Abstract
The Severe Acute Respiratory Syndrome-related Coronavirus 2 (COVID-19 or SARS-CoV-2) epidemic is professed as world disaster producing a worrying increasing mortality, particularly amongst vulnerable humans worldwide. Whether COVID-19 has a strong ability for acceptable genetic flexibility that amended for breaking immune responses quickly, it is critical to understand the adaptation mechanism between viruses and hosts that allows individuals to follow viral development. This can contribute to finding the appropriate treatment to combat the epidemic. However, the present information about viral adaptation mechanisms in hosts is still insufficient, and future investigations may reveal the unknown. Mutations and genetic variations are naturally occurring; however, the current knowledge about their mechanism and pathways still has many secrets. The present review also provides insights into the immune system, immunological memory, and the development of the COVID-19 vaccine. Other fighting methods against COVID-19 are also highlighted. The potential of antibodies, natural metabolites, and current suggest vaccines were applied to the face of this new threat.
Keywords: SARS-COV-2, COVID-19, Immune system, Vaccine
1. Introduction
Currently, viral infections have become one of the fundamental health problems. Viruses are known as submicroscopic non-living organisms causing several human diseases, consisting of DNA or RNA strains. Different viral diseases are commonly known, including the dengue virus, herpes simplex, human immunodeficiency virus (HIV), influenza, and hepatitis [1]. From December 2019, a different kind of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV-2) has been discovered; the primary pathogen of the present pandemic disease termed coronaviruses (COVID-19/SARS-CoV-2) [2]. The symptoms related to human coronaviruses have evolved from the communal cold to the Middle East and, in most cases, led to sudden death owing to the severe acute respiratory syndrome. It also caused massive economic losses and social burdens in several fields. In a few months, coronaviruses become the most significant danger threatened the world. The search for a cure has become a constant concern and attention of scientists worldwide to save humanity from this pandemic. The main attention concerning this kind of disease is connected by the inherent features of viruses, including effects of replication mechanism and mutation capacity. The virus's proliferation in a living agent is dependent on viral inoculation in healthy cells, after which replication occurs through the host cell's mechanisms [3]. As a result, it can drastically alter the organism's functionalities and metabolism, and thus important responsibilities may fail.
On the other hand, mutations could happen as a result of biotic and abiotic properties related to the host. The development of antiviral vaccines becomes a tricky and urgent need that relies on pathobiology studies. The virus inoculation in host health cells develops a crisis in finding effective and safe drugs without side effects [1], [4].
Though it is possible to establish a virus genome in a few days, developing vaccines and drugs is time-consuming. The two ways need further analysis for safety and efficiency that may cause the whole process to continue for at least a year [5]. This is what happened with the second wave of the virus when the first wave witnessed numerous human death because of vaccines shortage. In this vein, the current review discusses replication of the virus and their inoculation. The binding mechanism with the host cell was explained and the immunological activity that may help to find the suitable treatment.
2. Description of coronaviruses
Until now seven species of human coronaviruses have been recognized counting HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, Middle East respiratory syndrome coronavirus, severe acute respiratory syndrome coronavirus (SARS-CoV), and SARS-CoV-2 or COVID-19 evolving in Wuhan, China [5].
2.1. Structure of COVID-19
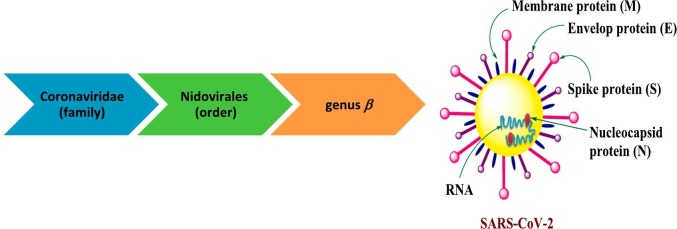
Today coronaviruses are considered one of the most enveloped RNA viruses containing many surface spikes and belong to Nidovirales order (Fig. 1 ) [6]. Coronaviruses have a large genome of single-stranded positive-sense RNA (30+ kb) encoding for numerous open reading edges. The first edge (S protein) encodes the spike protein. This is known as class I fusion protein which enhanced the correlation between virus and receptors on the surface of the cells and after then uptake into endosomes (for several types of coronaviruses). Proteolytic cracking of the S protein, viral fusion, and endosomal membranes; all contribute in viral RNA release into the cytosol [6]. Several 30 poly (A) tail and 50 cap units are included in RNA which permits and helped in replication process that is encoded roughly by two-thirds of the genome. The final three codes are associated with structural and accessory proteins. The replicase can be recognized as two polyproteins: pp1a and pp1ab; each of them contains up to 16 nonstructural proteins. These structures are formed by 2–3 viral proteases encoded within the replicase processing pp1a and pp1ab. Several nonstructural proteins accumulate into replicase-transcriptase complex which in the host cell cytosol generates new viral genome anti-sense genome and subgenomic RNA that applied as mRNA. Matrix proteins (M), structural spike proteins (S) and envelope proteins (E) are then created that passed into the endoplasmic reticulum through secretory pathway of the endoplasmic reticulum-Golgi intermediate compartment. In few cases coronaviruses are encoding a hemagglutinin esterase as well. In majority of coronaviruses, the structural protein is split to S1 and S2 subunits via furin-like proteases usually. The RNA genome attached with nucleoprotein for generating virus particles and buds into Golgi. After aggregation, virions are transferred via vesicles to the surface of the cells and are exocytosis. Many accessory proteins are expressed as well which become significant for pathogenesis, however some of them still not functionality characterized [7].
Fig. 1.
Schematic diagram of the COVID-19 structure.
2.2. COVID-19 inoculation and replication
The SARS-CoV-2 genomic sequence can be used to identify the basic enzymes and proteins required in the virus's inoculation and replication processes [3]. Its genomic sequence is occasionally similar to one strain of coronaviruses that is only infectious in bats with a percentage of 96% and SARS-CoV-1 (79.5%). Genomic data revealed that three proteins are primarily involved in inoculation and replication of COVID-19 in the host (human beings). These including papain-like protease (ACE2), the spike protein (TMPRSS2), and the three chymotrypsin-like protease (3CLpro) [4], [8], [9]. TMPRSS2 and ACE2 are considered a piece of the host cell.
The angiotensin-converting-enzyme type II (ACE2) consider the basic functional receptor of COVID-19, causing its attachment with bat and human cells and consequently its replication [10]. ACE2 is an analogue of angiotensin converting enzyme type I (ACE) and a renin–angiotensin system component, which regulates blood pressure. Lots of studies that target for development of anti-COVID-19 are basically searching on the ACE2 inhibition [4], [11], however, reactions with this central pressure control system may not be the efficient method.
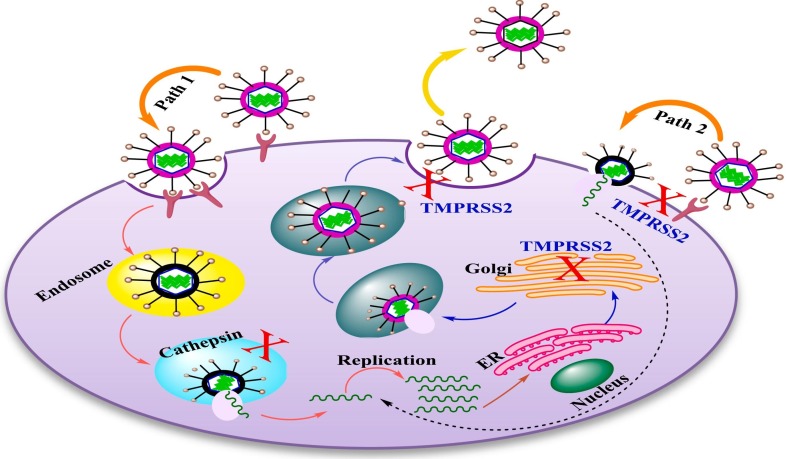
Several studies have shed light on the importance of TMPRSS2 in the proteolytic activation of COVID-19 [12]. It was found that SARS-COV can pass to the cell in two ways. The first pathway is mediated at the cell's surface by TMPRSS2 while the second is achieved by cathepsin L/B in the endosome as represented in Fig. 2 [13], [14], [15]. The studies revealed that coronaviruses can attached with the cellular receptor, causing an uptake of virions into endosomes (path A), whereas cathepsin used to activate the spike protein. The decreasing pH in the endosome allows the viral envelope to pass through the endosomal membrane and the viral genetic material to enter the cytosol. After then RNA transcription, replication and transcription occurred [12]. Newly synthetized RNA molecules are then transported to the ERGIC, where viral assembly take place. Virus particles bud into vesicles, which inserted to the cell surface for release. Instead, the spike protein activates on the cell's surface, which is responsible for fusion of the viral membrane with the plasma membrane (path 2). Spike is created as a precursor, which is then cleaved by host proteases. Throughout the virus's life cycle, spike cleavage can occur in various positions and at multiple times. Spike cleavage by cathepsin occurs in the endosome, whereas TMPRSS2 cleavage occurs in the Golgi or plasma membrane, during assembly or attachment and release [12].
Fig. 2.
The replication cycle of coronaviruses in the host and proteolytic cleavage of proteases.
The serine protease inhibitor camostat can successfully defend mice infected with the otherwise lethal SARS-CoV from morbidity and mortality; nevertheless, the inhibitor treatment containing both cathepsin and serine has not showed activity for developing survival significantly over that occurred by camostat only [16]. This is explained that SARS-CoV pathogenesis and propagation is mediated by TMPRSS2 rather than cathepsin in vivo. In addition, it was displayed that in the infected cells, cis-cleavage occurred when SARS-CoV S separated to several parts upon expression of TMPRSS2 besides the relation between TMPRSS2-expressing cells (trans-cleavage) and SARS-CoV S-expressing cells. Cis-cleavage produced from a wide spreading of SARS-CoV S particles into the cellular supernatant that could mixed with antibody-mediated neutralization, when Trans-cleavage leads to activation of SARS-CoV S on the target cell, caused an efficient SARS-CoV S-driven viral fusion [17]. Additionally, SARS-CoV activation by TMPRSS2 hinders the SARS-CoA S inhibition by interferon known as interferon-induced transmembrane proteins (IFITMs). The findings of relevant studies emphasized the importance of TMPRSS2 in SARS-CoV infection [18], [19].
On the other hand, 3CLpro leads to proteolytic damage of virus polypeptide in 11 non-structural proteins accountable for its replication. Consequently, the absence of 3CLpro from the host cell and existence only within SARS-CoV-2 represent a possible key for the potential treatment of COVID-19. The similarity between SARS-CoV-1 and SARS-CoV-2 serves as a basis for COVID-19 specific treatment progress, in addition to a “common” coronaviruses infections treatment [20].
2.3. COVID-19 binding mechanism
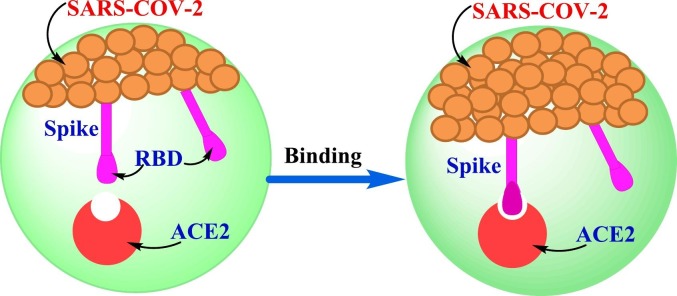
There is a lot of interest on the binding mechanism of coronaviruses with the target cell that will help researchers for developing an effective therapy. The strong ability explained the mechanism for attachment between the spike of COVID-19 and ACE2 used as an entry receptor to attack the target cell as displayed in Fig. 3 [21]. The spike protein on the surface of coronaviruses enhanced their passing into host cell. COVID-19 spike protein having a receptor-binding domain (RBD) that distinguishes clearly as its receptor ACE2 [22], [23]. The ACE2/RBD binding affinity has been recognized as a key principal of SARS-CoV pathogenesis and cross-species infections [24], [25]. This communication will provoke conformational variations of the C-terminal S2 subunit (causing fusion of virus-cell membrane) of the spike protein. The formed complex (ACE2/S protein) is then proteolytically handled by TMPRSS2 causing ACE2 cleavage and thus viral passes into the host cell as explained above [26].
Fig. 3.
The schematic shape of the COVID-19 binding mechanism.
Several natural mutations can occur around RBD sites. Two virus-binding sites were found on the surface of ACE2 which play significant role for binding of SARS-CoV-2 S. This is increased the infection and pathogenesis between human and species [25], [27].
3. Adaptive immune system
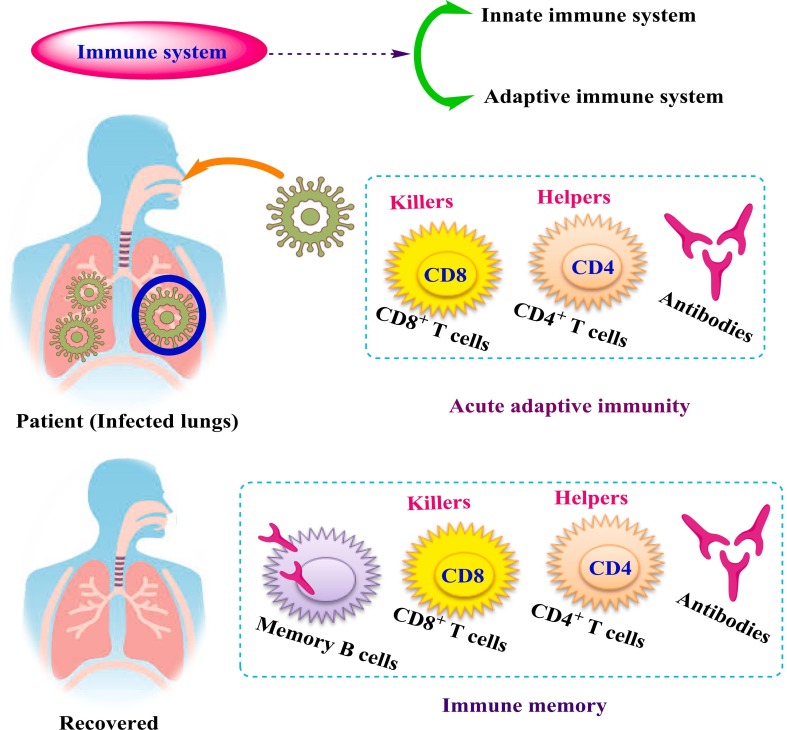
The immune system is generally divided into two parts: adaptive and innate. Despite their strong connection, the two parts are made up of different types of cells that perform different functions [28]. Deep investigations revealed that the adaptive immune system consists of three basic cell types; B cells, CD8+ T cells, and CD4+ T cells, as shown in Fig. 4 . It was found that B cells are the main sources for antibodies production and CD4+ have got different helper and effector functionalities, whereas CD8+ has strong ability to destroy the infected cells. The adaptive immune responses play an important role in control viral infections and in combination with the immune memory can control successfully of all vaccines [29]. COVID-19 is obviously unusually effective at evading the activation of early innate immune responses, such as type 1 interferons (IFNs) [28], [30], [31].
Fig. 4.
The fundamental elements of adaptive immunity are present in viral immune responses.
Several studies have confirmed the role of the innate immune system in detecting viral infections and triggering the alarm bells of type I IFN expression and related molecules [32], [33]. This action takes approximately 2 h to complete. The innate immune response continues to serve three primary components, including viral replication inhibition for infected cells, antiviral production in the local tissue, recruitment of innate immune system effector cells, and staining the adaptive immune response. The first two functions of the innate immune system reduce viral replication and spread, while the third is critical for activating the adaptive immune response. [28], [34], [35].
4. Mutated coronaviruses
In the current time, genetic variation of COVID-19 becomes one of the most critical topics in the area of research. Several viruses, especially RNA viruses, can adapt quickly with variations in their hosts because of high mutation rates in these viruses. Mutations are naturally selected however the available knowledge about their mechanism and pathways still contain a lot of mystifications. Additionally, the definite mechanisms of SARS-CoV adapted with several host receptors lack well understanding, revealing evolutionary and epidemic conundrums [25]. Several mutations of different SARS-CoV strains have been recognized in RBD. For example; regarding the response of antibodies, there are COVID-19 mutations which can disturb neutralizing antibody epitopes in humans. RBD is the best receptor for neutralizing antibodies and it is characterized by large sufficient surface area. This feature support mutation-selection as no single viral mutation is predicted to hinder neutralization via polyclonal human serum [36], [37]. These mutations may enhance the preferable interactions or decrease undesirable reactions with virus-binding hot spots on ACE2. Consequently, such mutations considered viral adaptations to ACE2 that possess enormous threats to human health [25]. The global well known spike D614G variant which is strongly bind with ACE2 and more transmissible [38], [39] is neutralized by plasma from the infected substances with original D614 virus [39]. Furthermore, despite the spike N439 variant's strong binding capability with ACE2, it is strongly neutralized by serum from the majority of COVID-19 patients [37].
Even though specific response to any coronavirus is still unknown, COVID-19 mutations are extremely unlikely to evade T cell immunity. This may occur due to the high rate of SARS-CoV-2 epitopes which recognized in humans composed of CD8+ T and CD4+ cell responses to more than 10 epitopes dispersed through genome of COVID-19, and changed from person to another [40], [41]. Agerer et al. have recently investigated mutations in major histocompatibility complex-I (MHC-I) restricted CD8+ cell epitopes which studied the sequencing of 747 COVID-19 virus isolate [42]. The study revealed that the mutant peptides demonstrated in vitro reduced cell-free MHC-I binding capacity linked with diminished IFN-γ production, cytotoxic activity and proliferation of CD8+ T cells isolated from COVID-19 patient. The results spot the light on the significant activity of COVID-19 to destroy CD8+ T cell surveillance via mutation points in MHC-I-restricted viral epitopes [42].
Though it might be critical to monitor the evolution of COVID-19, it is surprising that the virus can rapidly evolve escape variants that evade the majority of cellular and humoral immune memory in COVID-19 vaccine recipients or COVID-19 cases [28], [43].
5. Fighting methods against COVID-19
As the COVID-19 mortality and morbidity rates are still unknown due to difficulties in tracking cases and different containment protocols used by different countries, the disease's unprecedented rapid spread worldwide creates an urgent need for a treatment to be developed [3]. The rapid detection and diagnosis of the new coronavirus COVID-19 are highly appreciated for helping in an effective therapy. Although some new promising results with antiviral treatments, it is critical to exploit such investigations since the death worldwide provoked by COVID- 19 are continuously increasing every day. Different approaches have been applied for treatment and massive efforts are being done to develop and produce an effective vaccine [7], [44]. Despite the great efforts consuming for clinical trials (more than 200), investigating these drugs and others, their activity against COVID-19 infection is still unclear.
5.1. Currently available vaccines in the market
The current stage witnesses a massive revolution in vaccine development, but this effort is too late for controlling the first wave of a destructive epidemic. However, significant points can be accepted and applied during vaccine development against this quickly emerging virus disease. Significantly, COVID-19 vaccines could decrease mortality and morbidity even if the virus stabilizes people [7]. Many papers are recently discussed the development of most suggested vaccines against COVID-19 and clinical trials have been investigated worldwide, as displayed in Fig. 5 [45], [46].
Fig. 5.
The most suggested vaccines against outbreak of COVID-19 pandemic.
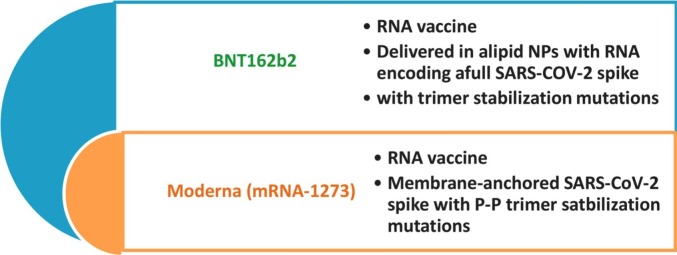
Phase 3 clinical trials (phase 3) Moderna and Pfizer/BioNtech RNA COVID-19 for example have been widely released. The effective vaccine would provoke long-lasting antibody titers and could deliver immunity to avert clinical disease and diffusion [28]. These vaccines achieved significant efficiency (95%) against COVID-19 disease [47]. 20,000 cases were injected with two dosages without any side effects and approved safety for the clinical trial at phase 1 [47], [48]. Moderna (mRNA-1273) and BNT 162b2 are considered RNA vaccine and applied in the form of lipid nanoparticles as membrane anchored with P—P trimer stabilization mutations (Fig. 6 ) [48], [49].
Fig. 6.
Characterization of currently applied vaccines against COVID-19.
The Moderna RNA vaccine was tested with 196 cases and proved protection activity with percent 94% against COVID-19. Phase 3 trial of this vaccine was released one week after Pfizer at 16th November [50]. The vaccine was displayed very successful results with severe cases.
On the other side, it was observed that efficacy of BNT162b2 vaccine was not depend on infections number during investigation but on the basis of symptomatic cases. The clinical trials appeared high activity for the tested vaccine which estimated by more than 90% however another vaccines such as measles and smallpox showed potent activity that close to 100% [28]. In the short term, Pfizer BNT162b2 (phase 1) clinical trial caused a successful detection of neutralizing antibody titers [48]. The data of T cell were not discussed for BNT162b2, whereas phase 1 trial data related to BNT162b1 vaccine revealed strong responses of the cells CD4+ T and CD8+ T cell [51]. Furthermore, the immunogenicity of the Pfizer BNT162b2 vaccine in different cases older than 65 years old was investigated, but the results were scarce. The efficacy of phase 3 trials demonstrated equivalent protective activity in younger and older age groups (estimated by 94% and 95%) [47].
5.2. Naturally occurring metabolites against COVID-19
The continuous fear and dread of the epidemic with the lack of resources, especially in developing countries, made people resort to natural resources, that characterized by safety, availability and may become the magic solution to strength the immune system. It plays a significant role in the treatment of several diseases. Many people who do not contribute to modern doctors depend on home therapies and folk medicine for treatment [5]. Natural compounds and their molecular frameworks have a long traditional use and contribute greatly to the history and landscape of new molecular entities. It showed high activity as antiviral agents so it could be an available, safe and cost effective candidate against coronavirus infectious diseases [52], [53], [54]. The World Health Organization (WHO) encourages utilization of scientifically proven traditional medicines for COVID-19 infection. Several natural sources have been employed as a valuable target for developing antiviral drugs such as plant species, vinegar and honeybees as displayed in Fig. 7 [44], [55].
Fig. 7.
Different natural sources are used against coronaviruses infections.
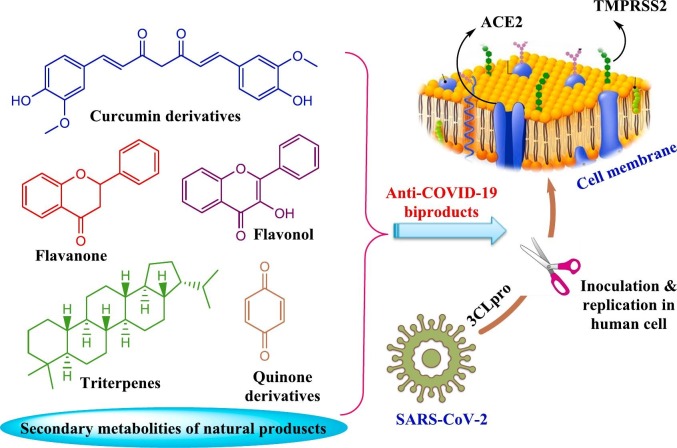
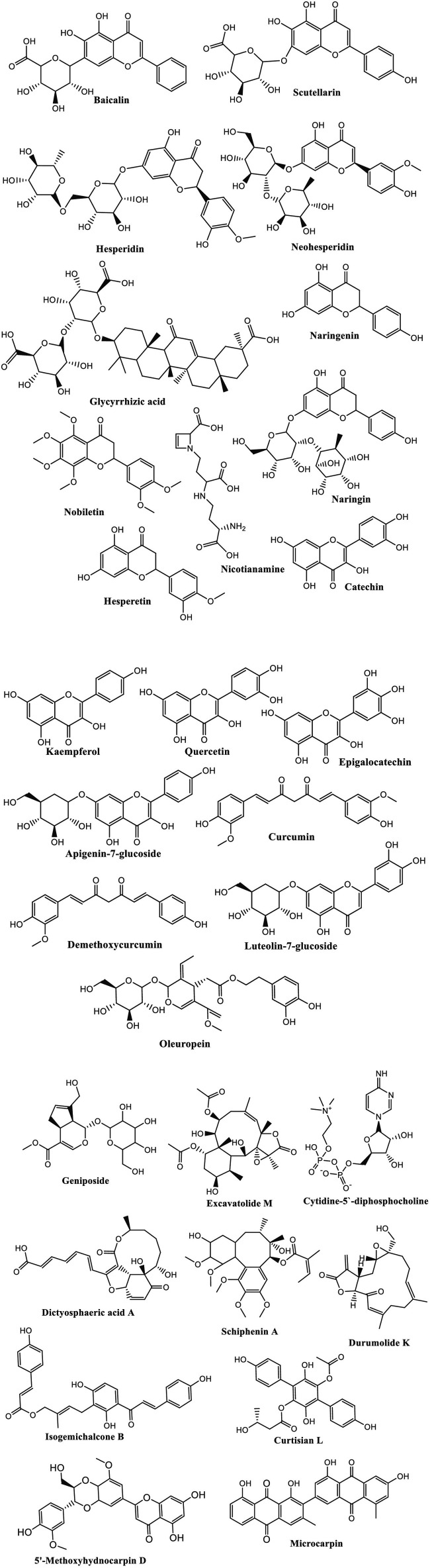
In China, for example, the herbal medicine has been used for treatment of COVID-19 since the beginning of this epidemic. The results were extremely encouraging as these traditional medicines caused 90% recovery of 214 patients treated. In addition, turmeric, garlic, ginger, black pepper cinnamon, and honey are being described as home therapies against COVID-19 in Pakistan [56]. A mixture of hot water with ginger, black cumin seeds, and clove extracts is reported for frightening coronaviruses' symptoms and, fruits and honey with vitamin C [5]. After 54 days of breakout COVID-19 as pandemic, several naturally occurring compounds from different classes revealed significant results on virtual molecular docking. Molecular docking of the identified compounds in the species and some others of antiviral drugs was investigated to evaluate binding capability with COVID-19 toward different molecular targets. Several plant species have investigated and induced activity for SARS-COVID. For example, N. sativa reported as an effective candidate against COVID-19. The results confirmed that the plant species is rich by different molecules with high activity against COVID-19 proteins and enzymes, including α-hederin, nigelledine, hederagenin, thymoquinone, and thymohydroquinone [2]. The roots of Scutellaria baicalensis and Oroxylum indicum seeds are considered the major source of baicalein which examined as TMPRSS-2 expression down-regulators and ACE2 inhibitor [57]. The relevant studies displayed a high diversity of natural chemical entities belonging to different chemical classes such as flavones, terpenes, quinones, fatty acids, alkaloids, and flavonoids, which applied as potential substances against coronaviruses disease as induced in Fig. 8 [2]. The phenolic compounds such as quercetin glycosylated derivates and myricetin have the ability to cause inhibition for ACE2. This action may occur due to the closely related active sites that are distinct basically in terms of the smaller intramolecular size of ACE2 sites [58], [59]. The natural metabolites that depress COVID-19 through ACE2 inhibition are characterized by low molecular weight (less than 500 Da), containing lower than five hydrogen bond donors and long P under 5 [60], [61]. In addition, Rahman et al. have reported the top 12 chemical compounds that have significant effectiveness to bind with TPMRSS2 and, hence, could be applied as potential inhibitors against COVID-19 [57]. These natural metabolites were extracted from different natural sources including plants (e.g., Camellia sinensis, Asphodelus ramosus), algae (e.g., Shisandra sphenanthera) and soft corals (e.g. Formosan gorgonian). Geniposide was found to be the most active substance as anti-coVID-19 [57]. Khaerunnisa and his group employed molecular docking to investigate different natural compounds able to inhibit 3Cro of COVID-19 for instance demethoxycurcumin, luteolin-7-glucoside, catechin, kaempferol, quercetin, apigenin 7-glucoside, naringenin, oleuropein, epigallocatechin and curcumin (Scheme 1 ) [62]. It is worth to mention that most of bioactive natural candidates belong to flavonoid however the other classes should take sufficient attention. It was observed that glycosylated compounds more active than their respective aglycone. Extracts and fractions showed more significant activity than the isolated molecules [54].
Fig. 8.
Key points for the effect of natural secondary metabolites against coronaviruses; anti-COVID-19.
Scheme 1.
Possible bioactive naturally occurring compounds used as COVID-19 inhibition.
However the development of bioactive natural metabolites against several diseases, including COVID-19, is quicker than vaccine improvement, it has remained a difficult process owing to the high variety of natural products, their extraction and sophisticated of their chemical structures. The plant species also lack of clinical trials on human coronavirus cases till now. Subsequently, we highly recommended deep and further studies either clinical or preclinical trials for curing coronavirus diseases especially COVID-19 [2].
5.3. Cellular and humeral immunity
The viruses causing diseases in human beings have a strong ability to evade the immune system and have one suggested mechanism for a trick. The viruses will be inoffensive without their evasion capacity against the immune system. The precise knowledge about viral immune evasion is an urgent request to evaluate the pathogenesis of the virus, obstacles that face the adaptive immune system, and any suggested vaccine then hence could control the disease [28]. The adaptive immune system is imperative to fight most viral infections. There are three essential parts of the adaptive immune system as described in Fig. 4. Cytotoxic CD8+ T-lymphocytes (CTLs) have significant effect in preserving immune memory and protection against viral pathogens [63], [64]. CTLs have a strong ability to kill infected cells based on the identification of viral epitopes demonstrated on the cell surface related to class I major histocompatibility complex proteins (MHC-I) [42]. The effect of CTL has described deeply in COVID-19 infected patients [65]. In case of high level of COVID-19 infection, CTLs displayed high cytotoxicity because of their significant molecules including perforin, granzyme B, and IFN-γ [28].
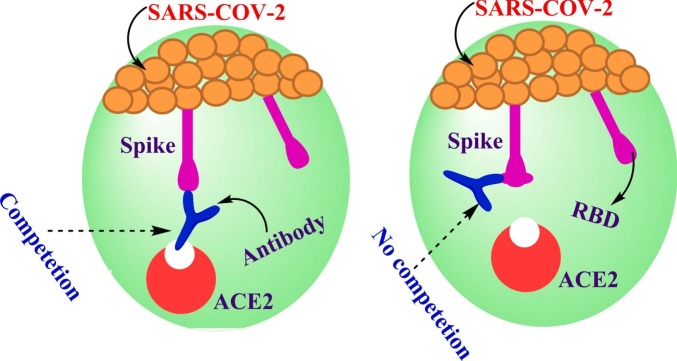
On the other side, infection of COVID-19 provokes expansive activation of the innate and adaptive arms of immunity [66]. Neutralizing antibodies are one of the basic correlates of protection that underwent a delicate development under the influence of coronaviruses. Antibodies are well-defined as glycoproteins created by plasma cells which act as adaptive immune response helping in an infection removing from the body. Much more attention has been paid for developing therapeutic antibodies to control COVID-19. Antibodies play a significant role in infection appearance in several ways. For example; agglutination of microbes, opsonization of pathogens to assist phagocytosis, stimulation of the complement system, and neutralization of toxins and viruses [21]. The mechanistic effect were explained as the antibodies prevent the viruses from insertion into the target cell receptor which hinder binding between them as represented in Fig. 9 . It is worth mentioning that antibody could attach on the surface cell at different positions, which occurs by several mechanisms but has the same effect. For instance, the first mechanism takes place through direct attaching of antibodies with site of COVID-19-RBD, causing competition between the specific receptor ACE2 with antibody. The different mechanism could happen via binding of antibody with other position on RBD but there is no vie with the targeted receptor (Fig. 9). The antibody/RBD complex at another position other than where receptor binds leads RBD structure modification and causing hindrance between key and lock attachment of RBD to ACE2 [21].
Fig. 9.
Schematic diagram of the attachment mechanism of antibodies and RBD complex.
A massive efforts have been paid, however deep studies have still required. It is observed that very few trials have done to investigate antibodies for example; one trial is recently examined the significant neutralization of COVID-19 through attaching with S glycoprotein of RBD [9]. The study recommended different types of antibodies as mixture enhancing neutralization effect on COVID-19 [9]. Previously, the results of convalescent patients who used antibodies for treatment infection of SARS-CoV have induced adverse effects in the cases like Antibody-Dependent Enhancement producing proliferation viral contamination and other side effects on the immune responses [67]. Based on several studies which paid for developing of anti-COVID-19 vaccines, it was observed that the chances of materialization the efforts for COVID-19 are meager. Consequently, natural products and traditional herbals characterized by high availability and safe consumption and ingestion by individuals may be one of the most effective candidates against COVID-19 [21].
6. Conclusion and future prospective
Coronaviruses become the biggest enemy of humanity in the current time, causing the global economy and loss of human lives. Today, the target for the most scientific groups worldwide is to search around the viruses to find an appropriate therapy that can effectively control this virus. Initially to achieve the goal; it is critical to understand the nature of these viruses and how it can contact the host and their binding mechanism. The mutation and genetic variations are very important to discuss but there is still limited. The current review also reported about the immune system and the adaptive immune response; however many secrets are present and further deep investigations are highly recommended. Currently, most researchers are keeping investigating the control methods. Natural products may offer an effective way to impair the viral infection cycle and treatment of this pandemic. In the light of use nanotechnology in the synthesis of nanoparticles that have anti-bacterial properties [69], [70], [71], [72], [73], [74], [75], [76], and based on previous studies on antibacterial nanoparticles, nanotechnology can be used in the diagnosis or treatment of COVID-19. Recently, nanotechnology tools can play a pivotal role in advancing COVID-19 treatment and vaccine development. The COVID-19 vaccine development journey has been very impressive, involving high-tech platforms such as viral vectors, antigen carriers, and delivery technology. Clinical trials for an mRNA-based vaccine delivered by lipid nanoparticles (LNPs) are already underway [77].
Declaration of competing interest
The author reports no conflicts of interest in this work.
Acknowledgments
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program to support publication in the top journal (Grant no. 42-FTTJ-36).
References
- 1.Denaro M., et al. Antiviral activity of plants and their isolated bioactive compounds: an update. Phyther. Res. 2020;34(4):742–768. doi: 10.1002/ptr.6575. [DOI] [PubMed] [Google Scholar]
- 2.Koshak D.A.E., Koshak P.E.A. Nigella sativa L as a potential phytotherapy for coronavirus disease 2019: a mini review of in silico studies. Curr. Ther. Res. - Clin. Exp. 2020;93 doi: 10.1016/j.curtheres.2020.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonio A.D.S., Wiedemann L.S.M., Veiga-Junior V.F. Natural products’ role against COVID-19. RSC Adv. 2020;10(39):23379–23393. doi: 10.1039/d0ra03774e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bambini S., Rappuoli R. 2020. The use of genomics in microbial vaccine development. no. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nur M., Azam K., Mahamud R.Al, Hasan A., Jahan R., Rahmatullah M. Some home remedies used for treatment of COVID-19 in Bangladesh. J. Med. Plants Stud. 2020;8(4):27–32. www.plantsjournal.com [Google Scholar]
- 7.Maier H.J., Bickerton E., Britton P. Coronaviruses: methods and protocols. Coronaviruses Methods Protoc. 2015;1282(1):1–282. doi: 10.1007/978-1-4939-2438-7. [DOI] [PubMed] [Google Scholar]
- 8.Amanat F., Krammer F. Perspective SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.BS, Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARSCoV antibody. Nature. 2020;383:290–295. doi: 10.1080/14756366.2019.1690480. [DOI] [PubMed] [Google Scholar]
- 11.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(March) doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meneguzzo F., Ciriminna R., Zabini F., Pagliaro M. Review of evidence available on hesperidin-rich products as potential tools against COVID-19 and hydrodynamic cavitation-based extraction as a method of increasing their production. Processes. 2020;8(5):1–19. doi: 10.3390/PR8050549. [DOI] [Google Scholar]
- 13.Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116(February):76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glowacka I., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/jvi.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang I.C., et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1) doi: 10.1371/journal.ppat.1001258. no. January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim Y., Ng Y., Tam J., Liu D. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4(4):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agostini M.L., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9(2):1–15. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abduljauwad S.N., Habib T., Ahmed H. 2020. Nano-clays as Potential Pseudo-antibodies for COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(May) doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 24.Ciulla M.M. Coronavirus uses as binding site in humans angiotensin - converting enzyme 2 functional receptor that is involved in arterial blood pressure control and fibrotic response to damage and is a drug target in cardiovascular disease. Is this just a phylogenet. J. Med. Virol. 2020:1–2. doi: 10.1002/jmv.25774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheahan T., et al. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 2008;82(5):2274–2285. doi: 10.1128/jvi.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of Host Receptor Adaptation by Severe Acute Respiratory Syndrome Coronavirus. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabi F.A., Zoubi M.S.Al, Kasasbeh G.A., Salameh D.M., Al-Nasser A.D. SARS-CoV-2 and Coronavirus Disease 2019: What We Know So Far. 2020. pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Infections C., Li F. Structural Analysis of Major Species Barriers between Humans and Palm Civets for Severe Acute Respiratory Syndrome. 2008;82(14):6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florindo H.F., et al. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15(8):630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boechat J.L., Chora I., Morais A., Delgado L. Vol. 27. ELSEVIER; 2021. The immune response to SARS-CoV-2 and COVID-19; pp. 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahmoodi M., et al. The effect of white vinegar on some blood biochemical factors in type 2 diabetic patients. J. Diabetes Endocrinol. 2013;4(1):1–5. doi: 10.5897/JDE12.015. [DOI] [Google Scholar]
- 33.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16(4):343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osamu Takeuchi S.A. Innate immunity to virus infection. Immunol. Rev. 2017;227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dogra P., et al. Innate immunity plays a key role in controlling viral load in COVID-19: mechanistic insights from a whole-body infection dynamics model. ACS Pharmacol. Transl. Sci. 2021;4(1):248–265. doi: 10.1021/acsptsci.0c00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J. Innate Immun. 2020;12(1):4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Q., et al. Article the impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity ll ll article the impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson E.C., et al. 2020. The circulating SARS-CoV-2 spike variant N439K maintains fitness while evading antibody-mediated immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou Y.J., et al. SARS-CoV-2 D614G variant exhibits enhanced replication ex vivo and earlier transmission in vivo. bioRxiv. 2020;1468(December):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korber B., et al. Article Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID- ll Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. 2020. pp. 812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grifoni A., Weiskopf D., Ramirez S., Diego S., Mateus J. 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. no. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarke A., et al. Article comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases ll ll comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Reports Med. 2021;2(2) doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agerer B. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8 + T cell responses. Vol. 6461. 2021. pp. 17–22. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah V.K., Firmal P., Alam A., Ganguly D., Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11(August):1–17. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benarba B., Pandiella A. Medicinal plants as sources of active molecules against COVID-19. Front. Pharmacol. 2020;11(August):1–16. doi: 10.3389/fphar.2020.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klasse P.J., Nixon D.F., Moore J.P. Immunogenicity of clinically relevant SARS-CoV-2 vaccines in nonhuman primates and humans. Sci. Adv. 2021 doi: 10.1126/sciadv.abe8065. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 48.Food and Drug Administration . Vaccines and Related Biological Products Advisory Committee Meeting FDA Briefing Document. Pfizer-BioNTech COVID-19 Vaccine. Fda; 2020. pp. 1–53. [Google Scholar]
- 49.Walsh E.E., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383(25):2439–2450. doi: 10.1056/nejmoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. bioRxiv. 2020;1263(March):1260–1263. doi: 10.1101/2020.02.11.944462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baden L.R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384(5):403–416. doi: 10.1056/nejmoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sahin U., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 53.Ashraf S., et al. Honey and Nigella sativa against COVID-19 in Pakistan (HNS-COVID-PK): a multi-center placebo-controlled randomized clinical trial. medRxiv. 2020 doi: 10.1101/2020.10.30.20217364. Available: [DOI] [PubMed] [Google Scholar]
- 54.Yoshimoto J., Ono C., Tsuchiya Y., Kabuto S., Kishi M., Matsuura Y. Virucidal effect of acetic acid and vinegar on SARS-CoV-2. 2020. pp. 3–7. [DOI] [Google Scholar]
- 55.Zakaryan H., Arabyan E., Oo A., Zandi K. Flavonoids: promising natural compounds against viral infections. Arch. Virol. 2017;162(9):2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhuiyan F.R., Howlader S., Raihan T., Hasan M. Plants metabolites: possibility of natural therapeutics against the COVID-19 pandemic. Front. Med. 2020;7(August):1–26. doi: 10.3389/fmed.2020.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rukh L., Technology I., Mohammad N. 2020. The use of home remedies for covid-19 in Pakistan: A Review. no. June. [DOI] [Google Scholar]
- 58.Rahman N., Basharat Z., Yousuf M., Castaldo G., Rastrelli L., Khan H. Virtual screening of natural products against type II virtual screening of natural products against type II transmembrane serine protease. Molecules. 2020;2:1–12. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice G.I., Thomas D.A., Grant P.J., Turner A.J., Hooper N.M. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem. J. 2004;383(1):45–51. doi: 10.1042/BJ20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daskaya-Dikmen C., Yucetepe A., Karbancioglu-Guler F., Daskaya H., Ozcelik B. Angiotensin-I-converting enzyme (ACE)-inhibitory peptides from plants. Nutrients. 2017;9(4) doi: 10.3390/nu9040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joshi T., et al. In silico screening of natural compounds against COVID-19 by targeting mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci. 2020;24(8):4529–4536. doi: 10.26355/eurrev_202004_21036. [DOI] [PubMed] [Google Scholar]
- 62.Guan L., et al. ADMET-score-a comprehensive scoring function for evaluation of chemical drug-likeness. Medchemcomm. 2019;10(1):148–157. doi: 10.1039/C8MD00472B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S. Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. 2020. pp. 1–14. Preprints, no. March. [DOI] [Google Scholar]
- 64.Goulder P.J.R., Watkins D.I. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 2008;8(8):619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thimme R., et al. CD8+ T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 2003;77(1):68–76. doi: 10.1128/jvi.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Braun J., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 67.Zhang X., et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 69.Salama A., Diab M.A., Abou-Zeid R.E., Aljohani H.A., Shoueir K.R. Crosslinked alginate/silica/zinc oxide nanocomposite: a sustainable material with antibacterial properties. Compos. Commun. 2018;7(November):7–11. doi: 10.1016/j.coco.2017.11.006. [DOI] [Google Scholar]
- 70.Fouda M.M.G., Ajarem J.S., Maodaa S.N., Allam A.A., Taher M.M., Ahmed M.K. Carboxymethyl cellulose supported green synthetic features of gold nanoparticles: antioxidant, cell viability, and antibacterial effectiveness. Synth. Met. 2020;(October) doi: 10.1016/j.synthmet.2020.116553. [DOI] [Google Scholar]
- 71.Ajarem J.S., Maodaa S.N., Allam A.A., Taher M.M., Khalaf M. Benign synthesis of cobalt oxide nanoparticles containing red algae extract: antioxidant, antimicrobial, anticancer, and anticoagulant activity. J. Clust. Sci. 2021;9 doi: 10.1007/s10876-021-02004-9. [DOI] [Google Scholar]
- 72.Al-Ahmed Z.A., et al. Electrospun nanofibrous scaffolds of ϵ-polycaprolactone containing graphene oxide and encapsulated with magnetite nanoparticles for wound healing utilizations. Mater. Res. Express. 2021;8(2) doi: 10.1088/2053-1591/abe42b. [DOI] [Google Scholar]
- 73.Hassan A.A., et al. Polycaprolactone based electrospun matrices loaded with Ag/hydroxyapatite as wound dressings: morphology, cell adhesion, and antibacterial activity. Int. J. Pharm. 2021;(December) doi: 10.1016/j.ijpharm.2020.120143. [DOI] [PubMed] [Google Scholar]
- 74.Al Jahdaly B.A., et al. Selenium nanoparticles synthesized using an eco-friendly method: dye decolorization from aqueous solutions, cell viability, antioxidant, and antibacterial effectiveness. J. Mater. Res. Technol. 2021;11(January):85–97. doi: 10.1016/j.jmrt.2020.12.098. [DOI] [Google Scholar]
- 75.Shaban N.Z., et al. Chitosan-based dithiophenolato nanoparticles: preparation, mechanistic information of DNA binding, antibacterial and cytotoxic activities. J. Mol. Liq. 2020;(November) doi: 10.1016/j.molliq.2020.114252. [DOI] [Google Scholar]
- 76.Al-Ahmed Z.A., Al-Radadi N.S., Ahmed M.K., Shoueir K., El-Kemary M. Dye removal, antibacterial properties, and morphological behavior of hydroxyapatite doped with Pd ions. Arab. J. Chem. 2020;13(12):8626–8637. doi: 10.1016/j.arabjc.2020.09.049. [DOI] [Google Scholar]
- 77.Chauhan G., Madou M.J., Kalra S., Chopra V., Ghosh D., Martinez-Chapa S.O. Nanotechnology for COVID-19: therapeutics and vaccine research. ACS Nano. 2020;14(7):7760–7782. doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]