Abstract
The COVID-19 pandemic has resulted in a worldwide health crisis. Rapid diagnosis, new therapeutics and effective vaccines will all be required to stop the spread of COVID-19. Quantitative evaluation of serum antibody levels against the SARS-CoV-2 virus provides a means of monitoring a patient's immune response to a natural viral infection or vaccination, as well as evidence of a prior infection. In this paper, a portable and low-cost electrochemical immunosensor is developed for the rapid and accurate quantification of SARS-CoV-2 serum antibodies. The immunosensor is capable of quantifying the concentrations of immunoglobulin G (IgG) and immunoglobulin M (IgM) antibodies against the SARS-CoV-2 spike protein in human serum. For IgG and IgM, it provides measurements in the range of 10.1 ng/mL − 60 μg/mL and 1.64 ng/mL − 50 μg/mL, respectively, both with an assay time of 13 min. We also developed device stabilization and storage strategies to achieve stable performance of the immunosensor over 24-week storage at room temperature. We evaluated the performance of the immunosensor using COVID-19 patient serum samples collected at different time points after symptom onset. The rapid and sensitive detection of IgG and IgM provided by our immunosensor fulfills the need of rapid COVID-19 serological testing for both point-of-care diagnosis and population immunity screening.
Keywords: Electrochemical biosensing, COVID-19, Serological test, Immunity screening, Point-of-care diagnosis
1. Introduction
Since the onset of the coronavirus disease 2019 (COVID-19) pandemic, this global health crisis has caused over 146 million infections and over 3 million fatalities (as of April 25th, 2021) (World Health Organization, 2021). To mitigate the impact of the pandemic, effective prevention of the spread of COVID-19 is urgently needed. It has been demonstrated in many countries that the COVID-19 pandemic could be controlled with a series of measures, such as rapid diagnosis, infection and contact tracing, large-scale vaccination, and immunotherapy (Bhalla et al., 2020; Carter et al., 2020; Cheng et al., 2020a; Ji et al., 2020; Jin et al., 2020; Qin et al., 2020; Ravi et al., 2020; Udugama et al., 2020). Serological assays for determining antibody responses against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) provide ammunition for these pandemic control tactics. The role of serological testing in clinical diagnostics and public health measures has been debated ever since the beginning of the pandemic (Tré-Hardy et al., 2020). For instance, it has been argued that the serological testing could serve as an alternative diagnostic method in countries and regions with limited access to molecular testing (Peeling et al., 2020). It can also be used as a complement to a polymerase chain reaction (PCR)-based diagnosis (Udugama et al., 2020). Another widely recognized use of serological testing is to determine the past infection history of individuals, allowing for longitudinal immunity tracking.
As the gold-standard diagnostic method for COVID-19, reverse-transcription PCR (RT-PCR) detects conserved regions of the SARS-CoV-2 RNA genome. However, it has a relatively high cost per test and requires costly equipment and well-trained laboratory personnel (Chaimayo et al., 2020; Corman et al., 2020; Moore et al., 2020). This makes the RT-PCR test less accessible in many developing countries (Giri and Rana, 2020; Mannan and Nseluka, 2020). Other diagnostic methods have been developed as alternatives to RT-PCR, including rapid viral antigen testing, loop-mediated isothermal amplification (LAMP)-based RNA testing, and serological testing (Ahmadivand et al., 2021; Fabiani et al., 2021; Lee et al., 2021; Liu et al., 2021; Raziq et al., 2021; Torrente-Rodríguez et al., 2020; Vabret et al., 2020; Yousefi et al., 2021). Because of its short turnaround time and low cost, serological testing has been recommended as an effective method for COVID-19 diagnosis, especially in countries/regions with limited capacity for large-scale molecular testing (Peeling et al., 2020). In addition, serological testing can also be used for population screening and contact tracing (Mathur and Mathur, 2020), as well as long-term population surveillance that could provide a reference for setting/adjusting pandemic control measures (Winter and Hegde, 2020). Since antibody levels can persist for months after a SARS-CoV-2 infection, serological testing is also suitable for longitudinal immunity assessment (Isho et al., 2020; Yongchen et al., 2020).
A variety of immunoassay platforms have been developed, by both academic laboratories and industrial companies, for the detection of SARS-CoV-2 antibodies. A widely adopted commercial immunoassay platform for COVID-19 serological testing is the lateral flow test (LFT) strip. Many LFT products have received regulatory approvals in different countries for COVID-19 diagnosis. Despite their merits such as ease of operation, short assay time, and low cost, these strips usually provide relatively low clinical sensitivity and specificity (Wu et al., 2020; C. Zhang et al., 2021). The conventional laboratory enzyme-linked immunosorbent assay (ELISA) is so far the most promising serology testing method for COVID-19 diagnosis because of its high sensitivity and specificity (Adams et al., 2020; GeurtsvanKessel et al., 2020). However, it requires laboratory infrastructure and equipment and takes hours for a single run (Kasetsirikul et al., 2020; Roy et al., 2020; Tan et al., 2020a). Targeting rapid and sensitive COVID-19 serological testing at the point of care (POC), a variety of portable immunoassay platforms have been developed based on microfluidics and biosensor technologies (Fabiani et al., 2021; Kim et al., 2021; Lee et al., 2021; Li et al., 2021; Raziq et al., 2021; Roda et al., 2021; Tan et al., 2020a; Torrente-Rodríguez et al., 2020; Yakoh et al., 2021; Zakashansky et al., 2021; C. Zhang et al., 2021; Z. Zhang et al., 2021; Zhao et al., 2021).
In this paper, we present a portable Serological testing Platform for rapid ElectrochEmical Detection of SARS-CoV-2 antibodies (SPEEDS). It is based on a low-cost electrochemical immunosensor that uses ELISA to quantify serum immunoglobulin G (IgG) or immunoglobulin M (IgM) antibodies against the SARS-CoV-2 spike protein (S-protein). The SPEEDS platform only takes 13 min for a complete assay and the electrochemical immunosensor can be batch-fabricated at low cost. Through simple device packaging, the prepared ready-to-use immunosensor chips can be stored at room temperature without performance deterioration for at least 24 weeks. We achieved wide measurement ranges of 10.1 ng/mL − 60 μg/mL and 1.64 ng/mL − 50 μg/mL for human monoclonal anti-SARS-CoV-2 IgG and IgM, respectively, which cover typical antibody levels in convalescent sera, as well as the sera of patients with both mild and severe COVID-19 infections. Using the SPEEDS platform, we performed serological testing of 30 patient samples and demonstrated satisfactory clinical performance. The SPEEDS platform provides a low-cost and reliable diagnostic tool for POC serological testing of COVID-19.
2. Materials and methods
2.1. Materials and reagents
Streptavidin (N7021S) was purchased from New England Biolabs (Whitby, ON, Canada). The blocking reagent for ELISA (11112589001), immunoassay stabilizer (s0950), alkaline phosphatase (ALP) conjugate stabilizer (76696), biotin (5-fluorescein) conjugate (53608), human serum (P2918), 10 × Tris-acetate-EDTA (TAE) buffer (T9650; used to prepare 1 × TAE buffer), K3[Fe(CN)6] and sulfuric acid (258105) were purchased from Sigma-Aldrich (Oakville, ON, Canada). ALP-conjugated goat anti-human IgG (ab97222) and goat anti-human IgM (ab97202) were purchased from Abcam (Toronto, ON, Canada). The molar ratio of the enzyme (ALP) to the second antibody protein is 1:1, and the samples were used as received. The electrochemical substrate p-aminophenyl phosphate (A-292-500) was purchased from Gold Biotechnology (St. Louis, MO, USA). Phosphate buffered saline (PBS) (10010023) and distilled water (15230162) were purchased from Thermo Fisher Scientific (Ottawa, ON, Canada). The ELISA diluent (3652-D2) was purchased from Mabtech (Cincinnati, OH, USA). The CR3022 IgG and IgM antibodies were constructed by cloning the CR3022 Fab into a human IgG1 and human IgM framework, respectively (Ter Meulen et al., 2006). Biotinylated SARS-CoV-2 S-protein receptor-binding domain (RBD), human monoclonal anti-SARS-CoV-2 CR3022 IgG and IgM were produced as previously described (Abe et al., 2020). Human total IgG (I4506) was purchase from Sigma-Aldrich. Patient serum samples (CoV-PosSet-S1) were purchased from RayBiotech (Peachtree Corners, GA, USA). The patient sample testing was approved by the research ethics boards at the University of Toronto (protocol number: 40357). Carbon ink (E3456) and Ag/AgCl ink (E2414-250G) for screen printing were purchased from Ercon Inc. (Wareham, MA, USA), and PDMS elastomer (SYLGARD™ 184) for hydrophobic line printing was purchase from Dow Corning (Midland, MI, USA). Blotting paper (Whatman Blotting Membranes, catalog #: 3030-6185, Cytiva) was used during the handling of fluids. Chip substrates, including Whatman Blotting Membranes (WHA3001861) and polyethylene terephthalate (PET) transparent film (CG7060), were purchased from Sigma-Aldrich and Amazon, respectively. Wax printing (Xerox 8560DX) was applied to pattern hydrophilic reaction zones on the sensor substrates. The protection film (3082307) for screen-printing was purchased from Dollarama, and airtight bags (B07QCM4MZ8) and desiccant (B00E880DYS) for device storage were purchased from Amazon.
2.2. Design of the SPEEDS platform
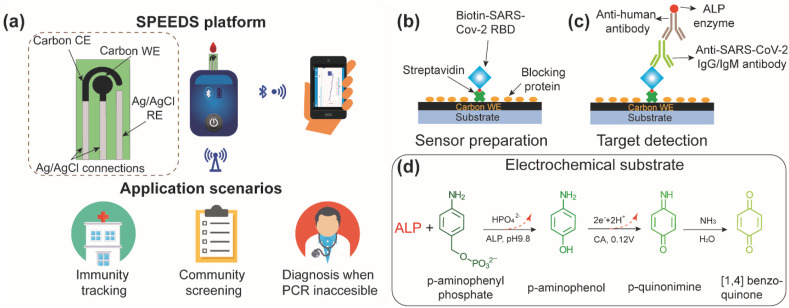
The SPEEDS platform, as shown in Fig. 1 a, consists of a custom-made electrochemical immunosensor and a commercial handheld potentiostat (EmStat3 Blue, PalmSens). The immunosensor includes three screen-printed electrodes (a carbon working electrode, a carbon counter electrode, and a Ag/AgCl reference electrode) on a PET transparent film, forming a three-electrode electrochemical cell. It can be directly inserted into a chip slot of the handheld potentiostat for electrochemical signal readout, and the testing data are transmitted, through a Bluetooth connection, to a smartphone (HUAWEI Mate20 X). The SPEEDS platform can address the urgent need for rapid COVID-19 serological testing in many settings including airports, customs/borders, long-term home cares, schools, and densely populated workplaces. By providing rapid and sensitive quantification of SARS-CoV-2 antibodies in serum samples, it can complement the PCR-based laboratory test and the rapid antigen COVID-19 test. It also allows for a retrospective study of COVID-19 infection via population immunity screening and tracking (Fig. 1a).
Fig. 1.
Design of the SPEEDS platform for detection of IgG and IgM antibodies against SARS-CoV-2 spike protein in human serum. (a) Schematic illustration of the SPEEDS platform and its application scenarios. The platform includes an electrochemical immunosensor and a handheld potentiostat. CE: counter electrode, WE: working electrode, and RE: reference electrode. The potentiostat can transmit testing results to a cell phone. (b) Schematic illustration of surface functionalization of the WE with biotinylated RBD protein as the capture probe. (c) Schematic illustration of capturing anti-SARS-CoV-2 IgG or IgM on the WE and subsequently labelling it with ALP-conjugated detection antibody. (d) Oxidation of the electrochemical substrate (pAPP) during chronoamperometry (CA) and production of the CA current.
The detection of the SARS-CoV-2 antibodies on the immunosensor is based on an electrochemical ELISA, as illustrated in Fig. 1b and c. Streptavidin was first immobilized, through physical absorption, on the working electrode (WE) of the immunosensor followed by the immobilization of the capture probe via streptavidin/biotin binding (Fig. 1b). The biotinylated SARS-CoV-2 spike receptor-binding domain (RBD) protein was used as the capture probe that provides high specificity to SARS-CoV-2 IgG and IgM antibodies. During each test, the SARS-CoV-2 IgG or IgM antibodies in the sample were captured on the WE by the RBD capture probe (Fig. 1c). Then, alkaline phosphatase (ALP)-labeled anti-human detection antibody, specifically against IgG or IgM antibody, was added to bind to the captured IgG or IgM antibody. Lastly, the electrochemical substrate of ALP, p-aminophenyl phosphate (pAPP), was added to the immunosensor to react with the ALP for chronoamperometric (CA) measurement (Fig. 1d). A higher concentration of the IgG or IgM antibody leads to a higher density of the immobilized ALP on the WE and thus a higher CA current. Therefore, the concentration of SARS-CoV-2 IgG or IgM antibody can be quantified based on the CA current.
2.3. Fabrication and preparation of the immunosensor
2.3.1. Fabrication of the immunosensor
The electrochemical immunosensor, as shown in Fig. 1a and S1a, contains three electrodes (carbon WE, carbon CE, and Ag/AgCl RE) screen-printed on a PET substrate, and all three electrodes are confined in a hydrophilic reaction zone patterned through solid wax printing. The screen-printing protocol for patterning carbon and Ag/AgCl electrodes has been reported previously (Zhao and Liu, 2016). Details of the electrode patterning are presented in the supporting information.
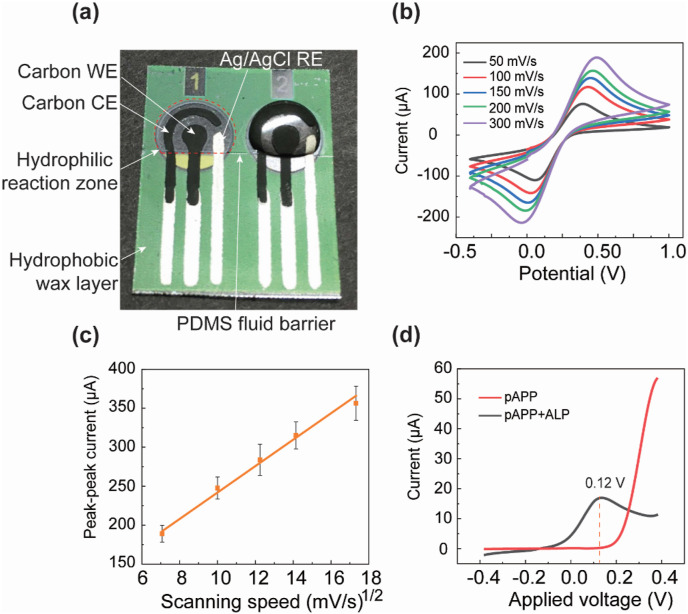
To prevent reagent solutions added to the WE from wicking out of the circular WE reaction zone through the WE tail connection (Fig. 2 a), a thin PDMS barrier was printed onto the WE top surface at the interconnection of the circular WE reaction zone and the tail connection through contact printing (Fig. S1a). To print the PDMS barrier, a fishing line of 100 μm in diameter coated with PDMS precursor (w/w ratio of the base and curing agent: 10:1) was brought into contact with the WE top surfaces of eight curved immunosensors attached on a 3D-printed bending mold (10 cm in diameter), as shown in Fig. S1a. The PDMS barrier can efficiently confine solutions added to the reaction zone of a WE, leading to consistent performance of each immunosensor. Surface functionalization and stabilization of capture proteins on the WE (see the next section for protocols) were conducted on the bare immunosensor chips to allow electrochemical ELISA of SARS-CoV-2 antibodies. The chips were then dried in air and stored in nitrogen-filled airtight bags with desiccant for long-term storage (see Fig. S2a). The ready-to-use immunosensors can be stored at room temperature.
Fig. 2.
Fabrication and characterization of the electrochemical immunosensor. (a) A photograph of the screen-printed three-electrode immunosensor loaded with sample solution in the reaction zone. The reaction zone is defined by a wax-printed cycle and a thin PDMS line barrier printed at the “tail” of the electrode. (b) Cyclic voltammetry (CV) measurements, conducted at scan rates of 50 mV/s, 100 mV/s, 150 mV/s, 200 mV/s, and 300 mV/s. (c) The peak-peak current of the CV curve versus the square root of the scan rate (n = 5). (d) The oxidation current-voltage signals measured from two electrochemical immunosensors loaded with the electrochemical substrate pAPP and a mixture of pAPP and ALP-conjugated detection antibody, respectively, both with a hydrodynamic linear sweeping voltage in the range of −0.4 V and 0.4 V with a scanning speed of 100 mV/s.
2.3.2. Activation, biofunctionalization, stabilization and long-term storage of the immunosensor
Before surface functionalization of the WE with capture probes, the reaction zone of the WE was loaded with 100 μL of 0.18 M sulfuric acid (diluted in 1 × PBS) and then activated for 3 min with a 1.3 V anodic voltage (against RE). This activation process creates abundant active carboxylic groups on the WE carbon surface and thus improves the sensitivity of the device (Díaz-González et al., 2005). After washing with DI water and then 1× PBS, the WE underwent a series of surface functionalization steps including i) immobilization of streptavidin (SA), ii) hybridization of biotinylated RBD, and iii) blocking of the void locations of the WE with blocking proteins (Fig. 1b). Finally, the biofunctionalized WE surface was then stabilized with immunoassay stabilizer. Unless otherwise stated, all the washing steps in the WE biofunctionalization process were finished by dispensing 50 μL buffer (1 × PBS or DI water) to the reaction zone, removing the solution by pipetting, and absorbing the residual buffer with blotting paper.
The immobilization of the RBD capture probe on the WE relies on the widely adopted biotin-streptavidin (SA) interaction (Diamandis and Christopoulos, 1991; Feng et al., 2021). First, the SA was immobilized on the WE by physical absorption. For this step, we investigated the absorption efficiency of SA on the WE with different incubation durations and studied the effect of washing cycles on the stability of SA immobilization. In our experiments, 5 μL of 1 mg/mL SA was added on the WE reaction zone and incubated at 4 °C for different durations in the range of 1–24 h. After incubation, the residual SA was washed off from the WE by PBS, and 5 μL of 20 μM biotin (5-fluorescein) conjugate was added on the WE and incubated for 10 min to examine the efficiency of SA immobilization. All the WE surfaces were imaged under a fluorescence microscope (under 20× objective), and the average fluorescence intensities of the WE surfaces were analyzed in ImageJ. Note that the fluorescence imaging method only provides qualitative analysis of the immobilized capture probes. Protein oscillation imaging could be employed to accurately quantify the binding kinetics of the immobilized capture proteins on the electrode surface (Ma et al., 2020a), with the immunosensor preparation always following the same protocol, we were able to establish a relationship between the input known concentration of antibody and the output electric signal.
Fig. S3b-f show typical fluorescent images of the WE surfaces after 1, 4, 8, 12 and 24 h incubation in the SA solution and then 3 times of PBS washing, respectively. One can observe uniform SA coating on the WE surface for all five SA incubation durations. Fig. S3g illustrates the average fluorescence intensity of the WE surface as a function of the incubation time. One can see there is an obvious increasing trend of the amount of immobilized SA with the incubation time, indicating longer incubation time improves the immobilization efficiency. To investigate the stability of the SA immobilization on the WE, the fluorescence intensities of the WE surfaces, which were modified with 12 h SA incubation and then washed by 50 μL of PBS for two, three or five times, were analyzed. The results (Fig. S3h) indicate that the average fluorescence intensity of the WE surface after five times of PBS washing is only 5.6% lower than that of the WE after twice of PBS washing, suggesting highly stable immobilization of SA on the WE through physical absorption. Based on the above data, the modification condition of 24-h SA absorption and three times of PBS washing were adopted for preparing all the immunosensors for the following experiments.
After the SA immobilization, the immunosensors were grafted with SARS-CoV-2 spike RBD protein. Details of the functionalization, blocking and stabilization are presented in the supporting information. All the incubation and drying steps were performed in air at room temperature (21 °C). The dried immunosensors were stored in a nitrogen-filled airtight bag with desiccant for long-term storage. The total material cost of an immunosensor chip is USD $2.10 in small quantity (see itemized material costs in Table S1).
2.4. Serological test procedure
For each serological test, the ready-to-use immunosensor was initially activated by adding 30 μL of immunoassay stabilizer to the WE reaction zone for 1-min incubation followed by washing with 50 μL of PBS twice. To minimize nonspecific binding of serum interference proteins on the capture antibody, a dilution factor of 5:1 (in ELISA diluent) was applied to all human serum samples tested in this work (see Fig. S5 for more information). 5 μL of diluted serum sample was pipetted on a freshly reactivated WE and incubated for 1 min to enable the capture of the target antibody on the WE, followed by washing with PBS for three times. Then, 5 μL of ALP-labeled anti-human antibody (20 μg/mL, diluted in the ALP conjugate stabilizer) was added to the WE and incubated for 3 min, which was followed by another three times of PBS washing. As the last step, 40 μL of pAPP (6 mM, diluted in 1 × TAE buffer) was added on the WE and incubated for 5 min to reach reaction equilibrium. The ALP catalyzes the oxi-reductive reaction of pAPP on the WE. The CA measurement was finally conducted and the stabilized faradaic current was measured 2 min after the stepwise CA potential was applied. For each test, the whole process took approximately 13 min.
2.5. Patient sample testing
A cohort of 20 COVID-19 positive serum samples (confirmed by RT-PCR) and 10 COVID-19 negative serum samples (collected before the pandemic) were tested using our immunosensors. Before the patient sample testing experiments, all the serum samples were treated with 0.5% Triton X-100 for 10 min to inactivate any potential infectious viruses. For every serum sample, both IgG and IgM tests were repeated twice each. All the data are listed in Table S3.
3. Results and discussion
3.1. Electrochemical characterization of the immunosensor
To verify the electrochemical performance of the three-electrode immunosensor, 40 μL of 10 mM K3[Fe(CN)6] (in 1 M KCl) was added to the WE of an immunosensor, and cyclic voltammetry (CV) measurements were conducted at different scan rates. As shown in Fig. 2b, the measured cyclic voltammograms reveal typical reversible electrochemical reactions at all scan rates. The peak-peak current of the cyclic voltammogram is linearly proportional to the square root of the scan rate, further confirming that the immunosensor is a reversible electrochemical system. For testing serum samples, we chose CA measurement for its lower signal-to-noise ratio and thus higher sensitivity over CV measurement (Dungchai et al., 2009; Zhao et al., 2013). To determine the optimal oxidation voltage of pAPP for CA measurement, hydrodynamic linear sweeping was performed on pure pAPP solution (6 mM in 1× TAE buffer) and pAPP plus ALP-labeled detection antibody solution (20 μg/mL in ALP conjugate stabilizer) at a 100 mV/s scan rate in the range of −0.4 V and 0.4 V. As shown in Fig. 2d, the highest oxidation peak was obtained at 0.12 V. Therefore, 0.12 V was adopted as the excitation voltage for CA testing of the serum samples.
3.2. Calibration of the immunosensor
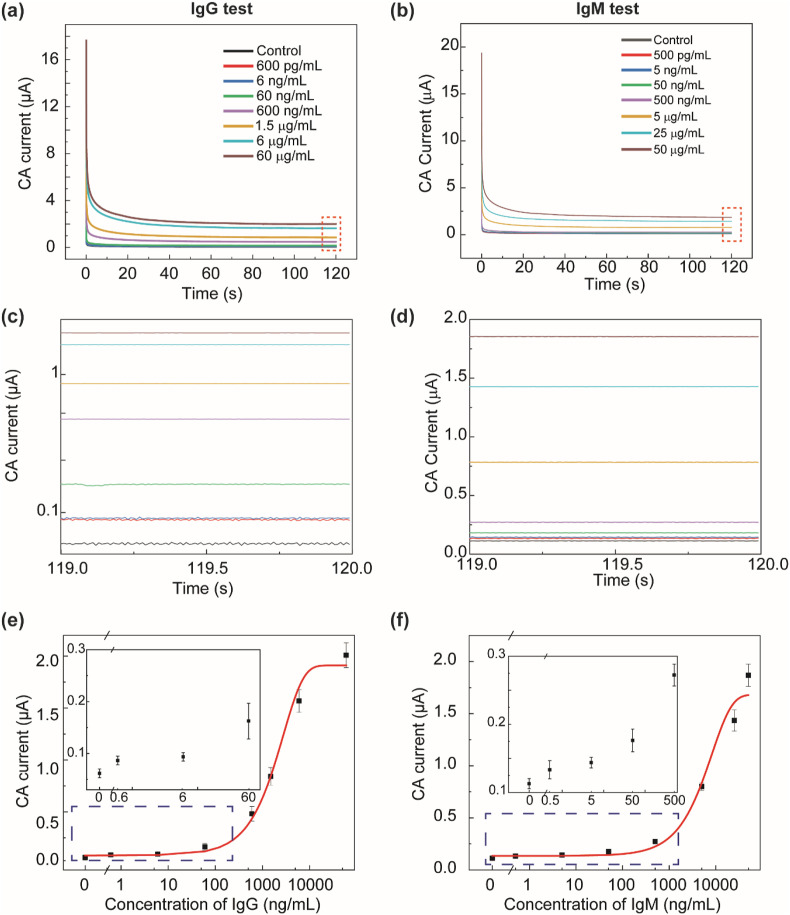
For device calibration, CR3022 IgG or IgM (concentration range: 500 pg/mL to 60 μg/mL) spiked in five-fold-diluted human serum was tested. Fig. 3 a and b show the typical CA current curves measured from IgG-spiked and IgM-spiked serum samples at different concentrations. To minimize errors caused by non-Faradaic current, the steady-state current value at ∼120 s after applyng the excitation voltage was collected as the immunosensor output (zoomed-in views of the CA curves shown in Fig. 3c and d). Using IgG as the model analyte, we also investigated the effect of biotinylated RBD capture protein concentration (in the range of 10–100 μg/mL) on the performance of the immunosensor. Fig. S4 shows the calibration curves of IgG detection with different RBD concentrations (concentrations of all other reagents are the same as described in Section 2.3), suggesting that low concentrations of RBD (10 and 25 μg/mL) would result in unsatisfactory sensitivity when quantifying the target antibody at low concentrations while a high RBD concentration (100 μg/mL) improves the testing sensitivity but also raises the reagent cost. Considering the assay sensitivity and the manufacturing cost, 50 μg/mL biotinylated RBD was adopted for all the experiments of device calibration and patient sample testing. Fig. 3e and f illustrate the calibration curves for the detection of CR3022 IgG and IgM in five-fold-diluted human serum, respectively. The LOD values of the immunosensor for IgG and IgM detection are 10.1 ng/mL and 1.64 ng/mL, respectively.
Fig. 3.
Calibration of the immunosensor for detecting anti-SARS-CoV-2 IgG and IgM in five-fold-diluted serum samples. (a)(b) Representative CA current curves measured on five-fold-diluted serum spiked with (a) CR3022 IgG antibody and (b) CR3022 IgM antibody at different concentrations. The quasi-steady-state current at 120 s was used as the signal readout for device calibration. (c)(d) The zoomed-in views of the CA current curves in (a)(b) during 119–120 s, respectively. (e) Calibration curve of the immunosensor for IgG detection at 600 pg/mL, 6 ng/mL, 60 ng/mL, 600 ng/mL, 1.5 μg/mL, 6 μg/mL and 60 μg/mL. (f) Calibration curve of the immunosensor for IgM detection at 500 pg/mL, 5 ng/mL, 50 ng/mL, 500 ng/mL, 5 μg/mL, 25 μg/mL, and 50 μg/mL. For both IgG and IgM calibration, n = 7 for current data at non-zero concentrations, and n = 10 for current data (background signals) at the zero concentration.
The IgG concentration in different individuals during and after SARS-CoV-2 infection varies significantly. Fortunately, there have been many studies revealing that the peak concentrations of antibodies against the SARS-CoV-2 RBD in COVID-19 patient serum are typically below 10's μg/mL, with only a few cases reaching 100 μg/mL (Ibarrondo et al., 2020; Iyer et al., 2020; H. Ma et al., 2020; Terpos et al., 2020; Torrente-Rodríguez et al., 2020). Therefore, we chose to set the measurement amplitude of our immunosensor to be up to 60 μg/mL. Monoclonal antibody CR3022 was chosen in device calibration for its ability to bind the SARS-CoV-2 S-protein RBD. The calibration curves show the typical sigmoidal relationship between the CA current and the antibody concentration. Because of the stable complex of the capture probe and the target antibody and the strong reaction realized under our final assay conditions, wide detection ranges have been achieved for both IgG (10.1 ng/mL – 60 μg/mL) and IgM (1.64 ng/mL to 50 μg/mL) antibodies. As mentioned above, the antibody concentrations among patients differ significantly, emphasizing the need for a wide measurement range. Our device responds to both antibody isotypes over wide ranges with LODs at the ng/mL level, making it suitable for COVID-19 diagnosis and long-term quantification of SARS-CoV-2 antibodies.
3.3. Cross-reactivity testing
Cross-reactivity testing of a biosensor is necessary when complex samples are involved. Herein, the potential interference of total human IgG to anti-SARS-CoV-2 IgG and the cross-reactivity between CR3022 IgG and IgM were tested. In the experiments, interference antibodies were spiked in targeting samples, and the mixtures were tested by following the testing procedure described above. For each cross-reactivity testing condition, the experiment was repeated for at least 5 times on freshly prepared chips.
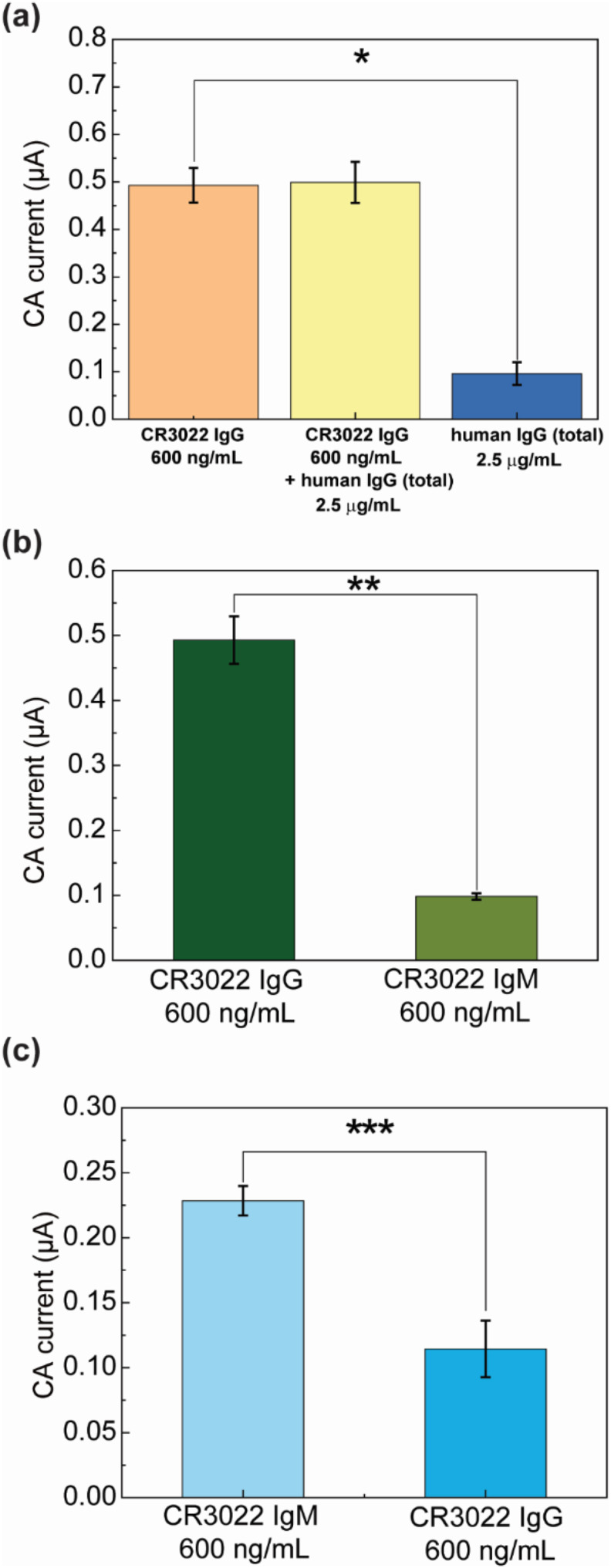
Firstly, the cross-reactivity between total human IgG and anti-SARS-CoV-2 IgG was tested. Since IgG is the most abundant antibody isotype in blood and persists for months while IgM is induced earlier and decays rapidly, we mainly focused on the cross-reactivity between anti-SARS-CoV-2 IgG and total human IgG. We used total human IgG produced before the COVID-19 pandemic as the interference antibody. Experiments were conducted with antibodies spiked in five-fold-diluted human serum, and the results are presented in Fig. 4 a: i) only CR3022 IgG (600 ng/mL), ii) CR3022 IgG (600 ng/mL) and total human IgG (2.5 μg/mL), and iii) only total human IgG (2.5 μg/mL). In all cases, ALP-labeled anti-human IgG antibody was used as the labelling antibody. All experiments were repeated 5 times on freshly prepared immunosensors. It should be noted that the incubation time was intentionally increased to 5 min to further amplify signals from any possible cross-reaction. For the serum samples (Fig. 4a), the average CA current of the “CR3022 IgG + total human IgG” sample (0.499 μA) was similar to that of the pure “CR3022 IgG” sample (0.493 μA), but both types of sample yielded average output current much larger than the pure “total human IgG” sample (0.0962 μA). Experiments on antibody-spiked PBS generated consistent results (Fig. S6a). These results show that our immunosensor for CR3022 IgG detection has negligible cross-reaction with non-specific total human IgG.
Fig. 4.
The cross-reactivity testing of the immunosensor for detection of anti-SARS-CoV-2 IgG and IgM. (a) The cross-reactivity data of CR3022 IgG detection using total human IgG antibody as the interference protein. *p = 3.58 × 10−8. (b) The cross-reactivity data of CR3022 IgG detection using CR3022 IgM as the interference protein. **p = 1.01 × 10−8. (c) The cross-reactivity data of CR3022 IgM detection using CR3022 IgG as the interference protein. ***p = 6.27 × 10−6. In (a–b), ALP-labeled anti-human IgG was used as the detection antibody. In (c), ALP-labeled anti-human IgM was used as the detection antibody. For all the testing data, n = 5.
The cross-reactivity between CR3022 IgG and IgM was also tested on our device in both diluted human serum (Fig. 4b and c) and PBS (Fig. S6). In the IgG assay, ALP-labeled anti-human IgG was used as the detection antibody, CR3022 IgG (600 ng/mL) and CR3022 IgM (600 ng/mL) were spiked in five-fold-diluted human serum and detected separately (mean currents were 0.493 μA and 0.0984 μA, respectively; see results in Fig. 4b). In the IgM assay, ALP-anti-human IgM was used as the detection antibody, CR3022 IgM (600 ng/mL) and CR3022 IgG (600 ng/mL) were spiked in diluted human serum and detected separately (mean currents were 0.228 μA and 0.114 μA, respectively; see results in Fig. 4c). All experiments were repeated five times on different immunosensors. From Fig. 4b and c, one can observe that the cross-reaction between the CR3022 IgG and IgM assays on our immunosensor is not significant (p-values: 1.01 × 10−8 and 6.27 × 10−6, respectively). The negligible cross-reactivity is due to the proper assay design as well as the high specificity of the secondary antibodies to IgG or IgM. Choosing RBD as the capture protein also proves to be beneficial since it has been found that S-protein-based serological tests showed less cross-reactivity than nucleocapsid protein (N-protein)-based assays (Amanat et al., 2020; Cheng et al., 2020b; Okba et al., 2020).
3.4. Long-term storage testing
The device stability over long-term storage is an important factor in the practical use of immunosensors. The possible degradation of capture proteins on the immunosensor could result in testing performance decline. The stability of our immunosensors was investigated over a 24-week storage period. All the immunosensors (with immunoassay stabilizers) were prepared in the same batch, packed in nitrogen-infused airtight bags with desiccant, and stored in either laboratory environment (21 °C) or a refrigerator (4 °C). CR3022 IgG diluted in PBS at 6 μg/mL was used to test the performance stability of the stored immunosensors.
The testing data (Fig. S7) show that for both room temperature and refrigerated storage conditions, no significant variation in the immunosensor output was observed over 24 weeks. These results confirm the high storage stability and performance reproducibility of our laboratory-made immunosensor, further demonstrating the high feasibility of our platform for practical COVID-19 serological testing. Comparing to the recently published reports on COVID-19 serological biosensors (Kasetsirikul et al., 2020; Tan et al., 2020b; Torrente-Rodríguez et al., 2020) with limited data on storage condition investigation, our study provides a practical method for long-term protein stabilization and room-temperature device storage. Our immunosensor shows a room-temperature shelf life comparable to that of existing commercial COVID-19 serological tests such as the Abbott BinaxNOW Ag Card Tests (6 months) and cPass™ SARS-CoV-2 Neutralization Antibody Detection Kit (6 months).
3.5. Patient sample testing
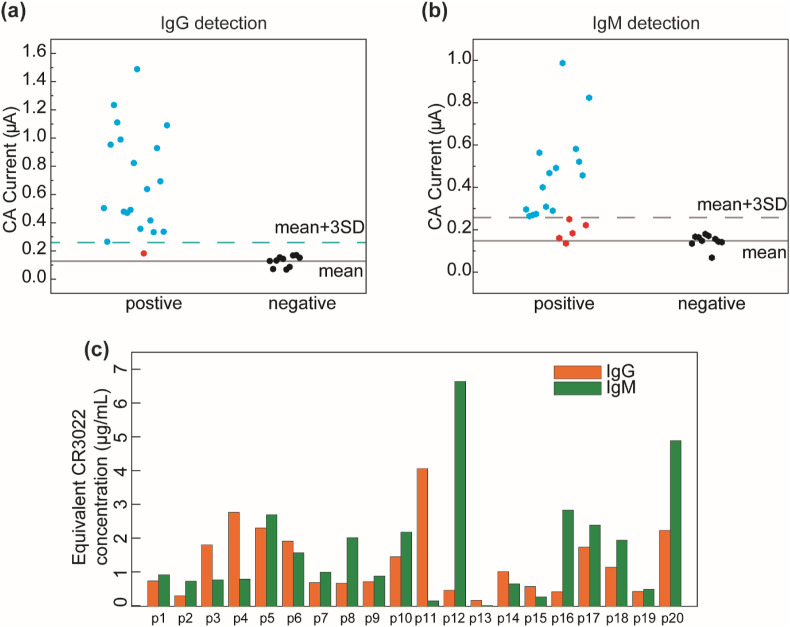
A cohort of 20 PCR-tested positive (CoV2+) patient serum samples and 10 pre-pandemic negative (CoV2−) serum samples were tested using our immunosensor, and the experimental setup is shown in Fig. S2. The CoV2+ serum samples were collected at different time points (1-35 days; see Table S3) after the onset of COVID-19 symptoms. Fig. 5 a and Fig. 5b show the immunosensor current outputs (values listed in Table S3) for detecting IgG and IgM of the 20 CoV2+ and 10 CoV2− samples, respectively. The cut-off values for IgG and IgM detection were determined to be 0.259 μA and 0.258 μA, which is defined as the average plus three times the standard deviation of the current outputs measured from the 10 CoV2− samples (Fig. 5a and b). Based on the calibration curves for CR3022 IgG and IgM detection (Fig. 3e and f), the equivalent CR3022 IgG and IgM concentrations in the five-fold-diluted patient samples were calculated and converted (Fig. 5c and Table S3). Noted that for the consistency of the testing protocol and the effectiveness of the calibration curves, the patient samples were all five-fold diluted before tests. The original antibody concentration in each patient sample was also caclulated by multiplying the concentration value in Fig. 5c with the dilution factor five, as shown in Table S3.
Fig. 5.
Patient sample testing of the immunosensor. (a)(b) Testing results of (a) IgG and (b) IgM of a cohort of 20 SARS-CoV-2 positive samples and 10 pre-pandemic negative samples. (c) The equivalent CR3022 IgG and IgM concentrations in the five-fold-diluted patient sera calculated based on the corresponding calibration curves.
From the patient sample testing results, one can see that most of the CoV2+ samples exhibited CA current signals higher than the cut-off values (IgG+, IgM+) within 1–35 days after symptom onset, indicating the active stage of infection. Only one CoV2+ sample (p13 in Table S3) generated both IgG and IgM signals lower than the cut-off values (IgG− and IgM−), which is due to the insufficient immune response of the patient (sample collected on the first day of symptom onset). It is reported that the IgM level generally increases rapidly with time at the early stage after symptom onset and decreases after long-term infection (Iyer et al., 2020). For example, the IgM levels in p12 and p18 samples (collected 8 and 9 days after symptom onset) are higher than the IgM cut-off value (indicating IgM+). The IgM levels in p11 and p15 samples (collected 33 or 34 days after symptom onset) become lower than the cut-off value (IgM−), but their IgG levels are still much higher than the cut-off value (IgG+). Detection results corresponding to 30, 33, 33, 34 days after symptom onset in p19, p2, p15, and p11 show positive IgG (IgG+) and negative IgM (IgM−) levels, indicating a rapid decay of IgM in these patient samples. Note that the immune systems of different individuals respond to viral infections quite differently; thus, the IgG and IgM concentrations vary from sample to sample. The binding activity of all antibodies against SARS-CoV-2 RBD could be quantified by the equivalent concentration of the monoclonal CR3022 antibody (Ibarrondo et al., 2020). For our immunosensor, the equivalent IgG and IgM concentrations of a patient sample can be easily converted from the measured CA currents based on the calibration curves; thus, our IgG and IgM tests are effective for the longitudinal assessment of individual patients. Our immunosensor can serve as a useful tool for immunology studies of COVID-19 including immunity tracking, immunotherapy and immunodiagnostics, where the quantitative evaluation of antibody levels is required.
Due to the limited human sera sample set (20 CoV2+ samples and 10 CoV2− samples) and the non-ideal distribution of the sample collection time points (16 out of 20 CoV2+ samples were collected during 30–35 days after the symptom onset), we did not calculate sensitivity and specificity of the immunosensor from the current patient sample data. Instead, we compare our SPEEDS platform with other portable biosensors reported in the literature for COVID-19 serological tests, in terms of analytical performance and other technical aspects (Table S4). One can see that our SPEEDS platform displays excellent performance in all aspects. In particular, our long-term device stability under room-temperature storage facilitates device transportation, storage, and field application.
4. Conclusion
We have successfully developed a low-cost and portable SPEEDS platform for the quantitative detection of IgG and IgM antibodies against the SARS-CoV-2 S-protein in human sera. The platform can be used for on-site COVID-19 serological tests and provides accurate results in 13 min. The electrochemical immunosensor can be batch-fabricated at a low material cost, and it can be stored at room temperature for at least 24 weeks without performance deterioration. Wide measurement ranges of 10.1 ng/mL − 60 μg/mL and 1.64 ng/mL − 50 μg/mL, have been achieved for IgG and IgM detection, respectively. The immunosensor showed negligible cross-reactivity with non-specific total human IgG and negligible cross-reactivity between CR3022 IgG and CR3022 IgM. A retrospective study on a cohort of 20 COVID-19 positive patient samples and 10 negative samples was conducted to demonstrate the practical use of the platform. We believe that the SPEEDS platform will find important applications in rapid and sensitive serological testing of COVID-19. It can be also extended to other types of serological tests and disease antigen tests.
CRediT authorship contribution statement
Ran Peng: Investigation, Formal analysis, Conceptualization, Methodology, Writing-Original Draft, Writing-Reviewing and Editing. Yueyue Pan: Investigation, Formal analysis, Conceptualization, Methodology, Writing-Original Draft, Writing-Reviewing and Editing. Zhijie Li: Conceptualization, Resources, Writing-Reviewing and Editing. Zhen Qin: Validation, Investigation. James M. Rini: Resources, Writing-Reviewing and Editing, Funding acquisition. Xinyu Liu: Conceptualization, Writing-Reviewing and Editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the Toronto COVID-19 Action Initiative Program (grant number: 508801), the National Sciences and Engineering Council of Canada (grant numbers: RGPIN-2017-06374 and RGPAS-2017-507980), the Canada Foundation for Innovation (grant number: JELF-38428), and the University of Toronto (through the Percy Edward Hart Professorship to Xinyu Liu).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113762.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., Budylowski P., Dupuis A.P., Girardin R.C., Rathod B., Wang J.H., Barrios-Rodiles M., Colwill K., McGeer A.J., Mubareka S., Gommerman J.L., Durocher Y., Ostrowski M., McDonough K.A., Drebot M.A., Drews S.J., Rini J.M., Gingras A.C. bioRxiv. 2020:1–14. doi: 10.1101/2020.07.10.197913. [DOI] [Google Scholar]
- Adams E.R., Ainsworth M., Anand R., Andersson M.I., Auckland K., Baillie J.K., Barnes E., Beer S., Bell J., Berry T., Bibi S., Carroll M., Chinnakannan S., Clutterbuck E., Cornall R.J., Crook D.W., De Silva T., Dejnirattisai W., Dingle K.E., Dold C., Espinosa A., Eyre D.W., Farmer H., Fernandez Mendoza M., Georgiou D., Hoosdally S.J., Hunter A., Jeffrey K., Klenerman P., Knight J., Knowles C., Kwok A.J., Leuschner U., Levin R., Liu C., Lopez-Camacho C., Martinez Garrido J.C., Matthews P.C., McGivern H., Mentzer A.J., Milton J., Mongkolsapaya J., Moore S.C., Oliveira M.S., Pereira F., Perez Lopez E., Peto T., Ploeg R.J., Pollard A., Prince T., Roberts D.J., Rudkin J.K., Sanchez V., Screaton G.R., Semple M.G., Skelly D.T., Slon-Campos J., Smith E.N., Sobrino Diaz A.J., Staves J., Stuart D., Supasa P., Surik T., Thraves H., Tsang P., Turtle L., Walker A.S., Wang B., Washington C., Watkins N., Whitehouse J. medRxiv. 2020 doi: 10.1101/2020.04.15.20066407. [DOI] [Google Scholar]
- Ahmadivand A., Gerislioglu B., Ramezani Z., Kaushik A., Manickam P., Ghoreishi S.A. Biosens. Bioelectron. 2021;177:112971. doi: 10.1016/j.bios.2021.112971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Pan Y., Yang Z., Payam A.F. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaimayo C., Kaewnaphan B., Tanlieng N., Athipanyasilp N., Sirijatuphat R., Chayakulkeeree M., Angkasekwinai N., Sutthent R., Puangpunngam N., Tharmviboonsri T., Pongraweewan O., Chuthapisith S., Sirivatanauksorn Y., Kantakamalakul W., Horthongkham N. Virol. J. 2020;17:177. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.P., Yansouni C.P., Basta N.E., Desjardins M., Kanjilal S., Paquette K., Caya C., Semret M., Quach C., Libman M., Mazzola L., Sacks J.A., Dittrich S., Papenburg J. Ann. Intern. Med. 2020;173:450–460. doi: 10.7326/M20-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.P., Yansouni C.P., Basta N.E., Desjardins M., Kanjilal S., Paquette K., Caya C., Semret M., Quach C., Libman M., Mazzola L., Sacks J.A., Dittrich S., Papenburg J. Ann. Intern. Med. 2020;173:450–460. doi: 10.7326/M20-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. 25. [DOI] [Google Scholar]
- Diamandis E.P., Christopoulos T.K. Clin. Chem. 1991;37:625–636. doi: 10.1093/clinchem/37.5.625. [DOI] [PubMed] [Google Scholar]
- Díaz-González M., Hernández-Santos D., González-García M.B., Costa-García A. Talanta. 2005;65:565–573. doi: 10.1016/j.talanta.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Dungchai W., Chailapakul O., Henry C.S. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Su J., Xu Y., He G., Wang C., Wang X., Pan T., Ding X., Mi X. Microsyst. Nanoeng. 2021;7:33. doi: 10.1038/s41378-021-00258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J., van den Akker J.P.C., van Kampen J.J.A., van der Eijk A.A., van Binnendijk R.S., Haagmans B., Koopmans M. Nat. Commun. 2020;11:1–5. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri A.K., Rana D.R. Biosaf. Health. 2020;2:53–56. doi: 10.1016/j.bsheal.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., Pu A., Christie-Holmes N., Gervais C., Ceccarelli D., Samavarchi-Tehrani P., Guvenc F., Budylowski P., Li A., Paterson A., Yun Y.F., Marin L.M., Caldwell L., Wrana J.L., Colwill K., Sicheri F., Mubareka S., Gray-Owen S.D., Drews S.J., Siqueira W.L., Barrios-Rodiles M., Ostrowski M., Rini J.M., Durocher Y., McGeer A.J., Gommerman J.L., Gingras A.C. Sci. Immunol. 2020;5:1–21. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F., Astudillo M., Yang D., Miller T.E., Oliver E., Fischinger S., Atyeo C., Iafrate A.J., Calderwood S.B., Lauer S.A., Yu J., Li Z., Feldman J., Hauser B.M., Caradonna T.M., Branda J.A., Turbett S.E., LaRocque R.C., Mellon G., Barouch D.H., Schmidt A.G., Azman A.S., Alter G., Ryan E.T., Harris J.B., Charles R.C. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T., Liu Z., Wang G.Q., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P., Fang C., Huang D., Huang L.Q., Huang Q., Han Y., Hu B., Hu F., Li B.H., Li Y.R., Liang K., Lin L.K., Luo L.S., Ma J., Ma L.L., Peng Z.Y., Pan Y.B., Pan Z.Y., Ren X.Q., Sun H.M., Wang Y., Wang Yun Yun, Weng H., Wei C.J., Wu D.F., Xia J., Xiong Y., Xu H.B., Yao X.M., Yuan Y.F., Ye T.S., Zhang X.C., Zhang Y.W., Zhang Y.G., Zhang H.M., Zhao Y., Zhao M.J., Zi H., Zeng X.T., Wang Yong Yan, Wang X.H. Mil. Med. Res. 2020;7:1–23. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasetsirikul S., Umer M., Soda N., Sreejith K.R., Shiddiky M.J.A., Nguyen N.-T. Analyst. 2020;145:7680–7686. doi: 10.1039/D0AN01609H. [DOI] [PubMed] [Google Scholar]
- Kim S., Hao Y., Miller E.A., Tay D.M.Y., Yee E., Kongsuphol P., Jia H., McBee M., Preiser P.R., Sikes H.D. 2021. ACS Sensors. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Choi M., Jung Y., Lee S.K., Lee C.S., Kim Jung, Kim Jongwoo, Kim N.H., Kim B.T., Kim H.G. Biosens. Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Qin Z., Fu H., Li T., Peng R., Li Z., Rini J.M., Liu X. Biosens. Bioelectron. 2021;177:112672. doi: 10.1016/j.bios.2020.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan J., Yan X. Biosens. Bioelectron. 2021;173:112817. doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Wan Z., Yang Y., Zhang P., Wang S., Tao N. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18547-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zeng W., He H., Zhao D., Yang Y., Jiang D., Qi P.Y., He W., Zhao C., Yi R., Wang X., Wang B., Yang Y.Y., Kombe Kombe A.J., Ding C., Xie J., Gao Y., Cheng L., Li Y., Ma X., Jin T. medRxiv. 2020 doi: 10.1101/2020.04.17.20064907. [DOI] [Google Scholar]
- Mannan D., Nseluka C. J. Prev. Rehabil. Med. 2020;2:5–7. doi: 10.21617/jprm2020.212. [DOI] [Google Scholar]
- Mathur G., Mathur S. Am. J. Clin. Pathol. 2020;154:1–3. doi: 10.1093/ajcp/aqaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore N.M., Li H., Schejbal D., Lindsley J., Hayden M.K. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J., Sall A., Tanuri A., Heymann D.L. Lancet Infect. Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z., Peng R., Baravik I.K., Liu X. Matter. 2020;3:628–651. doi: 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., Syritski V. Biosens. Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Biosens. Bioelectron. 2021;172:112765. doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy V., Fischinger S., Atyeo C., Slein M., Loos C., Balazs A., Luedemann C., Astudillo M.G., Yang D., Wesemann D., Charles R., Lafrate A.J., Feldman J., Hauser B., Caradonna T., Miller T.E., Murali M.R., Baden L., Nilles E., Ryan E., Lauffenburger D., Beltran W.G., Alter G., Alter G. J. Immunol. Methods. 2020;484–485:112832. doi: 10.1016/j.jim.2020.112832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Krel M., Dolgov E., Park S., Li X., Wu W., Sun Y.L., Zhang J., Khaing Oo M.K., Perlin D.S., Fan X. Biosens. Bioelectron. 2020;169:112572. doi: 10.1016/j.bios.2020.112572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Lin C., Zhang J., Khaing Oo M.K., Fan X. bioRxiv. 2020 doi: 10.1101/2020.04.20.052233. 2020.04.20.052233. [DOI] [Google Scholar]
- Ter Meulen J., Van Den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., Van Deventer E., Preiser W., Doerr H.W., Chow V.T., De Kruif J., Peiris J.S.M., Goudsmit J. PLoS Med. 2006;3:1071–1079. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos E., Mentis A., Dimopoulos M.A. N. Engl. J. Med. 2020;383:1693–1694. doi: 10.1056/nejmc2028468. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tré-Hardy M., Blairon L., Wilmet A., Beukinga I., Malonne H., Dogné J.M., Douxfils J. J. Infect. 2020;81:e91–e92. doi: 10.1016/j.jinf.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Pia L., Risson E., Saffern M., Salomé B., Esai Selvan M., Spindler M.P., Tan J., van der Heide V., Gregory J.K., Alexandropoulos K., Bhardwaj N., Brown B.D., Greenbaum B., Gümüş Z.H., Homann D., Horowitz A., Kamphorst A.O., Curotto de Lafaille M.A., Mehandru S., Merad M., Samstein R.M., Agrawal M., Aleynick M., Belabed M., Brown M., Casanova-Acebes M., Catalan J., Centa M., Charap A., Chan A., Chen S.T., Chung J., Bozkus C.C., Cody E., Cossarini F., Dalla E., Fernandez N., Grout J., Ruan D.F., Hamon P., Humblin E., Jha D., Kodysh J., Leader A., Lin M., Lindblad K., Lozano-Ojalvo D., Lubitz G., Magen A., Mahmood Z., Martinez-Delgado G., Mateus-Tique J., Meritt E., Moon C., Noel J., O'Donnell T., Ota M., Plitt T., Pothula V., Redes J., Reyes Torres I., Roberto M., Sanchez-Paulete A.R., Shang J., Schanoski A.S., Suprun M., Tran M., Vaninov N., Wilk C.M., Aguirre-Ghiso J., Bogunovic D., Cho J., Faith J., Grasset E., Heeger P., Kenigsberg E., Krammer F., Laserson U. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A.K., Hegde S.T. Lancet Infect. Dis. 2020;20:758–759. doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 World Health Organization [WWW Document]
- Wu J.L., Tseng W.P., Lin C.H., Lee T.F., Chung M.Y., Huang C.H., Chen S.Y., Hsueh P.R., Chen S.C. J. Infect. 2020;81:435–442. doi: 10.1016/j.jinf.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J., Chen Y., Gu B. Emerg. Microb. Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., McGeer A., Christie N., Gray-Owen S., Cochrane A., Rini J.M., Sargent E.H., Kelley S.O. J. Am. Chem. Soc. 2021 doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- Zakashansky J.A., Imamura A.H., Salgado D.F., Romero Mercieca H.C., Aguas R.F.L., Lao A.M., Pariser J., Arroyo-Currás N., Khine M. Anal. Methods. 2021;13:874–883. doi: 10.1039/d1ay00041a. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zheng T., Wang H., Chen W., Huang X., Liang J., Qiu L., Han D., Tan W. Anal. Chem. 2021;93:3325–3330. doi: 10.1021/acs.analchem.0c05059. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang X., Wei X., Zheng S.W., Lenhart B.J., Xu P., Li J., Pan J., Albrecht H., Liu C. Biosens. Bioelectron. 2021;181:113134. doi: 10.1016/j.bios.2021.113134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Che C., Wang W., Li N., Cunningham B.T. Talanta. 2021;225:122004. doi: 10.1016/j.talanta.2020.122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Liu X. Biomicrofluidics. 2016;10 doi: 10.1063/1.4945311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Thuo M.M., Liu X. Sci. Technol. Adv. Mater. 2013;14:54402. doi: 10.1088/1468-6996/14/5/054402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.