Abstract
We report transmission of an azole-resistant, isogenic strain of Candida albicans in a human immunodeficiency virus (HIV)-infected family of two children with symptomatic oropharyngeal candidiasis and a mother with asymptomatic colonization over a 5-year period. These findings were confirmed by three different molecular epidemiology methods: interrepeat PCR, Southern hybridization with a C. albicans repetitive element 2 probe, and electrophoretic karyotyping. This study contributes to an evolving understanding of the mode of transmission of C. albicans, particularly in children, and underscores the importance of monitoring specimens from family members of HIV-infected patients.
Transmission of genetically indistinguishable strains of Candida albicans between human immunodeficiency virus (HIV)-infected adult partners has been reported previously (1, 3, 4–6, 18). However, little is known about the transmission of azole-resistant C. albicans between children and within families (16). We report the transmission of an isogenic C. albicans strain within an HIV-infected family. All isolates were biotyped by interrepeat PCR (IR-PCR), Southern blot hybridization with a C. albicans repetitive element 2 (CARE-2) probe, and electrophoretic karyotyping (EK) in order to demonstrate the degree of genetic relatedness.
(This work was presented in part at the 5th Candida and Candidiasis Conference of the American Society for Microbiology, 1999 [11].)
Two brothers and their mother were monitored in a prospective study for 5 years in the Pediatric Oncology Branch of the National Cancer Institute (Table 1). The brothers had acquired HIV vertically and had a history of recurrent symptomatic oropharyngeal candidiasis (OPC), which had been treated with courses of clotrimazole (CLT), ketoconazole (KTC), itraconazole (ITC), fluconazole (FLC), amphotericin B (AMB), and cyclodextrin itraconazole (CD ITC). During the 5-year observation period, the mother was not treated for asymptomatic colonization with antifungal agents. A total of 13 oral surveillance cultures from the oral mucosa were obtained from the three patients between September 1993 and April 1998. Patient isolates were identified by the Clinical Microbiology Laboratory of the Warren Grant Magnuson Clinical Center of the National Institutes of Health. The 20C Analytic Profile Index strip (Biomerieux, Marcy l’Etoile, France) was used to identify C. albicans. Candida dubliniensis was distinguished from C. albicans by its differential growth at 42 and 45°C. All isolates were stored on potato dextrose agar (PDA) at −70°C and tested for antifungal susceptibilities at a later time. Stock solutions of AMB (Bristol-Myers Squibb, Princeton, N.J.) and FLC (Pfizer, Groton, Conn.) were prepared by using RPMI-1640 buffered with 0.165 M morpholinepropanesulfonic acid (MOPS) to pH 7.0 (BioWhittaker, Walkersville, Md.). Polyethylene glycol 400 was used to solubilize ITC and KTC (Janssen Pharmaceutica, Piscataway, N.J.). Serial twofold dilutions were further performed with the appropriate diluent. The final concentrations were 0.03 to 16 μg/ml for AMB, KTC, and ITC and 0.125 to 64 μg/ml for FLC. Broth microdilution testing was performed according to reference method M27-A of the National Committee for Clinical Laboratory Standards (Table 2) (12). All MICs at 24 and 48 h were determined at least three times for each isolate.
TABLE 1.
Brief clinical profiles of family members with azole-resistant C. albicans
| Patient | Transmission of HIV infection | CD4 count | HIV therapya | OPC | Antifungal history |
|---|---|---|---|---|---|
| Sibling 1 | Vertical | <800 | 3TC, ddI, D4T, ritonavir | Chronic | CLT, KTC, FLC, AMB, ITC |
| Sibling 2 | Vertical | <100 | AZT, ddI | Chronic | CLT, KTC, FLC, AMB, ITC, CD ITC |
| Mother | Horizontal | 200 | Nelfinavir, D4T, nevirapine | None | None |
3TC, lamivudine; ddI, dideoxyinosine; D4T, didehydrodideoxythymidine; AZT, zidovudine.
TABLE 2.
MICs of antifungal compounds against Candida spp. recovered from oral mucosa of family members
| Source and isolate no. | Species | Date of isolationa | MIC (μg/ml) of:
|
|||
|---|---|---|---|---|---|---|
| KTC | FLC | ITC | AMB | |||
| Sibling 1 | ||||||
| 4256 | C. albicans | 9/02/93 | 0.5 | 16 | 0.25 | 1 |
| 3961 | C. albicans | 2/26/94 | 0.5 | 64 | >16 | 0.25 |
| 1378 | C. albicans | 3/8/95 | 1 | 64 | 1 | 0.125 |
| 7582 | C. albicans | 5/23/95 | 0.5 | >64 | 2 | 0.125 |
| 2687 | C. albicans | 7/17/95 | 0.125 | 16 | 0.5 | 0.125 |
| 3312 | C. albicans | 7/08/96 | 0.5 | 64 | 16 | 0.125 |
| 6765 | C. albicans | 7/10/97 | 0.5 | >64 | 4 | 0.125 |
| 3146 | C. albicans | 4/06/98 | 0.125 | 32 | 1 | 0.125 |
| Sibling 2 | ||||||
| 4054 | C. albicans | 12/12/94 | 1 | 64 | 1 | 0.125 |
| 1379 | C. albicans | 3/8/95 | 0.125 | 64 | 1 | 0.125 |
| 7600 | C. albicans | 5/23/95 | 0.5 | 64 | 1 | 0.125 |
| Mother | ||||||
| 2823 | C. albicans | 7/17/95 | 0.125 | 64 | 0.5 | 0.5 |
| 3147 | C. dubliniensis | 4/6/98 | ≤0.03 | 0.25 | ≤0.03 | ≤0.03 |
Shown as month/day/year.
Molecular biotyping of the Candida strains was performed by using two DNA fingerprinting methods, IR-PCR and Southern hybridization with a CARE-2 probe, as well as EK. All typing was repeated at least three times to ensure reproducibility. For the IR-PCR assay, genomic DNA from Candida isolates was extracted with the DNeasy kit (Qiagen, Chatsworth, Calif.). The oligonucleotide pair 1245 (5′ AAGTAAGTGACTGGGGTGAGCG 3′) and 1246 (5′ ATGTAAGCTCCTGGGGATTCAC 3′) was used under the following conditions (21): a final amplification buffer of 6.25 mM MgCl2, 83 mM KCl, 16.7 mM Tris-HCl, 0.001% (wt/vol) gelatin, 1.33 mM deoxynucleoside triphosphates (Boehringer Mannheim, Indianapolis, Ind.), 0.01 μg of each primer/ml, 1.25 U of Taq DNA polymerase (Boehringer Mannheim), and 25 to 50 ng of genomic DNA/μl. Amplification conditions were as follows: 95°C for 5 min, 95°C for 1 min, 25°C for 1 min, and 74°C for 2 min for 35 cycles, followed by 74°C for 5 min. Amplification products were visualized on an ethidium bromide-stained 1.5% agarose gel. For the CARE-2 analysis, genomic DNA was extracted by using phenol-chloroform as described by Millon et al. (10). A 954-bp CARE-2 fragment was amplified by PCR as described previously (7). Ten micrograms of chromosomal DNA was digested with EcoRI, electrophoresed, and vacuum blotted onto a nylon membrane (Pall, Portsmouth, England) (17). After UV cross-linking, the filter was hybridized with 200 ng of enzyme chemiluminescence-labeled (ECL) (Amersham, Braunschweig, Germany) CARE-2 DNA. Detection of the chemiluminescent signal was performed according to the manufacturer’s directions. DNA was extracted for EK from agarose-embedded cells exposed to enzymatic digestion with minor modifications as described previously (19), by using the Bio-Rad (Hercules, Calif.) contour-clamped homogeneous electric field (CHEF) DR-III device. DNA was run on a 0.9% SeaKem Gold Agarose gel (FMC Bioproducts, Rockland, Maine). The gel was run at 14°C and 4.5 V/cm with a 120° angle. Running time was 36 h with a 60- to 300-s linear ramp (8). We analyzed isolate 8621 as a standard control for IR-PCR, CARE-2, and EK, and we analyzed four more unrelated clinical isolates (9329, 4389, 1672, and 2771) of C. albicans as controls for IR-PCR and CARE-2.
The isolates of C. albicans from both children demonstrated resistance to FLC and ITC (Table 2). One Candida isolate obtained from the mother was identified as C. albicans, with a MIC profile similar to those for the isolates from the two children, and the other isolate from the mother was characterized as a C. dubliniensis strain susceptible to FLC.
Molecular analysis of each isolate demonstrated that the same azole-resistant isogenic strain of C. albicans was shared among the two brothers and the mother. The three molecular typing methods also clearly distinguished between isolates of C. albicans and C. dubliniensis (Fig. 1 through 3). The separation of chromosomes in the EK demonstrated that within the index strain of C. albicans there were three variants differing at the level of chromosome rearrangement (Fig. 3). This chromosomal rearrangement may be a form of microevolution under antifungal azole pressure. As recommended by several investigators (2, 9, 13, 14, 20), we used other molecular methods to further determine the genotypic relatedness of these variant isolates. The evidence from a combination of a PCR-based method (IR-PCR) and Southern hybridization with CARE-2 supports the hypothesis that these isolates are highly related.
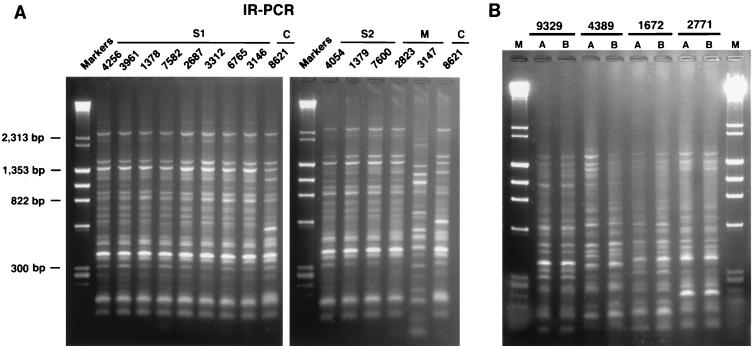
FIG. 1.
(A) IR-PCR banding patterns generated from genomic DNA of patients’ isolates by using the primer pair 1245 and 1246. S1, sibling 1; S2, sibling 2; M, mother; C, control lab isolate. (B) Distinct IR-PCR banding patterns generated from genomic DNA of unrelated clinical isolates of C. albicans: isolates 9329, 4389, 1672, and 2771. Each of these isolates was run in duplicate in parallel lanes (lanes A and B) to document the reproducibility of the gel pattern for each strain.
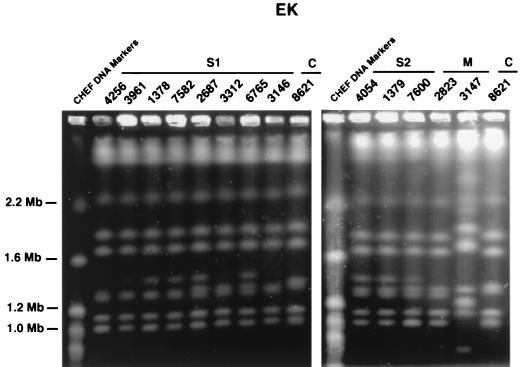
FIG. 3.
Electrophoretic karyotypes of chromosomal DNA of patients’ isolates obtained by pulsed-field gel electrophoresis. S1, sibling 1; S2, sibling 2; M, mother; C, control lab isolate. The banding pattern is similar for all isolates of the index strain. Note that there are three types of variants evidenced by alterations in chromosomes.
Azole-resistant OPC in HIV-infected patients develops after long-term exposure to azoles (15, 22, 23). However, our findings also reveal that transmission from symptomatic to asymptomatic family members is possible and perhaps represents a previously underappreciated factor in families with HIV infection, among whom more than one family member is at risk for serious and chronic complications of OPC. Asymptomatic family members, who have not received antifungal therapy, may also be colonized with azole-resistant C. albicans. The mode of transmission of azole-resistant C. albicans among siblings and parents may be the exchange of contaminated fomites, which commonly occurs in the sharing of food, utensils, and toys. The acquisition of an azole-resistant strain of C. albicans by an asymptomatic HIV-infected patient has important clinical implications and may result in de novo presentation of OPC refractory to initial azole therapy. The consequence is earlier use of AMB, a more toxic and inconvenient treatment alternative. Other families with children suffering from immunodeficiencies such as severe combined immunodeficiency, Wiskott-Aldrich syndrome, or chronic granulomatous disease carry similar risks for intrafamilial transmission of azole-resistant C. albicans and should be included in future infection control programs.
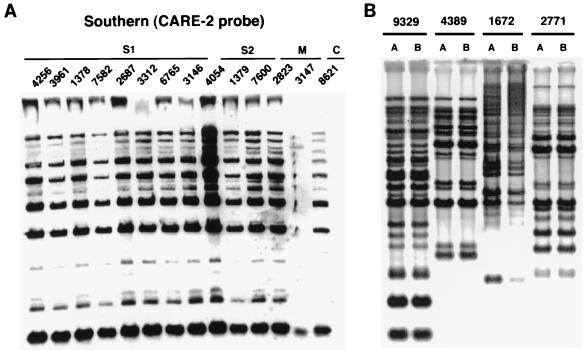
FIG. 2.
(A) Southern hybridization of EcoRI-digested genomic DNA of patients’ isolates probed with a CARE-2 probe. S1, sibling 1; S2, sibling 2; M, mother; C, control lab isolate. (B) Distinct CARE-2 probe banding patterns generated from genomic DNA of unrelated clinical isolates of C. albicans: isolates 9329, 4389, 1672, and 2771. Each of these isolates was run in duplicate in parallel lanes (lanes A and B) to document the reproducibility of the gel pattern for each strain.
Acknowledgments
We are grateful to Beatrice B. Magee and Paul T. Magee for the discussion of chromosome variants.
F.-M. C. Müller was supported by a grant from the Bundesministerium für Bildung und Forschung (BMBF).
REFERENCES
- 1.Barchiesi F, Hollis R J, Poeta M D, McGough D A, Scalise G, Rinaldi M G, Pfaller M A. Transmission of fluconazole-resistant Candida albicans between patients with AIDS and oropharyngeal candidiasis documented by pulsed-field gel electrophoresis. Clin Infect Dis. 1995;21:561–564. doi: 10.1093/clinids/21.3.561. [DOI] [PubMed] [Google Scholar]
- 2.Barton R C, van Belkum A, Scherer S. Stability of karyotype in serial isolates of Candida albicans from neutropenic patients. J Clin Microbiol. 1995;33:794–796. doi: 10.1128/jcm.33.4.794-796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerlin P, Addo M, Boerlin-Petzold F, Durussel C, Pagani J L, Chave J P, Bille J. Transmission of oral Candida albicans strains between HIV-positive patients. Lancet. 1995;345:1052–1053. doi: 10.1016/s0140-6736(95)90793-9. [DOI] [PubMed] [Google Scholar]
- 4.Cartledge J D, Midgley J, Gazzard B G. Transmission of fluconazole-resistant Candida strains between HIV-positive patients and their sexual partners. AIDS. 1998;12:1249–1251. doi: 10.1097/00002030-199810000-00022. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Guerra T M, Martinez-Suarez J V, Laguna F, Valencia E, Rodriguez-Tudela J L. Change in fluconazole susceptibility patterns and genetic relationship among oral Candida albicans isolates. AIDS. 1998;12:1601–1610. doi: 10.1097/00002030-199813000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Dromer F, Improvisi L, Dupont B, Eliaszewicz M, Pialoux G, Fournier S, Feuillie V. Oral transmission of Candida albicans between partners in HIV-infected couples could contribute to dissemination of fluconazole-resistant isolates. AIDS. 1997;15:1095–1101. doi: 10.1097/00002030-199709000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Lasker B A, Page L S, Lott T J, Kobayashi G S. Isolation, characterization and sequencing of Candida albicans repetitive element 2. Gene. 1992;116:51–57. doi: 10.1016/0378-1119(92)90628-3. [DOI] [PubMed] [Google Scholar]
- 8.Magee, B., and P. T. Magee. Personal communication.
- 9.Merz W G. Candida albicans strain delineation. Clin Microbiol Rev. 1990;3:321–334. doi: 10.1128/cmr.3.4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, Michel-Briand Y. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J Clin Microbiol. 1994;32:1115–1118. doi: 10.1128/jcm.32.4.1115-1118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller F M C, Kasai M, Francesconi A, Rishforth B, McEvoy M, Peter J, Chanock S J, Walsh T J. Abstracts of the 5th Candida and Candidiasis Conference of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1999. Transmission of azole-resistant isogenic Candida albicans strains among HIV-infected family members with oropharyngeal candidiasis, abstr. C36; pp. 58–59. [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. NCCLS M27-A. 17, no. 9. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 13.Odds F C, Brawner D L, Staudinger J, Magee P T, Soll D R. Typing of Candida albicans strains. J Med Vet Mycol. 1992;1:87–94. [PubMed] [Google Scholar]
- 14.Pfaller M A. Epidemiology of fungal infections: the promise of molecular typing. Clin Infect Dis. 1995;20:1535–1539. doi: 10.1093/clinids/20.6.1535. [DOI] [PubMed] [Google Scholar]
- 15.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruhnke M, Grosch-Wörner I, Steinmüller A, Neubauer A. Molecular epidemiology of Candida infections in HIV-infected mothers and their children. Wien Med Wochenschr. 1997;147:446–449. [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Sangeorzan J A, Bradley S F, He X, Zarins L T, Ridenour G L, Tiballi R N, Kauffman C A. Epidemiology of oral candidiasis in HIV-infected patients: colonization, infection, treatment, and emergence of fluconazole resistance. Am J Med. 1994;97:339–346. doi: 10.1016/0002-9343(94)90300-x. [DOI] [PubMed] [Google Scholar]
- 19.Scherer, S. 15 April 1999, posting date. [Online.] Candida albicans information. http://alces.med.umn.edu/Candida.html. [10 July 1999, last date accessed.]
- 20.Taylor J W, Geiser D M, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Belkum A, Melchers W, de Pauw B E, Scherer S, Quint W, Meis J F. Genotypic characterization of sequential Candida albicans isolates from fluconazole-treated neutropenic patients. J Infect Dis. 1994;169:1062–1070. doi: 10.1093/infdis/169.5.1062. [DOI] [PubMed] [Google Scholar]
- 22.vanden Bossche H, Dromer F, Improvisi I, Lozano-Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36:119–128. [PubMed] [Google Scholar]
- 23.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]