Abstract
Conventional CD8+ memory T cells develop upon stimulation with foreign antigen and provide increased protection upon re-challenge. Over the past two decades, new subsets of CD8+ T cells have been identified that acquire memory features independently of antigen exposure. These antigen-inexperienced memory T cells (TAIM) are described under several names including innate memory, virtual memory, and memory phenotype. TAIM cells exhibit characteristics of conventional or true memory cells, including antigen-specific responses. In addition, they show responsiveness to innate stimuli and have been suggested to provide additional levels of protection toward infections and cancer. Here, we discuss the current understanding of TAIM cells, focusing on extrinsic and intrinsic molecular conditions that favor their development, their molecular definitions and immunological properties, as well as their transcriptional and epigenetic regulation.
Keywords: CD8+ T cell, Antigen-inexperienced memory, Virtual memory, Innate memory
Introduction
T lymphocytes (T cells) are key players in cellular immune responses. Each T cell is equipped with a unique, clonotypic T cell receptor (TCR) capable of recognizing a number of small protein fragments (peptides) in complex with MHC molecules (pMHC) that are displayed on the surface of antigen-presenting cells. This ensures that, on a population level, T cells can respond to almost any infection. After a primary T cell-mediated immune response, a subset of antigen-specific T cells persists as long-lived memory T cells in lymphoid organs. Upon antigen re-challenge, memory cells are pre-programmed to differentiate rapidly into effector cells warranting effective secondary immune responses [1, 2].
In addition to these ‘true’ memory T cells (TTM), other, non-conventional memory T cells are being identified and characterized, as documented by an increasing number of independent studies. Specifically, CD8+ T cells with a memory phenotype can arise in an antigen-inexperienced manner, i.e., without previously being activated [3–5]. For simplicity, we here refer to these cells as antigen-inexperienced memory T cells (TAIM). TAIM cells constitute around 10–40% of total peripheral CD8+ T cells, indicating their potential importance in the immune system [6–13]. Two major CD8+ TAIM cell types have been described: innate memory and virtual memory cells. Innate memory (TIM) cells arise in the thymus, where they mostly depend on IL-4 [5, 14]. Virtual memory (TVM) cells develop in the periphery and depend on IL-15 [15]. However, the distinction between and origin of the two types of TAIM cells is still under debate [3, 16].
Functionally, TAIM cells partially resemble conventional antigen-experienced TTM cells by mounting a rapid immune response upon antigen stimulation [3, 5]. The immune response of TAIM cells from young animals is particularly fast, suggesting that they provide extra protection in young animals when the immune system is not fully established [17]. In aged mice, TAIM cells are major contributors to the anti-influenza response [18]. Besides mediating a TCR-dependent immune response, TAIM cells have also been shown to provide antigen non-specific bystander protection in response to innate stimuli [8, 9, 19]. In addition, a potential role for TAIM cells in the anti-tumor response has been suggested: TAIM cells are able to infiltrate tumors and express high densities of the inhibitory receptor PD-1, a common target for immunotherapy [12, 20]. Furthermore, chemotherapeutic treatment of tumor cells increases the abundance of TAIM cells [21]. These TAIM cells can inhibit tumor growth in an MHC-I independent manner [21]. This unique responsiveness of TAIM cells to different stimuli in combination with the substantial number of TAIM cells warrants detailed investigation into this ‘neglected’ CD8+ T cell compartment.
Another type of memory-phenotype cells are those induced by homeostatic proliferation in a lymphopenic environment [22]. It has been suggested that under lymphopenic conditions, these homeostatic proliferation-induced memory-phenotype cells (THP) arise from naïve T cells exposed to high levels of the homeostatic cytokine IL-7 [22]. The sum of these insights reveals that the T cell memory pool is very heterogenous and that the acquisition of T-cell memory features does not follow a single trajectory. Increasing our knowledge about TAIM cells will help to better understand the role of these diverse memory T cell subsets in controlling specific immune responses. In this review we will not discuss THP (for a detailed review see Sprent et al. [22]), but focus on TAIM cells that arise independent of experimentally induced lymphopenia. This review focuses on the origin of TAIM cells and how their numbers and functions are regulated under specific physiological and immunological conditions. Furthermore, we will discuss transcriptional and (epi)-genetic regulation underlying the generation of TAIM cells.
Markers of TAIM cells
How are CD8+ TAIM cells identified and distinguished from TTM cells? Several definitions of TAIM, TVM and TIM are being used in the literature, defining the subsets based on surface markers, functionality and antigen-specificity [3–5]. The basic definition of a TAIM cell is a T cell that has not been exposed to foreign antigen but has memory-like features. This antigen-inexperienced nature of TAIM cells can be defined in several ways. One option is tetramer staining to identify CD8+ T cells that are specific for defined pMHC class I complexes they have not been challenged with (e.g., the chicken ovalbumin OVA and vaccinia virus B8R epitopes) [9, 13, 15, 23–25]. Accordingly, one expects that CD8+ T cells capable of binding the OVA and B8R tetramers are naïve, since they have not been previously challenged with the same pMHCs. Even though a single TCR can detect many different pMHCs, the presence of memory-phenotype cells among these tetramer-binding T cells in unchallenged mice indicates that these cells are not TTM cells, but instead represent TAIM cells. Another option to guarantee that cells are antigen inexperienced is to use CD8+ T cells with a transgenic (tg) TCR, ideally on a background that prevents expression of endogenous TCRs, such as a RAG1/2 knock-out background [9, 25, 26]. When all CD8+ T cells express the same tgTCR that only recognizes a synthetic peptide (e.g., the OT-I TCR recognizing the SIINFEKL epitope derived from OVA), the memory-phenotype cells identified without exposure to that specific antigen are TAIM cells. Exposing the cells to the specific synthetic antigen will result in the formation of TTM cells. To identify TAIM-specific markers TAIM cells have been compared to naïve (TN) and TTM cells. These markers are now broadly applied to determine whether memory-phenotype cells in unchallenged mice are TAIM cells. Awareness of the TAIM subset and the specific markers to detect them is important to determine whether memory-phenotype cells in unchallenged mice are TTM or TAIM. Unchallenged mice housed under specific pathogen-free (SPF) conditions typically have a memory-phenotype population, of which the majority (80–90%) are TAIM cells [6, 27, 28]. Many studies in the past did not use TAIM-specific markers and as a result were regarded all as memory-phenotype cells, including TAIM, as TTM. The current knowledge about the heterogeneity of the memory-phenotype population in unchallenged mice warrants a re-evaluation of our understanding of conventional T-cell differentiation [4]. Precise identification of the different memory subsets is thus required to gain further insight into the heterogenous process of memory T cell differentiation.
TAIM cells are generally defined by several combinations of surface markers. They express many of the surface markers of conventional or true memory cells, including CD44 and CD62L. However, they can be distinguished from TTM cells by low expression of CD49d, a marker of previous antigen experience [13, 15], high surface levels of CD122, part of the IL-2 and IL-15 receptor [13, 15, 29] and increased surface levels of CD5 [9, 30]. Furthermore, NKG2D, an activating cell surface receptor, is upregulated on a subset of TVM cells, but not on all IL-4-induced TAIM cells [9, 24, 31]. Besides these surface markers, the intracellular expression of the transcription factors T-bet and EOMES is also used to distinguish TAIM from conventional memory cells. In C57Bl/6 mice, TVM cells express high levels of both T-bet and EOMES, whereas TTM are either T-bet+ or EOMES+ depending on being effector memory or central memory, respectively [26]. In BALB/c mice, TIM cells both in thymus and periphery are T-bet−EOMES+ [14, 32]. In the thymus, TIM cells are often described as CD44+CD122+EOMES+ CD8+CD4− thymocytes that express the chemokine receptor CXCR3 [14, 33] and have reduced levels of CD24, a feature of more mature CD8+ thymocytes [34]. A subset of the peripheral TAIM population also expresses CXCR3 [35]. Based on CD44 and CXCR3 expression, CD8+ T cells can be divided into naïve (TN; CD44−CXCR3−), intermediate (CD44+CXCR3−), and memory phenotype (CD44+CXCR3+) [31, 32]. Whether TIM and TVM are different stages of one TAIM subset or different subsets is still the topic of ongoing discussion [4, 16, 36]. It has recently been proposed that CD122 expression can function as marker to distinguish TIM and TVM in the periphery, with TIM cells having lower expression of CD122 than TVM but higher than TN [11]. A detailed overview of TAIM surface markers is provided in Table 1.
Table 1.
Markers of TAIM cells
| Marker | Function | Subset | Reference |
|---|---|---|---|
| CD122high |
Part of the IL-2 and IL-15 receptor [29]. Expression of CD122 is induced by EOMES and T-bet [37]. Lower CD122 expression in TIM compared to TVM [11]. |
TIM and TVM | [13, 15] |
| CD49dlow |
α4β-Integrin; Part of the very late antigen-4 (VLA4) integrin that is involved in homing of effector T cells to the site of infection. CD49d is highly expressed on antigen-activated effector memory (TEM) and effector (TEFF) CD8+ T cells [38]. CD49d upregulation is suppressed by IL-4 [38]. |
TIM and TVM | [13, 15] |
| CD5high |
Negative regulator of TCR signaling [39–41]. CD5 surface levels function as a proxy for TCR-binding affinity of the thymus [39–41]. |
TVM | [9, 30] |
| NKG2Dlow/high |
Natural killer group 2 member D is a receptor expressed on activated CD8 T cells in mice and constitutively on CD8 T cell in human [42]. It functions as a co-stimulatory receptor [42]. NKG2D expression can be downregulated by IL-4 [31]. |
High in a subset of TVM | [9, 24, 31] |
| CXCR3high | Chemokine receptor involved in migration of T cells to peripheral site of infection [43]. | TIM and TVM | [14, 31–33, 35] |
Mechanisms inducing differentiation toward TAIM
The precise mechanisms that induce differentiation toward innate and virtual memory development are not completely understood, but two major components have been identified: quality of the TCR signal and cytokine signaling in the developmental niche.
TCR signaling in TAIM cell differentiation
The quality of the TCR signal critically contributes to the decision making of developing and differentiating T cells. During thymic maturation, thymocytes expressing a TCR with very weak or strong affinity for self-peptides die by neglect or negative selection, respectively, and only thymocytes expressing intermediate affinity TCR are positively selected and migrate into the periphery [44]. In the periphery, T cells require tonic TCR signals for their survival [22]. Strong binding of an antigen to the TCR results in activation of the peripheral T cell and differentiation toward memory and effector cells [45, 46], but only when critical co-stimulatory signals are provided; otherwise the cells will become anergic, a long-lasting stage of T cell non-responsiveness [47]. The strength of TCR signaling in the thymus and periphery thus determines the fate of a T cell.
One characteristic of TVM cells is their increased CD5 surface level [9, 30]. High CD5 serves as a proxy for high TCR affinity during thymic selection and negatively regulates TCR signaling in the thymus, thereby increasing the positive selection window by preventing negative selection [39–41]. The relative level of CD5 correlates with the percentage of TVM cells in different mouse models [30]. Furthermore, CD5high naïve cells acquire a TVM phenotype more often after adoptive cell transfer than CD5low naïve cells [9]. This suggests that cells with higher affinity for self-antigens or with higher TCR signaling are more prone to become TAIM. This is corroborated in a mouse model with increased binding of the tyrosine kinase Lck to the CD8 co-receptor resulting in supraphysiological TCR signaling: mice with this increased TCR signaling have an increased number of TVM [30]. The importance of signaling through the CD8 co-receptor is further demonstrated in a mouse model with constitutive expression of CD8αβ. This results in increased numbers of TIM cells in the thymus, partly caused by extrinsic factors involving other thymic cell subsets and partly through cell-intrinsic mechanism [48]. Furthermore, mice deficient for DOCK2, which contributes to cellular signaling events by activating small G proteins, are more sensitive to TCR stimulation with low-affinity antigens and have increased numbers of TVM cells [10]. This also suggests that increased sensitivity for tonic/self-peptide signaling induces TAIM differentiation. A role for TCR quality was further supported by a study using mice with a transgenic TCRα and TCRβ chain. When these tgTCR mice are bred on a Rag-KO background, recombination of endogenous TCR genes is prevented and all T cells express the exact same TCR (OT-I) with the same affinity. Under these Rag-deficient conditions, the percentage of virtual memory cells is low (< 5%) and remains low during aging [25]. However, CD8+ T cells from mice on a Rag-proficient background, which are able to recombine the endogenous TCRα chain, generated higher TVM cells at higher frequency and number, both of which increased with age (> 50%) [25]. This indicates that some TCRs are more likely to support the gain of a TAIM phenotype than others.
The TCR repertoire of unchallenged TAIM cells is distinct from that of naïve CD8+ T cells (TN) [12, 30]. Several TCRs have been identified as TAIM-TCRs or TN-TCRs, both in mice with an endogenous TCR and in mice expressing a transgenic TCRβ chain [12, 30]. Retroviral expression of transgenic TAIM-TCRs contributes to an increased number of TAIM cells in vivo [12, 30]. Cells expressing a TAIM-TCR also respond stronger to stimulation with self-peptides by MHC-I on dendritic cells [12]. Furthermore, cells with a TAIM-TCR are already EOMES+ in the thymus and have increased surface CD5 expression, indicating that TCR-induced TAIM differentiation already starts in the thymus [12, 20]. This increased TCR affinity also correlates with the higher convergence score reported for memory-phenotype (CD44+CD122+) cells [10]. The convergence score is the number of unique TCR sequences that encode for the same TCR and a higher score is indicative of selection by self-antigens [10]. Interestingly, co-culture of naïve CD8+ T cells with chemotherapeutically treated tumor cells results in an increase of TVM-phenotype cells independent of MHC-I [21], suggesting that other signaling pathways might overrule TCR signaling in certain conditions. Taken together, this data suggests a model in which TAIM cells arise as a result of an intrinsically higher TCR signaling potential.
Given the increased TCR signaling in TAIM cells, Drobek et al. studied if TAIM cells are more likely to respond strongly to self-peptides and induce auto-immune pathology [30]. They found that TAIM cells do not break self-tolerance and are less effective in inducing auto-immune pathology compared to TTM cells [30]. This suggests that TAIM differentiation might function as an escape mechanism for self-reactive CD8+ T cells to avoid auto-immunity [30, 49]. Alternatively, when TAIM cells recognize self-peptides, they might ‘not properly’ be activated/primed and as a result of this have reduced cytotoxic capacities compared to properly activated TTM cells. In light of this, it is relevant to note that regulatory CD8+ T cells also have a CD122+CD49d− memory phenotype [50], although to our knowledge the immune-regulatory function of TAIM cells has not been studied.
Cytokine signaling in TAIM cell differentiation
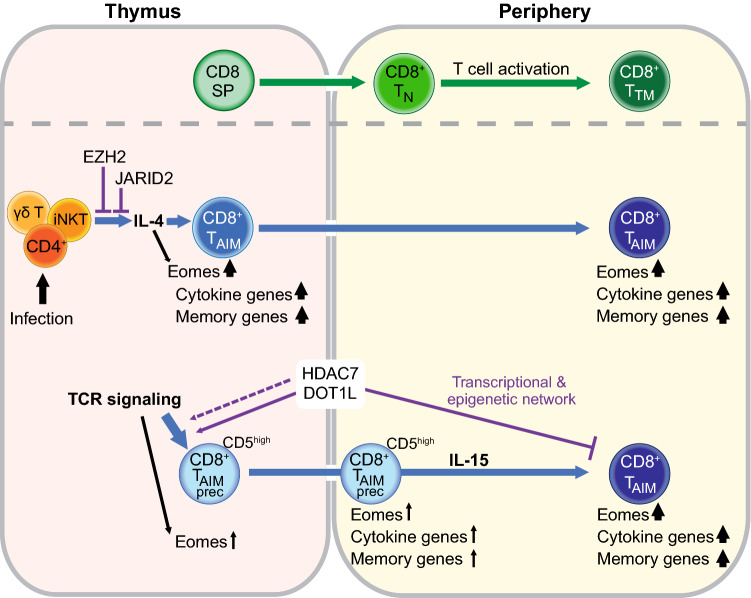
Cytokine signaling is critical to ensure that activated T cells follow the right trajectory of differentiation. The two main cytokines involved in TAIM development are IL-4 for TIM and IL-15 for TVM cells. IL-4 is mainly produced by invariant NKT (iNKT) and γδ-T cells in the thymus [14], although other thymic populations have also been reported as a source for IL-4 [51]. Mouse models in which the number of IL-4-producing iNKT cells, γδ-T cells or CD4 single positive T cells is increased have more innate memory cells [14, 52–55] as reviewed in [56]. Furthermore, the number of iNKT cells is higher in BALB/c mice compared to C57Bl/6 mice, correlating with a higher number of TIM cells [14, 57–59]. When IL-4 signaling is reduced in mouse models with increased IL-4 production, either by ablating iNKT cells, deleting the IL-4 receptor or by blocking IL-4 with antibodies, the TIM population is diminished, indicating that TIM cells require IL-4 [53, 60]. On the other hand, in vivo treatment with IL-4/anti-IL4 antibody complex results in increased number of CXCR3+ CD8+ T cells that have increased EOMES expression, indicating that increased IL-4 levels contribute to TIM differentiation [34, 61]. The exact mechanism of how increased IL-4 signaling stimulates TIM differentiation is not known. It has been reported that IL-4 drives EOMES expression through Stat6- and AKT-dependent pathways [32, 62]. However, TIM cells do not depend on increased IL-4 production under all conditions. There are also reports of TIM cells arising independently of excess IL-4 production [63]. Taken together, IL-4 is important, and in most cases, required for TIM differentiation in the thymus.
Although increased levels of IL-4 can contribute to an increased percentage and number of TVM in the periphery [19, 23, 57, 64], IL-15 is the most important cytokine for peripheral TAIM cells [3]. IL-15 is a key driver of homeostatic CD8+ T cell proliferation under steady-state conditions and regulates memory effector functions [65]. Several lines of evidence reveal that in the periphery, TAIM cells depend on IL-15. First, ablation of IL-15 production or abrogation of IL-15 signaling completely inhibits TAIM differentiation and survival [9, 15, 66, 67]. Second, TAIM differentiation in IL-15 depleted mice can be rescued by injecting IL-15/Rα complexes [9, 15]. Third, CD122, part of the IL-15 receptor complex, is more highly expressed on TVM compared to naïve and TTM [15]. Furthermore, a recent study suggests that regulatory CD4 T cells regulate TVM differentiation by inhibiting IL-15 trans-presentation by CD11b+ dendritic cells (DCs) [68]. How increased IL-15 signaling leads to the formation of TAIM cells has not been fully described yet. It has been suggested that IL-15 signaling results in homeostatic expansion of TVM cells that are already present in the periphery [15]. However, IL-15 signaling might also contribute toward TAIM differentiation itself. The source of IL-15 for TAIM cells are CD8α + DCs in peripheral lymphoid tissues [15]. This suggests that peripheral TAIM mostly depend on naturally occurring IL-15.
TAIM cell differentiation: a delicate balance of multiple signals
Taken together, the current understanding of signals involved in TAIM differentiation suggests a model in which TAIM differentiation is steered by a very delicate balance of at least two external signals mediated by the TCR and cytokine receptors and integrated intracellularly (Fig. 1). These signals lead to altered transcriptional output, which is further modulated by epigenetic mechanisms (see below). Natural fluctuations in gene expression of key genes that regulate TCR signaling or cytokine signaling or their downstream targets may render cells more or less susceptible to become TAIM cells. This is in line with the current notion that fluctuations in gene expression occur over time within cell populations and that these fluctuations can have long term impact on features of the original cell and its offspring [69, 70]. Among the genes whose fluctuation might affect TAIM differentiation is CD5, a negative regulator of TCR signaling. Fluctuations in CD5 expression may render cells more sensitive to tonic signals provided by self-peptide MHC complexes, making these clones more prone to inappropriate priming and differentiation, especially under pro-proliferative conditions like lymphopenia. In this context, the tyrosine kinase ITK, a mediator of TCR signaling, is also of interest. ITK plays an important role in TAIM differentiation. ITK ablation in the T cell lineage results in increased numbers of TAIM cells [66]. This skewed differentiation is IL-4 dependent and requires an external source of IL-4 [51, 56]. However, Huan and colleges suggest a cell-intrinsic role for ITK in TAIM differentiation [51]. While in vitro TCR activation in combination with IL-4 results in reduced expression of the IL-4 receptor (IL-4R) and EOMES and a decrease in the percentage of TAIM cells in WT CD8 single positive thymocytes, ITK-ablated CD8 single positive thymocytes are less sensitive to TCR activation and maintain higher levels of IL-4R and EOMES and a higher percentage of TAIM cells [51]. This suggests that ITK tunes the response to IL-4 signaling cell intrinsically [51]. Given the role of ITK as a mediator of TCR signaling, TCR signaling might no longer lead to reduced IL-4 receptor expression on CD8 single positive thymocytes in the absence of ITK, thereby rendering the cells more sensitive to IL-4. Cross talk between TCR signaling and cytokine signaling is also shown by the effect of TCR signaling on IL-4-induced expression of EOMES in vivo, where IL-4-induced upregulation of EOMES is stronger upon weak TCR signaling compared to strong TCR signaling [62]. This all suggests a model in which a delicate balance of TCR signaling and cytokine signaling is required to keep cells in a naïve state or induce TAIM differentiation.
Fig. 1.
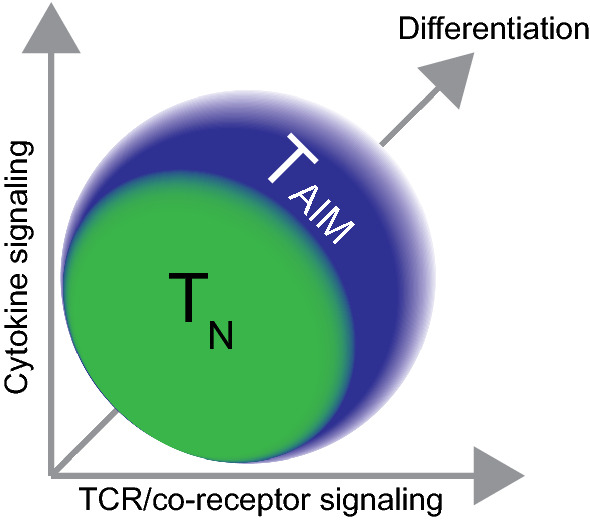
Antigen-inexperienced memory T cell (TAIM) differentiation: A delicate balance of integrated signaling. The quality of TCR/co-receptor and cytokine signals are central to TAIM differentiation. Cytokines most important for TAIM cells are IL-4 in the thymus and IL-15 in the periphery. Ablation of these cytokines or their receptors leads to a strong reduction in the number of TAIM cells. The balance between cytokine and TCR signals determines whether a CD8+ T cell remains naïve (TN) or differentiates into a TAIM cell. The TCR and cytokine signals lead to altered transcriptional output, which is further modulated by epigenetic mechanisms. The extracellular signals, and fluctuations in expression of genes involved in these signaling pathways due to transcription dynamics, as well as differentiation and proliferation together affect TAIM differentiation. The net result is a dynamic population of TAIM cells in mouse and humans
Physiological regulation of TAIM during development and infection
TAIM cells are observed in unchallenged wild-type mice and also in humans. Increased numbers of TAIM cells have been described in several genetically engineered mouse models (GEMM). The number of TAIM cells is also regulated during normal physiological development and it changes during life. Most studies on TAIM are performed in adult mice, which have a small but substantial population of TAIM cells (~ 20% of total CD8+ T cells) [6–13]. In young (infant) mice, there is a larger fraction of TAIM cells (~ 35%) [23]. Using an elegant approach to time stamp T cells produced at different ages, Smith et al. mapped the fate of cells from different developmental origins [17]. They showed that in adult mice the CD8+ T cells with a fetal origin have around five times more TVM cells than CD8+ T cells with an adult origin. This increased TVM frequency in CD8+ T cells with a fetal origin is not caused by lymphopenia, but rather is an intrinsic feature of CD8+ T cells of early developmental origin [17]. Physiologically, the early-developed cells contribute more rapidly to an immune response compared to late-developed T cells, as shown by a rapidly increased number of early time-stamped cells at the beginning of the immune response. Early-developed TVM cells respond in a more innate-like manner and differentiate predominantly toward short-lived effector cells, whereas late-developed TVM cells give preferentially rise to long-lived memory cells [17]. This suggests that the TVM cells developed during early stages of development might give increased rapid innate-like protection that is needed in young animals with a developing immune system. Importantly, this data shows that the TAIM pool is not homogenous, but that TAIM cells developed during different stages are differentially programmed and have distinct functions.
In aged mice, the percentage and absolute number of TAIM is again increased [9, 11, 28, 71]. The precise mechanism that contributes to this increase is unknown, but also in older individuals TAIM cells depend on a combination of TCR and cytokine signaling. Aged mice that have a transgenic TCR but that are also able to undergo recombination of the endogenous TCRα have increased numbers of TAIM, similar to younger mice [25]. This again demonstrates that the differentiation of TAIM cells requires a specific level of TCR signaling. Furthermore, the surface levels of CD5 are increased in unchallenged CD8+ T cells in aged mice [71], suggesting a selective advantage due to increased sensitivity for self-antigens. Consequently, in aged mice CD8+ T cells expressing TCRs with high affinity for self-pMHC and strong signaling potential increase, favor TAIM differentiation and with time TAIM cells outgrow other CD8+ T cells. Similar to TAIM in young mice, TAIM cells in aged mice still depend on IL-15, as was demonstrated by the absence of TVM in aged IL-15 KO mice [6]. Furthermore, expression of CD122, part of the IL-15 receptor, is increased on a subset of TAIM cells in aged BALB/c mice compared to young mice [11]. Taken together, this suggests that TAIM cells that accumulate in older animals require similar TCR and cytokine signals as early-developed TAIM cells.
Functionally, TAIM cells are major contributors to an immune response against influenza in aged mice and gain a TCM CD49d+ phenotype afterward [18]. However, when aged TVM and naïve CD8+ T cells are transferred together, the TVM cells are outperformed by the naïve cells [18]. Furthermore, aged TVM are more monofunctional, secreting only one effector cytokine, whereas naïve cells are more polyfunctional, secreting several different cytokines [7]. Aged TVM also have a reduced proliferative capacity [25] and they have a senescent phenotype characterized by resistance to apoptosis, increased γH2Ax indicating accumulated DNA damage and increased expression of Bcl-2 family members that control apoptosis [7]. The overall impaired immune response in aged individuals might among other changes also relate to reduced numbers of naïve CD8+ T cells and increased numbers of later developed TAIM cells.
There are several indications that the number of TAIM cells is also actively regulated during specific immunological challenges. During helminth infection, the concentration of IL-4 in the thymus increases and this correlates with an increased TVM pool [8, 19]. These newly expanded TVM cells are able to mount a more effective immune response against lytic gammaherpesvirus infection [8] and bacteria [19]. Infection with trypanosomes results in cellular composition changes in the thymus, leading to increased production of IL-4 and IL-15 and an increased percentage of memory-phenotype cells in the thymus although these cells are CD49d+ [72]. These TIM-phenotype cells offer increased protection against trypanosome infection in an antigen-independent manner [72]. TIM-phenotype cells (CD44+CXCR3+CD49d+) are also seen after treatment with metformin, a drug used in treatment of type 2 diabetes, and they can contribute to increased protection against infection with M. Tuberculosis [73]. However, the origin of these metformin-induced TIM-phenotype cells is not known [73]. Different infections can thus affect the immunological environment and thereby impact TAIM differentiation. Increased numbers of TAIM cells during certain immunological challenges are also seen in humans (discussed later); however, the exact mechanism is not always known. Given this recurrent correlation between infections and TAIM development, further research into how infections promote the generation of TAIM and the potential advantage of this increased TAIM population in establishing immunity is crucial.
Most studies on TAIM cells have been performed in mice housed under SPF conditions. This raises the question whether increased or decreased pathogen exposure affects TAIM differentiation. Several studies have shown that TAIM cells are also present in mice housed under germ-free conditions in a similar frequency as in SPF mice [11–13]. The presence of TAIM cells in ‘dirty’ feral mice has only recently been investigated. Feral mice have an increased frequency of TAIM cells compared to laboratory mice [11]. The frequency of TAIM cells in feral mice is even higher than in F1 offspring of feral mice born in captivity [11]. This indicates that environmental factors may be involved in TAIM differentiation, but the exact factors are not known. Co-housing of feral animals with laboratory animals affects the microbiome of the laboratory animals, but has very limited effect on TAIM cells [11]. Further studies are thus required to determine which environmental factors might impact TAIM differentiation.
Altogether, the number of TAIM cells is not static but physiologically regulated. Both the age at which TAIM cells are formed and the immunological challenges they encounter determine their number and function.
Transcriptional and epigenetic regulation of TAIM differentiation
To better understand how TAIM cells are regulated, we first need to define their developmental state. Are TAIM cells fully differentiated memory T cells or do they represent a semi-differentiated subset? To answer this question, several studies have looked beyond the small number of surface markers detected by flow cytometry and determined the full transcriptome of TAIM cells. Drobek et al. compared RNA-seq expression profiles of TN (CD44−CD62L+), TVM (CD44+CD62L+CD49d−) and TTM (Kb-OVA+CD44+CD62L+, after infection with Listeria monocytogenes-OVA) [30]. Based on publicly available datasets of TN and TTM they defined naïve and memory gene signatures. TVM cells are more enriched for memory signature genes than TN, but less than TTM. For naïve gene signatures TVM are also in between TN and TTM. A similar intermediate phenotype is seen for expression of cytokines and chemokines [30]. This suggests that TAIM cells indeed have an intermediate TN→TTM differentiation phenotype. However, this does not mean that they are just TN cells that are only differentiated halfway. When compared with TN and TTM, TVM cells have their own unique transcriptome features [7, 30]. This is shown by TVM cells clustering away from TN an TTM based on principal component analysis or multi-dimensional scaling analysis [7, 30]. Taken together, TAIM cells are a unique T cell subset with an intermediate differentiation phenotype as well as a unique transcriptome.
Further comparison of TN and TVM shows that TVM cells have high expression of genes related to cell killing, inflammatory cytokines, granzymes, and genes related to NK cell receptors, and reduced expression of CD49d [7]. Furthermore TVM cells have increased expression of genes involved in cytokine sensing, including IL-15 signaling mediators, and of the transcription factor EOMES [36]. A similar trend is seen for thymic TIM cells, which are also enriched for cytokine receptors and memory signature genes compared to naïve CD8 single positive thymocytes [34]. These changes correlate with a strong increase in histone H3K27ac on the promotors of genes upregulated in TIM cells. This histone mark is associated with active transcription and active enhancers [74]. The strongest epigenetic changes between TN and TIM are found in enhancers. Further comparison of H3K27ac peaks between TIM and TTM shows that many of the epigenetic changes in TIM cells compared to CD8 single positive thymocytes are similar to the changes observed between TN and TTM [34]. However, not all changes observed in TTM also occur in TIM [34]. This suggests that TIM cells use similar epigenetic programs as conventional antigen-experienced TTM cells, but also have TIM-specific changes in enhancer activity.
Interestingly, CD5high TN cells (CD44−CD62L+), which are more prone to differentiate toward TVM compared to CD5low TN cells, already show upregulation of key TAIM genes prior to memory differentiation [9]. CD5high TN cells also already show increased expression of cytokine genes associated with homeostatic cytokine responsiveness (Il2rb, Eomes, Il4r) compared to CD5low TN cells [9]. Furthermore, these cells show increased expression of gene clusters related to cell division and genes expressed in late effector and memory stages [67]. This upregulation of key TAIM genes in TAIM precursors seen in the peripheral CD5high cells, is in line with increased protein levels of EOMES in thymic TVM- precursors [12]. These findings suggest that transcriptomic changes are not merely a result of having a more differentiated phenotype, but that they lie at the basis of antigen-inexperienced memory differentiation.
As mentioned before, TAIM cells are a heterogeneous population, and this is also reflected in their transcriptome. Using the time-stamping technique Smith et al. isolated TN (CD8+CD44low) and TVM (CD8+CD44high) cells that developed during different time-points. Compared to TVM cells produced during adulthood, TVM cells produced early in life express more genes typically observed in short-term and late-effector CD8+ T cells after in vivo stimulation [17]. This also correlates with their chromatin landscape, with early-developed TVM cells having increased accessibility at binding sites for transcription factors associated with effector differentiation. On the other hand, transcription factors involved in repression of effector differentiation have increased binding in chromatin more accessible in late-developed TVM [17]. This suggests that the differential regulome of TVM cells with different developmental origins affects their responsiveness to key differentiation transcription factors [17]. TVM from aged mice have a more senescent phenotype that correlates with increased expression of anti-apoptotic transcripts including Bcl2 and downregulation of exhaustion markers, contributing to the accumulation of TVM over time [7]. Thus, the developmental origin of TAIM cells determines their transcriptome and epigenome. The transcriptomic and epigenomic data for TAIM cells described here has been acquired by bulk sequencing of TAIM populations. Given the heterogeneity of the TAIM pool, single-cell sequencing will be important to further understand how TAIM differentiation relates to regulation of the transcriptome and epigenomes.
Key epigenetic and transcriptional regulators in TAIM differentiation
EOMES
One of the key transcription factors expressed in TAIM cells is EOMES [36]. EOMES is a member of the T-Box family of transcription factors and is upregulated during conventional memory differentiation/antigen exposure [75]. EOMES is more highly expressed in TVM developed in the fetal stage compared to TVM developed during adulthood, but in all TVM subsets its expression is higher than in TN cells [17]. EOMES is very important in TAIM differentiation, as is shown by the strong reduction of TAIM cells in thymus and spleen in Eomes-KO mice [15, 53]. This was further confirmed in a model with transgenic overexpression of Eomes in thymocytes, resulting in an increase in the percentage of TIM cells [34].
EOMES is required for TAIM differentiation at several levels. EOMES has been shown to bind directly to the promotor and internal regions of Bcl2 and drive Bcl2 expression (pro-survival protein), thereby giving memory cells a survival advantage compared to the strongly stimulated effector cells [76]. Besides driving pro-survival signals, EOMES is also involved in upregulation of CD122, which is part of the IL-15 receptor and upregulated in TAIM cells [15, 29, 34, 37]. EOMES shows several interactions with other key regulators [34]. One potentially interesting interactor is RUNX3 because Runt motifs are slightly enriched in enhancer regions that have increased activity in TIM vs TN [34]. It has been suggested that EOMES is recruited toward regions where RUNX3 has bound [34]. In addition, BRG1, part of the SWI/SNIF chromatin remodeling complex, is critical to induce the EOMES-dependent program [34]. EOMES is thus a key transcriptional regulator in TAIM cells that is embedded in a broader network of transcriptional and epigenetic regulators.
EOMES can be induced by several upstream factors including TCR stimulation and cytokines. IL-4 is one of the main drivers of EOMES expression in TAIM cells. IL-4R knock-out (KO) mice show reduced EOMES levels in CD8+ T cells, including TAIM cells [32], while in vivo injection with IL-4 leads to increased EOMES expression in splenic TAIM cells [61]. In vitro, IL-4 is also sufficient to induce EOMES expression in CD8 single positive thymocytes [62]. Interestingly, this induction is stronger under submaximal TCR stimulation [62]. Also, in vivo submaximal TCR affinity results in the highest EOMES upregulation and drives central memory formation [76]. Besides IL-4, type 1 interferons (IFNs) can also induce EOMES expression as is shown by the reduced EOMES levels in TAIM cells lacking the receptor or signaling pathway required for type 1 IFN pathway. Loss of the type 1 IFN pathway correlates with a reduction in the number of TAIM cells. Mimicking type 1 IFN signaling by PolyI:C results in increased EOMES levels and increased TAIM numbers in vivo [77]. Several signals involved in TAIM differentiation are thus also involved in upregulation of the master TAIM transcription factor EOMES.
Epigenetic modulators
The epigenetic mechanisms involved in TAIM differentiation are only starting to be unraveled, but several epigenetic factors have already been linked to TAIM cells in different mouse models. The role of these epigenetic factors is mostly studied in conditional knock-out mouse models where the factor of interest is deleted in T cells during early T cell development. Among the epigenetic factors tested in this way is EZH2. EZH2 provides the catalytic part of the Polycomb repressive complex that is required for the generation of the repressive H3K27me3 mark [78–80]. Mice with conditional Ezh2 KO in T cells have reduced H3K27me3 on the EZH2 targets, including the Zbtb16 (PLZF) locus in iNKT cells. As a result, Ezh2-KO mice have aberrant iNKT differentiation and increased numbers of IL-4 producing iNKT cells, leading to increased numbers of TAIM cells [81]. A similar mechanism is seen in mice lacking JARID2, a component of three lysine methyltransferase complexes that are involved in transcriptional repression, Polycomb repressive complex 2 (PRC2) that methylates histone 3 lysine 27 (H3K27), and the GLP-G9a and SETDB1 complexes that methylate H3K9. One of the targets of JARID2 is PLZF. Deletion of Jarid2 results in reduced H3K9me3 on Zbtb16 resulting in increased PLZF expression. This leads to an increase in IL-4 producing PLZFhigh iNKT cells and as a result thereof more TIM cells in the thymus [33]. Thus, deletion of EZH2 or JARID2 contributes to TAIM differentiation at least in part by affecting thymic IL-4 production. Since EZH2 and JARID2 are major epigenetic regulators that affect the many aspects of the epigenome, it is likely that there are also other genes differentially expressed in these mouse models that contribute to TAIM differentiation. T cell-specific deletion of the histone acetyltransferase CREB binding protein (CBP) also results in increased TIM cells in a cell-extrinsic manner [14, 82]. This has been shown by a mixed bone marrow chimera experiment. When bone marrow cells from WT and Crebbp-KO were co-injected into the same mouse WT cells acquired the same phenotype as KO cells, indicating that CD8 cell-extrinsic factors are involved [14, 82].
In contrast to the above-described epigenetic regulators that affect TAIM differentiation in a cell-extrinsic manner, other epigenetic regulators impact TAIM differentiation cell intrinsically. The histone methyltransferase DOT1L is responsible for methylation of H3K79, which is associated with active transcription in gene bodies [83, 84]. Deletion of Dot1L in the T cell lineage results in a strong increase in the number of CD8+ TAIM cells. This increase in TAIM cells reflects a cell-intrinsic role of DOT1L as has been shown by mixed bone marrow chimeras [27]. When bone marrow cells from WT and Dot1L-KO were co-injected into the same recipient mouse, the WT cells remained mostly naïve, whereas Dot1L-KO cells still differentiated to TAIM cells [27]. The onset of TAIM differentiation in Dot1L-KO mice starts in the thymus where CD8 single positive thymocytes have increased expression of memory genes [27]. DOT1L-ablated T cells have aberrant expression of TCR signaling genes and reduced surface levels of the TCR complex [27]. This suggests that altered TCR signaling might be involved in TAIM differentiation in Dot1L-KO mice. H3K79me2 in WT CD8+ T cells marks active genes and loss of DOT1L activity results in downregulation of a subset of H3K79-methylated genes, whereas other H3K79-methylated genes remain highly expressed [27]. What determines which genes are sensitive to loss of H3K79me2 is still unclear, but the downregulated H3K79me2 marked genes include genes encoding TCR, co-stimulatory components, and TCR signaling components [27]. Dot1L-KO CD8+ T cells also show a group of upregulated genes that lack H3K79me2 in WT cells, suggesting that DOT1L controls expression of other transcriptional regulators, in particular repressors that indirectly regulate these genes. EZH2 was identified as a candidate negative regulator that is controlled by DOT1L and that can account for at least part of the indirect effects [27]. Taken together, DOT1L normally prevents TAIM differentiation by controlling TCR signaling and expression, and by regulating a network of other transcriptional and epigenetic regulators. How DOT1L maintains this network, through its catalytic activity toward H3K79 or also by other functions, remains to be determined [85–88]. It would also be interesting to understand what the role is of putative H3K79 methyl ‘reader’ proteins and DOT1L binding partners in TAIM differentiation, since these factors affect DOT1L output and activity [83, 89]. Finally, epigenetic regulators that impact DOT1L activity, including the histone deacetylase HDAC1 and the H2B ubiquitination machinery [83, 90–92], are also candidate factors affecting TAIM differentiation.
Another epigenetic modulator that has a central role in TAIM differentiation is the histone deacetylase HDAC7 [93]. HDAC7 a class IIa HDAC that functions as a transcriptional repressor [94]. The concentration of HDAC7 in the nucleus is actively regulated during T cell development in the thymus [95]. Deletion of Hdac7 results in reduced numbers of iNKT cells and a general reduction in the number of thymocytes, but in an increased percentage of TIM cells [93, 95]. Using mixed bone marrow chimeras, it has been shown that this increase in TIM cells is cell intrinsic [93]. However, it is not clear how deletion of Hdac7 results in increased percentages of TIM cells. HDAC7 itself has very minimal deacetylase activity but performs most of its catalytic function through interaction with the co-repressive N‐CoR/SMRT/HDAC3 complex [96–98]. In mature T cells, HDAC7 is continually phosphorylated resulting in translocation of HDAC7 from the nucleus to the cytosol [99]. In the thymus, HDAC7 is located in the nucleus during early thymocyte development and exported to the cytosol during positive selection [95]. The majority of the transcriptional changes observed during Hdac7 deletion in thymocytes overlap with changes induced by positive selection [95]. This suggests that HDAC7 is a negative regulator of the downstream effects of TCR activation [95]. A possible mechanism for TAIM differentiation in Hdac7-KO mice might thus involve increased TCR signaling. An overview of the role of epigenetic factors in TAIM cells is outlined in Fig. 2.
Fig. 2.
A model of TAIM differentiation regulators. During conventional CD8+ T cell differentiation CD8 single positive (CD8 SP) thymocytes exit the thymus and enter the periphery as naïve (TN) CD8+ cells. Upon encountering a foreign antigen presented on the MHC class I complex of an antigen-presenting cells, TN cells become activated and differentiate into short-lived effector and long-lived true memory (TTM) cells. TTM cells provide a rapid immune response when re-activated, thereby ensuring an effective secondary immune response. In addition to the conventional TTM cells, memory-phenotype cells also arise in an antigen-independent manner. The differentiation of the antigen-inexperienced memory T cells (TAIM) is guided by two main signals: cytokines and T cell receptor (TCR) signaling. Increased IL-4 levels in the thymus, either as a result of infection, or strain specific, or caused by deletion of the epigenetic modulator EZH2 or JARID2, induces TAIM differentiation in CD8 SP thymocytes. These cells have increased expression of cytokine genes and memory genes, including the key transcription factor EOMES. EOMES expression can also be upregulated directly by IL-4. A specific level of heightened TCR signaling also affects EOMES expression and results in the upregulation of CD5 on naïve CD8 single positive thymocytes that are more prone to become TAIM cells. Upon migration to the periphery, these CD5high cells already express mildly increased levels of cytokine genes and memory genes. IL-15 signaling further drives these cells to become TAIM cells. The histone modifiers DOT1L and HDAC7 prevent TAIM differentiation by regulating transcriptional and epigenetic networks that keep cells in a naïve state. Furthermore, DOT1L and, possibly also, HDAC7 are involved in regulating TCR signaling
Given the important role of epigenetic regulators in the execution of developmental transcriptional programs, it is likely that other epigenetic factors are also involved in TAIM differentiation. Studying these regulators in conditional knock-out mouse models might prove difficult since other cells subsets are likely also affected in these mouse models. This is for example the case with EZH2. Upregulation of EZH2 target genes has been shown in Dot1L-KO TAIM cells [27]. However, deletion of Ezh2 in the T cell lineage has major effects on iNKT cell differentiation, thereby indirectly affecting TAIM differentiation [81]. This strong effect of EZH2 on iNKT cell differentiation might mask the possible cell-intrinsic effects of Ezh2 deletion in CD8+ T cells. More advanced conditional ablation models combined with detailed epigenomic and transcriptomic analysis, preferably on single-cell level, will help to further understand the epigenetic mechanisms behind TAIM differentiation.
TAIM cells in humans
Most studies on TAIM cells have been performed in mice. Recently some studies described TAIM-phenotype cells in humans. Their precise origin and function are not known, but they are present in umbilical cord blood and fetal spleen, suggesting their antigen-inexperienced nature, and in blood and liver from mature adults [9, 54, 100–102]. In young adults TAIM cells constitute around 5% of the peripheral CD8+ T cell population and up to 15% in aged individuals [7, 103]. Human TAIM cells are characterized by expression of the innate natural killer (NK) markers NKG2A or KIR and expression of the memory transcription factor EOMES [9, 101, 102]. Most putative human TAIM cells in cord blood and in adult peripheral blood mononuclear cells have an effector memory re-expressing CD45RA (TEMRA) phenotype [9, 102, 103]. Similar to TIM cells in mice, human putative TAIM cells most likely also arise in response to IL-4. This has been studied in the context of chronic myeloid leukemia, a condition where iNKT activity, a major source for IL-4 production, is reduced. The first indication for IL-4 being important for human TAIM differentiation is that the percentage of TAIM-phenotype cells is decreased in chronic myeloid leukemia, and that this decrease can be partially restored after complete remission of chronic myeloid leukemia [104]. Furthermore, stimulation with IL-4 in vitro results in an increased number of TAIM-phenotype CD8+ T cells [104]. The authors suggest that iNKT cells are the source of the TAIM-stimulating IL-4 since the level of EOMES on TAIM-phenotype CD8+ T cells correlates with the level of PLZF on iNKT cells [104]. IL-15 has also been suggested to play a role in the maintenance/differentiation of human TAIM-phenotype cells. KIR/NKG2A+EOMES+ TAIM-phenotype CD8+ T cells are preferentially expanded in HIV-infected patients [103]. Interestingly, HIV-infected untreated patients have increased concentrations of IL-15 in the lymph nodes and an increased population of bystander expanded memory-phenotype cells (defined as CD45RO+ CCR7−) [105]. The increase of TAIM-phenotype cells in HIV-infected patients might also be (partially) due to increased homeostatic proliferation as a result of reduced CD4+ T cell numbers, but this remains to be studied. These studies suggest that human TAIM-phenotype cells are physiologically regulated and can be expanded under certain conditions.
As with murine TAIM cells, the function of human TAIM-phenotype cells has not been completely described. In vitro, the TAIM-phenotype KIR+/NKG2A+ CD8+ T cells rapidly produce IFNγ upon innate-like IL-12 + IL-18 stimulation [102, 104]. Furthermore, it has been suggested that TAIM-phenotype cells may contribute to control of the HIV viral reservoir [103]. However, further research is required to validate the antigen-naïve state of these putative human TAIM cells and to further study their origin and functionality.
Concluding remarks
Antigen-inexperienced memory cells form a substantial part of the CD8+ T cell pool. They represent a unique memory cell subset with its own functions, transcriptome and epigenome. The number and function of TAIM cells increases with age and during infections. This indicates that TAIM cell formation play a dynamic role in the immune system. The mechanisms and dynamics of TAIM differentiation have only been studied in the past few years and it is likely that many conditions that impact TAIM differentiation and functionality are yet to be discovered. For example, besides infections with helminths or trypanosomes, other infections that affect thymic IL-4 levels might also impact TAIM differentiation.
The presence and heterogeneity of TAIM cells shows that memory differentiation is not driven by a single process. Memory differentiation is guided by a complex network of different stimuli, involving TCR signaling and cytokines. Based on the conditions of their stimulation TAIM cells have specific functional characteristics. On a transcriptomic level, TAIM precursors already show increased expression of some key memory genes and fully differentiated TAIM cells have their own unique transcriptome. These transcriptomic changes correlate with changes in the epigenome but further studies are required to fully understand how epigenetic regulators contribute to TAIM differentiation by direct and indirect mechanisms.
In conclusion, CD8+ TAIM cells are a unique and intriguing subset of memory cells that help us to understand the complexity of memory cell differentiation and how different types of memory cells each contribute in unique ways to the immune response.
Author contributions
EMK-M, HJ and FvL all reviewed the literature and wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the Dutch Cancer Society (NKI2018-1 /11490 and NKI2014-7232) and ZonMW (Top91218022 and Top91213018).
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 2.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 3.White JT, Cross EW, Kedl RM. Antigen-inexperienced memory CD8+ T cells: where they come from and why we need them. Nat Rev Immunol. 2017;17:391–400. doi: 10.1038/nri.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiele D, La Gruta N, Nguyen A, Hussain T. Hiding in plain sight: virtually unrecognizable memory phenotype CD8+ T cells. Int J Mol Sci. 2020;21:8626. doi: 10.3390/ijms21228626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jameson SC, Lee YJ, Hogquist KA. Innate memory T cells. Adv Immunol. 2015;126:173–213. doi: 10.1016/bs.ai.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn KM, Hussain T, Kraus F, et al. Metabolic characteristics of CD8+ T cell subsets in young and aged individuals are not predictive of functionality. Nat Commun. 2020;11:2857. doi: 10.1038/s41467-020-16633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinn KM, Fox A, Harland KL, et al. Age-related decline in primary CD8+ T cell responses is associated with the development of senescence in virtual memory CD8+ T Cells. Cell Rep. 2018;23:3512–3524. doi: 10.1016/j.celrep.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 8.Rolot M, Dougall AM, Chetty A, et al. Helminth-induced IL-4 expands bystander memory CD8+ T cells for early control of viral infection. Nat Commun. 2018;9:4516. doi: 10.1038/s41467-018-06978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JT, Cross EW, Burchill MA, et al. Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat Commun. 2016;7:11291. doi: 10.1038/ncomms11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahajan VS, Demissie E, Alsufyani F, et al. DOCK2 sets the threshold for entry into the virtual memory CD8+ T cell compartment by negatively regulating tonic TCR triggering. J Immunol. 2020;204:49–57. doi: 10.4049/jimmunol.1900440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moudra A, Niederlova V, Novotny J, et al. Phenotypic and clonal stability of antigen-inexperienced memory-like T cells across the genetic background, hygienic status, and aging. J Immunol. 2021;206:2109–2121. doi: 10.1101/2020.09.01.277004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller CH, Klawon DEJ, Zeng S, et al. Eomes identifies thymic precursors of self-specific memory-phenotype CD8+ T cells. Nat Immunol. 2020;21:567–577. doi: 10.1038/s41590-020-0653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haluszczak C, Akue AD, Hamilton SE, et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sosinowski T, White JT, Cross EW, et al. CD8+ dendritic cell trans presentation of IL-15 to naive CD8+ T Cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J Immunol. 2013;190:1936–1947. doi: 10.4049/jimmunol.1203149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pribikova M, Moudra A, Stepanek O. Opinion: virtual memory CD8 T cells and lymphopenia-induced memory CD8 T cells represent a single subset: homeostatic memory T cells. Immunol Lett. 2018;203:57–61. doi: 10.1016/J.IMLET.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Smith NL, Patel RK, Reynaldi A, et al. Developmental origin governs CD8+ T cell fate decisions during infection. Cell. 2018;174:117–130.e14. doi: 10.1016/j.cell.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Lanzer KG, Cookenham T, Reiley WW, Blackman MA. Virtual memory cells make a major contribution to the response of aged influenza-naïve mice to influenza virus infection. Immun Ageing. 2018;15:17. doi: 10.1186/s12979-018-0122-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JS, Mohrs K, Szaba FM, et al. Virtual memory CD8 T cells expanded by helminth infection confer broad protection against bacterial infection. Mucosal Immunol. 2019;12:258–264. doi: 10.1038/s41385-018-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniels MA, Teixeiro E. Forget ‘ME’ not virtual memory T cells. Nat Immunol. 2020;21:499–500. doi: 10.1038/s41590-020-0668-7. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Waschke BC, Woolaver RA, et al. MHC class I-independent activation of virtual memory CD8 T cells induced by chemotherapeutic agent-treated cancer cells. Cell Mol Immunol. 2020;18:723–734. doi: 10.1038/s41423-020-0463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sprent J, Surh CD. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat Immunol. 2011;12:478–484. doi: 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akue AD, Lee J-Y, Jameson SC. Derivation and maintenance of virtual memory CD8 T cells. J Immunol. 2012;188:2516–2523. doi: 10.4049/jimmunol.1102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grau M, Valsesia S, Mafille J, et al. Antigen-induced but not innate memory CD8 T cells express NKG2D and are recruited to the lung parenchyma upon viral infection. J Immunol. 2018;200:3635–3646. doi: 10.4049/jimmunol.1701698. [DOI] [PubMed] [Google Scholar]
- 25.Renkema KR, Li G, Wu A, et al. Two separate defects affecting true naive or virtual memory T cell precursors combine to reduce naive T cell responses with aging. J Immunol. 2014;192:151–159. doi: 10.4049/jimmunol.1301453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JY, Hamilton SE, Akue AD, et al. Virtual memory CD8 T cells display unique functional properties. Proc Natl Acad Sci USA. 2013;110:13498–13503. doi: 10.1073/pnas.1307572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwesi-Maliepaard EM, Aslam MA, Alemdehy MF, et al. The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc Natl Acad Sci USA. 2020;117:20706–20716. doi: 10.1073/pnas.1920372117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu B-C, Martin BE, Stolberg VR, Chensue SW. Cutting edge: central memory CD8 T cells in aged mice are virtual memory cells. J Immunol. 2013;191:5793–5796. doi: 10.4049/jimmunol.1302509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 30.Drobek A, Moudra A, Mueller D, et al. Strong homeostatic TCR signals induce formation of self-tolerant virtual memory CD T cells. EMBO J. 2018;37:e98518. doi: 10.15252/embj.201798518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventre E, Brinza L, Schicklin S, et al. Negative regulation of NKG2D expression by IL-4 in memory CD8 T cells. J Immunol. 2012;189:3480–3489. doi: 10.4049/jimmunol.1102954. [DOI] [PubMed] [Google Scholar]
- 32.Renkema KR, Lee JY, Lee YJ, et al. IL-4 sensitivity shapes the peripheral CD8+ T cell pool and response to infection. J Exp Med. 2016;213:1319–1329. doi: 10.1084/jem.20151359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira RM, Martinez GJ, Engel I, et al. Jarid2 is induced by TCR signalling and controls iNKT cell maturation. Nat Commun. 2014;5:4540. doi: 10.1038/ncomms5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Istaces N, Splittgerber M, Lima Silva V, et al. EOMES interacts with RUNX3 and BRG1 to promote innate memory cell formation through epigenetic reprogramming. Nat Commun. 2019;10:3306. doi: 10.1038/s41467-019-11233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oghumu S, Dong R, Varikuti S, et al. Distinct populations of innate CD8+ T cells revealed in a CXCR3 reporter mouse. J Immunol. 2013;190:2229–2240. doi: 10.4049/jimmunol.1201170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussain T, Quinn KM. Similar but different: virtual memory CD8 T cells as a memory-like cell population. Immunol Cell Biol. 2019;97:675–684. doi: 10.1111/imcb.12277. [DOI] [PubMed] [Google Scholar]
- 37.Intlekofer AM, Takemoto N, Wherry EJ, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 38.Sackstein R, Schatton T, Barthel SR. T-lymphocyte homing: an underappreciated yet critical hurdle for successful cancer immunotherapy. Lab Investig. 2017;97:669–697. doi: 10.1038/labinvest.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azzam HS, Grinberg A, Lui K, et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgueño-Bucio E, Mier-Aguilar CA, Soldevila G. The multiple faces of CD5. J Leukoc Biol. 2019;105:891–904. doi: 10.1002/JLB.MR0618-226R. [DOI] [PubMed] [Google Scholar]
- 41.Voisinne G, Gonzalez de Peredo A, Roncagalli R. CD5, an undercover regulator of TCR signaling. Front Immunol. 2018;9:2900. doi: 10.3389/fimmu.2018.02900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prajapati K, Perez C, Rojas LBP, et al. Functions of NKG2D in CD8+ T cells: an opportunity for immunotherapy. Cell Mol Immunol. 2018;15:470–479. doi: 10.1038/cmi.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 45.Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev. 2010;236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 48.Kojo S, Ohno-Oishi M, Wada H, et al. Constitutive CD8 expression drives innate CD8+ T-cell differentiation via induction of iNKT2 cells. Life Sci Alliance. 2020;3:e202000642. doi: 10.26508/lsa.202000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truckenbrod EN, Jameson SC. The virtuous self-tolerance of virtual memory T cells. EMBO J. 2018;37:e99883. doi: 10.15252/embj.201899883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akane K, Kojima S, Mak TW, et al. CD8+CD122+CD49dlow regulatory T cells maintain T-cell homeostasis by killing activated T cells via Fas/FasL-mediated cytotoxicity. Proc Natl Acad Sci. 2016;113:2460–2465. doi: 10.1073/pnas.1525098113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang W, Huang F, Kannan AK, et al. ITK tunes IL-4-induced development of innate memory CD8 + T cells in a γδ T and invariant NKT cell-independent manner. J Leukoc Biol. 2014;96:55–63. doi: 10.1189/jlb.1ab0913-484rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prince AL, Kraus Z, Carty SA, et al. Development of innate CD4 + and CD8 + T cells in Itk-deficient mice is regulated by distinct pathways. J Immunol. 2014;193:688–699. doi: 10.4049/jimmunol.1302059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordon SM, Carty SA, Kim JS, et al. Requirements for eomesodermin and promyelocytic leukemia zinc finger in the development of innate-like CD8 + T cells. J Immunol. 2011;186:4573–4578. doi: 10.4049/jimmunol.1100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sook Min H, Jeong Lee Y, Kyung Jeon Y, et al. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8 + T cells. J Immunol. 2011;186:5749–5757. doi: 10.4049/jimmunol.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phalke SP, Huang Y, Rubtsova K, et al. γδ T cells shape memory-phenotype αβ T cell populations in non-immunized mice. PLoS ONE. 2019;14:e0218827. doi: 10.1371/journal.pone.0218827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 lineage. Trends Immunol. 2011;32:50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tripathi P, Morris SC, Perkins C, et al. IL-4 and IL-15 promotion of virtual memory CD8+ T cells is determined by genetic background. Eur J Immunol. 2016;46:2333–2339. doi: 10.1002/eji.201646404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai D, Zhu J, Wang T, et al. KLF13 sustains thymic memory-like CD8+ T cells in BALB/c mice by regulating IL-4-generating invariant natural killer T cells. J Exp Med. 2011;208:1093–1103. doi: 10.1084/jem.20101527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee YJ, Holzapfel KL, Zhu J, et al. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee A, Park SP, Park CH, et al. IL-4 induced innate CD8+ T cells control persistent viral infection. PLoS Pathog. 2015;11:e1005193. doi: 10.1371/journal.ppat.1005193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park HJ, Lee A, Il LJ, et al. Effect of IL-4 on the development and function of memory-like CD8 T cells in the peripheral lymphoid tissues. Immune Netw. 2016;16:126–133. doi: 10.4110/in.2016.16.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carty SA, Koretzky GA, Jordan MS. Interleukin-4 regulates eomesodermin in CD8+ T cell development and differentiation. PLoS ONE. 2014;9:e106659. doi: 10.1371/journal.pone.0106659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gugasyan R, Horat E, Kinkel SA, et al. The NF-κB1 transcription factor prevents the intrathymic development of CD8 T cells with memory properties. EMBO J. 2012;31:692–706. doi: 10.1038/emboj.2011.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurzweil V, LaRoche A, Oliver PM, et al. Increased peripheral IL-4 leads to an expanded virtual memory CD8+ population. J Immunol. 2014;192:5643–5651. doi: 10.4049/jimmunol.1301755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nolz JC, Richer MJ. Control of memory CD8+ T cell longevity and effector functions by IL-15. Mol Immunol. 2020;117:180–188. doi: 10.1016/j.molimm.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atherly LO, Lucas JA, Felices M, et al. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 67.Fulton RB, Hamilton SE, Xing Y, et al. The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8 + T cells to respond to foreign antigens. Nat Immunol. 2015;16:107–117. doi: 10.1038/ni.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Da Costa AS, Graham JB, Swarts JL, Lund JM. Regulatory T cells limit unconventional memory to preserve the capacity to mount protective CD8 memory responses to pathogens. Proc Natl Acad Sci USA. 2019;116:9969–9978. doi: 10.1073/pnas.1818327116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsabar M, Lovitch SB, Jambhekar A, Lahav G. Connecting timescales in biology: can early dynamical measurements predict long-term outcomes? Trends in Cancer. 2021;7:301–308. doi: 10.1016/j.trecan.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaffer SM, Emert BL, Reyes Hueros RA, et al. Memory sequencing reveals heritable single-cell gene expression programs associated with distinct cellular behaviors. Cell. 2020;182:947–959.e17. doi: 10.1016/j.cell.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinn KM, Zaloumis SG, Cukalac T, et al. Heightened self-reactivity associated with selective survival, but not expansion, of naïve virus-specific CD8+ T cells in aged mice. Proc Natl Acad Sci USA. 2016;113:1333–1338. doi: 10.1073/pnas.1525167113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baez NS, Cerbán F, Savid-Frontera C, et al. Thymic expression of IL-4 and IL-15 after systemic inflammatory or infectious Th1 disease processes induce the acquisition of “innate” characteristics during CD8 + T cell development. PLoS Pathog. 2019;15:e1007456. doi: 10.1371/journal.ppat.1007456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Böhme J, Martinez N, Li S, et al. Metformin enhances anti-mycobacterial responses by educating CD8+ T-cell immunometabolic circuits. Nat Commun. 2020;11:5225. doi: 10.1038/s41467-020-19095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kavazović I, Han H, Balzaretti G, et al. Eomes broadens the scope of CD8 T-cell memory by inhibiting apoptosis in cells of low affinity. PLoS Biol. 2020;18:e3000648. doi: 10.1371/journal.pbio.3000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinet V, Tonon S, Torres D, et al. Type i interferons regulate eomesodermin expression and the development of unconventional memory CD8 + T cells. Nat Commun. 2015;6:7089. doi: 10.1038/ncomms8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Mierlo G, Veenstra GJC, Vermeulen M, Marks H. The complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019;29:660–671. doi: 10.1016/j.tcb.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 80.Piunti A, Shilatifard A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol. 2021;22:326–345. doi: 10.1038/s41580-021-00341-1. [DOI] [PubMed] [Google Scholar]
- 81.Dobenecker M-W, Kim JK, Marcello J, et al. Coupling of T cell receptor specificity to natural killer T cell development by bivalent histone H3 methylation. J Exp Med. 2015;212:297–306. doi: 10.1084/jem.20141499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fukuyama T, Kasper LH, Boussouar F, et al. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol. 2009;29:3894–3904. doi: 10.1128/mcb.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vlaming H, van Leeuwen F. The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma. 2016;125:593–605. doi: 10.1007/s00412-015-0570-5. [DOI] [PubMed] [Google Scholar]
- 84.Steger DJ, Lefterova MI, Ying L, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao K, Ugarenko M, Ozark PA, et al. DOT1L-controlled cell-fate determination and transcription elongation are independent of H3K79 methylation. Proc Natl Acad Sci U S A. 2020;117:27365–27373. doi: 10.1073/pnas.2001075117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee S, Oh S, Jeong K, et al. Dot1 regulates nucleosome dynamics by its inherent histone chaperone activity in yeast. Nat Commun. 2018;9:240. doi: 10.1038/s41467-017-02759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stulemeijer IJE, Pike BL, Faber AW, et al. Dot1 binding induces chromatin rearrangements by histone methylation-dependent and -independent mechanisms. Epigenetics Chromatin. 2011;4:2. doi: 10.1186/1756-8935-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Welsem T, Korthout T, Ekkebus R, et al. Dot1 promotes H2B ubiquitination by a methyltransferase-independent mechanism. Nucleic Acids Res. 2018;46:11251–11261. doi: 10.1093/nar/gky801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wood K, Tellier M, Murphy S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules. 2018;8:11. doi: 10.3390/biom8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vlaming H, McLean CM, Korthout T, et al. Conserved crosstalk between histone deacetylation and H3K79 methylation generates DOT1L-dose dependency in HDAC1-deficient thymic lymphoma. EMBO J. 2019;38:e101564. doi: 10.15252/embj.2019101564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wille CK, Neumann EN, Deshpande AJ, Sridharan R (2020) Dot1L interaction partner AF10 safeguards cell identity during the acquisition of pluripotency. https://www.biorxiv.org/content/10.1101/2020.12.17.423347v1 [DOI] [PMC free article] [PubMed]
- 92.Uğurlu-Çimen D, Odluyurt D, Sevinç K et al (2021) AF10 (MLLT10) prevents somatic cell reprogramming through regulation of H3K79 methylation. Epigenetics Chromatin 14:32. 10.1186/s13072-021-00406-7 [DOI] [PMC free article] [PubMed]
- 93.Kasler HG, Lee IS, Lim HW, Verdin E. Histone deacetylase 7 mediates tissue-specific autoimmunity via control of innate effector function in invariant Natural Killer T cells. Elife. 2018;7:e32109. doi: 10.7554/eLife.32109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 95.Kasler HG, Young BD, Mottet D, et al. Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes. J Immunol. 2011;186:4782–4793. doi: 10.4049/jimmunol.1001179. [DOI] [PubMed] [Google Scholar]
- 96.Lahm A, Paolini C, Pallaoro M, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fischle W, Dequiedt F, Fillion M, et al. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 98.Desravines DC, Serna Martin I, Schneider R, et al. Structural characterization of the SMRT corepressor interacting with histone deacetylase. Sci Rep. 2017;7:3678. doi: 10.1038/s41598-017-03718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Navarro MN, Goebel J, Feijoo-Carnero C, et al. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol. 2011;12:352–362. doi: 10.1038/ni.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van Kaer L. Innate and virtual memory T cells in man. Eur J Immunol. 2015;45:1916–1920. doi: 10.1002/eji.201545761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Warren HS, Rana PM, Rieger DT, et al. CD8 T cells expressing killer Ig-like receptors and NKG2A are present in cord blood and express a more naive phenotype than their counterparts in adult blood. J Leukoc Biol. 2006;79:1252–1259. doi: 10.1189/jlb.0905536. [DOI] [PubMed] [Google Scholar]
- 102.Jacomet F, Cayssials E, Basbous S, et al. Evidence for eomesodermin-expressing innate-like CD8+ KIR/NKG2A+ T cells in human adults and cord blood samples. Eur J Immunol. 2015;45:1926–1933. doi: 10.1002/eji.201545539. [DOI] [PubMed] [Google Scholar]
- 103.Jin J-H, Huang H-H, Zhou M-J, et al. Virtual memory CD8+ T cells restrain the viral reservoir in HIV-1-infected patients with antiretroviral therapy through derepressing KIR-mediated inhibition. Cell Mol Immunol. 2020;17:1257–1265. doi: 10.1038/s41423-020-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jacomet F, Cayssials E, Barbarin A, et al. The hypothesis of the human iNKT/innate CD8+ T-cell axis applied to cancer: Evidence for a deficiency in chronic myeloid leukemia. Front Immunol. 2017;7:16. doi: 10.3389/fimmu.2016.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Younes S-A, Freeman ML, Mudd JC, et al. IL-15 promotes activation and expansion of CD8+ T cells in HIV-1 infection. J Clin Invest. 2016;126:2745–2756. doi: 10.1172/JCI85996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability
Not applicable.
Code availability
Not applicable.
Not applicable.