ABSTRACT
Commensal microbes in animal guts often help to exclude bacterial pathogens. In honey bees, perturbing or depleting the gut microbiota increases host mortality rates upon challenge with the opportunistic pathogen Serratia marcescens, suggesting antagonism between S. marcescens and one or more members of the bee gut microbiota. In laboratory culture, S. marcescens uses a type VI secretion system (T6SS) to kill bacterial competitors, but the role of this T6SS within hosts is unknown. Using infection assays, we determined how the microbiota impacts the abundance and persistence of S. marcescens in the gut and visualized colocalization of S. marcescens with specific community members in situ. Using T6SS-deficient S. marcescens strains, we measured T6SS-dependent killing of gut isolates in vitro and compared the persistence of mutant and wild-type strains in the gut. We found that S. marcescens is rapidly eliminated in the presence of the microbiota but persists in microbiota-free guts. Protection is reduced in monocolonized and antibiotic-treated bees, possibly because different symbionts occupy distinct niches. Serratia marcescens uses a T6SS to antagonize Escherichia coli and other S. marcescens strains but shows limited ability to kill bee symbionts. Furthermore, wild-type and T6SS-deficient S. marcescens strains achieved similar abundance and persistence in bee guts. Thus, an intact gut microbiota offers robust protection against this common pathogen, whose T6SSs do not confer the ability to compete with commensal species.
IMPORTANCE Bacteria living within guts of animals can provide protection against infection by pathogens. Some pathogens have been shown to use a molecular weapon known as a T6SS to kill beneficial bacteria during invasion of the mouse gut. In this study, we examined how bacteria native to the honey bee gut work together to exclude the opportunistic pathogen Serratia marcescens. Although S. marcescens has a T6SS that can kill bacteria, bee gut bacteria seem resistant to its effects. This limitation may partially explain why ingestion of S. marcescens is rarely lethal to insects with healthy gut communities.
KEYWORDS: Apis mellifera, Serratia marcescens, T6SS, colonization resistance, microbiota
INTRODUCTION
Serratia marcescens is an environmental bacterium that frequently acts as an opportunistic pathogen of insects (1, 2) and as a nosocomial pathogen of humans (3). In insects, S. marcescens is typically lethal only when injected into the hemolymph, but in some hosts (4–6) it can infect orally, escape from the gut into the body cavity, and ultimately kill the host. S. marcescens likely interacts with the native gut microbiota as it passes through the gut, suggesting that competition with commensal bacteria could modulate pathogenicity.
Several S. marcescens strains were recently identified as opportunistic pathogens of honey bees (6). These strains are capable of causing lethal infections when introduced orally, with greater mortality rates among bees whose gut microbiota has been perturbed by exposure to antibiotics or agrochemicals (7, 8). In honey bees, 10 core taxa comprise >95% of the gut microbiota (9–12). This community contributes to host health by promoting weight gain (13), assisting in breakdown of otherwise indigestible carbohydrates (14–16), and conferring colonization resistance to certain pathogens (17). Symbiont biofilms line regions of the gut (18) and may act as a barrier against pathogen invasion. Also, several symbionts contain type VI secretion systems (T6SSs) (19), which may be used to kill potential competitors, including pathogens. However, some pathogens may overcome these defenses by producing a chemical arsenal to kill gut bacteria and disrupt the microbiota. For example, T6SSs are increasingly recognized as important mediators of antagonism between Gram-negative bacteria (20). These protein complexes, which are used to deliver a variety of toxins through membranes of adjacent cells, are common in Gram-negative bacteria (20–22). T6SS-mediated killing of bacterial competitors has been demonstrated in vitro in a variety of species (23–27), including S. marcescens Db11, a close relative of S. marcescens strains isolated from honey bees (24, 28, 29). It is not known whether S. marcescens uses a T6SS to compete with commensal bacteria during infection of hosts. However, T6SSs have been shown to mediate interactions between commensal gut bacteria and other pathogens in mice (30–32) and fruit flies (33), suggesting that T6SSs may be a viable mechanism for opportunistic pathogens to overcome colonization resistance provided by native microbiotas.
In this study, we examined the effects of the microbiota on S. marcescens persistence after oral exposure. We observed rapid elimination of S. marcescens from bees previously colonized by gut symbionts and competition for space between S. marcescens and commensals in the ileum. We also tested the hypothesis that this common opportunistic pathogen, which has a well-studied antibacterial T6SS (24, 28, 29), would antagonize gut commensals during infection. When grown in culture, S. marcescens does use a T6SS to antagonize Escherichia coli and other S. marcescens strains, as well as the bee symbiont Gilliamella. However, inactivating the T6SS did not alter S. marcescens success in the gut, under any conditions tested. Overall, our results indicate that multiple symbiont species contribute to colonization resistance toward an opportunistic pathogen. Furthermore, the T6SSs of this pathogen do not enable it to overcome this community-conferred resistance and are instead likely to be deployed in some other ecological context.
RESULTS
The microbiota drives elimination of S. marcescens from the bee gut.
Bees with a perturbed gut microbiota have reduced survival rates after challenge with S. marcescens, compared to bees with a healthy microbiota (7, 8), but whether the microbiota contributes to host survival by killing S. marcescens is unknown. To determine whether S. marcescens abundance is reduced in bees with a healthy microbiota, we exposed bees with a conventional microbiota (CV), CV bees treated with tetracycline (Tet), or microbiota-free (MF) bees to S. marcescens kz11, a pathogenic isolate from bees (6, 8). Tet treatment was included to account for differences in metabolism, weight gain, and immune development between MF and CV bees (13, 34). Treatment with Tet reduces the abundance of gut bacteria and increases susceptibility to lethal infections but does not eliminate the commensal microbiota (8). One day after exposure, 100% of MF bees were infected, with an average of 1.85 × 105 S. marcescens CFU per gut, while 83.3% of CV bees contained no living S. marcescens cells (Fig. 1A). Bees in the Tet treatment group exhibited an intermediate and highly variable phenotype, with 4.10 × 106 S. marcescens CFU per gut on average and elimination of S. marcescens in 27.8% of bees. Loss of colonization resistance in Tet bees suggests that the gut microbiota may directly contribute to exclusion of S. marcescens from the gut, rather than acting through an indirect mechanism such as priming of the host immune system (6). Sucrose consumption is reduced in MF bees relative to CV bees (13), but differences in feeding behavior do not affect S. marcescens levels, as CV, Tet, MF, and MF Tet bees (MF bees treated with Tet) contained similar numbers of S. marcescens cells immediately after exposure (see Fig. S1A in the supplemental material), with differences between CV bees and other treatment groups developing over the next 24 h (see Fig. S1B and C). Additionally, protection from S. marcescens may vary without perturbation, due to variation in strains and relative abundances of gut taxa or due to host conditions. We observed a 1,000-fold difference in average S. marcescens CFU per gut between age-controlled bees from the same hive inoculated with conventional communities from two different hives (see Fig. S2).
FIG 1.
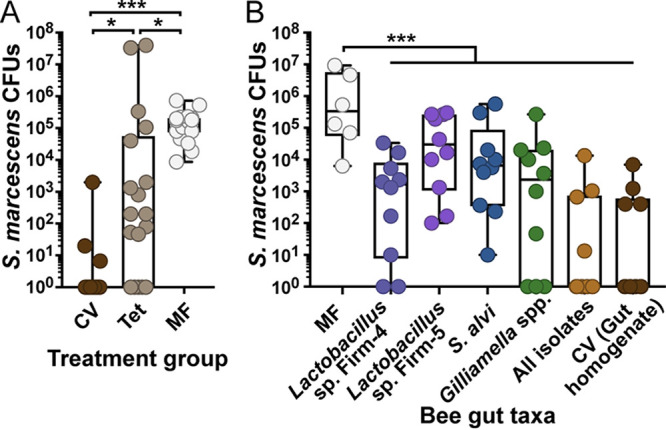
Serratia marcescens is eliminated from the guts of bees colonized by commensal species. (A) Total S. marcescens kz11 abundance in the midgut and hindgut of bees with a conventional, perturbed, or absent gut microbiota. Newly emerged bees from a single hive were inoculated with a conventional gut community (CV), inoculated with a conventional community and later treated with Tet (Tet), or kept microbiota free (MF). *, P < 0.05; ***, P < 0.005, Kruskal-Wallis test with Dunn’s multiple-comparison test. (B) S. marcescens abundance in bees colonized by individual gut taxa. MF bees were inoculated with representative strains of core gut taxa (Lactobacillus sp. Firm-4 DSM 26254 and DSM 26255, Lactobacillus sp. Firm-5 wkB8 and wkB10, S. alvi wkB2, G. apicola wkB1 and PEB0154, and G. apis PEB0162 and PEB0183), all isolates in combination, or homogenized gut of a bee collected from the hive. Bees were exposed for 1 day to 4 × 108 S. marcescens cells/ml in sugar syrup and dissected 1 day after the end of exposure. S. marcescens abundance was quantified by counting CFU. Box and whisker plots show the minimum, first quartile, median, third quartile, and maximum. ***, P < 0.005, one-way ANOVA with Tukey’s multiple-comparison test.
Multiple species contribute to elimination of S. marcescens.
Increased persistence of S. marcescens in Tet-treated bees suggests that microbiota composition is important for colonization resistance. To determine whether individual symbionts are capable of suppressing S. marcescens, we inoculated MF bees with representative isolates of the four most abundant core gut taxa, namely, Lactobacillus Firm-4 (DSM 26254 and DSM 26255), Lactobacillus Firm-5 (wkB8 and wkB10), Snodgrassella alvi (wkB2), or Gilliamella spp. (wkB1, PEB0154, PEB0162, and PEB0183). We also inoculated bees with all 9 isolates in combination or with homogenized hindgut from a hive bee (CV). After 5 days, the time required for microbiota establishment (18), bees were fed sucrose syrup containing S. marcescens at an optical density (OD600) of 0.5 for 1 day and then returned to a diet of sterile sucrose solution. Serratia marcescens abundance was quantified 1 day later. Serratia marcescens CFU were reduced in bees inoculated with any of the core gut taxa, relative to MF bees (P < 0.005, one-way analysis of variance [ANOVA] with Tukey’s multiple-comparison test) (Fig. 1B). MF bees were infected with an average 2.5 × 106 S. marcescens CFU per gut, in contrast to bees inoculated with Firm-4, Firm-5, S. alvi, or Gilliamella spp., which contained an average of 6.1 × 103, 1.1 × 105, 9.1 × 104, and 3.3 × 104 S. marcescens CFU per gut, respectively. The combined effect of these four taxa exceeded that of any taxon alone, with 1.5 × 103 S. marcescens CFU per gut and only 40% of bees still infected. These numbers resemble those in bees inoculated with gut homogenate (40% infected, with 9.0 × 102 CFU per gut). Therefore, antagonism of S. marcescens by the bee gut microbiota is likely to be a combined effect of multiple species.
Commensal bacteria may exclude S. marcescens from regions of the gut.
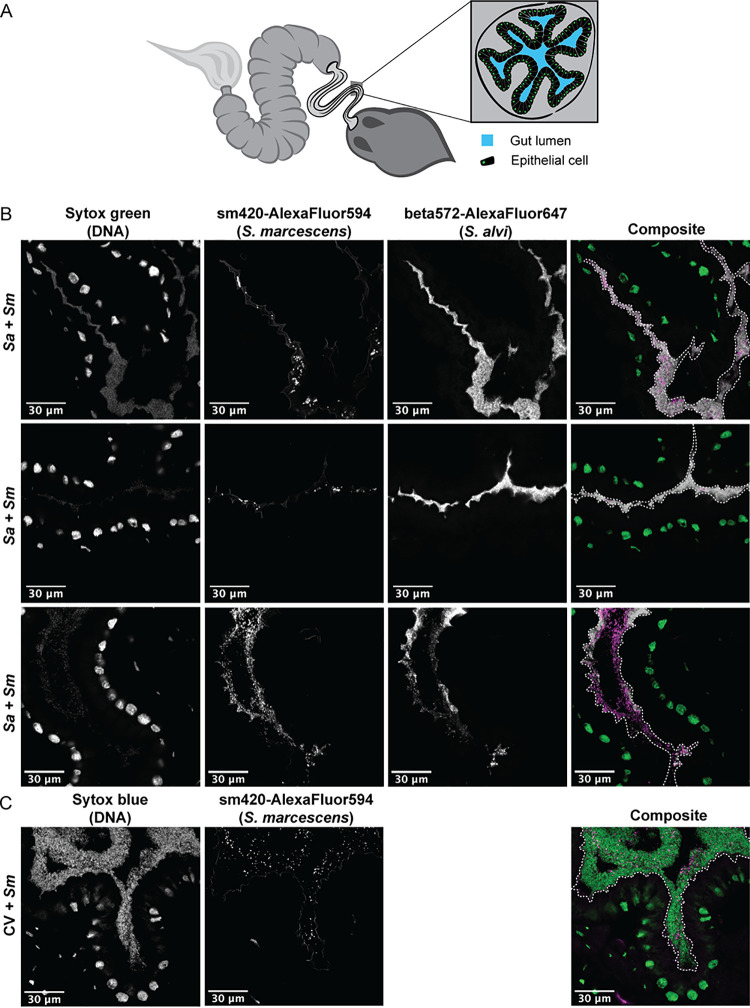
To determine whether S. marcescens physically interacts with gut symbionts, we used fluorescence in situ hybridization (FISH) microscopy to visualize the location of S. marcescens within the ilea of CV bees and bees monocolonized by S. alvi. The ileum is bordered by multiple folds of the epithelial layer, creating narrow grooves that are usually filled with gut symbionts (Fig. 2A). S. alvi colonizes the surface of the host epithelium and fills the lumen only in narrow regions of the ileum (18, 35). In MF bees, S. marcescens colonizes the same niche (see Fig. S3). When bees monocolonized by S. alvi are infected with S. marcescens, the pathogen infiltrates this space, forming small clusters of cells that are frequently separated from the host epithelium by a layer of symbiotic bacteria (Fig. 2B). Occasionally, larger populations of S. marcescens cells colonize the ilea of monoinoculated bees, suggesting that S. marcescens displaces S. alvi or exploits niches left open by the absence of other members of the microbiota (Fig. 2; also see Fig. S3). In CV bees 1 day after exposure, S. marcescens cells are scattered throughout a dense layer of commensal bacteria but do not seem to form aggregates, as in monocolonized bees (Fig. 2C). Thus, a diverse gut community appears capable of more fully exploiting niches within the gut, allowing greater exclusion of S. marcescens.
FIG 2.
Gut commensals compete with Serratia marcescens for space. (A) Diagram of the honey bee gut, showing a cross-section of the ileum. (B) Representative cross-sections of ilea from bees colonized by S. alvi (white) and S. marcescens (magenta). MF bees were inoculated with S. alvi wkB2 and exposed to S. marcescens after 5 days. S. alvi and S. marcescens were visualized using fluorescent probes that hybridize to 16S rRNA, while Sytox green stain indicates the presence of host nuclei and bacterial cells (green). (C) Cross-section of the ileum of a bee inoculated with gut homogenate (CV) and then exposed to S. marcescens (magenta). Sytox blue stain was used to label host nuclei and bacterial cells (green). Dashed white lines on the composite images outline the lumen of the gut.
Serratia marcescens kz11 contains two T6SSs.
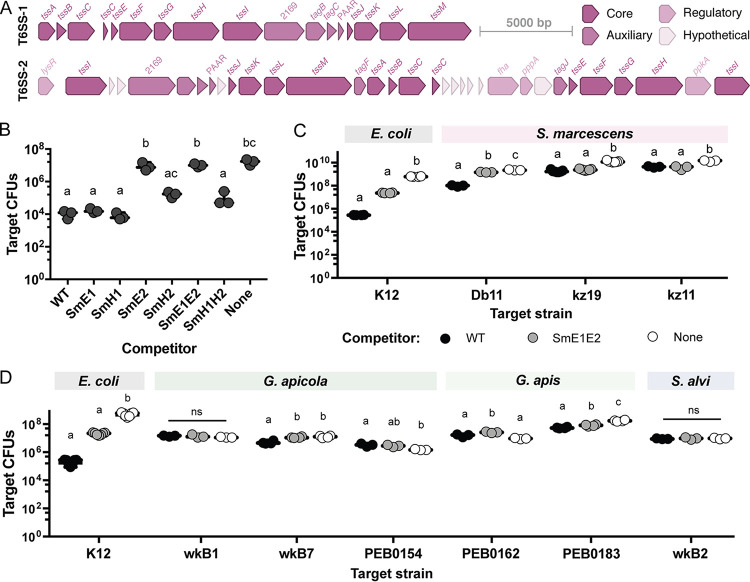
Serratia marcescens kz11 contains two complete sets of genes, each encoding the 13 required structural components of T6SSs, which are grouped into two loci (Fig. 3A). These loci share ≥90% nucleotide identity (≥70% coverage) with T6SS loci present in other S. marcescens strains. Both T6SSs are widespread within the genus, but T6SS-2 (which is also found in S. marcescens Db11) is more common among sequenced genomes (492 of 670 genomes) than T6SS-1 (189 of 670 genomes). We constructed in-frame replacements of genes encoding required structural components (tssE or tssH) of the two T6SSs (SmE1, SmH1, SmE2, and SmH2), as well as double replacements (SmE1E2 and SmH1H2). Strains lacking structural components of T6SS-2 had reduced ability to antagonize E. coli K-12 during in vitro competition assays, with deletion of tssE2 having a greater effect (Fig. 3B). In contrast, deletion of T6SS-1 genes had no effect on T6SS-mediated killing, indicating that T6SS-1 is not used to target E. coli or is not active under the tested conditions.
FIG 3.
Serratia marcescens kz11 uses a T6SS to antagonize other S. marcescens strains and one bee commensal. (A) Open reading frame (ORF) map of the T6SS loci in S. marcescens kz11, with structural genes in dark pink, auxiliary genes (such as adapters and effectors) in medium pink, genes for posttranscriptional regulation in light pink, and hypothetical genes encoding proteins of unknown function in white. (B) Recovery of E. coli K-12 Tn7-GmR after 4 h of coculture with the indicated S. marcescens strains. Competitions began with approximately 107 E. coli cells and a 1:4 ratio of E. coli to S. marcescens. (C) Recovery of E. coli K-12 and S. marcescens strain Db11, kz19, and kz11 CFU after 4 h of coculture with WT S. marcescens kz11 (black), SmE1E2 (gray), or buffer (white). (D) Recovery of E. coli K-12 and gut commensal G. apicola wkB1, wkB7, and PEB0154, G. apis PEB0162 and PEB0183, and S. alvi wkB2 CFU after coculture with S. marcescens. Target CFU were measured through plate counts on selective media. Letters indicate significant differences between treatment groups (one-way ANOVA with Tukey’s multiple-comparison test, P < 0.05). ns, not significant.
Serratia marcescens T6SSs have a limited target range.
To determine whether kz11 uses a T6SS to kill other S. marcescens strains, we set up pairwise competitions in vitro with a starting ratio of 10:1 attacker (wild-type [WT] or T6SS-deficient S. marcescens kz11) to target (S. marcescens Db11, kz19, or rifampin [Rif]-resistant kz11) (Fig. 3C). Fewer CFU were recovered from competitions than from competitor-free (phosphate-buffered saline [PBS]) controls for all three strains. More Db11 CFU were recovered after coculture with T6SS-SmE1E2 than with WT S. marcescens. While there was no significant difference in the number of kz19 CFU after coculture with either competitor, kz19 represented a larger percentage of the CFU recovered from coculture with SmE1E2 (see Fig. S4A). As expected, there was no difference in the number of WT kz11 CFU recovered after coculture with the kz11-derived mutants. As for E. coli K-12, antagonism of Db11 and kz19 appeared dependent on T6SS-2, but not T6SS-1, under the tested conditions (see Fig. S4B). Additionally, S. marcescens strains vary in susceptibility to T6SS-2, with Db11, an isolate from Drosophila, being more susceptible than kz19, an isolate from honey bees (8), to antagonism by kz11.
We tested the ability of S. marcescens to antagonize Gilliamella spp. and S. alvi, the two most abundant Gram-negative taxa in the honey bee gut. We recovered more Gilliamella apicola wkB7, Gilliamella apis PEB0162, and Gilliamella apis PEB0183 CFU after coculture with S. marcescens SmE1E2, relative to coculture with the WT strain, but found no difference in recovery of S. alvi wkB2, G. apicola wkB1, and G. apicola PEB0154 CFU (Fig. 3C). When fewer target CFU were recovered after coculture with WT S. marcescens, the target strain also represented a smaller proportion of the total recovered CFU (see Fig. S4). However, while statistically significant, the effect of the S. marcescens T6SSs on Gilliamella spp. in vitro was less than the effect on E. coli K-12 and S. marcescens Db11. Furthermore, there was no difference in Gilliamella or S. alvi abundance, based on 16S rRNA gene copies, for CV bees infected with WT S. marcescens versus SmE1E2 (see Fig. S5), suggesting that S. marcescens T6SSs do not affect the abundance of these species in vivo. We also found no difference in Gilliamella or S. alvi abundance between bees in which S. marcescens had been eliminated and bees in which it persisted.
Serratia marcescens T6SSs do not affect fitness in the bee gut.
To determine whether T6SSs produced by S. marcescens kz11 facilitate colonization, we exposed MF and CV bees to WT or SmE1E2 S. marcescens. In MF bees, WT and SmE1E2 CFU increased after exposure and persisted at high abundance over 4 days (Fig. 4A and B). In contrast, the percentage of CV bees infected and S. marcescens abundance in infected bees decreased over time (Fig. 4C and D). However, we observed no difference in the abundance of WT and SmE1E2 strains, indicating that T6SSs are not required to colonize or persist in MF bees and, further, provide no detectable advantage in CV bees. This result holds true over a range of tested conditions. In a similar experiment, we observed no difference in fitness between WT, SmE1, SmE2, and SmE1E2 strains in MF or CV bees (see Fig. S6). Furthermore, the WT strain does not appear to be more fit than the SmE1E2 strain in monoinoculated bees (see Fig. S7A), and a T6SS-deficient strain (SmC1H1) does not appear to be less fit than the WT strain in CV bees treated with Tet (see Fig. S7B).
FIG 4.
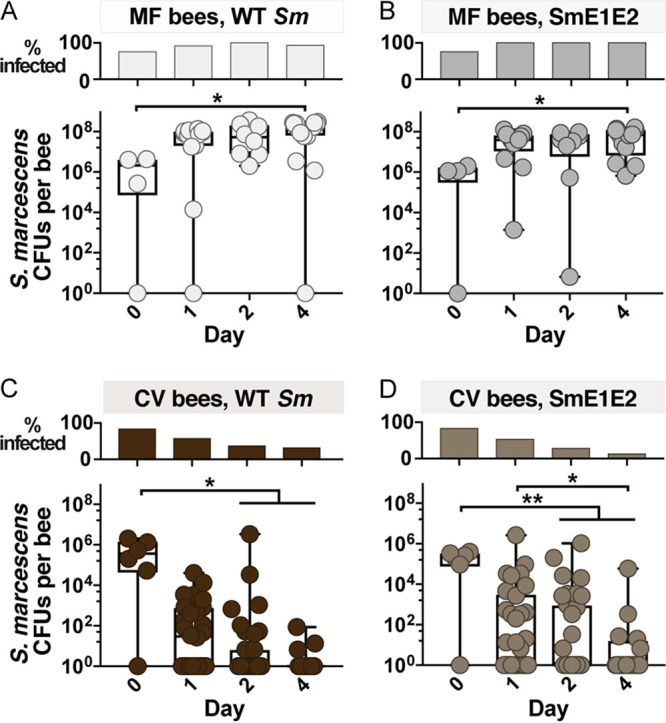
T6SS-deficient Serratia marcescens strains do not differ from the WT strain in fitness within the bee gut. Percentages of sampled bees infected with S. marcescens (top) and abundance of S. marcescens in the midguts and hindguts of individuals (bottom) 1 to 4 days after exposure are shown. MF bees were exposed to WT S. marcescens (A) or to SmE1E2 (B). Conventionalized (CV) bees were exposed to WT S. marcescens (C) or to SmE1E2 (D). Bees were exposed to 4 × 108 S. marcescens cells/ml in sugar syrup for 1 day. Abundance was quantified by counting CFU. Box and whisker plots show range, first and third quartiles, and median. *, P < 0.05; **, P < 0.005, Kruskal-Wallis test with Dunn’s multiple-comparison test.
DISCUSSION
Serratia marcescens is a pathogen of many animals, including bees, and is frequently found at low levels in honey bee hives (2, 8, 36). Although deadly when introduced directly into the bee hemolymph, many S. marcescens strains are not lethal when ingested (2). However, S. marcescens has been detected in the bee hemolymph after oral exposure, suggesting that some strains can travel from the gut to the hemolymph, through an unknown route (6). Furthermore, mortality rates are increased in bees whose native gut community has been perturbed by exposure to antibiotics (6, 8) or agrochemicals (7). Therefore, it seems likely that the bee gut microbiota protects hosts by directly antagonizing S. marcescens and/or by reducing access to host tissues.
In this study, S. marcescens kz11, a pathogenic isolate from bees, persisted in guts of MF bees and bees with a perturbed gut community but rapidly declined in guts of CV bees, indicating that the gut microbiota substantially contributes to elimination of this pathogen. Because honey bees do not defecate in captivity (37), reduced abundance is a result of S. marcescens cells dying, rather than leaving the system. We observed elimination of S. marcescens from CV bees within 1 day after exposure in summer (July to September) but only after ≥4 days in fall (October to November), suggesting that seasonal differences in host biology (38) or microbiota composition (39) have some effect. Stimulation of the host immune system by the commensal community may contribute to elimination of S. marcescens. Antimicrobial peptides (AMPs) apidaecin and hymenoptaecin are upregulated in the guts of CV bees or bees monocolonized with S. alvi, relative to MF bees (34, 40), but expression of these AMPs does not differ between CV bees and CV bees exposed to S. marcescens (6), suggesting that this pathway is not part of a host response to S. marcescens infection.
While S. marcescens abundance was reduced in bees colonized by single gut taxa, multiple species were required to attain the level of colonization resistance seen in CV bees, suggesting that bee symbiont species contribute to colonization resistance through different mechanisms. For instance, the microbiota alters bee gut chemistry by reducing oxygen concentration, pH, and redox potential within the gut (13, 41). S. alvi and G. apicola contain T6SSs (19) and may directly antagonize S. marcescens in the ileum. Lactobacillus spp. are abundant in the bee rectum (18) and are likely to produce antimicrobial molecules (41–43). Although this trait is not well characterized in bee isolates, Lactobacillus species are common probiotics for prevention of Clostridium difficile infections in humans (44). The participation of multiple species in colonization resistance is an important consideration for future investigation of the mechanisms involved. S. alvi appears to compete with S. marcescens for space in the ileum but colonizes only spaces adjacent to the host epithelium (18). Most bee commensals colonize specific regions of the gut (18), which may also explain why monoinoculation does not recapitulate the ability of the natural community to exclude S. marcescens. Intriguingly, CV communities from different bee colonies differed in the level of colonization resistance conferred (see Fig. S2 in the supplemental material), suggesting that the particular combination of species and strains determines the extent of protection, as observed in studies with bumble bees (45–47). Bee guts can also contain yeasts, which are acquired from environmental sources and vary seasonally and between hives (48, 49). These fungi may affect host immune function (50). Serratia marcescens secretes antifungal, as well as antibacterial, T6SS effectors (51) and may interact with yeasts in the bee gut. However, the core gut microbiota, even in the absence of yeasts and low-abundance bacteria, seems to be sufficient to provide colonization resistance.
Because the microbiota provides an obstacle to infection by S. marcescens, we hypothesized that T6SS-mediated antagonism of gut commensals might be important for colonization and pathogenicity, as for some pathogens in mice (30, 32). However, neither T6SS of S. marcescens kz11 contributes to its persistence in the bee gut. The T6SS-1 locus is present in many Serratia genomes but did not affect the ability to colonize the gut or to antagonize a range of bacterial competitors under the conditions we tested. In contrast, we observed T6SS-2-mediated killing of G. apicola wkB7, G. apis PEB0162, G. apis PEB0183, E. coli, and other S. marcescens strains in vitro but detected no effect on colonization of the gut. The antibacterial properties of T6SS-2 are consistent with previous studies of T6SS structure and function in the related strain S. marcescens Db11 (24). In Db11, the T6SS is constitutively active (52), and the lack of T6SS-mediated killing of bee symbionts by S. marcescens kz11 is unlikely to reflect a lack of T6SS activity. Horizontal transfer of immunity genes, which confer resistance to their cognate toxins, occurs within the gut microbiota of humans (53, 54) and bees (19, 55) and could explain symbiont survival. Alternatively, bee symbionts may be immune to S. marcescens toxins. For example, some toxins secreted by S. marcescens Db11 contain disulfide bonds and depend on a DsbA protein produced by the target cell for proper folding (56). Such toxins are likely to affect other S. marcescens strains but may not be functional within cells of more distantly related organisms. The utility of T6SSs may thus depend on the competitors involved, implying varied and dynamic roles within ecological communities.
CONCLUSION
Antagonistic interactions between pathogens and commensal microbiota can affect the severity of infection within hosts. We found that the opportunistic pathogen S. marcescens rapidly declines within honey bee guts containing the normal microbiota. Multiple symbiont taxa contribute to colonization resistance, possibly reflecting different niches and mechanisms of inhibition. T6SSs are likely to be important in many bacterial interactions; some pathogens use T6SSs to compete with commensals during infection (30, 32). Serratia marcescens possesses an antibacterial T6SS, but this T6SS does not appear to antagonize the bee gut microbiota. Instead, the S. marcescens T6SSs likely function in competition with close relatives and may have other roles that are still unexplored. These findings are relevant for understanding the ecology of S. marcescens and other opportunistic pathogens, which may often be poorly adapted for competition with host-associated bacteria.
MATERIALS AND METHODS
Detailed protocols are available in the supplemental material.
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are listed in Table S1 in the supplemental material. E. coli and S. marcescens strains were grown on solid LB plates at 37°C or in liquid LB medium at 37°C at 225 rpm. Bee isolates were grown at 35°C in 5% CO2, S. alvi and Gilliamella spp. on heart infusion agar supplemented with 5% sheep’s blood (blood HIA) and Lactobacillus spp. in MRS broth. When necessary, media were supplemented with antibiotics, i.e., 50 μg/ml kanamycin (Km), 20 μg/ml chloramphenicol (Cm), 60 μg/ml spectinomycin (Sp), 100 μg/ml carbenicillin (Cb), 12.5 μg/ml gentamycin (Gm), 100 μg/ml Rif, 15 μg/ml Tet, or 1 μg/ml anhydrotetracycline (ATc), or with 2,6-diaminopimelic acid (DAP) (0.3 mM).
Construction of T6SS-deficient mutants.
To create isogenic strains of S. marcescens kz11 with inactive T6SSs (see Table S1), genes encoding required components were replaced with antibiotic resistance markers Kmr or Cmr. Kmr was also inserted downstream of T6SS-1 to generate a Kmr strain with a WT phenotype. Plasmids used for allelic exchange contained antibiotic resistance markers flanked by ∼1-kb sequences homologous to regions on either side of the target gene, as well as a Tet-inducible toxin (Tse2) for counterselection. Plasmids were constructed using Golden Gate assembly (see the supplemental material). Plasmids were transformed into the donor strain E. coli MFDpir (a DAP auxotroph) through electroporation and then transferred into the recipient strain through conjugation. Transconjugants were selected for and then streaked on LB medium with Tet, ATc, and Km or Cm for selection. PCR was used to verify replacement.
Competition assays.
Competition assays were used to measure T6SSs-mediated antagonism of E. coli, other S. marcescens strains, or bee gut isolates by S. marcescens kz11. E. coli K-12-Tn7-Gmr and S. marcescens WT, SmE1, SmH1, SmE2, SmH2, SmE1E2, and SmH1H2 strains were mixed in a 1:4 ratio, and 25-μl droplets (∼106 E. coli cells) were spotted on LB medium and incubated for 4 h. For all other competition assays, target and attacker were mixed at a 1:10 ratio, and 25-μl droplets (∼107 target cells) were spotted on blood HIA and incubated for 4 h at 35°C in 5% CO2. E. coli K-12 was included as a target strain in each set of competition assays, as a control for T6SS activity. Cells from competitions with E. coli and S. marcescens target strains were collected and suspended in 500 μl PBS. Some Gilliamella strains were difficult to recover from agar; therefore, agar plugs were excised for competitions with E. coli, S. alvi, and Gilliamella target strains, placed in 500 μl PBS, and vortex-mixed to recover the cells. Serial dilutions (1:10) were prepared, and 10 μl of each dilution was spotted in triplicate on selective and nonselective media. Colonies were counted and data visualization and statistical analyses were performed with Prism 7.
Honey bee experiments.
MF bees were obtained by removing pupae, which naturally lack gut symbionts, from hives maintained by the laboratory, as described previously (57). CV bees and bees with defined communities were obtained by feeding pollen soaked in a mixture of sucrose-PBS (25% sucrose (w/v), 0.5X PBS) and either nurse gut homogenate or 8 × 108 bacterial cells from culture to MF bees within 2 days after emergence. Control MF bees were given pollen soaked in sterile sucrose-PBS. Tet-treated bees were fed 450 μg/ml Tet in sucrose syrup for 4 days, starting 5 days after inoculation. Bees were provided with 1 ml of S. marcescens at an OD600 of 0.5 in 1:1 sucrose or sterile sucrose 5 days after inoculation with gut bacteria or 1 day after the end of Tet treatment. After 1 day, feeding tubes containing S. marcescens were replaced with sterile sucrose. Preliminary experiments indicated that this approach did not increase variability in S. marcescens abundance, relative to hand-feeding a defined number of cells to each bee. To quantify S. marcescens, midguts and hindguts of individual bees were extracted and homogenized in 200 μl PBS. Serial dilutions were prepared, and 10 μl of each dilution was spotted in triplicate on LB agar supplemented with antibiotics. Colonies were counted to estimate S. marcescens CFU per gut.
Quantification of bee gut symbionts.
DNA was extracted from gut homogenate using the cetyltrimethylammonium bromide (CTAB) method, and quantitative PCR (qPCR) for absolute quantification of Gilliamella spp. and S. alvi 16S rRNA gene copies was performed as described by Powell et al. (57), using primers listed in Table S2 in the supplemental material. Quantification was performed in triplicate for each biological replicate.
FISH microscopy.
Bees were dissected 1 or 3 days after S. marcescens exposure. Guts were fixed in Carnoy’s solution, embedded in paraffin, and cut into 7-μm-thick sections. Fluorescent probes specific to 16S rRNA sequences of S. alvi and S. marcescens (see Table S2) were hybridized to sections, as described previously (18, 58). Sytox blue (50 mM) or green (0.5 mM) dyes were used to label host and microbial DNA. Images were acquired using a Zeiss 710 confocal microscope, except where noted in the figure legends, and processed using ImageJ (59, 60).
ACKNOWLEDGMENTS
We thank the following individuals for providing plasmids or strains: Herbert Schweizer, Harry Mobley, John Dueber, Jeff Barrick, Peng Geng, Kasie Raymann, and Sarah Coulthurst. We thank Ryuichi Koga, Julie Perreau, Bryan Davies, and Marvin Whiteley for useful discussions, Kim Hammond for maintaining honey bee hives, and Dan Deatherage for sequencing plasmids constructed for this study. This study utilized resources of the Texas Advanced Computing Center (TACC) at the University of Texas at Austin.
This work was funded by the National Science Foundation (grant DEB1701430 to M.I.S.) and the National Institutes of Health (grants RO1GM108477 and R35GM131738 to N.A.M. and grant F31DK115104 to M.I.S.).
Footnotes
Supplemental material is available online only.
Contributor Information
Nancy A. Moran, Email: nancy.moran@austin.utexas.edu.
Soo Chan Lee, University of Texas at San Antonio.
REFERENCES
- 1.Grimont PAD, Grimont F, Lysenko O. 1979. Species and biotype identification of Serratia strains associated with insects. Curr Microbiol 2:139–142. doi: 10.1007/BF02605870. [DOI] [Google Scholar]
- 2.Grimont PAD, Grimont F. 1978. The genus Serratia. Annu Rev Microbiol 32:221–248. doi: 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- 3.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, Katsura K, Kurokawa K, Oshima K, Hattori M, Parkhill J, Sebaihia M, Coulthurst SJ, Gotoh N, Thomson NR, Ewbank JJ, Hayashi T. 2014. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol 6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon RJ, Vennard CT, Buckling A, Charnley AK. 2005. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8:1291–1298. doi: 10.1111/j.1461-0248.2005.00828.x. [DOI] [Google Scholar]
- 5.Flyg C, Kenne K, Boman HG. 1980. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J Gen Microbiol 120:173–181. doi: 10.1099/00221287-120-1-173. [DOI] [PubMed] [Google Scholar]
- 6.Raymann K, Coon KL, Shaffer Z, Salisbury S, Moran NA. 2018. Pathogenicity of Serratia marcescens strains in honey bees. mBio 9:e01649-18. doi: 10.1128/mBio.01649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motta EVS, Raymann K, Moran NA. 2018. Glyphosate perturbs the gut microbiota of honey bees. Proc Natl Acad Sci USA 115:10305–10310. doi: 10.1073/pnas.1803880115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raymann K, Shaffer Z, Moran NA. 2017. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol 15:e2001861. doi: 10.1371/journal.pbio.2001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. 2011. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 11.Moran NA, Hansen AK, Powell JE, Sabree ZL. 2012. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabree ZL, Hansen AK, Moran NA. 2012. Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS One 7:e41250. doi: 10.1371/journal.pone.0041250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. 2017. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci USA 114:4775–4780. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel P, Moran NA. 2013. Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis. Gut Microbes 4:60–65. doi: 10.4161/gmic.22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI, Moran NA. 2016. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio 7:e01326-16. doi: 10.1128/mBio.01326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng H, Perreau J, Powell JE, Han B, Zhang Z, Kwong WK, Tringe SG, Moran NA. 2019. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc Natl Acad Sci USA 116:25909–25916. doi: 10.1073/pnas.1916224116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz RS, Moran NA, Evans JD. 2016. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc Natl Acad Sci USA 113:9345–9350. doi: 10.1073/pnas.1606631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol 78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steele MI, Kwong WK, Whiteley M, Moran NA. 2017. Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. mBio 8:e01630-17. doi: 10.1128/mBio.01630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coulthurst S. 2019. The type VI secretion system: a versatile bacterial weapon. Microbiology (Reading) 165:503–515. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 21.Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner’s guide. Curr Opin Microbiol 11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. 2014. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz S, West TE, Boyer F, Chiang W-C, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. 2014. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. 2016. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog 12:e1005735. doi: 10.1371/journal.ppat.1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. 2012. New secreted toxins and immunity proteins encoded within the type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol 86:921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ. 2017. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21:769–776.e3. doi: 10.1016/j.chom.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Wardenburg JB. 2016. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 17:1281–1291. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci USA 113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fast D, Kostiuk B, Foley E, Pukatzki S. 2018. Commensal pathogen competition impacts host viability. Proc Natl Acad Sci USA 115:7099–7104. doi: 10.1073/pnas.1802165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong WK, Mancenido AL, Moran NA. 2017. Immune system stimulation by the native gut microbiota of honey bees. R Soc Open Sci 4:170003. doi: 10.1098/rsos.170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell JE, Leonard SP, Kwong WK, Engel P, Moran NA. 2016. Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc Natl Acad Sci USA 113:13887–13892. doi: 10.1073/pnas.1610856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Sanousi SM, El Sarag MSA, Mohamed SE. 1987. Properties of Serratia marcescens isolated from diseased honeybee (Apis mellifera) larvae. Microbiology 133:215–219. doi: 10.1099/00221287-133-1-215. [DOI] [Google Scholar]
- 37.Weiss MR. 2006. Defecation behavior and ecology of insects. Annu Rev Entomol 51:635–661. doi: 10.1146/annurev.ento.49.061802.123212. [DOI] [PubMed] [Google Scholar]
- 38.Steinmann N, Corona M, Neumann P, Dainat B. 2015. Overwintering is associated with reduced expression of immune genes and higher susceptibility to virus infection in honey bees. PLoS One 10:e0129956. doi: 10.1371/journal.pone.0129956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kešnerová L, Emery O, Troilo M, Liberti J, Erkosar B, Engel P. 2020. Gut microbiota structure differs between honeybees in winter and summer. ISME J 14:801–814. doi: 10.1038/s41396-019-0568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horak RD, Leonard SP, Moran NA. 2020. Symbionts shape host innate immunity in honeybees. Proc Biol Sci 287:20201184. doi: 10.1098/rspb.2020.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer-Young EC, Raffel TR, McFrederick QS. 2019. pH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitology 146:380–388. doi: 10.1017/S0031182018001555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellegaard KM, Brochet S, Bonilla-Rosso G, Emery O, Glover N, Hadadi N, Jaron KS, van der Meer JR, Robinson-Rechavi M, Sentchilo V, Tagini F, Engel P, SAGE Class 2016–17, Engel P. 2019. Genomic changes underlying host specialization in the bee gut symbiont Lactobacillus Firm5. Mol Ecol 28:2224–2237. doi: 10.1111/mec.15075. [DOI] [PubMed] [Google Scholar]
- 43.Forsgren E, Olofsson TC, Vásquez A, Fries I. 2010. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41:99–108. doi: 10.1051/apido/2009065. [DOI] [Google Scholar]
- 44.McFarland LV, Ship N, Auclair J, Millette M. 2018. Primary prevention of Clostridium difficile infections with a specific probiotic combining Lactobacillus acidophilus, L. casei, and L. rhamnosus strains: assessing the evidence. J Hosp Infect 99:443–452. doi: 10.1016/j.jhin.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Koch H, Schmid-Hempel P. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA 108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch H, Schmid-Hempel P. 2012. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett 15:1095–1103. doi: 10.1111/j.1461-0248.2012.01831.x. [DOI] [PubMed] [Google Scholar]
- 47.Mockler BK, Kwong WK, Moran NA, Koch H. 2018. Microbiome structure influences infection by the parasite Crithidia bombi in bumble bees. Appl Environ Microbiol 84:e02335-17. doi: 10.1128/AEM.02335-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Callegari M, Crotti E, Fusi M, Marasco R, Gonella E, De Noni I, Romano D, Borin S, Tsiamis G, Cherif A, Alma A, Daffonchio D. 2021. Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. NPJ Biofilms Microbiomes 7:42. doi: 10.1038/s41522-021-00212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludvigsen J, Andersen Å, Hjeljord L, Rudi K. 2020. The honeybee gut mycobiota cluster by season versus the microbiota which cluster by gut segment. Vet Sci 8:4. doi: 10.3390/vetsci8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tauber JP, Nguyen V, Lopez D, Evans JD. 2019. Effects of a resident yeast from the honeybee gut on immunity, microbiota, and Nosema disease. Insects 10:E296. doi: 10.3390/insects10090296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trunk K, Peltier J, Liu Y-C, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, Coulthurst SJ. 2018. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol 3:920–931. doi: 10.1038/s41564-018-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerc AJ, Diepold A, Trunk K, Porter M, Rickman C, Armitage JP, Stanley-Wall NR, Coulthurst SJ. 2015. Visualization of the Serratia type VI secretion system reveals unprovoked attacks and dynamic assembly. Cell Rep 12:2131–2142. doi: 10.1016/j.celrep.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross BD, Verster AJ, Radey MC, Schmidtke DT, Pope CE, Hoffman LR, Hajjar AM, Peterson SB, Borenstein E, Mougous JD. 2019. Human gut bacteria contain acquired interbacterial defence systems. Nature 575:224–228. doi: 10.1038/s41586-019-1708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E. 2017. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 22:411–419. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwong WK, Engel P, Koch H, Moran NA. 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci USA 111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mariano G, Monlezun L, Coulthurst SJ. 2018. Dual role for DsbA in attacking and targeted bacterial cells during type VI secretion system-mediated competition. Cell Rep 22:774–785. doi: 10.1016/j.celrep.2017.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Powell JE, Martinson VG, Urban-Mead K, Moran NA. 2014. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol 80:7378–7387. doi: 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engel P, James RR, Koga R, Kwong WK, McFrederick QS, Moran NA. 2013. Standard methods for research on Apis mellifera gut symbionts. J Apicult Res 52:1–24. doi: 10.3896/IBRA.1.52.4.07. [DOI] [Google Scholar]
- 59.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00394-21_Supp_1_seq7.pdf, PDF file, 1.6 MB (1.6MB, pdf)