Abstract
The H/ACA small nucleolar RNAs (snoRNAs) are involved in pseudouridylation of pre-rRNAs. They usually fold into a two-domain hairpin-hinge-hairpin-tail structure, with the conserved motifs H and ACA located in the hinge and tail, respectively. Synthetic RNA transcripts and extracts from HeLa cells were used to reconstitute human U17 and other H/ACA ribonucleoproteins (RNPs) in vitro. Competition and UV cross-linking experiments showed that proteins of about 60, 29, 23, and 14 kDa interact specifically with U17 RNA. Except for U17, RNPs could be reconstituted only with full-length H/ACA snoRNAs. For U17, the 3′-terminal stem-loop followed by box ACA (U17/3′st) was sufficient to form an RNP, and U17/3′st could compete other full-length H/ACA snoRNAs for assembly. The H/ACA-like domain that constitutes the 3′ moiety of human telomerase RNA (hTR), and its 3′-terminal stem-loop (hTR/3′st), also could form an RNP by binding H/ACA proteins. Hence, the 3′-terminal stem-loops of U17 and hTR have some specific features that distinguish them from other H/ACA RNAs. Antibodies that specifically recognize the human GAR1 (hGAR1) protein could immunoprecipitate H/ACA snoRNAs and hTR from HeLa cell extracts, which demonstrates that hGAR1 is a component of H/ACA snoRNPs and telomerase in vivo. Moreover, we show that in vitro-reconstituted RNPs contain hGAR1 and that binding of hGAR1 does not appear to be a prerequisite for the assembly of the other H/ACA proteins.
The nucleolus of eukaryotic cells harbors a large population of small nucleolar RNAs (snoRNAs) that play critical roles in pre-rRNA processing and modification (for reviews, see references 43, 63, 64, 67, 71, 74, and 75). Two major classes of snoRNAs, the box C/D and box H/ACA snoRNAs, have been identified based on conserved sequence elements (3, 22, 68). The RNA subunit of RNase MRP is involved in pre-rRNA processing in yeast (14, 42, 60), but this snoRNA does not contain any of the common motifs (boxes C, D, H, or ACA) and therefore is not a member of these families. The RNase P RNA (RPR1 in yeast) may also be considered a snoRNA since a point mutation in RPR1 affects pre-rRNA processing (11). RNase MRP and RNase P RNAs are structurally similar (21) and share common proteins (12).
The box H/ACA snoRNAs direct the site-specific pseudouridylation of pre-rRNA (23, 53). A few members of this class are involved in cleavage reactions; snR10 and snR30 in yeast (51, 66) and U17/E1, E2, and E3 in vertebrates (18, 46). The H/ACA snoRNAs generally fold into a hairpin-hinge-hairpin-tail structure, with box H (consensus sequence ANANNA) located in the hinge region and the ACA triplet (box) positioned 3 nucleotides (nt) upstream from the 3′ end (3, 22). The hairpins are usually interrupted by an internal loop, the pseudouridylation pocket, which contains short sequences complementary to rRNA that allow selection of the target uridine to be converted to pseudouridine (for the detailed mechanism, see reference 23).
The snoRNAs function in the form of ribonucleoproteins (RNPs). Each class of snoRNAs is associated with distinct nucleolar proteins (reviewed in references 71 and 74). To date, four common H/ACA proteins (Gar1p, Cbf5p, Nhp2p, and Nop10p), have been identified in Saccharomyces cerevisiae, and all are essential for growth. Depletion of Gar1p (25 kDa) inhibits 18S rRNA production and pseudouridylation of pre-rRNA; however, it does not affect accumulation of H/ACA snoRNAs (8, 26), suggesting that Gar1p is not necessary to trigger assembly of H/ACA snoRNPs. Gar1p is very highly conserved among members of the domain Eucarya (2, 72; F. Dragon, V. Pogačić, and W. Filipowicz, unpublished data); it is characterized by the presence of glycine- and arginine-rich (GAR) domains that flank a central core domain, which is necessary and sufficient for function in vivo (27). In vitro-translated Gar1p, and more specifically its core domain, can bind directly to snR10 and snR30 transcripts (2). The 65-kDa protein Cbf5p is the putative pseudouridine synthase (32, 38, 39, 77). Its depletion impairs pre-rRNA pseudouridylation and 18S rRNA production and also affects the accumulation of H/ACA snoRNAs and Gar1p (39). Homologues of Cbf5p have been identified in various species; it is named NAP57 in rat, dyskerin in humans, and MFL (minifly) or Nop60B in Drosophila (25, 30, 44, 54). Interestingly, mutated forms of dyskerin were found in patients suffering from dykeratosis congenita, an X-linked recessive disease causing bone marrow failure and other disorders (30), and mutations in the minifly gene cause growth and developmental defects in Drosophila (25). More recently, Nhp2p (22 kDa) and Nop10p (10 kDa) were also shown to be required for H/ACA snoRNP stability since genetic depletion of either protein results in defects similar to those observed upon depletion of Cbf5p (31, 72). Nhp2p contains a putative RNA-binding domain shared by some ribosomal proteins (references 31 and 72 and references therein). The protein Sbp1p (formerly SSB1) was shown to be associated with some H/ACA snoRNAs in yeast (15); however, Sbp1p is not essential for growth, and it is not clear whether it is associated with all H/ACA snoRNAs.
Telomerase is an RNP that synthesizes telomeres at the ends of eukaryotic chromosomes. This reverse transcriptase uses a short sequence in its RNA subunit as a template to synthesize sequence repeats that form telomeres (reviewed in references 5 and 6). Mammalian telomerase RNAs contain a 3′-terminal extension that structurally resembles an H/ACA snoRNA (47). The H/ACA-like domain is required for the accumulation and function of human telomerase RNA (hTR) in vivo (47) but not in vitro (1, 4). Telomerase RNAs in lower eukaryotes do not contain H/ACA-like domains; ciliate RNAs are relatively short and do not bear any 3′-terminal extension (55), while yeast telomerase RNA contains a 3′-terminal domain that binds Sm proteins of the spliceosomal snRNPs (62). It is not known whether the H/ACA domain of hTR acts as a pseudouridylation guide; however, the fact that a fraction of hTR is localized in the nucleolus makes this an intriguing possibility (47).
In this report, we describe the use of an in vitro system to characterize the components and the assembly of human H/ACA snoRNPs. We show that U17 and other H/ACA snoRNPs can be assembled in HeLa cell extracts using synthetic snoRNAs as substrates. Competition and UV cross-linking experiments indicated that four proteins of about 60, 29, 23, and 14 kDa interact specifically with U17 RNA. The 3′-terminal stem-loop fragment of U17 RNA could form an RNP and could compete full-length snoRNAs for assembly, whereas equivalent portions of other H/ACA snoRNAs could not. The H/ACA-like domain of hTR and its 3′-terminal stem-loop also formed RNPs in vitro and competed with H/ACA snoRNAs for assembly, showing that telomerase contains H/ACA proteins. Antibodies specific for human GAR1 (hGAR1) could immunoprecipitate H/ACA snoRNAs and hTR from HeLa cell extracts, as well as the in vitro-reconstituted RNPs.
MATERIALS AND METHODS
Preparation of extracts.
HeLa cells were grown in Joklik medium supplemented with 5% fetal bovine serum (Gibco BRL) and harvested at a density of 5 × 105 cells/ml. Nuclear extracts (NE) and whole cell extracts (WCE) used for in vitro reconstitution of RNPs were prepared as described in references 19 and 45, respectively. Some experiments were performed with NE that had been heat denatured (5 min at 95°C) or pretreated with proteinase K (0.5 mg/ml for 20 min at 30°C) or with microccocal nuclease (1.5 U/μl for 15 min at 30°C in the presence of 4 mM CaCl2, then quenched with 8 mM EGTA).
Plasmids.
Unless otherwise stated, plasmids used in this study were pUC19 derivatives containing an EcoRI-BamHI insert with a T7 promoter downstream of the EcoRI site. Plasmid pHU17(E-B) contains the human U17a (207 nt [33]) DNA sequence preceded by 2 additional bp and terminated by an NaeI restriction site to linearize the template DNA; U17 RNA transcripts (209 nt) contain two additional G residues at the 5′ end. Truncated derivatives of pHU17(E-B) were pHU17/3′D and pHU17/3′st, which respectively encode U17 positions 117 to 207 and 135 to 207 preceded by an additional G residue. Derivatives of these plasmids were generated to produce RNA fragments bearing a mutated ACA motif (deletion of ACA or A→G transition of the first A). Plasmid pHU17/3′stΔ23 was derived from pHU17/3′st and codes for an RNA fragment missing positions 158 to 180 (replaced by a UUCG tetraloop). Plasmid pHU19 contains human U19 (200 nt [36]) DNA sequence; BamHI-linearized DNA template produced U19 RNA transcripts (210 nt) with two additional G residues at the 5′ end and an extra GGGGGAUC sequence at the 3′ end. Truncated derivatives of pHU19 were pHU19/3′D and pHU19/3′st, which respectively encode positions 74 to 200 and 142 to 200 preceded by two and one additional G, respectively. Plasmid pHU64 contains human U64 (134 nt [22]) DNA sequence preceded by 3 additional bp and terminated by a BsaHI restriction site to linearize the template DNA; U64 RNA transcripts (138 nt) contain three additional G residues at the 5′ end and an extra G residue at the 3′ end. Plasmid pHU64/3′D, a truncated version of pHU64, encodes positions 74 to 134 with two additional G's at the 5′ end. The DNA sequence of hTR was PCR amplified from genomic DNA by using a 5′ primer bearing a T7 promoter tailing sequence and a 3′ primer containing an FspI restriction site preceded by a tailing T3 promoter sequence; hTR-complementary sequences of the 5′ and 3′ primers were designed as described in references 20 and 76, respectively. The EcoRI-BamHI PCR fragment was cloned in pUC19 to generate phTR1. T7 transcription of FspI-linearized phTR1 produces an RNA transcript of 451 nt (as reported by Mitchell et al. [47]) with no extra residue. Truncated versions of phTR1 were generated that encode positions 148 to 451 with three additional G's at the 5′ end (phTR148), positions 206 to 451 (phTR206), positions 268 to 451 (phTR268), positions 308 to 451 (phTR308), and positions 379 to 451 with two extra G's at the 5′ end (phTR/3′st). Plasmid phTR/3′stA→G bears an A-to-G transition at position 446. DNA fragments encoding human E2 and E3 snoRNAs (57) were inserted between the KpnI and HindIII restriction sites of pBluescript KS(−) to generate pHE2 and pHE3, respectively. Plasmid pHU14.5 encoding human U14 contains intron 5 of the HSC70 gene cloned between the XbaI and EcoRI sites of pBluescript KS(−). A DNA fragment encoding human RNase P RNA (H1) was cloned between the EcoRI and XbaI sites of pBluescript KS(−). Plasmids encoding human U2, U3, U4, U13, U15, U22, and 7-2/MRP RNAs have been described previously (33, 34, 69, 70).
In vitro transcription.
Except for carrier tRNAs (Boehringer Mannheim), all competitor RNAs used in this study were gel purified as described elsewhere (16) after in vitro transcription with a Megashortscript kit (Ambion). Radiolabeled RNAs were synthesized in a mixture (10 μl) containing 1 μg of template DNA, 40 mM Tris-HCl (pH 7.9), 7.5 mM MgCl2, 10 mM dithiothreitol, 10 mM NaCl, 2 mM spermidine, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 5 μM UTP, 20 U of RNasin (Promega), 10 U of T7 RNA polymerase (Promega), and 15 μCi of [α-32P]UTP (800 Ci/mmol; NEN).
In vitro reconstitution.
Reconstitution reactions were performed in 25 μl containing 20 mM HEPES (pH 7.9), 120 mM KCl, 2 mM MgCl2, 1.5 mM ATP, 40 U of RNasin, 2.5 μg of tRNAs, 5 to 10 μl of NE (or WCE), and 10 fmol of radiolabeled RNA. The mixtures were incubated at 30°C for 60 to 120 min, then 5 μl of heparin (30 mg/ml; Sigma) was added, and the mixtures were further incubated 10 min at 30°C. In competition experiments, the unlabeled competitor RNAs were usually tested at three different concentrations (10-, 100-, and 1,000-fold molar excess).
Sucrose gradients and RNase A/T1 mapping.
HeLa cell nucleoli were prepared as described elsewhere (68) and fractionated by centrifugation through 5 to 20% linear sucrose gradients in an SW41 rotor at 4°C for 16 h at 25,000 rpm. Twenty fractions were collected from the top of the gradient and subjected to RNase A-RNase T1 (RNase A/T1) mapping with a U17 antisense RNA probe (35). Protected fragments were analyzed on a 6% sequencing gel. Radiolabeled RNPs reconstituted in vitro were analyzed similarly except that gradient fractions were directly analyzed on a 6% sequencing gel after phenol-chloroform extraction and ethanol precipitation.
Native gel analysis and UV cross-linking.
In vitro-reconstituted RNPs were analyzed by nondenaturing polyacrylamide gel electrophoresis (PAGE) in Tris-glycine buffer as described elsewhere (37). Typically, 1/10 of the reconstitution mixture was analyzed on native gel, and the rest of the mixture was subjected to UV light irradiation. Droplets were deposited on a Parafilm sheet covering an aluminum block placed on ice. Samples were irradiated about 3 cm from the UV source for 20 min in a Stratalinker (Stratagene) set at 120 mJ. The samples were then incubated with RNases A (1 mg/ml) and T1 (1000 U/ml) at 37°C for 30 min, precipitated with 20% trichloroacetic acid in the presence of deoxycholate (0.8 mg/ml) for 20 min on ice, and centrifuged at room temperature for 15 min. Pellets were washed with 3 volumes of acetone for 10 min and centrifuged at room temperature, then dried on ice for 10 min, and resuspended in 6 μl of sodium dodecyl sulfate (SDS) loading buffer (59). Samples were fractionated by SDS-PAGE.
Cloning of hGAR1 cDNA.
Database searches identified a human expressed sequence tag (GenBank AA071104) that encodes most of the core domain and the C-terminal GAR domain of hGAR1. The PCR-amplified core domain portion of this clone served as a template for random priming (59) to generate a probe that was used to screen a plasmid cDNA library from human Namalwa (Burkitt lymphoma) cell line (65). The longest positive clones were sequenced with an ABI automated sequencer.
Ab production.
Using the PHD program (56) of the PredictProtein server (www.embl-heidelberg.de/predictprotein/), two regions of hGAR1 core domain that could potentially be immunogenic were chosen. The corresponding keyhole limpet hemocyanin-conjugated peptides P795 (CTTDENKVPYFNAPV) and P796 (RFLPRPPGEKGPPC) were used to immunize rabbits; note that the C in P796 does not belong to hGAR1 sequence. Both peptides antibodies (Abs) were affinity purified using Sulfo-Link (Pierce); anti-P795 and anti-P796 Abs were used for Western blotting in a multiscreen apparatus (Bio-Rad) and detected by enhanced chemiluminescence (Amersham Pharmacia Biotech). Only anti-P796 Abs (hereafter named anti-hGAR1) were suitable for immunoprecipitation experiments.
Immunoprecipitation of snoRNP particles from HeLa cells.
Protein A-Sepharose (PAS) beads (Amersham Pharmacia Biotech) were incubated with anti-hGAR1 Abs or antifibrillarin monoclonal Ab (MAb) 72B9 (a kind gift from J. A. Steitz and K. T. Tycowski, Yale University School of Medicine, and K. M. Pollard, The Scripps Research Institute) in NET-2 buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Nonidet P-40) with gentle agitation overnight at 4°C. HeLa cells were grown in suspension as described above, washed three times with phosphate-buffered saline, and resuspended in NET-2 buffer supplemented with the protease inhibitor Complete cocktail (Boehringer Mannheim). WCE prepared by sonication (3 times for 30 s each at 50 W; Braun sonicator) were spun at 15,000 × g for 10 min. For each immunoprecipitation, the lysate of 5 million cells was used. The lysate (500 μl) was mixed with the Ab-coated beads, and the mixtures were incubated with gentle agitation for 1 h at 4°C. The immunoprecipitates were washed five times with NET-2 buffer containing 150, 250, or 400 mM NaCl. RNAs were recovered by extraction with phenol-chloroform containing 0.5% SDS followed by ethanol precipitation in the presence of 40 μg of glycogen. 3′-end labeling of precipitated RNAs (17) was performed with [5′-32P]pCp (3,000 Ci/mmol; Amersham Pharmacia Biotech), and labeled RNAs were fractionated on 8% sequencing gels. Individual RNA species were identified by RNase A/T1 mapping (28) using antisense RNA probes to U3, U17, and U19 snoRNAs prepared as described previously (35, 36). The antisense RNA probe to hTR was prepared similarly by in vitro transcription with T3 RNA polymerase of plasmid phTR1 linearized by EcoRI. Protected RNA fragments were fractionated on 6% sequencing gels.
Immunoprecipitation of reconstituted RNPs.
In vitro-reconstituted RNPs were incubated for 2 h at 25°C with PAS beads alone (background control) or PAS beads that had been precoated with anti-hGAR1 Abs in the absence or presence of the immunizing peptide. Reactions were performed in reconstitution buffer (see above). Immunoprecipitates were washed five times with reconstitution buffer before phenol-chloroform extraction and ethanol precipitation. RNAs were resuspended in formamide loading buffer and analyzed on 6% sequencing gels.
In vitro reconstitution with hGAR1-depleted NE.
Depletion of hGAR1 was performed by two rounds of incubation of the NE with PAS beads coated with anti-hGAR1 Abs for 2 h at 4°C. For controls, NE was mock depleted (incubated with uncoated PAS beads) or incubated with PAS beads coated with immunoglobulin Gs (IgGs) from preimmune serum. In vitro reconstitution of U17 snoRNP was performed as described above. An aliquot (1/10) of the mixtures was analyzed on native gels; the rest of the samples were subjected to UV cross-linking and processed as described above.
Nucleotide sequence accession number.
The sequence of the hGAR1 cDNA has been deposited in the EMBL database under accession no. AJ276003.
RESULTS
U17 RNA can form an RNP in vitro.
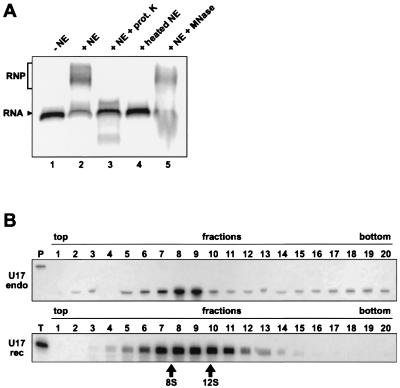
It has been shown previously that nuclear (splicing) extract preparations do not contain detectable amounts of snoRNAs (68). However, the snoRNPs are presumed to be assembled in the nucleoplasm (31, 58), and we reasoned that the NE could contain protein components of H/ACA snoRNPs. Radiolabeled transcripts corresponding to U17a snoRNA (33) were incubated with NE from HeLa cells, and the mixtures were fractionated by electrophoresis through native polyacrylamide gels (Fig. 1A). U17 RNA incubated with NE could form a larger complex, as shown by its retarded electrophoretic mobility (Fig. 1A, compare lanes 1 and 2). This mobility shift was not observed when we used NE that had been pretreated with proteinase K or heat denatured (lanes 3 and 4, respectively). Preincubation of the NE with micrococcal nuclease did not interfere with complex formation (lane 5). RNPs of similar mobility were also formed with U19 and U64 H/ACA RNAs (see below). Analysis of reconstituted U17 RNPs by centrifugation through 5 to 20% sucrose gradients indicated that they cosediment with native U17 monoparticles isolated from HeLa cell nucleoli at about 10S (Fig. 1B; for additional details, see the figure legend). Altogether these results demonstrate that U17 RNA can form an RNP having sedimentation properties similar to those of the endogenous particles when incubated with NE.
FIG. 1.
U17 RNA can form an RNP in vitro. (A) Gel mobility shift assay with radiolabeled U17 RNA. The labeled RNA was incubated in the absence (lane 1) or presence (lane 2) of NE or with NE preparations that had been pretreated with proteinase K (prot. K; lane 3), heat denatured (lane 4), or preincubated with micrococcal nuclease (MNase; lane 5). The arrowhead points to free RNA, and the bracket indicates the RNP complex. (B) Sedimentation of native (endo) and reconstituted (rec) U17 RNPs. HeLa cell nucleoli (top) and the in vitro reconstitution mixture with radiolabeled U17 RNA (bottom) were fractionated by centrifugation through 5 to 20% linear sucrose gradients. To detect native U17 monoparticles (top), each fraction was subjected to RNase A/T1 mapping (note that fraction 4 was lost during sample preparation). Reconstituted U17 RNPs were analyzed in parallel (bottom), together with sedimentation markers of 8S and 12S that correspond to fragments of Escherichia coli 16S rRNA, i.e., the 3′ domain and the 5′ and central domains, respectively. Fractions are numbered from top to bottom of the gradient. Lanes P and T correspond to undigested antisense probe and U17 transcript, respectively. Cerenkov scintillation counting of reconstituted RNPs indicated that fractions 8 and 9 correspond to peak fractions (not shown). The wider peak observed with reconstituted RNPs could result from loss of certain proteins during centrifugation (smaller particles) and association of some nonspecific proteins (larger particles).
U17 RNP contains proteins of the H/ACA class.
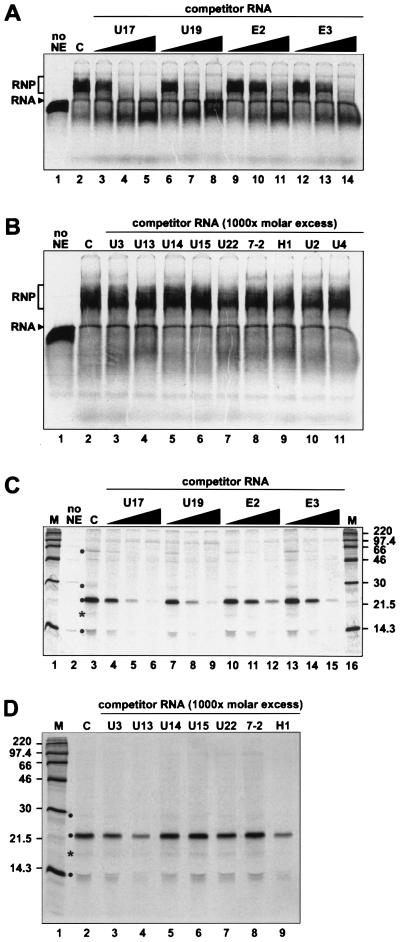
To determine whether U17 RNA interacts specifically with H/ACA proteins in vitro, competition experiments were performed with various classes of RNAs. Addition of increasing amounts of unlabeled competitor RNAs of the H/ACA class (U17, U19, E2, and E3) resulted in progressive disappearance of the radiolabeled U17 RNP complex (Fig. 2A). In contrast, RNAs belonging to other classes, such as the box C/D snoRNAs (U3, U13, U14, U15, and U22), RNAs of RNase MRP (7-2) and RNase P (H1), and spliceosomal RNAs (U2 and U4) could not compete for U17 RNP assembly (Fig. 2B). These results strongly suggest that in vitro-reconstituted U17 RNPs contain proteins of the H/ACA class.
FIG. 2.
In vitro-assembled U17 RNP contains a specific subset of proteins. (A) Gel mobility shift assay with competitor RNAs of the H/ACA class. Radiolabeled U17 RNA was incubated in the absence (lane 1) or presence (lane 2) of NE. Increasing amounts (10-, 100-, and 1,000-fold molar excess) of unlabeled competitor RNAs U17 (lanes 3 to 5), U19 (lanes 6 to 8), E2 (lanes 9 to 11), or E3 (lanes 12 to 14) were added to the assays. (B) Gel mobility shift assay with other classes of competitor RNAs. Radiolabeled U17 RNA was incubated in the absence (lane 1) or presence (lane 2) of NE. Box C/D RNAs (lanes 3 to 7), RNAs of RNase MRP (7-2; lane 8) and RNase P (H1; lane 9), and spliceosomal RNAs U2 (lane 10) and U4 (lane 11) were added as competitor at 1,000-fold molar excess. (C and D) UV cross-linking of proteins interacting with U17 RNA. Reconstituted U17 RNPs were subjected to UV light irradiation and RNase A/T1 digestion, and the cross-linked proteins were fractionated by SDS-PAGE. (C) UV cross-linking in the presence of competitor RNAs of the H/ACA class. Radiolabeled U17 RNA was incubated in the absence (lane 2) or presence (lane 3) of NE. Increasing amounts (10-, 100-, and 1,000-fold molar excess) of unlabeled competitor RNAs U17 (lanes 4 to 6), U19 (lanes 7 to 9), E2 (lanes 10 to 12), or E3 (lanes 13 to 15) were added to the assays. (D) UV cross-linking in the presence of nonspecific competitors. Box C/D RNAs (lanes 3 to 7), 7-2 RNA (lane 8), and RNA H1 (lane 9) were added as unlabeled competitors at 1,000-fold molar excess. Lane 2, cross-links with radiolabeled U17 RNA in the absence of the competitor. Cross-links that progressively disappeared upon increasing the concentration of specific competitor RNAs are indicated by a dot. The ∼17-kDa cross-link, likely to result from the proteolysis of the p23 cross-link, is marked with an asterisk. The masses of protein markers (lanes M) are indicated in kilodaltons.
Reconstituted RNPs were subjected to UV light irradiation (Fig. 2C and D), and the cross-linked proteins were analyzed by SDS-PAGE. Four proteins of about 60, 29, 23, and 14 kDa (hereafter called p60, p29, p23 and p14) could be specifically cross-linked to U17 RNA. Similar results were obtained when WCE instead of NE were used for reconstitution experiments (data not shown). Using different RNA preparations, we consistently observed that only those four cross-links progressively disappeared upon increasing the concentration of specific competitors, i.e., the H/ACA RNAs, whereas some nonspecific cross-links were not affected (Fig. 2C). It should be noted that in some experiments the bands corresponding to the p60 and p29 cross-links were very weak; it is possible that some RNA preparations did not fold optimally to allow proper binding of p60 and p29. The cross-linking pattern of reconstituted U17 RNP was not affected when competition was performed with RNAs of other classes (Fig. 2D); the p60 cross-link is not visible in Fig. 2D, but other experiments have shown that p60 binding is not impaired by RNAs other than those of the H/ACA class (data not shown). The ∼17-kDa cross-link marked with an asterisk also appears as H/ACA specific in the experiments shown in Fig. 2C and D. However, this cross-link was not seen in most other analyses (e.g., see Fig. 3C and 9B); most probably, it represents a breakdown product of the intense p23 cross-link. Therefore, we conclude that at least four proteins interact specifically with U17 RNA in the in vitro reconstitution system.
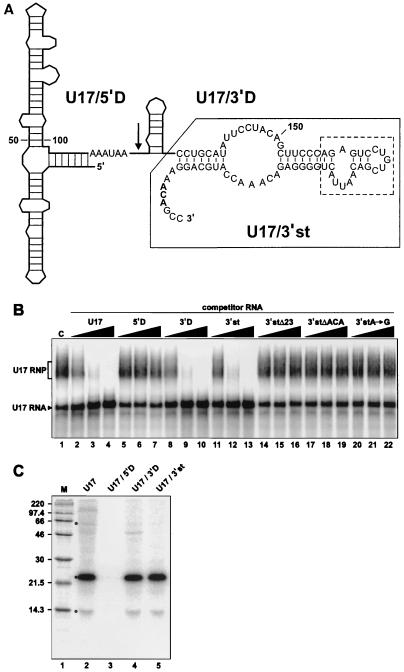
FIG. 3.
The 3′ domain RNA fragments of U17 can compete for RNP assembly. (A) Secondary structure model of human U17a RNA. Boxes H (AAAUAA) and ACA are shown in boldface. The arrow indicates the end of U17/5′D (positions 1 to 117), the 5′ domain RNA fragment that contains box H. The 3′ domain RNA fragment U17/3′D (positions 117 to 207) contains the ACA motif. The nucleotide sequence of the fragment corresponding to the 3′-terminal stem-loop of U17 (U17/3′st) is shown and boxed with a solid line, and the deleted portion of this fragment (23 nts replaced by a UUCG tetraloop) is indicated by a dashed line. The secondary structure model was adapted from references 10 and 61. (B) Gel mobility shift assay with U17 RNA and its derived RNA fragments used as competitors. As a control (C), radiolabeled U17 RNA was incubated with NE in the absence of competitor RNA (lane 1). Increasing amounts (10-, 100-, and 1,000-fold molar excess) of unlabeled competitor RNAs U17 (lanes 2 to 4), U17/5′D (lanes 5 to 7), U17/3′D (lanes 8 to 10), U17/3′st (lanes 11 to 13), and its derived mutated versions U17/3′stΔ23 (lanes 14 to 16), U17/3′stΔACA (lanes 17 to 19), and U17/3′stA→G (lanes 10 to 22) were added to the assays. (C) UV cross-linking with U17 RNA fragments. Radiolabeled U17 (lane 2) and its derived RNA fragments U17/5′D (lane 3), U17/3′D (lane 4), and U17/3′st (lane 5) were separately incubated with NE. The mixtures were subjected to UV light irradiation and RNase A/T1 digestion, and cross-linked proteins were fractionated by SDS-PAGE. The previously identified specific cross-links (Fig. 2C) are indicated by a dot. Positions of size markers (lane M) are indicated in kilodaltons.
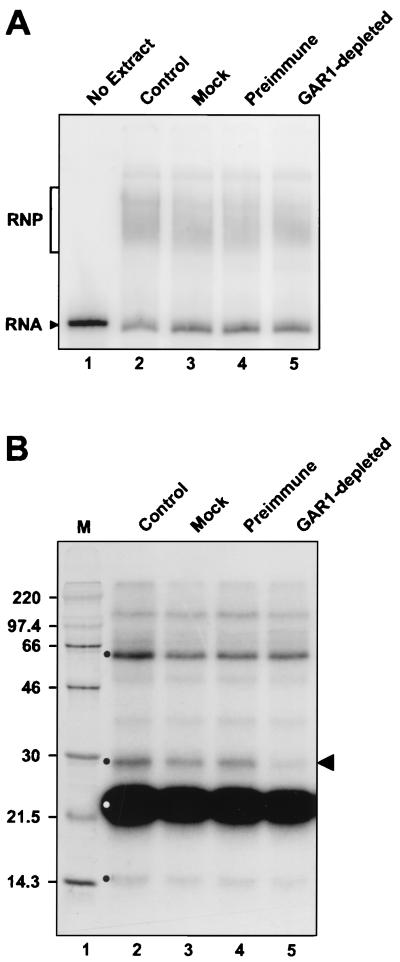
FIG. 9.
Assembly of U17 RNP does not require hGAR1. (A) Gel mobility shift assay with radiolabeled U17 RNA. Radiolabeled U17 RNA was incubated in the absence (lane 1) or presence (lane 2) of NE, mock-depleted NE (lane 3), NE preincubated with PAS beads coated with IgGs from preimmune serum (lane 4), or NE preincubated with PAS beads coated with anti-hGAR1 Abs. (B) UV cross-linking of proteins interacting with U17 RNA. In vitro reconstituted U17 RNPs were subjected to UV light irradiation and RNase A/T1 digestion, and the cross-linked proteins were fractionated by SDS-PAGE. Four H/ACA-specific proteins of about 60, 29, 23, and 14 kDa (indicated by a dot) could be cross-linked to U17 RNA incubated with NE (control lane 2), mock-depleted NE (lane 3), or NE pretreated with preimmune serum (lane 4). Only the cross-link corresponding to the 29-kDa protein (indicated by the arrowhead) was greatly diminished with the GAR1-depleted NE (lane 5). Preincubation with anti-hGAR1 Abs resulted in depletion of ∼70% of hGAR1, as verified by Western analysis (data not shown). Sizes of markers (M) are indicated in kilodaltons.
Molecular dissection of U17 snoRNA.
The U17 snoRNA is conserved among vertebrates, and secondary structure models of U17 have been proposed (10, 61). To delineate regions of human U17 RNA that are important for RNP assembly in vitro, different fragments of U17 were generated (Fig. 3A). First, U17 RNA was divided in two fragments: a 5′ domain fragment (U17/5′D; nt 1 to 117) that terminates 2 nt downstream of box H, and a 3′ domain fragment (U17/3′D; nt 117 to 207) containing box ACA. U17/5′D was unable to compete with U17 RNA for assembly (Fig. 3B, lanes 5 to 7). Addition of a 5′-cap structure and extension of the U17/5′D RNA to position 135, which places a stable hairpin at its 3′ end, to prevent potential degradation of the RNA did not improve its ability to compete for assembly (data not shown). In contrast to U17/5′D, U17/3′D competed as efficiently as the complete U17 RNA (lanes 8 to 10). U17/3′D was further trimmed by deleting its 5′ hairpin (Fig. 3A). This fragment (U17/3′st; nt 135 to 207) also proved to be a very potent competitor (lanes 11 to 13). Further trimming of U17/3′st by deleting 23 nt in its apical part (U17/3′stΔ23) produced a fragment that could no longer compete with U17 RNA (lanes 14 to 16). Note that the deleted portion was replaced by a very stable UUCG tetraloop (13) in order to favor the formation of a helix above the putative pseudouridylation pocket. Mutations in the ACA motif of U17/3′st (deleting it or changing its first A into G) also generated fragments that were unable to compete (lanes 17 to 19 and 20 to 22, respectively). Identical results were obtained when the ACA motif was mutated in the U17/3′D RNA fragment or full-length U17 (data not shown). In contrast, deletion of box H in U17 had no effect on its ability to compete (data not shown). Particles formed with different U17 subfragments were also analyzed by native gel electrophoresis and UV cross-linking. Native gel analysis revealed that consistent with the results of competition experiments, U17/3′D and U17/3′st but not U17/5′D could form an RNP in vitro (data not shown). UV light irradiation of complexes formed with U17/3′D and U17/3′st RNAs indicated that these fragments efficiently cross-link to p23 and p14, and also weakly to p60 (Fig. 3C, lanes 4 and 5). Taken together, these results indicate that the 3′-terminal hairpin of U17 can interact with H/ACA-specific proteins and that the ACA motif is a critical determinant of the assembly in vitro.
Similar molecular dissections were performed with other H/ACA snoRNAs. Competition experiments with fragments of U19 and U64 RNAs revealed that their 3′ domains, nt 70 to 200 of U19 (36) and nt 74 to 134 of U64 (22), or the 3′-terminal stem-loop with box ACA of U19 (residues 142 to 200), are unable to compete with full-length snoRNAs for assembly. Competition of different fragments for the assembly of radiolabeled U19 RNA is shown in Fig. 4. RNAs U17 (lanes 2 to 4), U17/3′st (lanes 5 to 7), U19 (lanes 8 to 10), and U64 (lanes 17 to 19) were effective competitors; however, fragments U19/3′D (lanes 11 to 13) and U19/3′st (lanes 14 to 16) could not compete for assembly of the parent U19 RNA. Similarly, a U64 RNA fragment encompassing its 3′ domain (U64/3′D), which consists of a single hairpin followed by the ACA motif, was unable to inhibit formation of U19 RNP (lanes 20 to 22). Identical results were obtained when the same set of RNAs was used in competition experiments with radiolabeled U17 or U64 RNAs (data not shown). Fragments corresponding to the 3′ domain of E2 and E3 snoRNAs were also tested in competition experiments, but none of them could compete for assembly (data not shown). Providing a 5′-cap structure to these noncompeting 3′ domain RNA fragments did not improve their ability to compete (data not shown). Taken together, these results suggest that the 3′-terminal stem-loop of U17 may have some structural advantage for assembly over the corresponding fragments of other H/ACA snoRNAs or that a U17-specific protein interacts with this region of U17 (see Discussion).
FIG. 4.
RNA fragments of U19 and U64 cannot compete for snoRNP assembly. The gel mobility shift assay was performed with radiolabeled U19 RNA, which was incubated with NE in the absence of competitor RNA (control [c]; lane 1) or in the presence of increasing concentrations (10-, 100-, and 1,000-fold molar excess) of unlabeled competitor RNAs U17 (lanes 2 to 4), U17/3′st (lanes 5 to 7), U19 (lanes 8 to 10), U19 3′ domain fragment (U19/3′D; lanes 11 to 13), U19 3′-terminal stem-loop fragment (U19/3′st; lanes 14 to 16), U64 (lanes 17 to 19), and U64 3′ domain fragment (U64/3′D; lanes 20 to 22).
The RNA subunit of telomerase is a specific H/ACA competitor.
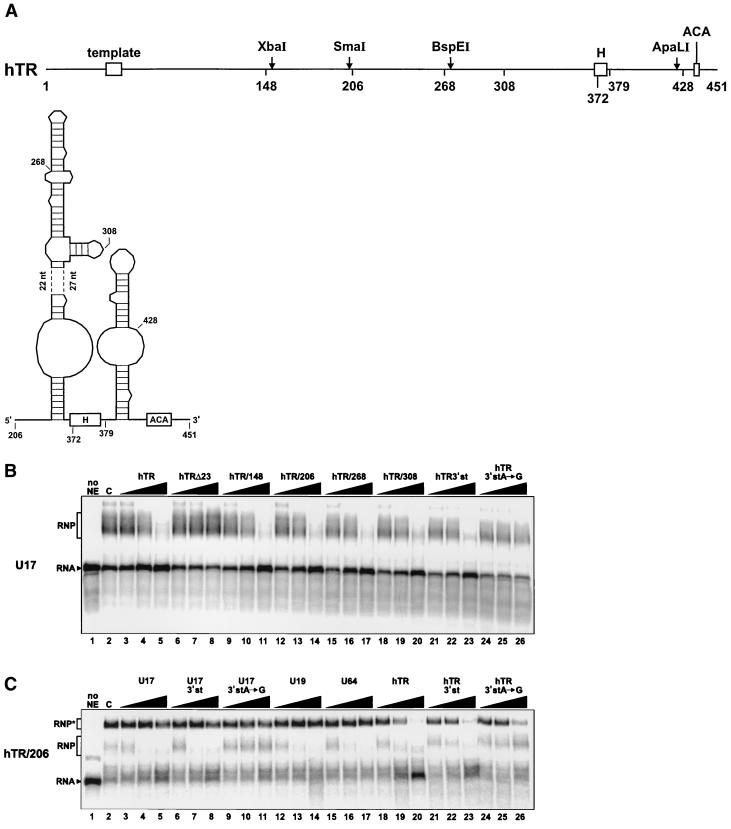
hTR contains a 3′-terminal domain that structurally resembles an H/ACA snoRNA (47) (Fig. 5A). We tested whether hTR or its derived RNA fragments could compete for U17 RNP assembly in vitro. For these experiments, we used WCE because active telomerase can be reconstituted from such extracts (1, 49). As shown in Fig. 5B (lanes 3 to 5), hTR proved to be a potent competitor for U17 assembly, suggesting that H/ACA proteins interact with hTR. Deletion of 23 nt at the 3′ end of hTR abolished its ability to compete (hTRΔ23; lanes 6 to 8). In contrast, truncations from the 5′ end of hTR had no effect. Indeed, RNA fragments hTR/148 (lanes 9 to 11), hTR/206 (lanes 12 to 14), hTR/268 (lanes 15 to 17), and hTR/308 (lanes 18 to 20) were able to compete for U17 assembly. A fragment encompassing the 3′-terminal stem-loop that follows box H (hTR/3′st; nt 379 to 451) could also compete for U17 RNP assembly (lanes 21 to 23). However, an A-to-G transition in the ACA motif of hTR/3′st abolished its ability to compete (lanes 24 to 26). Thus, hTR/3′st behaves similarly to U17/3′st, as it can bind H/ACA proteins and form an RNP in vitro (see below) and its ACA motif is critical for assembly.
FIG. 5.
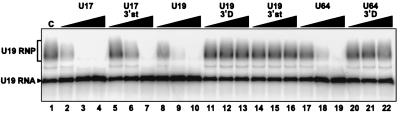
hTR and its derived 3′-terminal fragments can compete for U17 assembly. (A) Schematic structure of hTR RNA (nt 1 to 451) and secondary structure model of its H/ACA-like domain. Open boxes represent the template region and the H and ACA motifs. Relevant restriction sites are indicated by an arrow. Numbers indicate the first nucleotides of 5′-truncated hTR RNA fragments. The secondary structure model was adapted from reference 47. (B) Gel mobility shift assay in the presence of competitor RNAs hTR and its derived RNA fragments. Radiolabeled U17 RNA was incubated in the absence (lane 1) or presence (lane 2) of WCE. Increasing amounts (10-, 100-, and 1,000-fold molar excess) of unlabeled competitor RNAs hTR (lanes 3 to 5), hTRΔ23 (lanes 6 to 8), hTR/148 (lanes 9 to 11), hTR/206 (lanes 12 to 14), hTR/268 (lanes 15 to 17), hTR/308 (lanes 18 to 20), hTR/3′st (nt 378 to 451; lanes 21 to 23), and hTR/3′stA→G (lanes 24 to 26), which contains an A-to-G mutation in the ACA motif, were added to the assays. (C) Gel mobility shift assay with radiolabeled hTR/206. The labeled RNA was incubated in the absence (lane 1) or presence (lane 2) of WCE and could form two RNP complexes, designated RNP and RNP∗ for the faster- and the slower-migrating complexes, respectively. Unlabeled competitor RNAs U17 (lanes 3 to 5), U17/3′st (lanes 6 to 8), U17/3′stA→G (lanes 9 to 11), U19 (lanes 12 to 14), U64 (lanes 15 to 17), hTR (lanes 18 to 20), hTR/3′st (lanes 21 to 23) and hTR/3′stA→G (lanes 24 to 26) were added in increasing concentrations (10-, 100-, and 1,000-fold molar excess).
To determine whether the 3′-terminal domain of hTR behaves like a genuine H/ACA snoRNA, in vitro reconstitution experiments with radiolabeled hTR/206 RNA fragment, which corresponds to the H/ACA-like domain of hTR (47) (Fig. 5A), were performed in the presence of diverse unlabeled competitor H/ACA snoRNAs (Fig. 5C). When incubated with WCE, the RNA fragment hTR/206 could form two different complexes, as judged by gel mobility shift assay (Fig. 5C, lane 2). Competition experiments with H/ACA snoRNAs U17 (lanes 3 to 5), U19 (lanes 12 to 14), and U64 (lanes 15 to 17) showed that only the fast-migrating complex (RNP in Fig. 5C) progressively disappeared with increasing concentrations of competitor RNAs. Moreover, the RNA fragment U17/3′st (lanes 6 to 8), but not its derived mutant U17/3′stA→G (lanes 9 to 11), could compete for assembly of hTR/206 RNP. Addition of these competitor RNAs had no significant effect on the formation of the slow-migrating complex of hTR/206 (RNP∗ in Fig. 5C). In contrast, competition experiments with hTR (lanes 18 to 20) and its derived RNA fragment hTR/3′stem (lanes 21 to 23) revealed that both complexes (hTR/206 RNP and RNP∗) progressively disappeared with increasing concentrations of competitor RNAs. Furthermore, the mutant RNA fragment hTR/3′stA→G (lanes 24 to 26) had no effect on the formation of the fast-migrating complex RNP but had a noticeable effect on the formation of the slow-migrating complex RNP∗. Virtually identical results were obtained when radiolabeled hTR/3′st fragment was used in reconstitution experiments (data not shown). Taken together, these results indicate that the H/ACA-like domain of hTR interacts specifically with H/ACA proteins. Moreover, the fact that only hTR and hTR/3′st could compete for the formation of hTR/206 RNP∗ suggests that the 3′-terminal hairpin of hTR may, in addition, bind some hTR-specific protein(s). We have attempted to identify proteins interacting with hTR, or its fragments, using the UV cross-linking approach. Although in some experiments the p23 and p14 cross-links were clearly visible (results not shown), patterns of radiolabeled proteins were not reproducible enough to allow conclusive interpretation of the data.
Cloning of hGAR1 and generation of Abs.
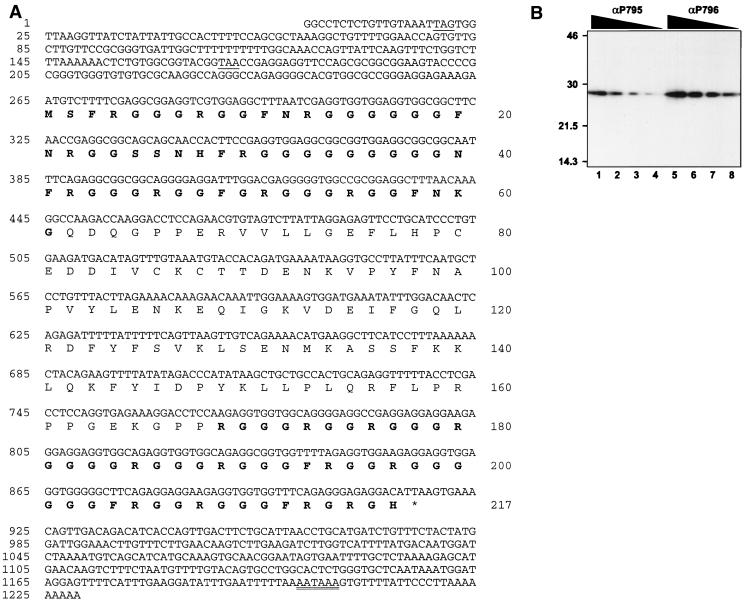
The nucleolar protein GAR1 was first identified in S. cerevisiae (26). Yeast Gar1p is specifically associated with H/ACA snoRNAs in vivo (3, 22, 26). This protein is characterized by the presence of GAR domains that flank a highly conserved core domain (2, 72). We have cloned a cDNA encoding hGAR1 (see Materials and Methods). The cDNA and predicted amino acid sequences of hGAR1 are presented in Fig. 6A. hGAR1 is composed of 217 amino acids; it has a pI of 10.9 and a predicted molecular mass of 22.3 kDa. Its core domain is flanked by GAR domains that account for half of the protein sequence. Two regions of the core domain were chosen to raise peptide Abs. As judged by Western blotting, both affinity-purified Abs recognize a single polypeptide of about 28 kDa (Fig. 6B). As for yeast Gar1p (26), the gel mobility of hGAR1 is slower than would be predicted from the calculated mass (∼28 kDa versus 22.3 kDa). This discrepancy could be due to the highly basic charge of hGAR1 or to posttranslational modifications, such as methylation of arginines in the GAR domains (24).
FIG. 6.
(A) hGAR1 cDNA and predicted protein sequences. Nucleotide positions are given on the left, and amino acids are numbered on the right. The open reading frame (nt 265 to 915) encodes a protein of 217 amino acids. Sequences of the GAR domains are in bold. Two in-frame termination codons of the 5′ untranslated region are underlined. A putative polyadenylation signal is double underlined. (B) Increasing dilutions of affinity-purified antipeptide Abs αP795 and αP796 were used for Western blotting of HeLa cell extracts in a multiscreen apparatus. Sizes are indicated in kilodaltons.
Immunoprecipitation of human H/ACA snoRNPs.
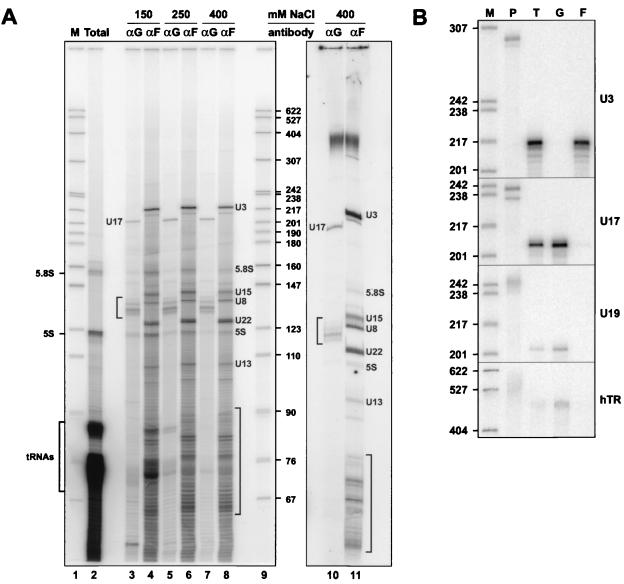
In yeast, Gar1p is associated with the H/ACA snoRNAs (3, 22), whereas Nop1p (fibrillarin) interacts with the box C/D snoRNAs (3, 22, 40, 68). Immunoprecipitation experiments were performed with WCE using the anti-hGAR1 Abs and the antifibrillarin MAb 72B9; the immunoprecipitated RNAs were labeled at the 3′ end with [5′-32P]pCp and fractionated on a sequencing gel. As shown in Fig. 7A, the immunoprecipitates are enriched in different population of RNAs. This is more apparent in the immunoprecipitation reactions carried out in the presence of 400 mM salt (lanes 7, 8, 10, and 11). Most of the anti-hGAR1 precipitated RNAs migrate in the range of 130 to 140 nt, with the exception of a very strong band at about 200 nt. This is in agreement with the size of known H/ACA snoRNAs: U17 (205 to 207 nt [33]), U19 (200 nt [36]), E2 and E3 (154 and 135 nt, respectively [57]), and U64 to U72 (about 130 to 140 nt [22]). The antifibrillarin MAb precipitated the box C/D snoRNAs that vary much more in size, such as U3 (217 nt), U8 (135 nt), U13 (105 nt), U15 (146 to 148 nt), U22 (125 nt), and some unidentified species in addition to the short antisense snoRNAs (60 to 90 nt).
FIG. 7.
Anti-hGAR1 Abs immunoprecipitate human H/ACA snoRNPs. (A) 3′-end-labeled RNAs isolated from WCE (an aliquot) (Total; lane 2) and from anti-hGAR1 (αG; lanes 3, 5, 7, and 10) and antifibrillarin (αF; lanes 4, 6, 8, and 11) immunoprecipitates were fractionated on an 8% sequencing gels. Sizes of DNA markers obtained from MspI-digested pBR322 (M; lanes 1 and 9) are indicated in nucleotides. Immunoprecipitates were washed with NET-2 buffer containing different concentrations of NaCl (indicated above the lanes). [5′-32P]pCp-labeled RNAs in lanes 10 and 11 were separated on a gel without prior ethanol precipitation. (B) RNase mappings. Antisense RNA probes (lanes P) specific for U3, U17, U19, and hTR were used for RNase A/T1 mapping of RNA isolated from WCE (lanes T) and from anti-hGAR1 (lanes G) and antifibrillarin (lanes F) immunoprecipitates.
To ascertain that the anti-hGAR1 immunoprecipitates correspond to H/ACA snoRNAs, RNAse A/T1 mapping of individual RNAs was performed with antisense RNA probes (Fig. 7B). Each probe was used to map the snoRNA from anti-hGAR1 and antifibrillarin immunoprecipitates (lanes G and F, respectively), as well as total cell extract (lanes T). The anti-U3 probe gave a strong signal in the antifibrillarin precipitate, but no signal was detected in the anti-hGAR1 immunoprecipitate. To the contrary, the H/ACA snoRNAs U17 and U19 were detected only with the anti-hGAR1 precipitates, thus showing that the anti-hGAR1 Abs are specific for the H/ACA class of snoRNPs. Moreover, hTR could be detected in the anti-hGAR1 but not antifibrillarin immunoprecipitate. These results demonstrate that hGAR1 is a protein subunit of H/ACA snoRNPs and telomerase in vivo.
Reconstituted RNPs contain hGAR1.
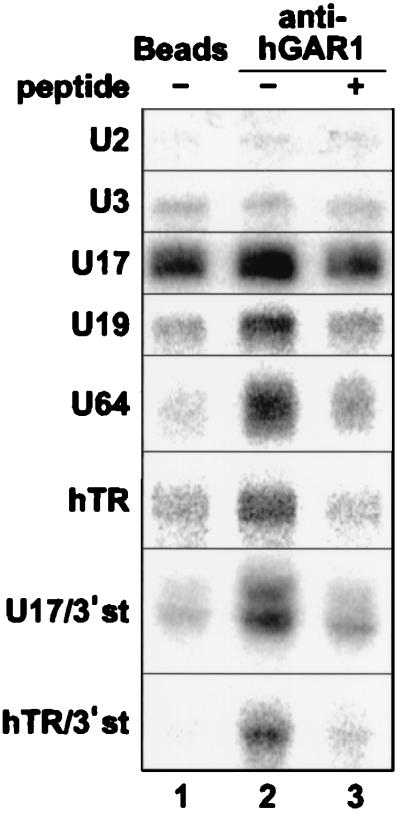
Immunoprecipitation experiments were also performed with radiolabeled H/ACA RNPs reconstituted in vitro with WCE (Fig. 8). For controls, radiolabeled U2 spliceosomal snRNA and U3 box C/D snoRNA, which should not interact with hGAR1, were treated similarly in parallel. Neither U2 nor U3 RNA could be immunoprecipitated with anti-hGAR1 Abs. In contrast, all other RNAs tested (U17, U19, U64, hTR, U17/3′st, and hTR/3′st) could be specifically immunoprecipitated with anti-hGAR1 Abs: the radioactive signals increased when resulting RNPs were incubated with anti-hGAR1 Abs (compare lanes 1 and 2) but fell to background levels when the anti-hGAR1 Abs were preincubated with the immunizing peptide (lane 3). U17 RNPs gave a strong background when incubated with beads alone (lane 1); nevertheless, PhosphorImager quantification (not shown) indicated that the signal increased about twofold when incubated with anti-hGAR1 Abs (lane 2) and reverted to the background level upon addition of the immunizing peptide (lane 3). These results demonstrate that hGAR1 is a component of in vitro-reconstituted RNPs.
FIG. 8.
Reconstituted RNPs contain hGAR1. Radiolabeled RNAs listed on the left were individually incubated with WCE, and aliquots of the mixtures were later incubated with PAS beads (lanes 1; background control) or PAS beads that had been precoated with anti-hGAR1 peptide Abs in the absence (lanes 2) or presence (lanes 3) of the immunizing peptide.
Reconstitution of U17 RNPs with hGAR1-depleted NE.
In yeast, depletion of Gar1p impairs pseudouridylation of the 35S pre-rRNA and formation of 18S rRNA but has no effect on the accumulation of H/ACA snoRNAs (8, 26), suggesting that Gar1p is not a primary binding protein required for the assembly of the other H/ACA proteins but is necessary for the activity or proper localization of H/ACA snoRNPs. We have found that immunodepletion of hGAR1 from the NE does not impair the ability of U17 RNA to associate with other proteins and form an RNP (Fig. 9A, compare lanes 2 and 5). Similar mobility of complexes formed in the immunodepleted and control extracts may be due to the small size of hGAR1 or the difference in the conformation of the hGAR1-free particle, resulting in its slower migration in a gel. When the reconstitution mixtures were subjected to UV light irradiation, only the 29-kDa cross-link (Fig. 9B) was altered when the hGAR1-depleted NE was used (compare lanes 2 and 5), while other cross-links were not affected. We conclude that, as observed in vivo in yeast (8, 26), hGAR1 is not required for the assembly of other H/ACA proteins and that the 29-kDa cross-link most probably corresponds to hGAR1.
DISCUSSION
Eukaryotic cells contain two major families of snoRNAs, the box C/D and the box H/ACA RNAs, involved in 2′-O-ribose methylation and pseudouridylation of pre-rRNA, respectively. In vertebrates, the C/D and H/ACA snoRNAs are usually encoded within introns of pre-mRNAs and, with few exceptions (9), are processed from the debranched introns by an exonucleolytic mechanism (35, 73). Intronic sequences that flank the snoRNA sequence can be changed without affecting the production of mature snoRNPs, indicating that the snoRNA itself contains all of the signals necessary for its processing and packaging into RNPs.
Experiments described in this work show that in vitro-transcribed human H/ACA snoRNAs, and also the H/ACA-like domain of hTR, can be assembled into RNPs when incubated with extracts prepared from HeLa cells. The assembly was competed by an excess of different H/ACA snoRNAs but not by other classes of small RNAs, or H/ACA snoRNAs containing mutations in the ACA motif, demonstrating that reconstituted RNPs contain H/ACA-specific proteins. In addition, analysis of the reconstituted U17 RNPs on sucrose gradients indicated that they cosediment with the endogenous U17 snoRNPs at about 10S.
We used the U17 snoRNA as a model in most of the reconstitution experiments. U17 is one of the very few H/ACA snoRNAs that are involved in cleavage of the pre-rRNA (18, 46). The 3′ domain of U17 RNA has the structural features of a pseudouridylation guide RNA, but its target remains unknown (23). Experiments combining UV cross-linking and competition assays have revealed that U17 interacts specifically with four proteins of about 60, 29, 23, and 14 kDa, termed p60, p29, p23, and p14, respectively. Since UV light irradiation generates zero-length cross-links, our data indicates that p60, p29, p23, and p14 are in contact with U17 RNA, most probably with single-stranded regions of the molecule. The apparent molecular masses of these four proteins resembles those of the four known yeast H/ACA proteins: the putative pseudouridine synthase Cbf5p of 65 kDa (32, 38, 39, 77), the 25-kDa protein Gar1p, which is required for snoRNP function (8, 26), and two recently identified proteins of 22 and 10 kDa termed Nhp2p and Nop10p, respectively (31, 72). Four tightly associated proteins of 65, 25 (Gar1p), 23, and 10 kDa were also identified in the yeast snR30 RNP (41).
We have cloned a full-length cDNA encoding hGAR1, the human homologue of yeast Gar1p. Like its counterparts in other eukaryotes (3, 72; Dragon et al., unpublished data), hGAR1 is composed of a highly conserved core region flanked by two GAR domains that account for approximately half of the protein. Abs raised against hGAR1 were found to immunoprecipitate H/ACA snoRNAs from HeLa cell extracts (Fig. 7), as well as RNPs reconstituted in vitro with radiolabeled H/ACA snoRNAs (Fig. 8), demonstrating that hGAR1 associates specifically with these RNAs in vivo and in vitro. It is very likely that the 29-kDa cross-linked protein corresponds to hGAR1 because p29 and hGAR1 have similar electrophoretic mobilities and, more importantly, immunodepletion of hGAR1 from the HeLa nuclear extract specifically decreases the amount of the 29-kDa cross-link seen with the reconstituted U17 RNP (Fig. 9). Since immunodepletion of hGAR1 does not impair the ability of other H/ACA proteins to assemble with U17 RNA, our results suggest that, as in yeast, association of hGAR1 with the H/ACA RNAs is not essential for binding of other proteins and may be a late assembly event.
It is likely that the 60-kDa cross-linked protein seen with the reconstituted U17 RNP corresponds to the 58-kDa protein dyskerin, the human homologue of yeast Cbf5p (30, 48). It is also probable that p23 and p14 represent human homologues of yeast Nhp2p (22 kDa) and Nop10p (10 kDa), respectively (31, 72). Interestingly, Nhp2p has sequence similarity to ribosomal protein L30 (formerly L32) and contains a putative RNA-binding domain (references 31 and 72 and references therein), and human p23 is always the most efficiently cross-linked protein. We have recently cloned cDNAs encoding the human homologues of Nhp2p and Nop10p. The human proteins, with apparent molecular masses of 22 and 12 kDa, respectively, associate with the H/ACA snoRNAs, as demonstrated by coimmunoprecipitation experiments performed with extracts from transfected mammalian cells (Pogačić et al., unpublished data). In summary, the in vitro reconstitution system seems to mimic the in vivo situation and provides a tool to study the assembly of human H/ACA snoRNPs.
Using reconstitution, competition, and UV cross-linking assays, we have delineated regions of U17 that are important for RNP assembly in vitro. The 5′-domain fragment that includes box H (U17/5′D) was found to be inactive in all these assays. In contrast, fragments corresponding to the entire 3′ domain (U17/3′D) or the 3′-terminal stem-loop (U17/3′st), both containing box ACA, competed for assembly as efficiently as the full-length U17 RNA. Deletion of the apical part of the 3′-terminal stem-loop or mutation of the ACA motif generated fragments that could no longer compete for U17 assembly. UV cross-linking experiments showed that p23 and p14 cross-linked efficiently to fragments U17/3′D and U17/3′st. In addition, we have shown that anti-hGAR1 Abs immunoprecipitate RNPs assembled with U17/3′st. Altogether, these results indicated that the 3′-terminal stem-loop binds proteins that are critical for U17 assembly in vitro and that the integrity of this stem-loop and box ACA are required for RNP formation.
hTR contains a 3′ domain that structurally resembles an H/ACA snoRNA (47) (Fig. 5A). This domain is essential for hTR stability and function in vivo (47), but is not required for telomerase activity in vitro (1, 4). Experiments carried out in this work indicate that hTR binds proteins specific to the H/ACA class of snoRNAs. 5′-terminal fragments of hTR that maintain telomerase activity in vitro (1, 4) were unable to compete for U17 assembly. In contrast, 3′-terminal fragments of hTR were very good competitors. Most notably, a short fragment corresponding to the 3′-terminal stem-loop of hTR followed by the ACA motif (hTR/3′st) competed as well as full-length hTR, but changing its ACA motif to GCA (hTR/3′stA→G) completely abolished its ability to compete (Fig. 5B). The conclusion that the H/ACA-like domain of hTR and its 3′-terminal stem-loop fragment associate with H/ACA-specific proteins was further corroborated by immunoprecipitation experiments. We have found that anti-hGAR1 Abs can immunoprecipitate endogenous hTR RNA from HeLa cell extracts and also radiolabeled hTR or its 3′-terminal stem-loop assembled into RNPs in vitro (Fig. 7 and 8). Association of the H/ACA-like domain of hTR with H/ACA proteins is further supported by the recent findings of Narayanan et al. (52). These authors have found that in microinjected Xenopus oocytes, hTR is targeted to the nucleolus by a box ACA-dependent mechanism. Moreover, Mitchell et al. (48) have shown that dyskerin is a component of H/ACA snoRNPs and telomerase in vivo. Our analysis of RNP formation with the H/ACA-like domain of hTR (fragment hTR/206) revealed that it forms two sets of complexes, designated RNP and RNP∗ (Fig. 5C). H/ACA snoRNA competitors could only inhibit formation of the fast-migrating RNP complexes, and not the slow-migrating RNP∗, while hTR and hTR/3′st competitor RNAs could inhibit formation of both complexes (Fig. 5C). These results suggest that the 3′-terminal stem-loop of hTR interacts with specific protein(s) in addition to common H/ACA proteins.
Specific structural features are required for the accumulation and function of H/ACA snoRNAs in vivo. Boxes H and ACA, and the integrity of both 5′ and 3′ hairpins, in particular the helical stems that bracket pseudouridylation pockets, are critical for the stability and function of H/ACA snoRNAs in S. cerevisiae (3, 7, 22, 23, 53). In microinjected Xenopus oocytes, a single pseudouridylation stem-loop of the human snoRNA U65 is targeted to the nucleolus when flanked by boxes H and ACA (52). Our findings that U17/3′st and hTR/3′st RNA fragments, lacking the entire upstream domain and box H, can form stable hGAR1-containing RNPs in vitro and can compete for assembly of full-length H/ACA snoRNAs does not correlate with the aforementioned studies performed with other H/ACA snoRNAs. It appears that the 3′-terminal stem-loops of U17 and hTR have some specific features that distinguish them from equivalent segments of other H/ACA snoRNAs. This suggestion is supported by our in vitro experiments with several other H/ACA snoRNAs (U19, U64, E2, and E3), indicating that their 3′-terminal fragments cannot compete for assembly of full-length snoRNAs (Fig. 4 and data not shown). Hence, the in vitro assembly of U17/3′st and hTR/3′st into RNPs indeed seems to reflect special properties of these RNAs. It is possible that U17/3′st and hTR/3′st are structurally favored and able to fold properly in the absence of the 5′ domain. Alternatively, they could bind some additional specific protein(s), allowing them to form a stable RNP in vitro. The latter possibility may apply to hTR/3′st, which forms two different RNP complexes in vitro, one being hTR specific. In conclusion, it appears that the 3′-terminal stem-loops of U17 and hTR act as independent assembly units. This view is further supported by the observation that hybrid U17 RNA molecules bearing the 3′-terminal stem-loop of U19 (U17/19 RNA) or U64 (U17/64 RNA) could not compete for U17 assembly (Dragon et al., unpublished data), indicating that the 3′-terminal hairpin of U17 is critical for its packaging into an RNP. Our findings that single hairpin domains of at least some H/ACA snoRNAs can form RNPs containing hGAR1 (Fig. 8), and possibly also other H/ACA proteins (Fig. 3C), is in line with the proposed three-dimensional model of H/ACA snoRNPs wherein a full set of proteins would bind to each of the two domains and form a U-shaped bipartite particle (72).
In yeast, GAR1 is a functional constituent of H/ACA snoRNPs; i.e., its depletion prevents pseudouridylation without affecting the stability and, most likely, the assembly of snoRNAs. Since, as shown in this work, hGAR1 is associated with U17 RNAs and hTR, it is tempting to speculate that their 3′-terminal stem-loops could function in pseudouridylation. Indeed, the 3′-terminal domains of both RNAs appear to fulfill the structural requirements to direct pseudouridylation but show no obvious complementarity to rRNA or any other potential targets (23, 47). Both U17 RNA and the H/ACA-like domain of hTR contain a 5′ domain that is much larger than those found in other human H/ACA snoRNAs (22). As in the case of the 3′-terminal stem-loop, it is not known whether these domains are involved in pseudouridylation or another reaction, such as pre-rRNA processing (18, 46). Refinement of the structural models of these RNAs by molecular probing techniques (49) and an approach involving the use of minigenes expressing potential targets for U17 and hTR (29) may prove useful in answering these intriguing questions.
ACKNOWLEDGMENTS
We thank Kazio Tycowski, Joan Steitz, and Michael Pollard for gifts of materials, and we thank Susan Baserga, Christian Grimm, Magda Konarska, Tom Meier, and Pawel Pelczar for experimental advice and protocols. We are grateful to Susan Baserga for critical reading of the manuscript and to our FMI colleagues Peter Müller for oligonucleotide synthesis, Herbert Angliker for DNA sequencing, Fritz Fischer for peptide synthesis, and Mike Rothnie for artwork.
V.P. was supported by the International Association for the Exchange of Students for Technical Experience.
REFERENCES
- 1.Autexier C, Pruzan R, Funk W D, Greider C W. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 2.Bagni C, Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J Biol Chem. 1998;273:10868–10873. doi: 10.1074/jbc.273.18.10868. [DOI] [PubMed] [Google Scholar]
- 3.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 4.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn E H. Telomerase. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 609–635. [Google Scholar]
- 6.Blackburn E H, Greider C W. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 7.Bortolin M-L, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bousquet-Antonelli C, Henry Y, Gélugne J-P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 10.Cecconi F, Mariottini P, Loreni F, Pierandrei-Amaldi P, Campioni N, Amaldi F. U17XS8, a small nucleolar RNA with a 12 nt complementarity to 18S rRNA and coded by a sequence repeated in the six introns of Xenopus laevis ribosomal protein S8 gene. Nucleic Acids Res. 1994;22:732–741. doi: 10.1093/nar/22.5.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamberlain J R, Pagán-Ramos E, Kindelberger D W, Engelke D R. An RNase P RNA subunit mutation that affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chastain M, Tinoco I., Jr Structural elements in RNA. Prog Nucleic Acid Res Mol Biol. 1991;41:131–177. doi: 10.1016/S0079-6603(08)60008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu S, Archer R H, Zengel J M, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark M W, Yip M L, Campbell J, Abelson J. SSB-1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J Cell Biol. 1990;111:1741–1751. doi: 10.1083/jcb.111.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragon F, Brakier-Gingras L. The interaction of Escherichia coli ribosomal protein S7 with 16S rRNA. Nucleic Acids Res. 1993;21:1199–1203. doi: 10.1093/nar/21.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.England T E, Bruce A G, Uhlenbeck O C. Specific labeling of 3′ termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- 18.Enright C A, Maxwell E S, Eliceiri G L, Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996;2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 19.Eperon I C, Krainer A R. Splicing of mRNA precursors in mammalian cells. In: Higgins S J, Hames B D, editors. RNA processing. A practical approach. Oxford, United Kingdom: IRL Press; 1994. pp. 57–101. [Google Scholar]
- 20.Feng J, Funk W D, Wang S-S, Weinrich S L, Avilion A A, Chiu C-P, Adams R R, Chang E, Allsopp R C, Yu J, Le S, West M D, Harley C B, Andrews W H, Greider C W, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 21.Foster A C, Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 22.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 23.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 24.Gary J D, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 25.Giordano E, Peluso I, Senger S, Furia M. minifly, a Drosophila gene required for ribosome biogenesis. J Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girard J-P, Bagni C, Caizergues-Ferrer M, Amalric F, Lapeyre B. Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J Biol Chem. 1994;269:18499–18506. [PubMed] [Google Scholar]
- 28.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 29.Hadjiolova K V, Normann A, Cavaillé J, Soupène E, Mazan S, Hadjiolov A A, Bachellerie J-P. Processing of truncated mouse or human rRNA transcribed from ribosomal minigenes transfected into mouse cells. Mol Cell Biol. 1994;14:4044–4056. doi: 10.1128/mcb.14.6.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heiss N S, Knight S W, Vulliamy T J, Klauck S M, Wiemann S, Mason P J, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 31.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gélugne J-P, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang W, Middleton K, Yoon H J, Fouquet C, Carbon J. An essential yeast protein, Cbf5p, binds in vitro to centromeres and microtubules. Mol Cell Biol. 1993;13:4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss T, Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- 35.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 36.Kiss T, Bortolin M-L, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol Cell Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konarska M M. Analysis of splicing complexes and small nuclear ribonucleoprotein particles by native gel electrophoresis. Methods Enzymol. 1989;180:442–453. doi: 10.1016/0076-6879(89)80116-8. [DOI] [PubMed] [Google Scholar]
- 38.Koonin E V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lischwe M A, Ochs R L, Reddy R, Cook R G, Yeoman L C, Tan E M, Reichlin M, Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr=34,000; pI 8.5) rich in NG, NG-dimethylarginine. J Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- 41.Lübben B, Fabrizio P, Kastner B, Lührmann R. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J Biol Chem. 1995;270:11549–11554. doi: 10.1074/jbc.270.19.11549. [DOI] [PubMed] [Google Scholar]
- 42.Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic pre-rRNA by RNase MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 43.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 44.Meier U T, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller K G, Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981;27:165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- 46.Mishra R K, Eliceiri G L. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc Natl Acad Sci USA. 1997;94:4972–4977. doi: 10.1073/pnas.94.10.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell J R, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell J R, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 49.Moine H, Ehresmann B, Ehresmann C, Romby P. Probing RNA structure and function in solution. In: Simons R W, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 77–115. [Google Scholar]
- 50.Morin G B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 51.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narayanan A, Lukowiak A, Jády B E, Dragon F, Kiss T, Terns R M, Terns M P. Nucleolar localization signals of Box H/ACA small nucleolar RNAs. EMBO J. 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 54.Phillips B, Billin A N, Cadwell C, Buchholz R, Erickson C, Merriam J R, Carbon J, Poole S J. The Nop60B gene of Drosophila encodes an essential nucleolar protein that functions in yeast. Mol Gen Genet. 1998;260:20–29. doi: 10.1007/s004380050866. [DOI] [PubMed] [Google Scholar]
- 55.Romero D P, Blackburn E H. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 56.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 57.Ruff E A, Rimoldi O J, Raghu B, Eliceiri G L. Three small nucleolar RNAs of unique nucleotide sequences. Proc Natl Acad Sci USA. 1993;90:635–638. doi: 10.1073/pnas.90.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif direct nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Schmitt M E, Clayton D A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selvamurugan N, Joost O H, Haas E S, Brown J W, Galvin N J, Eliceiri G L. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 1997;25:1591–1596. doi: 10.1093/nar/25.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seto A G, Zaug A J, Sobel S G, Wolin S L, Cech T R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 63.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 64.Sollner-Webb B, Tycowski K T, Steitz J A. Ribosomal RNA processing in eukaryotes. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, gene expression and function in protein synthesis. Boca Raton, Fla: CRC Press; 1996. pp. 469–490. [Google Scholar]
- 65.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 66.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 68.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tycowski K T, Shu M-D, Steitz J A. A small nucleolar RNA is processed from an intron from the human gene encoding ribosomal protein S3. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 70.Tycowski K T, Shu M-D, Steitz J A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 71.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 72.Watkins N J, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watkins N J, Leverette R D, Xia L, Andrews M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 74.Weinstein L B, Steitz J A. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 75.Yu Y-T, Scharl E C, Smith C M, Steitz J A. The growing world of small nuclear ribonucleoproteins. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 487–524. [Google Scholar]
- 76.Zaug A J, Linger J, Cech T R. Method for determining RNA 3′ ends and application to human telomerase RNA. Nucleic Acids Res. 1996;24:532–533. doi: 10.1093/nar/24.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zebarjadian Y, King T, Fournier M J, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]