et al. 2017). Although it is unclear how the melting of the DNA helix is accomplished in Sulfolobales after assembly of the replisome at the replication origin, the involvement of a replication-dedicated DNA-dependent RNA polymerase responsible for primer synthesis has been established. This DNA primase initiates leading strand synthesis, or the synthesis of Okazaki fragments for lagging strand synthesis, and is recruited by interacting with the GINS complex (Marinsek et al. 2006), with the primase in Sa. islandicus being a heterotrimer PriSLX (Liu et al. 2015). Finally, the enzyme responsible for DNA synthesis, DNA polymerase, has also been shown to be eukaryote-like. Crenarchaeota possess three different B-family DNA polymerases, with PolB1 being essential and PolB2 and PolB3 shown not to be required for cell viability and hypothesized to be involved in DNA damage repair (Greci and Bell 2020). Prior to elongation a sliding clamp, constituted by a heterotrimeric PCNA protein (Dionne et al. 2003a), is loaded onto the DNA by replication factor C (RFC) and forms a ring-shaped structure. This clamp functions as a molecular platform to recruit the DNA polymerase and other replication-associated enzymes. In contrast to bacteria, which harbor site-specific mechanisms, replication termination appears to be mediated by passive-fork collision taking place halfway between the active replication origin(s) in a site-unspecific manner (Lundgren et al. 2004; Duggin, Dubarry and Bell 2011; Samson et al. 2013).
Sulfolobales have an organized cell cycle (see the section 'Sulfolobus cell division'), with well-defined gap phases and in which the process of DNA replication is temporally separated from the process of chromosome segregation (Bernander and Poplawski 1997). Following the S phase in which the chromosome is replicated, there is a significant post-replicative period (G2 phase) in which sister chromatids remain bound together to form hemicatenane structures (Robinson and Bell 2007). Next, chromosome segregation is accomplished by a bacterial-like ParAB-like system; in Sa. solfataricus, this system consists of SegA, an ortholog of the bacterial, Walker-type ParA ATPase protein and an archaea-specific DNA-binding protein named SegB (Kalliomaa-Sanford et al. 2012).
Transcription and its regulation
Basal transcription machinery in the Sulfolobales
The small genomes of archaea share their genetic organization with bacteria, with an operonic transcription unit structure that is dense and characterized by short intergenic regions. Similar to other information processing steps, mechanisms of basal transcription have been extensively studied in archaeal species belonging to Sulfolobales, ranging from focused biochemical studies with in vitro reconstituted transcription systems (Qureshi, Bell and Jackson 1997; Bell et al. 1999; Blombach et al. 2019) to high-resolution mapping of the transcriptome (Wurtzel et al. 2010; Cohen et al. 2016; Dar et al. 2016). Not long after the isolation of the first thermoacidophilic crenarchaeal isolate S. acidocaldarius, Wolfram Zillig performed a biochemical analysis of its RNA polymerase, which is the key enzyme of transcription, thereby concluding that its subunit pattern resembles that of eukaryotic RNA polymerase (Zillig, Stetter and Janekovic 1979). Much later, structural analysis of the Sa. solfataricus and Sa. shibatae RNA polymerases confirmed that they are complexes consisting of 13-subunit proteins that display an evolutionary relationship with the eukaryotic RNA polymerase II (Hirata, Klein and Murakami 2008; Korkhin et al. 2009). Transcription initiation requires a set of additional general transcription factors that are also homologous to eukaryotic factors: TATA-binding protein (TBP), transcription factor B (TFB) and transcription factor E (TFE) (Fig. 4D). A typical promoter region in Sa. solfataricus is characterized by a core TATA-box region of which the center is located ∼26 base pairs (bps) upstream of the transcription initiation site, which is directly preceded by a purine-rich factor B recognition element (BRE) (Wurtzel et al. 2010). Besides these canonical archaeal promoter elements, an additional 6-bp, AT-rich, conserved promoter element was identified in Sa. islandicus just upstream of the transcription start site (TSS), named initiator (Inr) (Ao et al. 2013).
Initiation of transcription proceeds by the stepwise assembly of the different components in the pre-initiation complex (PIC), which was first studied with Sa. shibatae (Qureshi, Bell and Jackson 1997; Bell et al. 1999). First, the highly symmetrical TBP binds the TATA box region followed by the association of TFB to the TBP-DNA complex. By specific recognition of the BRE promoter element, TFB determines the correct orientation of the PIC (Bell et al. 1999). Next, the RNA polymerase as well as TFE are recruited, with TFE being a heterodimeric protein consisting of TFEα and TFEβ subunits in Sulfolobales (Blombach et al. 2015). Although TFE is not absolutely required for in vitro transcription reactions to proceed, it stabilizes the PIC and facilitates DNA melting in the Inr promoter region, thereby assisting the formation of an open complex during the transitioning from the initiation to the elongation phase (Bell et al. 2001; Blombach et al. 2015). During elongation, TBP and TFB dissociate from the RNA polymerase, which is assisted by different transcription elongation factors. Most of these factors, such as the transcript cleavage factor TFS and the processivity factors Spt4/5 and Elf1, are also evolutionarily related to eukaryotic elongation factors (Fouqueau et al. 2017; Blombach et al. 2019). Finally, although transcription termination remains understudied in archaea, a transcriptome-wide Term-seq approach enabled the mapping of all RNA 3′ termini in S. acidocaldarius, revealing a widespread occurrence of multiple terminators. This leads to alternative 3′ isoforms, with U-rich terminator motifs retrieved for 53% of all transcription units (Dar et al. 2016).
To some extent, components of the basal transcription machinery are capable of mediating a global regulation of the transcription initiation process. Certain archaea harbor multiple paralogs of TBP and TFB and it is hypothesized that these are employed for global gene regulation in a similar way as alternative sigma factors in bacteria (Facciotti et al. 2007). Sulfolobales typically harbor a single TBP and three TFB paralogs, with TFB3 being a truncated form. The latter functions as a transcriptional activator in a trans-dependent manner on the canonical TFB1 (Paytubi and White 2009). TFB3 activates the expression of Ups pili and the Ced DNA import system in response to UV irradiation (Paytubi and White 2009; Feng et al. 2018; Schult et al. 2018). On the other hand, TFE might be involved in global regulation in response to oxidative and heat shock stress, as the cellular protein levels were depleted under these stress conditions (Iqbal and Qureshi 2010; Blombach et al. 2015).
Regulatory transcription factors in the Sulfolobales
The observation of extensive transcriptome-wide differential gene expression in response to stress conditions or shifts in nutritional conditions (see the sections 'Extreme thermoacidophily and stress response' and 'Metabolism') indicates that transcription initiation is highly susceptible to regulation. It can be assumed that regulatory transcription factors (TFs) play an important role in this regard. In contrast to the eukaryotic basal transcription machinery, archaeal regulatory TFs resemble bacterial regulators pointing to a shared ancestry (Aravind 1999). One-component regulators are characterized by two domains: an N-terminal DNA-binding domain, with a wHTH or an HTH motif, and a C-terminal ligand-binding domain. The structural resemblance between TFs and wHTH-containing NAPs sometimes complicates their distinction, and dual-function DNA-binding proteins can be found within the entire spectrum between a specifically acting regulatory TF and a globally acting chromatin protein (Karr et al. 2017; Dorman et al. 2020). This is illustrated by the archaea-specific Lrs14 family of DNA-binding proteins, which is widespread in Sulfolobales and shown to bind DNA non-specifically and to regulate biofilm formation and motility in S. acidocaldarius (Orell et al. 2013a) (see the section 'Regulation of biofilm processes'). Given the complete absence of typical bacterial two-component systems in Crenarchaeota including the Sulfolobales (Galperin et al. 2018), these organisms are solely reliant on one-component regulators. Usually, TFs bind in the vicinity of the promoter elements of transcription units and interact with the different components of the PIC, thereby either repressing or activating transcription initiation (Peeters, Peixeiro and Sezonov 2013) (Table 2). In some cases, a single TF can have a dual function, depending on the target gene or in a concentration-dependent manner, as has been shown for the Sa. solfataricus TF Ss-LrpB (Peeters, Peixeiro and Sezonov 2013).
Table 2.
Overview of characterized transcription regulators in thermoacidophilic archaea and their viruses.
| Name | Family | Microbial or viral species | Gene number | Physiological role | Regulatory action | Reference |
|---|---|---|---|---|---|---|
| C68 | AbrB | S. islandicus plasmid–virus pSSVx | ORFC68 | (CRISPR-mediated) virus–host interactionsa | Activationa | (Contursi et al. 2011) |
| MerR | ArsR | S. solfataricus | SSO2688 | Mercury resistance | Repression | (Schelert et al. 2006) |
| IdeR | DtxR | T. acidophilum | TA0872 | Iron uptake and homeostasis | Repression | (Yeo et al. 2012; Yeo, Park and Lee 2014) |
| ArnA | FHA | S. acidocaldarius | Saci_1210 | Motility | Repression | (Duan and He 2011; Reimann et al. 2012) |
| S. tokodaii | ST0829 | |||||
| YtrA | GntR | S. acidocaldarius | Saci_1851 | Expression of membrane proteins | Repression | (Lemmens et al. 2019b) |
| BarR | Lrp | S. acidocaldarius | Saci_2136 | β-Alanine metabolism | Activation | (Liu et al. 2014a) |
| S. tokodaii | ST1115 | |||||
| Lrp | Lrp | S. acidocaldarius | Saci_1588 | Global regulation of metabolism and physiology | Dual | (Enoru-Eta et al. 2000; Vassart et al. 2013) |
| S. solfataricus | SSO0606 | |||||
| LysM | Lrp | S. acidocaldarius | Saci_0752 | Amino acid transport and metabolism | Activation | (Brinkman et al. 2002; Song et al. 2013) |
| S. solfataricus | SSO0157 | |||||
| LrpB | Lrp | S. solfataricus | SSO2131 | Regulation of pyruvate ferredoxin oxidoreductase and permeases | Dual | (Peeters et al. 2009; Peeters, Peixeiro and Sezonov 2013) |
| AbfR1 | Lrs14 | S. acidocaldarius | Saci_0446 | Biofilm formation and motility | Dual | (Orell et al. 2013a; Li et al. 2017) |
| Sta1 | Lrs14 | S. solfataricus | SSO0048 | Regulation of SIRV1 viral gene expression | Activation | (Kessler et al. 2006) |
| Lrs14 | Lrs14 | S. solfataricus | SSO1101 | N.A. | Repression | (Bell and Jackson 2000) |
| Csa3a | MarR | S. islandicus | SiRe_0764 | CRISPR spacer acquisition | Activation | (Liu et al. 2015) |
| BldR | MarR | S. solfataricus | SSO1352 | Detoxification of aromatic compounds | Activation | (Fiorentino et al. 2007) |
| BldR2 | MarR | S. solfataricus | SSO1082 | Stress response to aromatic compounds | N.A. | (Fiorentino et al. 2011) |
| N.A. | MarR | S. tokodaii | ST1710 | N.A. | N.A. | (Kumarevel et al. 2009) |
| MLPTv | MarR | T. volcanium | BAB59904b | N.A. | N.A. | (Liu, Walton and Rees 2010) |
| RbkR | MarRb | T. acidophilum | Ta1064 | Riboflavin biosynthesis | Activationa | (Rodionova et al. 2017) |
| M. yellowstonensis | EHP68448.1d | |||||
| FadR | TetR | S. acidocaldarius | Saci_1107 | Fatty acid and lipid metabolism | Repression | (Wang et al. 2019c) |
| HhcR | TrmB | M. yellowstonensis | H2C8P4d | Autotrophic metabolism | N.A. | (Leyn et al. 2015) |
| MalR | TrmB | S. acidocaldarius | Saci_1161 | Maltose transport and metabolism | Activation | (Wagner et al. 2014) |
| ArnB | vWA | S. acidocaldarius | Saci_1211 | Motility | Repression | (Reimann et al. 2012) |
| XylR | N.A. | S. acidocaldarius | Saci_2116 | Arabinose/xylose transport and metabolism | Activation | (van der Kolk et al. 2020) |
| ArnR | N.A. | S. acidocaldarius | Saci_1180 | Motility; type IV pili surface structures | Activation | (Lassak et al. 2013; Bischof, Haurat and Albers 2019) |
| ArnR1 | N.A. | S. acidocaldarius | Saci_1171 | Motility; type IV pili surface structures | Activation | (Lassak et al. 2013; Bischof, Haurat and Albers 2019) |
| CopR/CopT | N.A. | S. solfataricus | SSO2652 | Copper homeostasis | Repression | (Ettema et al. 2006; Villafane et al. 2009) |
| Fur | N.A. | T. volcanium | TVN0292 | Oxidative stress | N.A. | (Minoshima et al. 2014) |
| SvtR | N.A.e | S. islandicus rod-shaped virus 1 (SIRV1) | ORF56b | Viral development | Repression | (Guillière et al. 2009) |
| RIP | N.A.e | Acidianus two-tailed virus (ATV) | ORF145 | Global regulation of host transcription | Repression | (Sheppard et al. 2016) |
| Stf76 | N.A.e | S. islandicus plasmid–virus pSSVx | ORF76 | N.A. | N.A. | (Contursi et al. 2014) |
| F55 | N.A.e | Sulfolobus spindle-shaped virus 1 | Tlys | Viral lysogeny and UV induction | Repression | (Fusco et al. 2015) |
N.A. = not applicable (unknown based on published information).
Hypothesized;
UNIPROT number;
Multifunctional protein with enzymatic and transcription regulatory domains;
GenBank accession number;
Viral regulators are often difficult to classify into a family because of a lack of homology.
The functional understanding of the TFs in thermoacidophilic archaea is still limited and based on a relatively small number of case studies for individual TFs in model species, such as Sa. solfataricus, Sa. islandicus and S. acidocaldarius (Table 2). These TFs are involved in the regulation of various metabolic and physiological processes, such as motility, hetero- or autotrophic growth, metal resistance and detoxification mechanisms, typically in response to interactions with small molecules, e.g. metabolites (see the sections 'Regulation of biofilm processes', 'Extreme thermoacidophily and stress response' and 'Metabolism'). Unfortunately, a system-level approach for mapping TF-mediated gene regulatory networks in relation to common environmental stresses, similar to how it has been performed for the euryarchaeal model organism Halobacterium salinarum (Bonneau et al. 2007), has not yet been undertaken for a thermoacidophilic archaeal species. An understanding of these networks would be valuable to gain insights into the physiology and stress adaptation of thermoacidophilic archaea and could be exploited for the engineering of metabolism for biotechnological purposes. Most of the characterized TFs in Sulfolobales belong to the dominant TF families, Lrp/AsnC and MarR, which together encompass >50% of all TFs in Crenarchaeota (Perez-Rueda et al. 2018; Lemmens et al. 2019a). TFs belonging to the Lrp/AsnC family are responsive to amino acids or related small molecules and display either a global or specific regulatory function of central metabolic pathways (Vassart et al. 2013; Liu et al. 2014a). BldR and BldR2 of Sa. solfataricus are prototypical MarR-family TFs involved in the detoxification of aromatic compounds (Di Fiore et al. 2009; Fiorentino et al. 2011). Finally, members of archaea-specific TF families are also found in Sulfolobales, such as the TrmB family that is typically involved in the regulation of sugar metabolism (Wagner et al. 2014).
Intriguingly, archaeal genomes are predicted to harbor a lower fraction of TF-encoding genes as compared with bacterial genomes (Pérez-Rueda and Janga 2010), and Crenarchaeota typically have even lower numbers of TFs than Euryarchaeota (Coulson, Touboul and Ouzounis 2007). These observations raise the question as to how thermoacidophilic Crenarchaeota are capable of efficiently regulating their transcriptome with a limited repertoire of TFs. While this might be partially explained by an extensive specialization to living in niche habitats, alternative mechanisms are hypothesized to exist; for example, cross-interactions exist between paralogous TFs that lead to a combinatorial use of a limited set of regulators, as has been shown for members of the Lrp/AsnC family of TFs in Sa. solfataricus and S. acidocaldarius (Nguyen-Duc et al. 2013; Liu et al. 2016). Furthermore, additional layers of regulation might exist, for example at the post-transcriptional level (see below the section 'Translation and its regulation'), or by means of PTMs of TFs (see the section 'Protein phosphorylation in S. acidocaldarius'; Fig. 6). In this context, it is notable that phosphoproteomic studies have indicated the widespread occurrence of phosphorylation of TFs in S. acidocaldarius and Sa. solfataricus (Esser et al. 2012; Reimann et al. 2012). In S. acidocaldarius, phosphorylation has been shown to directly affect DNA binding of the Lrs14-type biofilm regulator AbfR1 (Li et al. 2017), or ligand interaction in the case of the acyl-CoA-responsive TetR-family regulator FadR (Maklad et al. 2020) (see the section 'Protein phosphorylation in S. acidocaldarius').
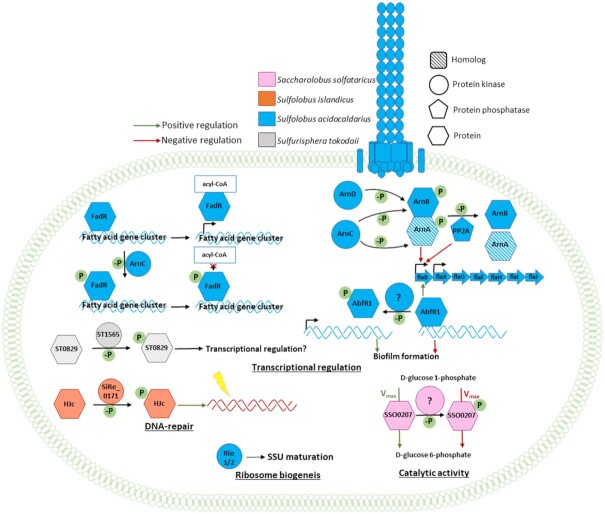
Figure 6.
Representation of known phosphorylation-based regulatory networks in different Sulfolobales strains (Sa. solfataricus in pink, Sa. islandicus in orange, S. acidocaldarius in blue and Sulfuri. tokodaii in gray) with their physiological function. From upper left to right: The FadR transcriptional regulator represses transcription of the fatty acid gene cluster and dissociates from the DNA upon binding to acyl-CoA. Phosphorylation of FadR by the ePK ArnC (Saci_1196) prevents acyl-CoA binding and thus hinders transcription of the gene cluster (Maklad et al. 2020). The archaellum regulatory network consists of the gene cluster arlBXGFHIJ (flaBXGFHIJ), which encodes the motility structure, the archaellum, and is under the control of two promoters, one upstream of arlB (flaB) being induced under starvation and one weak promoter upstream of arlX (flaX). The two negative regulators, ArnA (Saci_1210) and ArnB (Saci_1211), were shown to be phosphorylated by the ePKs ArnC and ArnD (Saci_1694) and dephosphorylated by the PP PP2A (Saci_0884). Deletion of the PP2A led to a hypermotile phenotype suggesting a negative influence on the gene cluster (Reimann et al. 2012; Hoffmann et al. 2017). The DNA-binding protein AbfR1 (Saci_0446) is a positive regulator of the arlB (flaB) promoter (Orell et al. 2013a). Phosphorylation of AbfR1 inhibits DNA binding and thus regulates biofilm formation and motility (Li et al. 2017). The FHA domain containing protein ST0829 was shown to interact and be phosphorylated by the ePK ST1565 indicating a role in transcription regulation (Duan and He 2011). The Holliday junction resolvase (Hjc) (SiRe_1431) is phosphorylated by the aPK SiRe_0171 facilitating DNA repair (Huang et al. 2019). The phosphohexomutase (SSO0207) exhibited a decreased Vmax value after being phosphorylated (Ray et al. 2005). The Rio kinases (Saci_0796 and Saci_0965) were shown to play a role in the ribosome maturation of the small subunit (Knüppel et al. 2018).
Translation and its regulation
Post-transcriptional regulatory mechanisms in the Sulfolobales
An alternative explanation for the compensation of the rather limited repertoire of TFs in Sulfolobales is the existence of gene regulatory mechanisms at alternative levels of information processing, such as the post-transcriptional level (Lemmens et al. 2019a). RNA-based regulation is supported by a widespread occurrence of small non-coding RNAs (ncRNAs) in Sa. solfataricus and, to a lesser extent, in S. acidocaldarius (Tang et al. 2005; Zago, Dennis and Omer 2005; Wurtzel et al. 2010; Cohen et al. 2016). More than 300 ncRNAs were identified in Sa. solfataricus, 60% of which are cis-acting antisense transcripts (Wurtzel et al. 2010). Possibly, these small RNAs assist in the stabilization of mRNA by RNA duplex formation, which is relevant given the thermophilic lifestyle of the organism (Gomes-Filho and Randau 2019). Nevertheless, given that most of these ncRNAs are conserved in closely related Sa. islandicus genomes (Reno et al. 2009), they likely have functional roles and these antisense ncRNAs may regulate translation in a similar manner as RNA silencing mechanisms in eukaryotes. Besides the observation that they are overrepresented in coding regions of genes involved in ion transport and metabolism (Wurtzel et al. 2010), the function of ncRNA-mediated regulation is still unclear as very few ncRNAs have been characterized thus far. A good example is RrrR in S. acidocaldarius, an antisense ncRNA that targets two mRNAs, including one that encodes a hypothetical membrane protein that was shown to influence biofilm formation (Orell et al. 2018) (see the section 'Regulation of biofilm processes'). In Sa. solfataricus, a small ncRNA has been shown to interact with the 3′-untranslated region (UTR) of its target mRNA in a phosphate-responsive gene regulatory process (Märtens et al. 2013). This is a logical regulatory site given that most transcripts in thermoacidophilic Crenarchaeota are leaderless and lack a 5′-UTR (Brenneis et al. 2007; Wurtzel et al. 2010), which is the preferred site of small RNA-mediated post-transcriptional regulation in bacteria.
There is a huge variation in the small RNAs found in transcriptomes of Sulfolobales; these can have lengths as small as 20–25 nucleotides (nts) or up to 500 nts (Wurtzel et al. 2010; Xu et al. 2012). Besides the antisense ncRNAs that have a classic regulatory role, Sa. solfataricus harbors transposon-associated ncRNAs involved in transposition, CRISPR-associated small RNAs (see the section 'CRISPR-Cas mediated viral defense in thermoacidophiles'), C/D box small nucleolar (sno) RNA that guide methylation sites in rRNAs and tRNAs (Zago, Dennis and Omer 2005), and small RNAs associated with TTSs (TTSaRNAs) (Zaramela et al. 2014). Different RNA-binding proteins act as chaperones of ncRNAs, such as members of the L7As/L30 protein family and the Sm superfamily. These proteins are not only involved in the biogenesis and functioning of small RNAs, but also in other aspects of processing of rRNA, tRNA and mRNA species (Gomes-Filho and Randau 2019).
Basal translation machinery in the Sulfolobales
Like replication and transcription, the machinery of translation in archaea shows striking similarities to those in eukaryotes. Studies of translation initiation have been mainly performed in Sa. solfataricus as a model for thermoacidophilic archaea (La Teana et al. 2013). Two different mechanisms have been discerned: (i) translation is initiated based on a canonical Shine–Dalgarno (SD)/anti-SD interaction analogous to bacteria at internal cistrons of polycistronic mRNAs or (ii) direct pairing of the start codon with the anticodon of the initiator methionine-tRNA occurs at leaderless transcripts devoid of a SD sequence (Benelli, Maone and Londei 2003). The latter mechanism is the most prevalent one, given the observation that most monocistronic mRNAs and the proximal cistrons of polycistronic mRNAs in Sulfolobales are leaderless, lacking SD-harboring 5′-UTRs (Brenneis et al. 2007; Wurtzel et al. 2010). The translation initiation machinery has a complexity reminiscent of their eukaryotic counterpart and consists of multiple translation initiation factors (IFs), some of which have eukaryotic but no bacterial homologs (La Teana et al. 2013). A crucial IF, especially for the translation initiation of leaderless transcripts, is aIF2, which is a heterotrimeric protein that forms a ternary complex with GTP and the methionine-loaded initiator tRNA, and binds the small ribosomal subunit. Other initiation factors are aIF1, aIF1a, aIF5b and aIF6. Despite the eukaryotic nature of the machinery, the operational steps in the translation initiation process in Sa. solfataricus resemble those in bacteria (La Teana et al. 2013).
POST-TRANSLATIONAL MODIFICATION BY REVERSIBLE PROTEIN PHOSPHORYLATION
All living cells, including thermoacidophiles, respond and adjust their cellular processes to a multitude of external and internal signals/cues. Information processing from gene to protein is also time and energy intensive. Therefore, post-translational-modifications (PTMs) are an efficient way to rapidly adjust protein function and coordinate the cellular response to changing needs. One of the best studied PTMs is the reversible protein phosphorylation mediated by protein kinases (PKs) and protein phosphatases (PPs) (Kennelly 2003, 2014; Esser et al. 2016; Papon and Stock 2019). Phosphoproteins were first found in the Euryarchaeon Halobacterium salinarum (Spudich and Stoeckenius 1980) but, despite this early report, little is still known about reversible phosphorylation in Archaea.
Two distinct phosphorylation systems exist. The two-component system (TCS) involves a histidine sensor kinase (HisK) and response regulator (RR); the covalent modification, i.e. addition of a phosphate group, takes place on histidine (His) and aspartate (Asp) residues, respectively (Galperin et al. 2018). It was originally thought that TCS were specific for Bacteria, but it has since been shown that TCS occurs in all three domains of life (Loomis, Shaulsky and Wang 1997; Kim and Forst 2001; Schaller, Shiu and Armitage 2011). The first TCS reported in Archaea was CheA and CheY in the Euryarchaeon Halobacterium salinarium (Rudolph et al. 1995). In thermoacidophiles, TCS have only been identified in the Thermoplasmata within the Euryarchaeota, although their function is still unknown (Galperin et al. 2018). In other archaeal phyla, i.e. the Crenarchaeota, the Korarchaeota and the Nanoarchaeota, TCSs are largely absent and the organisms solely rely on canonical Hanks-type protein kinases for signal transduction (Hanks 2003; Esser et al. 2016; Galperin et al. 2018). In these cases, autophosphorylation of the PK takes place on the amino acids Ser and Thr (eSTPKs) or tyrosine (Tyr). Often multiple PKs are interconnected, leading to a signal transduction cascade with a continuous hierarchical network structure. Hanks-type protein kinases can be broadly split into two groups, which have been well established in Archaea (Leonard, Aravind and Koonin 1998; Kennelly 2014; Esser et al. 2016; Hoffmann et al. 2017): the conventional Hanks-type protein kinases (ePKs) and the non-canonical, atypical Hanks-type protein kinases (aPKs). The largest group, the ePKs, all share a conserved catalytic domain consisting of twelve subdomains (Stancik et al. 2018). The other, smaller group, the aPKs, are distant members of the ePKs superfamily and share only some of the conserved subdomains (Esser et al. 2016). Although Tyr phosphorylation has been demonstrated, so far only eSTPKs (phosphorylation on Ser/Thr residues) have been identified and characterized in Archaea, with the responsible tyrosine kinases yet unknown (Smith, Kennelly and Potts 1997; Kennelly 2014).
The counterparts to PKs are PPs, which can remove the covalently linked phosphate residue, thereby making the process reversible. PPs can be classified into different subgroups, depending on their substrate specificity. In archaea, several phosphatases have been reported that differ in substrate specificity and cofactor requirement (Shi 2009). Ser/Thr phosphatases (PPPs) and Mg2+- or Mn2+-dependent protein phosphatases (PPM) act on pSer and/or pThr, whereas protein tyrosine phosphatases (PTPs) are active on pTyr (Shi 2009). Notably, dual activity of the PPM on pTyr and pSer/Thr was demonstrated in vitro in the thermoacidophile Thermoplasma volcanium (Dahche et al. 2009).
Among the thermoacidophilic archaea, the protein phosphorylation pathways of the Sulfolobales are the best characterized. When considering protein phosphorylation in the Sulfolobales, it is important to know which PKs and PPs have been characterized so far, which proteins are targeted, and, most importantly, what is the physiological and cellular impact of reversible protein phosphorylation. First, homology searches (BlastXP) were performed based on the PKs and PPs identified in S. acidocaldarius and Sa. solfataricus (Leng et al. 1995; Shi, Potts and Kennelly 1998; Lower, Bischoff and Kennelly 2000; Lower and Kennelly 2002, 2003; Lower, Potters and Kennelly 2004; Kennelly 2014; Ray et al. 2015),Sa. islandicus (Huang et al. 2017, 2019) and Sulf. tokodaii (Wang et al. 2010a) (Fig. 5). In surveying the Sulfolobales genomes, Sulf. tokodaii has 15 PKs, followed by S. acidocaldarius with 13 PKs, and Sa. islandicus and Sa. solfataricus both with 11 PKs. All strains harbor one typical ePK with an additional tetratricopeptide repeat (TPR-motif) that is known to mediate protein–protein interactions. In Sa. islandicus, the ePK was shown to act as a master PK, phosphorylating other PKs in vivo. Also, the homolog in S. acidocaldarius (ArnC) phosphorylates a variety of different target enzymes (Hoffmann et al. 2017; Huang et al. 2017; Knüppel et al. 2018; Maklad et al. 2020). Several PKs have an additional trans-membrane domain, implicating membrane-bound localization. In all Sulfolobales genomes, two Rio-like PKs (aPKs) are present. Rio kinases are an ancient conserved family that can be found in all three domains of life and are known to play a role in ribosome biogenesis (Esser and Siebers 2013; LaRonde 2014). All Sulfolobales are known to harbor a set of Rio B and Rio 2 PKs, except S. acidocaldarius, where Rio 1 and Rio 2 can be found (Esser and Siebers 2013). Rio 1 and Rio 2 are quite similar, except that Rio 2 possesses an additional winged helix-turn-helix domain (wHTH), a structural motif enabling binding of DNA. Recently, further evidence for the role that Rio kinases play in the synthesis of the ribosomal small subunit in archaea was uncovered (Knüppel et al. 2018). Furthermore, one piD261-like aPK, of which SSOPK5 is the only one studied in the Sulfolobales, one ABC1-like and one AQ578 aPK, both putative, can be found in Sa. solfataricus, Sulf. tokodaii, Sa. islandicus and S. acidocaldarius (Fig. 5). Notably, in all of these genomes, only two PPs were found: one annotated as Tyr-phosphatase and one as Ser/Thr-phosphatase.
Figure 5.
Predicted protein kinase and protein phosphatase homologs in the four different Sulfolobales species: S. acidocaldarius, Sa. islandicus, Sa. solfataricus and Sulfuri. tokodaii. Depicted are the different canonical and non-canonical Hanks-type protein kinases and protein phosphatases with their correspondent domain structure. (Lower Bischoff and Kennelly 2000; Lower and Kennelly 2002, 2003; Lower, Potters and Kennelly 2004; Haile and Kennelly 2011; Esser and Siebers 2013; Ray et al. 2015; Esser et al. 2016; Hoffmann et al. 2017; Huang et al. 2017).
Protein phosphorylation in Sa. solfataricus
The first evidence of protein phosphorylation in the Sulfolobales was reported in Sa. solfataricus (Kennelly et al. 1993). PP activity was detected in soluble extracts of Sa. solfataricus using 32P-casein as the substrate and allowed the isolation of the PP from the soluble fraction. Since no activity could be detected with p-Tyr labeled substrates, it was concluded that the PP was a Ser/Thr phosphatase. The amino acid sequence of the SSO-PP was similar to the eukaryotic PP1/2A/2B superfamily (Leng et al. 1995).
Sa. solfataricus was also the first Sulfolobales species in which PKs were characterized and now include four ePKs and one aPK (SSO-PK1, SSo-PK2, SSO-PK3, SSO-PK4 and SSO-PK5). Their activity on ‘artificial’ substrates, such as histone, myelin basic protein, p53, casein, reduced carboxyamidomethylated and maleylated lysozyme, in addition to their cofactor dependence, and inhibition by typical ePK inhibitors, have been analyzed (Lower Bischoff and Kennelly 2000; Lower and Kennelly 2002, 2003; Lower, Potters and Kennelly 2004; Haile and Kennelly 2011; Kennelly 2014; Ray et al. 2015; Esser et al. 2016). SSO-PK4 (encoded by SSO3182), like its eukaryotic homologs, was shown to phosphorylate the Sa. solfataricus eukaryotic translation initiation factor (eIF2α) homolog, aIF2α, in vitro, but not on the conserved phosphorylation sites known for eIF2α (Ray et al. 2015). Inhibition was observed in the presence of 3′,5′-cAMP in vitro, whereas a concentration-dependent activation occurred in the presence of oxidized CoA, an indicator of oxidative stress in archaea. Additionally, the aPK of the pID261/Bud32 kinase family (SSO-PK5) was characterized (Haile and Kennelly 2011). SSO-PK5 (encoded by SSO0433) phosphorylated artificial substrates, like p53 and casein, on Ser residues. Autophosphorylation of SSO-PK5 was shown to take place on both Ser and Thr residues and activation was observed in the presence of ADP-ribose (Haile and Kennelly 2011). Phosphohexomutase (SSO0207), first identified in tryptic digests, was shown to be phosphorylated. Site-directed mutagenesis (S309D) was used to mimic the presence of a phosphoryl group, which drastically decreased the Vmax value of the enzyme. Therefore, it was suggested that protein phosphorylation is used in vivo to regulate the phosphohexomutase activity, but the corresponding PKs are yet unknown (Fig. 6) (Ray et al. 2005). This appears to be the first report on regulation of central metabolic enzymes by reversible protein phosphorylation in not only the Sulfolobales, but Archaea in general.
In 2012, Esser et al. performed the first phosphoproteome study in Sa. solfataricus where they compared the phosphoproteome of cells grown on d-glucose vs cells grown on tryptone (Esser et al. 2012; Dopson 2016). Using a precursor acquisition independent from ion count (PAcIFIC) approach, 540 phosphoproteins in 21 out of 26 arCOGs were found, highlighting the importance of regulation by reversible protein phosphorylation in Sa. solfataricus. Interestingly, the phosphorylation profile was dependent on the respective carbon source and a high amount of Tyr phosphorylation (Ser/Thr/Tyr ratio of 26/21/54%) was detected. This was rather unexpected since, so far, no PKs with Tyr-phosphorylation activity had been reported. Due to the significant changes in the phosphoproteome in response to carbon source, it was concluded that reversible protein phosphorylation plays a major role in the regulation of central carbon metabolism (CCM) in Sa. solfataricus (Dopson 2016).
Protein phosphorylation in Sulf. tokodaii
Detailed characterization of the ePK STK_15650, which comprises all signatures of a canonical Hanks-type kinase in Sulf. tokodaii, showed that the important catalytic residues are all located in, or close to, the major functional domains of eSTKs (Wang et al. 2010a). It was the first time in Archaea that the interaction between an ePK and a target protein was characterized (Wang et al. 2010a). The interaction partner that is phosphorylated is the forkhead-associated (FHA) domain-containing protein (STK_00829). FHA domains are known to act as phosphorylation-dependent protein–protein interaction modules that can bind to pThr residues in their targets (England et al. 2009). Additionally, specific interactions of both proteins were demonstrated in vivo. Important residues for the protein–protein interaction were identified. It was proposed that STK_00829 might be a transcriptional regulator and, therefore, phosphorylation might play a role in transcriptional regulation in Archaea (Fig. 6) (Wang et al. 2010a; Duan and He 2011). Subsequently, the homolog of STK_00829 ArnA was shown to be a repressor of archaellum expression and part of the archaellum regulatory network in S. acidocaldarius (see Table 2) (Reimann et al. 2012).
Protein phosphorylation in Sa. islandicus
Autophosphorylation and cross-phosphorylation activities of the eleven PKs (three ePKs and eight aPKs) from S. islandicus REY15A revealed insights into the hierarchy of regulatory networks (Huang et al. 2017). The seven PKs (SiRe_0101KD, SiRe_0171, SiRe_0181, SiRe_1570, SiRe_1810, SiRe_2030 and SiRe_2056KD) exhibited autophosphorylation activities, with the highest activity shown for the ePK SiRe_2056KD. Dephosphorylation assays revealed that autophosphorylation mainly proceeds on Ser/Thr residues (Huang et al. 2017). To gain more insight into the cross-phosphorylation and cross-talk among the PKs, inactive ePKs were generated. Among the ePKs, SiRe_2030 and the truncated SiRe_2056KD were most active on phosphorylation of the other PKs. Next, to address the importance of ePKs, the effect of PK overexpression was evaluated. Only for SiRe_1531 and SiRe_2056 was there an obvious phenotype detectable, i.e. growth retardation (Huang et al. 2017). Therefore, SiRe_2056 and SiRe_2030 are the master PKs, and SiRe_0101 is an accessory kinase at the apex of the phosphorylation hierarchy in Sa. islandicus that transduced the signal toward the other substrate kinases. However, more physiological information is needed to elucidate the complex signaling pathways in Sa. islandicus (Huang et al. 2017).
With regard to targets in Sa. islandicus REY15A, phosphorylation of a conserved Holliday junction resolvase (Hjc), an enzyme employed in homologous recombination repair (HRR), by the PKs SiRe_0171 (Rio 1-like aPK), SiRe_2030 and SiRe_2056 (ePKs) was investigated (Huang et al. 2019). These PKs phosphorylated different residues in vitro and the analysis of the respective phosphorylation-mimic mutants revealed that the phosphorylation of S34 (phosphorylated by SiRe_0171) and S9 (phosphorylated by SiRe_2030) have a strong impact on Hjc activity. To elucidate the in vivo significance of Hjc protein phosphorylation, strains expressing the different phosphorylation-mimic mutants were tested for their sensitivity toward DNA damaging agents. The strain expressing S34E (mimicking phosphorylated Hjc) was less sensitive toward high doses of DNA-damaging agents (i.e. UV or cisplatin) indicating a higher DNA repair capability. In addition, deletion of the respective Rio 1 homolog SiRe_0171 (and thus preventing phosphorylation of the S34 residue) resulted in a strain with higher sensitivity toward DNA damaging agents, thus indicating that phosphorylation of S34 in Hjc enhances the DNA repair capability (Fig. 6).
Protein phosphorylation in S. acidocaldarius
Protein phosphorylation in S. acidocaldarius was initially identified when several proteins were found to be phosphorylated in the presence of [γ-32]ATP in vivo (Skórko 1984). Three PKs, ArnC, ArnD and ArnS, are involved in the regulation of the best studied complex signal transduction system in archaea, the archaellum regulatory network (Arn) (Fig. 6). The genes encoding for the archaeal motility structure, the archaellum, are arranged in an operon consisting of seven genes with two promoters. The main promoter upstream of the gene arlB (f. flaB) was induced upon starvation, whereas the second promoter upstream of arlX (f. flaX) revealed weak constitutive activity (Lassak et al. 2012). The transcriptional regulators, ArnA and ArnB, repress arlB (f. flaB) expression. arnA harbors a zincfinger (ZnF) and a forkhead-associated (FHA) domain and is a homolog to the previously identified regulator STK_00829 in Sulf. tokodaii, while ArnB possesses a von Willebrand type A domain (vWA). Deletion of either one of the two genes showed a hypermotile phenotype in vivo and protein levels of the archaellin ArlB (f. FlaB) were strongly enhanced compared with the wild-type strain (Reimann et al. 2012). The ePKs ArnC (Saci_1193), ArnD (Saci_1694) and ArnS (Saci_1181), and the Ser/Thr PP (Saci_0884) (Saci_PP2A), regulate the archaellum at the post-translational level by reversible phosphorylation. ArnC is able to phosphorylate both regulators ArnA and ArnB and deletion of the ePK resulted in reduced motility in vivo. In contrast, ArnD is only able to phosphorylate ArnB and its deletion resulted in a hypermotile phenotype. These divergent effects on motility suggested that the two PKs have different roles in the regulatory network. Both regulators were dephosphorylated by the addition of the Saci_PP2A (Reimann et al. 2012). Since ArnA and ArnB interact with each other in a phosphorylation-dependent manner, protein phosphorylation seems to be the key for their regulatory function in the Arn (Reimann et al. 2012). The deletion of the starvation induced ePK, ArnS, which is also located close to the archaellum operon, resulted in reduced motility, indicating an essential role of this ePK as well (Haurat et al. 2017). Finally, the deletion of the Ser/Thr PP PPP2A also revealed a hypermotile phenotype, suggesting a negative regulation, although the respective relay mechanism and target protein(s) are still unknown. Another player of the Arn is the transcriptional regulator of the Lrs14 family, the archaeal biofilm regulator 1 (AbfR1) (Orell et al. 2013a; Liu et al. 2017). AbfR1 binds to its own promoter, as well as the arlB, arlX (f. flaB, flaX, respectively) promoter, and has non-specific DNA-binding activity suggesting a general chromatin structuring function. In the non-phosphorylated state, it binds to DNA and increases motility by expression of the archaellum operon and negatively regulates biofilm formation by decreasing the production of extracellular polymeric substance (EPS). Phosphorylation of two residues, S87 and Y84, in the wHTH domain impair stable protein-DNA contacts in vivo (Li et al. 2017). Thus, AbfR1 phosphorylation promotes the transition from a motile, planktonic growth to a sessile lifestyle in S. acidocaldarius (Fig. 6). The PK(s) involved in phosphorylation of AbfR1 are still unknown.
The two Rio kinases, Rio 1 and Rio 2, of S. acidocaldarius exhibited ATP hydrolysis activity in a concentration-dependent manner (Knüppel et al. 2018) and are non-essential in S. acidocaldarius, as demonstrated by creation of single deletion mutants (Hoffmann et al. 2017). Both PPs in S. acidocaldarius were characterized in detail and the physiological role was addressed by comparison of the phosphoproteomes of the parent strain MW001 and the two PPs deletion mutants ΔSaci_ptp, ΔSaci_pp2a (Reimann et al. 2013). Saci-PTP is a dual-specific phosphatase (active with pSer/pThr and pTyr), whereas Saci-PP2A exhibited specific pSer/pThr activity and could be inhibited by okadaic acid (Reimann et al. 2013). The study revealed major differences in the phosphorylation and gene expression patterns of the two deletion strains, suggesting important roles for both phosphatases in signal transduction pathways (Reimann et al. 2013). Interestingly, the ratio of pSer/pThr/pTyr, varied slightly between the different strains and revealed a high amount of pTyr, also seen in Sa. solfataricus (Reimann et al. 2013; Dopson 2016).
A very recent study highlighted the physiological effect of protein phosphorylation on central metabolism (Fig. 6) (Maklad et al. 2020). The transcription factor, FadRSa, represses a 30-kb gene cluster encoding enzymes involved in the lipid/fatty acid degradation in S. acidocaldarius (Wang et al. 2019c). Acyl-CoAs act as inducers leading to DNA dissociation of FadRSa and thus transcription of the gene cluster. FadRSa repressed the gene cluster by only four binding sites; further studies revealed that FadRSa might have an additional function in organization of local chromatin architecture, as indicated by the interplay of FadRSa with the chromosome structuring factor coalescing (Takemata, Samson and Bell 2019). In previous phosphoproteome studies, FadRSa was found to be phosphorylated on three different residues (Y133, T134 and T135) (Reimann et al. 2013). Notably, all three residues are located within the binding pocket of acyl-CoA. In vitro phosphorylation studies with different PKs confirmed phosphorylation of FadR by ArnC and Saci_1041, whereas ArnD and Saci_0965 were not active. The constructed triple phosphomimetic mutant FadRSa (Y133D-T134E-T135E) was less sensitive for acyl-CoA. This implies that phosphorylation of FadRSa may act as an additional control mechanism that keeps the gene cluster in a condensed state and allows transcriptional repression of lipid/fatty acid metabolism in the presence of acyl-CoAs in S. acidocaldarius (Maklad et al. 2020).
CELL CYCLE AND MODES OF GROWTH
The planktonic lifestyle
The archaellum and other surface structures
Archaea form a multitude of different surface structures that play important roles in diverse processes, such as motility, adhesion, biofilm formation, DNA transfer and probably many more (Chaudhury, Quax and Albers 2018). Many of these structures are type IV pili or type IV pili-like, e.g. the archaellum, the motility structure of archaea (Albers and Pohlschröder 2009; Jarrell and Albers 2012; Makarova, Koonin and Albers 2016). Archaeal type IV pili are similar to those of bacteria in that both pilin precursor proteins exhibit a class III signal peptide at their N-terminus, which is processed by a type IV prepilin signal peptidase (PibD in Sulfolobales and Haloarchaea) (Albers, Szabó and Driessen 2003; Tripepi, Imam and Pohlschröder 2010). Only after removal of the signal peptide can the pilins be assembled into the pilus filament by the assembly system which is formed by an integral membrane protein and an ATPase (Fig. 7). The basic assembly mechanism of the archaellum resembles that of the type IV pilus. However, the associated accessory proteins (ArlFGH) enable the archaellum filament to rotate and therefore propel the cells forward (Jarrell and Albers 2012; Albers and Jarrell 2015).
Figure 7.
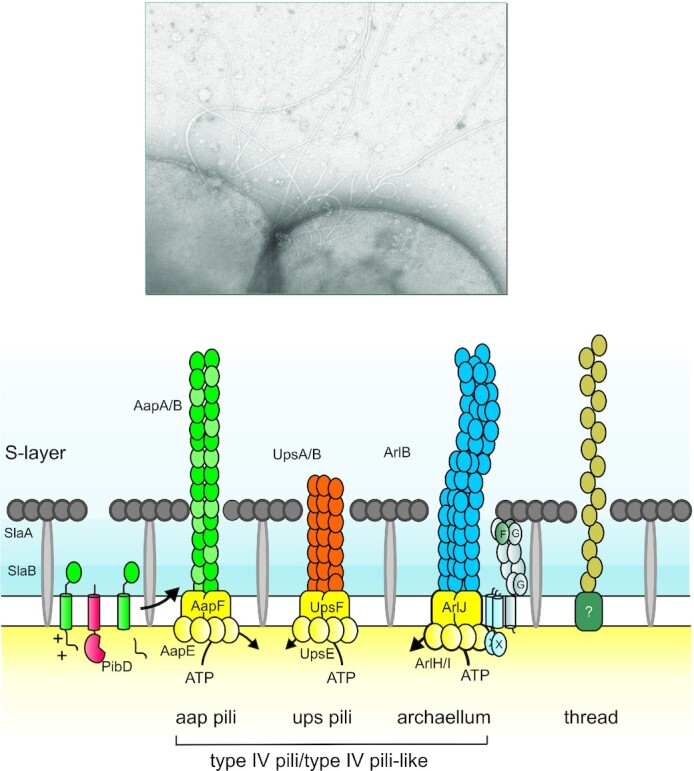
Archaeal cell surface structures involved in planktonic and biofilm growth. Upper image: electronic microscopy image from a S. acidocaldarius cell where archaellum and pilus can be seen. The upper image is reproduced from Albers and Meyer 2011. Lower image: schematic model with all proposed cell surface appendages in the Sulfolobales: the Aap pili (archaeal adhesive pili), the Ups pili (UV-induced pili), the archaellum and the threads. Also depicted is the S-layer and its proteins, SlaA and SlaB.
Among the Sulfolobales, S. acidocaldarius is the best studied with respect to surface structures as it exhibits a variety of them (Fig. 7 EM, Model): the Ups pili (UV-induced pili), the Aap pili (archaeal adhesive pili), the threads and the archaellum (Fröls et al. 2008; Henche et al. 2012a,b; Tsai et al. 2020). The threads, the only non-type IV pilus structure on the S. acidocaldarius surface, are formed by unknown proteins, but are used as binding sites for viruses (see the section 'Viruses and CRISPR systems of thermoacidophiles') (Hartman et al. 2019). The Ups pili are assembled after DNA double-strand breakages and lead to species-specific cell aggregation. During aggregation DNA is exchanged and subsequently used for DNA repair by homologous recombination (Fröls et al. 2007; van Wolferen et al. 2020). The species specificity is ensured by binding of the pilin subunits to the N-glycan trees on the S-layer protein that differ among species like Sa. solfataricus, Sulf. tokodaii and S. acidocaldarius (see the section 'S-layer') (van Wolferen et al. 2020). The Aap pili are important for adhesion of S. acidocaldarius to surfaces and biofilm formation (see the section 'Biofilms').
Biofilms
Biofilms are the most common form of microbial life. They consist of cells that are attached to a surface (which may or may not serve as a source of nutrients for the microorganism), embedded in extracellular polymeric substances (EPS) that form a matrix produced by the microbial population. This matrix consists of different types of polymers: lipids, polysaccharides, extracellular nucleic acids and proteins (Fröls 2013; van Wolferen, Orell and Albers 2018). Biofilms can also be formed at air-liquid and liquid-solid interfaces.
Biofilms are dynamic communities where cells can leave the biofilm structure and swim free in a planktonic lifestyle or attach to another surface to colonize it (Koechler et al. 2015). The first biofilm described for archaea was that of the extremely thermophilic euryarchaeon Thermococcus litoralis (Rinker and Kelly 1996). Similarly to bacterial biofilms, biofilms protect archaea against diverse kinds of stress, such as changes in pH, desiccation, UV radiation, and high salt and metal concentrations (Laplagia and Hartzell 1997; Fröls 2013). Biofilms also provide an advantage to cells because they form a microenvironment where they share nutrients, water channels, etc. (Petrova and Sauer 2012). In Sulfolobales species, stress factors like changes in pH and temperature induce biofilm formation (Koerdt et al. 2010), suggesting a role in protection against unfavorable conditions. A method to grow Sulfolobales biofilms in microtitrate plates has been standardized (Koerdt et al. 2010, 2011). The method consists of growing the species in Brock media pH 3 in microtiter plates covered with a gas permeable membrane at ∼75°C for 3–6 days without agitation. Using this method, biofilm from three Sulfolobales species were studied (Sa. solfataricus, S. acidocaldarius and Sulf. tokodaii). Also, some Sulfolobales have been studied in the context of acid mine drainage (AMD) biofilms, for example Sulfura. metallicus (Zhang et al. 2015b, 2019b) and Acidianus spp. (Zhang et al.2015b) on elemental sulfur and pyrite.
Biofilm formation process
There are three stages in biofilm formation: attachment of cells to the surface, formation of microcolonies and biofilm maturation, and finally, dispersion. In each of these stages, different structures are involved, and diverse morphologies had been described for Sulfolobales species (summarized in Table 3). The first stage for biofilm formation is the attachment of cells to a surface. Different cell surface structures play a role in this process, such as type IV pili, in the later stages, the archaellum, and depending on the archaeal species, hami and fimbria are involved (van Wolferen, Orell and Albers 2018). Saccharolobus solfataricus mutants that lack ArlB (formerly FlaB), the structural component of the archaellum, or either one of the Ups pili components, are defective in adhesion to surfaces like glass or pyrite (Zolghadr et al. 2010). Likewise, the deletion of Ups pili lead to less biofilm formation after 3 days of growth (Koerdt et al. 2010). The Aap pili are absent in Sa. solfataricus but present in S. acidocaldarius and, along with the Ups pili, are important for surface adhesion (Henche et al. 2012b). However, the archaellum does not play a role in adhesion in S. acidocaldarius, but seems to be involved in biofilm maturation (Henche et al. 2012b).
Table 3.
Biofilms of the Sulfolobales.
| Organism | Adhesion | Biofilm morphology | EPS components | Dispersion | References |
|---|---|---|---|---|---|
| Sa. solfataricus | Archaella and Ups pili | Carpet-like structure, low density of cells (20–30 mm thick) | Glucose, galactose, mannose and N-acetyl-glucosamine residues | Unknown | (Koerdt et al. 2010, 2012) |
| S. acidocaldarius | Aap and Ups pili | Dense biofilm with tower-like structures (25–35 mm thick) | Glucose, galactose, mannose and N-acetyl-glucosamine residues, eDNA and proteins | Depends on archaella | (Koerdt et al. 2010, 2012; Henche et al. 2012b) |
| Sulfuri. tokodaii | Unknown | Carpet-like structure with towers (25–35 mm thick) | Glucose, galactose, mannose and N-acetyl-glucosamine residues | Unknown | (Koerdt et al. 2010) |
| Sa. metallicus | Unknown | Micro- and macrocolonies on elemental sulfur | Galactose, mannose and N-acetyl-glucosamine residues, eDNA and proteins | Unknown | (Zhang et al. 2015a) |
Once attached to the surface, cells start to divide and produce EPS. In S. acidocaldarius, microcolony formation was seen after 36 h of inoculation. In Sa. solfataricus, S. acidocaldaricus and Sulfura. tokodaii during the first 3 days of growth, the secreted exopolysaccharides residues were mainly glucose and mannose, while from day five onward galactose and N-acetyl glucosamine were predominant (Koerdt et al. 2010). The complete path of exopolysaccharides biosynthesis is still unknown. However, an α-mannosidase encoded by SSO3006 in Sa. solfataricus affects EPS production (Koerdt et al. 2012). The mutant strain Sa. solfataricus PBL2025, which lacks a 50 kB region including SSO3006, produced more EPS than the wild-type strain and reverted back to the wild-type phenotype when complemented with SSO3006 (Koerdt et al. 2012). In Sulfura. metallicus DSM 6482(T), biofilm can form on elemental sulfur with microcolonies and cells clusters of up to 100 µm in diameter. This biofilm was rich in proteins and nucleic acids, in contrast to capsular EPS from planktonic cells, where the EPS mainly contained carbohydrates and proteins (Zhang et al. 2015a).
Mature biofilms formed by S. acidocaldarius, Sa. solfataricus and Sulf. tokodaii are morphologically different from each other. Saccharolobus solfataricus forms biofilms with a carpet-like structure, with 20–30 µm thick covering the whole surface but with a low density of cells. Sulf. tokodaii forms biofilms of 25–35 µm thick and also exhibits a carpet-like structure, but with high cell density and, occasionally, cell aggregates. Finally, S. acidocaldarius forms 25–35 µm thick biofilms that contained a high density of cells and large aggregates, forming towering structures above the surface of attached cells (Koerdt et al. 2010). Deletion mutant studies in S. acidocaldarius showed that the Ups pili and the Aap pili have profound impacts on the morphology of the biofilms (Henche et al. 2012b). While deletion of the Aap pilus led to a dense biofilm that was thinner than the wild type, the deletion of the Ups pili led to large aggregates of cells within a ‘fluffy’ biofilm characterized by a single dense layer of cells at the surface. Clearly, both pili play an essential role in the optimal layering and distancing of the cells in the wild-type biofilm (Henche et al. 2012b). Cell–cell connections were also seen extensively for S. acidocaldarius and Sulf. tokodaii, and to a lesser degree in Sa. solfataricus (Koerdt et al. 2010). Besides exopolysaccharides, proteins and extracellular (eDNA) can also be found in S. acidocaldarius biofilms. Composition analyses revealed several enzyme activities in EPS extracts, but most of them were cytoplasmic proteins (Jachlewski et al. 2015), probably derived from cell lyses as these species secrete only small amounts of proteins (Ellen, Albers and Driessen 2010).
Biofilm maturation was followed for several days in S. acidocaldarius (Fig. 8, biofilm). From days 3 to 6, an increase in cell density was observed, and in days 7 and 8 dispersion of the cells was seen (Henche et al. 2012b; Koerdt et al. 2012). In a ΔarlJ mutant, however, cell dispersion from the biofilm was decreased, showing that the archaellum is important for cells to leave the biofilm (Henche et al. 2012b). It is unknown what triggers dispersion of cells, but this process is important for colonizing other sites along the surface.
Figure 8.
Confocal laser microscopy images from static biofilm from S. acidocaldarius in days 3 to 7 of growth. Cells (DNA) stained with 4′,6-diamidino-2-phenylindole (DAPI; blue); extracellular glucose and mannose residues stained with fluorescently labeled concanavalin A (conA; green); and N-acetyl-d-glucosamine residues stained with fluorescently labeled lectin IB4 (yellow). Scale bars: 20 μm. Reproduced from Koerdt et al.2010.
Regulation of biofilm processes
The biofilm and planktonic lifestyles differ, as inferred from transcriptomic and proteomic studies comparing both populations (Koerdt et al. 2011), with the change from one to another depending on different regulators. In bacteria, it is known that secondary messenger and quorum sensing mechanisms are important for the regulation of biofilm formation, however these have not been described for the Sulfolobales. Nonetheless, high throughput proteomics and transcriptomics allowed for a first glance at the factors that might be involved in biofilm formation in three Sulfolobales species and a species-specific response was found (Koerdt et al. 2011). Among the common differentially regulated proteins were the archaea-specific Lrs14-like regulators (Leucine-responsive Regulator of Sulfolobus) (Koerdt et al. 2011). Later, deletion mutants confirmed that some of these Lrs14-like proteins are involved in biofilm regulation in S. acidocaldarius (see the section 'Regulatory transcription factors in Sulfolobales'; Table 2). Knockout mutants for Saci_1223 were impaired in biofilm formation, suggesting that this regulator promotes biofilm formation (Orell et al. 2013a). Furthermore, the deletion mutant of Saci_0446 produced more EPS than the wild type, and also showed a non-motile phenotype, where expression levels of the archaellum were downregulated and expression levels of Aap were increased. Therefore, it is thought to act as repressor of biofilm formation and named AbfR1 for Archaeal Biofilm Regulator 1 (see signal transduction in the section 'Post-translational modification by reversible protein phosphorylation'; Fig. 5) (Orell et al. 2013a). AbfR1 functions by stimulating motility through the induction of the archaellum by binding to the arlB promoter region, and repressing EPS production. Finally, detailed studies have shown that AbfR1 is also regulated by phosphorylation through unknown mechanisms, and it cannot bind DNA when phosphorylated (Li et al. 2017).
Some non-coding RNAs (ncRNA) also regulate biofilm formation. Sequencing of ncRNAs expressed in planktonic and biofilm cells was performed, and 29 ncRNA were differentially regulated in the latter (Orell et al. 2018). One in particular, ncRNA239, named RrrR (RNAse-resistant RNA), was abundant in planktonic cells but further upregulated in the biofilm. Moreover, deletion of this ncRNA led to impairment of biofilm formation. RrrR is a double stranded ncRNA, located in the intergenic region between Saci_1004 and Saci_1005. The sense transcript of this RNA interacts with RNA-binding Lsm proteins, and its antisense RNA binds two mRNAs. The antisense transcript of this RrrR seems to stabilize the sense transcript (Orell et al. 2018).
Polyphosphates (PolyP) are polymers of orthophosphate with roles in many cellular functions, including bacterial biofilm formation and related phenomena (Rashid et al. 2000; Shi, Rao and Kornberg 2004; Grillo-Puertas et al. 2012; Drozd, Chandrashekhar and Rajashekara 2014; Albi and Serrano 2016). In Escherichiacoli, PolyP is involved in biofilm formation by triggering type II autoinducers (AI-II) synthesis in the stationary phase of growth through PolyP degradation (Grillo-Puertas et al. 2012). In archaea, experiments in Sa. solfataricus and S. acidocaldarius demonstrated that this polymer is also involved in biofilm formation, adhesion and motility (unpublished results), although the mechanism is still unclear.
Cell envelope of Sulfolobales
S-layer
The architecture of archaeal cell envelopes can be very diverse (Albers and Meyer 2011; Klingl, Pickl and Flechsler 2019). However, studied Sulfolobales species mainly have a cytoplasmic membrane surrounded by a proteinaceous coat, called the S-layer. In contrast to other archaea, which also have an S-layer as the main cell wall component, most Sulfolobales have two proteins that form the S-layer: SlaB, which is the membrane anchor, and SlaA forming the outer layer on top of the cell (Grogan 1996; Veith et al. 2009). As S-layers are ordered in 2D lattices, they are excellent targets for structural studies. In 1982, Amos and colleagues used electron microscopy to study the structure of the S. acidocaldarius S-layer (Taylor, Deatherage and Amos 1982) and related studies then showed that the architecture of the S-layers is species-specific (Prüschenk, Baumeister and Zillig 1987). The S. acidocaldarius S-layer was found to be arranged in a conserved lattice with p3 symmetry, with 4.5 nm triangular and 8 nm hexagonal pores (Taylor, Deatherage and Amos 1982). By differential solubilization, the SlaB subunits could be detached from the SlaA lattice, allowing cryo-EM to pinpoint the placement of both subunits in the S-layer lattice (Fig. 9) (Gambelli et al. 2019).
Figure 9.
Model of the Sulfolobales S-layer. (A) The Sulfolobales S-layer consists of the two protein subunits. SlaA dimers (red, orange, yellow) form the outer S-layer canopy. Each SlaA protein is predicted to be rich in β-strands. The SlaA dimer has a boomerang-like shape, the angle of which determines the S-layer unit cell size. SlaB trimers (gray) form the membrane anchors of the S-layer. Each SlaB is predicted to consist of an N-terminal transmembrane domain (TMD), a coiled-coil domain (CC) and two to three C-terminal β-sandwich domains (β). SlaA and SlaB proteins are highly glycosylated (green). (B) Electron microscopy image from negatively stained isolated S-layer from S. acidocaldarius. (C)SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) of the isolation of S-layer from S. acidocaldarius cells using detergent buffers. Lane 1: pellet after incubating the cells with the detergent buffer the first time, the second time (lane 2) and the third time (lane 3). SlaB is being successively washed off and in the last wash a pure SlaA prep is obtained. (D) Subtomogram average of fully assembled S-layer. (E) Subtomogram average of SlaB-depleted S-layer. (F) Difference map (pink) overlaid with the complete S-layer visualizes location of SlaB. (Scale bars, and C–E, 20 nm). (Figure adapted from Gambelli et al. 2019).
As the only cell wall component, the S-layer provides stability to the cell and it has long been assumed that the S-layer is an essential component for Sulfolobales cells. However, a transposon library screen in Sa. islandicus indicated that both S-layer proteins can be deleted (Zhang et al. 2018). When SlaB was deleted in Sa. islandicus, the cells were still able to assemble partial SlaA containing S-layer lattices. However, cells lacking SlaA were not only deformed and sometimes very large, but they also had an aberrant number of chromosomes, indicating that the coordinated assembly of the S-layer is important for cell division (Zhang et al. 2019a). Furthermore, the S-layer in Sulfolobales plays a role in anchoring other surface structures, such as the bindosome, involved in sugar binding and uptake (Zolghadr et al. 2011), and the archaellum, where it is absolutely essential for torque generation (Tsai et al. 2020). The S-layer also acts as a sieve and as a surface for recognition that is conveyed by extensive N-glycosylation (discussed below).
Glycosylation
Many of the extracellular proteins of Archaea are glycosylated (this can be either O-glycosylation or N-glycosylation). Whereas we know very little about how proteins are O-glycosylated, the N-glycosylation pathway has been deciphered in halophiles, methanogens and S. acidocaldarius (Jarrell et al. 2014). Many archaeal extracellular proteins have a so-called ST-linker, which is a stretch of many serine (Ser) and threonine (Thr) residues in a row (Albers et al. 2004). In the Halobacteria S-layer, these residues were found to be O-glycosylated (Lechner and Sumper 1987; Sumper et al. 1990) and, thus, it is thought that other extracellular proteins, like SlaB or the sugar-binding proteins that also have these ST-linkers, are also O-glycosylated.
A number of proteins have been reported to be glycosylated in the Sulfolobales: the S-layer proteins (Peyfoon et al. 2010), cytochromes (Zähringer et al. 2000), sugar- and peptide-binding proteins (Albers, Konings and Driessen 1999; Elferink et al. 2001; Gogliettino et al. 2010), pilins (Wang et al.2019a), the archaellum (Meyer, Birich and Albers 2015) and many hypothetical proteins (Palmieri et al. 2013). Whereas proteins in other archaea have different glycans or the composition of the glycan changes due to environmental conditions, the Sulfolobales have only one kind of glycan decorating all glycosylated proteins in one species. All determined N-glycans of the Sulfolobales are in their basic structure quite similar to the eukaryotic N-glycan, as the glycans always start with two N-acetylglucosamines and are branched (Fig. 10) (Zähringer et al. 2000; Palmieri et al. 2013; van Wolferen et al. 2020). Interestingly, they all contain a sulfated saccharide called 6-sulfoquinovose, a sugar otherwise only found in chloroplast and membranes of photosynthetic bacteria (Meyer et al. 2011). However, except for this basic setup, the N-glycans of the different Sulfolobus species are diverse, as different numbers of mannose and glucose sugars are added to create a species-specific sugar labeling of extracellular proteins (see Fig. 10). This is used for species-specific recognition during DNA exchange (Fröls et al. 2008). During the pilin-dependent DNA exchange observed after double strand DNA breakages, the Sulfolobales exchange homologous DNA, by a sugar-specific binding of the pilin subunit to the S-layer of the aggregating cells (van Wolferen et al. 2020). Although the differences are minor between the N-glycans of Sa. solfataricus, Sulf. tokodaii and S. acidocaldarius (see Fig. 10), this is enough to ensure that aggregation and subsequent DNA exchange only happens with the same species (van Wolferen et al. 2020).
Figure 10.
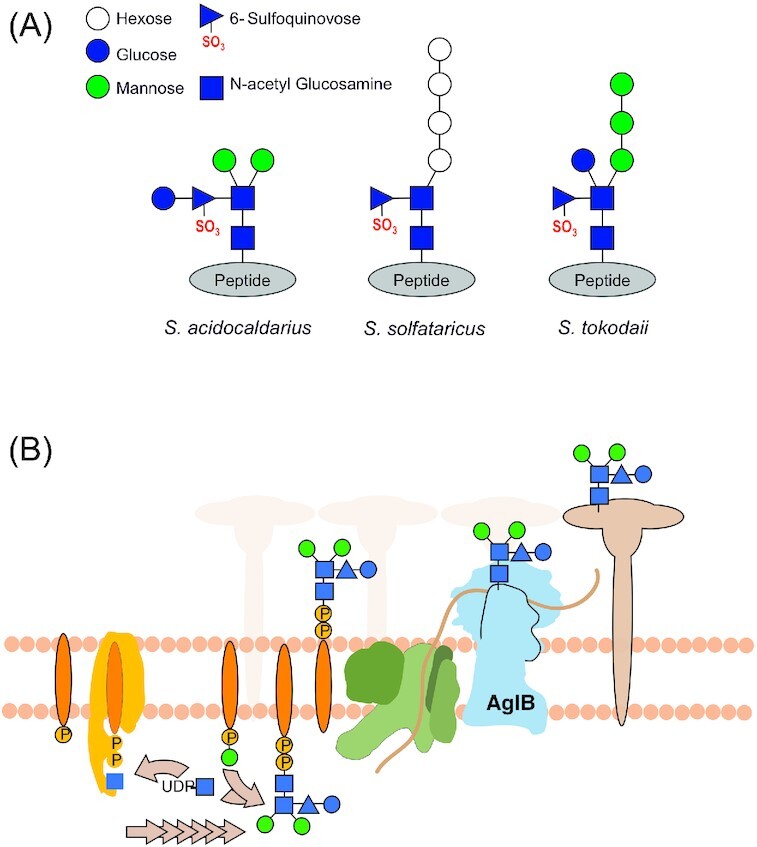
Glycosylation. (A) Comparison of the N-glycan trees of three different Sulfolobus/Saccharolobus species. (B) The current understanding of the N-glycosylation pathway in S. acidocaldarius. The N-glycan biosynthesis is initiated by adding nucleotide-activated monosaccharides sequentially to the lipid carrier dolichol phosphate on the cytoplasmic side of the membrane. The fully assembled dolichol pyrophosphate-linked N-glycan (hexasaccharide) is translocated across the membrane and then transferred by AlgB on the specific N-glycosylation sequons in secreted proteins. Sugar code is shown in (A).
In contrast to methanogens and halophiles (Jarrell et al. 2014), N-glycosylation seems to be essential in Sa. islandicus and S. acidocaldarius, as the enzyme attaching the N-glycan to the modified protein, the oligosaccharyl transferase AglB, cannot be deleted (Meyer and Albers 2014; Zhang et al. 2018). N-glycosylation is initiated by the assembly of the hexasaccharide N-glycan on short dolichol pyrophosphate carriers in S. acidocaldarius by AglH (Guan et al. 2016; Meyer, Shams-Eldin and Albers 2017). Deletion of the glycosyltransferase agl16 led to a removal of a terminal hexose from the N-glycan, whereas deletion of agl3, the sulfoquinovose synthase, reduced the N-glycan to a trisaccharide (Meyer et al. 2011, 2013). Both mutants had difficulties in adjusting to growth in media with elevated salt concentration. The N-glycosylation pathway of S. acidocaldarius is depicted in Fig. 10.
Sulfolobus cell division
The most investigated archaeal cell division system is the ESCRT-III-based system in the Sulfolobales. The proteins involved in cell division (Cdv) are organized in two groups. One group comprises the proteins CdvA, -B and -C organized in a gene cluster, while the other group consists of three CdvB paralogs located at different positions in the genome (Lindas et al. 2008; Samson et al. 2008, 2011). CdvA is the only protein of these two groups that does not share homologies to the eukaryotic ESCRT-system or any known protein family. CdvA can bind membranes and polymerizes into helical filaments on the outside of liposomes composed of tetraether lipids isolated from S. acidocaldarius. Interestingly, the presence of lipids was necessary for CdvA polymerization in vitro (Dobro et al. 2013). Transcriptional analysis of the cdv operon showed that cdvA is upregulated around 30 min before the cdvB genes are transcribed. Immunofluorescence with antibodies raised against CdvA displayed a ring-like localization pattern at midcell, but also outside the midcell region, before the genome was segregated (Lindas et al. 2008; Samson et al. 2011). These results suggest that CdvA is the earliest cell division protein localizing at the future division plane. However, it is unknown how CdvA is positioned. Remarkably, CdvA from Metallosphaera sedula, which was heterologously expressed and purified from E. coli, formed extended double-helical filaments that strongly interacted with the DNA of E. coli (Moriscot et al. 2011). This suggests that CdvA participates in the chromosome segregation processes or functions as an inverse nucleoid-occlusion type localization mechanism for the archaeal divisome (Caspi and Dekker 2018). However, the exact function of its DNA binding remains unknown. CdvA can recruit CdvB to preformed liposomes by direct interaction, resulting in extensively deformed vesicles. The β-strand forming C-terminal ESCRT-III-binding (E3B) peptide of CdvA interacts with the C-terminal incomplete winged-helix (wH) domain of CdvB. As such, the E3B peptide complements the ‘broken’ part of the CdvB wH domain, repairing the domain to a wH-related architecture. Importantly, the wH domain of CdvB, necessary for interaction with CdvA and the recruitment of CdvB to the membrane, is not present in the other three ESCRT-III homologs found in Sulfolobales species. Also, no interaction of the other CdvB paralogs with CdvA has been reported (Samson et al. 2011).
Interestingly, the CdvB paralogs can interact with themselves and the two other paralogs (Samson et al. 2008). A recent study implied that CdvB does not constrict in vivo but functions as a scaffold for CdvB1 and CdvB2 in S. acidocaldarius. The latter two proteins localize at the cell division plane, forming a ring-like structure after CdvB has already been recruited to midcell. Subsequent proteasomal degradation of CdvB allowed the formed CdvB1/B2 ring to constrict, completing cell division (Risa et al. 2019). Furthermore, CdvB1/B2 were shown to be involved in other membrane-remodeling events in the Sulfolobales, as they were found in secreted membrane vesicles (Ellen et al. 2009). Deletion of cdvB1 affected growth at 75°C only modestly and resulted in occasional failures in cell division. Remarkably, the ΔcdvB1 strain was severely impaired at growth at 65°C, suggesting a more important role under stress conditions (Pulschen et al. 2020). Additionally, deletion of cdvB2 showed a loss of cell division symmetry and was severely affected in cell growth (Pulschen et al. 2020). Analysis of cell division in the Sulfoscope (an inverted fluorescent microscope with a heated chamber) showed that the CdvB and CdvB1 rings were correctly assembled in the ΔcdvB2 strain, while after proteasome-mediated degradation of CdvB, CdvB1 rings were found at variable positions. This asymmetric division plane resulted in both ghost cells without DNA and cells with a double amount of DNA. This suggested that CdvB2 fixes the position of the ring after the loss of the CdvB scaffold. It was proposed that CdvB2 and CdvB1 are recruited to the cell center by the CdvB ring and that, after proteasome mediated degradation of CdvB, they hold the division ring in position while it constricts (Pulschen et al. 2020).
When cdvB3 was deleted, growth was delayed, colonies on plates were very small and cells had a division defect. In the cdvB3 deletion strain, CdvB was not localized at midcell in a ring-like structure. Instead, a diffuse CdvB signal was detected in the cytoplasm. Furthermore, CdvA was mislocalized and formed distinct foci at the cell membrane of enlarged cells (Yang and Driessen 2014). On the other hand, in Sa. islandicus, deletion of cdvB1 and cdvB2 was not successful. The reduction of intracellular CdvB1 protein levels led to a chain-like morphology of the cells, indicating that CdvB1 is important for the final abscission during cell division. In contrast to S. acidocaldarius,cdvB3 deletion in Sa. islandicus led to neither a cell division defect nor growth retardation. However, after infection with spindle-shaped virus 2 (STSV2), the ΔcdvB3 Sa. islandicus strain no longer developed viral buds on the cell surface (Liu et al. 2017). These findings indicate that although all Sulfolobus species have four ESCRT-III homologs, their specific function is different among the Sulfolobales. Yet, CdvB seems to be the key cell division protein of the four ESCRT-III homologs, providing a platform for later cell division proteins. Recently, ESCRT-I and -II homologs were identified in Lokiarchaeota. Though the function of these other ESCRT homologs is unknown, they may be involved in vesicle formation rather than in cell division as ESCRT-III homologs are also present in Lokiarchaeota (Caspi and Dekker 2018).
The third gene of the cdv operon encodes an ATPase (CdvC) that is homologous to the eukaryotic AAA-type (ATPase associated with various activities) ATPase Vps4 (Hobel et al. 2008). In HeLa cell lines, Vps4 was observed to localize at the cell center during cytokinesis simultaneously with ESCRT-III. It was shown that Vps4 is important for the dynamic assembly and disassembly of ESCRT-III filaments, leading to the formation and constriction of intercellular bridges. As such, the dynamic turnover rate of ESCRT-III was dependent on the Vps4 ATPase activity (Mierzwa et al. 2017; Caillat et al. 2019). With regard to archaea, it is assumed that CdvC also remodels CdvB filaments and is thereby responsible for membrane invagination.
Coinciding with the other two Cdv proteins, CdvC localizes in a band-like structure to midcell during cytokinesis (Lindas et al. 2008). Biochemical characterization of the CdvC protein from Sa. solfataricus showed (Samson et al. 2008; Caspi and Dekker 2018) that it assembles into single hexameric rings, comparable to Vps4 from Saccharomyces cerevisiae (Monroe et al. 2014). Furthermore, it was shown that CdvC directly interacts with CdvB (Obita et al. 2007). Important for this interaction is the N-terminal conserved MIT (microtubule interacting and trafficking) domain of CdvC that recognizes a C-terminal peptide motif of CdvB, called MIM2 (MIT domain interacting motif). The MIM2 domain is characterized by a proline-rich end part that is not present in the MIM2 domains of CdvB1 and CdvB2, resulting in a very weak binding affinity of the latter proteins to CdvC. Moreover, CdvB3 completely lacks the MIM2 domain and, hence, does not interact with CdvC (Samson et al. 2008; Caspi and Dekker 2018). This indicates that CdvB is the only ESCRT-III-like protein in S. acidocaldarius, recruiting CdvC to midcell. In contrast, a yeast two-hybrid assay with ESCRT-III homologs and CdvC from Sa. islandicus showed interaction between CdvC and CdvB1/B2 (Liu et al. 2017). Possibly, the lack of the proline-rich strand in CdvB1 and B2 is compensated by an additional interaction of the CdvB1/B2 protein with the MIT domain of CdvC (Caspi and Dekker 2018). Moreover, the different interaction pattern of CdvC with the respective CdvB paralogs in S. acidocaldarius and Sa. islandicus might explain their different influence on cytokinesis. However, the mechanism of how CdvC is involved in cell division is still unknown. See Fig. 11 for the model of cell division in S. acidocaldarius.
Figure 11.
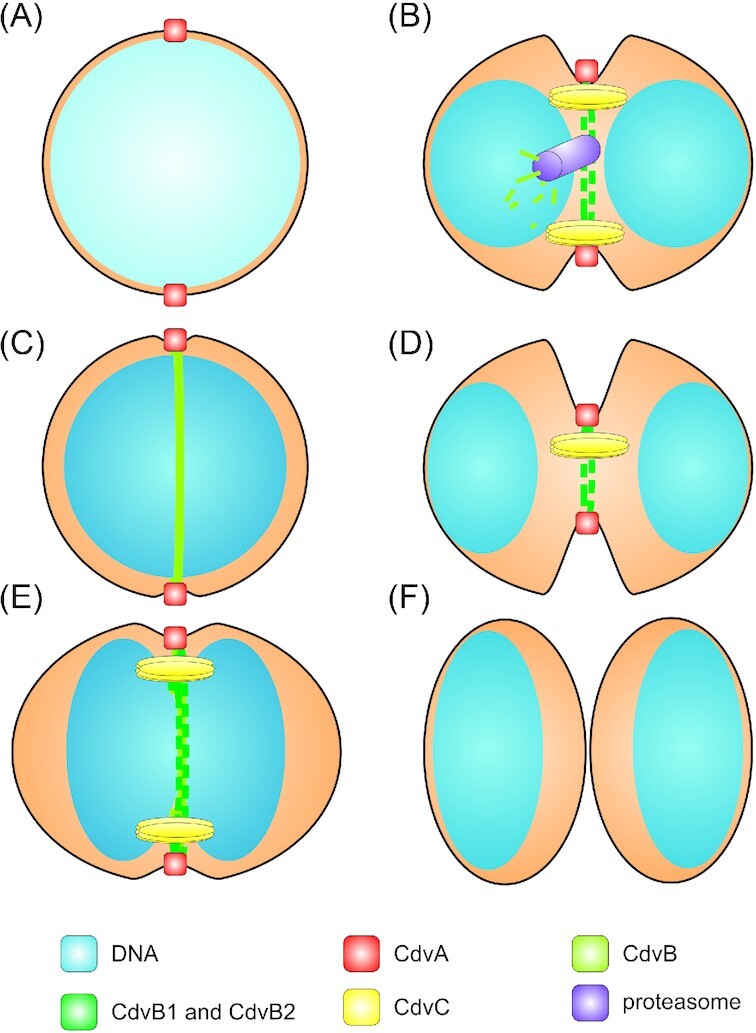
Schematic model of the cell division process in Sulfolobus acidocaldarius. (A)CdvA (red) is the first protein of the S. acidocaldarius cell division machinery that arrives at the future site of cell division, before DNA (light blue) segregation starts. (B) During nucleoid condensation (blue), CdvB (light green) forms a ring-like structure at midcell that is anchored to the membrane by CdvA. (C)CdvB provides a scaffold for CdvB1 and B2 (green) that are positioned at the cell center in ring-like structures. Additionally, CdvC (yellow), a homolog of the hexameric ATPase Vps4, localizes at the septum while nucleoid segregation and initial membrane invagination start. (D) After nucleoid segregation, the CdvB-ring undergoes proteasomal (purple) degradation. (E)Upon CdvB removal, CdvB1 and B2 constrict, leading to the final division of the cell. (F) Directly after fission, the new born cells have an oval shape that rapidly changes to the typical coccoid shape of S. acidocaldarius cells. CdvA and CdvC are organized in a ring-like structure as well. However, for a better overview the organization of both proteins at midcell was only indicated in the figure.
EXTREME THERMOACIDOPHILY AND STRESS RESPONSE
Thermoacidophily
Thermoacidophiles have specialized mechanisms to deal with stressors associated with their unusual temperature and pH optima and the fact that some thermal acidic environments are characterized by metal deposits. Although thermoacidophilic archaea largely do not have unique thermal or pH stress response mechanisms compared with thermophiles or acidophiles, some mechanisms play dual protective roles. Here, the major contributors to thermophily and acidophily will be discussed, with particular emphasis on where these stress responses overlap.
Because G–C bonds in DNA are more heat stable, it was initially hypothesized that thermophiles would have a higher G+C content then their mesophilic counterparts. Surprisingly, evaluation of DNA sequences from both thermophiles and mesophiles demonstrated that this was not the case (Tekaia, Yeramian and Dujon 2002; Wang, Susko and Roger 2006; Zeldovich, Berezovsky and Shakhnovich 2007). However, thermophiles have reduced mutation rates and have DNA repair mechanisms to combat biological damage at high temperatures. Though higher temperatures are correlated with an increase in mutations, base substitutions occur less frequently (Drake 2009), or at an equivalent rate (Grogan, Carver and Drake 2001) in thermophiles compared with mesophiles. Furthermore, DNA repair can also occur by homologous recombination and DNA uptake through type IV pili (reviewed in the section 'Cell cycle and modes of growth').
Even though DNA stability does not depend on G+C content as it does not correlate with higher growth temperatures, thermophiles do have codon/amino acid preferences to support enzyme thermal stability. For instance, thermophiles favor codons AGG and AGA over CGN for arginine (Farias and Bonato 2003; Singer and Hickey 2003) and ATA for isoleucine, instead of ATC or ATT (Singer and Hickey 2003). In addition, thermophiles have a nucleotide bias for the more heat-stable purines A and G; these bases are more frequently neighbors in thermophile compared with mesophile genomes (Zeldovich, Berezovsky and Shakhnovich 2007).
Not only do thermophiles have nucleotide preferences, but they also favor the use of certain amino acid residues. In general, thermophiles prefer amino acids that are charged or hydrophobic, while avoiding polar uncharged residues (Tekaia, Yeramian and Dujon 2002). Intuitively, thermophilic proteins also avoid heat labile amino acids, such as histidine, glutamine and threonine (Tekaia, Yeramian and Dujon 2002; Singer and Hickey 2003). Additionally, one study looking at the proteome amino acid composition of a variety of genomes discovered that the ratio (Glu + Lys)/(Gln + His) was lowest in mesophiles, higher in moderate thermophiles and highest in extreme thermophiles (Topt ≥ 70°C). This ratio was higher in thermostable chaperonins and DNA ligases in mesophiles compared with the rest of the proteome (Farias and Bonato 2003). Furthermore, another study demonstrated that the occurrence of certain amino acids (IVYWREL) correlated with microbial optimal growth temperature and, thus, was a predictor of thermophily (Zeldovich, Berezovsky and Shakhnovich 2007). Not only is the codon usage different in thermophiles but also their proteins averaged 283 aa compared with the 340 aa in mesophiles, possibly increasing the thermostability of the protein (Tekaia, Yeramian and Dujon 2002).
Another thermal adaptation is the use of a reverse DNA gyrase, a DNA topoisomerase that introduces positive supercoiling to DNA strands, thereby increasing its heat stability. When the reverse DNA gyrase was deleted in Thermococcus kodakarensis, this extreme thermophile was more sensitive to higher temperatures, although the mutation was not shown to be lethal (Atomi, Matsumi and Imanaka 2004). However, though multiple methods were used to mutate the gyrase genes in Sa. islandicus, no viable mutants were generated (Zhang et al. 2013b), indicating that this mutation can be lethal. In addition, a comparative genomic study showed that the only common gene in all extreme thermophiles sequenced at the time was a DNA reverse gyrase, lending further support to this enzyme's importance to thermophily (Forterre 2002).
In extreme thermophiles, the thermal stress response involves the thermosome (a molecular chaperone also referred to as the rosettasome, or archeaosome), a large HSP60-like protein complex originally discovered in S. shibitae (Trent, Osipiuk and Pinkau 1990, Trent et al. 1991). In archaea, the thermosome is composed of multiple α and β subunits that form ring structures with either 8- or 9-fold symmetry (Kagawa et al. 1995). A third γ subunit has been described in some extreme thermophiles that is downregulated during heat shock (Archibald, Logsdon and Doolittle 1999; Kagawa et al. 2003; Tachdjian and Kelly 2006). The thermosome is a Group II chaperonin, conserved among archaea (Kagawa et al. 1995), that shares structural and functional similarities to the bacterial Group I chaperonin GroEL complex (Trent et al. 1991). In both heat shock complexes, cytosolic proteins are sequestered inside an internal cavity where they can refold. As demonstrated for the Sa. solfataricus thermosome, complexes are formed with different subunit compositions in a temperature-dependent manner, which supposedly have different substrate specificities (Chaston et al. 2016). However, unlike the GroEL chaperonin of bacteria that has GroES to act as a lid for the complex, the thermosome does not have a separate subunit cap to close its internal cavity. Instead, the thermosome goes through conformational changes to close the apertures at each end (Schoehn et al. 2000). These conformational changes are correlated with the binding and hydrolysis of ATP (Bigotti and Clarke 2005). The apo-chaperonin is naturally in an open conformation and binding of ATP allows for the expansion of the thermosome entrance regions. The hydrolysis of bound ATP causes a conformational change to enclose a denatured protein. Finally, the release of ADP and/or Pi liberates the folded protein and reopens the thermosome (Gutsche et al. 2000; Gutsche, Mihalache and Baumeister 2000). The archaeal thermosomes are very similar in sequence to the cytosolic TCP1 eukaryotic chaperonins, the major difference being the number of subunits that compose each chaperonin's ring structures (Kagawa et al. 1995).
In addition to the thermosome (∼60 kDa subunits), thermoacidophiles also have HSP20 small heat shock proteins (sHSP), composed of 12–43 kDa subunits that also serve as chaperones during thermal stress response. HSP20 are common heat shock response proteins that are represented in every phylogenetic kingdom (Haslbeck et al. 2005). These small proteins form larger structures of up to 50 subunits that can bind and stabilize proteins upon exposure to supra-optimal temperatures preventing protein aggregation (Wang et al. 2010b; Li et al. 2012; Baes et al. 2020). Similar to the thermosome, the sHSP can also refold denatured proteins.
If denatured or damaged proteins cannot be refolded via chaperones, they will be degraded by the proteasome. In eukaryotes, ubiquitin and the ubiquitin-related modifier-1 (Urm1) mark proteins for degradation by the proteasome. Likewise, in thermoacidophilic archaea, an Urm1-like molecule is covalently attached to proteins by an E1-like enzyme that is reliant on the hydrolysis of ATP. This urmylation designates a protein for degradation by the archaeal proteasome. The archaeal Urm1 protein is also degraded in the process, unlike the Urm1 signal in eukaryotic systems (Anjum et al. 2015). The archaeal proteasome consists of two components: the 19S proteasome-activating nucleotidase (PAN) and the 20S proteolytic core particle. PANs are AAA+ ATPase regulatory particles that unfold proteins in preparation for degradation by the core particle. The 20S core particle of the proteasome is a cylindrical stack of four heptameric rings, which consists of two β rings flanked by two α rings. Within the core particle is an internal channel that contains the proteolytic site for protein degradation located within the two β rings. PAN is a homohexomeric ring that associates with the outside α rings of the core particle. Comprehensive reviews of proteasomes and AAA+ ATPases are available (Bar-Nun and Glickman 2012; Maupin-Furlow 2012).
Another possible way thermophiles handle thermal stress is through compatible solutes that were originally described only as an osmoprotectant (Brown and Simpson 1972). These organic molecules can accumulate intracellularly without interfering with cell metabolism to protect cytosolic components from other stressors, such as heat. For instance, Mycobacterium smegmatis mutants, unable to generate the compatible solute trehalose, exhibited sensitivities to elevated temperatures (Woodruff et al. 2004). Though trehalose is present in all domains of life, it has been found in large quantities in several thermophilic archaea, such as Sa. solfataricus (Nicolaus et al. 1988). On the other hand, the organic solutes di-myo-inositol-phosphate (DIP), di-mannosyl-di-myo-inositol-phosphate, di-glycerol-phosphate, mannosylglycerate and mannosylglyceramide are only observed in thermophilic organisms (reviewed in Santos and Da Costa 2002). Also, cellular quantities of DIP have been shown to increase in response to increasing temperature in the thermophiles Pyrococcus furiosus (Martins and Santos 1995) and Thermotoga neapolitana (Martins et al. 1996). Additionally, di-mannosyl-di-myo-inositol-phosphate also increases in T. neapolitana due to thermal stress (Martins et al. 1996). Although the exact mechanism of thermal protection by compatible solutes has not been determined, these compounds clearly have a part in the thermal stress response.
Some thermoacidophiles have pH optima near 0 (Schleper et al. 1995), but most have cytosols with a near neutral pH, thereby resulting in a large pH differential across the membrane. Even P. oshimae, previously mentioned to have an internal pH of ∼4.6, has a cytoplasmic pH several pH units above its environmental pH optima of <1. Acidophiles (both thermophilic and mesophilic) must have ways to maintain their cytoplasm near neutral and still generate a proton motor force for energy generation. For instance, acidophiles have a reversed membrane potential, where the intracellular membrane is positively charged and the extracellular is negatively charged. This reverse membrane potential is driven by the active transport of potassium ions into the cell and prevents passive proton transport. Despite the large ΔpH across the membrane, acidophiles still generate ATP by coupling the influx of protons to the phosphorylation of ADP through a membrane-bound ATP synthase (Moll and Schäfer 1988). However, this necessary influx of protons must be countered to maintain the intracellular pH of the cell. Proton pumps that are uniporters, symporters and antiporters are part of the energy generating respiratory chain that prevents the acidification of the intracellular environment by exporting internalized protons. These proton pumps are dependent on maintenance of the inversed membrane potential, which can be accomplished by pumping cations, such as potassium ions, into the cell (Schäfer 1996). In fact, potassium ions have been shown to be a vital part of proton pumping in the respirations chain of S. acidocaldarius (Moll and Schäfer 1988).
Alternatively, if protons do reach the cytoplasm, the cell must have ways to prevent the acidification of the cytosol. Intracellular molecules and proteins have buffering capacities that will neutralize internalized protons (Baker-Austin and Dopson 2007). For example, basic amino acids, such as histidine, arginine and lysine, can add to the buffering capacity of the cytoplasm. As discussed in the section 'Degradation of proteins and amino acids', members of the Sulfolobales have amino acid decarboxylases that may produce polyamines that are known contributers to cytoplasmic buffering. In fact, reduction of amino acids in continuous cultures of M. sedula caused a decrease in internal pH and an increase in thermosome protein levels (Peeples and Kelly 1995). Furthermore, protonated organic acids can permeate the cell and release a proton once inside the neutral cytoplasm. Heterotrophic acidophiles could possibly degrade organic acids without the production of a free proton to prevent the acidification of the cytoplasm (Baker-Austin and Dopson 2007). As is the case for thermal stress, damaged proteins due to low pH can either be repaired by microbial chaperones or be marked for degradation by the proteasome.
Membrane composition and thermoacidophily
Thermoacidophiles maintain their intracellular pH by having membranes that are more impermeable to protons than neutralophiles (Konings et al. 2002). The ether-linked lipids of archaeal membranes are more resistant to acid hydrolysis than the ester-linked lipids of bacterial membranes. Liposomes comprised of ether-linked lipids are more resistant to leakage due to thermal stress (Choquet et al. 1994). Furthermore, tetraether lipids that span the microbial membrane make a monolayer that reduces the fluidity of the archaeal membrane, thereby decreasing its permeability to proton penetration and increasing heat tolerance. These tetraether lipids can also have multiple cyclopentyl rings that will further increase the packing of these lipids, making the membrane even less permeable to the diffusion of protons and more resistant to elevated temperatures. This has been demonstrated in liposomes derived from the membranes of S. acidocaldarius (Elferink et al. 1994; Komatsu and Chong 1998) and P. oshimae (van de Vossenberg et al. 1998). The most abundant tetraether lipids are the glycerol dibiphytanyl glycerol tetraethers (GDGTs) that can have up to 4 cyclopentyl rings on each chain. The number of ring structures within these tetraether lipids increased with increasing temperature in Thermoplasma acidophilum (Uda et al. 2001), and with both decreasing pH and increasing temperature in P. torridus (Feyhl-Buska et al. 2016) and Acidilobus sulfurireducens (Boyd et al. 2011). These studies exemplify the importance of archaeal tetraether lipids with these cyclic moieties to survival in hot acid environments. However, only thermoacidophiles of the Sulfolobales have calditol, a cyclopentyl head group, ether linked to their GDGTs. Recently, the importance of this distinctive head group to acid resistance has been demonstrated in S. acidocaldarius as deletion of a calditol synthase resulted in a sensitivity to low pH (Zeng et al. 2018).
It is clear that thermoacidophiles have several different mechanisms to persist in elevated temperatures and low pH, including the thermosome and proteasome to manage the impact of damaged proteins in the cytosol, membranes made up of tetraether lipids with a reverse potential, and proton pumps to maintain intracellular pH homeostasis (Fig. 12).
Figure 12.
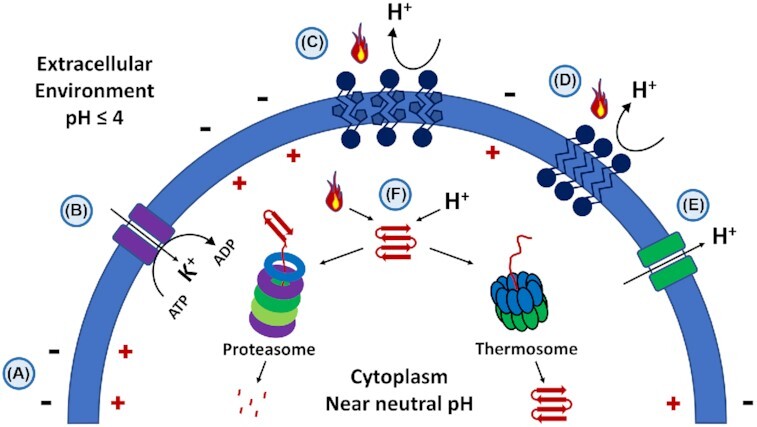
Major mechanisms of thermoacidophily. (1) Thermoacidophiles have an inverted membrane potential with a positive charge on the inside of the cellular membrane and a negative charge on the outside to prevent the acidification of the cytoplasm by the passive diffusion of protons. (2) The inverted membrane potential is maintained by transporting cations such as K+ into the cytoplasm. (3) Cyclopentyl ring moieties on tetraether lipids increase packing of the tetraether lipids decreasing the permeability of the membrane by protons and increasing cellular heat stability. (4) Tetraether lipids make a monolayer that is less permeable to protons and more heat stable than diether lipids. (5) Proton pumps export protons from the cytoplasm to prevent the acidification of the cytoplasm. (6) Heat-damaged or protonated proteins can either be degraded via the proteasome or properly refolded by the thermosome.
Thermal stress response
While the importance of the thermosome, sHSPs and proteasome to thermoacidophile heat tolerance is well known, the transcriptional thermal stress response of their associated genes is somewhat varied in the Sulfolobales. Although some sHSPs in other organisms are constitutively expressed, in Sa. solfataricus and S. acidocaldarius two sHSP are transcriptionally upregulated during thermal stress (Tachdjian and Kelly 2006; Li et al. 2012; Baes et al. 2020). However, the expression of the α and β subunits of the thermosome is unchanged during thermal stress in Sa. solfataricus (Tachdjian and Kelly 2006), while upregulated in Sa. shibatae and S. acidocaldarius (Kagawa et al. 2003; Baes et al. 2020). Also, as previously mentioned, transcriptional expression of the γ subunit of the thermosome decreases during thermal stress in most studied species (Archibald, Logsdon and Doolittle 1999; Kagawa et al. 2003; Tachdjian and Kelly 2006), with the exception of S. acidocaldarius (Baes et al. 2020). Although the proteasome plays a vital role in the heat shock response, the transcriptional expression of the 20S core components of the proteasome was unaffected by thermal stress in Sa. solfataricus, while the PAN regulatory component was significantly downregulated under these conditions (Tachdjian and Kelly 2006). However, post-transcriptional or -translational regulation, which has not yet been studied in detail, may also play a role in the thermosome's and proteasome's response to elevated temperatures.
The transcription of genes encoding components of the thermosome and proteasome are not significantly impacted by thermal stress, but this is not the case for much of the genome. In Sa. solfataricus, one-third of the genome, including sHSPs, was transcriptionally responsive to a 10°C temperature shift from 80°C to 90°C (Tachdjian and Kelly 2006). Of note, many type II TA loci, which encode a ribonucleolytic toxin and a corresponding antitoxin protein, were also upregulated. In particular, when the genes for one of these TA loci were deleted, the resulting Sa. solfataricus mutant became heat shock labile (Tachdjian and Kelly 2006; Cooper Charlotteet al. 2009; Maezato et al. 2011).
Thermoacidophiles can also use motility to seek out cooler conditions when thermal stress is encountered. Specifically, a temperature shift from 50°C to 80°C in S. acidocaldarius resulted in an increase in swimming speed and run time. Also, when exposed to a temperature gradient, cells migrated away from the higher temperature to regions closer to their optimum temperature. However, a similar response was not seen when the pH was shifted from 2 to 4, indicating that this was a temperature specific stress response for the thermoacidophile (Lewus and Ford 1999). Overall, thermoacidophiles mount a complex response to thermal stress that is varied among the members of the order Sulfolobales.
Metal stress response
Not only do thermoacidophiles have to combat the deleterious effects of life in hot acid, they also often encounter toxic heavy metals in their biotopes. In general, certain metal ions are essential for proper function of cells. Metals participate in many cellular processes, including as cofactors bound to proteins, as catalysts of redox reactions and to transport electrons in the respiratory chain. In mesophilic organisms, the focus is on acquiring biologically important heavy metals that are scarce in their environment. But, in the case of acidophilic organisms, especially in mining environments, the aim is to avoid their influx of heavy metals into the cell or at least to reduce the effective intracellular concentration. Mesophilic bacteria do this by using active and passive systems to remove the metal ions from the cell, form complexes with these metals or convert them into a less toxic form. It is also important to note that there are mechanisms associated with metabolic response to the reactive oxygen species (ROS) generated inside the cell after metal uptake (reviewed in Lemire et al. 2017). Most of the studies on heavy metal resistance in Crenarchaeota have been done in Sa. solfataricus and M. sedula, although many resistance mechanisms are still unknown, especially in species like Sulfura. metallicus, which has unusually high metal resistance levels.
Uranium and type II toxin–antitoxin systems
Some extreme thermoacidophiles have adapted to toxic metals, such as uranium, through unusual resistance mechanisms. Metallosphaera sedula, more so than a very similar species, Metallosphaera prunae, catalyzes the oxidization of U3O8 to soluble U(VI), mediated by Fe3+ generated from Fe2+ under chemolithoautotrophic conditions. However, the U(VI) so generated is toxic to the microbe (Mukherjee et al. 2012). As an adaptation to uranium U(VI) stress, M. prunae activates type II TA systems to degrade cellular rRNA during uranium stress, thereby inducing a population-wide dormancy. This response is not seen in M. sedula. This resistance mechanism allows M. prunae to withstand the toxic effects of living in the uranium-rich environment from which it was initially isolated (Mukherjee et al. 2012, 2017).
Active transporters for metal resistance
Active transport efflux is one of the most common approaches used for cellular metal resistance and the Cop system, which is involved in copper (Cu) resistance, is the best studied example in archaea. It includes a transcriptional regulator (CopT), a metal-binding chaperone (CopM) and a Cu-transporting ATPase (CopA) (Fig. 13) (Villafane et al. 2011). The genes for this system form a cluster in the Sa. solfataricus genome (She et al. 2001; Ettema et al. 2003, 2006), which is also present in M. sedula, S. acidocaldarius, Sulf. tokodaii (Martínez-Bussenius, Navarro and Jerez 2017) and Sulfura. metallicus (Orell et al. 2013b; Martínez-Bussenius, Navarro and Jerez 2017). Furthermore, in Sulfura. metallicus, the cop cluster is duplicated in the genome, although one cluster has a lower transcriptional response to copper. This duplication likely relates to the high heavy metal resistance of this organism (Orell et al.2013b).
Figure 13.
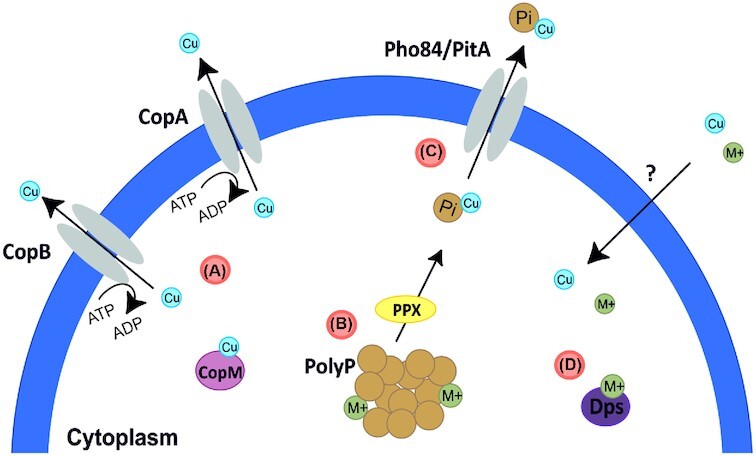
Model of heavy metal resistance in the Sulfolobales. (1) CopA and CopB export Cu outside the cell with ATP consumption. CopM is a metal chaperone that forms part of the Cop system that also includes a transcriptional factor called CopT (not shown). (2)PolyP can sequester cations via its negatively charged surface. (3) PolyP can also be degraded by PPX into inorganic phosphate to be exported outside the cell along with cations via PitA or Pho84 transporters. (4) Some proteins also act by sequestering metal ions, for example Dps. The mechanism for which metals enter the cell is still unknown.
Besides CopA, there is another copper-exporting ATPase, CopB, which contributes to Cu resistance and is present in some thermoacidophile genomes, including Sa. solfataricus. While CopA responds to intracellular Cu levels, CopB is constitutive (Völlmecke et al. 2012). The Cop system is also responsive to cadmium (Cd), but apparently not to silver (Ag), as seen in Sa. solfataricus (Ettema et al. 2006) and Sulfura. metallicus (Orell et al. 2013b). The Sulfura. metallicus Cop system also responds to chalcopyrite (CuFeS2) (Orell et al. 2013b) and likely reinforces its Cu resistance in mining environments.
Polyphosphate
Polyphosphates (PolyP) are ubiquitous molecules that play many cellular roles, including heavy metal resistance. These polymers are made of hundreds to thousands of inorganic orthophosphate (Pi) residues, linked by high-energy bonds, similar to those in ATP. There are two major enzymes involved in metabolism of this molecule: polyphosphate kinase (PPK) and exopolyphosphatase (PPX). The first enlarges the PolyP chain, adding a Pi to the end in a reversible reaction, using ATP as substrate, while PPX catalyzes the reverse reaction, hydrolyzing PolyP starting from the terminal Pi residue (there are endo-polyphosphatases as well, that cleaves internal PolyP bonds) (Kornberg, Rao and Ault-Riché 1999; Albi and Serrano 2016):
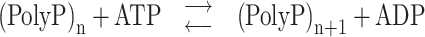 |
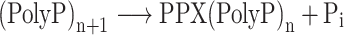 |
In the Crenarchaeota, the enzyme that synthesizes PolyP has not been identified, since neither PPK1 nor PPK2 has been identified. Several archaea, especially in the Sulfolobales, possess a PPX enzyme (Martínez-Bussenius, Navarro and Jerez 2017; Wang et al.2019d). Meanwhile, in Euryarchaeotes, a putative PPX was recently identified via bioinformatic analyses (Paula et al. 2019). High concentrations of intracellular polyP are related to higher metal resistance in Sulfolobales species (Remonsellez, Orell and Jerez 2006). Also, a Sa. solfataricus strain lacking PolyP due to overexpression of PPX, exhibited less copper resistance compared with the wild-type strain (Soto et al. 2019). Moreover, a spontaneous mutant with a functional PitA transporter, called M. sedula CuR1, was more resistant to copper and arsenic than the wild-type strain that carries a truncated pitA (McCarthy et al. 2014). PitA and Pho84 transporters are related to the uptake of Pi, and the exportation of Pi-Cu outside the cell (Fig. 13). In M. sedula, Pho84-like transporters responded to the presence of Cu and are also proposed to be involve in Pi-Cu uptake (Rivero et al. 2018). There are two ways in which PolyP helps directly to decrease intracellular effective concentration of metals. Since PolyP has negative charges, metal cations are attracted and complexed in the surface of the granule (Fig. 13). On the other hand, the increase in metal concentration triggers PolyP degradation, and Cu associated with Pi is exported outside the cell by PitA or Pho84 transporters (Fig. 13) (Keasling 1997; Remonsellez, Orell and Jerez 2006; Orell et al. 2012; Grillo-Puertas et al. 2014).
As mentioned, PolyP plays many roles in archaea and bacteria related to stress response and protein aggregation, the latter as an inorganic chaperone (Gray and Jakob 2015). PolyP can also sequester metals, like Fe, preventing the Fenton reaction and generating more free radicals (Gray and Jakob 2015). Proteomic studies in the Sa. solfataricus PolyP (-) mutant revealed upregulation of stress-related proteins when compared with the wild-type strain (Soto et al. 2019). Proteins, like peroxidases, were upregulated, and other enzymes from various metabolic pathways exhibited changes that collectively reassembled metabolic oxidative stress response (Soto et al. 2019).
Metal sequestration and transformation
Besides sequestration by PolyP granules, there are other molecules, such as DNA-binding proteins from starved cells (Dps) (Fig. 13), that are proposed to bind to metals, thereby avoiding deleterious effects as seen in Sa. solfataricus (Wiedenheft et al. 2005). Saccharolobus solfataricus also contains a mer operon, required for Hg (II) reduction. The operon contains a regulator, MerR, that controls production of a mercuric reductase (MerA) along with other components (Table 2). MerR acts as both a repressor and a metal-responsive activator of mercury resistance genes by binding an operator sequence (merO) that also forms part of the operon (Schelert et al. 2004, 2006).
Metal resistance and bioleaching
The use of mesoacidophiles and thermoacidophiles in biomining applications, that have reduced environmental impact, has driven interest in understanding heavy metal resistance in these organisms (Martínez-Bussenius, Navarro and Jerez 2017) (see the section 'Potential and current uses of thermoacidophiles in biotechnological applications'). The relationship between metal resistance and lithoautotrophy was seen in M. sedula when copA was disrupted, which lowered copper resistance and consequently chalcopyrite bioleaching (Maezato et al. 2012). In contrast, a spontaneous mutant of M. sedula, strain CuR1, also showed increased bioleaching capability, associated with its higher Cu resistance (Maezato et al. 2012). These examples illustrate that understanding metal resistance can lead to new strategies for bioleaching, where the Sulfolobales show great potential (see the section 'Potential and current uses of thermoacidophiles in biotechnological applications').
METABOLISM
Heterotrophic metabolism
As mentioned in the section 'The diversity of thermoacidophilic life', microorganisms adapted to both high temperature and low pH, so called thermoacidophiles, have been identified mainly in the domain of Archaea and most of them belong to the two orders, the euryarchaeal Thermoplasmatales, including the genera Thermoplasma, Picrophilus and the crenarchaeal Sulfolobales with the genera Caldivirga, Thermocladium, Acidianus, Desulfurolobus, Metallosphaera, Sulfolobus, Saccharolobus and Sulfurococcus (Bertoldo, Dock and Antranikian 2004; Zaparty and Siebers 2011; Sakai and Kurosawa 2018). Most of these organisms were isolated from solfataric fields or hot springs and are able to grow heterotrophically, using a variety of substrates as carbon and energy sources. Some have been also described as facultative heterotrophs that are also able to grow autotrophically by sulfur, metal or hydrogen oxidation. Among these species, the Crenarchaeota Sa. solfataricus and S. acidocaldarius have been most extensively analyzed with respect to the heterotrophic lifestyle and, therefore, represent archaeal model organisms for metabolic network reconstruction.
Saccharolobus solfataricus shows high metabolic versatility and is able to utilize a broad spectrum of substrates, including monosaccharides (e.g. d-glucose, d-galactose, l-fucose, d-fructose, d/l-arabinose and d-xylose), disaccharides (e.g. cellobiose, maltose, sucrose, trehalose and lactose), oligo- and polysaccharides (e.g. β-glucans, starch and dextrin), amino acids (e.g. glutamate), peptides and proteinaceous substrates (e.g. tryptone), and alcohols including aromatics (e.g. ethanol, phenol) (Quehenberger et al. 2017; Schocke, Bräsen and Siebers 2019). In contrast, S. acidocaldarius has a much narrower substrate spectrum; this could be attributed to its relatively smaller genome, which lacks numerous transport systems for substrate uptake. The metabolism of S. acidocaldarius is limited to a smaller range of substrates that include d-glucose, l-arabinose, d-xylose, sucrose, maltotriose, dextrin, starch, wheat bran, several fatty acids, and peptides and amino acids (Grogan 1989; Wang et al. 2019c). Over the past several decades, the metabolic network, especially the central carbohydrate metabolism, in these two model organisms has been studied intensely, and is characterized by the presence of unusual and/or unique pathways and enzymes (Bräsen et al. 2014; Quehenberger et al. 2017). As such, we focus on these two model organisms to provide a prospective on heterotrophy in thermoacidophilic Archaea (see Fig. 14 for central metabolism overview).
Figure 14.
An overview of the central metabolism in Sulfolobales. Dashed arrows indicate pathways, which have not yet been experimentally demonstrated. Abbreviations: F6P, fructose 6-phosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; D-KDG, 2-keto-3-deoxy-d-gluconate; D-KDGal, 2-keto-3-deoxy-d-galactonate; D-KDA, 2-keto-3-deoxy-d-arabinoate; L-KDA, 2-keto-3-deoxy-l-arabinoate; D-KDX, 2-keto-3-deoxy-d-xylonate; AA, amino acid; ED, Entner–Doudoroff pathway; EMP, Embden–Meyerhof–Parnas pathway; RuMP, reversed ribulose monophosphate pathway; TCA, tricarboxylic acid cycle; 3HP/4HB, 3-hydroxyproprionate/4-hydroxybutyrate cycle; ABC, ATP-binding cassette transporters; RC, respiratory chain.
Carbohydrates metabolism
Sugar catabolism: The two model organisms, Sa. solfataricus and S. acidocaldarius, can grow on a range of monosaccharides, disaccharides and also polysaccharides (Table 4). A variety of polysaccharides-degrading enzymes have been studied in Sa. solfataricus, such as cellulase (Maurelli et al. 2008), glucanase (Limauro et al. 2001; Huang et al. 2005; Girfoglio, Rossi and Cannio 2012), glucoamylase (Kim et al. 2004), β-glycosidase (Noh and Oh 2009), α-glucosidase (Rolfsmeier et al. 1998; Wagner et al. 2014), β-glucosidase (Moracci et al. 1995; Shin et al. 2013), α-amylase (Haseltine, Rolfsmeier and Blum 1996; Wagner et al. 2014), xylanase (Cannio et al. 2004; Maurelli et al. 2008) and xylosidase (Moracci et al. 2000). The degradation products, i.e. mono-, di- and/or oligosaccharides, are taken up into the cytoplasm primarily by various ATP-binding cassette (ABC) transporters (Albers et al. 2004; Choi, Hwang and Cha 2013; Wagner et al. 2017). The further processing of the monosaccharides follows a general scheme: the sugars are first oxidized to the corresponding lactone by sugar dehydrogenases, which in most cases belong to the medium chain dehydrogenase/reductase (MDR) superfamily (Lamble et al. 2003; Brouns et al. 2006; Nunn et al. 2010; Haferkamp et al. 2011; Wolf et al. 2016; Reinhardt, Johnsen and Schönheit 2019), and in only some cases to the short chain dehydrogenase/reductase (SDR) superfamily (Yasutake et al. 2007; Kim, Paek and Lee 2012). The sugar lactones are then hydrolyzed either enzymatically via lactonases or spontaneously in a non-enzymatic reaction favored by the extremely thermophilic conditions. The resulting sugar acids are subsequently dehydrated to the key intermediates, the 2-keto-3-deoxy sugar acids. These reactions are catalyzed by dehydratases mainly from the enolase superfamily. The further degradation then varies with respect to phosphorylation and (aldol) cleavage. In the modified branched ED pathway, the 2-keto-3-deoxy sugar acids, 2-keto-3-deoxy gluconate (KDG) and 2-keto-3-deoxy galactonate (KDGal) are either first phosphorylated by KDG kinase to KDPG/KDPGal and then cleaved to glyceraldehyde 3-phosphate (GAP) and pyruvate in the semi-phosphorylative (sp) ED branch, or they are directly cleaved by the same aldolase to yield glyceraldehyde (GA) and pyruvate in the non-phosphorylative (np) ED branch (Ahmed et al. 2005; Lamble et al. 2005). GAP is then directly oxidized to 3-phosphoglycerate (3PG) by a non-phosphorylating GAP dehydrogenase (GAPN), without coupling the oxidation to ATP generation via substrate level phosphorylation. 3PG is then further converted to a second molecule of pyruvate through the reaction sequence of the common lower Embden–Meyerhof–Parnas (EMP) pathway. In the npED branch, GA is first oxidized in a ferredoxin-dependent manner to glycerate by GA:ferredoxin oxidoreductase and then phosphorylated to 2-phosphoglycerate (2PG) by means of 2-phosphoglycerate kinase. 2PG again enters the lower common shunt of the EMP. The 2-keto-3-deoxy-l-fuconate derived from the 6-deoxy hexose l-fucose is cleaved by the same aldolase to lactaldehyde, and pyruvate and the lactaldehyde are further oxidized in two consecutive steps to lactate and finally to a second molecule of pyruvate (Wolf et al. 2016). Also, pentose degradation initially follows the same reaction sequence of oxidation and dehydration, finally leading to the corresponding 2-keto-3-deoxy acids, which are then further processed either in an aldolase-dependent manner, called the Dahms pathway, or in an aldolase-independent manner, referred to as the Weimberg pathway (Nunn et al. 2010). The aldol cleavage in the Dahms pathway yields pyruvate and glycolaldehyde, which is then oxidized in two steps to glyoxylate, converted with acetyl-CoA to malate via malate synthase, and finally enters the tricarboxylic acid (TCA) cycle as malate. In the Weimberg pathway, the 2-keto-3-deoxy pentanoates are converted via a second dehydration catalyzed by the 2-keto-3-deoxy xylonate dehydratase (KDXD) to 2-ketoglutarate semialdehyde (KGSA) and then oxidized through KGSA dehydrogenase (KGSADH) to 2-ketoglutarate entering the TCA cycle (Nunn et al. 2010; Wagner et al. 2017). While d-arabinose is only used as carbon source in Sa. solfataricus and degraded via the Weimberg pathway (Brouns et al. 2006), l-arabinose and d-xylose are converted by both pathways to the same extent in Sa. solfataricus and S. acidocaldarius (Nunn et al. 2010). However, deletion mutant analyses in S. acidocaldarius MW001 demonstrated that the Dahms pathway is dispensable, whereas the Weimberg pathway is essential for d-xylose degradation (Wagner et al. 2017). In contrast to the other hexoses, another 6-deoxy sugar, i.e. l-rhamnose, was also proposed to be degraded in an aldolase-independent manner via the so called 2,4-di-keto pathway. The 2-keto-3-deoxy rhamnoate generated by the common initial reactions is then oxidized at C4 to 2,4-keto-3-deoxy-rhamnoate, which is cleaved by a hydrolase to yield lactate and pyruvate (Reinhardt, Johnsen and Schönheit 2019). The lactate is then subsequently oxidized to a second molecule of pyruvate.
Table 4.
An overview of the reported substrates and degradation pathways in Sa. solfataricus and S. acidocaldarius.
| Substrate | Organism | Reference | |
|---|---|---|---|
| Carbohydrates | Cellulose | Sa. solfataricus | (Girfoglio, Rossi and Cannio 2012) |
| Xylan | Sa. solfataricus | (Cannio et al. 2004) | |
| Glucose | Sa. solfataricus,S. acidocaldarius | (De Rosa et al. 1984; Ahmed et al. 2005) | |
| Galactose | Sa. solfataricus | (Lamble et al. 2003) | |
| d-Arabinose | Sa. solfataricus | (Brouns et al. 2006) | |
| l-Arabinose | Sa. solfataricus,S. acidocaldarius | (Nunn et al. 2010; Wagner et al. 2017) | |
| d-Xylose | Sa. solfataricus,S. acidocaldarius | (Nunn et al. 2010; Wagner et al. 2017) | |
| l-Fucose | Sa. solfataricus | (Wolf et al. 2016) | |
| Proteins/peptides/amino acids | Proteins/peptides | Sa. solfataricus,S. acidocaldarius | (Lin and Tang 1990; Gogliettino et al. 2014) |
| Glutamate, methionine, leucine, phenylalanine, isoleucine, threonine, alanine, asparagine, glycine, tyrosine and serine | Sa. solfataricus | (Stark et al. 2017) | |
| Lipids/fatty acids | Olive oil, corn oil, p-nitrophenyl (PNP)-butyrate, PNP-caprylate, PNP-palmitate | Sa. solfataricus | (Choi et al. 2016) |
| Tributyrin, tricaproin | S. acidocaldarius | (Zweerink et al. 2017) | |
| Butyrate, hexanoate | S. acidocaldarius | (Wang et al. 2019c) | |
| Other substrates | Ethanol | Sa. solfataricus | (Chong et al. 2007a) |
| Phenol | Sa. solfataricus | (Izzo et al. 2005) |
An interesting feature of the sugar degradation routes in the Sulfolobales is the pronounced substrate promiscuity of the enzymes involved, especially in the upper part of the pathways catalyzing analogous reactions. This has first been described by Danson and co-workers, who found that the glucose dehydrogenase, the gluconate dehydratase and the KD(P)G aldolase from Sa. solfataricus also accept the d-galactose derivatives as substrates (Lamble et al. 2003, 2004, 2005). Subsequently, it was shown that the same sugar dehydrogenase is also responsible for the oxidation of the pentoses d-xylose and l-arabinose (Nunn et al. 2010). However, d-arabinose and l-fucose demonstrated the opposite stereochemistry on C2–C4 compared with l-arabinose oxidized by another dehydrogenase, which also accepted l-rhamnose as a substrate (Brouns et al. 2006; Wolf et al. 2016; Reinhardt, Johnsen and Schönheit 2019). Interestingly, the dehydratase's conversion of l-rhamnonate is different from the conversion of d-arabonate and l-fuconate, indicating that the promiscuity of the dehydratases is less pronounced than that of the dehydrogenases (Reinhardt, Johnsen and Schönheit 2019). This has also been proposed for the gluconate/galactonate dehydratase, which presumably does not accept the pentose derivatives as substrates suggesting the presence of an alternative dehydratase, although this has not been confirmed (Nunn et al. 2010). The KD(P)G aldolase, however, shows by far the most marked promiscuity, accepting 2-keto-3-deoxy acids derived from hexoses and pentoses, i.e. d-glucose, d-galactose, d-xylose, l-arabinose, d-arabinose and l-fucose, as well as the phosphorylated derivatives KDPG and KDPGal (Ahmed et al. 2005; Lamble et al. 2005; Nunn et al. 2010; Wolf et al. 2016). Thus, this enzyme plays a central role in sugar degradation via the branched ED pathway, comprising the npED and spED branch, as well as the Dahms pathway, both of which are involved in the breakdown of a wide variety of naturally occurring sugars in Sulfolobales.
The end products of the glycolytic pathways are pyruvate and the TCA cycle intermediates, α-ketoglutarate and malate. As in all Archaea, the pyruvate produced is then oxidatively decarboxylated to acetyl-CoA by ferredoxin oxidoreductase (Kerscher, Nowitzki and Oesterhelt 1982; Zhang et al. 1996; Yan et al. 2016). The resulting acetyl-CoA is then completely oxidized to CO2 in the TCA cycle (Danson 1988). Also, the oxidative decarboxylation of α-ketoglutarate is carried out by a ferredoxin-dependent oxidoreductase. NAD(P)+-dependent dehydrogenase complexes (i.e. pyruvate dehydrogenase complex), known from bacteria and eukaryotes, are not operative in the Sulfolobales or, for that matter, Archaea in general (Payne, Hough and Danson 2010).
Sugar anabolism: Sugar degradation in aerobic Archaea, including the Sulfolobales, proceeds via a modified branched ED pathway and is, at least in some ways, analogous to the Dahms and/or Weimberg pathways. Notably, for a complete EMP pathway, only a functional phosphofructokinase (PFK) is missing, which highlights the exclusively gluconeogenic function of the EMP in the Sulfolobales (Bräsen et al. 2014). In addition to the common lower shunt enzymes, gluconeogenesis is initiated by the phosphoenolpyruvate synthetase, bypassing the irreversible pyruvate kinase (PK) reaction (Haferkamp et al. 2019), and also involves the classical glyceraldehyde-3-phosphate (GAPDH)/phosphoglycerate kinase (PGK) couple, which bypasses the irreversible GAPN (Kouril et al. 2013b). Perhaps, the most striking difference to classical gluconeogenesis is the presence of the bifunctional fructose-bisphosphate aldolase/phosphatase (FBPA/ase), characterized among others from the Sulfolobales M. sedula and Sa. solfataricus, replacing the classical fructose-1,6-bisphosphate aldolase and F1,6BP phosphatase couple of the gluconeogenesis route in Bacteria and Eukarya (Say and Fuchs 2010). This enzyme catalyzes the conversion of GAP and DHAP to F6P without liberating the F1,6BP intermediate.
All of these modifications, both in sugar catabolism and anabolism, have been discussed as a mechanism of metabolic thermoadaptation, since the formation of extremely thermolabile triose phosphates, e.g. GAP, DHAP and 1,3BPG, is avoided (Say and Fuchs 2010; Kouril et al. 2013b; Figueiredo et al. 2017; Zhang et al. 2017). Also, the altered regulatory properties of the pathways and enzymes could contribute to thermoadaptation, since the classical control points, e.g. HK, PFK and PK, are missing or changed in Archaea, including in the Sulfolobales. For example, the main glycolytic control point is the GAPN, instead of the sugar(phosphate) kinases being activated by glucose 1-phosphate (Ettema et al. 2008; Kouril et al. 2017). Also, the PKs so far characterized, including those from Sulfolobus spp., show divergent regulation compared with the enzymes from the classical bacterial and eukaryotic pathways (Haferkamp et al. 2019; Johnsen et al. 2019).
The phosphoglucose isomerase (PGI) interconverting F6P and G6P and the phosphoglucomutase (PGM) present in the Sulfolobales represent the branch point to the synthesis of the storage compound glycogen and trehalose as the only compatible solutes in Sulfolobus spp. described so far (Martins et al. 1997; Bräsen et al. 2014). The additional presence of a hexokinase suggests that these biosynthesis reactions can also be directly initiated from the substrate molecule glucose. However, the spED branch, particularly the KDG kinase as its key enzyme, may be involved in gluconeogenesis by providing triose phosphates for anabolic purposes by directing the flux from the upper branched ED to the hexose phosphate synthesis (Kouril et al. 2013a).
The pentose metabolism in the Sulfolobales and Thermoplasmatales are also characterized by a missing oxidative pentose phosphate pathway (OPPP). Furthermore, the non-oxidative pentose phosphate pathway (NOPPP) is only partially present in the Sulfolobales lacking a ribulose-5-phosphate epimerase and a transaldolase, but is entirely present in the Thermoplasmatales (Thermoplasma and Picrophilus) (Bräsen et al. 2014). However, the pentose precursor ribulose-5-phosphate, as in most Archaea, is provided by the ribulose monophosphate (RuMP) pathway converting F6P to Ru5P and formaldehyde via d-arabinohexulose 6-phosphate. Ribose 5-phosphate (R5P) and erythrose 4-phosphate (E4P) are precursors of nucleotide and aromatic amino acid biosynthesis, respectively, and are then provided by the remaining reaction of the NOPPP. Thus, F6P is the main source of pentose/tetrose phosphates in most Archaea in general and, particularly, thermoacidophiles (Bräsen et al. 2014).
Degradation of proteins and amino acids
Although thermoacidophilic Archaea are routinely grown on proteinaceous substrates, surprisingly little is known about the breakdown of amino acids in these organisms. It appears that certain Sulfolobales species grow well on these complex protein substrates, such as yeast extract, casein hydrolysate, N-Z-Amine, and on mixtures of amino acids, but hardly if at all on single amino acids as a sole carbon, energy and nitrogen source (Grogan 1989; Stark et al. 2017; Quehenberger et al. 2019). The reason for this remains unclear so far. However, Sa. solfataricus, when growing on casein hydrolysate, prefers certain amino acids like glutamate, methionine, leucine, phenylalanine and isoleucine and, to a lesser extent, threonine, alanine, aspartate, glycine and tyrosine (Stark et al. 2017). Growth of Sa. solfataricus on glucose was stimulated most by glutamate and, to a lesser extent, by aspartate (Stark et al. 2017). Additionally, growth of S. acidocaldarius on glucose is enhanced by glutamate, followed by aspartate, arginine and lysine (Quehenberger et al. 2019). For the breakdown of (poly)peptides though, many proteinase and peptidase encoding genes are present in the genomes of the Sulfolobales, but few have been studied in detail (Cannio et al. 2010; Gogliettino et al. 2010). Additionally, ABC transporters for di/oligopeptides have been identified in Sa. solfataricus,Sulf. tokodaii and T. acidophilum (Albers et al. 2004).
There are generally three mechanisms known by which amino acid degradation is initiated: decarboxylation, transamination and (oxidative) deamination. Decarboxylation leads to biogenic amines and there are some reports in Sa. solfataricus for amino acid decarboxylases, e.g. arginine decarboxylase, which might play a role in the biosynthesis of spermidine and putrescine (Giles and Graham 2008; Esser et al. 2013). However, the more common mechanism for amino acid breakdown in Archaea, which was intensively studied for Thermococcales, is the transamination with 2-oxoacids, mainly alpha-ketoglutarate, and/or the oxidative deamination using amino acid dehydrogenases, most importantly the glutamate dehydrogenase liberating ammonia and concomitantly reducing NAD(P)+ (Yokooji et al. 2013; Awano et al. 2014; Scott, Poole and Adams 2014). Both mechanisms ultimately lead to the formation of the corresponding 2-keto acids that are subsequently oxidatively decarboxylated by ferredoxin-dependent 2-oxoacid:Fd oxidoreductases, which are also well known and characterized from Sulfolobales species, Sulf. tokodaii in particular (Kerscher, Nowitzki and Oesterhelt 1982; Zhang et al. 1996; Park et al. 2006; Yan et al. 2016). The reaction products are the corresponding CoA esters of the carboxylate backbone of the amino acids. In aerobes, these CoA esters are then completely oxidized to CO2 by channeling these compounds into the central energy metabolic pathways via pyruvate, PEP, acetyl-CoA and the TCA cycle intermediates. These pathways have also been identified in Sa. solfataricus by in silico metabolic reconstructions (Ulas et al. 2012). In anaerobic archaea, most well studied in Thermococcales, the CoA esters play pivotal roles in fermentative energy generation by being the sole source of net ATP via substrate level phosphorylation. The key enzymes are the ADP-forming acyl-CoA synthetases coupling the CoA ester hydrolysis to the ATP formation from ADP and Pi (Awano et al. 2014; Scott, Poole and Adams 2014; Weiße et al. 2016). However, these ADP-forming acyl-CoA synthetases are also present in aerobic archaea, including the Sulfolobales. Specifically, during Sa. solfataricus growth on casein hydrolysate carboxylic acids, mainly isovalerate, are excreted into the medium (Stark et al. 2017). Although such product formation appears unusual under aerobic conditions, it is well documented in bacterial model organisms, like E. coli and Bacillus subtilis (Bräsen and Schönheit 2004), and this so-called ‘overflow metabolism’ has also been observed in halophilic archaea (Bräsen and Schönheit 2004). However, under acidophilic conditions, carboxylic acids at elevated concentrations act as uncouplers/protonophores that lead to the acidification of the cytosol and the breakdown of membrane gradients (Baker-Austin and Dopson 2007). Moreover, the excretion of products results in the loss of carbon, which may account for the less efficient growth of Sa. solfataricus on casein hydrolysate compared with glucose. Furthermore, in Sa. solfataricus pyroglutamate may form from glutamate at high temperature and low pH to inhibit growth of thermophilic archaea (Stark et al. 2017). However, in S. acidocaldarius, pyroglutamate is not inhibitory and even serves as a carbon source, making S. acidocaldarius a better thermoacidophilic platform organism for applications with glutamate-containing media (Vetter et al. 2019). Finally, it has not been determined whether the carboxylic acids formed during exponential growth can be re-used by Sulfolobus species, as it has been shown for other aerobic organisms.
Degradation of lipids and fatty acids
In general, the mechanisms for fatty acid and, fatty acid-based, (phospho)lipid metabolism are not well understood in Archaea. However, Sa. solfataricus P1 can (partially) degrade corn oil as well as olive oil (Choi et al. 2016), and S. acidocaldarius can cleave triacylglycerols (Zweerink et al. 2017). Moreover, several extracellular esterases/lipases have been identified from Sa. solfataricus, S. acidocaldarius and Sulf. tokodaii (Suzuki, Miyamoto and Ohta 2004; Choi et al. 2016; Zweerink et al. 2017). A TetR-family transcription factor (FadRSa) plays a role in regulation of putative fatty acid metabolism-related genes in S. acidocaldarius (Table 2) and growth of this organism on short-chain fatty acids, i.e. butyrate and hexanoate, as sole carbon sources has been demonstrated (Wang et al. 2019c).
Genomic analyses revealed that all the genes encoding homologs of the key enzymes involved in the bacterial-like β-oxidation are present in the genomes of several Archaea, including Sa. solfataricus and S. acidocaldarius (Dibrova, Galperin and Mulkidjanian 2014; Wang et al. 2019c), implying that fatty acids could be degraded in these organisms through classical β-oxidation, but it remains unproven. Nonetheless, S. acidocaldarius can grow on acetate (as also described for many other Archaea including methanogens, Pyrobaculum, and halophilic Archaea) as the sole carbon and energy source. The glyoxylate shunt is operative under these conditions, demonstrating that C2 units can be assimilated by this reaction sequence (Uhrigshardt et al. 2002).
As a product of lipid hydrolysis, glycerol can be utilized as a carbon and energy source by many bacteria. In Archaea, glycerol degradation has been examined in halophiles (Sherwood, Cano and Maupin-Furlow 2009; Rawls, Martin and Maupin-Furlow 2011; Williams et al. 2017). Glycerol is taken up either by simple diffusion or glycerol transporters (Richey and Lin 1972; Stroud et al. 2003; Anderson et al. 2011). The haloarchaea employ one of the bacterial-like mechanisms, first phosphorylating glycerol followed by sn-glycerol-3-phosphate oxidation to DHAP. So far, glycerol catabolism has not been studied in thermoacidophilic archaea, although there are some indications that Sulfolobales spp. do not utilize glycerol as carbon and energy source (Grogan 1989). In contrast, genome-scale metabolic network reconstruction and modeling suggested glycerol as the most efficient carbon source for Sa. solfataricus (Ulas et al. 2012).
Degradation of other substrates
In addition to the three major types of nutrients described above, other substrates also support heterotrophic growth of thermoacidophilic archaea. For instance, Sa. solfataricus grows on acetoin, citric acid, alcohols and phenol (Izzo et al. 2005; Chong et al. 2007a; Wolf et al. 2016). There are several alcohol dehydrogenases in Sa. solfataricus, allowing oxidation of alcohols into aldehydes. The second step is then the oxidation to the corresponding carboxylic acids and subsequent activation of the CoA esters for further degradation (Chong et al. 2007a,b). Phenol is degraded in Sa. solfataricus in a classic pathway, as reported in some bacteria, e.g. Burkholderia pickettii and Pseudomonas stutzeri OX1. It is first converted to catechol, which undergoes a series of ring cleavage reactions producing products that finally enter TCA cycle (Izzo et al. 2005).
In conclusion, thermoacidophilic (facultative) heterotrophs, especially from the Sulfolobales, utilize a variety of substrates for cell growth (reported substrates with degradation pathways are concluded in Table 4). This metabolic versatility includes the potential for autotrophic growth, combined with the thermoacidophilic lifestyle, making them ideal candidates for the development as platform strains for the production of added-value compounds from renewable (waste) materials like lignocellulosics (see the section 'Potential and current uses of thermoacidophiles in biotechnological applications').
Autotrophy and chemolithotrophy
Autotrophy
The inhospitable environments in which the Sulfolobales thrive often have a scarcity of organic carbon available. As a result, many of the species within this order rely on the autotrophic fixation of CO2 to support growth. At present, six mechanisms for CO2 fixation are known throughout the domains of life. Several of these mechanisms build carbon–carbon bonds by fixing CO2 using oxygen-sensitive carboxylases, or in the case of the Calvin–Bassham–Benson cycle, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo) (Hugler et al. 2003). While RuBisCo is not oxygen sensitive, it does have a highly detrimental side reaction with oxygen, and the prevalence of oxygen-reducing terminal oxidases throughout the Sulfolobales (anaerobes included) seems to indicate that oxygen is essential for the organisms’ ability to survive their extreme conditions. The autotrophic pathway in the Sulfolobales circumvents the dependence on RuBisCo by incorporating bicarbonate molecules rather than CO2 (Gong et al. 2019). This pathway, named the 3-hydropropionate/4-hydroxybutyrate (3-HP/4-HB) cycle after two of its prominent intermediates, generates one acetyl-CoA per cycle (Berg et al. 2007). Initially identified in M. sedula, the 3-HP/4-HB pathway shares the enzymatic route from succinyl-CoA to acetyl-CoA with the dicarboxylate/4-hydroxybutyrate autotrophy cycle. However, the generation of succinyl-CoA eschews the traditional ferredoxin-powered pyruvate synthase and instead relies on the sequential addition of bicarbonate molecules and coenzyme-A moieties (Fuchs 2011). One full rotation of the 3-HP/4-HB cycle costs the cell 4 ATP and 4 NADPH, making it only a moderately expensive route to carbon fixation (Gong et al. 2019).
Since the discovery of the 3-HP/4-HB pathway, the enzymes responsible for each step in the pathway have been identified and characterized (Kockelkorn and Fuchs 2009; Teufel et al. 2009; Ramos-Vera et al. 2011). Metallosphaera sedula has served as the model organism for many of these efforts, and part of the pathway characterization has involved linking the enzymes of the 3-HP/4-HB pathway to open reading frames within M. sedula. This has proved to be challenging, considering the promiscuity of enzymes causing overlapping catalytic activities. In the case of 4-hydroxybutyrate-CoA synthetase, five candidate genes were identified based on bioinformatic analysis. However, three of these genes showed no activity for the desired reaction (Ramos-Vera et al. 2011), while the remaining two did demonstrate this activity (albeit with orders of magnitude of difference in Vmax values). Further investigation into these two candidates determined that one was merely a promiscuous acyl-CoA synthetase rather than the genuine 4-hydroxybutyrate-CoA synthetase (Hawkins et al. 2013).
As greater understanding of the precise enzymatic path of the 3-HP/4-HB cycle has developed, modeling of the pathway has become a point of interest. This effort has been greatly aided by the quantification of thermodynamic and kinetic parameters associated with each step in the pathway (Ramos-Vera et al. 2011; Loder et al. 2016). A kinetic model of the system in M. sedula revealed differing degrees of rotations the cycle can undergo, resulting in a different distribution of products including acetyl-CoA and succinyl-CoA (Loder et al. 2016). This modeling supports previous exploration into the channeling of carbon into various biosynthetic pathways from the 3-HP/4-HB cycle. Based on this isotope distribution, it was determined that the majority of CO2 taken up during autotrophy generates succinyl-CoA, representing a half-turn or one-and-a-half turns of the full cycle. Acetyl-CoA (one full turn) is generated during this process, but only enough for amino acids directly synthesized from acetyl-CoA. Otherwise, succinyl-CoA dominates as the product of this cycle (Estelmann et al. 2011). Extension of 3-HP/4-HB cycle modeling was done to identify the effect of the pathway on carbon isotopes and demonstrated that the source of carbon for the cycle may not be extracellular bicarbonate. Instead, bicarbonate is formed intracellularly as CO2 is taken up by the cell (Pearson et al. 2019).
While this autotrophic cycle requires 16 steps to generate acetyl-CoA from two bicarbonate molecules, only 13 enzymes are involved in the pathway. This inconsistency points toward the unusual redundancy in enzyme function throughout this pathway and it manifests in a variety of ways (Fig. 15). For example, the two-step conversion of crotonyl-CoA to acetoacetyl-CoA is catalyzed by the crotonyl-CoA hydratase. The first step of this conversion, however, can also be catalyzed by 3-hydroxypropionyl-CoA dehydratase, which also serves a separate function of dehydrating 3-hydroxypropionyl-CoA into acryloyl-CoA. Other redundancies include the reduction of malonyl-CoA and succinyl-CoA by the aptly named malonyl-CoA/succinyl-CoA reductase and the catalysis of hydroxypropionate to 3-hydroxypropionyl-CoA by either 3-hydroxypropionyl-CoA synthetase or the promiscuous 4-hydroxybutyryl-CoA synthetase (Loder et al. 2016). Notably, these overlaps in function reflect the structural similarity in the 3-hydroxypropionate half of the pathway and the 4-hydroxybutyrate half of the pathway. In fact, the two halves can almost be viewed as the same sequence of reactions, with enzymes acting on the same functional groups and the substrates varying only in length of the carbon chain.
Figure 15.
The enzymatic pathway of the 3-hydroxypropionate/4-hydroxybutyrate (3-HP/4-HB) cycle as characterized in Metallosphaera sedula. Enzyme names are contained within the yellow oval with arrows indicating reactions for which they have known catalytic activity. Enzymes in pink have shown activity for only a single reaction in the 3-HP/4-HB cycle; enzymes in green exhibit activity on multiple steps in the cycle.
The 3-HP/4-HB cycle is highly conserved within the Sulfolobales. In fact, with the exception of Sulfo. acidophilus, all genome-sequenced members of the order possess homologs to the characterized enzymes from M. sedula (Counts, Willard and Kelly 2020). In spite of this conservation, not all Sulfolobales appear to be capable of carbon fixation. A prime example of this is S. acidocaldarius, which at the time of its isolation was reported to grow chemolithoautotrophically in the presence of elemental sulfur (Brock et al. 1972). More recently, it appears that the commonly used lab strain, S. acidocaldarius DSM 639, is a strict heterotroph (Zeldes et al. 2019). This example may be explained through an incomplete transcriptional regulation pathway. Recently, a conserved transcriptional regulator, HhcR, was proposed to be a universal autotrophy regulator in the Sulfolobales (Leyn et al. 2015) (Table 2). Several genes involved in the 3-HP/4-HB cycle appear to be lacking the binding motif for this regulator in S. acidocaldarius (Zeldes et al. 2019). Whether this lack of promoter-binding regions is a result of strain domestication by extensive heterotrophic growth in the lab remains to be determined. Further complicating the network of autotrophy in the Sulfolobales is the existence of all genes necessary for the dicarboxylate/4-hydroxybutyrate in several chemolithoautotrophs, including M. sedula. However, these genes are transcribed at a low level even during autotrophic growth conditions (Berg et al. 2010b). One area of interest for this pathway is its application in engineered organisms. A partial pathway was engineered into E. coli to generate both 3-hydroxypropionate and 4-hydroxybutyrate as a means of generating a block copolymer product (Meng et al. 2012). Parts of the cycle from M. sedula have been introduced into Pyrococcus furiosus in an effort to convert CO2 into 3-hydroxypropionic acid (Keller et al. 2013). Similarly, E. coli has again been host to 3-HP/4-HB enzymes in order to produce propionic acid and acrylic acid (Liu and Liu 2016).
Chemolithotrophy at the cell surface
In order to power an autotrophic lifestyle, members of the Sulfolobales tend to rely on metals and inorganic compounds that prevail in their primordial environments. The oxidation of ferrous iron (Fe2+) is one such source of energy. Despite a very positive reduction potential (+0.77 V for Fe3+/Fe2+), chemolithoautotrophs are still able to leverage iron oxidation to drive the electron transport chain (ETC) (Amend and Shock 2001). While this pathway has been studied most intensely in mesoacidophiles like Acidithiobacillus ferrooxidans, transcriptomic studies of several Sulfolobales members have shed light on enzymes responsible for thermoacidophilic iron oxidation (Bathe and Norris 2007; Auernik and Kelly 2008; Kozubal et al. 2011). Metallosphaera sedula is a prolific iron oxidizer and has served as the organism of study to further elucidate the mechanism for iron oxidation. These results indicate control of iron oxidation by a locus known as the fox cluster. The cluster appears to be membrane-bound, with iron oxidation taking place at the surface of the cell membrane and funneling electrons into the ETC.
A variety of sulfur species also persist in the Sulfolobales’ habitats. Anaerobic Sulfolobales reduce zero-valent sulfur to hydrogen sulfide (H2S) in conjunction with oxidizing diatomic hydrogen. Much like iron oxidation, sulfur reduction takes place entirely at the surface of the cell membrane, where a membrane-bound sulfur reductase acts on zero-valent sulfur (Fig. 16A) (Laska, Lottspeich and Kletzin 2003). The complex achieves this reduction by transferring electrons from reduced quinols (specifically Sulfolobus quinol). Quinone cycling links sulfur reduction to hydrogen oxidation, and it both regenerates the oxidized quinone pool and powers proton export (Kletzin et al. 2004). The dominant form of zero-valent sulfur under acidic conditions is cyclooctasulfur, but experimental evidence suggests sulfane sulfur from polysulfide chains may be the substrate for sulfur reductase (Blumentals et al. 1990).
Figure 16.
Current knowledge of the mechanism of sulfur oxidation and reduction in the Sulfolobales. (A) Sulfur reduction and (B) sulfur oxidation. Solid arrows indicate involvement in a reaction; dotted arrows represent transport of species; and dashed lines indicate that the function is suspected but has not been demonstrated experimentally in the Sulfolobales. Enzyme colors indicate general grouping of function: coupled to electron transport chain (blue), involved in transporting or trafficking sulfur species (yellow), transformation of sulfur species with no energy conservation (orange) and transformation of sulfur species directly coupled to energy-conserving biomolecules (green). Abbreviations: sulfur reductase (Sre), hydrogenase (Hyn), heterodisulfide reductase (Hdr), tetrathionate hydrolase (TetH), sulfide:quinone oxidoreductase (SQO), sulfite:acceptor oxidoreductase (SAOR), thiosulfate:quinone oxidoreductase (TQO), sulfur oxygenase reductase (SOR), adenosine-5′-phosphosulfate reductase (APSR), adenosine-5′-phosphosulfate (APS), adenylylsulfate:phosphate adenylyltransferase (APAT), ATP sulfurylase (ATPS), adenylate kinase (AK).
Chemolithotrophy within the cytoplasm
In contrast to iron oxidation and sulfur reduction, sulfur oxidation takes place largely in the cytoplasm, where it cycles through a number of different reduced inorganic compounds (RISCs) (Fig. 16B). The premier enzyme of sulfur oxidation in the Sulfolobales is the cytoplasmic sulfur oxygenase reductase (SOR). The 24-subunit homomeric enzyme disproportionates zero-valent sulfur into H2S and sulfite (SO32−) without the assistance of any cofactors (Kletzin 1989). An abiotic reaction of these two products generates thiosulfate (S2O32−) as a by-product (Kletzin 1992). These species are coupled to the ETC through a variety of membrane-bound oxidoreductases: sulfide:quinone oxidoreductase (SQR) for H2S (Brito et al. 2009), thiosulfate:quinone oxidoreductase (TQO) for S2O32− (Müller et al. 2004) and sulfite:acceptor oxidoreductase (SAOR) for SO32− (Zimmermann, Laska and Kletzin 1999). In the case of SAOR, activity of the enzyme has been detected in Acidianus ambivalens, but the enzyme has not been linked to an open reading frame in any of the Sulfolobales. Caldariellaquinone is the primary acceptor of electrons for these oxidoreductases.
While the product of SQR is a polysulfide chain that can be recycled to SOR and the product of SAOR is fully oxidized sulfate (SO42−), TQO generates tetrathionate (S4O62−) as a product (Müller et al. 2004). Recent studies have investigated the possibility of tetrathionate acting as the substrate for a set of highly conserved genes in the Sulfolobales, the hdr/dsr/tusA locus. DsrE3A and TusA both appear to be sulfur-trafficking proteins, which cleave the sulfur–sulfur bond of S4O62− to regenerate S2O32− and form an organic persulfide compound. The persulfide ultimately acts as the substrate for the membrane-bound heterodisulfide reductase (Hdr) complex, which generate SO42− (Liu et al.2014b). A tentative role of the Hdr complex is once again the reduction of quinones to conserve energy. However, recent studies in the dimethyl sulfide (DMS)-degrading Hyphomicrobium denitrificans have demonstrated the association of a lipoate-binding protein with the Hdr complex and the importance of the hdr/dsr/tusA locus for energy conservation from DMS (Koch and Dahl 2018). Homologs of these binding proteins have been identified in some Sulfolobales, and the reduction potential of lipoate is sufficient to reduce NAD+, thereby conserving energy (Cao et al. 2018b). While the exact acceptor molecule of Hdr has yet to be confirmed in the Sulfolobales, it seems clear that the complex and its traffickers play some role in conserving energy from tetrathionate. In addition, the complex provides a route to total oxidation of S2O32− to SO42− and may serve to detoxify the abiotic by-product of SOR.
A final avenue to energy conservation is the phosphorylating pathway of cytoplasmic SO32− oxidation. In this pathway sulfite is attached to AMP by adenyl-5′-phosphosulfate reductase (APSR) using an unknown electron acceptor. Adenylylsulfate:phosphate adenylyltransferase (APAT) then replaces the sulfite group with phosphate to generate ADP. Finally, two molecules of ADP are converted to ATP and AMP by adenylate kinase (AK), thereby generating ATP directly from SO32− oxidation (Kappler and Dahl 2001). Much like SAOR, activity of these enzymes has been detected in A. ambivalens cell extracts, but the enzymes have not been purified, characterized or linked to a sequence in the genome (Zimmermann, Laska and Kletzin 1999).
Sulfur transport into the cytoplasm remains something of a mystery in the Sulfolobales. No transporter for zero-valent sulfur has been identified yet in any organism from this order, although a possible sulfate transporter in Metallosphaera cuprina has been identified in transcriptomic data (Jiang et al. 2014). Evidence has been presented, however, for the passive diffusion of H2S through the membrane (Mathai et al. 2009). In addition, an extracellular tetrathionate hydrolase is expressed in the Acidianus spp., many of which are prolific sulfur oxidizers (Protze et al. 2011). The reaction generates thiosulfate extracellularly, which may undergo abiotic reactions in the acidic environment to generate the appropriate sulfur species for transport across the membrane. Given the wide distribution of sulfur substrates used within the cytoplasm, it is difficult to say what RISC might act as the ‘starting point’ of sulfur oxidation, and understanding sulfur transport is still a key area of investigation for chemolithotrophy in the Sulfolobales.
Terminal oxidases and the electron transport chain (ETC)
The ETC in the Sulfolobales deviates from the traditional four-complex structure. While homologs to complexes I and II (NADH dehydrogenase and succinate dehydrogenase, respectively) have been identified (Lemos, Gomes and Teixeira 2001; Melo, Bandeiras and Teixeira 2004), complexes III and IV appear to be combined into single quinone oxidoreductase complexes. In fact, cytochrome c has yet to be found in any of the Sulfolobales. As a result, these quinone oxidoreductases are responsible for the reduction of molecular oxygen and pH homeostasis through proton pumping. A diverse array of terminal oxidases is present throughout the Sulfolobales and appear to relate to the mode of growth for each organism (Table 5). The DoxBCE complex is conserved throughout the Sulfolobales, including in the obligate anaerobe Stygiolobus azoricus, and it appears to sometimes co-purify with the DoxDA subunits of TQO (Purschke et al. 1997). However, SoxABCDD'L and SoxEFGHIM are more varied in their distribution. SoxABCDD'L, which directly pumps protons out of the cell (Gleissner et al. 1997), seems to be associated with aerobic growth. Meanwhile, SoxEFGHIM is absent from known chemolithoautotrophic organisms such as the Acidianus spp., and it is associated with heterotrophic growth (Lubben, Castresana and Warne 1994). An additional quinol oxidase, SoxLN-CbsAB, has been identified in A. ambivalens and is highly similar to the complex III cytochrome bc1 (Bandeiras et al. 2009), but the electron acceptor of this complex has yet to be definitively established. There is a wide range of binding affinities and kinetic parameters for these structures (Schafer, Moll and Schmidt 2001), and it is possible that the kinetics of quinol oxidation plays a key role in energy conservation from inorganic substrates in the Sulfolobales.
Table 5.
Distribution of oxidase complexes in the Sulfolobales.
| Organism | SoxABCDD'L | SoxEFGHIM | DoxBCE | SoxLN-CbsAB |
|---|---|---|---|---|
| Acidianus ambivalens | X | X | ||
| Acidianus brierleyi | X | X | X | |
| Metallosphaera cuprina | X | X | X | X |
| Metallosphaera sedula | X | X | X | X |
| Saccharolobus islandicus | X | X | X | X |
| Saccharolobus solfataricus | X | X | X | |
| Stygiolobus azoricus | X | |||
| Sulfodiicoccus acidophilus | X | X | X | |
| Sulfolobus acidocaldarius | X | X | X | X |
| Sulfuracidifex metallicus | X | X | X | |
| Sulfurisphaera tokodaii | X | X | X | X |
POTENTIAL AND CURRENT USES OF THERMOACIDOPHILES IN BIOTECHNOLOGICAL APPLICATIONS
Challenges in establishing genetic systems in Sulfolobales species
A genetic toolbox is essential to study and understand the function of genes and proteins of an organism and it has to include three essential components: (i) a strain, which is able to take up foreign DNA, (ii) a vector system, allowing for the introduction of genetic material and (iii) a selection/screening system for identification of mutated cells. Much effort has been put into the implementation of established systems from different prokaryotic organisms into the Sulfolobales. The main challenge of this objective is the natural growth conditions of this group. The high temperature and acidic environment lead to degradation of antibiotics and their resistance-mediating enzymes, which are mainly used as selectable markers in mesophilic organisms. There have been numerous attempts to adapt the systems to these difficult conditions, but either the attempt failed (Cammarano et al. 1985; Grogan 1989, 1991; Aagaard et al. 1994; Sanz et al. 1994; Ruggero and Londei 1996; Hjort and Bernander 2001; Bini et al. 2002; Reilly and Grogan 2002) or positive results could not be reproduced (Aravalli and Garrett 1997; Cannio et al. 1998, 2001). Another more promising approach is the use of auxotrophic systems, where metabolically deficient strains are complemented with a functional gene, which becomes a selectable marker. Because these systems use the endogenous metabolic system for selection, they are not affected by the harsh environmental conditions. Therefore, most genetic systems in the Sulfolobales make use of these auxotrophies for mutant selection, which are discussed in this section.
Transformation
The fundamental prerequisite for the establishment of a genetic tool is a reliable transformation protocol, consisting of a transformable strain and a way to introduce DNA into it. The first transformation in Sulfolobales was carried out by Schleper et al. in 1992 using electroporation, to test the infectivity of S. shibatae virus 1 (SSV1) (see the section 'Viruses and CRISPR systems of thermoacidophiles') derived shuttle vector system in Sa. solfataricus P1 (Schleper, Kubo and Zillig 1992). This protocol has been transferred and improved over the years into various other related strains (Zillig et al. 1993; Arnold et al. 1999; Aucelli et al. 2006). Transformation efficiency was later improved by altering the electroporation procedure as well as introducing a regeneration treatment to the cells after the electric shock (Kurosawa and Grogan 2005; Albers and Driessen 2007).
S. acidocaldarius expresses a restriction-modification enzyme, Sua I (Prangishvili et al. 1985), cleaving GGCC sequences lacking N4-methylation on the first cytosine (Grogan 2003). Therefore, the transformation protocol for S. acidocaldarius was significantly improved by methylating these sites on the plasmid DNA (Berkner et al. 2007). Another possibility is to delete Sua1 and use the resulting strain as a host for further experiments (Suzuki and Kurosawa 2016). While methylation of transformed plasmid DNA in S. acidocaldarius is mandatory, it is strain dependent in Sa. islandicus and Sa. solfataricus, with unmodified transformation possible for Sa. solfataricus P1 and Sa. solfataricus 98/2 and their derived strains (Stedman et al. 1999; Albers and Driessen 2007). In any case, electroporation has proven to be the most efficient means for the transformation of species used for genetic systems in the Sulfolobales.
Genetic stability
The most studied Sulfolobales species regarding genetic manipulation are Sa. solfataricus, S. acidocaldarius and Sa. islandicus. In contrast to the other two, S. acidocaldarius exhibits only a small number of insertion elements (Grogan, Carver and Drake 2001). These are highly mobile constructs resulting in an elevated mutation frequency, which is problematic for genetic studies, as whole sections of the genome can be inverted or rearranged within a couple of generations (Redder and Garrett 2006). These elements are most abundant in Sa. solfataricus (Martusewitsch, Sensen and Schleper 2000), making it the most unstable genome. For Sa. islandicus, it depends on the strain, some of which have low mutation frequencies (Berkner and Lipps 2008), making them preferable for studies of this species.
Cryptic and virus-based shuttle vectors
A broad spectrum of different viruses and plasmids was discovered for the Sulfolobales (reviewed by Prangishvili, Stedman and Zillig 2001; Snyder et al. 2003; Prangishvili and Garrett 2004; Lipps 2006), some of which were used to create the first generation of Saccharolobus/Sulfolobus–E. coli shuttle vector systems. The first derived genetic tool, used in different laboratories, was pMJ03 that consisted of the virus DNA of SSV1 and parts of the bacterial pUC18 vector (Jonuscheit et al. 2003). An advantage of using SSV1 DNA is that no selectable markers are required, as the plasmid can transfect cultures independently. However, this is also the largest disadvantage, since viral infection puts the cells under severe stress, causing other problems down the line (e.g. growth retardation or contamination of other close cultures). But with no other selection system available at that time, using virus DNA was the only reliable method for vector spreading.
Another approach was the use of naturally occurring cryptic plasmids as scaffolds for vector systems. The basis is the cryptic plasmids pRN1 and pRN2, which were extracted from Sa. islandicus (Keeling et al. 1996, 1998), with pRN1 as the main plasmid backbone in S. acidocaldarius systems (Berkner et al. 2007), and pRN2 in Sa. islandicus (Deng et al. 2009). However, these systems require a selectable marker for propagation. As the development of these marker systems advanced, the existing system based on SSV1 was largely replaced by cryptic plasmids, which today form the basis for the most used systems in S. acidocaldarius and Sa. islandicus (Deng et al. 2009; Berkner et al. 2010; Wagner et al. 2012).
LacS: first directed mutants in Sulfolobus
The first targeted mutation in Sulfolobales was performed by Blum and co-workers (Worthington et al. 2003), based on a natural lacS deficient strain, Sa. solfataricus lacS::IS1217, that had an insertion in the endogenous lacS cluster making it unable to grow on lactose as a carbon source. Therefore, growth on lactose could be used as a selection after electroporation of linear fragments containing the lacS gene flanked by the region of the DNA where the lacS gene had to be inserted. Later, the strain Sa. solfataricus PBL2025 proved to be more efficient as the recipient strain. In this natural mutant, a 50 kB region spanning the locus SSO_3004 to SSO3050 were deleted, including the lacS gene (Schelert et al. 2004). Improved transformation protocols and a 7–14 days adaptation time after the electroporation in minimal lactose medium enabled the more frequent use of this strain for genetic studies (Albers and Driessen 2007). However, it only works for Sa. solfataricus and Sa. islandicus, since S. acidocaldarius is not able to grow on lactose as the sole carbon source. Therefore, other methods were sought for S. acidocaldarius on the basis of uracil auxotrophic strains and vector systems containing the pyrEF gene cassette as a selection marker.
Genetic system for S. acidocaldarius
The generation of markerless deletion mutants in archaea was first established in the Euryarchaeota Haloferax volcanii using uracil auxotrophy (Bitan-Banin, Ortenberg and Mevarech 2003) and Thermococcus kodakarensis using uracil and tryptophan auxotrophic mutants (Sato et al. 2005), as well as in the methanogens Methanosarcina acetivorans (Pritchett, Zhang and Metcalf 2004) and Methanococcus maripaludis (Moore and Leigh 2005). The genes pyrE and pyrF encode for the enzymes orotate phosphoribosyl transferase and orotidine-5′-monophosphate decarboxylase, respectively, which catalyze the last two steps of the uridine monophosphate synthesis pathway (Grogan and Gunsalus 1993). Upon deletion of one of these two genes, cells lose the ability to grow without uracil supplementation. This deletion can be induced by exposure to the analog substrate 5′-fluoroorotic acid (5-FOA), which is metabolized to cytotoxic products, forcing mutations in the pyrEF cluster and generating auxotrophic colonies (Grogan 1991; Kondo, Yamagishi and Oshima 1991). These generated deficient mutants were first used to test horizontal marker transfer and homologous recombination in S. acidocaldarius (Grogan 1991; Kurosawa and Grogan 2005). S. acidocaldarius is capable of recombining linear DNA fragments into its genome via site specific interactions during rapid growth phases (Grogan and Stengel 2008). The efficiency of the recombination rises proportionally with the length of the fragments used, with 10–30 nt as the minimum length (Kurosawa and Grogan 2005). Additionally, attachment of short flanking sequences to a selectable marker (e.g. pyrEF) could lead to integration of the marker into a gene of interest (Sakofsky, Runck and Grogan 2011), similar to a technique used in S. cerevisiae (Kelly, Lamb and Kelly 2001) or the one previously described to obtain Sa. solfataricus lacS::IS1217 (Worthington et al. 2003). Wagner et al. were able to generate a pyrE deficient mutant, called MW001, derived from S. acidocaldarius DSM639 using this approach (Wagner et al. 2012). S. acidocaldarius MW001 contains a deletion of 322 bp (91–412 bp) in the pyrE gene and only grows in medium supplemented with uracil. In contrast to the typically used 5-FOA or UV light treatment (Grogan 1991), this method ensures a low probability for additional mutations in the genome.
Generation of markerless deletion mutants in S. acidocaldarius
The general idea of this method is an integration of pyrEF into a deficient strain via homologous recombination of target sequences, which flank the marker cassette and can interact with the region around the gene of interest (GOI) (Fig. 17). Positive clones can then be isolated in uracil free medium. The pyrEF sequence is derived from Sa. solfataricus to avoid homologous recombination between the pyrEF in the genome and plasmid. In addition to the auxotrophy, lacS from Sa. solfataricus was introduced into the vector system as a selectable marker allowing for standard blue/white screening with X-Gal staining, with positive clones exhibiting a blue color.
Figure 17.
Mechanisms for the generation of markerless deletion mutants.(A)Plasmid integration occurs via single crossover, resulting in a merodiploidal form. After counterselection with 5-FOA, the pyrEF marker cassette is looped out, either with or without the GOI, resulting in a theoretical ration of one to one in mutated and wild-type cells. Double crossover is feasible by introducing a linearized vector. Depending on the experimental design, either parts of the GOI (B) or an upstream (US) region (C) are introduced for recombination. Counterselection with 5-FOA produces marker-free deletion mutants. (D) A plasmid containing a CRISPR array and a repair fragment with homologous sequences to the GOI are introduced into a recipient strain. Upon induction, crRNA is transcribed and forms a ribonucleoprotein complex with the endogenous Cas protein, scanning the genomic DNA for the spacer sequence and cutting it. Only colonies that conducted recombination with the repair fragment survive. GOI, gene of interest; US, upstream; DS, downstream; pyrEF, pyrEF marker cassette; crRNA, CRISPR RNA; crRNP, ribonucleoprotein complex consisting of crRNA and Cas protein.
Following this, colonies are treated with 5-FOA and uracil, which imposes selective pressure, leading to a loop out of the pyrEF marker to avoid the formation of toxic by-products. The successful marker deletion can again be additionally tested via blue/white staining, with successfully obtained mutants displaying a white coloring. Depending on the design of the experiment, the GOI can be deleted by two different approaches. The first is by cloning the upstream (US) and downstream (DS) regions around the GOI consecutively next to the pyrEF marker cassette. Single-crossover can then occur leading to an integration of the plasmid (Fig. 17A, intermediate state). Upon treatment with 5-FOA, single cross-over can occur again, now between either of the two US or DS regions, respectively, leading to either a deletion of the GOI or the regeneration of the wild-type genotype (Fig. 17A) (Wagner et al. 2009, 2012). In an alternative approach, a linearized plasmid or linear PCR fragment is used, which allows for a double-crossover. For the second crossover site, a part of the GOI sequence is used and cloned in front of pyrEF (Fig. 17B). As only the US is present in a merodiploid form, subsequent removal of pyrEF via loop out generates only mutated colonies (Wagner et al. 2009, 2012).
This strategy is useful to test if a gene and its product have essential functions in the cell and, therefore, cannot be deleted. Removal of the pyrEF marker cassette by loop out allows for reuse of the marker, resulting in the possibility of multiple gene deletions in a single mutant (Meyer et al. 2011; Henche et al. 2012; Wagner et al. 2012). The S. acidocaldarius MW001 strain has so far been used successfully in >100 studies.
Generation of markerless deletion mutants in Sa. islandicus
The starting point in Sa. islandicus was the generation of a pyrEF deletion mutant, Sa. islandicus E233 (She et al. 2009), as a recipient strain for the generation of markerless deletion mutants (Deng et al. 2009). Deng et al. were able to show that their genetic system worked by using the single-crossover (Fig. 17A) and double-crossover (Fig. 17B) strategies, as in S. acidocaldarius. Alternatively, the plasmid is introduced as a linear fragment again, with the US and DS region flanking the pyrEF marker on both sites, plus one additional US or DS site, leading to a double cross-over event. Substitution of the GOI with only pyrEF (Fig. 17C) leads to mutated cells after 5-FOA treatment (Deng et al. 2009). However, the use of pyrEF was not successful in other Sa. islandicus strains, as the background growth on solid medium is always high (Zhang and Whitaker 2012). Therefore, additional selectable markers had to be introduced to achieve higher selective pressure.
Zheng et al. (2012) showed that simvastatin, a thermostable antibiotic, inhibits the growth of Sa. islandicus. Shuttle vector systems were developed for Sa. islandicus 16.4 (Zhang and Whitaker 2012) and Sa. islandicus REY15A (Zheng et al. 2012), based on the resistance mediated by the overexpression of the 3-hydroxy-3-methylglutaryl coenzyme A reductase gene (hmgA). It became apparent, however, that simvastatin-resistant cultures had fitness issues that could only be counteracted by enriching the mutants in liquid medium, resulting in a time-consuming protocol. Therefore, it was suggested to use the simvastatin selection only as a last resort.
By deleting the argD gene encoding for the arginine decarboxylase, Zhang et al. were able to generate an agmatine auxotrophic system (Zhang et al. 2013a). The methodology follows the uracil auxotrophy strategy, as it is possible to recover growth by supplying agmatine or expressing argD in a vector system. With this positive selectable marker, the problems derived from using pyrEF could be solved for Sa. islandicus (Zhang et al. 2013a). In addition, a second counterselectable genetic marker was introduced. Through inactivation of a putative adenine phosphoribosyltransferase, resistance could be mediated to 6-methylpurine (6-MP), a purine analog (Zhang et al. 2016a). Like 5-FOA, the metabolism of 6-MP leads to toxic compounds, which forces the cell to mutate the gene of the catalyzing enzyme from the genome. This system has been successfully used to establish a transposon library, revealing the essential genome of Sa. islandicus (Zhang et al. 2018). This counterselection was also later successfully implemented in Thermococcus barophilus (Birien et al. 2018). Table 6 summarizes the most frequently cited and applied methods of gene disruption and deletion.
Table 6.
Most used knockout systems in Sulfolobales.
| Sulfolobus acidocaldarius | Markerless deletion mutants via crossover based on pyrEF/5-FOA counterselection (Wagner et al. 2012) |
| Saccharolobus solfataricus | Gene disruption via permanent insertion of lacS reporter gene via homologous recombination (Albers and Driessen 2007) |
| Saccharolobus islandicus | Markerless deletion mutants via crossover based on pyrEF/5-FOA counterselection improved with argD selection (Zhang et al. 2013a) |
| Addition of apt/6-MP counterselection (Zhang et al. 2016a) | |
| CRISPR-based gene knockout (Li et al. 2016) |
Expression vectors: promoters and tags
The combination of the development of selectable markers and recipient strains stimulated efforts to create vectors for homologous protein expression, using different promoters and protein tags. Several attempts were made, but none were useful for high-level expression of proteins or for the study of promoters using reporter genes (Aagaard et al. 1996; Elferink, Schleper and Zillig 1996; Aravalli and Garrett 1997; Cannio et al. 1998). The first stable system for homologous expression and tagging of proteins was developed in Sa. solfataricus by Albers and co-workers (Jonuscheit et al. 2003; Albers et al. 2006). As previously mentioned, pMJ03 was designed using the SSV1 virus and pUC18 from E. coli (Jonuscheit et al. 2003). In this work, the pyrEF complementation served as a selectable marker and the heat inducible promoter of the chaperonin tf55α gene was used. The reporter gene was lacS, that codes for a β-galactosidase, and the recipient strain was a double pyrEF/lacS Sa. solfataricus mutant. Under heat shock conditions (shift from 75°C to 88°C), an increase of greater than 10-fold gene expression was seen, measured by northern blot analyses and activity assays of the enzyme. The promoter also had a strong basal expression. This vector was stable for 40–60 generations when cells were maintained in uracil, but propagation of transformants without selective pressure led to loss of the plasmid (Jonuscheit et al. 2003).
Subsequently, this vector was modified. A sugar-inducible promoter (d-arabinose) was added instead of the tf55α promoter along with cloning sites that allowed for the exchange of lacS for a gene of interest. Different tags for protein purification and detection, such as 6× His or Strep, were also added, leading to the pSVA plasmids set (Table 7). The strength of the D-Ara promoter was tested using LacS as a reporter resulting in an increase in activity of 13-fold when d-arabinose was added to the medium. The amount of protein obtained was similar to that of the tf55α promoter, but without the previously seen basal expression levels. The low basal expression can prevent adverse effects of high expression of proteins on growth before induction. While the His tag resulted in a 99% homogeneity rate in purification, Strep-tagged proteins co-eluted with a carboxylase from Sa. solfataricus when low yields of the recombinant protein were obtained (Albers et al. 2006).
Table 7.
Most used expression vectors in Sulfolobales.
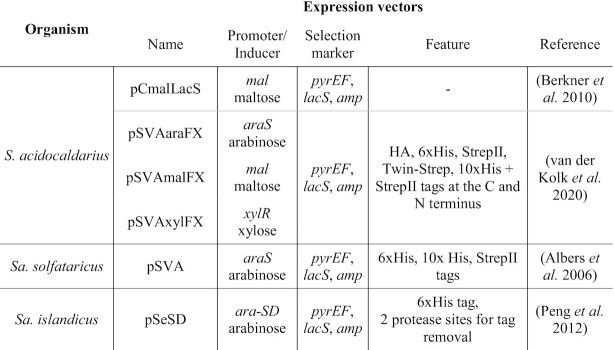
|
An advantage of pMJ03 and derived plasmids was that they were self-spreading, but the production of virus particles had an adverse effect on the transformed cells. A breakthrough came with the use of the plasmid pRN1 in S. acidocaldarius (Berkner et al. 2007). It was stable in different Sulfolobus species and could be easily selected for in S. acidocaldarius using the pyrEF marker cassette. Several promoters were tested including those from tf55α dps,lacS,mal,gdhA and sac7d (Berkner et al. 2010). The promoter from the maltose-binding protein (mal) showed low basal activity and increased expression in the presence of maltose or dextrin in the medium, leading to the plasmid pCmalLacS. The other promoters showed low (dps,lacS) or high constitutive expression (gdhA and sac7d) and were left aside. The promoter from copMA was also tested but exhibited even lower expression levels than the maltose promoter (Wagner et al. 2012).
In a recent study, a d-xylose/l-arabinose promoter was tested and showed less basal activity than the widely used maltose inducible promoter (van der Kolk et al. 2020). Furthermore, this promoter is also d-arabinose inducible, and can be used as a strong inducer similar to IPTG in E. coli since d-arabinose is not a growth substrate for S. acidocaldarius (van der Kolk et al. 2020). This study led to the availability of several expression plasmids for S. acidocaldarius. Using FX cloning (Geertsma 2013) for introduction of the desired gene, it is possible to choose between different promoters (d-xylose/l-arabinose/d-arabinose and maltose inducible) and several tags for proteins (StrepII, His, HA, His + Strep, etc.), either in the N-terminal or C-terminal region of the protein (Table 7) (van der Kolk et al. 2020).
Mutants via CRISPR-Cas and gene silencing
CRISPR systems and spacer acquisition are discussed in the section 'Viruses and CRISPR systems of thermoacidophiles'. CRISPR-Cas systems are classified into three groups so far: type I, associated with Cas3 protein; type II, associated with Cas9; and type III, associated with Cas10. Types I and II recognize DNA via PAM sequences in the protospacer in the target genome. Type III does not need a PAM sequence, but instead a seed sequence, corresponding to a mismatch in the 5′ end of the crRNA (van der Oost, Jackson and Wiedenheft 2014).
In the Sulfolobales, the idea of using the proper CRISPR-Cas system was first explored in Sa. islandicus. Li et al. (2016) used the native type IA and IIIB systems of this organism to generate mutants for lacS, so the phenotype could be easily tested with X-gal. The pSe-Rp plasmid was used as backbone and an artificial CRISPR array was introduced. The array consisted of two repeat sequences, flanking a spacer designed based on a protospacer sequence from the target gene. Also, the plasmid contained a donor region that does not exhibit DNA interference activity. Mutagenesis through deletion, insertion or point mutation is achieved via recombination of the donor region with the genomic DNA (Li et al. 2016).
The CRISPR RNA (crRNA) generated from the CRISPR array of the plasmid guides the native CRISPR-Cas system to self-targeting DNA from wild-type cells, killing them but not the mutants, which accomplished recombination with the donor DNA. Using a plasmid containing the donor sequence and the CRISPR array led to better results than performing co-transformation using a CRISPR plasmid and a short DNA fragment as donor sequence (Li et al. 2016).
In Sa. solfataricus, on the other hand, type III CRISPR-Cas was used to develop silencing via RNA interference (Zebec et al. 2016). Saccharolobus solfataricus has two type IIIB CRISPR complexes, both targeting RNA, and one of them also DNA (Zhang et al. 2016b). A plasmid containing a mini-CRISPR array with a spacer designed to target mRNA from β-galactosidase was used to demonstrate that 50% of gene silencing is possible to achieve using the native CRISPR-Cas system from Sa. solfataricus (Zebec et al. 2014). In later work, silencing up to 90% could be accomplished using a CRISPR array containing five different spacers from the same gene, in this case α-amylase (Zebec et al. 2016). Lower levels of silencing were achieved using one to three spacers, resulting in 35–82% gene silencing. Since one of the type IIIB complexes also targets DNA, the protospacers in the genome were chosen to have a flanking region that matches with the 5′-end handle of the crRNA. This inhibits DNA targeting since CRISPR-Cas system uses the repeat sequences to ‘protect itself’ by recognizing the cells' own genomic DNA (Manica and Schleper 2013).
CRISPR has also been used to edit rod-shaped virus 2 (SIRV-2) from Sa. islandicus, using the archaeon as a host and its endogenous CRISPR system as machinery (Mayo-Muñoz et al. 2018). This method was also used to investigate the core genome of the virus, generating knockout mutants, useful for probing the details of the infection process. In general, CRISPR-Cas systems are powerful genetic tools, which have had a huge impact in the life science community (van der Oost, Jackson and Wiedenheft 2014; Plagens et al. 2015; Mougiakos et al. 2016; Quehenberger et al. 2017), and also show promise for studying and engineering thermoacidophiles.
The potential of thermoacidophiles as metabolic engineering platforms
As outlined in the section 'Metabolism', representatives of the Sulfolobales have been examined for their autotrophic and heterotrophic lifestyles. They have enormous metabolic versatility that differs from species to species, with respect to their growth on a variety of carbon sources that include complex polymers (e.g. polysaccharides, proteins, lipids), monomers (e.g. carbohydrates, amino acids, fatty acids, alcohols, aldehydes) and CO2. In addition, comprehensive biochemical and functional genomics data are available for members of the Sulfolobales (Bräsen et al. 2014), including 13C NMR flux analysis for different carbon sources (Nunn et al. 2010). Furthermore, systems biology (www.sulfosys.com; Zaparty et al. 2009), genome-scale stoichiometric (FBA) (Ulas et al. 2012) and detailed kinetic models (e.g. for gluconeogenesis) (Kouril et al. 2013a,b, 2017) have been established for one of the model Sulfolobales, Sa. solfataricus.
Critical for metabolic engineering is the availability of advanced genetic tools, which are described above for several species in the Sulfolobales and include in-frame markerless deletion mutants, ectopic integration of foreign DNA, and a homologous expression system (Wagner et al. 2014) (see the section 'Metabolism'). Thus, S. acidocaldarius,Sa. islandicus and Sa. solfataricus are all a potential host ‘chassis’ on which to build biosynthetic designs of increasing complexity, although S. acidocaldarius’s genetic stability might be an advantage. Another important criterion for metabolic engineering and application in biotechnology is ease of cultivation under aerobic conditions. Complex and minimal media have been described, e.g. for S. acidocaldarius, and high cell density cultivation has been established for S. shibatae and S. acidocaldarius (Quehenberger et al. 2017, 2020; Schocke, Bräsen and Siebers 2019). The current genetic systems do require further improvement. In particular, the integration of larger gene clusters and the development of new regulatory strategies are needed to fully realize the biotechnological potential of these thermoacidophiles (Crosby et al. 2019).
Thermoacidophile biotechnology
As mentioned above, by their very nature, thermoacidophiles are robust microorganisms that can handle industrial processing conditions well and therefore offer potential advantages as metabolic engineering hosts over more established but less extremophilic species, such as E. coli and S. cerevisiae (Crosby et al. 2019). Their cytosolic enzymes are adapted to high temperature and neutral pH, whereas their extracellular enzymes, such as amylases, cellulases and lipases, are also adapted to low pH. Thus, the properties of thermoacidophilic biocatalysts are consistent with process schemes used in lignocellulosic biomass pre-treatments, which are typically done at high temperatures and low pH. The utilization of thermoacidophiles and their enzymes offers certain benefits for industrial biotechnology (Turner, Mamo and Karlsson 2007; Hess 2008; Zeldes et al. 2015; Straub et al. 2018): at high temperature, reaction rates increase and so does substrate accessibility for biopolymers such as starch and lignocellulosic carbohydrates, thereby enhancing biomass conversion. Since substrate solubility improves at higher temperatures, this enables mixing of otherwise viscous slurries. Furthermore, the energy input for cooling steps in bioreactors and thus production costs can be reduced. Under thermal conditions, volatile products can be removed through gas stripping and evaporation, facilitating product recovery. As such, expensive distillation steps as well as inhibition by toxic products can be minimized, allowing for novel design ‘one-pot’ strategies (Zeldes et al. 2018). Particularly important, microbial contamination is negligible at high temperatures and low pH, so that the use of antibiotics and the need for pharmaceutical-like processing can be avoided (Marhuenda-Egea and Bonete 2002; Champdore et al. 2007; Quehenberger et al. 2017; Cabrera and Blamey 2018).
There are also some disadvantages and challenges to overcome to fully realize the biotechnological potential of thermoacidophiles as industrial microorganisms. As previously described, central metabolic pathways (e.g. for lipid or glycerol degradation) are still not well understood and further work is needed to unravel the metabolic complexity of promising representatives of the Sulfolobales. In particular, networks with regulation at the gene and protein levels require further basic research, and only a few transcriptional regulators have been investigated, and regulation via post-translational modification is also not well understood (see the section 'Genetic mechanisms'). Processes that truly exploit thermoacidophily remain to be developed, although advances in thermoacidophile genetic motivate such efforts.
Biomining applications of thermoacidophiles
The importance of acidophilic organisms in the breakdown of sulfidic ore has been known for many years. Acid mine drainage is a by-product of acidophiles at work on pyritic mine waste and provides an environmental backdrop for studying the mechanism of biological oxidation (Schippers, Jozsa and Sand 1996). However, the same mechanism can be leveraged to extract base and precious metals from sulfidic ores through bioleaching operations. An important distinction to note is that bioleaching refers specifically to the dissolution of metals by bacteria and archaea, while biooxidation simply refers to the oxidation of metal and non-metal substrates by the same organisms.
At first glance, one might suspect that the acidophilic organisms directly attack the solid ore. However, closer inspection of the mechanism shows that the dissolution of both pyrite and other metal sulfides is an indirect result of biooxidation by acidophilic bacteria (Fig. 18). The ore undergoes electrophilic attack in the presence of ferric ions (Fe3+) and protons (Sand et al. 2001). Other ions have also been proposed to facilitate this initial attack, including copper and silver (Hiroyoshi et al. 2000; Zhao et al. 2019). The role of biooxidation in the bioleaching process is then the regeneration of ferrous ions into ferric ions that can again attack the ore and the total oxidation of the freed sulfur to sulfate (Li et al. 2013). Two distinct mechanisms for ore dissolution underscore the importance of the distribution of sulfur in the system. In the presence of pyrite, thiosulfate is directly generated during the electrophilic attack (Fig. 18A). This initiates a cycle of abiotic sulfur reactions, in which greater than 80% of the sulfur product is converted to sulfate (Sand et al. 2001). In contrast, the polysulfide mechanism applies to dissolution of metal sulfide ores, like chalcopyrite (Fig. 18B). Here, the sulfur product of the initial attack is hydrogen sulfide (H2S), which then undergoes a series of abiotic chain elongation reactions to form polysulfides. Ultimately, these polysulfides cyclize to form the thermodynamically stable and water-insoluble S8 ring (Steudel 1996), which accounts for 90–99% of the final sulfur product depending on the metal sulfide species (Sand et al. 2001). While pyrite is the dominant form of metal sulfide ores, mining streams generally contain a mixture of these various crystal structures (Neale et al. 2009), and thus these mechanisms exist simultaneously.
Figure 18.
Mechanisms of sulfidic ore dissolution. (A)Thiosulfate mechanism(B) and polysulfide mechanism. Green dashed arrows indicate biological steps; solid arrows indicate spontaneous abiotic reactions; blue dashed-dotted arrows indicate an overall transformation involving multiple reaction steps; yellow dashed-dotted arrows represent phase transition. Bold box around a species indicates that this is the dominant sulfur product of the dissolution process.
A key issue associated with the polysulfide mechanism generating solid cyclic sulfur is the passivation of the ore's surface (Zhao et al. 2019). Passivation is the formation of an inhibitory film on the surface of the ore that prevents further electrophilic attack (Klauber 2008). In the case of chalcopyrite, a passivating layer of solid sulfur coats the ore and significantly slows the dissolution rate. Another component of this passivating effect is the formation of jarosites, complex ferric sulfate compounds, that is only an issue with long leaching times. However, the slowed dissolution rate caused by passivating sulfur may allow for the accumulation of jarosites, and both modes of inhibition are seen in chalcopyrite bioleaching (Klauber 2008).
There are two primary formats for industrial-scale bioleaching. Heap leaching, as the name implies, is the open-air extraction of metals from large heaps of mine tailings. Leaching solution is percolated throughout the pile, with the metal-laden runoff collected for downstream processing (Schlitt 2006). The capital investment of these operations is generally low and uses low-grade ore to maximize yield from mining operations. These heaps are often self-inoculated and contain a consortium of acidophilic organisms, including mesophilic Acidithiobacillus spp., moderately thermophilic Sulfolobacillus spp., and extremely thermophilic Acidianus spp. and Metallosphaera spp. (Pradhan et al. 2008). Despite the open-air environment of the heaps, the highly exothermic oxidation of sulfur results in heap temperatures reaching as high as 81°C, which supports the growth of the Sulfolobales (Pradhan et al. 2008).
The second format is a traditional bioreactor. While the capital cost of this setup is much higher than that of heap leaching, the more controlled environment and improved contact area dramatically reduce the leaching time. While heap operations run for months to years (Schlitt 2006), bioreactor leaching has a retention time on the order of days (Neale et al. 2009). These bioreactors are often still self-inoculating and contain a complex microbial landscape. Notably, the acidic conditions provide a natural barrier to contamination, and this barrier is further enhanced when working at the high temperatures of the Sulfolobales.
Bioleaching at the high temperatures required by the Sulfolobales offers distinct advantages. Given the large exotherm associated with sulfur oxidation, the reactor does not need to be heated to maintain the temperatures necessary for Sulfolobales’ growth. In fact, cooling is necessary to maintain the constant temperatures (Neale et al. 2009), and at higher temperatures less cooling is necessary. Furthermore, chalcopyrite dissolution kinetics are significantly improved at higher temperatures (Watling 2006). In a 10-day lab-scale bioleaching reactor, around 70% copper recovery was achieved with the mesophilic Acidithiobacillus ferrooxidans and roughly 85% copper recovery was achieved with the moderate thermophile Sulfobacillus (Mousavi et al. 2005). At pilot scale, a thermoacidophile bioreactor containing Acidianus, Metallosphaera and Sulfolobus spp. achieved 95% copper recovery (Neale et al. 2009).
As understanding of the mechanisms for iron and sulfur oxidation improves, engineering an optimized bioleaching organism presents an intriguing possibility. In particular, this could overcome the obstacle of surface passivation caused by the accumulation of elemental sulfur and jarosites. Indeed, one means of controlling the passivation of ore is to lower the reduction potential of the reactor. This process controls the ratio of Fe3+/Fe2+ ions in order to limit the rate of elemental sulfur and jarosite formation. A variety of approaches to control the redox potential of the system have been explored, ranging from adding reagents to form new redox couples, controlling dissolved oxygen levels and adjusting the microbe composition (Zhao et al. 2019). This last option points toward the possibility of an optimized bioleaching organism that manages the redox potential through the relative rates of sulfur and iron oxidation. Efforts to generate an optimized strain of M. sedula through laboratory evolution had moderate success in this regard (McCarthy, Ai and Blum 2018). While natural evolution of the ability to breakdown chalcopyrite may provide novel insights, tailoring an organism to mitigate surface passivation may require a more controlled approach. As such, the genetic tools available to select members of the Sulfolobales provide a promising platform to expand their bioleaching capabilities.
CONCLUSIONS
The scientific and technological potential of the Sulfolobales has come a long way since their isolation more than a half century ago. During this time, molecular biology and genomics came of age, with all of the associated tools that can be brought to bear in understanding the microbiology of these thermoacidophiles. Here, the goal was to provide some historical perspective as well as to give an up-to-date overview of where the world of the Sulfolobales stands. Despite the length and breadth of this review, we have likely inadvertently left out important contributions to the field and thank all of those who have studied and reported on facets of these interesting microorganisms.
FUNDING
This work was supported by grants to RMK from the U.S. Air Force Office of Scientific Research (AFOSR) (FA9550-17-1-0268) and the U.S. National Science Foundation (CBET-1802939). JAC and DJW acknowledge support from the U.S. National Institutes of Health Biotechnology Traineeships (T32 GM008776-16, T32 GM133366-01). TEFQ was supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) with an Emmy Noether grant (411069969). AR and JB were supported by the grant HotAcidFactory (031B0848C) from the BMBF (Bundesministeriums für Bildung und Forschung; Federal Ministry of Education and Research, Germany). PN received funding from the Volkswagen Foundation on Momentum grant 94933. LS was supported by the Mercator Foundation with a MERCUR startup grant (Pr-2013-0010) and by the BMBF (Bundesministeriums für Bildung und Forschung; Federal Ministry of Education and Research, Germany) grant HotSysAPP (031L0078A) within the e:Bio2 funding initiative. LSch was supported by the BMBF (Bundesministeriums für Bildung und Forschung; Federal Ministry of Education and Research, Germany) grant HotAcidFactory (031B0848A). BS and CB acknowledge funding from the Volkswagen Foundation on the ‘Experiment?’ Lipid Divide grant (96725). EP was supported by the Research Foundation Flanders (Fonds Wetenschappelijk Onderzoek-Vlaanderen) (grant 1526418N and projects G021118 and G062820N).
Contributor Information
April M Lewis, Department of Chemical and Biomolecular Engineering, North Carolina State University. Raleigh, NC 27695, USA.
Alejandra Recalde, Institute for Biology, Molecular Biology of Archaea, University of Freiburg, 79104 Freiburg, Germany.
Christopher Bräsen, Department of Molecular Enzyme Technology and Biochemistry, Environmental Microbiology and Biotechnology, and Centre for Water and Environmental Research, University of Duisburg-Essen, 45117 Essen, Germany.
James A Counts, Department of Chemical and Biomolecular Engineering, North Carolina State University. Raleigh, NC 27695, USA.
Phillip Nussbaum, Institute for Biology, Molecular Biology of Archaea, University of Freiburg, 79104 Freiburg, Germany.
Jan Bost, Institute for Biology, Molecular Biology of Archaea, University of Freiburg, 79104 Freiburg, Germany.
Larissa Schocke, Department of Molecular Enzyme Technology and Biochemistry, Environmental Microbiology and Biotechnology, and Centre for Water and Environmental Research, University of Duisburg-Essen, 45117 Essen, Germany.
Lu Shen, Department of Molecular Enzyme Technology and Biochemistry, Environmental Microbiology and Biotechnology, and Centre for Water and Environmental Research, University of Duisburg-Essen, 45117 Essen, Germany.
Daniel J Willard, Department of Chemical and Biomolecular Engineering, North Carolina State University. Raleigh, NC 27695, USA.
Tessa E F Quax, Archaeal Virus–Host Interactions, Faculty of Biology, University of Freiburg, 79104 Freiburg, Germany.
Eveline Peeters, Research Group of Microbiology, Department of Bioengineering Sciences, Vrije Universiteit Brussel, 1050 Brussels, Belgium.
Bettina Siebers, Department of Molecular Enzyme Technology and Biochemistry, Environmental Microbiology and Biotechnology, and Centre for Water and Environmental Research, University of Duisburg-Essen, 45117 Essen, Germany.
Sonja-Verena Albers, Institute for Biology, Molecular Biology of Archaea, University of Freiburg, 79104 Freiburg, Germany.
Robert M Kelly, Department of Chemical and Biomolecular Engineering, North Carolina State University. Raleigh, NC 27695, USA.
Conflict of Interest
None declared.
REFERENCES
- Aagaard C, Leviev I, Aravalli RN et al. General vectors for archaeal hyperthermophiles: strategies based on a mobile intron and a plasmid. FEMS Microbiol Rev. 1996;18:93–104. [DOI] [PubMed] [Google Scholar]
- Aagaard C, Phan H, Trevisanato S et al. A spontaneous point mutation in the single 23S rRNA gene of the thermophilic arachaeon Sulfolobus acidocaldarius confers multiple drug resistance. J Bacteriol. 1994;176:7744–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed H, Ettema TJG, Tjaden B et al. The semi-phosphorylative Entner–Doudoroff pathway in hyperthermophilic archaea: a re-evaluation. Biochem J. 2005;390:529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S-V, Driessen AJM. Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea. 2007;2:145–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S-V, Jarrell KF. The archaellum: how Archaea swim. Front Microbiol. 2015;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S-V, Jonuscheit M, Dinkelaker S et al. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol. 2006;72:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S-V, Koning SM, Konings WN et al. Insights into ABC transport in archaea. J Bioenerg Biomembr. 2004;36:5–15. [DOI] [PubMed] [Google Scholar]
- Albers S-V, Konings WN, Driessen AJ. A unique short signal sequence in membrane-anchored proteins of Archaea. Mol Microbiol. 1999;31:1595–6. [DOI] [PubMed] [Google Scholar]
- Albers S-V, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–26. [DOI] [PubMed] [Google Scholar]
- Albers S-V, Pohlschröder M. Diversity of archaeal type IV pilin-like structures. Extremophiles. 2009;13:403–10. [DOI] [PubMed] [Google Scholar]
- Albers S-V, Szabó Z, Driessen AJM. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol. 2003;185:3918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albi T, Serrano A. Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J Microbiol Biotechnol. 2016;32:27. [DOI] [PubMed] [Google Scholar]
- Amend JP, Shock EL. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol Rev. 2001;25:175–243. [DOI] [PubMed] [Google Scholar]
- Anderson I, Scheuner C, Goker M et al. Novel insights into the diversity of catabolic metabolism from ten haloarchaeal genomes. PLoS One. 2011;6:e20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum RS, Bray SM, Blackwood JK et al. Involvement of a eukaryotic-like ubiquitin-related modifier in the proteasome pathway of the archaeon Sulfolobus acidocaldarius. Nat Commun. 2015;6:8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao X, Li Y, Wang F et al. The Sulfolobus initiator element is an important contributor to promoter strength. J Bacteriol. 2013;195:5216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravalli RN, Garrett RA. Shuttle vectors for hyperthermophilic archaea. Extremophiles. 1997;1:183–91. [DOI] [PubMed] [Google Scholar]
- Aravind L. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucl Acids Res. 1999;27:4658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM, Logsdon JM, Doolittle WF. Recurrent paralogy in the evolution of archaeal chaperonins. Curr Biol. 1999;9:1053–6. [DOI] [PubMed] [Google Scholar]
- Arnold HP, She Q, Phan H et al. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol Microbiol. 1999;34:217–26. [DOI] [PubMed] [Google Scholar]
- Arnold HP, Ziese U, Zillig W. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology. 2000;272:409–16. [DOI] [PubMed] [Google Scholar]
- Athukoralage JS, Graham S, Rouillon C et al. The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. eLife. 2020a;9:e55852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, McMahon SA, Zhang C et al. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020b;577:572–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, Rouillon C, Graham S et al. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature. 2018;562:277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atomi H, Matsumi R, Imanaka T. Reverse gyrase is not a prerequisite for hyperthermophilic life. J Bacteriol. 2004;186:4829–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aucelli T, Contursi P, Girfoglio M et al. A spreadable, non-integrative and high copy number shuttle vector for Sulfolobus solfataricus based on the genetic element pSSVx from Sulfolobus islandicus. Nucl Acids Res. 2006;34:e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auernik KS, Kelly RM. Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl Environ Microbiol. 2008;74:7723–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auernik KS, Maezato Y, Blum PH et al. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl Environ Microbiol. 2008;74:682–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano T, Wilming A, Tomita H et al. Characterization of two members among the five ADP-forming acyl coenzyme A (acyl-CoA) synthetases reveals the presence of a 2-(imidazol-4-yl)acetyl-CoA synthetase in Thermococcus kodakarensis. J Bacteriol. 2014;196:140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes R, Lemmens L, Mignon K et al. Defining heat shock response for the thermoacidophilic model crenarchaeon Sulfolobus acidocaldarius. Extremophiles. 2020;24:681–92. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C, Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007;15:165–71. [DOI] [PubMed] [Google Scholar]
- Bandeiras TM, Refojo PN, Todorovic S et al. The cytochrome ba complex from the thermoacidophilic crenarchaeote Acidianus ambivalens is an analog of bc(1) complexes. Biochim Biophys Acta. 2009;1787:37–45. [DOI] [PubMed] [Google Scholar]
- Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: structure and function. Biochim Biophys Acta. 2012;1823:67–82. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2:17092. [DOI] [PubMed] [Google Scholar]
- Bathe S, Norris PR. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl Environ Microbiol. 2007;73:2491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista MA, Black JA, Youngblut ND et al. Differentiation and structure in Sulfolobus islandicus rod-shaped virus populations. Viruses. 2017;9:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SD, Botting CH, Wardleworth BN et al. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–51. [DOI] [PubMed] [Google Scholar]
- Bell SD, Brinkman AB, Van Der Oost J et al. The archaeal TFIIEα homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2001;2:133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SD, Jackson SP. Mechanism of autoregulation by an archaeal transcriptional repressor. J Biol Chem. 2000;275:31624–9. [DOI] [PubMed] [Google Scholar]
- Bell SD, Kosa PL, Sigler PB et al. Orientation of the transcription preinitiation complex in Archaea. Proc Natl Acad Sci USA. 1999;96:13662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelli D, Maone E, Londei P. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol Microbiol. 2003;50:635–43. [DOI] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Buckel W et al. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–6. [DOI] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Ramos-Vera WH et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010a;8:447–60. [DOI] [PubMed] [Google Scholar]
- Berg IA, Ramos-Vera WH, Petri A et al. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiologyopen. 2010b;156:256–69. [DOI] [PubMed] [Google Scholar]
- Berkner S, Grogan D, Albers S-V et al. Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucl Acids Res. 2007;35:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner S, Lipps G. Mutation and reversion frequencies of different Sulfolobus species and strains. Extremophiles. 2008;12:263–70. [DOI] [PubMed] [Google Scholar]
- Berkner S, Wlodkowski A, Albers S-V et al. Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles. 2010;14:249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernander R, Poplawski A. Cell cycle characteristics of thermophilic Archaea. J Bacteriol. 1997;179:4963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoldo C, Dock C, Antranikian G. Thermoacidophilic microorganisms and their novel biocatalysts. Eng Life Sci. 2004;4:521–32. [Google Scholar]
- Bettstetter M, Peng X, Garrett RA et al. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology. 2003;315:68–79. [DOI] [PubMed] [Google Scholar]
- Bhoobalan-Chitty Y, Johansen TB, Di Cianni N et al. Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell. 2019;179:448–58.e411. [DOI] [PubMed] [Google Scholar]
- Bigotti MG, Clarke AR. Cooperativity in the thermosome. J Mol Biol. 2005;348:13–26. [DOI] [PubMed] [Google Scholar]
- Bini E, Dikshit V, Dirksen K et al. Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA. 2002;8:1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birien T, Thiel A, Henneke G et al. Development of an effective 6-methylpurine counterselection marker for genetic manipulation in Thermococcus barophilus. Genes. 2018;9:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof LF, Haurat MF, Albers SV. Two membrane-bound transcription factors regulate expression of various type-IV-pili surface structures in Sulfolobus acidocaldarius. PeerJ. 2019;7:e6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol. 2003;185:772–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bize A, Karlsson EA, Ekefjärd K et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106:11306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach F, Matelska D, Fouqueau T et al. Key concepts and challenges in archaeal transcription. J Mol Biol. 2019;431:4184–201. [DOI] [PubMed] [Google Scholar]
- Blombach F, Salvadori E, Fouqueau T et al. Archaeal TFEα/β is a hybrid of TFIIE and the RNA polymerase III subcomplex hRPC62/39. eLife. 2015;4:e08378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumentals II, Itoh M, Olson GJ et al. Role of polysulfides in reduction of elemental sulfur by the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl Environ Microbiol. 1990;56:1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum H, Zillig W, Mallok S et al. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology. 2001;281:6–9. [DOI] [PubMed] [Google Scholar]
- Bogdanova TI, Tsaplina IA, Kondrat'eva TF et al. Sulfobacillus thermotolerans sp. nov., a thermotolerant, chemolithotrophic bacterium. Int J Syst Evol Microbiol. 2006;56:1039–42. [DOI] [PubMed] [Google Scholar]
- Bolduc B, Shaughnessy DP, Wolf YI et al. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol. 2012;86:5562–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiologyopen. 2005;151:2551–61. [DOI] [PubMed] [Google Scholar]
- Bonneau R, Facciotti MT, Reiss DJ et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–65. [DOI] [PubMed] [Google Scholar]
- Borges AL, Davidson AR, Bondy-Denomy J. The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu Rev Virol. 2017;4:37–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Fecteau KM, Havig JR et al. Modeling the habitat range of phototrophs in Yellowstone National Park: toward the development of a comprehensive fitness landscape. Front Microbiol. 2012;3:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Jackson RA, Encarnacion G et al. Isolation, characterization, and ecology of sulfur-respiring Crenarchaea inhabiting acid-sulfate-chloride-containing geothermal springs in Yellowstone National Park. Appl Environ Microbiol. 2007;73:6669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Pearson A, Pi Y et al. Temperature and pH controls on glycerol dibiphytanyl glycerol tetraether lipid composition in the hyperthermophilic crenarchaeon Acidilobus sulfurireducens. Extremophiles. 2011;15:59–65. [DOI] [PubMed] [Google Scholar]
- Brenneis M, Hering O, Lange C et al. Experimental characterization of cis-acting elements important for translation and transcription in halophilic Archaea. PLoS Genet. 2007;3:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley CL, Brierley JA. A chemoautotrophic and thermophilic microorganism isolated from an acid hot spring. Can J Microbiol. 1973;19:183–8. [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Bell SD, Lebbinkt RJ et al. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J Biol Chem. 2002;277:29537–49. [DOI] [PubMed] [Google Scholar]
- Brito JA, Sousa FL, Stelter M et al. Structural and functional insights into sulfide: quinone oxidoreductase. Biochemistry. 2009;48:5613–22. [DOI] [PubMed] [Google Scholar]
- Brock TD, Brock KM, Belly RT et al. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. [DOI] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Walther J, Snijders APL et al. Identification of the missing links in prokaryotic pentose oxidation pathways. J Biol Chem. 2006;281:27378–88. [DOI] [PubMed] [Google Scholar]
- Brown AD, Simpson JR. Water relations of sugar-tolerant yeasts: the role of intracellular polyols. J Gen Microbiol. 1972;72:589–91. [DOI] [PubMed] [Google Scholar]
- Brown PB, Wolfe GV. Protist genetic diversity in the acidic hydrothermal environments of Lassen Volcanic National Park, USA. J Eukaryot Microbiol. 2006;53:420–31. [DOI] [PubMed] [Google Scholar]
- Brumfield SK, Ortmann AC, Ruigrok V et al. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus Sulfolobus turreted icosahedral virus. J Virol. 2009;83:5964–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräsen C, Esser D, Rauch B et al. Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol Mol Biol Rev. 2014;78:89–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräsen C, Schönheit P. Regulation of acetate and acetyl-CoA converting enzymes during growth on acetate and/or glucose in the halophilic archaeon Haloarcula marismortui. FEMS Microbiol Lett. 2004;241:21–6. [DOI] [PubMed] [Google Scholar]
- Bulaev AG, Kanygina AV, Manolov AI. Genome analysis of Acidiplasmasp. MBA-1, a polyextremophilic archaeon predominant in the microbial community of a bioleaching reactor. Microbiologyopen. 2017;86:89–95. [PubMed] [Google Scholar]
- Cabrera MÁ, Blamey JM. Biotechnological applications of archaeal enzymes from extreme environments. Biol Res. 2018;51:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillat C, Maity S, Miguet N et al. The role of VPS4 in ESCRT-III polymer remodeling. Biochem Soc Trans. 2019;47:441–8. [DOI] [PubMed] [Google Scholar]
- Cammarano P, Teichner A, Londei P et al. Insensitivity of archaebacterial ribosomes to protein synthesis inhibitors. Evolutionary implications. EMBO J. 1985;4:811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KM, Kouris A, England W et al. Sulfolobus islandicus meta-populations in Yellowstone National Park hot springs. Environ Microbiol. 2017;19:2334–47. [DOI] [PubMed] [Google Scholar]
- Cannio R, Catara G, Fiume I et al. Identification of a cell-bound extracellular protease overproduced by Sulfolobus solfataricus in peptide-rich media. Protein Pept Lett. 2010;17:78–85. [DOI] [PubMed] [Google Scholar]
- Cannio R, Contursi P, Rossi M et al. An autonomously replicating transforming vector for Sulfolobus solfataricus. J Bacteriol. 1998;180:3237–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannio R, Contursi P, Rossi M et al. Thermoadaptation of a mesophilic hygromycin B phosphotransferase by directed evolution in hyperthermophilic Archaea: selection of a stable genetic marker for DNA transfer into Sulfolobus solfataricus. Extremophiles. 2001;5:153–9. [DOI] [PubMed] [Google Scholar]
- Cannio R, Di Prizito N, Rossi M et al. A xylan-degrading strain of Sulfolobus solfataricus: isolation and characterization of the xylanase activity. Extremophiles. 2004;8:117–24. [DOI] [PubMed] [Google Scholar]
- Cao J, Wang Q, Liu T et al. Insights into the post-translational modifications of archaeal Sis10b (Alba): lysine-16 is methylated, not acetylated, and this does not regulate transcription or growth. Mol Microbiol. 2018a;109:192–208. [DOI] [PubMed] [Google Scholar]
- Cao X, Koch T, Steffens L et al. Lipoate-binding proteins and specific lipoate-protein ligases in microbial sulfur oxidation reveal an atpyical role for an old cofactor. eLife. 2018b;7:e37439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi Y, Dekker C. Dividing the archaeal way: the ancient Cdv cell-division machinery. Front Microbiol. 2018;9:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champdore Md, Staiano M, Rossi M et al. Proteins from extremophiles as stable tools for advanced biotechnological applications of high social interest. J R Soc Interface. 2007;4:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JJ, Smits C, Aragao D et al. Structural and functional insights into the evolution and stress adaptation of type II chaperonins. Structure. 2016;24:364–74. [DOI] [PubMed] [Google Scholar]
- Chaudhury P, Quax TEF, Albers S-V. Versatile cell surface structures of Archaea. Mol Microbiol. 2018;107:298–311. [DOI] [PubMed] [Google Scholar]
- Chen KY, Morris JC. Kinetics of oxidation of aqueous sulfide by oxygen. Environ Sci Technol. 1972;6:529–37. [Google Scholar]
- Chen L, Brügger K, Skovgaard M et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol. 2005;187:4992–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Hwang S, Cha J. Identification and characterization of MalA in the maltose/maltodextrin operon of Sulfolobus acidocaldarius DSM639. J Bacteriol. 2013;195:1789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y-H, Park Y-J, Yoon S-J et al. Purification and characterization of a new inducible thermostable extracellular lipolytic enzyme from the thermoacidophilic archaeon Sulfolobus solfataricus P1. J Mol Cataly B: Enzym. 2016;124:11–9. [Google Scholar]
- Chong PK, Burja AM, Radianingtyas H et al. Proteome and transcriptional analysis of ethanol-grown Sulfolobus solfataricus P2 reveals ADH2, a potential alcohol dehydrogenase. J Proteome Res. 2007a;6:3985–94. [DOI] [PubMed] [Google Scholar]
- Chong PK, Burja AM, Radianingtyas H et al. Translational and transcriptional analysis of Sulfolobus solfataricus P2 to provide insights into alcohol and ketone utilisation. Proteomics. 2007b;7:424–35. [DOI] [PubMed] [Google Scholar]
- Choquet CG, Patel GB, Sprott GD et al. Stability of pressure-extruded liposomes made from archaeobacterial ether lipids. Appl Microbiol Biotechnol. 1994;42:375–84. [DOI] [PubMed] [Google Scholar]
- Clore AJ, Stedman KM. The SSV1 viral integrase is not essential. Virology. 2007;361:103–11. [DOI] [PubMed] [Google Scholar]
- Cohen O, Doron S, Wurtzel O et al. Comparative transcriptomics across the prokaryotic tree of life. Nucleic Acids Res. 2016;44:W46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman DR, Poudel S, Hamilton TL et al. Geobiological feedbacks and the evolution of thermoacidophiles. ISME J. 2018;12:225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contursi P, D'Ambrosio K, Pirone L et al. C68 from the Sulfolobus islandicus plasmid–virus pSSVx is a novel member of the AbrB-like transcription factor family. Biochem J. 2011;435:157–66. [DOI] [PubMed] [Google Scholar]
- Contursi P, Farina B, Pirone L et al. Structural and functional studies of Stf76 from the Sulfolobus islandicus plasmid–virus pSSVx: a novel peculiar member of the winged helix-turn-helix transcription factor family. Nucleic Acids Res. 2014;42:5993–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper Charlotte R, Daugherty Amanda J, Tachdjian S et al. Role of vapBC toxin–antitoxin loci in the thermal stress response of Sulfolobus solfataricus. Biochem Soc Trans. 2009;37:123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram NJ, Rawlings DE. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40°C. Appl Environ Microbiol. 2002;68:838–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson RMR, Touboul N, Ouzounis CA. Lineage-specific partitions in archaeal transcription. Archaea. 2007;2:117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counts JA, Willard DJ, Kelly RM. Life in hot acid: a genome-based reassessment of the archaeal order Sulfolobales. Environ Microbiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counts JA, Zeldes BM, Lee LL et al. Physiological, metabolic and biotechnological features of extremely thermophilic microorganisms. Wiley Interdiscip Rev Syst Biol Med. 2017;9:1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M, Lindås AC. The DNA methylome of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. Front Microbiol. 2018;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby JR, Laemthong T, Lewis AM et al. Extreme thermophiles as emerging metabolic engineering platforms. Curr Opin Biotech. 2019;59:55–64. [DOI] [PubMed] [Google Scholar]
- Cunningham Dueber EL, Corn JE, Bell SD et al. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–3. [DOI] [PubMed] [Google Scholar]
- Dahche H, Abdullah A, Potters MB et al. A PPM-family protein phosphatase from the thermoacidophile Thermoplasma volcanium hydrolyzes protein-bound phosphotyrosine. Extremophiles. 2009;13:371. [DOI] [PubMed] [Google Scholar]
- Danson MJ. Archaebacteria: the comparative enzymology of their central metabolic pathways. Adv Microb Physiol. 1988;29:165–231. [DOI] [PubMed] [Google Scholar]
- Dar D, Prasse D, Schmitz RA et al. Widespread formation of alternative 3′ UTR isoforms via transcription termination in Archaea. Nat Microbiol. 2016;1:16143. [DOI] [PubMed] [Google Scholar]
- Darland G, Brock TD, Samsonoff W et al. A thermophilic, acidophilic mycoplasma isolated from a coal refuse pile. Science. 1970;170:1416–8. [DOI] [PubMed] [Google Scholar]
- Darland G, Brock TD. Bacillus acidocaldarius sp. nov., an acidophilic thermophilic spore-forming bacterium. J Gen Microbiol. 1971;67:9–15. [Google Scholar]
- Daum B, Quax TEF, Sachse M et al. Self-assembly of the general membrane-remodeling protein PVAP into sevenfold virus-associated pyramids. Proc Natl Acad Sci USA. 2014;111:3829–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellas N, Snyder JC, Bolduc B et al. Archaeal viruses: diversity, replication, and structure. Annu Rev Virol. 2014;1:399–426. [DOI] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA et al. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–99. [DOI] [PubMed] [Google Scholar]
- Deng L, He F, Bhoobalan-Chitty Y et al. Unveiling cell surface and type IV secretion proteins responsible for archaeal rudivirus entry. J Virol. 2014;88:10264–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Zhu H, Chen Z et al. Unmarked gene deletion and host–vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009;13:735. [DOI] [PubMed] [Google Scholar]
- De Rosa M, Gambacorta A, Nicolaus B et al. Glucose metabolism in the extreme thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem J. 1984;224:407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWerff SJ, Bautista MA, Pauly M et al. Killer Archaea: virus-mediated antagonism to CRISPR-immune populations results in emergent virus–host mutualism. mBio. 2020;11:e00404–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrova DV, Galperin MY, Mulkidjanian AY. Phylogenomic reconstruction of archaeal fatty acid metabolism. Environ Microbiol. 2014;16:907–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore A, Fiorentino G, Vitale RM et al. Structural analysis of BldR from Sulfolobus solfataricus provides insights into the molecular basis of transcriptional activation in Archaea by MarR family proteins. J Mol Biol. 2009;388:559–69. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Yu X, Rensen E et al. A virus that infects a hyperthermophile encapsidates A-form DNA. Science. 2015;348:914–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne I, Nookala RK, Jackson SP et al. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Cell. 2003a;11:275–82. [DOI] [PubMed] [Google Scholar]
- Dionne I, Robinson N, McGeoch A et al. DNA replication in the hyperthermophilic archaeon Sulfolobus solfataricus. Biochem Soc Trans. 2003b;31:674–6. [DOI] [PubMed] [Google Scholar]
- Dobro MJ, Samson RY, Yu Z et al. Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol Biol Cell. 2013;24:2319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopson M. Physiological and phylogenetic diversity of acidophilic Bacteria. In: Quatrini R, Barrie JD, (eds). Acidophiles: Life in Extremely Acidic Environments. Norfolk, UK: Caister Academic Press, 2016, 79–91. [Google Scholar]
- Dorman CJ, Schumacher MA, Bush MJ et al. When is a transcription factor a NAP?. Curr Opin Microbiol. 2020;55:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359:eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. Avoiding dangerous missense: thermophiles display especially low mutation rates. PLoS Genet. 2009;5:e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen RPC, Lin SN, Waterreus WJ et al. Diverse architectural properties of Sso10a proteins: evidence for a role in chromatin compaction and organization. Sci Rep. 2016;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozd M, Chandrashekhar K, Rajashekara G. Polyphosphate-mediated modulation of Campylobacter jejuni biofilm growth and stability. Virulence. 2014;5:680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, He Z-G. Characterization of the specific interaction between archaeal FHA domain-containing protein and the promoter of a flagellar-like gene-cluster and its regulation by phosphorylation. Biochem Biophys Res Commun. 2011;407:242–7. [DOI] [PubMed] [Google Scholar]
- Duggin IG, Dubarry N, Bell SD. Replication termination and chromosome dimer resolution in the archaeon Sulfolobus solfataricus. EMBO J. 2011;30:145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2008;105:16737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink MG, Albers SV, Konings WN et al. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol Microbiol. 2001;39:1494–503. [DOI] [PubMed] [Google Scholar]
- Elferink MG, Schleper C, Zillig W. Transformation of the extremely thermoacidophilic archaeon Sulfolobus solfataricus via a self-spreading vector. FEMS Microbiol Lett. 1996;137:31–5. [DOI] [PubMed] [Google Scholar]
- Elferink MGL, de Wit JG, Driessen AJM et al. Stability and proton-permeability of liposomes composed of archaeal tetraether lipids. Biochim Biophys Acta. 1994;1193:247–54. [DOI] [PubMed] [Google Scholar]
- Ellen AF, Albers S-V, Driessen AJ. Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles. 2010;14:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellen AF, Albers S-V, Huibers W et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles. 2009;13:67–79. [DOI] [PubMed] [Google Scholar]
- England P, Wehenkel A, Martins S et al. The FHA-containing protein GarA acts as a phosphorylation-dependent molecular switch in mycobacterial signaling. FEBS Lett. 2009;583:301–7. [DOI] [PubMed] [Google Scholar]
- Enoru-Eta J, Gigot D, Thia-Toong TL et al. Purification and characterization of Sa-Lrp, a DNA-binding protein from the extreme thermoacidophilic archaeon Sulfolobus acidocaldarius homologous to the bacterial global transcriptional regulator Lrp. J Bacteriol. 2000;182:3661–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser D, Hoffmann L, Pham TK et al. Protein phosphorylation and its role in archaeal signal transduction. FEMS Microbiol Rev. 2016;40:625–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser D, Kouril T, Talfournier F et al. Unraveling the function of paralogs of the aldehyde dehydrogenase super family from Sulfolobus solfataricus. Extremophiles. 2013;17:205–16. [DOI] [PubMed] [Google Scholar]
- Esser D, Pham T, Reimann J et al. Change of carbon source causes dramatic effects in the phospho-proteome of the archaeon Sulfolobus solfataricus. J Proteome Res. 2012;11:4823–33. [DOI] [PubMed] [Google Scholar]
- Esser D, Siebers B. Atypical protein kinases of the RIO family in archaea. Biochem Soc Trans. 2013;41:399–404. [DOI] [PubMed] [Google Scholar]
- Estelmann S, Hugler M, Eisenreich W et al. Labeling and enzyme studies of the central carbon metabolism in Metallosphaera sedula. J Bacteriol. 2011;193:1191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema TJG, Ahmed H, Geerling ACM et al. The non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) of Sulfolobus solfataricus: a key-enzyme of the semi-phosphorylative branch of the Entner–Doudoroff pathway. Extremophiles. 2008;12:75–88. [DOI] [PubMed] [Google Scholar]
- Ettema TJG, Brinkman AB, Lamers PP et al. Molecular characterization of a conserved archaeal copper resistance (cop) gene cluster and its copper-responsive regulator in Sulfolobus solfataricus P2. Microbiologyopen. 2006;152:1969–79. [DOI] [PubMed] [Google Scholar]
- Ettema TJG, Huynen MA, de Vos WM et al. TRASH: a novel metal-binding domain predicted to be involved in heavy-metal sensing, trafficking and resistance. Trends Biochem Sci. 2003;28:170–3. [DOI] [PubMed] [Google Scholar]
- Facciotti MT, Reiss DJ, Pan M et al. General transcription factor specified global gene regulation in Archaea. Proc Natl Acad Sci USA. 2007;104:4630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias ST, Bonato MC. Preferred amino acids and thermostability. Genet Mol Res. 2003;2:383–93. [PubMed] [Google Scholar]
- Feng X, Sun M, Han W et al. A transcriptional factor B paralog functions as an activator to DNA damage-responsive expression in Archaea. Nucleic Acids Res. 2018;46:7085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyhl-Buska J, Chen Y, Jia C et al. Influence of growth phase, pH, and temperature on the abundance and composition of tetraether lipids in the thermoacidophile Picrophilus torridus. Front Microbiol. 2016;7:1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo AS, Kouril T, Esser D et al. Systems biology of the modified branched Entner–Doudoroff pathway in Sulfolobus solfataricus. PLoS One. 2017;12:e0180331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino G, Del Giudice I, Bartolucci S et al. Identification and physicochemical characterization of BldR2 from Sulfolobus solfataricus, a novel archaeal member of the MarR transcription factor family. Biochemistry. 2011;50:6607–21. [DOI] [PubMed] [Google Scholar]
- Fiorentino G, Ronca R, Cannio R et al. MarR-like transcriptional regulator involved in detoxification of aromatic compounds in Sulfolobus solfataricus. J Bacteriol. 2007;189:7351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genetics. 2002;18:236–7. [DOI] [PubMed] [Google Scholar]
- Fouqueau T, Blombach F, Hartman R et al. The transcript cleavage factor paralogue TFS4 is a potent RNA polymerase inhibitor. Nat Commun. 2017;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröls S, Ajon M, Wagner M et al. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol Microbiol. 2008;70:938–52. [DOI] [PubMed] [Google Scholar]
- Fröls S, Gordon PM, Panlilio MA et al. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J Bacteriol. 2007;189:8708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröls S. Archaeal biofilms: widespread and complex. Biochem Soc Trans. 2013;41:393–8. [DOI] [PubMed] [Google Scholar]
- Fu C-Y, Wang K, Gan L et al. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure. 2010;18:1579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life?. Annu Rev Microbiol. 2011;65:631–58. [DOI] [PubMed] [Google Scholar]
- Fuchs T, Huber H, Teiner K et al. Metallosphaera prunae, sp. nov., a novel metal-mobilizing, thermoacidophilic archaeum, isolated from a uranium mine in Germany. Syst Appl Microbiol. 1995;18:560–6. [Google Scholar]
- Fusco S, She Q, Fiorentino G et al. Unravelling the role of the F55 regulator in the transition from lysogeny to UV induction of Sulfolobus spindle-shaped virus 1. J Virol. 2015;89:6453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer O, Angelov A, Liesegang H et al. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc Natl Acad Sci USA. 2004;101:9091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI et al. Phyletic distribution and lineage-specific domain architectures of archaeal two-component signal transduction systems. J Bacteriol. 2018;200:e00681–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambelli L, Meyer BH, McLaren M et al. Architecture and modular assembly of Sulfolobus S-layers revealed by electron cryotomography. Proc Natl Acad Sci USA. 2019;116:25278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AF, Bell SD, White MF et al. Protein–protein interactions leading to recruitment of the host DNA sliding clamp by the hyperthermophilic Sulfolobus islandicus rod-shaped virus 2. J Virol. 2014;88:7105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett RA, Shah SA, Vestergaard G et al. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem Soc Trans. 2011;39:51–7. [DOI] [PubMed] [Google Scholar]
- Geertsma ER. FX cloning: a versatile high-throughput cloning system for characterization of enzyme variants. Methods Mol Biol. 2013;978;133–48. [DOI] [PubMed] [Google Scholar]
- Giles TN, Graham DE. Crenarchaeal arginine decarboxylase evolved from an S-adenosylmethionine decarboxylase enzyme. J Biol Chem. 2008;283:25829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girfoglio M, Rossi M, Cannio R. Cellulose degradation by Sulfolobus solfataricus requires a cell-anchored endo-beta-1-4-glucanase. J Bacteriol. 2012;194:5091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleissner M, Kaiser U, Antonopoulos E et al. The archaeal SoxABCD complex is a proton pump in Sulfolobus acidocaldarius. J Biol Chem. 1997;272:8417–26. [DOI] [PubMed] [Google Scholar]
- Gogliettino M, Balestrieri M, Pocsfalvi G et al. A highly selective oligopeptide binding protein from the archaeon Sulfolobus solfataricus. J Bacteriol. 2010;192:3123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliettino M, Riccio A, Cocca E et al. A new pepstatin-insensitive thermopsin-like protease overproduced in peptide-rich cultures of Sulfolobus solfataricus. Int J Mol Sci. 2014;15:3204–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D et al. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golyshina OV, Yakimov MM, Lunsdorf H et al. Acidiplasma aeolicum gen. nov., sp. nov., a euryarchaeon of the family Ferroplasmaceae isolated from a hydrothermal pool, and transfer of Ferroplasma cupricumulans to Acidiplasma cupricumulans comb. nov. Int J Syst Evol Microbiol. 2009;59:2815–23. [DOI] [PubMed] [Google Scholar]
- Gomes-Filho JV, Randau L. RNA stabilization in hyperthermophilic Archaea. Ann NY Acad Sci. 2019;1447:88–96. [DOI] [PubMed] [Google Scholar]
- Gong F, Zhu H, Zhou J et al. Enhanced biological fixation of CO2 using microorganisms. In: Aresta M, Karimi IA, Kawi S (eds). An Economy Based on Carbon Dioxide and Water. Cham: Springer, 2019. [Google Scholar]
- Goto K, Tanimoto Y, Tamura T et al. Identification of thermoacidophilic bacteria and a new Alicyclobacillus genomic species isolated from acidic environments in Japan. Extremophiles. 2002;6:333–40. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Jakob U. Oxidative stress protection by polyphosphate—new roles for an old player. Curr Opin Microbiol. 2015;24:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greci MD, Bell SD. Archaeal DNA replication. Annu Rev Microbiol. 2020;74:65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo-Puertas M, Rintoul R, Villegas JM et al. Polyphosphate degradation in stationary phase triggers biofilm formation via LuxS quorum sensing system in Escherichia coli. PLoS One. 2012;7:e50368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo-Puertas M, Schurig-Briccio LA, Rodríguez-Montelongo L et al. Copper tolerance mediated by polyphosphate degradation and low-affinity inorganic phosphate transport system in Escherichia coli. BMC Microbiol. 2014;14:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan DW, Carver GT, Drake JW. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2001;98:7928–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan DW, Gunsalus RP. Sulfolobus acidocaldarius synthesizes UMP via a standard de novo pathway: results of biochemical-genetic study. J Bacteriol. 1993;175:1500–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan DW, Palm P, Zillig W. Isolate B12, which harbours a virus-like element, represents a new species of the archaebacterial genus Sulfolobus, Sulfolobus shibatae, sp. nov. Arch Microbiol. 1990;154:594–9. [DOI] [PubMed] [Google Scholar]
- Grogan DW, Stengel KR. Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli/lambda red and Sulfolobus acidocaldarius recombination systems. Mol Microbiol. 2008;69:1255–65. [DOI] [PubMed] [Google Scholar]
- Grogan DW. Cytosine methylation by the SuaI restriction-modification system: implications for genetic fidelity in a hyperthermophilic archaeon. J Bacteriol. 2003;185:4657–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan DW. Organization and interactions of cell envelope proteins of the extreme thermoacidophile Sulfolobus acidocaldarius. Can J Microbiol. 1996;42:1163–71. [Google Scholar]
- Grogan DW. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J Bacteriol. 1989;171:6710–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan DW. Selectable mutant phenotypes of the extremely thermophilic archaebacterium Sulfolobus acidocaldarius. J Bacteriol. 1991;173:7725–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Delago A, Nußbaum P et al. N-glycosylation in the thermoacidophilic archaeon Sulfolobus acidocaldarius involves a short dolichol pyrophosphate carrier. FEBS Lett. 2016;590:3168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillière F, Peixeiro N, Kessler A et al. Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1. J Biol Chem. 2009;284:22222–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Ding J, Guo R et al. Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea. J Biol Chem. 2014;289:1478–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Feng Y, Zhang Z et al. Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea. Nucleic Acids Res. 2008;36:1129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche I, Holzinger J, Rößle M et al. Conformational rearrangements of an archaeal chaperonin upon ATPase cycling. Curr Biol. 2000;10:405–8. [DOI] [PubMed] [Google Scholar]
- Gutsche I, Mihalache O, Baumeister W. ATPase cycle of an archaeal chaperonin. J Mol Biol. 2000;300:187–96. [DOI] [PubMed] [Google Scholar]
- Haferkamp P, Kutschki S, Treichel J et al. An additional glucose dehydrogenase from Sulfolobus solfataricus: fine-tuning of sugar degradation?. Biochem Soc Trans. 2011;39:77–81. [DOI] [PubMed] [Google Scholar]
- Haferkamp P, Tjaden B, Shen L et al. The carbon switch at the level of pyruvate and phosphoenolpyruvate in Sulfolobus solfataricus P2. Front Microbiol. 2019;10:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile JD, Kennelly PJ. The activity of an ancient atypical protein kinase is stimulated by ADP-ribose i n vitro. Arch Biochem Biophys. 2011;511:56–63. [DOI] [PubMed] [Google Scholar]
- Hampton HG, Watson BNJ, Fineran PC. The arms race between bacteria and their phage foes. Nature. 2020;577:327–36. [DOI] [PubMed] [Google Scholar]
- Han D, Krauss G. Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Letters. 2009;583:771–6. [DOI] [PubMed] [Google Scholar]
- Hanks SK. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 2003;4:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Rachel R, Peng X et al. Viral Diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J Virol. 2005;79:9904–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman R, Eilers BJ, Bollschweiler D et al. The molecular mechanism of cellular attachment for an archaeal virus. Structure. 2019;27:1634–46.e3. [DOI] [PubMed] [Google Scholar]
- Haseltine C, Rolfsmeier M, Blum P. The glucose effect and regulation of alpha-amylase synthesis in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 1996;178:945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D et al. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–6. [DOI] [PubMed] [Google Scholar]
- Haurat MF, Figueiredo AS, Hoffmann L et al. ArnS, a kinase involved in starvation-induced archaellum expression. Mol Microbiol. 2017;103:181–94. [DOI] [PubMed] [Google Scholar]
- Hawkes RB, Franzmann PD, O'Hara G et al. Ferroplasma cupricumulanssp. nov., a novel moderately thermophilic, acidophilic archaeon isolated from an industrial-scale chalcocite bioleach heap. Extremophiles. 2006;10:525–30. [DOI] [PubMed] [Google Scholar]
- Hawkins AS, Han Y, Bennett RK et al. Role of 4-hydroxybutyrate-CoA synthetase in the CO2 fixation cycle in thermoacidophilic archaea. J Biol Chem. 2013;288:4012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Bhoobalan-Chitty Y, Van LB et al. Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat Microbiol. 2018;3:461–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–66. [DOI] [PubMed] [Google Scholar]
- Henche A-L, Koerdt A, Ghosh A et al. Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ Microbiol. 2012b;14:779–93. [DOI] [PubMed] [Google Scholar]
- Henche AL, Ghosh A, Yu X et al. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ Microbiol. 2012a;14:3188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M. Thermoacidophilic proteins for biofuel production. Trends Microbiol. 2008;16:414–9. [DOI] [PubMed] [Google Scholar]
- Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008;451:851–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroyoshi N, Miki H, Hirajima T et al. A model for ferrous-promoted chalcopyrite leaching. Hydrometallurgy. 2000;57:31–8. [Google Scholar]
- Hjort K, Bernander R. Cell cycle regulation in the hyperthermophilic crenarchaeon Sulfolobus acidocaldarius. Mol Microbiol. 2001;40:225–34. [DOI] [PubMed] [Google Scholar]
- Hobel CFV, Albers SV, Driessen AJM et al. The Sulfolobus solfataricus AAA protein Sso0909, a homologue of the eukaryotic ESCRT Vps4 ATPase. Biochem Sci Trans. 2008;36:94–8. [DOI] [PubMed] [Google Scholar]
- Hocher A, Rojec M, Swadling JB et al. The DNA-binding protein HTa from Thermoplasma acidophilum is an archaeal histone analog. eLife. 2019;8:e52542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Schummer A, Reimann J et al. Expanding the archaellum regulatory network–the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius. Microbiologyopen. 2017;6:e00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Mayaka JBA, Zhong Q et al. Phosphorylation of the archaeal Holliday junction resolvase Hjc inhibits its catalytic activity and facilitates DNA repair in Sulfolobus islandicus REY15A. Front Microbiol. 2019;10:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Zhong Q, Mayaka J et al. Autophosphorylation and cross-phosphorylation of protein kinases from the Crenarchaeon Sulfolobus islandicus. Front Microbiol. 2017;8:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Krauss G, Cottaz S et al. A highly acid-stable and thermostable endo-β-glucanase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochem J. 2005;385:581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber G, Spinnler C, Gambacorta A et al. Metallosphaera sedula gen, and sp. nov. represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst Appl Microbiol. 1989;12:38–47. [Google Scholar]
- Huber G, Stetter KO. Sulfolobus metallicus, sp. nov., a novel strictly chemolithoautotrophic thermophilic archaeal species of metal-mobilizers. Syst Appl Microbiol. 1991;14:372–8. [Google Scholar]
- Hugler M, Huber H, Stetter KO et al. Autotrophic CO2 fixation pathways in archaea (Crenarchaeota). Arch Microbiol. 2003;179:160–73. [DOI] [PubMed] [Google Scholar]
- Hwang S, Maxwell KL. Meet the Anti-CRISPRs: widespread protein inhibitors of CRISPR-Cas systems. CRISPR J. 2019;2:23–30. [DOI] [PubMed] [Google Scholar]
- Häring M, Vestergaard G, Rachel R et al. Virology: independent virus development outside a host. Nature. 2005;436:1101–2. [DOI] [PubMed] [Google Scholar]
- Inskeep W, Jay Z, Tringe S et al. The YNP Metagenome Project: environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Front Microbiol. 2013;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Qureshi SA. Selective depletion of Sulfolobus solfataricus transcription factor E under heat shock conditions. J Bacteriol. 2010;192:2887–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Miura T, Sakai HD et al. Sulfuracidifex tepidarius gen. nov., sp. nov. and transfer of Sulfolobus metallicus Huber and Stetter 1992 to the genus Sulfuracidifex as Sulfuracidifex metallicus comb. nov. Int J Syst Evol Microbiol. 2020;70:1837–42. [DOI] [PubMed] [Google Scholar]
- Itoh T, Suzuki K, Sanchez PC et al. Caldisphaera lagunensisgen. nov., sp. nov., a novel thermoacidophilic crenarchaeote isolated from a hot spring at Mt Maquiling, Philippines. Int J Syst Evol Microbiol. 2003;53:1149–54. [DOI] [PubMed] [Google Scholar]
- Itoh T, Suzuki K, Sanchez PC et al. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int J Syst Evol Microbiol. 1999;49:1157–63. [DOI] [PubMed] [Google Scholar]
- Itoh T, Yoshikawa N, Takashina T. Thermogymnomonas acidicola gen. nov., sp. nov., a novel thermoacidophilic, cell wall-less archaeon in the order Thermoplasmatales, isolated from a solfataric soil in Hakone, Japan. Int J Syst Evol Microbiol. 2007;57:2557–61. [DOI] [PubMed] [Google Scholar]
- Izzo V, Notomista E, Picardi A et al. The thermophilic archaeon Sulfolobus solfataricus is able to grow on phenol. Res Microbiol. 2005;156:677–89. [DOI] [PubMed] [Google Scholar]
- Jachlewski S, Jachlewski WD, Linne U et al. Isolation of extracellular polymeric substances from biofilms of the thermoacidophilic archaeon Sulfolobus acidocaldarius. Front Bioeng Biotechnol. 2015;3:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, McKenzie RE, Fagerlund RD et al. CRISPR-Cas: adapting to change. Science. 2017;356:eaal5056. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Albers S-V. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–12. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Ding Y, Meyer BH et al. N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Molec Biol Rev. 2014;78:304–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay ZJ, Beam JP, Dlakic M et al. Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats. Nat Microbiol. 2018;3:732–40. [DOI] [PubMed] [Google Scholar]
- Jiang CY, Liu LJ, Guo X et al. Resolution of carbon metabolism and sulfur-oxidation pathways of Metallosphaera cuprina Ar-4 via comparative proteomics. J Proteomics. 2014;109:276–89. [DOI] [PubMed] [Google Scholar]
- Johnsen U, Reinhardt A, Landan G et al. New views on an old enzyme: allosteric regulation and evolution of archaeal pyruvate kinases. FEBS J. 2019;286:2471–89. [DOI] [PubMed] [Google Scholar]
- Johnson DB, Okibe N, Roberto FF. Novel thermo-acidophilic bacteria isolated from geothermal sites in Yellowstone National Park: physiological and phylogenetic characteristics. Arch Microbiol. 2003;180:60–8. [DOI] [PubMed] [Google Scholar]
- Johnson T, Payne S, Grove R et al. Methylation deficiency of chromatin proteins is a non-mutational and epigenetic-like trait in evolved lines of the archaeon Sulfolobus solfataricus. J Biol Chem. 2019;294:7821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonuscheit M, Martusewitsch E, Stedman KM et al. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol. 2003;48:1241–52. [DOI] [PubMed] [Google Scholar]
- Kagawa HK, Osipiuk J, Maltsev N et al. The 60 kDa heat shock proteins in the hyperthermophilic archaeon Sulfolobus shibatae. J Mol Biol. 1995;253:712–25. [DOI] [PubMed] [Google Scholar]
- Kagawa HK, Yaoi T, Brocchieri L et al. The composition, structure and stability of a group II chaperonin are temperature regulated in a hyperthermophilic archaeon. Mol Microbiol. 2003;48:143–56. [DOI] [PubMed] [Google Scholar]
- Kalichuk V, Béhar G, Renodon-Cornière A et al. The archaeal “7 kDa DNA-binding” proteins: extended characterization of an old gifted family. Sci Rep. 2016;6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomaa-Sanford AK, Rodriguez-Castañeda FA, McLeod BN et al. Chromosome segregation in Archaea mediated by a hybrid DNA partition machine. Proc Natl Acad Sci USA. 2012;109:3754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Barillà D. Combing chromosomal DNA mediated by the SMC complex: structure and mechanisms. BioEssays. 2018;40:1–8. [DOI] [PubMed] [Google Scholar]
- Kappler U, Dahl C. Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol Lett. 2001;203:1–9. [DOI] [PubMed] [Google Scholar]
- Karavaiko GI, Bogdanova TI, Tourova TP et al. Reclassification of ‘S ulfobacillus thermosulfidooxidans subsp.thermotolerans’ strain K1 as Alicyclobacillus tolerans sp. nov. and Sulfobacillus disulfidooxidans Dufresne et al. 1996 as Alicyclobacillus disulfidooxidans comb. nov., and emended description of the genus Alicyclobacillus. Int J Syst Evol Microbiol. 2005;55:941–7. [DOI] [PubMed] [Google Scholar]
- Karr EA, Isom CE, Trinh V et al. Transcription factor-mediated gene regulation in Archaea. In: RNA Metabolism and Gene Expression in Archaea, Vol. 32. Cham: Springer, 2017, 27–69. [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas Č et al. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–9. [DOI] [PubMed] [Google Scholar]
- Keasling JD. Regulation of intracellular toxic metals and other cations by hydrolysis of polyphosphate. Ann NY Acad Sci. 1997;829:242–9. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Klenk H-P, Singh RK et al. Complete nucleotide sequence of the Sulfolobus islandicus multicopy plasmid pRN1. Plasmid. 1996;35:141–4. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Klenk H-P, Singh RK et al. Sulfolobus islandicus plasmids pRN1 and pRN2 share distant but common evolutionary ancestry. Extremophiles. 1998;2:391–3. [DOI] [PubMed] [Google Scholar]
- Keller MW, Schut GJ, Lipscomb GL et al. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc Natl Acad Sci USA. 2013;110:5840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DS, Baross JA, Delaney JR. Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu Rev Earth Planetary Sci. 2002;30:385–491. [Google Scholar]
- Kelly DE, Lamb DC, Kelly SL. Genome-wide generation of yeast gene deletion strains. Comp Funct Genomics. 2001;2:236–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly P, Oxenrider K, Leng J et al. Identification of a serine/threonine-specific protein phosphatase from the archaebacterium Sulfolobus solfataricus. J Biol Chem. 1993;268:6505–10. [PubMed] [Google Scholar]
- Kennelly PJ. Archaeal protein kinases and protein phosphatases: insights from genomics and biochemistry. Biochem J. 2003;370:373–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly PJ. Protein Ser/Thr/Tyr phosphorylation in the Archaea. J Biol Chem. 2014;289:9480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher L, Nowitzki S, Oesterhelt D. Thermoacidophilic archaebacteria contain bacterial-type ferredoxins acting as electron acceptors of 2-oxoacid:ferredoxin oxidoreductases. Eur J Biochem. 1982;128:223–30. [DOI] [PubMed] [Google Scholar]
- Kessler A, Sezonov G, Guijarro JI et al. A novel archaeal regulatory protein, Sta1, activates transcription from viral promoters. Nucleic Acids Res. 2006;34:4837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-J, Forst S. Genomic analysis of the histidine kinase family in Bacteria and Archaea. Microbiologyopen. 2001;147:1197–212. [DOI] [PubMed] [Google Scholar]
- Kim MS, Park JT, Kim YW et al. Properties of a novel thermostable glucoamylase from the hyperthermophilic archaeon Sulfolobus solfataricus in relation to starch processing. Appl Environ Microbiol. 2004;70:3933–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Paek KH, Lee SB. Characterization of NADP+-specific l-rhamnose dehydrogenase from the thermoacidophilic archaeonThermoplasma acidophilum. Extremophiles. 2012;16:447–54. [DOI] [PubMed] [Google Scholar]
- Klauber C. A critical review of the surface chemistry of acidic ferric sulphate dissolution of chalcopyrite with regard to hindered dissolution. Int J Miner Process. 2008;86:1–17. [Google Scholar]
- Kletzin A, Urich T, Muller F et al. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J Bioenerg Biomembr. 2004;36:77–91. [DOI] [PubMed] [Google Scholar]
- Kletzin A. Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J Bacteriol. 1989;171:1638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzin A. Molecular characterization of the sor gene, which encodes the sulfur oxygenase/reductase of the thermoacidophilic archaeum Desulfurolobus ambivalens. J Bacteriol. 1992;174:5854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingl A, Pickl C, Flechsler J. Archaeal cell walls. Subcell Biochem. 2019;92:471–93. [DOI] [PubMed] [Google Scholar]
- Knüppel R, Christensen RH, Gray FC et al. Insights into the evolutionary conserved regulation of Rio ATPase activity. Nucl Acids Res. 2018;46:1441–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch T, Dahl C. A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 2018;12:2479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockelkorn D, Fuchs G. Malonic semialdehyde reductase, succinic semialdehyde reductase, and succinyl-coenzyme A reductase from Metallosphaera sedula: enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales. J Bacteriol. 2009;191:6352–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koechler S, Farasin J, Cleiss-Arnold J et al. Toxic metal resistance in biofilms: diversity of microbial responses and their evolution. Res Microbiol. 2015;166:764–73. [DOI] [PubMed] [Google Scholar]
- Koerdt A, Gödeke J, Berger J et al. Crenarchaeal biofilm formation under extreme conditions. PLoS One. 2010;5:e14104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerdt A, Jachlewski S, Ghosh A et al. Complementation of Sulfolobus solfataricus PBL2025 with an α-mannosidase: effects on surface attachment and biofilm formation. Extremophiles. 2012;16:115–25. [DOI] [PubMed] [Google Scholar]
- Koerdt A, Orell A, Pham TK et al. Macromolecular fingerprinting of Sulfolobus species in biofilm: a transcriptomic and proteomic approach combined with spectroscopic analysis. J Proteome Res. 2011;10:4105–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H, Chong PL-G. Low permeability of liposomal membranes composed of bipolar tetraether lipids from thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochemistry. 1998;37:107–15. [DOI] [PubMed] [Google Scholar]
- Kondo S, Yamagishi A, Oshima T. Positive selection for uracil auxotrophs of the sulfur-dependent thermophilic archaebacterium Sulfolobus acidocaldarius by use of 5-fluoroorotic acid. J Bacteriol. 1991;173:7698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrat'eva TF, Pivovarova TA, Tsaplina IA et al. Diversity of acidophilic chemolithotrophic microbial consortia in natural and anthropogenic ecosystems. Mikrobiologjia. 2012;81:3–27. [PubMed] [Google Scholar]
- Konings WN, Albers S-V, Koning S et al. The cell membrane plays a crucial role in survival of Bacteria and Archaea in extreme environments. Antonie van Leeuwenhoek. 2002;81:61–72. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Wolf YI. Evolutionary genomics of defense systems in Archaea and Bacteria. Ann Rev Microbiol. 2017;71:233–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhin Y, Unligil UM, Littlefield O et al. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol. 2009;7:e1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riché D. Inorganic polyphosphate: a molecule of many functions. Ann Rev Biochem. 1999;68:89–125. [DOI] [PubMed] [Google Scholar]
- Kouril T, Eicher JJ, Siebers B et al. Phosphoglycerate kinase acts as a futile cycle at high temperature. Microbiologyopen. 2017;163:1604–12. [DOI] [PubMed] [Google Scholar]
- Kouril T, Esser D, Kort J et al. Intermediate instability at high temperature leads to low pathway efficiency for an in vitro reconstituted system of gluconeogenesis in Sulfolobus solfataricus. FEBS J. 2013a;280:4666–80. [DOI] [PubMed] [Google Scholar]
- Kouril T, Wieloch P, Reimann J et al. Unraveling the function of the two Entner−Doudoroff branches in the thermoacidophilic Crenarchaeon Sulfolobus solfataricus P2. FEBS J. 2013b;280:1126–38. [DOI] [PubMed] [Google Scholar]
- Kozubal MA, Dlakic M, Macur RE et al. Terminal oxidase diversity and function in “Metallosphaera yellowstonensis”: gene expression and protein modeling suggest mechanisms of Fe(II) oxidation in the Sulfolobales. Appl Environ Microbiol. 2011;77:1844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubal MA, Romine M, Jennings R et al. Geoarchaeota: a new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J. 2013;7:622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarevel T, Tanaka T, Umehara T et al. ST1710-DNA complex crystal structure reveals the DNA binding mechanism of the MarR family of regulators. Nucleic Acids Res. 2009;37:4723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa N, Grogan DW. Homologous recombination of exogenous DNA with the Sulfolobus acidocaldarius genome: properties and uses. FEMS Microbiol. 2005;253:141–9. [DOI] [PubMed] [Google Scholar]
- Kurosawa N, Itoh YH, Iwai T et al. Sulfurisphaera ohwakuensis gen. nov., sp. nov., a novel extremely thermophilic acidophile of the order Sulfolobales. Int J Syst Evol Microbiol. 1998;48:451–6. [DOI] [PubMed] [Google Scholar]
- Lamble HJ, Heyer NI, Bull SD et al. Metabolic pathway promiscuity in the archaeon Sulfolobus solfataricus revealed by studies on glucose dehydrogenase and 2-keto-3-deoxygluconate aldolase. J Biol Chem. 2003;278:34066–72. [DOI] [PubMed] [Google Scholar]
- Lamble HJ, Milburn CC, Taylor GL et al. Gluconate dehydratase from the promiscuous Entner–Doudoroff pathway in Sulfolobus solfataricus. FEBS Lett. 2004;576:133–6. [DOI] [PubMed] [Google Scholar]
- Lamble HJ, Theodossis A, Milburn CC et al. Promiscuity in the part-phosphorylative Entner–Doudoroff pathway of the archaeon Sulfolobus solfataricus. FEBS Lett. 2005;579:6865–9. [DOI] [PubMed] [Google Scholar]
- Laplagia C, Hartzell PL. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl Environ Microbiol. 1997;63:3158–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde NA. The ancient microbial RIO kinases. J Biol Chem. 2014;289:9488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laska S, Lottspeich F, Kletzin A. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiologyopen. 2003;149:2357–71. [DOI] [PubMed] [Google Scholar]
- Lassak K, Neiner T, Ghosh A et al. Molecular analysis of the crenarchaeal flagellum. Mol Microbiol. 2012;83:110–24. [DOI] [PubMed] [Google Scholar]
- Lassak K, Peeters E, Wróbel S et al. The one-component system ArnR: a membrane-bound activator of the crenarchaeal archaellum. Mol Microbiol. 2013;88:125–39. [DOI] [PubMed] [Google Scholar]
- La Teana A, Benelli D, Londei P et al. Translation initiation in the crenarchaeonSulfolobus solfataricus: eukaryotic features but bacterial route. Biochem Soc T. 2013;41:350–5. [DOI] [PubMed] [Google Scholar]
- Laurens N, Driessen RP, Heller I et al. Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA. Nat Commun. 2012;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of Halobacteria. J Biol Chem. 1987;262:9724–29. [PubMed] [Google Scholar]
- Lemire J, Alhasawi A, Appanna VP et al. Metabolic defence against oxidative stress: the road less travelled so far. J Appl Microbiol. 2017;123:798–809. [DOI] [PubMed] [Google Scholar]
- Lemmens L, Maklad HR, Bervoets I et al. Transcription regulators in Archaea: homologies and differences with bacterial regulators. J Mol Biol. 2019a;431:4132–46. [DOI] [PubMed] [Google Scholar]
- Lemmens L, Tilleman L, De Koning E et al. YtrASa, a GntR-family transcription factor, represses two genetic loci encoding membrane proteins in Sulfolobus acidocaldarius. Front Microbiol. 2019b;10:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos RS, Gomes CM, Teixeira M. Acidianus ambivalens complex II typifies a novel family of succinate dehydrogenases. Biochem Biophys Res Commun. 2001;281:141–50. [DOI] [PubMed] [Google Scholar]
- Leng J, Cameron A, Buckel S et al. Isolation and cloning of a protein-serine/threonine phosphatase from an archaeon. J Bacteriol. 1995;177:6510–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in Bacteria and Archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 1998;8:1038–47. [DOI] [PubMed] [Google Scholar]
- Lewus P, Ford RM. Temperature-sensitive motility of Sulfolobus acidocaldarius influences population distribution in extreme environments. J Bacteriol. 1999;181:4020–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyn SA, Rodionova IA, Li X et al. Novel transcriptional regulons for autotrophic cycle genes in Crenarchaeota. J Bacteriol. 2015;197:2383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Sobrino C, Kot WP, Garrett RA. Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous CRISPR spacer acquisition. Mol Microbiol. 2016;99:719–28. [DOI] [PubMed] [Google Scholar]
- Lian H, Zeldes BM, Lipscomb GL et al. Ancillary contributions of heterologous biotin protein ligase and carbonic anhydrase for CO2 incorporation into 3-hydroxypropionate by metabolically engineered Pyrococcus furiosus. Biotechnol Bioeng. 2016;113:2652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D-C, Yang F, Lu B et al. Thermotolerance and molecular chaperone function of the small heat shock protein HSP20 from hyperthermophilic archaeon, Sulfolobus solfataricus P2. Cell Stress Chaper. 2012;17:103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Banerjee A, Bischof LF et al. Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius. Mol Microbiol. 2017;105:777–93. [DOI] [PubMed] [Google Scholar]
- Lillestøl RK, Shah SA, Brügger K et al. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–72. [DOI] [PubMed] [Google Scholar]
- Limauro D, Cannio R, Fiorentino G et al. Identification and molecular characterization of an endoglucanase gene, celS, from the extremely thermophilic archaeon Sulfolobus solfataricus. Extremophiles. 2001;5:213–9. [DOI] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT et al. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X-l, Tang J. Purification, characterization, and gene cloning of thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius. J Biol Chem. 1990;265:1490–5. [PubMed] [Google Scholar]
- Lipps G. Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus. Extremophiles. 2006;10:17–28. [DOI] [PubMed] [Google Scholar]
- Liu B, Ouyang S, Makarova KS et al. A primase subunit essential for efficient primer synthesis by an archaeal eukaryotic-type primase. Nat Commun. 2015;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Orell A, Maes D et al. BarR, an Lrp-type transcription factor in Sulfolobus acidocaldarius, regulates an aminotransferase gene in a β-alanine responsive manner. Mol Microbiol. 2014a;92:625–39. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang K, Lindås AC et al. The genome-scale DNA-binding profile of BarR, a β-alanine responsive transcription factor in the archaeon Sulfolobus acidocaldarius. BMC Genomics. 2016;17:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L-J, You X-Y, Guo X et al. Metallosphaera cuprina sp. nov., an acidothermophilic, metal-mobilizing archaeon. Int J Syst Evol. 2011;61:2395–400. [DOI] [PubMed] [Google Scholar]
- Liu LJ, Stockdreher Y, Koch T et al. Thiosulfate transfer mediated by DsrE. /TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina. J Biol Chem. 2014b;289:26949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ishino S, Ishino Y et al. A novel type of polyhedral viruses infecting hyperthermophilic Archaea. J Virol. 2017;91:e00589–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu T. Production of acrylic acid and propionic acid by constructing a portion of the 3-hydroxypropionate/4-hydroxybutyrate cycle from Metallosphaera sedula in Escherichia coli. J Ind Microbiol Biotechnol. 2016;43:1659–70. [DOI] [PubMed] [Google Scholar]
- Liu Z, Walton TA, Rees DC. A reported archaeal mechanosensitive channel is a structural homolog of MarR-like transcriptional regulators. Protein Sci. 2010;19:808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kawashima N, Li J et al. A review of the structure, and fundamental mechanisms and kinetics of the leaching of chalcopyrite. Adv Colloid Interface Sci. 2013;197−198:1–32. [DOI] [PubMed] [Google Scholar]
- Li Y, Pan S, Zhang Y et al. Harnessing type I and type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loder AJ, Han Y, Hawkins AB et al. Reaction kinetic analysis of the 3-hydroxypropionate/4-hydroxybutyrate CO2 fixation cycle in extremely thermoacidophilic Archaea. Metab Eng. 2016;38:446–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF, Shaulsky G, Wang N. Histidine kinases in signal transduction pathways of eukaryotes. J Cell Sci. 1997;110:1141–5. [DOI] [PubMed] [Google Scholar]
- Lower BH, Bischoff KM, Kennelly PJ. The archaeon Sulfolobus solfataricus contains a membrane-associated protein kinase activity that preferentially phosphorylates threonine residues in vitro. J Bacteriol. 2000;182:3452–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower BH, Kennelly PJ. Open reading frame sso2387 from the archaeon Sulfolobus solfataricus encodes a polypeptide with protein-serine kinase activity. J Bacteriol. 2003;185:3436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower BH, Kennelly PJ. The membrane-associated protein-serine/threonine kinase from Sulfolobus solfataricus is a glycoprotein. J Bacteriol. 2002;184:2614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower BH, Potters MB, Kennelly PJ. A phosphoprotein from the archaeon Sulfolobus solfataricus with protein-serine/threonine kinase activity. J Bacteriol. 2004;186:463–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubben M, Castresana J, Warne A. Terminal Oxidases of Sulfolobus: genes and proteins. Syst Appl Microbiol. 1994;16:556–9. [Google Scholar]
- Lundgren M, Andersson A, Chen L et al. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci USA. 2004;101:7046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Pathak C, Lee SJ et al. Alba from Thermoplasma volcanium belongs to α-NAT's: an insight into the structural aspects of Tv Alba and its acetylation by Tv Ard1. Arch Biochem Biophys. 2016;590:90–100. [DOI] [PubMed] [Google Scholar]
- Maezato Y, Daugherty A, Dana K et al. VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus. RNA. 2011;17:1381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezato Y, Johnson T, McCarthy S et al. Metal resistance and lithoautotrophy in the extreme thermoacidophile Metallosphaera sedula. J Bacteriol. 2012;194:6856–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai VQ, Chen X, Hong R et al. Small abundant DNA binding proteins from the thermoacidophilic archaeon Sulfolobus shibatae constrain negative DNA supercoils. J Bacteriol. 1998;180:2560–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV, Albers S-V. Diversity and evolution of type IV pili systems in Archaea. Front Microbiol. 2016;7:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2019;18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, van der Oost J et al. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct. 2009;4:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklad HR, Gutierrez GJ, Esser D et al. Phosphorylation of the acyl-CoA binding pocket of the FadR transcription regulator in Sulfolobus acidocaldarius. Biochimie. 2020;175:120–4. [DOI] [PubMed] [Google Scholar]
- Manica A, Schleper C. CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus. RNA Biol. 2013;10:671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhuenda-Egea FC, Bonete MaJ. Extreme halophilic enzymes in organic solvents. Curr Opin Biotech. 2002;13:385–9. [DOI] [PubMed] [Google Scholar]
- Marinsek N, Barry ER, Makarova KS et al. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7:539–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Yeats S, Janekovic D et al. SAV 1, a temperate UV-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 1984;3:2165–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LO, Carreto LS, Da Costa MS et al. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J Bactriol. 1996;178:5644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LO, Huber R, Huber H et al. Organic solutes in hyperthermophilic Archaea. Appl Environ Microbiol. 1997;63:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LO, Santos H. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl Environ Microbiol. 1995;61:3299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martusewitsch E, Sensen CW, Schleper C. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J Bacteriol. 2000;182:2574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Alvarez L, Deng L, Peng X. Formation of a viral replication focus in Sulfolobus cells infected by the rudivirus Sulfolobus islandicus rod-shaped virus 2. J Virol. 2017;91:e00486–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Bussenius C, Navarro CA, Jerez CA. Microbial copper resistance: importance in biohydrometallurgy. Microb Biotechnol. 2017;10:279–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathai JC, Missner A, Kugler P et al. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci USA. 2009;106:16633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat Rev Microbiol. 2012;10:100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli L, Giovane A, Esposito A et al. Evidence that the xylanase activity from Sulfolobus solfataricus Oα is encoded by the endoglucanase precursor gene (sso1354) and characterization of the associated cellulase activity. Extremophiles. 2008;12:689–700. [DOI] [PubMed] [Google Scholar]
- Mayo-Muñoz D, He F, Jørgensen JB et al. Anti-CRISPR-based and CRISPR-based genome editing of Sulfolobus islandicus rod-shaped virus 2. Viruses. 2018;10:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Ai C, Blum P. Enhancement of Metallosphaera sedula bioleaching by targeted recombination and adaptive laboratory evolution. Adv Appl Microbiol. 2018;104:135–65. [DOI] [PubMed] [Google Scholar]
- McCarthy S, Ai C, Wheaton G et al. Role of an archaeal pita transporter in the copper and arsenic resistance of Metallosphaera sedula, an extreme thermoacidophile. J Bacteriol. 2014;196:3562–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher M, Epling LB, Enemark EJ. DNA translocation mechanism of the MCM complex and implications for replication initiation. Nat Commun. 2019;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud VS, Pivovarova TA, Tourova TP et al. Sulfobacillus sibericus sp. nov., a new moderately thermophilic bacterium. MicrobiologyOpen. 2003;72:605–12. [PubMed] [Google Scholar]
- Melo AM, Bandeiras TM, Teixeira M. New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol Mol Biol Rev. 2004;68:603–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng DC, Shi ZY, Wu LP et al. Production and characterization of poly(3-hydroxypropionate-co-4-hydroxybutyrate) with fully controllable structures by recombinant Escherichia coli containing an engineered pathway. Metab Eng. 2012;14:317–24. [DOI] [PubMed] [Google Scholar]
- Merola A, Castaldo R, De Luca P et al. Revision of Cyanidium caldarium. Three species of acidophilis algae. G Bot Lt. 1982;116:189–95. [Google Scholar]
- Meyer BH, Albers S-V. AglB, catalyzing the oligosaccharyl transferase step of the archaeal N-glycosylation process, is essential in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. Microbiologyopen. 2014;3:531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BH, Birich A, Albers S-V. N-glycosylation of the archaellum filament is not important for archaella assembly and motility, although N-glycosylation is essential for motility in Sulfolobus acidocaldarius. Biochimie. 2015;118:294–301. [DOI] [PubMed] [Google Scholar]
- Meyer BH, Peyfoon E, Dietrich C et al. Agl16, a thermophilic glycosyltransferase mediating the last step of N-glycan biosynthesis in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J Bacteriol. 2013;195:2177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BH, Shams-Eldin H, Albers S-V. AglH, a thermophilic UDP-N-acetylglucosamine-1-phosphate: dolichyl phosphate GlcNAc-1-phosphotransferase initiating protein N-glycosylation pathway in Sulfolobus acidocaldarius, is capable of complementing the eukaryal Alg7. Extremophiles. 2017;21:121–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BH, Zolghadr B, Peyfoon E et al. Sulfoquinovose synthase: an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol Microbiol. 2011;82:1150–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa BE, Chiaruttini N, Redondo-Morata L et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol. 2017;19:787–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima H, Ikeda Y, Fujii M et al. Specificity of Fur binding to the oxidative stress response gene promoter in the facultative anaerobic archaeon Thermoplasma volcanium. Biol Pharm Bull. 2014;37:481–5. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. [DOI] [PubMed] [Google Scholar]
- Moll R, Schäfer G. Chemiosmotic H+ cycling across the plasma membrane of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1988;232:359–63. [Google Scholar]
- Monroe N, Han H, Gonciarz MD et al. The oligomeric state of the active Vps4 AAA ATPase. J Mol Biol. 2014;426:510–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol. 2005;187:972–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moracci M, Cobucci Ponzano B, Trincone A et al. Identification and molecular characterization of the first alpha-xylosidase from an archaeon. J Biol Chem. 2000;275:22082–9. [DOI] [PubMed] [Google Scholar]
- Moracci M, Nucci R, Febbraio F et al. Expression and extensive characterization of a beta-glycosidase from the extreme thermoacidophilic archaeon Sulfolobus solfataricus in Escherichia coli: authenticity of the recombinant enzyme. Enzyme Microb Technol. 1995;17:992–7. [DOI] [PubMed] [Google Scholar]
- Moriscot C, Gribaldo S, Jault JM et al. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS One. 2011;6:e21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougiakos I, Bosma EF, de Vos WM et al. Next generation prokaryotic engineering: the CRISPR-Cas toolkit. Trends Biotechnol. 2016;34:575–87. [DOI] [PubMed] [Google Scholar]
- Mousavi SM, Yaghmaei S, Vossoughi M et al. Comparison of bioleaching ability of two native mesophilic and thermophilic bacteria on copper recovery from chalcopyrite concentrate in an airlift bioreactor. Hydrometallurgy. 2005;80:139–44. [Google Scholar]
- Mukherjee A, Wheaton GH, Blum PH et al. Uranium extremophily is an adaptive, rather than intrinsic, feature for extremely thermoacidophilic Metallosphera prunae. Proc Natl Acad Sci USA. 2012;109:16702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Wheaton GH, Counts JA et al. VapC toxins drive cellular dormancy under uranium stress for the extreme thermoacidophile Metallosphaera prunae. Environ Microbiol. 2017;19:2831–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson-Mcgee JH, Peng S, Dewerff S et al. A virus or more in (nearly) every cell: ubiquitous networks of virus–host interactions in extreme environments. ISME J. 2018;12:1706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson-Mcgee JH, Snyder JC, Young MJ. Archaeal viruses from high-temperature environments. Genes. 2018;9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskhelishvili G, Palm P, Zillig W. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol Gen Genet. 1993;237:334–42. [DOI] [PubMed] [Google Scholar]
- Märtens B, Manoharadas S, Hasenöhrl D et al. Antisense regulation by transposon-derived RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO Rep. 2013;14:527–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller FH, Bandeiras TM, Urich T et al. Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol Microbiol. 2004;53:1147–60. [DOI] [PubMed] [Google Scholar]
- Neale JW, Robertson SW, Muller HH et al. Integrated piloting of a thermophilic bioleaching process for the treatment of a low-grade nickel-copper sulphide concentrate. J S Afr I Min Metall. 2009;109:273–93. [Google Scholar]
- Nguyen-Duc T, van Oeffelen L, Song N et al. The genome-wide binding profile of the Sulfolobus solfataricus transcription factor Ss-LrpB shows binding events beyond direct transcription regulation. BMC Genomics. 2013;14:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaus B, Gambacorta A, Basso AL et al. Trehalose in Archaebacteria. Sys Appl Microbiol. 1988;10:215–7. [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostøl JT et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–8. [DOI] [PubMed] [Google Scholar]
- Niu Y, Xia Y, Wang S et al. A prototypic lysine methyltransferase 4 from Archaea with degenerate sequence specificity methylates chromatin proteins Sul7d and Cren7 in different patterns. J Biol Chem. 2013;288:13728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh KH, Oh DK. Production of the rare ginsenosides compound K, compound Y, and compound Mc by a thermostable beta-glycosidase from Sulfolobus acidocaldarius. Biol Pharm Bull. 2009;32:1830–5. [DOI] [PubMed] [Google Scholar]
- Nordstrom KD, McCleskey BR, Ball JW. Sulfur geochemistry of hydrothermal waters in Yellowstone National Park: IV. Acid-sulfate waters. Appl Geochem. 2009;24:191–207. [Google Scholar]
- Norris PR, Clark DA, Owen JP et al. Characteristics of Sulfobacillus acidophilus sp. nov. and other moderately thermophilic mineral-sulphide-oxidizing bacteria. Microbiologyopen. 1996;142:775–83. [DOI] [PubMed] [Google Scholar]
- Nunn CEM, Johnsen U, Schönheit P et al. Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J Biol Chem. 2010;285:33701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–9. [DOI] [PubMed] [Google Scholar]
- Odling-Smee FJ, Laland KN, Feldman MW. Niche construction. Am Nat. 1996;147:641–8. [Google Scholar]
- Ogino H, Ishino S, Kohda D et al. The RecJ2 protein in the thermophilic archaeon Thermoplasma acidophilum is a 3′-5′ exonuclease that associates with a DNA replication complex. J Biol Chem. 2017;292:7921–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke M, Carter LG, Johnson KA et al. The Scottish structural proteomics facility: targets, methods and outputs. J Struct Funct Genomics. 2010;11:167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke M, Kerou M, Liu H et al. A dimeric rep protein initiates replication of a linear archaeal virus genome: implications for the rep mechanism and viral replication. J Virol. 2011;85:925–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orell A, Navarro CA, Rivero M et al. Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles. 2012;16:573–83. [DOI] [PubMed] [Google Scholar]
- Orell A, Peeters E, Vassen V et al. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. ISME J. 2013a;7:1886–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orell A, Remonsellez F, Arancibia R et al. Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus. Archaea. 2013b;2013:289236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orell A, Tripp V, Aliaga-Tobar V et al. A regulatory RNA is involved in RNA duplex formation and biofilm regulation in Sulfolobus acidocaldarius. Nucleic Acids Res. 2018;46:4794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann AC, Brumfield SK, Walther J et al. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008;82:6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri G, Balestrieri M, Peter-Katalinić J et al. Surface-exposed glycoproteins of hyperthermophilic Sulfolobus solfataricus P2 show a common N-glycosylation profile. J Proteome Res. 2013;12:2779–90. [DOI] [PubMed] [Google Scholar]
- Papon N, Stock AM. What do archaeal and eukaryotic histidine kinases sense? F1000Res. 2019;8:F1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Yoo CB, Choi SY et al. Purifications and characterizations of a ferredoxin and its related 2-oxoacid:ferredoxin oxidoreductase from the hyperthermophilic archaeon, Sulfolobus solfataricus P1. J Biochem Mol Biol. 2006;39:46–54. [DOI] [PubMed] [Google Scholar]
- Paula FS, Chin JP, Schnürer A et al. The potential for polyphosphate metabolism in Archaea and anaerobic polyphosphate formation in Methanosarcina mazei. Sci Rep. 2019;9:17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly MD, Bautista MA, Black JA et al. Diversified local CRISPR-Cas immunity to viruses of Sulfolobus islandicus. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KAP, Hough DW, Danson MJ. Discovery of a putative acetoin dehydrogenase complex in the hyperthermophilic archaeon Sulfolobus solfataricus. FEBS Lett. 2010;584:1231–4. [DOI] [PubMed] [Google Scholar]
- Payne S, McCarthy S, Johnson T et al. Nonmutational mechanism of inheritance in the Archaeon Sulfolobus solfataricus. Proc Natl Acad Sci USA. 2018;115:12271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paytubi S, White MF. The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol Microbiol. 2009;72:1487–99. [DOI] [PubMed] [Google Scholar]
- Pearson A, Hurley SJ, Elling FJ et al. CO2-dependent carbon isotope fractionation in Archaea, Part I: modeling the 3HP/4HB pathway. Geochim Cosmochim Acta. 2019;261:368–82. [Google Scholar]
- Peeples TL, Kelly RM. Bioenergetic response of the extreme thermoacidophile Metallosphaera sedula to thermal and nutritional stresses. Appl Environ Microbiol. 1995;61:2314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters E, Albers SV, Vassart A et al. Ss-LrpB, a transcriptional regulator from Sulfolobus solfataricus, regulates a gene cluster with a pyruvate ferredoxin oxidoreductase-encoding operon and permease genes. Mol Microbiol. 2009;71:972–88. [DOI] [PubMed] [Google Scholar]
- Peeters E, Driessen RP, Werner F et al. The interplay between nucleoid organization and transcription in archaeal genomes. Nat Rev Microbiol. 2015;13:333–41. [DOI] [PubMed] [Google Scholar]
- Peeters E, Peixeiro N, Sezonov G. Cis-regulatory logic in archaeal transcription. Biochem Soc T. 2013;41:326–31. [DOI] [PubMed] [Google Scholar]
- Peng N, Deng L, Mei Y et al. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl Environ Microbiol. 2012;78:5630–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Blum H, She Q et al. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology. 2001;291:226–34. [DOI] [PubMed] [Google Scholar]
- Perez-Rueda E, Hernandez-Guerrero R, Martinez-Nuñez MA et al. Abundance, diversity and domain architecture variability in prokaryotic DNA-binding transcription factors. PLoS One. 2018;13:e0195332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Sticky situations: key components that control bacterial surface attachment. J Bacteriol. 2012;194:2413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyfoon E, Meyer B, Hitchen PG et al. The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with chitobiose-linked N-glycans. Archaea. 2010;2010:754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilä MK, Demina TA, Atanasova NS et al. Archaeal viruses and bacteriophages: comparisons and contrasts. Trends Microbiol. 2014;22:334–44. [DOI] [PubMed] [Google Scholar]
- Pina M, Basta T, Quax TE et al. Unique genome replication mechanism of the archaeal virus AFV 1. Mol Microbiol. 2014;92:1313–25. [DOI] [PubMed] [Google Scholar]
- Pina M, Bize A, Forterre P et al. The archeoviruses. FEMS Microbiol Rev. 2011;35:1035–54. [DOI] [PubMed] [Google Scholar]
- Plagens A, Richter H, Charpentier E et al. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol Rev. 2015;39:442–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb JJ, Haddad CM, Gibson JaE et al. Acidianus sulfidivorans sp. nov., an extremely acidophilic, thermophilic archaeon isolated from a solfatara on Lihir Island, Papua New Guinea, and emendation of the genus description. Int J Syst Evol Microbiol. 2007;57:1418–23. [DOI] [PubMed] [Google Scholar]
- Poranen MM, Daugelavičius R, Bamford DH. Common principles in viral entry. Ann Rev Microbiol. 2002;56:521–38. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiologyopen. 2005;151:653–63. [DOI] [PubMed] [Google Scholar]
- Pradhan N, Nathsarma KC, Rao KS et al. Heap bioleaching of chalcopyrite: a review. Miner Eng. 2008;21:355–65. [Google Scholar]
- Prangishvili D, Arnold HP, Götz D et al. A novel virus family, the Rudiviridae: structure, virus–host interactions and genome variability of the Sulfolobus viruses SIRV1 and SIRV2. Genetics. 1999;152:1387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prangishvili D, Bamford DH, Forterre P et al. The enigmatic archaeal virosphere. Nat Rev Microbiol. 2017;15:724–39. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006;4:837–48. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Garrett RA. Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses. Biochem Soc Trans. 2004;32:204–8. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Koonin EV, Krupovic M. Genomics and biology of Rudiviruses, a model for the study of virus–host interactions in Archaea. Biochem Soc Trans. 2013;41:443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prangishvili D, Quax TE. Exceptional virion release mechanism: one more surprise from archaeal viruses. Curr Opin Microbiol. 2011;14:315–20. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Stedman K, Zillig W. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol. 2001;9:39–43. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Vestergaard G, Häring M et al. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol. 2006;359:1203–16. [DOI] [PubMed] [Google Scholar]
- Prangishvili D. Archaeal viruses: living fossils of the ancient virosphere?. Ann NY Acad Sci. 2015;1341:35–40. [DOI] [PubMed] [Google Scholar]
- Prangishvili DA, Vashakidze RP, Chelidze MG et al. A restriction endonuclease SuaI from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1985;192:57–60. [DOI] [PubMed] [Google Scholar]
- Pritchett MA, Zhang JK, Metcalf WW. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl Environ Microbiol. 2004;70:1425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokofeva MI, Kostrikina NA, Kolganova TV et al. Isolation of the anaerobic thermoacidophilic crenarchaeote Acidilobus saccharovorans sp. nov. and proposal of Acidilobales ord. nov., including Acidilobaceae fam. nov. and Caldisphaeraceae fam. nov. Int J Syst Evol Microbiol. 2009;59:3116–22. [DOI] [PubMed] [Google Scholar]
- Prokofeva MI, Miroshnichenko ML, Kostrikina NA et al. Acidilobus aceticus gen. nov., sp. nov., a novel anaerobic thermoacidophilic archaeon from continental hot vents in Kamchatcka. Int J Syst Evol Microbiol. 2000;50:2001–8. [DOI] [PubMed] [Google Scholar]
- Protze J, Müller F, Lauber K et al. An extracellular tetrathionate hydrolase from the thermoacidophilic archaeon Acidianus ambivalens with an activity optimum at pH 1. Front Microbiol. 2011;2:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüschenk R, Baumeister W, Zillig W. Surface structure variants in different species of Sulfolobus. FEMS Microbiol Lett. 1987;43:327–30. [Google Scholar]
- Pulschen AA, Mutavchiev DR, Culley S et al. Live imaging of a hyperthermophilic archaeon reveals distinct roles for two ESCRT-III homologs in ensuring a robust and symmetric division. Curr Biol. 2020;30:2852–9.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purschke WG, Schmidt CL, Petersen A et al. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J Bacteriol. 1997;179:1344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rueda E, Janga SC. Identification and genomic analysis of transcription factors in archaeal genomes exemplifies their functional architecture and evolutionary origin. Mol Biol Evol. 2010;27:1449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TE, Krupovič M, Lucas S et al. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology. 2010;404:1–4. [DOI] [PubMed] [Google Scholar]
- Quax TE, Lucas S, Reimann J et al. Simple and elegant design of a virion egress structure in Archaea. Proc Natl Acad Sci USA. 2011;108:3354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TEF, Daum B. Structure and assembly mechanism of virus-associated pyramids. Biophys Rev. 2017;10:551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TEF, Voet M, Sismeiro O et al. Massive activation of archaeal defense genes during viral infection. J Virol. 2013;87:8419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger J, Albersmeier A, Glatzel H et al. A defined cultivation medium for Sulfolobus acidocaldarius. J Biotechnol. 2019;301:56–67. [DOI] [PubMed] [Google Scholar]
- Quehenberger J, Pittenauer E, Allmaier G et al. The influence of the specific growth rate on the lipid composition of Sulfolobus acidocaldarius. Extremophiles. 2020;24:413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger J, Shen L, Albers S-V et al. Sulfolobus: a potential key organism in future biotechnology. Front Microbiol. 2017;8:2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ER, Lucas S, Daum B et al. First insights into the entry process of hyperthermophilic archaeal viruses. J Virol. 2013;87:13379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ERJ, Chlanda P, Sachse M et al. Eukaryotic-like virus budding in Archaea. mBio. 2016;7:e01439–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ERJ, Quax TEF. Archaeal viruses at the cell envelope: entry and egress. Front Microbiol. 2015;6:552–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi SA, Bell SD, Jackson SP. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Vera WH, Weiss M, Strittmatter E et al. Identification of missing genes and enzymes for autotrophic carbon fixation in Crenarchaeota. J Bacteriol. 2011;193:1201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid MH, Rumbaugh K, Passador L et al. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:9636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls KS, Martin JH, Maupin-Furlow JA. Activity and transcriptional regulation of bacterial protein-like glycerol-3-phosphate dehydrogenase of the haloarchaea in Haloferax volcanii. J Bacteriol. 2011;193:4469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray WK, Keith SM, DeSantis AM et al. A phosphohexomutase from the archaeon Sulfolobus solfataricus is covalently modified by phosphorylation on serine. J Bacteriol. 2005;187:4270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray WK, Potters MB, Haile JD et al. Activation of SsoPK4, an Archaeal eIF2α kinase homolog, by oxidized CoA. Proteomes. 2015;3:89–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redder P, Garrett RA. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J Bacteriol. 2006;188:4198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MS, Grogan DW. Biological effects of DNA damage in the hyperthermophilic archaeon Sulfolobus acidocaldarius. FEMS Microbiol Lett. 2002;208:29–34. [DOI] [PubMed] [Google Scholar]
- Reimann J, Esser D, Orell A et al. Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius. Mol Cel Proteomics. 2013;12:3908–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann J, Lassak K, Khadouma S et al. Regulation of archaella expression by the FHA and von Willebrand domain-containing proteins ArnA and ArnB in Sulfolobus acidocaldarius. Mol Microbiol. 2012;86:24–36. [DOI] [PubMed] [Google Scholar]
- Reinhardt A, Johnsen U, Schönheit P. l-Rhamnose catabolism in Archaea. Mol Microbiol. 2019;111:1093–108. [DOI] [PubMed] [Google Scholar]
- Remonsellez F, Orell A, Jerez CA. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiologyopen. 2006;152:59–66. [DOI] [PubMed] [Google Scholar]
- Reno ML, Held NL, Fields CJ et al. Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci USA. 2009;106:8605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A-L, Banta AB, Boone DR et al. Microbial essentials at hydrothemal vents. Nature. 2000;404:835. [DOI] [PubMed] [Google Scholar]
- Reysenbach AL, Liu Y, Banta AB et al. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–7. [DOI] [PubMed] [Google Scholar]
- Rice G, Tang L, Stedman K et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci USA. 2004;101:7716–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richey D, Lin E. Importance of facilitated diffusion for effective utilization of glycerol by Escherichia coli. J Bacteriol. 1972;112:784–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker KD, Kelly RM. Growth physiology of the hyperthermophilic archaeon Thermococcus litoralis: development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence of biofilm formation. Appl Environ Microbiol. 1996;62:4478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risa GT, Hurtig F, Bray S et al. Proteasome-mediated protein degradation resets the cell division cycle and triggers ESCRT-III-mediated cytokinesis in an archaeon. bioRxiv. 2019;774273. [Google Scholar]
- Rivero M, Torres-Paris C, Munoz R et al. Inorganic polyphosphate, exopolyphosphatase, and Pho84-like transporters may be involved in copper resistance in Metallosphaera sedula DSM 5348T. Archaea. 2018;2018:5251061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci USA. 2007;104:5806–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Dionne I, Lundgren M et al. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. [DOI] [PubMed] [Google Scholar]
- Rodionova IA, Vetting MW, Li X et al. A novel bifunctional transcriptional regulator of riboflavin metabolism in Archaea. Nucleic Acids Res. 2017;45:3785–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfsmeier M, Haseltine C, Bini E et al. Molecular characterization of the alpha-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 1998;180:1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostøl JT, Marraffini LA. Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR–Cas immunity. Nature Microbiol. 2019;4:656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild LJ, Mancinelli RL. Life in extreme environments. Nature. 2001;409:1092–101. [DOI] [PubMed] [Google Scholar]
- Rouillon C, Athukoralage JS, Graham S et al. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife. 2018;7:e36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland EF, Bautista MA, Zhang C et al. Surface resistance to SSVs and SIRVs in pilin deletions ofSulfolobus islandicus. Mol Microbiol. 2020;113:718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J, Tolliday N, Schmitt C et al. Phosphorylation in halobacterial signal transduction. EMBO J. 1995;14:4249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Londei P. Differential antibiotic sensitivity determined by the large ribosomal subunit in thermophilic archaea. J Bacteriol. 1996;178:3396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai HD, Kurosawa N. Complete genome sequence of the Sulfodiicoccus acidiphilus strain HS-1(T), the first crenarchaeon that lacks polB3, isolated from an acidic hot spring in Ohwaku-dani, Hakone, Japan. BMC Res Notes. 2019;12:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai HD, Kurosawa N. Saccharolobus caldissimus gen. nov., sp. nov., a facultatively anaerobic iron-reducing hyperthermophilic archaeon isolated from an acidic terrestrial hot spring, and reclassification of Sulfolobus solfataricus as Saccharolobus solfataricus comb. nov. and Sulfolobus shibatae as Saccharolobus shibatae comb. nov. Int J Syst Evol Microbiol. 2018;68:1271–8. [DOI] [PubMed] [Google Scholar]
- Sakai HD, Kurosawa N. Sulfodiicoccus acidiphilus gen. nov., sp. nov., a sulfur-inhibited thermoacidophilic archaeon belonging to the order Sulfolobales isolated from a terrestrial acidic hot spring. Int J Syst Evol Microbiol. 2017;67:1880–6. [DOI] [PubMed] [Google Scholar]
- Sakofsky CJ, Runck LA, Grogan DW. Sulfolobus mutants, generated via PCR products, which lack putative enzymes of UV photoproduct repair. Archaea. 2011;2011:864015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Abeyrathne PD, Bell SD. Mechanism of archaeal MCM helicase recruitment to DNA replication origins. Mol Cell. 2016;61:287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM et al. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Hodgson B et al. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41:186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Xu Y, Gadelha C et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 2013;3:485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand W, Gehrke T, Jozsa PG et al. (Bio)chemistry of bacterial leaching: direct vs. indirect bioleaching. Hydrometallurgy. 2001;59:159–75. [Google Scholar]
- Santos H, Da Costa MS. Compatible solutes of organisms that live in hot saline environments. Environ Microbiol. 2002;4:501–9. [DOI] [PubMed] [Google Scholar]
- Sanz JL, Huber G, Huber H et al. Using protein synthesis inhibitors to establish the phylogenetic relationships of the Sulfolobales order. J Mol Evol. 1994;39:528–32. [DOI] [PubMed] [Google Scholar]
- Sato T, Fukui T, Atomi H et al. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol. 2005;71:3889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Say RF, Fuchs G. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature. 2010;464:1077–81. [DOI] [PubMed] [Google Scholar]
- Schafer G, Moll R, Schmidt CL. Respiratory enzymes from Sulfolobus acidocaldarius. Methods Enzymol. 2001;331:369–410. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Shiu S-H, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21:R320–30. [DOI] [PubMed] [Google Scholar]
- Scheele U, Erdmann S, Ungewickell EJ et al. Chaperone role for proteins p618 and p892 in the extracellular tail development of Acidianus two-tailed virus. J Virol. 2011;85:4812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelert J, Dixit V, Hoang V et al. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol. 2004;186:427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelert J, Drozda M, Dixit V et al. Regulation of mercury resistance in the Crenarchaeote Sulfolobus solfataricus. J Bacteriol. 2006;188:7141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers A, Jozsa P, Sand W. Sulfur chemistry in bacterial leaching of pyrite. Appl Environ Microbiol. 1996;62:3424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C, Kubo K, Zillig W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci USA. 1992;89:7645–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C, Puehler G, Holz I et al. Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J Bacteriol. 1995;177:7050–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C, Puhler G, Klenk H-P et al. Picrophilus oshimae and Picrophilus torridusfam. nov., gen. nov., sp. nov., two species of hyperacidophilic, thermophilic, heterotrophic, aerobic archaea. J Syst Evol Microbiol. 1996;46:814–6. [Google Scholar]
- Schlitt WJ. Kennecott's million-ton test heap: the active leach program. Miner Metall Proc. 2006;23:1–16. [Google Scholar]
- Schocke L, Bräsen C, Siebers B. Thermoacidophilic Sulfolobus species as source for extremozymes and as novel archaeal platform organisms. Curr Opin Biotechnol. 2019;59:71–7. [DOI] [PubMed] [Google Scholar]
- Schoehn G, Quaite-Randall E, Jiménez JL et al. Three conformations of an archaeal chaperonin, TF55 from Sulfolobus shibatae. J Mol Biol. 2000;296:813–9. [DOI] [PubMed] [Google Scholar]
- Schult F, Le TN, Albersmeier A et al. Effect of UV irradiation on Sulfolobus acidocaldarius and involvement of the general transcription factor TFB3 in the early UV response. Nucleic Acids Res. 2018;46:7179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer G. Bioenergetics of the archaebacterium Sulfolobus. Biochim Biophys Acta. 1996;1277:163–200. [DOI] [PubMed] [Google Scholar]
- Scott JW, Poole FL, Adams MWW. Characterization of ten heterotetrameric NDP-dependent acyl-CoA synthetases of the hyperthermophilic archaeon Pyrococcus furiosus. Archaea. 2014;2014:176863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerer A, Neuner A, Kristjansson JK et al. Acidianus infernus gen. nov., sp. nov., and Acidianus brierleyi Comb. nov.: facultatively aerobic, extremely acidophilic thermophilic sulfur-metabolizing archaebacteria. Int J Syst Evol Microbiol. 1986;36:559–64. [Google Scholar]
- Segerer AH, Trincone A, Gahrtz M et al. Stygiolobus azoricus gen. nov., sp. nov. represents a novel genus of anaerobic, extremely thermoacidophilic archaebacteria of the order Sulfolobales. Int J Syst Bacteriol. 1991;41:495–501. [Google Scholar]
- Segerer ALT, Stetter KO. Thermoplasma acidophilum and Thermoplasma volcaniumsp. nov. from solfatara fields. Syst Appl Microbiol. 1988;10:161–71. [Google Scholar]
- Serre MC, Letzelter C, Garel JR et al. Cleavage properties of an archaeal site-specific recombinase, the SSV1 integrase. J Biol Chem. 2002;277:16758–67. [DOI] [PubMed] [Google Scholar]
- Sheppard C, Blombach F, Belsom A et al. Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP. Nat Commun. 2016;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Zhang C, Deng L et al. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem Soc Trans. 2009;37:92–6. [DOI] [PubMed] [Google Scholar]
- Sherwood KE, Cano DJ, Maupin-Furlow JA. Glycerol-mediated repression of glucose metabolism and glycerol kinase as the sole route of glycerol catabolism in the haloarchaeon Haloferax volcanii. J Bacteriol. 2009;191:4307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Potts M, Kennelly PJ. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol Rev. 1998;22:229–53. [DOI] [PubMed] [Google Scholar]
- Shima S, Suzuki K-I. Hydrogenobacter acidophilus sp. nov., a thermoacidophilic, aerobic, hydrogen-oxidizing bacterium requiring elemental sulfur for growth. Int J Syst Evol Microbiol. 1993;43:703–8. [Google Scholar]
- Shin KC, Oh HJ, Kim BJ et al. Complete conversion of major protopanaxadiol ginsenosides to compound K by the combined use of α-l-arabinofuranosidase and β-galactosidase from Caldicellulosiruptor saccharolyticus and β-glucosidase from Sulfolobus acidocaldarius. J Biotechnol. 2013;167:33–40. [DOI] [PubMed] [Google Scholar]
- Shi X, Rao NN, Kornberg A. Inorganic polyphosphate in Bacillus cereus: motility, biofilm formation, and sporulation. Proc Natl Acad Sci USA. 2004;101:17061–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–84. [DOI] [PubMed] [Google Scholar]
- Singer GAC, Hickey DA. Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content. Gene. 2003;317:39–47. [DOI] [PubMed] [Google Scholar]
- Skórko R. Protein phosphorylation in the archaebacterium Sulfolobus acidocaldarius. Eur J Biochem. 1984;145:617–22. [DOI] [PubMed] [Google Scholar]
- Smith SC, Kennelly PJ, Potts M. Protein-tyrosine phosphorylation in the Archaea. J Bacteriol. 1997;179:2418–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Brumfield SK, Peng N et al. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J Virol. 2011;85:6287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Stedman K, Rice G et al. Viruses of hyperthermophilic Archaea. Res Microbiol. 2003;154:474–82. [DOI] [PubMed] [Google Scholar]
- Song N, Duc TN, Van Oeffelen L et al. Expanded target and cofactor repertoire for the transcriptional activator LysM from Sulfolobus. Nucleic Acids Res. 2013;41:2932–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto DF, Recalde A, Orell A et al. Global effect of the lack of inorganic polyphosphate in the extremophilic archaeon Sulfolobus solfataricus: a proteomic approach. J Proteomics. 2019;191:143–52. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Stoeckenius W. Light-regulated retinal-dependent reversible phosphorylation of Halobacterium proteins. J Biol Chem. 1980;255:5501–3. [PubMed] [Google Scholar]
- Stancik IA, Šestak MS, Ji B et al. Serine/Threonine protein kinases from Bacteria, Archaea and Eukarya share a common evolutionary origin deeply rooted in the tree of life. J Mol Biol. 2018;430:27–32. [DOI] [PubMed] [Google Scholar]
- Stark H, Wolf J, Albersmeier A et al. Oxidative Stickland reactions in an obligate aerobic organism: amino acid catabolism in the Crenarchaeon Sulfolobus solfataricus. FEBS J. 2017;284:2078–95. [DOI] [PubMed] [Google Scholar]
- Stedman KM, Kosmicki NR, Diemer GS. Codon usage frequency of RNA virus genomes from high-temperature acidic-environment metagenomes. J Virol. 2013;87:1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman KM, Schleper C, Rumpf E et al. Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: construction and testing of viral shuttle vectors. Genetics. 1999;152:1397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steudel R. Mechanism for the formation of elemental sulfur from aqueous sulfide in chemical and microbiological desulfurization processes. Ind Eng Chem Res. 1996;35:7. [Google Scholar]
- Straub CT, Counts JA, Nguyen DMN et al. Biotechnology of extremely thermophilic archaea. FEMS Microbiol Rev. 2018;42:543–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud RM, Miercke LJ, O'Connell J et al. Glycerol facilitator GlpF and the associated aquaporin family of channels. Curr Opin Struct Biol. 2003;13:424–31. [DOI] [PubMed] [Google Scholar]
- Sumper M, Berg E, Mengele R et al. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J Bacteriol. 1990;172:7111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Kurosawa N. Disruption of the gene encoding restriction endonuclease SuaI and development of a host–vector system for the thermoacidophilic archaeon Sulfolobus acidocaldarius. Extremophiles. 2016;20:139–48. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Iwasaki T, Uzawa T et al. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles. 2002;6:39–44. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Miyamoto K, Ohta H. A novel thermostable esterase from the thermoacidophilic archaeon Sulfolobus tokodaii strain 7. FEMS Microbiol Lett. 2004;236:97–102. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ER et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014a;507:258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang Y et al. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. 2014b;21:743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachdjian S, Kelly RM. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2006;188:4553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi S, Kawasaki H, Sugimori K et al. Sulfolobus hakonensis sp. nov., a novel species of acidothermophilic archaeon. Int J Sys Evol Microbiol. 1996;46:377–82. [DOI] [PubMed] [Google Scholar]
- Takemata N, Samson RY, Bell SD. Physical and functional compartmentalization of archaeal chromosomes. Cell. 2019;179:165–79.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M et al. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–81. [DOI] [PubMed] [Google Scholar]
- Tansey MR, Brock TD. The upper temperature limit for eukaryotic organisms. Proc Nat Acad Sci USA. 1972;69:2426–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Deatherage JF, Amos LA. Structure of the S-layer of Sulfolobus acidocaldarius. Nature. 1982;299:840–2. [Google Scholar]
- Tekaia F, Yeramian E, Dujon B. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: a global picture with correspondence analysis. Gene. 2002;297:51–60. [DOI] [PubMed] [Google Scholar]
- Teufel R, Kung JW, Kockelkorn D et al. 3-hydroxypropionyl-coenzyme A dehydratase and acryloyl-coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in the Sulfolobales. J Bacteriol. 2009;191:4572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JD, Nimmesgern E, Wall JS et al. A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature. 1991;354:490–3. [DOI] [PubMed] [Google Scholar]
- Trent JD, Osipiuk J, Pinkau T. Acquired thermotolerance and heat shock in the extremely thermophilic archaebacterium Sulfolobus sp. strain B12. J Bacteriol. 1990;172:1478–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripepi M, Imam S, Pohlschröder M. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J Bacteriol. 2010;192:3093–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-L, Tripp P, Sivabalasarma S et al. The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility. Nat Microbiol. 2020;5:216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi K, Sakai HD, Nur N et al. Sulfurisphaera javensis sp. nov., a hyperthermophilic and acidophilic archaeon isolated from Indonesian hot spring, and reclassification of Sulfolobus tokodaii Suzuki et al. 2002 as Sulfurisphaera tokodaii comb. nov. Int J Syst Evol Microbiol. 2018;68:1907–13. [DOI] [PubMed] [Google Scholar]
- Turner P, Mamo G, Karlsson EN. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb Cell Fact. 2007;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda I, Sugai A, Itoh YH et al. Variation in molecular species of polar lipids from Thermoplasma acidophilum depends on growth temperature. Lipids. 2001;36:103–5. [DOI] [PubMed] [Google Scholar]
- Uhrigshardt H, Walden M, John H et al. Evidence for an operative glyoxylate cycle in the thermoacidophilic crenarchaeon S ulfolobus acidocaldarius. FEBS Lett. 2002;513:223–9. [DOI] [PubMed] [Google Scholar]
- Ulas T, Riemer SA, Zaparty M et al. Genome-scale reconstruction and analysis of the metabolic network in the hyperthermophilic archaeon Sulfolobus solfataricus. PLoS One. 2012;7:e43401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldahl KB, Jensen SB, Bhoobalan-Chitty Y et al. Life cycle characterization of Sulfolobus monocaudavirus 1, an extremophilic spindle-shaped virus with extracellular tail development. J Virol. 2016;90:5693–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbieta MS, Rascovan N, Castro C et al. Draft genome sequence of the novel thermoacidophilic archaeon Acidianus copahuensis strain ALE1, isolated from the Copahue volcanic area in Neuquén, Argentina. Genome Announc. 2014;2:e00259–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:316–23. [DOI] [PubMed] [Google Scholar]
- van der Kolk N, Wagner A, Wagner M et al. Identification of XylR, the activator of arabinose/xylose inducible regulon in Sulfolobus acidocaldarius and its application for homologous protein expression. Front Microbiol. 2020;11:1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost JWE, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vossenberg JLCM, Driessen AJM, Zillig W et al. Bioenergetics and cytoplasmic membrane stability of the extremely acidophilic, thermophilic archaeon Picrophilus oshimae. Extremophiles. 1998;2:67–74. [DOI] [PubMed] [Google Scholar]
- van Wolferen M, Orell A, Albers S-V. Archaeal biofilm formation. Nat Rev Microbiol. 2018;16:699–713. [DOI] [PubMed] [Google Scholar]
- van Wolferen M, Shajahan A, Heinrich K et al. Species-specific recognition of Sulfolobales mediated by UV-inducible pili and S-layer glycosylation patterns. mBio. 2020;11:e03014–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassart A, Van Wolferen M, Orell A et al. Sa-Lrp from Sulfolobus acidocaldarius is a versatile, glutamine-responsive, and architectural transcriptional regulator. Microbiologyopen. 2013;2:75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith A, Klingl A, Zolghadr B et al. Acidianus,Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol Microbiol. 2009;73:58–72. [DOI] [PubMed] [Google Scholar]
- Vestergaard G, Shah SA, Bize A et al. Stygiolobus rod-shaped virus and the interplay of crenarchaeal rudiviruses with the CRISPR antiviral system. J Bacteriol. 2008;190:6837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter AM, Helmecke J, Schomburg D et al. The impact of pyroglutamate: Sulfolobus acidocaldarius has a growth advantage over Saccharolobus solfataricus in glutamate-containing media. Archaea. 2019;2019:3208051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafane A, Voskoboynik Y, Ruhl I et al. CopR of Sulfolobus solfataricus represents a novel class of archaeal-specific copper-responsive activators of transcription. Microbiologyopen. 2011;157:2808–17. [DOI] [PubMed] [Google Scholar]
- Villafane AA, Voskoboynik Y, Cuebas M et al. Response to excess copper in the hyperthermophile Sulfolobus solfataricus strain 98/2. Biochem Biophys Res Commun. 2009;385:67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorontsov EA, Rensen E, Prangishvili D et al. Abundant lysine methylation and N-terminal acetylation in Sulfolobus islandicus revealed by bottom-up and top-down proteomics. Mol Cell Proteomics. 2016;15:3388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völlmecke C, Drees SL, Reimann J et al. The ATPases CopA and CopB both contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiologyopen. 2012;158:1622–33. [DOI] [PubMed] [Google Scholar]
- Wagner M, Berkner S, Ajon M et al. Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem Soc Trans. 2009;37:97–101. [DOI] [PubMed] [Google Scholar]
- Wagner M, Shen L, Albersmeier A et al. Sulfolobus acidocaldarius uptakes pentoses via a cut2-type ABC transporter and metabolizes them through the aldolase-independent Weimberg pathway. Appl Environ Microbiol. 2017;84:1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, van Wolferen M, Wagner A et al. Versatile genetic tool box for the Crenarchaeote Sulfolobus acidocaldarius. Front Microbiol. 2012;3:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Wagner A, Ma X et al. Investigation of the malE promoter and MalR, a positive regulator of the maltose regulon, for an improved expression system in Sulfolobus acidocaldarius. Appl Environ Microbiol. 2014;80:1072–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang S, Zhang L et al. Archaeal eukaryote-like serine/threonine protein kinase interacts with and phosphorylates a forkhead-associated-domain-containing protein. J Bacteriol. 2010a;192:1956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Cvirkaite-Krupovic V, Kreutzberger MA et al. An extensively glycosylated archaeal pilus survives extreme conditions. Nat Microbiol. 2019a;4:1401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liu Y, Su Z et al. A packing for A-form DNA in an icosahedral virus. Proc Natl Acad Sci USA. 2019b;116:22591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-C, Susko E, Roger AJ. On the correlation between genomic G+C content and optimal growth temperature in prokaryotes: data quality and confounding factors. Biochem Biophys Res Comm. 2006;342:681–4. [DOI] [PubMed] [Google Scholar]
- Wang K, Sybers D, Maklad HR et al. A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius. Nat Commun. 2019c;10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Q, Wu X et al. Bioinformatics analysis of metabolism pathways of archaeal energy reserves. Sci Rep. 2019d;9:1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu X, Wen Z et al. Isolation, purification, and properties of a novel small heat shock protein from the hyperthermophile Sulfolobus solfataricus. Appl Biochem Biotechnol. 2010b;162:476–85. [DOI] [PubMed] [Google Scholar]
- Ward L, Taylor MW, Power JF et al. Microbial community dynamics in Inferno Crater Lake, a thermally fluctuating geothermal spring. ISME J. 2017;11:1158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watling HR. The bioleaching of sulphide minerals with emphasis on copper sulphides: a review. Hydrometallurgy. 2006;84:81–108. [Google Scholar]
- Weinberger AD, Wolf YI, Lobkovsky AE et al. Viral diversity threshold for adaptive immunity in prokaryotes. mBio. 2012;3:e00456–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiße RH-J, Faust A, Schmidt M et al. Structure of NDP-forming acetyl-CoA synthetase ACD1 reveals a large rearrangement for phosphoryl transfer. Proc Natl Acad Sci USA. 2016;113:E519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Mosolf J, Willits D et al. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci USA. 2005;102:10551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, Allen M, Tschitschko B et al. Glycerol metabolism of Haloarchaea. Environ Microbiol. 2017;19:864–77. [DOI] [PubMed] [Google Scholar]
- Willkomm S, Oellig CA, Zander A et al. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat Microbiol. 2017;2:17035. [DOI] [PubMed] [Google Scholar]
- Wolf J, Stark H, Fafenrot K et al. A systems biology approach reveals major metabolic changes in the thermoacidophilic archaeon Sulfolobus solfataricus in response to the carbon source l-fucose versus d-glucose. Mol Microbiol. 2016;102:882–908. [DOI] [PubMed] [Google Scholar]
- Woodruff PJ, Carlson BL, Siridechadilok B et al. Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem. 2004;279:28835–43. [DOI] [PubMed] [Google Scholar]
- Worthington P, Hoang V, Perez-Pomares F et al. Targeted disruption of the α-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2003;185:482–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F et al. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Li Y, Zhao YT et al. Identification and characterization of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. PLoS One. 2012;7:e35306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gristwood T, Hodgson B et al. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proc Natl Acad Sci USA. 2016;113:13390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Driessen AJM. Deletion of cdvB paralogous genes of Sulfolobus acidocaldarius impairs cell division. Extremophiles. 2014;18:331–9. [DOI] [PubMed] [Google Scholar]
- Yan Z, Maruyama A, Arakawa T et al. Crystal structures of archaeal 2-oxoacid:ferredoxin oxidoreductases from Sulfolobus tokodaii. Sci Rep. 2016;6:33061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutake Y, Nishiya Y, Tamura N et al. Structural insights into unique substrate selectivity of Thermoplasma acidophilum d-aldohexose dehydrogenase. J Mol Biol. 2007;367:1034–46. [DOI] [PubMed] [Google Scholar]
- Yeo HK, Kang J, Park YW et al. Crystallization and preliminary X-ray diffraction analysis of the metalloregulatory protein DtxR from Thermoplasma acidophilum. Acta Crystallogr F. 2012;68:172–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo HK, Park YW, Lee JY. Structural analysis and insight into metal-ion activation of the iron-dependent regulator from Thermoplasma acidophilum. Acta Crystallogr D. 2014;70:1281–8. [DOI] [PubMed] [Google Scholar]
- Yokooji Y, Sato T, Fujiwara S et al. Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol. 2013;195:1940–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zago MA, Dennis PP, Omer AD. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:1812–28. [DOI] [PubMed] [Google Scholar]
- Zaparty M, Esser D, Gertig S et al. "Hot standards" for the thermoacidophilic archaeon Sulfolobus solfataricus. Extremophiles. 2009;14:119–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaparty M, Siebers B. Physiology, metabolism, and enzymology of thermoacidophiles. In: Horikoshi K (ed.). Extremophiles Handbook. Japan, Tokyo: Springer, 2011, 601–39. [Google Scholar]
- Zaramela LS, Vêncio RZN, Ten-Caten F et al. Transcription start site associated RNAs (TSSaRNAs) are ubiquitous in all domains of life. PLoS One. 2014;9:e107680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebec Z, Manica A, Zhang J et al. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014;42:5280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebec Z, Zink IA, Kerou M et al. Efficient CRISPR-Mediated post-transcriptional gene silencing in a hyperthermophilic archaeon using multiplexed crRNA expression. G3. 2016;6:3161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldes BM, Keller MW, Loder AJ et al. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front Microbiol. 2015;6:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldes BM, Loder AJ, Counts JA et al. Determinants of sulphur chemolithoautotrophy in the extremely thermoacidophilic Sulfolobales. Environ Microbiol. 2019;21:3696–710. [DOI] [PubMed] [Google Scholar]
- Zeldes BM, Straub CT, Otten JK et al. A synthetic enzymatic pathway for extremely thermophilic acetone production based on the unexpectedly thermostable acetoacetate decarboxylase from Clostridium acetobutylicum. Biotechnol Bioeng. 2018;115:2951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 2007;3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Liu X-L, Wei JH et al. Calditol-linked membrane lipids are required for acid tolerance in Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2018;115:12932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cooper TE, Krause DJ et al. Augmenting the genetic toolbox for Sulfolobus islandicuswith a stringent positive selectable marker for agmatine prototrophy. Appl Environ Microbiol. 2013a;79:5539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Phillips APR, Wipfler RL et al. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat Commun. 2018;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, She Q, Bi H et al. The apt/6-methylpurine counterselection system and its applications in genetic studies of the hyperthermophilic archaeon Sulfolobus islandicus. Appl Environ Microbiol. 2016a;82:3070–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Tian B, Li S et al. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem Soc Trans. 2013b;41:405–10. [DOI] [PubMed] [Google Scholar]
- Zhang C, Whitaker RJ. A broadly applicable gene knockout system for the thermoacidophilic archaeon Sulfolobus islandicus based on simvastatin selection. Microbiologyopen. 2012;158:1513–22. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wipfler RL, Li Y et al. Cell structure changes in the hyperthermophilic crenarchaeon Sulfolobus islandicus lacking the S-layer. mBio. 2019a;10:e01589–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Graham S, Tello A et al. Multiple nucleic acid cleavage modes in divergent type III CRISPR systems. Nucleic Acids Res. 2016b;44:1789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, White MF. Hot and crispy: CRISPR-Cas systems in the hyperthermophile Sulfolobus solfataricus. Biochem Soc Trans. 2013;41:1422–6. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Iwasaki T, Wakagi T et al. 2-Oxoacid:ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J Biochem. 1996;120:587–99. [DOI] [PubMed] [Google Scholar]
- Zhang R, Neu TR, Blanchard V et al. Biofilm dynamics and EPS production of a thermoacidophilic bioleaching archaeon. New Biotechnol. 2019b;51:21–30. [DOI] [PubMed] [Google Scholar]
- Zhang R, Neu TR, Zhang Y et al. Visualization and analysis of EPS glycoconjugates of the thermoacidophilic archaeon Sulfolobus metallicus. Appl Microbiol Biotechnol. 2015a;99:7343–56. [DOI] [PubMed] [Google Scholar]
- Zhang RY, Neu TR, Bellenberg S et al. Use of lectins to in situ visualize glycoconjugates of extracellular polymeric substances in acidophilic archaeal biofilms. Microb Biotechnol. 2015b;8:448–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kouril T, Snoep JL et al. The peculiar glycolytic pathway in hyperthermophylic archaea: understanding its whims by experimentation in silico. Int J Mol Sci. 2017;18:876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhan Z, Wang B et al. Archaeal chromatin proteins Cren7 and Sul7d compact DNA by bending and bridging. mBio. 2020;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhao M, Chen Y et al. Architectural roles of Cren7 in folding crenarchaeal chromatin filament. Mol Microbiol. 2019c;111:556–69. [DOI] [PubMed] [Google Scholar]
- Zhao HB, Zhang YS, Zhang X et al. The dissolution and passivation mechanism of chalcopyrite in bioleaching: an overview. Miner Eng. 2019;136:140–54. [Google Scholar]
- Zheng T, Huang Q, Zhang C et al. Development of a simvastatin selection marker for a hyperthermophilic acidophile, Sulfolobus islandicus. Appl Environ Microbiol. 2012;78:568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Klompe SE, Vlot M et al. Shooting the messenger: RNA-targetting CRISPR-Cas systems. Biosci Rep. 2018;38:BSR20170788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W, Kletzin A, Schleper C et al. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol. 1993;16:609–28. [Google Scholar]
- Zillig W, Stetter KO, Janekovic D. DNA-dependent RNA polymerase from the archaebacterium Sulfolobus acidocaldarius. Eur J Biochem. 1979;96:597–604. [DOI] [PubMed] [Google Scholar]
- Zillig W, Stetter KO, Wunderl S et al. The Sulfolobus-“Caldariella” group: taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch Microbiol. 1980;125:259–69. [Google Scholar]
- Zillig W, Yeats S, Holz I et al. Desulfurolobus ambivalens, gen. nov., sp. nov., an autotrophic archaebacterium facultatively oxidizing or reducing sulfur. Syst Appl Microbiol. 1986;8:197–203. [Google Scholar]
- Zimmermann P, Laska S, Kletzin A. Two modes of sulfite oxidation in the extremely thermophilic and acidophilic archaeon Acidianus ambivalens. Arch Microbiol. 1999;172:76–82. [DOI] [PubMed] [Google Scholar]
- Zolghadr B, Kling A, Koerdt A et al. Appendage-mediated surface adherence of Sulfolobus solfataricus. J Bacteriol. 2010;192:104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweerink S, Kallnik V, Ninck S et al. Activity-based protein profiling as a robust method for enzyme identification and screening in extremophilic Archaea. Nat Commun. 2017;8:15352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zähringer U, Moll H, Hettmann T et al. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur J Biochem. 2000;267:4144–9. [DOI] [PubMed] [Google Scholar]