ABSTRACT
The entry of antibiotic resistance genes (ARGs) into aquatic systems has been documented for large municipal wastewater treatment plants (WWTPs), but there is less study of the impact of smaller plants that are situated on small rural rivers. We sampled water metagenomes for ARGs and taxa composition from the Kokosing River, a small rural river in Knox County, Ohio, which has been designated an Ohio State Scenic River for retention of natural character. Samples were obtained 1.0 km upstream, 120 m downstream, and 6.4 km downstream from the effluent release of the Mount Vernon WWTP. ARGs were identified in metagenomes using ShortBRED markers from the comprehensive antibiotic resistance database (CARD) screened against UniPROT. Through all seasons, the metagenome just downstream of the WWTP effluent showed a substantial elevation of at least 15 different ARGs, including 6 ARGs commonly associated with Acinetobacter baumannii, such as msrE, mphE (macrolide resistance), and tet(39) (tetracycline resistance). The ARGs most prevalent near the effluent pipe persisted 6.4 km downriver. Using metagenomic phylogenetic analysis (MetaPhlAn2) clade-specific marker genes, the taxa distribution near the effluent showed elevation of reads annotated as Acinetobacter species as well as gut-associated taxa, Bacteroides and Firmicutes. The ARG levels and taxa prevalence showed little dependence on seasonal chlorination of the effluent. Nitrogen and phosphorus were elevated near the effluent pipe but had no consistent correlation with ARG levels. We show that in a rural river microbiome, year-round wastewater effluent substantially elevates ARGs, including those associated with multidrug-resistant A. baumannii.
IMPORTANCE Antibiotic resistance is a growing problem worldwide, with frequent transmission between pathogens and environmental organisms. Rural rivers can support high levels of recreational use by people unaware of inputs from treated wastewater, while wastewater treatment plants (WWTPs) can generate a small but significant portion of flow volume into a river surrounded by forest and agriculture. There is little information on the rural impacts of WWTP effluent on the delivery and transport of antibiotic resistance genes. In our study, the river water proximal to wastewater effluent shows evidence for the influx of multidrug-resistant Acinetobacter baumannii, an opportunistic pathogen of concern for hospitals but also widespread in natural environments. Our work highlights the importance of wastewater effluent in management of environmental antibiotic resistance, even in high quality, rural river systems.
KEYWORDS: Acinetobacter, antibiotic resistance, metagenome, microbiome, river, wastewater
INTRODUCTION
Environmental sources of antibiotic resistance increasingly threaten global public health (1–3). Antibiotics from clinical use and livestock husbandry can promote the development of resistant bacteria, and they readily pollute urban and rural waterways (4–6). Even very low concentrations of antimicrobial drugs select for resistance (7). Antibiotic resistance genes (ARGs) that enter environmental microbial communities have the potential for transfer to pathogenic bacteria (8). Yet, the public is rarely aware of the potential for exposure to ARG-carrying organisms in rural aquatic systems, particularly those designated for preservation by government agencies, such as the Ohio Scenic Rivers Program (https://ohiodnr.gov).
A major source of ARGs and antibiotics in aquatic systems is the effluent of wastewater treatment plants (WWTPs) (9–11). Wastewater treatment may actually select for increased antibiotic resistance of potential pathogens, such as Acinetobacter species (12, 13). It is important to understand the potential of WWTP to transfer ARGs as well as resistant microbes into rural streams where they may disturb autochthonous microbial communities and spread drug resistance to human microbiomes. We investigated the impact of WWTP effluent on the taxa distribution and ARG counts in the Kokosing River, a rural river designated a state “Scenic” river by the Ohio Department of Natural Resources (ODNR) that meets the criteria for Exceptional Habitat by the Ohio Environmental Protection Agency (Ohio EPA) due to its high species diversity and high ecological condition (14).
The river microbiome may be affected by WWTP effluent in various ways, including by elevation of phosphorus, nitrogen, and organic nutrients, by introduction of exogenous microbes and antibiotics, and by introduction of DNA, including ARGs. The WWTP in our study chlorinates effluent only during the months of May through October, so we compared both conditions. While chlorination effectively decreases bacterial biomass by three log units (15, 16), it does not fully remove ARGs from effluent. Some studies show partial decreases of ARGs by chlorine (17, 18), whereas others show that chlorination may increase the effluent content of ARGs and promote their conjugative transfer (19, 20). Various stress conditions in the WWTP can coselect antibiotic resistances and virulence properties (21). In some cases, the release of heavy metals, antibiotics, and other compounds into receiving rivers further propagates resistance by selecting for ARGs that encode multidrug efflux pumps (10, 22–24).
The establishment of antibiotic resistance in environmental microbial communities can be controlled when municipalities reduce antibiotic use (25). Therefore, understanding the impact of ARG pollutants on rural river resistomes is important for understanding the lasting potential of resistance in the environment. River resistomes offer the opportunity for surveillance of opportunistic pathogens that move between environment and human host, such as the ESKAPE pathogen Acinetobacter baumannii (26–28). The ESKAPE acronym comprises six leading hospital-acquired pathogens with multidrug resistance, including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (29). While A. baumannii is known for hospital transmission, recent reports indicate community acquisition of strains that carry ARGs on plasmids (30, 31). In the Kokosing River, we examined evidence for Acinetobacter ARGs, such as tet(39) (32, 33) and msrE and mphE (34).
To understand how WWTP effluent with secondary treatment might alter rural river microbial communities, we sampled sites upstream, just downstream, and further downstream of the effluent release of the Mount Vernon WWTP on the Kokosing River. The Kokosing river in east central Ohio flows 92 km into the Walhonding River, a part of the watershed of the Mississippi River (35). The Kokosing is included in Ohio’s Scenic Rivers Program; “scenic” designates “a waterway that retains much of its natural character for the majority of its length” (ODNR, https://ohiodnr.gov). The river is designated for exceptional warmwater habitat, agricultural water supply, industrial water supply, and primary contact recreation (14). The river is used regularly for recreation by the local residents, including students from an undergraduate college (approximately 1,800 students) situated at the downstream site reported by this study. Nevertheless, the Ohio EPA recognizes some localized impairment of the Kokosing’s warmwater habitat and use for recreational activities (https://mywaterway.epa.gov/).
Our study focused on a segment of the Kokosing in Knox County, proximal to the WWTP that serves the city of Mount Vernon (population 17,000). Mount Vernon includes surrounding suburban and rural homes as well as a 65-bed hospital. The WWTP system diagram is presented in Fig. S1 in the supplemental material. The design flow is 5.0 million gallons per day (MGD); actual discharge rates vary from 2.4 to 16.0 MGD (36). During our study dates, the discharge accounted for 2 to 7% of the river’s daily flow rate (Table S1). This fraction is small compared to the base flow contribution of municipal WWTP effluent to some rivers (37). Because it represents a small proportion of the river discharge, we asked whether the WWTP effluent would affect the microbiome of the system downriver of the plant. Small wastewater plants are situated approximately 25 km upstream (Village of Fredericktown, design flow 0.70 MGD) and 8 km downstream (Village of Gambier, 0.45 MGD). All of these plants disinfect their effluent by chlorination during 6 months of the year (May 1st through October 31st).
We focused our study on the river water microbiomes upstream, midstream (proximal to effluent pipe), and downstream of the Mount Vernon WWTP. We examined how ARG numbers are associated with the WWTP and how much ARG elevation may persist downstream of the effluent.
RESULTS
ARGs are elevated downstream of the WWTP effluent.
We sought to determine how the ARG distribution of the Kokosing River microbiomes was affected by the effluent from the Mount Vernon WWTP. Microbial samples were obtained from three sites on the Kokosing River, designated Upstream (1.0 km upstream of the WWTP), Midstream (120 m below the effluent pipe), and Downstream (6.4 km downstream of the effluent pipe) (Fig. 1). From all sites, the metagenomic DNA sequences were analyzed for ARG prevalence using the ShortBRED pipeline (38) applied to the CARD database (39). For each marker, the numbers of read hits were summed across all samples and dates, and the markers were ranked according to total hits (Tables S2 and S3 in the supplemental material). Results for the top 60 scoring markers are presented as a heat map (Fig. 2).
FIG 1.
Map of water sampling sites on the Kokosing River. The Upstream site is 1 km upstream of the Mount Vernon City wastewater treatment plant (WWTP). The Midstream site is located 120 m downstream of the WWTP. The Downstream site is 6.4 km downstream of the WWTP. The map was generated using the National Wild and Scenic Rivers System (2021).
FIG 2.
Heat map of relative abundance of the top 60 ARG marker hits. Read hit numbers are ranked in descending order by total hits across samples. Yellow represents highest abundance, while cyan represents lowest abundance.
Most of the top-scoring ARGs were elevated in the Midstream samples compared to samples from either Upstream or Downstream. The elevated ARGs include resistance determinants from several organisms that are of clinical concern. Most striking, six of the abundant ARGs are associated with the ESKAPE pathogen Acinetobacter baumannii and related strains: msrE, mphE, tet(39), cfxA6, oxa280, and aadA4 (26–28). The three top-ranked ARGs [msrE, mphE, and tet(39)] are found together on A. baumannii plasmid pS30-1 (34). Overall, the four top A. baumannii ARGs account for 37% of the total ARG hits found.
We tested whether the Downstream samples show evidence of carryover from the Midstream site. First, the numbers of ARG hit reads were re-sorted by Midstream site ARG totals (Table 1). For each of the top-ranked Midstream ARGs, we present the difference in ARG hits between Upstream and Downstream. The top 27 most abundant Midstream ARGs, including those associated with Acinetobacter, all show higher numbers at the Downstream site than at the Upstream location. In addition, we ranked Midstream ARGs separately for each of the six individual sampling dates and tested the top 20 ARGs for evidence of persistence downstream by using the Wilcoxon signed rank test (Fig. S2). Four of the six dates showed a significant increase of Midstream ARGs at the Downstream site compared to the Upstream site (P < 0.0083, with Bonferroni correction).
TABLE 1.
Top-ranked Midstream ARG hits: difference in hit numbers between Downstream and Upstream sample sites
| Mid rank | ARG familya | Host organism for ARG (example) | Reads: down-upb |
|---|---|---|---|
| 1 | msrE | Acinetobacter baumannii | 60 |
| 2 | mphE | Acinetobacter baumannii | 64 |
| 3 | tet(39) | Acinetobacter baumannii | 51 |
| 4 | CfxA6 | Acinetobacter baumannii | 70 |
| 5 | OXA-256 | Enterobacter cloacae | 28 |
| 6 | Mef(En2) | Bacteroides fragilis | 36 |
| 7 | AAC(3)-VIIa | Streptomyces rimosus | 34 |
| 8 | OXA-5 | Pseudomonas aeruginosa | 9 |
| 9 | QnrS6 | Aeromonas hydrophila | 25 |
| 10 | OXA-226 | Pseudomonas aeruginosa | 16 |
| 11 | ANT(4′)-Ib | Staphylococcus aureus | 38 |
| 12 | mtrA | Mycobacterium tuberculosis | 8 |
| 13 | sul1 | Vibrio fluvialis | 12 |
| 14 | OXA-280 | Acinetobacter johnsonii | 11 |
| 15 | tetE | Escherichia coli | 4 |
| 16 | EreA2 | Providencia stuartii | 3 |
| 17 | mefC | Photobacterium damselae | 7 |
| 18 | tetQ | Bacteroides fragilis | 9 |
| 19 | OXA-46 | Pseudomonas aeruginosa | 9 |
| 20 | MOX-5 | Aeromonas caviae | 11 |
| 21 | CbIA-1 | Bacteroides uniformis | 2 |
| 22 | ErmB | Enterococcus faecium | 17 |
| 23 | (aadA4) | Acinetobacter baumannii | 1 |
| 24 | mphG | Photobacterium damselae | 6 |
| 25 | ErmG | Bacteroides thetaiotaomicron | 4 |
| 26 | dfrF | Enterococcus faecalis | 2 |
| 27 | ErmF | Bacteroides fragilis | 14 |
ARGs shown represent the top 27 most abundant ARGs from Midstream sites. Gray shading indicates ARGs associated with Acinetobacter species.
Values indicate the difference between total Downstream reads and total Upstream reads that match the marker shown.
The overall percentage of reads that matched ARG markers ranged from 0.0015 to 0.0052% for Midstream samples and from 0.0002 to 0.0010% for Upstream and Downstream samples. These numbers indicate a roughly 5-fold elevation of ARG hits in the Midstream compared to the other two sites. We considered the possible effect of sample size, that is, whether the ARG hit numbers reflect the number of reads in our samples (Table S4). The read counts from individual samples deviated less than 20% from the mean. There was no significant difference in read numbers among the three collection sites Upstream, Midstream, and Downstream. Thus, the elevated number of ARGs near the effluent pipe was independent of the number of sequenced reads per sample.
Taxa profiles associated with WWTP effluent.
We investigated whether the elevation of ARGs by the WWTP was associated with specific microbial taxa. The taxa structure of our river metagenomes was determined using the pipeline metagenomic phylogenetic analysis (MetaPhlAn2) (40). The distribution of major bacterial phyla and classes in our samples is shown in Fig. 3A, with P values for Wilcoxon rank sum test displayed in Fig. 4A. Reads annotated to the genus Acinetobacter showed a striking prevalence in the Midstream, accounting for as high as 30% of predicted organisms (June sample, Midstream), and in some months elevated levels persisted downriver (December and April) (Fig. 3B). By comparison, through ShortBRED, ARGs associated with A. baumannii accounted for 37% of the total ARG hits. This result is striking, since the ShortBRED and MetaPhlAn2 pipelines use very different marker sets (ARGs versus core genome components). Thus, the two pipelines offer orthogonal evidence consistent with a high level of multidrug-resistant A. baumannii associated with the WWTP plant effluent.
FIG 3.
ARGs and bacterial taxa distribution across sampling sites and dates. (A) Percentages of phyla and of proteobacterial classes predicted by MetaPhlAn2. Taxa with prevalence too small to be quantified were grouped as “Other.” Samples are sorted by site then by sampling date. Horizontal black bars indicate dates when effluent was unchlorinated. (B) Percentage of Acinetobacter species predicted by MetaPhlAn2.
FIG 4.
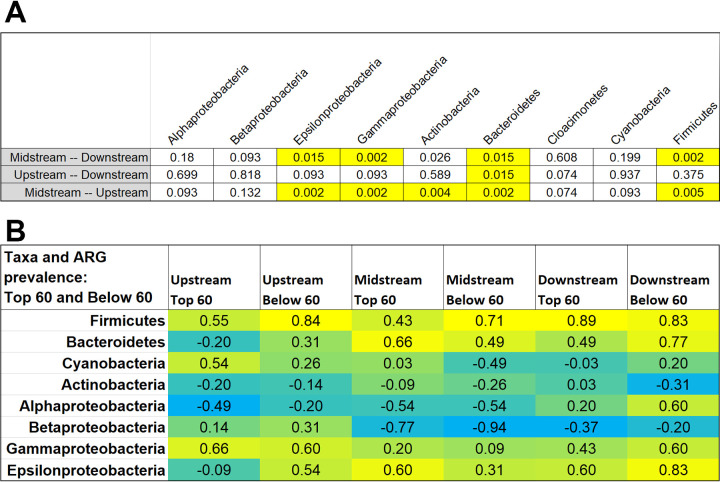
Taxa correlations across river sites. (A) Wilcoxon rank sum test P values for comparison of taxa percentages at different sites grouped across all dates (Fig. 3A). P values of ≤0.02 indicate significant differences (highlighted). (B) Spearman rank correlations between bacterial taxa and ARG abundance. Highlight color range indicates positive (yellow) versus negative (cyan) correlations.
The Midstream site showed significantly higher proportions of several major taxa than the Upstream site (Fig. 3A). The taxa with greater abundance include Bacteroidetes (P = 0.002), Epsilonproteobacteria (P = 0.002), Gammaproteobacteria (P = 0.002), and Firmicutes (P = 0.005). Downstream taxa appeared largely similar to those Upstream, with the exception of elevated abundance of Bacteroidetes (P = 0.015). These four taxa are consistent with a human fecal source during the period of effluent chlorination as well as during absence of chlorination. The Upstream and Downstream sites showed higher proportions of Actinobacteria relative to the Midstream. Alphaproteobacteria and Betaproteobacteria showed high prevalence across all three sites. High levels of Actinobacteria and Betaproteobacteria are consistent with metagenomic studies of freshwater oligotrophic lakes and rivers (41).
We considered whether the Midstream elevated ARGs might be associated with bacterial clades that were enriched in Midstream samples. A Spearman rank correlation was performed comparing ARG hits with the major taxa identified (Fig. 4B). ARGs were categorized as “Top 60” and “Below 60” based on overall rank prevalence (Fig. 2 and Table S3). The Top 60 were those ARG classes showing relative elevation at the Midstream site near the WWTP effluent, whereas ARGs Below 60 (ranked below the top 60 ARGs) more likely represent autochthonous genes commonly found in a relatively undisturbed river ecosystem. The number of ARG hits at Midstream and Downstream showed a positive correlation with Firmicutes and Epsilonproteobacteria, taxa that might be expected to arise from the WWTP effluent (Fig. 4B). Negative correlations were seen between ARGs and Betaproteobacteria, which are most likely native to the river.
If the source of Top 60 ARGs is the WWTP, are they carried by the genomes of effluent bacteria, or do they enter the river in the form of environmental DNA? The answer is unclear from our data. However, the occurrence of effluent chlorination (during the months of November, December, and April) shows no significant effect on the Midstream taxa profiles (Fig. 3A). If live bacteria are responsible for ARG elevation, significant numbers must be surviving chlorination.
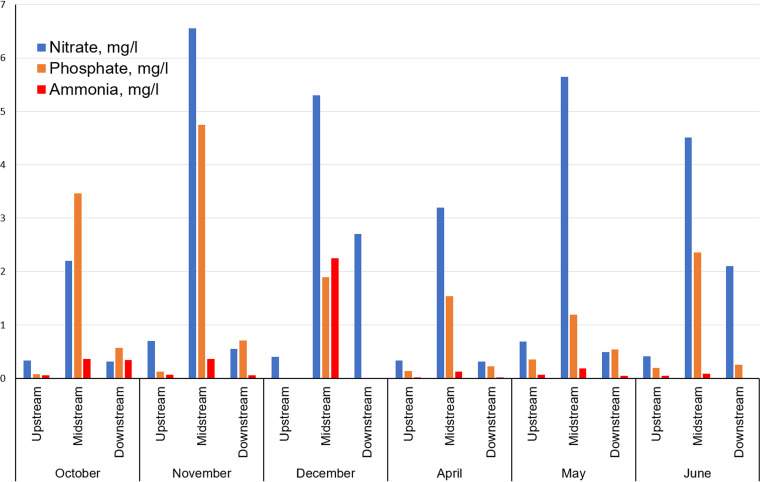
Nitrate, phosphate, and ammonia levels show no correlation with elevated ARGs.
The Mount Vernon WWTP effluent commonly includes total suspended solids (1 to 37 mg/liter), phosphorus (2.6 to 4.1 mg/liter), nitrate (5.86 to 28.9 mg/liter), and ammonia (0.107 to 5.77 mg/liter [summer] and 0.31 to 10.5 mg/liter [winter]) (14). Consistent with the above data, our Midstream water samples showed elevated levels of nitrate, phosphate, and ammonia relative to the Upstream and Downstream sites (Fig. 5 and Table S1). We therefore looked for possible correlations between water chemistry and ARG prevalence. Spearman rank correlations were performed for ARG levels and various chemical and physical factors (Table S6). Correlations were run separately for the sums of Top 60 ARG hits and for the sums of Below 60 ARG hits. We hypothesized that the Top 60 ARGs are dominated by the WWTP effluent and would therefore show stronger correlations with the Midstream chemistry.
FIG 5.
Nitrate, phosphate, and ammonia concentrations across sites and dates. Concentrations of nitrate, phosphorus, and ammonia were measured at each river site for each month. Full metadata are presented in Table S1 in the supplemental material, and metadata correlations with ARG abundance are in Table S6.
In fact, the nitrate, phosphate, and ammonia levels showed no consistent correlations with ARGs, either Top 60 or Below 60. This finding suggests that despite the higher concentration of these nutrients near the effluent pipe, the elevated levels of nitrogen and phosphorus are not correlated with the increased level of ARGs.
The plant effluent typically has a dissolved oxygen content (DO) of 5.3 to 10.2 mg/liter (14). In the Kokosing river samples, we observed DO values ranging from 8.22 to 12.60 mg/liter (Table S1). There was no significant correlation between river DO values and ARG prevalence (Spearman rank correlations) (Table S6).
Electrical conductivity (EC) was measured in the Kokosing samples, which has been shown to be an indirect indicator for dissolved organic carbon (DOC) (42–44). Previous studies have found connections between DOC and ARG abundance (45, 46). Over the course of our study, EC values ranged from 500 to 890 μS/cm (Table S1), but no significant correlation was found with site location or season nor with ARG levels (Table S6).
ARG numbers increased with pH and temperature.
The strongest correlations we saw between ARGs and water chemistry were for pH and temperature (Table S6). The range of pH values observed was pH 7.19 to 8.55 (Table S1). At Midstream and Downstream sites, pH showed positive correlations with ARG hits, particularly the Below 60 ARGs. These results suggest the possibility that low pH might select against ARGs that commonly occur in metagenomes of the undisturbed river. In laboratory evolution experiments on Escherichia coli, low pH and membrane-permeant aromatic acids select for loss of ARGs and ARG regulators (47, 48).
Temperature showed a strongly negative correlation with ARG levels, particularly those Below 60. This finding suggests the possibility of high-temperature selection against ARGs commonly found in the river community.
DISCUSSION
Past studies have investigated ARGs in urban waterways, but there has been relatively little research on the occurrence of ARGs in rural watersheds characterized by low human population density and agricultural land use. In addition, few studies have focused on rivers that are considered to be of exceptional quality, such as the Kokosing River investigated here. Forty-seven miles of the river are designated “scenic” by the state, and the river attracts members of the public for fishing, birding, and canoeing. Nonetheless, portions of the river are impacted by livestock and agriculture as well as pollution from a residential lakeside development (14). In 2010, portions of the watershed were reported to be impacted by gravel mining, erosion, and conversion to row crops.
Despite the overall high water quality of this river system and the relatively small contributions of WWTP effluent to stream discharge, we found substantially higher ARG abundance downriver of a WWTP than at the more agricultural portions of the watershed that lie upstream. We found substantially higher ARG abundance downriver of a WWTP than at the more agricultural portions of the watershed that lie upstream. The WWTP influx inputs a few percent of the total river flow rate (Table S1 in the supplemental material). Thus, a relatively small city WWTP (catchment population 17,000) may foster the spread of ARGs in a river that is in excellent ecological condition, as has been shown for anthropogenic contaminants in large urban centers (see, for example, reference 49).
The footprint of the WWTP effluent release was evident across our data, including shifts in ARG prevalence (Fig. 2; Table 1), microbial community taxa distribution (Fig. 3), and chemical indicators (Fig. 5). The top three ARGs for ShortBRED markers ranked in our metagenomes are known to occur together on A. baumannii plasmid pS30-1 (34). In addition, the MetaPhlAn2 taxonomic pipeline, with completely different markers, found high prevalence of A. baumannii near the WWTP effluent (Fig. 3B). It is possible that the multidrug-resistant A. baumannii actually comes from the WWTP. Wastewater treatment is known to increase the prevalence of multidrug resistance in A. baumannii from influent to the final effluent (12).
An effect of the wastewater effluent could be to increase community exposure to drug-resistant strains of this ESKAPE pathogen. It is also possible that the ARGs associated with A. baumannii have been acquired by other members of the native river microbial community. Nevertheless, the possibility of A. baumannii contamination should be followed up by further studies. Acinetobacter species of concern are emerging worldwide, especially in warmer climates, and their prevalence likely will increase with climate change (50–52). River levels of Acinetobacter species can be examined by targeted metagenomic analysis (53), amplicon assessment (54), and culture-based methods (55).
Rural rivers have substantial economic and cultural significance for local human communities. Nevertheless, the public is rarely aware of the potential impact of WWTP ARG exposure, with the common presence of WWTP plants along rural rivers. For example, 25 km upstream of the Mount Vernon plant is the Fredericktown WWTP, and just downstream of our sampled sites in Gambier, another small WWTP that releases effluent to the Kokosing. Further downstream from Gambier (20 km) lies the Danville WWTP (design flow 0.20 MGD).
We found evidence that detectable levels of ARGs persist in the river microbial community at least several kilometers past the effluent pipe. The Downstream site exhibited higher ARG counts than Upstream for the top 27 ARGs elevated at Midstream (Table 1). Thus, WWTP-associated ARGs persist and are transported in the environment at least 6.4 km downstream. Most of these ARGs are found in multiple species and may be transmitted by mobile elements (39). These ARGs might become established in the river microbial resistome and could propagate to pathogenic bacteria in the future, posing a risk to human health.
The WWTP-proximal site also showed substantial alteration of overall taxa distributions, such as increased prevalence of Bacteroidetes and Firmicutes (Fig. 4), findings that are consistent with previous studies (35, 56). The increase in Bacteroidetes persisted 6.4 km downstream. In addition, the WWTP-proximal site showed depletion of Actinobacteria, although the levels of this river group recovered downstream.
There is a need for future investigation regarding efficient methods of ARG control from WWTP in freshwater systems (57). In addition, the public should be more aware of the entry of wastewater into recreational waterways. Better awareness of the consequences of WWTP effluent release into rivers will improve our ability to sustain healthy microbial communities in our freshwater systems.
MATERIALS AND METHODS
Water sampling and metadata.
All water samples were obtained from the Kokosing River, Knox County, Ohio. Water samples were obtained at three sites on the river to yield data on water quality upstream of the WWTP effluent, just downstream of the WWTP in the mixing zone where wastewater is mixed with river water (Midstream site), and further downstream where plant effluent has been fully diluted (Fig. 1). The Upstream site (coordinates 40.38368, −82.47042) lies approximately 1.0 km upstream of the Mount Vernon WWTP effluent discharge. WWTP discharge rates and river flow rates on the dates of sample collection are presented in Table S1 in the supplemental material. The Midstream site, nearest the WWTP (40.378007, −82.467822) is located approximately 120 m downstream of the effluent release pipe, within the mixing zone of the plant where the effluent is initially mixed with river water. The Downstream site (40.376038, −82.40346) lies approximately 6.4 km downstream of the WWTP. The next nearest site where wastewater enters the Kokosing is the Fredericktown WWTP, a small plant (0.70 MGD) approximately 25 km upstream of the Mount Vernon WWTP. The three river sites were sampled using identical procedures at six dates throughout the year: 27 October, 3 November, and 3 December 2019 and 13 April, 28 May, and 25 June 2020. The WWTP effluent undergoes chlorination before discharge only from May 1st through October 31st; thus, only the October, May, and June samples occurred during the time that effluent was chlorinated.
On each sampling date, the three sites were sampled within a 2-h period. At each site, 400 ml of water was collected from the river using a dipper and was sealed in sterile Whirl-Pak bags. Within 24 h of sample collection, three 100-ml samples were vacuum-filtered through a sterile 0.22-μm filter 45 mm in diameter. Filter paper was folded using sterile forceps and deposited in centrifuge tubes, which were then frozen at −80°C to preserve microbial DNA. Water pH, conductivity, temperature, and dissolved oxygen (DO) were measured in the field using a Hannah pH/conductivity combination meter and a YSI Pro20 DO meter (Yellow Springs Instruments). Nutrient concentrations were analyzed using collected water samples within 24 h using a portable a Hach DR900 multiparameter portable colorimeter, including nitrate (NO3− -N), ammonia (NH3-N), and phosphate (PO4−-P; Table S1).
DNA isolation and sequencing.
Metagenomic DNA was isolated using a ZymoBIOMICS DNA miniprep kit. For control samples, 2 μg of the ZymoBIOMICS microbial community standard was processed under the same conditions. This community standard contains defined proportions of 10 microbes (5 Gram-positive bacteria, 3 Gram-negative bacteria, 2 fungal microbes).
Each filter was cut into small pieces and transferred to a ZR BashingBead lysis tube. ZymoBIOMICS lysis solution (650 μl) was added, and all tubes were processed on a Vortex Genie 2 for 40 min. The remainder of the preparation was performed according to the manufacturer’s protocol. Shotgun sequencing of DNA was performed by Admera Health (www.admerahealth.com). Libraries for sequencing were prepared using Illumina’s Nextera XT DNA library preparation kit following the manufacturer’s instructions. Final libraries were then pooled and sequenced on an Illumina HiSeq X sequencer for 150-bp read length in paired-end mode with an output of 40 million reads per sample.
ARG marker analysis.
Sequence reads were analyzed for ARG marker hits using ShortBRED, a computational pipeline from Huttenhower Biobakery (38). ShortBRED-Identify was used to create a database of short marker peptides specific to ARG protein families compiled from the comprehensive antibiotic resistance database (CARD) (39). From the ARG families, short consensus peptides were identified based on regions of amino acid sequence identity. To maintain high specificity, the set of peptides was then filtered against the universal protein database UNIREF90 (https://www.uniprot.org/uniref/) (data accessed 23 October 2019). This database was used to eliminate markers that match sequences outside a specific ARG. One additional marker, ARO_3002930 (vanRO, Rhodococcus hoagii) was removed from the marker set because it lacked specificity. The final list of markers used for our study (ShortBRED-2019) is presented in Table S2.
The ShortBRED-2019 marker list was used to screen metagenomic reads from each of the three river sites from six sampling dates (Table S3). Total read counts per sample were determined using Trimmomatic (58) (Table S4).
Taxa profiles.
The microbial taxa were profiled using the Huttenhower lab pipeline metagenomic phylogenetic analysis (MetaPhlAn2) (40, 59). MetaPhlAn2 assigns metagenomic reads to taxa using a set of clade-specific marker genes identified from approximately 17,000 microbial reference genomes. Taxa were grouped at the levels of phylum, class, order, family, and genus (Table S5). For a control, MetaPhlAn2 was also used to predict the taxa of ZymoBIOMICS microbial community standards that had been prepared concurrently with our experimental samples. For all preparation sets, MetaPhlAn2 consistently predicted the genera of the eight bacterial components and one fungal component of the standard (Table S7).
Data analysis.
To generate an ARG heat map from the ShortBRED data, we used R Studio 1.3.073. For one-way analysis of variance (ANOVA), we used JMP 14.2.0. One-way ANOVA was used to analyze the significance of ARG and taxa variances among the sites. ARG hits and metadata were correlated by the Spearman rank correlation using R (Table S6).
Data availability.
For all DNA sequences, FASTQ files are available at NCBI, SRA accession number PRJNA706754.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation award MCB-1923077. We acknowledge contributions of students in the Kenyon College course BIOL 239 Experimental Microbiology.
Footnotes
Supplemental material is available online only.
Contributor Information
Joan L. Slonczewski, Email: slonczewski@kenyon.edu.
Kathryn T. Elliott, College of New Jersey
REFERENCES
- 1.Bengtsson-Palme J, Larsson DGJ. 2015. Antibiotic resistance genes in the environment: prioritizing risks. Nat Rev Microbiol 13:396. doi: 10.1038/nrmicro3399-c1. [DOI] [PubMed] [Google Scholar]
- 2.Zhang T, Fukuda K, Topp E, Zhu YG, Smalla K, Tiedje JM, Larsson DGJ. 2020. Editorial: the environmental dimension of antibiotic resistance. FEMS Microbiol Ecol 96:fiaa130. doi: 10.1093/femsec/fiaa130. [DOI] [PubMed] [Google Scholar]
- 3.Yin X, Deng Y, Ma L, Wang Y, Chan LYL, Zhang T. 2019. Exploration of the antibiotic resistome in a wastewater treatment plant by a nine-year longitudinal metagenomic study. Environ Int 133:105270. doi: 10.1016/j.envint.2019.105270. [DOI] [PubMed] [Google Scholar]
- 4.Nesme J, Simonet P. 2015. The soil resistome: a critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ Microbiol 17:913–930. doi: 10.1111/1462-2920.12631. [DOI] [PubMed] [Google Scholar]
- 5.Pärnänen KMM, Narciso-Da-Rocha C, Kneis D, Berendonk TU, Cacace D, Do TT, Elpers C, Fatta-Kassinos D, Henriques I, Jaeger T, Karkman A, Martinez JL, Michael SG, Michael-Kordatou I, O’Sullivan K, Rodriguez-Mozaz S, Schwartz T, Sheng H, Sørum H, Stedtfeld RD, Tiedje JM, Della Giustina SV, Walsh F, Vaz-Moreira I, Virta M, Manaia CM. 2019. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci Adv 5:eaau9124. doi: 10.1126/sciadv.aau9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabri NA, Schmitt H, Van Der Zaan B, Gerritsen HW, Zuidema T, Rijnaarts HHM, Langenhoff AAM. 2020. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J Environ Chem Eng 8:102245. doi: 10.1016/j.jece.2018.03.004. [DOI] [Google Scholar]
- 7.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manaia CM. 2017. Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol 25:173–181. doi: 10.1016/j.tim.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D. 2013. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 10.Karkman A, Do TT, Walsh F, Virta MPJ. 2018. Antibiotic-resistance genes in waste water. Trends Microbiol 26:220–228. doi: 10.1016/j.tim.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Hultman J, Tamminen M, Pärnänen K, Cairns J, Karkman A, Virta M. 2018. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol Ecol 94:fiy038. doi: 10.1093/femsec/fiy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Marrs CF, Simon C, Xi C. 2009. Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci Total Environ 407:3702–3706. doi: 10.1016/j.scitotenv.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Higgins PG, Hrenovic J, Seifert H, Dekic S. 2018. Characterization of Acinetobacter baumannii from water and sludge line of secondary wastewater treatment plant. Water Res 140:261–267. doi: 10.1016/j.watres.2018.04.057. [DOI] [PubMed] [Google Scholar]
- 14.Strickland T, Korleski C, Korleski C. 2010. Biological and water quality study of the Kokosing River and selected tributaries 2007. Ohio EPA Technical Report EAS/2010-05-09. State of Ohio Environmental Protection Agency, Columbus, OH. [Google Scholar]
- 15.Owoseni MC, Olaniran AO, Okoh AI. 2017. Chlorine tolerance and inactivation of Escherichia coli recovered from wastewater treatment plants in the Eastern Cape, South Africa. Appl Sci 7:810. doi: 10.3390/app7080810. [DOI] [Google Scholar]
- 16.Bin Yuan Q, Guo MT, Yang J. 2015. Fate of antibiotic resistant bacteria and genes during wastewater chlorination: implication for antibiotic resistance control. PLoS One 10:e0119403. doi: 10.1371/journal.pone.0119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quach-Cu J, Herrera-Lynch B, Marciniak C, Adams S, Simmerman A, Reinke RA. 2018. The effect of primary, secondary, and tertiary wastewater treatment processes on antibiotic resistance gene (ARG) concentrations in solid and dissolved wastewater fractions. Water (Basel) 10:37. doi: 10.3390/w10010037. [DOI] [Google Scholar]
- 18.Zheng J, Su C, Zhou J, Xu L, Qian Y, Chen H. 2017. Effects and mechanisms of ultraviolet, chlorination, and ozone disinfection on antibiotic resistance genes in secondary effluents of municipal wastewater treatment plants. Chem Eng J 317:309–316. doi: 10.1016/j.cej.2017.02.076. [DOI] [Google Scholar]
- 19.Liu SS, Qu HM, Yang D, Hu H, Liu WL, Qiu ZG, Hou AM, Guo J, Li JW, Shen ZQ, Jin M. 2018. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res 136:131–136. doi: 10.1016/j.watres.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 20.Guo MT, Bin Yuan Q, Yang J. 2015. Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ Sci Technol 49:5771–5778. doi: 10.1021/acs.est.5b00644. [DOI] [PubMed] [Google Scholar]
- 21.Di Cesare A, Eckert EM, Corno G. 2016. Co-selection of antibiotic and heavy metal resistance in freshwater bacteria. J Limnol 75. doi: 10.4081/jlimnol.2016.1198. [DOI] [Google Scholar]
- 22.Seiler C, Berendonk TU. 2012. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 3:399. doi: 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P, Wu D, You X, Su Y, Xie B. 2021. Antibiotic and metal resistance genes are closely linked with nitrogen-processing functions in municipal solid waste landfills. J Hazard Mater 403:123689. doi: 10.1016/j.jhazmat.2020.123689. [DOI] [PubMed] [Google Scholar]
- 24.Bernier SP, Létoffé S, Delepierre M, Ghigo JM. 2011. Biogenic ammonia modifies antibiotic resistance at a distance in physically separated bacteria. Mol Microbiol 81:705–716. doi: 10.1111/j.1365-2958.2011.07724.x. [DOI] [PubMed] [Google Scholar]
- 25.Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons MN, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL. 2015. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 26.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha CJ, Jeong BC, Lee SH. 2017. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santajit S, Indrawattana N. 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chusri S, Chongsuvivatwong V, Silpapojakul K, Singkhamanan K, Hortiwakul T, Charernmak B, Doi Y. 2019. Clinical characteristics and outcomes of community and hospital-acquired Acinetobacter baumannii bacteremia. J Microbiol Immunol Infect 52:796–806. doi: 10.1016/j.jmii.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Salgado-Camargo AD, Castro-Jaimes S, Gutierrez-Rios RM, Lozano LF, Altamirano-Pacheco L, Silva-Sanchez J, Pérez-Oseguera Á, Volkow P, Castillo-Ramírez S, Cevallos MA. 2020. Structure and evolution of Acinetobacter baumannii plasmids. Front Microbiol 11:1283. doi: 10.3389/fmicb.2020.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agersø Y, Guardabassi L. 2005. Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J Antimicrob Chemother 55:566–569. doi: 10.1093/jac/dki051. [DOI] [PubMed] [Google Scholar]
- 33.Rumbo C, Gato E, López M, Ruiz De Alegría C, Fernández-Cuenca F, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual A, Bou G, Tomás M, Spanish Group of Nosocomial Infections and Mechanisms of Action and Resistance to Antimicrobials (GEIH-GEMARA), Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC), Spanish Network for Research in Infectious Diseases (REIPI). 2013. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackwell GA, Hall RM. 2017. The tet39 determinant and the msrE-mphE genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob Agents Chemother 61:e00780-17. doi: 10.1128/AAC.00780-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne JT, Millar JJ, Jackson CR, Ochs CA. 2017. Patterns of variation in diversity of the Mississippi river microbiome over 1,300 kilometers. PLoS One 12:e0174890. doi: 10.1371/journal.pone.0174890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Pollutant Discharge Elimination System (NPDES). 2019. Fact sheet regarding an NPDES permit to discharge to waters of the state of Ohio for Mount Vernon wastewater treatment plant (WWTP). Ohio Environmental Protection Agency, Columbus, OH. [Google Scholar]
- 37.Luthy RG, Sedlak DL, Plumlee MH, Austin D, Resh VH. 2015. Wastewater-effluent-dominated streams as ecosystem-management tools in a drier climate. Front Ecol Environ 13:477–485. doi: 10.1890/150038. [DOI] [Google Scholar]
- 38.Kaminski J, Gibson MK, Franzosa EA, Segata N, Dantas G, Huttenhower C. 2015. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput Biol 11:e1004557. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen ALV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monteiro MTF, Oliveira SM, Luizão FJ, Cândido LA, Ishida FY, Tomasella J. 2014. Dissolved organic carbon concentration and its relationship to electrical conductivity in the waters of a stream in a forested Amazonian blackwater catchment. Plant Ecol Divers 7:205–213. doi: 10.1080/17550874.2013.820223. [DOI] [Google Scholar]
- 43.Monteiro MTF, Tomasella J, Candido LA, Luizão F. 2015. Application of D-SEM to a catchment in Central Amazonia: calibration and validation of the carbon and nitrogen cycles. Ecohydrol Hydrobiol 15:192–207. doi: 10.1016/j.ecohyd.2015.07.001. [DOI] [Google Scholar]
- 44.Ye LL, Wu XD, Liu B, Yan DZ, Kong FX. 2015. Dynamics of dissolved organic carbon in eutrophic Lake Taihu and its tributaries and their implications for bacterial abundance during autumn and winter. J Freshw Ecol 30:129–142. doi: 10.1080/02705060.2014.939108. [DOI] [Google Scholar]
- 45.Garner E, Chen C, Xia K, Bowers J, Engelthaler DM, McLain J, Edwards MA, Pruden A. 2018. Metagenomic characterization of antibiotic resistance genes in full-scale reclaimed water distribution systems and corresponding potable systems. Environ Sci Technol 52:6113–6125. doi: 10.1021/acs.est.7b05419. [DOI] [PubMed] [Google Scholar]
- 46.Wang C, Hong P-Y. 2020. Genome-resolved metagenomics and antibiotic resistance genes analysis in reclaimed water distribution systems. Water (Basel) 12:3477. doi: 10.3390/w12123477. [DOI] [Google Scholar]
- 47.Moore JP, Li H, Engmann ML, Bischof KM, Kunka KS, Harris ME, Tancredi AC, Ditmars FS, Basting PJ, George NS, Bhagwat AA, Slonczewski JL. 2019. Inverted regulation of multidrug efflux pumps, acid resistance, and porins in benzoate-evolved Escherichia coli K-12. Appl Environ Microbiol 85:e00966-19. doi: 10.1128/AEM.00966-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He A, Penix SR, Basting PJ, Griffith JM, Creamer KE, Camperchioli D, Clark MW, Gonzales AS, Erazo JSC, George NS, Bhagwat AA, Slonczewski JL. 2017. Acid evolution of Escherichia coli K-12 eliminates amino acid decarboxylases and reregulates catabolism. Appl Environ Microbiol 83:e00442-17. doi: 10.1128/AEM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Posada-Perlaza CE, Ramírez-Rojas A, Porras P, Adu-Oppong B, Botero-Coy AM, Hernández F, Anzola JM, Díaz L, Dantas G, Reyes A, Zambrano MM. 2019. Bogotá River anthropogenic contamination alters microbial communities and promotes spread of antibiotic resistance genes. Sci Rep 9:11764. doi: 10.1038/s41598-019-48200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dekić S, Klobučar G, Ivanković T, Zanella D, Vucić M, Bourdineaud JP, Hrenović J. 2018. Emerging human pathogen Acinetobacter baumannii in the natural aquatic environment: a public health risk? Int J Environ Health Res 28:315–322. doi: 10.1080/09603123.2018.1472746. [DOI] [PubMed] [Google Scholar]
- 51.Kittinger C, Kirschner A, Lipp M, Baumert R, Mascher F, Farnleitner AH, Zarfel GE. 2018. Antibiotic resistance of Acinetobacter spp. isolates from the river Danube: susceptibility stays high. Int J Environ Res Public Health 15:52. doi: 10.3390/ijerph1501005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serwecińska L, Kiedrzyńska E, Kiedrzyński M. 2021. A catchment-scale assessment of the sanitary condition of treated wastewater and river water based on fecal indicators and carbapenem-resistant Acinetobacter spp. Sci Total Environ 750:142266. doi: 10.1016/j.scitotenv.2020.14226. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Garcera M, Touchon M, Brisse S, Rocha EPC. 2017. Metagenomic assessment of the interplay between the environment and the genetic diversification of Acinetobacter. Environ Microbiol 19:5010–5024. doi: 10.1111/1462-2920.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin F, Cai D, Sun Y, Guo D, Wu Z, Jiang D. 2014. Exploring the diversity of Acinetobacter populations in river water with genus-specific primers and probes. J Gen Appl Microbiol 60:51–58. doi: 10.2323/jgam.60.51. [DOI] [PubMed] [Google Scholar]
- 55.Benoit T, Cloutier M, Schop R, Lowerison MW, Khan IUH. 2020. Comparative assessment of growth media and incubation conditions for enhanced recovery and isolation of Acinetobacter baumannii from aquatic matrices. J Microbiol Methods 176:106023. doi: 10.1016/j.mimet.2020.106023. [DOI] [PubMed] [Google Scholar]
- 56.Price JR, Ledford SH, Ryan MO, Toran L, Sales CM. 2018. Wastewater treatment plant effluent introduces recoverable shifts in microbial community composition in receiving streams. Sci Total Environ 613–614:1104–1116. doi: 10.1016/j.scitotenv.2017.09.162. [DOI] [PubMed] [Google Scholar]
- 57.Barancheshme F, Munir M. 2017. Strategies to combat antibiotic resistance in the wastewater treatment plants. Front Microbiol 8:2603. doi: 10.3389/fmicb.2017.02603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible read trimming tool for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download Spectrum.00941-21-s0001.xlsx, XLSX file, 1.3 MB (1.4MB, xlsx) .
Data Availability Statement
For all DNA sequences, FASTQ files are available at NCBI, SRA accession number PRJNA706754.