Abstract
The global spread of antibiotic-resistant infections has meant that there is an urgent need to develop new antimicrobial alternatives. In this study, we developed a strategy to boost and/or synergize the activity of conventional antibiotics by combination with antimicrobial peptides tagged with the bulky non-natural amino acid β-naphthylalanine (Nal) to their N- or C-terminus. A checkerboard method was used to evaluate synergistic effects of the parent peptide and the Nal-tagged peptides. Moreover, boron-dipyrro-methene labeled vancomycin was used to characterize the synergistic mechanism of action between the peptides and vancomycin on the bacterial strains. These Nal-tagged antimicrobial peptides also reduced the antibiotic-induced release of lipopolysaccharide from Gram-negative bacteria by more than 99.95%. Our results demonstrate that Nal-tagged peptides could help in developing antimicrobial peptides that not only have enhanced antibacterial activities but also increase the synergistic effects with conventional antibiotics against antibiotic-resistant bacteria.
Keywords: antimicrobial peptides (AMP), antibiotic resistance, synergism, bulky non-natural amino acid, end-tagging
Introduction
The emergence of multidrug-resistant pathogens emphasizes the need for novel and alternative therapeutics to fight against infections (Marston et al., 2016). Some AMPs can work synergistically with conventional antibiotics to overcome the resistant problems and reduce the amount of antibiotics (Cassone and Otvos, 2010; Sierra et al., 2017; Martinez et al., 2019; Zharkova et al., 2019; Li et al., 2020). However, rules governing the design of AMPs with synergistic effects are still not clear (Lazar et al., 2018).
Tryptophan, owing to its membrane disruptive and lipid interface anchoring activities, has been found to play important roles in the design and development of Trp-rich AMPs (Mojsoska and Jenssen, 2015; Godballe et al., 2016; Arias et al., 2018). Previously, a Trp-rich peptide PEM-2-W5K/A9W (Ac-KKWRKWLKWLAKK-NH2) was developed based on the C-terminal region of Bothrops asper myotoxin II (Yu et al., 2010). PEM-2-W5K/A9W was found to possess strong activities against both bacteria and fungi even under high salt conditions (Yu et al., 2010). However, peptide S1 (Ac-KKWRKWLAKK-NH2), a shortened version of PEM-2-W5K/A9W, only had antimicrobial activities in the low salt LYM broth media (Chu et al., 2013). The antimicrobial activities of S1 were diminished at high salt concentrations.
Several problems such as salt resistance, proteolytic stability, as well as the lipopolysaccharide (LPS, endotoxin) outer membrane of bacteria may hinder the development of AMPs (Chu et al., 2013; Chih et al., 2015, 2019). Recent studies have indicated that the salt resistance, serum proteolytic stability, and LPS neutralizing activities can be increased by adding bulky non-natural amino acids to the termini of short AMPs such as peptide S1 (Chu et al., 2013; Chih et al., 2015). Nuclear magnetic resonance structures have shown that the two terminal β-naphthylalanine residues of S1-Nal-Nal were inserted into the hydrophobic lipid A motif of LPS micelles and enhanced membrane permeabilization and translocation of this peptide (Yu et al., 2017). Similar results were also observed by adding hydrophobic oligopeptide end tags to the termini of AMPs (Schmidtchen et al., 2009, 2011; Song et al., 2019).
Evidence has highlighted the synergistic effects of using AMPs in combination with conventional antibiotics against multidrug-resistant bacterial strains. For example, polymyxin B and its derivatives were found to function as “permeabilizers” or “potentiators” to sensitize bacteria to other antibiotics or potentiate the action of other antibiotics by interacting with anionic lipopolysaccharide (LPS) outer leaflet of Gram-negative bacteria (Vaara, 2019). It was also shown that multidrug-resistant bacteria frequently show collateral sensitivity to AMPs (Lazar et al., 2018). This finding can be used to develop peptide-antibiotic combinations that can fight against resistant bacteria (Lazar et al., 2018). Zharkova et al. (2019) also found that synergism mainly occurs between highly membrane-active AMPs and antibiotics with intracellular targets. Cytotoxicity of these combinations toward normal eukaryotic cells were rarely synergistic (Zharkova et al., 2019). In addition, a bulky non-natural amino acid substituted version of the antimicrobial peptide Bip-P-113 was found to act as a potentiator to sensitize vancomycin against Gram-negative bacteria such as Escherichia coli and to boost synergistic effects with vancomycin against drug-resistant Gram-positive bacteria (Wu et al., 2020).
The aim of this study was to test if the bulky non-natural amino acid end tagging strategy of short AMPs can be used in combination with various antibiotics to fight against antibiotic-resistant bacterial strains. These results could provide important insights into the synergistic mechanism between AMPs and traditional antibiotics.
Materials and Methods
Materials
S1, S1-Nal, and S1-Nal-Nal (Table 1) were purchased from Kelowna International Scientific Inc. (Taipei, Taiwan). The identity of the peptides was confirmed by electrospray mass spectroscopy and the purity (>95%) was assessed by high-performance liquid chromatography. Both identity and purity data were provided by Kelowna International Scientific Inc. (Taipei, Taiwan). Tetracycline, ciprofloxacin and vancomycin were purchased from Bio Basic Inc. (Toronto, ON, Canada). BODIPY-labeled Vancomycin was obtained from Thermo Fisher Scientific (Waltham, MA, United States). Mueller-Hinton broth (MHB), Tryptic soy broth (TSB) were purchased from Becton, Dickinson and Company (Franklin Lakes, NJ, United States). Calcein-AM dye was obtained from Sigma-Aldrich (St. Louis, MO, United States).
TABLE 1.
Sequences of S1, S1-Nal, and S1-Nal-Nal.
| Name | Sequencea | Molecular weight (Da) |
| S1 | Ac-KKWRKWLAKK-NH2 | 1,412.79 |
| S1-Nal | Ac-KKWRKWLAKK-Nal-NH2 | 1,609.98 |
| S1-Nal-Nal | Ac-KKWRKWLAKK-Nal-Nal-NH2 | 1,806.99 |
aNal, β-naphthylalanine.
Bacterial Strains and Culture Conditions
Enterococcus faecium (BCRC 15B0132, VRE), Acinetobacter baumannii strains including 14B0091, 14B0097, and 14B0100, and E. coli strains including BCRC 13B0198 and BCRC 13B0207 were purchased from Bioresources Collection & Research Center1 (BCRC, FIRDI, Hsinchu, Taiwan). Escherichia coli and A. baumannii strains were incubated in MHB, and E. faecium BCRC 15B0132 were incubated in TSB overnight at 37°C with 150 rpm shaking overnight. The turbidity of bacteria was measured by the absorbance of optical density at 600 nm (OD600 = 1, equal to approximately 108 CFU/mL) with UV/Visible spectrophotometer (Biochrom, Cambridge, United Kingdom).
Antimicrobial Activity Assay
The minimal inhibitory concentrations (MICs) of peptides and antibiotics were determined by using microbroth dilution technique, as described by the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (2015). The assay was conducted manually using the single pipette following the CLSI with some modifications. Briefly, E. coli and A. baumannii strains were incubated in MHB, and E. faecium BCRC 15B0132 were incubated in TSB overnight at 37°C. The cell cultures were regrown to mid-log phase and then diluted to a final concentration of 5 × 105 CFU/mL. 99 μl of each diluted microbes were transferred into each well of a 96-well plate, into which 1 μl of peptides or antibiotics has previously been added and the final concentration of the peptides and antibiotics would be 64–0.13 μg/mL. After incubation for about 16 h at 37°C, the MIC value of peptides or antibiotics was determined by inspecting the visible growth. The MIC value was defined as the lowest concentration of an antimicrobial that will inhibit the visible growth of a microorganism. All experiments were repeated three times independently.
Checkerboard Assay
The synergistic effects of peptides in combination with antibiotics was assessed by using the microbroth dilution checkerboard assay (Mataraci and Dosler, 2012; Mohamed et al., 2016). Each well containing the mixture of 1 μl peptide (¼ × MIC at final concentration) and 1 μl antibiotic (64–0.13 μg/mL at final concentration), and loaded 98 μl bacteria at the final concentration of 5 × 105 CFU/mL into each well of a 96-well plate. After incubation for about 16 h at 37°C, the MIC value of peptide/antibiotic combination was determined by inspecting the visible growth. MIC values obtained were used to evaluate the effects of combination between peptides and antibiotics by calculating the Fractional Inhibitory Concentration Index (FICI) according to the following formula:
The calculated FICI was interpreted as synergistic (FICI ≤ 0.5), no interaction (0.5 < FICI ≤ 4), or antagonistic (FICI > 4) (Odds, 2003). The experiments were repeated three times independently.
Confocal Laser Scanning Microscopy
The fluorescence experiments were performed with slight modification as described previously (Joshi et al., 2010). Briefly, the bacteria were diluted to 107 CFU/mL and incubated with S1, S1-Nal, and S1-Nal-Nal peptides at 0.5 × MIC for 30 mins. Then, 2 μg/mL BODIPY-labeled vancomycin were treated for 30 mins. Bacteria treated with BODIPY-labeled vancomycin only served as a control. After treatment, the cell pellet was obtained by centrifugation and washed with PBS to remove free BODIPY-labeled vancomycin. The pellet was resuspended and loaded on the glass slides (PolysineTM, Thermo Fisher Scientific, Waltham, MA, United States) and visualized under confocal laser scanning microscope (LSM 510 META, Carl Zeiss, Jena, Thüringen, Germany) equipped with 64 × oil objective lens (Carl Zeiss, Jena, Thüringen, Germany). All experiments were repeated three times independently.
Calcein Leakage Assay
Calcein acetoxymethyl ester (calcein-AM) is a non-fluorescent dye and enters into the bacteria through diffusing across the cell membrane, and hydrolyzed to fluorescent calcein (C30H26N2O13) by cytoplasmic esterases (Xiong et al., 2005). Briefly, the microbes were grown to mid-log phase and the pellet was obtained by centrifugation, washed with PBS, resuspended to OD600 of 1.0 with PBS containing 10% (vol/vol) broth. The microbes were incubated with 3 μM calcein-AM for 1 h at 37°C to form calcein-AM loaded cells. The calcein-AM loaded cells were collected by centrifugation (3,000 × g, 10 mins), and resuspended to 107 CFU/ml by PBS. Cobalt was added into cells and was used to quench the fluorescence of calcein released into the extracellular environment. The cells were treated with 0.5 × MIC of peptides in the sterile black-wall 96-well plate and measured the fluorescence intensity for 60 mins at an excitation wavelength of 485 nm and an emission wavelength of 510 nm on a fluorescence plate reader (VICTOR3, PerkinElmer, United States) (Essodaigui et al., 1998). Bacteria treated with H2O served as negative controls. The intensity of membrane permeabilization (%) was calculated as the absolute percent calcein leakage by peptides with respect to non-treated calcein-AM loaded cells (Koo et al., 2001). All experiments were repeated three times independently.
Anti-endotoxin Studies
Escherichia coli BCRC 13B0198 were cultured at the mid-log phase and diluted to 104 CFU/ml, and then treated with peptide alone at 1 × MIC or combination with vancomycin (both at 0.5 × MIC) at 37°C for 6 h. The cells were filtered by using a pyrogen-free 0.2 μm pore filter (Acrodisc, Pall Corporation, United States) and the endotoxin level was determined by limulus amebocyte lysate (LAL) PYROCHROME® test (Associates of Cape Cod, United States). The kinetic turbidity was analyzed by using a microplate reader (SpectraMax ABS, Molecular Devices, San Jose, CA, United States). The experiments were repeated three times independently.
Statistical Analysis
The statistical results are performed as the mean ± SEM and were analyzed using one-way ANOVA analysis of variance. Statistical analysis was performed using GraphPad Prism version 8.0 (San Diego, CA, United States), where p < 0.05 was considered to indicate a statistically significant difference.
Results
Antimicrobial Activity
The antibacterial activities of S1, S1-Nal, S1-Nal-Nal, vancomycin, ciprofloxacin, and tetracycline against Gram-positive bacterium including vancomycin-resistant E. faecium BCRC 15B0132 (VRE) and Gram-negative bacteria including A. baumannii BCRC 14B0091, A. baumannii BCRC 14B0097, A. baumannii BCRC 14B0100, E. coli BCRC 13B0198, and E. coli BCRC 13B0207 were assessed by MIC assay. As shown in Table 2, antibiotics, including vancomycin, ciprofloxacin, and tetracycline had limited or no activities against all bacterial strains except for E. coli BCRC 13B0207, which is sensitive to ciprofloxacin. S1 peptide also had limited or no antibacterial activities (>64 μg/ml). On the other hand, peptides S1-Nal and S1-Nal-Nal with the addition of bulky non-natural amino acid β-naphthylalanine at the C-termini demonstrated enhanced activities against all bacterial strains. S1-Nal had the MICs of 64 μg/ml against E. faecium, 8 μg/ml against A. baumannii, and 32 μg/ml against E. coli. S1-Nal-Nal had the MICs of 32 μg/ml against E. faecium, 2 μg/ml against A. baumannii BCRC 14B0091, 4 μg/ml against A. baumannii BCRC 14B0100, 8 μg/ml against A. baumannii BCRC 14B0097, and 16 μg/ml against E. coli. These results indicated that the addition of only one or two bulky non-natural amino acid end tags could boost the antimicrobial activity.
TABLE 2.
The minimal inhibitory concentrations (MICs) of S1, S1-Nal, S1-Nal-Nal, vancomycin, ciprofloxacin, and tetracycline.
| Bacterial strains | MICa (μg/ml) | |||||
| S1 | S1-Nal | S1-Nal-Nal | Vancomycin | Ciprofloxacin | Tetracycline | |
| Enterococcus faecium BCRC 15B0132 | >64 | 64 | 32 | >64 | >64 | 64 |
| Acinetobacter baumannii BCRC 14B0091 | >64 | 8 | 2 | >64 | 32 | >64 |
| Acinetobacter baumannii BCRC 14B0097 | >64 | 8 | 8 | >64 | >64 | >64 |
| Acinetobacter baumannii BCRC 14B0100 | >64 | 8 | 4 | >64 | 64 | >64 |
| Escherichia coli BCRC 13B0198 | 64 | 32 | 16 | >64 | 32 | >64 |
| Escherichia coli BCRC 13B0207 | >64 | 32 | 16 | >64 | 0.25 | >64 |
aMIC, minimum inhibitory concentration.
Bacterial Membrane Permeabilization
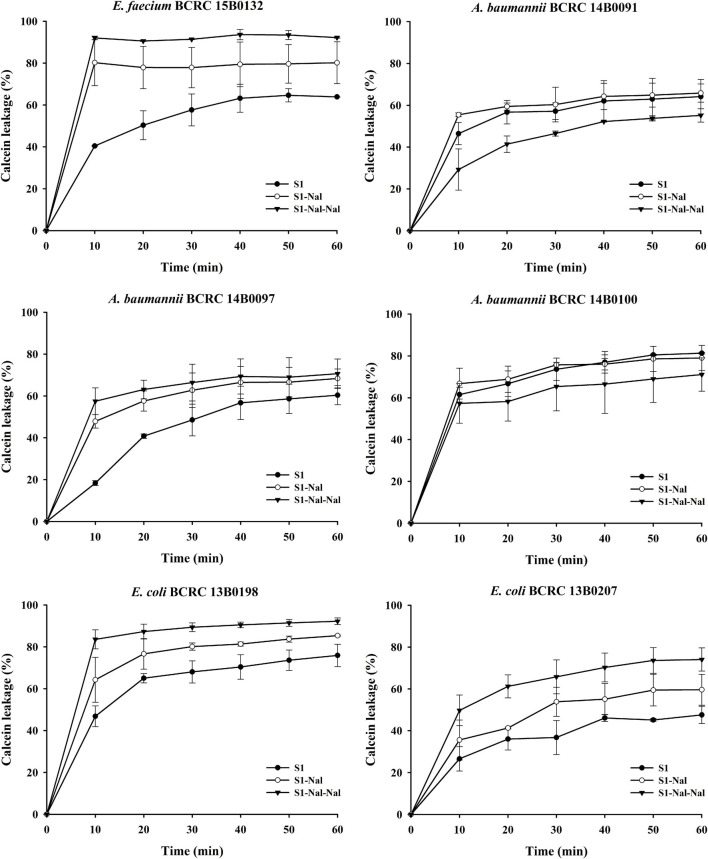
Permeabilities of S1, S1-Nal, and S1-Nal-Nal were measured by the peptide-induced leakage of the fluorescent dye calcein from bacterial cells (Figure 1). To determine the mechanism of antibacterial action of the AMPs, we evaluated the ability of peptides to permeabilize intact E. faecium, A. baumannii, and E. coli membranes by measuring the peptide-induced leakage of the fluorescent dye calcein from bacterial cells (Figure 1). In general, the permeabilization ability is in the order of S1-Nal-Nal > S1-Nal > S1. Addition of β-naphthylalanine to the termini of S1-Nal and S1-Nal-Nal can increase permeability from 60 to 90% for E. faecium, from 50 to 60% for A. baumannii BCRC 14B0097, from 65 to 90% for E. coli BCRC13B0198, and from 40 to 70% for E. coli BCRC13B0207. However, for bacterial strains A. baumannii BCRC 14B0091 and A. baumannii BCRC 14B0100, the order of the permeabilization ability is S1-Nal = S1 > S1-Nal-Nal. Nevertheless, All peptides showed substantial release of calcein from these six bacterial cells.
FIGURE 1.
The membrane permeabilization of Enterococcus faecium BCRC 15B0132, Acinetobacter baumannii BCRC 14B0091, A. baumannii BCRC 14B0097, A. baumannii BCRC 14B0100, Escherichia coli BCRC 13B0198, and E. coli BCRC 13B0207, by S1, S1-Nal, and S1-Nal-Nal for 60 min exposure. Calcein-AM loaded cells (107 CFU/ml) were resuspended by PBS, and the aliquots of 100 μl were added into a sterile black-wall 96-well plate, then treated with 0.5 × MIC of peptides in each well and measured the calcein leakage immediately. Each time point for the peptides was repeated three times independently.
Synergistic Effect With Vancomycin, Ciprofloxacin, and Tetracycline in the Presence of a Sub-Inhibitory Concentration (¼ × Minimal Inhibitory Concentrations) of Peptides
Based on the strong antimicrobial activity and bacterial membrane permeability, it is suggested that these peptides could be used to potentiate conventional antibiotics such as vancomycin, ciprofloxacin, and tetracycline against antibiotic-resistant Gram-positive and Gram-negative bacterial strains. Synergetic activities of S1, S1-Nal, and S1-Nal-Nal combined with vancomycin, ciprofloxacin, and tetracycline were then determined by the checkerboard assay (Table 3). For E. faecium BCRC 15B0132 and A. baumannii BCRC 14B0097, S1-Nal-Nal showed substantial synergy while combined with vancomycin, ciprofloxacin, and tetracycline. The order of the synergistic effects against E. faecium BCRC 15B0132 and A. baumannii BCRC 14B0097 is S1-Nal-Nal > S1-Nal > S1. For A. baumannii BCRC 14B0091 and A. baumannii BCRC 14B0100, S1-Nal demonstrated better synergistic effects than S1-Nal-Nal and S1. For E. coli BCRC 13B0198, all peptides had strong synergy while combined with ciprofloxacin and vancomycin, but these three peptides had only limited or no synergy while combined with tetracycline. For E. coli BCRC 13B0207, all peptides demonstrated strong synergy while combined with ciprofloxacin, but had only limited activity while combined with vancomycin and tetracycline.
TABLE 3.
Synergistic effects of S1, S1-Nal, and S1-Nal-Nal in combination with antibiotics against bacterial strains studied.
| Strains | Antibiotics (μg/mL) | AMP (μg/mL) (¼ × MIC) | |||||
| S1 | S1-Nal | S1-Nal-Nal | |||||
| + | FICIa | + | FICI | + | FICI | ||
| Enterococcus faecium BCRC 15B0132 | Vancomycin | >64 | 1.25 | 8 | 0.31 | 2 | 0.27 |
| Ciprofloxacin | 64 | 0.75 | 16 | 0.38 | 2 | 0.27 | |
| Tetracycline | 16 | 0.5 | 8 | 0.38 | 1 | 0.27 | |
| Acinetobacter baumannii BCRC 14B0091 | Vancomycin | 16 | 0.38 | 2 | 0.27 | >64 | 1.25 |
| Ciprofloxacin | 32 | 0.5 | 32 | 0.5 | 64 | 2.25 | |
| Tetracycline | 64 | 0.75 | 32 | 0.5 | >64 | 1.25 | |
| Acinetobacter baumannii BCRC 14B0097 | Vancomycin | 32 | 0.5 | 2 | 0.27 | 2 | 0.27 |
| Ciprofloxacin | >64 | 1.25 | 64 | 0.75 | 2 | 0.27 | |
| Tetracycline | 64 | 0.75 | 64 | 0.75 | 2 | 0.27 | |
| Acinetobacter baumannii BCRC 14B0100 | Vancomycin | 32 | 0.5 | 2 | 0.27 | 4 | 0.28 |
| Ciprofloxacin | 64 | 0.75 | 32 | 0.5 | 32 | 0.75 | |
| Tetracycline | 64 | 0.75 | 64 | 0.75 | >64 | 1.25 | |
| Escherichia coli BCRC 13B0198 | Vancomycin | 32 | 0.5 | 32 | 0.5 | 32 | 0.5 |
| Ciprofloxacin | 2 | 0.27 | 2 | 0.27 | 2 | 0.31 | |
| Tetracycline | >64 | 1.25 | 64 | 0.75 | 64 | 0.75 | |
| Escherichia coli BCRC 13B0207 | Vancomycin | 64 | 0.75 | >64 | 1.25 | 64 | 0.75 |
| Ciprofloxacin | 0.02 | 0.31 | 0.06 | 0.5 | 0.13 | 0.75 | |
| Tetracycline | 64 | 0.75 | 64 | 0.75 | 64 | 0.75 | |
aFICI, fractional inhibitory concentration index, FICI ≤ 0.5, synergy; 0.5 < FICI ≤ 4, no interaction; FICI > 4, antagonism.
+, the MIC of antibiotic in combination with ¼ × MIC antimicrobial peptides (AMPs). Bold values indicated synergistic effects.
Mechanism of the Synergistic Effects Studied by Boron-Dipyrro-Methene Labeled Vancomycin
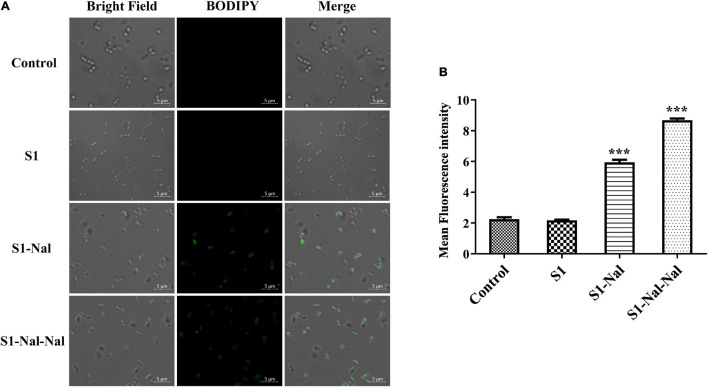
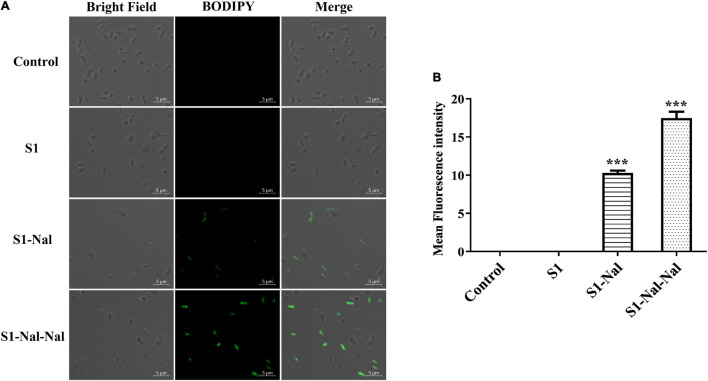
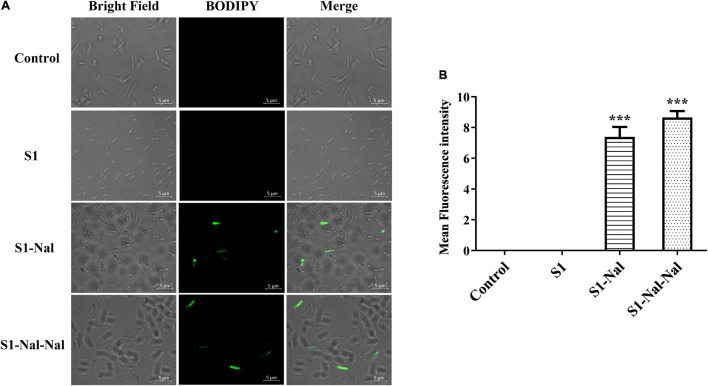
Boron-Dipyrro-Methene (BODIPY)-labeled vancomycin was used to study the mechanism of synergistic effects of S1, S1-Nal, and S1-Nal-Nal by measuring the uptake of vancomycin in E. faecium BCRC 15B0132, A. baumannii BCRC 14B0097, and E. coli BCRC 13B0207 strains (Chu et al., 2020). The results demonstrated that S1-Nal and S1-Nal-Nal with β-naphthylalanine end-tagging markedly enhanced the entry of BODIPY-labeled vancomycin into the Gram-positive E. faecium BCRC 15B0132 strain (Figure 2). Similar results were also found for the Gram-negative A. baumannii BCRC 14B0097, and E. coli BCRC 13B0207 strains (Figures 3, 4).
FIGURE 2.
S-Nal and S1-Nal-Nal increase the uptake of boron-dipyrro-methene (BODIPY)-labeled vancomycin in E. faecium. (A) Fluorescence images of 107 CFU/ml E. faecium BCRC 15B0132 treated with 0.5 × MIC of S1, S1-Nal and S1-Nal-Nal at 37°C for 30 min, then treated BODIPY-labeled vancomycin for 30 min (scale bar represents 5 μm). (B) Mean fluorescence intensity of S1, S1-Nal, and S1-Nal-Nal treated in E. faecium BCRC 15B0132. Samples treated with BODIPY-labeled vancomycin only served as a control. Results are presented as means ± SEM, ***P < 0.001 compared with control. n.s., no significant differences.
FIGURE 3.
S1-Nal and S1-Nal-Nal increase the uptake of BODIPY-labeled vancomycin in A. baumannii. (A) Fluorescence images of 107 CFU/ml A. baumannii BCRC 14B0097 treated with 0.5 × MIC of S1, S1-Nal, and S1-Nal-Nal at 37°C for 30 min, then treated BODIPY-labeled vancomycin for 30 min (scale bar represents 5 μm). (B) Mean fluorescence intensity of S1, S1-Nal, and S1-Nal-Nal treated in A. baumannii BCRC 14B0097. Samples treated with BODIPY-labeled vancomycin only served as a control. Results are presented as means ± SEM, ***P < 0.001 compared with control. n.s., no significant differences.
FIGURE 4.
S1-Nal and S1-Nal-Nal increase the uptake of BODIPY-labeled vancomycin in E. coli. (A) Fluorescence images of 107 CFU/ml E. coli BCRC 13B0207 treated with 0.5 × MIC of S1, S1-Nal, and S1-Nal-Nal at 37°C for 30 min, then treated BODIPY-labeled vancomycin for 30 mins (scale bar represents 5 μm). (B) Mean fluorescence intensity of S1, S1-Nal, and S1-Nal-Nal treated in E. coli BCRC 13B0207. Samples treated with BODIPY-labeled vancomycin only served as a control. Results are presented as means ± SEM, ***P < 0.001 compared with control.
S1-Nal and S1-Nal-Nal Attenuate Vancomycin-Induced Lipopolysaccharide Release
Antibiotic-treatment can cause the release of lipopolysaccharide (LPS, endotoxin) from Gram-negative bacteria into the bloodstream of the host and has been shown to be associated with the deterioration of the patients (Trautmann et al., 1998; Li et al., 2017). Herein, we used LAL assay to measure the concentration of LPS in supernatants induced by the treatment of vancomycin (Figure 5). Escherichia coli BCRC 13B0198 cells were treated with S1, S1-Nal, and S1-Nal-Nal or the combination of vancomycin and the peptides. The LPS concentration decreased tremendously with the S1-Nal, and S1-Nal-Nal treatment alone or the combination of S1-Nal, S1-Nal-Nal, and vancomycin treatment (Figure 5).
FIGURE 5.
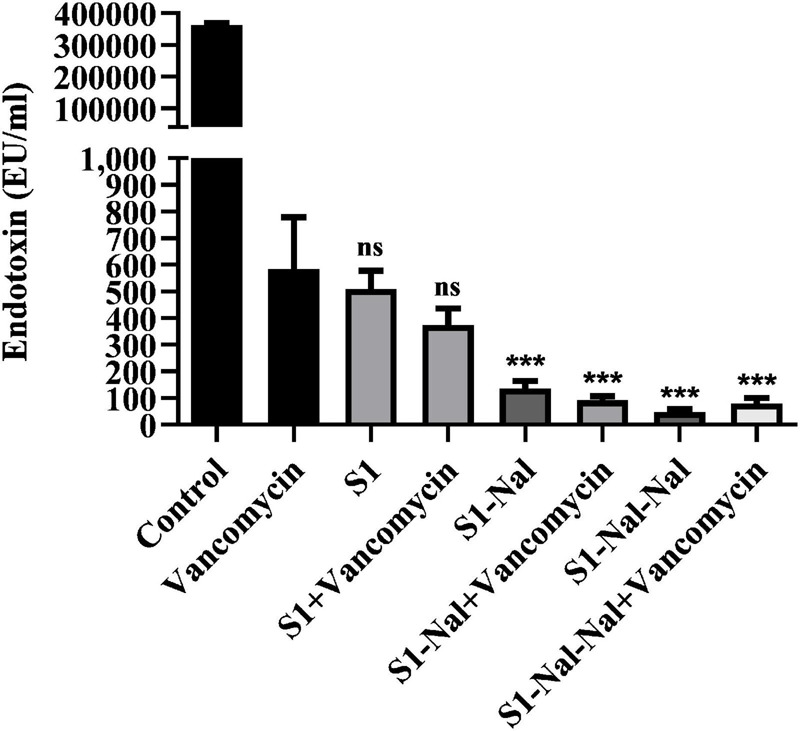
Attenuation of LPS release from E. coli. Escherichia coli BCRC 13B0198 were incubated at the mid-log phase (104 CFU/ml) and treated with peptide alone (at 1 × MIC) or combination with vancomycin (both at 0.5 × MIC) at 37°C for 6 h. The samples were filtered through 0.2 μm pore filter and the endotoxin level was detected by LAL assay. ***P < 0.001 compared with vancomycin only. ns, no significant differences compared with vancomycin only.
Discussion
End-tagging peptides with hydrophobic moieties have been developed to increase salt resistance and potency of AMPs. For example, tryptophan and/or phenylalanine stretch (Schmidtchen et al., 2009), as well as fatty acid, vitamin E, or cholesterol were added to the termini of short AMPs (Avrahami and Shai, 2002; Makovitzki et al., 2006; Serrano et al., 2009; Rosenfeld et al., 2010; Arnusch et al., 2012). This strategy was modified by adding only one or two β-naphthylalanine to the termini of short antimicrobial peptide to boost its salt resistance, serum proteolytic stability and antiendotoxin activities (Chu et al., 2013; Chih et al., 2015). There are several advantages to using β-naphthylalanine than using tryptophan/phenylalanine or fatty acid as end-tags. For example, a stretch of five tryptophan end-tags were needed to provide salt resistance to the antimicrobial peptide KNK10 under 150 mM NaCl (Pasupuleti et al., 2009). Only one β-naphthylalanine end-tag was needed to provide substantial salt resistance (Chu et al., 2013). Furthermore, β-naphthylalanine end-tags can provide superior serum stability due to its non-natural and bulky characteristics (Chu et al., 2013; Chih et al., 2015). This is particularly important for the development of short AMPs to lower the cost of synthesis. In this study, we have extended the β-naphthylalanine end-tagging strategy to boost the synergistic effects of S1-Nal and S1-Nal-Nal with conventional antibiotics against drug-resistant bacteria. Our results indicated that both S1-Nal and S1-Nal-Nal displayed synergistic effects in combination with conventional antibiotics such as vancomycin, ciprofloxacin, and tetracycline against the Gram-positive E. faecium BCRC15B0132 strain. S1-Nal or S1-Nal-Nal also displayed synergistic effects with vancomycin and ciprofloxacin against the Gram-negative A. baumannii BCRC14B0091, A. baumannii BCRC14B0097, A. baumannii BCRC14B0100, E. coli BCRC13B0198, and E. coli BCRC13B0207 strains. However, lack of synergy with tetracycline for all peptides was found against the A. baumannii BCRC14B0100, E. coli BCRC13B0198, and E. coli BCRC13B0207 strains.
Polymyxin B has been used as a potentiator to act synergistically with antibiotics against drug-resistant Gram-negative bacteria (Vaara, 2019). The mechanism of synergism was attributed to the increase of bacterial membrane permeabilization caused by polymyxin B (Vaara, 2019). AMP DP7 shows synergistic effects with the antibiotic, azithromycin, against the antibiotic-resistant strains including S. aureus and P. aeruginosa isolates (Wu et al., 2017). The synergistic effects may be attributed to the cell wall proteins reduced by azithromycin and cell wall disruption by DP7 peptide (Wu et al., 2017). Antimicrobial peptide CLP-19 displayed synergistic effects with antibiotics such as ampicillin, ceftazidime, and levofloxacin against E. coli ATCC 25922 and S. aureus ATCC 29213 (Li et al., 2017). In addition, the CLP-19 and CLP-19/antibiotic combination could induce the generation of hydroxyl radicals from bacteria through depleted NADH, which were related to the synergistic effects of CLP-19/antibiotic combination (Li et al., 2017). As can be seen from Figure 1 in this study, increases of membrane permeabilization of β-naphthylalanine end-tagging against the six bacterial strains studied were found. The results demonstrated that the increase of membrane permeabilization correlated well with synergistic effect with antibiotics against drug-resistant bacterial strains. Moreover, BODIPY-labeled vancomycin was used to study membrane permeabilization and uptake of vancomycin in E. faecium, A. baumannii, and E. coli strains (Figures 2–4). The results indicated that the antimicrobial peptide-induced fluorescent intensity changes of BODIPY-labeled vancomycin inside bacterial cells were concordant with the membrane permeability of the peptides. One interesting thing to note from bacterial membrane permeabilization studies is that β-naphthylalanine end-tags caused higher membrane permeabilization of the Gram-positive bacterial strain (from 60% calcein leakage of S1 to 90% calcein leakage of S1-Nal-Nal) than the Gram-negative bacterial strains (Figure 1). S1-Nal and S1-Nal-Nal both have excellent synergistic effects with vancomycin, ciprofloxacin, and tetracycline against the Gram-positive E. faecium BCRC 15B0132 strain (Table 3).
Lipopolysaccharide constitutes the major component of the outer leaflet of Gram-negative bacteria. It was shown that S1-Nal and S1-Nal-Nal possess high antiendotoxin activities (Chih et al., 2015). Furthermore, S1-Nal and S1-Nal-Nal can inhibit LPS-induced nitrite oxide and TNF-α production in murine macrophage cells and suppress TNF-α release in endotoxemia mouse model (Chih et al., 2015). NMR structural studies of S1-Nal-Nal and LPS micelles complex indicated that S1-Nal-Nal rotated its two terminal β-naphthylalanine residues into the hydrophobic lipid A motif of LPS micelles and blocked the LPS-induced inflammation (Yu et al., 2013). We and others have demonstrated that some AMPs may reduce antibiotic-induced release of LPS from Gram-negative bacteria (Li et al., 2017; Chu et al., 2020). In this study, we have found that S1-Nal and S1-Nal-Nal can reduce antibiotic-induced release of LPS from E. coli (Figure 5).
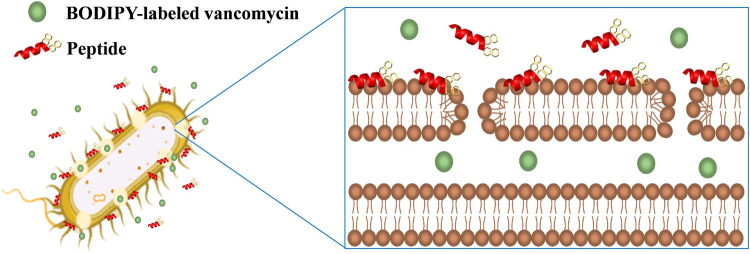
Vancomycin was shown to act synergistically with AMPs against vancomycin-persistent Gram-positive bacterial cells (Feng et al., 2015). However, vancomycin itself has no antibacterial effect against Gram-negative bacteria due to the LPS outer leaflet preventing large glycopeptide antibiotics such as vancomycin from being transported to intracellular targets. Recently, it was found that vancomycin can eradicate some E. coli cells under cold stress conditions through inhibition of peptidoglycan biosynthesis that is similar to the mechanism of action of vancomycin to Gram-positive bacteria (Stokes et al., 2016). Silver ion was also shown to increase membrane permeability of E. coli cells and potentiate vancomycin against E. coli (Morones-Ramirez et al., 2013). In this study, we have also demonstrated that the β-naphthylalanine end-tagged S1-Nal and S-Nal-Nal can be used with vancomycin to fight against Gram-negative bacterial infections (Figure 6).
FIGURE 6.
Schematic illustration of the synergistic mechanism for BODIPY-labeled vancomycin and peptides. S1-Nal-Nal (red) disturbs the bacterial outer membrane and enhances the entry of BODIPY-labeled vancomycin (green circle) into bacteria.
In conclusion, we describe a strategy to boost synergistic effects of short AMPs with conventional antibiotics against resistant bacterial strains by adding β-naphthylalanine (Nal) to one of the peptides’ termini. The described Nal-tagged peptides (S1-Nal and S1-Nal-Nal) enhanced the antibacterial activity of the plain peptides and generated a better synergistic effect when combined with conventional antibiotics. Increasing the membrane permeabilization against the bacterial strains was also observed. Additionally, these Nal-tagged AMPs reduced the antibiotic-induced release of LPS from Gram-negative bacteria by more than 99.95%. Only one or two Nal end-tags were needed to have these biological impacts on the peptides, making this a potential strategy for developing new antimicrobial agents.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
C-LW, Y-HC, and K-LP performed the experiments and analyzed the data. C-LW and J-WC wrote the manuscript. B-SY and J-WC planned the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Daniel Cheng for editing the manuscript.
Footnotes
Funding
This work was supported by grants from the Ministry of Science and Technology of Taiwan to J-WC (109-2113-M-007-009) and National Taiwan University Hospital Hsinchu Branch to B-SY.
References
- Arias M., Piga K. B., Hyndman M. E., Vogel H. J. (2018). Improving the activity of Trp-rich antimicrobial peptides by Arg/Lys substitutions and changing the length of cationic residues. Biomolecules 8:19. 10.3390/biom8020019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnusch C. J., Ulm H., Josten M., Shadkchan Y., Osherov N., Sahl H. G., et al. (2012). Ultrashort peptide bioconjugates are exclusively antifungal agents and synergize with cyclodextrin and amphotericin B. Antimcrob. Agents Chemother. 56 1–9. 10.1128/aac.00468-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami D., Shai Y. (2002). Conjugation of a magainin analogue with lipophilic acids controls hydrophobicity, solution assembly, and cell selectivity. Biochemistry 41 2254–2263. 10.1021/bi011549t [DOI] [PubMed] [Google Scholar]
- Cassone M., Otvos L. J. (2010). Synergy among antibacterial peptides and between peptides and small-molecule antibiotics. Expert Rev. Anti Infect. Ther. 8 703–716. 10.1586/eri.10.38 [DOI] [PubMed] [Google Scholar]
- Chih Y. H., Lin Y. S., Yip B. S., Wei H. J., Chu H. L., Yu H. Y., et al. (2015). Ultrashort antimicrobial peptides with antiendotoxin properties. Antimcrob. Agents Chemother. 59 5052–5056. 10.1128/aac.00519-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih Y. H., Wang S. Y., Yip B. S., Cheng K. T., Hsu S. Y., Wu C. L., et al. (2019). Dependence on size and shape of non-nature amino acids in the enhancement of lipopolysaccharide (LPS) neutralizing activities of antimicrobial peptides. J. Colliod Interface Sci. 533 492–502. 10.1016/j.jcis.2018.08.042 [DOI] [PubMed] [Google Scholar]
- Chu H. L., Chih Y. H., Peng K. L., Wu C. L., Yu H. Y., Cheng D., et al. (2020). Antimicrobial peptides with enhanced salt resistance and antiendotoxin properties. Int. J. Mol. Sci. 21:6810. 10.3390/ijms21186810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. L., Yu H. Y., Yip B. S., Chih Y. H., Liang C. W., Cheng H. T., et al. (2013). Boosting salt resistance of short antimicrobial peptides. Antimcrob. Agents Chemother. 57 4050–4052. 10.1128/aac.00252-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Essodaigui M., Broxterman H. J., Garnier-Suillerot A. (1998). Kinetic analysis of calcein and calcein-acetoxymethylester efflux mediated by the multidrug resistance protein and P-glycoprotein. Biochemistry 37 2243–2250. 10.1021/bi9718043 [DOI] [PubMed] [Google Scholar]
- Feng Q., Huang Y., Chen M., Li G., Chen Y. (2015). Functional synergy of a-helical antimicrobial peptides and traditional antibiotics against Gram-negative and Gram-positive bacteria in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis 34 197–204. 10.1007/s10096-014-2219-3 [DOI] [PubMed] [Google Scholar]
- Godballe T., Mojsoska B., Nielsen H. M., Jenssen H. (2016). Antimicrobial activity of GN peptides and their mode of action. Biopolymers 106 172–183. 10.1002/bip.22796 [DOI] [PubMed] [Google Scholar]
- Joshi S., Bisht G. S., Rawat D. S., Kumar A., Kumar R., Maiti S., et al. (2010). Interaction studies of novel cell selective antimicrobial peptides with model membranes and E coli ATCC 11775. Biochim. Biophys. Acta 1798 1864–1875. 10.1016/j.bbamem.2010.06.016 [DOI] [PubMed] [Google Scholar]
- Koo S. P., Bayer A. S., Yeaman M. R. (2001). Diversity in antistaphylococcal mechanisms among membrane-targeting antimicrobial peptides. Infect. Immun. 69 4916–4922. 10.1128/iai.69.8.4916-4922.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar V., Martins A., Spohn R., Daruka L., Grezal G., Fekete G., et al. (2018). Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3 718–731. 10.1038/s41564-018-0164-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Yang Y., Tian Z., Lv J., Sun F., Wang Q., et al. (2017). Synergistic antibiotic effect of looped antimicrobial peptide CLP-19 with bactericidal and bacteriostatic agents. Oncotarget 8 55958–55966. 10.18632/oncotarget.18124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Fernandez-Millan P., Boix E. (2020). Synergism between host defence peptides and antibiotics against bacterial infections. Curr. Top. Med. Chem. 20 1238–1263. 10.2174/1568026620666200303122626 [DOI] [PubMed] [Google Scholar]
- Makovitzki A., Avrahami D., Shai Y. (2006). Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. U.S.A. 103 15997–16002. 10.1073/pnas.0606129103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston H. D., Dixon D. M., Knisely J. M., Palmore T. N., Fauci A. S. (2016). Antimicrobial resistance. JAMA 316 1193–1204. [DOI] [PubMed] [Google Scholar]
- Martinez M., Goncalves S., Felicio M. R., Maturana P., Santos N. C., Semorile L., et al. (2019). Synergistic and antibiofilm activity of the antimicrobial peptide P5 against carbapenem-resistant Pseudomonas aeruginosa. Biochim. Biophys. Acta Biomembr. 1861 1329–1337. 10.1016/j.bbamem.2019.05.008 [DOI] [PubMed] [Google Scholar]
- Mataraci E., Dosler S. (2012). In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimcrob. Agents Chemother. 56 6366–6371. 10.1128/aac.01180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M. F., Abdelkhalek A., Seleem M. N. (2016). Evaluation of short synthetic antimicrobial peptides for treatment of drug-resistant and intracellular Staphylococcus aureus. Sci. Rep. 6:29707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojsoska B., Jenssen H. (2015). Peptides and peptidomimetics for antimicrobial drug design. Pharmaceuticals (Basel) 8 366–415. 10.3390/ph8030366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morones-Ramirez J. R., Winkler J. A., Spina C. S., Collins J. J. (2013). Silver enhances antibiotics activity against gram-negative bacteria. Sci. Transl. Med. 5:190ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F. C. (2003). Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52:1. [DOI] [PubMed] [Google Scholar]
- Pasupuleti M., Chalupka A., Morgelin M., Schmidtchen A., Malmsten M. (2009). Tryptophan end-tagging of antimicrobial peptides for increased potency against Pseudomonas aeruginosa. Biochim. Biophys. Acta 1790 800–808. 10.1016/j.bbagen.2009.03.029 [DOI] [PubMed] [Google Scholar]
- Rosenfeld Y., Lev N., Shai Y. (2010). Effect of the hydrophobicity to net positive charge ratio on antibacterial and anti-endotoxin activities of structurally similar antimicrobial peptides. Biochemistry 49 853–861. 10.1021/bi900724x [DOI] [PubMed] [Google Scholar]
- Schmidtchen A., Pasupuleti M., Morgelin M., Davoudi M., Alenfall J., Chalupka A., et al. (2009). Boosting antimicrobial peptides by hydrophobic oligopeptide end tags. J. Biol. Chem. 284 17584–17594. 10.1074/jbc.m109.011650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtchen A., Ringstad L., Kasetty G., Mizuno H., Rutland M. W., Malmsten M. (2011). Membrane selectivity by W-tagging of antimicrobial peptides. Biochim. Biophys. Acta 1808 1081–1091. 10.1016/j.bbamem.2010.12.020 [DOI] [PubMed] [Google Scholar]
- Serrano G. N., Zhanel G. G., Schweizer F. (2009). Antibacterial activity of ultrashort cationic lipo-b-Peptides. Antimcrob. Agents Chemother. 53 2215–2217. 10.1128/aac.01100-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra J. M., Fuste E., Rabanal F., Vinuesa T., Vinas M. (2017). An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 17 663–676. 10.1080/14712598.2017.1315402 [DOI] [PubMed] [Google Scholar]
- Song J., Wang J., Zhan N., Sun T., Yu W., Zhang L., et al. (2019). Therapeutic potential of Trp-Rich engineered amphiphiles by single hydrophobic amino acid end-tagging. ACS Appl. Mater. Interfaces 11 43820–43834. 10.1021/acsami.9b12706 [DOI] [PubMed] [Google Scholar]
- Stokes J. M., French S., Ovchinnikova O. G., Bouwman C., Whitfield C., Brown E. D. (2016). Cold stress makes Escherichia coli susceptible to glycopeptide antibiotics by altering outer membrane integrity. Cell Chem. Biol. 23 267–277. 10.1016/j.chembiol.2015.12.011 [DOI] [PubMed] [Google Scholar]
- Trautmann M., Heinemann M., Zick R., Moricke A., Seidelmann M., Berger D. (1998). Antibacterial activity of meropenem against Pseudomonas aeruginosa, including antibiotic-induced morphological changes and endotoxin-liberating effects. Eur. J. Clin. Microbiol. Infect. Dis 17 754–760. 10.1007/s100960050180 [DOI] [PubMed] [Google Scholar]
- Vaara M. (2019). Polymyxin derivatives that sensitize gram-negative bacteria to other antibiotics. Molecules 24:249. 10.3390/molecules24020249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. L., Hsueh J. Y., Yip B. S., Chih Y. H., Peng K. L., Cheng J. W. (2020). Antimicrobial peptides display strong synergy with vancomycin against vancomycin-reisstant E. faecium, S. aureus, and wild-type E. coli. Int. J. Mol. Sci. 21:4578. 10.3390/ijms21134578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li Z., Li X., Tian Y., Fan Y., Yu C., et al. (2017). Synergistic effects of antimicrobial peptide DP7 combined with antibiotics against multidrug-resistant bacteria. Drug Des. Devel. Ther. 11 939–946. 10.2147/dddt.s107195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y. Q., Mukhopadhyay K., Yeaman M. R., Adler-Moore J., Bayer A. S. (2005). Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob. Agents Chemother. 49 3114–3121. 10.1128/aac.49.8.3114-3121.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Y., Chen Y. A., Yip B. S., Wang S. Y., Wei H. J., Chih Y. H., et al. (2017). Role of b-naphthylalanine end-tags in the enhancement of antiendotoxin activities: solution structure of the antimicrobial peptide S1-Nal-Nal in complex with lipopolysaccharide. Biochim. Biophys. Acta 1859 1114–1123. 10.1016/j.bbamem.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Yu H. Y., Huang K. C., Yip B. S., Tu C. H., Chen H. L., Cheng H. T., et al. (2010). Rational design of tryptophan-rich antimicrobial peptides with enhanced antimicrobial activities and specificities. Chembiochem 11 2273–2282. 10.1002/cbic.201000372 [DOI] [PubMed] [Google Scholar]
- Yu H. Y., Yip B. S., Tu C. H., Chen H. L., Chu H. L., Chih Y. H., et al. (2013). Correlations between membrane immersion depth, orientation, and salt-resistance of tryptophan-rich antimicrobial peptides. Biochim. Biophys. Acta 1828 2720–2728. [DOI] [PubMed] [Google Scholar]
- Zharkova M. S., Orlov D. S., Golubeva O. Y., Chakchir O. B., Eliseev I. E., Grinchuk T. M., et al. (2019). Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics-a novel way to combat antibiotic resistance. Front. Cell. Infect. Microbiol. 9:128. 10.3389/fcimb.2019.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.