Abstract
SWI-SNF is an ATP-dependent chromatin remodeling complex that disrupts DNA-histone interactions. Several studies of SWI-SNF activity on mononucleosome substrates have suggested that remodeling leads to novel, accessible nucleosomes which persist in the absence of continuous ATP hydrolysis. In contrast, we have reported that SWI-SNF-dependent remodeling of nucleosomal arrays is rapidly reversed after removal of ATP. One possibility is that these contrasting results are due to the different assays used; alternatively, the lability of the SWI-SNF-remodeled state might be different on mononucleosomes versus nucleosomal arrays. To investigate these possibilities, we use a coupled SWI-SNF remodeling–restriction enzyme assay to directly compare the remodeling of mononucleosome and nucleosomal array substrates. We find that SWI-SNF action causes a mobilization of histone octamers for both the mononucleosome and nucleosomal array substrates, and these changes in nucleosome positioning persist in the absence of continued ATP hydrolysis or SWI-SNF binding. In the case of mononucleosomes, the histone octamers accumulate at the DNA ends even in the presence of continued ATP hydrolysis. On nucleosomal arrays, SWI-SNF and ATP lead to a more dynamic state where nucleosomes appear to be constantly redistributed and restriction enzyme sites throughout the array have increased accessibility. This random positioning of nucleosomes within the array persists after removal of ATP, but inactivation of SWI-SNF is accompanied by an increased occlusion of many restriction enzyme sites. Our results also indicate that remodeling of mononucleosomes or nucleosomal arrays does not lead to an accumulation of novel nucleosomes that maintain an accessible state in the absence of continuous ATP hydrolysis.
Eukaryotic chromatin has seen a rebirth of intense study over the past few years. Foremost among the biochemical reactions impinging on chromatin structure is ATP-dependent chromatin remodeling, which leads to an enhanced accessibility of nucleosomal DNA (for recent reviews, see references 18, 19, and 48). This reaction plays a key role in the regulation of transcription by RNA polymerase II, and it has been proposed to be a prerequisite for a variety of other cellular processes that require access to the chromatin template (for reviews, see references 35 and 48). In addition to ATP-dependent nucleosome remodeling, multisubunit complexes that can acetylate (12, 31, 39, 41) or methylate (6) histone and nonhistone proteins have the potential to directly modify chromatin structure and function.
A host of ATP-dependent chromatin remodeling complexes have been identified via biochemical fractionation of cell extracts, yeast genetics, or genome database mining (2, 5, 7, 17, 20, 23, 33, 44, 46, 47, 49, 50, 53, 55). A hallmark of these multisubunit complexes is that they contain a member of the SWI2/SNF2 subfamily of DNA-stimulated ATPases. Seventeen members of the SWI2/SNF2 family have been identified in the yeast genome (10, 38), and to date, four of these ATPases have been purified as subunits of distinct chromatin remodeling complexes (SWI-SNF [7]; RSC [5]; ISW1 and ISW2 [47]). Additional ATP-dependent remodeling complexes have been identified in Drosophila (ACF [17], CHRAC [49]), NURF ([4], and brm [33]), humans (hSWI-SNF [20], NURD [44, 53, 55], and RSF [25]), and frogs (Mi-2 [50]). Each of these complexes appears to catalyze a reaction in which the energy of ATP hydrolysis is used to weaken histone-DNA interactions which leads to an increase in nucleosomal DNA accessibility. In the case of the yeast SWI-SNF, Drosophila brm, and human SWI-SNF complexes, this reaction is required for transcriptional regulation of target genes in vivo (18).
Many of the in vitro studies that have focused on the mechanism of ATP-dependent remodeling have used mononucleosome substrates. In these cases, ATP-dependent remodeling is often scored as a disruption of the DNase I digestion pattern of rotationally phased nucleosomal DNA or an enhancement of transcription factor binding to nucleosomal sites. For yeast SWI-SNF, RSC, and human SWI-SNF complexes, the remodeled state of mononucleosomes is stable after removal of ATP or SWI-SNF (13, 16, 27, 42). Furthermore, this persistent disruption of mononucleosome structure is accompanied by formation of a novel, stably remodeled species that resembles a dinucleosome and retains a full complement of histones and DNA (27, 42; reviewed in references 18 and 45). In addition, under some conditions RSC can also generate persistent changes in mononucleosome accessibility by transferring the histone octamer onto acceptor DNA (28).
In contrast to studies with mononucleosomes, ATP-dependent remodeling of nucleosomal array substrates by yeast SWI-SNF and RSC leads to the formation of an unstable remodeled state that requires continuous ATP hydrolysis (24, 25; C. Logie, L. Boyer, and C. L. Peterson, unpublished data). In these studies, remodeling was monitored by quantifying the enhanced kinetics of restriction enzyme digestion of a unique SalI/HincII site located within the central nucleosome of a positioned nucleosomal array. Addition of a remodeling enzyme leads to a 30- to 40-fold increase in digestion kinetics, but removal of ATP results in almost immediate reversal of the SalI/HincII restriction site to the occluded state.
The apparent differences in stability of the remodeled state between mononucleosome and nucleosomal array substrates might be due to the different assays used to detect remodeling events; alternatively, remodeling of mononucleosome substrates may yield novel, stable products that do not occur on nucleosomal arrays. Here we have tested these two possibilities by directly comparing the labilities of remodeled mononucleosomes and nucleosomal arrays by using a quantitative restriction enzyme coupled remodeling assay. Using this assay, we confirm that the remodeling of mononucleosome substrates by yeast SWI-SNF leads to a persistent accessibility of DNA that was previously occluded by a nucleosome, whereas the enhanced restriction enzyme accessibility of nucleosomal arrays appears to be more labile. We then show that the stable, accessible state of mononucleosomes correlates with the SWI-SNF-dependent movement of the histone octamer to the DNA ends. Surprisingly, SWI-SNF and ATP also lead to a randomization of nucleosome positions on the array substrate which persists after removal of ATP. Thus, SWI-SNF remodeling leads to persistent changes in nucleosome positioning on both mononucleosome and nucleosomal array substrates. However, whereas the randomization of nucleosomes within the array persists in the absence of ATP, the enhanced accessibility of nucleosomal restriction enzyme sites does not persist. We propose that SWI-SNF and ATP can establish a dynamic state of continuous nucleosome mobilization only on nucleosomal arrays and that this fluid chromatin state is required for enhanced restriction enzyme accessibility.
MATERIALS AND METHODS
Plasmid constructions.
A partial EcoRI digestion was carried out on pCL7b (24) to release a DNA fragment encompassing five head-to-tail repeats of the 208-bp Lyetechinus variegatus 5S ribosomal DNA (rDNA) nucleosome positioning element. This EcoRI fragment was then cloned into the unique EcoRI site of pCL6 (24), to yield pCL113, where the last repeat bears the unique SalI/HincII site and is flanked by a unique PstI site distal to the NotI site of pBS-SKII (+). To generate pCL114 (which contains a total of seven 5S repeats), a single, blunt-ended, EcoRI 208-bp L. variegatus 5S rDNA nucleosome positioning element was subcloned into the filled-in XbaI site of pCL113 to introduce a wild-type 208-bp rDNA repeat between the NotI site and the modified SalI/HincII site-bearing repeat of pCL113. pCL115 contains a single 5S repeat and was generated by fill-in of the SalI site located in the polylinker of pCL6 so as to leave a unique SalI/HincII site at the predicted dyad axis of the single 5S repeat.
Reagent preparation and nucleosome reconstitutions.
SWI-SNF and histone octamers were purified as described elsewhere (25). Apyrase was from Sigma (A-6410) and was diluted to a concentration of 1 U/μl as described elsewhere (16). Nucleosomal array DNA templates (NotI-EcoRV fragments derived from pCL7b [24], pCL113, or pCL114) were labeled by the Klenow polymerase fill-in reaction using [α-32P]dCTP (6,000 μCi/mmol; Amersham). Linear nucleosomal arrays were reconstituted at a ratio of 1.0 to 1.3 octamers per 5S DNA repeat, and samples were characterized by EcoRI analysis as previously described (25). Circular minichromosomes were reconstituted as for the linear arrays, but in this case 2 μg of negatively supercoiled pCL115 and 2 μg of purified chicken histone octamers were assembled in 100-μl reactions (a histone/DNA ratio of ∼1 octamer/150 bp).
Mono- and dinucleosomes were obtained by digestion of the appropriate labeled nucleosomal arrays with PstI. The PstI digestions were carried out 3 μg of the corresponding labeled nucleosomal array (1.5 × 106 cpm), 100 U of PstI (New England Biolabs), 10 mM NaCl, 5 mM Tris-HCl, 1 mM MgCl2, and 0.1 mM dithiothreitol (DTT) in a final volume of 200 μl. After 2 h at 37°C, the reactions were loaded on top of a 15-ml 10 to 30% linear glycerol gradient containing 1% bovine serum albumin (BSA), 0.2% phenylmethylsulfonyl fluoride, 0.1% Tween 20, 1 mM DTT, 10 mM Tris-HCl (pH 8.0), and 125 mM NaCl. The gradients were centrifuged for 17 h at 28,000 rpm in an SW28 rotor. The gradients were fractionated into 500-μl fractions, the position of the labeled DNA was determined by scintillation counting, and 1/30 of the radioactive fractions (∼104 cpm) was analyzed on 4% native polyacrylamide gels. Mononucleosomes (216 bp) were detected in fractions 9 to 12; dinucleosomes (427 bp) were detected in fractions 14 to 19. Mononucleosomes reconstituted on the 427-bp DNA fragment were detected in fractions 9 to 12 of the dinucleosome gradient. For reconstitution of the 154-bp 5S mononucleosome, an NruI-BamHI fragment (154 bp) from pCL113 was used in a fast salt dilution reconstitution protocol (17). After the reconstitution procedure, mononucleosomes were purified through a 5-ml 5 to 30% linear glycerol gradient, and 150-μl fractions were collected, counted, and analyzed on a 4% native acrylamide gel.
Reaction conditions.
For the coupled SWI-SNF reactions, reconstituted arrays, mononucleosomes, or dinucleosomes (0.3 to 2 nM, final DNA concentration) were mixed with 3 nM SWI-SNF complex and 10 U of restriction enzyme in a buffer containing final concentrations of 125 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10 mM Tris-HCl (pH 7.9), 100 μg of BSA per ml, and 3% glycerol. Where indicated, ATP was added to a final concentration of 1 mM, and 0.5 U of apyrase was added per 50 μl of reaction mixture. The reactions were incubated at 37°C. Under these conditions, removal of ATP by apyrase was complete in <2 min. At the indicated time points, an aliquot of the reaction was vigorously mixed for 10 s with 25 μl of Tris-EDTA and 50 μl of a 1:1 solution of phenol-chloroform. After this extraction, samples were treated with 1 mg of proteinase K per ml for 1 h at 37°C. The purified DNA fragments were resolved either by nondenaturing agarose gel electrophoresis in the presence of ethidium bromide or on 4% native acrylamide gels. For SWI-SNF remodeling reactions containing the circular minichromosomes, DNA topoisomers were resolved on 20-cm 1.75% agarose gels in 40 mM Tris (pH 8.0)–30 mM NaPO4–1 mM EDTA at 40 V for 2 days (13), followed by Southern blotting and probing with pCL115 sequence. The fractions of topoisomers and of cut and uncut DNA were obtained by phosphorimager analysis using the ImageQuant software.
MNase digestion.
Reconstituted arrays at a concentration of 0.8 nM were digested with either 1, 2, or 4 U of micrococcal nuclease (MNase; Sigma) per ml in 50 mM NaCl–10 mM Tris-HCl–2.5 mM MgCl2–0.25 mM CaCl2–1 mM DTT–0.1 mg of BSA per ml at 37°C for 10 min. 32P-labeled DNA was digested under the same conditions at a concentration of 10 pM. For nucleosome protection experiments, 3 nM arrays was digested with 500 U of MNase. Purified DNA was analyzed on a 1% agarose gel or a 4% acrylamide gel (acrylamide-to-bisacrylamide ratio of 30:0.8).
RESULTS
SWI-SNF-dependent enhancement of restriction enzyme accessibility requires the continuous presence of ATP.
To quantify the accessibility of nucleosomal DNA in the context of nucleosomal arrays, we have developed a biochemical assay where nucleosome remodeling activity is coupled to restriction enzyme activity such that remodeling is revealed as an enhancement of restriction enzyme cleavage rates (24). In our previous studies, the central nucleosome of an 11-mer nucleosomal array contained a unique SalI/HincII site located at the predicted dyad axis of symmetry (24–26, 37, 54). Restriction enzyme kinetics are biphasic in this system; the first phase is rapid and reflects the fraction of restriction sites that are not occluded by a nucleosome (due primarily in our assays to nucleosomes that occupy minor translational positions; see Discussion and also references 24 to 26). The second phase is very slow and reflects a dynamic equilibrium between the occluded and open nucleosomal DNA states (Fig. 1B) (24, 37). Addition of SWI-SNF and ATP stimulates the second phase of SalI/HincII digestion 20- to 30-fold, but in contrast to the persistent remodeling of mononucleosome substrates, the enhancement of nucleosomal array digestion by SalI or HincII requires continuous ATP hydrolysis (24, 25).
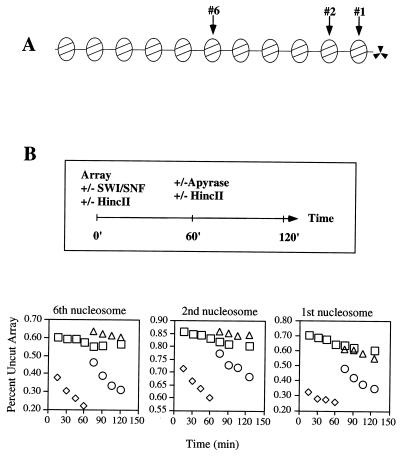
FIG. 1.
Remodeled nucleosomes do not accumulate in the context of linear nucleosomal arrays. (A) Schematic representation of the nucleosomal DNA templates used for the coupled restriction enzyme–SWI-SNF remodeling assay. Each template is composed of head-to-tail repeats of a 5S rDNA nucleosome positioning sequence from L. variegatus. The first, second, or sixth nucleosome is tagged by a unique SalI/HincII restriction site. (B) The nucleosomal arrays were incubated with HincII (□), SWI-SNF (○, ▵), or both (◊). After 1 h, HincII (○) or HincII and apyrase (▵) were added to the reaction to test for accumulation of remodeled template or for ATP dependence, respectively. Cleavage rates were quantified as described in Materials and Methods. Similar results were obtained in three separate experiments.
To investigate whether these kinetics of SWI-SNF remodeling are unique to the central nucleosome of an array, we constructed new DNA templates where the nucleosome positioning sequence marked by the SalI/HincII site was located within the last or second to last position of the array (Fig. 1A). We then reconstituted these new DNA templates into nucleosomal arrays and compared the kinetics of nucleosomal array remodeling by SWI-SNF with the rates of remodeling of our original array template (Fig. 1B). Addition of SWI-SNF to remodeling reactions resulted in a dramatic enhancement of restriction enzyme activity for all three nucleosomal array substrates; preincubation of the arrays with SWI-SNF for 1 h prior to restriction enzyme addition also resulted in enhanced cleavage rates; preincubation of the arrays with SWI-SNF for 1 h followed by coaddition of restriction enzyme and apyrase (to enzymatically remove ATP) resulted in cleavage kinetics that were identical to reactions where SWI-SNF was omitted from the reaction (Fig. 1B). These results indicate that novel, accessible nucleosomes do not accumulate during the preincubation with SWI-SNF and ATP regardless of their positions within the array.
Remodeling of closed circular nucleosomal arrays.
In contrast to our studies with linear nucleosomal arrays, Kingston and colleagues have reported that SWI-SNF-dependent remodeling of closed circular nucleosomal arrays, as assayed by a decrease in the number of constrained negative supercoils, does not require continuous ATP hydrolysis (i.e., a persistent change in minichromosome structure) (13, 16, 42). To test whether the topology of the nucleosomal array influences the stability of the remodeled state, as assayed by restriction enzyme digestion, we reconstituted an average of seven nucleosomes onto a ∼3-kb plasmid DNA template that contains a single 5S nucleosome positioning sequence harboring a unique SalI/HincII site (Fig. 2). This array was then subjected to a remodeling/reversal experiment identical to those displayed in Fig. 1. In the absence of SWI-SNF, we found that 57% of the plasmid was linearized during the 30-min HincII digestion, which reflects the fraction of HincII sites not occluded by histone octamers (Fig. 2B, lane 1). Addition of SWI-SNF and ATP resulted in 90% of the plasmid being cleaved (Fig. 2B, lane 2). However, preincubation of the circular nucleosomal array with SWI-SNF for 30 min, followed by removal of ATP with apyrase and subsequent exposure to HincII, resulted in only 49% cleavage (Fig. 2B, lane 3). Thus, as we observed for linear nucleosomal arrays, the SWI-SNF-dependent stimulation of HincII cleavage of circular nucleosomal arrays requires continuous ATP hydrolysis.
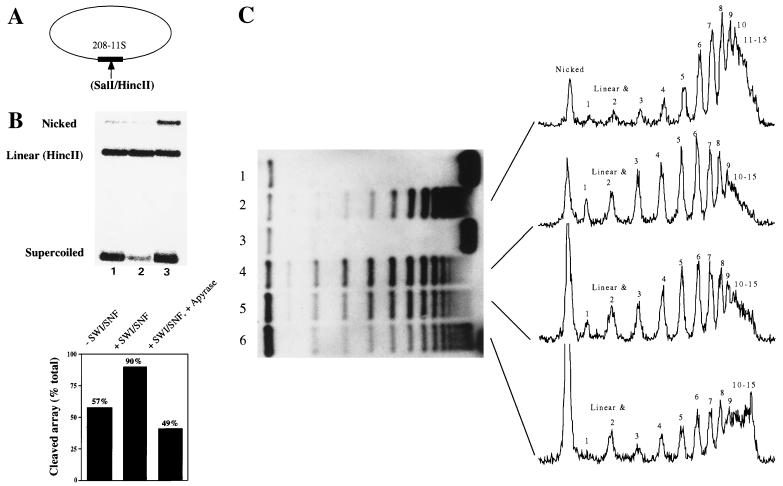
FIG. 2.
Remodeled nucleosomes do not accumulate on closed circular nucleosomal arrays. (A) Nucleosomal arrays were reconstituted on negatively supercoiled closed circular plasmids bearing a unique SalI/HincII site on a single 5S nucleosome positioning sequence. (B) The closed circular arrays were subjected to HincII digestion for 30 min followed by a 30-min incubation with (lane 2) or without (lane 1) SWI-SNF. Alternatively, the arrays were incubated for 30 min with SWI-SNF, followed by removal of ATP with apyrase and digestion with HincII for 30 min (lane 3). (C) A supercoiling assay was performed on the closed circular nucleosomal arrays (lane 1) in order to detect remodeling as changes in plasmid linking number. Addition of calf thymus topoisomerase I resulted in the appearance of approximately 15 bands after deproteination and agarose gel electrophoresis (lane 2). Incubation of the arrays with SWI-SNF and ATP for 30 min had no effect on topology (lane 3). Addition of SWI-SNF, ATP, and topoisomerase I resulted in a redistribution of the topoisomers (lane 4). Removal of ATP by addition of apyrase after 30 min of incubation in the presence of SWI-SNF plus topoisomerase did not affect the distribution of topoisomers (lane 5). Incubation of the arrays with SWI-SNF and ATP for 30 min followed by apyrase and then topoisomerase resulted in a topoisomer distribution similar to that in lane 2 (lane 6). Quantification of the autoradiograph indicates that for lanes 2 and 6, the predominant band corresponds to a linking number of 8; for lanes 4 and 5, the predominant band corresponds to a linking number of 6. Note that apyrase treatment of the circular array results in an increase in the nicked plasmid form (B, lane 3; C, lanes 5 and 6). Apyrase-induced plasmid nicking does not affect HincII activity or the relative distribution of topoisomers upon topoisomerase I treatment of the array (data not shown).
Previous studies with circular nucleosomal array substrates detected a persistent decrease in plasmid linking number due to a combined incubation of the arrays with SWI-SNF, ATP, and topoisomerase I (13, 16, 42). One possibility is that SWI-SNF-dependent changes in the topology of minichromosomes are persistent, whereas the enhanced accessibility of nucleosomal DNA within a single nucleosome requires continuous ATP hydrolysis. Alternatively, inclusion of topoisomerase I during the remodeling reaction may trap the remodeled state irreversibly (e.g., by inducing nucleosome loss). To investigate this latter possibility, we monitored the changes in minichromosome topology under conditions where we could temporally separate topoisomerase I and SWI-SNF activities. Figure 2C shows that addition of calf thymus topoisomerase I to our reconstituted minichromosomes resulted in the appearance of ∼15 discernible topoisomers after deproteination and agarose gel electrophoresis (Fig. 2C, lane 2). Quantification of the intensity of the topoisomers indicated that an average of seven nucleosomes had been reconstituted onto this 3.2-kb plasmid (peak topoisomer has a linking number of 8 [about 50% saturation]). This degree of saturation correlates well with the percentage of nucleosomal HincII sites (Fig. 2B). Incubation of the minichromosome with topoisomerase I, SWI-SNF, and ATP resulted in a redistribution of the topoisomers corresponding to a loss of about two constrained supercoils per plasmid (Fig. 2C, lane 4; the peak topoisomer has a linking number of 6). When apyrase was added after the incubation with SWI-SNF and topoisomerase I, the pattern of topoisomers was not greatly altered, indicating that the effect of SWI-SNF action was persistent in this assay as previously observed (Fig. 2C, lane 5) (13, 16, 42). However, if the minichromosomes were incubated for 30 min with SWI-SNF and ATP, and topoisomerase I was added after treatment of the SWI-SNF reaction with apyrase, then the topoisomer distribution became similar to that in the control reactions that lacked SWI-SNF (peak topoisomer has a linking number of 8) (Fig. 2C, lane 6). In fact, there is an increase in the proportion of topoisomers that appear to contain 11 to 15 constrained supercoils, which may reflect topoisomerase I-induced DNA knotting of stable SWI-SNF-nicked array DNA complexes (Fig. 2C, lane 6; see reference 9). Importantly, apyrase does not inhibit topoisomerase I activity (data not shown). We conclude that only the topoisomerase I-relaxed and remodeled state has a stable change in topology after removal of ATP. In contrast, in the absence of topoisomerase I, SWI-SNF-induced changes in minichromosome topology rapidly collapse (reverse) after removal of ATP.
Persistent remodeling of isolated di- and mononucleosomes.
In contrast to nucleosomal arrays, several studies have reported persistent alterations in mononucleosome accessibility due to SWI-SNF action (13, 16, 27, 42). Moreover, novel, “stably remodeled species” that resemble dinucleosomes were formed from mononucleosome substrates (27, 42). In light of the above results, we wished to study remodeling on mono- and dinucleosomes, using the coupled remodeling-restriction enzyme assay to test whether it could also detect a stable remodeled species. To obtain pure di- and mononucleosomes, we engineered nucleosomal array DNA templates where a PstI site flanks the ultimate or the penultimate (SalI/HincII-tagged) nucleosome positioning sequence (Fig. 3A and data not shown). Nucleosomal arrays were reconstituted on these templates and digested with PstI, and the products of the restriction reaction were fractionated on glycerol gradients to isolate homogeneous 216-bp mono- or 427-bp dinucleosome particles.
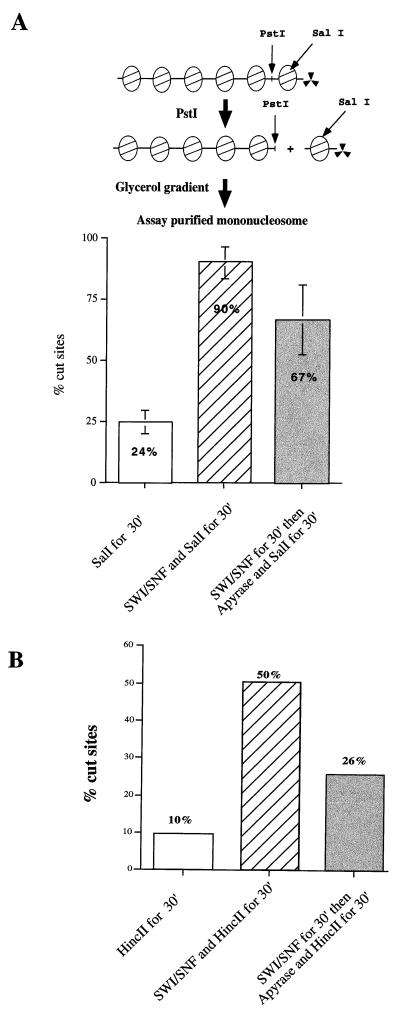
FIG. 3.
SWI-SNF remodeling of isolated mononucleosomes leads to persistent DNA accessibility. (A) Purified mononucleosomes, obtained following PstI digestion of a 6-mer nucleosomal array, were treated with SWI-SNF, SalI, and/or apyrase as indicated. (B) Mononucleosomes assembled on a 154-bp DNA fragment were used as substrates in the same assays as panel A. Similar results were obtained in two additional, independent experiments.
Isolated mono- or dinucleosomes were used as substrates in restriction enzyme digests in the presence or absence of SWI-SNF. In the absence of SWI-SNF complex, 24% of the mononucleosomes were rapidly cleaved by SalI, indicating that this population contains accessible SalI sites (Fig. 3A). The remaining 76% of the mononucleosomes were cleaved with the slow digestion kinetics diagnostic of nucleosomal SalI sites (data not shown). In contrast, when SWI-SNF and ATP were included in the reaction, 90% of the mononucleosomes were cleaved by SalI during a 30-min incubation. No stimulation of mononucleosome digestion was observed if ATP was omitted from the reactions (data not shown). In contrast, preincubation of the mononucleosomes with SWI-SNF and ATP, followed by coaddition of apyrase and SalI, resulted in 67% cleavage of the mononucleosomes (Fig. 3A). Identical results were obtained when, after the preincubation step, the binding of SWI-SNF to the mononucleosome substrate was competed for by addition of a 20-fold excess of chicken erythrocyte oligonucleosomes (data not shown). Similar results were also obtained with the dinucleosome substrates (data not shown). Thus, in contrast to our results with nucleosomal array substrates (Fig. 1 and 2), the majority of the SWI-SNF-dependent enhancement of mono- or dinucleosome accessibility, as assayed by SalI digestion, does not require continuous ATP hydrolysis.
Previous studies have used mononucleosome substrates assembled onto short DNA fragments (e.g., 154 bp [7, 16]), and thus we wished to confirm that our restriction enzyme assay would also be able to detect persistent remodeling of these types of mononucleosome substrates. To address this issue, we reconstituted 154-bp mononucleosomes using the reconstitution method described by Imbalzano et al. (16) (see Materials and Methods; Fig. 3B). In this case only 10% of the HincII sites were cleaved after 30 min of digestion in the absence of SWI-SNF. Incubation of these 154-bp mononucleosomes with SWI-SNF and ATP led to 50% cleavage after the 30-min incubation. Finally, if the mononucleosomes were preincubated with SWI-SNF and ATP, and then apyrase and HincII were added, 26% of the sites were cleaved (Fig. 3B). Although the remodeled state of the 154-bp mononucleosomes was clearly less stable than for the 216-bp mononucleosomes, much of the stimulation of HincII digestion due to SWI-SNF action was persistent in the absence of continuous ATP hydrolysis.
SWI-SNF remodeling of mononucleosomes results in a protection of the ends of the DNA.
Our data indicate that SWI-SNF-dependent remodeling of mononucleosomes leads to a more stable, accessible reaction product than does SWI-SNF remodeling of nucleosomal arrays. One possibility that we considered is that SWI-SNF action might lead to translational movements of histone octamers. A prediction of this hypothesis is that the stable, increased accessibility of the HincII site should be accompanied by an increased protection of DNA elsewhere on the template. To test this idea, we digested a 216-bp mononucleosome with two additional restriction enzymes, BamHI and NcoI, whose sites are located 9 and 23 bp, respectively, from the ends of the DNA template (Fig. 4A). In the absence of SWI-SNF, we find that 65 to 90% of the NcoI and BamHI sites were freely accessible, consistent with the known preferred position of the 5S nucleosome (Fig. 4B). Upon incubation of the mononucleosomes with SWI-SNF for 30 min, followed by addition of apyrase, we observe an increased protection of the NcoI and BamHI sites (25 and 50% increase in proportion of occluded sites, respectively [Fig. 4C]). This contrasts with the HincII sites, of which 25% were rendered more accessible by SWI-SNF action and subsequent removal of ATP (Fig. 3 and 4B). Similar results were observed if the binding of SWI-SNF to the mononucleosome substrate was competed for by addition of a 20-fold excess of chicken erythrocyte oligonucleosomes (data not shown). These experiments strongly imply that SWI-SNF remodeling can result in altered translational positioning of histone octamers on DNA. Furthermore, the dramatic increase in protected restriction sites near the ends of the DNA fragment indicate that DNA ends may act as sinks where SWI-SNF remodeled nucleosomes preferentially accumulate.
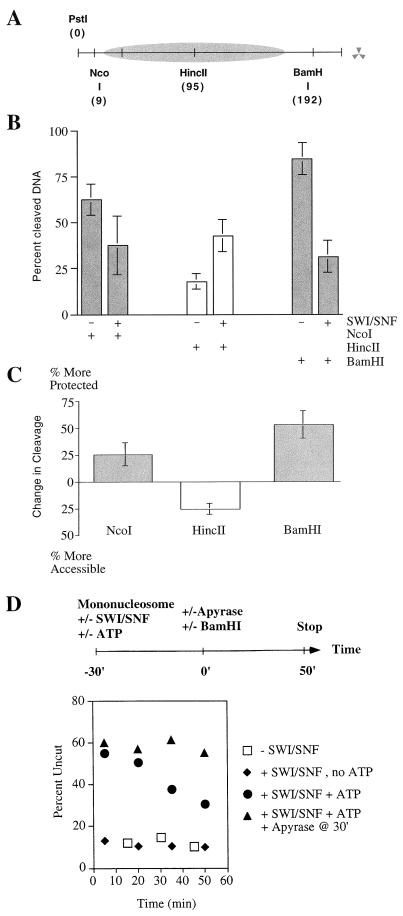
FIG. 4.
SWI-SNF action leads to increased protection of DNA ends. (A) Schematic of the 216-bp mononucleosome substrate. (B) The ends of the DNA fragment are more protected after a 30-min incubation with SWI-SNF. The graph represents the percentage of cleaved nucleosomal DNA after restriction enzyme digestion without SWI-SNF (−) or after a 30-min incubation with SWI-SNF followed by removal of ATP with apyrase (+). Error bars represent the standard deviation from at least three experiments. (C) Difference in percent nucleosomal DNA cleaved by the restriction enzyme in the absence (−) and presence (+) of SWI-SNF (see panel B). (D) Time course of BamHI DNA cleavage. Mononucleosomes were preincubated for 30 min in the absence of SWI-SNF (□), in the presence of SWI-SNF without ATP (⧫), or in the presence of SWI-SNF and ATP (●, ▴). Reactions containing SWI-SNF and ATP were then incubated with (▴) or without (●) apyrase, BamHI was added to all reactions, and the amount of cleavage was determined throughout a 50-min time course.
To further investigate the SWI-SNF-dependent protection of mononucleosomal DNA, we determined the kinetics of BamHI cleavage in the presence or absence of SWI-SNF and in the presence or absence of continued ATP hydrolysis. In the absence of SWI-SNF, 72% of the BamHI sites were rapidly cleaved, and only 18% of the BamHI sites were cleaved at a rate diagnostic of nucleosomal DNA (Fig. 4D). Thus, the majority of histone octamers do not appear to be positioned over the BamHI site. After a 30-min incubation with SWI-SNF and ATP, 60% of the mononucleosomes were more resistant to subsequent BamHI cleavage, reflecting the possible movement of the histone octamer (time zero; Fig. 4D). If apyrase was added after the 30-min preincubation to remove ATP, mononucleosomes were digested by BamHI at the low rate that is diagnostic of nucleosomal DNA; in contrast, if ATP hydrolysis was allowed to continue, SWI-SNF was able to remodel the newly occluded BamHI sites as assayed by the increased kinetics of BamHI digestion (Fig. 4D). These results support our view that SWI-SNF action can lead to movement of histone octamers to the DNA ends, and furthermore that these remodeled nucleosomes represent cannonical nucleosomes that can inhibit the accessibility of DNA as well as serve as new substrates for SWI-SNF remodeling.
SWI-SNF action alters the translational positioning of mononucleosomes.
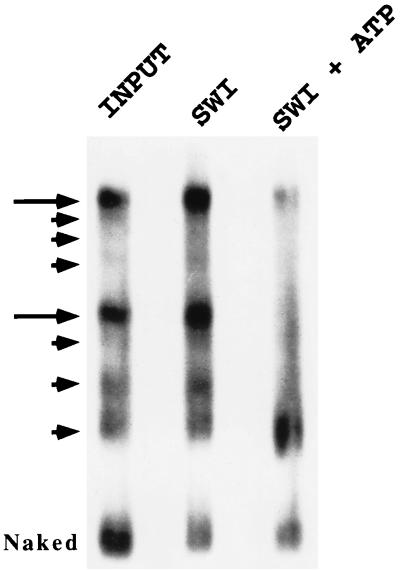
To verify that the persistent aspect of SWI-SNF-mediated mononucleosome remodeling is due to a repositioning of histone octamers, we used a nucleosome mobility assay where populations of mononucleosomes with heterogeneous translational positions are resolved by virtue of their different electrophoretic properties on native polyacrylamide gels (29). First, we used the strategy outlined in Fig. 3B to isolate mononucleosomes reconstituted onto a 427-bp DNA fragment that contains two 5S nucleosome positioning sequences. This substrate is nearly identical to the 416-bp mononucleosome described by Meersseman et al. (29); as this group previously observed (29), these purified mononucleosomes resolve into eight electrophoretically distinct species on 4% native acrylamide gels (Fig. 5, lane 1). Two of the species make up ∼60% of the total population (Fig. 5, lane 1), and the fastest-migrating species represent octamers located close to the DNA ends (29). In the absence of ATP, exposure to SWI-SNF had no effect on the distribution of species (Fig. 5, lane 2). In contrast, addition of SWI-SNF and ATP resulted in a quantitative switch of >90% of the total population to the faster-migrating species (Fig. 5, lane 3). Incubation of these mononucleosomes with SWI-SNF and ATP did not increase the amount of free DNA (Fig. 5, compare lanes 2 and 3), and these remodeled mononucleosomes still cosedimented in glycerol gradients with mononucleosomes that had not been remodeled (data not shown). Furthermore, prolonged incubation of the mononucleosomes (up to 2 h) resulted in the same pattern (data not shown), suggesting that the reaction rapidly reaches an equilibrium where the distribution of nucleosome translational positions is strongly biased towards histone octamers occupying end positions.
FIG. 5.
SWI-SNF action alters the translational positioning of a mononucleosome. Mononucleosomes reconstituted onto a 416-bp DNA fragment that contains two copies of a 5S nucleosome positioning sequence were electrophoresed on a 4% native polyacrylamide gel for 8 h. Arrows denote eight electrophoretically distinct particles that reflect different nucleosome translational positions. Mononucleosomes were incubated for 30 min at 37°C in the absence of SWI-SNF (lane 1), in the presence of SWI-SNF but in the absence of ATP (lane 2), and in the presence of both SWI-SNF and ATP (lane 3). Prior to loading of samples, all three reactions received a 100-fold molar excess of unlabeled chicken oligonucleosomes to compete for binding of SWI-SNF to the mononucleosome substrate. Note that SWI-SNF and ATP shifts the distribution of mononucleosome particles to the faster-migrating species without leading to an increase in the amount of free, naked DNA. Similar results were obtained in at least three additional experiments.
SWI-SNF action mobilizes histone octamers within positioned nucleosomal arrays.
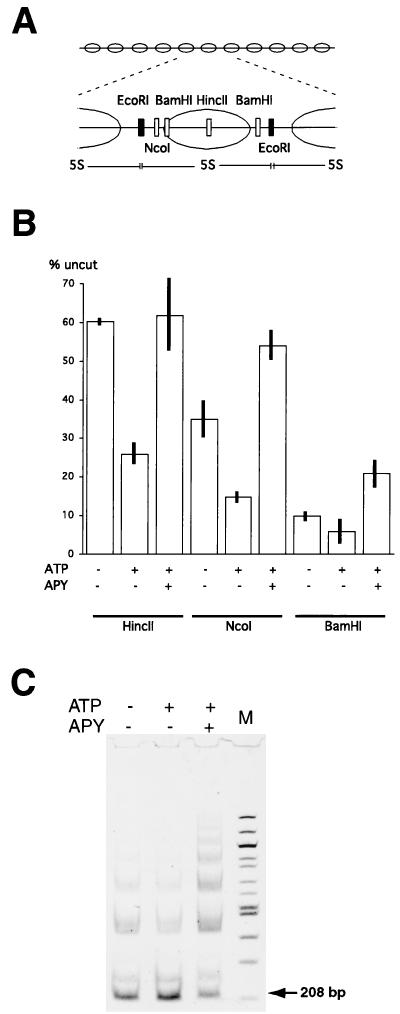
The observed difference in the stability of the enhanced restriction enzyme accessibility for mononucleosome and nucleosomal array substrates might be due to an inhibition of nucleosome mobility within an array context. In this case, the remodeled state on arrays may represent novel, accessible nucleosomal structures whose maintenance requires continuous ATP hydrolysis. On the other hand, the remodeling of nucleosomal arrays might also reflect the ATP-dependent movement of histone octamers. In this case, continuous ATP hydrolysis might be needed to maintain changes in nucleosome positions that create an accessible HincII/SalI site. To test this latter possibility, we first monitored the accessibility of the BamHI and NcoI restriction sites located adjacent to the SalI/HincII-marked central nucleosome (Fig. 6A). In the absence of SWI-SNF, 65% of the NcoI sites and 90% of the BamHI sites were accessible to restriction enzyme (Fig. 6B), which is consistent with the major translational frame of the 5S nucleosome and is similar to what we observed for the 216-bp 5S mononucleosome (Fig. 4). However, in contrast to our results with the mononucleosome substrates, addition of SWI-SNF and ATP led to a further increase in the accessibility of NcoI and BamHI sites (Fig. 6B). If the nucleosomal arrays were preincubated with SWI-SNF and ATP, and then apyrase was added with the restriction enzyme, an increased occlusion of the BamHI and NcoI sites (25% change for NcoI and 15% change for BamHI [Fig. 6B]) resulted. These results suggest that the enhanced accessibility of the HincII, BamHI, and NcoI sites requires continuous ATP hydrolysis, and furthermore, as we observed for mononucleosome substrates, that SWI-SNF may stably alter nucleosome positioning within the array.
FIG. 6.
SWI-SNF-dependent remodeling changes nucleosome positions within reconstituted arrays. (A) Schematic representation of the DNA template showing positions of restriction endonuclease sites unique to the central nucleosome (open bars). EcoRI sites (filled bars) are located in the linker region between nucleosomes in every 5S repeat. (B) Change in the accessibility of restriction endonuclease sites at the nucleosome dyad (HincII) and at the linker region (NcoI and BamHI) following SWI-SNF reaction. The bars represent percentages of uncut arrays after restriction endonuclease cleavage of reconstituted arrays in the presence of ATP and/or SWI-SNF as indicated. In apyrase (APY) experiments, the SWI-SNF reaction was stopped with apyrase and restriction enzyme was then added. Error bars represent the standard deviation from at least three experiments. (C) SWI-SNF reaction results in a decrease in accessibility of linker regions throughout nucleosomal arrays. The EcoRI digestion of reconstituted nucleosomal arrays was carried out as in panel B; DNA was isolated and analyzed in a 4% acrylamide gel. Gel was stained with Vistra Green (Amersham) and scanned with a Molecular Dynamics Storm scanner. The limit digestion product (208-bp 5S DNA) is marked with an arrow. The 232-bp product represents the HincII/SalI-marked nucleosome. Larger products represent partial digestion products. Lane M contains a 100-bp DNA ladder (New England Biolabs).
To investigate the accessibility of DNA adjacent to every nucleosome within the array, we analyzed the accessibility of the EcoRI sites that are located between each of the 11 5S DNA repeats (Fig. 6A). In the absence of SWI-SNF, EcoRI digestion of the nucleosomal array yields primarily the 208-bp 5S DNA limit product, as well as some di- and trinucleosome-size partial digestion products (Fig. 6C). This result is consistent with the majority of histone octamers occupying positions between EcoRI sites. When arrays are incubated with SWI-SNF and ATP, we observe a small increase in EcoRI cleavage which is best visualized by a decrease in the amount of di- and tri-nucleosome sized DNA fragments (Fig. 6C). However, when arrays are preincubated with SWI-SNF and ATP for 30 min, followed by addition of apyrase and EcoRI, accessibility of the EcoRI sites is dramatically reduced as visualized by a decrease in the amount of 208-bp DNA product and a large increase in the amount of partial digestion products (Fig. 6C). This SWI-SNF-dependent decrease in DNA accessibility following removal of ATP is similar to the changes in NcoI and BamHI accessibility at the central nucleosome and are consistent with the hypothesis that SWI-SNF might alter nucleosome positioning within the array.
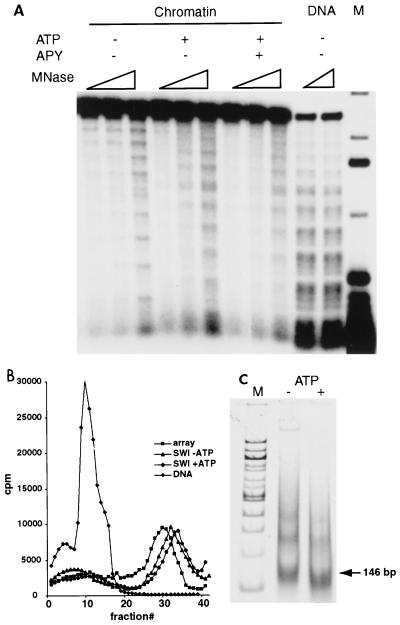
To further investigate persistent changes in nucleosome positioning within the array due to SWI-SNF action, we performed partial MNase digestions. MNase cleavages occur only within the linker regions between nucleosomes; thus, in the absence of SWI-SNF, MNase digestion reveals a repeating pattern of cleavages and protections indicative of a positioned array of 11 nucleosomes (Fig. 7A). Addition of SWI-SNF and ATP disrupts the positioned nucleosomal array, yielding digestion products that are nearly identical to those of the unassembled 5S DNA template (Fig. 7A; note that MNase cleavage of the 5S array DNA yields a repeating pattern of cleavages and protections that is the inverse of the nucleosomal pattern). Furthermore, this disruption of the positioned 5S array is persistent and does not require continuous ATP hydrolysis (Fig. 7A, +APY lanes). This effect of SWI-SNF remodeling represents changes in translational positioning of nucleosomes rather than nucleosome loss, as the remodeled arrays still cosediment with control arrays on glycerol gradients (Fig. 7B), and they still contain nucleosomes that protect ∼150 bp of DNA after extensive digestion with MNase (Fig. 7C). Thus, these results indicate that SWI-SNF remodeling of 5S arrays is associated with a dramatic randomization of nucleosome translational positions. In the presence of SWI-SNF and ATP, this randomized state is associated with an enhanced restriction enzyme accessibility throughout the array. In contrast, the subsequent inactivation of SWI-SNF remodeling activity leads to a randomized array where many restriction enzyme sites are persistently occluded.
FIG. 7.
Persistent randomization of nucleosome positions within reconstituted arrays as a result of SWI-SNF-dependent remodeling. (A) MNase digestion of reconstituted 11-mer arrays. [α-32P]dATP-labeled reconstituted nucleosomal arrays were digested with increasing amounts of MNase in the presence of SWI-SNF and either in the presence or absence of ATP before or after addition of apyrase (APY). DNA was isolated and analyzed on a 1% agarose gel. Lane M, 32P-labeled 1-kb DNA ladder (Gibco BRL). Note that 5S array DNA also exhibits a repeating pattern of MNase cleavages that is the inverse pattern of nucleosomal arrays. (B) SWI-SNF-dependent remodeling does not change the sedimentation properties of nucleosomal arrays. Shown are 10 to 40% linear glycerol gradient profiles of naked DNA and reconstituted nucleosomal arrays either before or after SWI-SNF-dependent remodeling. 32P-labeled reconstituted arrays were incubated in the presence of ATP and/or SWI-SNF for 30 min, loaded on top of the gradient, and centrifuged in an SW28 rotor (Beckman) at 33,000 × g for 16 h. Fractions of 0.4 ml were collected from the top of the gradient and counted by scintillation. (C) Nucleosomes within remodeled arrays protect ∼150 bp of DNA. Nucleosomal arrays were incubated in the presence of SWI-SNF with or without ATP, the reaction was stopped with apyrase, and remodeled arrays were digested with a high concentration of MNase. DNA was isolated, fractionated in a 4% acrylamide gel, and visualized by staining with Vistra Green. The position of the 146-bp nucleosomal DNA is marked with an arrow. Lane M, 100-bp DNA ladder.
DISCUSSION
In this study, we carried out experiments aimed at reconciling two seemingly contradictory experimental observations concerning the persistence of the remodeled nucleosome state induced by yeast SWI-SNF and related ATP-dependent remodeling complexes (8, 13, 16, 24, 27, 42). First, when the remodeling activity of SWI-SNF and RSC was analyzed on mononucleosome substrates, the remodeled, accessible state (detected by restriction enzyme cleavage, transcription factor binding, or DNase I digestion patterns) appeared to persist for over 30 min upon removal of ATP or SWI-SNF from the reaction (8, 13, 16, 27, 42). However, when SWI-SNF, RSC, NURF, CHRAC, or Mi-2 activity was analyzed on nucleosomal array substrates, remodeling (detected by restriction enzyme digestion) seemed to be reversible (24, 32; Logie et al., unpublished data). The experiments presented here suggest that the stable, accessible state that is detected on mononucleosomes is due to SWI-SNF-induced movement of histone octamers to the DNA ends. Furthermore, the apparent reversibility of nucleosomal array remodeling is a misnomer since SWI-SNF action leads to persistent randomization of nucleosome positions within the array. Our data are consistent with a recent study on yeast SWI-SNF by Whitehouse et al. (51) and with two studies which showed that chromatin remodeling complexes that contain the ISWI ATPase can induce nucleosome mobility (14, 21).
Nucleosome mobilization: mononucleosomes versus arrays.
Why does the SWI-SNF-dependent enhancement of restriction enzyme digestion appear to be reversible on nucleosomal array substrates? For each 5S repeat within an array, nucleosomes assume a major translational position that is present in at least 50% of the population. The remaining nucleosomes assume multiple, minor translational positions that differ from the major frame by multiples of 10 bp (34). Thus, the 5S array substrate is a heterogeneous population where 60 to 75% of the arrays contain a central nucleosome positioned at or near the major translational position (containing an occluded SalI/HincII site), and the remaining arrays contain a central nucleosome that occupies several different minor translational frames (characterized by an accessible SalI/HincII site). MNase analysis (Fig. 7) indicates that SWI-SNF and ATP rapidly redistribute these nucleosome positions such that nucleosomes are randomly positioned. In the presence of ATP, this randomized state is also associated with enhanced accessibility of restriction enzyme sites throughout the array. When ATP is subsequently removed from the reaction, nucleosomes remain randomly positioned as determined by MNase analysis, but there is an increased occlusion of restriction enzyme sites compared to arrays incubated with SWI-SNF and ATP. Thus, continual ATP hydrolysis is required to maintain enhanced restriction enzyme accessibility but not to maintain random nucleosome positions. This is in contrast to the 216-bp mononucleosome substrate, where SWI-SNF action leads to an accessible SalI/HincII site that persists after removal of ATP. We propose that SWI-SNF and ATP leads to a state of constant redistribution of nucleosome positions within an array and that this fluid chromatin state favors restriction enzyme accessibility. When ATP is removed from the array assay, the continual redistribution of nucleosomes terminates, and a random pattern of nucleosome positioning is frozen. In this case, the randomized array is associated with a decreased accessibility of restriction enzyme sites throughout the array. For restriction enzyme sites located within the minor translational frames of the starting substrate (EcoRI, NcoI, and BamHI), SWI-SNF action, followed by removal of ATP, leads to increased occlusion of these sites compared to the starting array. Furthermore, in the case of the SalI/HincII site, inactivation of SWI-SNF leads to a level of occlusion that is fortuitously similar to the starting substrate, making the remodeling reaction appear to be reversible. Furthermore, and most important, although SWI-SNF creates changes in nucleosome positions on all substrates which then persist in the absence of continuous ATP hydrolysis, SWI-SNF does not appear to create stable, novel nucleosomes that have enhanced DNA accessibility.
We note that in our studies we have monitored the persistence of the remodeled state for only short time periods (<30 min) following inactivation of SWI-SNF. Previous studies have indicated that the remodeled state of mononucleosomes is at least partially reversible after more extended incubations, and it is expected that nucleosomes will eventually reestablish the preferred translational positions within the 5S arrays (32). Thus, it is not clear whether any feature of the remodeled state is truly stable or if there are differences in the rate of reversibility between remodeled mononucleosomes and nucleosomal arrays.
Role of octamer mobilization in remodeling of 154-bp mononucleosomes.
Given that persistent disruption of mononucleosomal DNA (as assayed by restriction enzyme digestion) appears to correlate with the translational movement of the histone octamer, it seems surprising that SWI-SNF action can lead to a stable disruption of mononucleosomes that are reconstituted on very short DNA fragments (e.g., 154 bp). On these substrates the HincII site is located ∼75 bp from the nucleosomal edge, and thus a movement of the octamer to the end of the DNA fragment will still leave the HincII site buried within the nucleosome. One possibility is that SWI-SNF remodeling of 154-bp mononucleosomes yields novel reaction products that are not generated on nucleosomal arrays or on mononucleosomes with longer stretches of linker DNA. Alternatively, we favor a model in which SWI-SNF induces movement of the histone octamer off the end of the DNA fragment, leading to a histone octamer with less than 147 bp of DNA wrapped onto its surface. This type of reaction may not be favored; consistent with this view, remodeling of the 154-bp mononucleosomes does lead to fewer accessible HincII sites after SWI-SNF inactivation compared to the 216-bp mononucleosome (Fig. 3). Previous studies have shown that reconstitution of histone octamers onto a 145-bp DNA fragment can lead to the preferential assembly of only 128 bp of DNA (40). Furthermore, visualization of SWI-SNF remodeling by electron microscopy indicates that nearly 20 bp of DNA is lost from a remodeled nucleosome (3). If SWI-SNF generates remodeled mononucleosomes that contain a significant number of unoccupied DNA binding sites, then these particles may show a propensity to self-associate via histone-DNA interactions. This model may provide a simple explanation for the previously described novel reaction product that behaves biochemically as a dinucleosome (27, 42).
Is ATP-dependent remodeling equivalent to histone octamer mobilization?
Our results indicate that nucleosome remodeling by SWI-SNF and related enzymes leads to dramatic changes in nucleosome positioning. One simple model posits that remodeling is equivalent to octamer mobilization and that changes in nucleosome positions are responsible for the enhanced binding of transcription factors or activity of restriction enzymes. In this case, the energy of ATP hydrolysis might be used to directly move histone octamers, perhaps by “screwing” DNA over the octamer surface as suggested by Varga-Weisz and Becker (48). Furthermore, in this model the disruption of DNase I digestion patterns might not reflect changes in DNA path around the octamer as previously suggested (8), but disruption might be due to octamer movement and subsequent exposure of nucleosome-free DNA. In this case, disruption of the DNase I ladder would represent a mixture of species, where on average 50% of all sequences would be more or less nucleosome-free.
Alternatively, SWI-SNF-like enzymes might use the energy of ATP hydrolysis to generate a high-energy intermediate where DNA-histone contacts have been disrupted but the translational frame of the histone octamer on DNA has not yet been altered. This activated state may also constrain fewer negative supercoils due to the weakened histone-DNA interactions (see below). A similar activated intermediate consisting of weakened histone-DNA contacts has been proposed for remodeling catalyzed by ISWI-containing complexes (1, 17). This high-energy state might then decay into stable changes in nucleosome positions. In this model, the preferred stable outcome for mononucleosomes appears to be histone octamers at the DNA ends, but for nucleosomal arrays, where DNA ends do not flank each nucleosome, remodeling leads to a more random positioning of nucleosomes. Consistent with this high-energy-intermediate model, human SWI-SNF is still proficient at ATP-dependent remodeling of immobilized substrates which contain histone octamers cross-linked to nucleosomal DNA (22).
Stability of the remodeled state as a function of chromatin topology.
Kingston and colleagues have used a minichromosome topology assay to monitor the persistence of nucleosomal array remodeling by human SWI-SNF complex (13, 16, 42). In this remodeling assay, nucleosomes are reconstituted onto a closed circular DNA template in the presence of topoisomerase I, and these substrates are then incubated with SWI-SNF and ATP in the presence of topoisomerase I. In these reactions, ATP-dependent remodeling by human SWI-SNF results in the loss of a large number of constrained supercoils, and these topological changes persist for several hours after inactivation or removal of the remodeling enzyme. Based on these results, it was proposed that the stability of remodeled minichromosomes (as detected by these changes in topology) reflects a novel remodeled nucleosome species. In contrast, we hypothesized that the persistent nature of the remodeled product detected in the topology assay might be due to the combined incubation of minichromosomes with SWI-SNF and a topoisomerase, rather than a property inherent to the SWI-SNF remodeling reaction. To test this possibility, we designed a remodeling experiment where the activities of SWI-SNF and topoisomerase I were temporally uncoupled (Fig. 2). Whereas coincubation of SWI-SNF and topoisomerase I led to a change in minichromosome topology that persisted in the absence of ATP or SWI-SNF, incubation of the minichromosome with SWI-SNF and ATP, followed by removal of ATP and subsequent addition of topoisomerase I, eliminated the stable change in topology. Thus, persistent changes in minichromosome topology requires the combined action of SWI-SNF and topoisomerase I. Although the nature of this persistent change is not clear, it is possible that topoisomerase I traps a transient change in DNA topology that is induced during the remodeling reaction. In the absence of topoisomerase I, this change in topology would collapse, due either to the artificially high level of supercoiling on the minichromosomes or to an inherent reversibility of the remodeling reaction. Alternatively, the combined action of SWI-SNF and a topoisomerase might lead to more efficient eviction of histone octamers from circular chromatin.
Stability of the remodeled state in vivo.
Two studies have recently addressed the issue of continuous versus transient requirement of the yeast SWI-SNF complex in vivo (4, 43). In both cases, inactivation of SWI-SNF led to the rapid loss of gene expression, indicating that SWI-SNF is continuously required to maintain activated levels of gene expression. Previously it had been proposed that this continuous requirement for SWI-SNF observed in vivo might reflect the activity of other remodeling complexes (such as RSC) that might use the energy of ATP hydrolysis to reestablish a repressive chromatin structure (45). The data presented here, however, imply that there is no a priori need to invoke a balance between positively and negatively acting ATP-dependent remodeling complexes since termination of the nucleosome remodeling reaction leads to reappearance of bona fide, DNA-occluding nucleosomes.
Our data also provide a simple model to explain how SWI-SNF can be required for transcriptional repression of some genes in vivo (15). As we observe here, SWI-SNF might use the energy of ATP hydrolysis to alter the positions of nucleosomes surrounding cis-acting regulatory sites in vivo. In some cases, these movements might enhance DNA accessibility (either by continuous action of SWI-SNF or by placing DNA sites between nucleosomes), but in other instances SWI-SNF action might lead to the occlusion of DNA sites that are required for gene expression. In fact, SWI-SNF action might first act to promote accessibility of DNA to key transcription factors, but inactivation or loss of SWI-SNF from the target gene would help reestablish a repressed state by imposing inhibitory nucleosome positions.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank T. Imbalzano for assistance with the reconstitution of the 154-bp mononucleosomes as well as for insightful comments during the course of this work. We thank P. Becker for communicating his unpublished result that yeast SWI-SNF mobilizes nucleosomes to the ends of DNA fragments.
This work was supported by grant GM49650-07 from the NIH to C.L.P. and a fellowship from the Human Frontiers Science Program Organization to C.L.
REFERENCES
- 1.Alexiadis V, Varga-Weisz P D, Bonte E, Becker P B, Gruss C. In vitro chromatin remodelling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J. 1998;17:3428–3438. doi: 10.1093/emboj/17.12.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 3.Bazett-Jones D P, Cote J, Landel C C, Peterson C L, Workman J L. SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol Cell Biol. 1999;19:1470–1478. doi: 10.1128/mcb.19.2.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggar S R, Crabtree G R. Continuous and widespread roles for the SwiSnf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, Ma H, Hong H, Koh S S, Huang S M, Schurter B T, Aswad D W, Stallcup M R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 7.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF protein complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 8.Côté J, Peterson C L, Workman J L. Perturbation of nucleosome core structure by the SWI/SNF complex persists following its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean F B, Stasiak A, Koller T, Cozzarelli N R. Duplex DNA knots produced by Escherichia coli topoisomerase I. Structure and requirements for formation. J Biol Chem. 1985;260:4975–4983. [PubMed] [Google Scholar]
- 10.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elfring L K, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek S J, Waldrip W R, Daubresse G, DePace A, Kennison J A, Tamkun J W. Genetic analysis of brahma (brm): the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 13.Guyon J R, Narlikar G J, Sif S, Kingston R E. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol Cell Biol. 1999;19:2088–2097. doi: 10.1128/mcb.19.3.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamiche A, Sandaltzopoulos R, Gdula D A, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 15.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 16.Imbalzano A N, Schnitzler G R, Kingston R E. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 18.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2252. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 20.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 21.Langst G, Bonte E J, Corona D F, Becker P B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 22.Lee K M, Sif S, Kingston R E, Hayes J J. hSWI/SNF disrupts interactions between the H2A N-terminal tail and nucleosomal DNA. Biochemistry. 1999;38:8423–8429. doi: 10.1021/bi990090o. [DOI] [PubMed] [Google Scholar]
- 23.LeRoy G, Orphanides G, Lane W S, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 24.Logie C, Peterson C L. Catalytic nucleosome remodeling by the yeast SWI/SNF complex on nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logie C, Peterson C L. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 1999;304:726–741. doi: 10.1016/s0076-6879(99)04044-6. [DOI] [PubMed] [Google Scholar]
- 26.Logie C, Tse C, Hansen J, Peterson C L. The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry. 1999;38:2514–2522. doi: 10.1021/bi982109d. [DOI] [PubMed] [Google Scholar]
- 27.Lorch Y, Cairns B R, Zhang M, Kornberg R D. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 28.Lorch Y, Zhang M, Kornberg R D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 29.Meersseman G, Pennings S, Bradbury E M. Mobile nucleosomes—a general behavior. EMBO J. 1992;11:2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 32.Owen-Hughes T A, Utley R T, Cote J, Peterson C L, Workman J L. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 33.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 34.Pennings S, Meersseman G, Bradbury E M. Mobility of positioned nucleosomes on 5 S rDNA. J Mol Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 35.Peterson C L. Multiple SWItches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 36.Peterson C L. SWI/SNF complex: dissection of a chromatin remodeling machine. Cold Spring Harbor Symp Quant Biol. 1998;63:545–552. doi: 10.1101/sqb.1998.63.545. [DOI] [PubMed] [Google Scholar]
- 37.Polach K J, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 38.Pollard K J, Peterson C L. Chromatin remodeling: a marriage of two families? Bioessays. 1998;20:771–780. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 39.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramsay N, Felsenfeld G, Rushton B M, McGhee J D. A 145-base pair DNA sequence that positions itself precisely and asymmetrically on the nucleosome core. EMBO J. 1984;11:2605–2611. doi: 10.1002/j.1460-2075.1984.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saleh A, Lang V, Cook R, Brandl C J. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 42.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 43.Sudarsanam P, Cao Y, Wu L, Laurent B C, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodeling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 45.Travers A. An engine for nucleosome remodeling. Cell. 1999;96:311–314. doi: 10.1016/s0092-8674(00)80543-7. [DOI] [PubMed] [Google Scholar]
- 46.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 47.Tsukiyama T, Palmer J, Landel C C, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varga-Weisz P D, Becker P B. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 49.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 50.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 51.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 52.Winston F, Carlson M. Yeast SWI/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 53.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 54.Yudkovky N, Logie C, Hahn S, Peterson C L. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi-2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]