ABSTRACT
Protein phosphorylation is involved in several key biological roles in the complex life cycle of Trypanosoma cruzi, the etiological agent of Chagas disease, and protein kinases are potential drug targets. Here, we report that the AGC essential kinase 1 (TcAEK1) exhibits a cytosolic localization and a higher level of expression in the replicative stages of the parasite. A CRISPR/Cas9 editing technique was used to generate ATP analog-sensitive TcAEK1 gatekeeper residue mutants that were selectively and acutely inhibited by bumped kinase inhibitors (BKIs). Analysis of a single allele deletion cell line (TcAEK1-SKO), and gatekeeper mutants upon treatment with inhibitor, showed that epimastigote forms exhibited a severe defect in cytokinesis. Moreover, we also demonstrated that TcAEK1 is essential for epimastigote proliferation, trypomastigote host cell invasion, and amastigote replication. We suggest that TcAEK1 is a pleiotropic player involved in cytokinesis regulation in T. cruzi and thus validate TcAEK1 as a drug target for further exploration. The gene editing strategy we applied to construct the ATP analog-sensitive enzyme could be appropriate for the study of other proteins of the T. cruzi kinome.
IMPORTANCE Chagas disease affects 6 to 7 million people in the Americas, and its treatment has been limited to drugs with relatively high toxicity and low efficacy in the chronic phase of the infection. New validated targets are needed to combat this disease. In this work, we report the chemical and genetic validation of the protein kinase AEK1, which is essential for cytokinesis and infectivity, using a novel gene editing strategy.
KEYWORDS: CRISPR/Cas9-based mutagenesis, AGC kinase, ATP analog-sensitive, Trypanosoma cruzi, cytokinesis
INTRODUCTION
Trypanosoma cruzi, the causative agent of Chagas disease, is a parasitic protist that belongs to the order Kinetoplastida and exhibits a complex life cycle. This parasite has four major developmental stages, epimastigotes and metacyclic trypomastigotes in the insect vector and cell-derived trypomastigotes and amastigotes in the mammalian host, with two replicative stages: the epimastigote and amastigote forms.
Chagas disease affects 6 to 7 million people in the Americas, with over 10,000 deaths annually (https://www.who.int/chagas/en/), and although it is endemic in Latin America, migration has contributed to the globalization of the disease, with more than 300,000 infected people currently residing in the United States (1). There is no vaccine available and, for decades, the treatment of Chagas disease has been limited to two drugs (benznidazole and nifurtimox) with relatively high toxicity and low efficacy for the chronic phase of the infection, in which most of the patients are diagnosed (2–4). Moreover, natural drug resistance has been reported (5, 6).
The approaches to generate new treatments for Chagas disease involve new schemes or strategies for the administration of the available drugs to prevent side effects or searching for new therapeutic candidates to replace them (7). In humans, protein kinases have been a significant focus of drug discovery efforts due to their relevance in cancer, diabetes, inflammation, and neurodegeneration (8). Kinases represent almost 30% of the “druggable human genome,” and approximately 20 protein kinases in the human kinome are targets of FDA-approved inhibitors (8, 9).
Protein phosphorylation plays key roles in intracellular signal transduction pathways and in regulation of many biological processes such as the cell cycle, cell growth, and apoptosis. Phosphoproteomic studies indicate that T. cruzi protein phosphorylation is likely to have an important role in the regulation of the cell cycle, differentiation, metabolism, and survival (10–14). Therefore, protein kinases constitute an attractive class of molecular targets for drug discovery for the treatment of T. cruzi infection.
T. cruzi has approximately 190 eukaryotic protein kinases divided in six groups and an additional group classified as “other” (15). The AGC group of protein kinases, so named for its members cAMP-dependent protein kinase (PKA), cGMP-dependent protein kinase (PKG), and protein kinase C (PKC), includes more than 60 evolutionary related serine/threonine protein kinases in humans (16) and has been considered relatively underrepresented in trypanosomatid genomes. Thirteen genes encoding protein kinases belonging to the AGC group can be found in the T. cruzi genome, which represent about 7% of the kinases of T. cruzi (15, 17), although normalizing to the size of genome, trypanosomatids have about half the AGC kinases of humans (15). Some trypanosomatid AGC kinases, like AEK1 (named AGC essential kinase 1) (18), cannot be directly assigned to the AGC subfamilies conserved in higher organisms. AEK1 orthologs in Trypanosoma brucei and Leishmania mexicana, the causative agents of human African trypanosomiasis and cutaneous form of leishmaniasis, respectively, have been described as probably essential in large RNA interference (RNAi) and CRISPR/Cas9 screenings (19–21). Using conditional knockouts, Jensen et al. (18) confirmed that AEK1 is essential in T. brucei bloodstream forms in vitro and in vivo. However, no studies have been reported on AEK1 from T. cruzi.
Most Ser/Thr kinases have a large hydrophobic residue, named the gatekeeper residue, that limits the access to the ATP-binding pocket. This highly conserved amino acid (methionine, leucine, phenylalanine, threonine, etc.) can be mutated, naturally or artificially, to a small residue such as glycine or alanine, creating an extended space within the ATP-binding site (22). The resulting kinase is known as ATP analog-sensitive because the small gatekeeper residue allows that ATP analogs containing a bulky substituent (termed bumped kinase inhibitors or BKIs) complement the shape of the mutant ATP pocket and potentially and specifically inhibit the kinase (23–25). In addition, the ATP analog-sensitive approach has been used in T. brucei to perform functional studies with endogenously expressed small gatekeeper mutants of Polo-like kinase and CRK9 and with conditional knockouts (KOs) of TbAEK1 (18, 26, 27). To our knowledge, this methodology has not yet been applied to study T. cruzi protein kinases.
In this work, we demonstrate the cytosolic localization of TcAEK1 and the expression levels at different stages of the parasite’s life cycle by analyzing endogenously tagged and overexpressing TcAEK1 strains. We also generated single knockout mutants (TcAEK1-SKO) and used a CRISPR/Cas9-based editing strategy to substitute the TcAEK1 gatekeeper residue and generate mutants sensitive to known ATP-competitive small molecule inhibitors. ATP analog-sensitive kinase technology has been used as a tool to validate drug targets when no specific inhibitors were available (24). We demonstrate that TcAEK1 is essential for epimastigote proliferation, trypomastigotes host cell invasion, and amastigote replication. We also show that downregulation or inhibition of TcAEK1 causes a severe impairment of cytokinesis in the replicative epimastigote form. The engineered TcAEK1 gatekeeper mutants allowed us to confirm results obtained with TcAEK1-SKO cells and to validate TcAEK1 as a drug target.
RESULTS
Expression and localization of the AGC kinase TcAEK1.
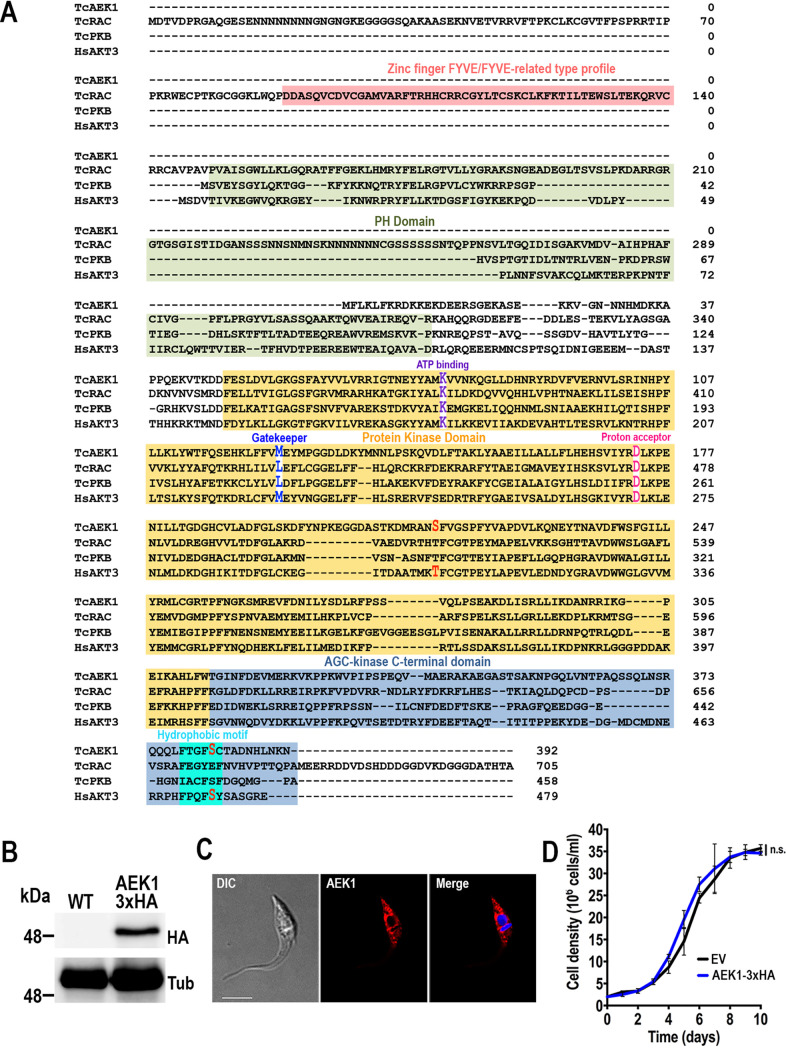
AEK1 has orthologs in all kinetoplastid species that were found in genome databases. The AEK1 ortholog of T. cruzi Y strain was found in the TriTryp genome database (http://tritrypdb.org/tritrypdb/, ID: TcYC6_0120630), and it is a single-copy gene composed of 1,179 nucleotides encoding 392 amino acids (∼44.8 kDa). TcAEK1 shares amino acid identity from 68% to 76% (84% to 87% similarity) with the orthologs found in T. brucei and Bodo saltans, respectively. Additionally, full-length TcAEK1 shows 33% identity (58.2% similarity) with human AKT3 (RAC-gamma serine/threonine kinase), reaching 40.7% (65.5% similarity) within the catalytic domain. Interestingly, the annotated ortholog in T. cruzi CL Brener strain genome (Trytrypdb: TcCLB.508479.150) is longer (1,425 nucleotides [nt]) than those found in other T. cruzi strains or kinetoplastid organisms. The open reading frame (ORF) of TcAEK1 in Y strain starts ∼250 nt downstream of that in CL Brener strain (Fig. S1A). The apparent loss of reading frame in that region occurs in areas of polypyrimidines, and in the absence of confirmation of the molecular weight of the AEK1 protein in CL Brener, the longer N terminus of this strain is likely an annotation error. TcAEK1 shares common features with other members of the AGC protein kinase family, such as the catalytic core and the C-terminal domain, which includes the hydrophobic motif (Fig. 1A). TcAEK1 is not predicted to have a Pleckstrin homology domain (PH) or other N-terminal domain involved in binding biological membranes. Putative phosphorylation sites, which are conserved in AGC kinase proteins to regulate their function, are also predicted in TcAEK1: the first is located in the kinase domain at the “activation loop” (S216, corresponding to HsAKT3 T305), and the second is located in the AGC kinase C-terminal domain (S382, corresponding to HsAKT3 S472) (Fig. 1A).
FIG 1.
TcAEK1 structure and localization. (A) Amino acid sequence alignment of AGC kinase subfamily members from T. cruzi, TcAEK1, TcRAC, and TcPKB (TriTrypDB IDs: TcYC6_0120630, TcYC6_0106530, TcYC6_0095770, respectively), and human HsAKT3 (GenBank accession number: NP_001357003). Important conserved domains and residues are highlighted. In red are putative conserved phosphorylated amino acid residues. PH, Pleckstrin homology domain. (B) TcAEK1-3×HA (AEK1-3×HA) overexpression was confirmed by Western blotting using anti-HA antibodies. Cells transfected with pTREX-n empty vector (EV) were used as the control cell line. Tubulin (Tub) was used as the loading control. (C) IFA showed subcellular localization of TcAEK1-3×HA detected with anti-HA antibodies (AEK1 [red]) in T. cruzi epimastigotes. Merge of red signal and DAPI staining (blue) and DIC (differential interference contrast) images is also shown. Scale bar is 5 μm. (D) Growth of control (EV) and TcAEK1 (AEK1-3×HA) epimastigotes in LIT medium. No significant (n.s.) differences in growth rates were found using one-way ANOVA with multiple comparisons (n = 3).
To determine TcAEK1 cellular localization, we generated a cell line overexpressing the full-length C-terminal triple hemagglutinin (3×HA)-tagged protein (TcAEK1-3×HA). Western blotting of epimastigote extracts using anti-HA antibodies showed a major protein band of ∼48 kDa, which is compatible with the predicted mass of the full-length protein plus the 3×HA tag (Fig. 1B). Fluorescence microscopy images showed a cytosolic subcellular localization of the overexpressed-tagged protein. After image deconvolution, TcAEK1 signals exhibited a punctate pattern with an apparent concentration near the nucleus and kinetoplast (Fig. 1C). TcAEK1-3×HA had the same growth rate as control cells transfected with the pTREX-n empty vector (Fig. 1D).
TcAEK1 expression level is higher in the replicative forms of T. cruzi.
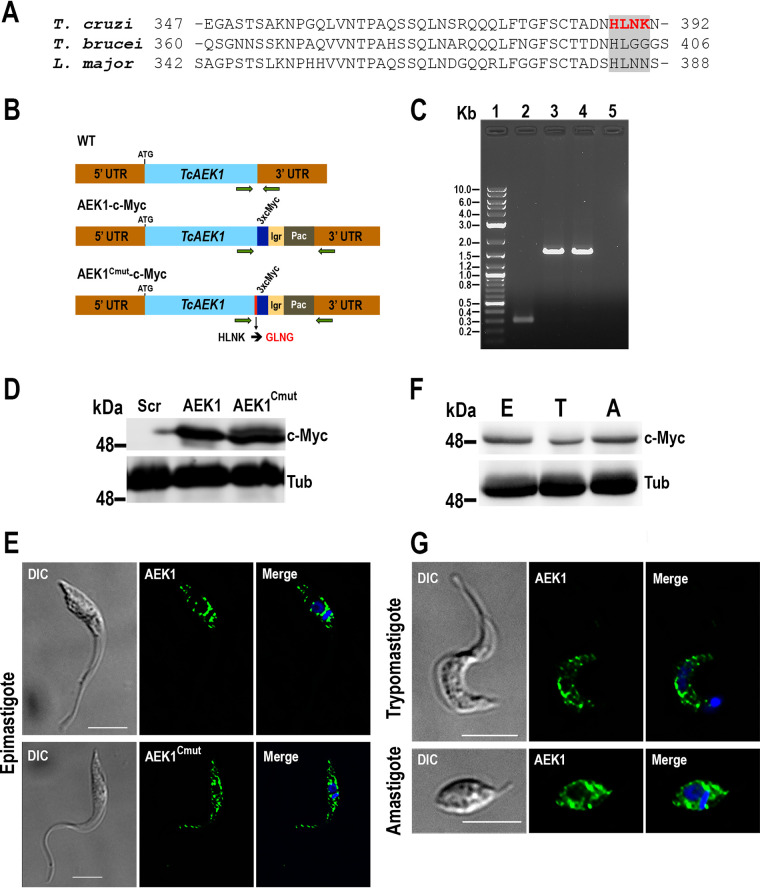
The WoLF PSORT tool predicted that the HLNK residues at the C terminus of TcAEK1 could be an endoplasmic reticulum (ER) membrane retention signal: KKXX-like (28) (Fig. 2A). Interestingly, this ER membrane retention signal was not predicted in orthologs from other kinetoplastid organisms. To examine this prediction, we used CRISPR/Cas9 gene editing to generate two endogenously tagged mutant cell lines: one wild-type TcAEK1 (TcAEK1-3xc-Myc) and one with a mutated C terminus (TcAEK1Cmut-3xc-Myc) (Fig. 2B). TcAEK1 tagging was confirmed by PCR (Fig. 2C) and also by Western blotting using anti-c-Myc antibodies (Fig. 2D). As shown in the immunofluorescence analysis (IFA) images in Figure 2E, both tagged proteins exhibited subcellular localization pattern similar to that of epimastigotes with the overexpressed TcAEK1 (Fig. 1C). TcAEK1Cmut-3xc-Myc epimastigotes did not show a growth defect (not shown). This result suggests that the predicted ER membrane retention signal does not determine the localization of TcAEK1. Furthermore, antibodies against ER (TbBIP) and reservosome (TcCruzipain) markers did not colocalize with TcAEK1 (Fig. S1B and C). Finally, to rule out the possibility that C-terminal tagging (overexpressed or endogenous) affected TcAEK1 localization (i.e., masking the KKXX-like motif), we generated an overexpression cell line with an internal N-terminal Flag tag (TcAEK1-Flag) (Fig. S1D and E), which exhibited cellular localization in epimastigote forms similar to that observed with the endogenously C-terminal TcAEK1-tagged versions.
FIG 2.
Analysis of expression of endogenously tagged TcAEK1-3xc-Myc. (A) Amino acid sequence alignment of the C-terminal region of AEK1 orthologs from T. cruzi, T. brucei, and L. major (TriTrypDB IDs: TcYC6_0120630, Tbg972.3.2430, and LmjF.25.2340, respectively). Enclosed in a gray box, a putative ER retention signal is highlighted in red text (T. cruzi): KKXX-like motif in the C terminus (HLNK) predicted by WoLF PSORT method (https://wolfpsort.hgc.jp/). (B) Schematic representation of TcAEK1 (WT), endogenously tagged TcAEK1-3xc-Myc (AEK1-c-Myc), and TcAEK1Cmut-3xc-Myc, which has the KKXX-like motif mutated from HLNK to GLNG amino acid residues (AEK1Cmut-c-Myc). UTR, untranslated region; Igr, tubulin intergenic region; Pac, puromycin N-acetyltransferase. Horizontal green arrows indicate primers used for tagging verification. (C) PCR verification of integration of DNA donor in the 3′ end of the endogenous TcAEK1 locus. TcAEK1 allele size: WT, 300 bp; 3xc-Myc-tagged, 1,581 bp. Lanes: 1, 1-kb plus ladder; 2, scrambled control; 3, TcAEK1-3xc-Myc; 4, TcAEK1Cmut-3xc-Myc; 5, PCR negative control. (D) TcAEK1 endogenous tagging was verified by Western blotting using anti-c-Myc antibodies (c-Myc). Tubulin (Tub) was used as a loading control. Scr, scrambled; AEK1, TcAEK1-3xc-Myc; AEK1Cmut, TcAEK1Cmut-3xc-Myc. (E) Immunofluorescence analysis (IFA) showed localization of endogenously tagged TcAEK1-3xc-Myc (upper panel) and TcAEK1Cmut-3xc-Myc (lower panel) epimastigotes detected with anti-c-Myc antibodies (green). Merge of green signal (AEK1) and DAPI staining (blue) and DIC images are also shown. Scale bars represent 5 μm. (F) Representative Western blotting of TcAEK1-3xc-Myc in total extracts of epimastigotes (column E), tissue culture cell-derived trypomastigotes (column T), and amastigotes (column A) using anti-c-Myc antibodies. Tubulin was used as the loading control. (G) IFA shows localization of endogenously tagged TcAEK1-3xc-Myc detected with anti-c-Myc antibodies (green) in trypomastigote and amastigote. Merge of green signal (AEK1) and DAPI staining (blue) and DIC images are also shown. Scale bars represent 5 μm.
We also used the endogenously tagged TcAEK1-3xc-Myc cell line to study the expression pattern of TcAEK1 in the other two developmental stages maintained under laboratory conditions. Initially, TcAEK1-3xc-Myc was detected in cell extracts of epimastigotes, cell-derived trypomastigotes, and amastigotes by Western blotting using anti-c-Myc antibodies (Fig. 2F). As was observed in epimastigotes (Fig. 2E), TcAEK1-3xc-Myc detected with anti-c-Myc antibodies was distributed as punctate pattern and larger aggregates throughout the cytoplasm but concentrated near the nucleus and kinetoplast, in both cell-derived trypomastigotes and amastigotes (Fig. 2G). Therefore, our results confirm the expression of TcAEK1 protein in the three main stages of the T. cruzi life cycle. Densitometry analyses of Western blot experiments, using anti-c-Myc antibodies (Fig. S2), normalized to tubulin protein levels suggest that endogenous TcAEK1 protein levels in cell-derived trypomastigotes were lower than those found in epimastigotes or amastigotes.
TcAEK1 knockdown affects growth of epimastigotes and amastigotes and trypomastigote invasion.
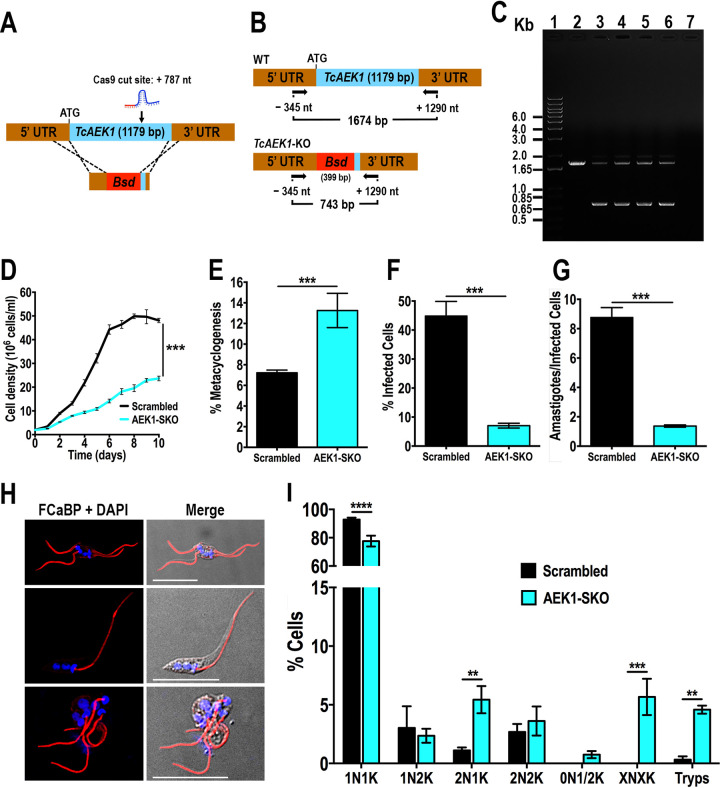
To explore the role of TcAEK1, we designed a CRISPR/Cas9 strategy to generate knockout mutants of this gene (Fig. 3A). The method, which has been successfully adapted recently to T. cruzi (29–33), involves the constitutive expression of Cas9 and specific single guide RNA (sgRNA) and the utilization of a template cassette to promote double-strand break repair by homologous-directed repair (HDR). After several attempts cotransfecting epimastigotes with the sgRNA/Cas9-harboring plasmid (using two different sgRNA separately) together with a DNA donor, we were unable to obtain drug-resistant stable transfectants. Therefore, we followed a two-step strategy: first, we obtained a cell line that constitutively expresses Cas9 and TcAEK1-sgRNA-787, and later, these epimastigotes were further transfected with the blasticidin resistance cassette. Subsequently, we obtained clonal populations from stable transfectants by limiting dilution. We confirmed by PCR that only one TcAEK1 allele was replaced by the DNA donor cassette at the specific loci (Fig. 3B and C) in all clonal populations obtained. Another attempt following the same approach but using a different DNA donor cassette also resulted in a single TcAEK1 allele eliminated (Fig. S3A to C).
FIG 3.
Generation and characterization of TcAEK1-SKO cells. (A) Schematic representation of the strategy designed to generate a TcAEK1-KO mutant by CRISPR/Cas9-induced homologous recombination. Cas9 introduced a DNA double-strand break at nt +787 of the TcAEK1 ORF (1,179 bp). DNA was repaired with a blasticidin S-deaminase (Bsd) cassette containing 100-bp homologous regions spanning from nt −342 to −243 and from nt +1,094 to +1,190 of the TcAEK1 locus. (B) PCR primers used to verify TcAEK1 ablation. Arrows indicate primers added to the reaction. The intact locus generates a PCR product of 1,674 bp, while the disrupted locus generates a DNA fragment of 743 bp. UTR, untranslated region. (C) Only one TcAEK1 allele was disrupted at its genomic locus in the SKO cell lines. Lanes: 1, 1-kb plus ladder; 2, WT; 3, TcAEK1-SKO G418/blasticidin resistant mixed population; 4, TcAEK1-SKO clone A3; 5, TcAEK1-SKO clone A6; 6, TcAEK1-SKO clone B3; 7, PCR negative control. (D) Growth of control (scrambled) and TcAEK1-SKO (clone A6) epimastigotes in LIT medium. Student’s t test was applied to growth rates calculated from each growth curve (n = 3; ***, P < 0.001). (E) Percentage of metacyclic trypomastigotes in epimastigote cultures after incubation in TAU 3AAG medium. Scrambled and TcAEK1-SKO (clone A6) epimastigote differentiation to metacyclic trypomastigotes was quantified by staining with DAPI to distinguish the position of the kinetoplast by fluorescence microscopy. (F and G) Scrambled and TcAEK1-SKO (clone A6) trypomastigote infection of Vero cells. There were significant differences in the percentage of infected Vero cells at 4 h postinfection (F) as well as in the number of intracellular amastigotes per infected host cell observed 48 h postinfection (G) by TcAEK1-SKO mutant cells. In panels E, F, and G, values are means ± SD (n = 3), ***, P < 0.001 by Student’s t test. (H) IFA of representative cytological abnormalities observed in TcAEK1-SKO epimastigote culture. Flagella were detected with monoclonal anti-TcFCaBP antibodies (red), and DAPI was used to stain kinetoplast and nuclear DNA (blue). DIC (differential interference contrast) images are also shown. Scale bars represent 15 μm. (I) Effect of single TcAEK1 gene KO on nucleus/kinetoplast patterns established after DNA labeling with DAPI. At least 200 cells were counted for each analysis. Data represent means, and error bars indicate SD from 3 independent experiments. XNXK, X > 2; Tryps, metacyclic trypomastigotes. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-way ANOVA with Sidak’s multiple-comparison test).
We then followed a third strategy to generate TcAEK1 null mutants, which was designed to disrupt the wild-type (wt) allele with a puromycin resistance cassette in a TcAEK1-SKO clonal population obtained in the second attempt (Fig. S4A). First, we confirmed that the protospacer and protospacer-adjacent motif (PAM) sequence for TcAEK1-sgRNA-787 was intact at the wt allele and that Cas9-GFP was expressed (Fig. S4B and C). After transfection and selection, genomic DNA (gDNA) of G418/blasticidin/puromycin-resistant stable populations were analyzed by PCR, revealing the presence of three bands whose sizes corresponded to those predicted for the wt TcAEK1 allele and those disrupted by the blasticidin and puromycin cassettes (Fig. S3D and E). This result suggests that TcAEK1-SKO cells probably amplified the TcAEK1 locus because null alleles can have lethal effects in epimastigotes.
We therefore studied the effects of the TcAEK1 hemizygous deletion or single knockout (SKO) on proliferation and infectivity of T. cruzi. TcAEK1-SKO epimastigotes exhibited a growth rate in liver infusion tryptose (LIT) medium significantly lower than that of parasites transfected with a scrambled sgRNA (Fig. 3D), providing evidence that AEK1 is important for epimastigote proliferation. Moreover, the ability of TcAEK1-SKO epimastigotes to differentiate in vitro into metacyclic trypomastigotes (metacyclogenesis in triatome artificial urine [TAU] 3AAG medium) was significantly higher compared to that of control cells (Fig. 3E). We also investigated the ability of trypomastigotes to infect tissue-cultured cells. Both invasion of host cells and replication of intracellular amastigotes were significant impaired in TcAEK1-SKO (Fig. 3F and G). These results together with the failure in obtaining TcAEK1 null mutants suggest that this gene is essential in T. cruzi.
TcAEK1-SKO epimastigotes with a severe growth defect were observed under fluorescence microscopy using anti-TcFCaBP antibodies to stain the flagella. Approximately 10 to 15% of TcAEK1-SKO cells were larger and showed morphological alterations, as well as an aberrant number of nuclei/kinetoplasts and flagella (Fig. 3H, see also Fig. S3D). To further examine the growth defect phenotype upon knockdown of TcAEK1, we quantified the number of nuclei and kinetoplasts per cell after DAPI (4′,6-diamidino-2-phenylindole) staining in control and TcAEK1-SKO epimastigotes cultured in LIT medium (Fig. 3I). Nuclear/kinetoplast content (N/K) in T. cruzi epimastigotes progresses during its normal cell cycle from 1N1K to 1N2K to 2N2K (34). The kinetoplast divides before the nucleus, and mitosis of 1N2K yields 2N2K cells that divide during cytokinesis into two 1N1K cells. In normal nonsynchronous, logarithmic-phase epimastigote cultures, most of the cells have 1N1K content (∼80 to 95%) (34). Compared with those of control cells, TcAEK1-SKO epimastigotes stained with DAPI exhibited an increase of binucleated cells, with cell numbers of 2N1K significantly higher. We also observed a significant accumulation of multinucleated cells (XNXK, X > 2), suggesting a defective cytokinesis. Interestingly, TcAEK1-SKO epimastigote cultures in LIT medium exhibited nearly 5% of cells (P < 0.01) with morphology compatible with that of metacyclic trypomastigotes.
Design and analysis of ATP analog-sensitive TcAEK1.
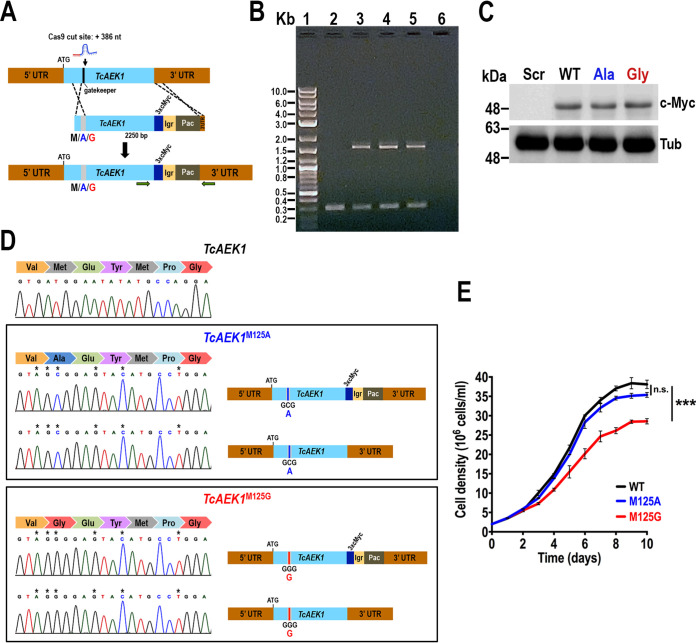
To gain more insight into the TcAEK1 protein function in vivo and to chemically validate it as a potential drug target, we generated TcAEK1 mutants specifically susceptible to ATP analogs that behave as BKIs. We designed a CRISPR/Cas9 mutagenesis strategy for mutation of the gatekeeper residue (methionine 125; M125) to a small residue (alanine or glycine) conferring specific sensitivity to BKIs. The method used consisted of cotransfecting T. cruzi wt epimastigotes with the sgRNA/Cas9-harboring plasmid plus a linear DNA donor (2,250 bp), which promotes HDR and introduces the gatekeeper mutation, a 3xc-Myc-tag to verify the expression of mutated TcAEK1 by Western blotting, and the Pac gene to allow selection of antibiotic-resistant mutants (Fig. 4A). After G418/puromycin selection, we obtained clonal populations for both TcAEK1 gatekeeper residue mutants (TcAEK1M125A and TcAEK1M125G). To confirm the TcAEK1 tagging, we extracted gDNA and performed PCRs with specific primers, and in all clones obtained we detected a single tagged allele (Fig. 4B). Expression of the TcAEK1 tagged allele in the three mutant cell lines (TcAEK1WT, TcAEK1M125A, and TcAEK1M125G) was verified by Western blotting using anti-c-Myc antibodies (Fig. 4C), and Sanger sequencing confirmed that both alleles had gatekeeper residue mutated TcAEK1M125A (two clones) and TcAEK1M125G (one clone), in addition to including the silent mutations introduced in the protospacer sequence (Fig. 4D). Thus, these results indicate that in TcAEK1M125A-3xc-Myc and TcAEK1M125G-3xc-Myc cell lines, the HDR mechanism repaired the TcAEK1 normal allele just in the region close to the Cas9/sgRNA cleavage site, probably using the mutated TcAEK1 allele (tagged) as the DNA donor. TcAEK1WT and TcAEK1M125A epimastigotes exhibited similar growth rates in a rich medium (LIT), whereas TcAEK1M125G revealed a growth rate significantly lower than that of the control (Fig. 4E).
FIG 4.
Generation of analog-sensitive TcAEK1. (A) Schematic representation of CRISPR/Cas9-mediated strategy for TcAEK1 gatekeeper residue knock-in and C-terminal endogenous tagging. Cas9 introduced a DNA double-stranded break at nt +386 of the TcAEK1 ORF. Homologous-directed repair was induced cotransfecting epimastigotes with the DNA donor cassette, containing homologous regions to TcAEK1 (light blue) and to the TcAEK1 3′ UTR (light brown). The HDR determines integration of 3xc-Myc and antibiotic resistance gene (Pac, puromycin N-acetyltransferase) at the 3′ end of TcAEK1, as well as the mutation of the TcAEK1 gatekeeper residue codon sequence, which is included in the protospacer, as well as other silent mutations in the protospacer sequence avoiding being targets of the constitutively expressed Cas9. UTR, untranslated region; Igr, tubulin intergenic region. We used 3 different DNA donor cassettes to maintain the methionine in the wild-type TcAEK1 gatekeeper residue (M125) or to mutate it to the smaller alanine (M125A) or glycine (M125G) residues. M, M125; A, M125A; G, M125G. Horizontal green arrows indicate primers used for tagging verification. (B) PCR verification of cassette integration into TcAEK1 locus. TcAEK1 allele size: WT, 300 bp; 3xc-Myc-tagged, 1,581 bp. Lanes: 1, 1-kb plus ladder; 2, scrambled control; 3, TcAEK1WT-3xc-Myc; 4, TcAEK1M125A-3xc-Myc; 5, TcAEK1M125G-3xc-Myc; 6, PCR negative control. (C) TcAEK1 endogenous tagging in gatekeeper residue mutants was verified by Western blotting using anti-c-Myc antibodies (c-My). Tubulin (Tub) was used as a loading control. Scr, scrambled, WT, TcAEK1WT-3xc-Myc; Ala, TcAEK1M125A-3xc-Myc; Gly, TcAEK1M125G-3xc-Myc. (D) TcAEK1 gatekeeper in situ-mutated nucleotide sequence was confirmed in PCR-amplified wild-type and tagged size DNA fragments. Electropherograms show that, despite that only one allele was tagged at its C terminus, both alleles were mutated in each TcAEK1 gatekeeper mutant cell line: TcAEK1M125A and TcAEK1M125G. Asterisks (*) indicate mutations generated in the protospacer sequence. (E) Growth of control TcAEK1WT (WT), TcAEK1M125A (M125A), and TcAEK1M125G (M125G) epimastigotes in LIT medium. One-way ANOVA with multiple comparisons was applied to growth rates calculated from each growth curve (n = 3, ***, P < 0.001).
Effects of BKIs on morphology and viability of ATP analog-sensitive TcAEK1 gatekeeper mutant epimastigotes and metacyclic trypomastigotes.
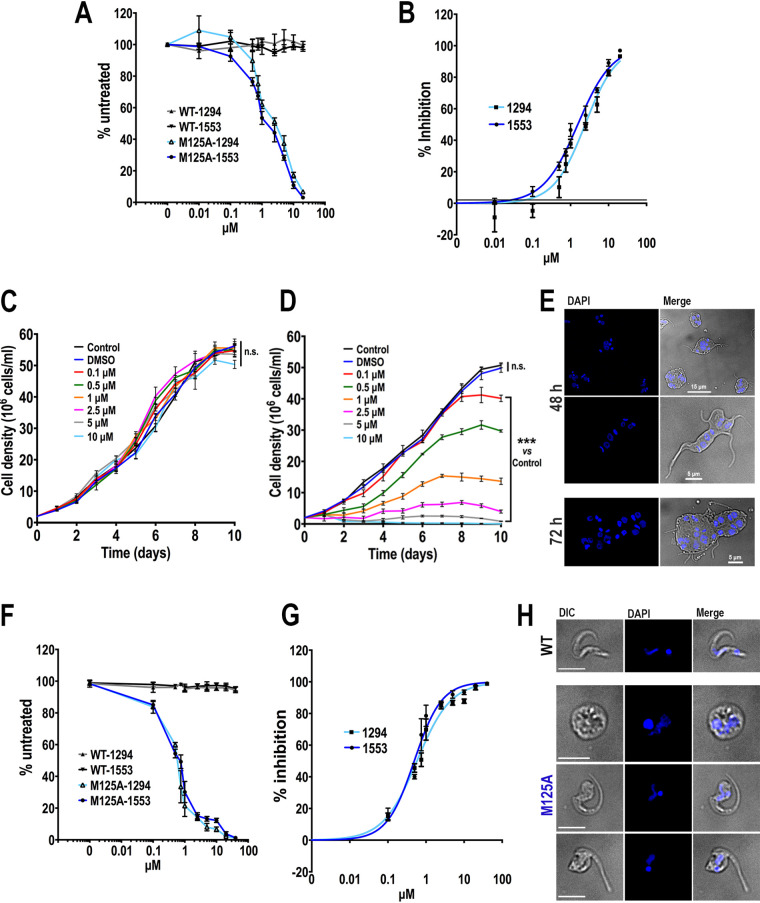
To explore the sensitivity of the ATP analog-sensitive TcAEK1 gatekeeper mutants to BKIs, we performed a pilot experiment growing cells in increasing concentrations of the BKI compounds 1294 and 1553 (Fig. S5A and B). BKI-1294 and its derivative BKI-1553 have been used to specifically inhibit in vitro and in vivo calcium-dependent protein kinase (CDPK) family members in apicomplexan parasites and the ATP analog-sensitive-AEK1 of T. brucei (18, 35–39). Cells were observed under light microscopy after they were incubated for 48 h in LIT medium. Rounded abnormal forms were present in BKI-1294- or BKI-1553-treated cultures of both mutant cell lines TcAEK1M125A (M125A) and TcAEK1M125G (M125G). At high concentrations of BKIs (>5 μM), cells were fragmented in a dose-dependent manner, while cells were morphologically intact in control TcAEK1WT (WT) incubated with BKIs and in M125A and M125G mutants incubated with 0.2% dimethyl sulfoxide (DMSO).
Based on the fact that the effect of the BKIs on both mutants is apparently similar but the TcAEK1M125G mutant grows more slowly, we focused on parasites expressing TcAEK1WT or the TcAEK1M125A mutant. The reduction of alamarBlue by metabolically active cells was used as a viability indicator. As seen in Figure 5A, parasites expressing wt TcAEK1 (WT) were not affected by up to 20 μM either BKI-1294 or BKI-1553. In contrast, survival of parasites expressing TcAEK1M125A mutant (M125A) was greatly decreased in the presence of BKI-1294 or BKI-1553 in a dose-dependent manner starting at 0.5 μM. The 50% inhibitory concentration (IC50)/48 h value of compounds BKI-1294 and BKI-1553 for TcAEK1M125A epimastigotes was 2.42 ± 0.35 μM and 1.55 ± 0.15 μM, respectively (Fig. 5B). BKI-1553 was chosen for further study because of its lower IC50 for TcAEK1M125A epimastigotes. A representative set of growth curves of wt and TcAEK1M125A epimastigotes, in the presence of various concentrations of BKI-1533 (100 nM to 10 μM), is shown in Figure 5C and D, respectively. No differences were found in the growth rates of TcAEK1WT cells in all ranges of concentrations compared with those of epimastigotes cultured without drug (control) (Fig. 5C). On the contrary, TcAEK1M125A cells exhibited a significant dose-dependent decrease of growth rates in the presence of BKI-1553, whereas parasites in the presence of solvent (0.1% DMSO) grew normally (Fig. 5D), confirming the importance of TcAEK1 for epimastigote proliferation. After 48 h in culture with 1.5 μM BKI-1553, DAPI-stained TcAEK1M125A cells were large, pleomorphic in shape, and multinucleated and had multiple attached flagella (Fig. 5E).
FIG 5.
Effect of BKI-1553 on TcAEK1 gatekeeper mutant epimastigotes and metacyclic trypomastigotes. (A) Cell viability of TcAEK1WT (WT) and TcAEK1M125A (M125A) epimastigotes incubated for 48 h with various drug (1294 or 1553) concentrations in LIT medium. Cell viability was monitored with the alamarBlue assay. Values were compared with the same cell line in the absence of the inhibitor (set as 100%). (B) Dose response of BKIs 1294 or 1553 against TcAEK1M125A epimastigotes. Values in panels A and B are means ± SD of 2 independent experiments, each done in triplicate. (C and D) Growth of TcAEK1WT (C) and TcAEK1M125A (D) epimastigotes in LIT medium incubated with different concentrations of compound 1553. One-way ANOVA with multiple comparisons was applied to growth rates calculated from each growth curve compared with that of the control (n = 3, n.s., not significant, ***, P < 0.001). (E) Representative images displaying aberrant cell morphology and DNA content of TcAEK1M125A epimastigotes incubated with 1.5 μM compound 1553 for 48 h and 72 h. DAPI staining (blue) and merge with DIC images are shown. Scale bars are shown. (F) Cell viability of TcAEK1WT (WT) and TcAEK1M125A (M125A) metacyclic trypomastigotes incubated for 24 h with various BKI (1294 or 1553) concentrations in LIT medium. Values were compared with the same cell line in the absence of the inhibitor (set as 100%). (G) Dose response of BKIs 1294 or 1553 against TcAEK1M125A metacyclic trypomastigotes. For panels F and G, number of parasites was counted in a Neubauer chamber and death percentage was estimated relative to control treatment (0.1% DMSO). Values plotted are means ± SD of 2 independent experiments, each done in triplicate. (H) Representative images of TcAEK1WT (WT) and TcAEK1M125A (M125A) metacyclic trypomastigotes incubated with 0.5 μM compound 1553 for 24 h. DAPI staining (blue) and merge with DIC images are shown. Scale bars represent 5 μm.
We were unable to test BKI compounds in host cell invasion assays because the TcAEK1 gatekeeper mutants have severely reduced ability to infect Vero cells, even though TcAEK1M125A cells exhibited no defects in in vitro metacyclogenesis compared with TcAEK1WT cells (Fig. S6).
We next incubated metacyclic trypomastigotes purified by DEAE cellulose columns with BKI-1294 or BKI-1553 (100 nM to 40 μM). Those expressing TcAEK1M125A showed a drastic reduction of viability of metacyclic trypomastigotes upon BKI treatment in a dose-dependent manner (Fig. 5F), while TcAEK1WT metacyclic cells remained fully viable in the presence of the BKIs. The estimated IC50 value (IC50/24 h) of BKI-1294 or BKI-1553 for TcAEK1M125A metacyclic trypomastigotes was 578 ± 72 nM and 421 ± 76 nM, respectively (Fig. 5G). As shown in Figure 5H, TcAEK1M125A metacyclic trypomastigotes incubated for 24 h with BKI-1553 exhibited severe morphological alterations. This last set of results indicates that TcAEK1 also plays an important role in this nonreplicative stage of T. cruzi.
Gatekeeper mutants treated with BKI confirmed that TcAKE1 is required for cytokinesis in the epimastigote forms of T. cruzi.
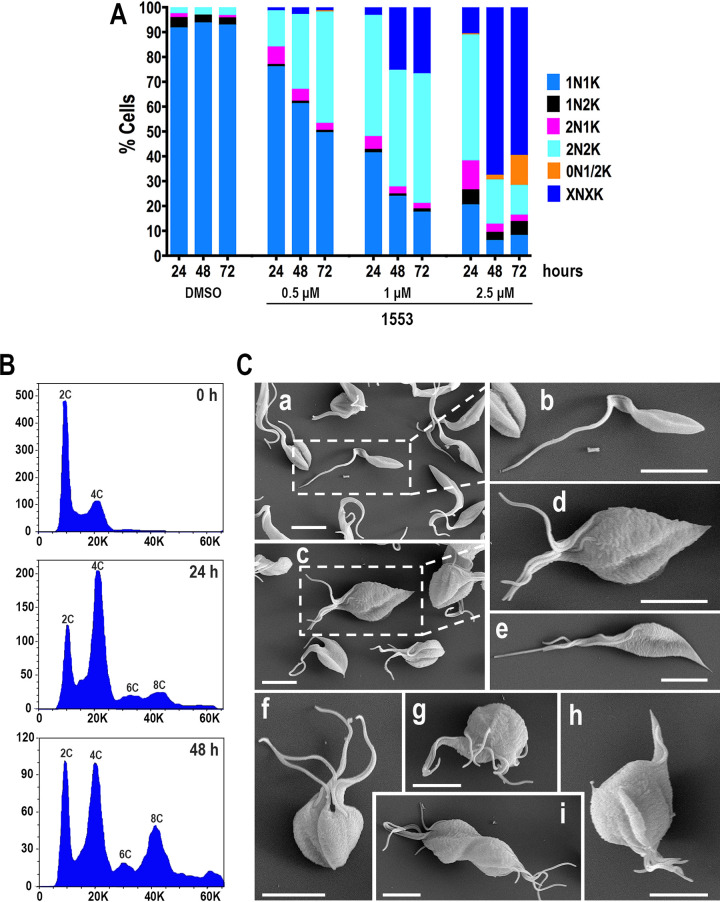
We also evaluated the N/K pattern of TcAEK1M125A epimastigotes cultured in the presence of BKI-1553. Figure 6A shows a decrease in 1N1K cells and an increase in binucleated (2N2K and 2N1K) and multinucleated cells (XNXK, X > 2) depending on the drug concentration and incubation time. These data are consistent with defective cytokinesis and suggest a failure in cell cycle progression in parasites with inhibited TcAEK1. Additionally, cells containing 1 or 2 kinetoplasts (termed a zoid) were observed at 48 and 72 h of incubation with the higher concentration of BKI-1553 tested (2.5 μM), suggesting that under these conditions the inhibition of TcAEK1M125A can impair the cell cycle in other points or cause nuclear mis-segregation or degradation. Cell DNA content in TcAEK1M125A epimastigotes was determined by flow cytometry at 0, 24, and 48 h of incubation with BKI-1553 (Fig. 6B). At the beginning of the experiment (0 h of incubation), TcAEK1M125A epimastigotes showed a typical cell cycle curve, having a predominant G0/G1 population (DNA content, 2C) with relatively fewer cells in S phase and G2/M (DNA synthesis and 4N DNA content, respectively) (Fig. 6B, upper panel). The number of tetraploid (4C, G2/M) and polyploid (6C and 8C) cells was abnormally elevated at 24 h of treatment (Fig. 6B, middle panel). After 48 h of incubation, TcAEK1M125A cells exhibited a predominance of polyploidy with an increase in the proportion of higher-DNA-content cells (Fig. 6B, bottom panel), which is consistent with a defect in cytokinesis.
FIG 6.
Incubation of ATP analog-sensitive TcAEK1M125A epimastigotes with BKI 1553 leads to cell cycle and morphological alterations. (A) Kinetoplast/nucleus configuration of TcAEK1M125A epimastigotes, which were treated with 0, 0.5, 1.0, or 2.5 μM compound 1553, was quantified by DAPI staining at 24, 48, and 72 h. Representative of three experiments (n = 200 cells). XNXK, X > 2. (B) Flow cytometry analysis of TcAEK1M125A epimastigotes incubated with 1.5 μM compound 1553. Analysis was made at 0 h (upper panel), 24 h (middle panel), and 48 h (bottom panel) of incubation with drug. The positions of cells having 2C DNA content (diploid), 4C, 6C, and 8C are indicated. (C) Representative images of scanning electron microscopy of control TcAEK1WT (a, b) and TcAEK1M125A parasites cultured in the presence of 1.5 μM compound 1553 for 48 h (c to i). Note that analog-sensitive TcAEK1M125A shows a multiflagellated and rosette-like morphology.
Electron microscopy analysis of ATP analog-sensitive TcAEK1 treated with BKIs confirms cell division defects.
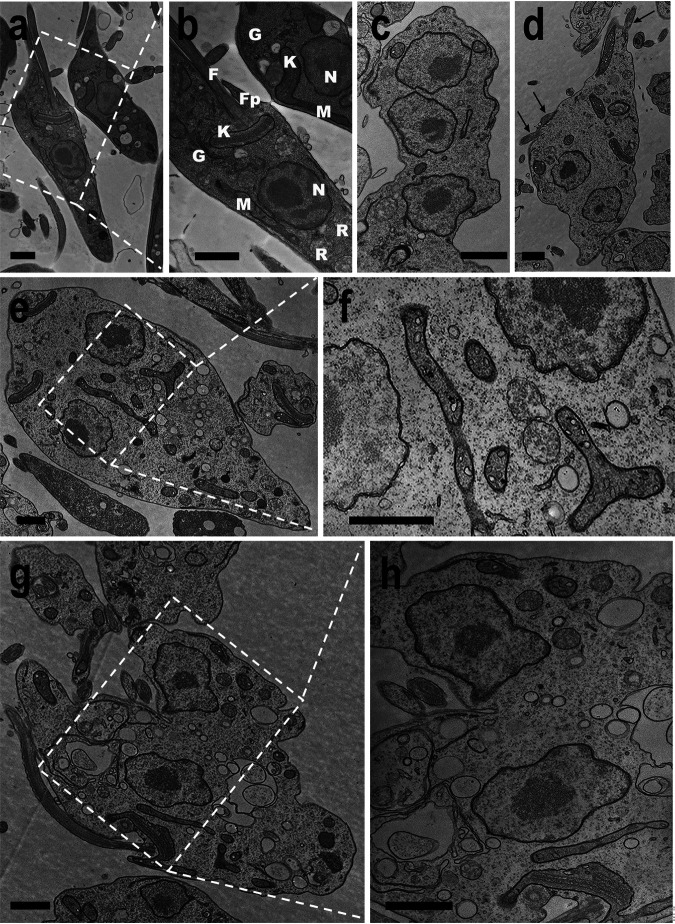
Figure 6C shows scanning electron microscopy (SEM) pictures of TcAEK1WT (panels a and b) and TcAEK1M125A cells (panels c to i) after treatment with BKI-1553 for 48 h. Most TcAEK1M125A cells are arrested in cytokinesis exhibiting a rosette-like or “pumpkin-like” morphology bearing several cell bodies connected. These aggregates of cells show multiple flagella attached, and in some cases (panels c, d, and f), each component still retains the shape of normal cells bound at the posterior end. Parasites did not have, or showed only an incipient, anterior cleavage furrow and longitudinal invagination, but these did not progress. These images are compatible with those observed under fluorescence microscopy (Fig. 5E) and also with DNA content analyzed by flow cytometry (Fig. 6B). Transmission electron microscopy (TEM) cross sections of TcAEK1M125A epimastigotes treated for 48 h confirmed that cell aggregates corresponded to a single massive undivided cell (Fig. 7c to h). In general, it is possible to distinguish the cell components of the multinuclear bodies and associated peripheral flagella. Large vacuoles with retracted content and double membrane in cytokinesis-arrested cells could correspond to autophagosomes (Fig. 7g and h). Some mitochondria of aggregate cells have dilated cristae (Fig. 7e and f). Therefore, we stained live TcAEK1M125A cells treated with BKI-1553 with MitoTracker. A net-like mitochondrial staining compatible with undivided cell bodies was observed (Fig. S7). Interestingly, MitoTracker fluorescence of treated TcAEK1M125A also suggests no obvious defect in mitochondrial membrane potential.
FIG 7.
Transmission electron microscopy of T. cruzi cells treated with BKI 1553. Control TcAEK1WT (a, b) and TcAEK1M125A parasites were treated with 1.5 μM compound 1553 for 48 h (c to h). Arrows indicate multiple flagella of rosette-like cells (d). Cytosolic vacuolization can be observed in g and h. N, nucleus; K, kinetoplast; F, flagellum; Fp, flagellar pocket; R, reservosome; G, Golgi complex; M, mitochondrion. Bars represent 1 μm.
DISCUSSION
Our studies genetically and chemically validated TcAEK1 as an essential protein in T. cruzi. We found that parasites hemizygous for AEK1 and those in which AEK1 was specifically inhibited through a chemical genetic approach showed similar phenotypes. A single KO of TcAEK1 resulted in slowed growth of the replicative stages of the parasite and inhibition of cell-derived trypomastigote invasion of host cells. Both TcAEK1-SKO and mutants susceptible to inhibition by BKIs showed similar effects on cytokinesis, as revealed by the appearance of multinucleated cells with several flagella. Metacyclic forms susceptible to BKIs also underwent a severe loss of viability when exposed to those inhibitors, suggesting that TcAEK1 is also important for the nonreplicative stages of the parasite.
In this work, we generated T. cruzi cell lines that exclusively express ATP analog-sensitive TcAEK1 mutants by adapting a CRISPR/Cas9-based mutagenesis strategy that we used previously (40). It is known that the space-creating mutation at the ATP-bonding pocket of kinases is often functionally silent. However, some kinases can suffer variable degrees of activity impairment (25, 41). It is known that there are no mammalian kinases with small gatekeeper residues (25), while that is not the case in apicomplexan parasites, and this has been exploited to perform functional studies and to test selective inhibitors (35, 37, 42–49). Interestingly, we found in the T. cruzi Y strain genome one protein kinase that possesses a small gatekeeper residue (TriTrypDB ID: TcYC6_0069280, residue A1141), which is annotated as STE/STE11 Ser/Thr-protein kinase, although no functional data are available on this putative kinase. The correlation in the phenotype between TcAEK1M125A in the presence of BKIs and the TcAEK1-SKO, combined with the lack of any phenotype in edited strains expressing tagged wild-type alleles upon exposure to BKI, indicates that there was a selective and specific effect of the inhibitors against the engineered TcAEK1.
Our data indicate that TcAEK1 appears to be required for epimastigote entry into cytokinesis after cells replicated their DNA and underwent mitosis. Cytokinesis is a critical step in the cell division cycle with multiple pathways and various degrees of redundancy that are monitored by specific signaling of checkpoints. This process in trypanosomatids possesses some complexities and peculiar features related to their complex life cycle, developmental stages which are morphologically different and highly polarized, and the need to replicate and segregate a series of single-copy organelles. Moreover, in these organisms, cytokinesis has been considered a microtubule-focused mechanism (50, 51). It has been described in T. brucei that two evolutionarily conserved protein kinases, the Polo-like kinase and the Aurora B kinase, and a group of trypanosome-specific proteins, including the cytokinesis initiation factors CIF1 to CIF4 and CIF1-interacting proteins FRW1 and FPRC, predominantly regulate control of cytokinesis initiation (52–57). Furthermore, Jones et al. (19) identified several protein kinases as essential for cytokinesis in bloodstream forms in their high-throughput RNAi screening.
Knowledge about cytokinesis and its regulation in T. cruzi lags far behind that in T. brucei, since only a few genes have been associated with this biological process in the first parasite. Thus, in T. cruzi, three Aurora kinase homologues were identified (TcAUK1 to 3), being TcAUK1 functional homologues to human Aurora B kinase and related to mitotic spindle assembling, chromosome segregation, and the initiation of kinetoplast duplication (58). Deletion of TcKMP-11 (kinetoplastid membrane protein-11) resulted in a substantial population of epimastigotes with cytokinesis defects (59). Furthermore, T. cruzi epimastigotes overexpressing high mobility group B protein (TcHMGB) exhibited impaired cytokinesis, suggesting a role in DNA replication and cell cycle progression control (60). Finally, inhibition of casein kinase 2, for which interaction with tubulin has been established, also triggered blockage of cytokinesis (61).
The essentiality and importance in cytokinesis of AEK1 were previously assessed in T. brucei. In a high-throughput RNAi-based phenotyping approach, bloodstream forms exhibited a significant loss of fitness after 6 days of RNAi targeting TbAEK1 (21). Moreover, Jones et al. (19), using a T. brucei kinome-wide RNAi library that included 190 predicted protein kinases, observed that knockdown of TbAEK1 induced a severe defect in cytokinesis and cell death in bloodstream forms. After induction of RNAi, cultures exhibited accumulation of cells with multiple nuclei and kinetoplasts (>2N > 2K cells). Essentiality of TbAEK1 in bloodstream forms was subsequently confirmed using conditional KOs and RNAi experiments (18), demonstrating that TbAEK1 knockdown caused reduced growth, defective cytokinesis, and cell death. Jensen et al. (18) also generated ATP analog-sensitive mutants in which the gatekeeper site was mutated to alanine or glycine, and they observed that BKI-1294 treatment of mice infected with bloodstream forms expressing TbAEK1 gatekeeper mutants (M138G) delayed parasitemia and death, with some mice being apparently cured of infection. Finally, the L. mexicana AEK1 ortholog has been also considered likely essential in promastigotes because no null mutants could be generated in a kinome-wide gene deletion library (20).
TcAEK1 null mutants could not be generated carrying out a methodology that has been previously used successfully for obtaining several gene KO cell lines (32). Instead, we obtained single allele KO parasites, which gives support to the essentiality of this gene for T. cruzi. In addition, our results also suggest that gene or chromosome duplication could have arisen when a second allele was targeted for disruption. Retention of essential genes following attempts at generating gene deletion cell lines is a common mechanism observed in Leishmania and is related to drug resistance, gene expression regulation, and adaptation to the host environment (20, 62–64). Variation in DNA copy number associated with drug selection and resistance has also been described for T. cruzi (6, 65).
The TcAEK1 amino acid sequence includes several features of the AGC kinase group of protein kinases, although the domain architecture outside the catalytic region is less conserved. In addition, no apparent AEK1 orthologs can be found in mammals. Many AGC kinases are activated by serine/threonine phosphorylation of two highly conserved motifs by the 3-phosphoinositide-dependent kinase-1 (PDK-1): the activation loop in the catalytic domain and the hydrophobic motif, C-terminal to the kinase domain (66). Phosphoproteomic analyses in T. brucei and T. cruzi indicate that phosphoserines are present at these two domains in AEK1 orthologs of both species (10, 14, 67). Additionally, a PDK-1 homolog is present in the T. cruzi genome (15, 17) (TriTrypDB ID: TcYC6_0028570 for T. cruzi Y C6 strain). Although functional evidence is absent, the data suggest a conserved activation/modulation mechanism for members of the AGC group of protein kinases in T. cruzi and for TcAEK1 in particular.
Interestingly, although tubulin may not be an optimal housekeeping protein for normalization when comparing protein expression levels between different T. cruzi developmental stages, our results suggest that the expression level of the endogenously tagged TcAEK1 protein was higher in the replicative stages (epimastigotes and amastigotes) than in the nondividing trypomastigote forms. The TcAEK1-SKO epimastigotes exhibited an increased in vitro metacyclogenesis, which correlates with the significant number of metacyclic trypomastigotes observed in mutant epimastigotes in the exponential growth phase in LIT medium. A potential explanation may be that the downregulation of TcAEK1 could lead epimastigotes to “escape” into a nonreplicative form. Similarly, T. cruzi epimastigotes exhibited an increased metacyclogenesis along with impairment of cytokinesis when they were cultured with an inhibitor of casein kinase 2 (61).
The defects observed in different T. cruzi developmental stages after knocking down or inhibiting TcAEK1 with BKIs suggest that this protein kinase is a pleiotropic player involved in crucial cellular processes in T. cruzi. In the absence of efficient inducible downregulation methods in T. cruzi, and lacking specific inhibitors, our work highlights the relevance of generating ATP analog-sensitive mutants in T. cruzi by CRISPR/Cas9-based strategies for functional studies of protein kinases and validation of therapeutic targets. Moreover, the use of selective and/or specific inhibitors permits an almost instantaneous loss of activity in a population of mutant cells, preventing a gradual reduction of proteins and adaptation of mutant cells.
In summary, by analyzing single allele deletion and engineered ATP analog-sensitive cell lines obtained using CRISPR/Cas9 technology, we demonstrated that TcAEK1 is critical for successful transit through multiple life cycle stages and the generation of pathogenic stages. It is essential for epimastigote proliferation, trypomastigote host cell invasion, and amastigote replication. We also demonstrated its requirement for cytokinesis in the epimastigote form. The use of BKIs against ATP analog-sensitive TcAEK1 allows us to consider this protein as a validated drug target, providing further impetus to develop and/or test inhibitor molecules in vitro and in vivo.
MATERIALS AND METHODS
Chemicals and reagents.
MitoTracker Deep Red FM, Alexa-conjugated secondary antibodies, Pierce ECL Western blotting substrate, and BCA protein assay kit were from Thermo Fisher Scientific Inc. Blasticidin S HCl was from AG Scientific (San Diego, CA). BLUelf prestained protein ladder was from FroggaBio (Wheatfield, NY). Puromycin was from Acros Organics (Fair Lawn, NJ). Anti-HA.11 epitope tag monoclonal antibody was from BioLegend (San Diego, CA). Alexa Fluor-conjugated secondary antibodies and horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Life Technologies. Benzonase nuclease was from Novagen (EMD Millipore, Billerica, MA). GoTaq DNA polymerase and T4 DNA ligase were from Promega (Madison, WI). Antarctic phosphatase, restriction enzymes, and Q5 high-fidelity DNA polymerase were from New England Biolabs (Ipswich, MA). Fluoromount-G was from SouthernBiotech (Birmingham, AL). Nitrocellulose membrane and alamarBlue were from Bio-Rad (Hercules, CA). DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coraville, IA.). Escherichia coli DH5α Mix & Go competent cells, ZymoPURE plasmid midiprep kit, and ZymoPURE plasmid miniprep kit were from Zymo Research (Irvine, CA). G418 sulfate was from KSE Scientific (Durham, NC). Anti-tubulin monoclonal antibody, anti-Flag antibody, mammalian cell protease inhibitor mixture (Sigma P8340), other protease inhibitors, and all other reagents of analytical grade were from Sigma (St. Louis, MO). Rabbit polyclonal antibody against cruzipain was a gift from Vanina Alvarez (Universidad Nacional de San Martin, Argentina). The pMOTag23M vector (68) was from Thomas Seebeck (University of Bern, Bern, Switzerland). Polyclonal antibody against TbBIP was from Jay Bangs (State University of New, York, Buffalo, NY). Stock solutions of bumped kinase inhibitors (BKI) 1294 (38) and 1553 (39) (20 mM) were kindly provided by Dustin Maly and Rama S.R. Vidadala (University of Washington).
Cell cultures.
T. cruzi Y strain epimastigotes were cultured in liver infusion tryptose (LIT) medium containing 10% heat-inactivated fetal bovine serum (FBS) at 28°C (69). CRISPR/Cas9 mutant cell lines were maintained in medium containing 250 μg/ml G418 and 10 μg/ml blasticidin or 5 μg/ml puromycin. Parasites from the control cell lines transfected with scrambled sgRNA (scrambled) or pTREX-n empty vector (EV) and overexpressing TcAEK1-sgRNA-787/Cas9, TcAEK1-3×HA, or TcAEK1-Flag were cultured in medium containing 250 g/ml G418. The growth rate of epimastigotes was determined by counting cells in a Neubauer chamber. Tissue culture cell-derived trypomastigotes were obtained from Vero cells infected with metacyclic trypomastigotes obtained as described below. T. cruzi trypomastigotes were collected from the culture medium of infected host cells as described previously (70). Vero cells were grown in RPMI supplemented with 10% fetal bovine serum and maintained at 37°C with 5% CO2.
In silico analysis.
Prediction of protein domains of TcAEK1 amino acid sequence was made using web servers https://www.ebi.ac.uk/interpro/ and https://prosite.expasy.org/scanprosite/. WoLF PSORT method (https://wolfpsort.hgc.jp/) was used to predict subcellular localization sites based on amino acid sequences. For in silico analyses for selection of protospacers for knockout and tagging of TcAEK1 gene, we used the CRISPR/Cas9 genome editing method for T. cruzi developed before (29, 71, 72). Selection of protospacers for both strategies was performed using EuPaGDT (eukaryotic pathogen CRISPR guide RNA/DNA design tool; http://grna.ctegd.uga.edu) (73).
TcAEK1 overexpression.
TcAEK1 open reading frame (ORF) (1,179 nt) was PCR amplified using T. cruzi Y strain genomic DNA as the template (primers 1 and 2; Table S1) and cloned into the pTREX-n/3×HA vector (31) by restriction sites XbaI/XhoI. Gene cloning was confirmed by PCR and sequencing, and constructs were subsequently used to transfect T. cruzi epimastigotes. TcAEK1 overexpression was confirmed by Western blotting using anti-HA antibodies.
TcAEK1 endogenous C-terminal tagging and mutation.
CRISPR/Cas9-mediated endogenous C-terminal tagging of TcAEK1 was done as described previously (71, 72). We chose a specific sgRNA sequence targeting the 3′ end of TcAEK1 gene (TriTrypDB ID: TcYC6_ 0120630). The construct TcAEK1-Ctag-sgRNA/Cas9/pTREX-n (primers 3 and 4; Table S1), together with a DNA donor cassette to induce homology-directed repair, was used to cotransfect T. cruzi epimastigotes and to insert a specific 3xc-Myc-tag sequence at the 3′ end of the gene. DNA donor cassette containing the 3xc-Myc-tag sequence and the puromycin resistance gene was amplified using the pMOTag23M vector (68) as the template (primers 5 and 7; Table S1). One additional DNA donor cassette was designed to generate amino acid substitutions, along with the insertion of the 3xc-Myc-tag, from HLNK to GLNG amino acid residues at the C terminus (residues 388 to 391) of TcAEK1 (primers 6 and 7; Table S1). Epimastigotes cotransfected with TcAEK1-Ctag-sgRNA/Cas9/pTREX-n plasmid and any of the DNA donor cassettes were cultured for 5 weeks with G418 and puromycin for selection of double-resistant parasites. Endogenous gene tagging was verified by PCR from gDNA using primers 8 and 9 (Table S1) and by Western blotting.
CRISPR-Cas9 mediated deletion of a single allele of TcAEK1.
We used the TcAEK1 sequence from T. cruzi Y C6 strain to design a single guide RNA (sgRNA), which is predicted to target DNA cleavage at nucleotide +787 of TcAEK1 (TcAEK1-sgRNA-787). TcAEK1-sgRNA-787 was PCR amplified (primers 4 and 10; Table S1) and cloned into Cas9/pTREX-n as described previously (29, 30) to generate TcAEK1-sgRNA-787/Cas9/pTREX-n plasmid. A scrambled sgRNA cloned in Cas9/pTREX-n was used as the control. In the three strategies (see Fig. 3A, Fig. S2A, and Fig. S3A) designed to try to generate a TcAEK1-KO cell line, 3 different linear DNA donor cassettes were obtained by PCR using a set of 120-nt primers (ultramers). In the first and second strategies, each one of the ultramers had 20 nt annealing on the blasticidin S-deaminase (Bsd) gene (primers 11, 12, and 14; Table S1), whereas for the third strategy, ultramers allowed the amplification of the puromycin N-acetyltransferase gene (Pac) (primers 13 and 15; Table S1). We first transfected epimastigotes with TcAEK1-sgRNA-787/Cas9/pTREX-n plasmid, and after obtaining a homogeneous G418-resistant population expressing Cas9-GFP in the nuclei of epimastigotes, the TcAEK1-sgRNA-787/Cas9 cells were transfected with a repair cassette fragment containing the Bsd resistance marker to promote the disruption of TcAEK1 ORF. Transfected cells were selected with G418 and blasticidin. For the third strategy, TcAEK1-sgRNA-787/Cas9 cells with one allele disrupted by Bsd cassette (TcAEK1-SKO) were transfected with a DNA donor cassette fragment containing the Pac resistance marker to promote the disruption of the WT remaining TcAEK1 allele. Stable transfected cells were obtained by selection with G418, blasticidin, and puromycin. TcAEK1 gene disruption was verified by PCR using primers 9 and 16 (Table S1).
Generation of ATP analog-sensitive TcAEK1.
To generate parasite cell lines with TcAEK1 mutated at the gatekeeper residue, we followed a CRISPR/Cas9-induced knock-in strategy based on that we used previously (40). The design provides a DNA donor molecule to induce homologous-directed repair, which introduces mutations at the gatekeeper residue M125 (to generate substitution to alanine or glycine) and silent mutations in the protospacer sequence to prevent the constitutively expressed sgRNA and Cas9 from continuing to cleave their targets. In addition, the repair cassettes also allowed tagging of the mutated gene to follow its expression by immunofluorescence analysis and Western blotting. We chose sgRNA-386 that directs DNA cleavage near codon 125 of TcAEK1 (primer 17; Table S1).
To generate donor DNA constructs, we performed a two-step-PCR strategy. In the first step, we amplified in two separate PCRs a 5′ portion of the gene including a 120-nt ultramer with the WT or mutated TcAEK1 gatekeeper residue M125 (primers 18 to 21; Table S1), while in the second step, the 3xc-Myc-tag sequence and the puromycin resistance gene were amplified using the pMOTag23M vector as the template (primers 7 and 22; Table S1) (68). Subsequently, we performed an overlap extension PCR including the two above-mentioned fragments as the templates (primers 7 and 18 to 20; Table S1). Finally, the resulting PCR amplicons were used as donor DNAs to generate these mutant cells: TcAEK1WT, TcAEK1M125A, and TcAEK1M125G. Epimastigotes cotransfected with TcAEK1-sgRNA-386/Cas9/pTREX-n and DNA donor were cultured for 5 weeks with G418 and puromycin for selection of double-resistant parasites and then cloned by limiting dilution. Endogenous gene tagging was verified by PCR from gDNA using specific primer sets (primers 8 and 9; Table S1) and by Western blotting. Mutation at the TcAKE1 gatekeeper residue was confirmed by amplifying gDNA with primers 9 and 23 (Table S1). Electrophoresed DNA fragments were gel-purified and submitted to determine their nucleotide sequence using primer 24 (Table S1).
Cell transfections.
T. cruzi Y strain epimastigotes were transfected as described previously (31). Briefly, T. cruzi epimastigotes in early exponential phase (4 × 107 cells) were washed with phosphate-buffered saline (PBS; pH 7.4) at room temperature (RT) and transfected in ice-cold CytoMix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4, 25 mM HEPES, 2 mM EDTA, 5 mM MgCl2 [pH 7.6]) containing 25 μg of each plasmid construct and 25 μg of donor DNA in 4-mm electroporation cuvettes with three pulses (1,500 V, 25 microfarads) delivered by a Gene Pulser Xcell electroporation system (Bio-Rad). Transfected epimastigotes were cultured in LIT medium supplemented with 20% heat-inactivated FBS until stable cell lines were obtained. When needed, the antibiotic used for drug selection and maintenance was 250 μg/ml G418, 10 μg/ml blasticidin, or 5 μg/ml puromycin. Parasite clones were obtained by limiting dilution.
Western blotting.
Western blotting was carried out as described previously (74). Briefly, parasites were harvested and washed twice in PBS and subsequently resuspended in radio-immunoprecipitation assay buffer (RIPA: 150 mM NaCl, 20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% SDS, 0.1% Triton X-100) plus a mammalian cell protease inhibitor mixture (diluted 1:250), 1 mM phenylmethylsulfonyl fluoride, 2.5 mM tosyl phenylalanyl chloromethyl ketone (TPCK), 100 M N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide (E64), and benzonase nuclease (25 units/ml of culture). The cells were then incubated for 1 h on ice and protein concentration was determined by bicinchoninic acid (BCA) protein assay. Thirty micrograms of protein from each cell lysate was mixed with Laemmli sample buffer (125 mM Tris-HCl [pH 7], 10% [wt/vol] β-mercaptoethanol, 20% [vol/vol] glycerol, 4.0% [wt/vol] SDS, 4.0% [wt/vol] bromphenol blue) before application to 10% SDS-polyacrylamide gels. Electrophoresed proteins were transferred onto nitrocellulose membranes with a Bio-Rad transblot apparatus. Membranes were blocked with 5% nonfat dried skim milk in PBS-T (PBS containing 0.1% vol/vol Tween 20) overnight at 4°C. Next, blots were incubated for 1 h at RT, with a primary antibody, i.e., monoclonal anti-HA (1:5,000), monoclonal anti-c-Myc-tag (1:100), rabbit anti-Flag (1:20,000), and monoclonal anti-tubulin (1:40,000). After three washes with PBS-T, blots were incubated with the secondary antibody (goat anti-mouse IgG or goat anti-rabbit IgG, HRP-conjugated antibody, diluted [1:10,000]). Membranes were washed three times with PBS-T, and then blots were incubated with Pierce ECL plus substrate and images were obtained and processed with a ChemiDoc Imaging System (Bio-Rad). Blot densitometry analyses were performed with ImageJ v1.48 software.
Immunofluorescence analysis.
Cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 1 h at RT. Cells were allowed to adhere to poly-l-lysine-coated coverslips and then permeabilized for 5 min with 0.1% Triton X-100. Permeabilized cells were blocked with PBS containing 3% bovine serum albumin (BSA), 1% fish gelatin, 50 mM NH4Cl, and 5% goat serum overnight at 4°C. Then, cells were incubated with a primary antibody (monoclonal anti-HA-Tag [1:50], monoclonal anti-c-Myc-tag [1:10], monoclonal anti-TcFCaBP [1:10], rabbit polyclonal anti-TcBIP [1:50], or rabbit polyclonal anti-cruzipain [1:100]) diluted in 1% BSA in PBS (pH 8.0) for 1 h at RT. Cells were washed three times with 1% BSA in PBS (pH 8.0) and then incubated for 1 h at RT in the dark with Alexa Fluor 488- or Alexa Fluor 546-conjugated goat anti-mouse secondary antibodies (1:1,000). Then, cells were washed and mounted on slides using Fluoromount-G mounting medium containing 5 μg/ml of 4,6-diamidino-2-phenylindole (DAPI) to stain DNA. Controls were done as described above but in the absence of a primary antibody. Differential interference contrast and fluorescence optical images were captured with a 100× objective (1.35-aperture) lens under nonsaturating conditions with an Olympus IX-71 inverted fluorescence microscope with a Photometrix CoolSnapHQ charge-coupled device camera driven by DeltaVision software (Applied Precision, Issaquah, WA); images were then deconvolved for 15 cycles using Sotwarx deconvolution software.
Metacyclogenesis.
Metacyclic trypomastigotes were obtained following the protocol described by Bourguignon et al. (75) with minor modifications. Epimastigotes were obtained after 4 days in LIT medium and incubated for 2 h in triatome artificial urine (TAU) medium (190 mM NaCl, 17 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 0.035% sodium bicarbonate, 8 mM phosphate [pH 6.9]) at room temperature. Then, parasites were incubated for 96 h in TAU 3AAG medium (TAU medium supplemented with 10 mM l-proline, 50 mM sodium l-glutamate, 2 mM sodium l-aspartate, and 10 mM glucose). Samples were harvested from the TAU 3AAG plus FBS-containing medium at day 4 of cultivation, washed with PBS, and fixed with 4% paraformaldehyde in PBS for 1 h at RT. Cells were adhered to poly-l-lysine-coated coverslips and mounted on slides using Fluoromount-G mounting medium containing 5 μg/ml of DAPI to stain DNA. For incubation with drugs, the metacyclic trypomastigotes were purified using anion-exchange chromatography in DEAE cellulose columns as described previously (76).
Invasion and replication assays.
Gamma-irradiated (2,000 radiation-absorbed doses) Vero cells (4.5 × 105 cells) were plated onto sterile coverslips in a 12-well plate and incubated overnight at 37°C with 7% CO2 in RPMI medium supplemented with 10% fresh FBS. Tissue culture-derived trypomastigote collections were incubated at 4°C overnight to allow amastigotes to settle from swimming trypomastigotes. Trypomastigotes from the supernatants of these collections were counted and used to infect the mammalian cells at a 10:1 ratio of parasites to host cells. At 4 h postinfection, coverslips were washed extensively with Hanks’ balanced salt solution, followed by PBS (pH 7.4), to remove any extracellular parasites. This protocol has been optimized with a multiplicity of infection (MOI) of 10 to ensure that at most one trypomastigote (Y strain) invades each irradiated host cell. Coverslips were immediately fixed in 4% paraformaldehyde in PBS (pH 7.4) at 4°C for 30 min and then washed once with PBS and counterstained with DAPI (15 μg/ml) in Fluoromount-G mounting medium on glass slides, which stains host and parasite DNA. Coverslips were analyzed on an Olympus BX60 microscope to quantify the number of host cells that contained intracellular parasites and the number of intracellular parasites per cell in 40 randomly selected fields. Four hundred host cells were counted per sample in three independent experiments. To quantify amastigote replication, Vero cells were infected at a 10:1 ratio of parasites to host cells, and after they were washed at 4 h postinfection as described above, coverslips were allowed to incubate for 48 h at 35°C with 7% CO2 prior to fixation and DAPI staining.
Drug solutions.
Stock solutions of bumped kinase inhibitors (BKIs) 1294 (38) and 1553 (39) (20 mM) were prepared in dimethyl sulfoxide (DMSO), with the final concentration of DMSO in the experiments never being higher than 0.2%.
Effects of BKIs in ATP analog-sensitive TcAEK1 mutants.
To calculate the concentration of BKI-1294 and BKI-1553 that inhibited 50% epimastigote growth (IC50), 100-μl log phase parasites (2.5 × 106/ml) were incubated at 28°C with different concentrations of drugs (0.01 to 20 μM) in biological triplicates for 48 h. After 48 h, 20 μl of alamarBlue (Bio-Rad) was added to each plate, allowing color changes in response to the chemical reduction by viable trypanosomes. Plates were incubated for an additional 8 h. Later, fluorescence was quantified at an excitation wavelength of 570 nm and an emission wavelength of 590 nm using a microplate reader (BioTek Instruments, Winooski, VT) driven by Gen5 software for data collection and analysis. To calculate the IC50 for metacyclic trypomastigotes, the parasites (2.5 × 106 cells/ml) were incubated with different concentrations of BKI-1294 and BKI-1553 (0.1 to 40 μM) in biological triplicates. After 24 h at 28°C, the parasite number was counted in a Neubauer chamber. In both epimastigotes and metacyclic trypomastigotes, the death percentage was estimated relative to the untreated control (LIT medium with 0.1% DMSO), generating dose-effect curves. GraphPad Prism software was used to calculate the IC50 values employing the death percentage for each BKI concentration. Each experiment was performed on three independent occasions.
Flow cytometry analysis.
For flow cytometry analysis, 1 × 106 cells were harvested by centrifugation, washed with PBS, and fixed in ice-cold 70% ethanol/30% PBS for 30 min. Then, they were washed once with PBS and resuspended in 1 ml of staining solution (20 μg/ml propidium iodide, 38 mM citrate buffer [pH 7.40], 20 μg/ml RNase, 0.3% NP-40). After the sample was inverted three times, the cell suspension was incubated at 37°C for 30 min. DNA content of propidium iodide-stained cells was analyzed using a CyAn ADP analyzer flow cytometer (Beckman Coulter, Hialeah, FL) at the CTEGD Cytometry Shared Resource Lab (UGA).
Scanning and transmission electron microscopy.
Cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate-HCl buffer (pH 7.25) and subsequently processed for transmission electron microscopy (TEM) and scanning electron microscopy (SEM) at the Electron Microscopy Facility of the Department of Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA. TEM images were obtained with a JEOL JEM1011 transmission electron microscope (JEOL Tokyo, Japan) equipped with a high-contrast 2k by 2k AMT midmount digital camera from Advanced Microscopy Techniques, Corp. (Woburn, MA). Parasites for SEM were viewed with a Teneo field-emission scanning electron microscope (ThermoFisher, Hillsboro, OR).
Statistical analysis.
Statistical analyses were performed with GraphPad Prism software (La Jolla, CA), version 6.0. Reported values are means ± standard deviation (SD) of n independent experiments, as indicated in the figure legends. The level of significance was evaluated by Student’s t test for comparisons between two cell lines, one-way analysis of variance (ANOVA) for comparisons between more than two cell lines, and two-way ANOVA with multiple-comparison tests for analyses of grouped data.
ACKNOWLEDGMENTS
We thank Marilyn Parsons (University of Washington) for useful discussions, Dustin J. Maly and Rama S.R. Vidadala for BKI inhibitors, Vanina Alvarez (University of San Martin, Argentina) for cruzipain antibodies, David Engman (Northwestern University) for TcFCaBP antibodies, Jay Bangs (State University of New, York, Buffalo, NY) for TbBIP antibodies, Thomas Seebeck (University of Bern, Bern, Switzerland) for the pMOTag23M vector, Noelia Lander for help in invasion assays and suggestions, Srinivasan Ramakrishnan and Mayara Bertolini (University of Georgia, GA) for help in the host invasion and replication assays, Julie Nelson from the CTEGD Cytometry Shared Resource Laboratory (UGA) for assistance in cell sorting, and Mary Ard for sample processing and cell imaging by SEM and TEM. Funding for this work was provided by the U.S. National Institutes of Health (grant AI140421 to R.D.). B.C.J. was supported by the U.S. National Institutes of Health (grant AI130685).
Footnotes
Supplemental material is available online only.
Contributor Information
Miguel A. Chiurillo, Email: chiurima@ucmail.uc.edu.
Roberto Docampo, Email: rdocampo@uga.edu.
Ayush Kumar, University of Manitoba.
REFERENCES
- 1.Montgomery SP, Parise ME, Dotson EM, Bialek SR. 2016. What do we know about Chagas disease in the United States? Am J Trop Med Hyg 95:1225–1227. doi: 10.4269/ajtmh.16-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urbina JA, Docampo R. 2003. Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol 19:495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Bern C. 2011. Antitrypanosomal therapy for chronic Chagas' disease. N Engl J Med 364:2527–2534. doi: 10.1056/NEJMct1014204. [DOI] [PubMed] [Google Scholar]
- 4.Andrade DV, Gollob KJ, Dutra WO. 2014. Acute chagas disease: new global challenges for an old neglected disease. PLoS Negl Trop Dis 8:e3010. doi: 10.1371/journal.pntd.0003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filardi LS, Brener Z. 1987. Susceptibility and natural resistance of Trypanosoma cruzi strains to drugs used clinically in Chagas disease. Trans R Soc Trop Med Hyg 81:755–759. doi: 10.1016/0035-9203(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 6.Campos MC, Phelan J, Francisco AF, Taylor MC, Lewis MD, Pain A, Clark TG, Kelly JM. 2017. Genome-wide mutagenesis and multi-drug resistance in American trypanosomes induced by the front-line drug benznidazole. Sci Rep 7:14407. doi: 10.1038/s41598-017-14986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez SJ, Romano PS, Engman DM. 2020. Precision health for Chagas disease: integrating parasite and host factors to predict outcome of infection and response to therapy. Front Cell Infect Microbiol 10:210. doi: 10.3389/fcimb.2020.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson FM, Gray NS. 2018. Kinase inhibitors: the road ahead. Nat Rev Drug Discov 17:353–377. doi: 10.1038/nrd.2018.21. [DOI] [PubMed] [Google Scholar]
- 9.Fabbro D. 2015. 25 years of small molecular weight kinase inhibitors: potentials and limitations. Mol Pharmacol 87:766–775. doi: 10.1124/mol.114.095489. [DOI] [PubMed] [Google Scholar]
- 10.Amorim JC, Batista M, da Cunha ES, Lucena ACR, Lima CVP, Sousa K, Krieger MA, Marchini FK. 2017. Quantitative proteome and phosphoproteome analyses highlight the adherent population during Trypanosoma cruzi metacyclogenesis. Sci Rep 7:9899. doi: 10.1038/s41598-017-10292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos Junior A, Melo RM, Ferreira BVG, Pontes AH, Lima CMR, Fontes W, Sousa MV, Lima BD, Ricart CAO. 2021. Quantitative proteomics and phosphoproteomics of Trypanosoma cruzi epimastigote cell cycle. Biochim Biophys Acta Proteins Proteom 1869:140619. doi: 10.1016/j.bbapap.2021.140619. [DOI] [PubMed] [Google Scholar]
- 12.Nakayasu ES, Gaynor MR, Sobreira TJ, Ross JA, Almeida IC. 2009. Phosphoproteomic analysis of the human pathogen Trypanosoma cruzi at the epimastigote stage. Proteomics 9:3489–3506. doi: 10.1002/pmic.200800874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchini FK, de Godoy LM, Rampazzo RC, Pavoni DP, Probst CM, Gnad F, Mann M, Krieger MA. 2011. Profiling the Trypanosoma cruzi phosphoproteome. PLoS One 6:e25381. doi: 10.1371/journal.pone.0025381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucena ACR, Amorim JC, de Paula Lima CV, Batista M, Krieger MA, de Godoy LMF, Marchini FK. 2019. Quantitative phosphoproteome and proteome analyses emphasize the influence of phosphorylation events during the nutritional stress of Trypanosoma cruzi: the initial moments of in vitro metacyclogenesis. Cell Stress Chaperones 24:927–936. doi: 10.1007/s12192-019-01018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons M, Worthey EA, Ward PN, Mottram JC. 2005. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6:127. doi: 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 17.Bachmaier S, Boshart M. 2013. Kinetoplastid AGC kinases. In Doerig C, Späth G, Wiese M (ed), Protein Phosphorylation in Parasites. Wiley-VCH Verlag GmbH & Co, Weinheim, Germany. [Google Scholar]
- 18.Jensen BC, Booster N, Vidadala RS, Maly DJ, Parsons M. 2016. A novel protein kinase is essential in bloodstream Trypanosoma brucei. Int J Parasitol 46:479–483. doi: 10.1016/j.ijpara.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones NG, Thomas EB, Brown E, Dickens NJ, Hammarton TC, Mottram JC. 2014. Regulators of Trypanosoma brucei cell cycle progression and differentiation identified using a kinome-wide RNAi screen. PLoS Pathog 10:e1003886. doi: 10.1371/journal.ppat.1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker N, Catta-Preta CMC, Neish R, Sadlova J, Powell B, Alves-Ferreira EVC, Geoghegan V, Carnielli JBT, Newling K, Hughes C, Vojtkova B, Anand J, Mihut A, Walrad PB, Wilson LG, Pitchford JW, Volf P, Mottram JC. 2021. Systematic functional analysis of Leishmania protein kinases identifies regulators of differentiation or survival. Nat Commun 12:1244. doi: 10.1038/s41467-021-21360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. 2011. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Bishop A, Witucki L, Kraybill B, Shimizu E, Tsien J, Ubersax J, Blethrow J, Morgan DO, Shokat KM. 1999. Structural basis for selective inhibition of Src family kinases by PP1. Chem Biol 6:671–678. doi: 10.1016/s1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 23.Bishop AC, Shokat KM. 1999. Acquisition of inhibitor-sensitive protein kinases through protein design. Pharmacol Ther 82:337–346. doi: 10.1016/s0163-7258(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 24.Lopez MS, Kliegman JI, Shokat KM. 2014. The logic and design of analog-sensitive kinases and their small molecule inhibitors. Methods Enzymol 548:189–213. doi: 10.1016/B978-0-12-397918-6.00008-2. [DOI] [PubMed] [Google Scholar]
- 25.Shah K, Kim H. 2019. The significant others: global search for direct kinase substrates using chemical approaches. IUBMB Life 71:721–737. doi: 10.1002/iub.2023. [DOI] [PubMed] [Google Scholar]
- 26.Lozano-Nunez A, Ikeda KN, Sauer T, de Graffenried CL. 2013. An analogue-sensitive approach identifies basal body rotation and flagellum attachment zone elongation as key functions of PLK in Trypanosoma brucei. Mol Biol Cell 24:1321–1333. doi: 10.1091/mbc.E12-12-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gosavi U, Srivastava A, Badjatia N, Gunzl A. 2020. Rapid block of pre-mRNA splicing by chemical inhibition of analog-sensitive CRK9 in Trypanosoma brucei. Mol Microbiol 113:1225–1239. doi: 10.1111/mmi.14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin J, Dunbrack RL, Jr, Lee S, Strominger JL. 1991. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc Natl Acad Sci USA 88:1918–1922. doi: 10.1073/pnas.88.5.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander N, Li ZH, Niyogi S, Docampo R. 2015. CRISPR/Cas9-induced disruption of paraflagellar rod protein 1 and 2 genes in Trypanosoma cruzi reveals their role in flagellar attachment. mBio 6:e01012. doi: 10.1128/mBio.01012-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander N, Chiurillo MA, Docampo R. 2019. Genome editing by CRISPR/Cas9 in Trypanosoma cruzi. Methods Mol Biol 1955:61–76. doi: 10.1007/978-1-4939-9148-8_5. [DOI] [PubMed] [Google Scholar]
- 31.Chiurillo MA, Lander N, Bertolini MS, Storey M, Vercesi AE, Docampo R. 2017. Different roles of mitochondrial calcium uniporter complex subunits in growth and infectivity of Trypanosoma cruzi. mBio 8. doi: 10.1128/mBio.00574-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lander N, Chiurillo MA. 2019. State-of-the-art CRISPR/Cas9 technology for genome editing in trypanosomatids. J Eukaryot Microbiol 66:981–991. doi: 10.1111/jeu.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiurillo MA, Lander N, Vercesi AE, Docampo R. 2020. IP3 receptor-mediated Ca2+ release from acidocalcisomes regulates mitochondrial bioenergetics and prevents autophagy in Trypanosoma cruzi. Cell Calcium 92:102284. doi: 10.1016/j.ceca.2020.102284. [DOI] [PubMed] [Google Scholar]
- 34.Elias MC, da Cunha JP, de Faria FP, Mortara RA, Freymuller E, Schenkman S. 2007. Morphological events during the Trypanosoma cruzi cell cycle. Protist 158:147–157. doi: 10.1016/j.protis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Van Voorhis WC, Doggett JS, Parsons M, Hulverson MA, Choi R, Arnold SLM, Riggs MW, Hemphill A, Howe DK, Mealey RH, Lau AOT, Merritt EA, Maly DJ, Fan E, Ojo KK. 2017. Extended-spectrum antiprotozoal bumped kinase inhibitors: a review. Exp Parasitol 180:71–83. doi: 10.1016/j.exppara.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doggett JS, Ojo KK, Fan E, Maly DJ, Van Voorhis WC. 2014. Bumped kinase inhibitor 1294 treats established Toxoplasma gondii infection. Antimicrob Agents Chemother 58:3547–3549. doi: 10.1128/AAC.01823-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ojo KK, Reid MC, Kallur Siddaramaiah L, Muller J, Winzer P, Zhang Z, Keyloun KR, Vidadala RS, Merritt EA, Hol WG, Maly DJ, Fan E, Van Voorhis WC, Hemphill A. 2014. Neospora caninum calcium-dependent protein kinase 1 is an effective drug target for neosporosis therapy. PLoS One 9:e92929. doi: 10.1371/journal.pone.0092929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson SM, Murphy RC, Geiger JA, DeRocher AE, Zhang Z, Ojo KK, Larson ET, Perera BG, Dale EJ, He P, Reid MC, Fox AM, Mueller NR, Merritt EA, Fan E, Parsons M, Van Voorhis WC, Maly DJ. 2012. Development of Toxoplasma gondii calcium-dependent protein kinase 1 (TgCDPK1) inhibitors with potent anti-toxoplasma activity. J Med Chem 55:2416–2426. doi: 10.1021/jm201713h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller J, Aguado-Martinez A, Balmer V, Maly DJ, Fan E, Ortega-Mora LM, Ojo KK, Van Voorhis WC, Hemphill A. 2017. Two novel calcium-dependent protein kinase 1 inhibitors interfere with vertical transmission in mice infected with Neospora caninum Tachyzoites. Antimicrob Agents Chemother 61. doi: 10.1128/AAC.02324-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiurillo MA, Lander N, Bertolini MS, Vercesi AE, Docampo R. 2019. Functional analysis and importance for host cell infection of the Ca2+-conducting subunits of the mitochondrial calcium uniporter of Trypanosoma cruzi. Mol Biol Cell 30:1676–1690. doi: 10.1091/mbc.E19-03-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garske AL, Peters U, Cortesi AT, Perez JL, Shokat KM. 2011. Chemical genetic strategy for targeting protein kinases based on covalent complementarity. Proc Natl Acad Sci USA 108:15046–15052. doi: 10.1073/pnas.1111239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lendner M, Bottcher D, Delling C, Ojo KK, Van Voorhis WC, Daugschies A. 2015. A novel CDPK1 inhibitor–a potential treatment for cryptosporidiosis in calves? Parasitol Res 114:335–336. doi: 10.1007/s00436-014-4228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, Sibley LD. 2010. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature 465:359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy RC, Ojo KK, Larson ET, Castellanos-Gonzalez A, Perera BG, Keyloun KR, Kim JE, Bhandari JG, Muller NR, Verlinde CL, White AC, Jr, Merritt EA, Van Voorhis WC, Maly DJ. 2010. Discovery of potent and selective inhibitors of calcium-dependent protein kinase 1 (CDPK1) from C. parvum and T. gondii. ACS Med Chem Lett 1:331–335. doi: 10.1021/ml100096t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keyloun KR, Reid MC, Choi R, Song Y, Fox AMW, Hillesland HK, Zhang Z, Vidadala R, Merritt EA, Lau AOT, Maly DJ, Fan E, Barrett LK, Van Voorhis WC, Ojo KK. 2014. The gatekeeper residue and beyond: homologous calcium-dependent protein kinases as drug development targets for veterinarian Apicomplexa parasites. Parasitology 141:1499–1509. doi: 10.1017/S0031182014000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winzer P, Muller J, Imhof D, Ritler D, Uldry AC, Braga-Lagache S, Heller M, Ojo KK, Van Voorhis WC, Ortega-Mora LM, Hemphill A. 2020. Neospora caninum: differential proteome of multinucleated complexes induced by the bumped kinase inhibitor BKI-1294. Microorganisms 8. doi: 10.3390/microorganisms8060801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jimenez-Melendez A, Ojo KK, Wallace AM, Smith TR, Hemphill A, Balmer V, Regidor-Cerrillo J, Ortega-Mora LM, Hehl AB, Fan E, Maly DJ, Van Voorhis WC, Alvarez GG. 2017. In vitro efficacy of bumped kinase inhibitors against Besnoitia besnoiti tachyzoites. Int J Parasitol 47:811–821. doi: 10.1016/j.ijpara.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 48.Ojo KK, Eastman RT, Vidadala R, Zhang Z, Rivas KL, Choi R, Lutz JD, Reid MC, Fox AM, Hulverson MA, Kennedy M, Isoherranen N, Kim LM, Comess KM, Kempf DJ, Verlinde CL, Su XZ, Kappe SH, Maly DJ, Fan E, Van Voorhis WC. 2014. A specific inhibitor of PfCDPK4 blocks malaria transmission: chemical-genetic validation. J Infect Dis 209:275–284. doi: 10.1093/infdis/jit522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidadala RS, Ojo KK, Johnson SM, Zhang Z, Leonard SE, Mitra A, Choi R, Reid MC, Keyloun KR, Fox AM, Kennedy M, Silver-Brace T, Hume JC, Kappe S, Verlinde CL, Fan E, Merritt EA, Van Voorhis WC, Maly DJ. 2014. Development of potent and selective Plasmodium falciparum calcium-dependent protein kinase 4 (PfCDPK4) inhibitors that block the transmission of malaria to mosquitoes. Eur J Med Chem 74:562–573. doi: 10.1016/j.ejmech.2013.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wheeler RJ, Scheumann N, Wickstead B, Gull K, Vaughan S. 2013. Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: implications for microtubule-based morphogenesis and mutant analysis. Mol Microbiol 90:1339–1355. doi: 10.1111/mmi.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farr H, Gull K. 2012. Cytokinesis in trypanosomes. Cytoskeleton (Hoboken) 69:931–941. doi: 10.1002/cm.21074. [DOI] [PubMed] [Google Scholar]
- 52.Hu H, An T, Kurasawa Y, Zhou Q, Li Z. 2019. The trypanosome-specific proteins FPRC and CIF4 regulate cytokinesis initiation by recruiting CIF1 to the cytokinesis initiation site. J Biol Chem 294:16672–16683. doi: 10.1074/jbc.RA119.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Hu H, Lun ZR, Li Z. 2019. Functional analyses of an axonemal inner-arm dynein complex in the bloodstream form of Trypanosoma brucei uncover its essential role in cytokinesis initiation. Mol Microbiol 112:1718–1730. doi: 10.1111/mmi.14385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu X, Kumar P, Li Z, Wang CC. 2006. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J Biol Chem 281:9677–9687. doi: 10.1074/jbc.M511504200. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Q, Gu J, Lun ZR, Ayala FJ, Li Z. 2016. Two distinct cytokinesis pathways drive trypanosome cell division initiation from opposite cell ends. Proc Natl Acad Sci USA 113:3287–3292. doi: 10.1073/pnas.1601596113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar P, Wang CC. 2006. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot Cell 5:92–102. doi: 10.1128/EC.5.1.92-102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammarton TC, Kramer S, Tetley L, Boshart M, Mottram JC. 2007. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol Microbiol 65:1229–1248. doi: 10.1111/j.1365-2958.2007.05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fassolari M, Alonso GD. 2019. Aurora kinase protein family in Trypanosoma cruzi: novel role of an AUK-B homologue in kinetoplast replication. PLoS Negl Trop Dis 13:e0007256. doi: 10.1371/journal.pntd.0007256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soares Medeiros LC, South L, Peng D, Bustamante JM, Wang W, Bunkofske M, Perumal N, Sanchez-Valdez F, Tarleton RL. 2017. Rapid, selection-free, high-efficiency genome editing in protozoan parasites using CRISPR-Cas9 ribonucleoproteins. mBio 8. doi: 10.1128/mBio.01788-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tavernelli LE, Motta MCM, Goncalves CS, da Silva MS, Elias MC, Alonso VL, Serra E, Cribb P. 2019. Overexpression of Trypanosoma cruzi High Mobility Group B protein (TcHMGB) alters the nuclear structure, impairs cytokinesis and reduces the parasite infectivity. Sci Rep 9:192. doi: 10.1038/s41598-018-36718-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Lima AR, Noris-Suarez K, Bretana A, Contreras VT, Navarro MC, Perez-Ybarra L, Bubis J. 2017. Growth arrest and morphological changes triggered by emodin on Trypanosoma cruzi epimastigotes cultivated in axenic medium. Biochimie 142:31–40. doi: 10.1016/j.biochi.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, de Doncker S, Maes I, Mottram JC, Quail MA, Rijal S, Sanders M, Schonian G, Stark O, Sundar S, Vanaerschot M, Hertz-Fowler C, Dujardin JC, Berriman M. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mannaert A, Downing T, Imamura H, Dujardin JC. 2012. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol 28:370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 64.Damianou A, Burge RJ, Catta-Preta CMC, Geoghegan V, Nievas YR, Newling K, Brown E, Burchmore R, Rodenko B, Mottram JC. 2020. Essential roles for deubiquitination in Leishmania life cycle progression. PLoS Pathog 16:e1008455. doi: 10.1371/journal.ppat.1008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardoso MS, Junqueira C, Trigueiro RC, Shams-Eldin H, Macedo CS, Araujo PR, Gomes DA, Martinelli PM, Kimmel J, Stahl P, Niehus S, Schwarz RT, Previato JO, Mendonca-Previato L, Gazzinelli RT, Teixeira SM. 2013. Identification and functional analysis of Trypanosoma cruzi genes that encode proteins of the glycosylphosphatidylinositol biosynthetic pathway. PLoS Negl Trop Dis 7:e2369. doi: 10.1371/journal.pntd.0002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frodin M, Antal TL, Dummler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM. 2002. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J 21:5396–5407. doi: 10.1093/emboj/cdf551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urbaniak MD, Martin DM, Ferguson MA. 2013. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res 12:2233–2244. doi: 10.1021/pr400086y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oberholzer M, Morand S, Kunz S, Seebeck T. 2006. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol 145:117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 69.Bone GJ, Steinert M. 1956. Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature 178:308–309. doi: 10.1038/178308a0. [DOI] [PubMed] [Google Scholar]
- 70.Moreno SN, Silva J, Vercesi AE, Docampo R. 1994. Cytosolic-free calcium elevation in Trypanosoma cruzi is required for cell invasion. J Exp Med 180:1535–1540. doi: 10.1084/jem.180.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lander N, Chiurillo MA, Vercesi AE, Docampo R. 2017. Endogenous C-terminal tagging by CRISPR/Cas9 in Trypanosoma cruzi. Bio Protoc 7. doi: 10.21769/BioProtoc.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lander N, Chiurillo MA, Storey M, Vercesi AE, Docampo R. 2016. CRISPR/Cas9-mediated endogenous C-terminal tagging of Trypanosoma cruzi genes reveals the acidocalcisome localization of the inositol 1,4,5-trisphosphate receptor. J Biol Chem 291:25505–25515. doi: 10.1074/jbc.M116.749655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng D, Tarleton R. 2015. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb Genom 1:e000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lander N, Bernal C, Diez N, Anez N, Docampo R, Ramirez JL. 2010. Localization and developmental regulation of a dispersed gene family 1 protein in Trypanosoma cruzi. Infect Immun 78:231–240. doi: 10.1128/IAI.00780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bourguignon SC, de Souza W, Souto-Padron T. 1998. Localization of lectin-binding sites on the surface of Trypanosoma cruzi grown in chemically defined conditions. Histochem Cell Biol 110:527–534. doi: 10.1007/s004180050314. [DOI] [PubMed] [Google Scholar]
- 76.de Sousa MA. 1983. A simple method to purify biologically and antigenically preserved bloodstream trypomastigotes of Trypanosoma cruzi using DEAE-cellulose columns. Mem Inst Oswaldo Cruz 78:317–333. doi: 10.1590/s0074-02761983000300009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00738-21_Supp_1_seq11.pdf, PDF file, 7.3 MB (7.5MB, pdf)