Abstract
The reverse transcription-polymerase chain reaction (RT-PCR) method has been adopted worldwide to diagnose severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Although this method has good sensitivity and specificity, there is a need to develop a more rapid diagnostic technology, given the virus’s rapid spread. However, the RT-PCR method takes a long time to diagnose SARS-CoV-2 because of the required thermocycling steps. Therefore, we developed a surface-enhanced Raman scattering (SERS)-PCR detection method using an AuNP-internalized Au nanodimple substrate (AuNDS) to shorten the diagnosis time by reducing the number of thermocycling steps needed to amplify the DNA. For the representative target markers, namely, the envelope protein (E) and RNA-dependent RNA polymerase (RdRp) genes of SARS-CoV-2, 25 RT-PCR thermocycles are required to reach a detectable threshold value, while 15 cycles are needed for magnetic bead-based SERS-PCR when the initial DNA concentration was 1.00× 105 copies/μL. However, only 8 cycles are needed for the AuNDS-based SERS-PCR. The corresponding detectable target DNA concentrations were 3.36 × 1012, 3.28 × 109, and 2.56 × 107 copies/μL, respectively. Therefore, AuNDS-based SERS-PCR is seen as being a new molecular diagnostic platform that can shorten the time required for the thermocycling steps relative to the conventional RT-PCR.
Keywords: surface-Enhanced Raman scattering, SERS-PCR, SARS-CoV-2, Au nanodimple substrate, Nanogap
1. Introduction
The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has had a huge political, social, and cultural impact (He et al., 2020; Ferriti et al., 2020; Zhu et al., 2019). Several different types of vaccines have been developed and distributed to control the spread to some extent; however, the world is suffering again due to the recent spread of mutations of the virus (Harvey et al., 2021; Chen et al., 2021a, Chen et al., 2021b). As the number of confirmed cases of SARS-CoV-2 increases, it is essential to identify and isolate infected persons quickly. The reverse transcription-polymerase chain reaction (RT-PCR) is currently used as a standard diagnostic method for SARS-CoV-2 (Smyrlaki et al., 2020). However, more rapid diagnostic technology is required to effectively respond to the virus’s rapid spread since the RT-PCR method takes approximately 3–4 h of diagnosis time, including viral RNA extraction, reverse transcription, thermocycling amplification, and fluorescence detection (Broughton et al. 2020; Long et al., 2020). An infection can be confirmed within 30 min of using an immunochromatographic rapid kit that relies on an antibody–antigen reaction. However, the diagnostic sensitivity for a recently infected or asymptomatic infected person is remarkably low (50% or less) (Vandenberg et al., 2021; Corman et al., 2021). Thus, there is a problem with the reliability of the diagnosis. In particular, the issue of “false-negative” results, whereby positive patients are diagnosed as negative, is the main reason this method cannot be adopted as a standard diagnostic method (Ghodake et al., 2021; Xu et al., 2020). Therefore, many researchers are striving to develop a new molecular diagnostic technology that can shorten the diagnostic time while maintaining the diagnostic accuracy of RT-PCR.
Fluorescence detection using TaqMan probes is part of the RT-PCR process, but a gene amplification process involving 30–40 thermocycles is essential because the detection sensitivity is limited (Hwang et al., 2021; Faulds et al., 2004). It would be possible to shorten the diagnosis time by reducing the number of thermocycling steps and using a more sensitive method than fluorescence detection. Recently, many studies have addressed the use of the surface-enhanced Raman scattering (SERS) detection method, which offers a better detection sensitivity than fluorescence detection (Sitjar et al., 2021; Chen et al., 2021a, Chen et al., 2021b, Zhang et al., 2021; Ramos-Guzman et al., 2021; Kim et al., 2021). If a biomarker molecule related to the change in the concentration of the target genes can exist in a nanogap, the localized surface plasmon resonance effect would greatly amplify any incident light (Baumberg et al., 2019; Chen et al., 2018; Wang et al., 2017). We recently reported on a novel nano-plasmonic detection platform that produces high-sensitivity SERS signals with regularly arranged gold nanoparticles (AuNPs) in the cavities of Au nanodimple substrates (AuNDSs) (Dang et al., 2021). The AuNPs were uniformly arranged over the AuNDS cavity surface by DNA hybridization. Thus, it is possible to attain a high level of sensitivity using strongly enhanced SERS signals for the specific gene markers generated in this nanogap.
In this study, we developed a new SERS-based SARS-CoV-2 diagnostic platform using previously reported AuNP-internalized AuNDS (Dang et al., 2021). This method can dramatically reduce the diagnostic time relative to conventional RT-PCR, which is currently used as a standard diagnostic method for SARS-CoV-2. The assay results for the envelope protein (E) and RNA-dependent RNA polymerase (RdRp) genes of SARS-CoV-2 were compared with those measured by RT-PCR and magnetic bead-based SERS-PCR. The AuNDS-based SERS-PCR system addressed in this work could detect bridge DNAs with fewer than ten thermocycling steps; therefore, it can dramatically reduce the diagnosis time relative to the currently used RT-PCR. Furthermore, we propose a SERS-PCR system that integrates a sampling module capable of automatically assaying samples and AuNPs into AuNDS with a portable Raman spectrophotometer, that is, a next-generation molecular diagnostic system capable of contributing to the rapid and accurate diagnosis of SARS-CoV-2.
2. Materials and methods
2.1. Materials and reagents
Gold chloride trihydrate (HAuCl4), trisodium citrate (Na3-citrate), tris (2-carboxyethyl) phosphine hydrochloride (TCEP), polyvinylpyrrolidone (PVP), and NaCl. Tris-EDTA buffer (TE, pH 7.0, 100), Tris-HCl buffer (pH 7.0, 10), and saline-sodium citrate buffer (SSC, pH 7.0, 20) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Malachite green isothiocyanate (MGITC) was purchased from Life Technologies (Eugene, OR, USA) and streptavidin-conjugated magnetic beads (Dynabeads MyOneTM) were purchased from Invitrogen (Carlsbad, CA, USA). MicroAmp 96-well tray/retainer sets (4379590) and support bases (4381850) were purchased from Applied Biosystems (Foster City, CA, USA). Premix EX TaqTM was purchased from Takara Bio Inc. (Shiga, Japan). The oligonucleotide DNA probes, SARS-CoV-2 primers, and TaqMan probe panels were purchased from Integrated DNA Technologies, Inc. (Coralville, IA, USA). Milli-Q water (18.2 MΩ cm) was obtained from an Arium comfort purification system (Göttingen, Germany).
2.2. Instrumentation
UV–visible absorption data were obtained using a Synergy H1 hybrid multi-mode reader (BioTek, Winooski, VT, USA). Dynamic light scattering (DLS) data were obtained using a Nano-ZS90 apparatus (Malvern Instruments, Malvern, UK). Scanning electron microscopy (SEM) images were obtained using a JSM-7800F Prime instrument (JEOL Ltd., Japan). Transmission electron microscopy (TEM) images were obtained using a JEM-1400 transmission electron microscope (JEOL Ltd., Japan). Raman measurements were performed using a confocal micro-Raman/PL mapping system (NOST, Korea) and an inVia Renishaw Raman microscope system (Renishaw, New Mills, UK). TaqMan-based RT-PCR assays were performed using a QuantStudioTM3 real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Preparation of detection DNA-conjugated SERS nanotags and capture DNA-conjugated 3D Au nanodimple substrate
Eighty nanometer-sized AuNPs were synthesized using the seeded growth method (Bastús et al., 2011). One milliliter of 0.1 nM AuNP was reacted with 10 μL of 1 μM MGITC for 1 h at room temperature. Thiolated DNA detection probes were then mixed with MGITC-labeled AuNPs at a molar ratio of 5000:1 in distilled water containing 0.05% PVP (Wu et al., 2020). The mixture was then incubated for 12 h at 4 °C. Next, a 2 M NaCl buffer containing 1TE and 0.05% PVP was slowly added to the mixture for 12 h until the final concentration was 0.2 M NaCl. Here, the salting process was carried out at 4 °C to avoid aggregation of AuNPs. After overnight incubation at 4 °C to induce a salt-aging process, the prepared SERS nanotags (detection DNA and MGITC-functionalized AuNPs) were redispersed in 1TE buffer for further use. The fabrication process of AuNDSs has been reported in detail elsewhere (Hao et al., 2020). The substrate (5 mm × 5 mm) was rinsed five times with distilled water and then dried in an oven at 70 °C. Next, the substrate was immersed in 0.25 mL thiolated capture DNA solution containing a 2 M NaCl buffer for 12 h. Then, the substrate was sequentially washed with 1TE buffer and Milli-Q water. After washing five times, the captured DNA-conjugated Au nanodimple substrate was dried in an oven at 70 °C and then cooled to room temperature.
2.4. SERS-PCR assays of SARS-CoV-2 using AuNP-inserted nanodimple substrates
First, 1 μL of RdRp or E target gene and 1 μL of corresponding bridge DNA was added to 23 μL of PCR mixture. The concentrations of forward and reverse primer pairs in the PCR mixture and assay conditions were the same as those described in a previous report (Corman et al., 2020). However, unlike the TaqMan probe, the bridge DNA probe, which does not contain either a fluorescent dye (FAM) or quencher at its 5’- and 3’- terminal positions, was used as a hydrolysis probe. After PCR amplification, 5 μL of the PCR product solution was extracted from the microtube for the SERS-based DNA hybridization assay. Second, a solution containing 5 μL of PCR product solution, 30 μL of 80-nm SERS nanotags, and 4 μL of a 20SSC buffer was dropped onto the surface of the Au nanodimple substrate. The substrate was then incubated for 40 min in a humid chamber to form DNA hybridization complexes between the SERS nanotags and the cavity surfaces of the Au nano-dimpled substrates. The DNA complexes were rinsed with 1SSC buffer and water five times and subsequently dried in an oven at 70 °C prior to the SERS measurements.
3. Results and discussion
Fig. 1 shows the principle of SERS-PCR assays using the internalization of AuNPs into AuNDS. Label-free bridge DNA was used as a hydrolysis agent instead of the TaqMan DNA, with a fluorescent dye attached to the terminal group, used for RT-PCR. As shown in Fig. 1a, the bridge DNA is broken into A, G, T, and C fragments by hybridization between the target DNA and the forward/reverse primers when a target DNA is present. As a result, the bridge DNA concentration decreases with an increase in the number of thermocycles (Wu et al., 2020). However, if the target gene is not present, there is no change in the concentration of bridge DNA because hybridization between the primer and the target DNA does not occur. Therefore, the concentration of the target DNA was determined by measuring the concentration of the remaining bridge DNA after PCR. In this study, the concentration of the remaining bridge DNAs was determined using AuNP-internalized AuNDS capable of ultrasensitive SERS detection, as shown in Fig. 1b. Herein, the large hot spot formed between the AuNP and Au dimple cavity surface induces strong localized surface plasmon effects, and a high-sensitivity SERS-PCR assay can be implemented using this area. The fabrication of AuNDS has been previously reported (Dang et al., 2021). Figs. S1a and S1b show a TEM image of 80-nm AuNPs and a SEM image of the AuNDS used in this study. Fig. 1c shows the series of processes for forming DNA hybridization complexes in the curved cavity of an AuNDS. First, Raman reporter MGITC molecules are labeled on the surface of the 80-nm AuNPs through electrostatic interaction. Then, the detection DNA with a thiol group attached to the terminal was immobilized on the AuNP surface through a covalent bond. Similarly, capture DNAs were bound to the curved cavity surface of the AuNDS through the covalent bond of the thiol group attached to its terminal position, as shown in Fig. 1c. Fig. 1d is a schematic representation of AuNP-internalized AuNDS formed through bridge DNA sandwich complexes and DNA hybridization.
Fig. 1.
Schematic illustration of SERS-PCR assays using the internalization of AuNPs into AuNDS. (a) PCR amplification process for bridge DNAs. (b) SERS detections for bridge DNAs. (c) Sequential process for the fabrication of detection DNA-conjugated SERS nanotags (top) and capture DNA probe-immobilized AuNDS (bottom). (d) Formation of sandwich DNA hybridization complexes with (left) and without (right) bridge DNAs.
Fig. S2 shows the finite-difference time-domain (FDTD) simulation results for the three-dimensional electric field distribution of AuNPs in an Au nanodimple cavity. Fig. S2a shows a three-dimensional image of AuNP-internalized AuNDS. Figs. S2b and S2c show two-dimensional electric field distribution images for the xz and xy planes, respectively. Linearly polarized light with a wavelength of 633 nm was directly illuminated onto the 80-nm AuNP in the cavity of AuNDS. The distance between the AuNPs and the curved cavity was fixed to 4 nm, based on the observed TEM image shown in Fig. S3, and the scale bar on the right is a color-decoding scheme that represents the electric field distribution intensity. The brightness of the color indicates the strength of the electric field distribution. Because the hot spot between AuNP and the curved cavity has a sizable volumetric area, a strong localized surface plasmon enhancement effect would occur. Fig. S3a shows a SEM image of AuNP-internalized AuNDS formed through bridge DNA sandwich complexes and DNA hybridization. This image shows that AuNPs can be uniformly distributed in the AuNDS cavity through DNA hybridization. Fig. S3b shows cross-sectional TEM images measured by FIB-TEM technology. From this image, the distance between the AuNPs and the cavity surface was estimated to be 4 nm. The DNA hybridization complex formed in wet conditions has 10–15 nm but is reduced to less than 5 nm in dry conditions. This reduction in DNA length under dry conditions has also been previously reported (Li et al., 2019; 19, 4314; Lim et al., 2009). We also confirmed the average distance for nanogaps between the AuNPs and the cavities from their TEM images. Figs. S3c and S3d show the FIB-TEM images for eight different nanogaps. The corresponding average distance and standard deviation were evaluated to be 3.89 nm and 0.36 nm, respectively.
We compared the Raman mapping images of the empty and AuNP-filled AuNDS in Fig. 2 . A 633-nm He–Ne laser was used as the light source, and Raman mapping images were measured for a total of 2500 pixels over an area of 250 μm × 250 μm at 5-μm intervals for each substrate, as shown in Fig. 2a. Fig. 2b and c shows the Raman mapping and corresponding SEM images of the empty and AuNP-filled AuNDS, respectively. The images show the results of the SERS-PCR assays for the RdRp gene, which is a representative biomarker of SARS-CoV-2. In this experiment, 105 copies/μL of the RdRp target gene was amplified over 25 PCR cycles, and then SERS detection was performed on the bridge DNA remaining in the solution for 0 (top) and 25 cycles (bottom). As shown in the SEM image (top) in Fig. 2c, when the PCR cycle was 0, the bridge DNAs remained in the PCR microtube without fragmentation, forming a hybridization complex with capture and detection DNAs in the curved cavities of the AuNDS.
Fig. 2.
Schematic illustration of acquisition of Raman mapping image. (a) Raman mapping was performed using a 633-nm He–Ne laser (laser power = 1.45 mW). The mapping area was 250 μm × 250 μm, and a total of 2500 pixels were imaged at 5-μm intervals (i.e., the size of one pixel was 5 μm × 5 μm). (b) Raman mapping images for remaining bridge DNAs before (top) and after (bottom) PCR amplification. (c) Corresponding magnified SEM images for one pixel (5 μm × 5 μm). The initial concentration of the target RdRp DNA was 105 copies/μL, and 25 PCR cycles were run.
In contrast, when the PCR cycle increased to 25, most of the bridge DNA was decomposed by hydrolysis, and AuNPs could not enter the AuNDS cavities through DNA hybridization as shown in the SEM image (bottom) in Fig. 2c.
We also examined the reproducibility of the Raman signals on the substrates. Fig. 3 a is a photograph of the Raman mapping images of five different samples of AuNP-internalized AuNDS, captured using a Raman microscope. First, we tested the reproducibility of five different substrate samples. As shown in Fig. 3b, these five pieces of substrate represent a uniform decoding color distribution; therefore, the substrate-to-substrate fluctuation of the Raman signal on the five substrates is minimal. Fig. 3c shows the Raman spectra for 2500 spots on the first substrate sample. Fig. 3d (left) shows the histogram for the average values of 2500 Raman intensities measured for 250 μm × 250 μm regions on each substrate sample. The Raman substrates exhibit excellent reproducibility, with RSD = 4.64% for the five 5 mm × 5 mm substrates. Second, the spot-to-spot reproducibility for a substrate was also investigated. Fig. 3d shows the histogram for the average values of 2500 Raman peak intensities measured for five randomly selected 250 μm × 250 μm regions on the first substrate. The relative standard deviation (RSD) for the spot-to-spot fluctuation was estimated to be 6.75%. Therefore, the spot-to-spot fluctuations of the Raman signal for different spots on the same substrate were also minimal.
Fig. 3.
Reproducibility tests of AuNP-internalized AuNDS. (a) Photograph of acquisition of Raman mapping images of five different AuNPs-internalized AuNDS samples. (b) Raman mapping images of five randomly selected 250 μm × 250 μm regions on first 5 mm × 5 mm substrate sample for the reproducibility test of spot-to-spot fluctuations. (c) Raman spectra for 2500 spots of the first substrate sample. (d) Histogram of average values of a total of 12,500 Raman intensities measured for five randomly selected 250 μm × 250 μm regions on five substrate samples for reproducibility test of substrate-to-substrate fluctuations.
Fig. 4 shows the results of the SERS-PCR assay for the SARS-CoV-2 E and RdRp genes. Table 1 lists the base sequences for the forward/reverse primer genes, capture/detection DNA probes, and bridge DNAs for the E and RdRp target genes used in the SARS-CoV-2 assays (Corman et al., 2020). In RT-PCR, the DNA concentration is amplified as the cycle number increases. When the fluorescence signal intensity exceeds the threshold level, the corresponding cycle number is called the cycle threshold (Ct) value, and at this time, it is determined as positive. Therefore, it is impossible to take the signal of the initial concentration as a reference or control because we cannot measure the fluorescence signal of the unamplified initial concentration because its fluorescence intensity is too weak to be detected by the current optical detection method. In SERS-PCR, Raman signal was also measured while increasing the number of PCR cycles, and a calibration curve was determined by measuring the Raman signal intensity for each process. Herein, label-free bridge DNA probes were used as hydrolysis agents instead of the fluorescent dye (FAM)-labeled TaqMan probes used in RT-PCR. Eight T-spacers were used in the capture/detection DNA terminals to quickly form a sandwich DNA hybridization complex between the SERS nanotag and the cavity surface. The concentrations of the capture DNA, SERS nanotag, bridge DNA, SSC buffer solution, and the reaction temperature and time were varied to determine the optimal reaction conditions for the DNA hybridization complex formed in the nanogap. Fig. S4 shows the obtained SERS measurements as histograms, with the optimal conditions for each being marked in red. Since the clinical trial results showed that the average concentration of E and RdRp viral RNA genes in patients undergoing the SARS-CoV-2 test is 105 copies/μL (Nadège et al., 2021; Jin et al., 2020), we set this concentration as the initial concentration of the target genes.
Fig. 4.
SERS spectra for remaining bridge DNAs measured by AuNP- internalized AuNDS. Initial concentrations of 105 copies/μL target DNAs for E gene (a) and RdRp gene (b) were sequentially amplified by varying the cycle count from 0 to 10.
Table 1.
Base sequences of forward/reverse primers, capture/detection DNA probes, and bridge DNAs for E and RdRp genes used in AuNDS-based SERS-PCR assays.
| Target genes and Probe DNAs | Base sequence (5'- 3') | Fragment size (bp) |
|---|---|---|
| E gene | NC_004718 (26141-26269) | 128 |
| Forward primer | ACAGGTACGTTAATAGTTAATAGCGT | 26 |
| Reverse primer | ATATTGCAGCAGTACGCACACA | 22 |
| Capture DNA probe | GATGGCTAGTGTTTTTTTTT-(CH2)3-SH | 20 |
| Detection DNA probe | SH-(CH2)6-TTTTTTTTCGAAGCGCAGTA | 20 |
| Bridge DNA |
iSP3-ACACTAGCCATCCTTACTGCGCTTCG-iSP3 |
26 |
| RdRp gene | MN908947 (15431-15531) | 100 |
| Forward primer | GTGARATGGTCATGTGTGGCGG | 22 |
| Reverse primer | CARATGTTAAASACACTATTAGCATA | 26 |
| Capture DNA probe | AGGTTCCACCTGTTTTTTTT-(CH2)3-SH | 20 |
| Detection DNA probe | SH-(CH2)6-TTTTTTTTGCATCTCCTGAT | 20 |
| Bridge DNA | iSP3-CAGGTGGAACCTCATCAGGAGATGC-iSP3 | 25 |
As shown in Fig. 4, 105 copies/μL target DNAs for the E gene (a) and RdRp gene (b) were sequentially amplified by varying the thermocycling number from 0 to 10, and the SERS spectra for the remaining bridge DNAs were measured using AuNP-internalized AuNDS. The SERS spectra shown on the right exhibit the characteristic Raman peak intensity at 1616 cm-1 of the Raman reporter MGITC as the cycling number increased. In both the E and RdRp genes, as the cycling number increased, the bridge DNA concentration decreased, and thus the SERS peak intensity also tended to decrease. In addition, Raman mapping images were obtained for the change in the number of thermocycles to confirm the reproducibility of the SERS measurement of the AuNDS for the difference in the concentration of the target E and RdRp gene concentrations. The results are shown in Fig. S5. As shown in this figure, the decoding color consistently changed from bright yellow to dark red with the increase in the cycling number for both the E and RdRp genes. Therefore, we can conclude that our SERS-PCR detection method for the target genes has consistent reproducibility.
Fig. S6 also shows the variation of Raman spectra when there was no target gene in the microtube. Variations in Raman signal intensity were observed while changing the PCR cycle number from 0 to 10 in the same way as when 105 copies/μL of the target gene was present. As expected, there was no change in bridge DNA concentration in the absence of target DNA, and no change in Raman signal intensity was observed.
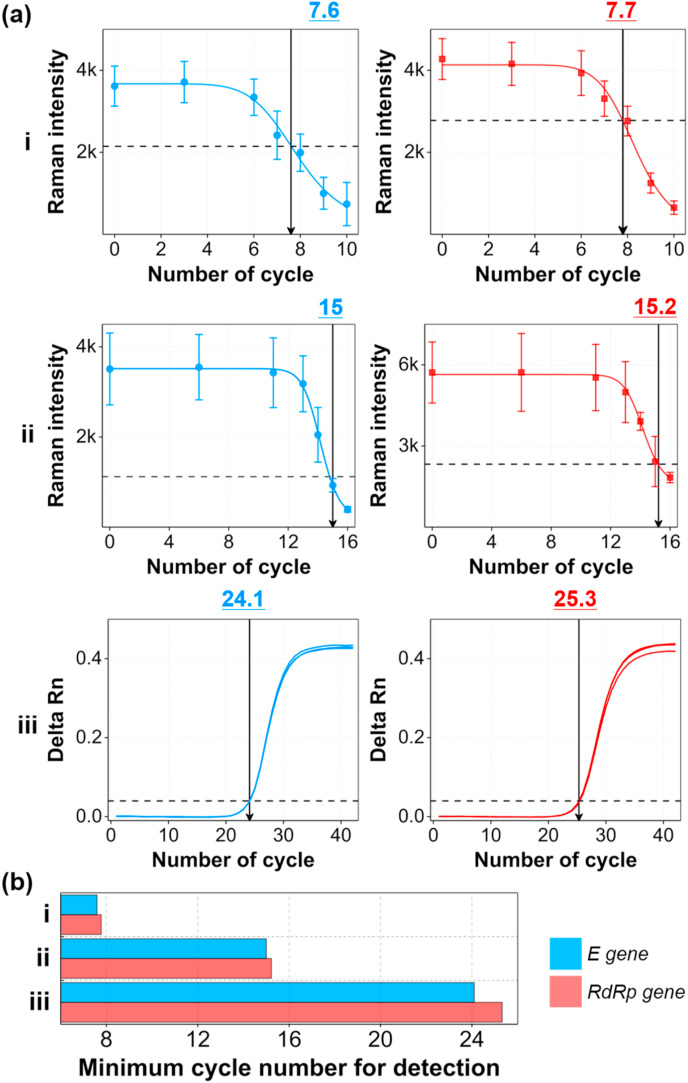
Based on the Raman spectral data shown in Fig. 4, we determined the SERS-PCR calibration curves of the E and RdRp genes for the change in the thermocycling number, and the results are shown in Fig. 5 a. In addition, the calibration curves determined by the RT-PCR method and the SERS-PCR method using magnetic beads for the same initial concentration of E and RdRp genes were compared, as shown in Fig. 5a, to compare their detection sensitivities. Fig. S7 is a schematic of the SERS-PCR assay using magnetic beads (a) and the conventional RT-PCR assay (b). A SERS-PCR assay using magnetic beads was recently reported (Wu et al., 2020; Wee et al., 2016). Herein, capture DNA-conjugated magnetic beads, and detection DNA-conjugated AuNP SERS nanotags were used as supporting substrates and detection probes, respectively. Capture and detection DNAs were designed to hybridize with a bridge probe in a sandwich format. After forming a sandwich hybridization complex for the remaining bridge DNA between the magnetic beads and SERS nanotags, the SERS signal was measured to determine the concentration of the target gene. (Fig. S7a). In contrast, the concentration of the target gene was determined by measuring the fluorescence signal of the dyes fragmented from the TaqMan probes along with the amplification of the target gene in the RT-PCR (Fig. S7b). The corresponding calibration curves for the SARS-CoV-2 E and RdRp genes, determined from two different types of SERS-PCR assays and RT-PCR assays, are displayed in Fig. 5a. In the magnetic bead-based SERS-PCR assays, error bars were determined by measuring the SERS intensity three times for each cycling count. On the other hand, error bars were determined from the Raman peak intensities for 2500 SERS data in the Raman mapping images in the AuNDS-based SERS-PCR. Unlike RT-PCR, the concentration of the bridge DNA decreases with the number of cycles in SERS-PCR; thus, the SERS intensity decreases. In addition, the threshold values corresponding to the Ct values in RT-PCR, which is a criterion for discriminating between a positive/negative SARS-CoV-2 infection, and the corresponding cycle counts were determined from the calibration curves.
Fig. 5.
SERS-PCR calibration curves for E and RdRp genes for different cycle counts. (a) Comparison of calibration curves determined by SERS-PCR using AuNDS (i), magnetic beads (ii), and RT-PCR (iii). The threshold lines in SERS-PCR were determined from the LoD of each target DNA. (b) Histogram of minimum cycle counts needed to reach the threshold value for E and RdRp DNA genes for two different types of SERS-PCR and RT-PCR assays. The initial concentration (cycle number = 0) for both E and RdRp genes was 105 copies/μL.
The threshold value in the SERS-PCR was determined from the limit of detection (LoD) of each target DNA. When this threshold value is reached at a small cycle count, sensitive detection is possible even with a small amount of the target DNA. Fig. 5b shows the minimum cycling count needed to attain the threshold value that can discriminate between positive and negative for E and RdRp DNA genes for two different types of SERS-PCR and RT-PCR assays. When the target DNA concentration was 105 copies/μL, the corresponding cycle counts needed to reach the threshold values for RT-PCR, magnetic bead-based SERS-PCR, and AuNDS-based SERS-PCR were 25, 15, and 8, respectively. In PCR assays, the concentration of the target gene is increased 2N times as the cycle number (N) increases. In the case of Au NDS-based SERS-PCR, the detection was possible after 8 cycles when the initial concentration of the target gene was 105 copies/μL, and the concentration of the target gene at that time was 105 × 28 = 2.56 × 107 copies/μL (8 cycles). When calculated for magnetic bead-based SERS-PCR and RT-PCR similarly, the values were 3.28× 109 (15 cycles) and 3.36 × 1012 (25 cycles) copies/μL, respectively. This means that the AuNDS-based SERS-PCR assay has a sensitivity that is 102 and 105 orders of magnitude better than magnetic bead-based SERS-PCR and RT-PCR, respectively.
On the other hand, the concentration at which patients begin to feel symptoms is 102 copies/μL. To confirm the diagnostic feasibility of our AuNDS-based SERS-PCR platform at this low concentration, we also evaluated the performance for (i) AuNDS-based SERS-PCR, (ii) magnetic bead-based SERS-PCR, and (iii) RT-PCR at this low concentration (Fig. S8a). The corresponding thermocycling numbers of the E and RdRp genes to reach the threshold value were estimated to be 18, 25∼26, and 34∼36, respectively (Fig. S8b). This result also demonstrates that the Au NDS-based SERS-PCR assay has a sensitivity much better than magnetic bead-based SERS-PCR and RT-PCR for the low SARS-CoV-2 concentration sample.
Therefore, we can dramatically reduce the diagnostic time required for SARS-CoV-2 in that we can distinguish between SARS-CoV-2 positive and negative samples by amplifying the target gene over only a small number of thermocycles. In addition to the shorter detection time, our Au NDS-based SERS-PCR assays is expected to improve accuracy than RT-PCR. Because the long PCR thermocycling steps increase the concentration of some non-specific genes as well as the concentration of the target gene, the intensity of the background noise increases. Since Au NDS-based SERS-PCR can reduce the cycle number for amplification, the signal-to-noise ratio of the target gene is also improved, which enables more accurate analysis. Nonetheless, it is difficult for the proposed Au NDS-based SERS-PCR platform to replace the RT-PCR in the current state. This work presented a proof-of-concept of reducing the overall diagnosis time by shortening the PCR amplification step, which takes the longest in RT-PCR assay. The current problems for the use of humid chambers and high-temperature conditions for the hybridization process might be solved using microfluidic technology in the future. We are developing a point-of-care diagnostic system that integrates a microfluidic device with a portable Raman spectrophotometer. Here, the virus pretreatment and PCR processes will be implemented into the chip using a microfluidic device. We expect that an automatic point-of-care assay system that can detect viruses rapidly and accurately in the field might be realized by integrating this microfluidic device into the portable Raman system.
4. Conclusions
In this study, we developed a new SARS-CoV-2 molecular diagnostic technology that combines Au nanoparticles synthesized by a bottom-up approach with AuNDS fabricated by a top-down approach. Strong electromagnetic effects were generated in the nanogaps formed by inserting 80-nm AuNPs into the AuNDS. The E and RdRp target genes were quantified by measuring the SERS signal of the hybridization complexes for the bridge DNA remaining after successive hybridization reactions. According to our SERS detection data, the number of cycles required to reach the threshold value that can discriminate between positive and negative patients were 25 cycles for RT-PCR, 15 cycles for magnetic bead-based SERS-PCR, and 8 cycles for AuNDS-based SERS-PCR when the initial DNA concentration was 1.00 × 105 copies/μL. The corresponding detection concentrations were estimated to be 3.36 × 1012, 3.28 × 109, and 2.56 × 107 copies/μL, respectively. That is, it is possible to distinguish between positive and negative patients after only eight cycles of amplification by using AuNDS-based SERS-PCR. Although the molecular diagnostic results obtained using the AuNDS described in this work are still in the proof-of-concept stage, it is seen as being a novel molecular diagnostic platform technology that should be able to dramatically shorten the diagnostic time relative to conventional RT-PCR.
CRediT authorship contribution statement
Yixuan Wu: Conceptualization, Validation, Formal analysis, Investigation, Writing – original draft. Hajun Dang: Validation, Formal analysis, Investigation. Sung-Gyu Park: Funding acquisition, Methodology, Investigation. Lingxin Chen: Conceptualization, Methodology, Writing – review & editing. Jaebum Choo: Funding acquisition, Conceptualization, Methodology, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the National Research Foundation of Korea (Grant Numbers 2019R1A2C3004375 and 2020R1A5A1018052) and by the Ministry of Trade, Industry and Energy (Grant Number: 20012405). National Natural Science Foundation of China of China (Grant Number 21976209) and Taishan Scholar Project Special Funding (Grant Number ts20190962) also supported this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113736.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bastús N.G., Comenge J., Puntes V. Langmuir. 2011;27:11098–11105. doi: 10.1021/la201938u. [DOI] [PubMed] [Google Scholar]
- Baumberg J.J., Aizpurua J., Mikkelsen M.H., Smith D.R. Nat. Mater. 2019;18:668–678. doi: 10.1038/s41563-019-0290-y. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang Y., Shih T.-M., Yang W., Hu S., Hu X., Li J., Ren B., Mao B., Yang Z., Tian Z. Nano Lett. 2018;18:2209–2216. doi: 10.1021/acs.nanolett.7b04385. [DOI] [PubMed] [Google Scholar]
- Chen H., Park S.-G., Choi N., Kwon H.-J., Kang T., Lee M.-K., Choo J. ACS Sens. 2021;6:2378–2385. doi: 10.1021/acssensors.1c00596. [DOI] [PubMed] [Google Scholar]
- Chen J., Gao K., Wang R., Wei G.-W. Chem. Sci. 2021;12:6929–6948. doi: 10.1039/d1sc01203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Euro Surveill. 2020;25:23–30. [Google Scholar]
- Corman V.M., Haage V.C., Bleicker T., Schmidt M.L., Mühlemann B., Zuchowski M., Jo W.K., Tscheak P., Möncke-Buchner E., Müller M.A., Krumbholz A., Drexler J.F., Drosten C. Lancet Microbe. 2021;2:e311–e319. doi: 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H., Park S.-G., Wu Y., Choi J.-Y., Yang S., Lee S., Joo S.-W., Chen L., Choo J. Adv. Funct. Mater. 2021;31:2105703. [Google Scholar]
- Faulds K., Barbagallo R.P., Keer J.T., Smith W.E., Graham D. Analyst. 2004;129:567–568. doi: 10.1039/b406423b. [DOI] [PubMed] [Google Scholar]
- Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L., Parker M., Bonsall D., Fraser C. Science. 2020;368 doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodake G.S., Shinde S.K., Kadam A.A., Saratale R.G., Saratale G.D., Syed A., Elgorban A.M., Marraiki N., Kim D.-Y. Biosens. Bioelectron. 2021;177:112969. doi: 10.1016/j.bios.2021.112969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock J., Robertson D.L. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hwang M.J., Jang A.S., Lim D.-K. Sensor. Actuator. B. 2021;329:129134. [Google Scholar]
- Jin A., Yan B., Hua W., Feng D., Xu B., Liang L., Guo C. Biosaf. Heal. 2020;2:104–111. doi: 10.1016/j.bsheal.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kashefi-Kheyrabadi L., Joung Y., Kim K., Dang H., Chavan S.G., Lee M.-H., Choo J. Sensor. Actuator. B. 2021;329:129214. doi: 10.1016/j.snb.2020.129214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Deng T.-S., Liu X., Dolan J.A., Scherer N.F., Nealey P.F. Nano Lett. 2019;19:4314–4320. doi: 10.1021/acs.nanolett.9b00792. [DOI] [PubMed] [Google Scholar]
- Lim D.-K., Jeon K.-S., Kim H.M., Nam J.-M., Suh Y.D. Nat. Mater. 2009;9:60–67. doi: 10.1038/nmat2596. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Néant N., Lingas G., Le Hingrat Q., Ghosn J., Engelmann I., Lepiller Q., Gaymard A., Ferré V., Hartard C., Plantier J.-C., Thibault V., Marlet J., Montes B., Bouiller K., Lescure F.-X., Timsit J.-F., Faure E., Poissy J., Chidiac C., Raffi F., Kimmoun A., Etienne M., Richard J.-C., Tattevin P., Garot D., Le Moing V., Bachelet D., Tardivon C., Duval X., Yazdanpanah Y., Mentré F., Laouénan C., Visseaux B., Guedj J. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2017962118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Guzman C.A., Ruiz-Pernia J.J., Tunon I. Chem. Sci. 2021;12:3489–3496. doi: 10.1039/d0sc04978f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitjar J., Liao J.-D., Lee H., Tsai H.-P., Wang J.-R., Liu P.-Y. Biosens. Bioelectron. 2021;181:113153. doi: 10.1016/j.bios.2021.113153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., de Sousa N.R., Papanicolaou N., Vondracek M., Aarum J., Safari H., Muradrasoli S., Rothfuchs A.G., Albert J., Högberg B., Reinius B. Nat. Commun. 2020;11:4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Nat. Rev. Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zong S., Wu L., Zhu D., Cui Y. Chem. Rev. 2017;117:7910–7963. doi: 10.1021/acs.chemrev.7b00027. [DOI] [PubMed] [Google Scholar]
- Wee E.J.H., Wang Y., Tsao S.C.-H., Trau M. Theranostics. 2016;6:1506–1513. doi: 10.7150/thno.15871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Choi N., Chen H., Dang H., Chen L., Choo J. Anal. Chem. 2020;92:2628–2634. doi: 10.1021/acs.analchem.9b04522. [DOI] [PubMed] [Google Scholar]
- Xu L., Li D., Ramadan S., Li Y., Klein N. Biosens. Bioelectron. 2020;170:112673. doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Li X., Pan J., Zhang Y., Zhang L., Wang C., Yan X., Liu X., Lu G. Biosens. Bioelectron. 2021;190:113421. doi: 10.1016/j.bios.2021.113421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. N. Engl. J. Med. 2019;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.