Abstract
Background
Pseudomonas aeruginosa is an important nosocomial infectious bacterium, more and more multidrug resistant P. aeruginosa have been isolated and posed severe challenges to clinical antibiotic treatment, bringing additional morbidity, mortality, and economic burden. Bacteriophages can lyse bacteria specificity and are feasible alternatives to antibiotics.
Methods
A Pseudomonas aeruginosa-infecting phage vB_PaeP_PA01EW was isolated. Phage plaque assays, transmission electron microscopy, host-range determination, infection assay analyses, whole-genome sequencing and annotation were performed for the phage. Mice pneumonia model using liquid aerosol-exposure Pseudomonas aeruginosa was established, and phage therapy was evaluated.
Results
vB_PaeP_PA01EW belongs to the family Podoviridae according to transmission electron microscopy and was identified as a Luz24likevirus according to the genome analysis. For the phage therapy, compared with the bacteria-infected group, the phage-rescue group has some characteristics. First, adventitial edema and diffuse infiltration of inflammatory cells in tissues were alleviated, Second, bronchial epithelial cell proliferation was reduced. Third, the bacterial burden was significantly decreased.
Conclusion
This study provided data support and theoretical basis for the clinical application of bacteriophages. It has important guiding significance and reference value for the application of bacteriophage therapy of other pathogenic bacteria.
Keywords: P. aeruginosa, bacteriophage, vB_PaeP_PA01EW, aerosol-exposure
Background
Bacterial resistance is a growing concern in the nosocomial environment in which Pseudomonas aeruginosa plays an important role due to their opportunism and metal-β-lactamase production.1 This bacterium has been considered an important emerging multidrug resistant (MDR) pathogen over the past two decades due to its frequently causing healthcare-associated infections, such as pneumonia, meningitis, urinary tract infections, surgical site infections and sepsis.2,3 Bloodstream infections with MDR P. aeruginosa have been associated with high mortality rates.4,5 P. aeruginosa is isolated as human clinical specimens from respiratory, urinary, blood, or gastrointestinal tract.6,7 It is a significant nosocomial pathogen and a common cause of iatrogenic bacteremia.8
The emergence of broad-spectrum antimicrobial-resistant P. aeruginosa isolates caused that no therapeutic option was available. Phage therapy is of great significance with the continuous emergence of drug-resistant pathogen.9 Bacteriophage treatment is particularly desirable because of the side effects and inefficacy associated with antibiotics and the emergence of new antibiotic-resistant strains.10 Antibacterial agents against the antibiotic resistance strains have been discovered.11,12 Conventional phage therapy has shown hopeful results in human clinical cases.13,14 In animal models and in vitro studies, phages are used as therapeutics.15,16 Previous studies of phage therapy for P. aeruginosa infection have been reported. Jault et al indicated that PP1131 (a cocktail of 12 phages) decreased bacterial burden in burn wounds infected by P aeruginosa.17 Fong et al demonstrated that CT-PA (a cocktail of 4 phages) significantly reduced biofilms formed in vitro by a range of P. aeruginosa isolates.18 Khairnar et al indicated that phage MBL effectively cured ulcerative lesions caused by MDR P. aeruginosa infection in Clarias gariepinus.19 Jeon et al demonstrated that two novel phages, Bφ-R656 and Bφ-R1836, improved survival in Galleria mellonella and mouse infected with extensively drug-resistant P. aeruginosa.20 Yang et al demonstrated that inhaled phage KPP10 exerted a significant protective effect against pneumonia caused by P. aeruginosa D4.21 Morello et al demonstrated that two different bacteriophages (PAK-P3 and P3-CHA) administered intranasally are effective in treating lung infections by two different P. aeruginosa strains.22 Forti et al demonstrated a broad-range bacteriophage cocktail (PYO2, DEV, E215, E217, PAK_P1 and PAK_P4) can reduce P. aeruginosa biofilms and treat acute infections in two animal models (Galleria larva and mice).23 Abd El-Aziz et al demonstrated intranasal administration of a single dose of phage MMI-Ps1 immediately after infection with P. aeruginosa provided a significant level of protection and increased the survival duration.24
The ideal therapeutic option for ventilator-associated pneumonia caused by carbapenem-resistant P. aeruginosa isolates was not defined.25 Respiratory tract infection in the form of aerosol was one of the most common ways of infection.26 There was little research evidence to determine if aerosol spread of infectious P. aeruginosa was possible.27 In this study, mouse models with pulmonary infection due to exposure to liquid aerosol of P. aeruginosa were established and phage therapy effects were assessed. Aerosol delivery has advantages: it delivers medication directly to where it is needed and it avoids the first-pass effect with minimum reduction of bioavailability.28,29 The inhaled route is increasingly developed to deliver locally acting or systemic therapies, and rodent models are used to assess tolerance before clinical studies.30
Given the severity of P. aeruginosa infection and the urgent need for better treatment options for multidrug resistant bacteria, alternative treatments for these infections are being sought to help solve this problem.31 In this study, we presented mouse models of lung infection with MDR P. aeruginosa, and the newly isolated lytic phage, vB_PaeP_PA01EW, was used to treat lung infection. Our results demonstrated that bacteriophage could be a promising treatment for lung infection caused by carbapenem-resistant P. aeruginosa in mice.
Methods
Bacterial Strain and Phage
Bacteriophage vB_PaeP_PA01EW (GenBank accession number MG589386) was isolated from a sewage wastewater sample from the Rocket Army General Hospital, Beijing, China, using a double agar overlay plaque assay, as described previously for the isolation of lytic phage.32–35
Electron Microscopy Examination
Phage preparations were stained with 2% potassium phosphotungstate (pH 7.0), and then examined using a Tecnai Spirit 120-kV transmission electron microscope (FEI Company, USA) at different magnitudes to determine the phage morphologies.
Characterization of vB_PaeP_PA01EW
The in vitro infection assay of phage vB_PaeP_PA01EW against P. aeruginosa was determined by optical densitometry (OD600). Briefly, the phages were added to bacterial cultures at different MOIs, SM buffer was added to the bacterial culture as a control. The mixture was incubated at 37°C for 6 h. The OD600 values were measured at 20 min intervals.36 Temperature stability of vB_PaeP_PA01EW was tested at 4 °C, 25 °C, 37 °C, 50 °C, 60 °C, 70 °C and 80 °C (pH 7.0) for 120 min. The phage titers were tested by the double-layer agar method.
Genomic DNA Purification and Sequencing
Phage DNA was extracted using a phenol-chloroform extraction method, as previously described.37 Genomic sequencing was performed by the CNRS sequencing facility in Gif sur Yvette (IMAGIF) using the Illumina platform (Illumina Genome Analyzer IIx). Assembly of short sequence reads was performed using BioNumerics tools (Applied Maths, Sint-Martens-Latem, Belgium) as described.38 The phage genome was annotated automatically using the BaSyS annotation tools.39 Bacterial genome contigs were annotated using BioNumerics annotation tools.38 Detailed methods are available on the website http://bacteriophages.igmors.u-psud.fr.
Mice and Intratracheal Delivery Device
Eight-week-old BALB/c mice (female) were purchased from Beijing Vital River Laboratory Animal Technology Company. Mice were maintained in ventilated cages in a specific pathogen-free room. An intratracheal delivery device (HRH-MAG4 microsprayer) was purchased from Beijing Huironghe Technology Co., Ltd. The microsprayer generates a fine plume of liquid aerosol throughout the lungs.
Mice Pneumonia Model Establishment
The BALB/c mice were randomly assigned to two groups and were anesthetized by intraperitoneal injection with 1% pentobarbital (50 mg/kg), and then inoculated by trans-oral insertion of a HRH-MAG4 rigid microsprayer into an endotracheal tube inserted into the trachea of the anesthetized mouse. An inoculum of 106 bacterial CFU was determined to be optimal and was used in the following experiments. To establish the mouse models of lung infection with MDR P. aeruginosa, 50 μL P. aeruginosa strain PA01 solution containing 0.05% poloxamer 188 was injected through the trachea and 50 μL physiological saline containing 0.05% poloxamer 188 was injected through the trachea 1 hpi (group B, n = 8). As the control group, 50 μL physiological saline containing 0.05% poloxamer 188 was injected through the trachea using an HRH-MAG4 microsprayer, 1 hour later, 50 μL physiological saline containing 0.05% poloxamer 188 was injected via the trachea (group C, n = 6).
Phage Treatment Against P. aeruginosa Infection in the Mouse Model
To evaluate the therapeutic effect of bacteriophage on lung infections induced by P. aeruginosa, the mice were randomly assigned to four groups: group A, control group, 50 μL physiological saline containing 0.05% poloxamer 188 was injected through the trachea using a HRH-MAG4 microsprayer, 1 hour later, 50 μL physiological saline containing 0.05% poloxamer 188 was injected via the trachea (n = 6); group B, phage group, 50 μL physiological saline containing 0.05% poloxamer 188 was injected through the trachea, 1 hour later, 50 μL phage solution (109 PFU) containing 0.05% poloxamer 188 was injected through the trachea (n = 6); group C, bacteria-infected group, 50 μL P. aeruginosa strain PA01 solution (106 CFU) containing 0.05% poloxamer 188 was injected through the trachea and 50 μL physiological saline containing 0.05% poloxamer 188 was injected through the trachea 1 hpi (n = 7); group D, phage-rescue group, 50 μL P. aeruginosa strain PA01 solution (106 CFU) containing 0.05% poloxamer 188 was injected through the trachea and 50 μL phage solution (109 PFU) containing 0.05% poloxamer 188 was injected through the trachea 1 hpi (n = 8).
Histopathology and Bacterial Burden of Lung Tissues
Mice were sacrificed (at 12 hpi, 24 hpi, 36 hpi and 48 hpi, respectively) by intraperitoneal injection with pentobarbital according to the guideline for euthanasia of the Ethics Committee of the Academy of Military Medical Sciences (SCXK-2007-004). The lung tissues were fixed in 4% (v/v) phosphate-buffered paraformaldehyde overnight. After fixation, the tissues were embedded in paraffin, sectioned, and stained with hematoxylin and eosin (HE) for the examination of histological changes by light microscopy (Microscope: NIKON. Eclipse. Ci, imaging system: NIKON.digital.sight.DS-FI2, MADE.IN.JAPAN, magnification: 100×, 200×). The remaining lung homogenate was diluted and plated onto Luria-Bertani agar to determine the colony formation unit.
Statistical Analysis
Statistical analysis of the results was performed by GraphPad Prism version 5 (GraphPad Software Inc., California, USA). Comparison of clinical parameters among control group, phage group, bacteria-infected group and phage-rescue group was calculated by non-parametric Mann–Whitney test (for continuous variables) or Pearson χ2 test on cross table (for categorical variables). For multiple groups, comparisons were made by two-way ANOVA or non-parametric Kruskal–Wallis test followed by multiple comparisons. P < 0.05 was considered to indicate a significant difference.
Ethics Statement
All protocols were approved by the Ethics Committee of the Academy of Military Medical Sciences (SCXK-2007-004), Beijing, China, and all experimental procedures were conducted per the European Community guidelines (Directive 2010/63/EU) and the International Council for Laboratory Animal Science (ICLAS).
Results
vB_PaeP_PA01EW Virulence Spectrum
The P. aeruginosa strain PA01 strain was used as an indicator for phage screening of sewage wastewater sample and a P. aeruginosa phage named vB_PaeP_PA01EW was isolated. A total of 10 P. aeruginosa strains and other species were tested for their susceptibility to the phage. vB_PaeP_PA01EW was responsible for complete lysis of 7 P. aeruginosa strains. For the other strains, no significant signs of lysis were detected (Table 1). Luria–Bertani (LB) broth or LB agar was used to culture the bacterium.
Table 1.
Host Range Infection of the Phage vB_PaeP_PA01EW
| Species | ID | Infection |
|---|---|---|
| P. aeruginosa | PA01 | + |
| P. aeruginosa | 28097 | + |
| P. aeruginosa | 4013 | + |
| P. aeruginosa | 4018 | + |
| P. aeruginosa | 10052 | – |
| P. aeruginosa | 27144 | – |
| P. aeruginosa | 27056 | + |
| P. aeruginosa | 15018 | + |
| P. aeruginosa | P104 | + |
| P. aeruginosa | 10224 | – |
| Stenotrophomonas maltophilia | 27056 | – |
| Staphylococcus aureus | 15058 | – |
| Stenotrophomonas maltophilia | 16107 | – |
| Klebsiella pneumoniae | 12056 | – |
| Klebsiella pneumoniae | 16019 | – |
| Escherichia coli | DH10B | – |
| Staphylococcus aureus | MU50 | – |
| Enterobacter cloacae | T5282 | – |
| Enterobacter aerogenes | 3-SP | – |
| Acinetobacter baumannii | N1 | – |
| Klebsiella pneumoniae | ATCC BAA-2146 | – |
| Shigella Soong | #1083 | – |
| Pseudomonas putida | P60 | – |
| Serratia marcescens | wk2050 | – |
| Vibrio parahaemolyticus | J5421 | – |
| Flavobacterium indoleans | SJJ19 | – |
| Streptococcus lactis | 5621 | – |
Notes: –Absent; +Present.
Morphology of Phage Particles
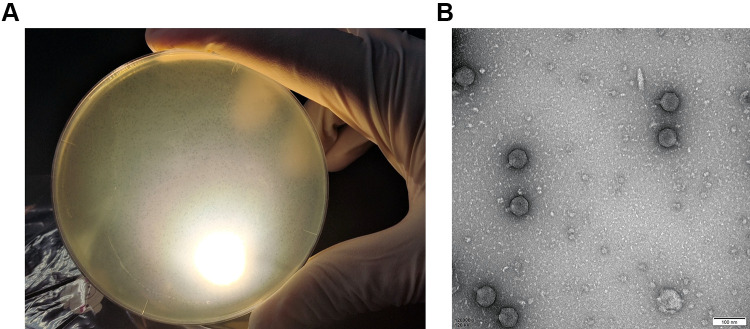
When cultured with P. aeruginosa PA01, vB_PaeP_PA01EW formed small, clear, and uniform plaques (Figure 1A). And then vB_PaeP_PA01EW was characterized under transmission electron microscopy, as shown in Figure 1B. vB_PaeP_PA01EW has an icosahedral head of approximately 45 nm diameter and a clearly visible short tail with no tail fibers being visible. According to the system of Ackermann classification,40 vB_PaeP_PA01EW is a member of the family Podoviridae (order, Caudovirales).
Figure 1.
(A) Plaques of phage vB_PaeP_PA01EW on P. aeruginosa PA01. (B) Morphology of phage vB_PaeP_PA01EW.
Characterization of Phage vB_PaeP_PA01EW
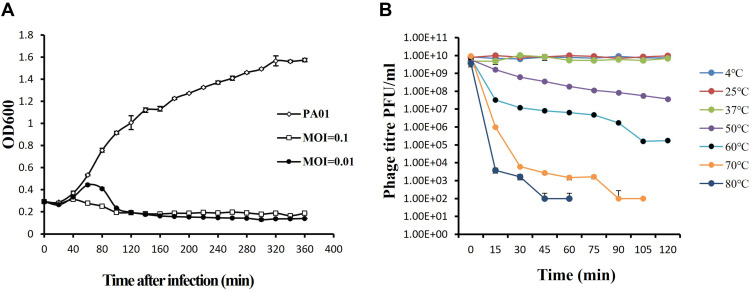
To assess the ability of vB_PaeP_PA01EW to lyse P. aeruginosa in vitro, we monitored the growth of P. aeruginosa PA01 in vB_PaeP_PA01EW’s presence. The infection assays of phage vB_PaeP_PA01EW against PAO1 was performed at 37°C. Our results showed that at an MOI of 0.01 and 0.1, phage vB_PaeP_PA01EW could effectively inhibit the growth of PAO1, keeping the optical density at 600 nm (OD600) at approximately 0.2 within 6 h (Figure 2A). We could conclude that phage vB_PaeP_PA01EW was able to effectively lyse P. aeruginosa PA01 in vitro. Temperature stability was an important influence factor of the therapeutic application of the phage in vivo. In this study, phage vB_PaeP_PA01EW could maintain good stability and high titer at 4 °C, 25 °C and 37 °C (conditions in the lungs in vivo) (Figure 2B).
Figure 2.
(A) Infection assay of phage vB_PaeP_PA01EW against P. aeruginosa in vitro. Each data point is a mean of three experiments. (B) Stability of phage vB_PaeP_PA01EW under different temperatures. Each assay was performed as three repetitions and the values represented are the means.
Open Reading Frames (ORFs) and Comparative Genomics
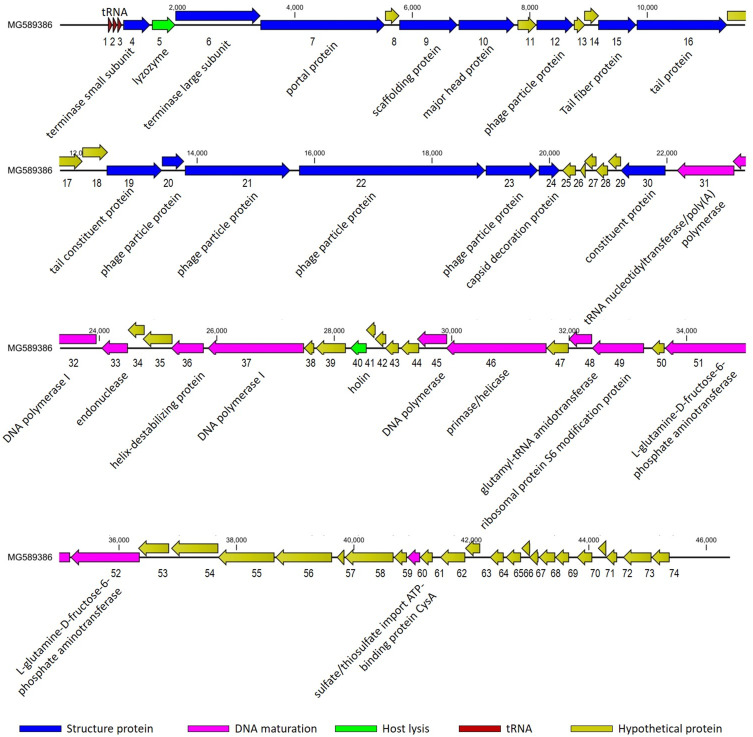
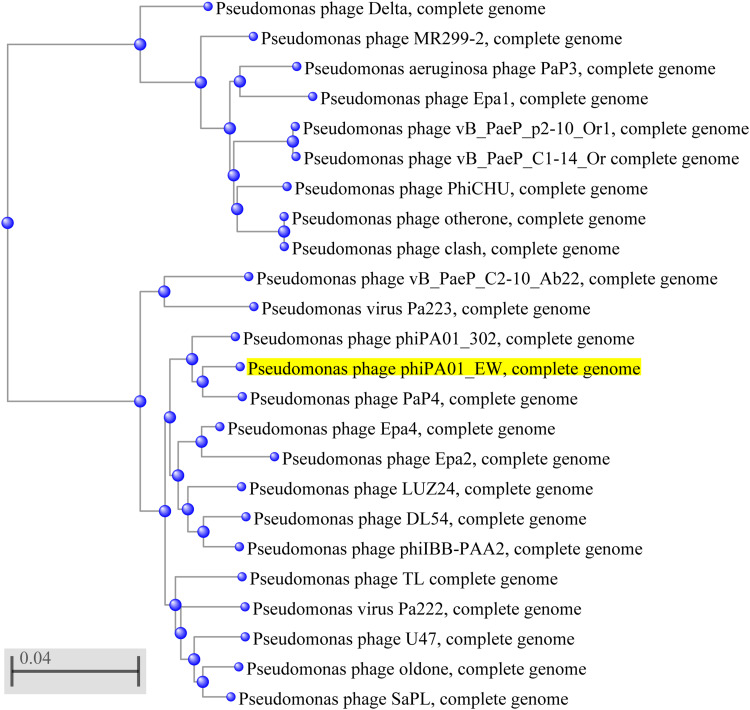
Analysis of the genomes of phages is essential to their safe use as alternative biocontrol agents.41 vB_PaeP_PA01EW has a double-stranded DNA genome consisting of 46,403 bp and 74 predicted ORFs, including three tRNAs genes, tRNA-Ile, tRNA-Asp and tRNA-Pro. However, functional predictions could only be made for 27 ORFs (38%) based on gene prediction and annotation (Figure 3). Coding sequences for phage structure and replication were identified in the genome of vB_PaeP_PA01EW (Figure 4). The shortest ORF encodes a putative protein of 31 amino acid residues (orf26), the longest encodes a putative protein of 1055 residues (orf22). At the nucleic acid level, vB_PaeP_PA01EW has a significant similarity to Pseudomonas phage PaP4 with an identity of 97.38% (94% coverage) and to Pseudomonas virus LUZ24 with an identity of 97.21% (91% coverage). Phylogenetic analysis revealed that the vB_PaeP_PA01EW was clustered in the same clade with Luz24 and was genetically like other Luz24likevirus (Figures 4 and 5). Most protein sequences showed high identity to proteins from the Luz24likevirus genus. vB_PaeP_PA01EW should be classified to Luz24likevirus according to the International Committee of Virus Taxonomy (ICTV) classification scheme based on morphology, biological characteristics, and genome organization (http://www.ictvonline.org/virusTaxonomy.asp). Twelve genes (ORFs 31–33, 36–37, 45–46, 48-49, 51–52, 60) involved in nucleotide biosynthesis and viral replication process were found in the vB_PaeP_PA01EW genome, named early genes. ORF31 encodes tRNA nucleotidyltransferase/poly(A) polymerase sharing 99.05% identity (96% coverage) with Pseudomonas phage U47. ORF32 encodes 5ʹ-3ʹ-exonuclease sharing 98.91% identity with phage Luz24. Fourteen genes (ORFs4, 6, 7, 9, 10, 15–16, 19–24, 30) involved in virion structure and assembly were also found in the vB_PaeP_PA01EW genome. ORF4 and ORF6 encode the small and large terminase subunits, which preform the translocation of viral genomic DNA into the capsid during the packaging process by ATP hydrolysis.41 ORF7 encodes the portal protein; ORF10 encodes the major head protein; and ORF9 encodes a scaffolding protein, which is a chaperone possibly related to viral particle assembly. ORFs 12, 19–24 encode particle/structural proteins; ORF15 encodes the tail fiber protein, which derived from the tail fibers of bacteriophages recognizing specific bacterial surface receptors. In the vB_PaeP_PA01EW genome, ORF5 encodes a lysozyme that is used in the process of host cell breakage through the lysis of the peptidoglycan layer,42 this protein shares 97.73% with Luz24. ORF40 encodes a holin with three transmembrane domains like those from the phage LUZ24, the holin protein share 97.83% identity with phage Luz24. The annotated vB_PaeP_PA01EW phage sequence has been deposited at NCBI GenBank under accession number MG589386. There are still many genes whose function is unknown or unannotated. The elucidation of the function of unknown phage proteins is one of the main challenges of phage molecular biology in the future.43
Figure 3.
Genomic map of vB_PaeP_PA01EW. The genome map was performed using the CLC Main Workbench, version 6.1.1 (CLC bio, Aarhus, Denmark). The genome is schematically presented with predicted ORFs indicated by arrows; the direction of the arrow represents the direction of transcription. Blue arrows, phage structural proteins; purple arrows, DNA regulation module; red arrows, tRNA; green arrows, host lysis, yellow arrows, hypothetical proteins.
Figure 4.
Phylogenetic tree of selected bacteriophages. The phylogenetic tree was constructed by the Neighbor-Joining method. Reference sequences used in the analysis were obtained from the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The highlighted mark indicates the phage vB_PaeP_PA01EW.
Figure 5.
Multiple genome alignment generated by Mauve software (http://asap.ahabs.wisc.edu/mauve/). Genome similarity is represented by the height of the bars, which correspond to the average level of conservation in that region of the genome sequence. Completely white regions represent fragments that were not aligned or contained sequence elements specific to a particular genome.
Histological Changes and Bacterial Burden
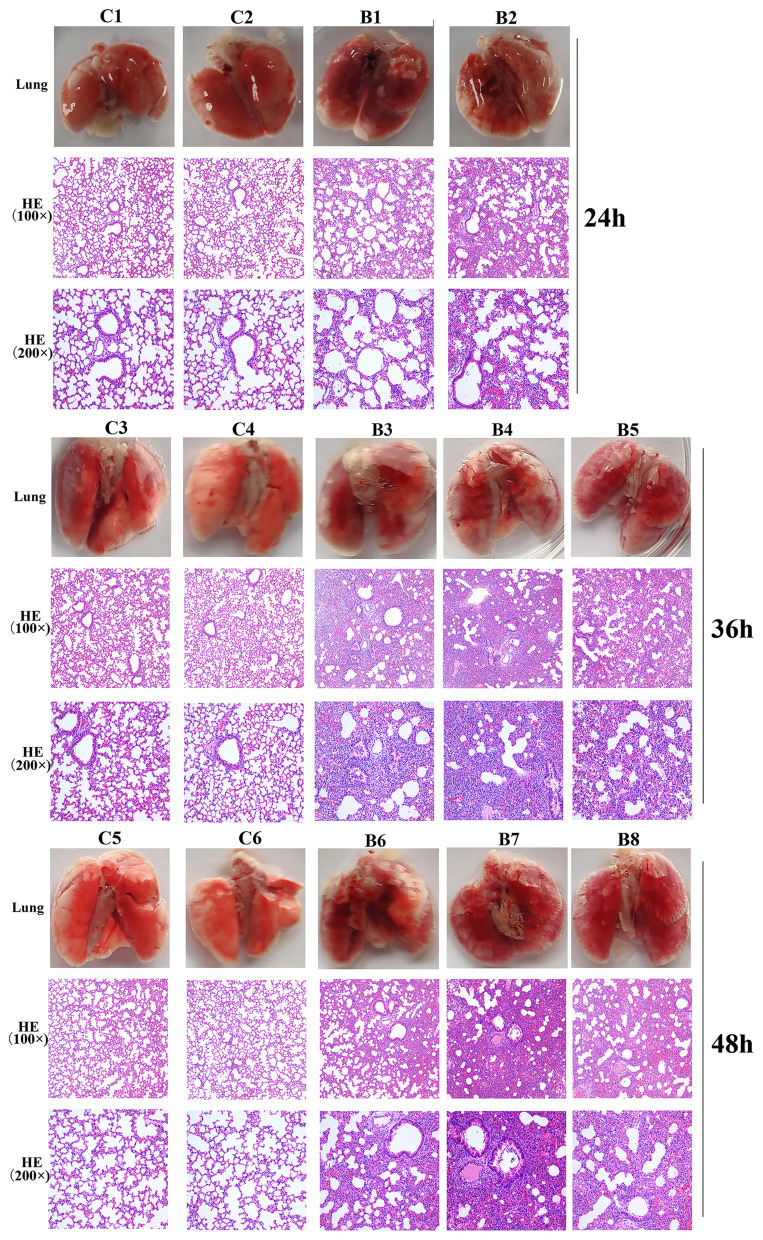
Lung tissue from bacteria-infected group showed severe thickening and congestion of the alveolar walls and marked inflammatory cell infiltration in the perivascular and peribronchial areas compared with the control group at 24 hpi (Figure 6). After 36 hours, local alveolar atrophy and disappearance were observed, and some bronchial epithelial cells proliferated and the wall thickened, accompanied by inflammatory cell infiltration compared with the control group. After 48 hours, the lung consolidation worsened, and red blood cells overflowed in some alveoli, inflammatory cell infiltration in lung tissue and bronchial lumen aggravated compared with the control group. This prompted that mice pneumonia model was successfully established using MDR P. aeruginosa PA01.
Figure 6.
Photos and HE staining of lung tissue of the control group (C) and bacteria infected group (B) at 24 hpi, 36 hpi and 48hpi.
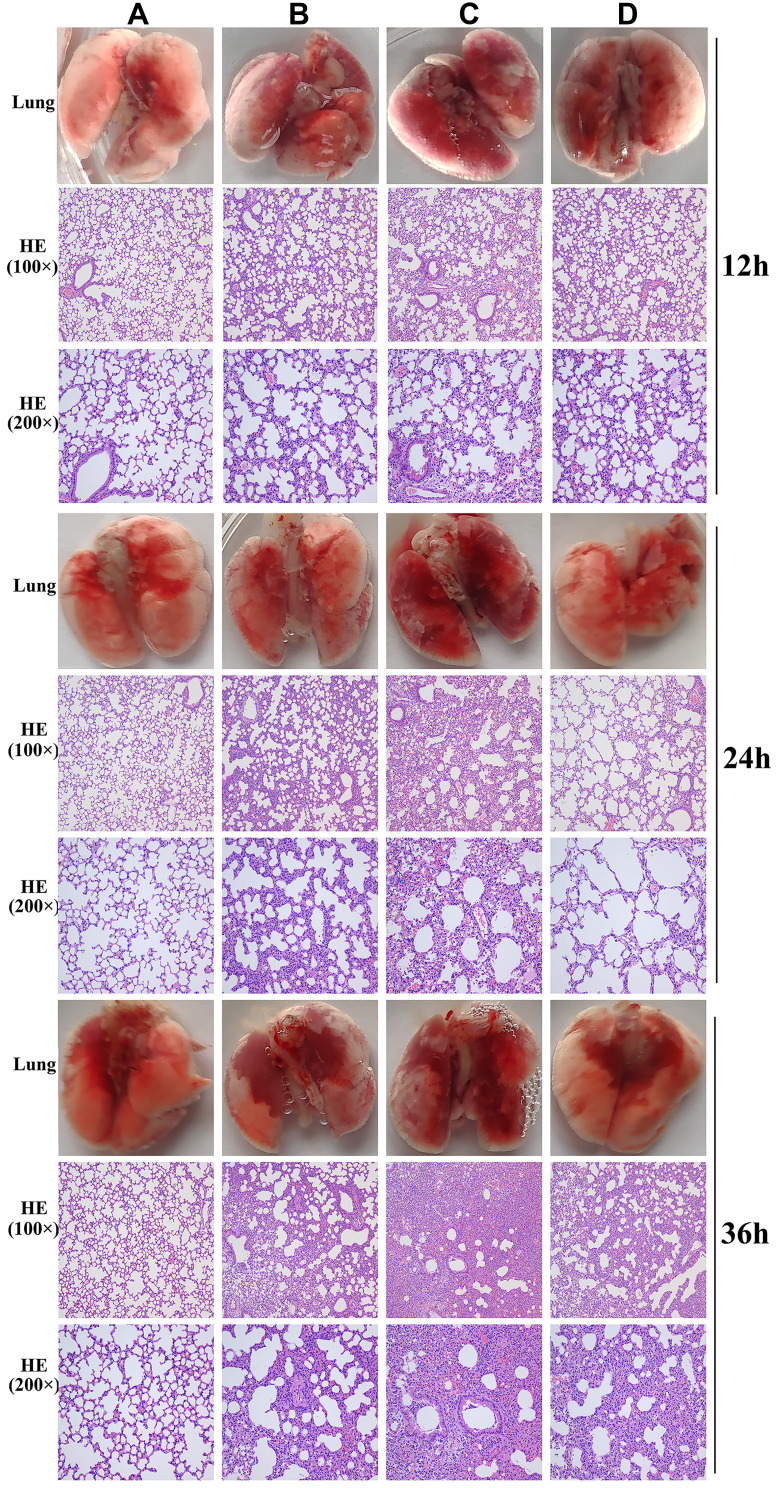
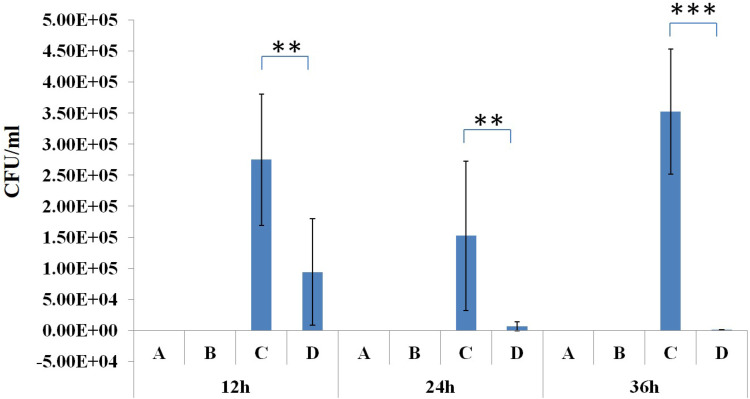
In the phage-rescue group, it was alleviated in both adventitial edema and diffuse infiltration of inflammatory cells in tissues compared with the bacteria-infected group, and bronchial epithelial cell proliferation was also reduced compared with the bacteria-infected group. However, it can still be seen that local alveolar epithelial cells proliferate, alveolar wall thickens, infiltration of inflammatory cells in some alveolar walls, as well as distribution of inflammatory cells in some bronchi (Figure 7). Furthermore, mouse lung homogenate was cultured, and colony formation units were detected. In the phage-rescue group, the number of bacteria was significantly reduced compared with that in the bacteria-infected group (P < 0.01, 12h; P < 0.01, 24h; P < 0.001, 36h) (Figure 8), indicating that the phage vB_PaeP_PA01EW can effectively kill P. aeruginosa PA01 in vivo.
Figure 7.
Photos and HE staining of lung tissue of the control group (A), phage group (B), bacteria-infected group (C) and phage rescued group (D) at 12 hpi; 24 hpi and 36 hpi.
Figure 8.
Bacterial load in the lung tissue of the control group (A), phage group (B), bacteria-infected group (C) and phage-rescue group (D) at 12 hpi; 24 hpi and 36 hpi. **P<0.01; ***P<0.001.
Discussion
P. aeruginosa is an indole-negative motile, non-spore forming, Gram-negative bacteria and classified as Pseudomonadaceae, Proteobacteria.6 This bacterium has taken on clinical significance as opportunistic bacteria and have emerged as nosocomial pathogens from intensive care patients.44,45 In order to successfully apply phage as an alternative to traditional antibiotic therapy, the discovery of novel phage, complete genome analysis, and careful evaluation of its therapeutic potential in vivo are essential before clinical application.14,26 In preliminary clinical trials, no adverse reactions were observed in the treatment of pulmonary infections by bacteriophage nebulization.46,47 In this study, we isolated a bacteriophage (vB_PaeP_PA01EW) which could improve the clinical syndrome of pneumonia caused by MDR P. aeruginosa in vivo. In addition, our study also implied the safety of phage vB_PaeP_PA01EW applied by liquid aerosol-exposure in mice. No obvious alterations were observed in lung tissue of the phage group compared to the control group, which provided a further safety evidence for the phage therapy by intratracheal aerosol delivery route and supported the feasibility of phage therapy in respiratory infection.
The in vitro study showed that vB_PaeP_PA01EW could effectively lyse P. aeruginosa with a large surge of release after a short incubation time. In addition, the phage vB_PaeP_PA01EW possessed good stability within physiological temperature ranges. Host range infection test suggested that phage PA01EW was highly specific and can only infect a single species. The bacterial burden was significantly decreased in the phage-rescue group compared with the bacteria-infected group, manifesting that the phage could effectively kill P. aeruginosa in vivo. However, in the phage-rescue group, there was still slight damage in the lungs of the mice, indicating that single phage treatment was difficult to completely cure pneumonia, but can be used as an auxiliary method, phage combined with antibiotics may achieve more significant therapeutic effects clinically. The previous report on the positive interaction between phage and antibiotic therapy for controlling PA01 infection in a cystic fibrosis zebrafish model have been provided,48 indicating that this combination is a useful treatment method reducing the doses and administration time of antibiotics. In this study, although the in vivo phage efficacy was promising, this experiment cannot fully resemble the real clinical situation, for such short time intervals are impossible in clinical practice. Thus, the ultimate applicability of phage vB_PaeP_PA01EW against P. aeruginosa infection still needs to be supported by clinical trials.
Funding Statement
This work received support from Beijing Municipal Natural Science Foundation Project (No. 5202027) and National Science and Technology Major Project (No. 2018ZX10712001-12-2).
Abbreviations
hpi, hours post-infection; MDR, multidrug resistant; ORF, open reading frame; MOI, multiplicity of infection; ICTV, International Committee of Virus Taxonomy.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author (Xiangna Zhao) on reasonable request.
Ethics Approval and Consent to Participate
All protocols were approved by the Ethics Committee of the Academy of Military Medical Sciences (SCXK-2007-004), Beijing, China, and all experimental procedures were conducted per the European Community guidelines (Directive 2010/63/EU).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Cai S, Chen Y, Song D, Kong J, Wu Y, Lu H. Study on the resistance mechanism via outer membrane protein OprD2 and metal beta-lactamase expression in the cell wall of Pseudomonas aeruginosa. Exp Ther Med. 2016;12(5):2869–2872. doi: 10.3892/etm.2016.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reza A, Sutton JM, Rahman KM. Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in gram-negative (ESKAPEE) bacteria. Antibiotics. 2019;8(4):229. doi: 10.3390/antibiotics8040229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crivaro V, Di Popolo A, Fau - Caprio A, et al. Pseudomonas aeruginosa in a neonatal intensive care unit: molecular epidemiology and infection control measures. BMC Infect Dis. 2009;9(1):1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aghamollaei H, Moghaddam MM, Kooshki H, Heiat M, Mirnejad R, Barzi NS. Detection of Pseudomonas aeruginosa by a triplex polymerase chain reaction assay based on lasI/R and gyrB genes. J Infect Public Health. 2015;8(4):314–322. doi: 10.1016/j.jiph.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 5.Ruiz-Azcona L, Santibanez M, Gimeno A, et al. Etiology of bloodstream infections at a population level during 2013-2017 in the Autonomous Community of Valencia, Spain. Revista espanola de quimioterapia. 2020;33(3):200–206. doi: 10.37201/req/024.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng YL, Lee HC, Yeung CY, Chan WT. Clinical significance in previously healthy children of Pseudomonas aeruginosa in the stool. Pediatr Neonatol. 2009;50(1):13–17. doi: 10.1016/S1875-9572(09)60024-3 [DOI] [PubMed] [Google Scholar]
- 7.Ohara T, Itoh K. Significance of Pseudomonas aeruginosa colonization of the gastrointestinal tract. Intern Med. 2003;42(11):1072–1076. doi: 10.2169/internalmedicine.42.1072 [DOI] [PubMed] [Google Scholar]
- 8.Chakravorty S, Arun P. Antibiotic/adjuvant combinations (ceftriaxone + sulbactam + adjuvant disodium edetate) as an alternative empiric therapy for the treatment of nosocomial infections: results of a retrospective study. Indian J Cancer. 2017;54(4):685–690. doi: 10.4103/ijc.IJC_364_17 [DOI] [PubMed] [Google Scholar]
- 9.Qadir MI, Sajjad S. Phage therapy against streptococcus pneumoniae: modern tool to control pneumonia. Crit Rev Eukaryot Gene Expr. 2017;27(4):289–295. doi: 10.1615/CritRevEukaryotGeneExpr.2017019527 [DOI] [PubMed] [Google Scholar]
- 10.Cao F, Wang X, Wang L, et al. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Research Support, Non-U.S. Gov’t. Biomed Res Int. 2015;2015:752930. doi: 10.1155/2015/752930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos CS, Silva AR. Current and alternative trends in antibacterial agents used in mammalian semen technology. Anim Reprod. 2020;17(1):e20190111. doi: 10.21451/1984-3143-AR2019-0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raimondi MV, Presentato A, Li Petri G, et al. New synthetic nitro-pyrrolomycins as promising antibacterial and anticancer agents. Antibiotics. 2020;9(6):292. doi: 10.3390/antibiotics9060292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miedzybrodzki R, Borysowski J, Weber-Dabrowska B, et al. Clinical aspects of phage therapy. Adv Virus Res. 2012;83:73–121. doi: 10.1016/B978-0-12-394438-2.00003-7 [DOI] [PubMed] [Google Scholar]
- 14.Abedon ST. Phage therapy: eco-physiological pharmacology. Scientifica. 2014;2014:581639. doi: 10.1155/2014/581639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brix A, Cafora M, Aureli M, Pistocchi A. Animal models to translate phage therapy to human medicine. Int J Mol Sci. 2020;21(10):3715. doi: 10.3390/ijms21103715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber-Dabrowska B, Zimecki M, Mulczyk M. Effective phage therapy is associated with normalization of cytokine production by blood cell cultures. Arch Immunol Ther Exp. 2000;48(1):31–37. [PubMed] [Google Scholar]
- 17.Jault P, Leclerc T, Jennes S, et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind Phase 1/2 trial. Lancet Infect Dis. 2019;19(1):35–45. doi: 10.1016/S1473-3099(18)30482-1 [DOI] [PubMed] [Google Scholar]
- 18.Fong SA, Drilling A, Morales S, et al. Activity of bacteriophages in removing biofilms of pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front Cell Infect Microbiol. 2017;7. doi: 10.3389/fcimb.2017.00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khairnar K, Raut MP, Chandekar RH, et al. Novel bacteriophage therapy for controlling metallo-beta-lactamase producing Pseudomonas aeruginosa infection in catfish. BMC Vet Res. 2013;9(1):264. doi: 10.1186/1746-6148-9-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon J, Yong D. Two novel bacteriophages improve survival in galleria mellonella infection and mouse acute pneumonia models infected with extensively drug-resistant Pseudomonas aeruginosa. Appl Environ Microbiol. 2019;85(9). doi: 10.1128/AEM.02900-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, Haque A, Matsuzaki S, Matsumoto T, Nakamura S. The efficacy of phage therapy in a murine model of pseudomonas aeruginosa Pneumonia and sepsis. Front Microbiol. 2021;12:1698. doi: 10.3389/fmicb.2021.682255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morello E, Saussereau E, Fau - Maura D, et al. Pulmonary bacteriophage therapy on Pseudomonas aeruginosa cystic fibrosis strains: first steps towards treatment and prevention. PLoS One. 2011;6(2):e16963. doi: 10.1371/journal.pone.0016963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forti F, Roach DR, Cafora M. et al. Design of a broad-range bacteriophage cocktail that reduces pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob Agents Chemoth. 2018;62(6):e02573–e02617. doi: 10.1128/AAC.02573-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abd El-Aziz AM, Elgaml A, Ali YM. Bacteriophage therapy increases complement-mediated lysis of bacteria and enhances bacterial clearance after acute lung infection with multidrug-resistant Pseudomonas aeruginosa. J Infect Dis. 2019;219(9):1439–1447. doi: 10.1093/infdis/jiy678 [DOI] [PubMed] [Google Scholar]
- 25.Tuon FF, Graf ME, Merlini A, et al. Risk factors for mortality in patients with ventilator-associated pneumonia caused by carbapenem-resistant Enterobacteriaceae. Braz J Infect Dis. 2017;21(1):1–6. doi: 10.1016/j.bjid.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berendt RF, Magruder RD, Frola FR. Treatment of Klebsiella pneumoniae respiratory tract infection of squirrel monkeys with aerosol administration of kanamycin. Am J Vet Res. 1980;41(9):1492–1494. [PubMed] [Google Scholar]
- 27.Moyano AJ, Mas CR, Colque CA, Smania AM. Dealing with biofilms of Pseudomonas aeruginosa and Staphylococcus aureus: in vitro evaluation of a novel aerosol formulation of silver sulfadiazine. Burns. 2020;46(1):128–135. doi: 10.1016/j.burns.2019.07.027 [DOI] [PubMed] [Google Scholar]
- 28.Hofstetter C, Flondor M, Hoegl S, et al. Aerosol delivery during mechanical ventilation to the rat. Exp Lung Res. 2004;30(7):635–651. doi: 10.1080/01902140490489126 [DOI] [PubMed] [Google Scholar]
- 29.Chatmongkolchart S, Schettino GP, Dillman C, Kacmarek RM, Hess DR. In vitro evaluation of aerosol bronchodilator delivery during noninvasive positive pressure ventilation: effect of ventilator settings and nebulizer position. Crit Care Med. 2002;30(11):2515–2519. doi: 10.1097/00003246-200211000-00018 [DOI] [PubMed] [Google Scholar]
- 30.Guillon A, Montharu J, Vecellio L, et al. Pulmonary delivery of dry powders to rats: tolerability limits of an intra-tracheal administration model. Int J Pharm. 2012;434(1–2):481–487. doi: 10.1016/j.ijpharm.2012.05.013 [DOI] [PubMed] [Google Scholar]
- 31.Hoyle N, Zhvaniya P, Balarjishvili N, et al. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: a case report. Res Microbiol. 2018;169(9):540–542. doi: 10.1016/j.resmic.2018.05.001 [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Wang K, Jiao N, Chen F. Genome sequences of two novel phages infecting marine roseobacters. Environ Microbiol. 2009;11(8):2055–2064. doi: 10.1111/j.1462-2920.2009.01927.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li E, Zhao J, Ma Y, et al. Characterization of a novel Achromobacter xylosoxidans specific siphoviruse: phiAxp-1. Sci Rep. 2016;6(1). doi: 10.1038/srep21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Germida JJ, Casida LE. Ensifer adhaerens predatory activity against other bacteria in soil, as monitored by indirect phage analysis. Appl Environ Microbiol. 1983;45(4):1380–1388. doi: 10.1128/aem.45.4.1380-1388.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latino L, Midoux C, Hauck Y, Vergnaud G, Pourcel C. Pseudolysogeny and sequential mutations build multiresistance to virulent bacteriophages in Pseudomonas aeruginosa. Microbiology. 2016;162(5):748–763. doi: 10.1099/mic.0.000263 [DOI] [PubMed] [Google Scholar]
- 36.Chen F, Cheng X, Li J, et al. Novel lytic phages protect cells and mice against pseudomonas aeruginosa infection. J Virol. 2021;95(8):e01832. doi: 10.1128/JVI.01832-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rittich B, Spanova A, Skalnikova M, Benes MJ. Chromatographic behaviour and purification of linear lambda phage and plasmid DNA molecules on 2-hydroxyethyl methacrylate-ethylene dimethacrylate-based supports. J Chromatogr A. 2003;1009(1–2):207–214. doi: 10.1016/s0021-9673(03)00241-3 [DOI] [PubMed] [Google Scholar]
- 38.Pedersen MK, Andersen AB, Folkvardsen DB, et al. Set-up and validation of mycobacterial interspersed repetitive unit-variable number of tandem repeat (MIRU-VNTR) analysis of Mycobacterium tuberculosis using BioNumerics software. PLoS One. 2018;13(10):e0205336. doi: 10.1371/journal.pone.0205336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Domselaar GH, Stothard P, Shrivastava S, et al. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33(WebServer issue):W455–9. doi: 10.1093/nar/gki593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ackermann HW. Phage Classification and Characterization. Humana Press: 2009. [DOI] [PubMed] [Google Scholar]
- 41.Eller MR, Vidigal PMP, Salgado RL, et al. UFV-P2 as a member of the Luz24likevirus genus: a new overview on comparative functional genome analyses of the LUZ24-like phages. BMC Genomics. 2014;15:7. doi: 10.1186/1471-2164-15-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo T, Xin Y, Zhang C, Kong J. A cytoplasmic antiholin is embedded in frame with the holin in a lactobacillus fermentum bacteriophage. Appl Environ Microbiol. 2018;84(6). doi: 10.1128/AEM.02518-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagemans J, Delattre AS, Uytterhoeven B, et al. Antibacterial phage ORFans of Pseudomonas aeruginosa phage LUZ24 reveal a novel MvaT inhibiting protein. Front Microbiol. 2015;6. doi: 10.3389/fmicb.2015.01242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanvazadeh F, Khosravi AD, Zolfaghari MR, Parhizgari N. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns. 2013;39(7):1409–1413. doi: 10.1016/j.burns.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 45.Vidal F, Mensa J, Martinez JA, et al. Pseudomonas aeruginosa bacteremia in patients infected with human immunodeficiency virus type 1. Eur J Clin Microbiol Infect Dis. 1999;18(7):473–477. doi: 10.1007/s100960050326 [DOI] [PubMed] [Google Scholar]
- 46.Hua Y, Luo T, Yang Y, et al. Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front Microbiol. 2017;8:2659. doi: 10.3389/fmicb.2017.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kvachadze L, Balarjishvili N, Meskhi T, et al. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb Biotechnol. 2011;4(5):643–650. doi: 10.1111/j.1751-7915.2011.00259.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cafora M, Deflorian G, Forti F, et al. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci Rep. 2019;9(1). doi: 10.1038/s41598-018-37636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]