Abstract
Purpose
Menin, encoded by the MEN1 gene, was recently reported to be involved in breast cancers, though the underlying mechanisms remain elusive. In the current study, we sought to further determine its role in mammary cells.
Methods
Menin expression in mammary lesions from mammary-specific Men1 mutant mice was detected using immunofluorescence staining. RT-qPCR and western blot were performed to determine the role of menin in ERα expression in human breast cancer cell lines. ChIP-qPCR and reporter gene assays were carried out to dissect the action of menin on the proximal ESR1 promoter. Menin expression in female patients with breast cancer was analyzed and its correlation with breast cancer subtypes was investigated.
Results
Immunofluorescence staining revealed that early mammary neoplasia in Men1 mutant mice displayed weak ERα expression. Furthermore, MEN1 silencing led to both reduced ESR1 mRNA and ERα protein expression in MCF7 and T47D cells. To further dissect the regulation of ESR1 transcription by menin, we examined whether and in which way menin could regulate the proximal ESR1 promoter, which has not been fully explored. Using ChIP analysis and reporter gene assays covering − 2500 bp to + 2000 bp of the TSS position, we showed that the activity of the proximal ESR1 promoter was markedly reduced upon menin downregulation independently of H3K4me3 status. Importantly, by analyzing the expression of menin in 354 human breast cancers, we found that a lower expression was associated with ER-negative breast cancer (P = 0.041). Moreover, among the 294 ER-positive breast cancer samples, reduced menin expression was not only associated with larger tumors (P = 0.01) and higher SBR grades (P = 0.005) but also with the luminal B-like breast cancer subtype (P = 0.006). Consistent with our clinical data, we demonstrated that GATA3 and FOXA1, co-factors in ESR1 regulation, interact physically with menin in MCF7 cells, and MEN1 knockdown led to altered protein expression of GATA3, the latter being a known marker of the luminal A subtype, in MCF7 cells.
Conclusion
Taken together, our data provide clues to the important role of menin in ERα regulation and the formation of breast cancer subtypes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10549-021-06339-9.
Keywords: Menin, ESR1, ERα, GATA3, Breast cancer, Luminal subtypes
Introduction
Breast cancers are among the most common malignancies worldwide and remain the leading cause of cancer-related mortality in women [1]. Previous receptor expression analyses enabled their classification into 4 major clinical subtypes, including luminal A, luminal B, HER2-enriched and triple-negative [2]. The luminal A subtype encompasses approximately 44% of breast cancers. This subtype is estrogen receptor (ER)-positive and/or progesterone receptor (PR)-positive and human epidermal growth factor receptor 2 (HER2)-negative, which displays a reduced expression of proliferation-related genes [3] and is sensitive to endocrine therapy with an overall favorable prognosis. The luminal B subtype represents around 20% of breast cancers and displays lower expression of ERα-related genes, a variable expression of (HER2), and a higher expression of proliferation-related genes [4]. This subtype harbors more genomic instability and has a poorer prognosis than the luminal A subtype [5]. The HER2-enriched subtype is ER-negative, PR-negative, HER2-positive, and highly sensitive to therapies targeting the HER2 receptor. The triple-negative breast cancer (TNBC) subtype is negative for all three receptors [6] and is the most aggressive with the worst prognosis.
Patients harboring MEN1 mutations are predisposed to multiple endocrine neoplasia type 1 (MEN1) syndrome, which is associated with multi-occurring endocrine tumors [7], as well as several types of non-endocrine tumors [8]. Numerous studies have revealed that menin is a multifaceted protein involved not only in the development and control of cell growth of endocrine cells but also in a variety of biological processes, including hematopoiesis and osteogenesis [9–11]. The wide range of biological functions regulated by menin results from its interaction with numerous proteins [12]. These menin-interacting proteins include transcription factors (the components of AP1, NFkB, and the TGFβ signaling pathways) and chromatin-modifying proteins (mixed lineage leukemia (MLL), Sin3A, and HDAC) [12, 13]. Notably, menin physically interacts with a range of nuclear receptors, including ERα and the androgen receptor (AR), to regulate their pathways [14–16].
Over the last few years, evidence has emerged, in vivo, to suggest that menin may play a role in breast cancers [17]. (1) Female heterozygous Men1 knockout mice develop cancers of mammary cells with a low frequency [18], and conditional mammary gland-specific Men1 disruption leads to the development of mammary intraepithelial neoplasia (MIN) in over 50% of female mutant mice [19]. (2) Importantly, an exhaustive analysis of several cohorts of MEN1 patients revealed a significant predisposition to breast cancer [20]. (3) Menin downregulation was detected in a substantial proportion of human sporadic breast cancer samples [19], and MEN1 mutations were found, although rarely in sporadic breast cancers, justifying its addition to the list of driver mutations/genes of this pathology [21, 22]. Of note, the abovementioned analyses all highlight the suppressive role of menin in mammary cell tumorigenesis. However, Imachi et al. found that, among 65 ERα + breast cancer samples treated with tamoxifen, menin-positive tumors (20 patients) had worse clinical outcome and were more resistant to tamoxifen than menin-negative tumors, suggesting that menin exerts oncogenic effects in these cases [15]. Interestingly, a recent publication, revealing the role of menin in regulating the enhancer of the ESR1 gene coding for ERα, suggests distinct functions for menin in primary normal mammary cells and in breast cancers [23]. The authors showed that, although menin possesses a crucial tumor-suppressive role in normal mammary cells, it acts as an oncogenic factor in ERα + breast cancer cell lines through an enhancer-mediated regulation of ESR1 transcription. Notably, they demonstrated, by the ZR75-1 breast cancer cell line which does not express menin, that the re-expression of menin leads to enhanced ERα expression. Given the heterogeneous nature of breast cancers, we sought to further investigate the regulation of ESR1 by menin and assess the putative relationship between ESR1 dysregulation due to menin inactivation and the occurrence of human breast cancer subtypes.
Results
Men1 deficiency in mice leads to ERα downregulation in early mammary lesions
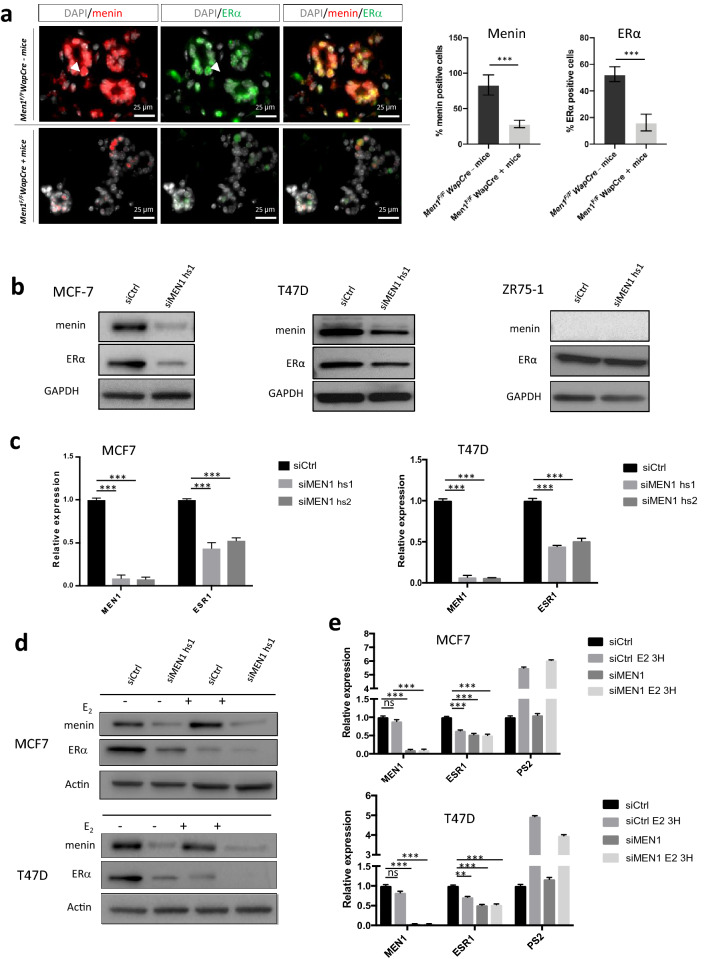
We previously reported the occurrence at a high incidence of mammary intraepithelial neoplasia (MIN) lesions, displaying weak ERα expression, in Men1 mammary conditional mutant mice [19]. To further determine the causative role of menin deficiency in reduced ERα expression, we carried out double IF analysis of menin and ERα expression in normal and young mutant mice with MIN lesions, before the development of breast cancer. Three mice per control or mutant group were analyzed. As shown in Fig. 1a, all of the mammary luminal cells in Men1F/F-WapCre− control mice expressed menin. Conversely, menin expression was lost in 71.1% of mammary cells in these young Men1F/F-WapCre+ mice (Fig. 1a). ERα was expressed in approximately 52.5% of luminal cells expressing menin in the former group (Fig. 1a, upper panel), whereas immunofluorescence revealed that ERα expression was nearly 3.2-fold lower in Men1-deficient cells in Men1F/F-WapCre+ mice (lower panel), compared to Men1F/F-WapCre− mice. The merged images of menin and ERα staining clearly highlight that ERα is less expressed specifically in the nuclei of menin-deficient luminal cells (Fig. 1a).
Fig. 1.
Reduced menin expression leads to a decrease in ERα expression. a Co-immunofluorescence against menin and ERα on mammary gland sections from Men1F/FWapCre− mice (upper panel) and Men1F/FWapCre+ mice (lower panel) at < 12 months of age. Quantification of IF signals for menin and ERα is shown on the right. b Western blot analyses using antibodies against menin and ERα in MCF7, T47D, and ZR75-1 cells treated with siRNA control (siCtrl) or siRNA targeting the MEN1 gene (siMEN1 hs1). c Quantitative RT-qPCR analyses of MEN1 and ESR1 expression in MCF7 and T47D cells treated with siCtrl or two different siMEN1 (hs1 or hs2). d Western blot analyses using antibodies against menin and ERα in MCF7 and T47D treated with siRNA control (siCtrl) or siMEN1 hs1 and then subjected to estradiol (E2) stimulation at a concentration of 10 nM. e Quantitative RT-qPCR analyses of the MEN1 and ESR1 expression in MCF7 and T47D cells treated with siCtrl or siMEN1 hs1 and then subjected to estradiol (E2) stimulation at a concentration of 10 nM. PS2 transcript was used as a positive control. All data are expressed as mean ± SEM, ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001
Menin downregulation in human ERα + mammary cells leads to reduced ERα expression
Next, we further dissected the regulation of ERα expression by menin using different approaches in ERα + breast cancer cell lines. To achieve this, we first performed MEN1 knockdown (KD) using a siRNA approach. As shown in Fig. 1b, MEN1 KD MCF7 and T47D cells displayed reduced ERα protein expression by western blot analysis, unlike the menin-negative ERα + cell line, ZR75-1. Moreover, MEN1 KD led to a twofold decrease in ESR1 mRNA levels by RT-qPCR analysis in MCF7 and T47D cell lines (Fig. 1c). We then verified the effects of MEN1 KD on ESR1 mRNA and ERα expression levels under estrogen (E2) stimulation. Western blot and RT-qPCR analyses showed that MEN1 silencing further abrogated ERα expression under E2 stimulation but had no additional effect on ESR1 transcription (Fig. 1d, e), most likely due to the fact that transcription had already reached its lowest level upon estrogen stimulation. All the above data thus confirm our in vivo analyses, indicating that menin is essential in maintaining ESR1 transcription and ERα expression.
Menin binds to the proximal region of the ESR1 promoter
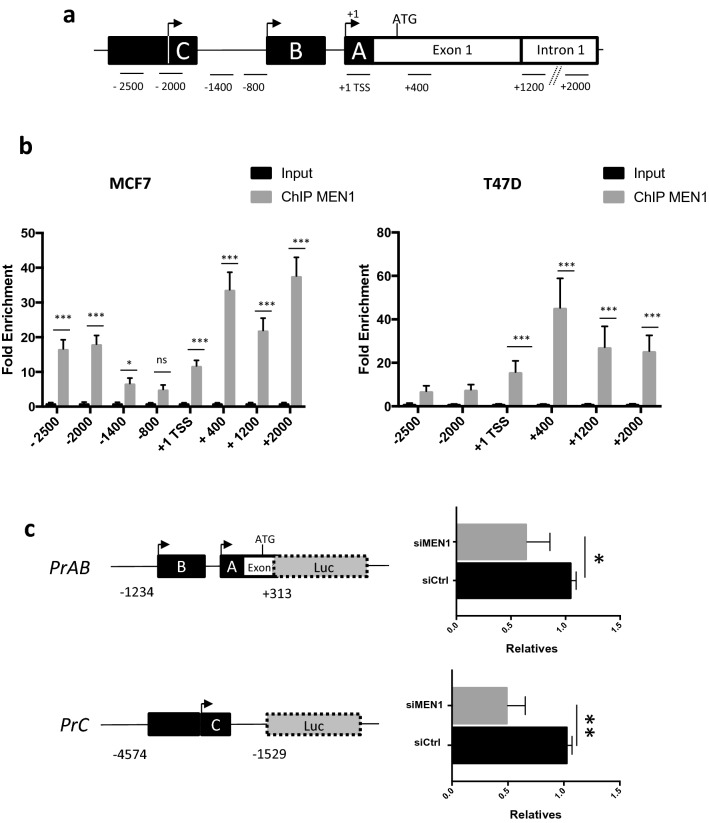
Dreijerink et al. reported that menin plays a crucial role in the regulation of ESR1 transcription in an enhancer-mediated way [23]. We noticed that, although the study revealed the binding of menin at the transcription start site (TSS) of the ESR1 promoter, no further analyses were reported on this region. We thus carried out ChIP analyses to evaluate the binding of menin to the − 2500 bp to + 2000 bp region around the TSS, defined based on previously reported works (Fig. 2a) [23, 24], to fully decipher the regulation of ESR1 transcription by menin at the proximal promoter region. Menin was significantly enriched in the ESR1 promoter region encompassing the TSS to + 2000 bp in MCF7 (Fig. 2b, left panel) and T47D (Fig. 2b, right panel) cells and more specifically in the promoter area C in MCF7 cells. Importantly, we confirmed by luciferase reporter assays that the transcriptional activity of the proximal ESR1 promoter region A/B and C was markedly reduced when MEN1 was knocked down (Fig. 2c).
Fig. 2.
Recruitment and effects of menin on the proximal ESR1 promoter. a Schematic diagram of the different regions of the ESR1 promoter studied, including A, B, and C regions of ESR1, that were cloned into luciferase reporter constructs (see c). b ChIP-Quantitative PCR analyses of menin binding on the − 2500 bp/ + 2000 bp area flanking the transcription start site (TSS) of ESR1 in MCF7 (left) and T47D (right) cells. c MCF7 cells were transfected with siCtrl or siMEN1 hs1 and pGL3-PrAB or pGL3-PrC luciferase reporter. After 48 h of transfection, luciferase activities were measured in triplicate for each condition. All the data are expressed as mean ± SEM, ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001
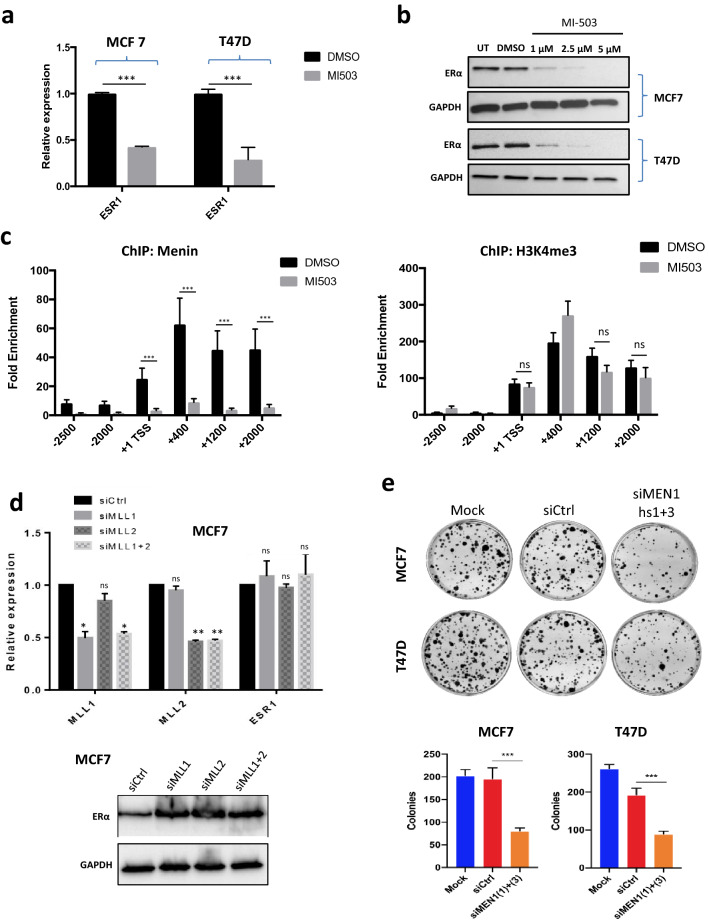
On considering the data published by Dreijerink et al. [23] showing that H3K4me3 marks are more abundant in the proximal part of the ESR1 promoter, we sought to investigate the involvement of the MLL complex, a major actor modifying H3K4me3 marks [13], in the regulation of the ESR1 promoter. By treating MCF7 and T47D cells with an inhibitor of the menin–MLL interaction, MI503, RT-qPCR analyses unveiled a more than twofold decrease in ESR1 transcription (Fig. 3a). Western blot analyses in MCF7 and T47D cells using the same inhibitor also revealed a decrease in ERα expression at the protein level (Fig. 3b). We then verified the potential alteration of H3K4me3 marks at this region upon inhibition of the MEN1/MLL complex. ChIP analysis with anti-H3K4me3 antibodies showed that, while MI503 treatment led to a markedly reduced binding of menin (Fig. 3c, left panel) at 48 h, H3K4me3 methylation was not altered upon menin/MLL inhibition in the tested region at this time point (Fig. 3c, right panel). We then performed the same analysis at 72 h and 96 h and found that at 72 h, one of the tested H3K4me3 marks significantly decreased, while other H3K4me3 marks slightly decreased but insignificantly (Fig. S1, upper panel). At 96 h, all H3K4me3 marks had decreased significantly, except for the one on the TSS site (Fig. S1, lower panel), whereas a substantial proportion of the MI503-treated cells stopped to grow. Furthermore, RT-qPCR analyses showed that neither siMLL1, nor siMLL2, nor their combination affected ESR1 transcription (Fig. 3d, left panel) and ERα expression (Fig. 3d, right panel). Taken together, our data provide evidence that menin regulates the proximal ESR1 promoter and raise the question of the involvement of factors other than the MLL complex in this regulation.
Fig. 3.
The regulation of the proximal ESR1 promoter by menin does not entirely rely on the MLL complex. a Quantitative RT-qPCR analyses of ESR1 expression in MCF7 and T47D cells treated or not with the inhibitor of menin–MLL interaction, MI503 at a concentration of 2 µM. b Western blot analyses of ERα expression in MCF7 and T47D cells untreated (UT) or treated with MI503 at concentrations of 1, 2.5, and 5 µM. c ChIP-qPCR analysis of the binding of the anti-menin antibody (left panel), or anti-H3K4me3 antibody (right panel) to the − 2500 bp/+ 2000 bp area flanking the transcription start site (TSS) on the ESR1 in MCF7 cells treated or not with MI503 at a concentration of 2 µM for 48 h. d Quantitative RT-qPCR (left panel) and western blot (right panel) analyses of the expression of MLL1 (KMT2A), MLL2 (KMT2B), and ESR1 in MCF7 cells treated with siCtrl, siMLL1, siMLL2, or combined siMLL1 and siMLL2. e Representative images of foci formation assay with MCF7 and T47D cells treated with siMEN1(1) + (3) or siCrtl. Quantification of foci formation assay is shown on the right. All the data are expressed as mean ± SEM, ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001
Having confirmed and extended the role of menin in regulating ESR1 transcription, we sought to further confirm its role in the growth of ER+ breast cancer cells, as previously reported [23]. To achieve this, we used colony formation assays to investigate cell growth behavior after MEN1 knockdown. As shown in Fig. 3e, MEN1 silencing in both MCF7 and T47D cells led to reduced colony formation, supporting that menin is needed for the growth of these ER+ breast cancer cells.
Lower menin expression is associated with luminal B-like and ER-negative breast cancer subtypes
Our observations prompted us to perform a thorough investigation of the levels of the menin protein in a cohort of breast cancer patients having undergone surgery at the Centre Léon Bérard (CLB) hospital from 2001 to 2003. Among 354 patients, 151 (42.7%) had a low menin expression, while 203 (57.3%) had a high expression. Among the 294 patients with ER+/HER2− tumors, 116 patients (39.5%) had a low nuclear menin expression and 178 patients (60.5%) had a high expression. In the cohort of 354 patients, we found that lower nuclear menin (H score ≤ 100) expression was significantly associated with ER-negative breast cancers (P = 0.041) and with the HER2-enriched subtype (P = 0.049, Table 1). Moreover, among the 294 ER+/HER2− patients, we observed that low menin expression was associated with the luminal B-like breast cancer subtype (P = 0.006), larger tumors (P = 0.016), and higher SBR grades (P = 0.005, Table 2).
Table 1.
Correlation between menin expression and the clinico-pathological factors of 354 breast cancer patients
| Variable | Menin low (≤ 100) | Menin high (> 100) | P* | ||
|---|---|---|---|---|---|
| No. 151 |
(%) (42.7) |
No. 203 |
(%) (57.3) |
||
| Age (year) | |||||
| Mean (± SD) | 59.2 | (± 13.3) | 58.6 | (± 11.2) | 0.081† |
| Age groups | |||||
| ≤ 50 years | 31 | (20.5) | 58 | (28.6) | 0.085 |
| > 50 years | 120 | (79.5) | 145 | (71.4) | |
| BMI | |||||
| ≤ 25 | 78 | (53.4) | 133 | (67.9) | 0.007 |
| > 25 | 68 | (46.6) | 63 | (32.1) | |
| T. size | |||||
| ≤ 2 cm | 72 | (47.7) | 131 | (64.5) | |
| > 2 cm | 79 | (52.3) | 72 | (35.5) | 0.002 |
| LN invasion | |||||
| No | 64 | (42.4) | 84 | (41.1) | |
| Yes | 87 | (57.6) | 119 | (58.6) | 0.85 |
| SBR grade | |||||
| Gr 1 | 16 | (10.6) | 46 | (22.7) | 0.001 |
| Gr 2 | 68 | (45.0) | 102 | (50.2) | |
| Gr 3 | 67 | (44.4) | 55 | (27.1) | |
| ER status | |||||
| Negative | 26 | (17.2) | 20 | (9.9) | 0.041 |
| Positive | 125 | (82.8) | 183 | (90.1) | |
| PR status | |||||
| Negative | 44 | (29.1) | 44 | (21.7) | 0.108 |
| Positive | 107 | (70.9) | 159 | (78.3) | |
| Her 2 status | |||||
| Negative | 84 | (89.6) | 100 | (95.2) | 0.049 |
| Over-expressed | 12 | (10.4) | 5 | (4.8) | |
Bold indicates statistically significant values
*Correlation by Pearson’s χ2 test unless otherwise specified
†Difference between means analyzed using the Student’s t test
Table 2.
Correlation between menin expression and the clinico-pathological factors of 294 ER+/HER2− breast cancer patients
| Variable | Menin low | Menin high | P* | ||
|---|---|---|---|---|---|
| No. 116 |
(%) (39.5) |
No. 178 |
(%) (60.5) |
||
| Age groups | |||||
| ≤ 50 years | 21 | (18.1) | 51 | (28.7) | 0.040 |
| > 50 years | 95 | (81.9) | 127 | (71.3) | |
| Menopausal status | |||||
| Premenopausal | 22 | (19.9) | 56 | (32.6) | 0.053 |
| Postmenopausal | 92 | (80.1) | 118 | (67.4) | |
| BMI | |||||
| ≤ 25 | 64 | (57.1) | 116 | (67.8) | 0.045 |
| > 25 | 48 | (42.9) | 55 | (32.2) | |
| T. size | |||||
| ≤ 2 cm | 62 | (53.4) | 120 | (67.4) | 0.016 |
| > 2 cm | 54 | (46.6) | 58 | (32.6) | |
| LN invasion | |||||
| No | 46 | (39.7) | 74 | (41.6) | 0.744 |
| Yes | 70 | (60.3) | 104 | (58.4) | |
| SBR grade | |||||
| Gr 1 | 16 | (13.8) | 46 | (25.8) | 0.005 |
| Gr 2 | 62 | (53.4) | 99 | (55.6) | |
| Gr 3 | 38 | (32.8) | 33 | (18.5) | |
| PR status | |||||
| Negative | 18 | (15.5) | 21 | (11.8) | 0.358 |
| Positive | 98 | (84.5) | 157 | (88.2) | |
| Breast Ca. subtype | |||||
| Luminal A | 67 | (57.8) | 130 | (73.0) | 0.006 |
| Luminal B | 49 | (42.2) | 48 | (27.0) | |
| Type of adjuvant hormonal | |||||
| Tamoxifen | 51 | (45.5) | 81 | (45.8) | 0.970 |
| AI | 61 | (54.5) | 96 | (54.2) | |
| Type of adjuvant chemo | |||||
| Anthracyclin | 51 | (96.2) | 77 | (88.5) | 0.509 |
| Anthra/Taxane | 2 | (3.8) | 9 | (10.3) | |
| Other | 0 | (0) | 1 | (1.1) | |
Bold indicates statistically significant values
*Correlation by Fisher’s exact test
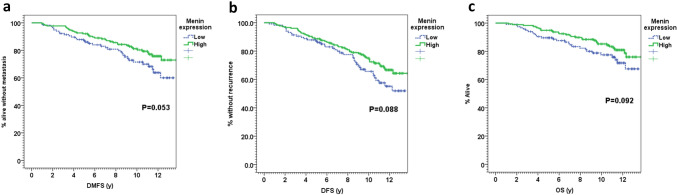
Interestingly, among the ER+/HER2− cohort, we found that low menin expression was associated with worse distant metastasis-free survival (DMFS), with a 10-year DMFS of 71.5% versus 81.2% in patients with high menin expression, P = 0.053 (Fig. 4a). Furthermore, low menin expression was also associated with a trend for worse disease-free survival (DFS), with a DFS of 65.7% at 10 years versus 75.0% in patients expressing high levels of menin (P = 0.088, Fig. 4b). Finally, lower expression of menin was also associated with a tendency toward worse overall survival (10-year OS of 77.5% versus 85.2%, P = 0.092, Fig. 4c).
Fig. 4.
Comparison of Kaplan–Meier estimates for a distant metastasis-free survival (DMFS); b disease-free survival (DFS); and c overall survival (OS) in the ER+/HER2− cohort of breast cancer patients included in the TMA of breast cancer patients, showing the survival curves of tumors with high (> 100) (green curve) or low menin nuclear staining (≤ 100) (blue curve)
The abovementioned data obtained in human patients, reminiscent of the observations made in Men1-deficient mutant mice, highlight a relationship between reduced menin expression and weaker ERα expression, suggesting that decreased ERα expression triggered by Men1 deficiency could be related to the occurrence of luminal B-like and ER-negative breast cancer subtypes.
Menin downregulation alters GATA3 and FOXA1 expression in ER+ breast cancer cells
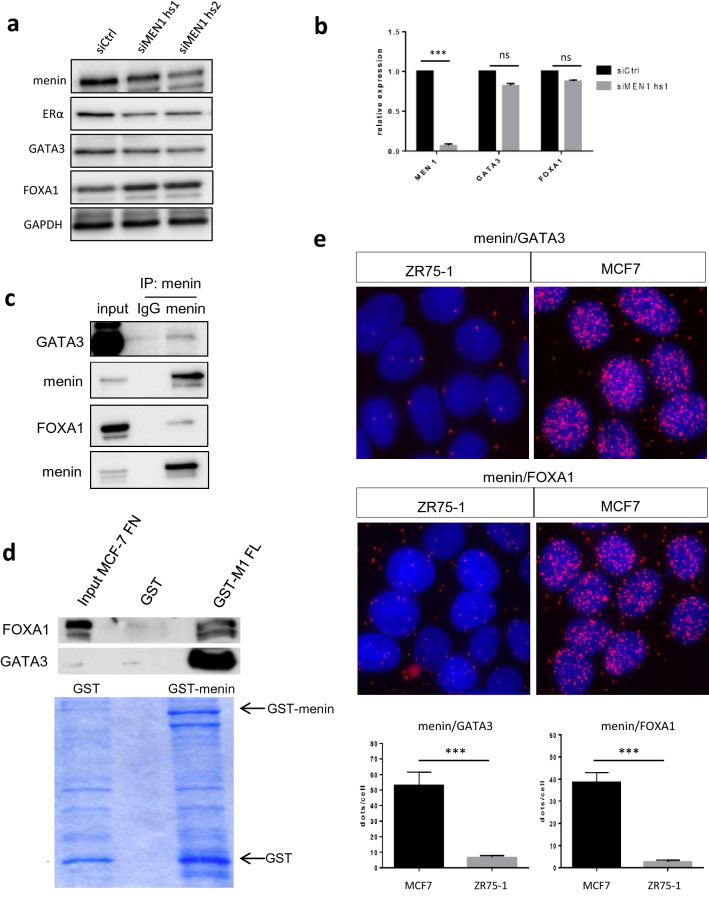
Having demonstrated a clinical correlation between menin inactivation and breast cancer subtypes, we wondered whether the factors important for luminal cell differentiation could be affected by menin in ER+ breast cancer cells. GATA3 is known to be a major factor involved in the regulation of ESR1 expression and is ubiquitously present in luminal A breast cancers [25]. Western blot analysis revealed that GATA3 expression was greatly reduced in both MCF7 and T47D cells at the protein level after MEN1 KD, although its level of mRNA was not impacted (Figs. 5a, b, S2). In parallel, we investigated the expression of FOXA1, which plays an important role in mammary cell differentiation and tumorigenesis, and found that its protein expression increased upon MEN1 KD in MCF7 cells, but remained unchanged in T47D cells, whereas no transcriptional alteration could be detected in both cell lines (Figs. 5a, b, S2). Since GATA3 has been reported to interact with menin in lymphocytes [26], and menin is known to interact with one member of the FOXA family, FOXA2 [27], we performed immunoprecipitation (IP) and PLA analyses to determine whether menin could interact with GATA3 and FOXA1 in breast cancer cells. The data obtained demonstrated that they interact with menin in MCF7 cells, as evidenced by IP at the endogenous level (Fig. 5c), by GST pull-down (Fig. 5d) and by PLA (Fig. 5e). Taken together, the current work revealed that menin interacts both with GATA3 and FOXA1 in ER + breast cancer cells. Moreover, its expression could be critically related to the expression of GATA3, a well-recognized marker of the luminal A subtype.
Fig. 5.
Menin interacts with GATA3 and FOXA1 and influences their expression. a Western blot analyses using antibodies against menin, GATA3, and FOXA1 in MCF7 cells treated with siCtrl or siMEN1 hs1. b Quantitative RT-qPCR analyses of the GATA3 and FOXA1 transcription in MCF7 cells treated with siCtrl or siMEN1 hs1. c Co-immunoprecipitation analyses were carried out by incubating nuclear lysates of MCF7 cells with either anti-IgG, or anti-GATA3 or FOXA1 antibodies and subjected to western blot analyses. d GST pull-down using GST-full-length (FL) menin and nuclear fraction of protein lysates of MCF7 cells, detected by western blot using the anti-GATA3 or FOXA1 antibodies. Coomassie blue-stained gel showing levels of recombinant GST proteins used in GST pull-down assay. e Upper: PLA analysis with anti-menin and anti-GATA3 antibodies in MCF7 and ZR75-1 cells, the latter expressing no menin. Under: the quantification of PLA analysis. All the data are expressed as mean ± SEM, ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
The current work provides both clinical and experimental data showing that menin is critically involved in ERα expression and that its inactivation in mammary cells is correlated with the occurrence of luminal B and ER-negative breast cancer subtypes. Our data highlighted cellular and molecular consequences of reduced menin expression in mammary cells, which may affect not only cell proliferation but also other hallmarks of cancer cells, in particular, cell differentiation.
We previously observed that the mammary lesions developing in mammary cell-specific Men1 mutant mice displayed low ERα expression [19]. Our current study further demonstrates that the decrease occurs in the precancerous lesions, suggesting that menin inactivation favors the tumorigenesis of mammary cells with weak ERα expression. Interestingly, by analyzing this expression in a large cohort of breast cancer patients, we found that reduced menin expression is significantly correlated with both ERα-negative and luminal B-like breast cancer subtypes. Consistently, low levels of menin were correlated with larger tumors, more advanced SBR grades, and worse prognosis, all of which are major features of these two breast cancer subtypes [4]. It is worth mentioning that these clinical data, together with those concerning reduced ERα expression in Men1-deficient mouse MIC lesions, further support the oncosuppressive role played by the MEN1 gene in the tumorigenesis of normal mammary gland cells [19, 23]. Moreover, while searching for luminal cell factors likely interacting with menin, we unveiled that menin binds physically to GATA3 and FOXA1 in mammary cells, and that MEN1 silencing reduces GATA3 expression in MCF7 and T47D cells. Of note, reduced GATA3 expression is often seen in the luminal B breast cancer subtype but not in luminal A [28]. However, the mechanisms leading to the occurrence of both luminal breast cancer subtypes remain elusive. The current work may provide useful insight and generate interest for further studies. In the meantime, considering the retrospective nature of the study and the heterogeneity of the therapies received by the patients included, the clinical analyses, which could be limited with the cutoff definition by IHC, should be confirmed in other cohorts, preferably through prospective studies.
Dreijerink et al. first described the capacity of menin to regulate ESR1 transcription by binding to the remote upstream part of regulatory sequences of ESR1, through an enhancer-mediated looping mechanism, involving GATA3 [23]. Moreover, the occupancy of this enhancer sequence by GATA3 has been reported to play an important role in the regulation of ERα expression upon estradiol stimulation [25]. Our findings provide complementary information related to the role of menin in ESR1 regulation through its proximal promoter. Intriguingly, our data showed retarded H3K4me3 methylation on the proximal ESR1 promoter with MI503 treatment, as well as a lack of clear ESR1 transcriptional alteration after single MLL1, MLL2, or their combined knockdown with siRNA. Since MI503 has been demonstrated not only to inhibit the interaction between menin and MLL1/MLL2 but also to reduce menin expression itself [29]; our data may suggest that factors other than the MLL complex may also participate in this regulation. It would be interesting in the future to identify the factors or cofactors that interact, positively or negatively, with menin to regulate this gene. In addition, our data seem to support the oncogenic role played by menin in ERα+ breast cancer cell lines, the proliferation of which is highly ERα-dependent. Therefore, by combining the data obtained from our experimental and clinical analyses, we consider that menin most likely acts as an oncogenic cofactor in the luminal A breast cancer subtype.
Conclusion
The emerging role for the MEN1 gene in mammary cell tumorigenesis appears to be multifaceted. Our current results provide further data showing that menin may play different, even opposite, roles in the development of different breast cancers, in agreement with the findings reported by Dreijerink et al. Taken together, these results may explain seemingly controversial data reported so far, in particular when comparing data obtained from naturally occurring tumors and those of cultured cancer cells. Furthermore, our findings may also raise awareness to the breast cancer subtypes selected when designing new therapeutic strategies involving the eventual use of menin and MLL inhibitors.
Materials and methods
Patients
We screened a total of 433 consecutive female patients with breast cancer who underwent surgery and (neo)/adjuvant therapy at the Centre Léon Bérard (CLB) between January 2001 and December 2003 (Additional file 1). Patients with complete data and with adequate samples assessable for menin by IHC were 354, among which 294 patients had ER+/HER2− tumors. The intrinsic subtypes of breast cancer were defined by the histological grade and IHC surrogates as per St Gallen 2013 consensus [30]. Patients were defined as luminal A-like if positive for ER and PR, negative for HER2 expression, and low proliferation (grade I or grade II with low Ki67 or mitotic index). Luminal B-like was defined as ER-positive and either: PR negativity, HER2 positivity or high proliferation. The study was conducted in accordance with the guidelines in the Declaration of Helsinki and the use of all patient tissues was approved by local IRB and carried out according to French laws and regulations.
TMA analysis of human breast cancers
Formalin-fixed paraffin-embedded breast cancers were prepared and processed for immunostaining as previously described [19]. Tissue micro-array (TMA) block preparation, menin nuclear expression assessment using IHC, and statistical analyses were performed as previously described [19]. The percentage of stained cells was multiplied by the intensity of staining to obtain the H score [31]. For the sake of correlations and survival analyses, the most discriminative cutoff in terms of DFS (as determined by Kaplan–Meier method) was chosen to divide the whole cohort of patients into high menin expression (H score > 100) and low menin expression (H score ≤ 100).
Animal breeding
Men1F/F-WapCre+ and Men1+/+-WapCre+ mice previously generated in our lab were used [19]. All animal experiments were conducted in accordance with accepted standards of animal care and were approved by the Animal Care and Use Committee of the University Lyon 1.
Cell culture, transfections, and luciferase assays
Three breast cancer cell lines expressing ERα, namely MCF7, T47D, and ZR75-1, were used in this study. Transient transfections were carried out in phenol red-free medium supplemented with 10% charcoal-stripped serum (Biowest) in order to remove steroid hormones (steroid depletion). Cells were transfected with 20 nM siRNA of, respectively, control siRNA (5 nmoles, Eurogentec), two different siRNA targeting human MEN1 transcript (siMEN1 hs1 (HSS106462) and hs2 (HSS181079), ThermoFisher Sci.), siRNA targeting human MLL1 (SiKMT2A: siRNA 107,890 ThermoFisher Sci.), siRNA targeting human MLL2 (SiKMT2B: siRNA s18833 ThermoFisher Sci.) using Jetprime® transfection reagent (Polyplus) for 72 h according to the manufacturer’s instructions. Inhibition of the menin–MLL interaction was achieved by MI503 (Active Biochem) at different concentrations. Prior to performing treatment with E2 and MI503, cells were grown in phenol red-free medium supplemented with 10% charcoal-stripped serum (Biowest) in order to remove steroid hormones (steroid depletion). Cells were then treated for 3 h with E2 (Sigma) 10−8 M and MI503 for 48 h. The treatment was repeated after 24 h due to the degradation of the inhibitor with time. Please also see Additional file 2—Supplemental Materials & Methods.
Foci formation assay
For foci formation assay, cells were seeded in 6-well culture plates at 5 × 102 cells for MCF7 and T47D. Cells were transfected with siRNA or treated with MI503 and cultured for 2 weeks. The ensuing colonies were stained with 0.05% crystal violet. The images of the plates were analyzed using ImageJ software. Each experiment was conducted in triplicate and statistical analyses were performed using the Prism software.
Construction of luciferase constructs
We used genomic DNA extracts from MCF7 cells to generate regions of the human ESR1 promoter, PrAB (genomic location: Chr6q25.1; 152127793-152129027) and PrC (genomic location: Chr6q25.1; 152124474-152127509) (Additional file 2). The resulting fragments of the proximal ESR1 promoter were cloned into the pGL3 Basic vector (Promega, Madison, WI).
Luciferase assays
For luciferase assays, MCF7 cells were cultured in 24-well plate. 48 h after transfection with 250 ng of the reporter plasmid PrAB or PrC, and 5 ng pRL-TK internal control vector, cell lysates were prepared and analyzed using a dual-luciferase reporter assay system (Promega, Madison, WI), as previously reported [27]. Comparisons between mean values were assessed using the two-tailed Student t test.
Real-time reverse transcription and qPCR analyses
RNAs were extracted using RNeasy-Kits (Qiagen, Valencia, USA). Real-Time PCR analyses were carried out on a Step-One RT-System (Applied Biosystem, France) using SYBR-Green (Life Technologies, France) and corresponding primers (Additional file 2). Results of each sample were normalized.
Protein extraction, immunoprecipitation, GST pull-down, and immunoblotting
Total protein extracts from cells and immunoprecipitation were prepared and analyzed as described previously [27]. For GST pull-down assays, 1.25 µg purified GST menin protein or GST control protein was incubated with 1 mg or 2 mg of nuclear cell extracts prepared from MCF7 cells, as previously described [27]. The co-sedimented proteins were detected by western blot using standard conditions.
Immunostaining
Tissue preparation, immunostaining, and statistical analyses were performed as previously described [19]. Briefly, endogenous peroxidases were quenched in 3% H2O2 solution for 30 min at room temperature. Heat-induced epitope retrieval was performed by immersion in antigen-unmasking solution (catalog no. H-3300; Vector Laboratories) in a microwave oven for 15 min. After blocking with antibody diluent (Dako), sections were incubated overnight with a primary antibody (Additional file 2). For immunofluorescence (IF) staining, signals were detected with a Cy3 or Cy5 tyramide amplification kit (PerkinElmer), with prior incubation with the appropriate biotinylated secondary antibody according to the manufacturer’s instructions. Images were acquired on an Eclipse-NiE NIKON microscope using the NIS-Elements Software.
Proximity ligation assay (PLA), image acquisition, and analysis
MCF7 cells were fixed in methanol for 5 min and washed twice in PBS and then treated and analyzed according to the manufacturer’s instructions (Duolink II Fluorescence, Olink Bioscience, Sweden). Images were acquired on an Eclipse NiE NIKON microscope using the NIS-Elements Software. For each sample, at least one hundred cells were counted. Analysis and quantification of these samples were performed using the ImageJ software (free access). PLA dots were quantified on 8-bit images using the ‘Analyse Particles’ command, while cells were counted using the cell counter plugin.
ChIP-qPCR assay
Chromatin for ChIP analysis was prepared from 5 million MCF7 or T47D cells. Briefly, cells were fixed in 1% formaldehyde for 10 min, nuclei were obtained and lysed in 300 μl ice-cold RIPA buffer prior to Chromatin-DNA shearing with a Diogene Bioruptor. ChIP was performed using 5 μg of primary antibodies. Dynabeads® Protein G (10003D, Life Technologies, France) was used to retrieve immunocomplexes according to manufacturers’ instructions.
Statistical analyses
For molecular biology experiments, statistical analyses were performed as described in the Fig. legends; unpaired Student’s t tests were used unless otherwise indicated. All analyses were conducted using the Prism5 software (GraphPad, USA); a P value of < 0.05 was considered to be significant. Results are expressed as means ± standard errors of the means (SEM). For the patient samples, numerical variables were compared using Student’s t test, while categorical variables were compared using χ2 test. Distant metastasis-free survival (DMFS) was defined as time from diagnosis to the date of distant metastasis or death or last follow-up. Disease-free survival (DFS), defined as the time from diagnosis to death or progression or to date of last follow-up (for censored patients), was also calculated. Survival rates were estimated using the Kaplan–Meier method, and comparisons between menin expression groups were performed using the log-rank test. All statistical tests were two-sided, and the P value was considered statistically significant if lower than 5%. Statistical analyses were performed using SPSS 20.0 statistics package.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Brigitte MANSHIP for her assistance in proofreading the manuscript.
Author contributions
RT and RAZ conducted the experiments and analyzed and interpreted the data, prepared figures, and manuscript; LK participated in the design of the clinical study, performed the statistical analysis, and participated in manuscript preparation; YL participated in the molecular and cellular studies and interpreted the data; LM participated in the molecular and cellular studies and interpreted the data; SG participated in the design of the molecular study and established methods; LC participated in the design of the molecular study and data interpretation and helped to draft the manuscript; PB provided technical and material support and participated in data interpretation and manuscript preparation; TB conceived the design of the clinical study and participated in clinical data collection and data interpretation; IT conceived the design of the clinical study; participated in clinical data collection, data interpretation, and manuscript preparation; and provided critical revision of the manuscript; MLR and CXZ conceived and supervised the study and manuscript preparation and obtained funding.
Funding
This study was supported by Epigenetics & Cancer Program (ASC14092CSA), the Fondation de l’Association pour la Recherche contre le Cancer (SFI20101201530), the Ligue Inter-régionale contre le Cancer (R19040CC), the Association: « Le Cancer du sein, parlons-en», and the Région Auvergne–Rhône-Alpes (SICORRA22425). R.T was the recipients of a PhD-fellowship from French Ministry of Higher Education and Research and of a 4th year doctoral fellowship from Société Française d’Endocrinologie and the Fondation ARC, France, respectively. R.A.Z was the recipient of a PhD-fellowship from Association "G04MEDIA S.A.R.L," Lebanon. Y.K was the recipient of a PhD-fellowship from China Scholarship Council.
Data availability
The data that support the findings of this study are available from the corresponding author, [CXZ for basic research and IT for clinical data analyses], upon reasonable request.
Declarations
Conflict of interest
Nothing to disclose.
Ethical approval
The specimens of patients with breast cancer were conducted according to French laws and regulations and written informed consent was obtained from each patient. All animal experiments were performed in accordance with the animal care guidelines of the European Union and were validated by the local Animal Ethic Evaluation Committee (CECCAPP: C2EA-15 agreed by the French Ministry of High School and Research, « Autorisation de projet» CLB-2012-053).
Informed consent
All of the authors approved the contents and the data in this manuscript and agreed for its submission for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Romain Teinturier and RazanAbou Ziki share the first authorship.
Contributor Information
Isabelle Treilleux, Email: isabelle.treilleux@lyon.unicancer.fr.
Chang Xian Zhang, Email: chang.zhang@lyon.unicancer.fr.
References
- 1.Tao Z, et al. Breast cancer: epidemiology and etiology. Cell Biochem Biophys. 2015;72:333–338. doi: 10.1007/s12013-014-0459-6. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai X, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 4.Creighton CJ. The molecular profile of luminal B breast cancer. Biologics. 2012;6:289–297. doi: 10.2147/BTT.S29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kittaneh M, Montero AJ. Biomarkers in cancer molecular profiling for breast cancer: a comprehensive review. Biomark Cancer. 2013;5:61–70. doi: 10.4137/BIC.S9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eliyatkın N, Yalçın E, Zengel B, Aktaş S, Vardar E. Molecular classification of breast carcinoma: from traditional, old-fashioned way to a new age, and a new way. J Breast Health. 2015;11:59–66. doi: 10.5152/tjbh.2015.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakker RV. Multiple endocrine neoplasia—syndromes of the twentieth century. J Clin Endocrinol Metab. 1998;83:2617–2620. doi: 10.1210/jcem.83.8.5045. [DOI] [PubMed] [Google Scholar]
- 8.Falchetti A, et al. Multiple endocrine neoplasia type 1 (MEN1): not only inherited endocrine tumors. Genet Med. 2009;11:825–835. doi: 10.1097/GIM.0b013e3181be5c97. [DOI] [PubMed] [Google Scholar]
- 9.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowa H, et al. Menin is required for bone morphogenetic protein 2- and transforming growth factor beta-regulated osteoblastic differentiation through interaction with Smads and Runx2. J Biol Chem. 2004;279:40267–40275. doi: 10.1074/jbc.M401312200. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, et al. Loss of menin in osteoblast lineage affects osteocyte-osteoclast crosstalk causing osteoporosis. Cell Death Differ. 2017;24:672–682. doi: 10.1038/cdd.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matkar S, Thiel A, Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci. 2013;38:394–402. doi: 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreijerink KMA, et al. Menin links estrogen receptor activation to histone H3K4 trimethylation. Can Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 15.Imachi H, et al. Menin, a product of the MENI gene, binds to estrogen receptor to enhance its activity in breast cancer cells: possibility of a novel predictive factor for tamoxifen resistance. Breast Cancer Res Treat. 2010;122:395–407. doi: 10.1007/s10549-009-0581-0. [DOI] [PubMed] [Google Scholar]
- 16.Malik R, et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med. 2015;21:344–352. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abou Ziki R, Luo Y, Vlaeminck-Guillem V, Le Romancer M, Zhang CX. Involvement of the MEN1 gene in hormone-related cancers: clues from molecular studies, mouse models, and patient investigations. Endocrines. 2020;1:58–81. doi: 10.3390/endocrines1020007. [DOI] [Google Scholar]
- 18.Bertolino P, Tong W-M, Galendo D, Wang Z-Q, Zhang C-X. Heterozygous men1 mutant mice develop a range of endocrine tumors mimicking multiple endocrine neoplasia type 1. Mol Endocrinol. 2003;17:1880–1892. doi: 10.1210/me.2003-0154. [DOI] [PubMed] [Google Scholar]
- 19.Seigne C, et al. High incidence of mammary intraepithelial neoplasia development in Men1-disrupted murine mammary glands. J Pathol. 2013;229:546–558. doi: 10.1002/path.4146. [DOI] [PubMed] [Google Scholar]
- 20.Dreijerink KMA, Goudet P, Burgess JR, Valk GD. Breast-cancer predisposition in multiple endocrine neoplasia type 1. N Engl J Med. 2014;371:583–584. doi: 10.1056/NEJMc1406028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong YJ, Oh HK, Bong JG. Multiple endocrine neoplasia type 1 associated with breast cancer: a case report and review of the literature. Oncol Lett. 2014;8:230–234. doi: 10.3892/ol.2014.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreijerink KM, et al. Article enhancer-mediated oncogenic function of the menin tumor suppressor in breast cancer. Cell Rep. 2017;18(10):2359–2372. doi: 10.1016/j.celrep.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kos M, Reid G, Denger S, Gannon F. Minireview: genomic organization of the human ERalpha gene promoter region. Mol Endocrinol. 2001;15:2057–2063. doi: 10.1210/mend.15.12.0731. [DOI] [PubMed] [Google Scholar]
- 25.Eeckhoute J, et al. Positive cross-regulatory loop ties GATA-3 to estrogen receptor? Expression in breast cancer. Can Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 26.Nakata Y, et al. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. J Am Soc Hematol. 2013;116:1280–1290. doi: 10.1182/blood-2009-05-223255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonnavion R, et al. Foxa2, a novel protein partner of the tumour suppressor menin, is deregulated in mouse and human MEN1 glucagonomas. J Pathol. 2017 doi: 10.1002/path.4885. [DOI] [PubMed] [Google Scholar]
- 28.Mair B, et al. Gain- and loss-of-function mutations in the breast cancer gene GATA3 result in differential drug sensitivity. PLoS Genet. 2016;12:1–26. doi: 10.1371/journal.pgen.1006279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, et al. Disruption of the menin-MLL interaction triggers menin protein degradation via ubiquitin-proteasome pathway. Am J Cancer Res. 2019;9:1682–1694. [PMC free article] [PubMed] [Google Scholar]
- 30.Goldhirsch A, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choucair A, et al. The arginine methyltransferase PRMT1 regulates IGF-1 signaling in breast cancer. Oncogene. 2019;38:4015–4027. doi: 10.1038/s41388-019-0694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, [CXZ for basic research and IT for clinical data analyses], upon reasonable request.