Abstract
We have identified an 86-kDa protein containing a single amino-terminal RNA recognition motif and two carboxy-terminal domains rich in serine-arginine (SR) dipeptides. Despite structural similarity to members of the SR protein family, p86 is clearly unique. It is not found in standard SR protein preparations, does not precipitate in the presence of high magnesium concentrations, is not recognized by antibodies specific for SR proteins, and cannot complement splicing-defective S100 extracts. However, we have found that p86 can inhibit the ability of purified SR proteins to activate splicing in S100 extracts and can even inhibit the in vitro and in vivo activation of specific splice sites by a subset of SR proteins, including ASF/SF2, SC35, and SRp55. In contrast, p86 activates splicing in the presence of SRp20. Thus, it appears that pairwise combination of p86 with specific SR proteins leads to altered splicing efficiency and differential splice site selection. In all cases, such regulation requires the presence of the two RS domains and a unique intervening EK-rich region, which appear to mediate direct protein-protein contact between these family members. Full-length p86, but not a mutant lacking the RS-EK-RS domains, was found to preferentially interact with itself, SRp20, ASF/SF2, SRp55, and, to a slightly lesser extent, SC35. Because of the primary sequence and unique properties of p86, we have named this protein SRrp86 for SR-related protein of 86 kDa.
Mammalian gene expression is dependent on the precise, efficient removal of intervening sequences from pre-mRNA. For alternatively spliced genes, the pattern of exon inclusion can result in the production of multiple functionally distinct protein products from the same gene (59, 70). While the overall mechanism of intron removal is likely to be RNA catalyzed, regulated splicing in higher eukaryotes depends on the action of multiple protein components (reviewed in reference 7). A family of proteins that plays a vital role in both constitutive and alternative splicing is characterized by the presence of one or two amino-terminal RNA recognition motifs (RRMs) and a carboxy-terminal domain rich in serine and arginine residues (SR proteins; reviewed in references 8, 20, and 40). Because many SR protein family members can individually complement splicing-deficient S100 extracts, it first appeared that they might be functionally redundant. However, more recent studies have shown that individual SR proteins display substrate specificity (19, 56, 68, 79) and targeted disruption of a single SR protein-encoding gene (ASF/SF2) is cell lethal (69). In addition, individual SR proteins have distinct functions in alternative splicing events and can even negatively regulate splicing (23, 29, 41, 42). Thus, overall regulation of alternative splicing is likely to be intimately tied to the concentration and/or activity of various SR proteins between different cell types (27, 56, 75).
Consistent with substrate-specific effects, individual SR proteins have been shown to display distinct RNA binding specificity (2, 28, 32, 57, 63, 64, 81). One of the first SR proteins to be identified, ASF/SF2, can bind to 5′ splice sites, leading to recruitment of U1 snRNP (32, 81). However, SR proteins also bind to purine-rich splicing enhancer sequences typically located within exons flanked by weak splicing signals (8, 40). The binding of SR proteins to enhancer elements leads to the activation of splicing through the formation of bridge complexes mediated by protein-protein interactions across either introns or exons (1, 3, 32, 60, 62, 71). These interactions have been observed mainly through two-hybrid and immunoprecipitation analyses and include interactions between SR superfamily members (2, 32, 71, 72, 79), between SR proteins and non-SR splicing factors (13, 26, 65), and between SR proteins and putative nuclear matrix components (4).
Despite the growing body of knowledge concerning the effects of SR proteins on alternative splicing, little is known about how these factors are themselves regulated. SR proteins are subject to phosphorylation, and it appears that their phosphorylation state changes as splicing proceeds (10, 30, 72, 76). Several kinases capable of phosphorylating SR proteins have been identified, but it remains to be determined exactly how these enzymes, and the level of SR protein phosphorylation, affect function (13, 16, 25, 26, 37, 51, 54, 61, 67). In addition to phosphorylation, other factors have been shown to inhibit SR protein function. The activity of ASF/SF2 can be counteracted by the effects of the hnRNP A/B proteins, but the mechanism of this inhibition is not clear (42, 43, 73). Furthermore, recent studies have identified p32 and RSF1 as inhibitors of SR protein function (38, 50). Originally found to copurify with ASF/SF2 (35), p32 has been shown to regulate splicing through direct inhibition of ASF/SF2. RSF1, a Drosophila melanogaster RNA binding protein, was shown to both antagonize ASF/SF2 in vitro and relieve the B52/SRp55 overexpression phenotype in flies. However, despite these advances, much about the regulation of SR proteins remains unknown.
Here we report the identification and characterization of a novel 86-kDa protein (SRrp86) containing a single consensus RRM at its amino terminus and two carboxy-terminal RS domains separated by an unusual glutamic acid-lysine (EK)-rich region. SRrp86 is a nuclear protein that closely resembles members of the SR protein family; however, despite structural similarity, it cannot complement splicing-deficient S100 extracts and can actually inhibit the rescue of splicing by SC35, ASF/SF2, and SRp55. In contrast, SRrp86 activates splicing in the presence of SRp20. Similarly, SRrp86 blocks the activation of alternative splice sites promoted by SC35, ASF/SF2, and SRp55 but does not block splice site selection in the presence of SRp20. When combined with SRp20, SRrp86 either has no effect or can modestly increase splice sites activated by SRp20, depending on the concentration of each protein. Thus, it appears that SRrp86 acts to regulate the activity of distinct SR protein family members. Such regulation is apparently due to direct protein-protein interaction mediated by the two RS domains and an intervening EK-rich region in SRrp86.
MATERIALS AND METHODS
Sequence analysis of SRrp86.
A rat intestinal cDNA expression library (a generous gift from Larry Chan) was screened with radiolabeled RNA probes as previously described (46). A clone containing a partial open reading frame (ORF) was isolated during the initial screen and then used to probe a rat brain cDNA library (Stratagene). One of the clones from this screen contained a full-length ORF. Additional cDNA clones were obtained from screening of rat cDNA libraries and from Genome Systems. The 5′ and 3′ ends were further mapped using rapid amplification of cDNA end protocols from rat and human RNA preparations (18). Sequencing was performed using fmole sequencing kits (Promega), and deduced amino acids and alignments were compiled and compared using Mac-DNASIS.
Protein expression and purification.
Using PCR mutagenesis, a HindIII site was inserted 33 nucleotides upstream of the initiation codon of a rat cDNA clone containing the full-length ORF of SRrp86. This ORF was cloned into pcDNA AMP at the HindIII site. The ΔRS mutant was generated by removing amino acids 204 to 455 using reverse PCR (14). The pcDNA AMP constructs were in vitro translated using the TnT system (Promega). For baculovirus expression, wild-type (WT) SRrp86 was cloned into the StuI site of pAcHLT-B and the ΔRS mutant was cloned into the XhoI site of pAcHLT-C (Pharmigen). ASF/SF2, SC35, SRp55, and SRp75 were each cloned into the EcoRI site of pAcHLT-A, and SRp20 was cloned into the EcoRI site of pAcHLT-C. The resulting plasmid constructs were then cotransfected into Sf9 cells with linearized BaculoGold, and recombinant viruses were generated and amplified. The splicing factor-expressing viruses, as well as a control virus expressing XylE (pAcHLT-XylE; Pharmingen), were used to infect Sf9 or Hi5 cells for recombinant protein expression. For protein purification, insect cells were infected with high-titer virus and incubated at 27°C for 48 to 72 h. Cells were harvested and resuspended in lysis buffer (10 mM Tris [pH 7.5], 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM NaPi, 10 mM NaPPi, 0.5 mM phenylmethylsulfonyl fluoride) on ice for 20 min and then sonicated twice for 30 s (each time). Lysates were cleared by centrifugation at 30,000 × g for 30 min and then incubated with Ni-nitrilotriacetic acid (NTA) resin at 4°C for 1 h. Bound proteins were washed with buffer containing 50 mM NaPO4 (pH 7.5), 300 mM NaCl, 10 mM imidazole, and 10% glycerol and then eluted with buffer containing 50 mM NaPO4 (pH 7.5), 300 mM NaCl, 10% glycerol, and 250 mM imidazole. Purified proteins were dialyzed against buffer E (20 mM Tris [pH 8], 100 mM KCl, 0.2 mM EDTA [pH 8], 0.5 mM dithiothreitol) with 5% glycerol and frozen at −70°C. Purified SR proteins were prepared from calf thymus as previously described (74). Prior to use, aliquots were centrifuged and resuspended in buffer E with 5% glycerol. Protein concentrations were determined using the Pierce BCA Protein Assay.
Antibody production.
Purified SRrp86 was isolated and used to immunize rabbits (East Acres Biologicals). Antibodies were affinity purified over a column of SRrp86 coupled to Affi-Gel 10 (Bio-Rad). Bound antibody was eluted by treatment with 100 mM glycine (pH 2.5), after which the pH was immediately neutralized by the addition of 1 M Tris (pH 8), followed by dialysis against buffer E with 20% glycerol.
Western analysis.
Proteins were separated by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore) in a semidry transfer cell (Bio-Rad). For detection of SRrp86, membranes were blocked for 1 h in buffer containing 5% nonfat dry milk, 10 mM Tris (pH 8), 0.9% NaCl, and 0.1% Tween 20 and then incubated for 1 h with anti-SRrp86 antibodies diluted 1:2,000 in blocking buffer. Membranes were washed with blocking buffer and incubated for 1 h in a 1:2,000 dilution (in blocking buffer) of horseradish peroxidase-conjugated anti-rabbit antibodies (Amersham). Blots were then washed several times in blocking buffer and then in phosphate-buffered saline (PBS) prior to visualization using ECL reagents (Amersham). SR proteins were detected using monoclonal antibodies mAB104 (55), mAB1H4 (44), anti-SC35 (21), and B1C8 (5) and the procedures described above, except that the blocking buffer was changed to 3% bovine serum albumin (BSA) and 10 mM β-glycerophosphate in TBST (50 mM Tris [pH 7.5], 150 mM NaCl, 0.05% Tween 20). Membranes were incubated with monoclonal antibodies for 1 h and then washed with TBST. Secondary horseradish peroxidase-conjugated anti-mouse serum (Amersham) was diluted 1:2,000 in blocking buffer and incubated with membranes for 30 to 40 min and then washed several times in TBST and PBS.
In vitro transcription and splicing assays.
Transcription and splicing of substrate RNAs were carried out as previously described (17, 47, 48). An adenovirus-derived substrate (17) was linearized with BamHI and transcribed with T7 RNA polymerase (Promega). The Cis-parent (15) and SP64 5′D-16X (53) templates were linearized with BamHI and transcribed with SP6 RNA polymerase. Products from reactions of both substrates were resolved on 8% denaturing gels, whereas those from splicing reactions using the adenovirus-derived substrate were resolved on 15% gels.
Both 5′ splice site selection (1.5 h) and 3′ splice site selection (1 h) reactions were spliced in nuclear extract supplemented with indicated recombinant proteins at 30°C. S100 complementation was carried out at 30°C for 1 h with adenovirus-derived substrates in the presence of purified SR proteins or SRrp86 as indicated. Proteins were incubated with RNA prior to addition of either nuclear or S100 extract.
For depletion studies, HeLa cell nuclear extracts were incubated with anti-SRrp86 antibodies on ice for 30 min. Extracts were then transferred to 2/3 volume of protein A-Sepharose beads equilibrated with buffer E containing 20% glycerol and incubated at room temperature with gentle rocking for 30 min. The beads were then pelleted, and the supernatants were tested for splicing ability.
Treatment of nuclear extract with immobilized SRrp86 was carried out by incubation of 1 volume of nuclear extract with 2 volumes of SRrp86 beads or mock beads at room temperature for 40 min. Following brief centrifugation, supernatants were removed and used for splicing of adenovirus-derived pre-mRNAs either alone or supplemented with purified SR proteins as described above. Pelleted beads were washed extensively with buffer E and resuspended in Laemmli loading buffer for subsequent Western analysis of bound proteins.
In vivo splicing.
The HindIII/BamHI fragment of 5′D-16X was cloned into pcDNA AMP to create an in vivo splicing substrate. HeLa cells grown to approximately 70% confluency were transfected using 20 μl of TransIT-LT2 (Pan Vera Corporation) with 1 μg of pcDNA 5′D-16X in conjunction with either 6 μg of control vector (pcDNA), pcDNA SRrp86, or the pcDNA ΔRS mutant. Cells were harvested 24 h after transfection, and total RNA was isolated using TRI REAGENT (Molecular Research Center, Inc.). Reverse transcription (RT)-PCR analysis was done as previously described (79). E1A transfections were similar, except that they contained 500 ng of pMTE1A (77) and 200 ng of pcDNA, pcDNA SRrp86, or pcDNA ASF/SF2. RT-PCR analysis was done as previously described (9). PCR was limited to 20 to 25 cycles, and products were resolved on 8% (5′D-16X) or 5% (E1A) denaturing gels and visualized using a PhosphorImager. Product bands were quantitated using IPLab Gel (Signal Analytics Corporation).
SRrp86 affinity chromatography.
Recombinant SRrp86 and the ΔRS mutant were purified as described above, except that HEPES (pH 7.6) was substituted for Tris in the lysis buffer and dialysis was performed against 0.1 M NaHCO3 and 0.5 M NaCl (binding buffer). For coupling, either an equivalent molar amount of one of the two proteins or buffer containing ethanolamine was incubated with CNBr-activated Sepharose 4B (Pharmacia) with gentle rocking at room temperature for 1 h. The beads were then pelleted and washed with 5 volumes of binding buffer; this was followed by blocking of the remaining active sites by incubation with 1 M ethanolamine (pH 8) for 2 h at room temperature. The coupled beads were washed with alternating cycles of 0.1 M acetate (pH 4) and 0.5 M NaCl or 0.1 M Tris (pH 8) and 0.5 M NaCl, followed by storage at 4°C in PBS containing 0.2% sodium azide. Prior to use, protein-coupled beads were equilibrated in buffer E with 5% glycerol.
Far-Western analysis.
Membrane-bound protein binding assays were carried out as previously described (78), with the following modifications. Briefly, Western blots containing purified SR proteins were blocked for 1 h in buffer containing 100 mM KCl, 20 mM Tris (pH 8), 1 mM dithiothreitol, 0.1 mM EDTA, 3 mM MgCl2, 10% glycerol, 5% BSA, and 0.1% Tween 20. Blots were then incubated in 10 ml of blocking buffer with 45 μl of in vitro-translated, 35S-labeled WT SRrp86 or the ΔRS mutant for 2 h at room temperature. Membranes were then washed four times for 5 to 10 min each in blocking buffer without BSA, dried, and exposed to PhosphorImager plates.
Nucleotide sequence accession number.
The sequence of the full-length ORF described here has been submitted to the GenBank database and assigned accession no. AF234765.
RESULTS
Identification of SRrp86.
In an effort to identify sequence-specific RNA binding proteins, we devised a method to screen cDNA expression libraries with short, radiolabeled RNA probes (46). During one of these screens, we isolated a clone that avidly bound both WT and mutant RNA probes. Despite the lack of obvious binding specificity, a portion of the cDNA was sequenced to determine its identity. From the partial cDNA sequence, the protein contained an amino-terminal RRM and two regions rich in SR dipeptides. Subsequently, full-length cDNA clones and/or overlapping expressed sequence tag sequences were obtained from rat, mouse, and human sources. By Northern blot analysis, a single mRNA species of approximately 4 kb was detected in all of the tissues examined (data not shown). The deduced amino acid sequence predicts a protein of 495 amino acids with a calculated molecular mass of 56.8 kDa and an isoelectric point of 10.90 (Fig. 1A). The single RRM is located between residues 71 and 127 and the two RS domains are located between residues 195 and 260 and residues 345 and 429, separated by a unique EK-rich region whose function is unknown. The EK region contributes extensively to the overall charge of the protein and, together with potential phosphorylation of the serine residues within the RS domain, likely accounts for its anomalous migration at 86 kDa on SDS-PAGE (Fig. 1B).
FIG. 1.
Amino acid sequence of SRrp86. (A) The amino acid sequence of rat SRrp86 is shown with the RRM shaded and the conserved RNP-2 and RNP-1 boxes in italics. The RS domains are in white with a gray background, while a unique region rich in glutamic acid and lysine (EK-rich region) is in white with a black background. The human and rat proteins are 86% identical, with the exception of a 16-amino-acid insertion in the human protein. (B) SRrp86 was in vitro translated (IVT) in the presence of [35S]methionine and subjected to SDS-PAGE. Molecular masses are in kilodaltons.
The full-length sequence exhibits little overall homology to any known protein at either the nucleotide or the amino acid level. The two most closely related proteins, with 25 and 18% identity over the full-length protein, respectively, are drosophila protein SRp54 and its human homolog, an arginine-rich nuclear protein, p54 (12, 31, 79). When comparisons were restricted to the RS domains, the percent identity increased for both SRp54 and p54 (36 and 33%, respectively). Despite relatively limited homology, the primary structure of p86 is similar to that of members of the SR protein family, suggesting that it is a new member of this growing superfamily. On the basis of its sequence and functional properties, we have named this protein SRrp86 for SR-related protein of 86 kDa, using nomenclature suggested by Fu (20).
Histidine-tagged recombinant SRrp86 was expressed in insect (Sf9 or Hi5) cells using the baculovirus system and subsequently purified over Ni-NTA agarose. The recombinant protein migrated on SDS-PAGE with an apparent molecular mass of 86 kDa (see Fig. 5A), which is substantially higher than the predicted molecular mass but consistent with the migration of in vitro-translated protein (Fig. 1B). A deletion mutant lacking the RS domains and the intervening EK-rich region (hereafter referred to as the ΔRS mutant) migrated at its expected size (26 kDa), in agreement with the hypothesis that these domains are primarily responsible for the anomalous migration of full-length SRrp86. Similar disparities between predicted molecular mass and observed migration on SDS-PAGE have been reported for other RS domain-containing proteins, including U1 70K, p54, and SRp75 (12, 52, 76).
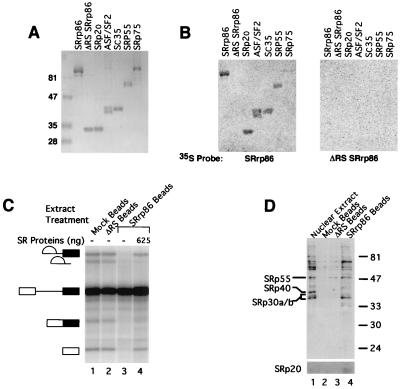
FIG. 5.
SRrp86 interacts with SR proteins. (A) Recombinant proteins were expressed in baculovirus-infected cells, purified by passage over Ni-NTA agarose, subjected to SDS-PAGE, and stained with Coomassie blue. Molecular masses in kilodaltons are shown on the left. (B) Recombinant proteins separated by SDS-PAGE as in panel A were transferred to PVDF membranes, and incubated with equivalent amounts of 35S-labeled SRrp86 or the 35S-labeled ΔRS mutant. (C) CNBr-activated Sepharose was coupled to WT SRrp86 or the ΔRS mutant or treated with ethanolamine (mock beads). Splicing extracts were incubated with the various resins, and flowthrough extracts were used in splicing reactions alone (lanes 1 to 3) or with the addition of purified SR proteins (lane 4). (D) Proteins retained on the resins in panel B were separated by SDS-PAGE, and Western blot analyses were performed with anti-SR antibody mAB1H4. The identities of known SR proteins are indicated on the left, and molecular masses are indicated on the right in kilodaltons. A longer exposure of the bottom portion of the gel shows the retention of SRp20.
Full-length recombinant SRrp86 was used to raise polyclonal antisera in rabbits. Western blot analyses using affinity-purified antibodies showed that anti-SRrp86 antibodies recognize a protein of approximately 86 kDa in HeLa nuclear extract, similar in size to the recombinant protein (Fig. 2A). Interestingly, SRrp86 is not present in either S100 extracts or the purified SR protein preparations. In addition, SRrp86 is not recognized by antibodies against SR proteins, including mAB104 (55), mAB1H4 (44), and anti-SC35 (21) (data not shown). Thus, despite structural similarity, SRrp86 is clearly different from canonical SR protein family members.
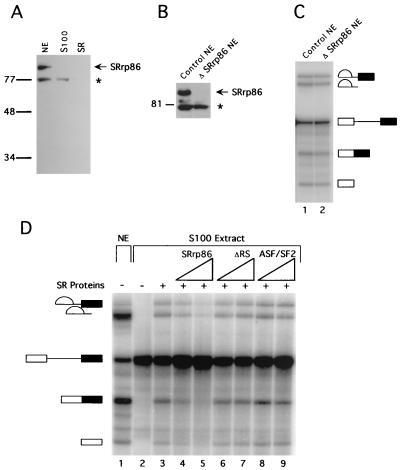
FIG. 2.
SRrp86 inhibits SR proteins but is not required for splicing. (A) Affinity-purified antibodies against SRrp86 were used to probe a Western blot of nuclear extract (NE), S100 extract, and purified SR proteins. The asterisk denotes a 77-kDa protein that cross-reacts with anti-SRrp86 antibodies only under denaturing conditions. (B) Nuclear extracts were immunodepleted of SRrp86 and subjected to Western blot analysis using antibodies against SRrp86. Note that the cross-reactive 77-kDa protein did not coimmunoprecipitate. Molecular masses in kilodaltons are shown to the left of panels A and B. (C) In vitro splicing of an adenovirus-derived substrate was carried out in control extracts or in extracts immunodepleted of SRrp86 (ΔSRrp86 NE). (D) In vitro splicing of the same substrate as in panel C was performed in splicing-deficient S100 extracts in the absence (lane 2) or presence (lane 3) of SR proteins purified from calf thymus (0.5 μg). For comparison, splicing in nuclear extract is shown with the products and intermediates of splicing as indicated. Increasing amounts of SRrp86 (1.5 to 3 μg) inhibited complementation by calf thymus SR proteins (lanes 4 and 5), whereas addition of the ΔRS mutant (1.5 to 3 μg) did not (lanes 6 and 7), nor did the addition of recombinant ASF/SF2 (1.85 to 3.7 μg; lanes 8 and 9).
SRrp86 is not required for pre-mRNA splicing.
To examine whether SRrp86 is required for splicing, nuclear extracts were immunodepleted of SRrp86 and then tested for the ability to splice an adenovirus-derived RNA substrate (Fig. 2C) or an α-tropomyosin (α-TM) substrate (data not shown). In both cases, depletion of SRrp86 did not affect splicing activity (Fig. 2C). Western blot analyses of the depleted extracts showed that SRrp86 had been reduced to undetectable levels, yet splicing was unaffected, suggesting that SRrp86 is not essential for splicing (Fig. 2B).
A well-established activity common to all of the core SR proteins is the ability to complement splicing-deficient cytoplasmic S100 extracts. This activity is observed with preparations containing all of the core SR proteins (74), as well as with individually purified and/or recombinant proteins (11, 34, 76, 79). Since SRrp86 is not found in S100 extracts (Fig. 2A), we sought to determine whether SRrp86 could complement in vitro splicing reactions performed with S100 extracts. In contrast to SR proteins, recombinant SRrp86 was unable to restore splicing (data not shown). However, when SR proteins and SRrp86 were simultaneously added to S100 extracts, the presence of increasing amounts of SRrp86 inhibited the ability of SR proteins to activate splicing activity (Fig. 2D). Deletion of the RS-EK-RS domains (ΔRS) abolished the ability of SRrp86 to inhibit SR activation of splicing (Fig. 2D), suggesting that the inhibition was specific to SRrp86. However, to control for the possibility that excess recombinant protein might nonspecifically block splicing, S100 extracts were supplemented with either recombinant ASF/SF2, which resulted in further activation of splicing (Fig. 2D) or a baculovirus-expressed 40-kDa control protein (XylE; Pharmingen), which had no inhibitory effect (data not shown). Thus, the inhibition of SR protein activity by SRrp86 appears to be specific. In addition, such inhibition does not appear to be substrate specific, as identical results were obtained with a β-globin pre-mRNA (data not shown). Together, the complementation and depletion indicate that SRrp86 is not required for splicing and, instead, raise the possibility that it might actually inhibit the function of canonical SR proteins.
Positive and negative regulation of splicing.
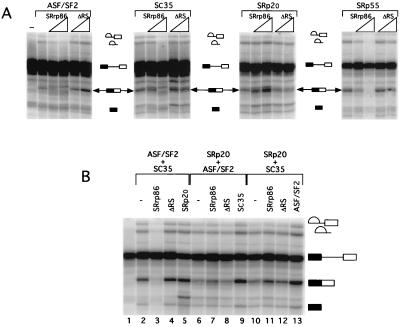
To more rigorously test the hypothesis that SRrp86 might inhibit SR proteins, individual recombinant SR proteins were expressed in Sf9 cells and used in the S100 complementation assay in the presence of either SRrp86 or the ΔRS mutant. In agreement with previous studies, recombinant SR proteins (ASF/SF2, SC35, SRp20, and SRp55) were able to rescue splicing in S100 extracts (35, 56) although the efficiency of complementation was significantly weaker with individual recombinant proteins than with bulk SR proteins purified from calf thymus. Consistent with the results observed in the presence of calf thymus SR proteins, SRrp86 was able to inhibit the rescue of splicing by ASF/SF2, SC35, and SRp55. In contrast, not only did SRrp86 not inhibit the rescue of splicing by SRp20, it actually activated splicing. This suggests that the inhibition of SR protein-dependent splicing by SRrp86 does not represent an overall nonspecific inhibition of splicing but rather a targeted inhibition of specific SR proteins.
Due to the inefficient rescue of splicing with single recombinant SR proteins (Fig. 3A), pairwise combinations of recombinant SR proteins were next used in the S100 complementation assay to improve the efficiency of splicing and to test the activity of SRrp86 (Fig. 3B). Under these conditions, SRrp86 was still able to inhibit the rescue of splicing in the presence of the combination of ASF/SF2 and SC35 (Fig. 3B, compare lanes 2 and 3) or the combination of ASF/SF2 with SRp55 (data not shown). In contrast, when SRp20 was present in combination with either ASF/SF2 or SC35, the addition of SRrp86 did not inhibit the rescue of splicing and, instead, resulted in a modest increase in spliced-product formation (lanes 6 and 7 and lanes 10 and 11, respectively). This increase was also observed in the presence of SRp55 and SRp20 (data not shown). As in Fig. 2, a variety of controls were performed to ensure that the inhibition of splicing was specific. First, the addition of the ΔRS mutant had no effect, consistent with the results shown above. Second, addition of a third SR protein purified from baculovirus-infected cells resulted in increased splicing efficiency, arguing against possible nonspecific inhibition of splicing. Thus, it appears that SRrp86 acts both positively and negatively to regulate splicing.
FIG. 3.
Specificity of SRrp86 for individual SR proteins. (A) Individual recombinant SR proteins were used to complement S100 extract splicing of an adenovirus-derived substrate. SRrp86 (0.75 to 1.5 μg) or an equimolar amount of the ΔRS mutant was added to S100 extracts in the presence of the indicated recombinant SR protein. The first lane in panel A shows splicing in unsupplemented S100 extracts. (B) Rescue of splicing in S100 extracts by pairwise combinations of recombinant SR proteins in the presence of SRrp86 (1.5 μg) or the ΔRS mutant. In each set, a third SR protein (as indicated) was also added to control for possible nonspecific inhibition by excess recombinant protein. The concentrations of SRrp86, ΔRS, and recombinant SR proteins used in this panel were determined in separate titration experiments.
Regulation of splice site selection by SRrp86.
In several cases, it has been demonstrated that both 5′ and 3′ in vitro splice site selection can be altered by varying the concentration of either bulk or specific SR proteins (22–24, 33, 41, 42). Therefore, we sought to determine if SRrp86 might play a role in controlling alternative splice site selection by performing in vitro splicing assays with substrates containing alternative 5′ or 3′ splice sites in the presence of SR proteins and increasing amounts of SRrp86 or the ΔRS mutant. As shown in Fig. 4A, addition of calf thymus-purified SR proteins to splicing reactions containing a β-globin-derived alternative splicing substrate (53) resulted in a dramatic increase in splicing efficiency and a modest increase in proximal splice site usage. However, the addition of increasing quantities of recombinant SRrp86 was able to inhibit the activation of proximal splicing, from 45 to 18% proximal splice site usage. Consistent with the S100 splicing assays, the RS-EK-RS domains were required to mediate this switch. Similarly, with an α-TM-derived substrate containing alternative 3′ splice sites, SR proteins again activated proximal splice site selection (Fig. 4B) and this effect could be counteracted by SRrp86, resulting in a decrease from 44 to 14% proximal splice site selection. As above, the addition of control protein preparations, including the ΔRS mutant, did not result in inhibition of proximal splice site selection (data not shown). Thus, it appears that SRrp86 can antagonize the function of canonical SR proteins in both 5′ and 3′ splice site selection.
FIG. 4.
Splice site selection by SR proteins affected by SRrp86. (A) In vitro splicing of a substrate derived from β-globin containing competing 5′ splice sites (5′D-16X) was performed in the presence of purified calf thymus SR proteins supplemented with the indicated amount of either WT SRrp86 or the ΔRS mutant. (B) Splicing of an α-TM-derived substrate containing competing 3′ splice sites was performed in the presence of purified calf thymus SR proteins supplemented as in panel A. (C) The 5′D-16X (left panel) and α-TM-derived substrates (right panel) were spliced in the presence of recombinant SR (rSR) protein ASF/SF2 (0.75 μg), SC35 (0.33 μg), SRp20 (0.6 μg), or SRp55 (1.1 μg) with or without SRrp86 (1.5 μg). As in Fig. 3B, the amounts of each of the proteins used in panel C were determined in separate titration experiments. The precursor and products of splicing for each substrate are diagrammed to the left of each gel.
To examine whether individual SR proteins would be affected by SRrp86 under alternative splicing conditions, splicing extracts were supplemented with individual recombinant SR proteins and incubated with alternative splicing substrates in the presence or absence of SRrp86 (Fig. 4C). As in Fig. 4A, the addition of either ASF/SF2 or SC35 to a β-globin-derived substrate with competing 5′ splice sites resulted in activation of splicing and an increase in proximal splice site selection (Fig. 4C). This activity could be counteracted by the addition of SRrp86. Similar results were observed upon the addition of SRp55 to the α-TM-derived substrate containing alternative 3′ splice sites (Fig. 4C). However, in this substrate, SRp20 strongly activated proximal 3′ splice site selection and no effect was detected upon the addition of SRrp86. With each of the four SR proteins tested, no changes were observed upon the addition of the ΔRS mutant (data not shown) and simple addition of SRrp86 to either of the substrates in the absence of additional SR proteins did not affect splicing activity. The lack of inhibition observed upon addition of SRrp86 to nuclear extracts, compared to that seen in S100 extracts (Fig. 2 and 3), likely reflects the overall ratio of SR proteins to SRrp86 in nuclear versus cytoplasmic S100 extracts.
SRrp86 interacts with SR proteins.
From the experiments described above, it appears that SRrp86 can alter the activity of SR proteins both positively and negatively. Among several possibilities, one mechanism to account for these differential effects could be direct interaction between SRrp86 and SR proteins. To test this hypothesis, coimmunoprecipitation reactions were performed using anti-SRrp86 antibodies. Potential association could be detected between SRrp86 and SRp55, SRp30a,b, and SRp20, but the interactions were fairly weak (data not shown). Under the conditions used, complete depletion of SRrp86 from nuclear extract was possible whereas quantitative removal of SR family members was not, consistent with the finding that extracts depleted of SRrp86 are active for splicing (Fig. 2C).
As an alternative to immunoprecipitation, far-Western blot analyses were then performed in which recombinant proteins were resolved by SDS-PAGE, transferred to PVDF membranes, and incubated with 35S-labeled SRrp86. As shown in Fig. 5B, SRrp86 associated with itself, SRp20, ASF/SF2, SC35, and SRp55 whereas no association was detected with the ΔRS mutant and only minimal interaction with SRp75 was detected. Despite the signals detected in Fig. 5B, repeat experiments have shown that the interaction between SRrp86 and SC35 is much weaker than the binding of SRrp86 to itself or any of the other SR proteins used in the experiment whose results are shown in Fig. 5B, except SRp75 (data not shown). For all of these interactions, the RS-EK-RS domains appear to play a vital role. Far-Western blots using equivalent levels of 35S-labeled ΔRS mutant protein did not result in the detection of any of the recombinant SR proteins or of WT SRrp86. Thus, it appears that SRrp86 preferentially interacts with a subset of SR proteins, including itself.
To attempt to confirm these interactions, a fourth assay was employed in which large amounts of SRrp86 were covalently coupled to Sepharose beads and incubated with HeLa cell nuclear extract, followed by determination of whether such extracts are active for splicing and whether SR proteins are retained by the SRrp86-coated resin. As shown in Fig. 5C, pretreatment of splicing extracts with SRrp86-coated beads severely inhibited splicing compared to mock-treated extracts or extracts incubated with resin containing the ΔRS mutant protein. However, the addition of purified SR proteins could partially restore splicing, especially the first step. To examine the composition of the retained proteins, antibodies against SR proteins (four different monoclonal antibodies) were used in Western blot analyses to identify whether specific SR proteins were being retained. As shown in Fig. 5D, three major SR proteins that migrate at approximately 30, 55, and 70 kDa were found to interact with SRrp86. Longer exposures also showed that a protein of approximately 20 kDa also interacted with SRrp86, but either the relatively low levels of this protein in nuclear extract or poor immunoreactivity limited detection. Additional minor bands were also detected, but the major conclusion is that SRrp86 interacts with a specific subset of SR proteins, as demonstrated by the change in the stoichiometry of SR proteins between untreated nuclear extracts and extracts exposed to SRrp86-coated resin. We do not know the exact identities of these proteins, but based on migration rates, it seems likely that they include SRp55, SRp20, and one or more of the proteins that comigrate around 30 kDa (ASF/SF2, SC35, and SRp30c). These interactions are apparently mediated by direct protein-protein interaction based on the results of the far-Western data (Fig. 5B) and the fact that pretreatment of the extracts with RNase as shown in Fig. 5D did not alter the pattern of proteins retained by the SRrp86-coupled resin (data not shown).
In vivo regulation of splice site selection by SRrp86.
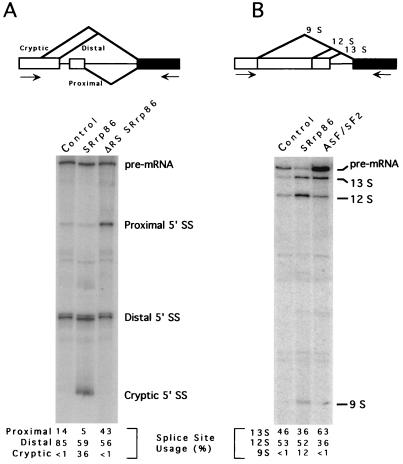
If SRrp86 interacts with a subset of SR proteins, modulating their activity, then overexpression of SRrp86 in cells should be able to alter the splicing patterns of alternatively spliced model substrates due to sequestration and/or activation of specific SR proteins. To test this, we examined the in vivo splicing patterns of two alternatively spliced substrates. The first substrate (Fig. 6A) is the same as that used in Fig. 4A, containing three alternative 5′ splice sites which compete for joining to the same 3′ splice site (53, 79). Two of these sites are derived from duplicated 5′ splice sites, whereas the third is a cryptic splice site that can be activated in vivo. In HeLa cells transfected with this substrate, most (85%) of the mRNA utilized the WT 5′ (distal) splice site with only a small amount of proximal 5′ splice site selection (Fig. 6A). However, in cells transfected with WT SRrp86, the use of the cryptic 5′ splice site was dramatically increased to greater than approximately 36% of the total mRNA. In contrast, transfection of the ΔRS mutant led to a nearly threefold increase in proximal 5′ splice site selection and no cryptic splice site selection was detectable. Similar results were obtained with an adenovirus E1A substrate (Fig. 6B) (79) which contains three alternative 5′ splice sites competing for the same 3′ splice site. Cotransfection of ASF/SF2 with this substrate caused activation of the most proximal 5′ splice site (13S) from 46 to 63%, consistent with previous results (9). In contrast, cotransfection of SRrp86 resulted in the activation of the most distal site (9S) from virtually undetectable levels in the control to approximately 12% of the total spliced products. While the absolute numbers changed from transfection to transfection, the same trend was observed in multiple independent transfections and different RT-PCRs for both substrates. Thus, it appears that overexpression of SRrp86 can alter the patterns of alternative splice site selection in vivo, consistent with the hypothesis that SRrp86 interacts with and modulates the activity of distinct SR protein family members.
FIG. 6.
In vivo regulation of 5′ splice site selection by SRrp86. HeLa cells were transfected with the in vivo splicing construct pcDNA 5′D-16X (A) or adenovirus E1A (B). Cells were cotransfected with either a control vector (pcDNA) or a vector expressing SRrp86, the ΔRS mutant, or ASF/SF2, as indicated. RNA was isolated 48 h after transfection, and RT-PCR was performed to amplify the three possible spliced products, which were quantitated by PhosphorImager analysis, as indicated. The positions of the primers used for RT-PCR are indicated by the arrows.
DISCUSSION
SRrp86 appears to be a unique member of the SR protein superfamily capable of regulating other SR protein family members. To our knowledge, this is the first example of an SR-related protein that regulates splice site selection by inhibiting the effects of some SR proteins while activating others, apparently through direct protein-protein interaction. Thus, in addition to differences in the concentration and/or the activity of SR proteins mediated by posttranslational modification, alternative splicing can also be controlled by regulatory proteins such as SRrp86 that interact with and regulate specific SR protein family members.
Protein characteristics.
Similar to SRp75 and U1 70K, SRrp86 contains two regions rich in serine-arginine dipeptides (52, 76). The region separating the two RS domains in SRrp86 contains 84 amino acids consisting of 60% lysine or glutamic acid residues, often found as EK or KE dipeptides. Combined, the two RS regions and the EK-rich region contribute extensively to the overall charge of SRrp86 and are likely responsible for the anomalous migration of SRrp86 on SDS-PAGE. One of the characteristics common to many SR proteins is the presence of domains that contain alternating positive and negative charges, typically consisting of arginine alternating with either glutamate (E) or aspartate (D) residues (36, 45). SRrp86 contains only a few RE or RD dipeptides, but the unique EK-rich region contains an extraordinarily long stretch of alternating positively and negatively charged residues. Since SRrp86 is not recognized by any of the anti-SR protein phosphoepitope antibodies, the RS domains in SRrp86 are different from those in other SR proteins, even though SRrp86 can be phosphorylated (data not shown). Whether regulated phosphorylation affects the function of SRrp86 remains to be determined, but preliminary evidence suggests that phosphorylation of SRrp86 does not affect its ability to interact with other SR proteins (data not shown).
SR protein regulation and splice site selection.
For splice site selection, antagonistic relationships between the hnRNP A/B proteins and ASF/SF2 and between PTB and U2AF have been well characterized (39, 42, 43, 49, 58, 66). Recently, the small number of proteins that can antagonize SR protein function has increased with the characterization of RSF1 (38) and p32 (50). In addition, individual SR proteins can differentially activate or repress specific splice sites within a variety of pre-mRNA substrates (27, 29, 56, 68, 75). Further, alternative splicing of pre-mRNAs encoding specific SR proteins can result in the production of isoforms with differential activity (29, 80). Thus, the overall levels and activity of SR proteins between tissues and cells can clearly influence splice site selection in a combinatorial manner (27, 75).
The initial observation that SRrp86 is able to inhibit the activity of a subset of SR proteins, both in the rescue of splicing-deficient S100 extracts and in alternative splice site selection, indicates that it may be another member of this group of inhibitory proteins yet unique in its resemblance to canonical SR proteins. Experiments conducted with individual SR proteins suggest that SRrp86 is different from other proteins that antagonize SR proteins in several ways. First, the antagonism is not directed against a single SR protein. Instead, SRrp86 is able to inhibit the activity of at least three SR proteins, including ASF/SF2, SC35, and SRp55. Further studies intended to determine if SRrp86 is able to inhibit the activity of other SR proteins are under way. The second and most striking point is that SRrp86 not only fails to inhibit SRp20 but, instead, causes activation of splicing. This activation is observed in experiments with both recombinant SRp20 alone (Fig. 4A) and in the presence of other SR proteins (Fig. 4B). This differential regulation of SR proteins indicates a potentially important role for SRrp86 in determining the relative activity of SR proteins involved in splicing. Variable SRrp86 concentration differences between cells and tissues could dramatically alter alternative splice site selection.
The results of transient-transfection assays support the notion that increases in SRrp86 levels can alter splice site selection in vivo. In cells containing only endogenous levels of SRrp86, as well as in cells transfected with the ΔRS mutant, little or no splicing to the most distal 5′ splice site of either the β-globin-derived or E1A substrate (the cryptic splice site and 9S, respectively) was observed. In contrast, in cells overexpressing WT SRrp86, 36% (β-globin derived) and 12% (E1A) of the spliced mRNA utilized this site. This is likely to be an underestimate of the change in splice site selection produced by overexpression of SRrp86, since the experiment requires cotransfection of both plasmids, and cells that take up only the vector will produce the pattern seen in the control lane. Regardless, in vivo overexpression of SRrp86 resulted in significant changes in splice site selection, consistent with the hypothesis that SRrp86 may act to regulate SR protein family members.
Two interesting results suggest a possible mechanism by which SRrp86 mediates these effects. First, in the substrates where we observed decreases in proximal splice site selection, corresponding increases in distal splice site selection were never observed. Second, supplementation of extracts with additional SRrp86 inhibited splicing in cytoplasmic S100 extracts but had no effect in nuclear extracts. Both of these results can potentially be explained by postulating that SRrp86 interacts with specific SR proteins, both positively and negatively, and the overall effects of SRrp86 depend on the stoichiometry between these different proteins and SRrp86. For example, SRp20 and ASF/SF2 act antagonistically to determine the splicing patterns of pre-mRNA transcripts encoded by the SRp20 gene itself; autoregulation of SRp20 depends on the ratio of its own concentration to that of ASF/SF2 (29). Using α-TM pre-mRNA substrates, we have also observed an antagonistic relationship between SRp20 and ASF/SF2 but observed that inclusion of SRrp86 further enhanced the pattern of splicing activated by SRp20 (unpublished data).
Protein-protein interaction.
Previous studies have shown that SR proteins are involved in a variety of protein-protein interactions and that the RS domains play a crucial role in these interactions (2, 32, 71, 72). In this paper, we have shown that specific SR proteins interact with SRrp86, dependent on the presence of the two RS domains and the intervening EK-rich region. The far-Western blots demonstrate that SRrp86 has the ability to interact with multiple RS domain-containing proteins with some specificity. In addition, when large amounts of SRrp86 were covalently coupled to Sepharose beads and incubated with splicing extracts, multiple SR proteins were retained. From all of the interaction assays, a common group of SR proteins were detected that interact with SRrp86, including SRp55, ASF/SF2, SC35, and SRp20. Furthermore, the affinity chromatography assay (Fig. 5D) showed a marked change in the concentration of a protein that migrates at 70 kDa and reacts with mAB104. Far-Western blots suggest that this protein is not SRp75. Based on migration, it could be SRp54 (79), but further studies are required to confirm this possibility. Since RNase treatment did not alter the pattern of retention on the affinity chromatography assay or the far-Western blots, it appears that the interaction between these proteins does not require RNA and is due to direct protein-protein interaction. We are currently using yeast two-hybrid screens to test the interaction between SRrp86 and specific SR proteins, as well as to identify other potential cellular targets. It will be interesting to eventually compare the levels of the interacting SR proteins and SRrp86 in various cell types to determine how the concentrations of these factors compare with the splicing patterns of alternatively spliced pre-mRNAs. Correlation of the changes in the ratios of these proteins with specific splicing patterns could provide further support for the idea that the regulation of alternative splicing is due to the relative levels of a variety of more widely expressed splicing factors rather than the existence of tissue- or cell-specific splicing regulators.
ACKNOWLEDGMENTS
We thank Jane Wu, Adrian Krainer, Brent Gravely, Tom Maniatis, Phil Sharp, and Mark Roth for their generous gifts of antibodies, expression constructs, and cDNA clones and Billy Dye for purified calf thymus SR proteins. Thanks also go to Sue Berget, Chris Smith, and Ron Emeson for comments and suggestions.
This work was supported by a grant from the NIH (GM50418). PhosphorImager analysis was made possible by a grant from the NSF (BIR-9419667).
REFERENCES
- 1.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 2.Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by transformer-2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 3.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 4.Blencowe B J, Issner R, Nickerson J A, Sharp P A. A coactivator of pre-mRNA splicing. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boche I, Fanning E. Nucleocytoplasmic recycling of the nuclear localization signal receptor α subunit in vivo is dependent on a nuclear export signal, energy, and RCC1. J Cell Biol. 1997;139:313–325. doi: 10.1083/jcb.139.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burge C B, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by spliceosomes. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 8.Caceres J F, Krainer A R. Mammalian pre-mRNA splicing factors. In: Krainer A R, editor. Eukaryotic mRNA processing. Oxford, England: IRL Press; 1997. pp. 174–212. [Google Scholar]
- 9.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1708. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 10.Cao W, Jamison S F, Garcia-Blanco M A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 11.Cavaloc Y, Popielarz M, Fuchs J-P, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhary N, McMahon C, Blobel G. Primary structure of a human arginine-rich nuclear protein that colocalizes with spliceosome components. Proc Natl Acad Sci USA. 1991;88:8189–8193. doi: 10.1073/pnas.88.18.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colwill K, Pawson T, Andrews B, Prasad J, Manley J L, Bell J C, Duncan P I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intracellular distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 14.Coolidge C J, Patton J G. Run-around PCR: a novel way to create duplications using polymerase chain reaction. BioTechniques. 1995;18:763–764. [PubMed] [Google Scholar]
- 15.Coolidge C J, Seely R J, Patton J G. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–896. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan P I, Stojdl D F, Marius R M, Bell J C. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol Cell Biol. 1997;17:5996–6001. doi: 10.1128/mcb.17.10.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dye B T, Buvoli M, Mayer S A, Lin C-H, Patton J G. Enhancer elements activate the weak 3′ splice site of α-tropomyosin exon 2. RNA. 1998;4:1523–1536. doi: 10.1017/s1355838298980360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frohman M A. RACE: rapid amplification of cDNA ends. In: Innis M A, Gelfand D A, Sninsky J J, White T J, editors. PCR protocols. Boston, Mass: Academic Press; 1990. pp. 28–38. [Google Scholar]
- 19.Fu X-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 20.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 21.Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 22.Fu X-D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:860–866. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallego M E, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge H, Manley J L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 25.Gui J-F, Lane W S, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 26.Gui J F, Tronchere H, Chandler S D, Fu X-D. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci USA. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 28.Heinrichs V, Baker B S. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jumaa H, Nielsen P J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy C F, Kramer A, Berget S M. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branch point. Mol Cell Biol. 1998;18:5425–5434. doi: 10.1128/mcb.18.9.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J. Protein-protein interactions and 5′ splice site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 33.Krainer A R, Conway G C, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 34.Krainer A R, Conway G C, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 35.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 36.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 37.Kuroyanagi N, Onogi H, Wakabayashi T, Hagiwara M. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem Biophys Res Commun. 1998;242:357–364. doi: 10.1006/bbrc.1997.7913. [DOI] [PubMed] [Google Scholar]
- 38.Labourier E, Bourbon H-M, Gallouzi I-E, Fostier M, Allemand E, Tazi J. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev. 1999;13:740–753. doi: 10.1101/gad.13.6.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin C-H, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 40.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 41.Mayeda A, Helfman D M, Krainer A R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 43.Mayeda A, Munroe S H, Caceres J F, Krainer A R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neugebauer K M, Roth M B. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- 45.Neugebauer K M, Stolk J A, Roth M B. A conserved epitope on a subset of SR proteins defines a larger family of pre-mRNA splicing factors. J Cell Biol. 1995;129:899–908. doi: 10.1083/jcb.129.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patton J G, Dye B T, Barnard D C, McAfee J G. Identification of pre-mRNA splicing factors and analysis of RNA-protein interaction. In: Richter J D, editor. Analysis of mRNA formation and function. San Diego, Calif: Academic Press; 1997. pp. 55–78. [Google Scholar]
- 47.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 48.Patton J G, Porro E B, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 49.Perez I, Lin C-H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of α-tropomyosin alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen-Mahrt S K, Estmer C, Ohrmalm C, Matthews D A, Russel W C, Akusjarvi G. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasad J, Colwill K, Pawson T, Manley J L. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Query C C, Bentley R C, Keene J D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 53.Reed R, Maniatis T. A role for exon sequences and splice site proximity in splice site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 54.Rossi F, Labourier E, Forne T, Divita G, Derancourt J, Riou J F, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- 55.Roth M B, Murphy C, Gall J G. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi H, Hoffman B E, Lis J T. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol Cell Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 59.Smith C W J, Patton J G, Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- 60.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanford J R, Bruzik J P. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stark J M, Bazett-Jones D P, Herfort M, Roth M B. SR proteins are sufficient for exon bridging across an intron. Proc Natl Acad Sci USA. 1998;95:2163–2168. doi: 10.1073/pnas.95.5.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tacke R, Chen Y, Manley J L. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tronchere H, Wang J, Fu X-D. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- 66.Valcarcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 67.Wang H-Y, Lin W, Dyck J A, Yeakley J M, Songyang Z, Cantley L C, Fu X-D. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Takagaki Y, Manley J L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y-C, Selvakumar M, Helfman D. Alternative pre-mRNA splicing. In: Krainer A R, editor. Eukaryotic mRNA processing. Vol. 17. New York, N.Y: IRL Press; 1997. pp. 242–279. [Google Scholar]
- 71.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 72.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 73.Yang X, Bani M-R, Lu S-J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 75.Zahler A M, Neugebauer K M, Lane W S, Roth M. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 76.Zahler A M, Neugebauer K M, Stolk J A, Roth M B. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zerler B, Moran B, Maruyama K, Moomaw J, Grodzicker T, Ruley E H. Adenovirus E1A coding sequences that enable ras and pmt oncogenes to transform cultured primary cells. Mol Cell Biol. 1986;6:887–899. doi: 10.1128/mcb.6.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang M, Zamore P D, Carmo-Fonseca M, Lamond A I, Green M R. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang W-J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zuo P, Manley J L. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zuo P, Manley J L. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc Natl Acad Sci USA. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]