ABSTRACT
The Mycobacterium avium complex (MAC) is one of the most prevalent causes of nontuberculous mycobacteria pulmonary infection in the United States, and yet it remains understudied. Current MAC treatment requires more than a year of intermittent to daily combination antibiotic therapy, depending on disease severity. In order to shorten and simplify curative regimens, it is important to identify the innate bacterial factors contributing to reduced antibiotic susceptibility, namely, antibiotic tolerance genes. In this study, we performed a genome-wide transposon screen to elucidate M. avium genes that play a role in the bacterium’s tolerance to first- and second-line antibiotics. We identified a total of 193 unique M. avium mutants with significantly altered susceptibility to at least one of the four clinically used antibiotics we tested, including two mutants (in DFS55_00905 and DFS55_12730) with panhypersusceptibility. The products of the antibiotic tolerance genes we have identified may represent novel targets for future drug development studies aimed at shortening the duration of therapy for MAC infections.
IMPORTANCE The prolonged treatment required to eradicate Mycobacterium avium complex (MAC) infection is likely due to the presence of subpopulations of antibiotic-tolerant bacteria with reduced susceptibility to currently available drugs. However, little is known about the genes and pathways responsible for antibiotic tolerance in MAC. In this study, we performed a forward genetic screen to identify M. avium antibiotic tolerance genes, whose products may represent attractive targets for the development of novel adjunctive drugs capable of shortening the curative treatment for MAC infections.
KEYWORDS: DNA sequencing, bioinformatics, mechanisms of action, molecular genetics, persistence, tolerance
INTRODUCTION
Nontuberculous mycobacteria (NTM) are found ubiquitously in the environment, and several species can cause disease especially in the elderly, those with preexisting lung disease, and the immunocompromised, including those infected with HIV (1–4). The Mycobacterium avium complex (MAC), a group of 12 related, slow-growing mycobacteria with Mycobacterium intracellulare and Mycobacterium avium as the most prevalent species, accounts for the majority of pulmonary infections due to NTM in the United States (5, 6). Although the true incidence of pulmonary MAC infections in the United States is not known, a study in Oregon reported 4.8 cases per 100,000 person-years in 2012 (7). Winthrop et al. estimated the annual incidence of NTM infections in the United States to be 4.73 cases per 100,000 person-years (8), and the NTM Network European Trials Group has reported that MAC accounts for 52% of NTM isolates in the United States and Canada (9), suggesting that the annual incidence of MAC in the United States may be closer to ∼2.5 per 100,000 person-years.
The current treatment for MAC comprises a combination of multiple antibiotics given for at least 12 months following the conversion of sputum cultures from positive to negative. Since sputum culture conversion occurs in ∼50% of cases after 5 months of antibiotic therapy, a typical patient receives a minimum of 15 to 18 months of treatment (10, 11). Macrolide-susceptible MAC infection is usually treated with at least three antibiotics, including a macrolide (azithromycin or clarithromycin), a rifamycin (rifampin or rifabutin), and ethambutol, either intermittently (three times weekly) or daily for severe fibronodular or cavitary disease. If the infecting MAC strain is macrolide-resistant or the patient is unable to take the first-line regimen, alternative antibiotics, such as moxifloxacin, clofazimine, or linezolid, are often used (11, 12).
The lengthy and complicated treatment course required to eradicate MAC infection has been attributed to the presence of persistent organisms, which exhibit reduced susceptibility, or tolerance, to antibiotics (13). Unlike antibiotic resistance, which results from a heritable genetic alteration permitting continued bacterial growth in the presence of antibiotic concentrations exceeding the MIC, antibiotic tolerance is a transient, nonheritable phenotype without associated change in the MIC. The term antibiotic tolerance was originally coined in 1970 by Tomasz et al. to denote the ability of bacteria to withstand the bactericidal activity of antibiotics, especially of cell wall-active agents, primarily by reducing their replication rate (14). In the intervening decades, additional mechanisms have been proposed to mediate bacterial antibiotic tolerance, including biofilm formation (15, 16), induction of the stringent response (17–20) or efflux pumps (21–23), and altered metabolism (24, 25). Following ingestion by macrophages, MAC members acquire an antibiotic-tolerant phenotype within the arrested phagosome (26). Moreover, various stress conditions, including nutrient starvation, low pH, and hypoxia, induce a nonreplicative, antibiotic-tolerant state (24), which is characterized by transcriptional changes (24, 25), leading to altered cell wall membrane permeability (3) and an increased expression of efflux pumps (21). This stress-induced adaptation of MAC is accompanied by dramatically reduced metabolism, with a shift to the glyoxylate shunt, stabilization of the mycolate pool, and a switch to transcription of only essential genes (25). Additionally, the glyoxylate shunt enzyme isocitrate lyase is critical for the long-term survival of the related pathogen Mycobacterium tuberculosis in host tissues (27) and can be used in a reductive amination pathway to produce NAD, which may serve as an alternative energy source in a nonreplicative state and under anaerobic conditions (28, 29). However, the molecular mechanisms driving antibiotic tolerance in MAC remain poorly understood.
Transposon insertion sequencing (Tn-seq) is a powerful technique for determining bacterial genotype-phenotype relationships, particularly specific bacterial genes that are required for growth and/or survival under controlled stress conditions (30–33). Modifications of this technique have been used to define essential genes for in vitro growth of M. tuberculosis (34) and M. avium (35). Xu et al. screened a saturated transposon mutant library in the presence of partially inhibitory concentrations of various antibiotics with diverse mechanisms of action to identify genetic determinants of intrinsic antibiotic susceptibility in M. tuberculosis (36). In the current study, we used a similar approach to identify genes responsible for the intrinsic tolerance of M. avium to the antibiotics clarithromycin (CLR), rifabutin (RFB), moxifloxacin (MOX), and ethambutol (EMB). The hits we have identified may serve as targets for the development of novel antibiotics, with the objective of shortening the duration of curative treatment for MAC.
RESULTS
Effects of antibiotics on bacterial growth at the population level.
To monitor the effects of individual antibiotics on the entire bacterial population, we measured CFU and optical density at 600 nm (OD600) during antibiotic exposure. CFU values obtained following antibiotic exposure for 0, 12, and 48 h are provided in Fig. S2 in the supplemental material. OD600 values are provided in Fig. S3 in the supplemental material. The same no-drug (vehicle) control data appear in all four plots (performed in triplicate). Notably, the no-drug control curve has an inflection at the 12-h time point. Truly logarithmic growth should appear as a straight line on this plot. Light microscopy of unstained samples of the no-drug control cultures revealed clumps of approximately 5 bacteria (data not shown), likely accounting for the inflection point. Bacterial clumping was likely also present in the antibiotic-containing tubes, although these were not specifically examined. Additionally, we observed that the OD curve increased for most samples from 0 h to 12 h, whereas the CFU remained stable or declined over this time. In our view, this discrepancy between OD and CFU is most likely accounted for by growth of new bacteria in the context of larger bacterial clumps, which scatter more light (increased OD) but may not increase the CFU (although viable bacterial cells per clump will increase). The presence of clumping is unlikely to affect the results of the screen, as there is no reason to suspect that individual transposon mutants were disproportionately distributed among the clumps.
Applying the set of criteria for selecting samples for processing as described in the Methods, we prepared libraries and sequenced both time points at the following concentrations: 0.54 and 5.4 μg/ml CLR, 0.1 and 1.0 μg/ml MOX, 0.063 and 0.63 μg/ml RFB, and 0.21 and 2.1 μg/ml EMB. The colored arrows in Fig. S2 and S3 indicate the time points sequenced.
Identification of mutants with altered antibiotic susceptibility.
A total of 161 mutants showed increased susceptibility and 32 mutants showed reduced susceptibility to at least 1 of the 4 antibiotics. A total of 46 mutants were hypersusceptible and 14 mutants were hypertolerant to CLR. Six mutants were found to be hypersusceptible to EMB, while no mutants were hypertolerant to this antibiotic. The MOX screen revealed 103 hypersusceptible and 2 hypertolerant mutants. A total of 84 mutants were found to be hypersusceptible and 108 mutants were hypertolerant to RFB. Effect sizes (after 48 hours of exposure) for mutants with significantly altered antibiotic susceptibility are plotted in Fig. 1 and summary data are provided in Tables S3 to S6 in the supplemental material.
FIG 1.
Bar chart showing the effect size of each statistically significant mutant. Each bar represents a single gene. A negative value represents a hypersusceptible mutant, while a positive value signifies that a mutant is less susceptible (hypertolerant) to the antibiotic. (A) Clarithromycin, (B) ethambutol, (C) moxifloxacin, (D) rifabutin.
We also evaluated for overlaps between the different drug classes tested. Our results of this analysis are summarized in Fig. 2 and Table S1 (hypersusceptible mutants) and Table S2 (hypertolerant mutants) in the supplemental material. Mutants hypersusceptible to multiple antibiotics may reflect genes with a role in more general bacterial persistence mechanisms, while mutants hypertolerant to multiple antibiotics may suggest genes promoting antibiotic susceptibility. Notably, no mutant was found to be hypersusceptible to one antibiotic but hypertolerant to another.
FIG 2.
Venn diagram of identified hypersusceptible (A) and hypertolerant transposon mutants (B). Note that in A, the set of ethambutol-hypersusceptible mutants has been partitioned into two sets (both in yellow). Partitioning in this way greatly simplifies the diagram. Gene names in each category can be found in Table S1 and S2.
Two mutants were hypersusceptible to all four antibiotics tested, namely, DFS55_00905 (annotated as an acyltransferase, homologous to M. tuberculosis Rv0111) and DFS55_12730 (hypothetical protein, homologous to Rv1836c). Ten mutants were hypersusceptible to CLR, MOX, and RFB, but not to EMB.
The following two mutants were identified as hypertolerant to CLR, MOX, and RFB (no mutant was hypertolerant to EMB): DFS55_10765 (annotated as a pyruvate kinase, homologous to Rv1617) and DFS55_20040 (DUF1707 domain-containing protein, homologous to Rv0966c). An additional 4 mutants were found to be hypertolerant to RFB and CLR only, but not to MOX. These mutants included DFS55_10660 (quinolinate synthase, homologus to Rv1594), DFS55_10665 (l-aspartate oxidase, homologous to Rv1595), DFS55_16845 (trigger factor, homologous to Rv2462c), and DFS55_21750 (hypothetical protein, homologous to Rv3489).
DISCUSSION
In this work, we utilized a genome-wide transposon mutant pool to screen for M. avium mutants with altered susceptibility to various clinically relevant antibiotics. Compared with the other antibiotics, exposure to MOX yielded the highest number of hypersusceptible mutants, highlighting the many potential targets which might synergize with this antibiotic. Also, given the strong effect sizes observed with MOX relative to the other antibiotics tested, MOX synthetic lethality may represent the greatest opportunity for novel treatment-shortening strategies.
Transposon insertions in several known virulence genes were found to enhance the susceptibility of M. avium to multiple antibiotics. For example, mutations in secA2 (DFS55_12665 or rv1821) conferred hypersusceptibility to both CLR and MOX. The Sec export pathway is conserved across bacteria and exports secreted proteins across the cytoplasmic membrane (37). Mycobacteria have two SecA proteins, namely, SecA1 and SecA2 (37). While SecA1 is essential and facilitates the transport of unfolded proteins though the SecYEG channel via its ATPase activity, the mechanism of export in the SecA2 pathway is less well understood (38). SecA2 is required for secretion of M. tuberculosis virulence proteins and arrest of phagosome maturation by preventing acidification, thereby facilitating M. tuberculosis growth within macrophages (39). In particular, the SecA2-secreted phosphatase SapM and the kinase PknG have been identified as effectors with direct roles in preventing phagosome maturation and promoting M. tuberculosis intracellular survival and replication (40). As CLR inhibits protein synthesis (41) and SecA2 disruption impairs the secretion of virulence-related proteins, these two alterations, which both dysregulate proteostasis, may have a synergistic or additive effect, leading to higher antibiotic susceptibility for this mutant. A similar but more indirect mechanism could be proposed for the sensitization of this mutant to MOX, which inhibits DNA gyrase and topoisomerase IV (41). Both of these enzymes are involved in the winding and unwinding of DNA and are necessary for DNA replication and RNA transcription (42, 43). MOX-induced reductions in mRNA transcripts may also dysregulate proteostasis in MAC, potentially explaining the similar phenotypes observed with CLR exposure.
RecA (DFS55_08530 or Rv2737c), which was found to be required for MAC tolerance to both MOX and RFB, plays a critical role in the mycobacterial DNA damage response, specifically in the repair of double-stranded breaks, as M. smegmatis cells lacking RecA are more sensitive to DNA damage (44). Specifically, after double-stranded breaks are resected by AdnAB, RecA is loaded onto the 3′ end of the DNA, helping to mediate a homology search and subsequent strand invasion (44). By inhibiting DNA topoisomerases, MOX promotes DNA damage and triggers a mutagenic SOS response, which can lead to the formation of persister cells (45). RecA activation promotes the self-cleavage of LexA leading to upregulation of the SOS regulon (46). In turn, removal of RecA leads to the suppression of the SOS regulon and decreased persister formation. Chemical inhibition of RecA with suramin in DNA gyrase-depleted cells has been shown to improve killing of M. tuberculosis by several anti-TB drugs, including rifampin and EMB (45). RFB, the rifamycin tested in our study, inhibits DNA-dependent RNA polymerase and suppresses RNA synthesis. Previous work showed that a recA-deficient M. tuberculosis mutant was unable to develop resistance to rifampin, possibly due to an inability to generate an SOS response (47). In short, it appears that inhibiting RecA, thereby suppressing the SOS response, could provide a means to decrease persister formation and improve killing of pathogenic mycobacteria, such as M. tuberculosis and MAC.
Interestingly, we identified two mutants as hypersusceptible to all four antibiotics tested in our study (Fig. 2). They included mutants with transposon insertions in DFS55_00905 (annotated as an acyltransferase, homologous to rv0111) and DFS55_12730 (hypothetical protein, homologous to rv1836c). Mutants in DFS55_00905 displayed particularly robust hypersusceptibility to MOX (effect size of −2.0 at 1 μg/ml and 48-h exposure) and EMB (effect size of −1.4 at 2.1 μg/ml, 48 h, which was the largest effect size we observed with this drug at 48 h). Mutants in DFS55_12730 were strongly hypersusceptible to CLR (effect size of-1.6 at 5.4 μg/ml and 48 h), MOX (−1.9 at 1.0 μg/ml and 48 h), and RFB (−1.5 at 0.63 μg/ml and 48 h). Future work should investigate the function of these gene products and their relationship to the pansusceptibility phenotype observed.
An additional 10 mutants were found to be hypersusceptible to CLR, MOX, and RFB, but not to EMB. They included mutants in sigE (DFS55_18590, rv1221) and an alpha-beta hydrolase gene (DFS55_15065, homologous to rv2224c, also known as caeA or hip1). Deficiency of sigma factor E (SigE) has been shown to confer increased susceptibility of M. tuberculosis to multiple drugs, including EMB and rifampin, but not to ciprofloxacin (48). These results differ somewhat from the results of our study. Upon closer examination of our results for EMB, we find that this mutant was barely outside our conservative thresholds for defining hypersusceptible mutants. While adjusted P values for EMB at both time points were below the cutoff (0.045 and 0.0001; cutoff, 0.05) (see Table S6), the corresponding log-fold changes were barely above our chosen thresholds (−0.47 and −1; cutoff, −0.5) (Table S6). Therefore, it is possible that our stringent cutoffs misclassified this mutant as displaying a similar EMB susceptibility as the wild type. On the other hand, the discrepancy regarding hypersusceptibility of sigE-deficient mycobacteria to fluoroquinolones may be due to differences between the two species (M. tuberculosis versus MAC, which have substantially different growth rates) and/or the experimental designs of the two studies (resazurin microtiter assay versus Tn-seq screen). Additional studies are required to further evaluate the impact of sigE deficiency on MAC susceptibility to EMB and fluoroquinolones. Consistent with our data in MAC, M. tuberculosis mutants in caeA/hip1/rv2224c have been shown to be hypersusceptible to rifamycins (rifampin) (36, 49) and macrolides (erythromycin) (49). Our data suggest that caeA (DFS55_15065) deficiency also confers enhanced susceptibility to fluoroquinolones (Table S1 and S4), which might be useful for designing novel therapies for M. tuberculosis.
Xu et al. screened a M. tuberculosis transposon mutant pool exposed to EMB, the only antibiotic shared with our study, and identified 45 hypersusceptible transposon mutants (q value of <0.05) (36). While the species of organism used and experimental design of our study differ from those of Xu et al., we were intrigued to discover that 3 of the 6 genes meeting our stringent cutoffs (DFS55_00120/rv0019c, DFS55_03885/rv0642c, and DFS55_12730/rv1836c) were shared by both lists of hypersusceptible mutants. This large fraction of genetic overlap suggests there may be some common genetic elements that lead to EMB hypersusceptibility in mycobacteria more generally.
We also identified two mutants with hypertolerance to three of the four antibiotics tested (CLR, MOX, and RFB), namely, DFS55_10765 (annotated as a pyruvate kinase, rv1617) and DFS55_20040 (DUF1707 domain-containing protein, rv0966c). Interestingly, Rv1617 deficiency is associated with a large growth defect in M. tuberculosis (34, 35), but the same phenotype is not observed in DFS55_10765-deficient M. avium (35). This result suggests that the metabolic impact of pyruvate kinase deficiency is remarkably different between MAC and M. tuberculosis. Pyruvate kinase catalyzes the transfer of a phosphate group from phosphoenolpyruvate to ADP (yielding pyruvate and ATP). Central metabolism may be disrupted in bacteria lacking this enzyme, possibly triggering the stringent response, which has been previously shown to protect bacteria from antibiotic-mediated killing (50, 51). Deletion of pyruvate kinase in M. tuberculosis causes allosteric inhibition of the trichloroacetic acid (TCA) cycle through accumulation of phosphoenolpyruvate (52). Disruption of the TCA cycle, especially alternate catabolism through the glyoxylate shunt, has been linked to antibiotic tolerance in multiple bacterial species, including Pseudomonas aeruginosa (53), Staphylococcus aureus (54, 55), and Staphylococcus epidermis (56), suggesting that this pathway may be a common mechanism for promoting antibiotic tolerance. In M. tuberculosis, downregulation of the malate synthase GlcB, one of the enzymes in the glyoxylate shunt, caused increased susceptibility to both rifampin and to nitrosative and oxidative stresses in vitro (57). Deficiency of isocitrate lyase, another glyoxylate shunt enzyme, also led to increased susceptibility of M. tuberculosis to antibiotics in vitro (58) and decreased survival in activated macrophages and mice (27). Thus, mycobacterial metabolism, and the TCA cycle in particular, clearly plays an important role in the development of antibiotic tolerance, although more work is necessary to fully elucidate its contributions. DFS55_20040 appears to lack an annotated function in the literature. Additional work is needed to understand the function of these two genes and determine their relationship to antibiotic tolerance in mycobacteria.
Our approach has several limitations. First, mutants in essential genes (or those without TA insertion sites) could not be screened, as they could not be recovered with our bacterial regrowth techniques. Therefore, we were unable to assess the potential role in antibiotic tolerance of genes essential for growth of M. avium in nutrient-rich medium. Second, gene disruptions leading to changes in secreted factors (e.g., extracellular proteins) may have been missed by our screen, as these factors may be complemented by factors produced by nondefective mutants present in the same culture. Third, we chose a conservative statistical approach (JT-test) and conservative thresholds for P values and log-fold changes (LFCs), which must be met at two different time points. It is likely that mutants with low numbers of insertion sites or somewhat weaker effect sizes were missed. Lastly, we have performed these screens in only a single strain of M. avium (MAC109), and it remains to be determined to what extent our data apply to other M. avium strains.
As we noted above, some clumping was observed in cultures at the 12-h time point, reducing the usefulness of the CFU measure but unlikely to impact the loss of hypersusceptible mutants. Tween 80 is a synthetic detergent known to greatly reduce clumping in some other mycobacterial species, such as M. tuberculosis. However, inclusion of this detergent is known to strongly impact antibiotic susceptibility in mycobacteria, including M. avium (59). Given that our preliminary culture work displayed limited clumping using medium without Tween 80, we decided to exclude it from our experiments (data not shown). It is possible that the addition of this detergent could reduce the clumping we observed at 12 h, although we did not test this possibility.
Previous studies have examined mycobacterial antibiotic hypersusceptibility in the context of very low antibiotic concentrations (36). In such an experimental setup, the entire bacterial population continues to grow during antibiotic exposure, and libraries are generated directly from bacterial cultures. In contrast, our approach here uses an additional regrowth step on solid agar after antibiotic exposure. This regrowth step produces sufficient material for library generation independent of whether the aggregate bacterial population is growing, stable, or dying. Therefore, our approach is more generally applicable to clinical scenarios in which higher doses of antibiotics may be used, inhibiting aggregate mycobacterial growth.
Mutations causing defects while the aggregate population declines or is static are interpreted in our screen as amplifying the killing effect of the antibiotic (given that the wild-type organisms can be assumed to be nongrowing). However, an observation of hypersusceptibility in the context of aggregate growth is more difficult to precisely resolve. Thus, it could be that the mutant is killed in the presence of the antibiotic, whereas the wild type is able to grow, or it is possible that the mutant is more inhibited by the antibiotic than the wild type but continues to grow, albeit at a lower rate. In particular, the overall population declined in the presence of 5.4 μg/ml CLR, suggesting that any defective mutants are killed more rapidly than the wild type. However, at 0.54 μg/ml CLR, the overall population increased, suggesting either that defective mutants could be killed more rapidly or merely that their growth is inhibited to a greater extent than that of wild type (see Fig. S2). Follow-up studies are needed in order to resolve the behavior of hypersusceptible mutants at this dose.
Our study represents a first step toward the development of novel, treatment-shortening strategies for MAC infections through identification of genes mediating antibiotic susceptibility. Biochemical characterization of the corresponding gene products might yield novel insights into the mechanisms of MAC antibiotic tolerance and lay the groundwork for the development of novel antibiotics, which might synergize with currently available drugs to kill tolerant organisms and shorten curative treatment for MAC infections more effectively. Future work is needed to validate the susceptibility phenotypes of individual gene-deficient mutants and their respective complemented strains in axenic cultures. Proof-of-concept studies could then be performed to demonstrate the treatment-shortening potential of candidate targets in a relevant animal model of pulmonary MAC disease (60, 61).
MATERIALS AND METHODS
Strains.
All experiments were performed using Mycobacterium avium subsp. hominissuis strain 109 (35).
Media and buffers.
To make 7H11 agar, 10.25 g of 7H11 without malachite green powder (catalog [cat] no. 511A; HiMedia) was added to 450 ml deionized water. A volume of 5 ml 50% glycerol was added and then the mixture was autoclaved. The agar was cooled to 55°C before addition of 50 ml oleic acid-albumin-dextrose-catalase (OADC) enrichment (Becton, Dickinson). To make 7H9-30% OADC, 2.35 g of 7H9 powder was added to 350 ml deionized water. After sterilization (by autoclaving at 121°C or passing through a 0.22-μm filter), 150 ml of OADC enrichment was added. Unless otherwise specified, no Tween 80 or glycerol was included in the media. To make phosphate-buffered saline with Tween 80 (PBS-T), 1.25 ml of filter-sterilized 20% Tween 80 was added to 500 ml of sterile PBS.
Overview of transposon screen setup.
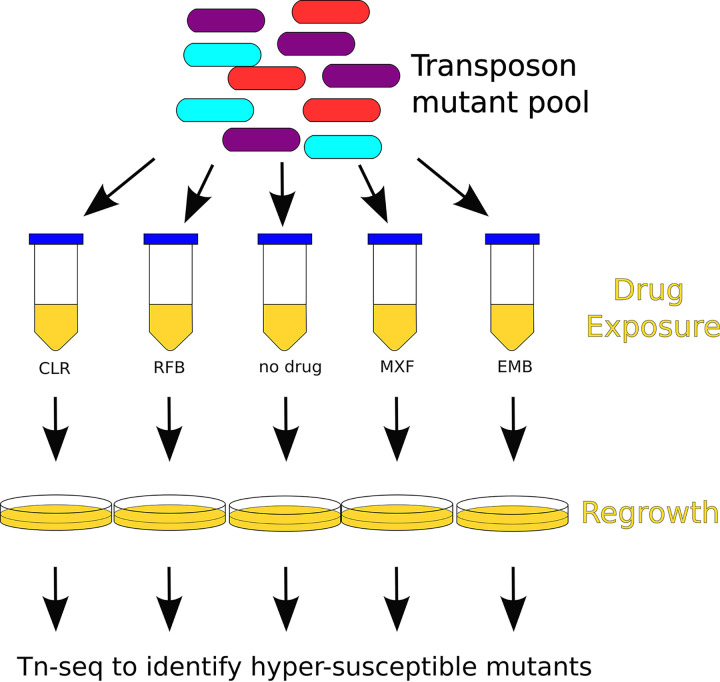
The setup for our genome-wide differential susceptibility screen is shown in Fig. 3. We inoculated two 1-ml frozen aliquots of a previously generated diverse Himar1 transposon mutant pool (consisting of approximately 1.2 × 106 unique mutants each) (35) into two 1.3-liter roller bottles each containing 250 ml of 7H9-30% OADC. The bottles was shaken overnight at 37°C to allow for some growth and reduce bacterial clumping (190 rpm, 0.75-in [1.905 cm] orbit). The optical density at 600 nm (OD600) was tracked until reaching 0.2 and 0.22. The bottles were then pooled, diluted to OD600 of 0.1 with cold 7H9-30% OADC, and aliquoted into 89 50-ml conical tubes (10 ml/tube). Tubes were incubated for 5 hours while shaking at 37°C to allow the bacteria to return to log-phase growth. Five tubes were randomly selected for processing at 0 hours. These samples were then processed for colony forming unit (CFU) enumeration and regrowth. The same tube was used for both operations. Following regrowth, bacterial samples were scraped, the DNA was extracted, and libraries were prepared for Tn-seq. Bacterial regrowth is required before Tn-seq library preparation in order to remove the contribution of DNA to the sequencing library from dead transposon mutants. After collection of enumeration and regrowth of samples at 0 hours, antibiotics (or dimethyl sulfoxide [DMSO] vehicle as a control) were then added to the remaining 84 tubes. Samples were collected at 12 and 48 hours after antibiotic exposure in triplicate and processed for CFU enumeration, bacterial regrowth, and Tn-seq library preparation in the same manner as the 0-hour samples.
FIG 3.
Schematic of transposon mutant screen to identify hypersusceptible mutants following exposure to multiple doses of various antibiotics in liquid culture. After antibiotic exposure, cultures were regrown on solid agar to enrich for surviving bacteria. After regrowth, DNA was extracted and prepared for Tn-seq analysis. Hypersusceptible mutants were identified using a nonparametric statistical approach.
Sample processing for CFU enumeration and regrowth.
The bacterial density (CFU/ml) was estimated by removing 400 μl of bacterial culture, centrifuging (2,000 × g for 5 minutes), and washing twice with PBS-T to remove the antibiotic. Washed samples were diluted 10-fold, and 50 μl of each dilution was plated onto 7H11 agar without malachite green. Malachite green, which is typically present in standard 7H10 and 7H11 agar formulations, was excluded to avoid potential issues with postantibiotic recovery in mycobacteria (62). T-shaped spreaders were used to spread liquid evenly across agar plates. CFUs were counted after 7 to 8 days.
For bacterial regrowth, the remainder of each tube (after removing samples for CFU enumeration) was centrifuged twice and washed (2,000 × g for 10 min) with 10 ml of PBS-T to remove the antibiotic. The samples were centrifuged once more, and the bacterial pellet was resuspended in 250 μl PBS-T. Fifty microliters of the washed transposon pool was plated onto each of four 7H11 agar plates and spread with 10 to 15 3-mm sterile glass beads to ensure even distribution of liquid across the plate. Samples were regrown for 7 to 8 days. Bacterial lawns from the four agar plates were scraped and pooled into 2-ml tubes. DNA was extracted from regrown samples, as described previously (short-read sequencing protocol) (63). DNA was processed for Tn-Seq, as described previously (35). Libraries were sequenced (2 × 75 bp) on an Illumina HiSeq 2500 instrument by the Johns Hopkins Genetic Resources Core Facility (GRCF) High Throughput Sequencing Center. A total of 59 samples (5 input-pool samples and 18 groups of output-pool triplicates) were sequenced, yielding between 2,333,295 and 7,193,522 reads per sample for a total of 269,324,560 paired-end reads.
Antibiotic selection.
Doses were selected to reflect antibiotic concentrations at the most common site of infection in noncompromised patients (the lungs) following standard antibiotic dosing. Based on a search of the pharmacokinetic literature, maximum achievable doses in lung tissues were taken to be 54 μg/ml for CLR (64), 0.63 μg/ml for RFB (65), 10.0 μg/ml for MOX (66), and 21.0 μg/ml for EMB (67) (based on nonhuman primate data). For each drug, a 10-fold dilution series of the estimated maximum achievable dose was performed to explore the impact of dose.
Before setting up the cultures used for this study, a preliminary calibration experiment (single replicate for each antibiotic, not processed for sequencing) was performed to estimate the bacterial viability at different antibiotic concentrations (data not shown) and to select the number of concentrations to include in the study (4 total concentrations for CLR, 3 concentrations for the other antibiotics in addition to drug-free controls).
Sample selection for sequencing from susceptibility screen.
A subset of samples from the differential susceptibility screen was chosen for Tn-seq library prep and sequencing. Samples were sequenced at two manually chosen concentrations for each drug at both available time points (12 h and 48 h). To help identify which should be processed further, objective criteria were established a priori, with the goal of identifying mutants with higher susceptibility to antibiotics but also reducing the likelihood that mutants were removed by chance due to low bacterial viability during antibiotic exposure. We considered the following 3 criteria for selecting samples:
Bacterial numbers must exceed 106 CFU/ml at all times during exposure. This ensures that the probability of losing a nondefective mutant is minimized, given the 60,129 possible thymine-adenine (TA) sites for the Himar1 transposon to insert across the MAC109 genome. Only CFU data were used, and we did not attempt to directly estimate the total number of bacterial cells, which can differ significantly from CFU.
There should be decreased bacterial viability after antibiotic exposure relative to drug-free controls (as measured by CFU). This ensures that the concentration of antibiotic is high enough to have bactericidal activity. Otherwise, the antibiotic concentration might be too low to select for or against mutants with growth phenotypes.
Drug concentrations near or below achievable serum concentrations of the drug after standard dosing are preferred. We assumed approximate serum values of 2.31 μg/ml for CLR, 4.42 μg/ml for MOX, 0.52 μg/ml RFB, and 2.27 μg/mL for EMB (68). In our view, this criterion makes the results more clinically relevant.
Identification of hypersusceptible mutants from sequencing data.
A schematic of the pipeline to process the data is provided in Fig. S1 in the supplemental material. Raw reads were mapped using the TRANSIT preprocessor (tpp) (69). Counts from tpp were then processed with a custom python script to produce a *.csv file to be read by pandas (version 0.24.1) used for downstream analyses.
Effect size/log-fold change calculation.
For calculation of the normalized read counts for each mutant, a pseudocount (70) of 4 was added to the raw count from all samples before dividing read counts by total read count:
where ˜xt,ir is the normalized read count; Xt,ir is the raw read count for transposon insertion site t, for antibiotic treatment group i, and for replicate r. α is the pseudocount (α = 4); and T is the number of transposon insertion sites. This pseudocount was added to stabilize the normalized count and was determined by manual examination of read counts in genes known to be essential. The pseudocount was set to be substantially larger than occasional background reads. The normalized read counts were then averaged over samples:
where μt,i is the average representation of each mutant across samples and ni is the number of replicates for treatment group i. An aggregate log-fold change (LFC) was used as a measure of effect size for differentially susceptible mutants. The aggregate LFC between treatment groups i and j was calculated as the median of the log-fold change at individual transposon insertion sites within a gene:
where Gg is the set of transposon insertion sites annotated to belong to gene g and LFCg,(i/j) is the log-fold change between treatment groups i and j for gene g. Although this formula is generally useful for comparing any pair of groups, for the work presented here, i always represents a drug-containing treatment group and j always represents the matching drug-free group at the same time point.
P value calculation.
For the calculation of P values, read counts for each sample were first normalized by dividing by the total read count in each sample (pseudocounts are unnecessary for the nonparametric test described below and thus were not used).
The Jonckheere-Terpstra (JT) test was then applied to the normalized read counts at each time point (71, 72). Briefly, the JT test is a nonparametric test of trend, which is more powerful than the more commonly used Kruskal-Wallis test when the alternative hypothesis assumes a monotonic trend of the treatment groups. In this case, we have three treatment groups for each drug at each time point, namely, no drug, low dose, and high dose. We postulated that if a mutant is hypersusceptible at a low dose of antibiotic, it will be even more hypersusceptible at a higher dose of that antibiotic. We define the alternative hypothesis as:
against a null hypothesis:
where θ1…θK are measures of a centrality parameter and K is the number of treatment groups for the drug at a particular time point (in this case K = 3 for each drug). The JT test statistic (Bt) at site t is defined as:
(where 1[] is the indicator function). P values at individual sites are computed via permutation test. Naively, this would result in nonuniformly distributed P values due to the discrete nature of the distribution. To ensure a truly uniform distribution, we performed a small correction on the permutation test P values by sampling from a uniform distribution bounded between adjacent values of the discrete permutation distribution. This process gives precisely uniform P values under the null hypothesis.
Pooling the P values within each gene (one P value calculated for each transposon insertion site) is then accomplished with the two-sided Stouffer’s method. Finally, adjusted P values are then computed using the Benjamini-Hochberg procedure.
Thresholds for defining differentially susceptible mutants.
Gene mutants are considered differentially susceptible to a drug if the absolute value of the LFC at the high dose of each drug (relative to no drug control) is greater than 0.5 and the adjusted P value is less than 0.05. These conditions must be met at both the 12-hours and 48-hour time points in order for the mutant to be defined as differentially susceptible to the drug. Mutants with negative LFC are predicted to be hypersusceptible to the drug, while a positive LFC indicates the mutant is hypertolerant.
Data availability.
The raw sequencing data (*.fastq) from this project can be found in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA559896. A Jupyter notebook and associated scripts to reproduce the data analysis (including figures) from the raw data are provided online at github (https://doi.org/10.5281/zenodo.4542412).
ACKNOWLEDGMENTS
This publication was made possible by funding from the following to P.C.K.: R21AI114507 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), and a pilot research grant from the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases, Division of Infectious Diseases of the Johns Hopkins University School of Medicine.
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID/NIH, Fisher Center or Johns Hopkins University School of Medicine.
W.M.M., H.P., C.D., L.H., and P.C.K. have no competing interests. J.S.B. is a founder, director, and equity holder of Neochromosome, Inc. Neochromosome is developing a yeast synthetic biology platform for pathway-level antimicrobial screens.
Footnotes
Supplemental material is available online only.
Contributor Information
Petros C. Karakousis, Email: petros@jhmi.edu.
Kileen L. Shier, Quest Dianostics Nicols Institute
REFERENCES
- 1.Falkinham JO. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev 9:177–215. doi: 10.1128/CMR.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolinsky E. 1979. Nontuberculous mycobacteria and associated diseases. Am Rev Respir Dis 119:107–159. doi: 10.1164/arrd.1979.119.1.107. [DOI] [PubMed] [Google Scholar]
- 3.Inderlied CB, Kemper CA, Bermudez LE. 1993. The Mycobacterium avium complex. Clin Microbiol Rev 6:266–310. doi: 10.1128/CMR.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito Y, Hirai T, Maekawa K, Fujita K, Imai S, Tatsumi S, Handa T, Matsumoto H, Muro S, Niimi A, Mishima M. 2012. Predictors of 5-year mortality in pulmonary Mycobacterium avium-intracellulare complex disease. Int J Tuber Lung Dis 16:408–414. doi: 10.5588/ijtld.11.0148. [DOI] [PubMed] [Google Scholar]
- 5.Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, Montes de Oca R, Shea YR, Seitz AE, Holland SM, Olivier KN. 2010. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med 182:970–976. doi: 10.1164/rccm.201002-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spaulding AB, Lai YL, Zelazny AM, Olivier KN, Kadri SS, Prevots DR, Adjemian J. 2017. Geographic distribution of nontuberculous mycobacterial species identified among clinical isolates in the United States, 2009–2013. Ann Am Thorac Soc 14:1655–1661. doi: 10.1513/AnnalsATS.201611-860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henkle E, Hedberg K, Schafer S, Novosad S, Winthrop KL. 2015. Population-based incidence of pulmonary nontuberculous mycobacterial disease in Oregon 2007 to 2012. Ann Am Thorac Soc 12:642–647. doi: 10.1513/AnnalsATS.201412-559OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winthrop KL, Marras TK, Adjemian J, Zhang H, Wang P, Zhang Q. 2020. Incidence and prevalence of nontuberculous mycobacterial lung disease in a large U.S. managed care health plan, 2008–2015. Ann Am Thorac Soc 17:178–185. doi: 10.1513/AnnalsATS.201804-236OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, Chimara E, Churchyard G, Cias R, Daza R, Daley CL, Dekhuijzen PNR, Domingo D, Drobniewski F, Esteban J, Fauville-Dufaux M, Folkvardsen DB, Gibbons N, Gómez-Mampaso E, Gonzalez R, Hoffmann H, Hsueh P-R, Indra A, Jagielski T, Jamieson F, Jankovic M, Jong E, Keane J, Koh W-J, Lange B, Leao S, Macedo R, Mannsåker T, Marras TK, Maugein J, Milburn HJ, Mlinkó T, Morcillo N, Morimoto K, Papaventsis D, Palenque E, Paez-Peña M, Piersimoni C, Polanová M, Rastogi N, Richter E, et al. , Nontuberculous Mycobacteria Network European Trials Group. 2013. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J 42:1604–1613. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 10.Griffith DE, Adjemian J, Brown-Elliott BA, Philley JV, Prevots DR, Gaston C, Olivier KN, Wallace RJ. 2015. Semiquantitative culture analysis during therapy for Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 192:754–760. doi: 10.1164/rccm.201503-0444OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Andrejak C, Böttger EC, Brozek J, Griffith DE, Guglielmetti L, Huitt GA, Knight SL, Leitman P, Marras TK, Olivier KN, Santin M, Stout JE, Tortoli E, van Ingen J, Wagner D, Winthrop KL. 2020. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Clin Infect Dis 71:905–913. doi: 10.1093/cid/ciaa1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffith DE. 2007. Therapy of nontuberculous mycobacterial disease. Curr Opin Infect Dis 20:198–203. doi: 10.1097/QCO.0b013e328055d9a2. [DOI] [PubMed] [Google Scholar]
- 13.Parker H, Lorenc R, Ruelas Castillo J, Karakousis PC. 2020. Mechanisms of antibiotic tolerance in Mycobacterium avium complex: lessons from related mycobacteria. Front Microbiol 11:573983. doi: 10.3389/fmicb.2020.573983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomasz A, Albino A, Zanati E. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 15.Liao J, Sauer K. 2012. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol 194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. 2010. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1->3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- 17.Thayil SM, Morrison N, Schechter N, Rubin H, Karakousis PC. 2011. The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One 6:e28076. doi: 10.1371/journal.pone.0028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutte CC, Crosson S. 2013. Bacterial lifestyle shapes stringent response activation. Trends Microbiol 21:174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang Y-M, Belchis DA, Karakousis PC. 2013. The polyphosphate kinase gene ppk2 is required for Mycobacterium tuberculosis inorganic polyphosphate regulation and virulence. mBio 4:e00039-13. doi: 10.1128/mBio.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang Y-M, Bandyopadhyay N, Rifat D, Rubin H, Bader JS, Karakousis PC. 2015. Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis. mBio 6:e02428. doi: 10.1128/mBio.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues L, Sampaio D, Couto I, Machado D, Kern WV, Amaral L, Viveiros M. 2009. The role of efflux pumps in macrolide resistance in Mycobacterium avium complex. Int J Antimicrob Agents 34:529–533. doi: 10.1016/j.ijantimicag.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues L, Machado D, Couto I, Amaral L, Viveiros M. 2012. Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect Genet Evol 12:695–700. doi: 10.1016/j.meegid.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Gupta AK, Katoch VM, Chauhan DS, Sharma R, Singh M, Venkatesan K, Sharma VD. 2010. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microb Drug Resist 16:21–28. doi: 10.1089/mdr.2009.0054. [DOI] [PubMed] [Google Scholar]
- 24.Rojony R, Martin M, Campeau A, Wozniak JM, Gonzalez DJ, Jaiswal P, Danelishvili L, Bermudez LE. 2019. Quantitative analysis of Mycobacterium avium subsp. hominissuis proteome in response to antibiotics and during exposure to different environmental conditions. Clin Proteomics 16:39. doi: 10.1186/s12014-019-9260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archuleta RJ, Yvonne Hoppes P, Primm TP. 2005. Mycobacterium avium enters a state of metabolic dormancy in response to starvation. Tuberculosis (Edinb) 85:147–158. doi: 10.1016/j.tube.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Sangari FJ, Parker A, Bermudez LE. 1999. Mycobacterium avium interaction with macrophages and intestinal epithelial cells. Front Biosci 4:582–588. doi: 10.2741/sangari. [DOI] [PubMed] [Google Scholar]
- 27.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 28.Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wayne LG, Lin KY. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun 37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langridge GC, Phan M-D, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA 106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matern WM, Jenquin RL, Bader JS, Karakousis PC. 2020. Identifying the essential genes of Mycobacterium avium subsp. hominissuis with Tn-Seq using a rank-based filter procedure. Sci Rep 10:1095. doi: 10.1038/s41598-020-57845-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, DeJesus MA, Rücker N, Engelhart CA, Wright MG, Healy C, Lin K, Wang R, Park SW, Ioerger TR, Schnappinger D, Ehrt S. 2017. Chemical genomic interaction profiling reveals determinants of intrinsic antibiotic resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e01334-17. doi: 10.1128/AAC.01334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braunstein M, Brown AM, Kurtz S, Jacobs WR. 2001. Two nonredundant SecA homologues function in mycobacteria. J Bacteriol 183:6979–6990. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ligon LS, Rigel NW, Romanchuk A, Jones CD, Braunstein M. 2013. Suppressor analysis reveals a role for secY in the secA2-dependent protein export pathway of mycobacteria. J Bacteriol 195:4456–4465. doi: 10.1128/JB.00630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan JT, Young EF, Mccann JR, Braunstein M. 2012. The Mycobacterium tuberculosis SecA2 system subverts phagosome maturation to promote growth in macrophages. Infect Immun 80:996–1006. doi: 10.1128/IAI.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zulauf KE, Sullivan JT, Braunstein M. 2018. The SecA2 pathway of Mycobacterium tuberculosis exports effectors that work in concert to arrest phagosome and autophagosome maturation. PLoS Pathog 14:e1007011. doi: 10.1371/journal.ppat.1007011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karakousis PC. 2009. Mechanisms of action and resistance of antimycobacterial agents, p 271–291. In Mayers DL (ed), Antimicrobial drug resistance. Humana Press, Totowa, NJ. [Google Scholar]
- 42.Dorman CJ. 2019. DNA supercoiling and transcription in bacteria: a two-way street. BMC Mol Cell Biol 20:26. doi: 10.1186/s12860-019-0211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pham TDM, Ziora ZM, Blaskovich MAT. 2019. Quinolone antibiotics. Medchemcomm 10:1719–1739. doi: 10.1039/c9md00120d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta R, Barkan D, Redelman-Sidi G, Shuman S, Glickman MS. 2011. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol Microbiol 79:316–330. doi: 10.1111/j.1365-2958.2010.07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choudhary E, Sharma R, Kumar Y, Agarwal N. 2019. Conditional silencing by CRISPRi reveals the role of DNA gyrase in formation of drug-tolerant persister population in Mycobacterium tuberculosis. Front Cell Infect Microbiol 9:70. doi: 10.3389/fcimb.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malik M, Chavda K, Zhao X, Shah N, Hussain S, Kurepina N, Kreiswirth BN, Kerns RJ, Drlica K. 2012. Induction of mycobacterial resistance to quinolone class antimicrobials. Antimicrob Agents Chemother 56:3879–3887. doi: 10.1128/AAC.00474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wipperman MF, Heaton BE, Nautiyal A, Adefisayo O, Evans H, Gupta R, van Ditmarsch D, Soni R, Hendrickson R, Johnson J, Krogan N, Glickman MS. 2018. Mycobacterial mutagenesis and drug resistance are controlled by phosphorylation- and cardiolipin-mediated inhibition of the RecA coprotease. Mol Cell 72:152–161.e7. doi: 10.1016/j.molcel.2018.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pisu D, Provvedi R, Espinosa DM, Payan JB, Boldrin F, Palù G, Hernandez-Pando R, Manganelli R. 2017. The alternative sigma factors SigE and SigB are involved in tolerance and persistence to antitubercular drugs. Antimicrob Agents Chemother 61:e01596-17. doi: 10.1128/AAC.01596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. 2009. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol 191:625–631. doi: 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dutta NK, Klinkenberg LG, Vazquez M-J, Segura-Carro D, Colmenarejo G, Ramon F, Rodriguez-Miquel B, Mata-Cantero L, Porras-De Francisco E, Chuang Y-M, Rubin H, Lee JJ, Eoh H, Bader JS, Perez-Herran E, Mendoza-Losana A, Karakousis PC. 2019. Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci Adv 5:eaav2104. doi: 10.1126/sciadv.aav2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehrt S, Schnappinger D, Rhee KY. 2018. Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat Rev Microbiol 16:496–507. doi: 10.1038/s41579-018-0013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. 2017. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol 24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zalis EA, Nuxoll AS, Manuse S, Clair G, Radlinski LC, Conlon BP, Adkins J, Lewis K. 2019. Stochastic variation in expression of the tricarboxylic acid cycle produces persister cells. mBio 10:e01930-19. doi: 10.1128/mBio.01930-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowe SE, Wagner NJ, Li L, Beam JE, Wilkinson AD, Radlinski LC, Zhang Q, Miao EA, Conlon BP. 2020. Reactive oxygen species induce antibiotic tolerance during systemic Staphylococcus aureus infection. Nat Microbiol 5:282–290. doi: 10.1038/s41564-019-0627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas VC, Chittezham Thomas V, Kinkead LC, Janssen A, Schaeffer CR, Woods KM, Lindgren JK, Peaster JM, Chaudhari SS, Sadykov M, Jones J, AbdelGhani SMM, Zimmerman MC, Bayles KW, Somerville GA, Fey PD. 2013. A dysfunctional tricarboxylic acid cycle enhances fitness of Staphylococcus epidermidis during β-lactam stress. mBio 4:e00437-13. doi: 10.1128/mBio.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh KS, Sharma R, Keshari D, Singh N, Singh SK. 2017. Down-regulation of malate synthase in Mycobacterium tuberculosis H37Ra leads to reduced stress tolerance, persistence and survival in macrophages. Tuberculosis (Edinb) 106:73–81. doi: 10.1016/j.tube.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 59.Masaki S, Sugimori G, Okamoto A, Imose J, Hayashi Y. 1990. Effect of Tween 80 on the growth of Mycobacterium avium complex. Microbiol Immunol 34:653–663. doi: 10.1111/j.1348-0421.1990.tb01041.x. [DOI] [PubMed] [Google Scholar]
- 60.Andréjak C, Almeida DV, Tyagi S, Converse PJ, Ammerman NC, Grosset JH. 2015. Characterization of mouse models of Mycobacterium avium complex infection and evaluation of drug combinations. Antimicrob Agents Chemother 59:2129–2135. doi: 10.1128/AAC.04841-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verma D, Stapleton M, Gadwa J, Vongtongsalee K, Schenkel AR, Chan ED, Ordway D. 2019. Mycobacterium avium infection in a C3HeB/FeJ mouse model. Front Microbiol 10:693. doi: 10.3389/fmicb.2019.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gelman E, McKinney JD, Dhar N. 2012. Malachite green interferes with postantibiotic recovery of mycobacteria. Antimicrob Agents Chemother 56:3610–3614. doi: 10.1128/AAC.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matern WM, Bader JS, Karakousis PC. 2018. Genome analysis of Mycobacterium avium subspecies hominissuis strain 109. Sci Data 5:180277. doi: 10.1038/sdata.2018.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fish DN, Gotfried MH, Danziger LH, Rodvold KA. 1994. Penetration of clarithromycin into lung tissues from patients undergoing lung resection. Antimicrob Agents Chemother 38:876–878. doi: 10.1128/AAC.38.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blaschke TF, Skinner MH. 1996. The clinical pharmacokinetics of rifabutin. Clin Infect Dis 22:S15–S22. doi: 10.1093/clinids/22.Supplement_1.S15. [DOI] [PubMed] [Google Scholar]
- 66.Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O'Brien P, Chen C, Kaya F, Weiner DM, Chen P-Y, Song T, Lee M, Shim TS, Cho JS, Kim W, Cho SN, Olivier KN, Barry CE, Dartois V. 2015. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med 21:1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liss RH, Letourneau RJ, Schepis JP. 1981. Distribution of ethambutol in primate tissues and cells. Am Rev Respir Dis 123:529–532. doi: 10.1164/arrd.1981.123.5.529. [DOI] [PubMed] [Google Scholar]
- 68.van Ingen J, Egelund EF, Levin A, Totten SE, Boeree MJ, Mouton JW, Aarnoutse RE, Heifets LB, Peloquin CA, Daley CL. 2012. The pharmacokinetics and pharmacodynamics of pulmonary Mycobacterium avium complex disease treatment. Am J Respir Crit Care Med 186:559–565. doi: 10.1164/rccm.201204-0682OC. [DOI] [PubMed] [Google Scholar]
- 69.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen SF, Goodman J. 1996. An empirical study of smoothing techniques for language modeling, p 310–318. In Proceedings of the 34th annual meeting on Association for Computational Linguistics. Association for Computational Linguistics, Santa Cruz, CA. [Google Scholar]
- 71.Jonckheere AR. 1954. A distribution-free k-sample test against ordered alternatives. Biometrika 41:133–145. doi: 10.2307/2333011. [DOI] [Google Scholar]
- 72.Terpstra TJ. 1952. The asymptotic normality and consistency of Kendall’s test against trend, when ties are present in one ranking. Indag Math 55:327–333. doi: 10.1016/S1385-7258(52)50043-X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00246-21_Supp_1_seq4.csv, CSV file, 0.02 MB (16.8KB, csv)
Supplemental material. Download SPECTRUM00246-21_Supp_2_seq5.csv, CSV file, 0.1 MB (3.3KB, csv)
Supplemental material. Download SPECTRUM00246-21_Supp_3_seq6.csv, CSV file, 0.5 MB (479.3KB, csv)
Supplemental material. Download SPECTRUM00246-21_Supp_4_seq7.csv, CSV file, 0.5 MB (483.9KB, csv)
Supplemental material. Download SPECTRUM00246-21_Supp_5_seq8.csv, CSV file, 0.5 MB (481.8KB, csv)
Supplemental material. Download SPECTRUM00246-21_Supp_6_seq9.csv, CSV file, 0.5 MB (482KB, csv)
Supplemental material. Download SPECTRUM00246-21_Supp_7_seq13.pdf, PDF file, 0.5 MB (490.7KB, pdf)
Data Availability Statement
The raw sequencing data (*.fastq) from this project can be found in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA559896. A Jupyter notebook and associated scripts to reproduce the data analysis (including figures) from the raw data are provided online at github (https://doi.org/10.5281/zenodo.4542412).