ABSTRACT
The influenza A virus (IAV) infection is usually restricted to the respiratory tract and only rarely enters the central nervous system (CNS) and causes neurological symptoms. However, the roles of host factors involved in IAV infection in the CNS remain largely undetermined. Therefore, we aimed to characterize the host responses to IAV infection in the brain. We isolated a strain of IAV H5N6, which is neurotoxic and highly pathogenic to mice. High-throughput RNA sequencing (RNA-seq) revealed 240 differentially expressed genes in IAV-infected brains. Among the significantly downregulated genes, we focused on the gene encoding progesterone receptor membrane component-1 (PGRMC1) and observed that IAV H5N6 infection clearly inhibited PGRMC1 in both neuroblastoma and glioma cells. Furthermore, treatment with AG205, a PGRMC1-specific inhibitor, or PGRMC1 knockout promoted H5N6 multiplication in vitro, while overexpression of PGRMC1 resulted in opposite effects. Furthermore, AG205 treatment or PGRMC1 knockout significantly inhibited the retinoic acid-inducible gene I (RIG-I)-mediated interferon beta (IFN-β) signaling pathway and reduced the levels of several antiviral proteins (Mx1 and ISG15). In addition, PGRMC1-mediated regulation of IFN signaling relied on inhibition of the expression and ubiquitination of RIG-I. The loss of PGRMC1 leads to an increased susceptibility of mice (brain and lung) to influenza A virus infection. Conclusively, our results show for the first time that IAV H5N6 downregulates PGRMC1 expression to contribute to virus proliferation by inhibiting RIG-I-mediated IFN-β production in the brain. These findings may offer new insights regarding the interplay between IAV and host factors that may impact IAV pathogenicity in the brain.
IMPORTANCE Central nervous system (CNS) disease is one of the most common extra-respiratory tract complications of influenza A virus (IAV) infections. However, there is still little knowledge about IAV regulating host responses in brain. In this study, we identified progesterone receptor membrane component-1 (PGRMC1) as a novel host factor involved in the replication and propagation of IAV H5N6 in the host brain. We also observed that PGRMC1 antagonism was required for viral evasion from the host immune response during IAV infection via inhibition of the retinoic acid-inducible gene I (RIG-I)-mediated interferon beta (IFN-β) signaling pathway and downstream antiviral gene expression. This study revealed a newly identified regulatory mechanism used by IAV H5N6 to ensure its life cycle in the CNS.
KEYWORDS: H5N6 virus, CNS, PGRMC1, RIG-I, virus replication
INTRODUCTION
Influenza A virus (IAV), the causative agent of flu, occasionally causes acute respiratory distress syndrome among humans and many mammalian and avian species. Novel IAVs have emerged via antigenic drift and reassortment events with other IAVs. The highly pathogenic avian influenza virus H5N1 (HPAIV/H5N1) was first detected in a domestic goose in Guangdong Province, China, in 1996 (1). Since then, HPAIV has caused devastating outbreaks repeatedly in wild birds and poultry, as well as sporadic human infections with high mortality. Since 2013, 5 H7N9 influenza epidemic waves have resulted in 1,344 cases, with 511 deaths in China (2). In addition, HPAIV/H5N6 has replaced H5N1 as one of the dominant IAV subtypes circulating in waterfowls and causing human infections in China (3). Therefore, the continuous evolution of the virus represents a long-term threat to public health and the poultry industry.
IAV requires the host cell machinery to support replication of its genome and for the production of new virions (4). Conversely, host cells produce various factors that target different steps in the virus life cycle to restrict virus infection and multiplication. Being part of the first line of defense against infections, type I interferons (IFNs) are key components of the host antiviral innate immune response and are modulators of the adaptive immune response. The innate immune response is triggered in an IAV-infected host to block virus replication and accelerate viral clearance (5). Host recognition of pathogens via pattern recognition receptors (PRRs) is the first step in triggering an immune response. The IAV RNA is recognized by various PRRs, including Toll-like receptors (TLR3, TLR7, and TLR8) retinoic acid-inducible gene I (RIG-I), and NOD-like receptor (NLRP3) (5). These sensors activate signaling cascades resulting in the expression of specific inflammatory cytokines and chemokines (6). Cytokines play various roles, such as direct inhibition of viral replication and activation of the cytolytic functions of T cells, whereas chemokines recruit innate immune cells, such as macrophages, neutrophils, natural killer (NK) cells, and inflammatory monocytes, to the lungs and airways (7). However, viruses have evolved sophisticated mechanisms to evade or counteract host innate immune responses. In particular, genes carried by viruses inhibit host innate antiviral responses by directly targeting IFN gene expression or/and IFN-induced host effector molecules. The nonstructural protein 1 (NS1) is the best characterized and the most important IFN antagonist protein of IAV. NS1 can block type I interferon signaling downstream of RIG-I or may block RIG-I ubiquitination (8). PB1-F2 from IAV strain A/Puerto Rico/8/1934 can interact with mitochondrial antiviral signaling (MAVS) to inhibit interferon production (9).
Central nervous system (CNS) disease is one of the most common extra-respiratory tract complications of IAV infections (10, 11). IAV has been associated with CNS disease since the 1918 H1N1 pandemic, and CNS disease has been observed during all subsequent pandemics, as well as during seasonal epidemics, with occasional detection of IAV in the CNS of humans (12, 13). Zoonotic IAV infects humans only sporadically, but when they do, they are linked frequently to severe and systemic disease (14). HPAIV H5N1 and H7N9 viruses, two recently identified zoonotic IAVs, are associated with CNS disease (14, 15). The HPAI H5N1 virus is possibly the most neurotropic IAV known and has been frequently associated with CNS disease in humans and in other naturally and experimentally infected mammalian species. IAV needs to infiltrate the CNS to infect, replicate, and spread throughout the CNS. IAV reaches the CNS via the olfactory, sympathetic, trigeminal, vagus, and possibly other cranial nerves (16, 17). However, there is still little knowledge about IAV regulation of host responses in the brain.
Progesterone receptor membrane component-1 (PGRMC1) is a 26–28 kDa membrane-spanning protein with a short extracellular domain, a single transmembrane region, and a cytoplasmic domain. The cytoplasmic segment contains a cytochrome P450 b5/heme binding sequence, which can bind to various pharmacological compounds (18). PGRMC1, also called Dap1, human progesterone receptor (HPR6.6), and IZA, is a membrane-associated progestin receptor (MAPR) occurring in different species (19). It is highly expressed in the CNS, where it exerts a neurotrophic effect (20). PGRMC1 has been shown to be involved in multiple cellular functions, such as cholesterol regulation, axonal guidance, endocytosis, and alteration of reproductive behaviors (21–24). However, how PGRMC1 affects pathogens, especially IAV, remains unclear.
In this study, we aimed to investigate the role of PGRMC1 in regulating antiviral defense response in brain tissue. Toward this aim, we used a mouse model of IAV infection and human neuroblastoma and glioma cell lines. High-throughput RNA sequencing (RNA-seq) was used to obtain an unbiased profile of the cellular response to IAV H5N6 infection in mice brain. We demonstrated that downregulation of PGRMC1 is required for IAV replication and propagation in both the human neuroblastoma cell line SK-N-SH and human brain glioma cell line U251. Furthermore, we showed that the PGRMC1-specific inhibitor AG205, or PGRMC1 knockout (KO), functions as an IFN antagonist in IAV infection by negatively regulating type I IFN induction and antiviral gene expression. Our findings reveal a novel mechanism via which IAV H5N6 inhibits host IFN response and emphasizes the vital role of PGRMC1 in modulating antiviral defenses in the brain.
RESULTS
Infection of BALB/c mice with the JX H5N6 virus.
Previously, we have demonstrated that HPAIV H5N6 (A/duck/Hubei/WH18/2015, abbreviated as JX) is neurotoxic and highly pathogenic to mice. To establish animal models of brain damage caused by IAV, mice (n = 11) were infected intranasally (i.n.) with 50 μl of 105 50% tissue culture infective dose (TCID50), and the mock-treated mice were used as control. Six mice in each group were sacrificed on 3 days postinfection (dpi), and their lung and brains were collected for further study. The remaining five mice in each group were observed daily for the signs of disease and death for 14 dpi. Mice infected with JX showed clearly detectable symptoms of the flu at 3 dpi and showed drastic weight loss (Fig. 1A) [F (1, 98) = 293.2, P < 0.0001]. Strikingly, these symptoms had progressed to neurological symptoms, such as balancing problems, hind limb weakness (Fig. 1B), and paralysis by 5 and 6 dpi, and mice began to die by 7 dpi. At the end of the experimental period, only two out of five mice survived (Fig. 1C) (Mantel-Cox log-rank test, P = 0.0511). Next, infectious virion titers in brain were determined, and immunohistochemistry (IHC) was performed to investigate whether the brain was successfully infected by the JX virus. Results suggested that the brains of mice were successfully infected. Low viral titers in brain were detected (Fig. 1D). Meanwhile, IHC staining for IAV viral antigen revealed virus-positive neurons in the brains of mice sacrificed on 3 dpi. Virus antigen was detected in multiple areas of the brain, including cerebral cortex, brainstem, and occipital lobe. For representative regions, an image of the cerebral cortex with a single staining of IAV antigen is shown (Fig. 1E). Brain tissue injury caused by the JX virus involves submeningeal hemorrhage, degeneration, and necrosis of neurons and local glial cell nodules (Fig. 1F to H). Collectively, these results demonstrated that the JX virus can infect and spread throughout the CNS, thereby successfully establishing a mouse model for further functional studies.
FIG 1.
Infection of BALB/c mice (n = 11) with H5N6 virus. Mice receiving PBS were used as controls. The mice were monitored for 14 days. (A) Body weight changes were depicted as the percentage of the starting weight of mice (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; using two-way ANOVA). (B) The mice developed neurological symptoms. (C) Survival of infected mice (Mantel-Cox log-rank test). (D) Infectious virion titers in lung and brain were calculated through TCID50. (E) Immunohistochemical analysis of the nucleoprotein antigen in brainstem. (F to H) Degeneration and necrosis of neurons and local glial cell nodules in the cerebral cortex. The data shown are representative of three independent experiments with similar results.
Brain transcription profile after JX virus infection.
To determine the differential responses of the host brain tissue to infection with JX and mock infection, RNA samples were prepared from whole brains and the complete gene expression profile was compared after whole-transcriptome sequencing (Fig. 2). Histopathological and immunohistochemical examinations showed neuron necrosis, proliferation of glial cells, and viral antigen positivity on 3 dpi. Therefore, brain samples were obtained on this day. A cutoff of >2.0 in fold change and a P value of <0.05 were used initially to examine the significantly differentially expressed genes that were up or downregulated following JX infection (Fig. 2). In total, 32,716 genes were detected in the brain, of which 240 were identified as differentially expressed mRNAs between the virus-infected and uninfected mice. Among these genes, 179 were upregulated with log2 fold change values ranging from 1.01 to 5.08, while 61 genes were downregulated with log2 fold change values ranging from 1 to 4.59. The top 10 upregulated genes listed were Cxx1b/Cxx1a, Scrt1, Bmyc, Pcp2, Car8, Arl4c, Fat2, Mybpc3, Rn7sk, and Irf2bp1 (Table 1). The top 10 most downregulated genes were Nudc-ps1, Glrx5, Pgrmc1, Rnf113a2, Irgq, Rgs4, Tmem97, Zik1, Zcchc3, and Gng4 (Table 2). To further assess the connection between gene expression pattern and IAV infection-induced biological processes in the brain, a functional classification of mRNA transcripts and a pathway analysis were performed. These analyses revealed that these genes were involved in regulating neuronal differentiation, ion transmembrane transport, and protein binding. Some of the significant pathways involving the differentially expressed genes were associated with herpes simplex infection, glutamatergic synapse, and chemokine signaling pathway. Among the significantly downregulated genes, we focused on PGRMC1 to evaluate the role in IAV infection in the brain.
FIG 2.
Analysis of the transcriptome profiles of brain tissue after infection with H5N6 virus. (A) Gene volcano plot. (B) Heat map. (C) Scatterplot showing Gene Ontology (GO) and (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis.
TABLE 1.
The top 10 most upregulated genes of virus-infected mice compared with control mice
| Symbol | Gene name | Log2FC | P value |
|---|---|---|---|
| Cxx1b;Cxx1a | CAAX box 1A;CAAX box 1B | 5.68 | 0.014 |
| Scrt1 | Scratch family zinc finger 1 | 4.47 | 0.021 |
| Bmyc | Brain expressed myelocytomatosis oncogene | 4.35 | 0.046 |
| Pcp2 | Purkinje cell protein 2 (L7) | 4.31 | 0.037 |
| Car8 | Carbonic anhydrase 8 | 4.25 | 0.044 |
| Arl4c | ADP-ribosylation factor-like 4C | 3.94 | 0.046 |
| Fat2 | FAT atypical cadherin 2 | 3.56 | 0.045 |
| Mybpc3 | Myosin binding protein C, cardiac | 3.51 | 0.038 |
| Rn7sk | RNA, 7SK, nuclear | 3.32 | 0.05 |
| Irf2bp1 | Interferon regulatory factor 2 binding protein 1 | 3.27 | 0.046 |
TABLE 2.
The top 10 most downregulated genes of virus-infected mice compared with that in the control mice
| Symbol | Gene name | Log2FC | P value |
|---|---|---|---|
| Nudc-ps1 | Nuclear distribution gene C homolog (Aspergillus), pseudogene 1 | 4.59 | 0.001 |
| Glrx5 | Glutaredoxin 5 | 3.87 | 0.043 |
| Pgrmc1 | Progesterone receptor membrane component 1 | 3.35 | 0 |
| Rnf113a2 | Ring finger protein 113A2 | 3.1 | 0.038 |
| Irgq | Immunity-related gtpase family, Q | 2.92 | 0.014 |
| Rgs4 | Regulator of G-protein signaling 4 | 2.6 | 0.007 |
| Tmem97 | Transmembrane protein 97 | 2.38 | 0.031 |
| Zik1 | Zinc finger protein interacting with K protein 1 | 2.16 | 0.034 |
| Zcchc3 | Zinc finger, CCHC domain containing 3 | 2.11 | 0.019 |
| Gng4 | Guanine nucleotide binding protein (G protein), gamma 4 | 2.1 | 0.014 |
H5N6 downregulates the expression of PGRMC1 in vitro.
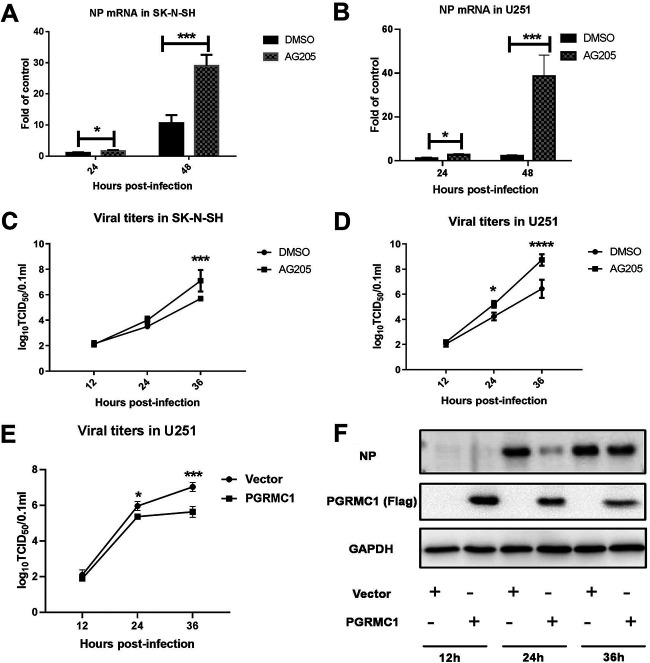
It is well known that the nervous tissue is composed of two primary cell types, namely, neurons and glial cells. Neurons transmit nerve messages, while the surrounding glial cells are in direct contact with neurons (24). To verify whether the H5N6 virus can downregulate PGRMC1 in vitro, both human neuroblastoma cell line SK-N-SH and human glioma cell line U251 were used. SK-N-SH or U251 cells were infected with JX at multiplicity of infection (MOI; PFU/cell) of 0.01. At 24 and 48 hours postinfection (hpi), PGRMC1 mRNA and protein levels were assessed using reverse transcription-quantitative PCR (qRT-PCR) and Western blotting, respectively (Fig. 3). Compared with the mock, JX significantly inhibited the production of PGRMC1 mRNA in SK-N-SH cells after 24 h, which was in agreement with the protein level (Fig. 3A and C) [F (1, 23) = 201.3, P < 0.0001]. At 48 hpi, the extent of PGRMC1 protein inhibition by the JX virus was lower than that at 24 hpi. Similar result was observed in U251 cells (Fig. 3B and D) [F (1, 20) = 87.57, P < 0.0001]. These results indicated that IAV JX significantly inhibit the expression of PGRMC1, which is in agreement with genes identified using RNA-seq.
FIG 3.
JX inhibited PGRMC1 expression in vitro. PGRMC1 expression level in SK-N-SH cells (A and C) and U251 cells (B and D). Cells were infected with JX at an MOI of 0.01. Samples were collected at 24 and 48 hpi, followed by qRT-PCR and Western blotting to determine the mRNA (A and B) and protein (C and D) levels of PGRMC1. For real-time PCR analysis, the mRNA level was normalized to the GAPDH level. For Western blotting, the band intensities were analyzed using ImageJ, and GAPDH was used as a control in each time point. The relative PGRMC1 levels (PGRMC1/GAPDH) are shown. The data are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; using two-way ANOVA).
PGRMC1 inhibits H5N6 replication in vitro.
To better understand the biological role of PGRMC1 in H5N6 replication, AG205, a putative PGRMC1-specific inhibitor (20), was used. SK-N-SH or U251 cells were seeded in 12-well plates and grown to 80% confluence. Then, the cells were treated with 15 μM AG-205 or vehicle (cell culture-grade dimethyl sulfoxide [DMSO]) for 24 h. Cells were infected with JX virus at an MOI of 0.01. Viral nucleoprotein (NP) mRNA levels and viral titers were evaluated at different time points (Fig. 4). NP mRNA levels in the two cell lines significantly increased in the AG205 group compared with that in the control (Fig. 4A) [F (1, 21) = 116.0, P < 0.0001] (Fig. 4B) [F (1, 19) = 62.26, P < 0.0001]. Next, we observed that AG205 significantly increased the viral titers in U251 24 h and 36 h postinfection (Fig. 4C) [F (1, 12) = 44.47, P < 0.0001] (Fig. 4D) [F (1, 12) = 37.78, P < 0.0001). However, viral titers significantly increased after AG205 treatment of SK-N-SH cells at 36 h, although no effect on virus replication was evident 24 h after AG205 treatment. Subsequently, we compared the effect of AG205 on viral proliferation in the two cell types and observed that AG205 consistently enhanced the viral titers of IAV JX in the U251 cells compared with that in the SK-N-SH cells. The effect of PGRMC1 overexpression on viral replication was also determined by transfecting U251 cells with Flag-PGRMC1. The efficiency of PGRMC1 overexpression was determined using Western blotting (Fig. 4F). As observed in the TCID50 assays, virus titers at 24 and 36 hpi were lower in the PGRMC1-overexpressing groups than in the control group (Fig. 4E) [F (1, 8) = 26.36, P = 0.0009]. Taken together, these findings indicated that PGRMC1 was an IAV JX restriction factor.
FIG 4.
Specific inhibition of PGRMC1 by AG205 increased JX H5N6 virus replication. SK-N-SH cells (A and C) and U251 cells (B and D) were pretreated with 15 μM AG205 or DMSO over 24 h. Cells were infected with JX at an MOI of 0.01. Samples were collected at different time points, and viral NP mRNA and viral titers were determined using qRT-PCR and TCID50, respectively. U251 cells (E and F) were transfected with Flag-PGRMC1 and empty vectors. After 24 h, the cells were infected with JX at MOI of 0.01 and expression was detected using Western blotting. TCID50 assays (E) for the virus titer of JX were performed to estimate virus multiplication at 12, 24, and 36 hpi. The expressions of PGRMC1 and H5N6 NP protein in U251 cells were detected by Western blot (F). The data are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; using two-way ANOVA).
AG205 inhibits the IAV-induced RIG-I-dependent antiviral response.
AG205 affects virus multiplication more in U251 cells than in SK-N-SH cells. Consistently, the innate immune surveillance is coordinated mainly by glial cells in the CNS (25). These CNS resident cells are assumed to orchestrate the immune response, which assists in combating infections in the brain. Hence, we hypothesized that AG205 may inhibit the IAV H5N6-mediated innate immune response. To test this hypothesis, we evaluated a series of immunological molecules in IAV JX-infected U251 cells, which were incubated with AG205 or DMSO (Fig. 5, Fig. 6). As expected, AG205 significantly downregulated IFN-β induced by IAV JX (Fig. 5A) [F (1, 8) = 126.1, P < 0.0001]. In addition, the expression of the downstream effectors of IFN-β, namely, ISG15, was suppressed (Fig. 6). We further observed that AG205 markedly inhibited poly(I·C)-mediated IFN-β induction (Fig. 5B to G). Overall, these observations suggested that AG205 inhibited the type I interferon response.
FIG 5.
AG205 negatively modulated the IFN-β-inducing pathway. U251 cells were pretreated with 15 μM AG205 or DMSO for 24 h. The cells were infected with JX at an MOI of 0.01 (A) or were stimulated with 100 ng of poly (I·C) (B to F) for 24 h. The mRNA levels of the target genes were detected and normalized to the GAPDH level, and the expression in the DMSO and poly (I·C)-unstimulated groups were set to 1. U251 cells were transfected with Flag-PGRMC1 and empty vectors (G). After 24 h, the cells were stimulated with 100 ng of poly (I·C) for 24 h and IFN-β mRNA levels were detected. The data are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; A was analyzed by repeated measures two-way ANOVA; B to G were analyzed by one-way ANOVA).
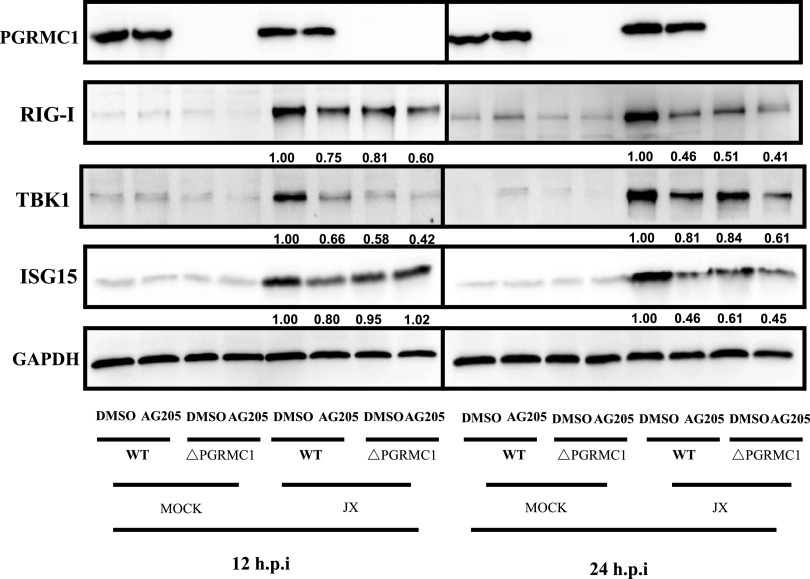
FIG 6.
AG205 or PGRMC1 knockout inhibited the RIG-I-mediated IFN-β signaling pathway. PGRMC1-KO U251 cells and wild-type cells were treated with 15 μM AG 205 or DMSO for 24 h. Cells were infected with JX at MOI of 0.01. Samples were collected at 12 and 24 h. IFN-β signaling molecules were detected using Western blotting. The band intensities were analyzed using ImageJ, and GAPDH was used as the control.
AG205 inhibits RIG-I-mediated IFN-β production.
The RIG-I signaling pathway is essential for IAV recognition. Once activated, RIG-I/MAVS induces the activation of transcription factor IRF3 and NF-κB, ultimately resulting in establishment of the host IFN-mediated antiviral response (5). To investigate how the PGRMC1 inhibitor AG205 regulates IFN-β, the IFN-β signaling pathway was analyzed using IFN-β luciferase activity assay (Fig. 7). AG205 or DMSO-pretreated 293 cells were cotransfected with the signal molecule expression plasmid, IFN-β promoter luciferase reporter plasmid, and internal control pRL-TK. AG205 markedly inhibited RIG-I-mediated IFN-β production (Fig. 7A) [F (1, 8) = 156.1, P < 0.0001]. Interestingly, it did not affect other molecules [MDA5, F (1, 8) = 3.410, P = 0.1020; TBK1, F (1, 8) = 0.08015, P = 0.7843; IKKξ, F (1, 8) = 1.774, P = 0.2196, IRF3, F (1, 7) = 3.168, P = 0.1183; IRF3-5D, F (1, 8) = 3.705, P = 0.0905] that stimulate IFN-β promoter luciferase activity (Fig. 7B to F). Notably, as shown in Fig. 7H (one-way analysis of variance [ANOVA], F = 46.24, P = 0.005), we observed a dose-dependent relationship between AG205 and IFN-β promoter luciferase activity. On the contrary, overexpression of PGRMC1 increased RIG-I-stimulated IFN-β luciferase activity in a dose-dependent manner (Fig. 7G) (one-way ANOVA, F = 78.66, P < 0.0001). Consequently, we concluded that downregulation or functional inhibition of PGRMC1 reduces the ability of RIG-I to induce IFN-β production in cells infected with IAV H5N6.
FIG 7.
AG205 inhibited IFN-β promoter activity induced by RIG-I-mediated IFN-β signaling molecules. Effect of AG205 or PGRMC1 on the activation of IFN-β promoter induced by RIG-I (A), MDA5 (B), IKKε (C) TBK1 (D), IRF3 (E), and IRF3-5D (F). HEK293T cells were treated with 15 μM AG 205 or DMSO for 24 h. Then, the cells were transfected with IFN-β-luc, pRL-TK, and the indicated expression plasmids of the signaling molecules. The effect of different doses of PGRMC1 (G) or AG205 (H) on the activation of IFN-β promoter induced by RIG-I was measured 24 h posttransfection using the luciferase activity assay. The expression level of each signaling molecule was detected using Western blotting with an anti-Flag antibody. The data are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; A to F were analyzed by repeated measures two-way ANOVA; G and H were analyzed by one-way ANOVA).
PGRMC1 knockout promotes IAV replication by inhibiting the RIG-I-mediated IFN-β signaling pathway.
As the PGRMC1 inhibitor AG205 has been shown to significantly promote IAV replication, we speculated whether PGRMC1 knockdown similarly affected the proliferation of IAV H5N6 in U251 cells. Toward this aim, PGRMC1 was knocked out using the lentiviral clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system containing a pair of suitable guide RNAs (gRNAs) driven by two independent U6 promoters on the same plasmid (Fig. 8). After CRISPR-Cas9 gene editing and isolation of individual clones, PGRMC1 knockout was analyzed using sequencing and Western blot analysis. Three PGRMC1 KO clones were selected randomly for further research. Our results suggested that CRISPR-Cas9 targeting resulted in the complete loss of the PGRMC1 protein (Fig. 8A). We evaluated the effect of PGRMC1 deficiency on IAV JX propagation using a TCID50 assay. Indeed, cells with PGRMC1 knockout produced more infectious viral particles than control U251 cells (Fig. 8B) [F (1, 12) = 70.53, P < 0.0001], which is in agreement with the results obtained in cells treated with AG205. Furthermore, the expression of RIG-I (protein level, Fig. 6) and IFN-β (mRNA, Fig. 8C) [F (3, 61) = 185.1, P < 0.0001] in JX-stimulated cells decreased. Alternatively, the IFN-β upstream factors (RIG-I, and TBK1) and IFN-β downstream effector (ISG15) were also significantly downregulated (Fig. 6). These results further confirmed that PGRMC1 knockout promoted virus replication by efficiently suppressing RIG-I-mediated IFN-β production.
FIG 8.
Effect of PGRMC1 knockout on IAV replication and the RIG-I-mediated IFN-β signaling pathway. Generation of PGRMC1-KO U251 cells using the CRISPR-Cas9 system. PGRMC1 knockout was confirmed using Western blotting (A). PGRMC1-KO U251 cells (ΔPGRMC1) or wild-type U251 cells (WT) were infected with JX H5N6 virus at MOI of 0.01. Viral titers were determined using TCID50 on MDCK cells (B). The mRNA levels of the IFN-β were detected and normalized to the GAPDH level (C) (means ± SD from three independent experiments; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; using two way ANOVA).
PGRMC1 knockout antagonizes RIG-I ubiquitination.
RIG-I exists in an inactive closed conformation under normal conditions. RIG activation requires ubiquitination induced by the tripartite motif 25 (TRIM25) ubiquitin E3 ligase. Hence, we next addressed whether PGRMC1 knockout specifically suppresses RIG-I ubiquitination (26). Toward this aim, we assessed RIG-I ubiquitination in PGRMC1-KO U251 cells or wild-type (WT) U251 cells transfected with the pCAGGS-HA-Ub vector or the empty vector along with or without p3×Flag-RIG-I, followed by infection with JX. Samples were subjected to anti-Flag immunoprecipitation and immunoblotting with an anti-hemagglutination (HA) tag antibody to monitor RIG-I ubiquitination. As shown in Fig. 9, PGRMC1 knockout markedly inhibited RIG-I ubiquitination, as shown by comparison of lanes 3 and lane 6. These results revealed collectively that the PGRMC1 knockout inhibited RIG-I ubiquitination in U251 cells, thereby suppressing RIG-I signal transduction.
FIG 9.

PGRMC1 knockout inhibited RIG-I ubiquitination. PGRMC1-KO U251 cells and wild-type cells were transfected with Flag-RIG-I and pUb-HA. After 24 h, the cells were infected with JX at an MOI of 0.01. After 24 h, the cells were subjected to anti-Flag immunoprecipitation and immunoblotting with anti-HA to monitor RIG-I ubiquitination.
PGRMC1 knockout promotes IAV replication in brain and lung of mice.
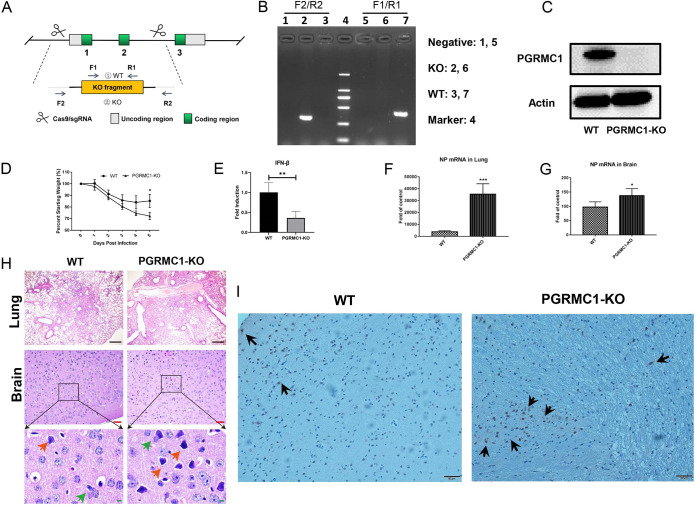
To further confirm the role of PGRMC1 in IAV infection in the brain, we utilized the PGRMC1 KO and wild-type (C57BL/6J) mice (Fig. 10). PGRMC1 KO mice were generated using the CRISPR-Cas9 technique (Fig. 10A) and were verified by PCR (Fig. 10B) and Western blotting (Fig. 10C). The PGRMC1 KO and WT mice were infected with JX IAV strain, and 5 days later, mice were sacrificed to collect the brain and the lung. Both KO and WT mice started losing weight from day 2 after virus inoculation (Fig. 10D). By day 5, WT mice had lost 15% of their body weight. In contrast, the KO mice lost a significantly higher amount of their body weight, namely, 28%, by day 5. The qRT-PCR analysis revealed that IFN-β mRNA level in KO mouse lung was significantly lower than that in wild-type mouse lung (Fig. 10E). More interestingly, the NP mRNA level in KO mouse lung and brain was significantly higher than that in WT mouse lungs (Fig. 10F and G). Histopathologic analysis showed that the lungs of PGRMC1 KO mice displayed more severe pathologies on day 5 postinfection than that of WT mice (Fig. 10H), including inflammatory cell infiltration, and deciduous cells in the bronchial lumen, even the alveoli, almost disappeared. In contrast to the brain in WT mice, more astrocytic hyperplasia, neuronophagia phenomenon, and neuronal cell necrosis were observed in the brain tissues from PGRMC1 KO mice. The immunohistochemical assays show a similar result in NP mRNA levels in the brain (Fig. 10I). These results further confirmed that PGRMC1 knockout promotes virus replication and leads to more serious damage in vivo.
FIG 10.
PGRMC1 knockout promoted IAV replication in mice. The PGRMC1 KO mice were made by using the CRISPR-Cas9 technique (A). The analyses of protein expressions in WT and PGRMC1 KO mice by PCR (B) and Western blotting using the antibody against PGRMC1 and actin (C). C57BL/6J WT and PGRMC1 KO female mice (n = 5) aged 4 to 6 weeks were infected intranasally with 50 μl of 105 TCID50 H5N6 virus. Body weights were monitored daily for 5 days (D), and three mice in each group were euthanized at 5 dpi. IFN-β mRNA in lung (E) and viral NP mRNA in lung (F) and brain (G) were determined through qRT-PCR. Hematoxylin and eosin (H&E) staining of lung and brain sections from WT and PGRMC1 KO mice infected H5N6 virus (H). Astrocytic hyperplasia, red arrow; neuronophagia, green arrow. (I) Viral NP proteins (black arrows) were detected in the cerebral cortex of WT and PGRMC1 KO mice. Scale bars (black), 200 μm. Scale bars (red), 50 μm. Scale bars (green), 10 μm. (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
IAV is known to target the respiratory system, while some particular subtypes of IAV can infect the brain and cause serious damage to the CNS. Multiple routes of IAV invasion into the CNS after intranasal exposure have been reported. In this study, we isolated a strain of HPAIV H5N6 virus from a duck, which showed high fatality and pathogenicity to mice. Some infected mice developed typical neurological symptoms, such as balancing problems, hind limb weakness, and paralysis. Surprisingly, although the mice survived until the completion of the experiment, hind limb paralysis did not improve. This observation was in accordance with the results of Hosseini et al., who reported that neuroinflammation caused by nonneurotropic and neurotropic IAV induces long-lasting impairments in hippocampal neuronal morphology and synaptic properties and cognitive function in adult animals (27). The viral loads in the brain of mice in the infected group were examined using a TCID50 assay. These results confirmed that the JX virus can infect, replicate, and spread throughout the CNS. This finding agrees with the observation that viruses of the HPAIV H5 subtype are associated more frequently with CNS disease in humans (28).
The molecular mechanisms underlying the involvement of host factors in IAV infection in CNS remain obscure. Here, we identified 240 genes in the IAV-infected mouse brains using a RNA-seq screen. These genes were involved in herpes simplex infection, glutamatergic synapse, and chemokine signaling pathway. Among these identified genes, H2-T23, H2-T24, Ifi1, Irf7, and Stat2 were involved in other virus infections, such as those caused by chikungunya virus, Duvenhage virus, and Zika virus. Zhao and coworkers observed that five virus-related proteins (TGTP, IFIT3, IFIT3-L, LCP1-1, and LCP1-2) were upregulated within 1 dpi in the lethal virus-inoculated group but did not show appreciable upregulation until 3 dpi in the nonlethal group. Their results suggest that proteomic approaches can distinguish the lethality of the influenza strain in a mouse model within 24 h of infection (29). Interestingly, the interferon-responsive gene Ifit3, which is reported to be involved in cognitive decline in aged mice (30), was specifically upregulated in the hippocampus of H7N7-infected mice and is expressed mainly in granule cells (31).
PGRMC1 participates in cellular processes relevant to CNS diseases, including neuroprotection, axonal migration, and mitochondrial protection. Furthermore, it is expressed highly in the brain. However, the relationship between PGRMC1 and virus infection in the brain remains uninvestigated. Here, we observed that HPAIV H5N6 significantly inhibited PGRMC1 expression in the brain. This phenomenon has further been demonstrated in SK-N-SH and U251 cells. Next, we investigated the effect of PGRMC1 on H5N6 and observed that AG205-mediated inhibition of PGRMC1 increased viral titers in these two cell lines. Although AG205 was advantageous for the proliferation of H5N6 in SK-N-SH and U251 cells, it promoted viral replication more in U251 cells than in SK-N-SH cells. U251 cells are derived from glial cells, which play a key role in regulating the immune responses in CNS. In addition, one of the first lines of host immunological defense against viruses involves the IFNs. Therefore, we hypothesized that AG205 treatment or PGRMC1 knockout may inhibit the H5N6-mediated IFN signaling pathway. As expected, AG205 treatment or PGRMC1 knockout inhibited the type I interferon response dramatically, implying that PGRMC1 is a host factor restricting IAV JX infection in the CNS.
IAV genomes consist of eight negative-sense single-stranded RNA segments (vRNA) that encode at least 14 viral proteins (32). Hence, we aimed to identify the gene(s) involved in PGRMC1 inhibition. To this end, eight viral gene segments were cloned individually in the pHW2000 expression vector. However, unfortunately, we observed that no single IAV JX gene segment was able to significantly downregulate PGRMC1 (Fig. S1). This finding indicated that a single gene or protein is not sufficient for inhibiting PGRMC1 and that downregulation of PGRMC1 is relevant for the biological characteristics of H5N6 virus.
During coevolution with hosts, IAV has developed multiple strategies to escape the host antiviral innate immunity, especially via blockade of the IFN system. Several IAV proteins have been well characterized as IFN antagonists, which antagonize the host IFN response at different steps. IAV antagonizes IFN response via various strategies, which involve viral proteins and host factors. For example, NS1 of IAV inhibits the activation of IRF3 and IFN-β transcription. In addition, PB2 and PB1-F2 interact with MAVS, which represses IFN-β expression (9). In this study, H5N6 downregulated PGRMC1 in the brain, which strongly suppressed the activation of the RIG-I-mediated IFN-β signaling pathway. Furthermore, AG205 treatment or PGRMC1 knockout inhibited not only RIG-I expression stimulated by the H5N6 virus but also RIG-I ubiquitination. These results suggest that IAV-induced downregulation of PGRMC1 is involved in controlling RIG-I signaling, which is beneficial for the virus infection in the CNS. However, the mechanisms by which downregulated PGRMC1 inhibits the expression and ubiquitination of RIG-I remain unclear. Possibly, downregulated PGRMC1 perturbs the interaction between RIG-I and its E3 ligase, TRIM25, which warrants further investigations. Previous studies have assumed that PGRMC1 can alter its subcellular location under certain circumstances. Peluso suggested that as a transcription factor, PGRMC1 is involved in the regulation of gene transcription (33).
Microglia and astrocytes are the CNS-resident cells involved most prominently in early innate responses to injury, autoimmune attack, or infection (25). Unfortunately, pathogens can induce devastating inflammatory damage to the CNS, leading to rapid mortality or long-term neurological disabilities. Nevertheless, some viruses may survive as latent infections in conducive environments, provided they become dormant and do not proliferate. Thus, the CNS “protects” these pathogens.
In summary, we identified PGRMC1 as a novel host factor involved in the replication and propagation of IAV H5N6 in the host brain. We also observed that PGRMC1 was required for evasion from the host immune response during IAV infection via inhibition of the expression and ubiquitination of RIG-I, thereby blocking activation of the RIG-I-mediated IFN-β signaling pathway and downstream antiviral gene expression. Overall, our study revealed a newly identified regulatory mechanism used by IAV H5N6 to ensure its life cycle in the CNS. However, further studies are necessary to explore more precisely the specific mechanism of the H5N6 virus that downregulates PGRMC1 expression.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were approved by the Research Ethics Committee, Huazhong Agricultural University, Hubei, China (HZAUMO-2016-022), and were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of the Research Ethics Committee, Huazhong Agricultural University, Hubei, China.
Cells and viruses.
The human neuroblastoma cell line SK-N-SH, human brain glioma cell line U251, and the Madin-Darby canine kidney cell line (MDCK) were cultured in Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (PAN Biotech) and 100 U/ml penicillin-streptomycin (Thermo Fisher Scientific, Inc.). HEK293T cells were cultured in RPMI 1640 medium (Invitrogen). The IAV strain used in this study was the A/duck/Hubei/WH18/2015(H5N6) (JX) (GenBank accession number KX652135) strain isolated in our laboratory from an infected duck lung. H5N6 was grown in 9-day-old embryonated eggs. The titers of IAV in culture media were determined using the 50% tissue culture infective dose (TCID50) and plaque assays in MDCK cells.
Mouse infection.
Female BALB/c mice aged 4 to 6 weeks were purchased from the Center for Animal Disease Control, Hubei Province, China. Mice (n = 11) were infected intranasally (i.n.) with 50 μl of 105 TCID50, while naive control mice were inoculated with phosphate-buffered saline (PBS). Six mice in each group were sacrificed on 3 dpi, and their brains were collected for further study. For pathogenicity analysis, three brains was split in half. The right hemisphere was collected in 1 ml of PBS supplemented with antibiotics (100 U/ml penicillin-streptomycin), and the left hemisphere was placed in formalin. For transcriptomic analysis, three other brains were harvested and stored in TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The remaining five mice in each group were observed daily for the signs of disease and death for 14 days postinfection (dpi).
Immunohistochemistry (IHC).
The fixed brains were paraffin embedded, sectioned, and stained with a rabbit anti-NP-specific monoclonal antibody (GeneTex). Goat anti-rabbit immunoglobulin conjugated to peroxidase (Maxim Bio, Fujian, China) was used as the secondary antibody. The sections were screened using an Olympus BX51 microscope coupled to a camera.
Plasmid construction.
The IFN-β-luc, RL-TK, RIG-I, MDA-5, TBK-1, IκB kinase ε (IKKε), IFN regulatory factor (IRF3), IRF3-5D, and pUb-HA plasmids were provided kindly by Zhengfan Jiang (Peking University) (4). PGRMC1 was cloned in the p3×Flag-CMV-14 vector. A lentiviral vector encoding EGFP-Cas9, the packaging plasmid pMD2.G, and the envelope plasmid psPAX2 were provided kindly by Lisheng Zhang (Huazhong Agricultural University). The PGRMC1 guide RNA was cloned in the lentiviral vector. Plasmids expressing Flag-RIG-I were amplified from HA-RIG-I and cloned into p3FLAG-CMV-14 vectors.
RNA sequencing and gene expression analysis.
Total RNA was isolated and purified using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s procedure and was dispatched to the LC-Bio Corporation (Hangzhou, Zhejiang, China) for microarray analysis. The reads were assembled using StringTie. Next, transcriptomes were merged to reconstruct a comprehensive transcriptome using perl scripts. In this study, StringTie was used to determine the expression level of mRNAs by calculating fragments per kilobase per million (FPKM). The differentially expressed genes were selected with log2 (fold change) of >1 or log2 (fold change) of <−1 and with statistical significance (P value of <0.05) by using R package Ballgown.
RNA extraction and reverse transcription-quantitative PCR (qRT-PCR).
Total RNA was extracted from brains or cells using TRIzol according to the manufacturer’s instructions. Next, the mixed genomic DNA was digested with DNase I and 1-μg RNA was reverse transcribed using the reverse transcriptase (TaKaRa Biotechnology, Dalian, China) from avian myeloblastosis virus with oligonucleotide (dT) 18. cDNA (0.5 μl) was used as the template for qRT-PCR. In addition, the reaction mixtures contained 5-μl 2× SYBR green master mix (Roche, Indianapolis, IN, USA), 0.25 μl of each primer (10 mM), and 4-μl ultrapure water. The assay was run on an ABIViiA 7 PCR system (Applied Biosystems, Waltham, MA, USA). The expression of each gene was normalized to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a control.
Luciferase reporter assays.
HEK293T cells cultured in 24-well plates were incubated with the PGRMC1 inhibitor AG205 (Sigma-Aldrich) or dimethyl sulfoxide (DMSO) for 24 h. The cells were transfected with IFN-β-luc (200 ng/well) and the internal control pRL-TK (10 ng/well). After 24 h, the cells were transfected with poly(I·C) (200 ng/ml) and incubated for 12 h, followed by analysis of cell lysates for luciferase activity with a dual luciferase reporter assay kit (Promega) according to the manufacturer’s instructions. HEK293T cells seeded in 24-well plates were cotransfected with the expression plasmid harboring PGRMC1 or the empty vector IFN-β-luc (200 ng/well) and the internal control pRL-TK (10 ng/well), together with the adaptor molecules (RIG-I, MDA-5, TBK-1, IKKε, IRF3, and IRF3-5D) of each signaling pathway. Luciferase activity was measured as mentioned above at 24 h posttransfection.
Western blotting.
The cells were washed with cold PBS and lysed on ice in Tris lysis buffer (Cell Signaling Technology, Danvers, MA, USA) containing 1% EDTA-free protease inhibitors (Roche). To remove cell debris, the cell lysates were centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was resuspended in 1× sodium dodecyl sulfate (SDS) loading buffer, was resolved using SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes (GE). The membrane was first probed using certain primary antibodies and then with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit secondary antibodies (Abcam; Cambridge, MA). Finally, a chemiluminescence imaging system (DNR, Neve Yamin, Israel) was used for the analysis, and images were acquired.
Generation of PGRMC1 knockout U251 cells.
PGRMC1-KO U251 cells were generated using the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system and lentiviral vectors as described previously (34). The single guide RNA sequence targeting the human PGRMC1 (5′-GCTCTACAAGATCGTGCGCG-3′) was cloned in the BsmBI site of the EGFP-Cas9 lentiviral vector. The EGFP-Cas9 lentiviral vector psPAX2 and pMD2.G were cotransfected in 293T cells. The cell supernatant was harvest 48 h posttransfection. Then, U251 cells were incubated with the viral supernatant or the empty vector lentivirus. Finally, the monoclonal cells were sorted using flow cytometry and seeded in a 96-well plate to generate clonal cell lines. PGRMC1 knockout was confirmed via sequencing and Western blotting.
Coimmunoprecipitation (co-IP) assay.
Wild-type (WT) or PGRMC1-KO U251 cells were transfected with pCAGGS-HA-Ub together with p3×Flag-RIG-I. At 48 h posttransfection, the cells were lysed with buffer for Western blotting and IP (Beyotime, Beijing, China) at 4°C for 30 min. The supernatants were collected and incubated overnight with anti-HA magnetic beads (Bimake, Beijing, China) at 4°C. Then, the beads were washed thrice with lysis buffer and eluted with 1× SDS loading buffer. Samples were tested using Western blotting.
H5N6 virus replication in WT and PGRMC1 KO mice.
The PGRMC1 KO mice were made by GemPharmatech Inc. by using the CRISPR-Cas9 technique. The gRNA sequences to deplete PGRMC1 are as follows: GGCGCTTAGGTGGCCGCGAA (gRNA1) and GGGCCCCCTATGTAGCCTCG (gRNA2). The 4,525-bp DNA fragment was depleted by these two gRNAs, resulting in a frameshift for the rest of the cDNA. For routine PCR genotyping of PGRMC1 KO mice, the following primer pair were designed to amplify a 414-bp (F1/R1) PCR product from wild-type mice and a 394-bp (F2/R2) product from mutant alleles: F1, CCTCGTCATCTGGTTGTATTTGC; R1, GCTAGCATCTACGACTGTGCTGTAC; F2, TGTCTGCGAGCTGCACTATGATC; and R2, TGCAGACACCATTGTTTGCTG.
C57BL/6J WT and PGRMC1 KO female mice (n = 5) aged 4 to 6 weeks were infected intranasally with 50 μl of 105 TCID50. Body weights were monitored daily for 5 days, and three mice in each group were euthanized at 5 dpi. Lungs and brains were collected for further studies. For lungs or brains, the right half was collected in 1-ml PBS supplemented with antibiotics (100 U/ml penicillin-streptomycin) and the left was placed in formalin for histopathology. Viral RNA copies in lungs and brains were determined with qRT-PCR.
Laboratory facility.
All experiments involving live viruses were performed in a biosafety level 3 (BSL3) facility in accordance with the institutional biosafety manual. The animals were housed in negative-pressure isolators with high-efficiency particulate air filters in the BSL3 facility.
Statistical analyses.
The results are expressed as means ± standard deviations (SD). Data were collected from triplicate experiments and are representative of at least three independent experiments. Groups were compared using one-way analysis of variance (ANOVA) in GraphPad Prism (San Diego, CA, USA). The level of significance was set at a P value of <0.05. Two-way ANOVA was used when appropriate. Mouse survival was compared using the Mantel-Cox version of the log-rank test.
Data availability.
The transcriptome data files were uploaded into the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) (BioProject identifier [ID] PRJNA720971).
ACKNOWLEDGMENTS
We thank the LC-Bio Corporation for assistance with RNA sequencing and gene expression analysis. We also thank Zhengfan Jiang from Peking University for providing the luciferase reporter constructs, including the IFNB promoter (IFNB-luc). We thank Min Cui for offering lots of advice and help. We thank all the members of the M.J. laboratory for helpful feedback.
This research was supported by National Natural Science Foundation of China (31820103015 and 31702255).
Footnotes
Supplemental material is available online only.
Contributor Information
Zhong Zou, Email: zz19841024@126.com.
Meilin Jin, Email: jml8328@126.com.
Abimbola O. Kolawole, Wright State University
REFERENCES
- 1.Jiao P, Song H, Liu X, Song Y, Cui J, Wu S, Ye J, Qu N, Zhang T, Liao M. 2016. Pathogenicity, transmission and antigenic variation of H5N1 highly pathogenic avian influenza viruses. Front Microbiol 7:635. doi: 10.3389/fmicb.2016.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding X, Luo J, Quan L, Wu A, Jiang T. 2017. Evolutionary genotypes of influenza A (H7N9) viruses over five epidemic waves in China. Infect Genet Evol 55:269–276. doi: 10.1016/j.meegid.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 3.Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 4.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. 2009. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavares LP, Teixeira MM, Garcia CC. 2017. The inflammatory response triggered by Influenza virus: a two edged sword. Inflamm Res 66:283–302. doi: 10.1007/s00011-016-0996-0. [DOI] [PubMed] [Google Scholar]
- 7.Shah M, Anwar MA, Kim JH, Choi S. 2016. Advances in antiviral therapies targeting Toll-like receptors. Expert Opin Invest Drugs 25:437–453. doi: 10.1517/13543784.2016.1154040. [DOI] [PubMed] [Google Scholar]
- 8.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. 2011. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog 7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Short KR, Veeris R, Leijten LM, van den Brand JM, Jong VL, Stittelaar K, Osterhaus A, Andeweg A, van Riel D. 2017. Proinflammatory cytokine responses in extra-respiratory tissues during severe influenza. J Infect Dis 216:829–833. doi: 10.1093/infdis/jix281. [DOI] [PubMed] [Google Scholar]
- 11.Toovey S. 2008. Influenza-associated central nervous system dysfunction: a literature review. Travel Med Infect Dis 6:114–124. doi: 10.1016/j.tmaid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Holtenius J, Gillman A. 2014. The Spanish flu in Uppsala, clinical and epidemiological impact of the influenza pandemic 1918–1919 on a Swedish county. Infect Ecol Epidemiol 4:1. doi: 10.3402/iee.v4.21528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheng ZM, Chertow DS, Ambroggio X, McCall S, Przygodzki RM, Cunningham RE, Maximova OA, Kash JC, Morens DM, Taubenberger JK. 2011. Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci USA 108:16416–16421. doi: 10.1073/pnas.1111179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korteweg C, Gu J. 2008. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am J Pathol 172:1155–1170. doi: 10.2353/ajpath.2008.070791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegers JY, van de Bildt MWG, Lin Z, Leijten LM, Lavrijssen RAM, Bestebroer T, Spronken MIJ, De Zeeuw CI, Gao Z, Schrauwen EJA, Kuiken T, van Riel D. 2019. Viral factors important for efficient replication of influenza A viruses in cells of the central nervous system. J Virol 93:e02273-18. doi: 10.1128/JVI.02273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leyva-Grado VH, Churchill L, Harding J, Krueger JM. 2010. The olfactory nerve has a role in the body temperature and brain cytokine responses to influenza virus. Brain Behav Immun 24:281–288. doi: 10.1016/j.bbi.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuiken T, Taubenberger JK. 2008. Pathology of human influenza revisited. Vaccine 26 :D59–D66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mifsud W, Bateman A. 2002. Membrane-bound progesterone receptors contain a cytochrome b5-like ligand-binding domain. Genome Biol 3:research0068.1. doi: 10.1186/gb-2002-3-12-research0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hand RA, Craven RJ. 2003. Hpr6.6 protein mediates cell death from oxidative damage in MCF-7 human breast cancer cells. J Cell Biochem 90:534–547. doi: 10.1002/jcb.10648. [DOI] [PubMed] [Google Scholar]
- 20.Teakel SL, Ludescher M, Thejer BM, Poschmann G, Forwood JK, Neubauer H, Cahill MA. 2020. Protein complexes including PGRMC1 and actin-associated proteins are disrupted by AG-205. Biochem Biophys Res Commun 524:64–69. doi: 10.1016/j.bbrc.2019.12.108. [DOI] [PubMed] [Google Scholar]
- 21.Nolte I, Jeckel D, Wieland FT, Sohn K. 2000. Localization and topology of ratp28, a member of a novel family of putative steroid-binding proteins. Biochim Biophys Acta 1543:123–130. doi: 10.1016/s0167-4838(00)00188-6. [DOI] [PubMed] [Google Scholar]
- 22.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA 101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura I, Yoshioka M, Konishi M, Miyake A, Itoh N. 2005. Neudesin, a novel secreted protein with a unique primary structure and neurotrophic activity. J Neurosci Res 79:287–294. doi: 10.1002/jnr.20356. [DOI] [PubMed] [Google Scholar]
- 24.Kimura I, Konishi M, Miyake A, Fujimoto M, Itoh N. 2006. Neudesin, a secreted factor, promotes neural cell proliferation and neuronal differentiation in mouse neural precursor cells. J Neurosci Res 83:1415–1424. doi: 10.1002/jnr.20849. [DOI] [PubMed] [Google Scholar]
- 25.Biswas S, Cottarelli A, Agalliu D. 2020. Neuronal and glial regulation of CNS angiogenesis and barriergenesis. Development 147:dev182279. doi: 10.1242/dev.182279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 27.Hosseini S, Wilk E, Michaelsen-Preusse K, Gerhauser I, Baumgartner W, Geffers R, Schughart K, Korte M. 2018. Long-term neuroinflammation induced by influenza A virus infection and the impact on hippocampal neuron morphology and function. J Neurosci 38:3060–3080. doi: 10.1523/JNEUROSCI.1740-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu J, Xie Z, Gao Z, Liu J, Korteweg C, Ye J, Lau LT, Lu J, Gao Z, Zhang B, McNutt MA, Lu M, Anderson VM, Gong E, Yu AC, Lipkin WI. 2007. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet 370:1137–1145. doi: 10.1016/S0140-6736(07)61515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao D, Liang L, Li Y, Liu L, Guan Y, Jiang Y, Chen H. 2012. Proteomic analysis of the lungs of mice infected with different pathotypes of H5N1 avian influenza viruses. Proteomics 12:1970–1982. doi: 10.1002/pmic.201100619. [DOI] [PubMed] [Google Scholar]
- 30.Bordner KA, Kitchen RR, Carlyle B, George ED, Mahajan MC, Mane SM, Taylor JR, Simen AA. 2011. Parallel declines in cognition, motivation, and locomotion in aging mice: association with immune gene upregulation in the medial prefrontal cortex. Exp Gerontol 46:643–659. doi: 10.1016/j.exger.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho H, Proll SC, Szretter KJ, Katze MG, Gale M, Jr, Diamond MS. 2013. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat Med 19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ampomah PB, Lim LHK. 2020. Influenza A virus-induced apoptosis and virus propagation. Apoptosis 25:1–11. doi: 10.1007/s10495-019-01575-3. [DOI] [PubMed] [Google Scholar]
- 33.Terzaghi L, Luciano AM, Dall'Acqua PC, Modina SC, Peluso JJ, Lodde V. 2018. PGRMC1 localization and putative function in the nucleolus of bovine granulosa cells and oocytes. Reproduction 155:273–282. doi: 10.1530/REP-17-0534. [DOI] [PubMed] [Google Scholar]
- 34.Zhao L, Zhang X, Wu Z, Huang K, Sun X, Chen H, Jin M. 2019. The downregulation of microRNA hsa-miR-340-5p in IAV-infected A549 cells suppresses viral replication by targeting RIG-I and OAS2. Mol Ther Nucleic Acids 14:509–519. doi: 10.1016/j.omtn.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00734-21_Supp_1_seq2.pdf, PDF file, 0.1 MB (142.3KB, pdf)
Data Availability Statement
The transcriptome data files were uploaded into the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) (BioProject identifier [ID] PRJNA720971).