Abstract
Extracellular vesicles (EVs) have demonstrated unique advantages in serving as nanocarriers for drug delivery, yet the cargo encapsulation efficiency is far from expectation, especially for hydrophilic chemotherapeutic drugs. Besides, the intrinsic heterogeneity of EVs renders it difficult to evaluate drug encapsulation behaviour. Inspired by the active drug loading strategy of liposomal nanomedicines, here we report the development of a method, named “Sonication and Extrusion‐assisted Active Loading” (SEAL), for effective and stable drug encapsulation of EVs. Using doxorubicin‐loaded milk‐derived EVs (Dox‐mEVs) as the model system, sonication was applied to temporarily permeabilize the membrane, facilitating the influx of ammonium sulfate solution into the lumen to establish the transmembrane ion gradient essential for active loading. Along with extrusion to downsize large mEVs, homogenize particle size and reshape the nonspherical or multilamellar vesicles, SEAL showed around 10‐fold enhancement of drug encapsulation efficiency compared with passive loading. Single‐particle analysis by nano‐flow cytometry was further employed to reveal the heterogeneous encapsulation behaviour of Dox‐mEVs which would otherwise be overlooked by bulk‐based approaches. Correlation analysis between doxorubicin auto‐fluorescence and the fluorescence of a lipophilic dye DiD suggested that only the lipid‐enclosed particles were actively loadable. Meanwhile, immunofluorescence analysis revealed that more than 85% of the casein positive particles was doxorubicin free. These findings further inspired the development of the lipid‐probe‐ and immuno‐mediated magnetic isolation techniques to selectively remove the contaminants of non‐lipid enclosed particles and casein assemblies, respectively. Finally, the intracellular assessments confirmed the superior performance of SEAL‐prepared mEV formulations, and demonstrated the impact of encapsulation heterogeneity on therapeutic outcome. The as‐developed cargo‐loading approach and nano‐flow cytometry‐based characterization method will provide an instructive insight in the development of EV‐based delivery systems.
Keywords: active cargo loading, extracellular vesicles, heterogeneity, nano‐flow cytometry, single particle analysis
1. INTRODUCTION
Extracellular vesicles (EVs) are biological nanoparticles with lipid‐bilayer enclosures and biomolecule contents, which are derived from most cell types to regulate intercellular communication (Jeppesen et al., 2019; Kalluri & LeBleu, 2020; Pegtel & Gould, 2019; van Niel et al., 2018). As naturally secreted nanovesicles, EVs have exhibited great potentials in drug delivery field, owing to the desired features such as inherent biocompatibility, appropriate size distribution, and intrinsic cell targeting capability (Agrahari et al., 2019; Colao et al., 2018; Gudbergsson et al., 2019; Wu et al., 2020). To date, there have been several EV‐based preclinical trials investigating the ability for delivering protein, RNA or other chemotherapeutic drugs to treat diseases (de Jong et al., 2019; Fais et al., 2016; Gilligan & Dwyer, 2017; Walker et al., 2019).
Despite the unique advantages on EVs therapeutics, some challenges remain to be addressed (Antimisiaris et al., 2018; Bunggulawa et al., 2018; Elsharkasy et al., 2020; Moller & Lobb, 2020; Zhao et al., 2020). To use EVs as delivery vehicles, stably encapsulating substantial quantities of therapeutic molecules is a prerequisite (S. P. Li, Lin, et al., 2018; Lu & Huang, 2020). Currently, the major drug loading approaches are divided into two categories, pre‐loading and post‐loading (Liao et al., 2018; C. Liu & Su, 2019; Vader et al., 2016). Pre‐loading is performed endogenously via co‐incubating small‐molecule agents or transfecting drug‐encoding DNA with parental cells, followed by isolating the secreted EVs naturally packed with therapeutic cargos (Batrakova & Kim, 2015; C. Liu & Su, 2019; Pascucci et al., 2014). However, this approach is limited to the culture cell derived EVs, which can hardly be applied to other sources such as blood plasma or food. For post‐loading approach, drugs can be incorporated into purified EVs upon passive or active stimuli such as incubation, sonication, freeze‐thaw and electroporation (Fuhrmann et al., 2015; Goh et al., 2017; Kamerkar et al., 2017; Kim et al., 2016). Nonetheless, compared to the artificial counterpart, all of the above‐mentioned methods encounter a common problem: poor cargo capacity, especially for the encapsulation of hydrophilic molecules (Donoso‐Quezada et al., 2020). Therefore, development of more advanced loading approach to facilitate encapsulation of therapeutic agents is a primary step in EV‐based drug delivery.
Besides, comprehensive evaluation of the cargo encapsulation behaviour is highly instructive to the development of novel drug loading approach, which is often overlooked (Ingato et al., 2016; S. Wang et al., 2020). Limited to the sensitivity of techniques, most current studies evaluate the loading capacity by extrapolating from bulk analysis rather than single‐particle basis (Nolan, 2015). For example, a commonly used parameter, encapsulation efficiency, was defined as the ratio of the overall nanocarrier‐incorporated cargo quantity to the initial input (Shen et al., 2017; J. Tang, Zhang, et al., 2018). However, EVs are naturally heterogeneous in particle size and molecular content, and often contaminated with co‐precipitated proteins and other impurities, which can lead to the unequal drug encapsulation (Gorgens & Nolan, 2020; Ramirez et al., 2018; Roy et al., 2019). Hence, the unknown heterogeneity necessitates quantitative characterization at the single‐particle level for evaluation and even purification of cargo‐loaded EVs (Ferguson & Nguyen, 2016; Shao et al., 2018).
Regarding to these challenges in EV‐based delivery systems, we proposed a highly‐efficient drug loading method named “Sonication and Extrusion‐assisted Active Loading” (SEAL) for encapsulation of ionizable hydrophilic agents. Using milk‐derived EVs (mEVs) and doxorubicin (DOX) as the model system, the drug encapsulation efficiency of SEAL was shown to be significantly higher than conventional methods. In addition, we developed a single‐particle methodology for the evaluation of loading behaviour, where a flow cytometer with nanoscale sensitivity (nFCM) was introduced to quantify encapsulation efficiency and reveal heterogeneity (Chen et al., 2019, 2017; Tian et al., 2018). Through multi‐parameter analysis, correlation between encapsulation heterogeneity and compositional variance of mEVs was revealed, further guiding the purification processes to remove the undesired components and increase purity.
2. RESULTS
2.1. Preparation of Dox‐mEVs by SEAL
Active drug loading based on transmembrane ion gradient represents one of the most high‐efficient methods for drug encapsulation in liposomal nanomedicines (Gubernator, 2011; T. Li, Cipolla, et al., 2018). Inspired from that, we proposed the SEAL method to increase the drug loading capacity of EVs (Figure 1a) by using mEVs and doxorubicin as the model system. The mEVs were resuspended and sonicated in ammonium sulfate ((NH4)2SO4) to temporarily permeabilize the membrane, facilitating a solution influx into the lumen to establish the transmembrane ammonium gradient required for active loading. Then, extrusion was carried out to downsize the large mEVs, homogenize particle size and reshape the nonspherical or multilamellar vesicles which are unfavourable for drug loading and delivery. Then the bulk solution was dialyzed against PBS, followed by doxorubicin incubation for active loading.
FIGURE 1.
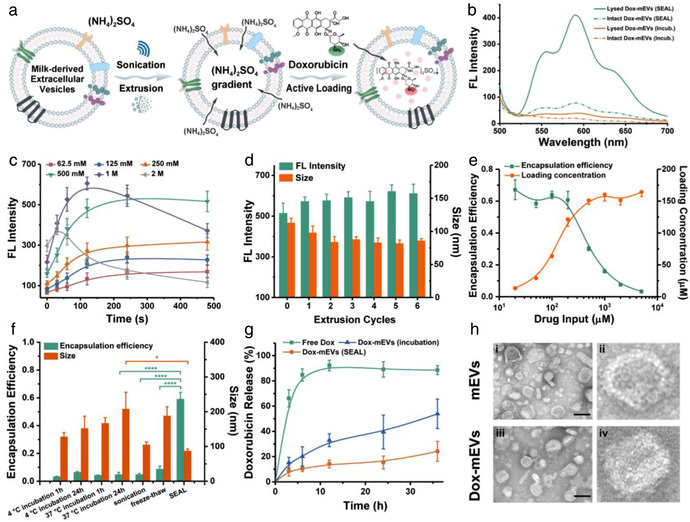
Preparation and analysis of Dox‐mEVs. (a) Schematic illustration representing the principle of SEAL. (b) Fluorescence analysis of Dox‐mEVs prepared by passive incubation and SEAL. (c) Fluorescence intensity of Dox‐mEVs at different ammonium sulfate concentration and sonication period (n = 3, mean ± SD). (d) Fluorescence intensity and particle size of Dox‐mEVs at various extrusion cycles (n = 3, mean ± SD). (e) Encapsulation efficiency and loading concentration of Dox‐mEVs at various drug input (n = 3, mean ± SD). (f) The encapsulation efficiency and particle size of Dox‐mEVs by various loading methods. p‐Values were calculated by t‐test. *p < 0.05, ****p < 0.0001 (n = 3, mean ± SD). (g) Drug release profile of free doxorubicin, as well as Dox‐mEVs prepared by SEAL and passive incubation, respectively (n = 3, mean ± SD). (h) TEM images of the unloaded mEVs (i) and SEAL‐prepared Dox‐mEVs (iii, scale bar = 100 nm). (ii) and (iv) depict a single mEV selected from the same image of (i) and (iii), respectively
To determine the efficiency of SEAL, the Dox‐mEV samples were lysed by Triton X‐100 to release the lumen‐aggregated doxorubicin, and the fluorescence signal was detected (Figure 1b). A 10.5‐fold increase in fluorescence intensity was observed compared to passive incubation, owing to the more efficient drug encapsulation. The significant fluorescence quenching, which was referred to as the fluorescence intensity ratio between released and mEV‐encapsulated doxorubicin, further confirmed that more drugs were actively accumulated in the lumen of SEAL‐prepared mEVs. Consistent results were obtained in a liposomal model where doxorubicin encapsulation was conducted using similar processes (Figure S1). The protein markers of CD81, CD63, CD9, TSG101, and HSP70 on mEVs before and after SEAL process were identified by western blot assays, addressing that sonication and extrusion treatment did not significantly compromise the protein content of mEVs (Figure S2).
Next, we sought to optimize the technical parameters of SEAL for improved drug loading. Firstly, it was found that the performance of SEAL process was highly dependent on both the salt concentration and sonication time, with optimal efficiency under conditions of 2–4 min sonication and 0.5–1 M ammonium sulfate solution (Figure 1c). Prolonged sonication time or more concentrated solution was unable to elevate the encapsulation efficiency without compromising the structure integrity. The impact of the extrusion operation was also investigated, showing that three extrusion cycles after sonication were sufficient to reduce the size of mEVs and increase the encapsulation efficiency (Figure 1d). An extra test using sole extrusion treatment without sonication further verified that the extrusion process did promote the solution exchange across the mEV membrane and facilitate the active loading process (Figure S3). Besides, the loading capacity with respect to loading concentration (mEV‐loaded doxorubicin concentration) and encapsulation efficiency (ratio of mEV‐encapsulated doxorubicin to the initial doxorubicin input) under different drug input was quantified, according to the fluorescence calibration curve of doxorubicin standard solutions with various concentrations (Figure S4). A distinct positive relationship between loading concentration and the drug input was observed when the initial concentration of doxorubicin was lower than 0.5 mM (Figure 1e). With continuously increased drug input, the loading concentration remained constant since additional doxorubicin could no longer be encapsulated limited to the cavity volume of mEVs. Correspondingly, a contrary trend was observed when calculating the drug encapsulation efficiency under various drug input. Except from the influence of the drug input, a positive correlation between mEVs concentration and the loading capacity was also revealed (Figure S5).
Furthermore, compared to other traditional methods including passive incubation, sonication, freeze‐thaw and electroporation, the much more excellent loading capacity of SEAL was verified (Figures 1f and S6). The particle size, polydispersity index and zeta potential of the Dox‐mEV samples were also measured by dynamic light scattering (DLS), illustrating that there was no significant aggregation or charge reverse upon drug loading (Figures 1f and S7). In addition, the drug release kinetics were analyzed using dialysis bag method (Byeon et al., 2016; Ponta et al., 2015). The delayed release profiles of the SEAL‐prepared Dox‐mEV samples verified the enhanced encapsulation stability compared to the traditional incubation method (Figure 1g).
Moreover, the morphological features of the Dox‐mEVs were examined by transmission electron microscopy (TEM, Figure 1h). Compared to the unloaded mEVs, an electron dense structure representing the doxorubicin precipitates was observed in the vesicular core for SEAL‐prepared Dox‐mEVs, which was consistent with the characteristic features of the liposomal counterparts. Meanwhile, TEM images also illustrated the coexistence of mEVs with other non‐vesicle particles. To address the universality of SEAL, a chemotherapeutic drug mitoxantrone (MXT) and a nucleic acid binding fluorescent dye acridine orange (AO) were also actively loaded into mEVs (Figure S8).
2.2. Single particle analysis of Dox‐mEVs by nFCM
Considering the drawbacks of TEM, which were the time‐consuming processes, low analysis throughputs and the potential imaging artefacts, an nFCM system was applied to more thoroughly reveal the heterogeneity and obtain a quantitative profile of the loading capacity of the Dox‐mEVs prepared by SEAL. As individual particles passing sequentially through the tightly focused laser beam of nFCM, the side scatter (SS) and doxorubicin fluorescence (FL‐Dox) signals were simultaneously detected. Figure 2a showed the representative SS and FL‐Dox burst traces of the Dox‐mEVs prepared by passive incubation and SEAL. For passive incubation samples, there were no particles loaded with sufficient amount of doxorubicin to emit detectable fluorescence for nFCM analysis (Figure 2a‐i&ii). For SEAL‐prepared samples, the concomitant detected peaks on both SS and FL‐Dox channels represented a subpopulation of actively‐loaded Dox‐mEVs that were encapsulated with large amounts of drugs. Meanwhile, particles without detectable FL‐Dox signals were also co‐existed, which were regarded as the un/poorly‐loaded subpopulation. In order to more quantitatively characterize the drug loading capacity from a single‐particle perspective, we introduced a parameter called “active loading fraction,” as a measure for describing the percentage of the actively‐loaded subpopulation. According to the bivariate dot‐plots of FL‐Dox versus SS derived from one minute's detection, the active loading fraction was calculated to be 18.2%, indicating that more than 80% of particles were unable to actively encapsulate doxorubicin (Figure 2b). In addition, “active loading efficiency,” which is defined as the average molecule number of doxorubicin encapsulated within an individual particle, was obtained via single particle enumeration by nFCM combined with the ensemble measurement of total drug loading content, which has been previously described in detail (Chen et al., 2017). For samples illustrated in Figure 2b, the particle concentration and total drug loading content were determined to be 9.0×1013 particles/mL and 162 μM, respectively. As a result, the active loading efficiency was calculated to be 1080 molecules per particle. Assumed that the doxorubicin associated with the un/poorly‐loaded subpopulation was negligible and only the actively‐loaded Dox‐mEVs were taken into account for calculation, the active loading efficiency was re‐estimated to be around 5934 on average. Moreover, the statistically representative histograms of the SS and FL‐Dox intensity distribution were illustrated in Figure 2c. The actively‐loaded subpopulation of Dox‐mEVs showed an enhanced light scattering compared to the un/poorly‐loaded counterparts, either due to the morphology change or the refractive index (RI) increase upon drug encapsulation. Besides, using silica nanoparticles as standards, the size profiles of the two subpopulations could be further deciphered (Figure S9). As for fluorescence detection, owing to the significant doxorubicin accumulation, the FL‐Dox histogram of the actively‐loaded Dox‐mEVs could be discriminated from the majority of the un/poorly‐loaded subpopulation (Figure 2c‐ii). Overall, by means of the nFCM‐based single particle analysis, more comprehensive drug loading behaviour could be revealed and quantified.
FIGURE 2.
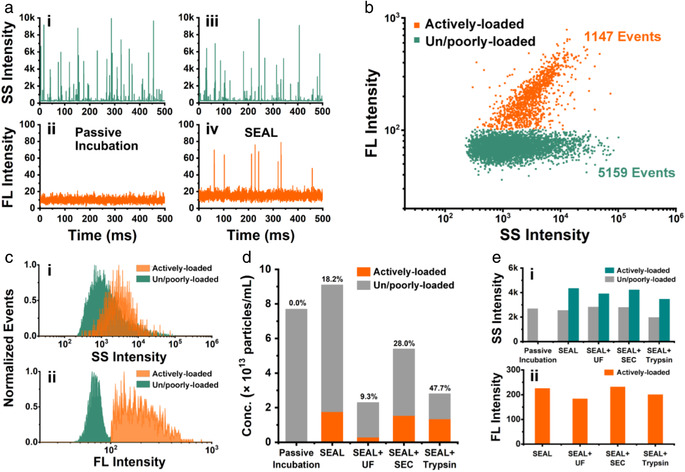
Single particle analysis of the drug loading capacity of SEAL‐prepared Dox‐mEVs by nFCM. (a) Representative SS and FL‐Dox burst traces of the Dox‐mEVs prepared by passive incubation (i, ii) and SEAL (iii, iv). (b) Bivariate dot plots of SS verse FL‐Dox of SEAL‐prepared Dox‐mEVs. (c) SS (i) and FL‐Dox (ii) intensity distribution histograms of the two subpopulations in SEAL‐prepared Dox‐mEVs. (d) Particle concentration and the active loading fraction of Dox‐mEVs prepared and purified by various methods. Note. The numbers on the above of each column indicated the active loading fractions. (e) The median SS (i) and FL‐Dox (ii) intensity of Dox‐mEVs prepared and purified by various methods
Considering the significant coexistence of un/poorly‐loaded particles, three EV purification methods, which were ultrafiltration (UF), size exclusion chromatography (SEC) and trypsin treatment, were applied to purify the Dox‐mEV samples and investigated the impact on the active loading fraction. The particle concentration of the two subpopulations were measured under various purification processes by nFCM, and the active loading fraction was calculated accordingly (Figure 2d). The results indicated that SEC and trypsin treatment were able to increase the active loading fraction, whereas UF purification caused a significant loss of the samples on contrary. The SS and FL‐Dox intensity were also recorded to confirm that there was no appreciable drug release or structure disruption during the purification processes (Figure 2e). Unless indicated otherwise, the Dox‐mEV samples used in the following studies were purified by SEC. The details about the determination of that the major fractions (7–11) containing mEVs were demonstrated in the supporting information (Figure S10).
2.3. Correlation analysis between encapsulation heterogeneity and lipid inclusion
To further clarify the relationship between the encapsulation heterogeneity and the structural features such as lipid inclusion, an upgraded version of nFCM with two lasers and three detection channels was adopted for multi‐parameter correlation analysis. By means of simultaneous scattering and fluorescence measurement, the lipid enclosed vesicles could be discriminated from other particles via fluorescence labelling with a lipophilic carbocyanine dye DiD (Figure 3a). From the dot‐plots of SS verse the fluorescence of DiD (FL‐DiD), it was determined that around 50% of the Dox‐mEV samples were DiD positive, representing the lipid‐enclosed vesicles (Figure 3b). Among them, a small fraction (∼5.9%) of DiD positive particles with only background level of SS intensity were also detected, which could be the dye aggregates or other small particles below the detection limit (∼40 nm; upper left panel in Figure 3b). The bivariate dot plots of FL‐DiD versus FL‐Dox indicated that almost all of the particles with detectable doxorubicin signals, thus the actively‐loaded Dox‐mEVs, were DiD positive (>95%, Figure 3c). Therefore, it was demonstrated that only the particles with lipid inclusion were actively loadable. Meanwhile, there were still 37.6% of the DiD positive particles without detectable FL‐Dox signals (upper left panel in Figure 3c). To more clearly demonstrate the heterogeneous nature of the sample, a Venn diagram with three subsets displaying the particle number of actively‐loaded, lipid‐enclosed and all detectable particles in Dox‐mEV samples was generated (Figure 3d). Besides, upon surfactant‐assisted disruption, a simultaneous reduction in event rates on both FL‐Dox and FL‐DiD channels was observed, further confirming the direct correlation between lipid inclusion and the active drug loading (Figure S11).
FIGURE 3.
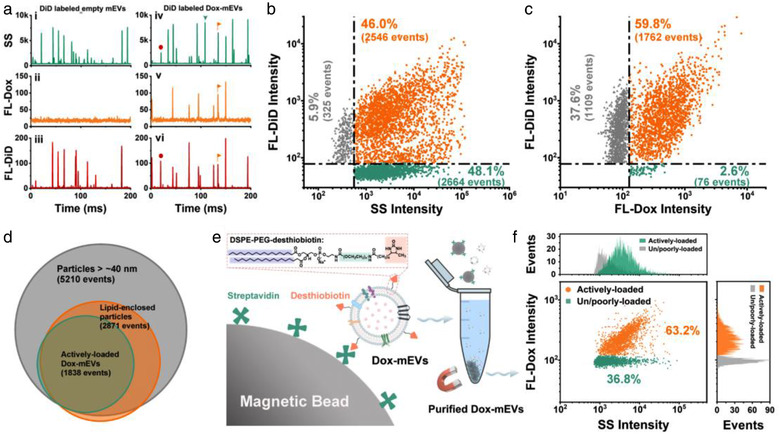
Multi‐parameter analysis of the correlation between drug loading capacity and lipid labelling pattern of Dox‐mEVs. (a) Representative SS, FL‐Dox, and FL‐DiD burst traces of Dox‐mEVs without (i, ii, iii) or with (iv, v, vi) DiD labelling. (b) Bivariate dot plots of SS verse FL‐DiD. (c) Bivariate dot plots of FL‐DiD verse FL‐Dox. (d) A Venn diagram showing the overlap of different subpopulations in SEAL‐prepared Dox‐mEVs. (e) Schematic illustration of the lipid‐probe based magnetic purification process. (f) Bivariate dot‐plots of SS versus FL‐Dox and the corresponding SS and FL‐Dox histograms of the LPMIT‐purified Dox‐mEV samples
2.4. Lipid‐probe mediated magnetic purification of Dox‐mEVs
Inspired from the above results, a lipid‐probe mediated magnetic isolation technique (LPMIT) was introduced to selectively remove the non‐lipid enclosed particles and increase the purity of the Dox‐mEV samples (Figure 3e) (Wan et al., 2017). The lipid‐probe, DSPE‐PEG‐desthiobiotin, was composed of a PEGylated lipid bearing two hydrophobic fatty acid tails for membrane insertion and a desthiobiotin tag for the subsequent affinity purification. Following the “post insertion” process, the desthiobiotin labelled particles could be enriched by streptavidin‐coated magnetic beads, and further released via the competitive binding of free biotin. The purified Dox‐mEV samples were then characterized by nFCM, showing an increased active loading fraction up to 63.2% (Figure 3f). Collectively, it was concluded that almost all of the actively loadable particles were lipid‐enclosed and as consequence, lipid‐probe mediated purification was feasible to purify Dox‐mEV samples and increase the active loading fraction.
2.5. Identification and removal of casein assemblies in Dox‐mEVs
Bovine milk contains large amounts of casein protein, which complicated mEV purification and application (Dalgleish & Corredig, 2012; de Kruif et al., 2012). Although the acid precipitation has been proved to be efficacious to remove the majority of monomeric caseins, the potential residual of the casein assemblies is hard to be completely eliminated (Chen et al., 2021; Yamauchi et al., 2019). Therefore, the LPMIT‐purified Dox‐mEV samples were immunolabeled with a fluorescent antibody against casein and analyzed by nFCM. Among the detectable events, the percentage of immunofluorescence‐positive particles was determined to be ∼33%, confirming the existence of casein assemblies (Figure 4a). Performing correlation analysis between FL‐Dox and the immunofluorescence of casein (FL‐Cas), we found that more than 85% of the FL‐Cas positive particles was unable to be actively loaded with doxorubicin (upper left panel in Figure 4b). Meanwhile, most of the actively loaded subpopulation was determined to be FL‐Cas negative (lower right panel in Figure 4b). By integrating the correlation information, a Venn diagram was generated to illustrate the heterogeneous composition of the LPMIT‐purified Dox‐mEV samples (Figure 4c). To remove the as‐observed casein assemblies and increase the active loading fraction, an immuno‐magnetic isolation technique (IMIT) was proposed (Figure 4d). Coupling magnetic beads with casein antibodies, the assemblies can be removed selectively without compromising the structure integrity of Dox‐mEVs. Finally, the active loading fraction of the purified Dox‐mEV samples was further increased to 83.6% (Figure 4e). It should be noted that although the active loading fraction was significantly increased, more than 70% of the actively loaded Dox‐mEVs was lost since far excessive Dox‐mEVs were applied during the LPMIT process in order to obtain as many purified samples as possible (Figure S12).
FIGURE 4.
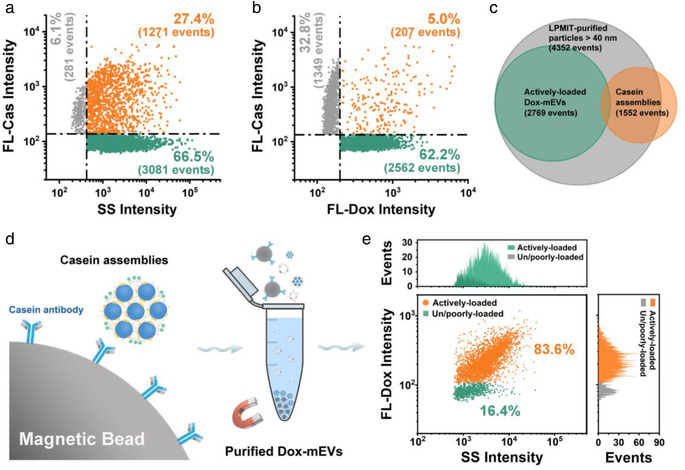
Correlation analysis between drug loading capacity and the immunofluorescence against casein protein of LPMIT‐purified Dox‐mEVs. (a) Bivariate dot plots of SS verse FL‐Cas. (b) Bivariate dot plots of FL‐Dox verse FL‐Cas. (c) A Venn diagram displaying the overlap of different subpopulations. (d) Schematic illustration of the IMIT process for the removal of casein assemblies. (e) Bivariate dot‐plots of SS versus FL‐Dox and the corresponding histograms of the purified Dox‐mEV samples
2.6. Cytotoxicity and cell internalization analysis of cargo‐loaded mEVs
To demonstrate the therapeutic potential of the cargo‐loaded mEVs prepared by SEAL and the impact of sample purity on the overall therapeutic performance, a series of intracellular experiments were further conducted. Firstly, CCK‐8 assays were performed to evaluate the cytotoxicity of the SEAL‐prepared Dox‐mEV samples. It was determined that blank mEVs showed negligible cytotoxicity, while the Dox‐mEVs exhibited a potent cytotoxic response which was more significant than the liposomal counterparts (Figure 5a). Due to the lack of active targeting moiety to promote the accumulation into HepG2 cells, the in vitro cytotoxicity of Dox‐mEVs and Dox‐LPs were lower than the free doxorubicin, which was consistent with the previous reports (Chen et al., 2021; Tian et al., 2014). However, EV formulations could have better therapeutic effects in vivo because of the nanoscale size distribution and the unique surface profile (Chen et al., 2021; J. Liu, Ye, et al., 2019; Zhang et al., 2020). To better exemplify the superiority of SEAL method, doxorubicin encapsulation was also achieved by various loading approaches under same drug input, and the cytotoxic effect was evaluated in parallel (Figure 5b). Using identical protein concentration, a commonly‐applied parameter to determine the dosage of EVs, Dox‐mEVs prepared by SEAL showed the most significant cytotoxicity compared to other methods. In addition, the viability of the cells treated with Dox‐mEVs purified by the above‐mentioned processes was lower than the unpurified samples, owing to the higher drug loading fraction.
FIGURE 5.
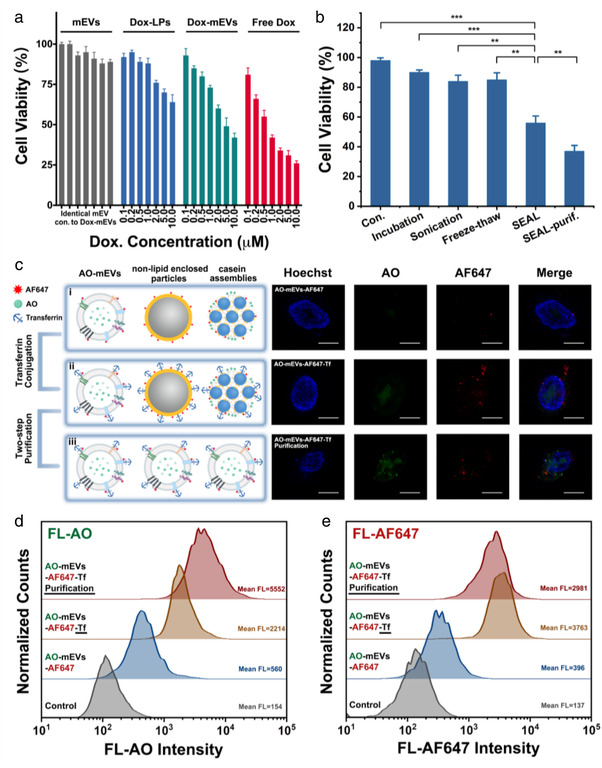
Analysis of the cell cytotoxicity and cell internalization ability of cargo‐loaded mEVs. (a) CCK‐8 based viability measurement of HepG2 cells incubated with blank mEVs, doxorubicin encapsulated liposomes (Dox‐LPs), Dox‐mEVs (purified) and free doxorubicin at various concentration ranging from 0.1 to 10 μM, respectively (n = 3, mean ± SD). Note: the particle concentration of blank mEVs was identical to Dox‐mEVs. (b) CCK‐8 based viability measurement of HepG2 cells incubated with mEVs and the Dox‐mEVs prepared by various methods. p‐Values were calculated by t‐test. **p < 0.01, ***p < 0.001 (n = 3, mean ± SD). (c) Confocal fluorescence microscopy analysis of the cellular internalization of AF647‐labeled AO‐mEVs (i, AO‐mEVs‐AF647), as well as the transferrin conjugated counterpart (AO‐mEVs‐AF647‐Tf) without (ii) and with the two‐step magnetic purification (iii). Note: HepG2 cells were stained with Hoechst 33342, and excited with 405, 488, and 638 nm lasers. Scale bars are 20 μm. (d, e) FCM analysis of the fluorescence intensity of AO (d) and AF647 (e) of HepG2 cells incubated with mEVs, AO‐mEVs‐AF647, AO‐mEVs‐AF647‐Tf without and with purification, respectively
To better analyze the cell internalization behaviour, mEVs were loaded with AO by SEAL, and further labelled with a fluorescence dye Alexa Fluor 647‐NHS via the NHS‐amine reaction (Morales‐Kastresana et al., 2017). After incubating HepG2 cells with the AO‐loaded and Alexa Fluor 647‐NHS labelled mEVs (AO‐mEVs‐AF647), green and red fluorescence could be observed using confocal fluorescence microscopy (CFM, Figure 5c‐i). Considering the unsatisfied cell targeting ability of natural mEVs, a widely used cancer targeting ligand transferrin was conjugated to the surface of AO‐mEVs by a protein‐based modification method developed previously to promote the internalization process (Chen et al., 2021; X. Wang et al., 2017). Compared to the unmodified mEVs, HepG2 cells treated with the transferrin conjugated samples (AO‐mEVs‐AF647‐Tf) displayed more intense fluorescence on both AO and AF647 channels (Figure 5c‐ii). The distinct subcellular localization of the fluorescence signal, where AO was observed in the nuclear fraction and AF647 was mainly located in the cytoplasm, indicated that the mEV‐encapsulated AO was released from the vesicular cavity after internalization. The enhanced cytotoxicity of Dox‐mEVs after transferrin conjugation further confirmed the ligand‐promoted internalization (Figure S13).
To demonstrate the impact of encapsulation heterogeneity on the cell internalization behaviour, the magnetic purified AO‐mEVs‐AF647‐Tf was prepared and analyzed in parallel (Figure 5c‐iii). As shown in CFM images, the AF647 fluorescence of the purified samples showed no observable difference to the unpurified counterpart, while the AO signals were much stronger when using identical particle concentration for cell treatment. The poorer AO delivery capability of unpurified samples was caused by the fact that the protein‐enriched nano‐contaminants were also readily to be modified with transferrin and internalized into the host cells through the receptor‐mediated pathway. Therefore, this receptor consumption effect would lead to the decreased uptake of the actively‐loaded subpopulation in a single transferrin internalization cycle.
Moreover, the flow cytometry (FCM) analysis represented a similar but more quantitative result (Figure 5d and e). Based on the cell fluorescence intensity, the AO delivery capability of the transferrin conjugated mEV formulations was about four times than the unmodified counterpart. After the magnetic purification, this difference was further enlarged (∼10 times). The transferrin conjugation also increased the AF647 intensity but showed a slight decline after magnetic purification, which could be derived from the inconsistent labelling degree between AO‐mEVs‐AF647‐Tf and other nano‐contaminants. Collectively, the integrated results proved the superior therapeutic performance of SEAL‐prepared mEV formulations, and revealed the potential impact of cargo encapsulation heterogeneity on the therapeutic outcome.
3. DISCUSSION
Highly efficient drug encapsulation is a prerequisite for successful application of EVs as nanomedicines, regardless the type of biological fluids employed as the EV sources. Although many different active cargo loading approaches have been developed to promote drug delivery of EVs, the efficiency is still insufficient and far inferior to the liposomal counterpart, especially for hydrophilic molecules. In addition, the encapsulated drugs are mainly existed as free form, which is unfavourable for long‐term preservation. A critical factor contributing to the clinical success of Doxil®, the first FDA‐approved nanomedicine, is the development of the active drug loading method driven by transmembrane (NH4)2SO4 gradient which ensures the stable and efficient doxorubicin encapsulation in liposomes. (Barenholz, 2012) Inspired from that, we conceive a remote loading approach, named “SEAL,” to actively load ionizable hydrophilic agents into EVs. Although sonication or extrusion has already been utilized for direct drug loading of EVs, the purpose of using these physical treatments here was to promote the influx of (NH4)2SO4 for constructing the transmembrane ion gradient required for remote drug loading. Consequently, compared to traditional passive incubation and other stimuli‐assisted active loading methods, SEAL shows much superior loading performance under identical mEV and drug input. The drug encapsulation efficiency for doxorubicin is increased to over 60%, while this value is lower than 40% by other methods reported so far (Figure 1f).
More importantly, the most commonly used parameters for the evaluation of loading performance, “encapsulation efficiency” and “loading efficiency,” are generally based on ensemble‐averaged analysis and only capable of measuring the overall cargo encapsulation amount or the averaged drug content per unit of delivered carrier (Rankin‐Turner et al., 2021; Walker et al., 2019). However, a large heterogeneity of EVs with regards to varying size, different composition and the co‐existence of non‐EV contaminants has been identified. which complicates the proper evaluation of the loading performance. In other word, there could be diverse subgroups of EVs with totally different drug loading outcome, regardless of the values of “encapsulation efficiency” or “loading efficiency.” Recently, a lot of techniques have been developed to reveal the intrinsic heterogeneity of EVs, including nFCM (Tian et al., 2018; S. Wang et al., 2020). Taking advantages of the superior sensitivity and multiparameter analysis merits of nFCM, the inhomogeneous drug loading behaviour of Dox‐mEVs was unravelled in present study. Counting the number of actively loaded and unloaded subpopulation, the active loading fraction was found to be around 20% which means that 80% of nFCM detectable nanoparticles are drug unloadable. The large population of empty carriers or other nano‐contaminants make no contribution to the therapeutic effect, even worse, might compromise the overall drug performance and lead to serious side‐effect. We also proved that SEC was applicable for further sample purification without significant loss of the desired population, which increased the active loading fraction. Whereas, utilization of another commonly used isolation method, UF, would lead to a significant loss of drug‐loaded vesicles.
For nano‐drug delivery application, EV formulations shall be as pure as possible. In order to increase the active loading fraction of cargo‐loaded EVs, the correlation between physiochemical properties and the drug encapsulation heterogeneity needs to be revealed. For Dox‐mEVs, it was found that almost all of the actively loaded Dox‐mEVs could be labelled with a lipophilic carbocyanine fluorescent dye DiD, whereas the other particles could not be fluorescently stained. Besides, casein protein was determined to be negatively related to active drug loading. It is worthy to mention that for EVs originated from different sources, the drug‐loading related features might vary largely depending on the structure and composition of the vesicles as well as the characteristic of the contaminants. Furthermore, taking advantage of the multiparameter analysis of nFCM, custom‐made purification approaches could be proposed to specifically extract the desired subpopulation with distinct features and remove the drug free contaminants to increase the purity of the drug‐loaded EVs. In our case, a series of magnetic separation methods were applied, leading to an increased active loading fraction higher than 80%. Meanwhile, it was found that more than 80% of the sample was lost during these purification steps. We speculated it might be because of that much more excessive Dox‐mEVs were applied than the magnetic beads could capture. However, the merits of high yield, low cost and easy accessibility of mEVs could counterbalance the drawback of low recovery rate (Figure S12). It should be noted that if the EVs were derived from less abundant or less accessible sources, higher number of magnetic beads or other purification methods could be applied to obtain an increased recovery rate. Thus, the balance between loading efficiency and recovery rate must be thoroughly optimized.
From the cell‐based assays, it was found that the therapeutic effect of Dox‐mEVs prepared by SEAL outperformed other conventional loading methods, owing to the superior drug encapsulation efficiency. Applying transferrin conjugated mEVs as a model, the impact of the drug‐loading heterogeneity on the active targeting behaviour was revealed. It was more convincingly confirmed that even if the great biosafety of EVs might allow for less pure formulations with a certain fraction of particles unloaded with drug, the occupation of the cell binding sites of these contaminants, would potentially compromise the therapeutic performance especially for the ligand coupled formulations.
4. CONCLUSION
Collectively, inspired from liposomal nanomedicines, we proposed the SEAL method for effective encapsulation of ionizable hydrophilic molecules in mEVs. The drug loading capacity was proved to be superior than conventional approaches. Moreover, an nFCM based single‐particle characterization strategy was developed to evaluate the heterogenous loading behaviour, and the “active loading fraction” was introduced to quantitatively reveal the co‐existence of actively‐loaded and un/poorly‐loaded subpopulations. Interpreting the correlation between compositional features and loading performance, a majority of the un/poorly‐loaded subpopulation was determined to be non‐lipid enclosed particles and casein assemblies. Accordingly, customized purification processes were proposed to selectively remove these nano‐contaminants, and purify the mEV formulations. Finally, the intracellular assessments demonstrated the superior therapeutic performance of drug‐loaded mEV formulations prepared by SEAL, and revealed the impact of encapsulation heterogeneity on the cargo‐delivery efficiency.
Although the practicability of SEAL was only validated by loading the ionizable and water‐soluble molecules here, the active encapsulation of the poorly ionizable or in‐soluble agents can be envisioned by means of the solvent or cyclodextrin assisted approaches (Sur et al., 2014; W. L. Tang, Tang, et al., 2018). It can also be conceived that in addition to Dox‐mEVs, vesicles isolated from other sources or with cargo loaded by various methods could easily be characterized using this nFCM‐based strategy. Doubtless, the loading efficiency and the major contaminants in the sample will be differed from SEAL‐prepared Dox‐mEVs, but custom designed purification approaches can also be inspired from the multiparameter analysis results of nFCM to improve the purity and therapeutic outcome of the EV formulations. Therefore, we believe that the as‐developed SEAL method will lower the primary hurdle in the development of EV‐based delivery systems, which is the unsatisfied cargo loading efficiency. More importantly, when it comes to another major challenge, which it that the natural and manufacturing heterogeneity of cargo‐loaded EV formulations significantly complicates the application, the nFCM‐based methodology can make a constructive contribution in evaluation of loading approach and assessment of quality.
5. EXPERIMENTAL SECTION
5.1. EVs isolation from raw milk
Milk derived EVs (mEVs) were isolated using ultracentrifugation‐based method with some modifications (Somiya et al., 2018; Yamauchi et al., 2019). Briefly, three litres of fresh unpasteurised milk was collected from a local dairy farm. The collected milk was aliquoted (500 mL) at −80°C and used within 3 days. The mEV samples were isolated by differential centrifugation. Firstly, raw milk was centrifuged at 2000 × g for 5 min at 4°C to remove milk fat globules and cells, and centrifuged again at 10,000 × g for 30 min to remove the fat residues and cellular debris. Then the supernatant was acidized to pH = 4.6 with acetic acid to aggregate the caseins, followed by 10 min incubation and 15 min centrifugation at 10,000 × g at 4°C. The supernatant was sequentially filtrated through 1, 0.45, and 0.22 μm polyethersulfone (PES) membranes to remove large particles, and centrifuged at 100,000 × g for 70 min to pellet EVs. Finally, the mEVs pellets were resuspended in PBS or ammonium sulfate for further experiments. A Beckman Coulter Avanti J‐26S XP centrifuge and a Beckman Coulter ultracentrifuge were used for the centrifugation throughout present study.
5.2. Preparation of cargo‐loaded mEVs
Sonication and Extrusion‐based Active Loading (SEAL) method was developed to prepare cargo‐loaded mEV formulations. Unless indicated otherwise, the experiment was performed as follows. Firstly, the ultracentrifuged mEV pellets were resuspended in 0.5 M ammonium sulfate solution, followed by probe sonication for 4 min (30 s on/off, on ice) at 20% amplitude by a Vibra Cell Ultrasonicator (Model VCX130) to promote the influx of ammonium sulfate into mEVs’ cavity. Then the sonicated mEVs were further extruded through 100 nm polycarbonate (PC) membranes for three cycles at room temperature (RT). The bulk solution was changed to PBS by 4°C dialysis overnight to generate the transmembrane ammonium gradient as the driving force for active loading. Finally, DOX, MXT or AO (Sigma‐Aldrich) was added to the dialyzed solution, and incubated at room temperature for 30 min for complete encapsulation. The mEV formulations were purified by gel filtration (Sephadex G‐25, GE Healthcare) to remove the free agents, and then subjected to DLS (ZetasizerNano ZS90, Malvern, UK) and TEM (HT‐7800, Hitachi, Japan) for size measurement and morphology observation, respectively.
Four traditional drug loading methods, passive incubation, sonication, freeze‐thaw and electroporation were also applied for doxorubicin encapsulation. For passive incubation, the mEVs were incubated with doxorubicin solution at 4°C or 37°C for 1 or 24 h after resuspended in PBS. In case of direct sonication treatment, mEVs were mixed with doxorubicin and sonicated using a Vibra Cell Ultrasonicator under the following settings: 20% amplitude, 30 s on/off for 4 min at room temperature. For freeze‐thaw method, the doxorubicin incubated mEVs were subjected to 3 freeze‐thaw cycles between −80°C and 37°C water bath. As for electroporation, doxorubicin was loaded into mEVs at 350 V and 150 μF in a 4 mm cuvette by Gene Pulser Electroporator (Bio‐Rad), according to previous reports with minor modification (Tian et al., 2014; Zhang et al., 2020). Gel filtration by Sephadex G‐25 column was used to separate Dox‐mEVs from excess drug.
5.3. Determination of drug loading capacity by fluorospectrophotometry
The drug loading capacity of Dox‐mEVs was determined by measuring the intrinsic fluorescence intensity using a spectrofluorometer (Agilent Cary Eclipse). Doxorubicin solutions with concentration of 0.25, 0.5, 1.0, 2.0, and 5.0 μM were used as standards. After drug loading and purification to remove the unloaded doxorubicin, the as‐fabricated Dox‐mEVs were pre‐treated with 1% Triton X‐100 (Sigma‐Aldrich) to disrupt the lipid membrane and release the encapsulated cargo and eliminate the inner filter effect (IFE)‐induced fluorescence quenching of doxorubicin. Then the fluorescence of this sample was detected, and the drug content was determined based on the calibration curve built upon the doxorubicin standard solutions. Finally, the loading efficiency was calculated by dividing the EV‐loaded doxorubicin content by the total drug input used in the assay.
5.4. Analysis of the drug release profiles of Dox‐mEVs
The drug release behaviour of Dox‐mEVs was performed by dialysis bag method. In brief, 1 mL of each sample was added in dialysis devices (Float‐A‐Lyzer G2, MWCO: 8 to 10 kDa, Repligen) suspended in external buffer of 1 L PBS at 4°C with gentle magnetic stirring. At scheduled intervals, 10 μL of the samples was collected for fluorescence measurement. The drug release profile was determined using the fluorescence intensity at different incubation time and the original value. The experiments were performed in triplicate.
5.5. Single‐particle analysis of SEAL‐prepared Dox‐mEVs by nFCM
A previously built nFCM system was used for single particle analysis of mEVs (Tian et al., 2018; Zhu et al., 2014). The mEVs with or without drug loading were diluted to a particle concentration at ∼109 particles/mL and loaded to the nFCM system. A 532 nm laser was used as the excitation source, and the side scatter and intrinsic fluorescence of doxorubicin emitted by single Dox‐mEVs were detected by two avalanche photodiodes (APDs), respectively. A bandpass filter (FF01‐582/75; Semrock Inc.) was set before the fluorescence detector. For multiparameter analysis, mEVs were fluorescent labeled with 50 μM lipophilic dye DiD (Sigma‐Aldrich) for 30 min at RT, and the unbound dye was removed by a Sephadex G‐25 column. Besides, a 60 mW, 642‐nm continuous‐wave solid‐state Nd:YAG laser was settled on nFCM as the second excitation source to trigger DiD fluorescence, which was further collected by a third APD. Regarding to the lysis experiments, 1% Triton X‐100 (Sigma‐Aldrich) was incubated with mEVs for 30 min at RT.
5.6. Dox‐mEVs purification by SEC, ultrafiltration and protease treatment
The Dox‐mEVs were further purified via different methods. For purification by SEC, a qEV2 / 70 nm column (#SP4, Izon Science Ltd, New Zealand) was used according to the instruction. Briefly, up to 2 mL Dox‐mEVs were loaded onto the PBS equilibrated column and 30 fractions (2 mL per fraction) were collected using PBS as elution buffer. The protein concentration of each fraction was measured by bicinchoninic acid assay (BCA assay, Thermo Scientific, USA) using a series of concentrations of bovine serum albumin (BSA, Sigma‐Aldrich) as standards. The mEV fractions (∼7–11) were used for further research. For UF process, 5 mL of mEVs were concentrated to 500 μL using a Vivaspin 20 Diafiltration device (Sartorius) with 300 kDa molecular weight cutoff (MWCO). Then PBS was added to reconstitute the concentrate to the original sample volume and the UF procedure was repeated twice. Finally, a pipettor was carefully inserted to the bottom of the device to withdraw the concentrated samples. For protease treatment, 0.25% trypsin (Sigma‐Aldrich) was added to the mEV samples and incubated at 37°C for 15 min.
5.7. Lipid‐probe mediated magnetic purification of mEVs
LPMIT was applied to purify the Dox‐mEVs. The lipid‐probe of DSPE‐PEG‐desthiobiotin was dissolved in chloroform and the solution was evaporated under vacuum for 3 h. The resulting film was rehydrated and sonicated in filtered PBS buffer for 30 min to form lipid micelles. Then DSPE‐PEG‐desthiobiotin micelles were added into the mEV suspension at a final lipid concentration of 0.1 mM. The samples were incubated at 37°C for 30 min and then mixed with streptavidin labeled magnetic beads (Dynabeads™, Thermo Scientific, USA) for 30 min. After rinsing twice with PBS to remove the non‐specific absorbed particles, mEVs were eluted using 10 mM free biotin.
5.8. Immuno‐magnetic isolation technique for the removal of casein assemblies
Briefly, casein antibody (Bioss Antibodies) was maleimide‐activated by sulfo‐SMCC (Sigma‐Aldrich) and conjugated to the thiol magnetic beads (MBs, PuriMag) according to the manufacturer's instructions. The unconjugated antibody was removed by washing with PBS for three times. For the removal of the casein assemblies, the LPMIT‐purified Dox‐mEV samples were mixed with the antibody conjugated MBs for 1 h at RT. The mixtures were then placed on a magnet for 1 min and the supernatant was transferred to a second tube of antibody conjugated MBs for another round of separation. The IMIT process was repeated three times for the effective removal of casein assemblies.
5.9. Cytotoxicity analysis of Dox‐mEVs
CCK‐8 assay was used to assess the cytotoxicity of Dox‐mEVs and doxorubicin encapsulated liposomes (Dox‐LPs). HepG2 cells were seeded in 96‐well plates and cultured for 6 h. Then Dox‐mEVs and Dox‐LPs with doxorubicin concentration ranging from 0.1 to 10 μM, and blank mEVs with identical particle concentration to Dox‐mEVs were added to the cells and cultured for another 16 h. Afterwards, the cells were further incubated with 10 μL CCK‐8 solution for 2 h, and the absorbance intensity at 450 nm was measured immediately. The measurements were carried out in triplicate. Besides, the cytotoxicity of Dox‐mEVs prepared by simple incubation, direct sonication and freeze‐thaw were also analyzed by CCK‐8.
5.10. Cellular internalization analysis of AO‐loaded mEVs
To better evaluate the cellular internalization, mEVs were loaded with 0.5 mM AO by SEAL, and labelled with Alexa fluor 647‐NHS (Thermo Scientific, USA) via the NHS‐amine reaction. As for the preparation of ligand conjugated mEVs, facile reduction of surface disulfides on mEVs was conducted followed by the click reaction with maleimide modified transferrin (Sigma‐Aldrich) (Chen et al., 2021). Then AO‐mEVs‐AF647, AO‐mEVs‐AF647‐Tf and the magnetic purified AO‐mEVs‐AF647‐Tf were incubated with HepG2 cells for 9 h, respectively. Once removing the uninternalized mEVs and washing with PBS, the cells were stained with Hoechst 33342 and imaged on a Leica SP8 confocal microscope. Regarding FCM analysis, three mEV formulations, as well as the blank mEVs, were incubated with HepG2 cells for 12 h. After removing the uninternalized mEVs, the fluorescence intensity of the cells was measured immediately using a flow cytometer (FACS Aria, BD).
5.11. Statistical analysis
Statistical results were reported as mean ± standard deviation (SD) values of three independent experiments conducted. Unpaired t test was performed to assess the difference between various groups: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 (n = 3).
CONFLICT OF INTEREST
The authors declare the following competing financial interest. X.Y. declares competing financial interest as a cofounder and shareholder of NanoFCM Inc., a company committed to commercializing the nFCM technology.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
This research was supported by the Start‐up Fund of Jimei University (4411/C618004), Natural Science Foundation of Fujian Province (2019J05097), and the National Natural Science Foundation of China (21627811 and 21934004).
Chen, C. , Sun, M. , Wang, J. , Su, L. , Lin, J. , & Yan, X. (2021). Active cargo loading into extracellular vesicles: Highlights the heterogeneous encapsulation behaviour. Journal of Extracellular Vesicles, 10,e12163. 10.1002/jev2.12163
Contributor Information
Chaoxiang Chen, Email: cxchen@jmu.edu.cn.
Xiaomei Yan, Email: xmyan@xmu.edu.cn.
REFERENCES
- Agrahari, V. , Agrahari, V. , Burnouf, P. A. , Chew, C. H. , & Burnouf, T. (2019). Extracellular microvesicles as new industrial therapeutic frontiers. Trends in Biotechnology, 37, 707–729. [DOI] [PubMed] [Google Scholar]
- Antimisiaris, S. G. , Mourtas, S. , & Marazioti, A. (2018). Exosomes and exosome‐inspired vesicles for targeted drug delivery. Pharmaceutics, 10, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenholz, Y. (2012). Doxil(R)—The first FDA‐approved nano‐drug: Lessons learned. Journal of Controlled Release, 160, 117–134. [DOI] [PubMed] [Google Scholar]
- Batrakova, E. V. , & Kim, M. S. (2015). Using exosomes, naturally‐equipped nanocarriers, for drug delivery. Journal of Controlled Release, 219, 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunggulawa, E. J. , Wang, W. , Yin, T. , Wang, N. , Durkan, C. , Wang, Y. , & Wang, G. (2018). Recent advancements in the use of exosomes as drug delivery systems. Journal of Nanbiotechnology, 16, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon, H. J. , le Thao, Q. , Lee, S. , Min, S. Y. , Lee, E. S. , Shin, B. S. , Choi, H. G. , & Youn, Y. S. (2016). Doxorubicin‐loaded nanoparticles consisted of cationic‐ and mannose‐modified‐albumins for dual‐targeting in brain tumors. Journal of Controlled Release, 225, 301–313. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Gao, K. , Lian, H. , Chen, C. , & Yan, X. (2019). Single‐particle characterization of theranostic liposomes with stimulus sensing and controlled drug release properties. Biosensors & Bioelectronics, 131, 185–192. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Sun, M. , Liu, X. , Wu, W. , Su, L. , Li, Y. , Liu, G. , & Yan, X. (2021). General and mild modification of food‐derived extracellular vesicles for enhanced cell targeting. Nanoscale, 13, 3061–3069. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Zhu, S. , Wang, S. , Zhang, W. , Cheng, Y. , & Yan, X. (2017). Multiparameter quantification of liposomal nanomedicines at the single‐particle level by high‐sensitivity flow cytometry. ACS Applied Materials & Interfaces, 9, 13913–13919. [DOI] [PubMed] [Google Scholar]
- Colao, I. L. , Corteling, R. , Bracewell, D. , & Wall, I. (2018). Manufacturing exosomes: A promising therapeutic platform. Trends in Molecular Medicine, 24, 242–256. [DOI] [PubMed] [Google Scholar]
- Dalgleish, D. G. , & Corredig, M. (2012). The structure of the casein micelle of milk and its changes during processing. Annual Review of Food Science and Technology, 3, 449–467. [DOI] [PubMed] [Google Scholar]
- de Jong, O. G. , Kooijmans, S. A. A. , Murphy, D. E. , Jiang, L. , Evers, M. J. W. , Sluijter, J. P. G. , Vader, P. , & Schiffelers, R. M. (2019). Drug delivery with extracellular vesicles: From imagination to innovation. Accounts of Chemical Research, 52, 1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kruif, C. G. , Huppertz, T. , Urban, V. S. , & Petukhov, A. V. (2012). Casein micelles and their internal structure. Advances in Colloid and Interface Science, 171–172, 36–52. [DOI] [PubMed] [Google Scholar]
- Donoso‐Quezada, J. , Ayala‐Mar, S. , & Gonzalez‐Valdez, J. (2020). State‐of‐the‐art exosome loading and functionalization techniques for enhanced therapeutics: A review. Critical Reviews in Biotechnology, 40, 804–820. [DOI] [PubMed] [Google Scholar]
- Elsharkasy, O. M. , Nordin, J. Z. , Hagey, D. W. , de Jong, O. G. , Schiffelers, R. M. , Andaloussi, S. E. , & Vader, P. (2020). Extracellular vesicles as drug delivery systems: Why and how? Advanced Drug Delivery Reviews, 159, 332–343. [DOI] [PubMed] [Google Scholar]
- Fais, S. , O'Driscoll, L. , Borras, F. E. , Buzas, E. , Camussi, G. , Cappello, F. , Carvalho, J. , Cordeiro da Silva, A. , Del Portillo, H. , El Andaloussi, S. , Ficko Trcek, T. , Furlan, R. , Hendrix, A. , Gursel, I. , Kralj‐Iglic, V. , Kaeffer, B. , Kosanovic, M. , Lekka, M. E. , Lipps, G. , … Giebel, B. (2016). Evidence‐based clinical use of nanoscale extracellular vesicles in nanomedicine. ACS Nano, 10, 3886–3899. [DOI] [PubMed] [Google Scholar]
- Ferguson, S. W. , & Nguyen, J. (2016). Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. Journal of Controlled Release, 228, 179–190. [DOI] [PubMed] [Google Scholar]
- Fuhrmann, G. , Serio, A. , Mazo, M. , Nair, R. , & Stevens, M. M. (2015). Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. Journal of Controlled Release, 205, 35–44. [DOI] [PubMed] [Google Scholar]
- Gilligan, K. E. , & Dwyer, R. M. (2017). Engineering exosomes for cancer therapy. International Journal of Molecular Sciences, 18, 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, W. J. , Lee, C. K. , Zou, S. , Woon, E. C. , Czarny, B. , & Pastorin, G. (2017). Doxorubicin‐loaded cell‐derived nanovesicles: An alternative targeted approach for anti‐tumor therapy. International Journal of Nanomedicine, 12, 2759–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgens, A. , & Nolan, J. P. (2020). Aiming to compare apples to apples: Analysis of extracellular vesicles and other nanosized particles by flow cytometry. Cytometry Part A, 97, 566–568. [DOI] [PubMed] [Google Scholar]
- Gubernator, J. (2011). Active methods of drug loading into liposomes: Recent strategies for stable drug entrapment and increased in vivo activity. Expert Opinion on Drug Delivery, 8, 565–580. [DOI] [PubMed] [Google Scholar]
- Gudbergsson, J. M. , Jonsson, K. , Simonsen, J. B. , & Johnsen, K. B. (2019). Systematic review of targeted extracellular vesicles for drug delivery—Considerations on methodological and biological heterogeneity. Journal of Controlled Release, 306, 108–120. [DOI] [PubMed] [Google Scholar]
- Ingato, D. , Lee, J. U. , Sim, S. J. , & Kwon, Y. J. (2016). Good things come in small packages: Overcoming challenges to harness extracellular vesicles for therapeutic delivery. Journal of Controlled Release, 241, 174–185. [DOI] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177, 428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri, R. , & LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science, 367, 640–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar, S. , LeBleu, V. S. , Sugimoto, H. , Yang, S. , Ruivo, C. F. , Melo, S. A. , Lee, J. J. , & Kalluri, R. (2017). Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature, 546, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. S. , Haney, M. J. , Zhao, Y. , Mahajan, V. , Deygen, I. , Klyachko, N. L. , Inskoe, E. , Piroyan, A. , Sokolsky, M. , Okolie, O. , Hingtgen, S. D. , Kabanov, A. V. , & Batrakova, E. V. (2016). Development of exosome‐encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine, 12, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. P. , Lin, Z. X. , Jiang, X. Y. , & Yu, X. Y. (2018). Exosomal cargo‐loading and synthetic exosome‐mimics as potential therapeutic tools. Acta Pharmacologica Sinica, 39, 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Cipolla, D. , Rades, T. , & Boyd, B. J. (2018). Drug nanocrystallisation within liposomes. Journal of Controlled Release, 288, 96–110. [DOI] [PubMed] [Google Scholar]
- Liao, W. , Du, Y. , Zhang, C. , Pan, F. , Yao, Y. , Zhang, T. , & Peng, Q. (2018). Exosomes: The next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomaterialia, 86, 1–14. [DOI] [PubMed] [Google Scholar]
- Liu, C. , & Su, C. (2019). Design strategies and application progress of therapeutic exosomes. Theranostics, 9, 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Ye, Z. , Xiang, M. , Chang, B. , Cui, J. , Ji, T. , Zhao, L. , Li, Q. , Deng, Y. , Xu, L. , Wang, G. , Wang, L. , & Wang, Z. (2019). Functional extracellular vesicles engineered with lipid‐grafted hyaluronic acid effectively reverse cancer drug resistance. Biomaterials, 223, 119475. [DOI] [PubMed] [Google Scholar]
- Lu, M. , & Huang, Y. (2020). Bioinspired exosome‐like therapeutics and delivery nanoplatforms. Biomaterials, 242, 119925. [DOI] [PubMed] [Google Scholar]
- Moller, A. , & Lobb, R. J. (2020). The evolving translational potential of small extracellular vesicles in cancer. Nature Reviews Cancer, 20, 697–709. [DOI] [PubMed] [Google Scholar]
- Morales‐Kastresana, A. , Telford, B. , Musich, T. A. , McKinnon, K. , Clayborne, C. , Braig, Z. , Rosner, A. , Demberg, T. , Watson, D. C. , Karpova, T. S. , Freeman, G. J. , DeKruyff, R. H. , Pavlakis, G. N. , Terabe, M. , Robert‐Guroff, M. , Berzofsky, J. A. , & Jones, J. C. (2017). Labeling extracellular vesicles for nanoscale flow cytometry. Scientific Reports, 7, 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, J. P. (2015). Flow cytometry of extracellular vesicles: Potential, pitfalls, and prospects. Current Protocols in Cytometry, 73, 13.14.1–13.14.16. [DOI] [PubMed] [Google Scholar]
- Pascucci, L. , Cocce, V. , Bonomi, A. , Ami, D. , Ceccarelli, P. , Ciusani, E. , Vigano, L. , Locatelli, A. , Sisto, F. , Doglia, S. M. , Parati, E. , Bernardo, M. E. , Muraca, M. , Alessandri, G. , Bondiolotti, G. , & Pessina, A. (2014). Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. Journal of Controlled Release, 192, 262–270. [DOI] [PubMed] [Google Scholar]
- Pegtel, D. M. , & Gould, S. J. (2019). Exosomes. Annual Review of Biochemistry, 88, 487–514. [DOI] [PubMed] [Google Scholar]
- Ponta, A. , Fugit, K. D. , Anderson, B. D. , & Bae, Y. (2015). Release, partitioning, and conjugation stability of doxorubicin in polymer micelles determined by mechanistic modeling. Pharmaceutical Research, 32, 1752–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, M. I. , Amorim, M. G. , Gadelha, C. , Milic, I. , Welsh, J. A. , Freitas, V. M. , Nawaz, M. , Akbar, N. , Couch, Y. , Makin, L. , Cooke, F. , Vettore, A. L. , Batista, P. X. , Freezor, R. , Pezuk, J. A. , Rosa‐Fernandes, L. , Carreira, A. C. O. , Devitt, A. , Jacobs, L. , … Dias‐Neto, E. (2018). Technical challenges of working with extracellular vesicles. Nanoscale, 10, 881–906. [DOI] [PubMed] [Google Scholar]
- Rankin‐Turner, S. , Vader, P. , O'Driscoll, L. , Giebel, B. , Heaney, L. M. , & Davies, O. G. (2021). A call for the standardised reporting of factors affecting the exogenous loading of extracellular vesicles with therapeutic cargos. Advanced Drug Delivery Reviews, 173, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S. , Lin, H. Y. , Chou, C. Y. , Huang, C. H. , Small, J. , Sadik, N. , Ayinon, C. M. , Lansbury, E. , Cruz, L. , Yekula, A. , Jones, P. S. , Balaj, L. , & Carter, B. S. (2019). Navigating the landscape of tumor extracellular vesicle heterogeneity. International Journal of Molecular Sciences, 20, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, H. , Im, H. , Castro, C. M. , Breakefield, X. , Weissleder, R. , & Lee, H. (2018). New technologies for analysis of extracellular vesicles. Chemical Reviews, 118, 1917–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, S. , Wu, Y. , Liu, Y. , & Wu, D. (2017). High drug‐loading nanomedicines: Progress, current status, and prospects. International Journal of Nanomedicine, 12, 4085–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somiya, M. , Yoshioka, Y. , & Ochiya, T. (2018). Biocompatibility of highly purified bovine milk‐derived extracellular vesicles. Journal of Extracellular Vesicles, 7, 1440132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur, S. , Fries, A. C. , Kinzler, K. W. , Zhou, S. , & Vogelstein, B. (2014). Remote loading of preencapsulated drugs into stealth liposomes. PNAS, 111, 2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J. , Zhang, R. , Guo, M. , Shao, L. , Liu, Y. , Zhao, Y. , Zhang, S. , Wu, Y. , & Chen, C. (2018). Nucleosome‐inspired nanocarrier obtains encapsulation efficiency enhancement and side effects reduction in chemotherapy by using fullerenol assembled with doxorubicin. Biomaterials, 167, 205–215. [DOI] [PubMed] [Google Scholar]
- Tang, W. L. , Tang, W. H. , Szeitz, A. , Kulkarni, J. , Cullis, P. , & Li, S. D. (2018). Systemic study of solvent‐assisted active loading of gambogic acid into liposomes and its formulation optimization for improved delivery. Biomaterials, 166, 13–26. [DOI] [PubMed] [Google Scholar]
- Tian, Y. , Li, S. , Song, J. , Ji, T. , Zhu, M. , Anderson, G. J. , Wei, J. , & Nie, G. (2014). A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials, 35, 2383–2390. [DOI] [PubMed] [Google Scholar]
- Tian, Y. , Ma, L. , Gong, M. , Su, G. , Zhu, S. , Zhang, W. , Wang, S. , Li, Z. , Chen, C. , Li, L. , Wu, L. , & Yan, X. (2018). Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano, 12, 671–680. [DOI] [PubMed] [Google Scholar]
- Vader, P. , Mol, E. A. , Pasterkamp, G. , & Schiffelers, R. M. (2016). Extracellular vesicles for drug delivery. Advanced Drug Delivery Reviews, 106, 148–156. [DOI] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19, 213–228. [DOI] [PubMed] [Google Scholar]
- Walker, S. , Busatto, S. , Pham, A. , Tian, M. , Suh, A. , Carson, K. , Quintero, A. , Lafrence, M. , Malik, H. , Santana, M. X. , & Wolfram, J. (2019). Extracellular vesicle‐based drug delivery systems for cancer treatment. Theranostics, 9, 8001–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , Cheng, G. , Liu, X. , Hao, S. J. , Nisic, M. , Zhu, C. D. , Xia, Y. Q. , Li, W. Q. , Wang, Z. G. , Zhang, W. L. , Rice, S. J. , Sebastian, A. , Albert, I. , Belani, C. P. , & Zheng, S. Y. (2017). Rapid magnetic isolation of extracellular vesicles via lipid‐based nanoprobes. Nature Biomedical Engineering, 1, 0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Khan, A. , Huang, R. , Ye, S. , Di, K. , Xiong, T. , & Li, Z. (2020). Recent advances in single extracellular vesicle detection methods. Biosensors & Bioelectronics, 154, 112056. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Wang, M. , Lei, R. , Zhu, S. F. , Zhao, Y. , & Chen, C. (2017). Chiral surface of nanoparticles determines the orientation of adsorbed transferrin and its interaction with receptors. ACS Nano, 11, 4606–4616. [DOI] [PubMed] [Google Scholar]
- Wu, P. , Zhang, B. , Ocansey, D. K. W. , Xu, W. , & Qian, H. (2020). Extracellular vesicles: A bright star of nanomedicine. Biomaterials, 269, 120467. [DOI] [PubMed] [Google Scholar]
- Yamauchi, M. , Shimizu, K. , Rahman, M. , Ishikawa, H. , Takase, H. , Ugawa, S. , Okada, A. , & Inoshima, Y. (2019). Efficient method for isolation of exosomes from raw bovine milk. Drug Development and Industrial Pharmacy, 45, 359–364. [DOI] [PubMed] [Google Scholar]
- Zhang, G. , Huang, X. , Xiu, H. , Sun, Y. , Chen, J. , Cheng, G. , Song, Z. , Peng, Y. , Shen, Y. , Wang, J. , & Cai, Z. (2020). Extracellular vesicles: Natural liver‐accumulating drug delivery vehicles for the treatment of liver diseases. Journal of Extracellular Vesicles, 10, E12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Wu, D. , Ma, X. , Wang, J. , Hou, W. , & Zhang, W. (2020). Exosomes as drug carriers for cancer therapy and challenges regarding exosome uptake. Biomedicine & Pharmacotherapy, 128, 110237. [DOI] [PubMed] [Google Scholar]
- Zhu, S. , Ma, L. , Wang, S. , Chen, C. , Zhang, W. , Yang, L. , Hang, W. , Nolan, J. P. , Wu, L. , & Yan, X. (2014). Light‐scattering detection below the level of single fluorescent molecules for high‐resolution characterization of functional nanoparticles. ACS Nano, 8, 10998–11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


