Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for the disease coronavirus-19 disease (COVID-19) has wreaked havoc on the health and economy of humanity. In addition, the disease is observed in domestic and wild animals. The disease has impacted directly and indirectly every corner of the planet. Currently, there are no effective therapies for the treatment of COVID-19. Vaccination to protect against COVID-19 started in December 2020. SARS-CoV-2 is an enveloped virus with a single-stranded RNA genome of 29.8 kb. More than two-thirds of the genome comprise Orf1ab encoding 16 nonstructural proteins (nsps) followed by mRNAs encoding structural proteins, spike (S), envelop (E), membrane (M), and nucleocapsid (N). These genes are interspaced with several accessory genes (open reading frames [Orfs] 3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10). The functions of these proteins are of particular interest for understanding the pathogenesis of SARS-CoV-2. Several of the nsps (nsp3, nsp4, and nsp6) and Orf3a are transmembrane proteins involved in regulating the host immunity, modifying host cell organelles for viral replication and escape and hence considered drug targets. In this paper, we report mapping the transmembrane structure of the nsps of SARS-CoV-2.
Keywords: coronavirus, COVID-19, double-membrane vesicle, endoplasmic reticulum, nonstructural proteins, nsp3, nsp4, nsp6, nsps, Orf1ab, Orf3a, SARS-CoV-2
1. INTRODUCTION
The coronavirus-19 disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019 is responsible for the pandemic and is a major public health concern. The disease has resulted in mortality, massive hospitalizations, lockouts, economic loss, employment layoffs, school and college closings, and have impacted all the countries. The SARS-CoV-2 virus not only infects humans, but also companion and wild animals. Even after a year, there are no effective therapies for the treatment of the disease (Thomas, 2020). Current effective strategies to decrease the incidence of the disease is social distancing, use of masks, and lockdowns in seriously affected areas. The first vaccines to protect against COVID-19 were rolled out in December 2020. All the vaccines currently available are based on the spike protein (S). However, there is a risk that the vaccines may not be effective against mutant strains of the virus. Recent reports suggest that the AstraZeneca vaccine do not provide complete protection against the South African strain (501Y.V2) of SARS-CoV-2.
People with diabetes are at risk of the disease. As yet we do not know why the virus is highly successful in causing the pandemic within 3 months of its first report (Thomas, 2020). Understanding the structure and function of the proteins of SARS-CoV-2 will lead to the development of effective vaccines and drugs to protect against the virus.
The structural proteins of SARS-CoV-2 include membrane glycoprotein (M), envelope protein (E), nucleocapsid protein (N), and the spike protein (S). SARS-CoV-2 contains a 29.8-kb single-stranded RNA genome wrapped in a helical nucleocapsid composed of multiple copies of N protein, which in turn is surrounded by an envelope containing S glycoprotein, M glycoprotein, and a small E protein. The viral gene order is similar to that in other known coronaviruses, with the first two open reading frames (Orfs) (1a and 1b) encoding the viral replicase and the downstream mRNAs encoding the structural proteins. Using bioinformatics, we previously showed that the M proteins of SARS-CoV-2 resemble the sugar transporters that may be involved in functions favorable to the virus (Thomas, 2020). The nonstructural proteins (nsps) of SARS-CoV-2 are involved in inhibiting innate immunity and also induce virus replication. Two overlapping Orfs, Orf1a and Orf1b, are translated from the positive-strand genomic RNA and generate continuous polypeptides, which are cleaved into a total of 16 nsps. The genes coding for the structural proteins are interspaced with several accessory genes, including the Orfs 3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10. The functions of these proteins are of particular interest for understanding the pathogenesis of SARS-CoV-2 (Gordon et al., 2020; Yoshimoto, 2020).
To evade detection by host innate immune sensors, viruses that replicate in the cytoplasm compartmentalize their genome transcription in organelle-like structures, thereby protecting the virus against host cell defenses and increasing the replication efficiency (Belov and van Kuppeveld, 2012). The transmembrane nsp3, nsp4, and nsp6 are known to rearrange endoplasmic reticulum (ER) membranes thereby inducing curvature of the ER membrane, essential for virus replication. Lack of antiviral therapies is the paucity of knowledge regarding the β-coronavirus–host cell interface (Ghosh et al., 2020). In this paper, we provide information on the transmembrane and lumen domains of the nsps of SARS-CoV-2.
2. MATERIALS AND METHODS
2.1. SARS-CoV-2 transmembrane nsp structure
The transmembrane nsp sequences of the SARS-CoV-2 were downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/protein/) protein database. The transmembrane nsps of SARS-COV-2 include nsp3 (accession no. YP_009725299), nsp4 (accession no. YP_009725300), nsp6 (accession no. YP_009725302), and Orf3a (accession no. BCI50534).
2.2. Protein modeling
For a comprehensive understanding of biological systems of how proteins complexes and networks operate a detailed description of the interactions of proteins and the overall quaternary structure is essential. Residue-based diagrams of proteins, also called snake diagrams or protein plots, are 2-D representations of a protein sequence that contain information about properties such as secondary structure (Skrabanek et al., 2003). To determine a snake diagram model of a protein we used Protter (http://wlab.ethz.ch/protter). Protter is an interactive and customizable web-based application that enables the integration and visualization of both annotated and predicted protein sequence features together with experimental proteomic evidence for peptides and post-translational modifications onto the transmembrane topology of a protein. It allows users to choose from numerous annotation sources, integrate their own proteomics data files, select the best-suited peptides for targeted quantitative proteomics applications, and export publication-quality illustrations (Omasits et al., 2014).
For the three-dimensional homology modeling we employed the iterative threading assembly refinement (I-TASSER) (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) with default settings. The protein sequences of SARS-CoV-2 was entered in FASTA format.
We used multiple bioinformatics software for the prediction of transmembrane regions of membrane proteins. Secondary structure prediction system of membrane proteins (SOSUI) discriminates membrane and soluble proteins and predicts transmembrane proteins based on the nature of the amino acids (Hirokawa et al., 1998) ( https://harrier.nagahama-i bio.ac.jp/sosui/sosui_submit.html). Transmembrane helix prediction (TMHMM) (transmembrane hidden Markov model) is a method for prediction of transmembrane helices based on a hidden Markov model (Krogh et al., 2001) (www.cbs.dtu.dk/services/TMHMM/). Subcellular Localization Predictive System (CELLO) is a multiclass support vector machines (SVMs) classification system. CELLO uses four types of sequence coding schemes: the amino acid composition, the dipeptide composition, the partitioned amino acid composition, and the sequence composition based on the physicochemical properties of amino acids. CELLO is a simple straightforward implementation of a single module (SVM) based on multiple n-peptide composition to predict subcellular localization (Yu et al., 2004) (http://cello.life.nctu.edu.tw/).
3. RESULTS
As yet there are no effective therapies and vaccines for COVID-19, to meet the increasing demand for the treatment of COVID-19 there is a need to accelerate novel antiviral drug development as quickly as possible. Target-based drug development may be a promising approach to achieve this goal (Liu et al., 2021). Identifying molecular targets could lead to development of medications that protect against SARS-CoV-2 virus.
Coronaviruses, including SARS-CoV-2 that replicate in the cytoplasm compartmentalize their genome transcription in organelle-like structures thereby protecting the virus against host cell defenses and increasing the replication efficiency (Santerre et al., 2020).
The nsps are critical elements of the replication and transcription complex (RTC), as well as immune system evasion. Through hijacking the ER membrane, nsps help the virus establish the RTC (Santerre et al., 2020). The structure and function of nsps of SARS-CoV-2 is similar to SARS-CoV. In SARS-CoV, the primary structures of the three nsps—nsp3, nsp4, and nsp6, contain hydrophobic stretches, and these proteins are predicted to be integral membrane proteins. Hence, they are likely to function in anchoring the replication complexes to the lipid bilayer (Oostra et al., 2007).
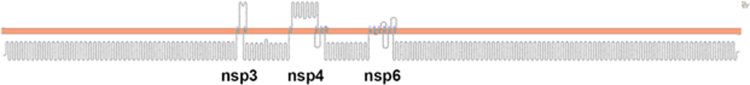
Using bioinformatics we mapped the transmembrane nsps of SARS-CoV-2. We initially mapped the Orf1ab. The Orf1ab codes for 16 nsps. The Orf1ab is cleaved into 16 nsps. Analysis of the Orf1ab by Protter showed the three transmembrane nsps—nsp3, nsp4, and nsp6 (Fig. 1).
FIG. 1. .
The topology of Orf1ab (snake diagram) determined using Protter. The Orf1ab is cleaved into 16 nsps. Analysis of the Orf1ab shows the three transmembrane nsps—nsp3, nsp4, and nsp6. The 12 transmembrane domains of the nsps are shown in Figures 2–4. nsps, nonstructural proteins.
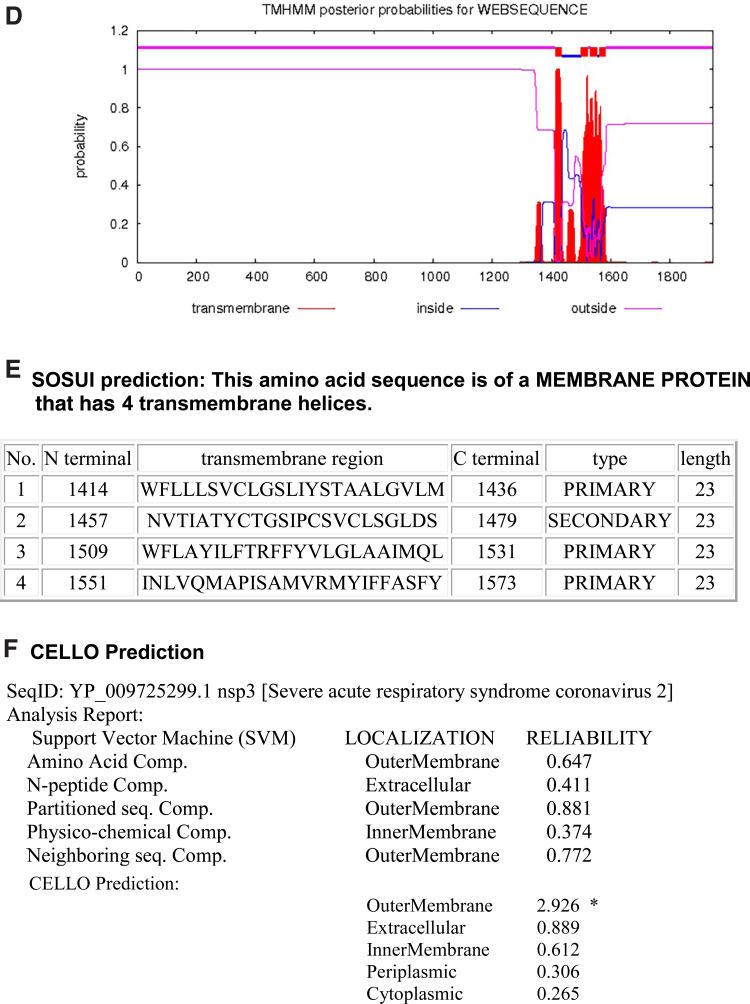
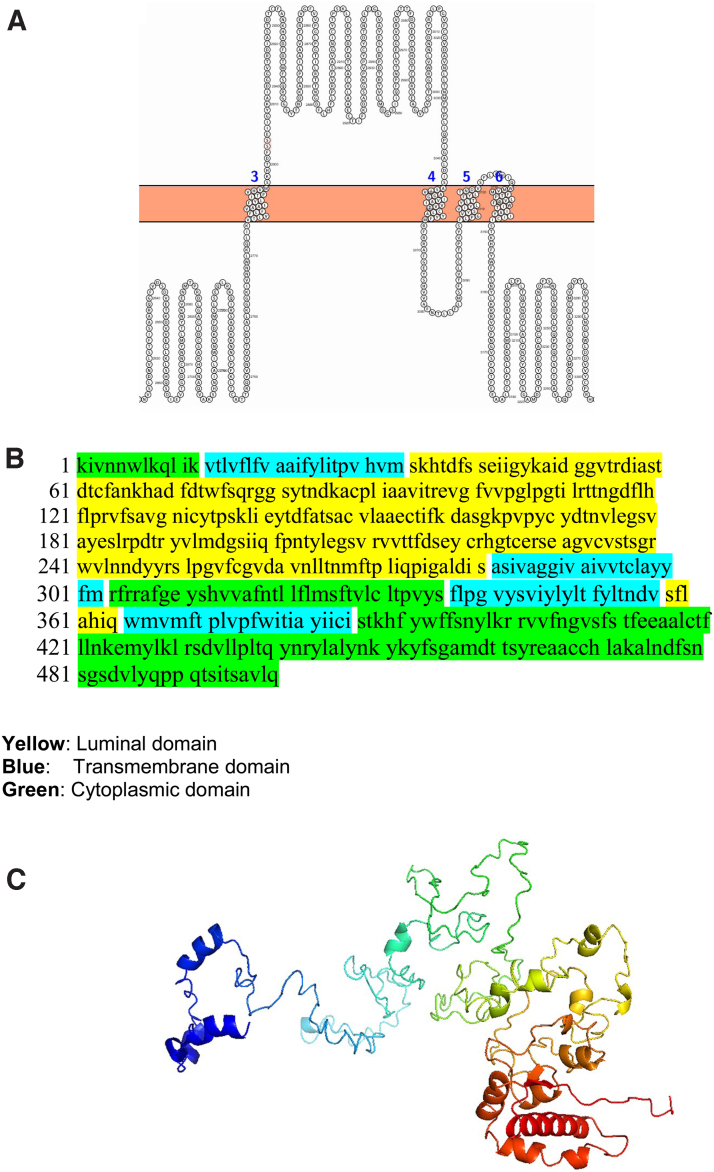
We modeled each of the transmembrane nsps. Luminal loops of the transmembrane nsps are essential for rearranging ER membranes thereby inducing curvature of the ER membrane. The nsp3 has two transmembrane domains and a luminal domain in the ER lumen. The nsp3 snake diagram, 3D model, and the various domains (cytoplasmic, transmembrane, and luminal) are shown in Figure 2. We used SOSUI, TMHMM, and CELLO bioinformatics software to determine the accuracy of the predicted transmembrane regions (Fig. 2D–F). The transmembrane regions predicted by all the bioinformatic software were similar confirming the accuracy.
FIG. 2.
The topology of nsp3 of SARS-CoV-2. (A) Membrane topology of nsp3 determined using Protter. (B) The domains (cytoplasmic, transmembrane, and luminal) of nsp3. (C) The predicted nsp3 protein structure of SARS-CoV-2 (ribbon diagram) determined using the software I-TASSER. The amino acid sequence from 1201 to 1945 of nsp3 is used for modeling the protein. The Orf1ab is cleaved into 16 nsps. The domains (1, 2) of nsp3 denote the first two transmembrane domain of Orf1ab. (D) TMHMM of nsp3. (E) SOSUI predicted the membrane has four transmembrane helices. (F) CELLO prediction of the nature of the nsp3 protein. CELLO, Subcellular Localization Predictive System; I-TASSER, iterative threading assembly refinement; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SOSUI, secondary structure prediction system of membrane proteins; TMHMM, transmembrane helix prediction.
The nsp4 is the largest transmembrane nsp and the second largest transmembrane protein of SARS-CoV-2 after the spike protein. The nsp4 protein has four transmembrane domains and a large luminal domain in the ER lumen between the first two transmembrane domains. Nsp4 also has a smaller luminal domain in the ER lumen between the third and fourth transmembrane domains (Fig. 3).
FIG. 3.
The topology of nsp4 of SARS-CoV-2. (A) Topology of nsp4 is determined using Protter. (B) The domains (cytoplasmic, transmembrane, and luminal) of nsp4. (C) The predicted nsp4 structure of SARS-CoV-2 (ribbon diagram) determined using the software I-TASSER. The Orf1ab is cleaved into 16 nsps. The domains (3–6) of nsp4 denote the transmembrane domain 3–6 of Orf1ab. (D) TMHMM of nsp4. (E) SOSUI predicted the membrane has four transmembrane helices. (F) CELLO prediction of the nature of the nsp4 protein.
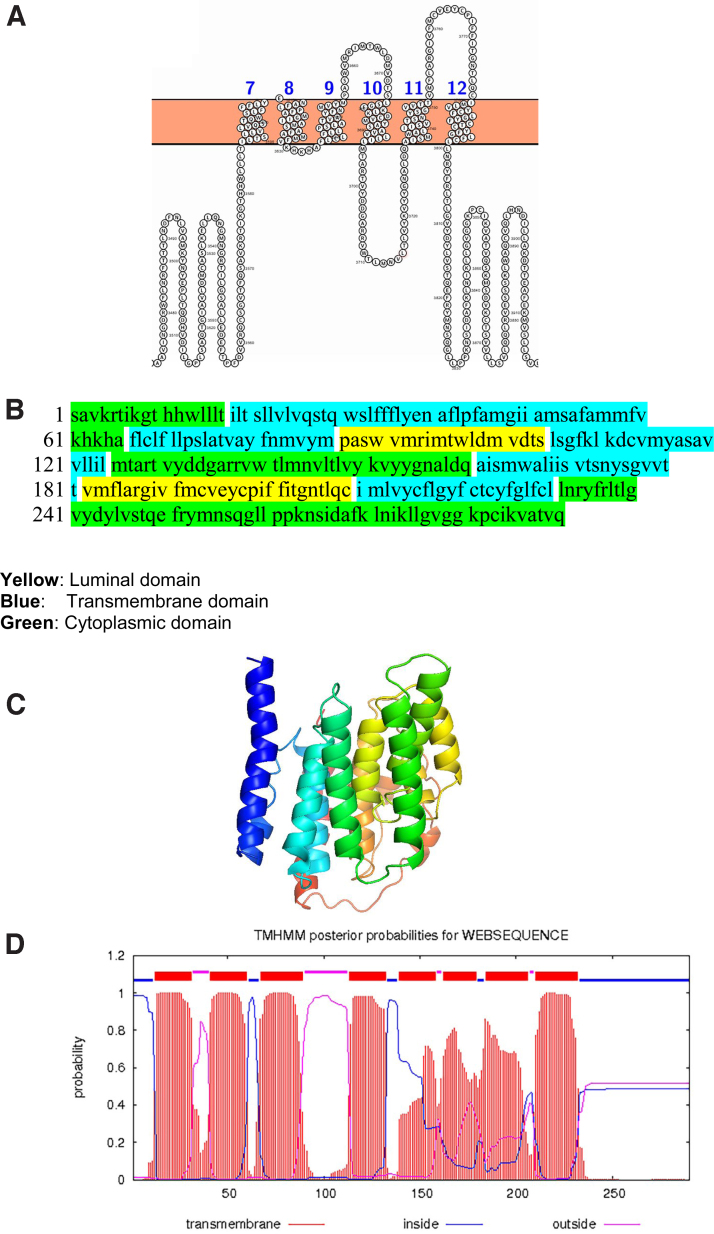
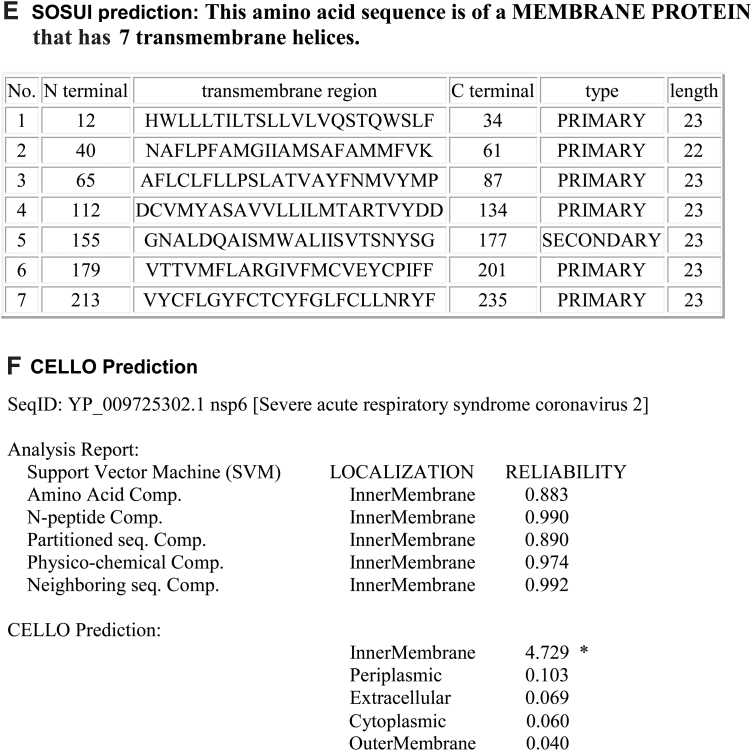
The nsp6 has six transmembrane domains and two small luminal domains between the third and fourth as well as fifth and sixth transmembrane in the ER lumen (Fig. 4).
FIG. 4.
The topology of nsp6 of SARS-CoV-2. (A) Topology of nsp6 is determined using Protter. (B) The domains (cytoplasmic, transmembrane, and ER lumen) of nsp6. (C) The predicted nsp6 protein structure of SARS-CoV-2 (ribbon diagram) determined using the software I-TASSER. The Orf1ab is cleaved into 16 nsps. The domains (7–12) of nsp6 denotes the transmembrane domain 7–12 of Orf1ab. (D) TMHMM of nsp6. (E) SOSUI predicted the membrane has seven transmembrane helices. (F) CELLO prediction of the nature of the nsp6 protein. ER, endoplasmic reticulum.
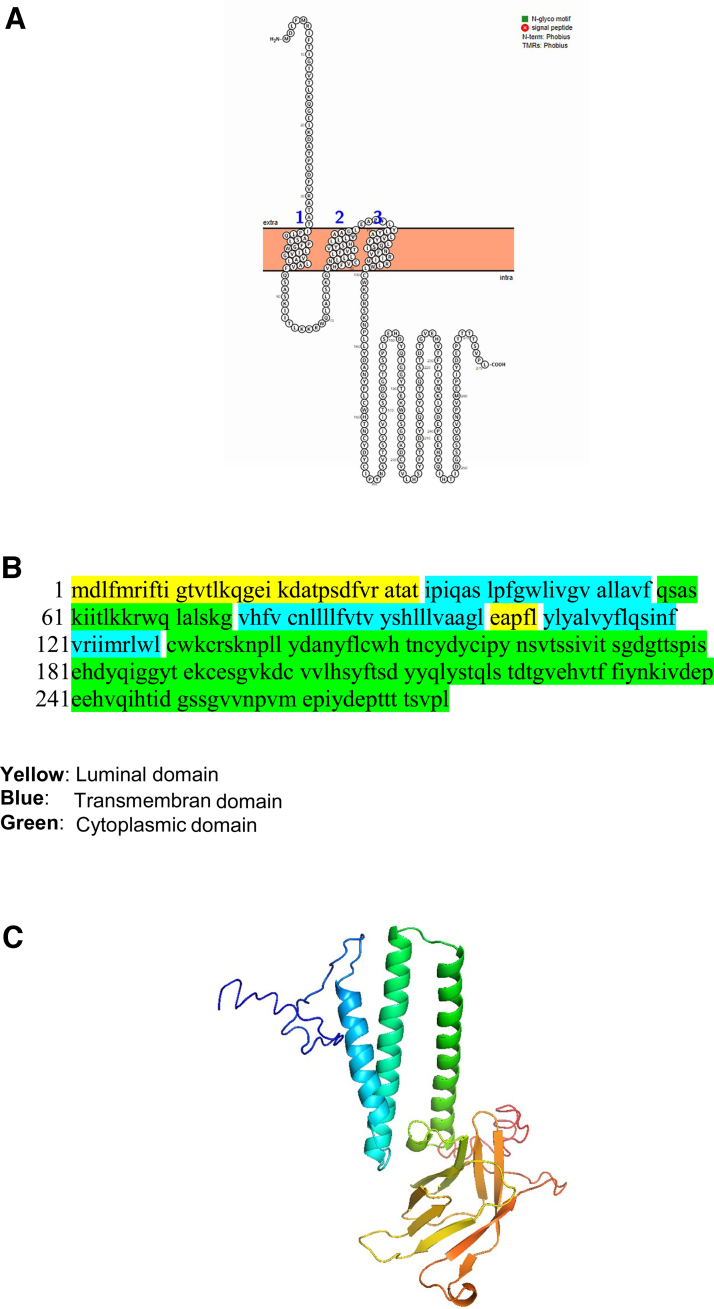
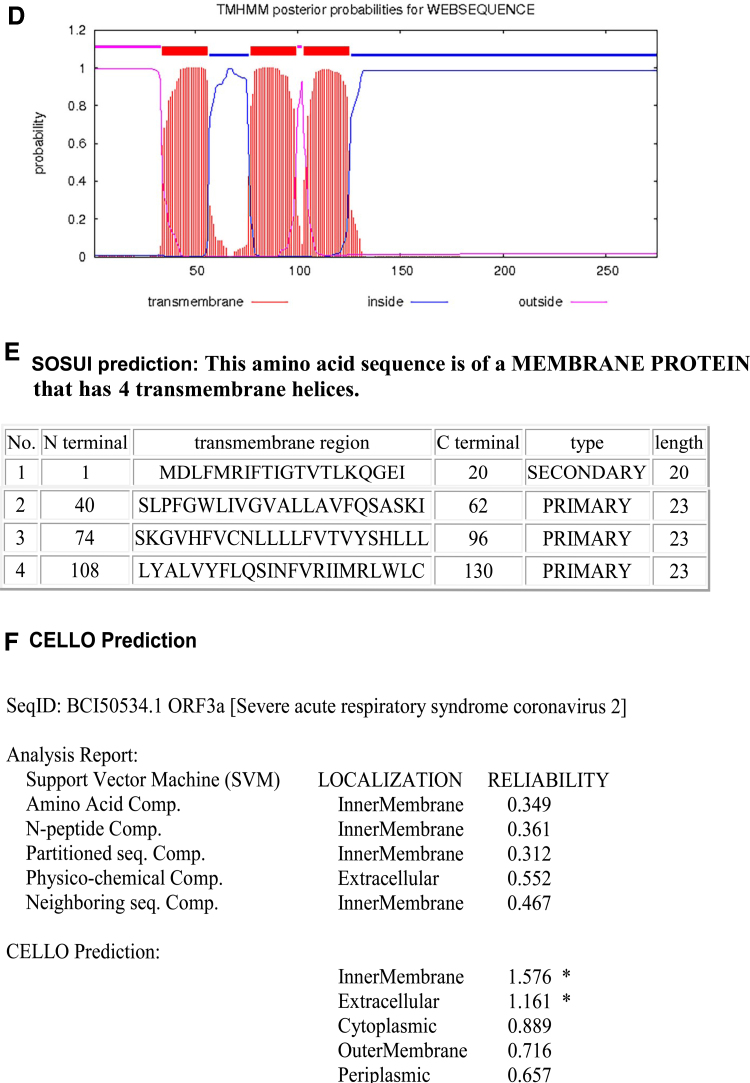
The ORF3a of SARS-CoV-2 is also a transmembrane protein. The Orf3a has three transmembrane domains and a long and short luminal domain jutting into the ER lumen (Fig. 5).
FIG. 5.
The topology of Orf3a of SARS-CoV-2. (A) The topology of Orf3a of SARS-CoV-2 determined using Protter. (B) The domains (cytoplasmic, transmembrane, and luminal) of Orf3a of SARS-CoV-2. (C) The predicted Orf3a protein structure of SARS-CoV-2 (ribbon diagram) determined using the software I-TASSER. (D) TMHMM of Orf3a. (E) SOSUI predicted the membrane has four transmembrane helices. (F) CELLO prediction of the nature of the Orf3a protein.
The transmembrane domains of the nsps of SARS-CoV-2 is responsible for inhibiting the host immunity as well as increasing the replication efficiency of the virus. In this paper, we have mapped the transmembrane nsps of SARS-CoV-2 and they could be used as a target to inhibit virus replication.
4. DISCUSSION
The COVID-19 pandemic caused by the SARS-CoV-2 virus has immobilized the world. It is the most severe pandemic of the twentieth century. As of the first week of December 2020, the virus has infected 63 million people, with 1.5 million deaths worldwide. The disease is more severe in old people compared with children and young adults. Currently there are no therapies and vaccines for the disease; hence, there is an urgent need to develop effective therapies and vaccines for the deadly disease.
The four structural proteins of SARS-CoV-2 are spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. In addition, the virus consists of a large polyprotein, Orf1ab that proteolytically cleave to form 16 nsps. There are also accessory proteins: Orf3a, Orf3b, Orf6, Orf7a, Orf7b, Orf8, Orf9b, Orf9c, and Orf10. Although accessory proteins have been viewed as dispensable for viral replication in vitro, some have been shown to play an important role in virus–host interactions in vivo (Gordon et al., 2020; Yoshimoto, 2020).
The entry of the SARS-CoV-2 into cells starts when the spike glycoprotein expressed on the viral envelope binds to ACE2 receptor of the host cell. The virus enters the cells through endocytosis process, which is possibly facilitated, through a pH-dependent endosomal cysteine protease cathepsins. Once inside the cells, SARS-CoV-2 exploits the endogenous transcriptional machinery of host cells to replicate and spread inside the cell. The virus activates or hijacks the intracellular pathways of the host in favor of its replication (Sureda et al., 2020).
Knowledge on the structure of the structural proteins of SARS-CoV-2 is essential to the development of vaccines. Most of the vaccines that protect against SARS-CoV-2 developed in the laboratory are based on the S protein (Gu et al., 2020; McKay et al., 2020; van Doremalen et al., 2020). Several commercial entities are also developing the vaccines based on the spike mRNA, adenovector, or recombinant protein.
In a previous article we showed the structures of the structural proteins of SARS-CoV-2. In silico analysis showed that the M protein of SARS-CoV-2 resembled the prokaryotic sugar transporter SemiSWEET (Thomas, 2020). The nsps are involved in inhibiting host innate immunity, inducing RNA replication and virus exit. They are potential drug targets. However, the transmembrane domains of these nsps are not clearly documented. In this article, we report the domains of the nsps that may be targets for potential drug candidates.
The first nsps encoded by Orf1a/Orf1ab are papain-like proteinase (PL proteinase, nsp3) and 3-chymotrypsin-like proteinase (3CLPro protease). The PL proteinase nsp3 cleaves nsps 1–3 and the 3CLPro proteinase slices the C-terminus from nsp4 to nsp16. Nsp3 is the largest element of the replication/transcription complex (RTC). In addition to cleaving, nsp3 alters cytokine expression to decrease the host innate immune response. Nsp3, nsp4, and nsp6 form a complex to induce double-membrane vesicles (DMVs) (Rohaim et al., 2020).
Replication and transcription of the virus happen within an RTC encoded by the virus with nsps as primary constituents. Nearly all RTC elements are encoded by the large replicase gene that consists of Orf1a and Orf1ab. The early formation of the RTC is an essential step in the SARS-CoV-2 life cycle to safeguard viral genome replication and to synthesize subgenomic mRNA (Santerre et al., 2020).
SARS-CoV-2 is a positive stranded RNA virus. The virus replication is supported by the transformed ER membranes of the host cell. The virus RNA synthesis is mediated in the DMV that acts as central hubs, offers a favorable microenvironment and may protect against innate immune sensors activated by dsRNA. The newly synthesized viral RNA is exported to the cytosol through the molecular pore spanning the DMVs. The components of the pore include nsp3, nsp4, and nsp6. Finally, the nucleocapsid (N) protein package the genome of the virus (Wolff et al., 2020). In this article, we map the ER luminal domain of nsp3, nsp4, and nsp6 that may be involved in DMV formation.
Interferons (IFNs) are cytokines with strong antiviral activities and is the first line of defense against invading pathogens. Multiple nsps are involved in inhibiting IFN-I production. The nsp6 binds TANK binding kinase 1 (TBK1) to suppress interferon regulatory factor 3 (IRF3) phosphorylation, nsp13 binds and blocks TBK1 phosphorylation, and Orf6 binds importin Karyopherin α 2 (KPNA2) to inhibit IRF3 nuclear translocation. SARS-CoV-2 nsp1 and nsp6 suppress IFN-I signaling more efficiently than SARS-CoV and Middle East respiratory syndrome coronavirus (Xia et al., 2020).
The Orf3a protein is expressed abundantly in infected and transfected cells, which localizes to intracellular and plasma membranes (Hassan et al., 2020). ORF3a induce apoptosis of cells mediated through caspase 3 (Ren et al., 2020). Issa et al. (2020) identified six functional domains (I–VI) in the SARS-CoV-2 Orf3a protein. The functional domains were linked to virulence, infectivity, ion channel formation, and virus release. Orf 3a may also be involved in vesicle trafficking (Gordon et al., 2020). Based on our analysis, the Orf3a has three transmembrane domains and a long and short luminal domain jutting into the ER lumen.
Overall, this article maps the structure of the nsps that modify the ER to DMVs so as to induce replication and further exit the host cell. Targeting the nsp transmembrane domains will lead to the development of therapies that treat COVID-19.
ACKNOWLEDGMENT
The author thanks Abraham Thomas Foundation for providing the resources for this study.
AUTHOR DISCLOSURE STATEMENT
The author declares there are no competing financial interests.
FUNDING INFORMATION
The author received no funding for this work.
References
- Belov, G.A., and van Kuppeveld, F.J.. 2012. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr. Opin. Virol. 2, 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., Dellibovi-Ragheb, T.A., Kerviel, A., et al. 2020. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 183, 1520.e14–1535.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D.E., Jang, G.M., Bouhaddou, M., et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H., Chen, Q., Yang, G., et al. 2020. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369,1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, S.S., Choudhury, P.P., Basu, P., et al. 2020. Molecular conservation and differential mutation on ORF3a gene in Indian SARS-CoV2 genomes. Genomics 112, 3226–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S.. 1998. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379. [DOI] [PubMed] [Google Scholar]
- Issa, E., Merhi, G., Panossian, B., et al. 2020. SARS-CoV-2 and ORF3a: Nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems 5, e00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., et al. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Liu, X.H., Zhang, X., Lu, Z.H., et al. 2021. Potential molecular targets of nonstructural proteins for the development of antiviral drugs against SARS-CoV-2 infection. Biomed. Pharmacother. 133, 111035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay, P.F., Hu, K., Blakney, A.K., et al. 2020. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 11, 3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits, U., Ahrens, C.H., Müller, S., et al. 2014. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. [DOI] [PubMed] [Google Scholar]
- Oostra, M., Lintelo, E.G., Deijs, M., et al. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: Involvement of the early secretory pathway in replication. J. Virol. 81, 12323–12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y., Shu, T., Wu, D., et al. 2020. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 17, 881–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaim, M.A., El Naggar, R.F., Clayton, E., et al. 2020. Structural and functional insights into non-structural proteins of coronaviruses. Microb. Pathog. 23, 104641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santerre, M., Arjona, S.P., Allen, C.N., et al. 2020. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 1–10. Online ahead of print, DOI: 10.1007/s00415-020-10197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrabanek, L., Campagne, F., and Weinstein, H.. 2003. Building protein diagrams on the web with the residue-based diagram editor RbDe. Nucleic Acids Res. 31, 3856–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureda, A., Alizadeh, J., Nabavi, S.F., et al. 2020. Endoplasmic reticulum as a potential therapeutic target for covid-19 infection management? Eur. J. Pharmacol. 882, 173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S. 2020. The Structure of the Membrane Protein of SARS-CoV-2 resembles the sugar transporter SemiSWEET. Pathog. Immun. 5, 342–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen, N., Lambe, T., Spencer, A., et al. 2020. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, G., Limpens, R.W.A.L., Zevenhoven-Dobbe, J.C., et al. 2020. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369, 1395–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., Cao, Z., Xie, X., et al. 2020. Evasion of type I interferon by SARS-CoV-2. Cell. Rep. 33, 108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, F.K. 2020. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 39, 198–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.S., Lin, C.J., and Hwang JK.. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]