ABSTRACT
There is a critical need for improved pharmacodynamic markers for use in human tuberculosis (TB) drug trials. Pharmacodynamic monitoring in TB has conventionally used culture or molecular methods to enumerate the burden of Mycobacterium tuberculosis organisms in sputum. A recently proposed assay called the rRNA synthesis (RS) ratio measures a fundamentally novel property, how drugs impact ongoing bacterial rRNA synthesis. Here, we evaluated RS ratio as a potential pharmacodynamic monitoring tool by testing pretreatment sputa from 38 Ugandan adults with drug-susceptible pulmonary TB. We quantified the RS ratio in paired pretreatment sputa and evaluated the relationship between the RS ratio and microbiologic and molecular markers of M. tuberculosis burden. We found that the RS ratio was highly repeatable and reproducible in sputum samples. The RS ratio was independent of M. tuberculosis burden, confirming that it measures a distinct new property. In contrast, markers of M. tuberculosis burden were strongly associated with each other. These results indicate that the RS ratio is repeatable and reproducible and provides a distinct type of information from markers of M. tuberculosis burden.
IMPORTANCE This study takes a major next step toward practical application of a novel pharmacodynamic marker that we believe will have transformative implications for tuberculosis. This article follows our recent report in Nature Communications that an assay called the rRNA synthesis (RS) ratio indicates the treatment-shortening of drugs and regimens. Distinct from traditional measures of bacterial burden, the RS ratio measures a fundamentally novel property, how drugs impact ongoing bacterial rRNA synthesis.
KEYWORDS: human, Mycobacterium tuberculosis, sputum, assay development, pharmacodynamics
INTRODUCTION
To achieve the World Health Organization End TB strategic goals, it will be necessary to develop new, shorter treatment regimens for both drug-susceptible and drug-resistant tuberculosis (TB) (1, 2). One key challenge for evaluation of new TB treatment regimens is the limitation of existing pharmacodynamic (PD) markers (3–5). There is an urgent need for new PD markers that maximize information gained from preclinical animal models and early-phase human clinical trials. More accurate PD markers would enable selection of the most efficacious regimens for testing in definitive phase III trials (6–8).
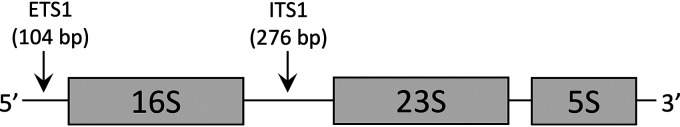
We recently proposed a new marker of TB treatment efficacy called the rRNA synthesis (RS) ratio (9). In Mycobacterium tuberculosis, the three rRNA sequences are transcribed on a single polycistronic precursor-rRNA (pre-rRNA) sequence with intervening short spacer sequences called internally transcribed spacer 1 (ITS1) and externally transcribed spacer 1 (ETS1) (Fig. 1). Since the spacer sequences are rapidly degraded, they serve as a marker of newly transcribed rRNA (10). The RS ratio estimates ongoing rRNA synthesis in M. tuberculosis populations by quantifying the relative abundance of ETS1 sequence relative to 23S rRNA sequence.
FIG 1.
Schematic of ribosomal operon in M. tuberculosis, illustrating rapidly degraded precursor spacer sequences (ETS1 and ITS1). The RS ratio is based on the abundance of ETS1 sequence relative to 23S rRNA sequence.
The RS ratio is unlike most conventional and investigational PD markers that enumerate the burden of M. tuberculosis, such as sputum smear grade, time to positivity (TTP) in liquid culture, GeneXpert MTB/RIF (Xpert) threshold cycle (CT) values, and M. tuberculosis rRNA burden. In contrast, the RS ratio measures the effect of drugs on the physiologic state of the pathogen. In principle, the RS ratio provides a different type of information than these existing measures of M. tuberculosis burden. Key insights from in vitro and murine studies are (i) drugs often affect the RS ratio and CFU burden differently and (ii) the RS ratio appears to indicate the sterilizing activity of drugs and regimens (9). This novel molecular approach has yet to be extensively investigated as a marker in humans.
As a preliminary evaluation of the performance of the RS ratio in human sputum, we evaluated pretreatment sputa from 38 Ugandan adults with drug-susceptible pulmonary TB. We tested the repeatability and reproducibility of the RS ratio in paired pretreatment sputa and evaluated the association of the RS ratio with conventional and investigational PD markers that enumerate M. tuberculosis burden. Our results suggest that the RS ratio is repeatable and reproducible and provide a distinct new type of information.
RESULTS
Study population characteristics.
Evaluation of 102 adults confirmed drug-susceptible pulmonary TB among 52 participants. After excluding 14 who declined to participate or were unable to produce additional sputa, 38 participants were included in this study (see Fig. S1 in the supplemental material). Table 1 provides participant characteristics.
TABLE 1.
Participant characteristicsa
| Variable | Value |
|---|---|
| Age in yrs, median (IQR) | 34 (26–38) |
| Wt in kg, median (IQR) | 53 (50–59) |
| Female (%) | 32 |
| HIV-uninfected (%) | 66 |
| Nonsmoking (%) | 82 |
| Smear grade (%) | |
| Negative | 3 |
| Positive 1+ | 32 |
| Positive 2+ | 32 |
| Positive 3+ | 26 |
| Other | 8 |
| TTP in days, median (IQR) | 6 (4–9) |
| Xpert CT cycles, median (IQR) | 18.2 (16.3–20.0) |
| 16S burden, median (IQR)b | |
| SS1 | 6.0 (4.9–7.6) |
| SS2 | 5.8 (4.8–7.3) |
| 23S burden, median (IQR)b | |
| SS1 | 5.4 (4.1–7.3) |
| SS2 | 5.2 (3.9–6.7) |
| RS ratio, median (IQR)b | |
| SS1 | 3.3 (3.0–3.5) |
| SS2 | 3.3 (3.2–3.6) |
IQR, interquartile range; CT, cycle threshold; TTP, time to culture positivity.
log10-transformed.
Repeatability and reproducibility of RS ratio in sputum.
In the paired pretreatment sputa, the RS ratio was quantifiable in 97% of first sputum samples (SS1) and 92% of second sputum samples (SS2). When SS1 samples were assayed in triplicate in a single experiment for assessment of repeatability, the intraclass correlation coefficient (ICC) was 0.99. When the RS ratio was conducted by two different lab workers using different instruments at a 12-month interval, interobserver reproducibility was high (ICC, 0.93). Repeatability and reproducibility results are summarized in Table 2.
TABLE 2.
Repeatability and reproducibility of the RS ratio in sputum
| Measure | Sample | Basis for assessment | Intraclass correlation coefficient |
|---|---|---|---|
| Repeatability in technical replicatesa | SS1 | Same sputum sample Same operator Same expt |
0.99 (0.99–1.00) |
| Interobserver reproducibilityb | SS1 | Same sputum sample Different operators Different instruments Different experiments |
0.93 (0.86–0.97) |
| Biological reproducibilityc | SS1 vs SS2 | Paired sputum samples Same operator Same instrument Same expt |
0.63 (0.37–0.79) |
Agreement among three replicate RS ratio results within a single experiment.
Agreement between RS ratio results in the same samples conducted by two different lab workers using different instruments at a 12-month interval.
Agreement between RS ratio results in two separate sputum samples collected within an hour of each other.
When the RS ratio was compared between two paired sputum samples from the same participant, the ICC was 0.63, indicating good biological reproducibility. The RS ratio estimates did not differ systematically between the first and second sputum sample (mean difference between RS ratios in SS1 and SS2, –0.11 [95% confidence interval (CI), –0.26 to 0.03]). The variability of the RS ratio between paired sputum samples (ICC, 0.63) was comparable to the variation observed in paired sputum samples in M. tuberculosis 16S rRNA (ICC, 0.64) and 23S rRNA (ICC, 0.64).
Association of RS ratio with markers of M. tuberculosis burden.
There was no significant relationship between RS ratios quantified in SS1 and M. tuberculosis rRNA burden, smear grade, TTP, or Xpert CT value (Table 3), reinforcing our understanding that the RS ratio provides a different type of information than markers of M. tuberculosis burden. Similar results were observed using RS ratios quantified in SS2 (Table 3). Conversely, with few exceptions, M. tuberculosis rRNA burden, smear grade, TTP, and Xpert CT values were significantly associated, despite the fact that some measurements were made on different sputum samples.
TABLE 3.
| Markers | RS ratiob | 16S rRNA burdenb | 23S rRNA burdenb | Smear grade | TTP in liquid culture | Xpert CT values |
|---|---|---|---|---|---|---|
| SS1 | ||||||
| RS ratiob |
ρ = –0.19 (P = 0.28) |
ρ = –0.20 (P = 0.24) |
ρ = –0.15 (P = 0.40) |
ρ = 0.22 (P = 0.21) |
ρ = 0.43 (P = 0.08) |
|
| 16S rRNA burdenb |
ρ = 0.98 (P < 0.01) |
ρ = 0.49 (P < 0.01) |
ρ = –0.64 (P < 0.01) |
ρ = –0.65 (P < 0.01) |
||
| 23S rRNA burdenb |
ρ = 0.45 (P < 0.01) |
ρ = –0.57 (P < 0.01) |
ρ = –0.59 (P = 0.01) |
|||
| Smear grade |
ρ = –0.72 (P < 0.01) |
ρ = –0.07 (P = 0.80) |
||||
| TTP in liquid culture | ρ = 0.62 | |||||
| (P < 0.01) | ||||||
| Markers | RS ratio c | 16S rRNA burden c | 23S rRNA burden c | Smear grade | TTP in liquid culture | Xpert CT values |
| SS2 | ||||||
| RS ratioc |
ρ = –0.05 (P = 0.75) |
ρ = –0.04 (P = 0.83) |
ρ = 0.12 (P = 0.50) |
ρ = –0.08 (P = 0.65) |
ρ = 0.10 (P = 0.68) |
|
| 16S rRNA burdenc |
ρ = 0.97 (P < 0.01) |
ρ = 0.48 (P < 0.01) |
ρ = −0.50 (P < 0.01) |
ρ = –0.37 (P = 0.13) |
||
| 23S rRNA burdenc |
ρ = 0.52 (P < 0.01) |
ρ = –0.50 (P < 0.01) |
ρ = –0.38 (P = 0.12) |
Spearman correlation coefficients (ρ) with P values (P) are provided. RS ratio and rRNA burden measurements were log10-transformed. CT, cycle threshold; TTP, time to culture positivity.
Using RS ratio, 16S and 23S rRNA from SS1.
Using RS ratio, 16S and 23S rRNA from SS2.
Sputum grade was converted to an ordinal scale with negative, positive 1+, positive 2+, and positive 3+ corresponding to 0, 1, 2, and 3, respectively.
Association of clinical and demographic factors with the RS ratio and markers of M. tuberculosis burden.
In general, there was no significant relationship between clinical and demographic factors, including sex, HIV status, and smoking, and the RS ratio or markers of M. tuberculosis burden. The only exception was for the relationship between Xpert CT values and sex, which had a P value of 0.03 (Table S1). After adjustment for multiple comparisons, this association was nonsignificant.
DISCUSSION
Using sputa from Ugandan adults with untreated TB, we evaluated the RS ratio in pretreatment sputa and compared the RS ratio with other conventional and investigational markers. We found that the RS ratio was highly repeatable in technical replicates and had high interobserver reproducibility. The sputum-to-sputum biological variability in the RS ratio approximated the variability observed in M. tuberculosis 16S and 23S rRNA burden. Comparison of the RS ratio with sputum smear, TTP, Xpert CT values and M. tuberculosis rRNA burden indicated that the RS ratio measures a distinct property that is independent of bacterial burden.
An important precursor to establishing a novel PD marker is understanding technical and biological variability of the assay (11–13). The sputum RS ratio demonstrated high technical consistency with high repeatability and interobserver reproducibility, consistent with the previously described performance of droplet digital PCR (ddPCR) in clinical samples (14, 15).
As anticipated, the biological variability exceeded the technical variability. The baseline sample is particularly important because clinical trials typically evaluate how changes in PD marker over time relative to baseline values relate to disease outcomes. Therefore, evaluating the repeatability and reproducibility of the RS ratio in paired baseline sputum samples helps us understand the source of variability. We suspect that the sources of biological variability differ between the RS ratio and markers that enumerate M. tuberculosis burden. Generally, production of greater sputum volume is associated with higher M. tuberculosis burden and correspondingly shorter TTP, lower Xpert values, and higher M. tuberculosis rRNA burden (16–18). The volume of sputum varies based on time of collection, participant effort to expectorate, and severity of lung disease (16). In contrast, the RS ratio is designed to be “self-normalizing” to bacterial burden because both the pre-rRNA numerator and the 23S rRNA denominator scale with change in M. tuberculosis burden. For the RS ratio, it is likely that sputum-to-sputum variation indicates biological variability in the M. tuberculosis populations present in samples originating from different regions of the lung (19, 20).
Consistent with our hypothesis that the RS ratio is not a marker of M. tuberculosis burden, we did not observe an association between this novel marker and conventional or investigational markers that enumerate M. tuberculosis burden. This is similar to findings from our in vitro and murine studies (9). Our findings suggest that the RS ratio is a feasible tool for evaluating human sputa that may complement markers of M. tuberculosis burden, providing novel insight into treatment response.
This study has several limitations. First, although our goal is a PD marker that can be used to monitor treatment effectiveness, here, we studied only pretreatment samples. Nevertheless, evaluation of technical and biological variability in pretreatment samples is an important preliminary step required for interpretation of longitudinal data. Second, by necessity, we used different sputum samples to evaluate different markers. For example, smear status and TTP were quantified in one sample, Xpert was quantified in another, and RS ratio and M. tuberculosis rRNA burden were quantified in separate paired samples. Nonetheless, all of the markers that enumerate M. tuberculosis burden were strongly associated.
In summary, this study determined that assaying the RS ratio in sputum is both repeatable and reproducible. The RS ratio may serve as a novel PD marker that offers a new physiologic perspective on TB treatment, distinct from existing assays of M. tuberculosis burden.
MATERIALS AND METHODS
Participant recruitment and specimen collection.
Participants were enrolled at Naguru Referral Hospital in Kampala, Uganda, from August 2018 to March 2019 as a component of a longitudinal observational cohort study of adults with pneumonia called the International HIV-Associated Opportunistic Pneumonias-Inflammation, Ageing, Microbes, and Obstructive Lung Disease study (21, 22). Participants were ≥18 years of age, with persistent cough, without signs of extrapulmonary TB, and without TB treatment within the past 2 years.
Each participant provided 4 spot sputum samples. The first and second sputum samples were processed for smear microscopy, liquid and solid cultures, and Xpert, as described in the supplemental material. Auramine O fluorescent smear microscopy used the direct method. Drug-susceptible pulmonary TB was confirmed by sputum smear microscopy, culture, and Xpert (23). Smear microscopy used a small amount of primary sputum. Culture and Xpert each used 1 ml of processed sputum. Two additional sputum samples were collected within a 1-h time interval for RNA-based assays in a guanidine thiocyanate (GTC)-based RNA preservative as described in the supplemental material. Both the parent and current study were approved by the institutional review boards in Uganda and the United States. All participants provided written informed consent for the use of their sputa and clinical data for a biomarker study.
RNA extraction and quantification of rRNA burden and RS ratio.
Total RNA was extracted from paired sputum samples using standard methods described in the supplemental material. Following reverse transcription, 16S and 23S rRNA transcripts were quantified in triplicate via reverse transcription quantitative PCR (RT-qPCR). Absolute copies were determined by reference to a standard DNA ladder. Employing methods similar to those used for the molecular bacterial load assay (24, 25), we used a spike-in to adjust for loss in RNA extraction (“retention percentage”) and normalized by sputum weight to estimate the burden of 16S or 23S rRNA in sputum (supplemental material). For the RS ratio assay, droplet digital PCR (ddPCR) was used to quantify the abundance of pre-rRNA relative to 23S rRNA, as previously described (9).
Evaluation of repeatability and reproducibility.
To understand the sources of variability in the RS ratio, we defined repeatability and reproducibility in three ways. Repeatability was the agreement among three replicate RS ratio results within a single experiment (i.e., technical replicates). Second, interobserver reproducibility was the agreement between RS ratio results in the same samples conducted by two different lab workers using different instruments at a 12-month interval. Finally, we defined a sputum-to-sputum biological reproducibility as the agreement between RS ratio results in two separate sputa (SS1 and SS2) collected within an hour of each other. We additionally evaluated variability in measurement of M. tuberculosis rRNA burden by qPCR, quantifying repeatability in technical replicates and sputum-to-sputum biological reproducibility. Repeatability and reproducibility were estimated based on the intraclass correlation coefficient (ICC), ranging from 0, indicating no agreement, to 1, indicating perfect agreement (26). We implemented the one-way random effects ICC framework (27), which assumes that each participant is measured by a different set of assays, using the function icc in the irr R package.
Comparison of RS ratio with existing and investigational markers of M. tuberculosis burden.
We tested the association of the RS ratio with sputum smear grade, TTP, Xpert, and rRNA burden using Spearman correlation tests. For the burden of 16S and 23S rRNA, we selected the median of triplicates for statistical analysis. P values of <0.05 were considered statistically significant. Relationships of the RS ratio and markers of M. tuberculosis burden with clinical and demographic factors were tested using two-sample Wilcoxon tests. Statistical analysis was conducted in R v 3.5.3 (R Development Core Team, Vienna, Austria).
Data availability.
All primary data are included in the supplemental material.
ACKNOWLEDGMENTS
We acknowledge the participants in this study; the staff and administration of Naguru Referral Hospital, Kampala-Uganda; the clinical, research, and administrative staff of the Infectious Diseases Research Collaboration, Catherine Nabakiibi and Yusuf Magezi; and the administrators of the Pulmonary Complications of AIDS Research Training (PART) program.
The MIND-IHOP study was funded by the IHOP grant (NIH R01 HL090335), Lung MicroCHIP grant (NIH U01 HL098964), and K24 grant (NIH K24 HL087713). These sources provided the funding to support participant enrollment and specimen collection. Emmanuel Musisi was supported by a scholarship from the Pulmonary Complications of AIDS Research Training Program (NIH D43 TW009607). N.D.W., R.M.S., J.L.D., and P.N. acknowledge funding from the U.S. National Institutes of Health (1R01AI127300-01A1). N.D.W. and M.I.V. acknowledge funding from the U.S. National Institutes of Health (1R21AI135652-01). N.D.W. acknowledges funding from Veterans Affairs (1IK2CX000914-01A1 and 1I01BX004527-01A1) and from the Doris Duke Charitable Foundation Clinical Scientist Development Award.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Emmanuel Musisi, Email: em303@st-andrews.ac.uk.
Christian Dide-Agossou, Email: christian.dide-agossou@cuanschutz.edu.
William Lainhart, University of Arizona/Banner Health.
REFERENCES
- 1.Uplekar M, Weil D, Lonnroth K, Jaramillo E, Lienhardt C, Dias HM, Falzon D, Floyd K, Gargioni G, Getahun H, Gilpin C, Glaziou P, Grzemska M, Mirzayev F, Nakatani H, Raviglione M. 2015. WHO’s new End TB Strategy. Lancet 385:1799–1801. doi: 10.1016/S0140-6736(15)60570-0. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JL, Hadad DJ, Dietze R, Noia Maciel EL, Sewali B, Gitta P, Okwera A, Mugerwa RD, Alcaneses MR, Quelapio MI, Tupasi TE, Horter L, Debanne SM, Eisenach KD, Boom WH. 2009. Shortening treatment in adults with noncavitary tuberculosis and 2-month culture conversion. Am J Respir Crit Care Med 180:558–563. doi: 10.1164/rccm.200904-0536OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horne DJ, Royce SE, Gooze L, Narita M, Hopewell PC, Nahid P, Steingart KR. 2010. Sputum monitoring during tuberculosis treatment for predicting outcome: systematic review and meta-analysis. Lancet Infect Dis 10:387–394. doi: 10.1016/S1473-3099(10)70071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips PPJ, Mendel CM, Burger DA, Crook AM, Nunn AJ, Dawson R, Diacon AH, Gillespie SH. 2016. Limited role of culture conversion for decision-making in individual patient care and for advancing novel regimens to confirmatory clinical trials. BMC Med 14:36. doi: 10.1186/s12916-016-0585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips PPJ, Fielding K, Nunn AJ. 2013. An evaluation of culture results during treatment for tuberculosis as surrogate endpoints for treatment failure and relapse. PLoS One 8:e63840. doi: 10.1371/journal.pone.0063840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartelink I, Zhang N, Keizer R, Strydom N, Converse P, Dooley K, Nuermberger E, Savic R. 2017. New paradigm for translational modeling to predict long-term tuberculosis treatment response. Clin Transl Sci 10:366–379. doi: 10.1111/cts.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahid P, Saukkonen J, Mac Kenzie WR, Johnson JL, Phillips PPJ, Andersen J, Bliven-Sizemore E, Belisle JT, Boom WH, Luetkemeyer A, Campbell TB, Eisenach KD, Hafner R, Lennox JL, Makhene M, Swindells S, Villarino ME, Weiner M, Benson C, Burman W, Centers for Disease Control and Prevention. 2011. Tuberculosis biomarker and surrogate endpoint research roadmap. Am J Respir Crit Care Med 184:972–979. doi: 10.1164/rccm.201105-0827WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dooley KE, Phillips PPJ, Nahid P, Hoelscher M. 2016. Challenges in the clinical assessment of novel tuberculosis drugs. Adv Drug Deliv Rev 102:116–122. doi: 10.1016/j.addr.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter ND, Born SEM, Robertson GT, Reichlen M, Dide-Agossou C, Ektnitphong VA, Rossmassler K, Ramey ME, Bauman AA, Ozols V, Bearrows SC, Schoolnik G, Dolganov G, Garcia B, Musisi E, Worodria W, Huang L, Davis JL, Nguyen NV, Nguyen HV, Nguyen ATV, Phan H, Wilusz C, Podell BK, Sanoussi ND, de Jong BC, Merle CS, Affolabi D, McIlleron H, Garcia-Cremades M, Maidji E, Eshun-Wilson F, Aguilar-Rodriguez B, Karthikeyan D, Mdluli K, Bansbach C, Lenaerts AJ, Savic RM, Nahid P, Vásquez JJ, Voskuil MI. 2021. Mycobacterium tuberculosis precursor rRNA as a measure of treatment-shortening activity of drugs and regimens. Nat Commun 12:2899. doi: 10.1038/s41467-021-22833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cangelosi GA, Meschke JS. 2014. Dead or alive: molecular assessment of microbial viability. Appl Environ Microbiol 80:5884–5891. doi: 10.1128/AEM.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thyagarajan B, Howard AG, Durazo-Arvizu R, Eckfeldt JH, Gellman MD, Kim RS, Liu K, Mendez AJ, Penedo FJ, Talavera GA, Youngblood ME, Zhao L, Sotres-Alvarez D. 2016. Analytical and biological variability in biomarker measurement in the Hispanic Community Health Study/Study of Latinos. Clin Chim Acta 463:129–137. doi: 10.1016/j.cca.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pletcher MJ, Pignone M. 2011. Evaluating the clinical utility of a biomarker: a review of methods for estimating health impact. Circulation 123:1116–1124. doi: 10.1161/CIRCULATIONAHA.110.943860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis KD, Aghaeepour N, Ahn AH, Angst MS, Borsook D, Brenton A, Burczynski ME, Crean C, Edwards R, Gaudilliere B, Hergenroeder GW, Iadarola MJ, Iyengar S, Jiang Y, Kong J-T, Mackey S, Saab CY, Sang CN, Scholz J, Segerdahl M, Tracey I, Veasley C, Wang J, Wager TD, Wasan AD, Pelleymounter MA. 2020. Discovery and validation of biomarkers to aid the development of safe and effective pain therapeutics: challenges and opportunities. Nat Rev Neurol 16:381–400. doi: 10.1038/s41582-020-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morisset D, Štebih D, Milavec M, Gruden K, Žel J. 2013. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One 8:e62583. doi: 10.1371/journal.pone.0062583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shehata HR, Li J, Chen S, Redda H, Cheng S, Tabujara N, Li H, Warriner K, Hanner R. 2017. Droplet digital polymerase chain reaction (ddPCR) assays integrated with an internal control for quantification of bovine, porcine, chicken and turkey species in food and feed. PLoS One 12:e0182872. doi: 10.1371/journal.pone.0182872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karinja MN, Esterhuizen TM, Friedrich SO, Diacon AH. 2015. Sputum volume predicts sputum mycobacterial load during the first 2 weeks of antituberculosis treatment. J Clin Microbiol 53:1087–1091. doi: 10.1128/JCM.02379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat J, Rao V, Muniyandi M, Yadav R, Karforma C, Luke C. 2014. Impact of sputum quality and quantity on smear and culture positivity: findings from a tuberculosis prevalence study in central India. Trans R Soc Trop Med Hyg 108:55–56. doi: 10.1093/trstmh/trt100. [DOI] [PubMed] [Google Scholar]
- 18.Yoon SH, Lee NK, Yim JJ. 2012. Impact of sputum gross appearance and volume on smear positivity of pulmonary tuberculosis: a prospective cohort study. BMC Infect Dis 12:172. doi: 10.1186/1471-2334-12-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radtke T, Böni L, Bohnacker P, Fischer P, Benden C, Dressel H. 2018. The many ways sputum flows: dealing with high within-subject variability in cystic fibrosis sputum rheology. Respir Physiol Neurobiol 254:36–39. doi: 10.1016/j.resp.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Ozkutuk A, Terek G, Coban H, Esen N. 2007. Is it valuable to examine more than one sputum smear per patient for the diagnosis of pulmonary tuberculosis? Jpn J Infect Dis 60:73–75. [PubMed] [Google Scholar]
- 21.Davis JL, Worodria W, Kisembo H, Metcalfe JZ, Cattamanchi A, Kawooya M, Kyeyune R, den Boon S, Powell K, Okello R, Yoo S, Huang L. 2010. Clinical and radiographic factors do not accurately diagnose smear-negative tuberculosis in HIV-infected inpatients in Uganda: a cross-sectional study. PLoS One 5:e9859. doi: 10.1371/journal.pone.0009859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang RJ, Moore J, Moisi D, Chang EG, Byanyima P, Kaswabuli S, Musisi E, Sanyu I, Sessolo A, Lalitha R, Worodria W, Davis JL, Crothers K, Lin J, Lederman MM, Hunt PW, Huang L. 2019. HIV infection is associated with elevated biomarkers of immune activation in Ugandan adults with pneumonia. PLoS One 14:e0216680. doi: 10.1371/journal.pone.0216680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musisi E, Matovu DK, Bukenya A, Kaswabuli S, Zawedde J, Andama A, Byanyima P, Sanyu I, Sessolo A, Seremba E, Davis JL, Worodria W, Huang L, Walter ND, Mayanja-Kizza H. 2018. Effect of anti-retroviral therapy on oxidative stress in hospitalized HIV-infected adults with and without TB. Afr Health Sci 18:512–522. doi: 10.4314/ahs.v18i3.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honeyborne I, McHugh TD, Phillips PPJ, Bannoo S, Bateson A, Carroll N, Perrin FM, Ronacher K, Wright L, van Helden PD, Walzl G, Gillespie SH. 2011. Molecular bacterial load assay, a culture-free biomarker for rapid and accurate quantification of sputum Mycobacterium tuberculosis bacillary load during treatment. J Clin Microbiol 49:3905–3911. doi: 10.1128/JCM.00547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honeyborne I, Mtafya B, Phillips PPJ, Hoelscher M, Ntinginya EN, Kohlenberg A, Rachow A, Rojas-Ponce G, McHugh TD, Heinrich N, Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics (PanACEA). 2014. The molecular bacterial load assay replaces solid culture for measuring early bactericidal response to antituberculosis treatment. J Clin Microbiol 52:3064–3067. doi: 10.1128/JCM.01128-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo TK, Li MY. 2016. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller R, Büttner P. 1994. A critical discussion of intraclass correlation coefficients. Stat Med 13:2465–2476. doi: 10.1002/sim.4780132310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00481-21_Supp_1_seq1.pdf, PDF file, 0.3 MB (341.7KB, pdf)
Supplemental material. Download SPECTRUM00481-21_Supp_2_seq2.xlsx, XLSX file, 0.02 MB (22.8KB, xlsx) .
Data Availability Statement
All primary data are included in the supplemental material.