ABSTRACT
Cigarettes and opium contain chemicals and particulate matter that may modify the oral microbiota. This study aimed to investigate the association between cigarette and opium use with the oral microbiota. A total of 558 participants were recruited from Iran between 2011 and 2015. Individuals were categorized as never cigarette nor opium users, ever cigarette-only smokers, ever opium-only users, and ever both cigarette and opium users. Participants provided saliva samples for 16S rRNA gene sequencing. Logistic regression, microbiome regression-based kernel association test (MiRKAT), and zero-inflated beta regression models were calculated. For every increase in 10 observed amplicon sequence variants (ASVs), the odds for being a cigarette-only smoker, opium-only user, and both user compared to never users decreased by 9% (odds ratio [OR] = 0.91; 95% confidence interval [95% CI] = 0.86 to 0.97), 13% (OR = 0.87; 95% CI = 0.75 to 1.01), and 12% (OR = 0.88; 95% CI = 0.80 to 0.96), respectively. The microbial communities differed by cigarette and opium use as indicated by MiRKAT models testing the three beta-diversity matrices (P < 0.05 for all). Three genera were less likely and one genus was more likely to be detected in cigarette-only smokers or opium-only users than in never users. The relative abundance of the phylum Actinobacteria (never, 14.78%; both, 21.20%) was higher and the phyla Bacteroidetes (never, 17.63%; both, 11.62%) and Proteobacteria (never, 9.06%; both, 3.70%) were lower in users of both cigarettes and opium, while the phylum Firmicutes (never, 54.29%; opium, 65.49%) was higher in opium-only users. Cigarette and opium use was associated with lower alpha-diversity, overall oral microbiota community composition, and both the presence and relative abundance of multiple taxa.
IMPORTANCE Cigarette smoking and opium use are associated with periodontal disease caused by specific bacteria such as Porphyromonas gingivalis, which suggests a link between cigarette smoking and opium use and the oral microbiota. Alterations of the oral microbiota in cigarette smokers compared to nonsmokers have been reported, but this has not been studied across diverse populations. Additionally, the association of opium use with the oral microbiota has not been investigated to date. We conducted this study to investigate differences in the oral microbiota between ever users of cigarettes only, opium only, and both cigarettes and opium and never users of cigarettes and opium in Iran. Lower alpha-diversity, distinct overall oral microbial communities, and the presence and relative abundance of multiple taxa have been found for users of cigarettes and/or opium.
KEYWORDS: cigarette, opium, oral microbiota, Iran
INTRODUCTION
The human oral cavity consists of distinctive structures of hard tissue and mucosa, including the tongue, tooth surface, buccal mucosa, tonsils, and soft and hard palate, which have varied microbiota compositions (1, 2) that may change due to environmental exposures (3). There are approximately 700 bacterial species reported to inhabit the human oral cavity, and these bacterial species are important in maintaining oral health (4). Changes in the oral microbiota are associated with oral diseases, such as dental caries, periodontal disease, endodontic lesions, and odontogenic infections (5, 6). In addition, oral bacteria have been found to be associated with certain systemic diseases of the digestive (7, 8), cardiovascular (9), and endocrine systems (10, 11).
Cigarettes and illicit drugs, such as opium, contain chemicals and particulate matter that may exert an effect on the oral microbiota and perturb the microbial ecology of the mouth (12–14). Cigarette and opium use are associated with periodontal disease (15–17), and because periodontal disease is caused by specific bacteria, including Porphyromonas gingivalis, this also suggests a link between cigarette and opium use and the oral microbiota. Previous studies have reported alterations of the oral microbiota in cigarette smokers compared to nonsmokers (18, 19), but this was studied predominantly in populations from the United States. Although not as prevalent as cigarette smoking, illicit drug use causes disease burden worldwide (20), and opium use is a major public health problem in Iran (21–23). High prevalence of oral health issues in people with drug addiction has been confirmed by a growing number of studies (24–26); however, no study has investigated the association between opium use and the oral microbiota to date.
Changes in the oral microbiota due to cigarettes and opium may help to understand how these exposures adversely affect the human body. To better understand their impact on the oral microbiota, we conducted this study to investigate differences in the oral microbiota using 16S rRNA gene sequencing between ever users of cigarettes only, opium only, and both cigarettes and opium and never users of cigarettes and opium in Iran.
RESULTS
A total of 558 individuals were included in the present analysis, of which 66.85% (n = 373) were never users of cigarettes and opium, 21.51% (n = 120) were ever cigarette-only smokers, 2.87% (n = 16) were ever opium-only users, and 8.78% (n = 49) were ever users of both cigarettes and opium. Demographic characteristics of the study population are shown in Table 1. Most participants had no formal education, lived in an urban area, and were in the normal body mass index (BMI) range. Participants who used cigarettes and opium were mostly male and less likely to drink alcohol.
TABLE 1.
Demographic characteristics of the study population by cigarette and/or opium use status
| Never cigarette nor opium |
Ever cigarette only |
Ever opium only |
Ever both cigarette and opium |
|||||
|---|---|---|---|---|---|---|---|---|
|
n = 373 |
n = 120 |
n = 16 |
n = 49 |
|||||
| Frequency | Percent | Frequency | Percent | Frequency | Percent | Frequency | Percent | |
| Age group | ||||||||
| <50 | 36 | 9.65% | 5 | 4.17% | 1 | 6.25% | 4 | 8.16% |
| 50–59 | 88 | 23.59% | 24 | 20.00% | 6 | 37.50% | 21 | 42.86% |
| 60–69 | 123 | 32.98% | 43 | 35.83% | 1 | 6.25% | 14 | 28.57% |
| 70–79 | 82 | 21.98% | 38 | 31.67% | 7 | 43.75% | 9 | 18.37% |
| ≥80 | 44 | 11.80% | 10 | 8.33% | 1 | 6.25% | 1 | 2.04% |
| Sex | ||||||||
| Male | 132 | 35.39% | 103 | 85.83% | 14 | 87.50% | 47 | 95.92% |
| Female | 241 | 64.61% | 17 | 14.17% | 2 | 12.50% | 2 | 4.08% |
| Body mass index (kg/m2) | ||||||||
| Underweight (<18) | 26 | 6.97% | 9 | 7.50% | 2 | 12.50% | 13 | 26.53% |
| Normal (18–24.9) | 176 | 47.18% | 78 | 65.00% | 8 | 50.00% | 27 | 55.10% |
| Overweight (25–29.9) | 121 | 32.44% | 31 | 25.83% | 2 | 12.50% | 7 | 14.29% |
| Obese (≥30) | 50 | 13.40% | 2 | 1.67% | 4 | 25.00% | 2 | 4.08% |
| Education | ||||||||
| No formal education | 169 | 45.31% | 37 | 30.83% | 6 | 37.50% | 14 | 28.57% |
| ≤5 y | 69 | 18.50% | 28 | 23.33% | 5 | 31.25% | 13 | 26.53% |
| 6–8 y | 44 | 11.80% | 14 | 11.67% | 2 | 12.50% | 2 | 4.08% |
| 9–12 y | 47 | 12.60% | 20 | 16.67% | 3 | 18.75% | 14 | 28.57% |
| Higher education | 44 | 11.80% | 21 | 17.50% | 0 | 0.00% | 6 | 12.24% |
| Residence | ||||||||
| Rural | 124 | 33.24% | 46 | 38.33% | 3 | 18.75% | 16 | 32.65% |
| Urban | 249 | 66.76% | 74 | 61.67% | 13 | 81.25% | 33 | 67.35% |
| Any alcohol consumption | ||||||||
| Yes | 364 | 97.59% | 106 | 88.33% | 13 | 81.25% | 33 | 67.35% |
| No | 9 | 2.41% | 14 | 11.67% | 3 | 18.75% | 16 | 32.65% |
| Pancreatic cancer | ||||||||
| Yes | 167 | 44.77% | 60 | 50.00% | 11 | 68.75% | 35 | 71.43% |
| No | 206 | 55.23% | 60 | 50.00% | 5 | 31.25% | 14 | 28.57% |
Compared to never users of cigarettes or opium, all of the cigarette and opium use categories had lower alpha-diversity (Fig. S1 in the supplemental material). The average number of observed amplicon sequence variants (ASVs) (± standard deviation) for cigarette-only smokers (82.13 ± 38.55), opium-only users (76.19 ± 40.71), and users of both cigarettes and opium (77.80 ± 42.83) were significantly lower than never users of cigarettes or opium (95.10 ± 44.03). Similar trends were observed for the Shannon index and Faith’s phylogenetic diversity (PD).
As shown in Table 2, after adjustment for age, sex, BMI, alcohol consumption, and case status, for every increase in 10 observed ASVs, the odds decreased by 9% (odds ratio [OR] = 0.91; 95% confidence interval [95%] CI = 0.86 to 0.97), 13% (OR = 0.87; 95% CI = 0.75 to 1.01), and 12% (OR = 0.88; 95% CI = 0.80 to 0.96) for being a cigarette-only smoker, opium-only user, and user of both cigarettes and opium, respectively, compared to never users. Decreased odds were also found for increases in one unit of the Shannon index and Faith’s PD. When the ORs were calculated by quartiles of alpha-diversity, the odds of being a cigarette and/or opium user decreased in the higher quartiles. For example, compared to the first quantile (Q1) of observed ASVs, individuals in the fourth quantile (Q4) were significantly less likely to be cigarette-only smokers (OR = 0.37; 95% CI = 0.18 to 0.74) and/or users of both cigarettes and opium (OR = 0.28; 95% CI = 0.09 to 0.90).
TABLE 2.
Association between alpha- and beta-diversity and cigarette and/or opium use status
| Ever cigarette onlya |
Ever opium onlya |
Ever both cigarette and opiuma |
||||
|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | OR | 95% CI | |
| Alpha-diversity | ||||||
| Observed ASVs | ||||||
| Continuous (10 ASVs per unit) | 0.91 | 0.86–0.97 | 0.87 | 0.76–1.01 | 0.88 | 0.80–0.96 |
| <57 (Q1) | Reference | Reference | Reference | |||
| 57–84 (Q2) | 0.75 | 0.39–1.44 | 0.48 | 0.12–1.90 | 1.22 | 0.44–3.37 |
| 85–117 (Q3) | 0.59 | 0.30–1.13 | 0.13 | 0.02–1.19 | 0.41 | 0.13–1.33 |
| ≥118 (Q4) | 0.37 | 0.18–0.74 | 0.30 | 0.07–1.26 | 0.28 | 0.09–0.90 |
| Shannon index | ||||||
| Continuous | 0.74 | 0.58–0.95 | 0.50 | 0.28–0.89 | 0.51 | 0.33–0.79 |
| <3.43 (Q1) | Reference | Reference | Reference | |||
| 3.43–4.09 (Q2) | 0.70 | 0.37–1.34 | 0.48 | 0.12–1.92 | 0.33 | 0.12–0.95 |
| 4.10–4.58 (Q3) | 0.83 | 0.43–1.59 | 0.82 | 0.23–3.02 | 0.39 | 0.14–1.09 |
| ≥4.59 (Q4) | 0.39 | 0.20–0.78 | NAb | NAb | 0.14 | 0.04–0.46 |
| Faith’s PD | ||||||
| Continuous | 0.87 | 0.78–0.96 | 0.80 | 0.62–1.02 | 0.85 | 0.73–1.00 |
| <5.13 (Q1) | Reference | Reference | Reference | |||
| 5.13–6.80 (Q2) | 0.90 | 0.47–1.73 | 1.15 | 0.30–4.36 | 1.18 | 0.42–3.31 |
| 6.81–8.64 (Q3) | 0.52 | 0.27–1.01 | 0.35 | 0.06–1.99 | 0.34 | 0.10–1.10 |
| ≥8.65 (Q4) | 0.47 | 0.24–0.93 | 0.36 | 0.07–1.85 | 0.46 | 0.15–1.36 |
| Beta-diversity | ||||||
| Bray-Curtis | ||||||
| PCoA1 (13.8%) | 0.72 | 0.56–0.92 | 0.59 | 0.33–1.01 | 0.31 | 0.19–0.48 |
| PCoA2 (10.5%) | 0.77 | 0.60–0.98 | 0.43 | 0.22–0.79 | 0.45 | 0.28–0.69 |
| PCoA3 (5.7%) | 1.18 | 0.94–1.50 | 0.93 | 0.58–1.62 | 1.35 | 0.91–2.08 |
| PCoA4 (4.7%) | 0.99 | 0.78–1.24 | 1.55 | 0.90–2.78 | 0.90 | 0.62–1.30 |
| PCoA5 (3.9%) | 1.08 | 0.85–1.37 | 1.44 | 0.78–2.83 | 1.29 | 0.89–1.92 |
| PCoA6 (3.4%) | 1.01 | 0.80–1.28 | 0.73 | 0.45–1.23 | 0.77 | 0.52–1.14 |
| Unweighted UniFrac | ||||||
| PCoA1 (24.4%) | 0.72 | 0.56–0.92 | 0.62 | 0.35–1.09 | 0.66 | 0.44–0.96 |
| PCoA2 (9.4%) | 1.05 | 0.83–1.32 | 2.05 | 1.17–3.90 | 2.06 | 1.40–3.12 |
| PCoA3 (4.6%) | 0.78 | 0.61–0.99 | 0.60 | 0.32–1.07 | 0.49 | 0.32–0.74 |
| PCoA4 (3.3%) | 1.06 | 0.84–1.33 | 1.09 | 0.62–1.88 | 1.00 | 0.68–1.45 |
| PCoA5 (3.0%) | 0.73 | 0.57–0.92 | 0.68 | 0.37–1.20 | 0.44 | 0.28–0.68 |
| PCoA6 (2.1%) | 0.94 | 0.74–1.19 | 0.72 | 0.40–1.25 | 0.91 | 0.63–1.30 |
| Weighted UniFrac | ||||||
| PCoA1 (31.2%) | 0.79 | 0.62–1.00 | 0.62 | 0.34–1.09 | 0.54 | 0.35–0.83 |
| PCoA2 (14.4%) | 0.86 | 0.67–1.07 | 0.48 | 0.23–0.90 | 0.43 | 0.25–0.69 |
| PCoA3 (6.8%) | 1.10 | 0.87–1.39 | 0.64 | 0.36–1.13 | 1.24 | 0.85–1.84 |
| PCoA4 (5.2%) | 0.92 | 0.73–1.17 | 0.55 | 0.35–0.86 | 0.79 | 0.55–1.14 |
| PCoA5 (3.9%) | 1.15 | 0.90–1.44 | 1.33 | 0.80–2.13 | 1.08 | 0.74–1.56 |
| PCoA6 (3.4%) | 1.36 | 1.07–1.74 | 0.92 | 0.55–1.60 | 1.58 | 1.04–2.51 |
Adjusted for age, sex, BMI, alcohol consumption, and case status.
NA, the 95% CI cannot be calculated because no individuals used opium in this quartile.
We next examined alpha-diversity differences between never (n = 389), former (n = 92), and current (n = 77) users of cigarettes and never (n = 493), former (n = 11), and current (n = 53) users of opium. In this analysis, the never cigarette smoker category could contain opium users and vice versa. Description of cigarette and opium use status is shown in Table S1. Alpha-diversity appeared to decrease according to smoking status, where current smokers/users had the lowest alpha-diversity, never smokers/users had the highest alpha-diversity, and former smokers/users were in the middle, although the difference between current smokers/users was only statistically significant (P < 0.05) compared to never smokers/users (Fig. S2). There was little evidence of a correlation between years since quitting for former cigarette smokers or opium users with alpha-diversity nor was there evidence of a correlation between pack-years of cigarettes and alpha-diversity (Table S2).
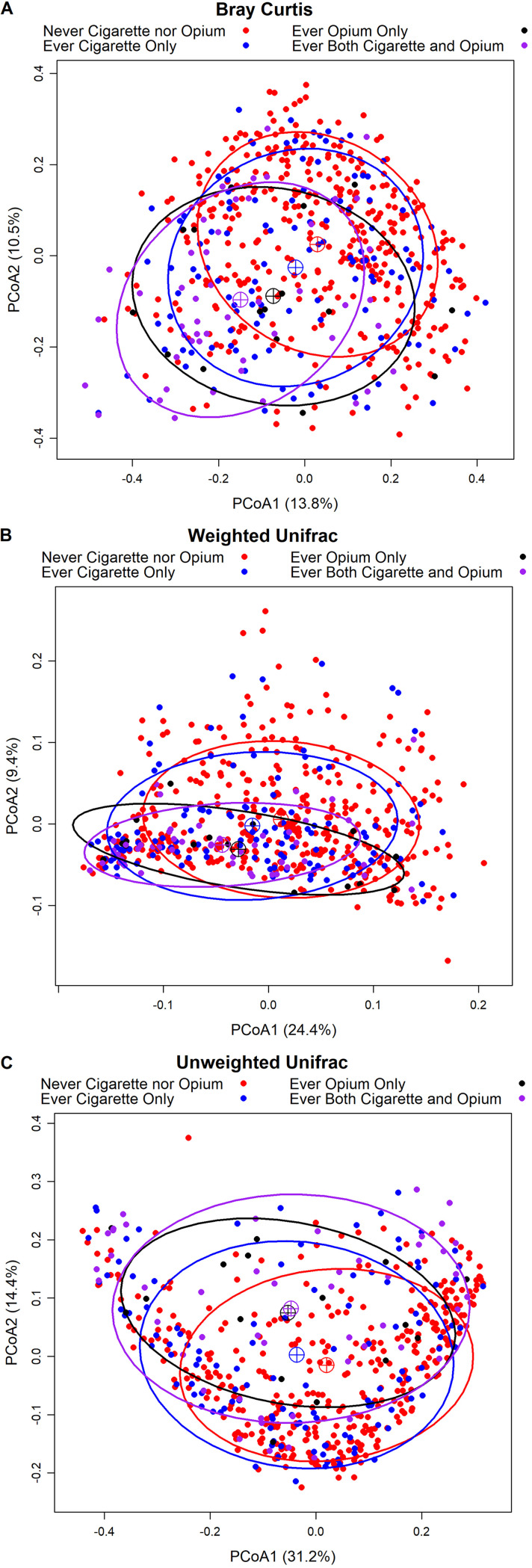
Compared to never users, overall beta-diversity for all three matrices significantly differed according to cigarette and opium use status as indicated by microbiome regression-based kernel association test (MiRKAT) models adjusted for age, sex, BMI, alcohol consumption, and case status (Table 3). However, no significant differences were found when comparing opium-only users to cigarette-only users (data not shown), which may be due to the limited sample size of cigarette- or opium-only users. When examining the first two principal coordinates analysis (PCoA) vectors, there was some indication for a shift by cigarette and opium use status (Fig. 1). The distance between the PCoA clustering centroids of never users and the centroids of users of both cigarettes and opium visually appeared to be larger than that observed between never users and cigarette-only smokers, or between never users and opium-only users. When considering the first six PCoA vectors from the three beta-diversity matrices, strong associations were observed for specific PCoA vectors in logistic regression models, especially for the first two vectors from all three beta-diversity matrices of users of both cigarettes and opium (Table 2). For example, for one standard deviation increase in the first PCoA vector of weighted UniFrac (31.2% of variability explained in the matrix), the odds of being a cigarette and opium user was 0.54 (95% CI = 0.35 to 0.83). In our analysis of associations of taxa presence or abundance with the first six PCoA vectors, multiple taxa were strongly and statistically significantly associated with the first six PCoA vectors (Table S3 and S4). The distance-based coefficient of determination, R2, for the three beta-diversity matrices for ever use cigarette only, ever use opium only, ever use both cigarettes and opium, and other demographic factors are shown in Fig. S3. Results indicated that ever use both cigarettes and opium explained a higher percentage of microbial variability than most demographic factors and was about 2-fold higher than cigarettes only or opium only, suggesting a potentially additive effect of cigarettes and opium.
TABLE 3.
MiRKAT test for association of beta-diversity matrices of cigarette and/or opium users compared to those of never users
| Analysis | Ever cigarette onlya | Ever opium onlya | Ever both cigarette and opiuma |
|---|---|---|---|
| Bray-Curtis | 0.0013 | 0.0118 | <0.0001 |
| Weighted UniFrac | 0.0417 | 0.0242 | 0.0002 |
| Unweighted UniFrac | 0.0076 | 0.0102 | 0.0003 |
Adjusted for age, sex, BMI, alcohol consumption, and case status.
FIG 1.
Principal coordinate analysis (PCoA) of Bray Curtis (A), weighted UniFrac (B), and unweighted UniFrac (C) distance according to cigarette and opium use status. Sixty-eight percent confidence ellipses were drawn, and the centroids represent the coordinate mean of PCoA1 and PCoA2 by group.
Table 4 presents the significant associations of bacterial taxa with cigarette and opium use from the zero-inflated beta regression analysis (results of all taxa are included in Table S5). For taxa presence, compared to never users of cigarettes or opium, the genus Lachnospiraceae G7 was detected in 14.17% of cigarette-only smokers, which was statistically significantly lower than the 20.11% detected in never users of cigarettes or opium, while an unclassified genus in the family Enterobacteriaceae was detected in 17.50% of cigarette-only smokers, significantly higher than the 13.40% detected in never users of cigarettes or opium. The genera Abiotrophia and Lautropia were detected in 50.00% and 18.75% of opium-only users, which was significantly lower than that detected in never users of cigarettes or opium (80.43% and 64.61%, respectively). The lower prevalence of Abiotrophia and Lautropia was also observed at order and family levels for users of opium only. The higher prevalence of the unclassified genus in the family Enterobacteriaceae was also observed at order and family levels for cigarette-only smokers. The relative abundance of the phylum Firmicutes was significantly higher in opium-only users (65.49%) than in never users of cigarettes or opium (54.29%). The phylum Actinobacteria had a significantly higher relative abundance in users of both cigarettes and opium (21.20%) than in never users of cigarettes or opium (14.78%), while the phyla Bacteroidetes (never 17.63%, both 11.62%) and Proteobacteria (never 9.06%, both 3.70%) had lower relative abundances in users of both cigarettes and opium. Similar significant associations were also found at the class, family, and genus levels for the taxa belonging to the phylum Firmicutes and at the class, order, and family levels of the taxa belonging to the phylum Actinobacteria. No significant associations were found for the relative abundance of any taxa and cigarette-only smokers.
TABLE 4.
Results from multivariable zero-inflated beta regression models detailing taxa presence and relative abundance significantly associated with different cigarette and/or opium use status
| Namec | Level | Proportion of presencea |
Relative abundancea |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Never cigarette nor opium | Cigarette only | P value | Adjusted P valueb | Opium only | P value | Adjusted P valueb | Both cigarette and opium | P value | Adjusted P valueb | Never cigarette nor opium | Cigarette only | P value | Adjusted P valueb | Opium only | P value | Adjusted P valueb | Both cigarette and opium | P value | Adjusted P valueb | ||
| Actinobacteria | Phylum | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.1478 | 0.1691 | 0.1615 | 1.0000 | 0.1536 | 0.3140 | 1.0000 | 0.2120 | 0.0000 | 0.0003 |
| Bacteroidetes | Phylum | 1.0000 | 1.0000 | 0.9498 | 1.0000 | 1.0000 | 0.9894 | 1.0000 | 0.9796 | 0.8917 | 1.0000 | 0.1763 | 0.1399 | 0.0148 | 0.1623 | 0.1492 | 0.0223 | 0.2448 | 0.1162 | 0.0027 | 0.0292 |
| Firmicutes | Phylum | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.5429 | 0.5820 | 0.0072 | 0.0793 | 0.6549 | 0.0003 | 0.0029 | 0.6098 | 0.0363 | 0.3990 |
| Proteobacteria | Phylum | 0.9920 | 0.9833 | 0.1908 | 1.0000 | 1.0000 | 0.7154 | 1.0000 | 1.0000 | 0.9990 | 1.0000 | 0.0906 | 0.0753 | 0.2922 | 1.0000 | 0.0231 | 0.0292 | 0.3214 | 0.0370 | 0.0001 | 0.0015 |
| Actinobacteria;Actinobacteria | Class | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.1365 | 0.1590 | 0.1095 | 1.0000 | 0.1404 | 0.3987 | 1.0000 | 0.2022 | 0.0000 | 0.0010 |
| Bacteroidetes;Flavobacteriia | Class | 0.9598 | 0.8750 | 0.4729 | 1.0000 | 0.7500 | 0.0047 | 0.1358 | 0.7551 | 0.0017 | 0.0487 | 0.0106 | 0.0057 | 0.3229 | 1.0000 | 0.0038 | 0.4711 | 1.0000 | 0.0029 | 0.0343 | 0.9949 |
| Firmicutes;Bacilli | Class | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.4316 | 0.4777 | 0.0093 | 0.2686 | 0.5696 | 0.0002 | 0.0062 | 0.5189 | 0.0205 | 0.5943 |
| Proteobacteria;Gammaproteobacteria | Class | 0.9330 | 0.9583 | 0.0544 | 1.0000 | 0.9375 | 0.4441 | 1.0000 | 0.8980 | 0.8651 | 1.0000 | 0.0476 | 0.0413 | 0.2430 | 1.0000 | 0.0108 | 0.0419 | 1.0000 | 0.0188 | 0.0006 | 0.0181 |
| Actinobacteria;Actinomycetales | Order | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.1264 | 0.1464 | 0.1303 | 1.0000 | 0.1318 | 0.5153 | 1.0000 | 0.1939 | 0.0001 | 0.0026 |
| Proteobacteria;Burkholderiales | Order | 0.6917 | 0.4750 | 0.6189 | 1.0000 | 0.2500 | 0.0000 | 0.0011 | 0.3673 | 0.0005 | 0.0264 | 0.0023 | 0.0012 | 0.7406 | 1.0000 | 0.0021 | 0.8640 | 1.0000 | 0.0029 | 0.4738 | 1.0000 |
| Proteobacteria;Enterobacteriales | Order | 0.1340 | 0.1750 | 0.0001 | 0.0034 | 0.1875 | 0.8211 | 1.0000 | 0.2449 | 0.9595 | 1.0000 | 0.0026 | 0.0011 | 0.5056 | 1.0000 | 0.0001 | 0.7952 | 1.0000 | 0.0025 | 0.9701 | 1.0000 |
| Actinobacteria;Micrococcaceae | Family | 0.9973 | 1.0000 | 0.9154 | 1.0000 | 1.0000 | 0.9549 | 1.0000 | 1.0000 | 0.9921 | 1.0000 | 0.1075 | 0.1302 | 0.1939 | 1.0000 | 0.1191 | 0.2704 | 1.0000 | 0.1725 | 0.0005 | 0.0407 |
| Firmicutes;Lactobacillales;NA | Family | 0.1180 | 0.1333 | 0.0260 | 1.0000 | 0.0625 | 0.7601 | 1.0000 | 0.1837 | 0.4162 | 1.0000 | 0.0001 | 0.0001 | 0.5593 | 1.0000 | 0.0420 | 0.0000 | 0.0000 | 0.0003 | 0.1777 | 1.0000 |
| Firmicutes;Aerococcaceae | Family | 0.8043 | 0.5917 | 0.2150 | 1.0000 | 0.5000 | 0.0001 | 0.0113 | 0.3469 | 0.0109 | 0.9783 | 0.0052 | 0.0034 | 0.2091 | 1.0000 | 0.0038 | 0.1694 | 1.0000 | 0.0011 | 0.3161 | 1.0000 |
| Proteobacteria;Burkholderiaceae | Family | 0.6461 | 0.3917 | 0.4916 | 1.0000 | 0.1875 | 0.0000 | 0.0002 | 0.2857 | 0.0007 | 0.0593 | 0.0021 | 0.0011 | 0.8800 | 1.0000 | 0.0021 | 0.5197 | 1.0000 | 0.0007 | 0.3734 | 1.0000 |
| Proteobacteria;Enterobacteriaceae | Family | 0.1340 | 0.1750 | 0.0001 | 0.0062 | 0.1875 | 0.8211 | 1.0000 | 0.2449 | 0.9595 | 1.0000 | 0.0026 | 0.0011 | 0.5056 | 1.0000 | 0.0001 | 0.7952 | 1.0000 | 0.0025 | 0.9701 | 1.0000 |
| Firmicutes;Lactobacillales;NA;NA | Genus | 0.1180 | 0.1333 | 0.0260 | 1.0000 | 0.0625 | 0.7601 | 1.0000 | 0.1837 | 0.4162 | 1.0000 | 0.0001 | 0.0001 | 0.5593 | 1.0000 | 0.0420 | 0.0000 | 0.0000 | 0.0003 | 0.1777 | 1.0000 |
| Firmicutes;Abiotrophia | Genus | 0.8043 | 0.5917 | 0.2150 | 1.0000 | 0.5000 | 0.0001 | 0.0219 | 0.3469 | 0.0109 | 1.0000 | 0.0052 | 0.0034 | 0.2091 | 1.0000 | 0.0038 | 0.1694 | 1.0000 | 0.0011 | 0.3161 | 1.0000 |
| Firmicutes;Lachnospiraceae G7 | Genus | 0.2011 | 0.1417 | 0.0001 | 0.0200 | 0.1250 | 0.0694 | 1.0000 | 0.2041 | 0.2609 | 1.0000 | 0.0000 | 0.0000 | 0.8958 | 1.0000 | 0.0000 | 0.8042 | 1.0000 | 0.0000 | 0.9640 | 1.0000 |
| Proteobacteria;Lautropia | Genus | 0.6461 | 0.3917 | 0.4916 | 1.0000 | 0.1875 | 0.0000 | 0.0003 | 0.2857 | 0.0007 | 0.1146 | 0.0021 | 0.0011 | 0.8800 | 1.0000 | 0.0021 | 0.5197 | 1.0000 | 0.0007 | 0.3734 | 1.0000 |
| Proteobacteria;Enterobacteriaceae;NA | Genus | 0.1340 | 0.1750 | 0.0001 | 0.0120 | 0.1875 | 0.8211 | 1.0000 | 0.2449 | 0.9595 | 1.0000 | 0.0026 | 0.0011 | 0.5041 | 1.0000 | 0.0001 | 0.7965 | 1.0000 | 0.0022 | 0.9504 | 1.0000 |
Adjusted for age, sex, BMI, alcohol consumption, and case status.
Adjusted P value was the P value that was adjusted using a Bonferroni correction.
NA, unclassified genus.
In the sensitivity analysis excluding pancreatic cancer cases, the results of the logistic regression models for alpha- and beta-diversity estimates were generally similar, although the confidence intervals were wider due to reduced sample size, and associations were often not statistically significant (Table S6). The P values of MiRKAT models were less statistically significant (Table S7). The prevalence and relative abundance of specific taxa were similar, but most significant results were no longer statistically significant (Table S8).
DISCUSSION
Cigarettes and opium, which contain chemicals and particulate matter that may alter the oral microbiota, are used globally, and the control of the use of these substances is urgent. Determination of changes in the oral microbiota due to cigarette and opium use can be a strategy to provide additional evidence regarding the health impacts of these exposures. In this study, we found that the oral microbiota of cigarette and opium users differed significantly from never users in Iran. Cigarette and opium users had lower alpha-diversity than never users. Additionally, cigarette and opium use were associated with differences in the overall microbial community as assessed using beta-diversity. There was also evidence of associations between the use of cigarettes or opium and both the presence and the relative abundance of specific taxa. These associations were more apparent within ever users of both cigarettes and opium, which may suggest a possible additive effect of these two exposures. To our knowledge, this is the first study investigating the association between opium use and the oral microbiota.
Previous research considering the oral microbiota of cigarette smokers has shown mixed results, potentially due to the small sample sizes in many studies. In some studies of the oral microbiota, assessed using samples from subgingival plaque, saliva, oropharynx swabs, or mouthwash, cigarette smokers tended to have higher alpha-diversity than nonsmokers (19, 27–31), while other studies did not detect a statistically significant difference between cigarette smokers and nonsmokers (32, 33). These results conflict with our study, which found lower diversity in the oral microbiota of cigarette and opium users. One possible explanation for the conflicting results may be due to heterogeneity of the human microbiota related to geography. The geographical areas in which individuals live has been shown to be associated with microbial diversity and to modify observed microbiota associations (34), but none of the previous studies of the association between cigarette smoking and the oral microbiota were conducted in Iran. Another possible explanation for our unique findings may be that the type of tobacco widely used in Iran differed from other populations (35) and that this tobacco has a different effect on the microbiota than tobacco used in the other countries. Recent studies showed that different types of tobacco and cigarette exposure can cause substantial changes in the structure and function of the general oral microbiota (31, 32, 36).
We found significant differences in overall microbial community composition by cigarette and opium use status, which has also been seen in previous studies comparing cigarette smokers and nonsmokers (18, 19, 27, 31, 32, 37). However, it is difficult to determine whether these community-level changes are similar across studies because commonly used beta-diversity measures are study specific. Exposure to cigarette smoking likely results in functional changes in the oral environment (38), which may impact the immune system and the competition between commensal and pathogenic bacteria. Studies have suggested that smoking may create a microenvironment that selects for a pathogen-rich, commensal-poor microbiota community (39). Opium and its components have been reported to induce gut microbial disruption (40) and have an effect on the nasopharyngeal microbial flora (41), while, for the first time, our study reported that opium use was associated with oral microbial communities, with or without cigarette smoking. In addition, the distance between the PCoA clustering centroids of never users and users of both cigarettes and opium appeared to be larger than that between never users and cigarette-only smokers or opium-only users. Use of both cigarettes and opium explained a 2-fold higher percentage of microbial variability than cigarettes only or opium only, suggesting that there may be an additive effect of cigarettes and opium, although this study is underpowered to statistically assess this potential interaction.
In this study, the relative abundance of the phylum Actinobacteria was significantly higher in users of both cigarettes and opium than in never users, while the phyla Bacteroidetes and Proteobacteria were lower in users of both cigarettes and opium, and Firmicutes was higher in opium-only users. Three genera Lachnospiraceae G7, Abiotrophia, and Lautropia were less prevalent in cigarette-only smokers or opium-only users, and an unclassified genus in the family Enterobacteriaceae was more prevalent in cigarette-only smokers. Previous studies have found that smoking was associated with depletion of aerobic taxa and enrichment with anaerobic microbiota (19, 32); however, the genus Abiotrophia is a facultative aerobe while the genus Lautropia is a facultative anaerobe, both of which were less prevalent in opium-only users in our study. The other taxa that were significantly associated with using both cigarettes and opium were a mixture of anaerobic and aerobic bacteria.
Although no study has previously considered the association of opium with the oral microbiota, studies have investigated associations between specific taxa in cigarette smokers and nonsmokers. In the United Arab Emirates Healthy Future Study of 33 cigarette smokers and 225 nonsmokers, the phyla Proteobacteria and Fusobacteria were depleted in cigarette smokers, while Firmicutes, Bacteroidetes, and Actinobacteria were enriched at all lower taxonomical levels in cigarette smokers (31). The depletion of Proteobacteria and enrichment of Firmicutes and Actinobacteria was consistent with our results, while the enrichment of Bacteroidetes was in the opposite direction compared to our results. Two studies in the United States had some consistent taxonomic results with our study at the phylum level. 16S rRNA gene sequencing of oral wash samples from 1,204 adults drawn from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial cohort (42) and the American Cancer Society Cancer Prevention Study II Nutrition cohort (43) showed a lower relative abundance of the phylum Proteobacteria (4.6%) among current smokers than among never smokers (11.7%) in addition to the genus Abiotrophia (18). In the New York City Health and Nutrition Examination study, the relative abundance of taxa in the phylum Proteobacteria was found to be lower in current cigarette smokers than in never smokers (32). In addition, oral bacterial taxa such as the genera Capnocytophaga, Corynebacterium, Neisseria, Haemophilus, Aggregatibacter, Porphyromonas, Prevotella, Leptotrichia, Peptostreptococcus, Selenomonas, Fusobacterium, Fretibacterium, Streptococcus, Veillonella, TM7, Filifactor, Parvimonas, Treponema, Prevotella, Campylobacter, and Bacteroides were also found to be associated with cigarette smoking in previous studies, but these associations were not replicated in our study (27, 28, 37, 44). We also found that the genus Lachnospiraceae G7 was less prevalent in cigarette-only smokers, but this was not seen in other studies.
Our study has several limitations. First, all participants of this analysis were from a pancreatic cancer case-control study. We adjusted for pancreatic cancer case status in all analyses and in supplementary analyses restricting to controls, and the results were similar. However, all individuals in the study, including controls, were individuals referred for endoscopic ultrasonography, and the effect of cigarette or opium use on the oral microbiota might be different from people in more optimal health. The sample size of ever opium-only users was relatively small in this study. To fully understand the association of opium with the oral microbiome will require a larger population of opium users. Additionally, there were only a limited number of female cigarette and opium users in our study. When stratified by sex, we found similar results in female users (data not shown), but it is possible that cigarette and opium exposure modifies the oral microbiota in females differently. Finally, the type of cigarettes or opium used may have unique effects on the oral microbiota; however, due to limited sample size, we were unable to assess differences by type of cigarettes or opium.
In conclusion, we found that cigarette and opium use was related to lower alpha-diversity, overall oral microbiota community composition, and the presence and relative abundance of specific taxa, including the phyla Actinobacteria, Proteobacteria, Bacteroidetes, and Firmicutes. Future studies should continue to investigate associations of cigarettes and opium with the oral microbiota in diverse populations and evaluate how the oral microbiota may mediate the associations between cigarette smoking and opium use with specific health outcomes.
MATERIALS AND METHODS
Study population.
Participants were drawn from a case-control study examining factors associated with pancreatic cancer in Iran, which has been described previously in detail (45). Both cases and controls were identified from patients at one of three tertiary hospitals or a specialty clinic in Tehran, and they were referred for endoscopic ultrasonography between January 2011 and January 2015 for suspicion of a mass or cyst in the pancreas or bile ducts, for assessment of submucosal lesions found during esophago-gastro-duodenal endoscopy, or to rule out bile duct stones. The inclusion and exclusion criteria of cases and controls have been described previously in detail (46). All cases and controls from the previous study were included in this study.
After providing informed consent, participants responded to a questionnaire and provided saliva samples, which were immediately stored at −70°C. The questionnaire has been described in detail previously (45). Briefly, questions related to cigarette and opium use included user status (never, former, current), age at initiation, duration of use, frequency of use and amount of use, type of cigarettes used (factory-made cigarettes with filters, factory-made cigarettes without filters, and hand-made cigarettes), and type of opium used (opium, heroin, burned opium, opium juice, crystal, crack, or cocaine).
DNA extraction, amplification, sequencing, and bioinformatic data processing.
Saliva samples were shipped on dry ice to the National Cancer Institute for processing. DNA extraction, PCR amplification, sequencing, and bioinformatic data processing were completed as described in detail previously (46). Briefly, the saliva samples were thawed at 4°C and extracted using the DSP DNA virus pathogen kit on a QIAsymphony instrument (Qiagen). The V4 region of the 16S rRNA gene was PCR amplified for 25 cycles, and 2 × 250 bp paired-end sequencing was performed on an Illumina MiSeq. Sequence data processing was performed with QIIME 2 2017.2 (47). Taxonomy was assigned to the resulting ASVs using q2-feature-classifier (48) and the Human Oral Microbiome Database, version 14.51 (49). Sequences were demultiplexed, and quality control and paired‐end read joining were performed with DADA2. The first 10 bases were trimmed from forward and reverse reads; forward reads were truncated at 225 bases, and reverse reads were truncated at 200 bases. The average read per study sample was 109,582. Taxonomy was assigned to the resulting ASVs using q2‐feature‐classifier and the Human Oral Microbiome Database version 14.51. ASVs not assigned at least to the phylum level were excluded. Taxonomic relative abundances from the phylum to genus level were generated. Alpha-diversity metrics, including observed ASVs, Shannon index, and Faith’s PD, and beta-diversity metrics, including Bray-Curtis dissimilarity and weighted and unweighted UniFrac, were computed with rarefaction at 40,000 sequences per sample. PCoA vectors were calculated from the three beta-diversity distance matrices. The quality control analysis of the sequencing data has been described previously in detail (46). In brief, a low false‐positive error rate and a low false‐negative detection rate were found when comparing the taxonomic composition of the oral artificial community samples to the known composition of the mock. In addition, both the oral artificial and chemostat communities displayed high levels of consistency across sequencing runs.
Statistical analysis.
Statistical analyses were conducted using R version 3.6.2. Participants who reported ever use of factory-made cigarettes with a filter, factory-made cigarettes without a filter, or hand-made cigarettes were categorized as ever smokers, while participants who reported ever use of opium, heroin, burned opium, opium juice, crystal, crack, or cocaine were considered ever opium users. According to the cigarette and opium use history, individuals were categorized as never users of cigarettes or opium, ever cigarette-only smokers, ever opium-only users, and ever users of both cigarettes and opium. Participants were also categorized as never, former, or current cigarette smokers and never, former, or current opium users where the never cigarette smoker category could contain opium users and vice versa. A former smoker/user was defined as a participant who had quit using the product for more than 1 year, and a current smoker/user was defined as using the product within the past year. Years since quitting for former users was calculated by subtracting the age the participant stopped using regularly from their current age. Pack-years for cigarette smokers was calculated as the product of smoking duration and number of cigarettes smoked per day divided by 20 (a standard pack of cigarettes). Descriptive characteristics and alpha-diversity estimates of the study population were described by cigarette and opium use status.
Logistic regression models were used to calculate prevalence odds ratios (OR) and 95% confidence intervals for the association between alpha-diversity (independent variable) and cigarette and opium use (dependent variable) with adjustment for age, sex, BMI, alcohol consumption, and case status (case or control). Alpha-diversity was modeled as a continuous variable and also using quartiles estimated from the distribution within the controls. Kruskal-Wallis and Dunn tests for multiple comparisons with adjusted P values by Bonferroni correction were used to compare alpha-diversity for current, former, or never smokers/users. Spearman correlation coefficients were used to estimate the correlations between alpha-diversity and years since quitting for former cigarette smokers and former opium users and pack-years for cigarette smokers.
For beta-diversity, the association between the overall beta-diversity matrices and cigarette and opium use was tested using MiRKAT (MiRKAT function, MiRKAT package) (50) with adjustment for age, sex, BMI, alcohol consumption, and case status. PCoA plots were generated using the first two PCoA vectors, which accounted for 9.4 to 31.2% of the overall variance, and were labeled according to cigarette and opium use status. Sixty-eight percent confidence ellipses, representing one standard deviation, were generated (xyplot function, latticeExtra package), and the centroids of the ellipses represent the coordinate mean of the first and second vectors. The first six PCoA vectors, which accounted for a total of 42 to 65% of the overall variance in the matrices, were modeled in independent logistic regression models. Each principal coordinate vector was standardized by subtracting the overall mean for the principal coordinate vector and then dividing by its standard deviation before modeling. Permutational multivariate analysis of variance (PERMANOVA) test for the beta-diversity matrices was used to calculate the distance-based coefficient of determination R2 (adonis function, vegan package) to quantify the percentage of microbiota variability explained by ever use of cigarettes only, opium only, or both cigarettes and opium, and the 95% CI was calculated using 10,000 bootstrap samples.
Restricting to taxa with an overall prevalence of at least 10%, a series of zero-inflated beta regression models was used to examine associations between the presence and relative abundance of specific taxa and cigarette and opium use with adjustment for age, sex, BMI, alcohol consumption, and case status (gamlss function, gamlss package). P values were adjusted using Bonferroni correction for the taxonomic analyses. Finally, a sensitivity analysis of the above associations excluding pancreatic cancer cases was conducted.
Data availability.
The sequencing data are available on the Sequence Read Archive (NCBI SRA) under BioProject ID PRJNA549488.
ACKNOWLEDGMENTS
This study was funded by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, USA, and the Digestive Disease Research Institute, Tehran University of Medical Sciences, Iran. Bioinformatics work performed by J.G.C. and N.B. was supported, in part, by the National Cancer Institute under the awards for the Partnership of Native American Cancer Prevention U54CA143924 (UACC) and U54CA143925 (NAU) by NSF Award 1565100 to J.G.C. The funders had no role in study design, data collection, and interpretation or the decision to submit the work for publication.
This work utilized the computational resources of the NIH high performance computing Biowulf cluster (http://hpc.nih.gov).
Footnotes
Supplemental material is available online only.
Contributor Information
Zeni Wu, Email: zeni.wu@nih.gov.
Akram Pourshams, Email: akrampourshams@gmail.com.
Jan Claesen, Lerner Research Institute.
REFERENCES
- 1.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verma D, Garg PK, Dubey AK. 2018. Insights into the human oral microbiome. Arch Microbiol 200:525–540. doi: 10.1007/s00203-018-1505-3. [DOI] [PubMed] [Google Scholar]
- 4.Krishnan K, Chen T, Paster BJ. 2017. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis 23:276–286. doi: 10.1111/odi.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson I, Witkowska E, Kaveh B, Lif Holgerson P, Tanner AC. 2016. The microbiome in populations with a low and high prevalence of caries. J Dent Res 95:80–86. doi: 10.1177/0022034515609554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Payne MA, Hashim A, Alsam A, Joseph S, Aduse-Opoku J, Wade WG, Curtis MA. 2019. Horizontal and vertical transfer of oral microbial dysbiosis and periodontal disease. J Dent Res 98:1503–1510. doi: 10.1177/0022034519877150. [DOI] [PubMed] [Google Scholar]
- 7.Kageyama S, Takeshita T, Takeuchi K, Asakawa M, Matsumi R, Furuta M, Shibata Y, Nagai K, Ikebe M, Morita M, Masuda M, Toh Y, Kiyohara Y, Ninomiya T, Yamashita Y. 2019. Characteristics of the salivary microbiota in patients with various digestive tract cancers. Front Microbiol 10:1780. doi: 10.3389/fmicb.2019.01780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn J, Chen CY, Hayes RB. 2012. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control 23:399–404. doi: 10.1007/s10552-011-9892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kholy KE, Genco RJ, Van Dyke TE. 2015. Oral infections and cardiovascular disease. Trends Endocrinol Metab 26:315–321. doi: 10.1016/j.tem.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Lindheim L, Bashir M, Munzker J, Trummer C, Zachhuber V, Pieber TR, Gorkiewicz G, Obermayer-Pietsch B. 2016. The salivary microbiome in polycystic ovary syndrome (PCOS) and its association with disease-related parameters: a pilot study. Front Microbiol 7:1270. doi: 10.3389/fmicb.2016.01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demmer RT, Jacobs DR Jr, Singh R, Zuk A, Rosenbaum M, Papapanou PN, Desvarieux M. 2015. Periodontal bacteria and prediabetes prevalence in ORIGINS: the oral infections, glucose intolerance, and insulin resistance study. J Dent Res 94:201S–211S. doi: 10.1177/0022034515590369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bougnoux ME, Dupont C, Turner L, Rouveix E, Dorra M, Nicolas-Chanoine MH. 1997. Mixed Candida glabrata and Candida albicans disseminated candidiasis in a heroin addict. Eur J Clin Microbiol Infect Dis 16:598–600. doi: 10.1007/BF02447924. [DOI] [PubMed] [Google Scholar]
- 13.Hutcherson JA, Scott DA, Bagaitkar J. 2015. Scratching the surface—tobacco-induced bacterial biofilms. Tob Induc Dis 13:1. doi: 10.1186/s12971-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Li M, Huang R. 2019. The effect of smoking on caries-related microorganisms. Tob Induc Dis 17:32. doi: 10.18332/tid/105913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leite FRM, Nascimento GG, Scheutz F, Lopez R. 2018. Effect of smoking on periodontitis: a systematic review and meta-regression. Am J Prev Med 54:831–841. doi: 10.1016/j.amepre.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Vogtmann E, Graubard B, Loftfield E, Chaturvedi A, Dye BA, Abnet CC, Freedman ND. 2017. Contemporary impact of tobacco use on periodontal disease in the USA. Tob Control 26:237–238. doi: 10.1136/tobaccocontrol-2015-052750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shekarchizadeh H, Khami MR, Mohebbi SZ, Ekhtiari H, Virtanen JI. 2019. Oral health status and its determinants among opiate dependents: a cross-sectional study. BMC Oral Health 19:5. doi: 10.1186/s12903-018-0691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, Ma Y, Purdue MP, Jacobs EJ, Gapstur SM, Li H, Alekseyenko AV, Hayes RB, Ahn J. 2016. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J 10:2435–2446. doi: 10.1038/ismej.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason MR, Preshaw PM, Nagaraja HN, Dabdoub SM, Rahman A, Kumar PS. 2015. The subgingival microbiome of clinically healthy current and never smokers. ISME J 9:268–272. doi: 10.1038/ismej.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peacock A, Leung J, Larney S, Colledge S, Hickman M, Rehm J, Giovino GA, West R, Hall W, Griffiths P, Ali R, Gowing L, Marsden J, Ferrari AJ, Grebely J, Farrell M, Degenhardt L. 2018. Global statistics on alcohol, tobacco and illicit drug use: 2017 status report. Addiction 113:1905–1926. doi: 10.1111/add.14234. [DOI] [PubMed] [Google Scholar]
- 21.Alinejad S, Aaseth J, Abdollahi M, Hassanian-Moghaddam H, Mehrpour O. 2018. Clinical aspects of opium adulterated with lead in Iran: a review. Basic Clin Pharmacol Toxicol 122:56–64. doi: 10.1111/bcpt.12855. [DOI] [PubMed] [Google Scholar]
- 22.Etemadi A, Khademi H, Kamangar F, Freedman ND, Abnet CC, Brennan P, Malekzadeh R, Golestan Cohort Study Team. 2017. Hazards of cigarettes, smokeless tobacco and waterpipe in a Middle Eastern population: a cohort study of 50 000 individuals from Iran. Tob Control 26:674–682. doi: 10.1136/tobaccocontrol-2016-053245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fallahzadeh MA, Salehi A, Naghshvarian M, Fallahzadeh MH, Poustchi H, Sepanlou SG, Gandomkar A, Malekzadeh R. 2017. Epidemiologic study of opium use in pars cohort study: a study of 9000 adults in a rural southern area of Iran. Arch Iran Med 20:205–210. [PubMed] [Google Scholar]
- 24.Yazdanian M, Armoon B, Noroozi A, Mohammadi R, Bayat AH, Ahounbar E, Higgs P, Nasab HS, Bayani A, Hemmat M. 2020. Dental caries and periodontal disease among people who use drugs: a systematic review and meta-analysis. BMC Oral Health 20:44. doi: 10.1186/s12903-020-1010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammadi TM, Hasheminejad N, Salari HR, Rostamizadeh MR, Najafipour H. 2017. Association between tooth loss and opium addiction: results of a community-based study on 5900 adult individuals in south east of Iran in 2015. J Int Soc Prev Community Dent 7:186–190. doi: 10.4103/jispcd.JISPCD_189_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shekarchizadeh H, Khami MR, Mohebbi SZ, Ekhtiari H, Virtanen JI. 2013. Oral health of drug abusers: a review of health effects and care. Iran J Public Health 42:929–940. [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar PS, Matthews CR, Joshi V, de Jager M, Aspiras M. 2011. Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect Immun 79:4730–4738. doi: 10.1128/IAI.05371-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon JH, Lee JH, Lee JY. 2015. Subgingival microbiome in smokers and non-smokers in Korean chronic periodontitis patients. Mol Oral Microbiol 30:227–241. doi: 10.1111/omi.12086. [DOI] [PubMed] [Google Scholar]
- 29.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, Jablonski K, Kleerup E, Lynch SV, Sodergren E, Twigg H, Young VB, Bassis CM, Venkataraman A, Schmidt TM, Weinstock GM, Lung HIV Microbiome Project. 2013. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeshita T, Kageyama S, Furuta M, Tsuboi H, Takeuchi K, Shibata Y, Shimazaki Y, Akifusa S, Ninomiya T, Kiyohara Y, Yamashita Y. 2016. Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Sci Rep 6:22164. doi: 10.1038/srep22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valles Y, Inman CK, Peters BA, Ali R, Wareth LA, Abdulle A, Alsafar H, Anouti FA, Dhaheri AA, Galani D, Haji M, Hamiz AA, Hosani AA, Houqani MA, Junaibi AA, Kazim M, Kirchhoff T, Mahmeed WA, Maskari FA, Alnaeemi A, Oumeziane N, Ramasamy R, Schmidt AM, Weitzman M, Zaabi EA, Sherman S, Hayes RB, Ahn J. 2018. Types of tobacco consumption and the oral microbiome in the United Arab Emirates Healthy Future (UAEHFS) Pilot Study. Sci Rep 8:11327. doi: 10.1038/s41598-018-29730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beghini F, Renson A, Zolnik CP, Geistlinger L, Usyk M, Moody TU, Thorpe L, Dowd JB, Burk R, Segata N, Jones HE, Waldron L. 2019. Tobacco exposure associated with oral microbiota oxygen utilization in the New York City Health and Nutrition Examination Study. Ann Epidemiol 34:18–25. doi: 10.1016/j.annepidem.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys MS, Ravel J, Ren Y, Caporaso NE. 2017. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome 5:3. doi: 10.1186/s40168-016-0226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, Chen MX, Chen ZH, Ji GY, Zheng ZD, Mujagond P, Chen XJ, Rong ZH, Chen P, Lyu LY, Wang X, Wu CB, Yu N, Xu YJ, Yin J, Raes J, Knight R, Ma WJ, Zhou HW. 2018. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med 24:1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 35.Taghavi S, Khashyarmanesh Z, Moalemzadeh-Haghighi H, Nassirli H, Eshraghi P, Jalali N, Hassanzadeh-Khayyat M. 2012. Nicotine content of domestic cigarettes, imported cigarettes and pipe tobacco in Iran. Addict Health 4:28–35. [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart CJ, Auchtung TA, Ajami NJ, Velasquez K, Smith DP, De La Garza R, II, Salas R, Petrosino JF. 2018. Effects of tobacco smoke and electronic cigarette vapor exposure on the oral and gut microbiota in humans: a pilot study. PeerJ 6:e4693. doi: 10.7717/peerj.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, Hwang J, Bushman FD, Collman RG. 2010. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One 5:e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brook I. 2011. The impact of smoking on oral and nasopharyngeal bacterial flora. J Dent Res 90:704–710. doi: 10.1177/0022034510391794. [DOI] [PubMed] [Google Scholar]
- 39.Hanioka T, Morita M, Yamamoto T, Inagaki K, Wang PL, Ito H, Morozumi T, Takeshita T, Suzuki N, Shigeishi H, Sugiyama M, Ohta K, Nagao T, Hanada N, Ojima M, Ogawa H. 2019. Smoking and periodontal microorganisms. Jpn Dent Sci Rev 55:88–94. doi: 10.1016/j.jdsr.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, Sindberg G, Wang F, Meng J, Sharma U, Zhang L, Dauer P, Chen C, Dalluge J, Johnson T, Roy S. 2016. Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal Immunol 9:1418–1428. doi: 10.1038/mi.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golshiri A, Mokhtaree MR, Shabani Z, Tabatabaee ST, Rahnama A, Moradi M, Sayadi AR, Faezi H. 2009. Effects of opium smoking cessation on the nasopharyngeal microbial flora. Addict Health 1:1–4. [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes RB, Reding D, Kopp W, Subar AF, Bhat N, Rothman N, Caporaso N, Ziegler RG, Johnson CC, Weissfeld JL, Hoover RN, Hartge P, Palace C, Gohagan JK, Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial Project. 2000. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin Trials 21:349S–355S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 43.Calle EE, Rodriguez C, Jacobs EJ, Almon ML, Chao A, McCullough ML, Feigelson HS, Thun MJ. 2002. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer 94:2490–2501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 44.Shchipkova AY, Nagaraja HN, Kumar PS. 2010. Subgingival microbial profiles of smokers with periodontitis. J Dent Res 89:1247–1253. doi: 10.1177/0022034510377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shakeri R, Kamangar F, Mohamadnejad M, Tabrizi R, Zamani F, Mohamadkhani A, Nikfam S, Nikmanesh A, Sotoudeh M, Sotoudehmanesh R, Shahbazkhani B, Ostovaneh MR, Islami F, Poustchi H, Boffetta P, Malekzadeh R, Pourshams A. 2016. Opium use, cigarette smoking, and alcohol consumption in relation to pancreatic cancer. Medicine (Baltimore) 95:e3922. doi: 10.1097/MD.0000000000003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogtmann E, Han Y, Caporaso JG, Bokulich N, Mohamadkhani A, Moayyedkazemi A, Hua X, Kamangar F, Wan Y, Suman S, Zhu B, Hutchinson A, Dagnall C, Jones K, Hicks B, Shi J, Malekzadeh R, Abnet CC, Pourshams A. 2020. Oral microbial community composition is associated with pancreatic cancer: a case-control study in Iran. Cancer Med 9:797–806. doi: 10.1002/cam4.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao N, Chen J, Carroll IM, Ringel-Kulka T, Epstein MP, Zhou H, Zhou JJ, Ringel Y, Li H, Wu MC. 2015. Testing in microbiome—profiling studies with MiRKAT, the microbiome regression-based kernel association test. Am J Hum Genet 96:797–807. doi: 10.1016/j.ajhg.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download Spectrum.00138-21-s0001.pdf, PDF file, 0.5 MB (575.4KB, pdf)
Supplemental material. Download Spectrum.00138-21-s0002.xlsx, XLSX file, 0.4 MB (390.6KB, xlsx)
Data Availability Statement
The sequencing data are available on the Sequence Read Archive (NCBI SRA) under BioProject ID PRJNA549488.